Abstract
Periodontitis is an inflammatory disease induced by local infection in tooth‐supporting tissue. Periodontitis is associated with systemic bone diseases, but little is known about the mechanism of the causal effect of periodontitis on systemic bone resorption. Bacteria‐derived extracellular vesicles (EVs) act as natural carriers of virulence factors that are responsible for systemic inflammation. In this study, we investigated the role of EVs derived from Filifactor alocis, a Gram‐positive, anaerobic periodontal pathogen, in systemic bone loss and osteoclast differentiation. F. alocis EVs accumulated in the long bones of mice after intraperitoneal administration. These EVs induced proinflammatory cytokines, osteoclastogenesis, and bone resorption via Toll‐like receptor 2 (TLR2). The phase separation of F. alocis EVs showed that amphiphilic molecules were responsible for the induced bone resorption and osteoclastogenesis. The osteoclastogenic effects of F. alocis EVs were reduced by lipoprotein lipase. Proteomic analysis of the amphiphilic molecules identified seven lipoproteins. Our results indicate that lipoprotein‐like molecules in F. alocis EVs may contribute to systemic bone loss via TLR2.
Keywords: bacterial extracellular vesicles, bone resorption, lipoproteins, osteoclastogenesis, TLR2
1. INTRODUCTION
Periodontitis is a chronic oral disease initiated by microbial complexes in the subgingival pocket and is characterized by gingival tissue destruction and bone resorption, leading to tooth loss. Periodontitis is also associated with systemic diseases, including cardiovascular diseases, Alzheimer's disease, rheumatoid arthritis, oral and colorectal carcinoma, pregnancy complications, pneumonia, and diabetes (Hajishengallis & Chavakis, 2021). Clinical studies have suggested that periodontitis is closely associated with osteoporosis and osteopenia, which are systemic skeletal diseases characterized by reduced bone mineral density (Passos et al., 2013; Xu et al., 2021). Compared to healthy individuals, patients with osteoporosis showed a twofold higher risk of periodontitis (Manjunath et al., 2019). Periodontitis and osteoporosis share risk factors, including aging, obesity, diabetes mellitus, menopause, and deficiency of vitamin D and calcium. Inflammation is also an important factor that is common to both diseases, indicating bidirectional relationships between the two diseases (Contaldo et al., 2020). Periodontitis may systemically activate osteoclastogenic bone resorption by releasing proinflammatory cytokines from local periodontal tissue inflammation (Pan et al., 2019). Proinflammatory cytokines such as IL‐1, IL‐6 and TNF‐α are known to be involved in both alveolar and osteoclastogenic bone resorption (Azuma et al., 2000; Ishimi et al., 1990; Jimi et al., 1999). In the infectious state, microbes or microbe‐derived bioactive molecules trigger proinflammatory responses to increase osteoclast activity and inhibit osteoblast activity, resulting in bone loss (Hienz et al., 2015).
Extracellular vesicles (EVs) are nano‐sized particles enclosed by lipid bilayers that are released by all living organisms, including bacteria, archaea, and eukaryotes (Gho & Lee, 2017). Bacterial EVs contain various molecules, including microbe‐associated molecular patterns (MAMPs), enzymes, and toxins, that can act as virulence factors in the host (Bitto et al., 2021; Kim et al., 2015). EVs are attracting great attention due to their ability to deliver their contents both locally and systematically by cell‐free intercellular communication (Yáñez‐Mó et al., 2015). In a patient with Neisseria meningitidis infection, EVs were observed in the patient's blood sample (Namork & Brandtzaeg, 2002). Helicobacter pylori EVs were observed in gastric juices from patients with gastric diseases, and orally administered H. pylori EVs were distributed in the stomach in a mouse model (Choi et al., 2017). EVs derived from Porphyromonas gingivalis, Tannerella forsythia, and Treponema denticola, which are so‐called ‘red‐complex’ periodontal pathogens, harbour various MAMPs (Cecil et al., 2016) and exert immunomodulatory effects through proinflammatory cytokine production (Cecil et al., 2017). EVs derived from Aggregatibacter actinomycetemcomitans, a periodontal pathogen, were able to cross the blood–brain barrier and induce TNF‐α production in the brain (Han et al., 2019). As EVs released from periodontal pathogens in the oral cavity can easily disseminate to other parts of the body, it is very intriguing to determine the role of EVs of periodontal pathogens in systemic bone metabolism.
Filifactor alocis is an asaccharolytic obligate anaerobe and a Gram‐positive periodontal pathogen. F. alocis has recently received attention as an important periodontal pathogen based on oral microbiome analysis (Griffen et al., 2012). F. alocis is highly detected in chronic and aggressive periodontitis, implantitis, and endodontic infections (Aruni et al., 2014). In patients with aggressive periodontitis, the presence of F. alocis with A. actinomycetemcomitans and Streptococcus parasanguinis in subgingival tissue is closely associated with bone loss (Fine et al., 2013). F. alocis was also found in extraoral sites such as brain abscesses (Hishiya et al., 2020) and thoracic empyema (Gray and Vidwans, 2019). F. alocis infections are characterized by the dysfunctional modulation of neutrophils (Edmisson et al., 2018), adhesion to and invasion of epithelial cells, resistance to oxidative stress (Aruni et al., 2011), and inhibition of complement cascades (Jusko et al., 2016). Recently, we reported that F. alocis EVs harboured lipoproteins, autolysins, and complement inhibitors, and induced as much immunostimulatory activity as whole bacterial cells (Kim, Lim, et al., 2020). Although EVs from pathogenic bacteria have been reported to have pathogenic potential (Kuipers et al., 2018; Park et al., 2010), no study has attempted to determine their roles in systemic bone resorption and osteoclastogenesis.
In this study, we investigated the role of F. alocis EVs in bone metabolism using a MAMP‐induced inflammatory bone loss model in mice to determine whether EVs derived from periodontal pathogens have the potential to cause osteoporosis. F. alocis EVs induced systemic bone loss and osteoclastogenesis via Toll‐like receptor (TLR) 2. Amphiphilic and lipoprotein lipase‐sensitive molecules are involved in this effect. This study provides new insights into the role of periodontal pathogen‐derived EVs in systemic bone resorption.
2. MATERIALS AND METHODS
2.1. Reagents and chemicals
Columbia broth, yeast extract and Bacto agar were purchased from BD Biosciences (San Jose, CA, USA). L‐cysteine, L‐arginine, resazurin, hemin, and vitamin K were purchased from Sigma (St. Louis, MO, USA). A vacuum filter system (0.22 μm pore size) was purchased from Corning (New York, NY, USA; cat#: 431098). A 100 kDa cut‐off centrifugal filter (cat#: UFC710008) and RIPA buffer were purchased from Merck (Darmstadt, Germany). RPMI 1640 medium and the phosphate‐buffered saline (PBS) were purchased from Welgene (Daegu, South Korea). Fetal bovine serum (FBS) and the penicillin/streptomycin solution were purchased from Gibco BRL (Paisley, UK). Pam2CSK4 (cat#: tlrl‐pm2s‐1), Pam3CSK4 (cat#: tlrl‐pms), Escherichia coli lipopolysaccharide (LPS) O111:B4 (cat#: tlrl‐eblps), ultrapure LPS (cat#: tlrl‐3pelps), G418 (cat#: ant‐gn), and hygromycin B (cat#: ant‐hg) were purchased from InvivoGen (San Diego, CA, USA). Recombinant mouse macrophage colony‐stimulating factor (M‐CSF; cat#: 315–02) and soluble mouse receptor activator of nuclear factor‐κB ligand (RANKL; cat#: 315–11) were purchased from PeproTech (Rocky Hill, NJ, USA). FITC anti‐human CD25 mouse monoclonal antibodies (cat#: 555431) and anti‐actin antibodies (cat#: 612656) were purchased from BD Biosciences. Anti‐NFATc1 mouse monoclonal antibodies (cat#: SC7294) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Anti‐c‐Fos rabbit polyclonal (cat#: 2250S), anti‐phospho‐ERK (Thr202/Tyr204; cat#: 9101S), anti‐phospho‐JNK (Thr182/Tyr185; cat#: 9251S), anti‐phospho‐p38 (Thr180/Tyr182; cat#: 9211S), anti‐phospho‐NF‐κB p65 antibodies (cat#: 3033S) and anti‐ERK (cat#: 9102S), JNK (cat#: 9252S), p38 (cat#: 9212S), NF‐κb p65 (cat#: 8242S) antibodies were purchased from Cell Signaling Technology (Beverly, MA, USA).
2.2. Bacteria and extracellular vesicles
F. alocis (ATCC 35896) was cultured at 37°C in an anaerobic atmosphere (10% CO2, 10% H2, 80% N2). F. alocis was grown in modified Columbia broth (5% yeast extract, 0.0025% resazurin, 5 μg/ml hemin, 1 μg/ml, vitamin K, 1 μg/ml L‐cysteine, 2 μg/ml L‐arginine) for 48 h until the late mid‐log phase was reached. Culture supernatants (2 L for one preparation) were harvested at an optical density (OD600nm) of 1.0–1.3 (colony‐forming units, ≥ 3.0 × 108 CFU/ml). EVs were purified and characterized as previously described with minor modifications (Kim, Lim et al., 2020). The supernatants were filtered using a 0.22 μm membrane filter and concentrated using a 100 kDa cut‐off centrifugal filter. The crude EVs were isolated by ultracentrifugation (160,000 × g, 4°C, 2 h) using a Type 45 Ti rotor (Beckman Coulter, Brea, CA, USA). Then, the crude EVs were dissolved in PBS (1 ml) and resuspended in 60% OptiPrep (2 ml, Sigma; cat#: D1556) to make 40% OptiPrep. The 40% crude EV/OptiPrep (3 ml) was overlaid with 35% OptiPrep (5 ml) and 10% OptiPrep (5 ml), and subjected to buoyant density gradient ultracentrifugation (100,000 × g, 4°C, 18 h) using an SW 40 Ti rotor (Beckman Coulter). Each fraction (1.3 ml) was obtained from the top of the gradient sample (fractions #1–10). Fractions 4 and 5 were pooled and EVs in Fractions 4 and 5 were harvested by ultracentrifugation (160,000 × g, 4°C, 2 h) using an SW 40 Ti rotor. Purified EVs were dissolved in 500 μL PBS. The number of EV particles per ml was measured by nanoparticle tracking analysis (NTA) using a NanoSight LM10 system and NTA 2.3 nanoparticle tracking and analysis software (Malvern Instruments Ltd, Worcestershire, UK). The absence of bacterial cells in the EV preparation was confirmed by transmission electron microscopy (TEM). The EV protein concentrations were measured by bicinchoninic acid (BCA) assays (Thermo Fisher Scientific Inc., Waltham, MA, USA; cat#: 23227). The yield of F. alocis EVs was approximately 150 μg of proteins or 1.8 × 1012 particles per 1 L of F. alocis culture supernatant. The EV particles per μg of protein was 1.2 × 1010, which was regarded as pure EVs as previously described (Webber & Clayton, 2013). For this study, at least 60 EV preparations were made. As a control, culture medium without bacteria was also subjected to EV isolation.
2.3. Isolation of the amphiphilic phase of F. alocis EVs using Triton X‐114
Amphiphilic phases from F. alocis EVs were isolated using a method for bacterial lipoprotein isolation as previously described with minor modifications (Armbruster & Meredith, 2018). Briefly, F. alocis EVs (2 mg proteins) were disrupted by ultrasonication (Vibra‐cell VCX500, Sonics & Materials, Inc., Newtown, CT, USA) with a protease inhibitor cocktail (Roche Diagnostics GmbH, Mannheim, Germany; cat#: 11836153001). The lysates were resuspended in 2% Triton X‐114 (Sigma; cat#: X114) and rotated at 4°C for 3 h. Then, the lysates were mixed with TBS and incubated at 37°C for 15 min. The amphiphilic phase and hydrophilic phase were separated by centrifugation (10,000 × g) at 37°C for 15 min. The amphiphilic phase was precipitated by methanol (Merck; cat#: 1.06009.1011) to remove residual chemicals and then dissolved in 10 mM octyl β‐D‐glucopyranoside (Sigma; cat#: O8001). The protein concentration of each phase was measured by BCA assays. Approximately 350 μg of amphiphilic phase proteins and 1.35 mg of hydrophilic phase proteins were obtained from 2 mg of F. alocis EV proteins with 300 μg of loss.
2.4. Lipoprotein identification of the amphiphilic phase of F. alocis EVs using LC–MS/MS
Four independently isolated amphiphilic phases of F. alocis EVs were used for proteomic analysis. In‐solution digestion and peptide analysis using liquid chromatography‐tandem mass spectrometry (LC–MS/MS) were performed as previously described (Lee et al., 2010). The amphiphilic phase of F. alocis EVs (50 μg of protein) was denatured and reduced in 8 M urea/5 mM dithiothreitol (Sigma), dissolved in 25 mM ammonium bicarbonate and incubated at 60°C for 1 h. After cooling, the sample was dissolved in 25 mM iodoacetamide/25 mM ammonium bicarbonate (Sigma) at room temperature for 1 h in the dark. The urea was diluted to a concentration of 1 M with 25 mM ammonium bicarbonate (Sigma), to which the 8‐fold diluted proteins were digested with MS grade Pierce trypsin protease (enzyme to substrate ratio = 1:50, Thermo Scientific; cat#: 90057) at 37°C overnight. The resulting peptide sample was desalted by Oasis HLB 1 cc extraction cartridges (Waters Corporation, MA, USA; cat#: WAT094225) and lyophilized. LC–MS/MS analysis was performed using a nano HPLC system (Agilent, Wilmington, DE, USA). A nanochip column (150 mm × 0.075 mm, Agilent; cat#: WK5065–9911) was used for peptide separation. Mobile phase A for LC separation was 0.1% formic acid in deionized water, and mobile phase B was 0.1% formic acid in acetonitrile. The chromatography gradient was designed for a linear increase from 3% B to 45% B in 70 min, 45% B to 95% B in 1 min, 95% B in 9 min, and 3% B in 10 min. The flow rate was maintained at 300 nL/min. Product ion spectra were collected in the information‐dependent acquisition mode and were analysed by Agilent 6530 Accurate‐Mass quadrupole time‐of‐flight (Q‐TOF) using continuous cycles of one full scan TOF MS from 300 to 2000 m/z (1.0 s) plus three product ion scans from 150 to 2000 m/z (1.5 s each). Precursor m/z values were selected starting with the most intense ion using a quadrupole selection with a resolution of 3 Da. The rolling collision energy feature was used, which determines collision energy based on the precursor value and charge state. The dynamic exclusion time for precursor ion m/z values was 60 s. Database searching was performed as previously described (Kim, Lim et al., 2020). Lipoproteins in the amphiphilic phase were identified using a database of bacterial lipoproteins (DOLOP, https://www.mrc‐lmb.cam.ac.uk/genomes/dolop/analysis.shtml) (Babu et al., 2006). Lipoprotein subcellular localization was predicted by PSORTb version 3.0.3 (https://www.psort.org/psortb/).
2.5. Enzyme treatment of F. alocis EVs
Enzyme treatment was performed as previously described with minor modifications (Hashimoto et al., 2006b; Seo & Nahm, 2009). F. alocis EVs (200 μg/200 μl PBS) were treated with lipoprotein lipase from the Pseudomonas species (0.5–50 μg/ml, Sigma; cat#: 62309), proteinase K (50 μg/ml, Roche; cat#: 3115879001), or platelet‐activating factor acetylhydrolase (PAF‐AH, 50 μg/ml, PeproTech; cat#: 140–10) at 37°C for 16 h. For the control, F. alocis EVs (200 μg/200 μl PBS) without enzyme were incubated under the same conditions. After the incubation period, F. alocis EVs were obtained by ultracentrifugation (160,000 × g, 4°C, 1 h) and resuspended in 200 μl PBS.
2.6. Animals
All animal experiments were approved by the Institutional Animal Care and Use Committee of Seoul National University (SNU‐190721–2–2). C57BL/6 mice were purchased from Orient Bio (Gyeonggi‐do, Korea). TLR2–/‐ mice (C57BL/6) were obtained from The Jackson Laboratory (Bar Harbour, ME, USA), and TLR4–/– mice (C57BL/6) were kindly provided by Dr Seung Hyun Han (Seoul National University, Seoul, Korea). All mouse experiments were performed using eight‐week‐old C57BL/6 male mice. Before experiments, mice were weighed, and all mice showed similar body weights.
2.7. In vivo tracking of F. alocis EVs
F. alocis EVs (500 μg of protein/1 ml PBS) were stained with Vybrant DiO dye (5 μl of 1 mM DiO dye, Invitrogen; cat#: V22886) at 37°C for 30 min. To exclude residual DiO dye, the DiO dye‐labeled F. alocis EV solutions were filtered using a 100‐kDa centrifugal filter. The retentate containing EVs were resuspended in PBS and concentrated using ultracentrifugation (160,000 × g) at 4°C for 1 h. Then, DiO dye‐labeled F. alocis EVs were resuspended in 1 ml of PBS. No pellets were visible when the same protocol was applied to the DiO dye solution without F. alocis EV. Eight‐week‐old male mice were intraperitoneally administered 50 μg of DiO dye‐labeled F. alocis EVs. At 3 or 12 h, the mice were euthanized, and the organs were collected and analysed within 5 min. The fluorescence intensity in the organs was visualized by a Fusion‐FX6 imaging system (Vilber Lourmat, Marne‐la‐Vallée, France).
2.8. In vivo bone loss model and micro‐computed tomography (μCT)
F. alocis EV‐induced bone loss experiments were performed as previously described (Chang et al., 2008) with minor modifications. Eight‐week‐old male mice were intraperitoneally administered the indicated stimuli (as protein, 50 μg) on Days 0 and 3. At Day 7, femurs were obtained from the mice and analysed with SkyScan 1172 (SkyScan, Aartselaar, Belgium; X‐ray output of 70 kV and 139 μA, 0.5 mm aluminium filter, 590 ms exposure, 6.94 μm‐pixel size resolution), and the data were reconstructed with NRecon software (SkyScan, version 1.7.4.2; beam hardening correction = 40%, ring artifact correction = 7). Trabecular bones were measured in the 1‐mm‐thick region of the distal femur starting 1 mm below the growth plate at a minimum threshold of 69 and a maximum of 255. Bone parameters were calculated by a CT‐analyser program (version 1.7; SkyScan) according to American Society for Bone and Mineral Research standard [Trabecular bone volume/total volume (Tb.BV/TV), trabecular number (Tb.N), trabecular separation (Tb.Sp), and trabecular thickness (Tb.Th)] (Dempster et al., 2013). Three‐dimensional images were reconstructed by CT‐volume software (version 2.2.3.0; SkyScan).
2.9. Histology
Histological measurements were performed as previously described (Chang et al., 2008). Briefly, femurs were fixed in 4% paraformaldehyde and decalcified with 12% ethylenediaminetetraacetic acid (EDTA) for 4 weeks. The decalcified femurs were dehydrated and embedded in paraffin. Five‐micrometre‐thick serial sections of the paraffin blocks were stained for TRAP (tartrate‐resistant acid phosphatase) using a TRAP staining kit (Sigma; cat#: 387A‐1KT) and haematoxylin (Sigma; cat#: H3136). The images were captured by bright field microscopy, and histomorphometric analysis was carried out with OsteoMeasure software (Osteometrics, Decatur, CA, USA). To determine the osteoclast parameters, the trabecular bone area and surface perimeters and the number of TRAP‐positive cells on the bone surfaces were measured in the same region below the growth plate.
2.10. Osteoclast differentiation
WT, TLR2–/–, or TLR4–/– bone marrow‐derived macrophages (BMMs) were prepared as previously described (Song et al., 2018). The osteoclastogenic effects of F. alocis EVs on committed osteoclast precursors (COCs) were determined as previously described (Zou & Bar‐Shavit, 2002) with minor modifications. To obtain COCs, BMMs (2 × 104/well) were plated on 48‐well culture plates with M‐CSF (30 ng/ml) for 1 day and cultured with M‐CSF (30 ng/ml) and RANKL (60 ng/ml) for 3 days. COCs were stimulated with F. alocis EVs (1, 3, or 10 μg/ml of protein; 0.6 ×, 1.8 ×, or 6 × 106 of EV particles/cell) in the presence of M‐CSF (30 ng/ml) for 2 days. COCs were also treated with lipoprotein lipase‐, proteinase K‐ and PAF‐AH‐treated F. alocis EVs (10 μg/ml of protein). For the TRAP staining, the cells were fixed and stained with a commercial TRAP staining kit according to the manufacturer's instructions. Representative images of TRAP‐positive MNCs (multinucleated cells) from triplicate samples were obtained by digital inverted microscopy (DS‐Ri2, Nikon, Tokyo, Japan). TRAP‐positive MNCs containing three or more nuclei in whole wells were counted from triplicate samples.
2.11. Resorption assay using calcium phosphate‐coated plates
Resorption assays were performed as previously described (Song et al., 2015). To obtain COCs, BMMs (2 × 104/well) were plated on calcium phosphate‐coated 48‐well plates and cultured with M‐CSF (30 ng/ml) and RANKL (60 ng/ml) for 3 days. Then, the COCs were stimulated with F. alocis EVs (1, 3, or 10 μg/ml of protein; 0.6 ×, 1.8 ×, or 6 × 106 of EV particles/cell) in the presence of M‐CSF (30 ng/ml) for 10 days. The plates were washed with distilled water to remove cells and subjected to von Kossa staining. For the staining, the plates were incubated with 5% silver nitrate solution for 30–60 min in the dark. The plates were then washed with distilled water and incubated in carbonate‐formaldehyde solution (5% Na2CO3, 9.25% formaldehyde) for 2 min before washing and drying. Representative images of the resorption area from triplicate samples were obtained by digital inverted microscopy (DS‐Ri2). The resorption area was measured by ImageJ software (National Institutes of Health, Bethesda, MD, USA) from representative images of triplicate samples.
2.12. Western blotting
BMMs (1 × 105/well) were plated on 12‐well culture plates with M‐CSF (30 ng/ml) for 1 day and then cultured with M‐CSF (30 ng/ml) and RANKL (60 ng/ml) for 3 days. Then, COCs were stimulated with F. alocis EVs for the indicated times. Then, cell lysates were prepared using RIPA buffer (10 mM Tris‐HCl, pH 7.5, 150 mM NaCl, 1% Triton X‐100) containing a protease inhibitor cocktail. The lysates (20 μg) were separated by sodium dodecyl sulphate‐polyacrylamide gel electrophoresis (12%) and transferred to polyvinylidene difluoride membranes (Millipore, Bedford, MA, USA). After the membranes were incubated with 5% skim milk in TBST (Tris‐buffered saline containing 0.1% Tween 20), they were incubated with specific antibodies against nuclear factor‐activated T cells c1 (NFATc1), c‐fos, p‐ERK, ERK, p‐p38, p38, p‐JNK, JNK, p‐NF‐κB p65, NF‐κB p65, or β‐actin at 4°C overnight. The membranes were washed with TBST 4 times and incubated with horseradish peroxidase‐conjugated secondary antibodies at room temperature for 1 h. The protein bands were visualized with SUPEX ECL solution (Neuronex, Pohang, Korea) using ChemiDoc MP (Bio–Rad, Hercules, CA, USA).
LTA in F. alocis EVs, amphiphilic phase, hydrophilic phase, and F. alocis cell lysates was detected using monoclonal antibody to Gram‐positive bacteria lipoteichoic acid (LTA, Invitrogen, Grand Island, NY, USA; cat#: MA1–7402). F. alocis cell lysates were prepared by ultrasonication of F. alocis cells. F. alocis EVs treated with lipoprotein lipase, proteinase K, or PAF‐AH were subjected to Western blotting with LTA antibody. Purified lipoteichoic acid (LTA) from Staphylococcus aureus (InvivoGen; cat#: tlrl‐pslta) was used as a positive control.
2.13. Real‐time reverse transcription‐polymerase chain reaction
BMMs (2 × 104/well) were plated on 48‐well culture plates with M‐CSF (30 ng/ml) for 1 day and then cultured with M‐CSF (30 ng/ml) and RANKL (60 ng/ml) for 3 days. Then, COCs were stimulated with F. alocis EVs in the presence of M‐CSF (30 ng/ml) for 12 h. Total RNA was isolated from the cells using TRIzol reagent (Invitrogen; cat#: 15596018). The RNA concentrations and purity were measured by 260/280 nm absorbance using a NanoDrop 2000 (Thermo Fisher Scientific Inc.). The 260/280 nm absorbance of the used RNA sample ranged 1.82–1.99. Complementary DNA was synthesized using an M‐MLV reverse transcriptase kit (Promega, Madison, WI, USA; cat#: M1701). Real‐time RT–PCR analysis of NFATc1, c‐fos, TRAP, Cathepsin K, dendritic cell‐specific transmembrane protein (DC‐STAMP), ATPase H+ transporting v0 subunit d2 (atp6v0d2), and glyceraldehyde dehydrogenase (GAPDH) was performed using Power SYBR Green master mix (Applied Biosystem, Warrington, UK; cat#: 4367659). Gene expression was calculated using the 2−ΔΔCt method. Average threshold cycles (Ct) for the gene of interest were obtained from triplicate samples and normalized by the Ct of GAPDH as a housekeeping gene. Then, the gene expression was calculated relative to the nontreated group. All experiments for real‐time RT–PCR were performed at least three times independently. The sequences of the primers used were as follows: NFATc1, forward 5′‐CCA GTA TAC CAG CTC TGC CA‐3′ and reverse 5′‐GTG GGA AGT CAG AAG TGG GT‐3′; c‐Fos, forward 5′‐GGG GAC AGC CTT TCC TAC TA‐3′ and reverse 5′‐CTG TCA CCG TGG GGA TAA AG‐3′; TRAP, forward 5′‐GCT CAC ATG ACC AAG GGG AG‐3′ and reverse 5′‐CCT CGC TCC TCT GAC ACT TAC‐3′; Cathepsin K, forward 5′‐GTA GCC ACG CTT CCT ATC CG‐3′ and reverse 5′‐GCC GAG AGA TTT CAT CCA CCT‐3′; DC‐STAMP, forward 5′‐CTG TTG CTT TGT GGC CTT CC‐3′ and reverse 5′‐AAG CGT TCC TAC CTT CAC GG‐3′; Atp6vod2, forward 5′‐CAG CCT GAG CAG TAT GCT TGA‐3′ and reverse 5′‐AGC CAG GAA GTT GCC ATA GTC‐3′; and GAPDH, forward 5′‐GTC GCC AGC CGA GCC‐3′ and reverse 5′‐TGA AGG GGT CAT TGA TGG CA‐3′.
2.14. Enzyme‐linked immunosorbent assay
For the in vitro experiments, BMMs (5 × 104/well) were plated on 96‐well culture plates with M‐CSF (30 ng/ml) for 1 day and then cultured with M‐CSF (30 ng/ml) and RANKL (60 ng/ml) for 3 days. Then, COCs were stimulated with F. alocis EVs in the presence of M‐CSF (30 ng/ml) for 24 h. Proinflammatory cytokines (TNF‐α, IL‐6 and IL‐1β) in the supernatants were analysed by an R&D ELISA kit (Minneapolis, MN, USA; TNF‐α, cat#: DY410; IL‐6, cat#: DY406; IL‐1β, cat#: DY401) according to the manufacturer's instructions. For the in vivo experiments, mice were systemically administered F. alocis EVs. At the indicated times, proinflammatory cytokines (TNF‐α, IL‐6 and IL‐1β) in the serum were analysed by R&D ELISA kits, and clinical markers of bone metabolism (C‐terminal telopeptide of type I collagen; CTX‐1 and N‐terminal propeptide of type I procollagen, P1NP) were analysed by RatLaps EIA kits (Immunodiagnostic Systems, Boldon, U.K.; CTX‐1, cat#: AC‐06F1, P1NP, cat#: AC‐33F1) according to the manufacturer's instructions.
2.15. Measurement of TLR2 and TLR4 activations
CHO/CD14/TLR2 and CHO/CD14/TLR4 cells were obtained from Douglas Golenbock (Boston Medical Centre, Boston, MA, USA). To measure TLR2 or TLR4 activation, CHO/CD14/TLR2 or CHO/CD14/TLR4 cells (3 ×105/well) were plated on 48‐well culture plates in the presence of G418 (1 mg/ml) and hygromycin B (0.4 mg/ml) for 20 h. Then, the cells were stimulated with the indicated conditions for 24 h and stained with specific antibodies against human CD25. The expression of CD25 was analysed by flow cytometry using a FACSCalibur (BD Biosciences) and FlowJo software (TreeStar, San Carlos, CA, USA). The gating strategy for flow cytometry is shown in Figure S1.
3. STATISTICAL ANALYSIS
All experiments were performed at least three times independently. The data were analysed using GraphPad Prism software. Unpaired, two‐tailed Student's t‐tests were used to determine statistical significance between two independent groups. One‐way analysis of variance (ANOVA) with Bonferroni's multiple comparison test was used to determine statistical significance among multiple groups with one independent variable. Two‐way ANOVA with Bonferroni's multiple comparison test was used to determine statistical significance among multiple groups with two independent variables. The data are shown as the mean values ± standard deviations. A p value of < 0.05 was considered statistically significant.
4. RESULTS
4.1. F. alocis EVs induce bone loss in vivo
F. alocis EVs were purified by the combination of ultracentrifugation and buoyant density gradient ultracentrifugation using OptiPrep reagent. After the buoyant density gradient ultracentrifugation, the particle numbers of each fraction were measured by NTA. As shown in Figure 1a, a distinct band was visible between fractions #4 and #5. Then, these fractions were pooled and subjected to EV preparation. Approximately, 150 μg protein (1.8 × 1012 particles) of F. alocis EVs was obtained from 1 L of F. alocis culture supernatant. The TEM images showed that F. alocis EVs had round vesicular structures without intact F. alocis cells, and the size was approximately 30–100 nm (Figure 1b). NTA data showed that the mean size and mode size of F. alocis EVs were 133.4 ± 5.5 nm and 113.0 ± 3.7, respectively (Figure 1c). NTA tends to measure biological EVs larger than that of TEM (Bachurski et al., 2019). The size of F. alocis EVs was similar to that of EVs derived from other Gram‐positive bacteria (Bitto et al., 2021; Kim, Lee et al., 2020b). No colonies were observed after plating F. alocis EVs on Columbia agar (data not shown). To examine the distribution of F. alocis EVs in vivo, F. alocis EVs were labelled with DiO dye and injected intraperitoneally into mice. Then, F. alocis EVs were tracked by measuring the fluorescence intensity in the brain, lung, heart, liver, kidney, spleen, tibiae, and femur. The fluorescence intensity of F. alocis EVs was strong in the liver, kidney, tibiae, and femur but weak in the brain, lung, heart and spleen at 1 h after ‐ administration (Figure 1d). The intensity was diminished after 12 h. Next, to investigate the effect of F. alocis EVs on long bones, F. alocis EVs were intraperitoneally administered to mice on Days 0 and 3 for 7 days according to a MAMP‐induced inflammatory bone loss model (Figure 1e, Chang et al., 2008). Micro‐CT images showed that the trabecular bone density was reduced by F. alocis EVs compared to that in the control group (Figure 1f). Quantitative μCT analysis showed that F. alocis EVs significantly reduced Tb.BV/TV and Tb.N, and increased Tb.Sp in the mouse femur (Figure 1g). There was no difference in Tb.Th. These results suggest that intraperitoneally administered F. alocis EVs accumulate in long bones and cause bone loss in vivo.
FIGURE 1.
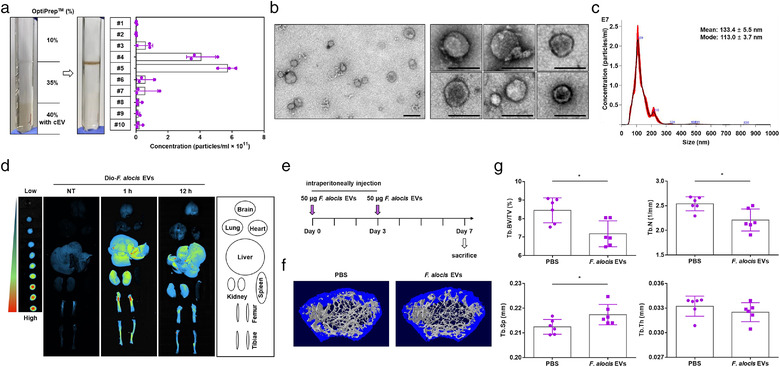
Bone loss in long bones in response to F. alocis EVs. (a) Representative image of F. alocis EV purification by buoyant density gradient ultracentrifugation using the OptiPrep reagent. The number of particles from each gradient fraction (#1–10) was analysed by NTA using nanoparticle tracking analysis software/instrument. Data are shown as the mean values ± standard deviations of three biological replicates. Fractions #4 and #5 were pooled and subjected to the isolation of F. alocis EVs. (b) F. alocis EVs were examined under TEM at both 10,000× magnification (left panel, scale bar: 100 nm) and 25,000× magnification (right panel, scale bar: 100 nm). (c) Size distributions of F. alocis EVs (1:1000 diluted sample) were analysed by NTA using nanoparticle tracking analysis software/instrument. For the TEM and NTA analysis, representative data from at least three biological replicates are shown. (d) Eight‐week‐old male mice were administered DiO‐labeled F. alocis EVs (50 μg of protein) for the indicated time periods. The brain, lung, heart, liver, kidney, spleen, femur and tibiae were isolated, and the distributions of DiO‐labeled F. alocis EVs were determined by an in vivo imaging system. (e‐g) Eight‐week‐old male mice were administered F. alocis EVs (50 μg of protein) on Days 0 and 3. On Day 7, the femurs were isolated, and trabecular bone was analysed by μCT (PBS, and F. alocis EVs: n = 6). Representative 3D reconstruction images of femurs from PBS‐treated or F. alocis EV‐treated mice (f). Trabecular bone volume/total volume (Tb.BV/TV), trabecular number (Tb.N), trabecular separation (Tb.Sp), and trabecular thickness (Tb.Th) were calculated with the μCT analysis program (g, n = 6). The graphs are shown as the mean values ± standard deviations. Statistical significance was determined by two‐tailed Student's t‐test. Representative data are shown for at least three biological replicates. * P < 0.05 compared to PBS group
4.2. F. alocis EVs induce osteoclast differentiation in vitro
Osteoclasts are multinucleated cells that originate from bone marrow‐derived hematopoietic precursor cells. Osteoclasts play an important role in the regulation of bone homeostasis by resorbing bone (Teitelbaum, 2000), and their differentiation and activation are mainly regulated by RANKL and M‐CSF (Khosla, 2001; Tanaka et al., 2005). To determine the role of F. alocis EVs in osteoclastogenesis in vitro, we prepared COCs from BMMs using RANKL and M‐CSF, as shown in Figure 2a. Since RANKL can activate COCs to mature osteoclasts (Boyle et al., 2003), COCs were stimulated with F. alocis EVs (1, 3, or 10 μg/ml of protein; 0.6 ×, 1.8 ×, or 6 × 106 of EV particles/ cell) for 2 days in the presence of M‐CSF without RANKL. After stimulation for 2 days, TRAP staining was performed, and TRAP‐positive multinucleated cells (MNCs) containing three or more nuclei were counted. As shown in Figure 2b, F. alocis EVs dose‐dependently increased the number of TRAP‐positive MNCs. Consistent with the TRAP staining results, F. alocis EVs dose‐dependently induced the osteoclastogenic cytokines TNF‐α, IL‐6, and IL‐1β in COCs (Figure 2c). Next, to determine the bone‐resorption activity of osteoclasts differentiated by F. alocis EVs, BMMs were seeded on calcium phosphate‐coated plates that mimic bone material (Dorozhkin & Epple, 2002) and cultured with M‐CSF and RANKL for 3 days to prepare COCs. Then, the cells were stimulated with F. alocis EVs in the presence of M‐CSF for 10 days. F. alocis EVs dose‐dependently increased the bone resorption areas in calcium phosphate‐coated plates (Figure 2d). To confirm that the osteoclastogenic activity was not due to a component derived from the bacterial culture medium used for EV isolation, the same protocol was used to perform EV isolation from bacterial culture media that was free of bacteria. The fraction isolated showed no activity (data not shown). These results suggest that F. alocis EVs are potent inducers of osteoclast differentiation and activation.
FIGURE 2.
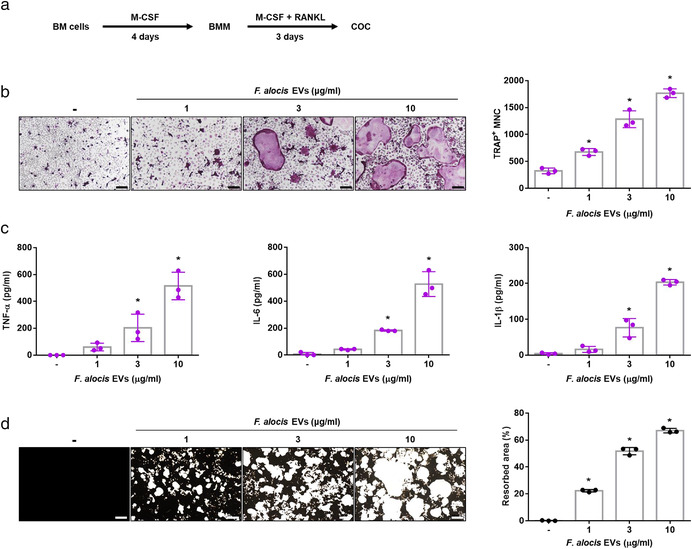
Osteoclast differentiation and activation by F. alocis EVs. (a) Schematic overview of the preparation of COCs from BM cells. (b) COCs were stimulated with F. alocis EVs (1, 3, or 10 μg/ml of protein; 0.6 ×, 1.8 ×, or 6 × 106 of EV particles/cell) in the presence of M‐CSF (30 ng/ml) for 2 days. Then, the cells were stained for TRAP activity, and the representative images of TRAP‐positive MNCs from triplicate samples are shown (left panel, scale bar: 200 μm). The number of TRAP‐positive MNCs was counted with triplicate samples (right panel). (c) COCs were stimulated with F. alocis EVs (1, 3, or 10 μg/ml of protein; 0.6 ×, 1.8 ×, or 6 × 106 of EV particles/cell) in the presence of M‐CSF (30 ng/ml) for 24 h. TNF‐α, IL‐6, and IL‐1β in the culture supernatants from triplicate samples were analysed using ELISA. (d) COCs were cultured in calcium phosphate‐coated plates and stimulated with F. alocis EVs (1, 3, or 10 μg/ml of protein; 0.6 ×, 1.8 ×, or 6 × 106 of EV particles/cell) in the presence of M‐CSF (30 ng/ml) for 10 days. Then, calcium phosphate was stained with Von Kossa reagents, and the representative images from triplicate samples are shown (left panel, Scale bar: 200 μm). The resorption area was measured from triplicate samples by ImageJ software (right panel). The graphs are shown as the mean values ± standard deviations. Representative data from three biological replicates are shown. Statistical significance was determined by one‐way ANOVA with Bonferroni's multiple comparison test. * P < 0.05 compared to the nontreatment group. (‐) denotes the nontreatment group
4.3. F. alocis EVs induce osteoclastogenesis via TLR2
TLRs are involved in the recognition of various MAMPs and the intracellular signalling and are expressed by osteoclast progenitors (Souza & Lerner, 2019). TLR2 is one of the major pattern recognition receptors that protects against infection by Gram‐positive pathogens (Takeuchi et al., 1999; Takeuchi et al., 2000). F. alocis activated p38 mitogen‐activated protein kinase (MAPK) in neutrophils through TLR2 signalling (Armstrong et al., 2016). We therefore analysed the involvement of TLR2 by stimulating the NF‐κB reporter cell lines CHO/CD14/TLR2 and CHO/CD14/TLR4 with F. alocis EVs. F. alocis EVs and F. alocis whole cells dose‐dependently activated TLR2 but not TLR4 (Figure 3a). To test whether TLR2 activation by F. alocis EVs is required for osteoclastogenesis, we prepared COCs from WT, TLR2–/– or TLR4–/– mice. As shown in Figure 3b, F. alocis EVs dose‐dependently induced TRAP‐positive MNCs in WT and TLR4–/– COCs but not in TLR2–/– COCs. As a control, E. coli LPS (a TLR4 ligand) and Pam2CSK4 (a TLR2 ligand) were used. In addition, F. alocis EVs dose‐dependently induced the mRNA expression of osteoclastogenic markers (TRAP, Cathepsin K, DC‐STAMP, and Atp6v0d2) in the WT COCs but not in the TLR2–/– COCs (Figure 3c). These results suggest that TLR2 is an essential receptor for F. alocis EV‐induced osteoclastogenesis I n vitro.
FIGURE 3.
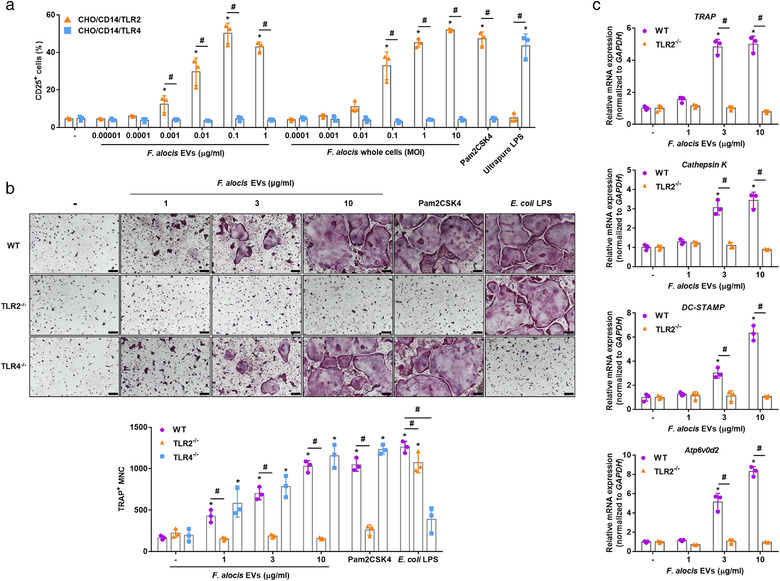
TLR2 signalling in osteoclasts induced by F. alocis EVs. (a) CHO/CD14/TLR2 or CHO/CD14/TLR4 cells were stimulated with the indicated treatments for 24 h. The expression of the TLR activation marker CD25 was measured by flow cytometry. Pam2CSK4 (100 ng/ml) and ultrapure LPS (100 ng/ml) were used as positive controls for TLR2 and TLR4 activation, respectively. Data are shown as the mean values ± standard deviations of triplicate samples. (b) WT, TLR2–/–, and TLR4–/– COCs were stimulated with F. alocis EVs (1, 3, or 10 μg/ml of protein; 0.6 ×, 1.8 ×, or 6 × 106 of EV particles/cell) in the presence of M‐CSF (30 ng/ml) for 2 days. Then, the cells were stained for TRAP activity, and the representative images of TRAP‐positive MNCs from triplicate samples are shown (upper panel, scale bar: 200 μm). The number of TRAP‐positive MNCs was counted with triplicate samples (lower panel). (c) WT and TLR2–/– COCs were stimulated with F. alocis EVs (1, 3, or 10 μg/ml of protein; 0.6 ×, 1.8 ×, or 6 × 106 of EV particles/cell) in the presence of M‐CSF (30 ng/ml) for 12 h and subjected to real‐time RT‐PCR to determine the mRNA expression levels of osteoclastogenic markers (TRAP, Cathepsin K, DC‐STAMP, and Atp6v0d2). The expression of osteoclastogenic markers was normalized to the expression of GAPDH. Relative expression of the genes was quantified between the nontreatment and treatment groups from the mean value of triplicate samples. The graphs are shown as the mean values ± standard deviations. Representative data from three biological replicates are shown. Statistical significance was determined by two‐way ANOVA with Bonferroni's multiple comparison test. * P < 0.05 compared to nontreatment group. # P < 0.05 compared to the indicated group. (‐) denotes the nontreatment group
4.4. F. alocis EVs activate transcription factors in committed osteoclast precursors via TLR2
TLR2 activation induces the expression of NFATc1 and c‐Fos (Kassem et al., 2016; Kim et al., 2013), which are master transcription factors for osteoclastogenesis in COCs (Lee & Kim, 2003). To confirm the involvement of TLR2 in the activation of osteoclastogenesis transcription factors by F. alocis EVs, WT or TLR2–/– COCs were stimulated with F. alocis EVs. In WT COCs, F. alocis EVs dose‐dependently induced the expression of NFATc1 and c‐Fos at both the protein and mRNA levels (Figure 4a and b). However, in TLR2–/– COCs, F. alocis EVs did not induce NFATc1 or c‐Fos. Likewise, F. alocis EVs markedly induced the phosphorylation of NF‐κB, ERK, JNK, and p38 in WT COCs but not in TLR2–/– COCs (Figure 4c). Inhibitors of ERK (U0126), JNK (SP600125), p38 (SB203580), or NF‐κB (Bay‐117082) reduced the expression of NFATc1 and c‐Fos in response to F. alocis EVs in COCs (Figure 4d). Furthermore, the inhibitors significantly reduced the number of TRAP‐positive MNCs induced by F. alocis EVs (Figure 4e). These results suggest that F. alocis EVs efficiently activate osteoclastogenesis transcription factors in COCs through the TLR2‐MAPKs/NF‐κB signalling pathway.
FIGURE 4.
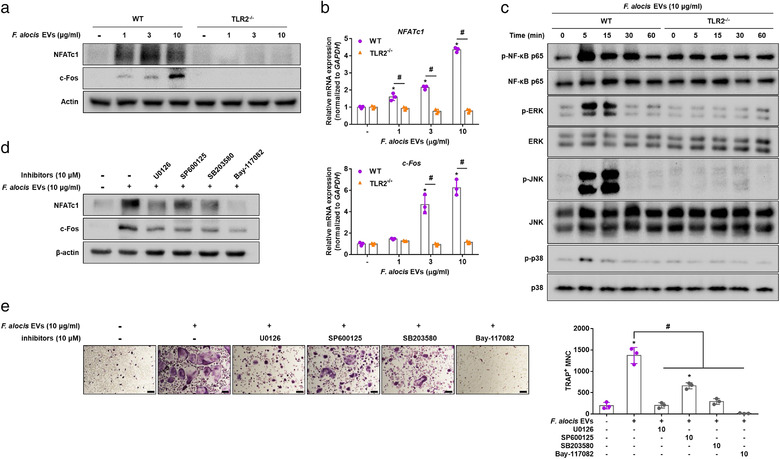
Activation of transcription factors by F. alocis EVs in COCs. (a, b) WT and TLR2–/– COCs were stimulated with F. alocis EVs (1, 3, or 10 μg/ml of protein; 0.12 ×, 0.36 ×, or 1.2 × 106 of EV particles/cell) in the presence of M‐CSF (30 ng/ml) for 12 h. The protein and mRNA expression levels of transcription factors (NFATc1 and c‐Fos) were measured by Western blotting (a) and real‐time RT‐PCR (b), respectively. (c) WT and TLR2–/– COCs were stimulated with F. alocis EVs (10 μg/ml of protein; 1.2 × 106 of EV particles/cell) for the indicated times and subjected to Western blotting to determine the phosphorylation levels of NF‐κB, ERK, p38 and JNK. (d, e) COCs were pre‐treated with U0126, SB203580, SP600125 or Bay‐117082 for 1 h. Then, the cells were stimulated with F. alocis EVs (10 μg/ml of protein; 1.2 × 106 of EV particles/cell) for 2 days. The protein expression levels of NFATc1 and c‐Fos were measured by Western blotting (d). The representative images of TRAP‐positive MNCs from triplicate samples are shown (e, left panel, scale bar: 200 μm). The number of TRAP‐positive MNCs was counted with triplicate samples (e, right panel). The graphs are shown as the mean values ± standard deviations. Representative data from the three biological replicates are shown. Statistical significances were determined by two‐way ANOVA with Bonferroni's multiple comparison test (b) or by one‐way ANOVA with Bonferroni's multiple comparison test (e). * P < 0.05 compared to the nontreatment group. # P < 0.05 compared to the indicated group. (‐) denotes the nontreatment group
4.5. F. alocis EVs induce bone resorption through TLR2 in vivo
Next, to examine the role of TLR2 in F. alocis EV‐induced bone resorption in vivo, we injected WT or TLR2–/– mice intraperitoneally with F. alocis EVs on Days 0 and 3 for seven days. As shown in Figure 5a and 5b, F. alocis EVs induced trabecular bone resorption in the femurs of WT mice but not TLR2–/– mice. Similarly, the histological analysis of femurs showed that F. alocis EVs significantly increased osteoclast surface/bone surface (Oc.S/BS) in WT mice but not in TLR2–/‐ mice (Figure 5c and d). The number of osteoclast/bone perimeter (N.Oc/B.Pm) showed a tendency to increase with F. alocis EVs treatment in WT mice (Figure 5d, p value = 0.0839), but not in TLR2–/‐ mice. We also measured serum levels of CTX‐1, a clinical marker of bone resorption, and P1NP, a clinical marker of bone formation (Kuo & Chen, 2017). F. alocis EVs increased serum levels of CTX‐1 in WT mice but not in TLR2–/‐ mice. P1NP expression was not affected by F. alocis EVs in either WT or TLR2–/‐ mice (Figure 5e). To further examine the role of TLR2 in F. alocis EV‐induced proinflammatory cytokine expression, we administered F. alocis EVs intraperitoneally to WT or TLR2–/– mice for 3 h. F. alocis EVs induced TNF‐α, IL‐6, and IL‐1β expression in the serum of WT mice but not TLR2–/‐ mice (Figure 5f). These results suggest that TLR2 stimulation by F. alocis EVs is a major cause of bone resorption, osteoclast differentiation, and bone‐resorptive cytokine expression in vivo.
FIGURE 5.
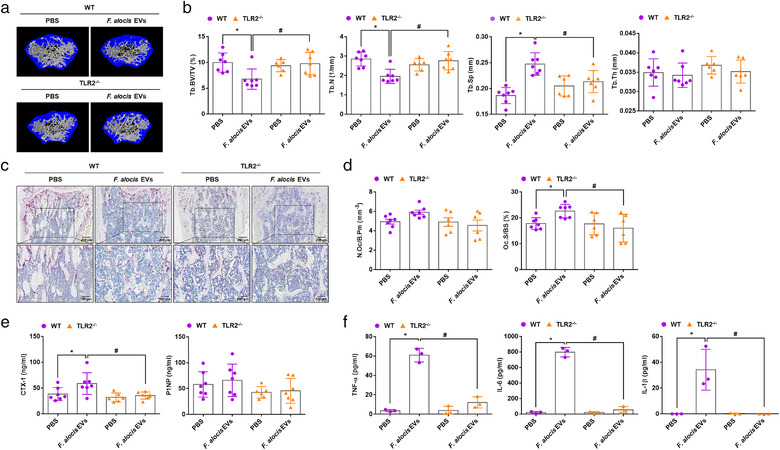
TLR2‐dependent bone resorption induced by F. alocis EVs. (a‐e) Eight‐week‐old male WT or TLR2–/– mice were intraperitoneally administered F. alocis EVs (50 μg of protein) on Days 0 and 3. On Day 7, the femurs were isolated, and trabecular bone was analysed by μCT. Representative 3D reconstruction images of femurs from PBS‐treated or F. alocis EV‐treated WT or TLR2–/– mice were obtained by μCT analysis (a). Tb.BV/TV, Tb.N, Tb.Sp, and Tb.Th were calculated by μCT analysis (b, n = 7 for WT‐PBS, WT‐F. alocis EVs, and TLR2–/–‐F. alocis EVs; n = 6 for TLR2–/–‐PBS). Decalcified femurs were embedded in paraffin, sectioned, and stained with haematoxylin and for TRAP (c). The number of osteoclast/bone parameter (N.Oc/B.Pm) and osteoclast surface/bone surface (Oc.S/BS) were measured in femoral sections (d, n = 7 for WT‐PBS and WT‐F. alocis EVs; n = 6 for TLR2–/–‐PBS and TLR2–/–‐F. alocis EVs). CTX‐1 and P1NP levels in mouse serum were analysed using ELISA (e). (f) Eight‐week‐old male mice (n = 3) were administered F. alocis EVs (50 μg of protein) for 3 h. TNF‐α, IL‐6, and IL‐1β in the serum were analysed using ELISA. The graphs are shown as the mean values ± standard deviations. Representative data from three biological replicates are shown. Statistical significance was determined by two‐way ANOVA with Bonferroni's multiple comparison test. * P < 0.05 compared to the PBS group. # P < 0.05 compared to the indicated group
4.6. Amphiphilic molecules in F. alocis EVs induce bone resorption
Various amphiphilic molecules from bacteria are known to be potent immune stimulators in the host environment (Chandler & Ernst, 2017). Among them, bacterial lipoproteins in Gram‐positive bacteria are known to be potent TLR2 stimulators (Oliveira‐Nascimento et al., 2012). To identify the active molecules in F. alocis EVs that induce TLR2 activation and bone resorption, F. alocis EVs were separated into amphiphilic and hydrophilic phases using Triton X‐114, a nonionic detergent that is used to enrich bacterial lipoproteins (Figure S2a). The molecular profiles of each fraction were analysed by silver staining (Figure S2b). As expected, the amphiphilic phase potently induced TLR2 activation in CHO/CD14/TLR2 reporter cells, whereas the hydrophilic phase did not (Figure S2c). Next, to evaluate the osteoclastogenic effects of the amphiphilic phase, COCs were stimulated with the amphiphilic phase or the hydrophilic phase. As shown in Figure 6a, only the amphiphilic phase dose‐dependently increased the number of TRAP‐positive MNCs, and the osteoclastogenic activity of the amphiphilic phase was dependent on TLR2. To further examine whether the amphiphilic phase of F. alocis EVs induced bone resorption in vivo, mice were intraperitoneally administered the F. alocis EV‐derived amphiphilic phase or hydrophilic phase on Days 0 and 3 for 7 days. Only the amphiphilic phase significantly induced trabecular bone resorption in vivo (Figure 6b). The amphiphilic phase significantly reduced the Tb.BV/TV and Tb.N in the mouse femurs. Interestingly, the hydrophilic phase significantly reduced Tb.Sp in the mouse femurs, but it did not affect Tb.BV/TV, Tb.N, or Tb.Th (Figure 6c). Next, to inactivate the bacterial lipoproteins in F. alocis EVs, F. alocis EVs were treated with lipoprotein lipase (0, 0.5, 5, or 50 μg/ml) at 37°C for 16 h. As shown in Figure 6d, F. alocis EVs treated with lipoprotein lipase exhibited significantly decreased osteoclastogenic potency. Pam3CSK4, a synthetic lipopeptide, treated with lipoprotein lipase also exhibited decreased osteoclastogenic potency.
FIGURE 6.
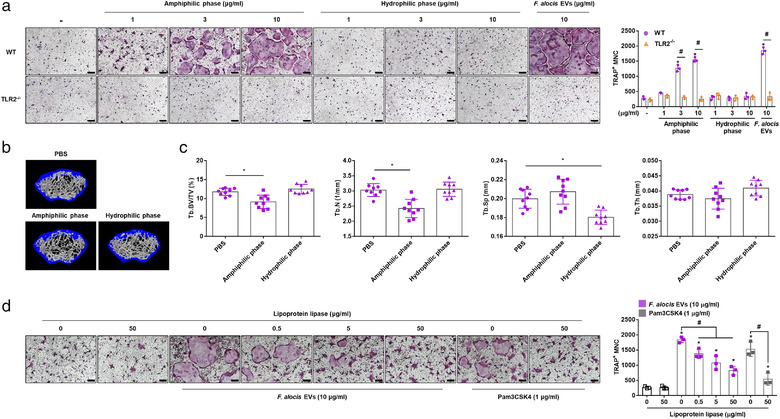
Bone resorption and osteoclastogenesis mediated by amphiphilic molecules in F. alocis EVs. (a) The WT and TLR2–/– COCs were stimulated with the indicated treatments in the presence of M‐CSF (30 ng/ml) for 2 days. Then, the cells were stained for TRAP activity, and representative images of TRAP‐positive MNCs from triplicate samples are shown (left panel, scale bar: 200 μm). The number of TRAP‐positive MNCs was counted with triplicate samples (right panel). (b, c) Eight‐week‐old male mice were administered the amphiphilic phase or hydrophilic phase (50 μg of protein) on Days 0 and 3. On Day 7, femurs were isolated, and trabecular bone was analysed by μCT. Representative 3D reconstruction images of femurs from PBS‐treated, amphiphilic phase‐treated or hydrophilic phase‐treated mice were analysed by μCT (b). Tb.BV/TV, Tb.N, Tb.Sp, and Tb.Th were calculated by μCT analysis (c, n = 9). (d) F. alocis EVs or Pam3CSK4 were incubated in the presence or absence of the indicated concentration of lipoprotein lipase at 37°C for 16 h. Then, COCs were stimulated with the indicated treatments in the presence of M‐CSF (30 ng/ml) for 2 days. Then, the cells were stained for TRAP activity, and representative images of TRAP‐positive MNCs from triplicate samples are shown (left panel, scale bar: 200 μm). The number of TRAP‐positive MNCs was counted with triplicate samples (right panel). The graphs are shown as the mean values ± standard deviations. Representative data from three biological replicates are shown. Statistical significance was determined by two‐way ANOVA with Bonferroni's multiple comparison test (a) or by one‐way ANOVA with Bonferroni's multiple comparison test (c, d). * P < 0.05 compared to the nontreatment group. # P < 0.05 compared to the indicated group. (‐) denotes the nontreatment group
LTA is also a TLR2 ligand with amphiphilic structure (Percy & Grundling, 2014). To determine the presence of LTA in F. alocis EVs, F. alocis EVs, hydrophilic phase, amphiphilic phase, and F. alocis cell lysates were analysed by Western blotting using anti‐Gram‐positive bacteria LTA antibody. F. alocis EVs and the amphiphilic phase harboured LTA (Figure S3a). Next, to identify the possible involvement of LTA in the osteoclastogenic effects of F. alocis EVs, we used PAF‐AH, which can specifically inactivate LTA by removing the acyl chain in the sn‐2 (nucleophilic substitution‐2) position of LTA without inactivation of bacterial lipoproteins (Seo & Nahm, 2009). We also used proteinase K and lipoprotein lipase, which can decrease the immunostimulatory potency of bacterial lipoproteins. As shown in Figure S3b, Western blotting showed that LTA disappeared from F. alocis EVs by PAF‐AH treatment. Lipoprotein lipase treatment also reduced the LTA amount, but proteinase K did not affect. Treatment with proteinase K significantly reduced the osteoclastogenic potency of F. alocis EVs to the same extent as treatment with lipoprotein lipase, whereas PAF‐AH treatment did not reduce the osteoclastogenic potency of F. alocis EVs (Figure S3c). Furthermore, we found seven lipoproteins in the amphiphilic phase of F. alocis EVs by LC–MS/MS (Table S1). The lipoproteins contain putative signal sequences composed of positively charged amino acid residues at the N‐terminus (n‐region), hydrophobic region (h‐region) and lipobox containing cysteine residue (c‐region; Babu et al., 2006). These results suggest that lipoproteins of F. alocis EVs may be the major TLR2 stimulators responsible for osteoclastogenesis and bone resorption.
5. DISCUSSION
Bacterial EVs have an advantage over the whole bacteria or soluble bacterial components released from bacteria in exerting physiological and pathological effects. EV components are protected by a lipid bilayer and can be transported for a long distance. Bacterial EVs can transfer bioactive molecules, including MAMPs and virulence factors, to distant host tissues or organs, leading to systemic proinflammatory responses. In the present study, we first demonstrated the role of periodontal pathogen‐derived EVs in systemic bone loss. F. alocis EVs increased osteoclastogenesis and bone resorption in vivo and in vitro. Bone integrity is maintained by continuous remodelling by bone‐forming osteoblasts and bone‐resorbing osteoclasts. In our previous study, F. alocis EVs exhibited immunostimulatory effects on human monocytes and oral keratinocytes similar to whole bacteria (Kim, Lim et al., 2020). We also reported that F. alocis EVs inhibited osteogenesis through TLR2 signalling in mouse bone‐derived mesenchymal stromal cells (Song et al., 2020). Taken together, these findings suggest that F. alocis EVs can accelerate bone loss by inhibiting bone formation and increasing bone resorption.
TLR2 signalling was common in reduction in osteogenesis and increase in osteoclastogenesis mediated by F. alocis EVs. F. alocis EVs accumulated in long bones after intraperitoneal administration in mice and increased serum levels of the osteoclast‐activating cytokines TNF‐α, IL‐6, and IL‐1β in vivo via TLR2 signalling. Lipoprotein lipase‐sensitive amphiphilic molecules in F. alocis EVs were responsible for osteoclastogenesis and bone resorption via TLR2. In this study, we identified seven lipoproteins in the amphiphilic phase of F. alocis EVs, of which four (TRAP transporter solute receptor, hypothetical protein HMPREF0389_01610, basic membrane protein, lipoprotein YaeC family) were also identified with whole F. alocis EVs in our previous study (Kim, Lim et al., 2020). Lipoproteins and LTA are amphiphilic molecules known to be representative TLR2 ligands in Gram‐positive bacteria. However, recent studies have shown that lipoproteins but not LTA are predominant TLR2 ligands that are responsible for immunostimulatory activity (Hashimoto et al., 2006b; Hashimoto et al., 2006a; Kim et al., 2018). Lipoprotein‐deficient S. aureus did not induce bone resorption, while the synthetic bacterial lipopeptides Pam2CSK4 and Pam3CSK4 efficiently induced bone resorption and osteoclast differentiation (Kim et al., 2013). EVs isolated from lipoprotein‐deficient S. aureus abrogated TLR2 signalling in human macrophages, resulting in reduction in IL‐1β, IL‐18, and IL‐6 expression levels (Wang et al., 2020). The EVs released from Mycobacterium tuberculosis were enriched in lipoproteins, and intratracheal injection of these EVs in mice evoked inflammatory responses in the lung in a TLR2‐dependent manner (Prados‐Rosales et al., 2011). Intratracheal injection of M. tuberculosis EVs before bacterial infection resulted in larger and more numerous granulomas than those in the controls not pre‐treated with EVs. We also observed that lipoprotein lipase and proteinase K treatment decreased the osteoclastogenic potency of F. alocis EVs, but PAF‐AH, an LTA inhibitor, did not. Based on these results, we hypothesize that lipoproteins in bacterial EVs are TLR2‐activating molecules primarily responsible for the induction of osteoclastogenesis and bone resorption.
The microbiota is closely associated with the homeostasis of host tissues, including bone (Sommer & Bäckhed, 2013). Microbiota‐derived molecules such as LPS, peptidoglycan, and short‐chain fatty acids (SCFAs) have been shown to directly modulate bone metabolism (Li et al., 2019). LPS, a representative TLR4 ligand derived from Gram‐negative bacteria, induced bone resorption and osteoclast differentiation (Mian et al., 2008). Peptidoglycan plays an important role in gut microbiota‐mediated TNF‐α and RANKL expression and bone resorption via nucleotide‐binding oligomerization domain 1 or 2 (Ohlsson et al., 2017). SCFAs, the main metabolites in the colon generated from bacterial fermentation of dietary fibres, increased bone mineral density by inhibiting osteoclast differentiation and bone resorption (Lucas et al., 2018). A recent study showed that EVs derived from gut microbiota enriched in Akkermansia muciniphila from healthy children showed suppressive effects on osteoclastogenesis and protective effects against osteoporosis in ovariectomized mice (Liu et al., 2021). A. muciniphila is a promising probiotic that also suppresses P. gingivalis‐induced osteoclast activation and bone resorption (Huck et al., 2020).
Periodontal pathogens include multiple species, and most of the species are Gram‐negative bacteria that release EVs called outer membrane vesicles (OMVs) through outer membrane blebbing. Recently, the immunostimulatory activities of periodontal pathogen‐derived EVs were demonstrated. ‘Red complex’ periodontal pathogens, which are known to be highly pathogenic and include P. gingivalis, T. denticola, and T. forsythia, release OMVs that efficiently activate TLRs (Cecil et al., 2016) and induce TNF‐α and IL‐1β in monocytes and macrophages (Cecil et al., 2017). A. actinomycetemcomitans OMVs contain proteases and leukotoxin (Nice et al., 2018), and intracardiac injection of OMVs showed penetration of the blood–brain barrier and induction of TNF‐α in the brains of mice (Han et al., 2019). Compared to Gram‐negative periodontal pathogens, Gram‐positive F. alocis has been relatively recently identified as an important periodontal pathogen. F. alocis showed increased invasion of HeLa cells and enhanced biofilm formation in the presence of P. gingivalis, a key pathogen (Aruni et al., 2011). In periodontitis, EVs derived from various oral bacteria may enter the bloodstream and have synergistic effects on systemic bone resorption. In the present study, we administered F. alocis EVs intraperitoneally to mice to mimic the systemic circulation of oral bacteria‐derived EVs. Therefore, further studies are needed to analyse the effects of EVs isolated from multiple bacteria, including pathogens and/or commensals, on systemic bone metabolism through various routes of administration, such as oral and intravenous administration, and to establish the causal relationship between periodontitis and systemic bone diseases such as osteoporosis.
In summary, we demonstrated that EVs derived from the Gram‐positive periodontal pathogen F. alocis significantly induced osteoclast differentiation and bone resorption in vivo and in vitro via TLR2. Amphiphilic molecules sensitive to lipoprotein lipase were primarily responsible for these activities via TLR2. Seven lipoproteins were identified in the amphiphilic phase from F. alocis EVs. For the first time, our results suggest that periodontal pathogens may affect systemic bone metabolism through circulating EVs in body fluids.
CONFLICT OF INTEREST
The authors have no competing financial interests to declare.
Supporting information
Supporting information.
ACKNOWLEDGEMENTS
This work was supported by grants from the National Research Foundation of Korea (NRF‐ 2021R1A2C1003952, NRF‐2020R1A2C2010082, and NRF‐2018R1A5A2024418) and the Dental Research Institute of Seoul National University.
Kim, H. Y. , Song, M.‐K. , Gho, Y. S. , Kim, H.‐H. , & Choi, B.‐K. (2021). Extracellular vesicles derived from the periodontal pathogen Filifactor alocis induce systemic bone loss through toll‐like receptor 2. Journal of Extracellular Vesicles, 10, e12157. 10.1002/jev2.12157
Hyun Young Kim and Min‐Kyoung Song contributed equally to this study.
Contributor Information
Hong‐Hee Kim, Email: hhbkim@snu.ac.kr.
Bong‐Kyu Choi, Email: bongchoi@snu.ac.kr.
REFERENCES
- Armbruster, K. M. , & Meredith, T. C. (2018). Enrichment of bacterial lipoproteins and preparation of N‐terminal lipopeptides for structural determination by mass spectrometry. Journal of Visualized Experiments, 21(135), 56842. 10.3791/56842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong, C. L. , Miralda, I. , Neff, A. C. , Tian, S. , Vashishta, A. , Perez, L. , Le, J. , Lamont, R. J. , & Uriarte, S. M. (2016). Filifactor alocis promotes neutrophil degranulation and chemotactic activity. Infection and Immunity, 84(12), 3423–3433. 10.1128/IAI.00496-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aruni, A. W. , Roy, F. , & Fletcher, H. M. (2011). Filifactor alocis has virulence attributes that can enhance its persistence under oxidative stress conditions and mediate invasion of epithelial cells by Porphyromonas gingivalis . Infection and Immunity, 79(10), 3872–3886. 10.1128/Iai.05631-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aruni, W. , Chioma, O. , & Fletcher, H. M. (2014). Filifactor alocis: The newly discovered kid on the block with special talents. Journal of Dental Research, 93(8), 725–732. 10.1177/0022034514538283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma, Y. , Kaji, K. , Katogi, R. , Takeshita, S. , & Kudo, A. (2000). Tumor necrosis factor‐α induces differentiation of and bone resorption by osteoclasts. Journal of Biological Chemistry, 275(7), 4858–4864. 10.1074/jbc.275.7.4858 [DOI] [PubMed] [Google Scholar]
- Babu, M. M. , Priya, M. L. , Selvan, A. T. , Madera, M. , Gough, J. , Aravind, L. , & Sankaran, K. (2006). A database of bacterial lipoproteins (DOLOP) with functional assignments to predicted lipoproteins. Journal of Bacteriology, 188(8), 2761–2773. 10.1128/Jb.188.8.2761-2773.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachurski, D. , Schuldner, M. , Nguyen, P. H. , Malz, A. , Reiners, K. S. , Grenzi, P. C. , Babatz, F. , Schauss, A. C. , Hansen, H. P. , Hallek, M. , & von Strandmann, E. P. (2019). Extracellular vesicle measurements with nanoparticle tracking analysis ‐ An accuracy and repeatability comparison between NanoSight NS300 and ZetaView. Journal of Extracellular Vesicles, 8(1). 1596016. 10.1080/20013078.2019.1596016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitto, N. J. , Cheng, L. , Johnston, E. L. , Pathirana, R. , Phan, T. K. , Poon, I. K. H. , O'Brien‐Simpson, N. M. , Hill, A. F. , Stinear, T. P. , & Kaparakis‐Liaskos, M. (2021). Staphylococcus aureus membrane vesicles contain immunostimulatory DNA, RNA and peptidoglycan that activate innate immune receptors and induce autophagy. Journal of Extracellular Vesicles, 10(6). e12080. 10.1002/jev2.12080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle, W. J. , Simonet, W. S. , & Lacey, D. L. (2003). Osteoclast differentiation and activation. Nature, 423(6937), 337–342. 10.1038/nature01658 [DOI] [PubMed] [Google Scholar]
- Cecil, J. D. , O'Brien‐Simpson, N. M. , Lenzo, J. C. , Holden, J. A. , Chen, Y. Y. , Singleton, W. , Gause, T. K. , Yan, Y. , Caruso, F. , & Reynolds, E. C. (2016). Differential responses of pattern recognition receptors to outer membrane vesicles of three periodontal pathogens. PLoS One, 11(4), e0151967. 10.1371/journal.pone.0151967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecil, J. D. , O'Brien‐Simpson, N. M. , Lenzo, J. C. , Holden, J. A. , Singleton, W. , Perez‐Gonzalez, A. , Mansell, A. , & Reynolds, E. C. (2017). Outer membrane vesicles prime and activate macrophage inflammasomes and cytokine secretion in vitro and in vivo. Frontiers in Immunology, 8, 1017. 10.3389/fimmu.2017.01017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler, C. E. , & Ernst, R. K. (2017). Bacterial lipids: Powerful modifiers of the innate immune response. F1000Research, 6(F1000 Faculty Rev), 1334. 10.12688/f1000research.11388.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, E. J. , Ha, J. , Oerlemans, F. , Lee, Y. J. , Lee, S. W. , Ryu, J. , Kim, H. J. , Lee, Y. , Kim, H. M. , Choi, J. Y. , Kim, J. Y. , Shin, C. S. , Pak, Y. K. , Tanaka, S. , Wieringa, B. , Lee, Z. H. , & Kim, H. H. (2008). Brain‐type creatine kinase has a crucial role in osteoclast‐mediated bone resorption. Nature Medicine, 14(9), 966–972. 10.1038/nm.1860 [DOI] [PubMed] [Google Scholar]
- Choi, H. I. , Choi, J. P. , Seo, J. , Kim, B. J. , Rho, M. , Han, J. K. , & Kim, J. G. (2017). Helicobacter pylori‐derived extracellular vesicles increased in the gastric juices of gastric adenocarcinoma patients and induced inflammation mainly via specific targeting of gastric epithelial cells. Experimental & Molecular Medicine, 49(5), e330. 10.1038/emm.2017.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contaldo, M. , Itro, A. , Lajolo, C. , Gioco, G. , Inchingolo, F. , & Serpico, R. (2020). Overview on osteoporosis, periodontitis and oral dysbiosis: The emerging role of oral microbiota. Applied Sciences, 10(17). 6000. 10.3390/app10176000 [DOI] [Google Scholar]
- Dempster, D. W. , Compston, J. E. , Drezner, M. K. , Glorieux, F. H. , Kanis, J. A. , Malluche, H. , Meunier, P. J. , Ott, S. M. , Recker, R. R. , & Parfitt, A. M. (2013). Standardized nomenclature, symbols, and units for bone histomorphometry: A 2012 update of the report of the ASBMR Histomorphometry Nomenclature Committee. Journal of Bone and Mineral Research, 28(1), 1–16. 10.1002/jbmr.1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorozhkin, S. V. , & Epple, M. (2002). Biological and medical significance of calcium phosphates. Angewandte Chemie‐International Edition, 41(17), 3130–3146. 10.1002/1521-3773(20020902)41:17<3130::Aid‐Anie3130>3.0.Co;2‐1 [DOI] [PubMed] [Google Scholar]
- Edmisson, J. S. , Tian, S. , Armstrong, C. L. , Vashishta, A. , Klaes, C. K. , Miralda, I. , Jimenez‐Flores, E. , Le, J. , Wang, Q. , Lamont, R. J. , & Uriarte, S. M. (2018). Filifactor alocis modulates human neutrophil antimicrobial functional responses. Cellular Microbiology, 20(6). e12829. 10.1111/cmi.12829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine, D. H. , Markowitz, K. , Fairlie, K. , Tischio‐Bereski, D. , Ferrendiz, J. , Furgang, D. , Paster, B. J. , & Dewhirst, F. E. (2013). A consortium of Aggregatibacter actinomycetemcomitans, Streptococcus parasanguinis, and Filifactor alocis is present in sites prior to bone loss in a longitudinal study of localized aggressive periodontitis. Journal of Clinical Microbiology, 51(9), 2850–2861. 10.1128/Jcm.00729-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gho, Y. S. , & Lee, C. (2017). Emergent properties of extracellular vesicles: A holistic approach to decode the complexity of intercellular communication networks. Molecular Biosystems, 13(7), 1291–1296. 10.1039/c7mb00146k [DOI] [PubMed] [Google Scholar]
- Gray, R. M. , & Vidwans, M. (2019). Mixed anaerobic thoracic empyema: The first report of Filifactor alocis causing extra‐oral disease. New Microbes and New Infections, 29. 100528. 10.1016/j.nmni.2019.100528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffen, A. L. , Beall, C. J. , Campbell, J. H. , Firestone, N. D. , Kumar, P. S. , Yang, Z. K. , Podar, M. , & Leys, E. J. (2012). Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME Journal, 6(6), 1176–1185. 10.1038/ismej.2011.191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis, G. , & Chavakis, T. (2021). Local and systemic mechanisms linking periodontal disease and inflammatory comorbidities. Nature Reviews Immunology. 28, 1–15. 10.1038/s41577-020-00488-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, E. C. , Choi, S. Y. , Lee, Y. , Park, J. W. , Hong, S. H. , & Lee, H. J. (2019). Extracellular RNAs in periodontopathogenic outer membrane vesicles promote TNF‐α production in human macrophages and cross the blood‐brain barrier in mice. FASEB Journal, 33(12), 13412–13422. 10.1096/fj.201901575R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto, M. , Tawaratsumida, K. , Kariya, H. , Aoyama, K. , Tamura, T. , & Suda, Y. (2006a). Lipoprotein is a predominant Toll‐like receptor 2 ligand in Staphylococcus aureus cell wall components. International Immunology, 18(2), 355–362. 10.1093/intimm/dxh374 [DOI] [PubMed] [Google Scholar]
- Hashimoto, M. , Tawaratsumida, K. , Kariya, H. , Kiyohara, A. , Suda, Y. , Krikae, F. , Kirikae, T. , & Götz, F. (2006b). Not lipoteichoic acid but lipoproteins appear to be the dominant immunobiologically active compounds in Staphylococcus aureus . Journal of Immunology, 177(5), 3162–3169. 10.4049/jimmunol.177.5.3162 [DOI] [PubMed] [Google Scholar]
- Hienz, S. A. , Paliwal, S. , & Ivanovski, S. (2015). Mechanisms of bone resorption in periodontitis. Journal of Immunological Research, 2015, 615486. 10.1155/2015/615486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hishiya, N. , Uno, K. , Amano, M. , Asada, K. , Masui, K. , Ishida, Y. , Suzuki, Y. , Hirai, N. , Nakano, A. , Nakano, R. , Kasahara, K. , Yano, H. , & Mikasa, K. (2020). Filifactor alocis brain abscess identified by 16S ribosomal RNA gene sequencing: A case report. Journal of Infection and Chemotherapy, 26(2), 305–307. 10.1016/j.jiac.2019.09.013 [DOI] [PubMed] [Google Scholar]
- Huck, O. , Mulhall, H. , Rubin, G. , Kizelnik, Z. , Iyer, R. , Perpich, J. D. , Haque, N. , Cani, P. D. , de Vos, W. M. , & Amar, S. (2020). Akkermansia muciniphila reduces Porphyromonas gingivalis‐induced inflammation and periodontal bone destruction. Journal of Clinical Periodontology, 47(2), 202–212. 10.1111/jcpe.13214 [DOI] [PubMed] [Google Scholar]
- Ishimi, Y. , Miyaura, C. , Jin, C. H. , Akatsu, T. , Abe, E. , Nakamura, Y. , Yamaguchi, A. , Yoshiki, S. , Matsuda, T. , & Hirano, T. (1990). IL‐6 is produced by osteoblasts and induces bone resorption. Journal of Immunology, 145(10), 3297–3303. http://www.jimmunol.org/content/145/10/3297 [PubMed] [Google Scholar]
- Jimi, E. , Nakamura, I. , Duong, L. T. , Ikebe, T. , Takahashi, N. , Rodan, G. A. , & Suda, T. (1999). Interleukin 1 induces multinucleation and bone‐resorbing activity of osteoclasts in the absence of osteoblasts/stromal cells. Experimental Cell Research, 247(1), 84–93. 10.1006/excr.1998.4320 [DOI] [PubMed] [Google Scholar]
- Jusko, M. , Miedziak, B. , Ermert, D. , Magda, M. , King, B. C. , Bielecka, E. , Riesbeck, K. , Eick, S. , Potempa, J. , & Blom, A. M. (2016). FACIN, a double‐edged sword of the emerging periodontal pathogen Filifactor alocis: A metabolic enzyme moonlighting as a complement inhibitor. Journal of Immunology, 197(8), 3245–3259. 10.4049/jimmunol.1600739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassem, A. , Lindholm, C. , & Lerner, U. H. (2016). Toll‐like receptor 2 stimulation of osteoblasts mediates Staphylococcus aureus induced bone resorption and osteoclastogenesis through enhanced RANKL. PLoS One, 11(6). e156708. 10.1371/journal.pone.0156708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosla, S. (2001). Minireview: The OPG/RANKL/RANK system. Endocrinology, 142(12), 5050–5055. 10.1210/en.142.12.5050 [DOI] [PubMed] [Google Scholar]
- Kim, H. Y. , Kim, A. R. , Seo, H. S. , Baik, J. E. , Ahn, K. B. , Yun, C. H. , & Han, S. H. (2018). Lipoproteins in Streptococcus gordonii are critical in the infection and inflammatory responses. Molecular Immunology, 101, 574–584. 10.1016/j.molimm.2018.08.023 [DOI] [PubMed] [Google Scholar]
- Kim, H. Y. , Lim, Y. , An, S. J. , & Choi, B. K. (2020a). Characterization and immunostimulatory activity of extracellular vesicles from Filifactor alocis . Molecular Oral Microbiology, 35(1), 1–9. 10.1111/omi.12272 [DOI] [PubMed] [Google Scholar]
- Kim, J. H. , Lee, J. , Park, J. , & Gho, Y. S. (2015). Gram‐negative and Gram‐positive bacterial extracellular vesicles. Seminars in Cell & Developmental Biology, 40, 97–104. 10.1016/j.semcdb.2015.02.006 [DOI] [PubMed] [Google Scholar]
- Kim, J. , Yang, J. , Park, O. J. , Kang, S. S. , Kim, W. S. , Kurokawa, K. , Yun, C. H. , Kim, H. H. , Lee, B. L. , & Han, S. H. (2013). Lipoproteins are an important bacterial component responsible for bone destruction through the induction of osteoclast differentiation and activation. Journal of Bone and Mineral Research, 28(11), 2381–2391. 10.1002/jbmr.1973 [DOI] [PubMed] [Google Scholar]
- Kim, W. , Lee, E. J. , Bae, I. H. , Myoung, K. , Kim, S. T. , Park, P. J. , Lee, K. , Quynh Pham, A. V. , Ko, J. , Oh, S. H. , & Cho, E. G. (2020b). Lactobacillus plantarum‐derived extracellular vesicles induce anti‐inflammatory M2 macrophage polarization in vitro. Journal of Extracellular Vesicles, 9(1), 1793514. 10.1080/20013078.2020.1793514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuipers, M. E. , Hokke, C. H. , Smits, H. H. , & Nolte‐’t Hoen, E. N. M. (2018). Pathogen‐derived extracellular vesicle‐associated molecules that affect the host immune system: An overview. Frontiers in Microbiology, 12(9), 2182. 10.3389/fmicb.2018.02182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo, T. R. , & Chen, C. H. (2017). Bone biomarker for the clinical assessment of osteoporosis: Recent developments and future perspectives. Biomarker Research, 18(5), 18. 10.1186/s40364-017-0097-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H. J. , Na, K. , Choi, E. Y. , Kim, K. S. , Kim, H. , & Paik, Y. K. (2010). Simple method for quantitative analysis of N‐linked glycoproteins in hepatocellular carcinoma specimens. Journal of Proteome Research, 9(1), 308–318. 10.1021/pr900649b [DOI] [PubMed] [Google Scholar]
- Lee, Z. H. , & Kim, H. H. (2003). Signal transduction by receptor activator of nuclear factor kappa B in osteoclasts. Biochemical and Biophysical Research Communications, 305(2), 211–214. 10.1016/S0006-291x(03)00695-8 [DOI] [PubMed] [Google Scholar]
- Li, L. S. , Rao, S. , Cheng, Y. , Zhuo, X. , Deng, C. , Xu, N. , Zhang, H. , & Yang, L. (2019). Microbial osteoporosis: The interplay between the gut microbiota and bones via host metabolism and immunity. Microbiologyopen, 8(8). e00810. 10.1002/mbo3.810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. H. , Chen, C. Y. , Liu, Z. Z. , Luo, Z. W. , Rao, S. S. , Jin, L. , Wan, T. F. , Yue, T. , Tan, Y. J. , Yin, H. , Yang, F. , Huang, F. Y. , Guo, J. , Wang, Y. Y. , Xia, K. , Cao, J. , Wang, Z. X. , Hong, C. G. , Luo, M. J. … Xie, H. (2021). Extracellular vesicles from child gut microbiota enter into bone to preserve bone mass and strength. Advanced Science. 8(9), 2004831. 10.1002/advs.202004831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas, S. , Omata, Y. , Hofmann, J. , Böttcher, M. , Iljazovic, A. , Sarter, K. , Albrecht, O. , Schulz, O. , Krishnacoumar, B. , Krönke, G. , Herrmann, M. , Mougiakakos, D. , Strowing, T. , Schett, G. , & Zaiss, M. M. (2018). Short‐chain fatty acids regulate systemic bone mass and protect from pathological bone loss. Nature Communications, 9(1), 55. 10.1038/s41467-017-02490-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjunath, S. H. , Rakhewar, P. , Nahar, P. , Tambe, V. , Gabhane, M. , & Kharde, A. (2019). Evaluation of the prevalence and severity of periodontal diseases between osteoporotic and nonosteoporotic subjects: A cross‐sectional comparative study. The Journal of Contemporary Dental Practice, 20(10), 1223–1228. https://www.thejcdp.com/doi/JCDP/pdf/10.5005/jp‐journals‐10024‐2717 [PubMed] [Google Scholar]
- Mian, A. H. , Saito, H. , Alles, N. , Shimokawa, H. , Aoki, K. , & Ohya, K. (2008). Lipopolysaccharide‐induced bone resorption is increased in TNF type 2 receptor‐deficient mice In vivo. Journal of Bone and Mineral Metabolism, 26(5), 469–477. 10.1007/s00774-007-0834-0 [DOI] [PubMed] [Google Scholar]
- Namork, E. , & Brandtzaeg, P. (2002). Fatal meningococcal septicaemia with “blebbing” meningococcus. Lancet, 360(9347), 1741. 10.1016/S0140-6736(02)11721-1 [DOI] [PubMed] [Google Scholar]
- Nice, J. B. , Balashova, N. V. , Kachlany, S. C. , Koufos, E. , Krueger, E. , Lally, E. T. , & Brown, A. C. (2018). Aggregatibacter actinomycetemcomitans leukotoxin is delivered to host cells in an LFA‐1‐indepdendent manner when associated with outer membrane vesicles. Toxins, 10(10). 414. 10.3390/toxins10100414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson, C. , Nigro, G. , Boneca, I. G. , Bäckhed, F. , Sansonetti, P. , & Sjögren, K. (2017). Regulation of bone mass by the gut microbiota is dependent on NOD1 and NOD2 signaling. Cellular Immunology, 317, 55–58. 10.1016/j.cellimm.2017.05.003 [DOI] [PubMed] [Google Scholar]
- Oliveira‐Nascimento, L. , Massari, P. , & Wetzler, L. M. (2012). The role of TLR2 in infection and immunity. Frontiers in Immunology, 18(3), 79. 10.3389/fimmu.2012.00079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, W. , Wang, Q. , & Chen, Q. (2019). The cytokine network involved in the host immune response to periodontitis. International Journal of Oral Science, 11(3), 30. 10.1038/s41368-019-0064-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, K. S. , Choi, K. H. , Kim, Y. S. , Hong, B. S. , Kim, O. Y. , Kim, J. H. , Yoon, C. M. , Koh, G. Y. , Kim, Y. K. , & Gho, Y. S. (2010). Outer membrane vesicles derived from Escherichia coli induce systemic inflammatory response syndrome. Plos One, 5(6). e11334. 10.1371/journal.pone.0011334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passos, J. S. , Vianna, M. I. P. , Gomes, I. S. , Cruz, S. S. , Barreto, M. L. , Adan, L. , Rösing, C. K. , Cerqueira, E. M. M. , Trindade, S. C. , & Coelho, J. M. F. (2013). Osteoporosis/osteopenia as an independent factor associated with periodontitis in postmenopausal women: A case‐control study. Osteoporosis International, 24(4), 1275–1283. 10.1007/s00198-012-2130-7 [DOI] [PubMed] [Google Scholar]
- Percy, M. G. , & Grundling, A. (2014). Lipoteichoic acid synthesis and function in Gram‐positive bacteria. Annual Review of Microbiology, 68, 81–100. 10.1146/annurev-micro-091213-112949 [DOI] [PubMed] [Google Scholar]
- Prados‐Rosales, R. , Baena, A. , Martinez, L. R. , Luque‐Garcia, J. , Kalscheuer, R. , Veeraraghavan, U. , Camara, C. , Nosanchuk, J. D. , Besra, G. S. , Chen, B. , Jimenez, J. , Glatman‐Freedman, A. , Jacobs, W. R. , Porcelli, S. A. , & Casadevall, A. (2011). Mycobacteria release active membrane vesicles that modulate immune responses in a TLR2‐dependent manner in mice. Journal of Clinical Investigation, 121(4), 1471–1483. 10.1172/Jci44261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, H. S. , & Nahm, M. H. (2009). Lipoprotein lipase and hydrofluoric acid deactivate both bacterial lipoproteins and lipoteichoic acids, but platelet‐activating factor‐acetylhydrolase degrades only lipoteichoic acids. Clinical and Vaccine Immunology, 16(8), 1187–1195. 10.1128/Cvi.00115-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer, F. , & Bäckhed, F. (2013). The gut microbiota‐masters of host development and physiology. Nature Reviews Microbiology, 11(4), 227–238. 10.1038/nrmicro2974 [DOI] [PubMed] [Google Scholar]
- Song, M. K. , Lee, Z. H. , & Kim, H. H. (2015). Adseverin mediates RANKL‐induced osteoclastogenesis by regulating NFATc1. Experimental & Molecular Medicine, 47(12), e199. 10.1038/emm.2015.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, M. K. , Kim, H. Y. , Choi, B. K. , & Kim, H. H. (2020). Filifactor alocis‐derived extracellular vesicles inhibit osteogenesis through TLR2 signaling. Molecular Oral Microbiology, 35(5), 202–210. 10.1111/omi.12307 [DOI] [PubMed] [Google Scholar]
- Song, M. K. , Park, C. , Lee, Y. D. , Kim, H. , Kim, M. K. , Kwon, J. O. , Koo, J. H. , Joo, M. S. , Kim, S. G. , & Kim, H. H. (2018). Gα12 regulates osteoclastogenesis by modulating NFATc1 expression. Journal of Cellular and Molecular Medicine, 22(2), 849–860. 10.1111/jcmm.13370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza, P. P. C. , & Lerner, U. H. (2019). Finding a toll on the route: The fate of osteoclast progenitors after toll‐like receptor activation. Frontiers in Immunology, 17(10), 1663. 10.3389/fimmu.2019.01663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi, O. , Hoshino, K. , & Akira, S. (2000). Cutting edge: TLR2‐deficient and MyD88‐deficient mice are highly susceptible to Staphylococcus aureus infection. Journal of Immunology, 165(10), 5392–5396. 10.4049/jimmunol.165.10.5392 [DOI] [PubMed] [Google Scholar]
- Takeuchi, O. , Hoshino, K. , Kawai, T. , Sanjo, H. , Takada, H. , Ogawa, T. , Takeda, K. , & Akira, S. (1999). Differential roles of TLR2 and TLR4 in recognition of Gram‐negative and Gram‐positive bacterial cell wall components. Immunity, 11(4), 443–451. 10.1016/S1074-7613(00)80119-3 [DOI] [PubMed] [Google Scholar]
- Tanaka, S. , Nakamura, K. , Takahasi, N. , & Suda, T. (2005). Role of RANKL in physiological and pathological bone resorption and therapeutics targeting the RANKL‐RANK signaling system. Immunological Reviews, 208, 30–49. 10.1111/j.0105-2896.2005.00327.x [DOI] [PubMed] [Google Scholar]
- Teitelbaum, S. L. (2000). Bone resorption by osteoclasts. Science, 289(5484), 1504–1508. 10.1126/science.289.5484.1504 [DOI] [PubMed] [Google Scholar]
- Wang, X. , Eagen, W. J. , & Lee, J. C. (2020). Orchestration of human macrophage NLRP3 inflammasome activation by Staphylococcus aureus extracellular vesicles. Proceedings of the National Academy of Sciences of the United States of America, 117(6), 3174–3184. 10.1073/pnas.1915829117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber, J. , & Clayton, A. (2013). How pure are your vesicles? Journal of Extracellular Vesicles, 10(2), 19861. 10.3402/jev.v2i0.19861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, S. , Zhang, G. , Guo, J. F. , & Tan, Y. H. (2021). Associations between osteoporosis and risk of periodontitis: A pooled analysis of observational studies. Oral Diseases, 27(2), 357–369. 10.1111/odi.13531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yáñez‐Mó, M. , Siljander, P. R. M. , Andreu, Z. , Zavec, A. B. , Borràs, F. E. , Buzas, E. I. , Buzas, K. , Casal, E. , Cappello, F. , Carvalho, J. , Colás, E. , Silva, Cordeiro‐da , A, Fais , S, Falcon‐Perez , J, M. , Ghobrial, I. M. , Giebel, B. , Gimona, M. , Graner, M. … De Wever, O. (2015). Biological properties of extracellular vesicles and their physiological functions. Journal of Extracellular Vesicles, 14(4), 27066. 10.3402/jev.v4.27066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, W. , & Bar‐Shavit, Z. (2002). Dual modulation of osteoclast differentiation by lipopolysaccharide. Journal of Bone and Mineral Research, 17(7), 1211–1218. 10.1359/jbmr.2002.17.7.1211 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.


