Abstract
The aim of this systematic review and meta-analysis was to identify a list of common, candidate genes associated with the three components of fitness, specifically cardiovascular fitness, muscular strength, and anaerobic power, and how these genes are associated with exercise response phenotype variability, in previously untrained participants. A total of 3,969 potentially relevant papers were identified and processed for inclusion. After eligibility and study selection assessment, 24 studies were selected for meta-analysis, comprising a total of 3,012 participants (male n = 1,512; females n = 1,239; not stated n = 261; age 28 ± 9 years). Meta-Essentials spreadsheet 1.4 (Microsoft Excel) was used in creating the forest plots and meta-analysis. IBM SPSS statistics V24 was implemented for the statistical analyses and the alpha was set at p ≤ 0.05. 13 candidate genes and their associated alleles were identified, which were associated with the phenotypes of interest. Analysis of training group data showed significant differential phenotypic responses. Subgroup analysis showed; 44%, 72% and 10% of the response variance in aerobic, strength and power phenotypes, respectively, were explained by genetic influences. This analysis established that genetic variability explained a significant proportion of the adaptation differences across the three components of fitness in the participants post-training. The results also showed the importance of analysing and reporting specific gene alleles. Information obtained from these findings has the potential to inform and influence future exercise-related genes and training studies.
Introduction
Current evidence shows that cardiovascular fitness, muscular strength, and anaerobic power are key in determining an individuals’ health-related fitness, well-being, and quality of life, as well as successful performance in many sporting events [1–5]. For example, the , is a key index of cardiovascular and cardiorespiratory fitness and an increase in , improves the ability of the body to both supply and utilise oxygen. This prolongs the time to exhaustion and the ability to sustain aerobic exercise for longer periods of time, at higher intensities [6]. For some, this could mean the difference between being able to walk upstairs easily, rather than with effort and associated discomfort. Below average, is also correlated to increased morbidity and mortality and improvement in can prevent early mortality [7]. There is a similar rationale for improving both strength and power as well. Strength refers to the force that can be generated during a voluntary muscle contraction and is required for everyday tasks and mobility [2,5]. Anaerobic power is the ability of the neuromuscular system to produce the greatest possible action in a given time period and is needed for quick bursts explosive movements and agility [8]. Hence, it is beneficial for individuals to improve these components, irrespective of their initial level of fitness and especially, for those classed as untrained [4–6,9–12].
Although it has always been a key objective of health and exercise sciences to improve these specific key components of fitness, Schutte et al. [13] and Sarzynski, Ghosh and Bouchard, [14] stress that an individuals’ responsiveness to exercise training varies significantly, depending on the precise exercise-stimuli given. In this connection, previous studies show that a genetic component, in the response to exercise training, can explain up-to 80% of the variability in aerobic, strength and power adaptations [2,15–17]. Such findings suggest that the current health and exercise guidelines, promoting generic fitness classes and group exercises, are of questionable value, without consideration of individuals’ genetic profile. Accordingly, several well-studied genes, show significant associations with exercise trainability and successful increases in performance, providing advantages in sports and athletic competitions [18,19]. Many other genes have been shown to influence all aspects of fitness, including, but not limited to, energy-pathways, metabolism, storage, cell growth, protein, hormonal, and enzyme interactions [13,19–21]. All such genes have been termed ‘candidate genes’ [14,22–25] and may be useful indicators in predicting and producing successful training responses, to a particular exercise intervention and maximising health benefits. However, the difficulty is identifying and selecting key genes from the extensive suite of candidate genes, shown to be associated with exercise responses [13,15,17,26]. Since most current research typically relies on studies that only investigate a limited number of genes and/or single genes and their alleles, and mainly in twins or elite and high-level performance, a literature review may well be more suited in identifying multiple genes and their alleles and their applications on the untrained [17,27,28]. Thus, Ahmetov et al. [29] and Williams and Folland, [30] reported that, regardless of the high heritability of exercise response, no single gene, or polymorphism, has been shown to be solely responsible for a particular physiological variable, due to the large number of genetic polymorphisms associated with aerobic, strength and power phenotypes.
Accordingly, the aim of this review is to identify candidate genes and their alleles that best define exercise phenotype responses to training interventions, with respect to the three components of fitness, in the untrained population.
Methods
This review was conducted in accordance with the Cochrane guidelines of systematic reviews and the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) [31–33]. This review was ethically approved by the Faculty of Science and Engineering Research Ethics Panel at Anglia Ruskin University, Cambridge, UK. A comprehensive literature search was performed using Scopus, Web of Science, PubMed and SportDiscus, between June and July 2020. Multiple database thesauruses and MeSH terms were employed to pool keywords for the systematic search. An email alarm system, on each database, was created for notifications on new publications related to the search terms. All relevant studies were exported and sorted within a bibliographic management system (Refworks, ProQuest, USA) and an automatic deduplication tool was applied. Studies’ inclusion was screened in accordance with title, abstract and full text. Potential studies that met the inclusion criteria were further inspected [34]. The reference list of studies and grey literature was also explored, any missing information was requested from study authors, via email, if possible, otherwise results could not be included.
Eligibility criteria
The following PICOS criteria (Population/Participant, Intervention, Comparison, Outcome and Study type) [35] were implemented. Firstly, a minimum effective intervention time-course of two weeks (six sessions) was identified from studies such as, Astorino and Schubert, [26] and Hautala et al. [2]. Relevant outcomes were defined as the common phenotype measurements across all studies [3,5,17].
Population: (a) health untrained, human male and female participants with no stated medical conditions; (b) aged between 18–55 years; (c) training status below the ACSM norm values for cycle ergometer cardio-respiratory fitness (), one repetition maximum (1RM) leg-press and cycle ergometer peak power output (PPO), ( ≤ 45 ml.kg-1.min-1 for Males; ≤ 40 for females. Lower body 1RM ≤ 1.91 x mass for males; ≤ 1.32 x mass for females. PPO ≤ 9.22 W/kg for males; ≤ 7.65 W/kg for females) [5].
Intervention: (d) training ≥ two weeks (minimum six sessions, three per-week); (e) include either: 1) continuous endurance/aerobic Interval training, 2) resistance, or weight training, or 3) interval/sprint/anaerobic training; (f) no dietary manipulation.
Comparison: (g) Pre vs Post changes in primary phenotypes; (h) participants grouped by gene, or genotype.
Outcome: (i) change in primary variable(s): , lower body 1RM or PPO.
Study type: (j) repeated measures method; (k) original research study; (l) English language.
Studies that did not meet all the above PICOS criteria were excluded from this review.
Quality and risk of bias assessment
A COnsensus-based Standards for the selection of health status Measurement INstruments (COSMIN) checklist was implemented to evaluate the transparency and risk of bias of the studies, by measuring methodological quality [36]. The COSMIN ‘worst score’ approach was set for all items at ≥ 3, to meet the acceptable requirement of study quality and study selection [31]. To confirm the consistency and reliability of the COSMIN tool, two reviewers (HC and DG) independently evaluated the studies for inclusion. Each COSMIN item for all categories was scored from 4–1 (31 items total), where 4 was low and 1 was high risk, respectively, and any disagreements from authors were resolved using the mean COSMIN scores. Studies were only included if ≥3. 95% Limits of Agreement (LoA) were calculated using the Bland and Altman approximate method [37]. At least two studies, or groups, were required to report the same gene of interest to establish a conclusive outcome measurement, otherwise this could not be evaluated, by meta-analysis [38,39].
Statistical analysis
Means, Standard Deviations (SD), Standard Errors (SE) and 95% Confidence Intervals (CI) were extracted from all studies and pooled for analysis. Pre-to-post intervention scores for each study were converted into Effect Sizes (ES) and Standardised Mean Differences (SMD), to express intervention effects and genotypic variance between groups in standardised units. Pooled standard deviation, SE, variance (s1), 95% Cl and the weighted Mean Cohen’s d, were also calculated: classifying d values as 0.20–0.49 small, 0.50–0.79 medium and ≥ 0.80 large effect [39–41]. Meta-Essentials spreadsheet 1.4 (Microsoft Excel 2016, Washington, USA) was used for the meta-analysis and creation of forest plots. IBM SPSS statistics version 24 (SPSS, Chicago, Illinois) was used for all other statistical testing, with alpha set at p ≤ 0.05. For the assessment of any genetic effects, a subgroup stratification analysis was implemented. Here the training groups in each study were combined and then split, based on genotypes, and then further analysed and compared. Normality and homogeneity of variance were calculated via Shapiro-Wilk test and Levene’s test, respectively. Where necessary, a non-parametric Kruskal-Wallis H test was used to determine if there were any significant differences between the gene groups. Partial Eta Squared and Mean Ranks were used to determine the variability within subgroup genetics and the estimation of gene and allelic variability and their contribution towards the change in training phenotypes.
Results
A comprehensive flow diagram representing the study retrieval process and exclusions was created (Fig 1). The figure also outlines the process of the PICOS and the COSMIN checklist between reviewers. 29 studies were initially included for meta-analysis, however a further five studies were excluded, as the genes in that study were only reported on one occasion [42–46]. The final 24 studies contained 89 groups (43 aerobic; 29 strength; 17 power), with a total of 3,012 participants, and 13 candidate genes and associated alleles.
Fig 1. Flow diagram.
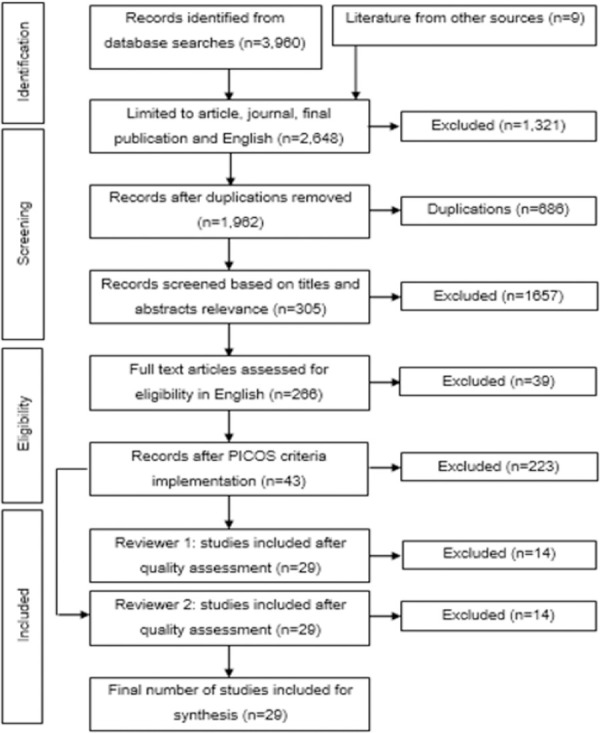
Step-by-step method of collecting and excluding studies at each stage for this review. Also, where other sources from unpublished material and grey literature were entered. This entire process was repeated twice.
Genes associated with cardiorespiratory fitness ()
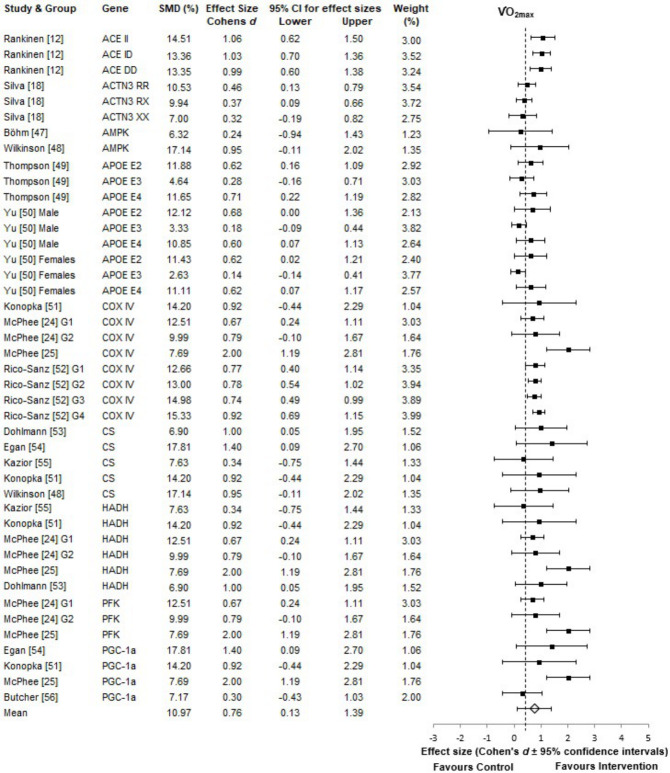
increased by 10.97 ± 3.80%, with aerobic training interventions, across all studies in this review. Forest plot analysis demonstrated that, irrespective of the genes, the overall results represented a medium to large effect-size change in , which was classed as a very highly significant improvement with training intervention alone (p < 0.001) (Fig 2). On average, these studies [12,18,24,25,47–56] used durations of 36 minutes, with intensities of 74% , or 77% HRmax, performed over 3-days a week and 12 weeks of training.
Fig 2. forest plot.
Effect sizes represent the change in post-intervention. For all plots the 95% confidence intervals were calculated and the overall mean effect size is represented with the diamond, whereas, the black squares are the individual effect size of each study. The weighting is adjusted for sample size, SD and variance. Where G1 = group 1 and G2 = group 2. Genes listed in alphabetical order.
Between subgroup analysis revealed non-normal distributions across the nine aerobic associated gene groups: D(43), .876, p < 0.05. Here, Kruskal-Wallis H testing found significant differences between the gene groups, H(8), 18.427, p = 0.018. Partial Eta Squared, calculated by Pearson’s χ2 tests, found that 44% of the total variability in the increase in post-training intervention was explained by nine gene subgroups (Table 1). Post-Hoc subgroup analysis shows the distribution of the variance in scores between the genetic groups.
Table 1. Candidate genes for cardio-respiratory fitness.
| Gene Groups | No. of Study groups | Total Group Sample Size | Mean Rank (Not adjusted for sample size) | Subgroup (Adjusted for sample size) % of weight | Mean Group Effect Size (d) |
|---|---|---|---|---|---|
| ACE | 3 | 188 | 35.33 | 14.24 | 1.02 |
| ACTN3 | 3 | 206 | 8.33 | 14.15 | 0.39 |
| AMPK/PRKAA2 | 2 | 18 | 17.25 | 7.84 | 0.62 |
| APOE | 9 | 437 | 10.44 | 13.67 | 0.41 |
| COX4I1 | 8 | 591 | 25.06 | 13.43 | 0.85 |
| CS | 5 | 46 | 28.00 | 12.12 | 0.90 |
| HADH | 6 | 106 | 25.25 | 10.58 | 0.96 |
| PFKM | 3 | 78 | 27.17 | 6.62 | 1.12 |
| PGC-1α | 4 | 52 | 28.25 | 7.36 | 1.14 |
| Total | 43 | 1,722 |
The group with the highest mean rank shows the greatest number of high scores in and subgroup % shows how much of the overall 44% variance is accounted for by each of the nine genes, adjusted for sample size, SE and within group variation in SMD.
Within this subgroup the ACE gene, regardless of allele, had the greatest mean rank and weight on the increase in . ACE alleles (II, ID, DD) showed no significant difference in effect sizes. Other “aerobic” genes (COX4I1, CS, HADH, PFKM and PGC-1α) showed no significant differences between mean rank scores and contributed equally to the increase in . However, these genes, which all showed a large effect size, had a higher mean rank than those of ACTN3, AMPK and APOE groups. Interestingly, for the APOE genotype, which showed medium effect sizes for certain alleles, the E3/E3 combination, in both males and females, showed the lowest effect size on , at 0.18 and 0.14, respectively (Fig 2).
Here an F test found that there were no significant differences in APOE alleles effect size scores across genders for E2, E3 and E4; p = 0.667, 0.488 and 0.776, respectively. However, further analysis revealed that, as a whole, there were highly significant differences between allele groups, regardless of gender F(2,6) = 59.52, p = 0.000. Paired samples t-test found significant differences when comparing E2/E3 allele scores with E3/E3 at p = 0.013, similarly when comparing E3/E4 with E3/E3, p = 0.002. There was, however, no significant difference when comparing E2 and E4 (p = 0.952). Post-hoc LSD test found that E3 allele was the least effective, compared to both E2 and E4, in terms of its effect size.
Genes associated with muscle strength (1RM)
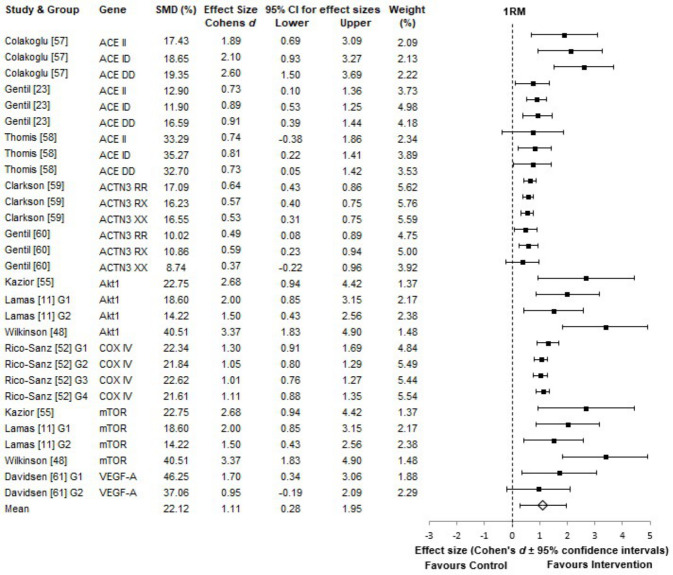
Strength training intervention studies [11,23,48,52,55,57–61] found an average increase in lower body 1RM of 22.12 ± 10.08% across the study groups, with an overall large effect size, which was very highly significant (p < 0.001) (Fig 3). Typically these studies did not report activity duration, rather number of repetitions performed, which on average was 174 reps, session at intensities of 75% 1RM, over 3-days per-week and 10 weeks of training.
Fig 3. 1RM forest plot.
The effect sizes are the change in 1RM post-intervention. For all plots the 95% confidence intervals were calculated and the overall mean effect size is represented with the diamond. The black squares represent the effect size of each study. Weighting was adjusted for sample size, SD and variance. Genes listed in alphabetical order.
Again, 1RM did not meet the requirements for parametric testing between the six strength gene subgroups when split, D(29), 0.886, p < 0.05. Kruskal-Wallis H test found very highly significant difference between the gene specific groups; H (5), 20.081, p = 0.001. Partial Eta Squared tests, revealed that 72% of the total variability in the increase of 1RM strength post-training intervention, was explained by the genetic subgrouping (Table 2).
Table 2. Candidate genes for strength.
| Gene Groups | No. of Study groups | Total Group Sample Size | Mean Rank (Not adjusted for sample size) | Subgroup (Adjusted for sample size) % of weight | Mean Group Effect Size (d) |
|---|---|---|---|---|---|
| ACE | 9 | 194 | 14.11 | 18.22 | 1.12 |
| ACTN3 | 6 | 743 | 3.50 | 22.96 | 0.57 |
| AKT1 | 4 | 39 | 24.00 | 11.71 | 2.27 |
| COX4I1 | 4 | 506 | 15.50 | 22.72 | 1.09 |
| mTOR | 4 | 39 | 24.00 | 11.71 | 2.27 |
| VEGF-A | 2 | 17 | 16.50 | 12.68 | 1.27 |
| Total | 29 | 1,538 |
The group with the highest mean rank shows the greatest number of high scores in 1RM. Subgroup % shows how much each of the six genes contribute to the 72% variance, adjusted for sample size, SE and within group variation in SMD.
All strength associated gene groups, identified, were found to have a large influence in the variability of lower body 1RM, with large effect-sizes, except for the ACTN3 group, which had a medium effect. This relative lack of effect was noted, regardless of ACTN3 genotype and allele (RR, RX, XX).
Genes associated with anaerobic power
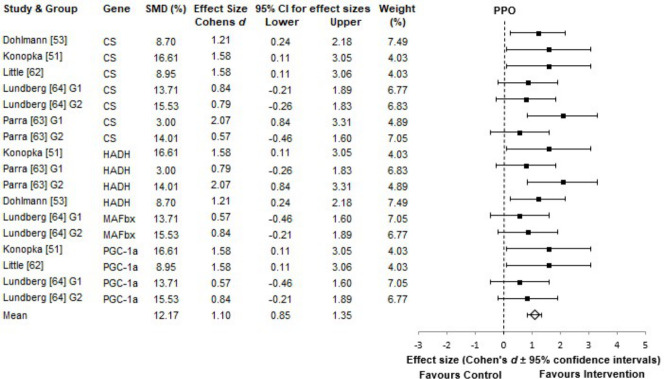
Here the analysis revealed a mean increase in peak power output of 12.17 ± 4.40%, irrespective of gene groups. Forest plot analysis found all studies [51,53,62–64] increased PPO by either a medium, or large effect-size (smallest ES 0.57), with a very highly significant improvement with training interventions (p < 0.001) (Fig 4). These studies performed between 4–12 cycle bouts, at average intensities of 90–110% or load of 0.075 per-kg bodyweight, over 3-days a week and 5 weeks of training.
Fig 4. PPO forest plot.
ES represents the change in PPO post-intervention. For all plots the 95% CI’s were calculated and the overall mean effect size is represented with the diamond, whereas the black squares are the effect size of each study. The weight is adjusted for sample size, SD and variance. Genes listed in alphabetical order.
As with previous data sets, PPO results were not normally distributed, D(17), .888, p < 0.05. However, there were no significant differences between gene subgroups, H(3), 1.592, p = 0.661 and partial Eta Squared found that only 10% of the variability in the 12.17% increase in PPO post-training, was explained by genetic sub-grouping (Table 3).
Table 3. Candidate genes for peak power output.
| Gene Groups | No. of Study groups | Total Group Sample Size | Mean Rank (Not adjusted for sample size) | Subgroup (Adjusted for sample size) % of weight | Mean Group Effect Size (d) |
|---|---|---|---|---|---|
| CS | 6 | 66 | 9.33 | 26.15 | 1.15 |
| HADH | 4 | 39 | 10.88 | 18.96 | 1.34 |
| MAFbx | 3 | 27 | 6.17 | 34.36 | 0.70 |
| PGC-1a | 4 | 34 | 8.75 | 20.53 | 1.02 |
| Total | 17 | 166 |
The group with the highest mean rank shows the greatest number of high scores in PPO and subgroups show how much of the 10% variance is accounted for by each of the four genes when normalised to 100%, adjusted for study sample size, SE and within group SD in SMD.
All genes showed a medium, or large effect size, with the HADH gene group showing the largest effect size score, (d = 1.34). However, HADH showed the lowest weighting, of 18.95%, when compared to other genes. MAFbx, with an effect size of 0.70, carried 34.36% of the weighting and explained about 1/3 of the total variability in the 10% power increase.
Discussion
The aim of this systematic review and meta-analysis was to identify candidate genes associated with the three key components of fitness. Additionally, to assess if these genes and their alleles, are associated with exercise response phenotype variability, in untrained human subjects, following an exercise training intervention. The results from this review are important, not just for the untrained, but all training populations. The inter-individual variation in the improvements in health-related components of fitness, identified in this study, for , strength and power can be explained genetics up to 44, 72 and 10% respectively. Such a finding emphasises the importance of assessing individuals’ genotype and planning training accordingly, thereby, making these findings relevant to the wider field of sport and exercise sciences.
13 candidate genes were identified that show significant associations with the fitness variables of interest. Overall, this review found that a genetic component for exercise responsiveness can explain between 10–72% of the variability in these key components. Such findings are consistent with previous studies, which reported variabilities of up to 80% in fitness phenotypes [2,13,15,17,27]. In this review, the subgroup analysis of the 13 candidate genes showed that nine were associated with cardiovascular fitness, six with muscular strength and four with anaerobic power phenotypes. Although studies, reported the ACE, ACTN3 and APOE allelic contribution, a key limitation with the majority of studies, was that they assessed individual genes, as a single variable, irrespective the gene’s allelic composition. Such an omission makes it difficult to assess the exact role of the gene(s) as the minor and major alleles often affect the phenotype differently, as shown in this study with APOE alleles [17,18]. Different genes also interact to produce the final phenotypic response [65], and here Genome Wide Association Studies (GWAS) will play an increasingly important role in identifying these and the variants. Thus, Williams et al. [65] identified a total of 97 genes that predicted trainability, and that the phenotype was dependent on several of these genotypes, which may contribute to approximately 50% of an individual’s trainability [65]. Further, a recent study by Al-Khelaifi et al. [66] also uncovered novel genes and associations using GWAS [66]. Using these studies that have identified the genes and potential associations to training this study identified 13 candidate genes, that provides a useful focus for future exercise intervention studies and how the variability of the phenotypes are affected.
In terms of cardiorespiratory fitness, all studies demonstrated an increased in response to aerobic exercise interventions. Here the well-studied ACE gene and its polymorphisms; II, ID, DD, showed the greatest influence on the phenotype, despite only having three groups in the analysis, with little differences between genotypes (188 participants out of 1,772, Table 1). Following this, COX4I1, CS and HADH genes also showed large influences on aerobic improvements. A possible explanation is that these genes code for key mitochondrial enzymes used in aerobic respiration. Thus, COX4I1 codes for cytochrome C oxidase, a key component of mitochondrial electron transport, whilst CS codes for citrate synthase, found in Krebs’ Cycle. Finally, HADH codes for 3-hydroxyacyl-CoA dehydrogenase, required for the oxidation of fatty acids [15,17,25,67,68]. PFKM and PGC1-α also displayed large influences and effect sizes, but only contributed 6.62 and 7.36% of the total 44% gene variability, respectively. A possible explanation is the combination of low study numbers and sample sizes (78 and 52 out of the 1,722 participants). AMPK had the smallest sample size, of 18 participants, hence it is difficult to draw firm conclusions on its effect on cardiorespiratory fitness, when compared to the other genes. However, AMPK has been found to directly affect PGC1- α, which is the independent master regulator of mitochondrial biogenesis and aerobic respiration [68,69]. Finally, APOE and ACTN3 results indicated no advantages and little influence on cardiorespiratory fitness. The APOE E3/E3 allele is very common, at 78.3%, worldwide [70] and is considered the neutral genotype, showing no effect on cardiorespiratory fitness, in agreement with the findings in this review. However, high to medium effect sizes were observed for the E2 and E4 alleles (Fig 2), concurring with the findings of Bernstein et al. [71] and Deeny et al. [72]. In this connection Obisesan et al. [73] found APOE genotypes explained significant variability in cardiorespiratory fitness, seen in training-induced increases after 24 weeks (p = 0.002). Further analysis in this review found that there was a highly significant difference when the E3 allele was compared to E2 and E4, in both males and females. This could explain the observed 44% genetic influence seen in this review, and why it was lower than reported rates in the literature, due to the confounding effect of APOE E3/E3 alleles [2,15,16,74,75]. Additionally, this would also emphasise the importance of examining the contributions of specific alleles of candidate genes, as opposed to whole gene analysis. These findings would also suggest that it is more advantageous to carry E2 and E4 genotypes, as opposed to E3, for improvements of post-training. Raichlen and Alexander, [70] stated, that these specific candidate genes and genotypes do not necessarily aid physical performance phenotypes, such as cardiorespiratory fitness, but instead, in the presence of physical activity, decrease health risks, such as coronary artery disease (CAD) and improve overall health status, therefore, when this is included in the meta-analysis it consequently decreases the associated gene variability [76].
Interestingly, the well-researched ACTN3 gene showed equivocal results in this review for the both and 1RM. Theoretically, homozygosity for the X (deletion) allele should abolish production of α actinin-3, leading to improved aerobic fitness, whilst the R allele should decrease aerobic fitness, due to increased α actinin-3 expression [18,27,60,61,77–79]. Additionally, Hogarth et al. [80] states that α-actinin-3 controls sarcomeric composition and muscle function in an allele dose-dependent fashion and promotes strength adaptations, but this was not observed in this review. However, the findings of this review agree with those of Gineviciene et al. [1], which directly assessed the influence of the ACTN3 R577X polymorphism on aerobic fitness and found no significant differences between alleles for the variability and trainability of the exercise phenotype [14]. Similarly, for the well-studied ACE genotypes, this review found no significant differences between ACE insertion (I) and deletion (D) alleles, as both alleles were associated with significant improvements in cardiorespiratory fitness. Here previous work has suggested that I allele is associated with greater increases in cardiorespiratory fitness and endurance, due to lower levels of ACE and increased maximal heart rate and improved blood circulatory role [58].
In this review, muscular strength phenotype, assessed by 1RM, showed the largest observed variability, in response to training interventions, with 72% accounted for by the six candidate genes. AKT1 and mTOR had equally large contributions to the phenotype response, displaying the largest mean rank and effect size. Previous studies [81–83] are consistent in showing interactions between Akt and mTOR regulation, which are activated through resistance exercise. Akt and its downstream signalling pathways, such as mTOR (Akt-mTOR pathway) is the central mediator of protein synthesis, associated with the control of skeletal muscle hypertrophy, muscle mass and strength [80–83]. However, due to the nature of the studies, reporting the findings from whole gene analysis, it is still unclear on the specific role of different alleles. Interestingly, the literature supports an upstream regulation in AMPK activation for endurance, suppressing Akt-mTOR, meaning the increased levels of AMPK may be detrimental to strength improvements [84–87]. Additionally, the literature suggests that mTOR polymorphism (rs2295080) alleles G = 45.2% and T = 54.8% also show different results [88]. The G-allele predominates in endurance athletes, whilst the T-allele frequency is greater in power and strength-oriented athletes [89]. Therefore, suggests the need to review this gene at an allele specific level.
ACE and VEGF-A made similar contributions to increased strength variability, despite the low number of participants (17 participants) in the VEGF-A study. The results further show that ACE and COX4I1 accounted for 40.9% of the 72% total variability found. Theoretically the ACE D allele results in increased ACE activity, which has been shown to be associated with strength performance and decreases in [23,29,90], however no such effect was noted in this study. Finally, ACTN3 had the lowest mean rank score, and the lowest effect size, when compared to the other genes. However, it is important to note that the increases in strength, associated with ACTN3, were still significant, which is consistent with previous findings [27,59,91]. Moreover, when sub-group analysis and sample size were accounted for, ACTN3 was the largest contributor to overall variability in the strength phenotype. However, this may simply reflect the high proportion of participants within this group (743 out of the 1,538).
PPO displayed the lowest genetic influence (10% variability) but was still significantly improved by the training intervention. Here one new candidate gene was identified, by this review, that had not already been linked with another component of fitness, the MAFbx (FBXO32), or Atrogin-1 gene. Despite low study numbers (27 participants), MAFbx accounted for 34.36% of the total 10% variability. MAFbx has previously been found to be associated with muscle strength gains during exercise induced muscle hypertrophy [65,92–94]. In agreement, Mascher et al. [93] found that this gene is involved in muscle breakdown and atrophy, and that resistance training reduced its expression, hence on this basis, might be associated with strength. Such findings reflect a paucity of investigations into candidate genes for the PPO phenotype, as only five studies were included in this review. Moreover, the current analysis also revealed three of the identified genes, were also associated with cardiorespiratory fitness. Again, the allele compositions of these genes might have been more informative but were not reported. Previous work [1] has identified that PPARGC1A gene (PGC-1α) is associated with power variables, as has this review.
It is important to note that, one possible reason why the anaerobic power candidate genes may be associated with different phenotype responses and low variability rates, could be due to studies incorrectly measuring anaerobic power and rather, measuring metabolic power, which is a mixture of energy sources. This may be a significant flaw in many power assessment studies [10]. In this connection, it is very common, when measuring power, to use 30 second Wingate tests (WAnT). Here energy from anaerobic phosphagenic, glycolytic and aerobic mitochondrial respiration metabolism, contributes to 31.1%, 50.3% and 18.6%, respectively [8,95]. Hence, many studies are misinterpreting power phenotypes, making the assessment more difficult. Therefore, we would recommend studies that include, all-out burst and brief sprints with durations of up to 10 seconds, where the initial energy source is primarily drawn from anaerobic metabolism only, and reported as peak power output, rather than mean power output over time [10,96,97].
A major strength of this meta-analytical was the ability to compare all studies, regardless of intervention, by grouping studies and assessing them by the genotype sub-groups. For example, in this review, all studies that assessed the same genes and alleles were combined, any effect on the phenotype was averaged and the overall variability assessed. This was then compared with the influences of the other candidate genes following the same method, rather than directly comparing studies against each other using different interventions. Such an approach helps account for the variation caused by the training intervention and other external influences, such as the environment on the phenotype. Another key strength of this review was that the analysis compared the contribution of multiple genes towards the total variance of the phenotypes, rather than a more restricted, single gene analysis approach. It is also important to note that the genetic make-up, alone, does not determine the phenotype, only the potential for expression of the phenotype in response to a particular lifestyle, environment, and intervention [13,17].
The main limitation to this review was the lack of allele specific analysis in the included studies. Another limitation is the possible exclusion of other candidate genes, potentially reducing the number of candidate genes identified. Such limitations reflect a generic problem with a systematic review, that it is limited to current published information and the requirement to ensure the comparability of different studies. Another factor to consider, is that baseline-training status also affects the adaptation responses to exercise. Although, this review attempted to address this by specifically selecting studies in which all participants were classed as untrained, according to widely accepted norms, it is clear there were still baseline differences between individuals. This is reflected in this review by the within-groups non-normal distributions and heterogeneity [2,4]. Moreover, the predisposition of the genetic heritability for advantageous baseline phenotypes, shows genes and specific alleles heavily influence adaptations and trainability, even before training interventions are implemented [14]. Finally, for a number of particular genes in this review, due to the low group sample size, it is not clear, nor possible to draw firm conclusions for the precise role of these genes on the phenotype and further investigation is required. Nevertheless, this review has identified 13 candidate genes, which explain a significant proportion of the variability and their contribution in the phenotype responses to trainability for the three components of fitness.
Practical applications
This review demonstrates that the candidate genes provide valuable information regarding genotype-specific training and variability in the phenotype responses. In theory a possible practical application of this could be to identify a person’s genotype and tailor a specific individual training intervention programme, based on their genotype. This would be more advantageous than implementing generic training programmes, which may provide relatively little value in terms of phenotype gains and improvements. These inferences also support and strengthen the evidence, that genes have on training variability suggested in the research literature.
Supporting information
The results of both reviewers using the quality assessment tool is mapped as the difference in scores against the average score (Bias). The 95% LoA are also calculated and represented as the upper and lower 1.96 dashed lines. The confidence intervals for the 95% LoA were calculated using Bland Altman’s approximate method.
(PDF)
Search terms implemented for all the databases and the number of results shown by hits.
(PDF)
The average score between reviewers was taken for the final inclusion. Power threshold was based at 0.8 (80%).
(PDF)
(PDF)
(PDF)
Acknowledgments
This meta-analytical review is non-profited and non-funded research. This work was supported by Anglia Ruskin University, the staff from library services and the Faculty of Science and Engineering (FSE). All data is available at the Cambridge Centre for Sport & Exercise Sciences, Anglia Ruskin University, UK.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
The author(s) received no specific funding for this work.
References
- 1.Gineviciene V, Jakaitiene A, Aksenov MO, Aksenova AV, Astratenkova AD, Egorova ES, et al. Association analysis of ACE, ACTN3 and PPARGC1A gene polymorphisms in two cohorts of European strength and power athletes. Biology of sport. 2016. Sep;33(3):199. doi: 10.5604/20831862.1201051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hautala A.J., Kiviniemi A.M., Mäkikallio T.H., Kinnunen H., Nissilä S., Huikuri H.V., et al. , 2006. Individual differences in the responses to endurance and resistance training. European journal of applied physiology, 96(5), pp.535–542. doi: 10.1007/s00421-005-0116-2 [DOI] [PubMed] [Google Scholar]
- 3.Metzl J. ACSM Blogs. 2017 May 16. [Cited 1 December 2020] In: ACSM Blogs [internet]. American college of sports and exercise 2020 - [Exercise guidelines] Available from: https://www.acsm.org/all-blog-posts/acsm-blog/acsm-blog/2017/05/16/science-of-exercise.
- 4.Peterson MD, Rhea MR, Alvar BA. Applications of the dose-response for muscular strength development: areview of meta-analytic efficacy and reliability for designing training prescription. The Journal of Strength & Conditioning Research. 2005. Nov 1;19(4):950–8. [DOI] [PubMed] [Google Scholar]
- 5.Ratamess NA. ACSM’s foundations of strength training and conditioning. Wolters Kluwer Health/Lippincott Williams & Wilkins; 2012.
- 6.Midgley AW, McNaughton LR, Polman R, Marchant D. Criteria for determination of maximal oxygen uptake. Sports medicine. 2007. Dec 1;37(12):1019–28. doi: 10.2165/00007256-200737120-00002 [DOI] [PubMed] [Google Scholar]
- 7.Blair SN, Kampert JB, Kohl HW, Barlow CE, Macera CA, Paffenbarger RS, et al. Influences of cardiorespiratory fitness and other precursors on cardiovascular disease and all-cause mortality in men and women. Jama. 1996. Jul 17;276(3):205–10. [PubMed] [Google Scholar]
- 8.Cometti G, Maffiuletti NA, Pousson M, Chatard JC, Maffulli N. Isokinetic strength and anaerobic power of elite, subelite and amateur French soccer players. International journal of sports medicine. 2001;22(01):45–51. doi: 10.1055/s-2001-11331 [DOI] [PubMed] [Google Scholar]
- 9.Beneke R, Pollmann CH, Bleif I, Leithäuser R, Hütler M. How anaerobic is the Wingate Anaerobic Test for humans?. European journal of applied physiology. 2002. Jan 1;87(4–5):388–92. doi: 10.1007/s00421-002-0622-4 [DOI] [PubMed] [Google Scholar]
- 10.Bundle MW, Weyand PG. Sprint exercise performance: does metabolic power matter?. Exercise and sport sciences reviews. 2012. Jul 1;40(3):174–82. doi: 10.1097/JES.0b013e318258e1c1 [DOI] [PubMed] [Google Scholar]
- 11.Lamas L, Aoki MS, Ugrinowitsch C, Campos GE, Regazzini M, Moriscot AS, et al. Expression of genes related to muscle plasticity after strength and power training regimens. Scandinavian journal of medicine & science in sports. 2010. Apr;20(2):216–25. doi: 10.1111/j.1600-0838.2009.00905.x [DOI] [PubMed] [Google Scholar]
- 12.Rankinen T, Pérusse L, Gagnon J, Chagnon YC, Leon AS, Skinner JS, et al. Angiotensin-converting enzyme ID polymorphism and fitness phenotype in the HERITAGE Family Study. Journal of Applied Physiology. 2000. Mar 1;88(3):1029–35. doi: 10.1152/jappl.2000.88.3.1029 [DOI] [PubMed] [Google Scholar]
- 13.Schutte NM, Nederend I, Hudziak JJ, Bartels M, de Geus EJ. Twin-sibling study and meta-analysis on the heritability of maximal oxygen consumption. Physiological Genomics. 2016. Mar;48(3):210–9. doi: 10.1152/physiolgenomics.00117.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sarzynski MA, Ghosh S, Bouchard C. Genomic and transcriptomic predictors of response levels to endurance exercise training. The Journal of Physiology. 2017. May 1;595(9):2931–9. doi: 10.1113/JP272559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bouchard C. Genomic predictors of trainability. Experimental physiology. 2012. Mar;97(3):347–52. doi: 10.1113/expphysiol.2011.058735 [DOI] [PubMed] [Google Scholar]
- 16.Huygens W, Thomis MA, Peeters MW, Vlietinck RF, Beunen GP. Determinants and upper-limit heritabilities of skeletal muscle mass and strength. Canadian Journal of Applied Physiology. 2004. Apr 1;29(2):186–200. doi: 10.1139/h04-014 [DOI] [PubMed] [Google Scholar]
- 17.Spurway N, Wackerhage H. Genetics and molecular biology of muscle adaptation. Elsevier Health Sciences; 2006. [Google Scholar]
- 18.Silva MS, Bolani W, Alves CR, Biagi DG, Lemos JR, da Silva JL, et al. Elimination of influences of the ACTN3 R577X variant on oxygen uptake by endurance training in healthy individuals. International journal of sports physiology and performance. 2015. Jul 1;10(5):636–41. doi: 10.1123/ijspp.2014-0205 [DOI] [PubMed] [Google Scholar]
- 19.Vancini RL, Pesquero JB, Fachina RJ, dos Santos Andrade M, Borin JP, Montagner PC, et al. Genetic aspects of athletic performance: the African runners phenomenon. Open access journal of sports medicine. 2014;5:123. doi: 10.2147/OAJSM.S61361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prud’Homme D, Fontaine E. of maximal aerobic power to training is genotype-dependent. Medicine and science in sports and exercise. 1984;16(5):459–93. doi: 10.1249/00005768-198410000-00012 [DOI] [PubMed] [Google Scholar]
- 21.Zambon AC, McDearmon EL, Salomonis N, Vranizan KM, Johansen KL, Adey D, et al. Time-and exercise-dependent gene regulation in human skeletal muscle. Genome biology. 2003. Oct 1;4(10):R61. doi: 10.1186/gb-2003-4-10-r61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cięszczyk P., Zarębska A., Jastrzębski Z., Sawczyn M., Kozakiewicz-Drobnik I., Leońska-Duniec A., et al. , 2016. Does the MTHFR A1298C Polymorphism Modulate the Cardiorespiratory Response to Training?. Journal of human kinetics, 54(1), pp.43–53. doi: 10.1515/hukin-2016-0055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gentil P, Lima RM, Pereira RW, Mourot J, Leite TK, Bottaro M. Lack of association of the ACE genotype with the muscle strength response to resistance training. European Journal of Sport Science. 2012. Jul 1;12(4):331–7. [Google Scholar]
- 24.McPhee JS, Perez-Schindler J, Degens H, Tomlinson D, Hennis P, Baar K, et al. HIF1A P582S gene association with endurance training responses in young women. European journal of applied physiology. 2011. Sep 1;111(9):2339–47. doi: 10.1007/s00421-011-1869-4 [DOI] [PubMed] [Google Scholar]
- 25.McPhee JS, Williams AG, Perez-Schindler J, Degens H, Baar K, Jones DA. Variability in the magnitude of response of metabolic enzymes reveals patterns of co-ordinated expression following endurance training in women. Experimental physiology. 2011. Jul 1;96(7):699–707. doi: 10.1113/expphysiol.2011.057729 [DOI] [PubMed] [Google Scholar]
- 26.Astorino TA, Schubert MM. Individual responses to completion of short-term and chronic interval training: a retrospective study. PLoS One. 2014. May 21;9(5):e97638. doi: 10.1371/journal.pone.0097638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Del Coso J, Hiam D, Houweling P, Pérez LM, Eynon N, Lucía A. More than a ‘speed gene’: ACTN3 R577X genotype, trainability, muscle damage, and the risk for injuries. European journal of applied physiology. 2019. Jan;119(1):49–60. doi: 10.1007/s00421-018-4010-0 [DOI] [PubMed] [Google Scholar]
- 28.Yvert T, Miyamoto-Mikami E, Murakami H, Miyachi M, Kawahara T, Fuku N. Lack of replication of associations between multiple genetic polymorphisms and endurance athlete status in Japanese population. Physiological reports. 2016. Oct 1;4(20). doi: 10.14814/phy2.13003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahmetov II, Egorova ES, Gabdrakhmanova LJ, Fedotovskaya ON. Genes and athletic performance: an update. InGenetics and Sports 2016. (Vol. 61, pp. 41–54). Karger Publishers. doi: 10.1159/000445240 [DOI] [PubMed] [Google Scholar]
- 30.Williams AG, Folland JP. Similarity of polygenic profiles limits the potential for elite human physical performance. The journal of physiology. 2008. Jan 1;586(1):113–21. doi: 10.1113/jphysiol.2007.141887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Int J Surg. 2010. Jan 1;8(5):336–41. doi: 10.1016/j.ijsu.2010.02.007 [DOI] [PubMed] [Google Scholar]
- 32.Needleman IG. A guide to systematic reviews. Journal of clinical periodontology. 2002. Dec;29:6–9. doi: 10.1034/j.1600-051x.29.s3.15.x [DOI] [PubMed] [Google Scholar]
- 33.Terwee CB, Mokkink LB, Knol DL, Ostelo RW, Bouter LM, de Vet HC. Rating the methodological quality in systematic reviews of studies on measurement properties: a scoring system for the COSMIN checklist. Quality of life research. 2012. May 1;21(4):651–7. doi: 10.1007/s11136-011-9960-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kitchenham B, Brereton P, Li Z, Budgen D, Burn A. Repeatability of systematic literature reviews. In15th Annual Conference on Evaluation & Assessment in Software Engineering (EASE 2011) 2011 Apr 11 (pp. 46–55). IET.
- 35.Methley AM, Campbell S, Chew-Graham C, McNally R, Cheraghi-Sohi S. PICO, PICOS and SPIDER: a comparison study of specificity and sensitivity in three search tools for qualitative systematic reviews. BMC health services research. 2014. Dec;14(1):579. doi: 10.1186/s12913-014-0579-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mokkink LB, Terwee CB, Patrick DL, Alonso J, Stratford PW, Knol DL, et al. The COSMIN checklist for assessing the methodological quality of studies on measurement properties of health status measurement instruments: an international Delphi study. Quality of life research. 2010. May 1;19(4):539–49. doi: 10.1007/s11136-010-9606-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. International journal of nursing studies. 2010. Aug 1;47(8):931–6. [Google Scholar]
- 38.Suurmond R, van Rhee H, Hak T. Introduction, comparison, and validation of Meta-Essentials: A free and simple tool for meta-analysis. Research synthesis methods. 2017. Dec;8(4):537–53. doi: 10.1002/jrsm.1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valentine JC, Pigott TD, Rothstein HR. How many studies do you need? A primer on statistical power for meta-analysis. Journal of Educational and Behavioral Statistics. 2010. Apr;35(2):215–47. [Google Scholar]
- 40.Cohen J. Statistical power analysis for the behavioral sciences, 2nd edn. Á/L.
- 41.Suresh KP, Chandrashekara S. Sample size estimation and power analysis for clinical research studies. Journal of human reproductive sciences. 2012. Jan;5(1):7. doi: 10.4103/0974-1208.97779 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Ahtiainen JP, Hulmi JJ, Kraemer WJ, Lehti M, Nyman K, Selänne H, et al. Heavy resistance exercise training and skeletal muscle androgen receptor expression in younger and older men. Steroids. 2011. Jan 1;76(1–2):183–92. doi: 10.1016/j.steroids.2010.10.012 [DOI] [PubMed] [Google Scholar]
- 43.Bouchard C, Sarzynski MA, Rice TK, Kraus WE, Church TS, Sung YJ, et al. Genomic predictors of the maximal O2 uptake response to standardized exercise training programs. Journal of applied physiology. 2011. May 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Erskine RM, Williams AG, Jones DA, Stewart CE, Degens H. Do PTK2 gene polymorphisms contribute to the interindividual variability in muscle strength and the response to resistance training? A preliminary report. Journal of applied physiology. 2012. Apr 15;112(8):1329–34. doi: 10.1152/japplphysiol.01137.2011 [DOI] [PubMed] [Google Scholar]
- 45.He ZH, Hu Y, Wang HY, Li YC, Lu YL, Zhang L, et al. Are calcineurin genes associated with endurance phenotype traits?. European journal of applied physiology. 2010. Jun 1;109(3):359–69. doi: 10.1007/s00421-010-1361-6 [DOI] [PubMed] [Google Scholar]
- 46.Rankinen T, Pérusse L, Borecki I, Chagnon YC, Gagnon J, Leon AS, et al. The Na+-K+-ATPase α2 gene and trainability of cardiorespiratory endurance: the HERITAGE Family Study. Journal of applied physiology. 2000. Jan 1;88(1):346–51. doi: 10.1152/jappl.2000.88.1.346 [DOI] [PubMed] [Google Scholar]
- 47.Böhm A, Hoffmann C, Irmler M, Schneeweiss P, Schnauder G, Sailer C, et al. TGF-β contributes to impaired exercise response by suppression of mitochondrial key regulators in skeletal muscle. Diabetes. 2016. Oct 1;65(10):2849–61. doi: 10.2337/db15-1723 [DOI] [PubMed] [Google Scholar]
- 48.Wilkinson SB, Phillips SM, Atherton PJ, Patel R, Yarasheski KE, Tarnopolsky MA, et al. Differential effects of resistance and endurance exercise in the fed state on signalling molecule phosphorylation and protein synthesis in human muscle. The journal of physiology. 2008. Aug 1;586(15):3701–17. doi: 10.1113/jphysiol.2008.153916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson PD, Tsongalis GJ, Seip RL, Bilbie C, Miles M, Zoeller R, et al. Apolipoprotein E genotype and changes in serum lipids and maximal oxygen uptake with exercise training. Metabolism. 2004. Feb 1;53(2):193–202. doi: 10.1016/j.metabol.2003.09.010 [DOI] [PubMed] [Google Scholar]
- 50.Yu B, Chen W, Wang R, Qi Q, Li K, Zhang W, et al. Association of apolipoprotein E polymorphism with maximal oxygen uptake after exercise training: a study of Chinese young adult. Lipids in health and disease. 2014. Dec 1;13(1):40. doi: 10.1186/1476-511X-13-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Konopka AR, Suer MK, Wolff CA, Harber MP. Markers of human skeletal muscle mitochondrial biogenesis and quality control: effects of age and aerobic exercise training. Journals of Gerontology Series A: Biomedical Sciences and Medical Sciences. 2014. Apr 1;69(4):371–8. doi: 10.1093/gerona/glt107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rico-Sanz J, Rankinen T, Rice T, Leon AS, Skinner JS, Wilmore JH, et al. Quantitative trait loci for maximal exercise capacity phenotypes and their responses to training in the HERITAGE Family Study. Physiological genomics. 2004. Jan 15;16(2):256–60. doi: 10.1152/physiolgenomics.00035.2003 [DOI] [PubMed] [Google Scholar]
- 53.Dohlmann TL, Hindsø M, Dela F, Helge JW, Larsen S. High-intensity interval training changes mitochondrial respiratory capacity differently in adipose tissue and skeletal muscle. Physiological Reports. 2018. Sep;6(18):e13857. doi: 10.14814/phy2.13857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Egan B, O’connor PL, Zierath JR, O’gorman DJ. Time course analysis reveals gene-specific transcript and protein kinetics of adaptation to short-term aerobic exercise training in human skeletal muscle. PloS one. 2013. Sep 12;8(9):e74098. doi: 10.1371/journal.pone.0074098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kazior Z, Willis SJ, Moberg M, Apró W, Calbet JA, Holmberg HC, et al. Endurance exercise enhances the effect of strength training on muscle fiber size and protein expression of Akt and mTOR. PLoS One. 2016. Feb 17;11(2):e0149082. doi: 10.1371/journal.pone.0149082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Butcher LR, Thomas A, Backx K, Roberts A, Webb R, Morris K. Low-intensity exercise exerts beneficial effects on plasma lipids via PPARγ. Medicine & Science in Sports & Exercise. 2008. Jul 1;40(7):1263–70. doi: 10.1249/MSS.0b013e31816c091d [DOI] [PubMed] [Google Scholar]
- 57.Colakoglu M, Cam FS, Kayitken B, Cetinoz F, Colakoglu S, Turkmen M, et al. ACE genotype may have an effect on single versus multiple set preferences in strength training. European Journal of Applied Physiology. 2005. Sep 1;95(1):20–6. doi: 10.1007/s00421-005-1335-2 [DOI] [PubMed] [Google Scholar]
- 58.Thomis MA, Huygens W, Heuninckx S, Chagnon M, Maes HH, Claessens AL, et al. Exploration of myostatin polymorphisms and the angiotensin-converting enzyme insertion/deletion genotype in responses of human muscle to strength training. European journal of applied physiology. 2004. Jul 1;92(3):267–74. doi: 10.1007/s00421-004-1093-6 [DOI] [PubMed] [Google Scholar]
- 59.Clarkson PM, Devaney JM, Gordish-Dressman H, Thompson PD, Hubal MJ, Urso M, et al. ACTN3 genotype is associated with increases in muscle strength in response to resistance training in women. Journal of Applied Physiology. 2005. Jul;99(1):154–63. doi: 10.1152/japplphysiol.01139.2004 [DOI] [PubMed] [Google Scholar]
- 60.Gentil P, Pereira RW, Leite TK, Bottaro M. ACTN3 R577X polymorphism and neuromuscular response to resistance training. Journal of Sports Science & Medicine. 2011. Jun;10(2):393. [PMC free article] [PubMed] [Google Scholar]
- 61.Davidsen PK, Gallagher IJ, Hartman JW, Tarnopolsky MA, Dela F, Helge JW, et al. High responders to resistance exercise training demonstrate differential regulation of skeletal muscle microRNA expression. Journal of applied physiology. 2011. Feb 1. doi: 10.1152/japplphysiol.00901.2010 [DOI] [PubMed] [Google Scholar]
- 62.Little JP, Safdar A, Wilkin GP, Tarnopolsky MA, Gibala MJ. A practical model of low-volume high-intensity interval training induces mitochondrial biogenesis in human skeletal muscle: potential mechanisms. The Journal of physiology. 2010. Mar 15;588(6):1011–22. doi: 10.1113/jphysiol.2009.181743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Parra J, Cadefau JA, Rodas G, Amigo N, Cusso R. The distribution of rest periods affects performance and adaptations of energy metabolism induced by high-intensity training in human muscle. Acta Physiologica Scandinavica. 2000. Jun;169(2):157–65. doi: 10.1046/j.1365-201x.2000.00730.x [DOI] [PubMed] [Google Scholar]
- 64.Lundberg TR, Fernandez-Gonzalo R, Tesch PA. Exercise-induced AMPK activation does not interfere with muscle hypertrophy in response to resistance training in men. Journal of applied physiology. 2014. Mar 15;116(6):611–20. doi: 10.1152/japplphysiol.01082.2013 [DOI] [PubMed] [Google Scholar]
- 65.Williams CJ, Williams MG, Eynon N, Ashton KJ, Little JP, Wisloff U, et al. Genes to predict VO 2max trainability: a systematic review. BMC genomics. 2017. Nov;18(8):81–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Al-Khelaifi F., Yousri N.A., Diboun I., Semenova E.A., Kostryukova E.S., Kulemin N.A., et al. , 2020. Genome-wide association study reveals a novel association between MYBPC3 gene polymorphism, endurance athlete status, aerobic capacity and steroid metabolism. Frontiers in genetics, 11, p.595. doi: 10.3389/fgene.2020.00595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ma W, Sung HJ, Park JY, Matoba S, Hwang PM. A pivotal role for p53: balancing aerobic respiration and glycolysis. Journal of bioenergetics and biomembranes. 2007. Jun 1;39(3):243–6. doi: 10.1007/s10863-007-9083-0 [DOI] [PubMed] [Google Scholar]
- 68.Popov DV, Lysenko EA, Butkov AD, Vepkhvadze TF, Perfilov DV, Vinogradova OL. AMPK does not play a requisite role in regulation of PPARGC1A gene expression via the alternative promoter in endurance-trained human skeletal muscle. Experimental physiology. 2017. Mar 1;102(3):366–75. doi: 10.1113/EP086074 [DOI] [PubMed] [Google Scholar]
- 69.Lantier L, Fentz J, Mounier R, Leclerc J, Treebak JT, Pehmøller C, et al. AMPK controls exercise endurance, mitochondrial oxidative capacity, and skeletal muscle integrity. The FASEB Journal. 2014. Jul;28(7):3211–24. doi: 10.1096/fj.14-250449 [DOI] [PubMed] [Google Scholar]
- 70.Raichlen DA, Alexander GE. Exercise, APOE genotype, and the evolution of the human lifespan. Trends in neurosciences. 2014. May 1;37(5):247–55. doi: 10.1016/j.tins.2014.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bernstein MS, Costanza MC, James RW, Morris MA, Cambien F, Raoux S, et al. Physical activity may modulate effects of ApoE genotype on lipid profile. Arteriosclerosis, thrombosis, and vascular biology. 2002. Jan 1;22(1):133–40. doi: 10.1161/hq0102.101819 [DOI] [PubMed] [Google Scholar]
- 72.Deeny SP, Poeppel D, Zimmerman JB, Roth SM, Brandauer J, Witkowski S, et al. Exercise, APOE, and working memory: MEG and behavioral evidence for benefit of exercise in epsilon4 carriers. Biological psychology. 2008. May 1;78(2):179–87. doi: 10.1016/j.biopsycho.2008.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Obisesan TO, Ferrell RE, Goldberg AP, Phares DA, Ellis TJ, Hagberg JM. APOE genotype affects black-white responses of high-density lipoprotein cholesterol subspecies to aerobic exercise training. Metabolism. 2008. Dec 1;57(12):1669–76. doi: 10.1016/j.metabol.2008.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Klissouras VA. Heritability of adaptive variation. Journal of Applied Physiology. 1971. Sep;31(3):338–44. doi: 10.1152/jappl.1971.31.3.338 [DOI] [PubMed] [Google Scholar]
- 75.Komi PV, Viitasalo JH, Havu M, Thorstensson A, Sjödin B, Karlsson J. Skeletal muscle fibres and muscle enzyme activities in monozygous and dizygous twins of both sexes. Acta Physiologica Scandinavica. 1977. Aug;100(4):385–92. doi: 10.1111/j.1365-201X.1977.tb00001.x [DOI] [PubMed] [Google Scholar]
- 76.Richardson JT. Eta squared and partial eta squared as measures of effect size in educational research. Educational Research Review. 2011. Jan 1;6(2):135–47. [Google Scholar]
- 77.Keiller DR, Gordon DA. The plateau at V˙ O2max is associated with anaerobic alleles. Journal of science and medicine in sport. 2020. May 1;23(5):506–11. doi: 10.1016/j.jsams.2019.11.012 [DOI] [PubMed] [Google Scholar]
- 78.Pickering C, Kiely J. ACTN3: more than just a gene for speed. Frontiers in physiology. 2017. Dec 18;8:1080. doi: 10.3389/fphys.2017.01080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Seto JT, Quinlan KG, Lek M, Zheng XF, Garton F, MacArthur DG, et al. ACTN3 genotype influences muscle performance through the regulation of calcineurin signaling. The Journal of clinical investigation. 2013. Oct 1;123(10):4255–63. doi: 10.1172/JCI67691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hogarth MW, Garton FC, Houweling PJ, Tukiainen T, Lek M, Macarthur DG, et al. Analysis of the ACTN3 heterozygous genotype suggests that α-actinin-3 controls sarcomeric composition and muscle function in a dose-dependent fashion. Human molecular genetics. 2016. Mar 1;25(5):866–77. doi: 10.1093/hmg/ddv613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Léger B, Cartoni R, Praz M, Lamon S, Dériaz O, Crettenand A, et al. Akt signalling through GSK-3β, mTOR and Foxo1 is involved in human skeletal muscle hypertrophy and atrophy. The Journal of physiology. 2006. Nov 1;576(3):923–33. doi: 10.1113/jphysiol.2006.116715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Møller AB, Vendelbo MH, Rahbek SK, Clasen BF, Schjerling P, Vissing K, Jessen N. Resistance exercise, but not endurance exercise, induces IKKβ phosphorylation in human skeletal muscle of training-accustomed individuals. Pflügers Archiv-European Journal of Physiology. 2013. Dec 1;465(12):1785–95. doi: 10.1007/s00424-013-1318-9 [DOI] [PubMed] [Google Scholar]
- 83.Song Z, Moore DR, Hodson N, Ward C, Dent JR, O’Leary MF, et al. Resistance exercise initiates mechanistic target of rapamycin (mTOR) translocation and protein complex co-localisation in human skeletal muscle. Scientific reports. 2017. Jul 10;7(1):1–4. doi: 10.1038/s41598-016-0028-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Altamirano F, Oyarce C, Silva P, Estrada M. Testosterone activates mTOR/S6K1 pathway through intracellular calcium and ERK in cardiomyocytes. 2008: 742–4. [Google Scholar]
- 85.Brandauer J, Andersen MA, Kellezi H, Risis S, Frøsig C, Vienberg SG, et al. AMP-activated protein kinase controls exercise training-and AICAR-induced increases in SIRT3 and MnSOD. Frontiers in physiology. 2015. Mar 24;6:85. doi: 10.3389/fphys.2015.00085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Metcalfe RS, Koumanov F, Ruffino JS, Stokes KA, Holman GD, Thompson D, et al. Physiological and molecular responses to an acute bout of reduced-exertion high-intensity interval training (REHIT). European journal of applied physiology. 2015. Nov 1;115(11):2321–34. doi: 10.1007/s00421-015-3217-6 [DOI] [PubMed] [Google Scholar]
- 87.Tannerstedt J, Apró W, Blomstrand E. Maximal lengthening contractions induce different signaling responses in the type I and type II fibers of human skeletal muscle. Journal of applied physiology. 2009. Apr;106(4):1412–8. doi: 10.1152/japplphysiol.91243.2008 [DOI] [PubMed] [Google Scholar]
- 88.National Library of Medicine. Reference SNP (rs) Report; 2021 [cited 2021 Jul 6] Database: NLM [Internet]. Available from: https://www.ncbi.nlm.nih.gov/snp/rs2295080#frequency_tab.
- 89.Drozdovska S, Oleshko V. Association of gene FRAP1 Т/G (rs2295080) polymorphism with power-oriented athlete’s status. Sporto mokslas, 2016, nr. 3, p. 59–65. 2016. [Google Scholar]
- 90.Montgomery HE, Clarkson P, Dollery CM, Prasad K, Losi MA, Hemingway H, et al. Association of angiotensin-converting enzyme gene I/D polymorphism with change in left ventricular mass in response to physical training. Circulation. 1997. Aug 5;96(3):741–7. doi: 10.1161/01.cir.96.3.741 [DOI] [PubMed] [Google Scholar]
- 91.Bustamante-Ara N, Santiago C, Verde Z, Yvert T, Gómez-Gallego F, Rodríguez-Romo G, et al. ACE and ACTN3 genes and muscle phenotypes in nonagenarians. International journal of sports medicine. 2010. Apr;31(04):221–4. doi: 10.1055/s-0030-1247529 [DOI] [PubMed] [Google Scholar]
- 92.Lagirand-Cantaloube J, Offner N, Csibi A, Leibovitch MP, Batonnet-Pichon S, Tintignac LA, et al. The initiation factor eIF3-f is a major target for atrogin1/MAFbx function in skeletal muscle atrophy. The EMBO journal. 2008. Apr 23;27(8):1266–76. doi: 10.1038/emboj.2008.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mascher H, Tannerstedt J, Brink-Elfegoun T, Ekblom B, Gustafsson T, Blomstrand E. Repeated resistance exercise training induces different changes in mRNA expression of MAFbx and MuRF-1 in human skeletal muscle. American Journal of Physiology-Endocrinology and Metabolism. 2008. Jan;294(1):E43–51. doi: 10.1152/ajpendo.00504.2007 [DOI] [PubMed] [Google Scholar]
- 94.Seaborne RA, Hughes DC, Turner DC, Owens DJ, Baehr LM, Gorski P, et al. UBR5 is a novel E3 ubiquitin ligase involved in skeletal muscle hypertrophy and recovery from atrophy. The Journal of physiology. 2019. Jul;597(14):3727–49. doi: 10.1113/JP278073 [DOI] [PubMed] [Google Scholar]
- 95.Baker JS, McCormick MC, Robergs RA. Interaction among skeletal muscle metabolic energy systems during intense exercise. Journal of nutrition and metabolism. 2010. Oct;2010. doi: 10.1155/2010/905612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Driss T, Vandewalle H. The measurement of maximal (anaerobic) power output on a cycle ergometer: a critical review. BioMed research international. 2013. Oct;2013. doi: 10.1155/2013/589361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Silva MLD, Ferreira RCA. Anaerobic power analysis and training methods in professional soccer athletes. Int Phys Med Rehab J. 2019;4(4):198–202. doi: 10.15406/ipmrj.2019.04.00198 [DOI] [Google Scholar]