ABSTRACT
The complete genome sequence of Streptococcus pneumoniae strain Rx1, a Hex mismatch repair-deficient standard transformation recipient, was obtained by combining Nanopore and Illumina sequencing technologies. The genome consists of a 2.03-Mb circular chromosome, with 2,054 open reading frames and a GC content of 39.72%.
ANNOUNCEMENT
Streptococcus pneumoniae is a human pathogen and the most important model organism for studying bacterial genetics and genomics. Widely used laboratory strains include type 2 Avery’s strain D39 and its derivatives Rx1 and R6, which are standard transformation recipients (1, 2). We characterized the complete genome sequence of Rx1, a highly transformable and Hex mismatch repair system-deficient strain. To track the genomic changes that gave rise to Rx1, we also sequenced the genome of its unencapsulated parental strain R36A (Table 1). Strains, which were obtained from the Guild laboratory collection (3), were grown in tryptic soy broth at 37°C for 4 h until they reached an optical density at 590 nm (OD590) of 0.8. Pneumococcal cells were harvested by centrifugation (5,000 × g for 30 min at 4°C), and the cell pellet was dry vortex-mixed and lysed in 0.1% deoxycholate-0.008% SDS. High-molecular-weight DNA was purified three times with 1 volume of chloroform-isoamyl alcohol (24:1 [vol/vol]), precipitated in 0.6 volumes of ice-cold isopropanol, and spooled on a glass rod. DNA was resuspended in 10× saline-sodium citrate (SSC) buffer (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and then adjusted to 1× SSC and maintained at 4°C. The DNA solution was homogenized using a rotary mixer. Oxford Nanopore Technologies MinION and Illumina HiSeq 2500 instruments were used for DNA sequencing. DNA was not sheared; size selection was obtained with 0.8 volumes of AMPure XP beads (Beckman Coulter). The Nanopore sequencing library was prepared using the SQK-LSK108 kit (Oxford Nanopore Technologies) following the manufacturer’s instructions, and the sample was sequenced using an R9.4 flow cell (FLO-MIN106). Postsequencing high-accuracy base calling and adapter trimming of raw Nanopore reads were performed using Guppy v4.0.11 with configuration dna_r9.4.1_450bps_hac, and base-called reads were analyzed with NanoPlot v1.18.2 (4). Illumina sequencing was performed at MicrobesNG (University of Birmingham) using the Nextera XT library preparation kit (Illumina Inc.), followed by paired-end sequencing. Illumina reads were trimmed using Trimmomatic v0.30 (5) and analyzed with FastQC v0.11.5 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). Nanopore and Illumina sequencing generated 3,892 long reads (26,780,859 bp [N50, 18.3 kbp]) and 86,582 read pairs (2 × 250 bp), respectively, for Rx1, whereas 4,771 long reads (27,433,219 bp [N50, 16.9 kbp]) and 278,462 read pairs were obtained for R36A. Sequence coverage was 31.6× for Rx1 and 67.0× for R36A. A hybrid assembly of Nanopore and Illumina reads was obtained using Unicycler v0.4.712 (6). Assembly completeness and quality were assessed using Bandage v.0.8.1 (7) and Ideel (https://github.com/mw55309/ideel), respectively. Annotation was performed with the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) v5.1 (8). Default parameters were used for all tools unless otherwise specified. The Rx1 genome consists of a 2,030,186-bp single circular chromosome containing 2,054 open reading frames (ORFs), of which 1,813 have a predicted function. The 2,039,955-bp circular chromosome of R36A contains 2,059 ORFs, of which 1,834 have a putative function. Both genomes have (i) a GC content of 39.72%, (ii) 58 tRNA genes, 3 rRNA operons, and 3 structural RNAs, (iii) a 36.6-kb pneumococcal pathogenicity island 1 (PPI1) (9), (iv) prophage remnants, and (v) remnants of the integrative and conjugative element Tn5253 (10–12). Rx1 and R36A capsule loci are schematized in Fig. 1. Rx1 harbors type I restriction-modification system SpnD39III variant C, while R36A harbors variant D (13). In Rx1, g.168,614C>A, g.1,979,527G>A, and g. 1,629,603delA nucleotide changes introduce premature termination codons in hexB, pspc3.1, and dpnC, respectively.
TABLE 1.
Genealogy of the S. pneumoniae Rx1 strain
| Strain | Descriptiona | Relevant propertiesb | GenBank accession no. (year)a |
|---|---|---|---|
| D39 | Avery’s strain, clinical isolate (1916); type 2, virulent (3, 19–23) | pDP1+, Hex+, DpnI+, comC1-comD1, pspC3.1 | CP000410.1 (2007) (24) |
| R36 | D39 passaged 36 times in anti-type 2 serum (1944); rough, avirulent (3, 21, 22) | pDP1+, Hex+, DpnI+, comC1-comD1, pspC3.1 | Not available |
| R36A | Highly transformable R36 colony morphology variant (1944); rough, avirulent (3, 20, 23, 25) | pDP1−, Hex+, DpnI+, comC1-comD1, pspC3.1 | CP079922 (2021) (this study) |
| R6 | Highly transformable R36A single-colony isolate (1962); rough, avirulent (3, 26, 27) | pDP1−, Hex+, DpnI+, comC1-comD1, pspC3.1 | AE007317.1 (2001) (16) |
| A66 | Avery’s strain, clinical isolate (1949); type 3, virulent (23, 25) | Hex+, DpnI, comC2-comD2, pspC11.4 | LN847353.1 (2015) (28) |
| SIII-N | R36A transformed with A66 DNA (1949); type 3, virulent (20, 23, 25, 29) | comC1-comD1, pspC3.1 | Not available |
| Rx | Spontaneous rough derivative of R36A (1959); reduced type 3 capsule production, avirulent (3, 17, 23, 30) | pDP1−, Hex− (HexB−), comC1-comD1, pspC3.1 | Not available |
| Rx1 | Highly transformable derivative of Rx (1959); reduced type 3 capsule production (Ugd mutant), avirulent (3, 31) | pDP1−, Hex− (HexB−), DpnI− (DpnC−), comC1-comD1, pspC3.1’ | CP079923 (2021) (this study) |
The year of the first strain description (except for the D39 isolation year) or of the sequence release is reported in parentheses.
pDP1 is a 3,161-bp cryptic plasmid (32). Hex is the DNA mismatch repair system encoded by hexA and hexB (33). DpnI is a restriction system composed of the DpnI/DpnC endonuclease and DpnD (34). comC-comD competence genes encode the competence-stimulating peptide (CSP) and its ComD receptor (35–38). pspC encodes the virulence surface protein PspC (39, 40).
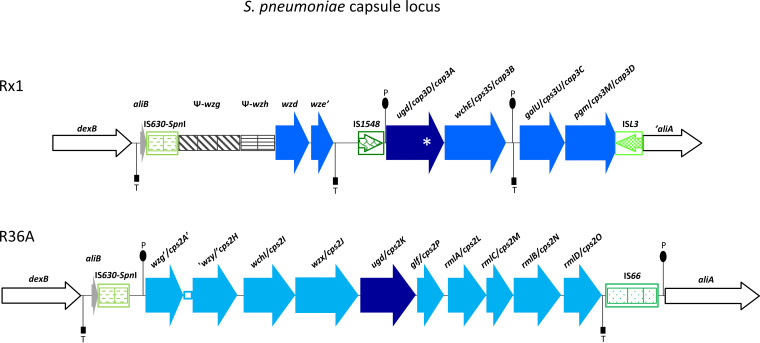
FIG 1.
S. pneumoniae capsule locus. Rx1 harbors a type 3 capsule locus acquired by A66 DNA through a double crossover between IS630-SpnI and aliA. At the 3′ end, recombination produced the insertion of an ISL3 transposase and a 950-bp deletion of the aliA 5′ end, as in the A66 capsule locus. IS1548 identifies (i) a 5′ fragment, common to all serotypes (14), that contains wzg and wzh pseudogenes and wzd and wze genes and is not involved in type 3 capsular synthesis (15) and (ii) a 3′ fragment containing ugd/cap3D/cap3A UDP-glucose dehydrogenase gene, wchE/cps3S/cap3B synthase gene, galU/cps3U/cap3C, and pgm/cps3M/cap3D genes involved in UDP-glucose biosynthesis (15–17). The nucleotide change g.317,495C>T in ugd/cps3A/cps3D (indicated with an asterisk) causes p.R320C in the UDP-glucose dehydrogenase UDP-binding domain. The type 2 capsule locus of R36A harbors a 7,505-bp deletion involving the 3′ end of wzg/cps2A, seven genes (namely, wzh/cps2B, wzd/cps2C, wze/cps2D, wchA/cps2E, wchF/cps2T, wchG/cps2F, and wchH/cps2G), and the 5′ end of wzy/cps2H (18). The deletion event left an inverted 25-bp fragment (indicated with an open box) belonging to the lost wzg/cps2A 3′ end.
Data availability.
The complete genome sequences of R36A and Rx1 are available under GenBank accession no. CP079922 and CP079923, respectively. The sequencing project is available under NCBI BioProject accession no. PRJNA748391. Nanopore and Illumina sequencing reads are available under Sequence Read Archive (SRA) accession no. SRR15216323 and SRR15216322, respectively, for R36A and SRA accession no. SRR15216380 and SRR15216379, respectively, for Rx1.
ACKNOWLEDGMENTS
This work was supported by the Italian Ministry of Education, University, and Research (MIUR-Italy), under grant 20177J5Y3P (Progetti di Ricerca di Rilevante Interesse Nazionale, Bando 2017). Illumina genome sequencing was provided by MicrobesNG (http://www.microbesng.uk).
Contributor Information
Francesco Iannelli, Email: francesco.iannelli@unisi.it.
Steven R. Gill, University of Rochester School of Medicine and Dentistry
REFERENCES
- 1.Pearce BJ, Iannelli F, Pozzi G. 2002. Construction of new unencapsulated (rough) strains of Streptococcus pneumoniae. Res Microbiol 153:243–247. doi: 10.1016/s0923-2508(02)01312-8. [DOI] [PubMed] [Google Scholar]
- 2.Santoro F, Iannelli F, Pozzi G. 2019. Genomics and genetics of Streptococcus pneumoniae. Microbiol Spectr 7:GPP3-0025-2018. doi: 10.1128/microbiolspec.GPP3-0025-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith MD, Guild WR. 1979. A plasmid in Streptococcus pneumoniae. J Bacteriol 137:735–739. doi: 10.1128/jb.137.2.735-739.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Coster W, D’Hert S, Schultz DT, Cruts M, Van Broeckhoven C. 2018. NanoPack: visualizing and processing long-read sequencing data. Bioinformatics 34:2666–2669. doi: 10.1093/bioinformatics/bty149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31:3350–3352. doi: 10.1093/bioinformatics/btv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. 2016. NCBI Prokaryotic Genome Annotation Pipeline. Nucleic Acids Res 44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown JS, Gilliland SM, Spratt BG, Holden DW. 2004. A locus contained within a variable region of pneumococcal pathogenicity island 1 contributes to virulence in mice. Infect Immun 72:1587–1593. doi: 10.1128/IAI.72.3.1587-1593.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santoro F, Oggioni MR, Pozzi G, Iannelli F. 2010. Nucleotide sequence and functional analysis of the tet(M)-carrying conjugative transposon Tn5251 of Streptococcus pneumoniae. FEMS Microbiol Lett 308:150–158. doi: 10.1111/j.1574-6968.2010.02002.x. [DOI] [PubMed] [Google Scholar]
- 11.Iannelli F, Santoro F, Oggioni MR, Pozzi G. 2014. Nucleotide sequence analysis of integrative conjugative element Tn5253 of Streptococcus pneumoniae. Antimicrob Agents Chemother 58:1235–1239. doi: 10.1128/AAC.01764-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santoro F, Romeo A, Pozzi G, Iannelli F. 2018. Excision and circularization of integrative conjugative element Tn5253 of Streptococcus pneumoniae. Front Microbiol 9:1779. doi: 10.3389/fmicb.2018.01779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manso AS, Chai MH, Atack JM, Furi L, De Ste Croix M, Haigh R, Trappetti C, Ogunniyi AD, Shewell LK, Boitano M, Clark TA, Korlach J, Blades M, Mirkes E, Gorban AN, Paton JC, Jennings MP, Oggioni MR. 2014. A random six-phase switch regulates pneumococcal virulence via global epigenetic changes. Nat Commun 5:5055. doi: 10.1038/ncomms6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bentley SD, Aanensen DM, Mavroidi A, Saunders D, Rabbinowitsch E, Collins M, Donohoe K, Harris D, Murphy L, Quail MA, Samuel G, Skovsted IC, Kaltoft MS, Barrell B, Reeves PR, Parkhill J, Spratt BG. 2006. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet 2:e31. doi: 10.1371/journal.pgen.0020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arrecubieta C, Garcia E, López R. 1995. Sequence and transcriptional analysis of a DNA region involved in the production of capsular polysaccharide in Streptococcus pneumoniae type 3. Gene 167:1–7. doi: 10.1016/0378-1119(95)00657-5. [DOI] [PubMed] [Google Scholar]
- 16.Hoskins J, Alborn WE, Arnold J, Blaszczak LC, Burgett S, DeHoff BS, Estrem ST, Fritz L, Fu DJ, Fuller W, Geringer C, Gilmour R, Glass JS, Khoja H, Kraft AR, Lagace RE, LeBlanc DJ, Lee LN, Lefkowitz EJ, Lu J, Matsushima P, McAhren SM, McHenney M, McLeaster K, Mundy CW, Nicas TI, Norris FH, O'Gara M, Peery RB, Robertson GT, Rockey P, Sun PM, Winkler ME, Yang Y, Young-Bellido M, Zhao G, Zook CA, Baltz RH, Jaskunas SR, Rosteck PR, Skatrud PL, Glass JI. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J Bacteriol 183:5709–5717. doi: 10.1128/JB.183.19.5709-5717.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prudhomme M, Martin B, Mejean V, Claverys JP. 1989. Nucleotide sequence of the Streptococcus pneumoniae hexB mismatch repair gene: homology of HexB to MutL of Salmonella typhimurium and to PMS1 of Saccharomyces cerevisiae. J Bacteriol 171:5332–5338. doi: 10.1128/jb.171.10.5332-5338.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iannelli F, Pearce BJ, Pozzi G. 1999. The type 2 capsule locus of Streptococcus pneumoniae. J Bacteriol 181:2652–2654. doi: 10.1128/JB.181.8.2652-2654.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffith F. 1928. The significance of pneumococcal types. J Hyg (Lond) 27:113–159. doi: 10.1017/s0022172400031879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Avery OT, Macleod CM, McCarty M. 1944. Studies on the chemical nature of the substance inducing transformation of pneumococcal types: induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J Exp Med 79:137–158. doi: 10.1084/jem.79.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacLeod CM, Krauss MR. 1947. Stepwise intratype transformation of pneumococcus from R to S by way of a variant intermediate in capsular polysaccharide production. J Exp Med 86:439–452. doi: 10.1084/jem.86.6.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Austrian R. 1953. Morphologic variation in pneumococcus. I. An analysis of the bases for morphologic variation in pneumococcus and description of a hitherto undefined morphologic variant. J Exp Med 98:21–34. doi: 10.1084/jem.98.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ravin AW. 1959. Reciprocal capsular transformations of pneumococci. J Bacteriol 77:296–309. doi: 10.1128/jb.77.3.296-309.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lanie JA, Ng W-L, Kazmierczak KM, Andrzejewski TM, Davidsen TM, Wayne KJ, Tettelin H, Glass JI, Winkler ME. 2007. Genome sequence of Avery’s virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J Bacteriol 189:38–51. doi: 10.1128/JB.01148-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor HE. 1949. Additive effects of certain transforming agents from some variants of pneumococcus. J Exp Med 89:399–424. doi: 10.1084/jem.89.4.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ottolenghi E, Hotchkiss RD. 1962. Release of genetic transforming agent from pneumococcal cultures during growth and disintegration. J Exp Med 116:491–519. doi: 10.1084/jem.116.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomasz A, Hotchkiss RD. 1964. Regulation of the transformability of pneumococcal cultures by macromolecular cell products. Proc Natl Acad Sci USA 51:480–487. doi: 10.1073/pnas.51.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hahn C, Harrison EM, Parkhill J, Holmes MA, Paterson GK. 2015. Draft genome sequence of the Streptococcus pneumoniae Avery strain A66. Genome Announc 3:e00697-15. doi: 10.1128/genomeA.00697-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Austrian R, Bernheimer HP, Smith EE, Mills GT. 1959. Simultaneous production of two capsular polysaccharides by pneumococcus. II. The genetic and biochemical bases of binary capsulation. J Exp Med 110:585–602. doi: 10.1084/jem.110.4.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dillard JP, Vandersea MW, Yother J. 1995. Characterization of the cassette containing genes for type 3 capsular polysaccharide biosynthesis in Streptococcus pneumoniae. J Exp Med 181:973–983. doi: 10.1084/jem.181.3.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guild WR, Shoemaker NB. 1974. Intracellular competition for a mismatch recognition system and marker-specific rescue of transforming DNA from inactivation by ultraviolet irradiation. Mol Gen Genet 128:291–300. doi: 10.1007/BF00268517. [DOI] [PubMed] [Google Scholar]
- 32.Oggioni MR, Iannelli F, Pozzi G. 1999. Characterization of cryptic plasmids pDP1 and pSMB1 of Streptococcus pneumoniae. Plasmid 41:70–72. doi: 10.1006/plas.1998.1364. [DOI] [PubMed] [Google Scholar]
- 33.Claverys JP, Lacks SA. 1986. Heteroduplex deoxyribonucleic acid base mismatch repair in bacteria. Microbiol Rev 50:133–165. doi: 10.1128/mr.50.2.133-165.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lacks SA, Mannarelli BM, Springhorn SS, Greenberg B. 1986. Genetic basis of the complementary DpnI and DpnII restriction systems of S. pneumoniae: an intercellular cassette mechanism. Cell 46:993–1000. doi: 10.1016/0092-8674(86)90698-7. [DOI] [PubMed] [Google Scholar]
- 35.Havarstein LS, Coomaraswamy G, Morrison DA. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc Natl Acad Sci USA 92:11140–11144. doi: 10.1073/pnas.92.24.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pozzi G, Masala L, Iannelli F, Manganelli R, Havarstein LS, Piccoli L, Simon D, Morrison DA. 1996. Competence for genetic transformation in encapsulated strains of Streptococcus pneumoniae: two allelic variants of the peptide pheromone. J Bacteriol 178:6087–6090. doi: 10.1128/jb.178.20.6087-6090.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pestova EV, Håvarstein LS, Morrison DA. 1996. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol Microbiol 21:853–862. doi: 10.1046/j.1365-2958.1996.501417.x. [DOI] [PubMed] [Google Scholar]
- 38.Iannelli F, Oggioni MR, Pozzi G. 2005. Sensor domain of histidine kinase ComD confers competence pherotype specificity in Streptococcus pneumoniae. FEMS Microbiol Lett 252:321–326. doi: 10.1016/j.femsle.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 39.Iannelli F, Oggioni MR, Pozzi G. 2002. Allelic variation in the highly polymorphic locus pspC of Streptococcus pneumoniae. Gene 284:63–71. doi: 10.1016/S0378-1119(01)00896-4. [DOI] [PubMed] [Google Scholar]
- 40.Iannelli F, Chiavolini D, Ricci S, Oggioni MR, Pozzi G. 2004. Pneumococcal surface protein C contributes to sepsis caused by Streptococcus pneumoniae in mice. Infect Immun 72:3077–3080. doi: 10.1128/IAI.72.5.3077-3080.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The complete genome sequences of R36A and Rx1 are available under GenBank accession no. CP079922 and CP079923, respectively. The sequencing project is available under NCBI BioProject accession no. PRJNA748391. Nanopore and Illumina sequencing reads are available under Sequence Read Archive (SRA) accession no. SRR15216323 and SRR15216322, respectively, for R36A and SRA accession no. SRR15216380 and SRR15216379, respectively, for Rx1.