Abstract
Expansion microscopy enables conventional diffraction limit microscopy to achieve super-resolution imaging. However, the enlarged tissue lacks an objective lens with sufficient working distance that can image tissues with whole-brain-scale coverage. Here, we present expansion tomography (ExT) to solve this problem. We have established a modified super-absorbent hydrogel (ExT gel) that possesses high mechanical strength and enables serial sectioning. ExT gel enables tissue and cell imaging and is compatible with various fluorescent labeling strategies. Combining with the high-throughput light-sheet tomography (HLTP) system, we have shown the capability of large volume imaging with nanoscale resolution of mouse brain intact neuronal circuits. The ExT method would allow image samples to support super-resolution imaging of intact tissues with virtually unlimited axial extensions.
1. Introduction
Mapping the architecture of neural circuits with both high resolution (synaptic-level) and wide view (whole-brain-scale) is one of the long-sought goals of modern neuroscience [1–3]. However, the optical diffraction limit prevents the conventional light microscopy from accessing structures smaller than ∼ 250 nm [4], resulting that many subcellular organelles and supramolecular complexes such as presynaptic terminals, postsynaptic spines elude visualization as an individual configuration [5,6]. Over the past few decades, electron microscopy (EM) has bypassed the diffraction limit to provide unparalleled details for neuronal circuit structures [7–10]. However, the labor-intensive nature makes EM unsuitable for high throughput imaging experimentation with large-scale tissues. Another imaging technique super-resolution (SR) microscopy [11–14], breaking the diffraction limit, allows many previously unnoticed molecular structures of biological systems to be directly visualized [15–19]. However, its slow imaging speed and rapid photobleaching are still major obstacles to image large tissues. Besides, the requirements of sophisticated equipment and specialized expertise are also limitations in widespread use of SR [20,21].
Recently, expansion microscopy (ExM) [22], a new super-resolution technique, enables nanoscale resolution imaging on various conventional diffraction-limited microscopes. More importantly, ExM is feasible for thick samples imaging. Since the pioneering work by Boyden lab first introduced the principle of ExM [22], a number of ExM protocols have been developed, allowing for nanoscale imaging of proteins [23–25], RNAs [26,27], or human clinical specimens [28]. Moreover, the development of iterative expansion microscopy (iExM) [29] and X10 expansion microscopy [21,30] have boosted the image resolution to ∼ 25-30 nm, which are comparable to those of the highest performing forms of super-resolution light microscopy [29]. However, the maximum imaging depth of ExM is determined by the maximum working distance of the objective lens. Even with the extremely long working distance objective lens, ExM only can obtain super-resolution data for a small piece of the whole organ, because the expanded sample can become rather bulky, at least ∼100-fold its original volume [31].
Cross-sectional tomography adopts sequentially alternating imaging and sectioning to increase the detection depth of optical microscopes [32]. Sequential sectioning imaging of the expanded tissue will be beneficial to obtain the larger volume of super-resolution data. However, current expansive hydrogels for tissue-embedding are too soft and fragile to ensure continuous, complete and long-term stable sectioning imaging of the swollen specimens, even if a vibratome suitable for the soft tissue sectioning is employed. On the other hand, too soft samples will cause serious deformation in the process of sectioning, which will bring great difficulties to the subsequent data reconstruction [33,34]. Therefore, it is necessary to develop a new swellable hydrogel with high mechanical strength for the tomography imaging.
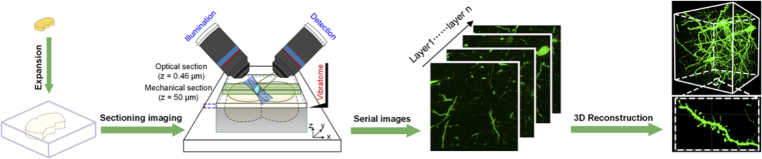
Here, we have developed a modified super-absorbent hydrogel with high mechanical strength that enables alternating imaging and sectioning with a cross-sectional tomography system. We termed the imaging strategy expansion tomography (ExT) (Fig. 1) and the super-absorbent hydrogel ExT gel. ExT gel has passed evaluation of various properties regarding ExM for cells and tissues, including that enables linearly expanding the tissue by ∼ 4×, achieving an effective resolution of ∼ 60 nm on a diffraction limited microscope, and having excellent fluorescence retention for fluorescent secondary antibodies of different wavelengths and chromophores after expansion treatment with various labeling strategies. Combined with high-throughput light sheet tomography (HLTP) system [35], ExT has obtained 3D super-resolution dataset of a mouse brain tissue with a volume of ∼ 20 mm3 (∼103 mm3 after expansion), achieving detailed characterization of the individual synaptic structure morphology spanning a size continuum multiple orders of magnitude in terms of volume.
Fig. 1.
Expansion tomography (ExT) strategy.
2. Materials and methods
2.1. Hydrogel preparation
The gel monomers were sodium 2-acryla-mido-2-methyl-1-propanesulfonic acid (AMPS-Na, Gelest, GEL-CXSO009) and acrylamide (AM, Aladdin, A108465). The crosslinking co-monomer was N, N'-methylenebisacrylamide (MBA, Macklin, M6024). The initiator was ammonium persulfate (APS, Macklin, A6295) and the accelerator was N, N, N’, N'-tetramethylethylenediamine (TEMED, Sigma-Aldrich, 176141). All materials were used as received. All the gels were prepared in deionized (DI) water.
A series of gels were prepared by changing the initial gel monomer concentration, and the corresponding concentrations were shown in Table S1. The gel was prepared as follows: gel monomers were dissolved in DI water according to the corresponding concentration (all concentrations were mass concentrations). Then, the monomer solution was bubbled for 30 min with N2 or Ar to remove oxygen from the solution to prevent oxygen inhibit the polymerization reaction. Immediately afterward, 0.05 wt% APS and 0.01 wt% TEMED were added for gelation.
2.2. Mechanical strength measurement
The mechanical properties of hydrogels were measured on the electronic dynamic and static fatigue testing machine (E1000, Instron corporation). Before measurement, all the polymerized hydrogels were in a state of fully swollen at room temperature. All the stress-strain tests were performed using a compression rate of 2 mm·min−1. The samples were compressed until failure. The compressive modulus was determined from the linear slope of the stress-strain curve at low strains (10%-15%). The cylindrical gel samples were tested diameter and height by a vernier caliper.
2.3. Surface flatness measurement
The surface flatness measurement was performed with Profilometer (Dektak XT), whose measuring principle was shown in Fig. S1. The gel after slicing was placed in Profilometer with the probe in slight contact with the surface of gel. The probe would shift along the Z direction with the fluctuation of the gel surface, and the displacement data would be recorded by the Profilometer. Surface fluctuation in different directions could be measured by placing the gel blocks in different directions. In this study, X direction (the direction of sectioning) and Y direction (perpendicular to the direction of sectioning) were chose to measure to describe the flatness of the gel sectioning surface.
2.4. Specimen preparation
(1) Animal
Animals were housed in a specific-pathogen-free (SPF) animal house under a 12/12 h light/dark cycle with unrestricted access to food and water. Wild-type (C57BL/6J, 8-12 weeks old) and thy1-GFP-M (8-12 weeks old) were used in this study. For sampling, adult mice were deeply anaesthetized with a mixture of 2% α-chloralose and 10% urethane (8 ml/kg) and were transcardially perfused with 0.01 M phosphate buffered saline (PBS, Sigma, P3813) followed by 4% paraformaldehyde (PFA, Sigma-Aldrich, 158127) in PBS for fixation. The brain was excised from the perfused animal head. All harvested samples were post-fixed 24 h in 4% PFA at 4°C. Coronal brain sections (50 μm, 100 μm, and 1 mm) were sliced using a vibration microtome (Leica VT 1200 s, Germany).
All animal experiments protocols were performed under the Experimental Animal Management Ordinance of Hubei Province, P. R. China, and the guidelines from the Huazhong University of Science and Technology. These protocols were approved by the Institutional Animal Ethics Committee of Huazhong University of Science and Technology.
(2) Cell
U-2 OS cells were grown in McCoy's 5A (Modified) Medium (Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco) and 1% penicillin (10,000 IU/mL)/streptomycin (10,000 μg/mL) at 37°C with 5% CO2. Before imaging, cells were seeded on 35-mm glass-bottom dishes overnight.
2.5. Immunostaining
(1) Immunostaining of cultured cells
U-2 OS cells were washed three times with PBS pre-warmed to 37 °C and fixed with 3% paraformaldehyde, 0.05% glutaraldehyde and 0.2% Triton X-100 (Sigma, T8787) in PBS for 10 min, followed by brief washing in PBS. After washing, samples were incubated with blocking/permeabilization buffer (3% bovine serum albumin (BSA, Solarbio, A8020) and 0.05% Triton X-100 in PBS) for 1 h with gently rocking. Cells were then incubated with primary antibodies (monoclonal mouse anti–α-tubulin, Sigma T5168, 1:500 diluted in the blocking buffer) at room temperature for 1.5 h, washed three times with PBS, and incubated with Alexa Fluor 546 goat anti-mouse IgG (Thermo Fisher, A-11003, 1:500 diluted in blocking buffer) for 1 h. Finally, samples were washed three times with PBS and stored in PBS at 4 °C until proceeding to the next step.
(2) Immunostaining of mouse tissues
For 50∼100 µm mouse brain slices, samples were first incubated in blocking/permeabilization buffer (5% BSA and 0.3% Triton X-100 in PBS (PBST)) for 2 h at room temperature, and then incubated in primary antibody diluted into blocking/permeabilization buffer for at least 24 h at 4 °C with gently rocking and was then washed five times in PBST (10 min each). Brain slices were then incubated with secondary antibody diluted into PBST for 3-6 h at 37 °C and afterwards were washed three times with PBST (10 min each) and two times with PBS (10 min each).
For 1-mm-thick mouse brain blocks, samples were immunostained using iDISCO [36] protocols with minor modifications. The brain blocks were initially pretreated: first, they were immersed in 30 wt% sucrose in PBS until the brain blocks sinking to the bottom of the container (50 ml centrifugal tubes were used in this experiment), refreshing the sucrose solution once after sinking and immersing until sinking again. Taken out tissues from the sucrose solution, samples were placed in liquid nitrogen for five minutes, then placed in air until recovering to room temperature. Samples were washed with PBS three times. Then, samples were treated with 25%-30% CUBIC agent 1 [37] 4 h for decoloring followed by washing three times in PBS. Next, the samples were dehydrated, bleached, and rehydrated (all steps were done at 4℃). They were incubated in 20%, 40%, 60%, 80% methanol (Sinopharm Chemical Reagent, 10014108) in DI water for 30 min at each step and then in 100% methanol for 30 min two times. Samples were then bleached with 5% H2O2 (Sinopharm Chemical Reagent, 10011208) in 20% DMSO (Sigma, D8418)/methanol (1 volume 30% H2O2 /1 volume DMSO/4 volume methanol). After bleaching, sections were washed in methanol for 45 min three times; then in 80%, 60%,40%, 20% methanol for 30 min each step; then in PBS for 30 min three times. For the immunostaining step, pretreated sections were incubated in PBS/0.2% Triton X-100/20% DMSO/0.3 M glycine (Sigma, G8898) at 37 °C (1-mm-thick for 24 h, 2-mm-thick for 48 h), then blocked in PBS/0.2% Triton X-100/10% DMSO/6% BSA at 37 °C for 1 d, washed in PBS/0.2% Tween-20 (Sigma, P9416) with 10 mg/ml heparin (PTwH) overnight and then incubated with primary antibody dilutions in PTwH/5% DMSO/3% bovine serum at 37°C with gentle shaking on an oscillator for 3–5 d. Sections were then washed for 1 h with PTwH four times and then incubated with secondary antibodies diluted in PTwH/3% goat serum at 37 C° with gentle shaking on an oscillator for 3–5 d. Sections were finally washed in PTwH 30 min five times and in PBS 30 min three times.
2.6. Expansion workflow
All the experimental steps were carried out according to the method of Jo. Vaughan’s group [23] and Ed. Boyden’s group [25,38,39]. For samples of different thickness, we made some adjustments in the processing time.
(1) Anchoring
Immunostained samples were incubated overnight at room temperature in 1 mM methacrylic acid N-hydroxysuccinimidyl ester (MA-NHS, Sigma-Aldrich, A8060) in PBS for mouse brain slices and 25 mM for cells. For 1-2 mm thick mouse brain blocks, samples was incubated in 5 mM MA-NHS in 2-(N-morpholino) ethanesulfonic acid (MES)-based saline (100 mM MES, 150 mM NaCl) at 4℃ for 48 h, refreshing the MA-NHS solution once after 24 h, and afterwards were washed three times with PBS (30 min each).
(2) Gelation
In the final decision, 10 wt% AM, 15 wt% AMPS-Na and 0.1wt% MBA were used. Immunostained samples were incubated with the monomer solution plus APS/TEMED at 4°C for 10 minute, 60 minutes or 120 minutes for cultured cells, 50 µm, 100 µm mouse brain slices respectively. For 1-2 mm tissues blocks, samples were incubated with the monomer solution plus APS/TEMED at 4°C for 6 h, exchanging fresh solution every two hours. Note when the monomer solution plus APS/TEMED was placed at 4°C, it did not polymerize for 24 hours without the addition of an inhibitor.
After incubation in monomer solution, specimens were transferred to a humidified 37°C incubator for three hours. For cells and ≤100 µm thick brain slices, the procedures for cross-linking and polymerization of gels and tissues could be referred to the previous reported literatures [25,38,39].
For polymerizing 1-2 mm thick brain blocks with gels, we printed 3D molds (Fig. S17) that could be used for brain block and gel polymerization. The size of the mold in XY direction is 19 mm×19 mm, and the size in Z direction can be adjusted arbitrarily according to the thickness of the samples. For gelation, a coverslip (18×18 mm) was placed in the bottom of mold and the tissue was placed above the coverslip, then the monomer solution plus APS/TEMED was fill in the mold. Next, a larger coverslip (24×24 mm) covered the top of the mold. Note that there must be no bubbles in the gel solution. Specimens were transferred to a humidified 37° C incubator for six hours.
(3) Digestion and expansion
For cells and ≤100 µm thick brain slices, proteinase K (New England Biolabs) was diluted 1:100 to 8 units/mL in digestion buffer (50 mM Tris (pH 8), 1 mM EDTA, 0.5% Triton X-100, 0.8 M guanidine HCl) and applied directly to gels in at least ten times volume excess. The gels were then incubated in digestion buffer for at least 12 hours. Digested gels were next placed in excess volumes of doubly DI water for 1 hour to expand, with longer times for thicker gels. This step was repeated 3-5 times in fresh water, until the size of the expanding sample plateaued.
For 1-2 mm thick brain blocks, after gels were polymerized, they were removed from the mold and the excess gel was removed from the edges of tissues. Digestion of the samples was carried out in the digestion buffer 37 ℃ for 24 hours and then in fresh digestion buffer for another 24 hours until the tissue became transparent. After digestion, tissues expanded in DI water with repeatedly refreshing until it reached its maximum volume.
2.7. Expansion factor and distortion measurement
The expansion factor was measured by direct comparison of images before and after ExT gel expansion. First, the physical distances between landmark positions in the pre-ExT images were measured, then the physical distances of the same landmark positions in the corresponding post-ExT images were measured, and the final expansion factor obtained as the distance after expansion divided by the distance before expansion. At least 10 such distance comparisons for each experiment was performed and the average was used as the expansion factor.
The distortion measurements were performed by the method descripted by Jo. Vaughan’s group [23]. We chose the same fields of view for imaging before and after expansion. Post-expansion images were first registered to the corresponding pre-expansion images by rotation, translation and uniform scaling with Fiji. These scaled images were treated with Gaussian blur to achieve a resolution similar to that of the pre-bloated images. These scaled images were then B-spline registered to the pre-expansion images, and the registration was performed with the method descripted by Jo. Vaughan’s group, and all the analysis was done using a Mathematica script they wrote. It should be noted that due to the limited resources of our institution, the distortion result in this study was calculated from images photographed by a confocal microscope, which couldn’t reflect the nanoscale disorder.
2.8. Evaluation of fluorescence retention
Quantification of fluorescent retention of fluorochromes during ExT gel treatment: fluorescent secondary antibodies were purchased from commercial vendors (Table S2). The retention was quantified via before and after expansion imaging mouse brain slices as described below. Thy1-GFP brain slices (50-µm-thick) were stained with anti-GFP primary antibody (rabbit anti GFP: Abcam, ab290; goat anti GFP: Abcam, ab6673; 1:500 diluted), and different secondary antibodies (1:500 diluted) described in Table S2. Confocal microscope (ZEISS, LSM710) images of brain slices were taken with 10 × 0.42 NA objective pre-gelation. Following ExT gel gelation and digestion, the brain slices were washed extensively with PBS (3 × 30 min), and confocal microscope images of the slice were taken again with identical imaging conditions. Cell somas in mouse pyramidal neurons (30 somas were counted for each fluorescent secondary antibody) were used to determine the loss of fluorescence during ExT gel expansion processing. Intensity measurements before and after sample processing were normalized by segmented area to account for fluorophore dilution.
2.9. HLTP imaging
For sectioning imaging, an ExT-embedded sample after expansion was fixed on a metal base and installed in a plastic chamber under the HLTP system. Imaging and sectioning procedures were all conducted in DI water. HLTP system used a customized vibratome [35]. The vibrating frequency of the knife is 60 Hz, and the moving speed of the stage along the x-axis during sectioning is 10 mm/min. The dataset was acquired with a 50-μm sectioning thickness and the 55 μm imaging thickness during the periodic sectioning imaging (Fig. S14e). Therefore, the last 5 μm of the upper layer and the first 5 μm of the next layer would overlap to a certain extent. The advantage of this imaging method is that, on the one hand, it avoids the signal loss caused by uneven sectioning. On the other hand, the same signal of upper and next layers could be used as landmarks for the following registration. The sCMOS operating in 1 × 1 binning mode (voxel sizes: 0.65 × 0.65 × 0.46 μm). The 1-mm-thickness half brain slice imaging had taken ∼24 hours.
2.10. 3D image processing and reconstruction
After imaging, the raw imaging data was preprocessed for alignment between layers. The workflow of image alignment in axial direction was shown in Fig. S14. The strict spatial alignment of 3D data method was performed with a custom script written by our group. In brief, first, the similar signal feature points (such as, the mouse brain contour edges and somas) were extracted from the last image of the upper layer and the first image of the next layer in the adjacent two layers, and then the dislocation value between layers was calculated for translation registration (bilinear interpolation was used for translation), and then the accuracy of the translated image was calculated according to the SSIM function which was used to judge the image similarity [40]. If the translation was accurate, the registered image was output. If the translation was not accurate, the iterative operation was performed until the accurate translation image was obtained. After preprocessing, the data was imported into Imaris software (Bitplane, Zurich, Switzerland 7.4) for visualization analysis. To reconstruct the morphology of dendritic spines, we have tracked the dendrites of individual neurons. Due to the large amount of fine data, which caused pressure to the server operation in the process of reconstruction, we chose to segment each dendrite as a single filament unit for reconstruction. After reconstruction, we classified the dendritic spines. The criteria for the classification of dendritic spines referred to the parameters in the Imaris software.
3. Results
3.1. Developing the high mechanical strength superabsorbent hydrogel
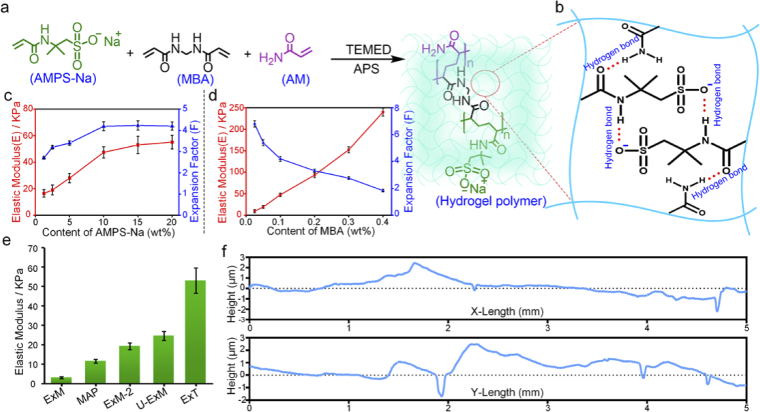
Various reported ExM recipes share similar gel monomer components, comprising the superabsorbent reagent sodium acrylate (SA), the cross-linker N-N’-methylenebisacrylamide (MBA) and the comonomer acrylamide (AM). However, these hydrogels are soft and fragile when expanded up to ∼ 4×. Increasing the concentration of MBA can improve their mechanical strength, but the expansion factor will decrease, resulting in decreased imaging resolution [41,42]. We considered to establish a modified hydrogel formula that could offer high mechanical strength to ensure continuous sectioning, meanwhile maintain high expansion factor to satisfy super-resolution imaging. Sodium 2-acryla-mido-2-methyl-1-propanesulfonic acid (AMPS-Na) (Fig. 2(a)), as a water absorbent agent for the synthesis of the superabsorbent hydrogel, has great potential to achieve this goal. AMPS-Na has a sulfonic acid group with a high ionization constant [43]. Furthermore, the interaction of the polyelectrolytes in the gel network may provide a rigid skeleton owing to the strong hydrogen bonding and the repulsion between ionic groups (Fig. 2(b)) [44,45].
Fig. 2.
The establishment of ExT gel. (a) The schematic representation of synthesis of ExT hydrogel. (b) Possible hydrogen-bonding structures in the swollen ExT hydrogel. (c) The elastic modulus and expansion factor of expansion hydrogels with different AMPS-Na concentration (mean ± s.d., n = 3 samples each). (d) The elastic modulus and expansion factor of expansion hydrogels with different MBA concentration (mean ± s.d., n = 3 samples each). (e) The compression modulus of the expansion hydrogels with various expansion recipes (ExM [22,25], MAP [24], U-ExM [46,47], ExM-2 is defined as an expansion recipe which has the same molar concentration as the corresponding monomer component in ExT gel solution) (mean ± s.d., n = 3 samples each). (f) Measurements of surface evenness of ExT hydrogel after continuous sectioning with a vibratome. X is the direction of sectioning and Y is perpendicular to the direction of sectioning. The Root Mean Square (RMS) values in X direction and Y direction are (948.11 ± 35.62) nm and (953.51 ± 51.74) nm respectively (n = 3).
To obtain the optimal performance of the expanded hydrogel, we conducted extensive testing and fine-tuning in the concentration of each component. According to our measuring results, the mechanical strength and expansion factor increased with the increase of AMPS-Na contents. When the content of APMS-Na reached 15%, the mechanical strength and expansion factor almost did not change (Fig. 2(c), Fig. S2a and Table S1). In addition, the content of MBA is also a major influence on mechanical strength and expansion factor. When the content of MBA was controlled below 0.1%, the expansion factor could be more than 4-fold (Fig. 2(c), Fig. S2b and Table S1). Comprehensive considering between high enough expansion factor for imaging and sufficiently stiff mechanical performance for sectioning, we confirmed the ExT hydrogel recipe as 15 wt% AMPS-Na, 0.1 wt% MBA and 10 wt% AM. In this recipe, ExT gel can expand to 4.0∼4.5 folds and the elastic modulus was obtained as ∼53.0 kPa which is much higher than expended hydrogels that have been reported (Fig. 2(e) and Fig. S3). To verify the sectioning performance of ExT gel, the expanded ExT gel was performed serial sectioning with a vibratome. ExT gel surface fluctuation was measured to be about ∼1 μm (Fig. 2(f)), providing superior sectioning performance compared to other expanded gels (Fig. S4)
3.2. Evaluation of the ExT hydrogel in super-resolution imaging
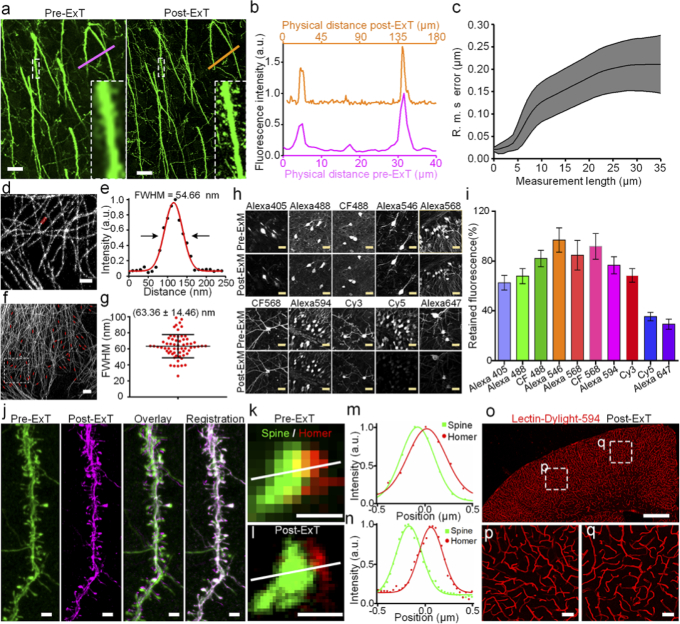
To measure the accurate expansion factor after the hydrogel cross-linked with biological tissues, immunostained Thy1::GFP-M line mouse brain slices processed by pre- versus post-ExT gel were performed fluorescence imaging. By measuring physical distance of distinctive landmarks in pre-ExT and post-ExT fluorescence images, the expansion factor could be obtained as 4.46 in this example (Fig. 3(a, b)). In addition, it was obvious that detailed structures becomes richer and finer after expansion (Insets of Fig. 3(a, b)). The distortions of architectures introduced by ExT gel during expansion was estimated by quantifying the root-mean-square (RMS) error from the deformation vector field between the images via a non-rigid registration process [22,23], and the result demonstrated that the errors in length were ∼ 1% of distance (Fig. 3(c), Fig. S5-S7), which indicated excellent expansion fidelity of ExT gel. Next, ExT gel in cultured cells was performed to further demonstrate its super-resolution capability. The cross-sectional profile of expanded microtubules were measured and an average Gaussian-fitted full width at half maximum (FWHM) of (63.36 ± 14.46 nm) was presented (mean ± s.d., n = 63) (Fig. 3(d-g)), suggesting an apparent lateral resolution of ∼ 60 nm. Comparison of images before and after expansion, distinct separated microtubules were observed after expansion that were originally clustered together in the unexpanded specimens (Fig. S8).
Fig. 3.
ExT gel enables biological tissue and cells imaging. (a) Confocal images of the immunostained GFP mouse brain slice before and after expansion treatment. (b) An exemplary measurement for the expansion factor. A line scan was drawn over corresponding regions before and after expansion, as indicated in panel (a) and (b) by the colored lines. The expansion factor in this example is 4.46-fold. (c) An analysis of the root mean square error (RMSE) of the distortions between aligned pre- and post-expansion images. (d) Imaging of immunostained α-tubulin in U-2 OS cells with Alexa 546 after ExT gel expansion (F=4.34). (e) The full width at half maximum (FWHM) measurement of the red region in (d) as an indication of resolution. FWHM is obtained by a best Gaussian curve fitting and its value is 54.66 nm in this example. (f-g) Numbers of statistical measurements of FWHM in microtubules. The FWHM average value is (63.36 ± 14.46) nm (mean ± s.d., n = 63 microtubules). (h) Representative images of various conventional fluorescent secondary antibody immunostained brain slides before and after ExT hydrogel expansion. (i) Fluorescence retention for these dyes after ExT hydrogel workflow treatment in immunolabeled mouse brain slices (mean ± s.d., n = 30 somas from four brain slices of each group). (j) Image of dendritic spines in the GFP brain slice without immunostaining, streptavidin conjugated Alexa 647(SA-Alexa647) staining after expansion, overlay pre-ExT and post-ExT images using similarity transformation in a local region, registration pre-ExT and post-ExT image used B-spline registration (F=4.42). (k-l) Synaptic structure resolved with a post-synaptic marker, homer1, and dendritic spines in a cortical region. Homer1 was stained with Alexa 568, and dendritic spines used GFP-positive signal as the reporter which stained with Alexa 488 (F=4.45). (m-n) Intensity plot along the axis of the synapse in (n) and (o). The lines represent the Gaussian curve fitting. (o-q) Images of blood vessels stained with lectin-DyLight 594 after ExT processing (F=4.29). All distances and scale bars correspond to pre-expansion dimensions. Scale bar: (a) 10 μm, (d) 5 μm, (f) 2 μm, (h) 20 μm, (j) 2 μm, (k-l) 500 nm, (o) 500 μm, (p-q) 20 μm.
The fluorescence retention was systematically measured for various commercial fluorescent dyes in the expansion workflow. Following gelation and digestion, mouse brain slices immunolabeled with secondary antibodies conjugating with small-molecule fluorophores of various fluorescence wavebands retained more than 60% of their initial brightness except cyanine dyes Cy5 and Alexa 647(30%-40%) (n = 30 somas from four brain slices; Fig. 3(h, i), Table S2). Compared with ExM gel, ExT gel has better fluorescence retention for dyes [25]. To give cyanine dyes a better image contrast in ExT gel, post-expansion staining was performed by adding streptavidin conjugated dyes to the expanded specimen that had been labeled with a biotin-conjugated secondary antibody (Fig. 3(j)) [23]. The result showed that this post-expansion staining made brighter signal and better details of dendritic spines.
The expression of fluorescent proteins in biological tissues by transgenic or viral injection is another kind of labeling strategy for biological research. The most commonly used transgenetically labeled Thy1-GFP mice were tested fluorescence retention after expansion. We found that GFP fluorescence became misty even disappeared over a few hours of imaging in deionized (DI) water (Fig. S9a). This phenomenon was consistent with the fluorescence retention of fluorescent protein in the ExM hydrogel [41]. Surprisingly, when the expanded tissues were transferred to PBS solution from DI water, the disappeared fluorescence was recovered and this process was reversible (Fig. S9a). This result implied that GFP might not be denatured or degraded in DI water. Although the buffered environment (PBS) prevents GFP fluorescence from dim, the tissue can only expand 1.7-fold in PBS solution, which contribute little to the imaging resolution improvement. The gradual dilution of 1×PBS solution could increase the expansion factor, but the fluorescence intensity of GFP also decreased correspondingly (Fig. S9a, b). Similar results were also observed in another fluorescent protein, mClover3, which expressed in mice by viral injection (Fig. S10). To obtain higher expansion factors and better image signal-to-noise ratio, we suggested immunostaining for fluorescent protein labeled tissues, as antibodies were currently available for all major fluorescent proteins [30].
To verify ability of ExT to image more elaborate neural architectures, dendritic spines and postsynaptic protein Homer1 were immunostained to GFP slices and processed with ExT. It was obvious that the junctions between synapses and dendritic spines were clearly observable after expansion (Fig. 3(k-n)). Although the fluorescence signal was diluted after expansion, all synaptic punctum could be found in the same corresponding position in the pre-expansion image could be clearly identified by individual (Fig. S11). In addition to antibodies labeling, blood vessels stained with lectin-DyLight 594 also could be expanded and the structure of the vascular system was also successfully acquired (Fig. 3(o-q)).
3.3. 3D super-resolution imaging with high-throughput light sheet tomography (HLTP)
High-throughput light-sheet tomography platform (HLTP) [35], which combines a vibratome and an inclined light sheet microscopy to enable successive block-face imaging, was used to implement the task of 3D super-resolution imaging of the swollen tissues. HLTP has nearly isotropic 3D resolution and ultra-fast imaging speed. Following expansion, optically inhomogeneous organic components (such as lipids and protein fragments) that are not anchored to the gel are sufficiently removed [41] so that the expanded gel has a refractive index nearly indistinguishable from water to be perfectly matched with water immersion objectives in HLTP system.
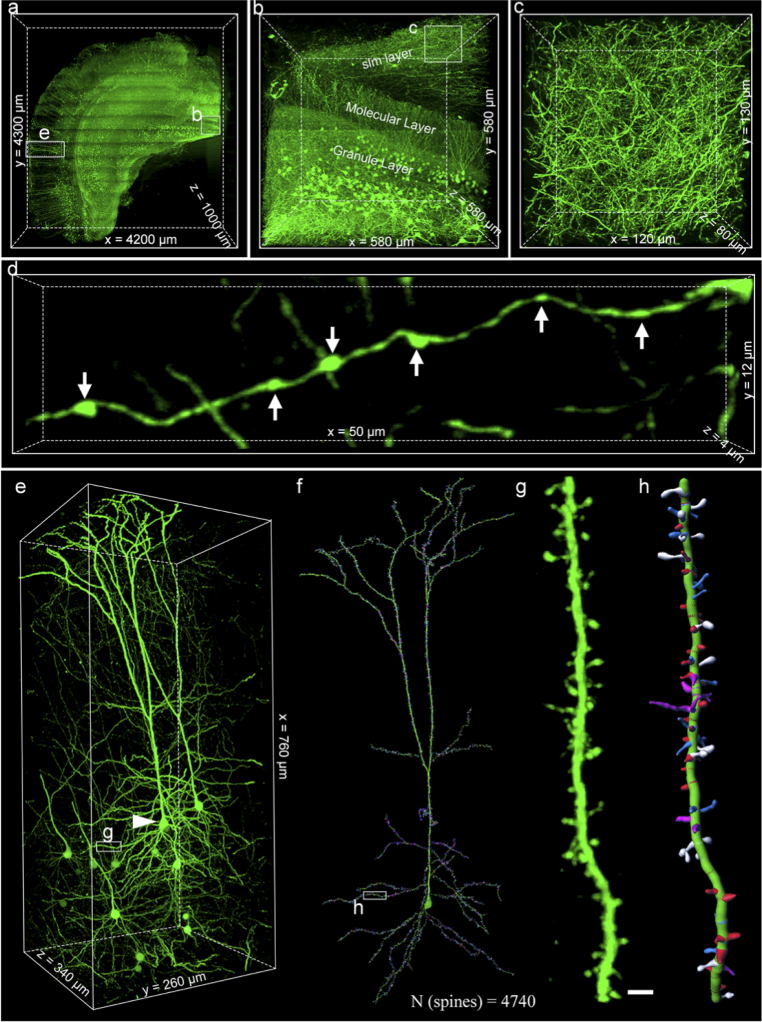
We imaged a volume of the adult Thy1-GFP-M mouse brain (immunostaining with Alexa 488) spanning 4.6- by 4.2- by 1.0-mm (original tissue slice sizes; the volume was extended to 20.7- by 18.9- by 4.5-mm after ExT gel expansion, F = 4.51). The voxel resolution of HLTP imaging system (650 × 650 × 460 nm3), divided by the expansion factor, yields an actually estimated voxel resolution of 144 × 144 × 101 nm3. The data acquisition took ∼ 24 h, and the raw dataset was larger than 20 terabytes (TB). The fluorescence intensity of the photographed thickness range was uniform and does not decay with increasing depth due to the completely transparent sample (Fig. S12). In addition, HLTP system used the field of view stitching strategy to extend the field of view. For every stitched panel, the illumination cannot be homogeneous, which introduced periodical strips in the raw data. Thus, periodic stitching artifacts were shown in Fig. 4(a) and 4(b). However, these stitches have no effect on the continuity of nervous architectures (Fig. S13). The mouse brain embedded in ExT gel was able to perform such continuous, long-term, and periodic sectioning imaging, meaning that ExT gel had satisfactory stability. By 3D reconstruction of the dataset (Fig. S14), intact and detailed structures in various brain regions could be clearly identified (Fig. 4(a), Visualization 1 (19.5MB, mp4) ). Figure 4(b), as one of the representatives, has exhibited the distributions of neuronal morphology in the granular layer of the dentate gyrus (GrDG) brain region of the hippocampus dentate gyrus (DG), including granule cells in the granular layer, hilum fibers in the molecular layer, and mossy fibers in the CA1 stratum lacunosum moleculare (slm) layer (Fig. 3(b)). In the dense fibers network of slm layer (Fig. 3(c)), the fine morphological characteristics of axon boutons structures could be clearly distinguished (Fig. 4(d)).
Fig. 4.
Scalable 3D super-resolution imaging of mouse brain tissue with HLTP system. (a) Volume rendering of a half coronal plane containing cortex and hippocampus. (b) Volume rendering of granule cells and fibers in dentate gyrus (DG). (c) Enlargements of insets (white boxes) from (b). (d) Magnified segmented views of the morphology of axonal boutons. (e) Volume rendering of a group of integral layer V pyramidal neurons in cortex. (f) Neural tracing and dendritic spine reconstruction across the mouse primary visual cortex. (g-h) Magnified segmented views of the morphology of a basal dendrite and its spines. All the scale bar was on a biological scale. Scale bars: (e-f) 2 μm.
To illustrate the nanoscale features of ExT strategy in multiscale imaging, we reconstructed a group of integral layer V pyramidal neuron in the mouse brain cortex (Fig. 4(e)). Focusing on the dendrite of the neuron, we obtained 4740 spines with different morphologies (Fig. 4(f), Visualization 2 (17.9MB, mp4) and Visualization 3 (6MB, mp4) ). Zooming into the raw data set, we observed all four types of dendritic spines with ultrahigh resolution: thin (blue), stubby (red), mushroom (white), and filopodium (magenta) spines (Fig. 4(g, h), Fig. S15 and Visualization 3 (6MB, mp4) ). These results indicate that the ExT strategy enables imaging and visualization of nanoscale features across multiple orders of magnitude scales, facilitating a systematic understanding of neural circuit connections.
4. Discussion and conclusion
We herein describe ExT, a strategy combining the tissue expansion and the tomography system for systematic interrogation of large volume tissues with nanoscale precision and molecular information. ExT hydrogel has high mechanical strength to ensure continuous and stable sectioning imaging, meanwhile maintaining its high expansion factor. Similar to other expansion hydrogels, ExT hydrogel enables to be compatible with nanoscale imaging of biological tissue labeled with off-the-shelf fluorescence tags. ExT hydrogel combined with HLTP system provides high-speed and large field-of-view volumetric imaging at nanoscale resolution with an organ-scale perspective. Most importantly, the sequential sectioning imaging is independent of the working distance of objective lens and can be appropriate for the expanded tissue with any thickness.
Many efforts have been made to obtain super-resolution data of large expanded tissues, and the most common method is to combine the light-sheet fluorescence microscopy (LSFM) with ExM [48–50]. However, LSFM is still limited by the working distance of the objective lens and can only perform tissue imaging up to 2-mm-thick (4× expansion), even the maximum working distance of the objective lens (WD; 8-mm XLPLN-series 10× and 25× Olympus lenses) is employed [51]. Sequential sectioning imaging can solve the limitation of the working distance of the objective lens. HTLP has an extremely high imaging throughput, and when combined with high mechanical strength ExT gel, it can quickly obtain super-resolution data for large samples, breaking the limitation of super-resolution imaging on sample size. In our results, it took ∼24 h to obtain a 1-mm-thick slice. As such an imaging speed, it will takes only ∼ 8-10 days to obtain the super-resolution data of a whole mouse brain, which is nearly 100-fold faster than the reported speed of LSFM [48].
The mechanical strength of the expanded hydrogel is an important factor for the sequential sectioning imaging of expanded samples. The low mechanical strength of the original ExM gel may be due to the low monomer concentration, because high amounts of acrylamide and acrylate significantly improved the mechanical strength of the gels (Fig. 2(e)), even though they did not have good sectioning properties (Fig. S3). Therefore, extensive adjustment of the monomer solution concentration of the original ExM gel recipe may be another way to study the expansion gel with high mechanical strength. TREx gels [52] with high amounts of acrylamide and acrylate may provide a new guidance for high rigidity gel recipes. Preliminary tests of TREx (4×) recipe have found that it might have similar mechanical properties to ExT gel (Fig. S16), and TREx (4×) may have great potential to be compatible with the tomography system.
Super elaborate data of large biological tissues brings huge storage space. 1-mm-thick half coronal section imaging has obtained ∼20 TB raw data, while accessing to the super resolution data of the whole brain or whole organs will bring hundreds of TB or even greater amounts of data. Therefore, subsequent data registration processing and data analysis have higher requirements on computer hardware and visualization software. It is believed that the technological update is continuous, and it is inevitable to realize the visualization of the whole mouse organs with higher precision.
Acknowledgements
We acknowledge the support of Analytical and Testing Center of Huazhong University of Science and Technology for mechanical strength measurement of hydrogels. We also thank the support of optical bio-imaging core facility of Britton Chance Center for Biomedical Photonics for confocal imaging. We also thank Ying Wu and Ting Cheng for the HLTP imaging.
Funding
National Natural Science Foundation of China10.13039/501100001809 (61475059); National Key Research and Development Program of China10.13039/501100012166 (2015CB755603).
Disclosures
The authors declare no conflict of interest.
Data availability
Data underlying the results presented in this paper are not publicly available at this time but may be obtained from the authors upon reasonable request.
Supplemental document
See Supplement 1 (10.1MB, pdf) for supporting content.
References
- 1.DeFelipe J., “From the connectome to the synaptome: an epic love story,” Science 330(6008), 1198–1201 (2010). 10.1126/science.1193378 [DOI] [PubMed] [Google Scholar]
- 2.Helmstaedter M., “Cellular-resolution connectomics: challenges of dense neural circuit reconstruction,” Nat. Methods 10(6), 501–507 (2013). 10.1038/nmeth.2476 [DOI] [PubMed] [Google Scholar]
- 3.Oh S. W., Harris J. A., Ng L., Winslow B., Cain N., Mihalas S., Wang Q., Lau C., Kuan L., Henry A. M., Mortrud M. T., Ouellette B., Nguyen T. N., Sorensen S. A., Slaughterbeck C. R., Wakeman W., Li Y., Feng D., Ho A., Nicholas E., Hirokawa K. E., Bohn P., Joines K. M., Peng H., Hawrylycz M. J., Phillips J. W., Hohmann J. G., Wohnoutka P., Gerfen C. R., Koch C., Bernard A., Dang C., Jones A. R., Zeng H., “A mesoscale connectome of the mouse brain,” Nature 508(7495), 207–214 (2014). 10.1038/nature13186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abbe E., “Beiträge zur Theorie des Mikroskops und der mikroskopischen Wahrnehmung,” Arch. Mikrosk. Anat. 9(1), 413–468 (1873). 10.1007/BF02956173 [DOI] [Google Scholar]
- 5.Shapira M., Zhai R. G., Dresbach T., Bresler T., Torres V. I., Gundelfinger E. D., Ziv N. E., Garner C. C., “Unitary assembly of presynaptic active zones from Piccolo-Bassoon transport vesicles,” Neuron 38(2), 237–252 (2003). 10.1016/S0896-6273(03)00207-1 [DOI] [PubMed] [Google Scholar]
- 6.Sudhof T. C., “The synaptic vesicle cycle,” Annu. Rev. Neurosci. 27(1), 509–547 (2004). 10.1146/annurev.neuro.26.041002.131412 [DOI] [PubMed] [Google Scholar]
- 7.Scheffer L. K., Xu C. S., Januszewski M., Lu Z., Takemura S. Y., Hayworth K. J., Huang G. B., Shinomiya K., Maitlin-Shepard J., Berg S., Clements J., Hubbard P. M., Katz W. T., Umayam L., Zhao T., Ackerman D., Blakely T., Bogovic J., Dolafi T., Kainmueller D., Kawase T., Khairy K. A., Leavitt L., Li P. H., Lindsey L., Neubarth N., Olbris D. J., Otsuna H., Trautman E. T., Ito M., Bates A. S., Goldammer J., Wolff T., Svirskas R., Schlegel P., Neace E., Knecht C. J., Alvarado C. X., Bailey D. A., Ballinger S., Borycz J. A., Canino B. S., Cheatham N., Cook M., Dreher M., Duclos O., Eubanks B., Fairbanks K., Finley S., Forknall N., Francis A., Hopkins G. P., Joyce E. M., Kim S., Kirk N. A., Kovalyak J., Lauchie S. A., Lohff A., Maldonado C., Manley E. A., McLin S., Mooney C., Ndama M., Ogundeyi O., Okeoma N., Ordish C., Padilla N., Patrick C. M., Paterson T., Phillips E. E., Phillips E. M., Rampally N., Ribeiro C., Robertson M. K., Rymer J. T., Ryan S. M., Sammons M., Scott A. K., Scott A. L., Shinomiya A., Smith C., Smith K., Smith N. L., Sobeski M. A., Suleiman A., Swift J., Takemura S., Talebi I., Tarnogorska D., Tenshaw E., Tokhi T., Walsh J. J., Yang T., Horne J. A., Li F., Parekh R., Rivlin P. K., Jayaraman V., Costa M., Jefferis G. S., Ito K., Saalfeld S., George R., Meinertzhagen I. A., Rubin G. M., Hess H. F., Jain V., Plaza S. M., “A connectome and analysis of the adult Drosophila central brain,” eLife 9, e57443 (2020). 10.7554/eLife.57443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Motta A., Berning M., Boergens K. M., Staffler B., Beining M., Loomba S., Hennig P., Wissler H., Helmstaedter M., “Dense connectomic reconstruction in layer 4 of the somatosensory cortex,” Science 366(6469), eaay3134 (2019). 10.1126/science.aay3134 [DOI] [PubMed] [Google Scholar]
- 9.Anderson J. R., Jones B. W., Watt C. B., Shaw M. V., Yang J. H., Demill D., Lauritzen J. S., Lin Y., Rapp K. D., Mastronarde D., Koshevoy P., Grimm B., Tasdizen T., Whitaker R., Marc R. E., “Exploring the retinal connectome,” Mol. Vis. 17, 355–379 (2011). 10.3390/MOLECULES17010355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson J. R., Jones B. W., Yang J. H., Shaw M. V., Watt C. B., Koshevoy P., Spaltenstein J., Jurrus E., U. R. V. K., Whitaker T., Mastronarde D., Tasdizen T., Marc R. E., “A computational framework for ultrastructural mapping of neural circuitry,” PLoS Biol 7(3), e1000074 (2009). 10.1371/journal.pbio.1000074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hell S. W., Wichmann J., “Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy,” Opt. Lett. 19(11), 780–782 (1994). 10.1364/OL.19.000780 [DOI] [PubMed] [Google Scholar]
- 12.Betzig E., Patterson G. H., Sougrat R., Lindwasser O. W., Olenych S., Bonifacino J. S., Davidson M. W., Lippincott-Schwartz J., Hess H. F., “Imaging intracellular fluorescent proteins at nanometer resolution,” Science 313(5793), 1642–1645 (2006). 10.1126/science.1127344 [DOI] [PubMed] [Google Scholar]
- 13.Rust M. J., Bates M., Zhuang X., “Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM),” Nat. Methods 3(10), 793–796 (2006). 10.1038/nmeth929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hess S. T., Girirajan T. P., Mason M. D., “Ultra-high resolution imaging by fluorescence photoactivation localization microscopy,” Biophys. J. 91(11), 4258–4272 (2006). 10.1529/biophysj.106.091116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu K., Zhong G., Zhuang X., “Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons,” Science 339(6118), 452–456 (2013). 10.1126/science.1232251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoopmann P., Punge A., Barysch S. V., Westphal V., Bückers J., Opazo F., Bethani I., Lauterbach M. A., Hell S. W., Rizzoli S. O., “Endosomal sorting of readily releasable synaptic vesicles,” Proc. Natl. Acad. Sci. U. S. A. 107(44), 19055–19060 (2010). 10.1073/pnas.1007037107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Westphal V., Rizzoli S. O., Lauterbach M. A., Kamin D., Jahn R., Hell S. W., “Video-rate far-field optical nanoscopy dissects synaptic vesicle movement,” Science 320(5873), 246–249 (2008). 10.1126/science.1154228 [DOI] [PubMed] [Google Scholar]
- 18.Willig K. I., Rizzoli S. O., Westphal V., Jahn R., Hell S. W., “STED microscopy reveals that synaptotagmin remains clustered after synaptic vesicle exocytosis,” Nature 440(7086), 935–939 (2006). 10.1038/nature04592 [DOI] [PubMed] [Google Scholar]
- 19.Kittel R. J., Wichmann C., Rasse T. M., Fouquet W., Schmidt M., Schmid A., Wagh D. A., Pawlu C., Kellner R. R., Willig K. I., Hell S. W., Buchner E., Heckmann M., Sigrist S. J., “Bruchpilot promotes active zone assembly, Ca2+ channel clustering, and vesicle release,” Science 312(5776), 1051–1054 (2006). 10.1126/science.1126308 [DOI] [PubMed] [Google Scholar]
- 20.Engerer P., Fecher C., Misgeld T., “Super-resolution microscopy writ large,” Nat. Biotechnol. 34(9), 928–930 (2016). 10.1038/nbt.3669 [DOI] [PubMed] [Google Scholar]
- 21.Truckenbrodt S., Sommer C., Rizzoli S. O., Danzl J. G., “A practical guide to optimization in X10 expansion microscopy,” Nat. Protoc. 14(3), 832–863 (2019). 10.1038/s41596-018-0117-3 [DOI] [PubMed] [Google Scholar]
- 22.Chen F., Tillberg P. W., Boyden E. S., “Optical imaging: expansion microscopy,” Science 347(6221), 543–548 (2015). 10.1126/science.1260088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chozinski T. J., Halpern A. R., Okawa H., Kim H. J., Tremel G. J., Wong R. O., Vaughan J. C., “Expansion microscopy with conventional antibodies and fluorescent proteins,” Nat. Methods 13(6), 485–488 (2016). 10.1038/nmeth.3833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ku T., Swaney J., Park J. Y., Albanese A., Murray E., Cho J. H., Park Y. G., Mangena V., Chen J., Chung K., “Multiplexed and scalable super-resolution imaging of three-dimensional protein localization in size-adjustable tissues,” Nat. Biotechnol. 34(9), 973–981 (2016). 10.1038/nbt.3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tillberg P. W., Chen F., Piatkevich K. D., Zhao Y., Yu C. C., English B. P., Gao L., Martorell A., Suk H. J., Yoshida F., DeGennaro E. M., Roossien D. H., Gong G., Seneviratne U., Tannenbaum S. R., Desimone R., Cai D., Boyden E. S., “Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies,” Nat. Biotechnol. 34(9), 987–992 (2016). 10.1038/nbt.3625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsanov N., Samacoits A., Chouaib R., Traboulsi A. M., Gostan T., Weber C., Zimmer C., Zibara K., Walter T., Peter M., Bertrand E., Mueller F., “smiFISH and FISH-quant - a flexible single RNA detection approach with super-resolution capability,” Nucleic Acids Res 44(22), e165 (2016). 10.1093/nar/gkw784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen F., Wassie A. T., Cote A. J., Sinha A., Alon S., Asano S., Daugharthy E. R., Chang J. B., Marblestone A., Church G. M., Raj A., Boyden E. S., “Nanoscale imaging of RNA with expansion microscopy,” Nat. Methods 13(8), 679–684 (2016). 10.1038/nmeth.3899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao Y., Bucur O., Irshad H., Chen F., Weins A., Stancu A. L., Oh E. Y., DiStasio M., Torous V., Glass B., Stillman I. E., Schnitt S. J., Beck A. H., Boyden E. S., “Nanoscale imaging of clinical specimens using pathology-optimized expansion microscopy,” Nat. Biotechnol. 35(8), 757–764 (2017). 10.1038/nbt.3892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang J. B., Chen F., Yoon Y. G., Jung E. E., Babcock H., Kang J. S., Asano S., Suk H. J., Pak N., Tillberg P. W., Wassie A. T., Cai D., Boyden E. S., “Iterative expansion microscopy,” Na. methods 14(6), 593–599 (2017). 10.1038/nmeth.4261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Truckenbrodt S., Maidorn M., Crzan D., Wildhagen H., Kabatas S., Rizzoli S. O., “X10 expansion microscopy enables 25-nm resolution on conventional microscopes,” EMBO Rep. 19(9), e45836 (2018). 10.15252/embr.201845836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao R., Asano S. M., Boyden E. S., “Q&A: Expansion microscopy,” BMC Biol. 15(1), 50 (2017). 10.1186/s12915-017-0393-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng T., Feng Z., Wang X., Jiang T., Jin R., Zhao P., Luo T., Gong H., Luo Q., Yuan J., “Review of micro-optical sectioning tomography (MOST): technology and applications for whole-brain optical imaging [Invited],” Biomed. Opt. Express 10(8), 4075–4096 (2019). 10.1364/BOE.10.004075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Economo M. N., Clack N. G., Lavis L. D., Gerfen C. R., Svoboda K., Myers E. W., Chandrashekar J., “A platform for brain-wide imaging and reconstruction of individual neurons,” eLife 5, e10566 (2016). 10.7554/eLife.10566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou C., Zheng T., Luo T., Yan C., Sun Q., Ren M., Zhao P., Chen W., Ji B., Wang Z., Li A., Gong H., Li X., “Continuous imaging of large-volume tissues with a machinable optical clearing method at subcellular resolution,” Biomed. Opt. Express 11(12), 7132–7149 (2020). 10.1364/BOE.405801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang X., Zhang Q., Huang F., Bai K., Guo Y., Zhang Y., Li N., Cui Y., Sun P., Zeng S., Lv X., “High-throughput light sheet tomography platform for automated fast imaging of whole mouse brain,” J. Biophotonics 11(9), e201800047 (2018). 10.1002/jbio.201800047 [DOI] [PubMed] [Google Scholar]
- 36.Renier N., Wu Z., Simon D. J., Yang J., Ariel P., Tessier-Lavigne M., “iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging,” Cell 159(4), 896–910 (2014). 10.1016/j.cell.2014.10.010 [DOI] [PubMed] [Google Scholar]
- 37.Susaki E. A., Tainaka K., Perrin D., Kishino F., Tawara T., Watanabe T. M., Yokoyama C., Onoe H., Eguchi M., Yamaguchi S., Abe T., Kiyonari H., Shimizu Y., Miyawaki A., Yokota H., Ueda H. R., “Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis,” Cell 157(3), 726–739 (2014). 10.1016/j.cell.2014.03.042 [DOI] [PubMed] [Google Scholar]
- 38.Asano S. M., Gao R., Wassie A. T., Tillberg P. W., Chen F., Boyden E. S., “Expansion microscopy: protocols for imaging proteins and RNA in cells and tissues,” Curr. Protoc. Cell Biol. 80(1), e56 (2018). 10.1002/cpcb.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang C., Kang J. S., Asano S. M., Gao R., Boyden E. S., “Expansion microscopy for beginners: visualizing microtubules in expanded cultured HeLa cells,” Curr. Protoc. Neurosci. 92(1), e96 (2020). 10.1002/cpns.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Z., Bovik A. C., Sheikh H. R., Simoncelli E. P., “Image quality assessment: from error visibility to structural similarity,” IEEE Trans. Image Process 13(4), 600–612 (2004). 10.1109/TIP.2003.819861 [DOI] [PubMed] [Google Scholar]
- 41.Wassie A. T., Zhao Y., Boyden E. S., “Expansion microscopy: principles and uses in biological research,” Nat. Methods 16(1), 33–41 (2019). 10.1038/s41592-018-0219-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu Y., Chu X., Chen T. T., Pan Q., Liu C., Yi J., Chu X., “Improving resolving ability of expansion microscopy by varying crosslinker concentration,” Chem. Commun. 56(30), 4176–4179 (2020). 10.1039/D0CC00052C [DOI] [PubMed] [Google Scholar]
- 43.Meng Y., Ye L., “Synthesis and swelling property of superabsorbent starch grafted with acrylic acid/2-acrylamido-2-methyl-1-propanesulfonic acid,” J. Sci. Food Agric. 97(11), 3831–3840 (2017). 10.1002/jsfa.8247 [DOI] [PubMed] [Google Scholar]
- 45.Zhao J., Liu P., Liu Y., “Adjustable tribological behavior of glucose-sensitive hydrogels,” Langmuir 34(25), 7479–7487 (2018). 10.1021/acs.langmuir.8b01388 [DOI] [PubMed] [Google Scholar]
- 44.Getachew B. A., Kim S. R., Kim J. H., “Self-healing hydrogel pore-filled water filtration membranes,” Environ. Sci. Technol. 51(2), 905–913 (2017). 10.1021/acs.est.6b04574 [DOI] [PubMed] [Google Scholar]
- 46.Gambarotto D., Zwettler F. U., Le Guennec M., Schmidt-Cernohorska M., Fortun D., Borgers S., Heine J., Schloetel J. G., Reuss M., Unser M., Boyden E. S., Sauer M., Hamel V., Guichard P., “Imaging cellular ultrastructures using expansion microscopy (U-ExM),” Nat. Methods 16(1), 71–74 (2019). 10.1038/s41592-018-0238-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gambarotto D., Hamel V., Guichard P., “Ultrastructure expansion microscopy (U-ExM),” Methods Cell Biol. 161(1), 57–81 (2021). 10.1016/bs.mcb.2020.05.006 [DOI] [PubMed] [Google Scholar]
- 48.Burgers J., Pavlova I., Rodriguez-Gatica J. E., Henneberger C., Oeller M., Ruland J. A., Siebrasse J. P., Kubitscheck U., Schwarz M. K., “Light-sheet fluorescence expansion microscopy: fast mapping of neural circuits at super resolution,” Neurophotonics 6(01), 1 (2019). 10.1117/1.NPh.6.1.015005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.During D. N., Rocha M. D., Dittrich F., Gahr M., Hahnloser R. H. R., “Expansion light sheet microscopy resolves subcellular structures in large portions of the songbird brain,” Front. Neuroanat. 13, 2 (2019). 10.3389/fnana.2019.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mascheroni L., Scherer K. M., Manton J. D., Ward E., Dibben O., Kaminski C. F., “Combining sample expansion and light sheet microscopy for the volumetric imaging of virus-infected cells with super-resolution,” Biomed. Opt. Express 11(9), 5032–5044 (2020). 10.1364/BOE.399404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murakami T. C., Mano T., Saikawa S., Horiguchi S. A., Shigeta D., Baba K., Sekiya H., Shimizu Y., Tanaka K. F., Kiyonari H., Iino M., Mochizuki H., Tainaka K., Ueda H. R., “A three-dimensional single-cell-resolution whole-brain atlas using CUBIC-X expansion microscopy and tissue clearing,” Nat. Neurosci. 21(4), 625–637 (2018). 10.1038/s41593-018-0109-1 [DOI] [PubMed] [Google Scholar]
- 52.Damstra H., Mohar B., Eddison M., Akhmanova A., Kapitein L., Tillberg P., “Visualizing cellular and tissue ultrastructure using Ten-fold Robust Expansion Microscopy (TREx),” bioRxiv 2021.02.03.428837 (2021). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data underlying the results presented in this paper are not publicly available at this time but may be obtained from the authors upon reasonable request.