Abstract
Despite their significance for mammalian embryogenesis, the molecular mechanisms that regulate placental growth and development have not been well defined. The Esx1 homeobox gene is of particular interest because it is among the few regulatory genes that have specific expression and function in the placenta during murine development. In addition, the ESX1 protein contains several notable features that are not often associated with homeoproteins, including an atypical homeodomain of the paired-like class, a proline-rich region that contains an SH3 binding motif, and a novel repeat region consisting of prolines alternating with phenylalanines or asparagines that we term the PF/PN motif. We have found that the ESX1 protein is expressed in the labyrinth layer of the placenta in vivo, where its subcellular localization is primarily cytoplasmic. Our results suggest that this unexpected subcellular localization is conferred by the PF/PN motif, which inhibits nuclear localization of ESX1 in cell culture, as well as its DNA binding activity in vitro. Finally, we show that the proline-rich region of ESX1 mediates interactions in vitro with the c-abl SH3 domain as well as with certain WW domains. We propose that the PF/PN motif provides a novel mechanism for regulating nuclear entry and that the essential function of ESX1 during placental development is mediated by its ability to couple cytoplasmic signal transduction events with transcriptional regulation in the nucleus.
During mammalian embryogenesis, the placenta provides a physical connection between the embryo and the mother, supplying nutrients that are critical for embryonic growth and survival. The multiple steps involved in the formation of the chorioallantoic placenta are poorly understood at the molecular level, yet they represent fundamental aspects of embryogenesis that are essential for development to term (13). Indeed, defects of implantation and placental development account for approximately one-third of spontaneous abortions in humans, and even minor defects in placentation can have severe consequences for embryo viability (reviewed in reference 15).
Among the candidate regulatory genes that have been implicated in placental development are the divergent homeobox gene Pem (28, 52), the zinc finger gene Rex-1 (40), the basic helix-loop-helix transcription factor Mash-2 (19), and the nuclear hormone receptor ERR-β (29, 34). In addition, we and others have been investigating Esx1 (also known as Spx1), which is a murine homeobox gene of the “paired-like” class (5, 26; Y.-T. Yan, S. Stein, P. Sciavolino, L. Yang, H. Wang, C. Abate-Shen, and M. M. Shen, unpublished observations). Esx1 is an X-linked gene that is chromosomally imprinted and is expressed specifically in extraembryonic tissues during development, as well as in the adult testis (5, 25, 26). Consistent with its restricted expression pattern, loss of function of Esx1 through targeted gene disruption leads to overgrowth and defective morphogenesis of the labyrinth layer of the placenta (25). These defects are consistent with an essential role for Esx1 in establishing and/or maintaining the maternal-fetal interface.
In addition to its restricted expression and function in extraembryonic tissues, the Esx1 homeobox gene is of particular interest because its predicted protein sequence includes several motifs that are generally not associated with homeoproteins. In this study, we have examined the expression pattern of Esx1 transcripts and corresponding protein product during placental development in the mouse. Unexpectedly, we found that ESX1 protein was localized primarily to the cytoplasm in vivo and in transfected cells and that a novel motif that we termed the PF/PN domain inhibited its nuclear localization. Furthermore, we showed that ESX1 contains a proline-rich SH3 binding motif that mediates interactions with the c-abl SH3 domain in vitro. These findings raise the possibility that ESX1 links cytoplasmic signaling events with nuclear transcriptional regulation during extraembryonic development.
MATERIALS AND METHODS
ES cell culture and cDNA library screening.
Culture of D3 ES cells (16) and differentiation of embryoid bodies were carried out as previously described (43, 44). To identify novel paired-like homeodomains, we designed an 1,152-fold degenerate oligonucleotide 5′ C(G/T)(G/C/T)C(G/T)(A/G)TT(C/T)T(T/G)(G/A)AACCA(G/C/A)AC(T/C)TG 3′ as a probe to recognize the amino acid sequence QVWF(Q/K)NRR. Screening of a primary cDNA library constructed from embryoid bodies on day 5 of differentiation was performed in the presence of 3 M tetramethylammonium chloride (Aldrich) (8). Screening of 200,000 primary phages resulted in the identification of a single homeobox-containing clone that corresponds to Esx1. We subsequently identified six additional Esx1 clones through rescreening of the embryoid-body cDNA library with a 5′ fragment of the original clone (see Fig. 1B, probe 2).
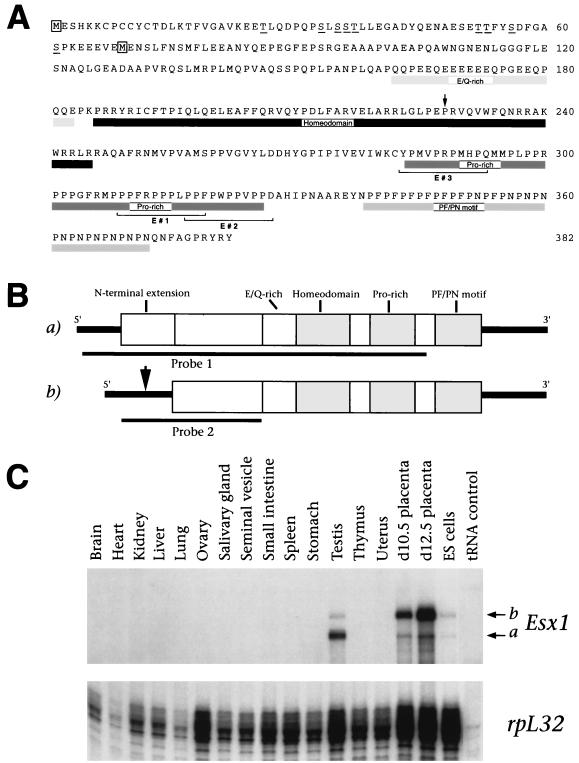
FIG. 1.
Differential expression of alternative Esx1 transcripts in the placenta and testes. (A) Amino acid sequence encoded by the longest Esx1 transcript (transcript A) (5). The putative initiator methionines for the products of transcripts A and B are boxed (transcript B) (26). Protein domains are indicated by boxes, the proline-rich peptides (E#1, E#2, and E#3) are bracketed, potential phosphorylation sites in the N-terminal extension are underlined, and the proline at position 43 of the homeodomain is indicated by an arrow. (B) Schematic diagram of the protein products of transcripts A and B. The protein regions depicted are the N-terminal extension, the glutamic acid- and glutamine-rich region (E/Q-rich), the homeodomain, the proline-rich region (Pro-rich), and the PF/PN motif. The arrow indicates the position in transcript B where its nucleotide sequence diverges at the 5′ end from transcript A. Probes for in situ hybridization (probe 1) and RNase protection assays (probe 2) are indicated. (C) RNase protection analysis of Esx1 transcripts. Each hybridization mixture contained 15 μg of total RNA (or 50 μg of yeast tRNA) and antisense riboprobes for Esx1 (probe 2) and ribosomal protein L32 (rpL32) as an internal standard (43). The positions of protected fragments corresponding to transcripts A and B are indicated.
Analysis of Esx1 mRNA and protein expression.
Intact embryos and extraembryonic membranes were dissected on days 7.5 through 12.5 post coitum, where 0.5 day post coitum is defined as noon of the day of the vaginal plug. RNase protection assays were carried out as described previously with total RNA prepared from embryonic or adult tissues (43). Nonradioactive in situ hybridization with digoxygenin-labeled riboprobes was performed on whole mounts or cryosections of embryos and extraembryonic tissues as described previously (42, 44). No specific staining was detected when a corresponding sense riboprobe for Esx1 was used (data not shown).
To generate polyclonal anti-ESX1 antisera, the homeodomain region of ESX1 (amino acids 183 to 248) was subcloned into plasmid pQE-9 (Qiagen) and the resulting hexahistidine fusion protein was purified by nickel affinity chromatography as described previously (9). The recombinant ESX1(183–248) protein was used as antigen to generate antisera in rabbits (Cocalico Biologicals). Affinity-purified immunoglobulin G (IgG) was prepared by chromatography of the anti-ESX1 antisera on an affinity resin in which the purified ESX1 antigen had been covalently cross-linked to CNBr-activated Sepharose (Pharmacia). For immunohistochemistry, cryosections (12 μ) were fixed in 4% paraformaldehyde and blocked with 3% hydrogen peroxide in methanol and normal goat serum. The sections were then incubated with affinity-purified anti-ESX1 IgG (1:100) followed by biotin-conjugated secondary antibody (1:200) and solutions A and B provided with the VectaStain Elite ABC kit (Vector Laboratories). Staining was visualized with 3,3′-diaminobenzidine tetrahydrochloride enhanced with nickel chloride (Vector Laboratories) and was followed by counterstaining with nuclear fast red (Vector Laboratories). No specific staining was detected following blocking of anti-ESX1 IgG by preincubation with purified ESX1 protein or in the absence of anti-ESX1 IgG (data not shown).
Production of ESX1 proteins and expression in mammalian cells.
Sequences corresponding to the full-length ESX1 coding region or various truncated derivatives were isolated by PCR amplification from a cDNA corresponding to transcript A (see Fig. 1), using primers containing EcoRI and XhoI restriction sites to facilitate cloning into pcDNA3 (Invitrogen). The 5′ PCR primers also contained sequences encoding an initiator methionine and a single Myc epitope, to ensure uniformity at the 5′ end. To produce ESX1 proteins in mammalian cells, these pcDNA3-Esx1 plasmids were transiently transfected into COS-1 cells. For Western blot analysis, transfected COS-1 cells were lysed in sodium dodecyl sulfate (SDS) sample buffer, cell lysates were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) and proteins were detected with anti-ESX1 antisera (1:5,000), followed by chemiluminescence detection (Amersham). For indirect immunofluorescence, transfected COS-1 cells were fixed in 3% paraformaldehyde and incubated with anti-ESX1 antisera (1:300). The cells were then incubated in fluorescein-conjugated goat anti-rabbit IgG (1:200) (Vector Laboratories) and subjected to nuclear staining with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI; 30 nM). Antibody specificity was verified in control experiments with pre-immune IgG or secondary antisera alone.
In vitro DNA binding assays and GST interaction assays.
Electrophoretic mobility shift assays were performed as described previously (9) using proteins produced by in vitro transcription and translation from the pcDNA3-Esx1 plasmids. The DNA sites were (top strand shown) 5′-ACACTAATTGGAGGC-3′ (site 6 of reference 9), 5′-ACAATAATTGGAGGC-3′ (site 6-5 of reference 9), 5′-ACACTAATGGAGGC-3′ (site 6-11 of reference 9), 5′-ACACTACTTGGAGGC-3′ (site 6-19 of reference 9), 5′-GATAATTGATTATC-3′ (P2 site of reference 53), and 5′-ACTAATTGAATTAGC-3′ (PRDQ9 site of reference 45).
For protein interaction assays, 11-mer peptide sequences from the ESX1 proline-rich domain (E#1, E#2, and E#3 [see Fig. 1A]), the SH3 domains from c-abl, c-fyn, and N-src (10), and the WW domains of formin-binding protein 11 (FBP-11WW) and formin-binding protein 30 (FBP-30WW) (3) were cloned into pGEX-2TK for production of glutathione S-transferase (GST) fusion proteins (23). Far-Western protein interaction assays were performed as described previously (10) with 32P-labeled GST-SH3 domain fusion proteins to probe total-cell lysates containing the GST ESX1 fusion peptides (GST E#1, GST E#2, and GST E#3). GST interaction assays were performed as described previously (55) with 35S-labeled ESX1 proteins obtained by in vitro transcription-translation and the purified GST-SH3 or GST-WW domain fusion proteins.
Nucleotide sequence accession numbers.
Transcripts A and B have been deposited in GenBank under accession no. AF017735 and AF017734, respectively.
RESULTS
Two distinct Esx1 transcripts have differing expression patterns.
We have previously used the differentiation of embryonic stem (ES) cells in culture as a model system for identifying genes involved in embryonic and extraembryonic development (43, 44). ES cells are pluripotent progenitors that spontaneously form cell aggregates termed embryoid bodies in suspension culture. These embryoid bodies differentiate into a wide range of embryonic derivatives, as well as extraembryonic derivatives such as visceral and parietal extraembryonic endoderm (16).
To identify novel homeobox genes expressed during ES cell differentiation, we screened a cDNA library constructed on day 5 of embryoid-body culture, which is prior to the appearance of many markers of terminal differentiation (43). Our screen was focused on isolating members of the paired-like class of homeobox genes, which are relatively underrepresented in sequence databases (7). Members of this class of homeobox genes display overall similarity to the Drosophila paired homeobox, but unlike prototypic paired/Pax genes, they lack sequences encoding a second DNA binding region (the paired domain). In addition, paired-like homeobox genes encode a glutamine at position 50 of the homeodomain instead of the serine characteristic of paired/Pax homeobox genes (7), which is notable since the residue at position 50 is an important determinant of DNA binding specificity (20, 50). Consequently, we designed a degenerate oligonucleotide to recognize the sequence QVWF(Q/K)NRR, which is representative of many members of the paired-like homeodomain subclass. Among the genes isolated by this approach was the paired-like homeobox gene Esx1 (5, 26).
Sequence analysis of seven independent Esx1 cDNA clones revealed the presence of two distinct transcripts that differ at their 5′ ends (transcripts A and B [Fig. 1A and B]). We examined the expression of these two Esx1 transcripts by RNase protection analysis with an antisense probe that produces two protected bands corresponding to transcripts A and B (Fig. 1B and C). We found that Esx1 expression was limited to extraembryonic tissues and to the adult testis (Fig. 1C), consistent with previous reports (5, 26). Furthermore, the relative abundance of the two Esx1 transcripts varies between different tissues, with transcript A being more abundant in the testis, transcript B being more abundant in the placenta, and equivalent levels being found in undifferentiated ES cells (Fig. 1C).
Because their 5′ sequences are distinct, the protein product of transcript B is predicted to begin at an internal methionine and to lack 68 residues at the N terminus of the product of transcript A (Fig. 1A and B). Aside from their differing N-terminal regions, the products of transcripts A and B are otherwise identical and contain several notable features: (i) a divergent paired-like homeodomain; (ii) a proline-rich region that contains a putative SH3 binding motif; (iii) a region consisting almost exclusively of glutamic acids and glutamines (E/Q-rich region), which may correspond to a transcriptional activation domain; and (iv) an unusual repeat sequence consisting of prolines alternating with phenylalanines or asparagines, that we termed the PF/PN motif, which is not found in known proteins in sequence databases (Fig. 1A and B). Therefore, the predicted ESX1 protein displays a complex modular organization, since alternative transcripts encode proteins with distinct N-terminal regions that have features of transcription factors (the homeodomain and glutamic acid/glutamine-rich regions) and signaling molecules (putative SH3 binding domains), as well as other novel regions (the PF/PN motif).
The subcellular distribution of ESX1 protein is primarily cytoplasmic in the developing placenta.
A notable feature of the Esx1 expression pattern during development is that it is restricted to extraembryonic tissues, as shown by published in situ hybridization studies (26) and by our RNase protection analysis (Fig. 1C). We now show that Esx1 is specifically expressed during early development in extraembryonic tissues that form the placenta and subsequently continues to be expressed in the labyrinth layer of the placenta, where its protein product is localized primarily to the cytoplasm (Fig. 2 and 3).
FIG. 2.
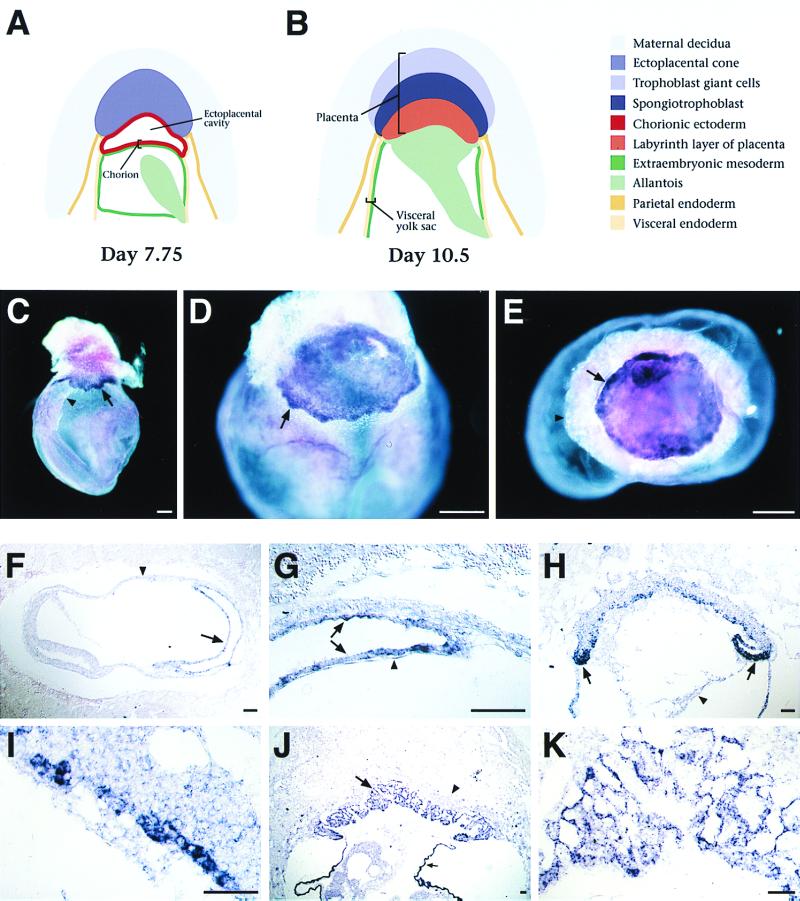
Esx1 expression in extraembryonic tissues. (A and B) Schematic depiction of stages in extraembryonic development using nomenclature from a previous report (24). (C to E) Whole-mount in situ hybridization analysis of intact egg cylinders. (C) Expression of Esx1 in the chorion (arrow) on day 8.0 (2 somites), just prior to fusion with the allantois (arrowhead). (D) Punctate staining in the developing placenta (arrow) on day 8.5, shortly after chorioallantoic fusion. (E) View of the day 8.5 placenta from below with the embryo removed; the ectoplacental cone and visceral yolk sac are beneath the plane of focus. Esx1 expression appears highest around the edges of the placenta (arrow). (F to K) Section in situ hybridization to cryosections of egg cylinders on days 7.5 through 9.5 of gestation. (F) Sagittal section through a day 7.5 egg cylinder (head-fold stage); the ectoplacental cone is to the right. Esx1 is expressed in the chorion (arrow) but not in the visceral yolk sac (arrowhead). (G) Higher-power view of panel F, showing staining of the chorionic ectoderm (arrows) but not of the chorionic mesoderm (arrowhead). (H) Sagittal section on day 8.5 of gestation, with scattered Esx1 expression in the newly formed placenta, with the highest levels at the inverted lateral margins (laminae) of the ectoplacental plate (arrows); no expression is observed in the allantois (arrowhead). (I) Higher-power view of the ectoplacental plate from panel H. (J) Low-power view of the placenta on day 9.5 of gestation. Expression is found in the developing labyrinth layer (arrow) but not in the spongiotrophoblast layer (arrowhead). Expression is also evident in the visceral endoderm of the yolk sac (small arrow); note that this is consistent with the expression of Esx1 during differentiation of ES embryoid bodies, which produce visceral endoderm. (K) Higher-power view of the labyrinth layer. In panels C to E, bars represent 200 μm; in panels F to K, bars represent 50 μm.
FIG. 3.
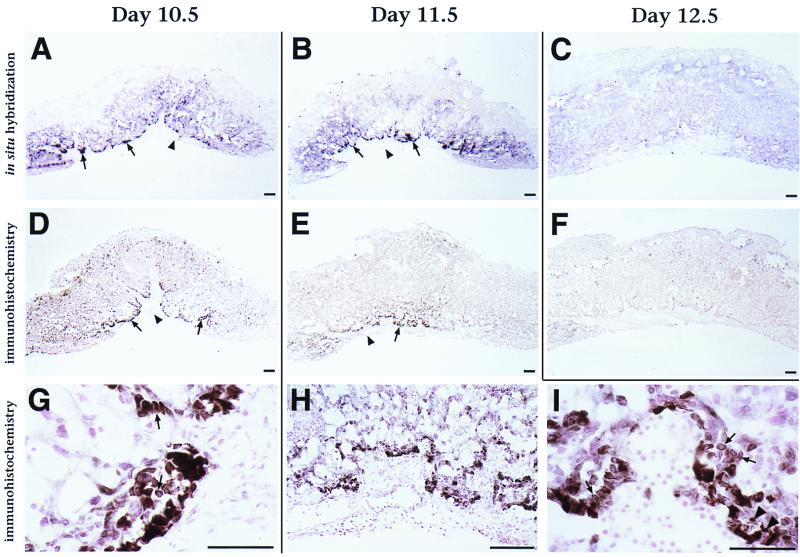
ESX1 protein is localized primarily to the cytoplasm of extraembryonic tissues. (A to C) Section in situ hybridization analysis of Esx1 expression in the placenta on days 10.5, 11.5, and 12.5 of gestation. Arrows point to intense punctate expression at the edge of the labyrinth layer of the placenta on days 10.5 and 11.5; note the lack of expression in the adjacent allantoic plate (arrowheads). (D to I) Immunohistochemical staining of ESX1 protein expression in the placenta on days 10.5, 11.5, and 12.5. Note that the darker staining (brown) corresponds to ESX1 immunoreactivity whereas the light color (pink) corresponds to the counterstain with nuclear fast red. (D to F) Low-power views of ESX1 protein expression in the labyrinth layer of the placenta; note the overall similarity to the in situ hybridization results. As in panels A to C, arrows indicate expression at the edge of the labyrinth layer and arrowheads indicate the adjacent allantoic plate. (G to I) High-power views show intense ESX1 expression in a subpopulation of the labyrinth cells on day 10.5 (G) and on day 11.5 (H and I), with localization predominantly to the cytoplasm (arrows) and with scattered nuclear staining (arrowheads). Bars, 25 μm.
In the mouse, the placenta develops from a complex association of tissue layers that are derived from several distinct extraembryonic lineages, as shown in Fig. 2A and B, which schematically display these tissue layers prior to and following placenta formation. Unlike other tissues, the placenta is derived from both the trophectoderm and the inner cell mass of the blastocyst (reviewed in reference 15). The trophectoderm gives rise to the chorionic ectoderm, which will contribute to the labyrinth layer of the placenta, and it also gives rise to the ectoplacental cone, which ultimately forms the trophoblast giant cells and spongiotrophoblast layers of the placenta. The inner cell mass, which also gives rise to the embryo proper, generates extraembryonic mesoderm and the allantois, as well as the extraembryonic parietal and viceral endoderm. Placenta formation occurs around embryonic day 8.5 when the allantois fuses with the chorion, with the chorionic tissue subsequently forming the labyrinth layer of the placenta (24, 33). Chorioallantoic fusion allows nutrient exchange between the mother and the fetus, with the labyrinth layer providing the primary maternal-fetal interface.
To examine the distribution of Esx1 transcripts during development, we performed in situ hybridization analysis from days 7.5 to 12.5 of mouse gestation by using digoxygenin-labeled riboprobes, which afford a high degree of cellular and anatomical resolution. Our in situ analysis, which confirms and extends previous studies (26), shows that Esx1 was expressed during late gastrulation (on day 7.75) in the chorionic ectoderm, prior to its fusion with the allantois (Fig. 2C and F). At this stage, expression was undetectable in any other extraembryonic tissue, including the ectoplacental cone and the visceral endoderm (Fig. 2C, F, and G). Following chorioallantoic fusion, Esx1 was expressed in the ectoplacental plate (on day 8.5) and the developing labyrinth layer of the placenta (on day 9.5) (Fig. 2D, E, and H to K). Interestingly, Esx1 was expressed in a punctate fashion within the ectoplacental plate and was most highly expressed in its inverted lateral margins (laminae), as is evident by both whole-mount and section in situ hybridization (Fig. 2E and H). Esx1 expression continued in the labyrinth layer of the placenta from day 9.5 onward, with a peak of expression on days 10.5 and 11.5 but with decreased expression on day 12.5 (Fig. 2J and K; Fig. 3A to C).
To investigate the distribution of ESX1 protein, we performed immunohistochemical analysis of placental tissues by using affinity-purified anti-ESX1 IgG (Fig. 3D to I). We found that the distribution of ESX1 protein in the placenta mirrored that of its corresponding mRNA (compare Fig. 3A to C with Fig. 3D to F). Thus, anti-ESX1 staining is restricted to the labyrinth layer of the placenta, where it is concentrated in the outer margin and appears to have a punctate distribution. The anti-ESX1 staining was detectable in the placenta on day 9.5, becoming most intense on days 10.5 and 11.5 and decreasing in intensity by day 12.5 (Fig. 3D to F) (data not shown), which is similar to the expression of Esx1 transcripts. Notably, the peak expression of Esx1 transcripts and protein occurring on day 11.5 corresponds temporally to the initial appearance of the placental defects in Esx1 knockout mice (25).
High-power views revealed an unexpected feature of the anti-ESX1 staining pattern, namely, that it was primarily cytoplasmic in a majority of the immunostained cells (Fig. 3G to H). In contrast, only a subset of the cells displayed the nuclear staining that would be anticipated for a putative transcription factor. We have also observed a cytoplasmic distribution of anti-ESX1 staining in the adult testis (data not shown), another major site of Esx1 expression (Fig. 1C) (5). The observed cytoplasmic localization of ESX1 protein has interesting implications for its potential function and/or mode of regulation during placental development.
The PF/PN motif confers cytoplasmic localization of ESX1.
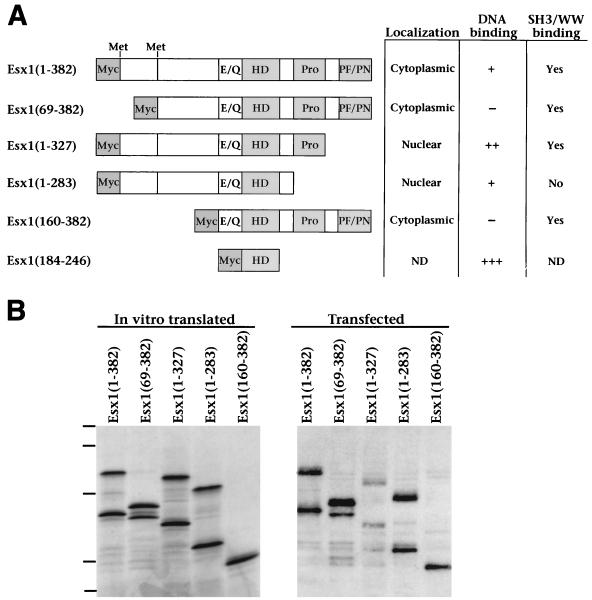
To investigate the contribution of the various protein regions of ESX1 to its subcellular localization and other biochemical activities, we produced a series of truncated ESX1 proteins, which contained the homeodomain but lacked different portions of the N- or C-terminal regions (Fig. 4A). Since ESX1 has two alternative transcripts that differ at their 5′ ends (Fig. 1A and B), we designated the putative initiator methionine of the longer form (transcript A) amino acid 1 [ESX1(1–382)] and that of the shorter form (transcript B) as amino acid 69 [ESX1(69–382)] (Fig. 1A, boxed). Each of the truncated ESX1 proteins was stably expressed following in vitro translation or transient transfection in mammalian cells (Fig. 4B).
FIG. 4.
Analysis of ESX1 protein motifs. (A) Diagram of the full-length and truncated ESX1 proteins. Shown are the glutamic acid- and glutamine-rich region (E/Q), the homeodomain (HD), the proline-rich region (Pro), and the PF/PN region. The numbering of amino acid sequences (indicated in parentheses) is based on the product of transcript A, such that ESX1(1–382) corresponds to the product of transcript A and ESX1(69–382) corresponds to the product of transcript B. Also indicated are the positions of the two alternative initiator methionines for transcripts A and B at amino acids 1 and 69, respectively. The truncated ESX1 proteins also contain an N-terminal Myc epitope as indicated. The table on the right summarizes the subcellular localization, DNA binding properties, and SH3/WW domain interactions for each ESX1 protein; primary data are shown in Fig. 5 to 7. Symbols: +++, strong DNA binding activity; ++ and +, weaker binding activity; −, no detectable binding activity; ND, not determined. (B) Comparison of ESX1 proteins produced by in vitro transcription-translation or by transfected mammalian cells. For in vitro transcription-translation (left), pcDNA3-Esx1 plasmids encoding the indicated proteins were used to synthesize the corresponding mRNA and this was followed by translation with rabbit reticulyocyte lysate in the presence of [35S]methionine. In vitro-translated proteins (3 μl) were resolved by SDS-PAGE (12% polyacrylamide) and visualized by autoradiography. For expression in mammalian cells (right), these pcDNA3-Esx1 plasmids were transfected into COS1 cells and cellular proteins were extracted in SDS lysis buffer. Cell lysates (15 μl) were resolved by SDS-PAGE (12% polyacrylamide) and visualized by Western blot analysis with anti-ESX1 antisera. Dashes correspond to molecular mass standards of 110, 68, 45, 31, and 20 kDa.
The ESX1 derivatives containing the first 68 amino acids (the N-terminal extension) [ESX1(1–382), ESX1(1–327), and ESX1(1–283)] generated two polypeptides of approximately equal intensity, which differed significantly in their mobility following in vitro translation or transient transfection (Fig. 4B). Because both polypeptides were recognized by anti-ESX1 antisera, while only the lower-mobility ESX1 polypeptides (the upper bands) were recognized by anti-Myc antisera (Fig. 4B) (data not shown), we infer that the upper bands initiate at amino acid 1 while the higher-mobility polypeptides (the lower bands) initiate at the downstream methionine (amino acid 69). Therefore, the downstream methionine at amino acid 69, which is presumed to be the initiator methionine for transcript B (Fig. 1A and B), is efficiently utilized in vitro as well as in mammalian cells. Inspection of the amino acid sequence of the N-terminal extension reveals the presence of multiple serine and threonine residues that are putative phosphorylation sites (Fig. 1A). Furthermore, we have observed that phosphatase treatment significantly alters the mobility of the upper bands (data not shown). Therefore, the N-terminal extension is likely to be extensively modified by phosphorylation, accounting for the relatively low mobility of ESX1 proteins containing this region.
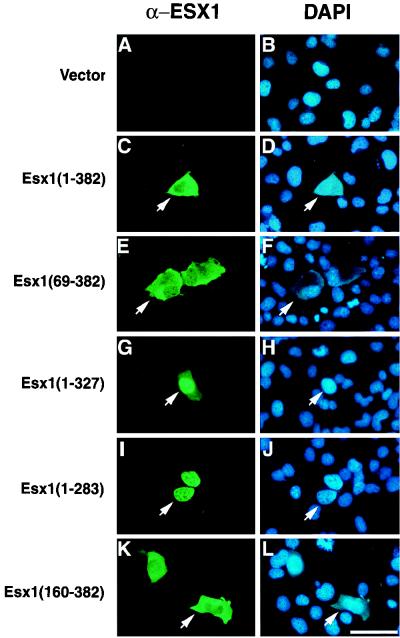
To examine the subcellular localization of the full-length and truncated ESX1 polypeptides, we transiently transfected the corresponding expression plasmids into COS-1 cells and examined their distribution by indirect immunofluorescence with anti-ESX1 antisera (Fig. 5). As we observed for endogenous ESX1 (Fig. 3G to I), the full-length protein [ESX1(1–382)] was localized primarily to the cytoplasm of transfected cells and did not overlap with the DAPI nuclear staining (Fig. 5C and D). The N-terminal extension or other N-terminal sequences did not influence the subcellular localization of ESX1, since polypeptides lacking these N-terminal regions [ESX1(69–382) and ESX1(160–382)] were also primarily cytoplasmic (Fig. 5E, F, K, and L). In contrast, ESX1 polypeptides lacking C-terminal sequences, in particular the PF/PN motif [ESX1(1–327) and ESX1(1–283)], exhibited nuclear localization, which overlaps with the DAPI staining (Fig. 5G to J). Taken together, these findings suggest that the PF/PN motif masks or inhibits the nuclear localization signal(s) located in other regions of the ESX1 protein.
FIG. 5.
The PF/PN domain inhibits nuclear localization of ESX1 in transfected cells. A control plasmid (Vector) or pcDNA3-Esx1 plasmids encoding the indicated proteins were transfected into COS1 cells, and the ESX1 proteins were visualized by indirect immunofluorescence with anti-ESX1 antisera (α-Esx1) (A, C, E, G, I, and K), while cell nuclei were visualized by DAPI staining (B, D, F, H, J, and L). For each pair of images, arrows point to the same cell showing the anti-ESX1 antiserum staining (green) or the DAPI staining (blue). Note that ESX1 proteins containing the PF/PN domain [ESX1(1–382), ESX1(69–382), and ESX1(160–382)] are primarily cytoplasmic whereas those lacking this domain [ESX1(1–327) and ESX1(1–283)] are primarily nuclear. Bar, 25 μm.
The PF/PN motif inhibits the DNA binding activity of ESX1.
To date, few members of the paired-like class of homeodomains have been characterized in terms of their DNA binding properties. Moreover, although the ESX1 homeodomain displays the defining sequence features of the paired-like class, it has only 62% amino acid identity to the most closely related homeodomains (7) and does not contain a conserved region (the paired-tail motif) that defines a large subclass of paired-like proteins (32). Furthermore, inspection of the ESX1 sequence reveals an unusual feature of its homeodomain, namely, a proline at position 43 (Fig. 1A), which is rarely found in homeodomains (7) and would be expected to truncate the DNA recognition helix by three residues. Therefore, we performed electrophoretic mobility shift assays to examine the DNA binding properties of the full-length and truncated ESX1 polypeptides produced by in vitro translation (Fig. 6). For this purpose, we used DNA sites that are known to interact with members of several classes of homeodomains (sites 6, 6-5, and 6-11) (9) or with members of the paired (site P2) (53) or paired-like (site PRDQ9) (45) classes of homeodomains, or a control site lacking the TAAT core that is required for homeoprotein-DNA binding (site 6-19) (9).
FIG. 6.
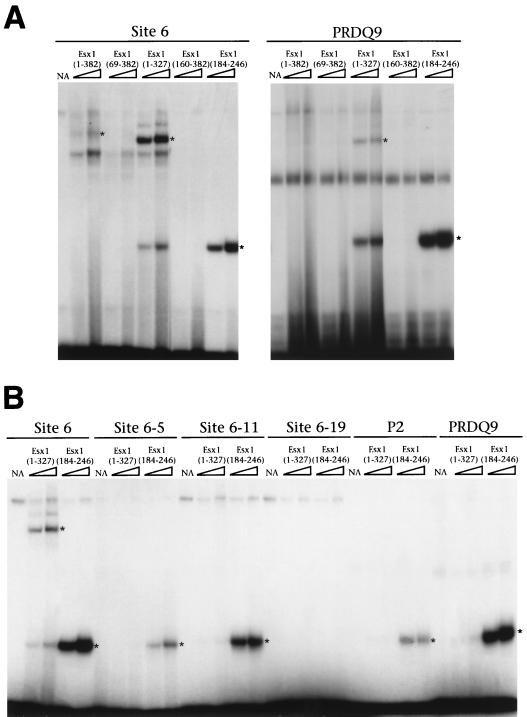
The PF/PN domain inhibits DNA binding of ESX1 proteins. Electrophoretic mobility shift assays were performed as described previously (9), using increasing amounts (3 or 6 μl; triangle) of the indicated ESX1 proteins obtained by in vitro translation. The DNA sites used are indicated; their sequences are provided in Materials and Methods. The specific protein-DNA complexes are indicated by asterisks. NA, unprogrammed reticulocyte lysate.
In general, the DNA binding activity of full-length ESX1 [ESX1(1–382)] was relatively weak and was detectable only with DNA site 6 (Fig. 6A). Moreover, no DNA binding activity was observed for an ESX1 protein lacking the N-terminal extension [ESX1(69–382)] or one lacking most of the N-terminal region [ESX1(160–382] (Fig. 6A). However, an ESX1 protein lacking the PF/PN domain [ESX1(1–327)] bound avidly to DNA while retaining a preference for binding to site 6 (Fig. 6). A truncated polypeptide corresponding to the homeodomain [ESX1(184–246)] bound to DNA with greater avidity although with broader specificity, since it interacted to various degrees with all of the DNA sites except the control site lacking the TAAT core (site 6-19).
Taken together, these data reveal several notable features of the DNA binding properties of ESX1. First, these findings indicate that ESX1 interacts specifically with DNA, albeit with low affinity, and with a distinct binding specificity from that of other paired-like homeoproteins, since it binds preferentially to site 6 rather than to sites P2 and/or PRDQ9. Second, N-terminal protein regions of ESX1 appear to contribute to its overall binding specificity and affinity for DNA. Third, ESX1 appears to bind to DNA as a monomer, in contrast to other paired-like homeodomains, which bind as dimers (45, 53), as is apparent in Fig. 6 and confirmed by more extensive mixing experiments (data not shown). Finally, and most notably, our results reveal a second potential function for the PF/PN motif, which is to inhibit the DNA binding activity of ESX1.
The ESX1 protein contains an SH3 binding motif.
The SH3 domain is a modular protein interaction domain found in a variety of proteins, including many involved in signal transduction (reviewed in references 12 and 47). This domain recognizes short proline-rich sequences that consist of a PXXP core flanked by other residues, frequently prolines and basic amino acids, which confer binding specificity (18, 27, 36, 47, 54). Inspection of the ESX1 proline-rich region revealed one sequence with an excellent match to the c-abl SH3 binding consensus (E#1, PPFRPPPLPPF) (36), as well as two other sequences that contain a PXXP core but whose flanking amino acids do not appear to correspond to SH3 binding motifs (E#2, PPFPWPPVPPD; E#3, YPMVPRPMHPQ) (Fig. 1A and B). Notably, the sequence of ESX1 peptide E#1 is similar to that of the SH3 binding domain of 3BP-1 (APTMPPPLPP), which was isolated on the basis of its interaction with c-abl (11, 36); however, ESX1 peptide E#1 lacks a leucine at position 6 that is strongly preferred for binding to the c-fyn and c-src SH3 domains (37, 46). Moreover, ESX1 peptide #1 has an arginine at position 4, which is frequently found in SH3 binding peptides that bind in the class I orientation, and it has been suggested to interact with a corresponding conserved aspartic acid residue in the SH3 domain (18, 27). While the sequence determinants of SH3 binding specificity are not yet completely defined, it is clear that nonproline residues confer specific recognition of distinct SH3 domains (12). In particular, polyproline stretches such as those found in many transcription factors do not bind SH3 domains in vitro (36).
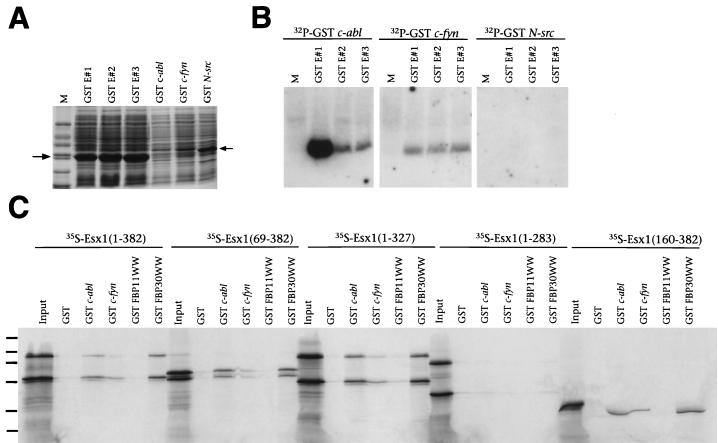
To investigate whether these proline-rich ESX1 peptides bind to SH3 domains in vitro, we performed Far-Western interaction experiments with GST fusion proteins containing E#1, E#2, and E#3 and radiolabeled SH3 domains from c-abl, c-fyn, or N-src (Fig. 7A and B). We found that ESX1 peptide E#1, but not E#2 or E#3, binds strongly in vitro to the c-abl SH3 domain, weakly to the c-fyn SH3 domain, and not at all to the N-src SH3 domain (Fig. 7B). We next asked whether the full-length or truncated ESX1 proteins exhibit SH3 binding activity by using GST interaction assays performed with the GST–c-abl and GST–c-fyn fusion proteins and 35S-labeled ESX1 proteins obtained by in vitro translation (Fig. 7C). We found that each of the ESX1 proteins that contain the proline-rich region, including peptide E#1 [ESX(1–382), ESX(69–382), ESX(1–327), and ESX(160–382)], interacted in vitro with the c-abl SH3 domain and to a lesser extent with the c-fyn SH3 domain (Fig. 7C). In contrast, an ESX1 protein lacking the proline-rich region [ESX1(1–283)] did not interact with these SH3 domains (Fig. 7C). Thus, the proline-rich region of ESX1 mediates specific interactions with SH3 domains in vitro.
FIG. 7.
The proline-rich region of ESX1 confers binding to SH3 and WW domains. (A) SDS-PAGE analysis of total-cell lysates containing GST ESX1 fusion peptides (GST E#1, GST E#2, or GST E#3) and GST SH3 domain fusion proteins (GST c-abl, GST c-fyn, or GST N-src), visualized by staining with Coomassie blue. Markers (M) are 66, 45, 36, 24, 20, and 14 kDa. (B) Far-Western analysis of the interaction of GST ESX1 fusion peptides with radiolabeled SH3 domains. Each lane contains equal amounts (15 μg) of a total-cell lysate containing GST ESX1 fusion peptides (GST E#1, GST E#2, or GST E#3) probed with the indicated radiolabeled GST SH3 domain fusion proteins. M denotes the lane containing molecular mass markers. (C) GST interaction assays with radiolabeled in vitro-translated ESX1 proteins (input), followed by incubation with GST alone, with GST fused to SH3 domains (c-abl or c-fyn), or with GST fused to WW domains (FBP11WW or FBP30WW). Proteins were visualized by SDS-PAGE followed by autoradiography. Molecular mass standards (130, 90, 66, 45, 36, and 24 kDa) are indicated by dashes.
The sequence determinants for binding to the c-abl SH3 domain are similar to those for binding to several proteins containing the WW domain, a protein interaction domain named for its characteristic two tryptophan residues (3, 47). Therefore, we also tested the ability of ESX1 to interact in vitro with selected WW domains by using a GST interaction assay. Since the binding specificities of WW domains have not been as well studied as those of SH3 domains, we used GST fusions with WW domains from FBP11 and FBP30, which have distinct binding specificities (3). We found that ESX1 polypeptides containing the proline-rich region [ESX1(1–382), ESX1 (69–382), ESX1(1–327), and ESX1(160–382)] interacted strongly with the GST-FBP30 WW fusion protein and weakly with the GST-FBP11 WW protein (Fig. 7C) whereas an ESX1 protein lacking the proline-rich region [ESX1(1–283)] did not interact with these WW domains (Fig. 7C). Taken together, these findings demonstrate that the proline-rich region of ESX1 modulates specific interactions with both SH3 and WW domains, raising the possibility that ESX1 is involved in signaling interactions mediated through such protein-protein interactions.
DISCUSSION
Esx1 is one of the few regulatory genes known to have specific expression and function in the placenta during development. The present study has revealed several novel features of the expression pattern and biochemical functions of the ESX1 protein. First, we have found that ESX1 is localized primarily to the cytoplasm in the labyrinth layer of the placenta in vivo. Second, we have shown that ESX1 contains a novel protein motif, termed the PF/PN domain, which inhibits both nuclear localization and DNA binding activity. Finally, we have found that ESX1 contains a proline-rich region that mediates specific interactions with the c-abl SH3 domain as well as certain WW domains. These findings suggest that the essential function of ESX1 in the placenta during embryogenesis may be mediated by its ability to relay signaling events in the cytoplasm to transcriptional activity in the nucleus and may be regulated through the inhibitory action of the PF/PN domain.
The subcellular localization of ESX1 suggests its regulated nuclear entry.
In contrast to the numerous studies that have examined the expression of homeobox genes during development, fewer studies have investigated the expression patterns and/or subcellular localization of their corresponding protein products. Most of the homeoproteins that have been examined are localized to the nucleus, consistent with their roles as putative transcription factors (see, e.g., references 14, 48, and 51). However, some notable exceptions include the murine Emx1 homeoprotein, which exhibits nuclear localization early in development but is localized to olfactory axons in the nervous system at later stages (6). Moreover, the vertebrate Engrailed homeoproteins are localized to nuclei as well as caveola-like vesicles, where they are thought to relay signals from axon terminals to the nucleus (22, 30). In addition, members of the Extradenticle/PBX class of homeoproteins are found in the cytoplasm as well as in the nucleus, due to competition between active nuclear export through a CRM-dependent mechanism and nuclear import by association with the essential cofactors homothorax and PREP1 (1, 4, 31, 38). In each of these cases, the extranuclear localization of these homeoproteins is presumed to reflect their regulated nuclear entry and subsequent function as transcription factors. On the other hand, certain homeoproteins are known to have functions in the cytoplasm for posttranscriptional control; in particular, Bicoid functions as a translational regulator through binding to mRNA, in addition to its activity as a transcription factor (17, 39).
Our observation that the ESX1 protein is primarily cytoplasmic in vivo most probably reflects regulated nuclear transport and subsequent transcriptional activity, since it is also localized to the nucleus in a subset of the labyrinth cells of the placenta. Furthermore, ESX1 contains a nuclear localization signal(s) that is masked or inhibited by the PF/PN domain in cell culture. However, we cannot rule out the possibility that the cytoplasmic localization reflects an alternative function of ESX1, other than (or in addition to) its presumed activity as a transcription factor. In this regard, it is notable that ESX1 is relatively inefficient at DNA binding and that it contains an SH3 binding motif, which is often found in signaling molecules. Although our data do not distinguish between retention of ESX1 in the cytoplasm and its active export from the nucleus, the inhibitory activity of the PF/PN domain suggests a novel mechanism for regulating nuclear entry.
Potential roles of the unusual protein motifs in ESX1.
ESX1 contains at least two protein motifs that are not generally found in homeoproteins or other transcription factors. The first unusual feature is the presence of an SH3 binding motif that can interact in vitro with the c-abl SH3 domain as well as certain WW domains. While the precise in vivo target(s) for this interaction remains to be identified, the ability of ESX1 to bind to the c-abl SH3 domain in vitro indicates its potential to interact with related SH3 domains in vivo. It is noteworthy that a proline-rich domain containing potential SH3 binding motifs has been noted in another paired-like homeoprotein, Shox, although it is unknown whether it binds to SH3 domains (35).
Both the well-studied SH3 domain and the more recently identified WW domain represent protein interaction modules that participate in a broad range of cellular signaling pathways. While SH3 domains are found predominantly in cytoplasmic proteins, they are occasionally encountered in proteins that reside in or transit into the nucleus, such as c-abl (41). Indeed, SH3 domains may be used to direct proteins to specific subcellular locations, as in the case of the adapter protein GRB-2 and phospholipase C-γ (2). Furthermore, it has been suggested that certain SH3 domain interactions can relay cytoplasmic signaling events to the nucleus, as is the case for the interaction between c-vav and heterogeneous ribonucleoprotein K (21).
A second unusual feature of ESX1 is the PF/PN motif, which we have not found in other proteins in sequence databases. This peculiar motif consists of an alternating repeat of proline and phenylalanine residues followed by proline and asparagine residues (Fig. 1A), which is predicted to assume a flexible coiled-coil secondary structure. The PF/PN motif may provide an effective means of regulating the transcriptional activity of ESX1, since it can inhibit both nuclear localization and DNA binding activity. We propose that the inhibitory role of the PF/PN motif may be mediated by intramolecular interactions with the homeodomain and other protein regions responsible for nuclear localization and DNA binding and/or by interference with SH3 or WW domain binding by the proline-rich region. In this regard, we speculate that binding to an SH3 or WW domain-containing protein may unmask nuclear localization signals, allowing transport of ESX1 to the nucleus and subsequent transcriptional activity.
In addition to its unusual protein motifs and atypical homeobox sequence, other novel properties of Esx1 suggest its recent evolutionary origin. Esx1 is an X chromosome imprinted gene (25), which is notable since genomic imprinting arose during mammalian evolution and is intimately associated with placental development (49). The recent evolution of chorioallantoic placentation may have allowed the utilization of novel molecular mechanisms and signaling pathways that have not been encountered in the better-conserved processes that underlie fetal development.
ACKNOWLEDGMENTS
We thank Philip Leder, in whose laboratory this work was initiated, for his advice and generous support. We also thank Mark Bedford and Philip Leder for the WW domain fusion proteins. We acknowledge the contributions of Lu Yang, Hongyu Wang, Peter Sciavolino, and David Chan during the initial stages of this work. We are also grateful to Thomas Bürglin for advice on library screening, Chaosu E for assistance with DNA sequencing, and Drew Vershon for helpful comments on the manuscript.
S.M.S. was supported by a predoctoral fellowship (96-2003-CCR-00) from the New Jersey Commission on Cancer Research. C.A.-S. was supported by NIH grant HD33362, and M.M.S. was supported by a Leukemia Society of America Special Fellowship and by NIH grant HL60212.
Y.-T.Y., S.M.S., and J.D. contributed equally to this work.
REFERENCES
- 1.Abu-Shaar M, Ryoo H D, Mann R S. Control of the nuclear localization of Extradenticle by competing nuclear import and export signals. Genes Dev. 1999;13:935–945. doi: 10.1101/gad.13.8.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bar-Sagi D, Rotin D, Batzer A, Mandiyan V, Schlessinger J. SH3 domains direct cellular localization of signaling molecules. Cell. 1993;74:83–91. doi: 10.1016/0092-8674(93)90296-3. [DOI] [PubMed] [Google Scholar]
- 3.Bedford M T, Chan D C, Leder P. FBP WW domains and the Abl SH3 domain bind to a specific class of proline-rich ligands. EMBO J. 1997;16:2376–2383. doi: 10.1093/emboj/16.9.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berthelsen J, Kilstrup-Nielsen C, Blasi F, Mavilio F, Zappavigna V. The subcellular localization of PBX1 and EXD proteins depends on nuclear import and export signals and is modulated by association with PREP1 and HTH. Genes Dev. 1999;13:946–953. doi: 10.1101/gad.13.8.946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Branford W W, Zhao G-Q, Valerius M T, Weinstein M, Birkenmeier E H, Rowe L B, Potter S S. Spx1, a novel X-linked homeobox gene expressed during spermatogenesis. Mech Dev. 1997;65:87–98. doi: 10.1016/s0925-4773(97)00058-0. [DOI] [PubMed] [Google Scholar]
- 6.Briata P, Di Blas E, Gulisano M, Mallamaci A, Iannone R, Boncinelli E, Corte G. EMX1 homeoprotein is expressed in cell nuclei of the developing cerebral cortex and in the axons of the olfactory sensory neurons. Mech Dev. 1996;57:169–180. doi: 10.1016/0925-4773(96)00544-8. [DOI] [PubMed] [Google Scholar]
- 7.Bürglin T R. A comprehensive classification of homeobox genes. In: Duboule D, editor. Guidebook to the homeobox genes. Oxford, United Kingdom: Oxford University Press; 1994. pp. 25–71. [Google Scholar]
- 8.Bürglin T R, Finney M, Coulson A, Ruvkun G. Caenorhabditis elegans has scores of homeobox-containing genes. Nature. 1989;341:239–243. doi: 10.1038/341239a0. [DOI] [PubMed] [Google Scholar]
- 9.Catron K M, Iler N, Abate C. Nucleotides flanking a conserved TAAT core dictate the DNA binding specificity of three murine homeodomain proteins. Mol Cell Biol. 1993;13:2354–2365. doi: 10.1128/mcb.13.4.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan D C, Bedford M T, Leder P. Formin binding proteins bear WWP/WW domains that bind proline-rich peptides and functionally resemble SH3 domains. EMBO J. 1996;15:1045–1054. [PMC free article] [PubMed] [Google Scholar]
- 11.Cicchetti P, Mayer B J, Thiel G, Baltimore D. Identification of a protein that binds to the SH3 region of Abl and is similar to Bcr and GAP-rho. Science. 1992;257:803–806. doi: 10.1126/science.1379745. [DOI] [PubMed] [Google Scholar]
- 12.Cohen G B, Ren R, Baltimore D. Modular binding domains in signal transduction proteins. Cell. 1995;80:237–248. doi: 10.1016/0092-8674(95)90406-9. [DOI] [PubMed] [Google Scholar]
- 13.Copp A J. Death before birth: clues from gene knockouts and mutations. Trends Genet. 1995;11:87–93. doi: 10.1016/S0168-9525(00)89008-3. [DOI] [PubMed] [Google Scholar]
- 14.Corsetti M T, Levi G, Lancia F, Sanseverino L, Ferrini S, Boncinelli E, Corte G. Nucleolar localisation of three Hox homeoproteins. J Cell Sci. 1995;108:187–193. doi: 10.1242/jcs.108.1.187. [DOI] [PubMed] [Google Scholar]
- 15.Cross J C, Werb Z, Fisher S J. Implantation and the placenta: key pieces of the development puzzle. Science. 1994;266:1508–1518. doi: 10.1126/science.7985020. [DOI] [PubMed] [Google Scholar]
- 16.Doetschman T C, Eistetter H, Katz M, Schmidt W, Kemler R. The in vitro development of blastocyst-derived embryonic stem cell lines: formation of visceral yolk sac, blood islands and myocardium. J Embryol Exp Morphol. 1985;87:27–45. [PubMed] [Google Scholar]
- 17.Dubnau J, Struhl G. RNA recognition and translational regulation by a homeodomain protein. Nature. 1996;379:694–699. doi: 10.1038/379694a0. [DOI] [PubMed] [Google Scholar]
- 18.Feng S, Chen J K, Yu H, Simon J A, Schreiber S L. Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions. Science. 1994;266:1241–1247. doi: 10.1126/science.7526465. [DOI] [PubMed] [Google Scholar]
- 19.Guillemot F, Nagy A, Auerbach A, Rossant J, Joyner A L. Essential role of Mash-2 in extraembryonic development. Nature. 1994;371:333–336. doi: 10.1038/371333a0. [DOI] [PubMed] [Google Scholar]
- 20.Hanes S D, Brent R. DNA specificity of the bicoid activation protein is determined by homeodomain recognition helix residue 9. Cell. 1989;57:1275–1283. doi: 10.1016/0092-8674(89)90063-9. [DOI] [PubMed] [Google Scholar]
- 21.Hobert O, Jallal B, Schlessinger J, Ullrich A. Novel signaling pathway suggested by SH3 domain-mediated p95vav/heterogeneous ribonucleoprotein K interaction. J Biol Chem. 1994;269:20225–20228. [PubMed] [Google Scholar]
- 22.Joliot A, Trembleau A, Raposo G, Calvet S, Volovitch M, Prochiantz A. Association of Engrailed homeoproteins with vesicles presenting caveolae-like properties. Development. 1997;124:1865–1875. doi: 10.1242/dev.124.10.1865. [DOI] [PubMed] [Google Scholar]
- 23.Kaelin W G, Krek W, Sellers W R, DeCaprio J A, Ajchenbaum F, Fuchs C S, Chittenden T, Li Y, Farnham P J, Blanar M A, Livingston D M, Flemington E K. Expression cloning of a cDNA encoding a retinoblastoma-binding protein with E2F-like properties. Cell. 1992;70:351–364. doi: 10.1016/0092-8674(92)90108-o. [DOI] [PubMed] [Google Scholar]
- 24.Kaufman M H. The atlas of mouse development. San Diego, Calif: Academic Press, Inc.; 1992. [Google Scholar]
- 25.Li Y, Behringer R R. Esx1 is an X-chromosome-imprinted regulator of placental development and fetal growth. Nat Genet. 1998;20:309–311. doi: 10.1038/3129. [DOI] [PubMed] [Google Scholar]
- 26.Li Y, Lemaire P, Behringer R R. Esx1, a novel X chromosome-linked homeobox gene expressed in mouse extraembryonic tissues and male germ cells. Dev Biol. 1997;188:85–95. doi: 10.1006/dbio.1997.8640. [DOI] [PubMed] [Google Scholar]
- 27.Lim W A, Richards F M, Fox R O. Structural determinants of peptide-binding orientation and of sequence specificity in SH3 domains. Nature. 1994;372:375–379. doi: 10.1038/372375a0. [DOI] [PubMed] [Google Scholar]
- 28.Lin T-P, Labosky P A, Grabel L B, Kozak C A, Pitman J L, Kleeman J, MacLeod C L. The Pem homeobox gene is X-linked and exclusively expressed in extraembryonic tissues during early murine development. Dev Biol. 1994;166:170–179. doi: 10.1006/dbio.1994.1305. [DOI] [PubMed] [Google Scholar]
- 29.Luo J, Sladek R, Bader J A, Matthyssen A, Rossant J, Giguere V. Placental abnormalities in mouse embryos lacking the orphan nuclear receptor ERR-beta. Nature. 1997;388:778–782. doi: 10.1038/42022. [DOI] [PubMed] [Google Scholar]
- 30.Maizel A, Bensaude O, Prochiantz A, Joliot A. A short region of its homeodomain is necessary for Engrailed nuclear export and secretion. Development. 1999;126:3183–3190. doi: 10.1242/dev.126.14.3183. [DOI] [PubMed] [Google Scholar]
- 31.Mann R S, Abu-Shaar M. Nuclear import of the homeodomain protein extradenticle in response to Wg and Dpp signalling. Nature. 1996;383:630–633. doi: 10.1038/383630a0. [DOI] [PubMed] [Google Scholar]
- 32.Mathers P H, Grinberg A, Mahon K A, Jamrich M. The Rx homeobox gene is essential for vertebrate eye development. Nature. 1997;387:603–607. doi: 10.1038/42475. [DOI] [PubMed] [Google Scholar]
- 33.Mossman H W. Vertebrate fetal membranes. New Brunswick, N.J: Rutgers University Press; 1987. [Google Scholar]
- 34.Pettersson K, Svensson K, Mattsson R, Carlsson B, Ohlsson R, Berkenstam A. Expression of a novel member of estrogen response element-binding nuclear receptors is restricted to the early stages of chorion formation during mouse embryogenesis. Mech Dev. 1996;54:211–223. doi: 10.1016/0925-4773(95)00479-3. [DOI] [PubMed] [Google Scholar]
- 35.Rao E, Weiss B, Fukami M, Rump A, Niesler B, Mertz A, Muroya K, Binder G, Kirsch S, Winkelmann M, Nordsiek G, Heinrich U, Breuning M H, Ranke M B, Rosenthal A, Ogata T, Rappold G A. Pseudoautosomal deletions encompassing a novel homeobox gene cause growth failure in idiopathic short stature and Turner syndrome. Nat Genet. 1997;16:54–63. doi: 10.1038/ng0597-54. [DOI] [PubMed] [Google Scholar]
- 36.Ren R, Mayer B J, Cicchetti P, Baltimore D. Identification of a ten-amino acid proline-rich SH3 binding site. Science. 1993;259:1157–1161. doi: 10.1126/science.8438166. [DOI] [PubMed] [Google Scholar]
- 37.Rickles R J, Botfield M C, Weng Z, Taylor J A, Green O M, Brugge J S, Zoller M J. Identification of Src, Fyn, Lyn, PI3K and Abl SH3 domain ligands using phage display libraries. EMBO J. 1994;13:5598–5604. doi: 10.1002/j.1460-2075.1994.tb06897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rieckhof G E, Casares F, Ryoo H D, Abu-Shaar M, Mann R S. Nuclear translocation of extradenticle requires homothorax which encodes an extradenticle-related homeodomain protein. Cell. 1997;91:171–183. doi: 10.1016/s0092-8674(00)80400-6. [DOI] [PubMed] [Google Scholar]
- 39.Rivera-Pomar R, Niessing D, Schmidt-Ott U, Gehring W J, Jäckle H. RNA binding and translational suppression by bicoid. Nature. 1996;379:746–749. doi: 10.1038/379746a0. [DOI] [PubMed] [Google Scholar]
- 40.Rogers M B, Hosler B A, Gudas L J. Specific expression of a retinoic acid-regulated, zinc-finger gene, Rex-1, in preimplantation embryos, trophoblast and spermatocytes. Development. 1991;113:815–824. doi: 10.1242/dev.113.3.815. [DOI] [PubMed] [Google Scholar]
- 41.Sawyers C L, McLaughlin J, Goga A, Havlik M, Witte O. The nuclear tyrosine kinase c-Abl negatively regulates cell growth. Cell. 1994;77:121–131. doi: 10.1016/0092-8674(94)90240-2. [DOI] [PubMed] [Google Scholar]
- 42.Sciavolino P J, Abrams E W, Yang L, Austenberg L P, Shen M M, Abate-Shen C. Tissue-specific expression of murine Nkx3.1 in the male urogenital system. Dev Dyn. 1997;209:127–138. doi: 10.1002/(SICI)1097-0177(199705)209:1<127::AID-AJA12>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 43.Shen M M, Leder P. Leukemia inhibitory factor is expressed by the preimplantation uterus and selectively blocks primitive ectoderm formation in vitro. Proc Natl Acad Sci USA. 1992;89:8240–8244. doi: 10.1073/pnas.89.17.8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen M M, Wang H, Leder P. A differential display strategy identifies Cryptic, a novel EGF-related gene expressed in the axial and lateral mesoderm during mouse gastrulation. Development. 1997;124:429–442. doi: 10.1242/dev.124.2.429. [DOI] [PubMed] [Google Scholar]
- 45.Sornson M W, Wu W, Dasen J S, Flynn S E, Norman D J, O'Connell S M, Gukovsky I, Carriere C, Ryan A K, Miller A P, Zuo L, Gleiberman A S, Andersen B, Beamer W G, Rosenfeld M G. Pituitary lineage determination by the Prophet of Pit-1 homeodomain factor defective in Ames dwarfism. Nature. 1996;384:327–333. doi: 10.1038/384327a0. [DOI] [PubMed] [Google Scholar]
- 46.Sparks A B, Rider J E, Hoffman N G, Fowlkes D M, Quillam L A, Kay B K. Distinct ligand preferences of Src homology 3 domains from Src, Yes, Abl, Cortactin, p53bp2, PLCgamma, Crk, and Grb2. Proc Natl Acad Sci USA. 1996;93:1540–1544. doi: 10.1073/pnas.93.4.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sudol M. From Src homology domains to other signaling molecules: proposal of the “protein recognition code”. Oncogene. 1998;17:1469–1474. doi: 10.1038/sj.onc.1202182. [DOI] [PubMed] [Google Scholar]
- 48.Tani M, Odenwald W F, Lazzarini R A, Friedrich V L., Jr Progressive restriction in the distribution of the Hox-1.3 homeodomain protein during embryogenesis. J Neurosci Res. 1989;24:457–469. doi: 10.1002/jnr.490240402. [DOI] [PubMed] [Google Scholar]
- 49.Tilghman S M. The sins of the fathers and mothers: genomic imprinting in mammalian development. Cell. 1999;96:185–193. doi: 10.1016/s0092-8674(00)80559-0. [DOI] [PubMed] [Google Scholar]
- 50.Treisman J, Gonczy P, Vashishtha M, Harris E, Desplan C. A single amino acid can determine the DNA binding specificity of homeodomain proteins. Cell. 1989;59:553–562. doi: 10.1016/0092-8674(89)90038-x. [DOI] [PubMed] [Google Scholar]
- 51.Wall N A, Jones C M, Hogan B L, Wright C V. Expression and modification of Hox 2.1 protein in mouse embryos. Mech Dev. 1992;37:111–120. doi: 10.1016/0925-4773(92)90073-s. [DOI] [PubMed] [Google Scholar]
- 52.Wilkinson M F, Kleeman J, Richards J, MacLeod C L. A novel oncofetal gene is expressed in a stage-specific manner in murine embryonic development. Dev Biol. 1990;141:451–455. doi: 10.1016/0012-1606(90)90400-d. [DOI] [PubMed] [Google Scholar]
- 53.Wilson D, Sheng G, Lecuit T, Dostatni N, Desplan C. Cooperative dimerization of paired class homeo domains on DNA. Genes Dev. 1993;7:2120–2134. doi: 10.1101/gad.7.11.2120. [DOI] [PubMed] [Google Scholar]
- 54.Yu H, Chen J K, Feng S, Dalgarno D C, Brauer A W, Schreiber S L. Structural basis for the binding of proline-rich peptides to SH3 domains. Cell. 1994;76:933–945. doi: 10.1016/0092-8674(94)90367-0. [DOI] [PubMed] [Google Scholar]
- 55.Zhang H, Hu G, Wang H, Sciavolino P, Iler N, Shen M M, Abate-Shen C. Heterodimerization of Msx and Dlx homeoproteins results in functional antagonism. Mol Cell Biol. 1997;17:2920–2932. doi: 10.1128/mcb.17.5.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]