Abstract
Background
Various reports have now established that postoperative endoscopy to examine and intervene in the process of anastomotic healing is both feasible and safe. Here we present our preliminary experience with serial postoperative endoscopy to determine its feasibility, patient acceptance and the ability to obtain and the utility of perianastomotic material for molecular analysis.
Methods
Patients undergoing LAR with ileostomy for rectal cancer were recruited for study to undergo routine serial endoscopic surveillance (SES) at three time points during the course of LAR: intraoperatively, before discharge (postoperative day 3–7) and at follow-up (postoperative day 10–28). At each endoscopy, images were captured, anastomotic tissues were lavaged and lavage fluid was retrieved. Fluid samples were analyzed using proteomics, zymography, ELISA and bacteria via 16S rRNA gene amplicon sequencing and culture of collagenolytic strains.
Results
SES is feasible and acceptable to this limited set of patients following LAR. Biologic analysis of perianastomotic fluids was able to detect the presence of proteins, microbiota and inflammatory mediators previously identified at anastomotic sites in animals with pathologic healing.
Conclusion
SES can be implemented in patients undergoing LAR with a high degree of patient compliance and capture of biologic information and imaging. Application of this approach has the potential to uncover, for the first time, the natural history of normal versus pathologic anastomotic healing in patients undergoing anastomotic surgery.
Introduction
Despite the fact that anastomotic leak (AL) remains the most morbid and lethal complication following gastrointestinal surgery, the standard approach to “wait and watch” for symptoms severe enough to elicit imaging and treatment might be reconsidered given that today, patients still become disabled from, and die of, anastomotic leak [1,2,3,4,5].
Surprisingly, the natural history of human anastomotic healing remains unknown as it would require direct serial visualization and a molecular-level analysis of anastomotic tissues as they heal. Although advanced techniques are being used to image other surgical sites such as the biliary tree [6] and to assess intestinal blood via endoscopy during surgery [7], reluctance remains toward postoperative endoscopy as such an approach is considered risky, invasive, uncomfortable, costly and unnecessary. This reluctance persists despite a growing body of literature suggesting that postoperative endoscopy to directly examine anastomotic tissues is safe and both diagnostically and therapeutically useful [8,9,10,11,12,13,14].
Here we hypothesized that routine serial endoscopic surveillance (SES) following low anterior resection (LAR) would be acceptable to patients and could yield information important to the understanding of anastomotic healing. We assessed the willingness of patients to undergo the procedure, the experience of patients in terms of its duration and discomfort and the ability to capture images and relevant biologic information from perianastomotic fluid samples.
The pathobiology of anastomotic leak likely involves multiple factors including technique, blood flow and molecular elements of inflammation and healing. Our laboratory has been interested in the role of bacteria on the pathogenesis of anastomotic leak, particularly those that produce the tissue destroying enzyme collagenase [15, 16]. While bacteria are invariably present at anastomotic sites, our work suggests that, beyond species identification, the phenotype bacteria express in vivo (i.e., collagenase production), the provocative “cues” that are present in vivo that activate them and the specific phenotype of inflammation they elicit (i.e., MMP9, plasminogen) is most deterministic of their causal role in anastomotic leak [17]. In summary, our work demonstrates that bacteria are necessary but alone are not sufficient to cause anastomotic leak (Fig. 1). Therefore, the aim of this study was to determine if serial endoscopy following LAR can be successfully carried out in a limited cohort of patients and to determine if anastomotic lavage fluids can be successfully assayed for mediators of inflammation and bacterial phenotypes.
Fig.1. Bacteria are necessary, but alone not sufficient to cause anastomotic leak.
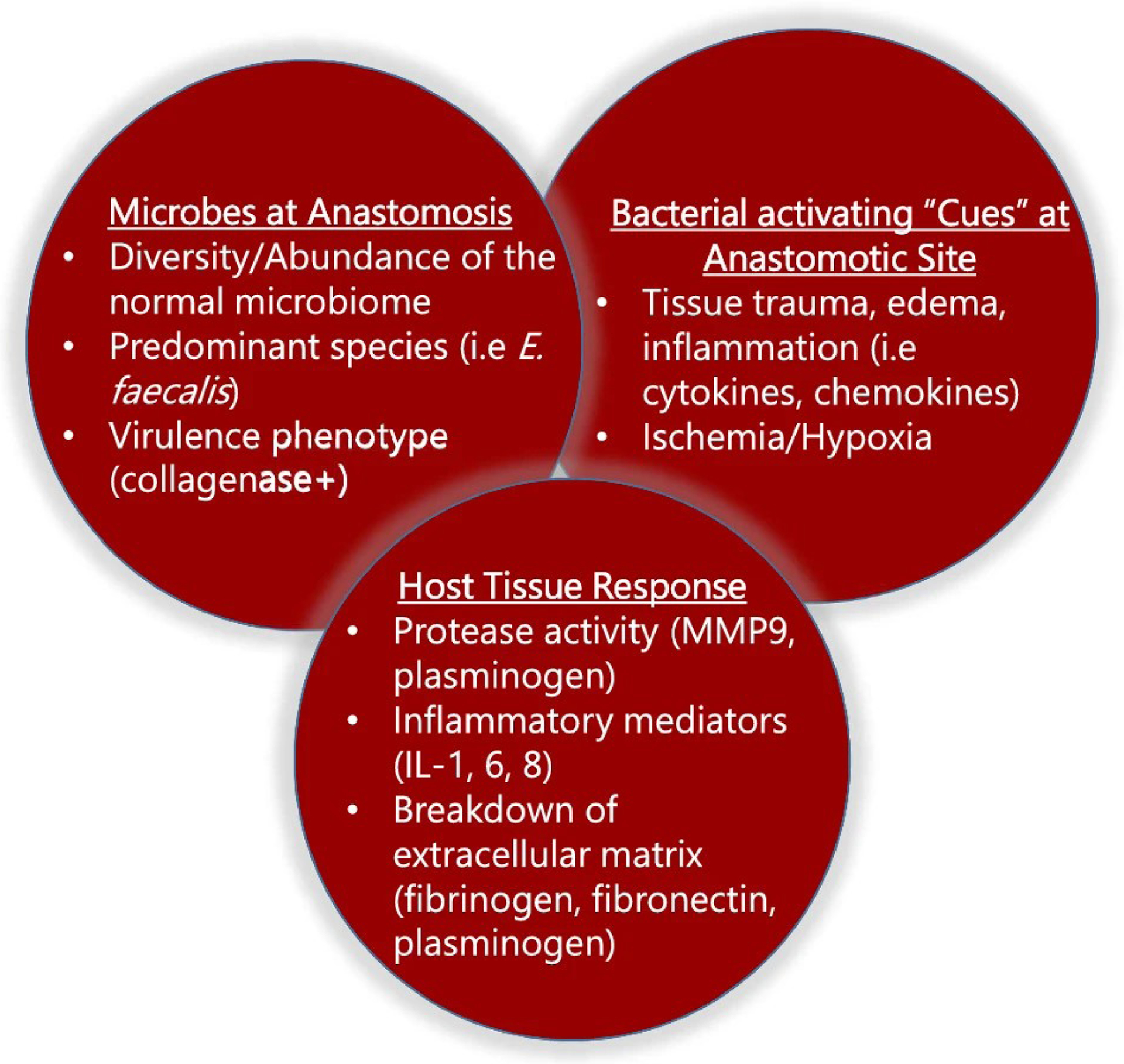
Previous work in animals demonstrates that the presence of collagenase-producing bacteria on anastomotic tissues is required for a clinically significant leak to occur. However, collagenolytic microbes alone are not sufficient to cause leak as “other factors” must be in play (loss of protective anaerobes, in vivo activation of collagenolytic activity by bacteria in response to host “cues,” shifts in tissue protease activity-MMP9, plasminogen, breakdown of extracellular matrix, etc.).
Materials and methods
Patients All patients aged 18 and over undergoing a low anterior resection with a primary anastomosis< 10 cm from the anal verge and diverting loop ileostomy were eligible for study inclusion. As a planned pilot study, 10 patients were included in this study, recruited from three institutions across Chicago. Patient characteristics (age, sex, use if bowel prep/antibiotics, tumor stage, etc.) are displayed in Table 1 for all patients. The study was approved by the Institutional Review Boards of each institution.
Table 1.
Patient characteristics. Patient s#7 was excluded from sample analysis later due to deviation of the protocol
| Patient | Age | Sex | Stoma | Cancer stage | Preop XRT | Cleaning prep | Oral antibiotics | Preparative antibiotic (IV) |
|---|---|---|---|---|---|---|---|---|
| 1 | 55 | M | Y | T3N1 | Y | Golytele | Erythomycin, Metronidazole | Gentamicin, Metronidazole |
| 2 | 53 | M | Y | T3N0 | Y | Golytele | Erythomycin, Metronidazole | Cefoxitin, Ampicillin |
| 3 | 66 | M | Y | T3N0 | Y | Mag Citrate | None | Cefoxitin |
| 4 | 49 | M | Y | T1N0 | N | Golytele | Erythomycin, Metronidazole | Cefoxitin, Ampicillin |
| 5 | 53 | F | Y | Anastomotic recurrence | N | MoviPrep | None | Cefoxitin |
| 6 | 65 | M | Y | T3bN1 | Y | Golytele | Erythomycin, Metronidazole | Cefoxitin, Ampicillin |
| 7 | 34 | M | Y | T3N1 | Y | MoviPrep | None | Cefoxitin |
| 8 | 50 | M | Y | T2N0 | N | Miralax, dulcolax | Neomycin, Metronidazole | Ertapenam |
| 9 | 68 | M | Y | T3N1 | Y | Miralax, dulcolax | Neomycin, Metronidazole | Ertapenam |
| 10 | 55 | M | Y | T3N1 | Y | Miralax, dulcolax | Neomycin, Metronidazole | Ertapenam |
Serial endoscopic surveillance (SES).
Using an adult flexible sigmoidoscope, patients underwent serial endoscopic surveillance at three different time points: in the operating room after completion of the anastomosis (SES-1) while under general anesthesia, while an inpatient during postoperative day 3–7 (SES-2), and as an outpatient at the initial follow clinic visit between postoperative day 10–28 (SES-3). All postoperative endoscopies were performed by the operating surgeon. No sedation was used for postoperative endoscopy and topical lidocaine gel was used at the discretion of the surgeon. Patients were closely monitored before, during and after the procedure by a study coordinator: length of the endoscopy, patient discomfort and satisfaction were recorded prospectively. Patient discomfort during postoperative endoscopy was scored on a scale of 1–5 (1: well tolerated, no discomfort 3: tolerated, moderate discomfort 5: procedure aborted). Pre- and post-operative management was otherwise not impacted and was determined by the practice of the operating surgeon.
Anastomosis imaging and anastomotic fluid sample collection.
During each endoscopy, select images of the anastomotic site were captured and stored. The anastomosis was then lavaged with 50–150 ml of normal saline under direct vision and samples retrieved using a Lukins trap. Samples were transported on dry ice, stored at − 80 °C for later analysis.
Anastomotic fluid analysis.
The collected fluids were assayed by zymography, proteomics and ELISA using standard techniques [18]. The matrix metalloproteinase 9 (MMP9) pro- and active forms were detected by zymography as previously described [15, 19]. LC–MS/MS analysis was performed as previously described [20, 21].
Microbial analysis by 16S rRNA.
DNA was isolated from anastomotic fluids using the DNeasy PowerSoil HTP 96 Kit (Qiagen). For library preparation, DNA was amplified using the barcoded 12-bp Golay primer set designed for the Earth Microbiome Project (EMP) [22]. The samples were run on an Illumina MiSeq at UIUC (150 bp × 2). 16S rRNA sequence data analysis was performed as previously described [23].
Microbial analysis by culture.
Microbial species were isolated by plating anastomotic fluids on selective culture plates (MacConkey II, Columbia CNA agar, Pseudomonas isolation agar, Enterococcosel agar). For the detection and isolation of collagenolytic bacteria, skim milk was incorporated as previously described [24]. Collagenolytic strains were analyzed for their ability to activate MMP9 using recombinant MMP9 (rMMP9) as a substrate as previously described [15]. Detection of MMP9-cleaving activity was assessed by zymography.
Quantitative PCR (qPCR)
Quantitative PCR (qPCR) was performed for quantitative analysis of Eubacteria (Universal 16S Eu-F 5′ACTCCTACGGGAGGCAGCAGT3′ and Eu-R 5′ ATTACCGCGGCTGCTGGC3′), E. faecalis (E.f.23S-F 5′CCTATCGGCCTGGGCTTAG’, E.f.23S-R 5′AGCGAAAGACAGGTGAGAATCC3′) and P. aeruginosa (P.a.-F 5′CCTGACCATCCGTCGCCACAAC3′, P.a.-R 5′CGCAGCAGGATGCCGACGCC3′) in anastomotic fluids.
Results
Patient demographics.
Of 19 consequent patients recruited for participation in the study, 11 were consented and enrolled. One patient was subsequently excluded due to a change in the operative plan leaving 10 patients for analysis. The demographics of the study cohort is displayed in Table 1. One patient (patient #7) was excluded due to deviation of the sampling protocol, rendering the fluid samples unusable. One patient (patient #1) elected to not complete the last endoscopic screening. Thus, the results of 9 patients are reported; among them, 8 patients completed all 3 endoscopic procedures and one patient completed two endoscopic screenings. Patient characteristics are summarized in Table 1.
Patient comfort.
The mean time to complete each SES episode was 2 min and 46 s and no sedation was required to complete any of the postoperative endoscopies. For SES-2 (the initial postoperative endoscopy), the mean comfort score was 1.5 (SD± 0.7), while for SES-3 every patient reported that it was ‘well tolerated’ (comfort score= 1). The patient who refused SES-3 experienced mild/moderate discomfort (comfort score= 3) during SES-2 and therefore declined to participate in SES-3. There was no difference in the comfort score between patients who received topical lidocaine vs those that did not.
Image Analysis.
Endoscopy images are displayed in Fig. 2. The patient #4 SES-1 image and the patient #10 SES images were not recovered after endoscopy. All patients’ SES-1 images (POD0) demonstrated no obvious pathology (Fig. 2). Edema on and extending beyond the staple line, ulceration and bruising were observed in SES-2 images (POD3–4) in several patients (Figs. 2, 3). Images from SES-3 demonstrated that in most patients, the edema had resolved, although white exudate could be observed on the staple line.
Fig.2.
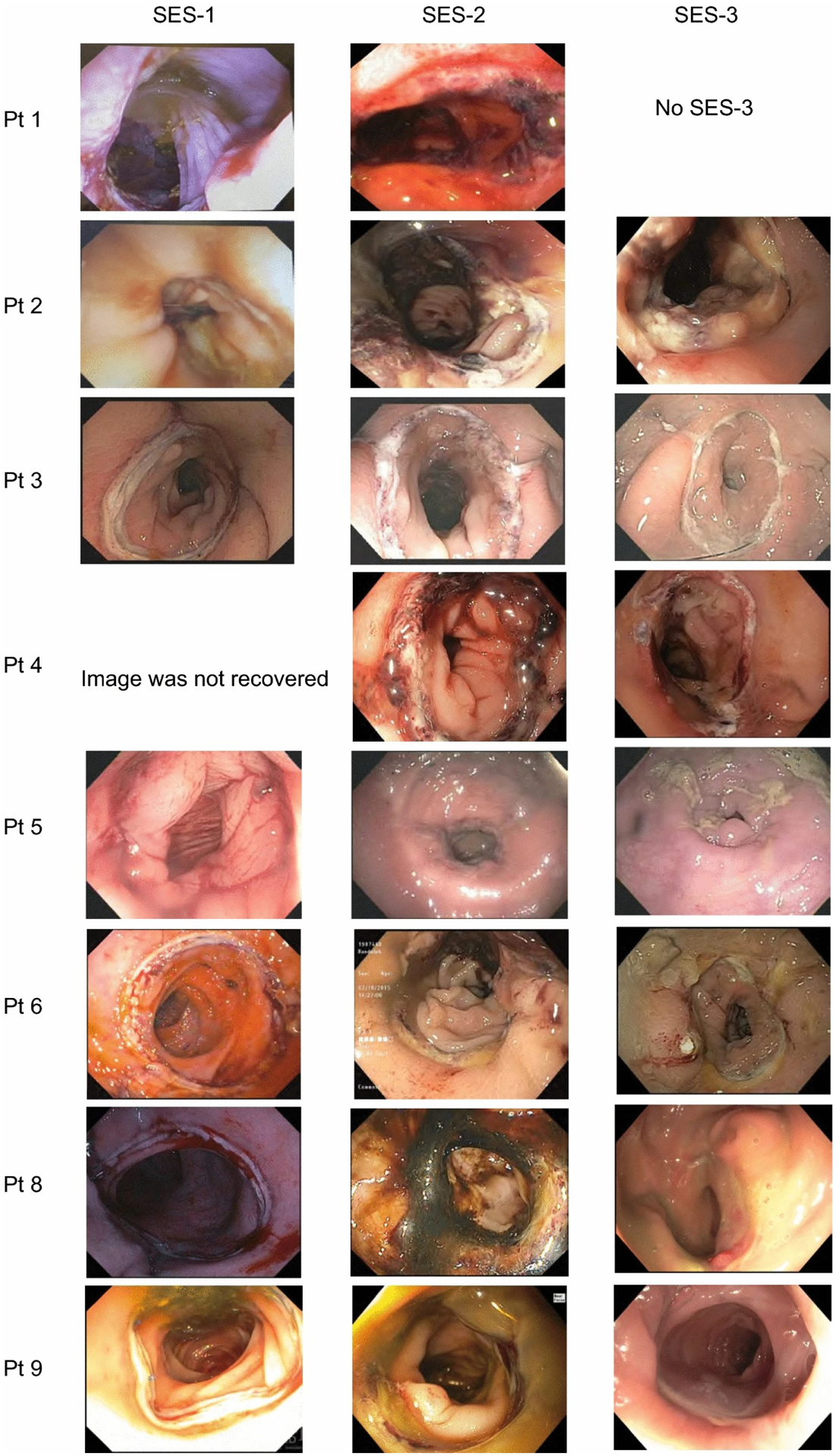
Serial endoscopic images of patients following low anterior resection.
Fig.3. SES-2 endoscopy images demonstrating visible complications.
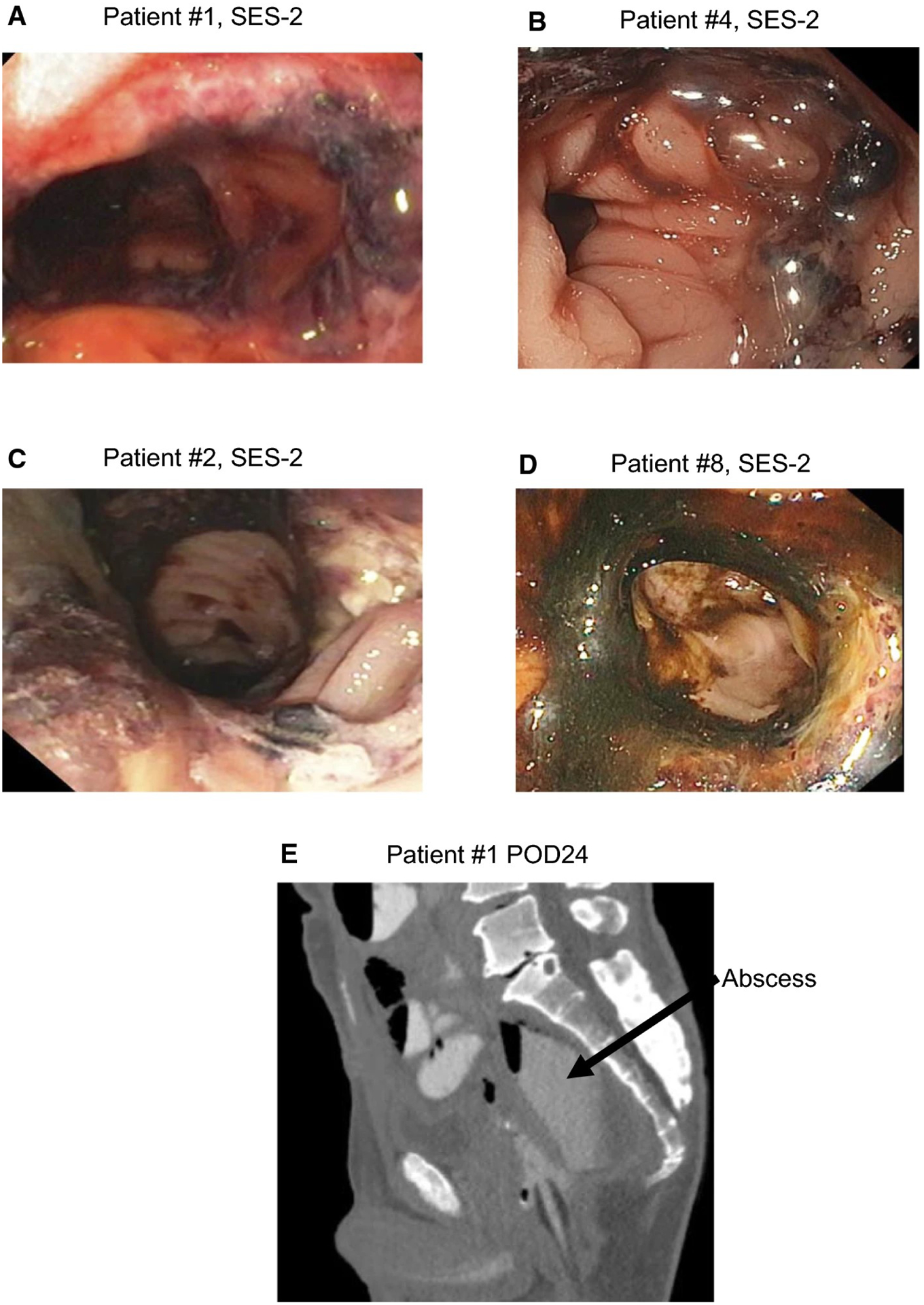
a, Edema on and extending the staple line in patient #1 at the SES-2 time point b, Edema on the staple line observed in patient #4 at SES-2; c, Ulceration observed on anastomotic line in patient #2 at SES-2; d, Edema on the staple line and ecchymosis patient #8 at SES-2; e, CT scan of patient #1 on postoperative day 24.
Postoperative complications.
Two patients developed superficial surgical site infections. Two of the nine patients were readmitted after discharge. Of these, one was for postoperative ileus, and one was subsequently diagnosed with anastomotic leak during the readmission on POD24 (patient #1). A CT scan and barium enema of patient #1 detected a posterior leak, which was drained percutaneously. The patient was mildly distended and reported mild/moderate discomfort on POD3 prior to the SES-2 procedure; at post-procedure, he was stable, not distended and in no distress. However, later he required nasogastric tube decompression for a persistent ileus, which eventually resolved prior to discharge on POD10. Noteworthy is that collagenolytic E. faecalis was cultured from the abscess material from the percutaneous drain on POD24.
Analyses of anastomotic lavage fluids.
A summary representation of selected results of the various parameters measured are displayed in Fig. 4. Anastomotic lavage fluids yielded the following results individually: Phase contrast microscopy performed on all patients’ lavage fluids to detect exfoliated epithelial cells revealed a massive amount of cells (Supplemental Fig. 1A) from fluid obtained from patient #1 on SES-2. Confocal microscopy with DAPI staining for visualization of nuclei (Supplemental Fig. 1B) confirmed the presence of live cells in the fluid. Proteomics analysis detected between 46 and 1724 human proteins in individual patients’ samples collected at SES-1, SES-2, and SES-3, totaling 2670 different proteins identified across all patient samples. The most abundant proteins belonged to actin family-related proteins, cytoskeletal proteins, hemoglobins, immunoglobulins, complement factors, mucin 2, myosins, extracellular matrix remodeling proteins, inhibitors of proteases (Supplemental Table S1, Supplemental Fig. 2A–B). Analysis of proteins revealed the presence of fibrinogen and fibronectin; proteins that function primarily to occlude blood vessels as part of the process of wound healing [25, 26] Supplemental Fig. 2A. The highest level of fibrinogen and fibronectin was observed in the patient #1 at SES-2 although a predominance of fibrinogen-fibronectin complex was observed in all patients (Supplemental Fig. 2B) compared to other proteins. MMP9 was also detected by proteomics.
Fig.4. Summary representation of microbial and tissue components assayed in endoscopic lavage fluids from selected samples.
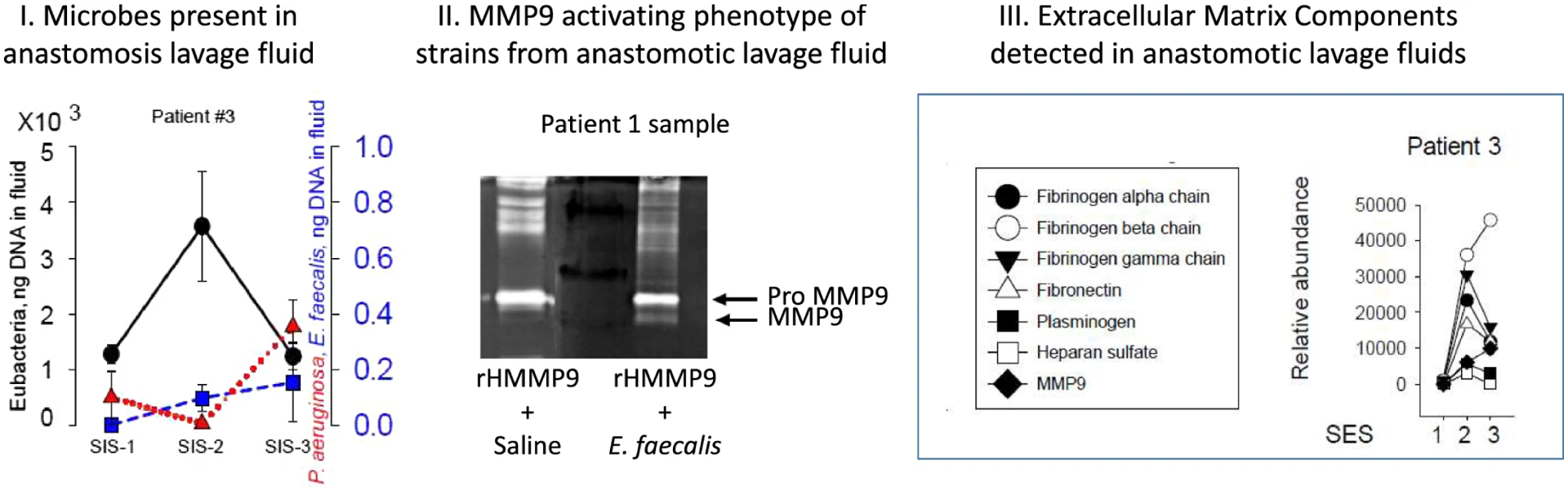
I.16 s rRNA microbial sequencing showing an increase in overall bacterial density (black dots of total Eubacteria) associated with suppressed pathogenic species (blue and red indicators of E. faecalis, P. aeruginosa. II. Biologic assays from patient #3 lavage samples demonstrating proteins can be detected that are relevant to extracellular matrix breakdown. III. Assay showing shift from pro-MMP9 (inactive form) to active MMP9 when recombinant human MMP9 (rHMMP9) is co-incubated with collagenase-producing E. faecalis isolated from an anastomotic lavage sample (patient #1)
A specific inhibitor of the MMP9, tissue inhibitor of metalloprotease (TIMP) is specifically designed to fine-tune the activity of metalloproteases through specific binding of MMPs [27]. Although TIMP was detected, its relative level was negligible. A high relative level of alpha-2-macroglobulin (α − 2 M) was detected in most of the patients (Supplemental Fig. 2C). Interestingly, a central role of α − 2 M has been suggested to function when TIMPs are deficient [28]. The α − 2 M acts as anti-protease and is able to inactivate an wide variety of proteinases including metalloproteases [29]. It also functions as an inhibitor of fibrinolysis [30]. Recent work by our lab has identified the plasmin/plasminogen system to be a key pathway involved in anastomotic leak pathogenesis [31]. Another potent inhibitor of plasmin and MMPs, the α−1-antitrypsin [32], was also found at relatively high abundance in anastomotic effluents.
The S100 family consists of proteins with anti-microbial function [33]. Among S100 family proteins, S100A8 and S100A9, were observed at the highest relative abundance, especially in patient #1 (Supplemental Fig. 2D). The complex of these two proteins is known as calprotectin and has potent antimicrobial activity [34]. Finally, a relatively high abundance of proteins that sequester iron (perhaps as an anti-microbial defense?) such as lactotransferrin, serotransferrin, and ceruloplasmin [35] were also detected in the patient #1 (Supplemental Fig. 2E).
Zymography was used to measure pro- and active forms of MMP9, a key mediator in the extracellular matrix remodeling and wound healing (Supplemental Fig. 3A–C). Both forms MMP9 were consistently non-detectable on POD0 (SES-1) and moderately elevated on POP3–4 (SES-2) in patients 2, 3 and 4. The exception was patient #1 who expressed a high level of pro- MMP9 on SES-2 (Supplemental Fig. 3B). Active MMP9 was detectable in lavage fluids in only three patients on SES-3. IL-1β, a proinflammatory cytokine, was observed to be elevated in patient #1 at SES-2 (Supplemental Fig. 3D) with IL-6 and IL-8 being among the highest in patients #1 and #10 (Supplemental Fig. 3E, F).
16S rRNA demonstrated significant inter-patient variability in bacterial composition at the family level among material retrieved from each endoscopic time-point (Supplemental Fig. 4A). Health-promoting (i.e., probiotic) bacteria belonging to the families Bacteroidaceae, Lachnospiraceae, Ruminococcaceae were dominant on SES-1 (Supplemental Fig. 4A).
Among all anastomotic lavage samples recovered by culture, Enterococcus faecalis and Pseudomonas aeruginosa were the most frequent (Supplemental Fig. 4B). E. faecalis was recovered from 4 patients, P. aeruginosa from 4 patients, and both E. faecalis and P. aeruginosa from one patient. Expression of the collagenolytic phenotype was most commonly identified with E. faecalis and P. aeruginosa. Collagenolytic Bacilli such as Virgibacillus proomii, Bacillus sp. and Bacillus licheniformis were second in frequency among cultured strains. Anaerobic collagenolytic bacteria Clostridia perfringes and Bacteroides fragilis were recovered from the one patient, patient #1. All recovered collagenolytic strains were tested for their ability to activate MMP9 using recombinant MMP9 as a substrate. Supplemental Fig. 4C represents an example of zymography demonstrating cleavage of MMP9 by an isolate of E. faecalis. P. aeruginosa and E. faecalis were predominant among strains expressing the collagenolytic (col +) and MMP9-activating (MMP9 +) phenotype (i.e., col + MMP9 +) Supplemental Fig. 4D).
qPCR was used to determine the abundance of collagenase positive (col +) and MM9 activating (MMP9 +) among E. faecalis and P. aeruginosa strains and was used to determine the total bacteria load in anastomotic fluids (Supplemental Fig. 5). qPCR analysis revealed dynamic changes in both total and col + MMP + bacteria that was observed in 5 of 6 patients that shifted in the same direction among samples (Supplemental Fig. 5). Curiously, in patient #1, a decrease in the total bacterial load (total bacterial DNA) was observed in the lavage sample from the SES-2 episode which was accompanied by an increase in col + MMP9 + E. faecalis (Supplemental Fig. 5).
Discussion
The present study demonstrates that it is feasible to serially track anastomotic healing via endoscopy and lavage perianastomotic tissues for molecular analysis. As a result of the limited dataset, neither the images nor the molecular analyses could be expected to be discriminative to predict anastomotic healing. However, the readouts generated from the lavage samples in this study were able to detect molecular markers we have previously identified in our mouse models such as the presence of collagenolytic bacteria as well as biomarkers involved in enhanced anastomotic proteolysis such as MMP9 and plasminogen [31, 36]. Obviously a much larger properly powered prospective study will be necessary to determine if it is possible to map a specific molecular signature to a wound healing score that can then be validated to be predictive of healing versus leak.
Given that anastomoses in the esophagus and rectum, two areas at highest risk for AL, are so easily reachable by endoscopy and given the result of the present study that demonstrate that a dense molecular analysis is possible with anastomotic lavage fluid, the reluctance to proceed with applying SES to high risk patients may be need to be reconsidered. Understanding which microbes re-occupy the anastomotic tissues as patients recover following exposure to antibiotics and hospital pathogens will require serial endoscopy and sampling [37].
There are several limitations to the current study. First of all, within this limited cohort, we have not established the full safety of the technique, although postoperative endoscopy and its use to control anastomotic bleeding has been established to be safe by others [9, 11]. Second, cost, both in terms of money and time, may be an issue in many centers thus prohibiting the application of SES. Thirdly, the choice of mechanical bowel prep, the use of oral antibiotics and the institutional standard intravenous antibiotic prophylaxis in this study could have influenced the microbiology and inflammatory mediator results. Yet, whether antibiotic use correlated to outcome in this study cannot be determined given the limited number of patients. Finally, whether the turnaround time for the microbial and immune markers will be clinically useful has not been established.
In summary, in this pilot study we demonstrate the promise of SES as a method to advance our understanding of anastomotic wound healing. Once both visual and molecular parameters indicating healthy versus pathologic healing are established, the ultimate goal of this line of inquiry is to predict, and potentially intervene in anastomotic leak before it becomes clinically evident.
Supplementary Material
Supplementary Figure 1. Microscopic image of anastomotic lavage fluid demonstrating the presence of shed cells in patient #1 at SES-2. (A) Phase contrast; (B) confocal microscopy of cells with DAPI staining for nuclei in fluid of the Patient #1 SES-2 sample (PDF 219 kb).
Supplementary Figure 2. Proteomic analysis demonstrating relative protein abundance in anastomotic effluents. (A), Family of extracellular matrix proteins; (B), Family of extracellular matrix proteins represented at different scale; (C), Inhibitors of proteases; (D) S100 family; (E), Iron binding proteins (PDF 89 kb).
Supplementary Figure 3. Extracellular matrix metalloproteases and pro-inflammatory cytokine analysis in anastomotic lavage fluids. (A), Representative zymography image with pro- and active MMP9 bands; (B, C), Quantification of zones of zpro-MMP9 (B) and its active form (C) by densitometry analysis using Image J software; (D-F), pro-inflammatory cytokines IL-1 beta (G),, IL-6 (H), and IL-8 (I) levels estimated by ELISA (PDF 67 kb).
Supplementary Figure 4. Composition and function of microbial populations in anastomotic lavage fluids. (A), 16S rRNA gene sequence analysis demonstrating the most predominant Families in anastomotic fluids; (B), Analysis of cultivable microorganisms under aerobic and anaerobic conditions. Species, demonstrating collagenolytic activity on skim milk plates, defined as “col+”. (C), Zymography image representing an example of rMMP9 cleavage by E. faecalis isolated from patient #1 at SES-2. Three other strains isolated from patient #1 did not demonstrate the capacity to cleave rMMP9. (D), Species, demonstrating both collagenolytic activity and the ability to activate MMP9 (Col+MMP9+ phenotype) using recombinant MMP9 (rMMP9) (PDF 110 kb).
Supplementary Figure 5. Quantitative analysis of total bacterial loading balanced against the bacterial load of Col+MMP9+ species in anastomotic lavage fluids as assessed by qPCR. The Y axes on the left show the dynamic changes in total bacterial load (as measured by using a universal primer that detects all bacterial DNA) on solid lines with circle symbols. The y axes on the right side show the dynamic changes in bacterial load of E. faecalis (Medium dash blue lines with square symbols) or P. aeruginosa (dotted red lines with triangle up symbols) (PDF 48 kb).
Supplementary Table: Relative abundance of proteins in anastomotic effluents. Lines 1–253: Relative spectra counts in 0.5 μg peptides. Lines 259–510: Relative abundance of proteins in anastomotic effluents (XLSX 168 kb)
Acknowledgements
The authors thank for Cameron Hunter, Cindy Bethel, Monika Krezalek MD, Konstantin Umanskiy MD, Joseph Muldoon MD, Bruce Orkin MD, Marc Singer MD for substantive contributions to the study.
Funding
Funded by Limited Project Grant from the American Society of Colon and Rectal Surgeons Research Foundation (LPG-098, JCA). Portions of this research were supported by the NIH National Institute of General Medical Sciences (GM103493), and the W.R. Wiley Environmental Molecular Science Laboratory (a national scientific user facility sponsored by the U.S. Department of Energy’s Office of Biological and Environmental Research and located at Pacific Northwest National Laboratory). Pacific Northwest National Laboratory is operated by Battelle Memorial Institute for the U.S. Department of Energy under contract DE-AC05-76RLO-1830.
Footnotes
Conflict of interests: None.
References
- 1.Wong NY, Eu KW (2005) A defunctioning ileostomy does not prevent clinical anastomotic leak after a low anterior resection: a prospective, comparative study. Dis Colon Rectum 48:2076–2079 [DOI] [PubMed] [Google Scholar]
- 2.Guel-Klein S, Biebl M, Knoll B et al. (2019) Anastomotic leak after transanal total mesorectal excision: grading of severity and management aimed at preservation of the anastomosis. Colorectal Dis 21:894–902 [DOI] [PubMed] [Google Scholar]
- 3.Angeramo CA, Dreifuss NH, Schlottmann F et al. (2020) Postoperative outcomes in patients undergoing colorectal surgery with anastomotic leak before and after hospital discharge. Updates Surg 72:463–468 [DOI] [PubMed] [Google Scholar]
- 4.Alanezi K, Urschel JD (2004) Mortality secondary to esophageal anastomotic leak. Ann Thorac Cardiovasc Surg 10:71–75 [PubMed] [Google Scholar]
- 5.Henneman D, van Leersum NJ, Ten Berge M et al. (2013) Failure-to-rescue after colorectal cancer surgery and the association with three structural hospital factors. Ann Surg Oncol 20:3370–3376 [DOI] [PubMed] [Google Scholar]
- 6.Quaresima S, Balla A, Palmieri L et al. (2020) Routine near infra-red indocyanine green fluorescent cholangiography versus intraoperative cholangiography during laparoscopic cholecystectomy: a case-matched comparison. Surg Endosc 34:1959–1967 [DOI] [PubMed] [Google Scholar]
- 7.Tsang YP, Leung LA, Lau CW et al. (2020) Indocyanine green fluorescence angiography to evaluate anastomotic perfusion in colorectal surgery. Int J Colorectal Dis 35:1133–1139 [DOI] [PubMed] [Google Scholar]
- 8.Chardavoyne R, Ratner LE, Jaume JC et al. (1989) Safety of endoscopy in the immediate postoperative period following gastric anastomosis. Surg Endosc 3:13–15 [DOI] [PubMed] [Google Scholar]
- 9.Amr MA, Alzghari MJ, Polites SF et al. (2014) Endoscopy in the early postoperative setting after primary gastrointestinal anastomosis. J Gastrointest Surg Official J Soc Surg Aliment Tract 18:1911–1916 [DOI] [PubMed] [Google Scholar]
- 10.Okada T, Kawada K, Nakajima Y et al. (2013) Internal pressure of the conduit during endoscopy on the day after esophagectomy. Dig Surg 30:183–189 [DOI] [PubMed] [Google Scholar]
- 11.Page RD, Asmat A, McShane J et al. (2013) Routine endoscopy to detect anastomotic leakage after esophagectomy. Ann Thorac Surg 95:292–298 [DOI] [PubMed] [Google Scholar]
- 12.Blumetti J, Chaudhry V, Prasad L et al. (2012) Delayed transanal repair of persistent coloanal anastomotic leak in diverted patients after resection for rectal cancer. Colorectal Dis Off J Assoc Coloproctol Great Br Irel 14:1238–1241 [DOI] [PubMed] [Google Scholar]
- 13.Dapri G (2020) Transanal minimally invasive surgery: a multi-purpose operation. Surg Technol Int 36 [PubMed] [Google Scholar]
- 14.Chen WT, Bansal S, Ke TW et al. (2018) Combined repeat laparoscopy and transanal endolumenal repair (hybrid approach) in the early management of postoperative colorectal anastomotic leaks: technique and outcomes. Surg Endosc 32:4472–4480 [DOI] [PubMed] [Google Scholar]
- 15.Shogan BD, Belogortseva N, Luong PM et al. (2015) Collagen degradation and MMP9 activation by Enterococcus faecalis contribute to intestinal anastomotic leak. Sci Transl Med 7:286ra68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olivas AD, Shogan BD, Valuckaite V et al. (2012) Intestinal tissues induce an SNP mutation in Pseudomonas aeruginosa that enhances its virulence: possible role in anastomotic leak. PLoS ONE 7:e44326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCarty SM, Percival SL (2013) Proteases and delayed wound healing. Adv Wound Care (New Rochelle) 2:438–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelly RT, Page JS, Luo Q et al. (2006) Chemically etched open tubular and monolithic emitters for nanoelectrospray ionization mass spectrometry. Anal Chem 78:7796–7801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Belogortseva N, Krezalek M, Guyton K et al. (2017) Media from macrophages co-incubated with Enterococcus faecalis induces epithelial cell monolayer reassembly and altered cell morphology. PLoS ONE 12:e0182825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maiolica A, Borsotti D, Rappsilber J (2005) Self-made frits for nanoscale columns in proteomics. Proteomics 5:3847–3850 [DOI] [PubMed] [Google Scholar]
- 21.Granholm V, Kim S, Navarro JC et al. (2014) Fast and accurate database searches with MS-GF+Percolator. J Proteom Res 13:890–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caporaso JG, Lauber CL, Walters WA et al. (2012) Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hyoju SK, Zaborin A, Keskey R, et al. (2019) Mice fed an obesogenic Western diet, administered antibiotics, and subjected to a sterile surgical procedure develop lethal septicemia with multidrug-resistant pathobionts. MBio 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guyton KL, Levine ZC, Lowry AC et al. (2019) Identification of collagenolytic bacteria in human samples: screening methods and clinical implications for resolving and preventing anastomotic leaks and wound complications. Dis Colon Rectum 62:972–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Carrim N, Ni H (2015) Fibronectin orchestrates thrombosis and hemostasis. Oncotarget 6:19350–19351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pereira M, Rybarczyk BJ, Odrljin TM et al. (2002) The incorporation of fibrinogen into extracellular matrix is dependent on active assembly of a fibronectin matrix. J Cell Sci 115:609–617 [DOI] [PubMed] [Google Scholar]
- 27.Arpino V, Brock M, Gill SE (2015) The role of TIMPs in regulation of extracellular matrix proteolysis. Matrix Biol 44–46:247–254 [DOI] [PubMed] [Google Scholar]
- 28.Chu CT, Howard GC, Misra UK et al. (1994) Alpha 2-macroglobulin: a sensor for proteolysis. Ann N Y Acad Sci 737:291–307 [DOI] [PubMed] [Google Scholar]
- 29.Rehman AA, Ahsan H, Khan FH (2013) Alpha-2-macroglobulin: a physiological guardian. J Cell Physiol 228:1665–1675 [DOI] [PubMed] [Google Scholar]
- 30.de Boer JP, Creasey AA, Chang A et al. (1993) Alpha-2-macroglobulin functions as an inhibitor of fibrinolytic, clotting, and neutrophilic proteinases in sepsis: studies using a baboon model. Infect Immun 61:5035–5043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jacobson RA, Williamson AJ, Wienholts K, et al. (2019) Prevention of anastomotic leak via local application of tranexamic acid to target bacterial-mediated plasminogen activation: a practical solution to a complex problem. Ann Surg [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stockley RA (2015) The multiple facets of alpha-1-antitrypsin. Ann Transl Med 3:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zackular JP, Chazin WJ, Skaar EP (2015) Nutritional immunity: S100 proteins at the host-pathogen interface. J Biol Chem 290:18991–18998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zygiel EM, Nolan EM (2018) Transition metal sequestration by the host-defense protein calprotectin. Annu Rev Biochem 87:621–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bonaccorsi di Patti MC, Cutone A, Polticelli F et al. (2018) The ferroportin-ceruloplasmin system and the mammalian iron homeostasis machine: regulatory pathways and the role of lactoferrin. Biometals 31:399–414 [DOI] [PubMed] [Google Scholar]
- 36.Jacobson RA, Wienholts K, Williamson AJ et al. (2020) Enterococcus faecalis exploits the human fibrinolytic system to drive excess collagenolysis: implications in gut healing and identification of druggable targets. Am J Physiol Gastrointest Liver Physiol 318:G1–G9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alverdy JC, Hyman N (2020) Bowel preparation under siege. Br J Surg 107:167–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Microscopic image of anastomotic lavage fluid demonstrating the presence of shed cells in patient #1 at SES-2. (A) Phase contrast; (B) confocal microscopy of cells with DAPI staining for nuclei in fluid of the Patient #1 SES-2 sample (PDF 219 kb).
Supplementary Figure 2. Proteomic analysis demonstrating relative protein abundance in anastomotic effluents. (A), Family of extracellular matrix proteins; (B), Family of extracellular matrix proteins represented at different scale; (C), Inhibitors of proteases; (D) S100 family; (E), Iron binding proteins (PDF 89 kb).
Supplementary Figure 3. Extracellular matrix metalloproteases and pro-inflammatory cytokine analysis in anastomotic lavage fluids. (A), Representative zymography image with pro- and active MMP9 bands; (B, C), Quantification of zones of zpro-MMP9 (B) and its active form (C) by densitometry analysis using Image J software; (D-F), pro-inflammatory cytokines IL-1 beta (G),, IL-6 (H), and IL-8 (I) levels estimated by ELISA (PDF 67 kb).
Supplementary Figure 4. Composition and function of microbial populations in anastomotic lavage fluids. (A), 16S rRNA gene sequence analysis demonstrating the most predominant Families in anastomotic fluids; (B), Analysis of cultivable microorganisms under aerobic and anaerobic conditions. Species, demonstrating collagenolytic activity on skim milk plates, defined as “col+”. (C), Zymography image representing an example of rMMP9 cleavage by E. faecalis isolated from patient #1 at SES-2. Three other strains isolated from patient #1 did not demonstrate the capacity to cleave rMMP9. (D), Species, demonstrating both collagenolytic activity and the ability to activate MMP9 (Col+MMP9+ phenotype) using recombinant MMP9 (rMMP9) (PDF 110 kb).
Supplementary Figure 5. Quantitative analysis of total bacterial loading balanced against the bacterial load of Col+MMP9+ species in anastomotic lavage fluids as assessed by qPCR. The Y axes on the left show the dynamic changes in total bacterial load (as measured by using a universal primer that detects all bacterial DNA) on solid lines with circle symbols. The y axes on the right side show the dynamic changes in bacterial load of E. faecalis (Medium dash blue lines with square symbols) or P. aeruginosa (dotted red lines with triangle up symbols) (PDF 48 kb).
Supplementary Table: Relative abundance of proteins in anastomotic effluents. Lines 1–253: Relative spectra counts in 0.5 μg peptides. Lines 259–510: Relative abundance of proteins in anastomotic effluents (XLSX 168 kb)


