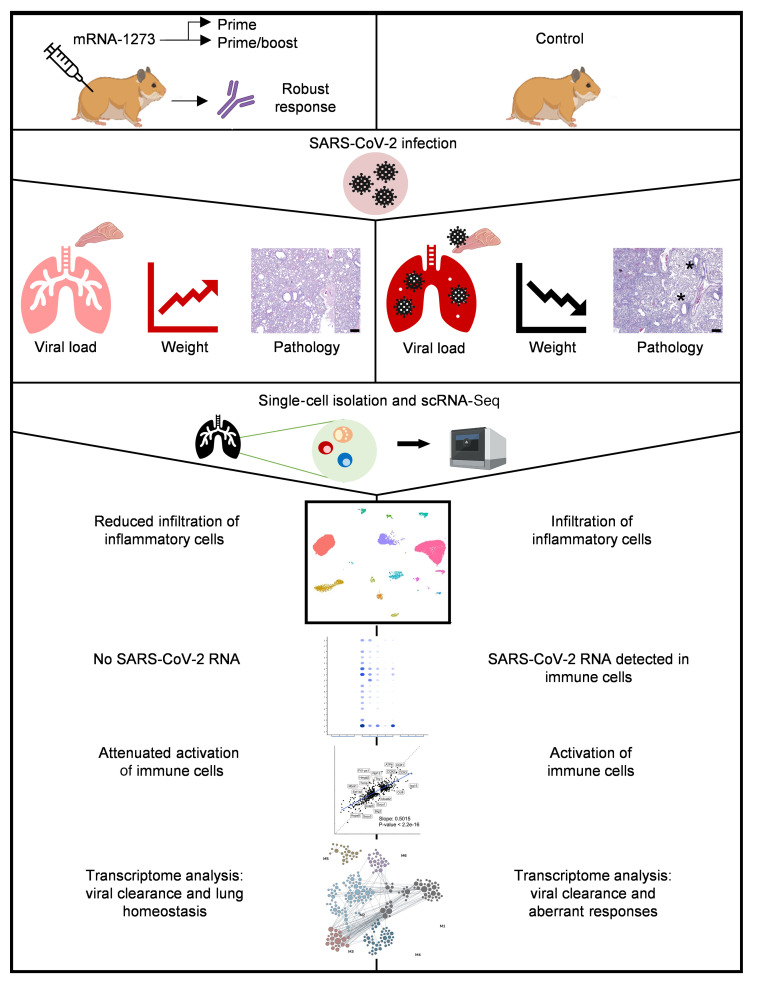
Abstract
The mRNA-1273 vaccine is effective against SARS-CoV-2 and was granted emergency use authorization by the FDA. Clinical studies, however, cannot provide the controlled response to infection and complex immunological insight that are only possible with preclinical studies. Hamsters are the only model that reliably exhibits severe SARS-CoV-2 disease similar to that in hospitalized patients, making them pertinent for vaccine evaluation. We demonstrate that prime or prime-boost administration of mRNA-1273 in hamsters elicited robust neutralizing antibodies, ameliorated weight loss, suppressed SARS-CoV-2 replication in the airways, and better protected against disease at the highest prime-boost dose. Unlike in mice and nonhuman primates, low-level virus replication in mRNA-1273–vaccinated hamsters coincided with an anamnestic response. Single-cell RNA sequencing of lung tissue permitted high-resolution analysis that is not possible in vaccinated humans. mRNA-1273 prevented inflammatory cell infiltration and the reduction of lymphocyte proportions, but enabled antiviral responses conducive to lung homeostasis. Surprisingly, infection triggered transcriptome programs in some types of immune cells from vaccinated hamsters that were shared, albeit attenuated, with mock-vaccinated hamsters. Our results support the use of mRNA-1273 in a 2-dose schedule and provide insight into the potential responses within the lungs of vaccinated humans who are exposed to SARS-CoV-2.
Keywords: Immunology, Vaccines
Keywords: Adaptive immunity, Bioinformatics, Cellular immune response
Introduction
When the World Health Organization declared severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) a global pandemic in March 2020, phase I clinical trials on the most promising vaccine candidates had already commenced. Nucleoside-modified mRNA, a relatively new addition to the arsenal of vaccine platforms, has shown promise against numerous viral infectious diseases in preclinical trials (1, 2) and phase I and II trials that are completed (3, 4) or currently under way (5–7). Previous preclinical work on a related betacoronavirus enabled the rapid development of mRNA-1273, a vaccine composed of a modified mRNA encoding a stabilized prefusion form of the SARS-CoV-2 spike (S) protein encapsulated in lipid nanoparticles (1). Vaccination with mRNA-1273 significantly reduced infection in the lungs and upper airways of mice (1) and rhesus macaques (8). In phase I (9, 10) and phase II studies the vaccine was found to be safe and to elicit robust neutralizing antibody responses. More recently, interim analysis of phase III data indicated that mRNA-1273 was 94.1% efficacious in prevention of COVID-19 disease and highly efficacious in protecting against severe disease (11). On December 18, 2020, the US Food and Drug Administration authorized the emergency use of mRNA-1273.
Nonhuman primate (NHP) (12–14), ferret (15–17), and mouse (18) models that have been developed to understand SARS-CoV-2 pathogenesis and test vaccine and therapeutics efficacies display a mild course of disease. The lack of overt symptoms in NHPs and ethical constraints due to the sheer numbers required to recapitulate low-frequency human cases with severe clinical manifestations limit the ability to understand all aspects of the disease and vaccine effectiveness. Hamsters are highly susceptible to SARS-CoV-2 and develop a severe pneumonia similar to that in COVID-19 patients (19–22). Hamster angiotensin-converting enzyme-2 (ACE2) receptors that enable SARS-CoV-2 binding and entry are analogous to the human receptor (20), and therefore virus adaptation (18) or transgenic modifications (23) are not required for permissiveness. Additionally, hamsters can support high levels of virus replication and transmission and demonstrate weight loss and lethality with high SARS-CoV-2 infectious doses (21), making them an ideal model for evaluating vaccines.
Here, we tested the efficacy of the mRNA-1273 vaccine with prime-only and 3 dose levels of prime-boost regimens using the stringent golden Syrian hamster model. Serological, virological, clinicopathological, and single-cell RNA sequencing (scRNA-Seq) analyses were conducted to characterize vaccine-mediated immunity before and after challenge. We show that vaccination induced robust virus-neutralizing antibody responses, attenuated virus replication, and mitigated the influx of inflammatory innate immune cells and the relative reduction of lymphocytes in the lungs after challenge, although activated immune cell populations were observed. Our data illustrate the viral, cellular, and immune dynamics within the lungs of vaccinated hamsters, which may offer a unique perspective into the events that occur within the lungs of vaccinated humans who are exposed to SARS-CoV-2.
Results
Vaccination induces potent antibody responses.
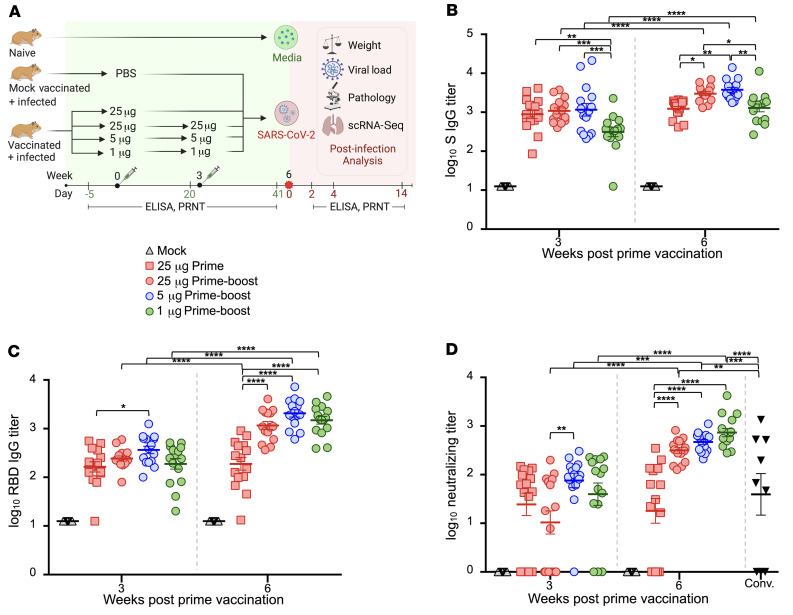
Three groups of outbred hamsters (n = 15) were vaccinated with 25 μg, 5 μg, and 1 μg of mRNA-1273 via the intramuscular (i.m.) route in a prime (week 0)–boost (week 3) regimen (Figure 1A). One group of hamsters (n = 15) received a prime-only dose of 25 μg at week 0, and another group (n = 15) were mock vaccinated as the study’s control. The humoral responses to vaccination were measured by ELISA specific for the SARS-CoV-2 S protein (Figure 1B) and its receptor-binding domain (RBD) (Figure 1C). Three weeks after the prime dose, higher S-specific IgG titers were detected in hamsters vaccinated with 25 μg and 5 μg doses compared with the 1 μg dose (Figure 1B). S-specific IgG titers were significantly augmented in all groups following the booster, but continued to be significantly higher in the 25 μg and 5 μg dose groups compared with the 1 μg dose group. RBD-specific IgG titers were comparable among hamsters in the prime-boost regimen groups after receipt of their first dose (Figure 1C). After a boost dose, RBD titers had increased significantly (P < 0.0001) and remained comparable among the prime-boost groups. RBD titers in the prime-boost vaccine groups were also significantly higher after the second dose in comparison with the 25 μg prime-only group (P < 0.0001), while titers remained unchanged for the 25 μg prime-only group between weeks 3 and 6 after vaccination. The ability of serum to neutralize live SARS-CoV-2 reporter virus was also determined (Figure 1D and Supplemental Methods; supplemental material available online with this article; https://doi.org/10.1172/JCI148036DS1). Most vaccinated hamsters produced neutralizing titers after the prime dose. As with RBD-binding titers, neutralizing titers significantly increased in all groups that received a booster dose, and the levels remained comparable among these hamster groups, but higher versus the prime-only vaccine group. The magnitude of neutralizing antibody titers in all booster-vaccinated groups was higher than in convalescent COVID-19 patients, while titers in the prime-only group were comparable to those seen in these subjects. Neutralizing titers significantly correlated with S-specific IgG titers (Supplemental Figure 1A) and RBD-specific titers (Supplemental Figure 1B), albeit the correlation was greater with RBD-specific IgG titers at both week 3 and week 6 after vaccination.
Figure 1. Serum antibody responses in vaccinated hamsters.
(A) Study design. Hamsters were prime-vaccinated via the i.m. route at week 0 and boosted at week 3, with 25 μg (n = 15), 5 μg (n = 15), and 1 μg (n = 15) of mRNA-1273. A group of hamsters (n = 15) received a prime dose only of 25 μg mRNA-1273, and a mock group received PBS (n = 15) at week 0. At week 6, animals were intranasally challenged with 105 PFU of SARS-CoV-2. On days 2, 4, and 14 after infection, hamsters (n = 5 per group) were euthanized for tissue collection. PRNT, plaque reduction neutralization test (B–D) Total serum SARS-CoV-2 S–specific (B) and RBD-specific (C) IgG titers in serum and neutralizing titers (D) in hamster groups prior to SARS-CoV-2 infection measured by ELISA and plaque reduction assays, respectively. Neutralizing titers were compared with a panel of human convalescent serum samples (Conv.). Bars denote group means ± SE. Significance was measured by ANOVA with Tukey’s or Šidák’s correction for multiple comparisons between vaccine groups or between time points, respectively (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001).
Prime-boost vaccination effectively protects against SARS-CoV-2 replication in the lungs.
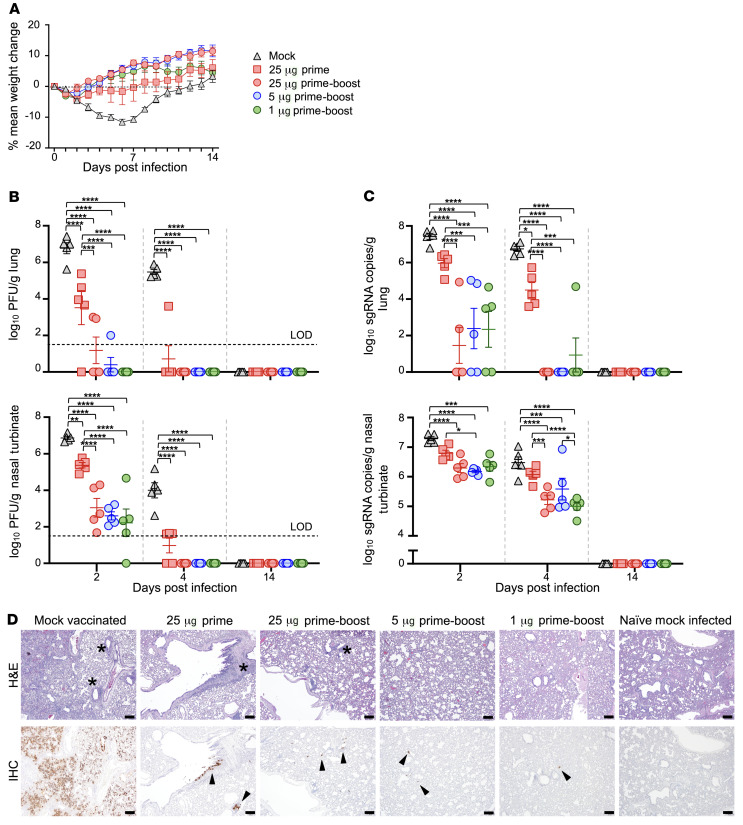
Six weeks after prime vaccination (3 weeks after the boost vaccination), all treatment-group hamsters were challenged intranasally with 105 plaque-forming units (PFU) of SARS-CoV-2. Hamsters were monitored daily for changes in body weight. At 2 and 4 days postinfection (dpi), 5 animals from each group were serially euthanized, and the viral load in the right lung and nasal turbinates was determined. The remaining hamsters were observed until the study’s endpoint at day 14. Mock-vaccinated hamsters lost an average maximum body weight of 12% by day 6 (Figure 2A and Supplemental Figure 2, A and B). The prime-boost regimen prevented significant weight loss in all but 1 hamster in the 5 μg prime-boost group; excluding animals necropsied at 2 and 4 dpi, the average maximum weight loss for the combined prime-boost dose vaccine groups was 2.25% over the 14-day infection period (Supplemental Figure 2B). The prime-only vaccinated group lost a maximal mean weight of 6.2%. Moderate inverse correlations were observed between maximum percentage weight loss and S-binding IgG and neutralizing antibody titers at week 6 (Supplemental Figure 3A).
Figure 2. Characteristics of clinical disease following SARS-CoV-2 challenge of vaccinated hamsters.
(A) Groups of 15 hamsters each vaccinated with 1 dose (25 μg) or 2 doses (25 μg, 5 μg, and 1 μg) of mRNA-1273 or PBS were monitored for mean percentage weight change over 14 days after challenge with SARS-CoV-2. (B) Viral load expressed in PFU per gram of tissue in the lungs and nasal turbinates at serial endpoint days (n = 5 per group euthanized at 2, 4, and 14 dpi). Dashed horizontal line indicates limit of detection (LOD). (C) Viral sgRNA was measured by qRT-PCR in the lungs and nasal turbinates at serial endpoint days (n = 5 per group euthanized at 2, 4, and 14 dpi). (B and C) Significance was measured by ANOVA with Tukey’s correction for multiple comparisons (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001). Error bars represent ± SE. (D) Lung sections from mock-vaccinated and prime-only– and prime-boost–vaccinated groups infected with SARS-CoV-2 and naive mock-infected groups at 4 dpi were stained with H&E. Representative photomicrographs (original magnification, ×4; scale bars, 200 μm) from each group with virus antigen (arrowheads) in lungs are shown, stained by immunohistochemistry. *Areas of perivascular-peribronchiolar inflammation (mostly mononuclear).
At 2 dpi, high viral loads were detected by plaque assay in the lungs and nasal turbinates of the mock-vaccinated group with a peak mean of 6.8 log10 PFU/g and 6.9 log10 PFU/g, respectively (Figure 2B). Markedly lower levels of virus were detected in the lungs of all vaccinated groups, while the 1 μg prime-boost dose group had no detectable virus in the lungs. The mean peak virus load in the nasal turbinates was similar among the prime-boost groups and 4.3 log10 lower in comparison with the mock-vaccinated group (P < 0.0001). At 4 dpi, there was no detectable virus in the lungs and nasal turbinates of all prime-boost recipients. Hamsters that received the prime-only vaccine dose had 4.8 log10 and 3.0 log10 reductions of peak virus load means, respectively, in the lungs and nasal turbinates compared with the control group. At 14 dpi, no virus was detected in all groups.
We measured viral subgenomic RNA (sgRNA) in these tissues by quantitative reverse transcription PCR (qRT-PCR) as a potential gauge for replicating virus. Peak sgRNA levels were detected in the mock group at 2 dpi, with geometric means of 7.4 log10 (95% CI, 7.0 log10 to 7.9 log10) copies/g in the lungs and 7.3 log10 (95% CI, 7.1 log10 to 7.5 log10) copies/g in nasal turbinates (Figure 2C). All prime-boost groups showed similar lower sgRNA levels compared with the mock vaccine group with peak geometric means of 2.1 log10 (95% CI, 0.86 log10 to 3.3 log10) sgRNA copies/g in lungs and 6.3 log10 (95% CI, 6.4 log10 to 6.1 log10) sgRNA copies/g in nasal turbinates. In the prime-only vaccine group, sgRNA levels were not significantly reduced in either of these tissues. In close agreement with viral load, no sgRNA was detected in the lungs of prime-boost groups by 4 dpi, except for 1 hamster in the 1 μg dose group. While viral load was not detected in the nasal turbinates of prime-boost recipients, sgRNA was detected, albeit at markedly reduced levels in comparison with the mock-vaccinated group. SARS-CoV-2 transmission has been shown to correlate with levels of infectious virus and not sgRNA (22). Therefore, the likelihood of onward transmission was highly reduced by mRNA-1273. The prime-only dose afforded a less robust protection according to sgRNA levels, particularly in the nasal turbinates. Neutralizing antibody titers at week 6 inversely correlated with viral load and sgRNA in both the lungs and nasal turbinates at 2 and 4 dpi (Supplemental Figure 3, B and C).
Fourteen days after challenge, the mock-vaccinated hamsters had measurable serum S-specific (Supplemental Figure 4A) and nucleocapsid-specific (NP-specific) (Supplemental Figure 4B) IgG titers and neutralizing titers (Supplemental Figure 4C). An anamnestic response was observed as soon as 4 dpi with the S-specific IgG increasing in recipients of the prime-only vaccine regimen. At 14 dpi, all vaccinated hamsters displayed an anamnestic S-specific IgG antibody response. This result contrasts with the unwavering virus-specific IgG levels observed after challenge in vaccinated NHPs (8). Furthermore, NP-specific IgG titers were detected in all groups at 14 dpi (Supplemental Figure 4B) confirming, together with the detection of sgRNA in the upper and lower respiratory tract (Figure 2C), replication of the challenge virus before clearance.
High-dose prime-boost vaccination protects against severe pathological changes in the lungs.
The lungs of hamsters were evaluated histologically after challenge with SARS-CoV-2 at 2-, 4-, and 14-day time points (Figure 2D and Supplemental Figures 5 and 6). A naive group of hamsters (n = 4), intranasally given media to mimic virus inoculum and euthanized 4 days later, was included as a control and presented with a moderately prominent alveolar interstitial hypercellularity. While these hypercellular areas may represent regions of atelectasis, the presence of Pasteurella multocida, a common respiratory commensal, was also detected by metagenomics analysis of all lung samples used in scRNA-Seq (Supplemental Table 1 and ref. 24). At 2 dpi, SARS-CoV-2 infection in mock-vaccinated animals caused mild interstitial inflammation in the lungs with some animals exhibiting a largely polymorphonuclear cellular infiltrate, composed predominantly of neutrophils/heterophils, in and around the lung airways (Supplemental Figure 6 and Supplemental Table 2). By 4 dpi, inflammation was largely associated with perivascular and peribronchiolar regions in a focally diffuse or multifocal distribution affecting, on average, 30%–50% of the lung that persisted until the study’s endpoint (Figure 2D, Supplemental Figure 5, and Supplemental Table 2). Over the 14-day infection course, prime-boost–vaccinated animals displayed mild to moderate inflammation mostly involving the pulmonary interstitium, characterized by expanded alveolar capillary profiles, diminished alveolar spaces, and influx of predominantly mononuclear cells (Figure 2D, Supplemental Figures 5 and 6, and Supplemental Table 2). In the 1 μg prime-boost group, the predominant inflammatory phenotype was mild to moderate interstitial and rarely perivascular-peribronchiolar (4 dpi) inflammation. However, 1 animal exhibited a more severe pulmonary inflammatory response at 2 dpi, characterized by mild to moderate edema and rare foci of hemorrhage and vascular congestion that were previously described in SARS-CoV-2–infected hamsters (19). Prime-only vaccinated animals generally exhibited reduced inflammation compared with mock-vaccinated animals at 4 dpi; 1 outlier hamster presented mild pulmonary edema and scant fibrin deposition that were not observed in the rest of the group. Irrespective of vaccination schedule and dose, the airway lumen remained clear of inflammatory cells and cellular debris.
Lung sections were also examined for SARS-CoV-2 antigen at 4 dpi by immunohistochemistry (Figure 2D, Supplemental Figure 5, Supplemental Table 2, and Supplemental Methods). Large amounts of viral antigen were found throughout the lungs of mock-vaccinated hamsters. Prime-boost vaccination limited the amount of detectable total viral antigen more than did prime-only vaccination; SARS-CoV-2 antigen was minimal or absent in the 25 μg and 5 μg prime-boost groups.
Notably, the 3 outlier hamsters with more severe histopathological phenotypes identified in the 25 μg prime-only group at 4 dpi and in the 1 μg prime-boost group at 2 and 4 dpi did not exhibit drastic weight loss. Their levels of infectious virus or sgRNA in the lungs or nasal turbinates were also comparable with those in other group members, with the exception of the single 1 μg recipient at 4 dpi that had detectable sgRNA in the lungs that was not detected in other prime-boost recipients at the same time point. Both 4 dpi outliers had the highest level of viral antigen among all vaccinated hamster samples. Interestingly, all 3 outliers did not produce neutralizing titers after prime vaccination, a finding that suggests the importance of vaccine dose level and regimen, and the evolving immune response following administration of a second dose.
Integrated single-cell analysis and cell type identification.
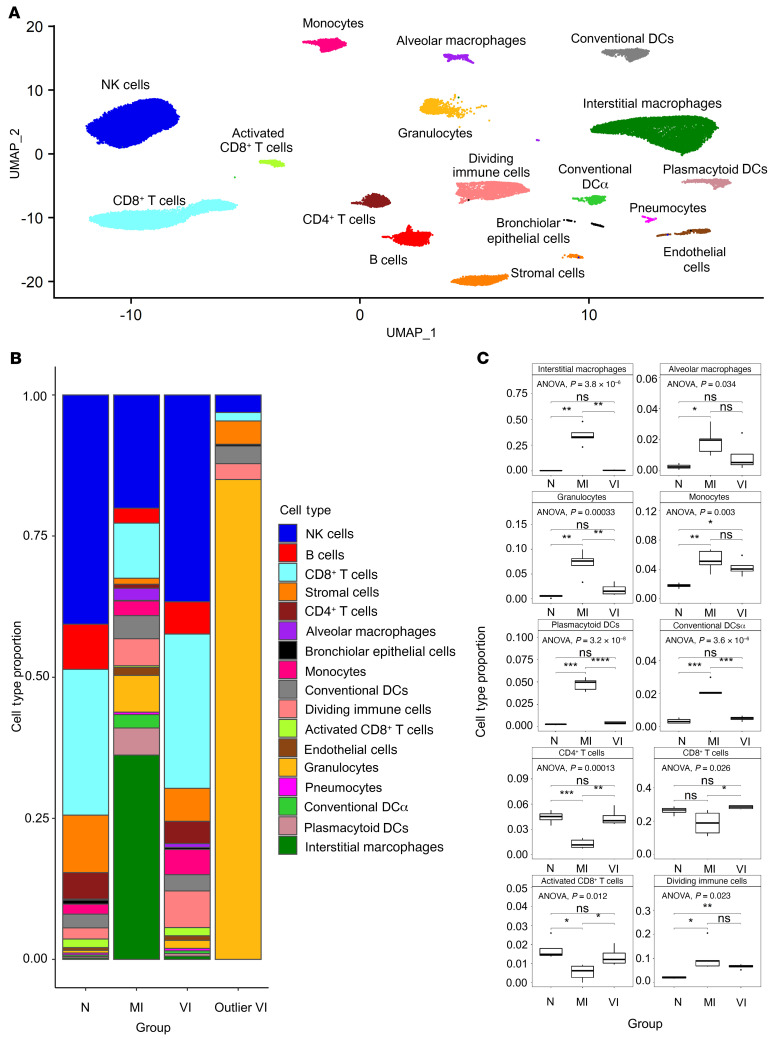
We performed scRNA-Seq on samples from the upper region of the left lung lobe from 3 of the study’s hamster groups that were euthanized at 4 dpi: the naive hamsters mock-infected with media (naive, N; n = 4) and the 5 μg prime-boost–vaccinated (vaccinated + infected, VI; n = 5) and mock-vaccinated (mock-vaccinated + infected, MI; n = 5) groups infected with SARS-CoV-2. Lung samples from the N group served as a baseline control for analysis. The scRNA-Seq data were aligned against the hamster and SARS-CoV-2 genomes and integrated using the Seurat scRNA-Seq analysis pipeline (25). Integration of the single-cell data identified 17 cell types, including multiple T cell, dendritic cell (DC), and macrophage subtypes (Figure 3A). The main markers used to distinguish the cell types are shown in Supplemental Figure 7A.
Figure 3. Single-cell RNA-Seq analysis of pulmonary tissues identifies cell population differences between N, MI, and VI animals.
(A) Integrated UMAP (uniform manifold approximation and projection) of all N (n = 4), MI (n = 5), and VI samples (n = 4) excluding one VI outlier sample. (B) Bar plot of cell-type proportions within N (n = 4), MI (n = 5), and VI (n = 4) groups and VI outlier. (C) Box plots comparing cell-type proportions observed in N (n = 4), MI (n = 5), and VI (n = 4) groups excluding one VI outlier. The limits of the box reflect the interquartile range (IQR: Q3–Q1) with median shown as horizontal bars. Whiskers extend to 1.5 times the IQR of the box. Outliers outside 1.5 × IQR are shown as individual points. ANOVA and pairwise comparisons by post hoc Tukey’s test are shown (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). DCα, activated DCs.
The expression profile of one VI animal (Supplemental Figure 8A) that showed excessive weight loss at 4 dpi (Supplemental Figure 2A) was very distinct and was not included in the integrated analysis. Granulocytes represented over 75% of the cells in this outlier sample but were fewer than 5% of cells in the other VI samples (Figure 3B). The gene expression pattern in the granulocytes from the outlier VI sample also differed from that in the granulocytes from the other VI samples (Supplemental Figure 8, B and C). Pathway analysis of the differentially expressed genes (DEGs) in granulocytes from the outlier showed upregulation of classical neutrophil processes associated with degranulation, activation, and proliferation, and downregulation of immune-suppressive interleukins (Supplemental Figure 8D).
The 3 groups showed distinct differences in cell-type proportions for many of the cell types (Figure 3, B and C, and Supplemental Figure 7B). Notably, while interstitial macrophages constituted a negligible fraction of the total cells in N hamsters, the proportion of interstitial macrophages in MI hamsters was greater than 30% of the total cells (Figure 3, B and C). Importantly, this increase in macrophages was not seen in VI hamsters. Significant increases in the proportions of alveolar macrophages, DC subtypes (plasmacytoid DC [pDC], conventional DC [cDC], and activated DC [DCα]), and granulocytes were also found in MI hamsters and not VI hamsters. In agreement with the observed cellular infiltration in MI hamsters, expression of biomarkers of inflammation including IL-6, MCP-1, and IL-2, measured in a multiplex assay (Supplemental Methods), was significantly greater in the lungs of MI hamsters compared with N hamsters. The expression of inflammatory markers in the lungs of VI hamsters was generally low and comparable with that in the lungs of the N hamsters (Supplemental Figure 9). We detected decreases in the proportions of CD4+, CD8+, and CD8+ activated T cells in MI hamsters and not VI hamsters (Figure 3, B and C). The MI and VI hamsters both showed increases in the proportion of the dividing immune cells and monocytes when compared with N hamsters. The moderate changes in the cell-type composition of VI hamsters and infiltration of inflammatory cells in lungs of MI hamsters are consistent with histology results described in the previous section.
Vaccination attenuates infection of immune cells and inflammation but enables recall responses.
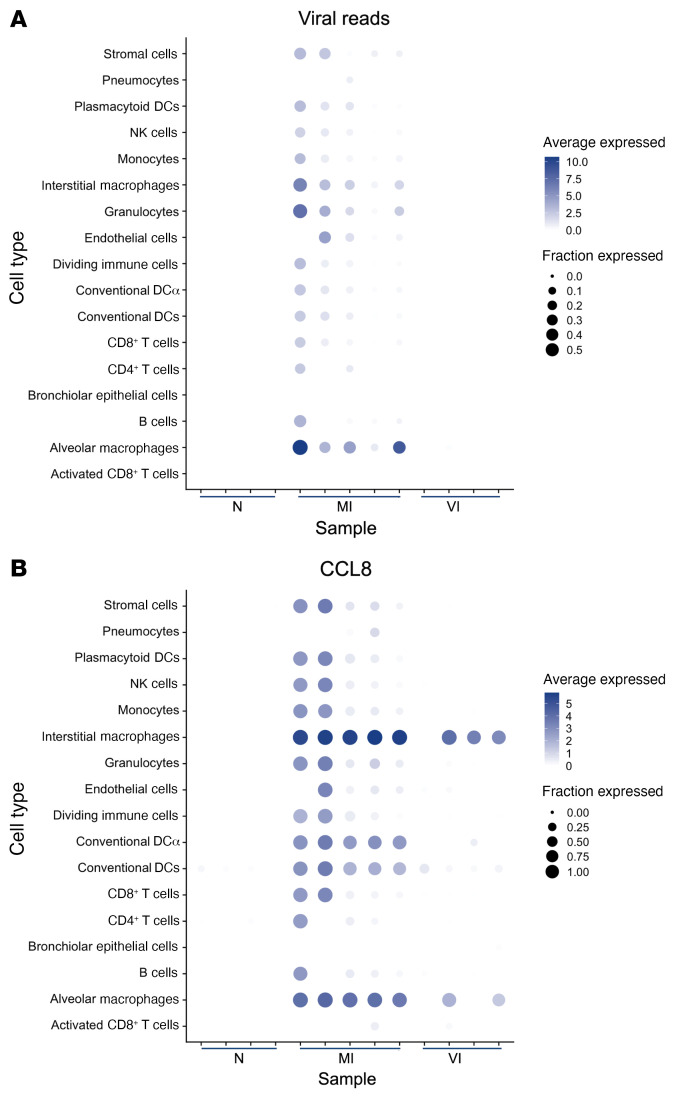
To identify the extent of infection of cells within the lungs, the scRNA-Seq reads were mapped to the SARS-CoV-2 genome. Viral gene reads were detected only in MI hamsters (Figure 4A and Supplemental Figure 10, A and B), consistent with the clearance of infection in the lungs of the VI hamsters that was observed by qRT-PCR (Figure 2C). Several MI samples with higher overall viral reads had virus gene expression detected in a broad selection of cell types in the lungs, including lymphoid and myeloid cells (Figure 4A), which is consonant with other studies sampling bronchoalveolar lavage fluid (BALF) (26), blood, or postmortem lung tissues (27, 28) from severe infections in humans. Macrophages and granulocytes had the highest average viral read counts (Supplemental Figure 10E). The relative levels of reads for viral gene segments within each lung sample were highly variable across animals (Supplemental Figure 10F). Interindividual variation in the relative levels of viral transcripts has been previously described in cells isolated from the BALF of COVID-19 patients (26) and lung tissues from NHPs (29).
Figure 4. Viral reads and CCL8 expression from lung tissue scRNA-Seq.
(A) Dot plot showing expression of all viral reads in each cell type in N (n = 4), MI (n = 5), and VI (n = 4) samples excluding one VI outlier sample. Cell type/sample pairs are only shown if there are more than 20 cells per cell type/sample. (B) Dot plot showing expression of CCL8 in each cell type in N (n = 4), MI (n = 5), and VI (n = 4) samples excluding one VI outlier sample. Cell type/sample pairs are only shown if there are more than 20 cells per cell type/sample.
Expression of CCL8, the chemoattractant for monocytes and macrophages reported to be significantly induced in response to SARS-CoV-2 infection in the ferret upper respiratory tract (17), was minimal in the lungs of N animals but highly upregulated in the MI hamsters (Figure 4B and Supplemental Figure 10C). The upregulation of CCL8 is consistent with macrophage infiltration of the lung observed in the MI group and in COVID-19 patients (30). The cellular distribution of CCL8 expression in MI lung mirrored that of the virus RNA, being highest in the interstitial and alveolar macrophage populations. In the VI group, increases in CCL8 expression were only observed in the macrophage populations in some of the samples studied, despite the absence of SARS-CoV-2 mRNA (Figure 4B and Supplemental Figure 10D). Overall, these data suggest that vaccination impedes SARS-CoV-2 infection of lymphoid and myeloid cells.
We assessed the activation states of nondividing lymphocyte cell clusters (Supplemental Figure 11) and their counterparts undergoing division (Supplemental Figure 12, A and B), and compared the transcriptional changes in expression markers between N, MI, and VI hamsters. All cell types assessed in both MI and VI hamsters underwent transcriptional changes signifying an activation state in comparison with N hamsters. The nondividing CD4+ T cells in VI hamsters had higher expression of genes representative of Th1 responses (Il2 and Anxa1) (31–33) and memory phenotypes (S100a4, Lgals1, and Anxa1) than did those in the MI and N groups (Supplemental Figure 11). Nondividing CD4+ T cells from MI hamsters had reduced levels of gene transcripts important for survival (Il7r), regulation (Socs2), and viral clearance (Ltb) (34) and increased levels of transcripts potentially driving proinflammatory effector functions (Ccl5, Isg15) (35) in comparison with VI and N hamsters. VI hamsters had a greater cluster of dividing CD4+ T cells compared with MI or N hamsters (Supplemental Figure 12C) that potentially enabled a rapid recall response within the lungs through augmentation of transcripts encoding cytotoxic molecules (36) and antibody binding Fc receptor (FcerIg; ref. 37) and repression of transcripts responsible for migration (S1pr1; Supplemental Figure 12D and ref. 38). Additionally, the transcriptional regulators Tbx21 and Zbtb32, which drive Th1 polarization (39), were increased in VI hamsters compared with MI and N hamsters, while Gata3, a Th2 marker, was reduced in comparison with N hamsters. These data are consistent with a Th1-polarized recall response to viral challenge in the VI animals and suggest that vaccine-induced CD4+ T cells contribute substantially toward virus clearance.
Nondividing B cells in VI hamsters were reduced in Ighm transcripts compared with those in MI and N hamsters, indicating they were differentially class-switched away from IgM production (Supplemental Figure 11). Dividing B cells in VI hamsters had more transcripts representative of memory or plasma cells (Ssr3 and Mzb1) than did those in MI and N hamsters (Supplemental Figure 12D). The transcription repressor Prmd1 was also greater in VI than in MI and N hamsters and, together with the Zbtb32 repressor (40), potentially enabled differentiation of memory cells into IgG-secreting plasma cells, thus coinciding with the observed amnestic antibody response induced after infection. Dividing B cells in MI hamsters had more transcripts associated with inflammation (Fos, Klf6, Zfp36, and S100a8) than did those in VI or N hamsters.
Both dividing and nondividing CD8+ T cells in VI hamsters possessed a lower effector and cytotoxic potential than those in MI hamsters (Supplemental Figure 11 and Supplemental Figure 12D). Transcripts encoding both stimulatory (Cd27, Tnfrsf4, and Tnfrsf18) and inhibitory (Lag3) molecules were increased in dividing CD8+ T cells in MI hamsters compared with VI and N hamsters, suggesting that these proliferating cells may be dysfunctional or exhausted (Supplemental Figure 12D and refs. 41, 42).
NK cells and their dividing counterparts in VI hamsters were enriched for FcerIg and Klrk and the cytotoxic Gzma transcripts and had reduced chemokine Xcl1 transcripts (43) compared with those in MI or N hamsters, suggesting that they were active or mature (Supplemental Figure 11 and Supplemental Figure 12D). Dividing and nondividing NK cells in MI hamsters were also augmented for transcripts driving effector functions (Ccl4 and Ccl5 chemokines and interferon response) in addition to transcripts encoding cytotoxic granules, indicating an enhanced cytolytic potential in comparison with VI hamsters.
Overall, the transcriptional changes in lymphocytes from VI hamsters that occurred after infection are likely to represent memory-driven responses from the adaptive arm of immunity established by mRNA-1273.
Following SARS-CoV-2 infection, similar transcriptional programs are regulated in lung immune cells from vaccinated and mock-vaccinated hamsters.
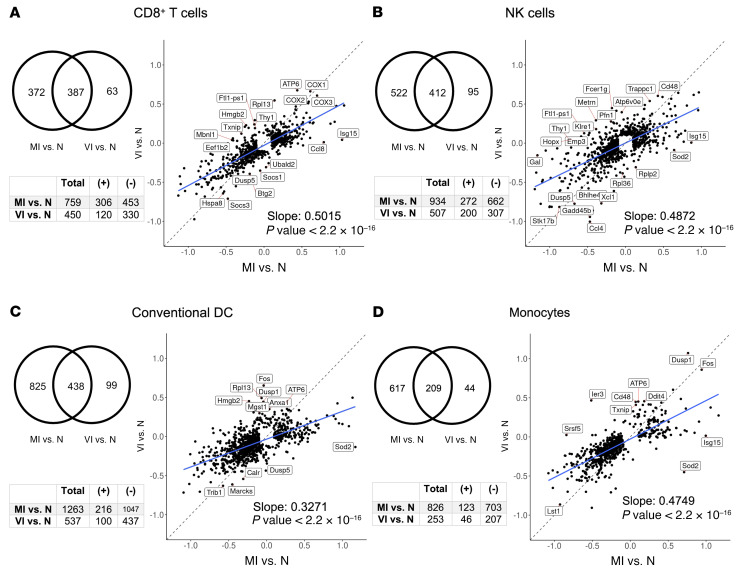
To determine whether immune cells in the lungs showed differences in their transcriptional states in the 3 groups studied, we focused on 2 lymphoid cell types critical for viral clearance during respiratory infections, CD8+ T cells and NK cells, which were relatively abundant across all samples. We also examined myeloid cells, including pDCs, cDCs, and monocytes. Differential expression analysis comparing the MI and VI groups for each cell type annotated for an increase in virus response and innate immune response pathways in all MI cell types (Supplemental Table 3). However, to more fully resolve the relationship of the pathways regulated in MI and VI cells, we subsequently performed differential expression analysis for each cell type (see Methods), comparing the MI samples with N samples and the VI samples with N samples. In all cell types, fewer transcripts were differentially expressed in the VI versus N comparison than in the MI versus N comparison; the DEGs in VI samples were predominantly a subset of the DEGs in MI samples (Figure 5). A scatterplot comparing the log fold changes of the set of transcripts significantly regulated (FDR < 0.05) in either MI or VI comparisons with N showed that changes of these transcripts were highly correlated for MI and VI samples (P < 2.2 × 10–16). The slope of the linear regression fit was less than 1, indicating that the gene expression changes were less pronounced in the VI samples. Overall, these results indicated that across all cell types analyzed, the transcriptome regulation in the VI group was similar but of lower magnitude in the MI group.
Figure 5. Differential gene expression analysis of MI versus N and VI versus N comparisons in CD8+ T cells, NK cells, conventional DCs, and monocytes.
(A–D) Venn diagrams of the union of DEGs of the 2 comparisons in corresponding cell types. The tables below the Venn diagrams indicate the total number of DEGs, number of upregulated DEGs (positive logFC), and number of downregulated DEGs (negative logFC) in each of the 2 comparisons. Scatterplots show the logFC of the union of DEGs of the 2 comparisons in corresponding cell types, i.e., the same group of genes as in the Venn diagram. For each comparison, DEGs were selected with thresholds FDR < 0.05 and absolute logFC > 0.1. Linear regression model was fitted to the scatterplot. Adjusted R2 for each of the cell types is 0.6687, 0.5147, 0.4042, and 0.5779, respectively. N, n = 4; MI, n = 5; VI, n = 4.
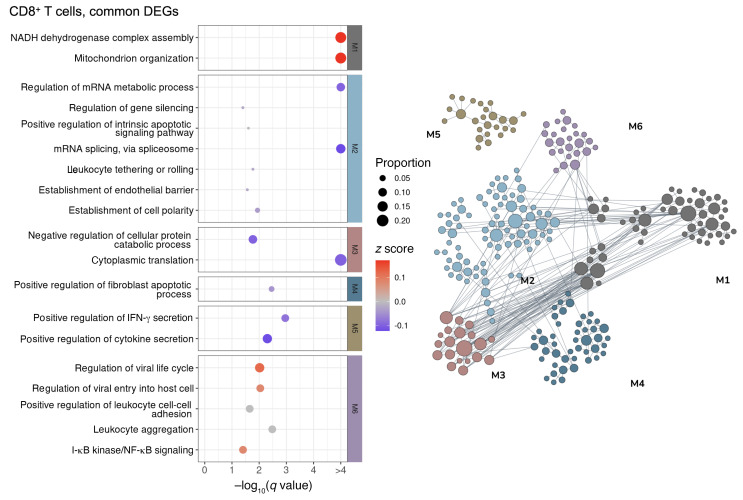
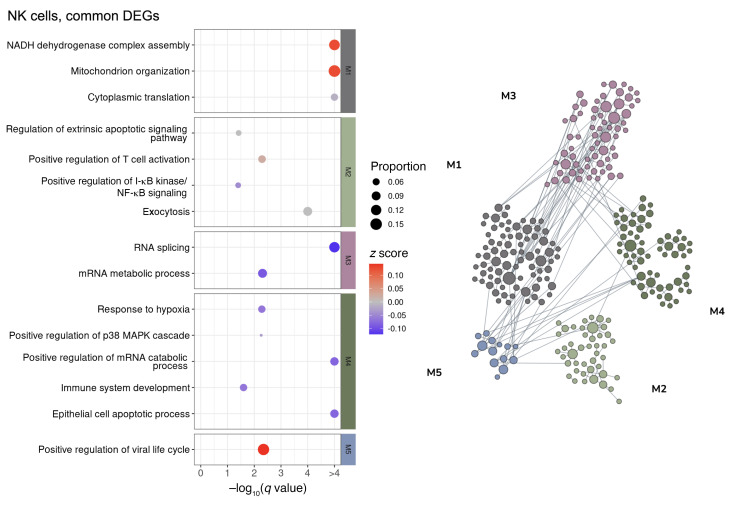
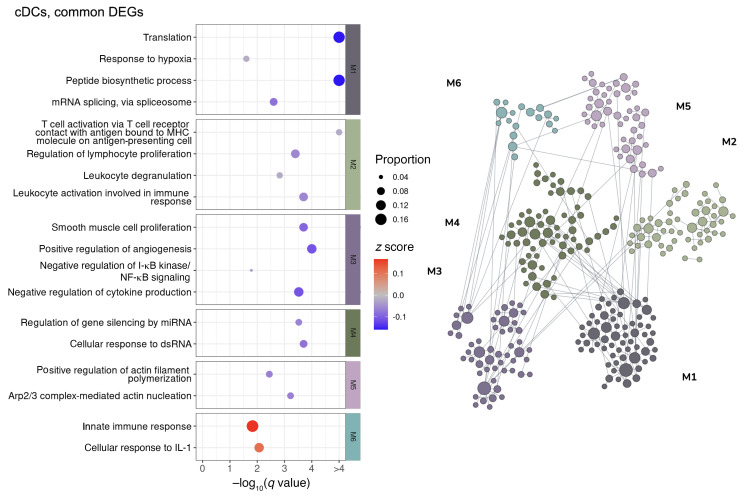
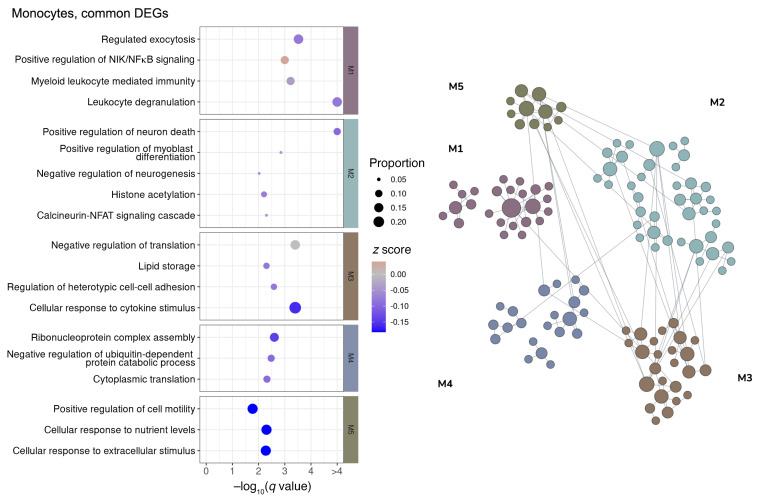
To elucidate the common pathways regulated in the MI and VI groups, we performed a network-based functional enrichment analysis that leverages genetic and epigenetic information from extensive public data sets prebuilt specifically for the lung tissue (44). DEGs were clustered into modules according to their functional relatedness in the lung using a community clustering algorithm to capture higher-order tissue-specific genetic functions. Among the modules that show an upregulated tendency, immune activation and viral response were noted across CD8+ T cells (Figure 6), NK cells (Figure 7), cDCs (Figure 8), and monocytes (Figure 9). The sensitivity for DEGs in pDCs (Supplemental Figure 13A) was limited by the number of cells captured in the N and VI samples (27 pDCs in N samples and 87 pDCs in VI samples). Common pathway enrichment analysis could not be performed for the pDCs because of the small number of DEGs identified in the VI group.
Figure 6. Network-based functional interpretation of common DEGs of MI versus N and VI versus N comparisons in CD8+ T cells.
The bubble plot (left) shows key pathways enriched in each functional module (M) within the network plot for DEGs common to both MI versus N and VI versus N comparisons (right) in CD8+ T cells. Left: The color of each bubble represents the z score for each pathway, which defines the directional tendency of regulation. A positive z score is represented by red for upregulation, and a negative z score is represented by blue for downregulation. Bubble size corresponds to the proportion of DEGs in each pathway relative to the total number of DEGs within M. Right: In the network plots, DEGs are clustered into M/gene communities based on functional relatedness in the lung tissue. Each DEG is represented by a node within M, and each edge represents a functional association between the pair of DEGs. A node with a larger size reflects a higher degree of connectivity in the network. N, n = 4; MI, n = 5; VI, n = 4.
Figure 7. Network-based functional interpretation of common DEGs of MI versus N and VI versus N comparisons in NK cells.
The bubble plot (left) shows key pathways enriched in each functional module (M) within the network plot for DEGs common to both MI versus N and VI versus N comparisons (right) in NK cells. Left: The color of each bubble represents the z score for each pathway, which defines the directional tendency of regulation. A positive z score is represented by red for upregulation, and a negative z score is represented by blue for downregulation. Bubble size corresponds to the proportion of DEGs in each pathway relative to the total number of DEGs within M. Right: In the network plots, DEGs are clustered into M/gene communities based on functional relatedness in the lung tissue. Each DEG is represented by a node within M, and each edge represents a functional association between the pair of DEGs. A node with a larger size reflects a higher degree of connectivity in the network. N, n = 4; MI, n = 5; VI, n = 4.
Figure 8. Network-based functional interpretation of DEGs of MI versus N and VI versus N comparisons in conventional DCs.
The bubble plot (left) shows key pathways enriched in each functional module (M) within the network plot for DEGs common to both MI versus N and VI versus N comparisons (right) in conventional DCs (cDCs). Left: The color of each bubble represents the z score for each pathway, which defines the directional tendency of regulation. A positive z score is represented by red for upregulation, and a negative z score is represented by blue for downregulation. Bubble size corresponds to the proportion of DEGs in each pathway relative to the total number of DEGs within M. Right: In the network plots, DEGs are clustered into M/gene communities based on functional relatedness in the lung tissue. Each DEG is represented by a node within M, and each edge represents a functional association between the pair of DEGs. A node with a larger size reflects a higher degree of connectivity in the network. N, n = 4; MI, n = 5; VI, n = 4.
Figure 9. Network-based functional interpretation of DEGs of MI versus N and VI versus N comparisons in monocytes.
The bubble plot (left) shows key pathways enriched in each functional module (M) within the network plot for DEGs common to both MI versus N and VI versus N comparisons (right) in monocytes. Left: The color of each bubble represents the z score for each pathway, which defines the directional tendency of regulation. A positive z score is represented by red for upregulation, and a negative z score is represented by blue for downregulation. Bubble size corresponds to the proportion of DEGs in each pathway relative to the total number of DEGs within M. Right: In the network plots, DEGs are clustered into M/gene communities based on functional relatedness in the lung tissue. Each DEG is represented by a node within M, and each edge represents a functional association between the pair of DEGs. A node with a larger size reflects a higher degree of connectivity in the network. N, n = 4; MI, n = 5; VI, n = 4.
We also performed the same type of network-based functional analysis on the DEGs that were specific to either the MI or the VI group for each cell type analyzed. Similar pathways were modulated in MI-specific CD8+ T cell DEGs and NK cell DEGs; viral response, migration, regulation of apoptotic signaling, and cellular responses to IFN-γ and oxidative stress were upregulated, while homeostasis and cellular maintenance were downregulated (Supplemental Figure 14, A and B). However, the proliferation process was upregulated in CD8+ T cells and negatively regulated in NK cells. Type I interferon production and viral response pathways were upregulated and cellular maintenance pathways were downregulated in MI-specific cDC DEGs and monocyte DEGs (Supplemental Figure 15, A and B). Adaptive immune response, including T cell activation, was modulated in both MI-specific cDC DEGs and MI-specific pDC DEGs (Supplemental Figure 15A and Supplemental Figure 13B). Finally, additional viral response pathways were upregulated in MI-specific pDC DEGs (Supplemental Figure 13B). Since fewer DEGs were identified as VI-specific, few regulated pathway modules were detected (Supplemental Figure 14, C and D, and Supplemental Figure 15, C and D).
Overall, these data show that SARS-CoV-2 infection induced largely similar transcriptional programs in immune cells of both VI and MI animals, including many viral response and immune activation pathways. In MI-specific transcriptional programs, positive regulation of cytokine signaling stood out as enriched across all cell types studied, while cellular maintenance functions were typically downregulated. On the other hand, in the VI animals, the magnitude of gene expression was lower and fewer differential genes were detected, resulting in few regulated pathway modules.
Discussion
In humans, COVID-19 can progress to severe clinical disease that manifests as pneumonia. mRNA-1273 was previously shown to be efficacious in NHPs (8), in mice (1), and, recently, in a large phase III trial (11). Hamsters consistently exhibit the hallmarks of severe COVID-19 disease and are therefore an important and stringent model for preclinical vaccine efficacy studies. We show that 2 doses of mRNA-1273 reduced viral load in the upper and lower airways of hamsters and protected against weight loss, while a prime-only vaccination provided partial protection. Neutralizing titers were not dependent on dosage in a prime-boost schedule, yet the highest prime-boost dose of 25 μg provided better protection against lung injury and weight loss. Despite a strong inverse correlation between neutralizing antibody titers and virus load in the respiratory tract, the inverse correlation between neutralizing or binding antibody titers and weight loss was only moderate. Therefore, other antibody- or cell-mediated mechanisms may be required for complete protection against SARS-CoV-2 disease in this model. This emphasizes the importance of a qualitative, not just quantitative, immune response, which may be influenced by the vaccine dose and regimen. The efficacy of mRNA-1273 in hamsters is distinguished from efficacy studies with NHP and mouse models, as protection was afforded against severe pathological phenotypes and clinical disease, including high levels of virus replication, weight loss, and extensive pneumonia, and immunity was more permissive to infection.
Gene expression functional pathways in vaccinated hamsters appeared to promote pulmonary homeostasis and a less proliferative and migratory response following infection while supporting virus clearance. While enriched gene expression pathways associated with immune effector functions were shared by both VI and MI hamsters, pathways regulating cell migration were generally unique to MI hamsters. Granulocytes and inflammatory macrophages were abundant in the lungs of MI hamsters, features that are also observed in patients with severe COVID-19 (31, 45). High viral RNA reads within these cells suggest acquisition of infected cells by phagocytosis or a susceptibility to infection. Vaccination averted proinflammatory responses and the influx of these innate immune cells. The dominant CCL8 response across all immune cells in MI hamsters was restricted to macrophages in VI hamsters and may represent a signal for regulated cellular trafficking as opposed to inflammation. Similar levels of monocytes infiltrated the lungs of both MI and VI hamsters. While monocytes from both MI and VI hamsters possessed DEGs compared with N samples that modulated their responses to external stimulus, the monocytes from MI, but not VI, hamsters exhibited upregulated inflammatory characteristics that are observed in severe COVID-19 cases (46). The controlled responses in VI monocytes suggest a supportive role in immune regulation. Upregulated adaptation to oxidative stress in monocytes, NK cells, and CD8+ T cells was unique to MI hamsters, indicating that vaccination controlled exacerbated responses to infection that promote tissue damage and hypoxic respiratory failure in patients with severe disease (47). Certain NK cell phenotypes were associated with COVID-19–related hyperinflammation and similar to the phenotypes of the NK cell populations in the BALF from patients with severe disease (48); the cytotoxic functions of NK cells in MI hamsters were enriched, while their proliferation processes were negatively regulated. NK cells in VI hamsters were functioning in a similar, albeit attenuated, capacity to those in MI hamsters, which suggests a reduced ability to clear virus-infected cells. NK cells maintained immune homeostasis in the lungs upon infection of mice vaccinated against influenza, rather than realizing their killing capacity, to enable virus clearance by the adaptive immune system with minimal pathology (49). We found that the transcriptional program induced by infection in lymphoid and myeloid cells is comparable but of much reduced amplitude in VI compared with MI hamsters, and the VI hamsters also show an attenuated CCL8 induction across immune cells. Overall, we interpret this as indicative of a generally lower state of innate immune cell activation in response to infection in the vaccinated hamsters. mRNA-1273 vaccination may enable a controlled cellular antiviral response conducive to preserving the lung milieu.
Patients with severe COVID-19 display aberrant T cell activation and differentiation and lymphopenia (50, 51) and generally have more proliferative T cells but lower CD8+ T cell proportions with limited clonal diversity in BALF (30). Conversely, non-hospitalized, recovered individuals have virus-specific T cell memory (52, 53). In the lungs of MI hamsters, T cell proportions declined, indicating that extravasation to affected tissues, a suggested cause of lymphopenia in severe patients, was not occurring. Instead, pathways supportive of cell cycling and survival during activation of naive T cells or environmental stresses, including macroautophagy and telomere maintenance, were downregulated in CD8+ T cells from MI hamsters (54, 55), even though their responsiveness to oxidative stress and cytokine stimulus was upregulated. Therefore, CD8+ T cells in MI hamsters may develop an effector phenotype but are replicative senescent, susceptible to depletion, or unable to form functional memory T cells (56, 57). Exhaustion of CD8+ T cells from hyperactivation or augmented expression of proapoptotic molecules has been linked to their depletion in severe COVID-19 cases (58, 59). Vaccination prevented a reduction in T cell frequencies in the lungs following infection. Rather, greater proportions of dividing CD4+ T cells were found in VI hamsters compared with MI hamsters, which appeared to represent a Th1-polarized recall response. While CD8+ T cells in VI hamsters were less enriched for activation and proliferative pathways compared with MI hamsters, they sustained effector function pathways. This suggests that the CD8+ T cell response in VI hamsters was either from memory effectors established after vaccination, to facilitate viral clearance, or from clonally diverse, tissue-resident cells with an effector phenotype after SARS-CoV-2 challenge, as seen in the BALF from moderate but not from severe COVID-19 patients (30).
Aberrant T cell responses or T cell depletion in severe COVID-19 may be indirectly attributed to DCs that are impaired in maturation and T cell activation (60). DC subsets infiltrated the lungs of MI hamsters but remained unaltered in VI hamsters. As in acute-phase coronavirus infections (61), infiltrating pDCs may provide rapid antiviral responses through the upregulation of cytokine production pathways. However, pDCs were not necessary for influenza A virus clearance and were shown to have a deleterious effect on CD8+ T cells during lethal infection of mice (62, 63). In MI hamsters, the lymphoid organ development pathway of pDCs was downregulated, suggesting that their role in T cell differentiation was compromised. The low numbers of pDCs in VI hamsters and their limited DEGs suggest that they were not involved in vaccine-mediated responses to infection. Dysfunctional T cell activation in COVID-19 patients has been associated with the downregulation of MHC and costimulatory molecules on antigen-presenting cells (60, 64, 65). Immune effector pathways and chemotactic cues from IL-1 to migrate to the lung-draining lymph nodes were upregulated in cDCs from both MI and VI hamsters, while DEGs involved in the activation of T cells via MHC molecules were paradoxically downregulated. Despite this remarkable similarity in MI- and VI-induced cDC pathways, disparities in DEG expression magnitude, cDC numbers, and external stimulus from the lung milieu may account for different cDC-mediated outcomes. Diminished MHC-dependent T cell activation by cDCs from MI hamsters likely impaired the adaptive immune response, while in VI hamsters, the controlled cell contact–dependent activation by cDCs may be sufficient for recall responses or for sustaining the effector functions of CD8+ T cells (66). mRNA-1273 promoted a controlled response to SARS-CoV-2 infection that prevented desynchrony between the innate and adaptive immune arms, which can exacerbate inflammation and disease severity.
The incomplete annotation of the hamster genome prevented identification of highly specific subtypes within immune cell populations. In particular, we were unable to identify resident memory cells, which are important for recall responses and durable immunity. Cell type identification partially based on mouse data and the pathway analysis based on human gene ontology terms did permit detailed profiling of cellular responses. While this study examined the response to infection directly in the lungs of VI and MI hamsters rather than BALF, scRNA-Seq analysis was limited to the upper region of the left lung lobe, and responses to an infection may be spatially distinct. As a result, we captured the functional dynamics of neutrophils/heterophils within a potential focal inflammatory lesion, formed after infection of a low-vaccine-dose VI hamster. This finding, along with excessive weight loss exhibited by this outlier hamster, may underscore the importance of dose in achieving complete protection against severe disease.
mRNA-1273 provided hamsters with infection-permissive immunity that, following SARS-CoV-2 infection, reduced virus load and severe disease by promoting humoral responses in the periphery and antiviral cellular responses in the lung. The protection demonstrated by mRNA-1273 is predominantly achieved through the induction of antibodies, and cellular responses may act to support complete clearance of the virus. Importantly, the safety of mRNA-1273 was indicated by the absence of aberrant cellular pathways in VI hamsters after challenge. Transient, low-level virus replication in VI hamsters did trigger the regulation of cellular programs in some immune cells that were strikingly similar to those in MI hamsters. However, the responses were attenuated and lacked the atypical manifestations that contribute to inflammation and lung injury in critical COVID-19 cases. The surprising commonalities in transcriptional pathway regulation between vaccinated and mock-vaccinated recipients during acute infection warrant investigation into how immunity induced by other vaccines is regulated following infection to provide the foundation for successful vaccination.
Methods
Preclinical mRNA-1273 and lipid nanoparticle production process.
Preclinical mRNA-1273 is a purified mRNA transcript encoding the prefusion-stabilized SARS-CoV-2 S-2P protein and encapsulated by a lipid nanoparticle. The process for mRNA synthesis, purification, and encapsulation was described previously (1).
Hamster study.
Three groups of 6- to 7-week-old female golden Syrian hamsters (Envigo) (n = 15 per group) were vaccinated with mRNA-1273 diluted to 25 μg, 5 μg, and 1 μg in PBS for prime-boost vaccine regimens. An additional group (n = 15) was prime-vaccinated only with 25 μg of mRNA-1273. A mock-vaccinated control group (n = 15) received PBS only. Formulations were administered by i.m. injection to each hind leg (50 μL per dose site). At week 3, prime-boost groups received their second vaccine dose. At week 6 (day 42), all animals were infected via the intranasal route with 100 μL of 2019-nCoV/USA-WA01/2020 (GenBank MN985325.1; courtesy of World Reference Center for Emerging Viruses and Arboviruses, University of Texas Medical Branch) at 1 × 105 PFU (approximately 50 μL per nostril). Over the course of the study, hamsters were monitored daily for weight changes. On each serial endpoint day (days 2, 4, and 14 after challenge), lungs, nasal turbinates, and serum were collected from 5 hamsters per group.
Analysis of viral load by qRT-PCR.
Replicating viral RNA was determined in the lungs and nasal turbinates by measurement of subgenomic SARS-CoV-2 E gene RNA by qRT-PCR using previously described primers, probe, and cycle conditions (67). Briefly, RNA was extracted from lung and nasal turbinate homogenates using TRIzol LS (Thermo Fisher Scientific) and Direct-zol RNA Microprep kit (Zymo Research). RNA (500 ng) was reverse-transcribed using SuperScript IV (Thermo Fisher Scientific) according to the manufacturer’s recommendations. Quantitative real-time PCR was performed using TaqMan Fast Advanced Master Mix (Thermo Fisher Scientific), and primers and a FAM-ZEN/Iowa Black FQ labeled probe sequence (Integrated DNA Technologies) on the QuantStudio 6 system (Applied Biosystems). An Ultramer DNA oligonucleotide (Integrated DNA Technologies) spanning the amplicon was used to generate standard curves to calculate the sgRNA copies per gram of tissue.
Single-cell isolation and RNA-Seq.
Left lung sections were taken at the time of necropsy at 4 dpi from the 5 μg prime-boost group (VI, n = 5), the mock-vaccinated group (MI, n = 5), and a naive group of hamsters (N, n = 4) that were mock-infected via the intranasal route with 100 μL of media inoculum. Lung samples were enzymatically digested and homogenized using the Lung Dissociation kit, mouse (Miltenyi Biotec), and GentleMACS Dissociator (Miltenyi Biotec) according to the manufacturer’s protocol. After 30 minutes of digestion, the samples were filtered through a 70 μm filter, and RBC lysis was performed (Thermo Fisher Scientific). After 2 washes in PBS containing 0.05 mM EDTA and 0.04% BSA, the cell pellet was resuspended in 1 mL of buffer and filtered through a 40 μm Flowmi Cell strainer (Bel-Art). The cell viability and concentration were determined on a TC20 cell counter (Bio-Rad). Dead cell removal was performed if the viability was lower than 80% using Dead Cell Removal Kit (Miltenyi Biotec) per the manufacturer’s recommendations.
Seven thousand cells were targeted for generation of barcoded gel bead emulsions using Chromium Single Cell 3′ version 3.0 chemistry (10x Genomics). After reverse transcription, the cDNA was amplified and purified using SPRISelect magnetic beads (Beckman Coulter). The purified cDNA was precipitated in 80% ethanol, removed from biosafety level 4 containment, and tested for quality on a Bioanalyzer (Agilent), and 3′ gene expression libraries were prepared per the manufacturer’s instructions. Libraries were quantified, and pooled libraries were submitted for sequencing (~140,000 reads per sample) on a NovaSeq S1 flow cell (New York Genome Center).
Processing of scRNA-Seq data.
scRNA-Seq data were processed using the Cell Ranger pipeline v4.0.0 (10× Genomics). Alignment was done against the Mesocricetus auratus Ensembl genome (MesAur1.0). STARsolo was used to quantify reads associated with the viral genome by aligning against the SARS-CoV-2 genome (ASM985889v3). The 5′ leader sequence and 3′-UTR of the SARS-CoV-2 genome were included as reference GTF entries. Resulting count tables were then analyzed using Seurat (v3.1.5; ref. 68). Cells with abnormally high mitochondrial gene content (greater than 20% of unique molecular identifiers per cell) and/or cells containing fewer than 300 unique genes were removed. A single sample in the VI group was a clear outlier (Supplemental Figure 8) and was removed from all further analysis. For each group (MI, n = 5; VI, n = 4; and N, n = 4), all samples were merged in an unbiased manner. The 3 resulting data sets (1 per group) were then integrated using the IntegrateData() function of Seurat, to account for potential batch effects. Finally, the principal component analysis (PCA) and UMAP (uniform manifold approximation and projection) (69) projections were calculated for the integrated sample set using the top 2000 highly variable genes, as determined by Seurat, and significant cell clusters were identified.
Cell type identification for scRNA-Seq data.
Integration of the data from 13 samples comprising 50,694 cells identified 17 cell types. Single-cell Mouse Cell Atlas (scMCA; ref. 70) was used to guide cell type identification. Reference gene markers were initially determined from the MCA adult lung data set. For each cell type, marker genes were defined as genes satisfying 2 conditions. Then, the average log fold change (logFC) of each marker gene was greater than 1.0 in comparison with the other cell types. Second, the fraction of cells expressing the marker gene in the cell type was higher than the fraction of cells expressing the marker gene in all other cell types by at least 0.4. Using the defined marker gene lists, the correlation of the average gene expression in each cell type in the MCA adult lung was computed with each cell in our data set, annotating each cell with the most highly correlated cell type. Additional marker sets and prior knowledge (71) were used for some cell types that were ambiguous or missing from the reference data (granulocytes, activated CD8+ T cells, DC subtypes). Cell subtypes in some instances were aggregated (pneumocytes, bronchiolar epithelial cells, DC subtypes) . Activated CD8+ T cells were identified by the coexpression of CD8+ T cell markers and NK cell markers, along with too low read count to be considered doublets. To identify clusters consisting of cells undergoing rapid cell division, scMCA’s labeling of dividing cells was used. These clusters were combined into a single population named “Dividing Immune Cells,” as cells within this population expressed markers for both lymphocytes and myeloid cells. This cluster annotation was also corroborated with the CellCycleScoring() function in Seurat, where 97.7% of the cells in the dividing immune cell cluster were predicted to be in G2/M (59.4%) or S (38.3%) phase. To account for invading cell types that were only present in MI samples, the interstitial macrophages and alveolar macrophages were removed from the cell-type proportion calculations of the remaining cell types.
Cell type–specific comparisons of MI with N and VI with N.
DEGs were identified in scRNA-Seq data for MI and VI comparisons with N in CD8+ T cells, NK cells, monocytes, and DC subtypes (pDCs and cDCs) using the nonparametric Wilcoxon’s rank sum test. The groups of activated DCs were incorporated into cDCs during DEG identification. Differential expression P values were adjusted for multiple hypothesis testing using the Benjamini-Hochberg procedure (44). DEGs with FDR less than 0.05 and absolute logFC greater than 0.1 for each comparison were selected as significant. We note that the threshold used for single-cell differential expression analysis within the same cell type accounted for fold-change distribution of DEGs and maintained recovery of biological signal (72–78).
To study gene function and pathway enrichment among DEGs, golden hamster genes were mapped to homolog genes in the Homo sapiens species using BioMart (https://www.ensembl.org/biomart/martview/). Selected genes were used to construct a functional network built by HumanBase (https://hb.flatironinstitute.org) specifically of the lung tissue (44). Neighboring genes that form a community are clustered into functional modules using the graph-based Louvain clustering algorithm based on the network-predicted functional similarity between genes in the lung tissue. Gene Ontology term enrichment was performed for genes in each module, while the direction of regulation for enriched pathways was elicited using a z score, defined as z = (Nup – Ndown)/(Nup + Ndown), where Nup represents the number of upregulated DEGs (positive logFC) in that module and Ndown represents the number of downregulated DEGs (negative logFC). Significantly enriched terms with adjusted P value (q value) less than 0.05 were selected. Each module was annotated with distinct and representative gene ontology terms. For CD8+ T cells (Figure 6), NK cells (Figure 7), cDCs (Figure 8), and monocytes (Figure 9), gene functional clustering and enrichment analysis was performed for DEGs that were common to the MI and VI comparisons with N and DEGs that were specific to either the MI or the VI comparison with N (Supplemental Figures 14 and 15). For pDCs, however, functional clustering and enrichment analysis was only performed on MI versus N given the insufficient number of DEGs in VI versus N (Supplemental Figure 13B).
Data availability.
RNA-Seq data were deposited in the NCBI’s Gene Expression Omnibus database and are accessible through GEO Series accession number GSE163838.
Statistics.
Statistical analysis was performed using GraphPad Prism software version 6. Two-way ANOVA with Tukey’s or Šidák’s correction, respectively, was performed for multiple comparisons between vaccine groups or between time points. Significance of difference between pre- and postinfection antibody titers was measured by multiple t tests with Holm-Šidák correction for multiple comparisons (Supplemental Figure 4). ANOVA and pairwise comparisons by post hoc Tukey’s test were used to determine the significance of scRNA-Seq–based cell-type proportion changes (Figure 3 and Supplemental Figure 7B). Correlations were determined by 2-sided Spearman’s rank tests. For lymphocyte marker comparisons (Supplemental Figure 11 and Supplemental Figure 12D), markers were selected for known functional activity, and for exhibiting both a statistically significant change (P < 0.05) and expression in at least 10% of the cells in a cell type.
Study approval.
All animal protocols were approved by the Institutional Animal Care and Use Committee at the University of Texas Medical Branch (UTMB).
Author contributions
M Meyer, DE, AC, and AB conceived and designed the study. GBESJ co-designed the vaccine construct. CEM performed the SARS-CoV-2 challenge experiment and animal procedures. M Meyer and CP processed hamster samples. CH, AW, and LM performed ELISAs. M Meyer performed and analyzed SARS-CoV-2–based virology and serology assays. PR performed single-cell RNA-Seq. YW, GRS, ABR, XC, YG, and WSC analyzed single-cell RNA-Seq data. KWB, M Minai, and BMN performed histopathology and immunohistochemistry. INM and SP analyzed the histopathology data. INM analyzed the immunohistochemistry data. BSG contributed to data interpretation. PYS contributed a new reagent. M Meyer, YW, GRS, ABR, XC, and INM prepared manuscript figures and tables. M Meyer, YW, GRS, ABR, XC, WSC, INM, IR, OGT, EZ, SCS, and AB wrote the manuscript. DE, EZ, OGT, AC, SCS, and AB supervised the research. All authors reviewed, edited, and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank members of the UTMB Animal Resource Center for technical assistance with the hamster experiment in ABSL-2 and ABSL-4 and husbandry support. We thank Steve Widen and the UTMB Next Generation Sequencing Core for quantification and pooling of scRNA-Seq libraries for submission to the New York Genome Center. We thank the Anatomic Pathology Core facility at UTMB for embedding, sectioning, and staining lung tissues for histopathology. The challenge virus used in this publication was generously provided by World Reference Center for Emerging Viruses and Arboviruses at UTMB. This work was supported by Moderna Inc.
Version 1. 08/27/2021
In-Press Preview
Version 2. 10/15/2021
Electronic publication
Funding Statement
This study was funded by Moderna, Inc.
Footnotes
Conflict of interest: DE, CH, AW, LM, GBESJ, and AC are employees of Moderna Inc.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(20):e148036.https://doi.org/10.1172/JCI148036.
Contributor Information
Michelle Meyer, Email: mnmeyer@utmb.edu.
Yuan Wang, Email: yuanwang@cs.princeton.edu.
Darin Edwards, Email: darin.edwards@modernatx.com.
Gregory R. Smith, Email: gregory.smith@mssm.edu.
Aliza B. Rubenstein, Email: aliza.rubenstein@mssm.edu.
Palaniappan Ramanathan, Email: paramana@UTMB.EDU.
Chad E. Mire, Email: chmire@UTMB.EDU.
Colette Pietzsch, Email: capietzs@utmb.edu.
Xi Chen, Email: xchen@flatironinstitute.org.
Yongchao Ge, Email: yongchao.ge@mssm.edu.
Wan Sze Cheng, Email: wansze.cheng@mssm.edu.
Carole Henry, Email: carole.henry@modernatx.com.
Angela Woods, Email: angela.woods@modernatx.com.
LingZhi Ma, Email: lingzhi.ma@modernatx.com.
Guillaume B.E. Stewart-Jones, Email: Guillaume.stewart-jones@modernatx.com.
Kevin W. Bock, Email: kevin.bock@nih.gov.
Mahnaz Minai, Email: minaim@niaid.nih.gov.
Bianca M. Nagata, Email: Bianca.nagata@nih.gov.
Sivakumar Periasamy, Email: siperias@utmb.edu.
Pei-Yong Shi, Email: peshi@UTMB.EDU.
Barney S. Graham, Email: bgraham@mail.nih.gov.
Ian N. Moore, Email: ian.moore@nih.gov.
Irene Ramos, Email: irene.ramos-lopez@mssm.edu.
Olga G. Troyanskaya, Email: ogt@cs.princeton.edu.
Elena Zaslavsky, Email: elena.zaslavsky@mssm.edu.
Andrea Carfi, Email: andrea.carfi@modernatx.com.
Stuart C. Sealfon, Email: stuart.sealfon@mssm.edu.
Alexander Bukreyev, Email: abukreye@utmb.edu.
References
- 1.Corbett KS, et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586(7830):567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyer M, et al. Modified mRNA-based vaccines elicit robust immune responses and protect guinea pigs from Ebola Virus Disease. J Infect Dis. 2018;217(3):451–455. doi: 10.1093/infdis/jix592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feldman RA, et al. mRNA vaccines against H10N8 and H7N9 influenza viruses of pandemic potential are immunogenic and well tolerated in healthy adults in phase 1 randomized clinical trials. Vaccine. 2019;37(25):3326–3334. doi: 10.1016/j.vaccine.2019.04.074. [DOI] [PubMed] [Google Scholar]
- 4.Aliprantis AO, et al. A phase 1, randomized, placebo-controlled study to evaluate the safety and immunogenicity of an mRNA-based RSV prefusion F protein vaccine in healthy younger and older adults. Hum Vaccin Immunother. 2021;17(5):1248–1261. doi: 10.1080/21645515.2020.1829899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. ClinicalTrials.gov. A Phase 1, Randomized, Observer-Blind, Placebo-Controlled, Dose-Ranging Study to Evaluate the Safety, Reactogenicity, and Immunogenicity of Cytomegalovirus Vaccines mRNA-1647 and mRNA-1443 When Administered to Healthy Adults. https://clinicaltrials.gov/ct2/show/NCT03382405 Updated January 15, 2021. Accessed August 26, 2021.
- 6. ClinicalTrials.gov. A Phase 1, Randomized, Observer-Blind, Placebo-Controlled, Dose-Ranging Study to Evaluate the Safety, Reactogenicity, and Immunogenicity of mRNA-1653, a Combined Human Metapneumovirus and Human Parainfluenza Virus Type 3 Vaccine, When Administered to Healthy Adults. https://clinicaltrials.gov/ct2/show/NCT03392389 Updated February 6, 2020. Accessed August 26, 2021.
- 7. ClinicalTrials.gov. A Phase 1, Randomized, Observer-Blind, Placebo-Controlled, Dose-Ranging Study to Evaluate the Safety, Tolerability, and Immunogenicity of Zika Vaccine mRNA-1893 in Healthy Flavivirus Seropositive and Seronegative Adults. https://clinicaltrials.gov/ct2/show/NCT04064905 Updated April 22, 2021. Accessed August 26, 2021.
- 8.Corbett KS, et al. Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. N Engl J Med. 2020;383(16):1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jackson LA, et al. An mRNA vaccine against SARS-CoV-2 — preliminary report. N Engl J Med. 2020;383(20):1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson EJ, et al. Safety and immunogenicity of SARS-CoV-2 mRNA-1273 vaccine in older adults. N Engl J Med. 2020;383(25):2427–2438. doi: 10.1056/NEJMoa2028436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baden LR, et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med. 2021;384(5):403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Munster VJ, et al. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature. 2020;585(7824):268–272. doi: 10.1038/s41586-020-2324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rockx B, et al. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science. 2020;368(6494):1012–1015. doi: 10.1126/science.abb7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woolsey C, et al. Establishment of an African green monkey model for COVID-19 and protection against re-infection. Nat Immunol. 2021;22(1):86–98. doi: 10.1038/s41590-020-00835-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi J, et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020;368(6494):1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim YI, et al. Infection and rapid transmission of SARS-CoV-2 in ferrets. Cell Host Microbe. 2020;27(5):704–709. doi: 10.1016/j.chom.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blanco-Melo D, et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181(5):1036–1045. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dinnon KH, 3rd, et al. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature. 2020;586(7830):560–566. doi: 10.1038/s41586-020-2708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imai M, et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc Natl Acad Sci U S A. 2020;117(28):16587–16595. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan JF, et al. Simulation of the clinical and pathological manifestations of Coronavirus Disease 2019 (COVID-19) in golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin Infect Dis. 2020;71(9):2428–2446. doi: 10.1093/cid/ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tostanoski LH, et al. Ad26 vaccine protects against SARS-CoV-2 severe clinical disease in hamsters. Nat Med. 2020;26(11):1694–1700. doi: 10.1038/s41591-020-1070-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sia SF, et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583(7818):834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Winkler ES, et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation impaired function. Nat Immunol. 2020;21(11):1327–1335. doi: 10.1038/s41590-020-0778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wood DE, et al. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20(1):257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Butler A, et al. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu T, et al. Differential expression of viral transcripts from single-cell RNA sequencing of moderate and severe COVID-19 patients and its implications for case severity. Front Microbiol. 2020;11:603509. doi: 10.3389/fmicb.2020.603509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. doi: 10.1101/2020.07.28.225912. Pontelli MC, et al. Infection of human lymphomononuclear cells by SARS-CoV-2 [preprint]. Posted on bioRxiv August 7, 2020. [DOI]
- 28.Ramos da Silva S, et al. Broad SARS-CoV-2 cell tropism and immunopathology in lung tissues from fatal COVID-19. J Infect Dis. 2021;223(11):1842–1854. doi: 10.1093/infdis/jiab195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Speranza E, et al. Single-cell RNA sequencing reveals SARS-CoV-2 infection dynamics in lungs of African green monkeys. Sci Transl Med. 2021;13(578):eabe8146. doi: 10.1126/scitranslmed.abe8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liao M, et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med. 2020;26(6):842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 31.Cano-Gamez E, et al. Single-cell transcriptomics identifies an effectorness gradient shaping the response of CD4+ T cells to cytokines. Nat Commun. 2020;11(1):1801. doi: 10.1038/s41467-020-15543-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Szabo PA, et al. Single-cell transcriptomics of human T cells reveals tissue and activation signatures in health and disease. Nat Commun. 2019;10(1):4706. doi: 10.1038/s41467-019-12464-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tibbitt CA, et al. Single-cell RNA sequencing of the T helper cell response to house dust mites defines a distinct gene expression signature in airway Th2 cells. Immunity. 2019;51(1):169–184. doi: 10.1016/j.immuni.2019.05.014. [DOI] [PubMed] [Google Scholar]
- 34.Koroleva EP, et al. Lymphotoxin in physiology of lymphoid tissues – implication for antiviral defense. Cytokine. 2018;101:39–47. doi: 10.1016/j.cyto.2016.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou Z, et al. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe. 2020;27(6):883–890. doi: 10.1016/j.chom.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oja AE, et al. Trigger-happy resident memory CD4+ T cells inhabit the human lungs. Mucosal Immunol. 2018;11(3):654–667. doi: 10.1038/mi.2017.94. [DOI] [PubMed] [Google Scholar]
- 37.Schoettler N, et al. Transcriptional programming and T cell receptor repertoires distinguish human lung and lymph node memory T cells. Commun Biol. 2019;2:411. doi: 10.1038/s42003-019-0657-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiong Y, et al. CD4 T cell sphingosine 1-phosphate receptor (S1PR)1 and S1PR4 and endothelial S1PR2 regulate afferent lymphatic migration. Sci Immunol. 2019;4(33):eaav1263. doi: 10.1126/sciimmunol.aav1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanhere A, et al. T-bet and GATA3 orchestrate Th1 and Th2 differentiation through lineage-specific targeting of distal regulatory elements. Nat Commun. 2012;3:1268. doi: 10.1038/ncomms2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoon HS, et al. ZBTB32 is an early repressor of the CIITA and MHC class II gene expression during B cell differentiation to plasma cells. J Immunol. 2012;189(5):2393–2403. doi: 10.4049/jimmunol.1103371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buchan S, et al. OX40- and CD27-mediated costimulation synergizes with anti-PD-L1 blockade by forcing exhausted CD8+ T cells to exit quiescence. J Immunol. 2015;194(1):125–133. doi: 10.4049/jimmunol.1401644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams JB, et al. The EGR2 targets LAG-3 and 4-1BB describe and regulate dysfunctional antigen-specific CD8+ T cells in the tumor microenvironment. J Exp Med. 2017;214(2):381–400. doi: 10.1084/jem.20160485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang C, et al. Heterogeneity of human bone marrow and blood natural killer cells defined by single-cell transcriptome. Nat Commun. 2019;10(1):3931. doi: 10.1038/s41467-019-11947-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Greene CS, et al. Understanding multicellular function and disease with human tissue-specific networks. Nat Genet. 2015;47(6):569–576. doi: 10.1038/ng.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chua RL, et al. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat Biotechnol. 2020;38(8):970–979. doi: 10.1038/s41587-020-0602-4. [DOI] [PubMed] [Google Scholar]
- 46.Zhang JY, et al. Single-cell landscape of immunological responses in patients with COVID-19. Nat Immunol. 2020;21(9):1107–1118. doi: 10.1038/s41590-020-0762-x. [DOI] [PubMed] [Google Scholar]
- 47.Berlin DA, et al. Severe Covid-19. N Engl J Med. 2020;383(25):2451–2460. doi: 10.1056/NEJMcp2009575. [DOI] [PubMed] [Google Scholar]
- 48.Maucourant C, et al. Natural killer cell immunotypes related to COVID-19 disease severity. Sci Immunol. 2020;5(50):eabd6832. doi: 10.1126/sciimmunol.abd6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mooney JP, et al. Natural killer cells dampen the pathogenic features of recall responses to influenza infection. Front Immunol. 2020;11:135. doi: 10.3389/fimmu.2020.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen G, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest. 2020;130(5):2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tan L, et al. Lymphopenia predicts disease severity of COVID-19: a descriptive and predictive study. Signal Transduct Target Ther. 2020;5(1):33. doi: 10.1038/s41392-020-0148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grifoni A, et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181(7):1489–1501. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Braun J, et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587(7833):270–274. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- 54.Lum JJ, et al. Autophagy in metazoans: cell survival in the land of plenty. Nat Rev Mol Cell Biol. 2005;6(6):439–448. doi: 10.1038/nrm1660. [DOI] [PubMed] [Google Scholar]
- 55.Pua HH, et al. Autophagy is essential for mitochondrial clearance in mature T lymphocytes. J Immunol. 2009;182(7):4046–4055. doi: 10.4049/jimmunol.0801143. [DOI] [PubMed] [Google Scholar]
- 56.Xu X, et al. Autophagy is essential for effector CD8(+) T cell survival and memory formation. Nat Immunol. 2014;15(12):1152–1161. doi: 10.1038/ni.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Akbar AN, Vukmanovic-Stejic M. Telomerase in T lymphocytes: use it and lose it? J Immunol. 2007;178(11):6689–6694. doi: 10.4049/jimmunol.178.11.6689. [DOI] [PubMed] [Google Scholar]
- 58.Mathew D, et al. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369(6508):eabc8511. doi: 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Laing AG, et al. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat Med. 2020;26(10):1623–1635. doi: 10.1038/s41591-020-1038-6. [DOI] [PubMed] [Google Scholar]
- 60.Zhou R, et al. Acute SARS-CoV-2 infection impairs dendritic cell and T cell responses. Immunity. 2020;53(4):864–877. doi: 10.1016/j.immuni.2020.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cervantes-Barragan L, et al. Control of coronavirus infection through plasmacytoid dendritic-cell-derived type I interferon. Blood. 2007;109(3):1131–1137. doi: 10.1182/blood-2006-05-023770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Langlois RA, Legge KL. Plasmacytoid dendritic cells enhance mortality during lethal influenza infections by eliminating virus-specific CD8 T cells. J Immunol. 2010;184(8):4440–4446. doi: 10.4049/jimmunol.0902984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wolf AI, et al. Plasmacytoid dendritic cells are dispensable during primary influenza virus infection. J Immunol. 2009;182(2):871–879. doi: 10.4049/jimmunol.182.2.871. [DOI] [PubMed] [Google Scholar]
- 64. doi: 10.1101/2020.07.16.20153437. Unterman A, et al. Single-cell omics reveals dyssynchrony of the innate and adaptive immune system in progressive COVID-19 [preprint]. Posted on medRxiv July 17, 2020. [DOI] [PMC free article] [PubMed]
- 65.Wilk AJ, et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat Med. 2020;26(7):1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McGill J, et al. IL-15 trans-presentation by pulmonary dendritic cells promotes effector CD8 T cell survival during influenza virus infection. J Exp Med. 2010;207(3):521–534. doi: 10.1084/jem.20091711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wolfel R, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 68.Stuart T, et al. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888–1902. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Becht E, et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol. 2019;37:38–44. doi: 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- 70.Sun H, et al. scMCA: a tool to define mouse cell types based on single-cell digital expression. Methods Mol Biol. 2019;1935:91–96. doi: 10.1007/978-1-4939-9057-3_6. [DOI] [PubMed] [Google Scholar]
- 71.Zilionis R, et al. Single-cell transcriptomics of human and mouse lung cancers reveals conserved myeloid populations across individuals and species. Immunity. 2019;50(5):1317–1334. doi: 10.1016/j.immuni.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sun S, et al. Hair cell mechanotransduction regulates spontaneous activity and spiral ganglion subtype specification in the auditory system. Cell. 2018;174(5):1247–1263. doi: 10.1016/j.cell.2018.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Popescu DM, et al. Decoding human fetal liver haematopoiesis. Nature. 2019;574(7778):365–371. doi: 10.1038/s41586-019-1652-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jiang Y, et al. Single cell RNA sequencing identifies an early monocyte gene signature in acute respiratory distress syndrome. JCI Insight. 2020;5(13):e135678. doi: 10.1172/jci.insight.135678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Soneson C, Robinson MD. Bias, robustness and scalability in single-cell differential expression analysis. Nat Methods. 2018;15(4):255–261. doi: 10.1038/nmeth.4612. [DOI] [PubMed] [Google Scholar]
- 76.Mou T, et al. Reproducibility of methods to detect differentially expressed genes from single-cell RNA sequencing. Front Genet. 2019;10:1331. doi: 10.3389/fgene.2019.01331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Luecken MD, Theis FJ. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol Syst Biol. 2019;15(6):e8746. doi: 10.15252/msb.20188746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yao C, et al. Cell-type-specific immune dysregulation in severely ill COVID-19 patients. Cell Rep. 2021;34(13):108943. doi: 10.1016/j.celrep.2021.108943. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-Seq data were deposited in the NCBI’s Gene Expression Omnibus database and are accessible through GEO Series accession number GSE163838.