ABSTRACT
Limited information is available in relation to surveillance, genotyping, genome sequences, and treatment outcomes for rare hepatitis C virus variants. Here, we have characterized a novel subtype of major hepatitis C virus genotype 1, which was deep sequenced before and after treatment failure with 4 weeks of glecaprevir and pibrentasvir.
ANNOUNCEMENT
Hepatitis C virus (HCV), which belongs to the genus Hepacivirus within the family Flaviviridae, causes liver cirrhosis and cancer (1). HCV is divided into 8 genotypes and >90 subtypes, and unassigned putative subtypes are still being detected, e.g., in Africa (2–4). By deep sequencing, we investigated an unknown subtype of HCV genotype 1 from a patient participating in the 4RIBC study (5) (EudraCT no. 2017-005179-21) who had been treated for 4 weeks with glecaprevir and pibrentasvir (Maviret). Treatment failed, and the patient was confirmed to be positive for the same virus at 12 weeks posttreatment. Subsequently, the patient was cured by 12 weeks of treatment with sofosbuvir, velpatasvir, and voxilaprevir (Vosevi). The viral load was 7.5 log IU/ml at baseline prior to treatment and 6.2 log IU/ml at failure, as quantified by the COBAS HCV assay (Roche). For sequencing, the RNA from the baseline sample (A106-Baseline) and the 12-week post-Maviret-treatment sample (A106-Post) was extracted from 100 μl of plasma with the TRIzol method (6). After RNA extraction, human rRNA was removed with the NEBNext rRNA depletion kit. Libraries were prepared with the NEBNext Ultra II directional RNA library preparation kit and sequenced with paired-end 150-bp reads on an Illumina NextSeq instrument (7). The human host reads (14,325,771 reads) were depleted by HISAT2 v.2.1.0 (8) mapping to the human genome hg37 (GenBank accession no. GCA_000001405.13). The unmapped reads (13,106,372 reads) were subjected to de novo assembly by IVA v.1.0.8 (9). Subsequent mapping and consensus calling were performed with BWA MEM and SAMtools with the single open reading frame (ORF) sequence (10). All tools were run with default parameters.
The ORF was annotated with Geneious v.10.2.3 (11) based on reference strain H77 (GenBank accession no. NC_004102) and had 9,036 bp, including the stop codon. No recombination sites were detected by RDP v.5.05 (12). The sequences of the 5′ and 3′ untranslated regions were omitted after assembly (5′ untranslated region, 207 bases; 3′ untranslated region, 101 bases). The genome coverage was ∼215,000×, and the genome had a G+C content of 58%.
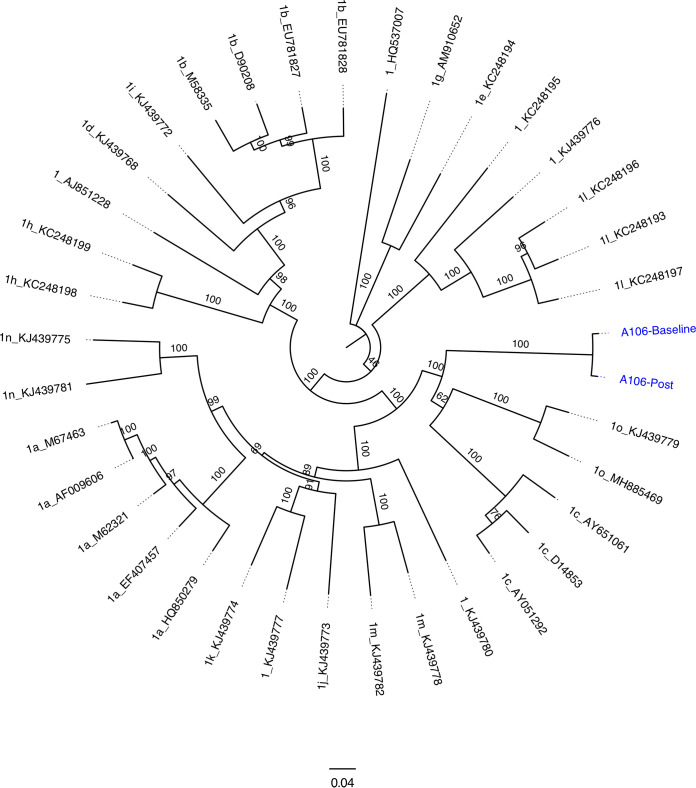
The genotype 1 reference sequences were obtained from the International Committee on Taxonomy of Viruses (ICTV) (13). The genotype 1 ORF nucleotide sequences and the A106-Baseline and A106-Post sequences were aligned with MUSCLE v.3.8.425 (14), and a maximum-likelihood phylogenetic tree was created with IQ-TREE v.1.6.8, with 1,000 bootstrap iterations (15), and visualized with FigTree v.1.4.3 (http://tree.bio.ed.ac.uk/software/figtree). The pretreatment and posttreatment samples create a distinct branch from the other subtypes (Fig. 1). After treatment (A106-Post), 172 nucleotide differences (1.9%) above 50% were detected in the consensus genome by mapping and variant calling; all except 1 could be detected at baseline (A106-Baseline) as minor variants above 0.5%. Resistance-associated substitutions (RASs) were determined with HCV-GLUE v.0.1.63 (16). Before treatment initiation, multiple RASs were detected in the nonstructural protein 3 (NS3) protease region and NS5A (NS3, 56F and 170I; NS5A, 24K, 28M, 30Q, 31M, and 37L), while none was found in the NS5B polymerase region. All RASs were found in 96 to 100% of the genome population reads and remained after treatment failure, without further additions.
FIG 1.
Phylogenetic tree with the genotype 1 reference sequences from the ICTV, with the two novel sequences, A106-Baseline and A106-Post, indicated in blue. The sequences were aligned with MUSCLE v.3.8.425; a maximum-likelihood phylogenetic tree was generated with IQ-TREE v.1.6.8, with 1,000 bootstrap iterations, and visualized with FigTree v.1.4.3. Each branch is labeled with the genotype number, subtype letter, and NCBI GenBank accession number. Internal branches are labeled with bootstrap support. The bar indicates substitutions per site.
The closest reference strain was 1o_KJ439779 from Africa (GenBank accession no. KJ439779.1), with a consensus sequence identity of ∼80% at the nucleotide level. The viral genomes in this study originated from a patient who had immigrated from Africa to Denmark in 1996, with known potential prior exposure to HCV in Nigeria, and rare genotype 1 subtypes in Africa have been found to have lower sustained virologic response (SVR) rates (2). The genome sequence presented and the appertaining information are important for future care of patients infected with HCV subtypes that are not commonly detected.
Data availability.
The genome sequences have been deposited in GenBank with accession no. MZ541883 and MZ541884 for A106-Baseline and A106-Post, respectively. The human read-depleted sequencing reads have been deposited in the NCBI database under BioProject no. PRJNA745515.
ACKNOWLEDGMENTS
This work was supported by the Danish Regions and a Ph.D. grant from the University of Southern Denmark (SDU). Additional support was from the Novo Nordisk Foundation, the Danish Cancer Society, and the Weimann Foundation. The funders had no influence on the study, data, or manuscript preparation.
Contributor Information
Jens Bukh, Email: jbukh@sund.ku.dk.
Kenneth M. Stedman, Portland State University
REFERENCES
- 1.Bukh J. 2016. The history of hepatitis C virus (HCV): basic research reveals unique features in phylogeny, evolution and the viral life cycle with new perspectives for epidemic control. J Hepatol 65(Suppl):S2–S21. doi: 10.1016/j.jhep.2016.07.035. [DOI] [PubMed] [Google Scholar]
- 2.Childs K, Davis C, Cannon M, Montague S, Filipe A, Tong L, Simmonds P, Smith D, Thomson EC, Dusheiko G, Agarwal K. 2019. Suboptimal SVR rates in African patients with atypical genotype 1 subtypes: implications for global elimination of hepatitis C. J Hepatol 71:1099–1105. doi: 10.1016/j.jhep.2019.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Niebel M, Singer JB, Nickbakhsh S, Gifford RJ, Thomson EC. 2017. Hepatitis C and the absence of genomic data in low-income countries: a barrier on the road to elimination? Lancet Gastroenterol Hepatol 2:700–701. doi: 10.1016/S2468-1253(17)30257-1. [DOI] [PubMed] [Google Scholar]
- 4.Pawlotsky JM, Negro F, Aghemo A, Berenguer M, Dalgard O, Dusheiko G, Marra F, Puoti M, Wedemeyer H. 2020. EASL recommendations on treatment of hepatitis C: final update of the series. J Hepatol 73:1170–1218. doi: 10.1016/j.jhep.2020.08.018. [DOI] [PubMed] [Google Scholar]
- 5.Madsen LW, Christensen PB, Fahnøe U, Pedersen MS, Bukh J, Øvrehus A. 2021. Inferior cure rate in pilot study of four‐week glecaprevir/pibrentasvir treatment with or without ribavirin of chronic hepatitis C. Liver Int doi: 10.1111/liv.14991. [DOI] [PubMed] [Google Scholar]
- 6.Fahnøe U, Bukh J. 2019. Full-length open reading frame amplification of hepatitis C virus. Methods Mol Biol 1911:85–91. doi: 10.1007/978-1-4939-8976-8_5. [DOI] [PubMed] [Google Scholar]
- 7.Pedersen MS, Mollerup S, Nielsen LG, Jenssen H, Bukh J, Schønning K. 2019. Genome sequence of an unknown subtype of hepatitis C virus genotype 6: another piece for the taxonomic puzzle. Microbiol Resour Announc 8:e01030-19. doi: 10.1128/MRA.01030-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hunt M, Gall A, Ong SH, Brener J, Ferns B, Goulder P, Nastouli E, Keane JA, Kellam P, Otto TD. 2015. IVA: accurate de novo assembly of RNA virus genomes. Bioinformatics 31:2374–2376. doi: 10.1093/bioinformatics/btv120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jensen SB, Fahnøe U, Pham LV, Serre SBN, Tang Q, Ghanem L, Pedersen MS, Ramirez S, Humes D, Pihl AF, Filskov J, Sølund CS, Dietz J, Fourati S, Pawlotsky JM, Sarrazin C, Weis N, Schønning K, Krarup H, Bukh J, Gottwein JM. 2019. Evolutionary pathways to persistence of highly fit and resistant hepatitis C virus protease inhibitor escape variants. Hepatology 70:771–787. doi: 10.1002/hep.30647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. 2015. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol 1:vev003. doi: 10.1093/ve/vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simmonds P, Becher P, Bukh J, Gould EA, Meyers G, Monath T, Muerhoff S, Pletnev A, Rico-Hesse R, Smith DB, Stapleton JT. 2017. ICTV virus taxonomy profile: Flaviviridae. J Gen Virol 98:2–3. doi: 10.1099/jgv.0.000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robert C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quang B, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, Von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol 37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singer JB, Thomson EC, McLauchlan J, Hughes J, Gifford RJ. 2018. GLUE: a flexible software system for virus sequence data. BMC Bioinformatics 19:532. doi: 10.1186/s12859-018-2459-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome sequences have been deposited in GenBank with accession no. MZ541883 and MZ541884 for A106-Baseline and A106-Post, respectively. The human read-depleted sequencing reads have been deposited in the NCBI database under BioProject no. PRJNA745515.