Abstract
The genomes of the O3:K6 strains of Vibrio parahaemolyticus which abruptly emerged in Calcutta, India, in February 1996 and which demonstrated an unusual potential to spread and an enhanced propensity to cause infections were examined by different molecular techniques to determine clonality. No restriction fragment length polymorphism (RFLP) in the gene encoding the thermostable direct hemolysin was observed among the O3:K6 isolates of V. parahaemolyticus. Clonal diversity among the O3:K6 strains became evident by examining the RFLPs of the rrn operons and by the use of pulsed-field gel electrophoresis. Five ribotypes were distinguished among the O3:K6 strains examined, with ribotype R4 constituting the major type. Strains of O3:K6 isolated between June and August 1996 showed different pulsotypes compared to the pulsotypes of strains isolated before and after this period, indicating genetic reassortment among these strains, but those isolated between August 1996 and March 1998 showed identical or nearly similar pulsotypes. It is clear that there is a certain degree of genomic reassortment among the O3:K6 clones but that these strains are predominantly one clone.
Vibrio parahaemolyticus is a gram-negative marine bacterium that can cause seafood-borne gastroenteritis, but not all strains have the same pathogenic potential. Infections caused by V. parahaemolyticus are usually associated with diverse serovars. However, recent studies have shown the emergence of a unique serovar, serovar O3:K6, which appears to have the potential to spread and to be associated with infections more often than other serovars (19). Strains belonging to the O3:K6 serovar abruptly appeared in Calcutta, India, in February 1996 and accounted for 50 to 80% of the strains of V. parahaemolyticus (19). Furthermore, strains belonging to the same serovar have now been isolated from other Southeast Asian countries (19), from travelers at a quarantine station in Japan (6), and from a food-borne outbreak in the United States (3).
Although the pathogenic mechanism of V. parahaemolyticus is poorly understood, it has long been known that one of the hemolysins detected on a special blood agar medium, Wagatsuma agar, is a major virulence determinant (16). Most strains isolated from patients demonstrate this hemolytic activity, which is known as the positive Kanagawa phenomenon (KP) (13, 20). The hemolysin produced by KP-positive strains is thermostable in nature, and the hemolytic activity is not enhanced by the addition of lecithin, which indicates that it has direct action on erythrocytes (21) and which is thus called thermostable direct hemolysin (TDH). Additionally, a TDH-related hemolysin (TRH) produced by a KP-negative strain was reported in the last decade (4, 5). The tdh and trh genes, which encode TDH and TRH, respectively, share 70% nucleotide sequence identity (17). Molecular epidemiological studies revealed that not only strains that carry the tdh gene but also strains that carry the trh gene, or strains that carry both genes, are strongly associated with gastroenteritis (7, 23). Recently, a strong correlation between the presence of the trh gene and urease production, which is an unusual trait for V. parahaemolyticus, has been observed (11, 16, 25).
Intense investigations of the O3:K6 strains isolated in Calcutta in 1996 revealed that all 61 strains examined shared identical traits (tdh positive, trh negative, and urease negative), with only 2 strains having an antibiogram different from those of the other strains (19). In addition, the representative O3:K6 strains were shown to be genetically indistinguishable by arbitrarily primed PCR analysis and were therefore considered to be a single clone (19). The present report describes the further examination of the genomes of the O3:K6 strains of V. parahaemolyticus isolated in Calcutta by a variety of molecular typing methods to determine clonality among the strains. These additional molecular studies indicate that this collection of strains is composed of multiple clonal populations.
A total of 30 clinical V. parahaemolyticus O3:K6 strains isolated between February 1996 and June 1998 from patients admitted to the Infectious Diseases Hospital, Calcutta, were included in this study. The strains were selected to represent strains from different periods of isolation subsequent to the initial isolation of the O3:K6 strains in February 1996. The strains numbers of the selected strains (with the date of isolation in parentheses) are as follows: VP45 (5 February 1996), VP53 (28 February 1996), VP59 (21 March 1996), VP61 (22 March 1996), VP72 (12 April 1996), VP77 (17 April 1996), VP79 (23 April 1996), VP86 (10 May 1996), VP96 (30 May 1996), VP100 (4 June 1996), VP122 (9 July 1996), VP136 (2 August 1996), VP138 (6 August 1996), VP144 (17 August 1996), VP151 (20 September 1996), VP155 (8 October 1996), VP159 (4 November 1996), VP165 (12 March 1997), VP170 (17 April 1997), VP176 (30 April 1996), VP178 (26 May 1997), VP186 (12 June 1997), VP187 (1 June 1997), VP199 (10 July 1997), VP208 (8 August 1997), VP210 (29 August 1997), VP218 (16 September 1997), VP233 (25 March 1998), VP238 (16 June 1998), and VP239 (18 June 1998). Strains were identified and characterized, and their O:K serovars were determined with specific antisera as described previously (18). V. parahaemolyticus strains were grown in a gyratory shaker at 37°C in Luria broth containing 3% NaCl and were maintained in nutrient agar with 3% NaCl at room temperature.
Genomic DNA extraction and Southern hybridization were carried out as described by Sambrook et al. (22). Restriction enzymes BglI and HindIII (Boehringer Mannheim GmbH, Mannheim, Germany) were used for ribotyping and tdh genotyping of the O3:K6 strains of V. parahaemolyticus, respectively. Enzyme-digested genomic DNA fragments were electrophoresed, UV irradiated, and transferred to a nylon membrane (Hybond-N+; Amersham Life Science, Buckinghamshire, England), followed by hybridization with specific gene probes. A 7.5-kb BamHI fragment of the recombinant plasmid pKK3535 containing an rRNA operon of Escherichia coli (2) was used as the rrn gene probe for ribotyping. The gene probe for tdh was a 415-bp PstI fragment of pCVD518 representing 71% of the structural gene coding for TDH (15). Labeling of the probes, hybridization conditions, washing conditions for the filters, and detection of bands were performed with the ECL detection system (Amersham Life Science).
To perform pulsed-field gel electrophoresis (PFGE), the genomic DNAs of the various V. parahaemolyticus strains were prepared in agarose plugs as described previously (1, 27). Agarose blocks containing genomic DNA were equilibrated in restriction enzyme buffer for 1 h at room temperature and were cleaved in fresh buffer at the appropriate incubation temperature. For complete digestion of the DNAs, 50 U of NotI was used. PFGE of NotI-digested inserts was performed by the contour-clamped homogeneous electric field method on a CHEF Mapper system (Bio-Rad, Richmond, Calif.) in 0.5× TBE buffer (44.5 mM Tris-HCl, 44.5 mM boric acid, 1.0 mM EDTA [pH 8.0]) for 40 h 24 min. A DNA size standard (bacteriophage λ ladder; Bio-Rad) was used as the molecular mass standard, and a minichiller (model 1000; Bio-Rad) was used to maintain the temperature of the buffer at 14°C. Run conditions were generated by the autoalgorithm mode of the CHEF Mapper PFGE system by using a size range of 20 to 300 kb. CeuI-digested DNAs were electrophoresed on 1% agarose (Bio-Rad) with the Pulsaphor Plus System (Pharmacia, Uppsala, Sweden), with the pulse time interpolated between 10 and 50 s for 24 h at 10 V/cm at 4°C. Phage λ multimeric DNA (Bio-Rad) and yeast chromosomal DNA (New England Biolabs, Boston, Mass.) were used as molecular mass markers. After electrophoresis, the gel was stained by placing it in ethidium bromide (1 μg/ml) for 30 min and was destained by placing it in water for 15 min twice. The DNA bands were visualized with UV transilluminator and were photographed with a Polaroid photo apparatus.
The tdh gene probe hybridized with two HindIII fragments of 2.5 and 1.3 kb for all strains examined (data not shown). No restriction fragment length polymorphism (RFLP) of the genes encoding TDH was observed among the O3:K6 strains examined in this study. Most of the KP-positive strains carry two copies of the tdh gene in their genomes, and since HindIII cleaves DNA sequences outside the tdh structural genes, the number of copies of the tdh gene can be determined (15). For clinical strains of V. parahaemolyticus isolated in Thailand, the HindIII restriction fragment patterns of tdh and trh genes could be classified into five and four types, respectively, and a strong association between the restriction fragment patterns of tdh and trh was observed (24).
For ribotyping of V. parahaemolyticus strains, we selected the BglI restriction enzyme because this enzyme has successfully been used to develop a ribotyping scheme for Vibrio cholerae (8). Five ribotypes were distinguished among the 28 V. parahaemolyticus strains tested, and the ribotypes were designated R1 through R5 (Fig. 1). The ribotypes obtained in this study exhibited stable and reproducible patterns consisting of 9 to 11 bands ranging from approximately 23 to 4 kb (Fig. 1). The most common ribotype was ribotype R4 (78.6%), while the other ribotypes were uncommon but were distinct from ribotype R4. Ribotype R4 was found throughout the study period, while ribotype R1 was found in May 1996 and June 1997, ribotype R5 was found in May 1996, ribotype R2 was found in July 1997, and ribotype R3 was found in August 1997.
FIG. 1.
Representative results for the five ribotypes of the O3:K6 V. parahaemolyticus strains encountered in this study. R1, strain VP86; R2, strain VP199; R3, strain VP208; R4, strain VP155; and R5, strain VP96. Numbers on the right are molecular sizes (in kilobases).
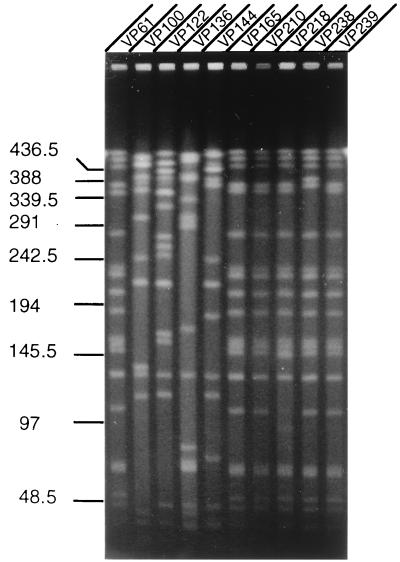
To further examine the clonality of O3:K6 isolates, the genomic DNAs of representative O3:K6 strains isolated during different time periods were analyzed by PFGE following digestion with NotI. The enzyme NotI produced several well-separated fragments in all the V. parahaemolyticus strains examined (Fig. 2). By using phage λ multimeric DNA and yeast chromosomal DNA as molecular mass markers, the NotI fragment sizes were estimated. The results showed several distinct RFLPs among the O3:K6 isolates (Fig. 2). O3:K6 strain VP61, which was isolated on 22 March 1996, had a pulsotype which resembled the pulsotypes of the strains isolated after August 1996. However, the O3:K6 strains isolated between June and August 1996 showed different pulsotypes, indicating genomic reassortment among these strains during these few months. The NotI profiles of the genomes of O3:K6 strains isolated between August 1996 and June 1998 were identical or nearly similar. The PFGE results suggest that genetic differences did exist between O3:K6 isolates, but only for a brief period.
FIG. 2.
PFGE of NotI-digested genomic DNAs of V. parahaemolyticus O3:K6 strains. The gel was stained with ethidium bromide. Numbers indicate the sizes of the molecular size markers (in kilobases). The RFLP patterns of strains isolated between February and August 1996 (strains VP61, VP100, VP122, and VP136) compared to the RFLP patterns of strains isolated between August 1996 and March 1998 (strains VP144, VP165, VP210, and VP218) are distinct. No RFLP was observed between the strains isolated in the later phase (June 1998) of the outbreak (strains VP238 and VP239).
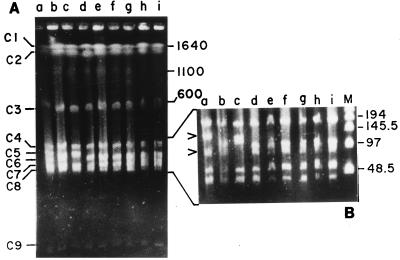
CeuI is encoded by a class I mobile intron (12), and CeuI digestion followed by PFGE is very useful for determination of intra- and interspecies genomic rearrangements and elucidation of the numbers of rrn operons in the genomes of prokaryotes. Since the enzyme cleaves genomic DNA only in a 19-bp sequence in the 23S rRNA gene of the rrn operon (10), it has also been successfully used to determine the number of rrn operons in the genomes of many organisms including V. cholerae, the type species of the genus Vibrio. When the genomes of representative V. parahaemolyticus O3:K6 strains were digested with CeuI and pulsed-field gel electrophoresed, nine fragments were detected in the ethidium bromide-stained gel, indicating that there are at least nine rrn operons in the V. parahaemolyticus genome (operons C1 to C9; Fig. 3). It is noteworthy that, as in the V. parahaemolyticus genome, there are 9 to 10 rrn operons in the V. cholerae genome (14). Apart from detecting the rrn operon in the genomes of prokaryotes, CeuI can serve as a tool for the rapid examination of the organizational variations in the genomes among various strains within a species (10). When we analyzed the CeuI digestion profiles of O3:K6 strains, RFLPs between O3:K6 strains were observed (Fig. 3). The CeuI-based RFLPs found between the O3:K6 strains matched those for the NotI-digested genomes of respective V. parahaemolyticus isolates (Fig. 2 and 3). These results confirm that there is clonal diversity among the O3:K6 strains, and genetic rearrangements were evident in strains isolated during a brief interim period after the genesis of this unique O3:K6 clone of V. parahaemolyticus.
FIG. 3.
PFGE of CeuI-digested genomic DNAs of V. parahaemolyticus O3:K6 isolates and identification of number of rrn operons. The enzyme-digested DNAs were electrophoresed, with pulse times interpolated between 20 and 200 s for 24 h at 10 V/cm at 4°C to separate CeuI fragments (operons C1 to C9) (A), and for better resolution of CeuI operons C4 to C8, the pulse time was interpolated between 5 and 100 s for 24 h at 10 V/cm at 4°C (the region for operon C4 to C8 is expanded in panel B. Since CeuI has sites only in rrn operons in the bacterial genomes examined so far, it appears that there are nine CeuI sites in the genome of V. parahaemolyticus and hence probably nine rrn operons. Lanes: a, VP100; b, VP122; c, VP136; d, VP144; e, VP165; f, VP165; g, VP210; h, VP218; i, VP238; and M, bacteriophage λ multimeric DNA as marker. Numbers indicate the sizes of the molecular size markers (in kilobases). The arrowhead indicates RFLP between initial isolates only in operons C5 and C6 (lanes a to d). Strains isolated from the later phase of the outbreak had identical CeuI pulsotypes (lanes e to i).
The BglI-generated ribotypes obtained from this study also indicate that there are at least nine or more rrn operons in the genome of V. parahaemolyticus, because all strains tested had 9 to 11 bands in the autoradiogram. It seems that most of the rrn operons of V. parahaemolyticus do not have a BglI site in the coding sequence. However, this needs further investigation to establish clearly the exact copy number of the rrn operon in this bacterium. rrn-mediated recombination may be one of the mechanisms by which pathogenic bacteria may maintain their genomic plasticity or diversity, and the possibility of genome rearrangements is greater when there are more rrn operons in the genome of any organism (9).
Intraspecies characterization of pathogenic bacteria for epidemiological purposes can now be achieved by several approaches. The development of molecular techniques has allowed the comparison of strains by examination of their genomes. We applied three such techniques to determine the clonality of the O3:K6 V. parahaemolyticus strains which abruptly appeared in Calcutta in February 1996 and which we previously defined as clonal (19). We used NotI and CeuI for PFGE analysis of the genomes of O3:K6 strains, and to our knowledge, this is the first report to describe an analysis of the genome of V. parahaemolyticus with these two enzymes. Previously, Wong et al. (26) used SfiI to distinguish a large number of V. parahaemolyticus strains isolated from different outbreaks in Taiwan, and they grouped all the strains into 14 PFGE types. From the present study it appears that the NotI enzyme can also be used to differentiate various V. parahaemolyticus strains because it produces a small number of large fragments with a uniform distribution of sizes separable by PFGE.
Although Southern analysis with the tdh gene as a probe failed to differentiate the strains, it confirmed that there are two chromosomal gene copies in strains showing a typical hemolysin-positive phenotype, as reported earlier (16). However, clonal diversity among O3:K6 isolates of V. parahaemolyticus became evident when we used the PFGE technique and the ribotyping method. The infection caused by the O3:K6 clone of V. parahaemolyticus can be categorized as an emerging infectious disease, considering the extent of its geographical spread. At this point we are not certain where this clone originated, but chronologically, it would appear that the clone abruptly established itself as an entity in Calcutta in February 1996 (19). Subsequently, the clone appears to have spread to other areas in Southeast Asia and now seems to have crossed over to the North American continent. This study indicates that the strains isolated during a brief interim period showed genetic instability, but over a period of time the clone stabilized. The global spread of this clone of V. parahaemolyticus needs to be carefully monitored.
Acknowledgments
This work was supported, in part, by the Japan International Cooperation Agency (JICA/NICED Project 054-1061-E-O).
REFERENCES
- 1.Bhadra R K, Roychoudhury S, Banerjee R K, Kar S, Majumdar R, Sengupta S, Chatterjee S, Khetawat G, Das J. Cholera toxin (CTX) genetic element in Vibrio cholerae O139. Microbiology. 1995;141:1977–1983. doi: 10.1099/13500872-141-8-1977. [DOI] [PubMed] [Google Scholar]
- 2.Brosius J, Ullrich A, Raker M A, Gray A, Dull T J, Gutell R R, Noller H F. Construction and fine mapping of recombinant plasmids containing the rrnB ribosomal RNA operon of E. coli. Plasmid. 1981;6:112–118. doi: 10.1016/0147-619x(81)90058-5. [DOI] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention (Atlanta, Ga.). Unpublished data.
- 4.Honda S, Goto I, Minematsu I, Ikeda N, Asano N, Ishibashi M, Kinoshita Y, Nishibuchi M, Honda T, Miwatani T. Gastroenteritis due to Kanagawa negative Vibrio parahaemolyticus. Lancet. 1987;i:331–332. doi: 10.1016/s0140-6736(87)92062-9. [DOI] [PubMed] [Google Scholar]
- 5.Honda T, Ni Y, Miwatani T. Purification and characterization of a hemolysin produced by a clinical isolate of Kanagawa phenomenon-negative Vibrio parahaemolyticus and related to the thermostable direct hemolysin. Infect Immun. 1988;56:961–965. doi: 10.1128/iai.56.4.961-965.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ishibashi, M. Unpublished data.
- 7.Kishishita M, Matsuoka N, Mukagai K, Yamasaki S, Takeda Y, Nishibuchi M. Sequence variation in the thermostable direct hemolysin-related hemolysin (trh) gene of Vibrio parahaemolyticus. Appl Environ Microbiol. 1992;58:2449–2457. doi: 10.1128/aem.58.8.2449-2457.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koblavi S, Grimont F, Grimont P A D. Clonal diversity of Vibrio cholerae O1 evidenced by rRNA gene restriction patterns. Res Microbiol. 1990;141:645–657. doi: 10.1016/0923-2508(90)90059-y. [DOI] [PubMed] [Google Scholar]
- 9.Krawiec S, Riley M. Organization of the bacterial genome. Microbiol Rev. 1990;54:502–539. doi: 10.1128/mr.54.4.502-539.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu S-L, Sanderson K E. I-CeuI reveals conservation of the genome of independent strains of Salmonella typhimurium. J Bacteriol. 1995;177:3355–3357. doi: 10.1128/jb.177.11.3355-3357.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Magalhães M, Takeda Y, Magalhães V, Tateno S. Brazilian urease-positive strains of Vibrio parahaemolyticus carry genetic potential to produce the TDH-related hemolysin. Mem Inst Oswaldo Cruz. 1992;87:167–168. doi: 10.1590/s0074-02761992000100027. [DOI] [PubMed] [Google Scholar]
- 12.Marshall P, Lemieux C. Cleavage pattern of the homing endonuclease encoded by the fifth intron in the chloroplast large subunit rRNA-encoding gene of Chlamydomonas eugametos. Gene. 1991;104:241–245. doi: 10.1016/0378-1119(91)90256-b. [DOI] [PubMed] [Google Scholar]
- 13.Miyamoto Y, Kato T, Obara Y, Aliyama S, Takizawa K, Yamai S. In vitro hemolytic characteristic of Vibrio parahaemolyticus: its close correlation with human pathogenicity. J Bacteriol. 1969;100:1147–1149. doi: 10.1128/jb.100.2.1147-1149.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nandi S, Khetawat G, Sengupta S, Majumder R, Kar S, Bhadra R K, Roychoudhury S, Das J. Rearrangements in the genomes of Vibrio cholerae strains belonging to different serovars and biovars. Int J Syst Bacteriol. 1997;47:858–862. doi: 10.1099/00207713-47-3-858. [DOI] [PubMed] [Google Scholar]
- 15.Nishibuchi M, Kaper J B. Duplication of the thermostable direct hemolysin (tdh) gene in Vibrio parahaemolyticus. Mol Microbiol. 1990;4:87–99. doi: 10.1111/j.1365-2958.1990.tb02017.x. [DOI] [PubMed] [Google Scholar]
- 16.Nishibuchi M, Kaper J B. Thermostable direct hemolysin gene of Vibrio parahaemolyticus: a virulence gene acquired by a marine bacterium. Infect Immun. 1995;64:2093–2099. doi: 10.1128/iai.63.6.2093-2099.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishibuchi M, Taniguchi T, Misawa T, Khaeomanee-iam V, Honda T, Miwatani T. Cloning and nucleotide sequence of the gene (trh) encoding the hemolysin related to the thermostable direct hemolysin of Vibrio parahaemolyticus. Infect Immun. 1989;57:2691–2697. doi: 10.1128/iai.57.9.2691-2697.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okuda J, Ishibashi M, Abbott S L, Janda J M, Nishibuchi M. Analysis of the thermostable direct hemolysin (tdh) gene and the tdh-related hemolysin (trh) genes in the urease-positive strains of Vibrio parahaemolyticus isolated on the West Coast of the United States. J Clin Microbiol. 1997;35:1965–1971. doi: 10.1128/jcm.35.8.1965-1971.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okuda J, Ishibashi M, Hayakaya E, Nishino T, Takeda Y, Mukhopadhyay A K, Garg S, Bhattacharya S K, Nair G B, Nishibuchi M. Emergence of a unique O3:K6 clone of Vibrio parahaemolyticus in Calcutta, India, and isolation of strains from the same clonal group from Southeast Asian travelers arriving in Japan. J Clin Microbiol. 1997;35:3150–3155. doi: 10.1128/jcm.35.12.3150-3155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sakazaki R, Tamura K, Kato T, Obara Y, Yamai S, Boho K. Studies of the enteropathogenic, facultatively halophilic bacteria, Vibrio parahaemolyticus. III. Enteropathogenicity. Jpn J Med Sci Biol. 1968;21:325–331. doi: 10.7883/yoken1952.21.325. [DOI] [PubMed] [Google Scholar]
- 21.Sakurai J, Matsuzaki A, Miwatani T. Purification and characterization of thermostable direct hemolysin of Vibrio parahaemolyticus. Infect Immun. 1973;8:775–780. doi: 10.1128/iai.8.5.775-780.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 23.Shirai H, Ito H, Hirayama T, Nakamoto Y, Nakabayashi N, Kumagai K, Takeda Y, Nishibuchi M. Molecular epidemiologic evidence for association of thermostable direct hemolysin (TDH) and TDH-related hemolysin of Vibrio parahaemolyticus with gastroenteritis. Infect Immun. 1990;58:3568–3573. doi: 10.1128/iai.58.11.3568-3573.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suthienkul O, Iida T, Park K-S, Ishibashi M, Supavej S, Yamamoto K, Honda T. Restriction fragment length polymorphism of the tdh and trh genes in clinical Vibrio parahaemolyticus strains. J Clin Microbiol. 1996;34:1293–1295. doi: 10.1128/jcm.34.5.1293-1295.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suthienkul O, Ishibashi M, Iida T, Nettip N, Supavej S, Eampokalam B, Makino M, Honda T. Urease production correlates with possession of the trh gene in Vibrio parahaemolyticus strains isolated in Thailand. J Infect Dis. 1995;172:1405–1408. doi: 10.1093/infdis/172.5.1405. [DOI] [PubMed] [Google Scholar]
- 26.Wong H C, Lu K T, Pan T M, Lee C L, Shih D Y C. Subspecies typing of Vibrio parahaemolyticus by pulsed-field gel electrophoresis. J Clin Microbiol. 1996;34:1535–1539. doi: 10.1128/jcm.34.6.1535-1539.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamasaki S, Nair G B, Bhattacharya S K, Yamamoto S, Kurazono H, Takeda Y. Cryptic appearance of a new clone of Vibrio cholerae serogroup O1 biotype El Tor in Calcutta, India. Microbiol Immunol. 1997;41:1–6. doi: 10.1111/j.1348-0421.1997.tb01165.x. [DOI] [PubMed] [Google Scholar]