Abstract
Background:
Glutamate (Glu) is of great interest in biomedical research. It is considered a biomarker in diabetes, which may potentially contribute to the development of autism in genetically vulnerable human populations, and it is found in relation to advanced glycation end products (AGEs) [1]. Additionally, Glu plays an active role in the function of ligand-gated ion channel glutamate receptors, chloride channels capable of filtering glutamate, as well as Potassium (K+)-channel [2]. Glu attains α [3] and β [4] crystal forms and Cβ-CH2 show asymmetric 1H signal pattern in NMR spectra.
Objectives:
The current study was undertaken to understand the signal patterns of Cβ-CH2 in Glu of the smallest dipeptide, Glycylglutamic Acid (GlyGlu), as well as the order, and planarity of the amide bond in the molecule.
Materials and Methods:
NMR spectra of GlyGlu were measured in D2O to deduce 1H and 13C chemical shifts and coupling constants. GlyGlu was crystallized from MeOH and the structure was determined by single crystal X-ray diffraction techniques.
Results:
The sidechain of Glu in the dipeptide dissimilates the β form. The amino group of Gly (Glycine) is protonated and exhibits hydrogen bonding with the main chain carboxylate group of a symmetry-related Glu that is deprotonated in the crystal packing of GlyGlu. The deprotonated main chain carboxylate of Glu is also in hydrogen-bonding distance from the side chain carboxylic acid group that is in the protonated form of a symmetry-related Glu of the dipeptide. The Cβ-CH2 geminal protons on the side chain of Glu have different chemical shifts and splitting pattern in 1H NMR reflecting their dissymmetric environment.
Conclusion:
The results reported will be useful for monitoring changes that Glu and/or molecules in connection to Glu may undergo in in vivo, in situ, and in vitro conditions. This provides a valuable metric which will enable the examination of the metabolites relevant to the detection and diagnosis of disease or developmental conditions, as well as scrutinizing the effectiveness of treatment options.
Keywords: Autism, Biomarker, Diabetes, Dipeptide, Structure, AGE, Derivatization
1. Introduction
Glu is an important metabolite because its accumulation causes neurotoxicity and is considered a novel biomarker in the development of type 2 diabetes. Gardener et al. (2009) reported that of maternal factors linked to autism, gestational diabetes was linked with a twofold increase in the incidence of autism [5–7]. Also the PI3K/Tor pathway is predicted to be activated by Insulin signaling through a mechanism that is comparable to the genetic changes as described by Scott et al. (1998) [8, 9]. Moreover, in neurons, the PI3K/Tor signaling pathway alters a form of synaptic plasticity that has been involved in autism [10, 11]. Furthermore, Glutamate, a ligand for metabotropic glutamate receptors (mGluR) facilitates synaptic plasticity known as long term depression. Insulin signaling has also been predicted to contribute to the development of autism in genetically susceptible individuals [12–14].
Diabetes Mellitus is a disease of impaired energy metabolism, characterized by damaged glucose metabolism and insulin resistance. Chronically elevated blood sugar levels lead to the rise of advanced glycation end products (AGEs) which may cause significant physiological complications which are the hallmarks of the disease [1]. This occurs when Glutamate catalyzes and/or participates in the glycation of susceptible proteins [15, 16] when located less than 5.0 Å [17] from AGE-modification sites in these proteins. Amino groups with lower pKa values are expected to be more reactive toward glycation due to their greater nucleophilicity [18, 19].
The acidic pK values for the α-amino, α-carboxyl and δ-carboxyl groups of Glu are 9.47, 2.10 and 4.07, respectively [20, 21]. Also Glu is known to exist in two defined α and β crystal forms. The amide C-N double bond strength increases and is accompanied by a transfer of electron density from the amide nitrogen to oxygen in Glycylglycine when in the zwitterionic state [22, 23]. The resonance-stabilized planar amide bond exhibits approximately 40% partial double-bond strength makes one of the highly stable and least reactive functional groups.
The dipeptide, GlyGlu, is chosen as a model system to study because: 1) the amino and carboxylate/carboxylic acid groups are covalently connected by a framework with a single amide moiety; 2) it contains β methylene geminal protons of Glu; and 3) will not introduce additional chiral centers. Characterization of the structural properties of this molecule will provide the required information about the amide moiety in it. What would happen to the diastereotopic signal pattern seen in 1H NMR pattern of Glu when it is a peptide? Here we present the crystal structure determination and NMR studies of GlyGlu. Since proteins and peptides are polymers of amide/peptide groups their properties are documented and their automated synthesis is considerably advanced for large scale production. Hence, the ability to generate derivatives with highly diverse functional substitutions provides a large collection of peptidomimetic analog variations for a given amide fragment based inhibitor of interest.
2. Materials and Methods
Glycylglutamic acid (GlyGlu) –
GlyGlu (CAS Number: 7412–78-4) was dissolved in 0.7 mL of D2O to produce a 0.105 M solution. NMR spectra were recorded using tertramethylsilane (TMS) as the standard on a Bruker Advance II 400 MHz NMR spectrometer with an indirect detection probe. Chemical shifts of the signals were reported in parts per million (ppm) and peak patterns defined as singlet (s), doublet (d), triplet (t), multiplet (m), doublet of doublets (dd) with coupling constants (J) in Hertz (Hz) indicated in NMR spectra.
The sample in D2O that was used for NMR studies was then expended for crystallization. Crystals were obtained by vapor diffusion technique using methanol as the solvent. Data were collected at 100 K using a Bruker SMART APEX CCD single crystal X-ray diffractometer with graphite monochromated Mo Kα radiation (λ=0.71073 Å) with a crystal to detector distance of 4.00 cm. Bruker Apex2 [24] and SAINT software packages [25] were used for data collection and integration. Collected data were corrected for absorption and other systematic errors using SADABS [26] by multi-scan methods based on the Laue symmetry using equivalent reflections. SHELXTL-PLUS [27–29] was used to determine the solution and perform the refinement of the structure. The structures were determined by direct methods and refined by full matrix least-squares refinement by minimizing while non-hydrogen atoms were refined anisotropically. All hydrogen atoms were located from difference Fourier maps and were refined freely using isotropic thermal parameters following the procedure accepted previously [29, 30]. Complete listings of geometrical parameters, positional and isotropic displacement coefficients for hydrogen atoms and anisotropic displacement coefficients for the non-hydrogen atoms are deposited with the Cambridge Crystallographic Data Centre as (CCDC 2090883). These data can be obtained from The Cambridge Crystallographic Data Centre www.ccdc.cam.ac.uk/data_request/cif.
3. Results
Single crystal structure determination
Single-crystal X-ray structure of GlyGlu (C7H12N2O5) (Figure 1) was determined and all hydrogen atoms were located in Difference Fourier maps. In the crystal structure (Figure 2) the molecule is in the zwitterionic form. The N-terminal amine group is protonated, the C-terminal carboxylic acid group is deprotonated, and the side chain carboxylic acid is protonated. The crystal packing is dominated by a multitude of N-H•••O and O-H•••O hydrogen bonds that involve the ammonium and amide groups. In the GlyGlu structure, these hydrogen bonds assemble the molecules into intricately connected three dimensional networks in the crystalline state.
Figure 1.

Chemical structure of GlyGlu is shown with atom numbering schemes based on that used in the crystal structure determination. Not all hydrogen atoms are labeled for clarity.
Figure 2.
View down the amide bond of the crystal structure of GlyGlu in the Thermal Ellipsoid Plot representation. Atom labels and numbering shown are the same as what was deposited in the CSD and used in the text. Atoms C4, C3, C2 and C1 correspond to Cα, Cβ, Cγ and Cδ respectively. The peptide bond torsion angle C7-C6-N1-C4 is 171.01° in the crystal structure.
The angle and distances associated with the amide moiety in GlyGlu are −6.8 (3) °, 1.233 (3) Å, 1.327 (3) Å and 0.8800 Å for O5-C6-N1-C4, C6-O5, C6-N1 and N1-H1, respectively, reinforcing the existence of partial double bond nature. The structure of GlyGlu is densely packed (Figure 3) with no residual void within the lattice structure or bound solvent molecules located in its crystalline state. This confirms the chemical structure and molecular formula of the compound used for NMR analysis. The Dx, for GlyGlu is 1.51 Mg m−3. This falls within the category of hydrophilic structures with abundant hydrogen bonding groups of dipeptides that are with the predicted [31] Dx value to typically fall within the 1.40 – 1.60 Mg m−3 range. Hydrogen bonds are found between atoms N1H1-O3’, N2H2C-O3’, N2H2D-O5’, N2H2E-O1’, N2H2E-O5 and O2H2-O4’. The typical hydrogen bonding pattern which results in the dimerization between two COOH groups such as that observed in Fenofibric acid [30] is absent in GlyGlu. However, COOH•••−OOC type interactions found between atoms, C1O2H2•••O4’C5’ with corresponding distances (H-O, O-O) and angles (O-H-O) are 1.7Å, 2.6Å and 171.9° and C1O2H2•••O3’C5’ are 2.8Å, 3.3Å and 121.8° in the GlyGlu structure. The distances and angles of 2.6 Å (between H2 and H2CN2), 2.8 Å (O2 and H2CN2) and 95.6° between atoms of the side chain COOH and main chain NH3+ groups are noticed whereas the O2 of C1OOH is 2.7 Å away from H2EN2 and makes 111.4°. Also, the distances and angles of main chain O3 of COO− is 1.9Å, 2.7Å and 161.3° from H2C, N2 of NH3+ symmetry related molecule and O4 of COO− is 2.7 Å, 3.4 Å and 131.2° from H2C and N2 of NH3+ symmetry related molecule. In addition, the distances and angles of main chain O3 of COO− is 2.0 Å and 2.9 Å away from H1N1 and N1 and forms 172.2° with main chain NH group.
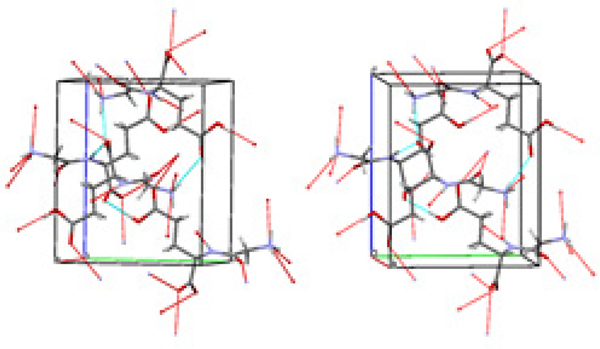
Figure 3.
The crystal packing diagram for GlyGlu. A unit cell with axes and hydrogen-bond pattern for molecules of a cell packing are shown for P212121 space group.
Glu side chain conformation in the crystalline state
A survey of CCDC (Ver. 2020.20.0) [32] for entries containing Glu as part of a mono-, di- or tripeptide structure were considered for comparison with that of GlyGlu. All the entries that met this prerequisite regardless of whether the Glu is the N or C terminal residue were included in the evaluation. Any entry of metal complex with Glu was excluded. Of the entries included, a superposition of 10 non-hydrogen atoms of Glu and the RMS deviation of their crystal structure coordinates from Glu of GlyGlu are shown along with the dihedral angles (Table 1, Figure 4). As shown in the (Figure 4) side chain conformations and as reflected by the RMS value of Glu in GlyGlu, it is very close to LGLUAC02 and Si2005 which are in α form but with and without bound HCl, however the conformation is not altered significantly.
Table 1.
Entries superimposed based on non-hydrogen atoms of Glu with GlyGlu and their RMS (Å) values.
| Entry | Polymorph | Torsion/Dihedral Angle (°) | Compound (Complex) | RMS (Å) | |||
|---|---|---|---|---|---|---|---|
| NCαCβCγ | CαCβCγCδ | CβCγCδOε(1) | CβCγCδOε(2) | ||||
| 2090883 | alpha | −58.15 | 176.61 | −1.73 | 179.14 | Glycyl-L-glutamic acid | 0.0 |
| LGLUAC02 | alpha | 178.43 | 68.83 | −104.92 | 73.99 | L-Glutamic acid | 0.137 |
| Si2005 | alpha | −69.92 | −171.72 | −166.60 | 14.87 | L-Glutamic acid Hydrochloride | 0.137 |
| ARGGLU10 | −56.83 | −172.05 | 179.91 | −2.58 | L-Arginine L-glutamate monohydrate | 0.138 | |
| BELCUQa | (C)−57.83 (N)60.31 |
−131.06 −174.55 |
36.02 174.20 |
−145.50 −4.20 |
α-L-Glutamyl-L-glutamic acid | 0.523 | |
| BOFZOL | 59.05 | 171.15 | −139.23 | 37.55 | α-L-Leucyl-L-glutamic acid | 0.634 | |
| CIJGUX | 178.46 | 174.31 | −179.62 | 0.30 | L-Valyl-L-glutamic acid | 0.773 | |
| LGLUAC11 | beta | −51.79 | −73.10 | −160.70 | 18.80 | L-Glutamic acid | 0.859 |
| DIYZIU | −171.14 | −179.19 | 119.71 | −59.71 | L-Arginyl-L-glutamic acid monohydrate | 1.031 | |
| LGLUAC12 | −52.12 | −73.15 | −160.72 | 19.34 | L-Glutamic acid | 1.053 | |
| LGLUAC01 | beta | −50.98 | −74.16 | −160.14 | 20.30 | L-Glutamic acid | 1.130 |
| BUDXUT | −170.26 | 70.26 | 175.00 | −5.67 | L-Prolyl-L-glutamic acid dihydrate | 1.131 | |
-C terminal residue 1, N terminal residue 2
Figure 4.
Superimposition of main chain atoms of Glu to GlyGlu. The C of GlyGlu-(Green); L-Glutamic acid Hydrochloride (SI2055-Pink); LULGAC11 (Maroon); LGLUAC12 (Yellow); and ARGGLU10 (Brown), N (Blue), O (Red) and H (White) atoms are shown with corresponding colors.
The most significant difference between the α and β forms are predominantly confined to the torsion angles defined by, N-Cα-Cβ-Cγ, Cβ-Cγ-Cδ-Oε(1) and Cβ-Cγ-Cδ-Oε(2), with the corresponding values, 178.2°, 74.2° and −104.6°, in α and −51.8°, 18.8° and −160.7° in β forms, respectively of Glu. In GlyGlu crystalline state these torsion angles of Glu are 58.1°, 1.8° and 179.1° which are closer to that displayed in the β form than that found in α form of Glu.
Glu side chain conformation in solution state
The 400 MHz 1D 1H NMR spectra of GlyGlu (Figure 5) were analyzed, and the spins were assigned, optimized and interpreted using the tools in the Guided Ideographic Spin System Model Optimization method [33]. The parametrization of spectra analysis of 1H NMR data reveal accurate spectral signatures of chemical shifts, coupling constants and splitting patterns (Table 2) for the proton signals of GlyGlu dissolved in D2O. The geminal Cβ methylene protons of Glu are chemically not alike and magnetically not equivalent when GlyGlu conformation/structure is in solution environment. There were no peaks downfield of 6ppm in 1H NMR which implies all the exchangeable protons are replaced by D from the solvent. Furthermore, 13C NMR (100 MHz, D2O) chemical shift (ppm) are: C1 – (Cδ Carboxyl) of Glu 178.22, C5 – (Cα Carboxyl) of Glu 177.49, C6 – (Cα Carboxyl)) of Gly 166.51, C4 – (Cα) of Glu 54.25, C7 – (Cα) of Gly 40.31, C2 – (Cγ) of Glu 30.95, C3 – (Cβ) of Glu 26.77.
Figure 5.

1D 400MHz 1H NMR spectrum of GlyGlu in D2O. Integration of peaks shown correspond to non-exchangeable protons of the dipeptide.
Table 2.
Chemical shift(s) and coupling constants for GlyGlu
| Label | Atom (Residue) | δ(ppm) | J(Hz)(coupling nuclei) | Splitting |
|---|---|---|---|---|
| H7B | αCH(Gly) | 3.88 | −16.06(H7B-H7A) | |
| αCH(Gly) | 3.88 | −16.05(H7A-H7B) | ||
| H4 | αCH(Glu) | 4.28 | 5.04(H4-H3A) | dd |
| 8.97(H4-H3B) | ||||
| H3A | βCH(Glu) | 2.17 | 5.04(H3A-H4) | ddd |
| −14.25(H3A-H3B) | ||||
| 8.35(H3A-H2B) | ||||
| 7.19(H3A-H2A) | ||||
| H3B | βCH(Glu) | 1.97 | 8.97(H3B-H4) | dd |
| −14.25(H3B-H3A) | ||||
| 5.45(H3B-H2B) | ||||
| 8.99(H3B-H2A) | ||||
| H2B | γCH(Glu) | 2.43 | 8.35(H2B-H3A) | dd |
| 5.45(H2B-H3B) | ||||
| −15.77(H2B-H2A) | ||||
| H2A | γCH(Glu) | 2.44 | 7.19(H2A-H3A) | |
| 8.98(H2A-H3B) | ||||
| −15.77(H2A-H2B) |
4. Discussion
In aqueous solution, the α-amino and the δ-carboxyl groups will be protonated and the proposed probability of finding a protonated α-carboxyl group is approximately two orders of magnitude less than that for the δ-carboxyl group [34] based on their pK values. Since the crystal structure reported here was produced in aqueous solution that is grown out of D2O, protonated state of the α-amino and δ-carboxyl and deprotonated α-carboxyl groups seen in the crystalline state are consistent with the NH3+, COOH and COO− charge form, respectively, in agreement with the pK values stated above. The protonated and deprotonated forms seen here are in agreement with corresponding 13C chemical shifts reported previously for Glu [35–37] and observed in the current solution state measurements of GlyGlu.
Görbitz reported that in the protonated state, the donor groups are engaged in two types of hydrogen bonds with significantly different mean N•••O distances, 2.644(17) and 2.730(17) Å [38]. Furthermore, studies by Görbitz [38] validated that peptides assemble head to tail in crystal structures due to hydrogen bonding interactions between the main chains as well as side chain atoms. Potential hydrogen bonding interactions could be divided into four categories: interaction between 1) two ionized moieties NH3+•••−OOC; 2) amide NH•••O=C; 3) NH3+•••O=C; and 4) amide NH•••−OOC of main chain atoms. Due to the electrostatic nature of the interaction between ionized moieties, category 1 is considered the strongest interaction. The presence of donor or acceptor groups in the side chain adds additional possibilities for hydrogen bonding interactions. Hydrogen bonding angles, defined by Donor-H•••Accepter are confined to be in the 180–170° and 150–110° ranges in previously reported peptide crystal structures [38]. Some of the Donor-H•••Accepter angles observed in GlyGlu are outside the range and may or may not be counted as standard hydrogen bonds.
Theoretically Glu may be able to attain over 103 possible conformations due to rotations around seven σ bonds. Major differences between the α and β forms of Glu originate from their torsion angles χ1: N-Cα-Cβ-Cγ (178.2° vs −51.8°) and χ3,4,2: Cβ-Cγ-Cδ-Oε(2) (−104.6° vs −160.7°). In the GlyGlu crystalline state the conformation of Glu is very close to the αpolymorph of L-Glutamic acid (LGLUAC02) and that in L-Glutamic acid Hydrochloride (SI2055) among the structures compared as reflected by the lowest RMS value of over 10 common non-hydrogen atoms. All the α-amino, α-carboxyl and δ-carboxyl groups in L-Glutamic acid Hydrochloride (SI2055) are in protonated form with a bound Cl− in the crystal structure. Noticeably the conformation of Glu found in GlyGlu deviates significantly from that of Glu in β polymorph of L-Glutamic acid (LGLUAC11, LGLUAC01).
Conformations of Glu, as seen in it or in molecules containing it, is not only limited to the above-mentioned entries but also may be relevant to specific functions the system is evolved to operate. Some examples of molecular assemblies that exploit such mechanics are: 1) In the vertebrate brain, ligand-gated ion channel glutamate receptors accommodate structural changes necessary to mediate excitatory synaptic transmissions [39, 40]. 2) Chloride Channels (ClC) selectively filter glutamate to clear the pathway for ion passage, thereby gating the pore [41–44] with a complementary structural mechanism. 3) Protonation of glutamate side chains mediated the activation of gate opening in Potassium (K+)-channel associated neural signal transduction, as it regulates selective conduction of K+ across biological membranes [2] which may have conformations of Glu controlling their biological functions.
In solution, vicinal J-coupling for Glu has been used previously to predict the ratio of different side chain conformers [45]. Moreover, it was suggested that glutamate exists predominantly in two conformations about Cα-Cβ bond, namely either gauche-gauche and gauche-trans or gauche-gauche and trans-gauche [45]. However, the relationship between the conformations defined by the angles reported previously for Glu and that specified by α, β forms are not clear.
The different proton chemical shifts and J coupling noticed for geminal protons in CβH2 group of Glu in the 1D 1H NMR of GlyGlu may bring about a significant signature. Though the plane of symmetry is present and chirality is absent, otherwise identical methylene geminal protons, CH2 of Glu are stereochemically and/or magnetically non-equivalent. This resultant splitting pattern seen in the NMR spectrum enables the identification of this metabolite uniquely. The information obtained from this study would provide detailed splitting patterns corresponding to the CβH2 in the detection of Glu and/or it’s metabolites’ signal and must be taken into account for the interpretation of measurements made in vivo and in situ real time dynamic conditions and may be useful in monitoring related biomarkers. Also, the information obtained on the conformations, protonation states and pK of substitutions of a single amide derivative will be helpful in the characterization of peptidomimetic analog studies. Moreover, the increased double bond character reflected in the amide bond reveals the stability of such functionality and may be incorporated in the fragment based drug design.
5. Conclusions
The benchmark signature of Glu as a dipeptide establishes a unique pattern which may be monitored in the cells under in vivo or in situ conditions that may occur due to natural development or changes caused by illness. As the accumulation of AGEs may be considered a significant marker for diagnosis, their proximity to Glu residues may be useful in identifying likely areas of protein modification/glycation sites. Furthermore, the pattern revealed by the dipeptide may become a helpful tool to monitor in vitro conversions in late-stage derivations of peptides at Glu and/or in its vicinity.
Acknowledgement
This work is supported by National Institutes of Health Grant. We thank Ray Hoff for all the technical assistance, Dr. Milo Westler of National Magnetic Resonance Facility at Madison for help with the NMR studies and Ohio Supercomputer Center, computing services, staff and for the access to the computing systems.
Abbreviations:
- CSD
Cambridge Structural Database
- CCD
Charge Coupled Device
- ClC
Chloride Channels
- Glu
Glutamic acid, Glutamate
- Gly
Glycine
- GlyGlu
Glycylglutamic acid
- 1D
One dimensional
- TMS
Tertramethylsilane
- ‘
symmetry related molecule
- s
singlet
- d
doublet
- t
triplet
- m
multiplet
- dd
doublet of doublets
- J
coupling constants
- RMS
root mean square
REFERENCES
- [1].Hayes K L, Pericytes in type 2 diabetes. Series “Pericytes in type 2 diabetes. vol. 1147: Springer.265–278, 2019. DOI: 10.1007/978-3-030-16908-4_12. [DOI] [Google Scholar]
- [2].Thompson A, Posson D, Parsa P, Nimigean C, Molecular mechanism of pH sensing in KcsA potassium channels. “Proc Natl Acad Sci USA,” vol. 105, no., 6900–5, 2008, DOI: 10.1073/pnas.0800873105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bernal JD, The crystal structure of the natural amino acids and related compounds. “Zeitschrift fuer Kristallographie, Kristallgeometrie, Kristallphysik, Kristallchemie,” vol. 78, no., 363–9, 1931, [Google Scholar]
- [4].Hirokawa S, A new modification of L-glutamic acid and its crystal structure. “Acta Crystallographica,” vol. 8, no., 637–41, 1955, DOI: 10.1107/S0365110X55001990 [DOI] [Google Scholar]
- [5].Frye RE, Vassall S, Kaur G, Lewis C, Karim M, Rossignol D, Emerging biomarkers in autism spectrum disorder: a systematic review. “Annals of Translational Medicine,” vol. 7, no. 23, 792, 2019, DOI: 10.21037/atm.2019.11.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Money KM, Barke TL, Serezani A, Gannon M, Garbett KA, Aronoff DM, Mirnics K, Gestational diabetes exacerbates maternal immune activation effects in the developing brain. “Molecular Psychiatry,” no., 2017, DOI: 10.1038/mp.2017.191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Gardener H, Spiegelman D, Buka SL, Prenatal risk factors for autism: comprehensive meta-analysis. “The British journal of psychiatry: the journal of mental science,” vol. 195, no. 1, 7–14, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Haeusler RA, McGraw TE, Accili D, Biochemical and cellular properties of insulin receptor signalling. “Nature Reviews Molecular Cell Biology,” vol. 19, no. 1, 31–44, 2018, DOI: 10.1038/nrm.2017.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Scott PH, Brunn GJ, Kohn AD, Roth RA, Lawrence JCJ, Evidence of insulin-stimulated phosphorylation and activation of the mammalian target of rapamycin mediated by a protein kinase B signaling pathway. “Proc. Natl. Acad. Sci. U.S.A,” vol. 95, no. 13, 7772–7, 1998, DOI: 10.1073/pnas.95.13.7772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Darnell RB, The Genetic Control of Stoichiometry Underlying Autism. “Annual Review of Neuroscience,” vol. 43, no., 509–533, 2020, DOI: 10.1146/annurev-neuro-100119-024851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Bear MF, Huber KM, Warren ST, The mGluR theory of fragile X mental retardation. “Trends Neurosci,” vol. 27, no. 7, 370–7, 2004, DOI: 10.1016/j.tins.2004.04.009 [DOI] [PubMed] [Google Scholar]
- [12].Savaheli S, Ahmadiani A, Obsessive-compulsive disorder and growth factors: A comparative review. “Behavioural Brain Research,” vol. 392, no., 111967, 2019, DOI: 10.1016/j.bbr.2019.111967 [DOI] [PubMed] [Google Scholar]
- [13].King BH, Promising forecast for autism spectrum disorders. “JAMA, the Journal of the American Medical Association,” vol. 313, no. 15, 1518–1519, 2015, DOI: 10.1001/jama.2015.2628 [DOI] [PubMed] [Google Scholar]
- [14].Stern M, Insulin signaling and autism. “Front. Endocrin,” vol. 2, no. Oct, 54, 2011, doi: 10.3389/fendo.2011.00054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Rabbani N, Thornalley PJ, Glycation of Proteins. Series “Glycation of Proteins,” ed. Griffiths JR, Unwin RD.307–332, 2017. DOI: 10.1002/9781119250906.ch8. [DOI] [Google Scholar]
- [16].Johansen MB, Kiemer L, Brunak S., Analysis and prediction of mammalian protein glycation. “Glycobiology,” vol. 16, no. 9, 844–53, 2006, DOI: 10.1093/glycob/cwl009 [DOI] [PubMed] [Google Scholar]
- [17].Bilova T, Paudel G, Shilyaev N, Schmidt R, Brauch D, Tarakhovskaya E, Milrud S, Smolikova G, Tissier A, Vogt T, Sinz A, Brandt W, Birkemeyer C, Wessjohann LA, Frolov A, Global proteomic analysis of advanced glycation end products in the Arabidopsis proteome provides evidence for age-related glycation hot spots. “J. Biol. Chem,” vol. 292, no. 38, 15758–76, 2017, DOI 10.1074/jbc.M117.794537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].18. Strickland SW, Campbell ST, Little RR, Bruns DE, Bazydlo LAL, Recognition of rare hemoglobin variants by hemoglobin A1c measurement procedures. “Clinica Chimica Acta,” vol. 476, no., 67–74, 2018, DOI: 10.1016/j.cca.2017.11.012 [DOI] [PubMed] [Google Scholar]
- [19].Bunn HF, Shapiro R, McManus M, Garrick L, McDonald MJ, Gallop PM, Gabbay KH, Structural heterogeneity of human hemoglobin A due to nonenzymatic glycosylation. “J. Bio. Chem,” vol. 254, no., 3892–98, 1979, DOI: 10.1016/S0021-9258(18)50671-2 [DOI] [PubMed] [Google Scholar]
- [20].Raum HN, Weininger U, Experimental pKa Value Determination of All Ionizable Groups of a Hyperstable Protein. “ChemBioChem,” vol. 20, no. 7, 922–930, 2019, DOI: 10.1002/cbic.201800628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Simms HS, The nature of the ionizable groups in proteins. “Journal of General Physiology,” vol. 11, no., 629–40, 1928, DOI: 10.1085/jgp.11.5.629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Olson ER, Gribble GW, 4-Fluoro-5-Methylacridine: In Search Of Long-Range “Lone-Pair Mediated” H-F And C-F Spin-Spin Coupling. “Organic Preparations and Procedures International,” vol. 53, no. 1, 100–104, 2021, DOI: 10.1080/00304948.2020.1851123 [DOI] [Google Scholar]
- [23].Irving CS, Lapido A, pH Dependence of the 15N and “0 Nuclear Magnetic Resonance Chemical Shifts of Glycylglycine. “J.C.S. CHEM. COMM,” vol. 2, no., 43, 1976, DOI: 10.1039/c39760000043 [DOI] [Google Scholar]
- [24].Bruker, APEX2, SAINT and SADABS, in Bruker AXS Inc.: Madison, Wisconsin, USA., 2013 [Google Scholar]
- [25].Bruker, Bruker Analytical X-Ray: Madison, WI, 2006. [Google Scholar]
- [26].Krause L, Herbst-Irmer R, Sheldrick GM, Stalke D, SADABS-2008/1. “J. Appl. Cryst,” vol. 48, no., 3–10, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Sheldrick GM, SHELXT - Integrated space-group and crystal-structure determination. “Acta Cryst,” vol. A71, no., 3–8, 2015, DOI: 10.1107/S2053273314026370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Sheldrick GM, Crystal structure refinement with SHELXL. “Acta Cryst,” vol. C71, no., 3–8, 2015, DOI: 10.1107/S2053229614024218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Sheldrick GM, A short history of SHELX. “Acta Crystallogr A,” vol. 64, no. Pt 1, 112–22, 2008, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18156677 [DOI] [PubMed] [Google Scholar]
- [30].Rath NP, Haq W, Balendiran GK, Fenofibric acid. “Acta Crystallogr C,” vol. 61, no. Pt 2, o81–4, 2005, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15695917 [DOI] [PubMed] [Google Scholar]
- [31].Görbitz CH, A new ‘hydrogen-bond rule’ applied to the structure of L-seryl-L-alanine and pairs of dipeptide retroanalogues. “Acta Cryst,” vol. C56, no. Part 4, 500–2, 2000 [DOI] [PubMed] [Google Scholar]
- [32].Groom CR, Bruno IJ, Lightfoot MP, Ward SC, The Cambridge Structural Database. “Acta Cryst,” vol. B72, no., 2016, DOI: 10.1107/S2052520616003954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Dashti H, Westler WM, Tonelli M, Wedell JR, Markley JL, Eghbalnia HR, Spin System Modeling of Nuclear Magnetic Resonance Spectra for Applications in Metabolomics and Small Molecule Screening. “Anal Chem,” vol. 89, no. 22, 12201–8, 2017, DOI: 10.1021/acs.analchem.7b02884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lehmann MS, Nunes AC, A short hydrogen bond between near identical carboxyl groups in the α-modification of L-glutamic acid. “Acta Crystallographica,” vol. B36, no. 7, 1621–5, 1980, DOI: 10.1107/S0567740880006711 [DOI] [Google Scholar]
- [35].Platzer G, Okon M, McIntosh LP, pH-dependent random coil (1)H, (13)C, and (15)N chemical shifts of the ionizable amino acids: a guide for protein pKa measurements. “J Biomol NMR,” vol. 60, no. 2–3, 109–29, 2014, doi: 10.1007/s10858-014-9862-y. Epub 2014 Sep 20. PMID: 25239571 [DOI] [PubMed] [Google Scholar]
- [36].Jameson CJ, Encyclopedia of NMR. Series “Encyclopedia of NMR,” ed. Harris RK, Wasylishen RE. vol. 4.2197–2214, 2012 [Google Scholar]
- [37].Servis KL, Domenick RL, Origin of deuterium isotope effects on carbon-13 chemical shifts. “J Am Chem Soc,” vol. 108, no. 9, 2211–4, 1986, PMID: 22175562 DOI: 10.1021/ja00269a015 [DOI] [PubMed] [Google Scholar]
- [38].Görbitz C H, Hydrogen-bond distances and angles in the structures of amino acids and peptides. “Acta Crystallographica,” vol. B45, no. 4, 390–5, 1989, DOI: 10.1107/S0108768189003666 [DOI] [Google Scholar]
- [39].Greger IH, Mayer ML, Structural biology of glutamate receptor ion channels: towards an understanding of mechanism. “Current Opinion in Structural Biology,” vol. 57, no. 185–195, 2019, DOI: 10.1016/j.sbi.2019.05.004 [DOI] [PubMed] [Google Scholar]
- [40].Meyerson J, Kumar J, Chittori S, Rao P, Pierson J, Bartesaghi A, Mayer M, Subramaniam S, Structural mechanism of glutamate receptor activation and desensitization. “Nature,” vol. 514, no. 7522, 328–34, 2014, DOI: 10.1038/nature13603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Jentsch TJ, Pusch M, CLC chloride channels and transporters: structure, function, physiology, and disease. “Physiological Reviews,” vol. 98, no. 3, 1493–1590, 2018, DOI: 10.1152/physrev.00047.2017 [DOI] [PubMed] [Google Scholar]
- [42].Dutzler R, Campbell E, MacKinnon R, Gating the selectivity filter in ClC chloride channels. “Science,” vol. 300, no., 108–12, 2003, DOI: 10.1126/science.1082708 [DOI] [PubMed] [Google Scholar]
- [43].Niemeyer M, Cid L, Zúñiga L, Catalán M, Sepúlveda F, A conserved pore-lining glutamate as a voltage- and chloride-dependent gate in the ClC-2 chloride channel. “J Physiol,” vol. 553, no. Part 3, 873–79, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Estévez R, Schroeder B, Accardi A, Jentsch T, Pusch M, Conservation of chloride channel structure revealed by an inhibitor binding site in ClC-1. “Neuron,” vol. 38, no. 1, 47–59, 2003, DOI: 10.1016/S0896-6273(03)00168-5 [DOI] [PubMed] [Google Scholar]
- [45].Govindaraju V, Basus VJ, Matson GB, Maudsley AA, Measurement of chemical shifts and coupling constants for glutamate and glutamine. “Magnetic Resonance in Medicine,” vol. 39, no. 6, 1011-3, 1998, DOI: 10.1002/mrm.1910390620 [DOI] [PubMed] [Google Scholar]