Abstract
Multisystem inflammatory syndrome in children (MIS-C) manifests as a severe and uncontrolled inflammatory response with multiorgan involvement, occurring weeks after SARS-CoV-2 infection. Here, we utilized proteomics, RNA sequencing, autoantibody arrays, and B cell receptor (BCR) repertoire analysis to characterize MIS-C immunopathogenesis and identify factors contributing to severe manifestations and intensive care unit admission. Inflammation markers, humoral immune responses, neutrophil activation, and complement and coagulation pathways were highly enriched in MIS-C patient serum, with a more hyperinflammatory profile in severe than in mild MIS-C cases. We identified a strong autoimmune signature in MIS-C, with autoantibodies targeted to both ubiquitously expressed and tissue-specific antigens, suggesting autoantigen release and excessive antigenic drive may result from systemic tissue damage. We further identified a cluster of patients with enhanced neutrophil responses as well as high anti-Spike IgG and autoantibody titers. BCR sequencing of these patients identified a strong imprint of antigenic drive with substantial BCR sequence connectivity and usage of autoimmunity-associated immunoglobulin heavy chain variable region (IGHV) genes. This cluster was linked to a TRBV11-2 expanded T cell receptor (TCR) repertoire, consistent with previous studies indicating a superantigen-driven pathogenic process. Overall, we identify a combination of pathogenic pathways that culminate in MIS-C and may inform treatment.
Keywords: COVID-19, Inflammation
Keywords: Autoimmune diseases, Cellular immune response, Cytokines
Introduction
While cases of severe coronavirus disease 2019 (COVID-19) occur relatively less frequently in children than adults (1, 2), a small proportion of SARS-CoV-2–infected children develop a novel pediatric febrile entity called multisystem inflammatory syndrome in children (MIS-C) within 2 to 5 weeks of initial SARS-CoV-2 exposure (3–9). MIS-C disproportionately affects male children and children of Hispanic or Black descent. It manifests as a severe and uncontrolled inflammatory response with multiorgan involvement (3–10). A hyperinflammatory state is evidenced by clinical makers that include high serum concentrations of C-reactive protein (CRP), ferritin, and D-dimers, and increased levels of proinflammatory cytokines (3, 6, 9, 11, 12). Children often present with persistent fever, severe gastrointestinal (GI) symptoms, cardiovascular manifestations, and respiratory and neurological symptoms (3, 4, 6–10). Cardiovascular manifestations include hypotension, shock, cardiac dysfunction, myocarditis, and pericardial effusion (5). In the United States, admission to the intensive care unit (ICU) occurs in approximately 58% of cases (5).
The immunological features of MIS-C involve perturbations in both innate and adaptive immune responses. MIS-C patients have a highly activated myeloid cell compartment (13–15) and elevated levels of cytokines and chemokines that mediate neutrophil and monocyte chemotaxis, differentiation, and activation (14). MIS-C is associated with severe T cell lymphopenia, high levels of T cell activation, and expansion of vascular patrolling CX3CR1+CD8+ T cells (13–16). We recently identified HLA class I alleles associated with expansion of TRBV11-2 T cells in severe MIS-C (17). Interestingly, in these cases TRBV11-2 expansion did not follow the typical CDR3-dependent clonal selection observed with conventional antigens (17). Instead, CDR3 regions of expanded TRBV11-2 T cells were diverse, similar to observations following superantigen-driven T cell expansion (17). This expansion correlated with disease severity and cytokine storm, suggesting TRBV11-2 T cell expansion may be an important mechanism driving severe MIS-C (17). B cell responses may also contribute to MIS-C pathogenesis, as plasmablast frequencies have been reported to be elevated during MIS-C (13, 18). In addition, most children with MIS-C have IgG and IgA anti–SARS-CoV-2 antibodies (14, 15, 19), including highly inflammatory anti–SARS-CoV-2 IgG antibodies involved in monocyte activation and Fc binding (19). A number of autoantibodies against immunomodulatory mediators and antigens from endothelial, cardiac, and GI tissues have been identified and may play a role in driving MIS-C pathogenesis (14, 16, 18).
Here, we sought to further our understanding of MIS-C immunopathogenesis through proteomics, RNA sequencing (RNA-seq), B cell receptor sequencing (BCR-seq), and autoantibody analysis of a unique cohort of MIS-C patients with severe and mild clinical courses. We identify a highly inflammatory MIS-C subset of patients that show both enhanced neutrophil activation and imprints of harmful adaptive immune responses. In addition to superantigenic T cell interactions, these patients show strong selection of autoreactive B cells that may be the cellular counterpart of the high autoantibody levels found in these patients.
Results
Severe MIS-C patients exhibit hyperinflammation and cytokine storm.
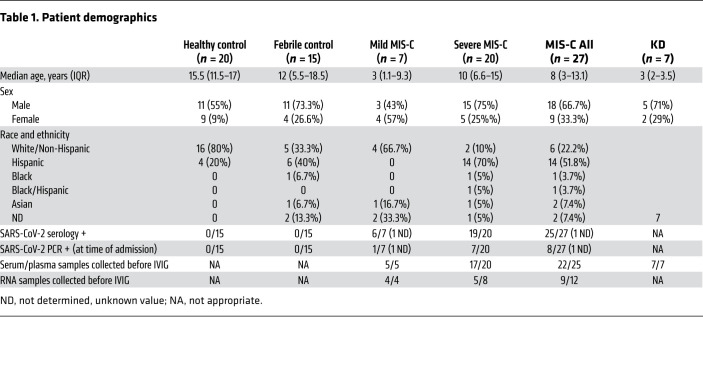
Our MIS-C cohort was composed of 7 patients with mild MIS-C and 20 patients with severe MIS-C (Table 1). MIS-C diagnosis was performed according to CDC guidelines, and patients who required treatment in the pediatric ICU were defined as severe MIS-C patients. We first compared circulating biomarkers of inflammation and heart failure, and cytokine profiles of severe and mild MIS-C patients. Both mild and severe MIS-C patients showed elevated levels of C-reactive protein (CRP), ferritin, fibrinogen, pro–B-type natriuretic peptide (proBNP), aspartate transaminase, alanine transaminase (ALT), D-dimers, and creatine compared with normal reference ranges (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI151520DS1). Severe MIS-C cases showed significantly higher levels of ferritin, proBNP, D-dimers, and creatine, and a trend toward increased levels of CRP, ALT, and fibrinogen compared with mild MIS-C patients (Supplemental Figure 1A).
Table 1. Patient demographics.
We next assessed and compared the circulating cytokine profiles of healthy controls, mild MIS-C, and severe MIS-C patients. MIS-C patients showed increased levels of IFN-γ, TNF-α, IL-6, IL-8, IL-10, and IL-1β, with severely ill patients showing stronger dysregulation than those with milder courses (Supplemental Figure 1B). Since some of the clinical symptoms of MIS-C, such as erythematous rashes, conjunctivitis, and inflammatory changes in the oral mucosa are suggestive of Kawasaki disease (KD), we also characterized the cytokine profiles of 7 KD patients who were recruited before the COVID-19 pandemic (Supplemental Figure 1B). KD diagnosis was performed according to the established American Heart Association guidelines (20). Except for IL-8, which appeared less dysregulated in KD, the overall cytokine pattern was similar in MIS-C and KD (Supplemental Figure 1B).
Proteomic analysis identifies a highly inflammatory proteomics profile in MIS-C.
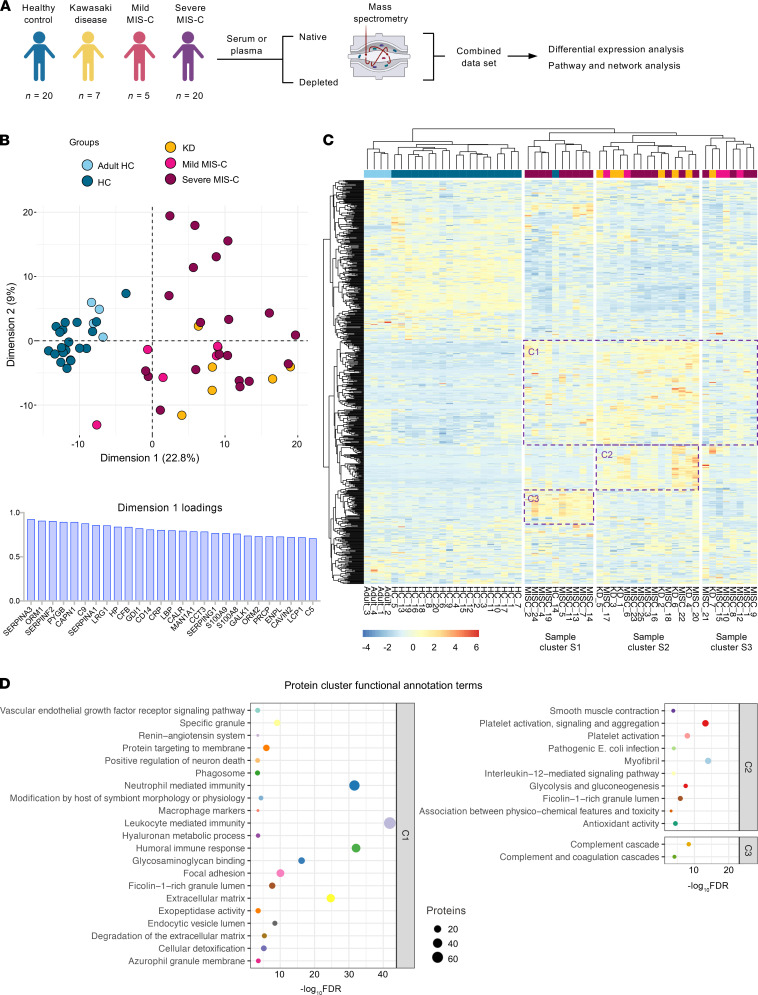
To assist in the elucidation of the pathogenesis of MIS-C and to identify proteins associated with the severe form of the disease, we performed proteomics analysis of serum or plasma samples from our study cohort (Figure 1A). We collected serum from healthy children (n = 20), mild MIS-C patients (non-ICU, n = 5), and severe MIS-C patients who required ICU treatment (n = 20). We also included analysis of plasma samples from KD patients that were collected prior to the pandemic (n = 7; Table 1). Healthy adult serum (n = 4) was used for reference range quality control. To obtain a high resolution of protein expression we performed discovery proteomics analysis on native and depleted (top 14 most abundant proteins) serum or plasma samples (ref. 21, Figure 1B, and Supplemental Figure 2A). Both data sets were integrated at the transition, peptide, and protein level. As plasma was used for KD samples, clotting factors (Supplemental Figure 2B) were removed from the data set for any downstream analysis involving the KD group. Principal component analysis (PCA) (Figure 1B) and hierarchical clustering (Figure 1C) showed that the MIS-C and KD proteomes clustered separately from healthy controls. Similar to our cytokine analysis, MIS-C and KD showed similar protein profiles, indicating that shared pathological pathways likely exist between the diseases (Figure 1, B and C). The major proteins contributing to dimension 1 of the PCA plot, which separates disease samples (MIS-C and KD) from the healthy controls, included inflammatory markers and alarmins such as SERPINA3, CRP, haptoglobin (HP)/zonulin, LPS-binding protein (LBP), CD14, S100A8 and S100A9 (Figure 1B and Supplemental 2C).
Figure 1. Proteomic profiling of MIS-C cases.
(A) Experimental design of native and depleted serum proteomics profiling of healthy controls (n = 20), mild MIS-C (n = 5), severe MIS-C (n = 20), and KD (n = 7) patients. (B) PCA of proteomics data and top proteins contributing to dimension 1 of the PCA plot. (C) Heatmap and hierarchical clustering of proteomics expression data revealed 3 protein sets (C1, C2, and C3) driving separation between 3 clades of MIS-C and KD patients (sample clusters S1, S2, and S3). (D) ClueGO ontology analysis via PINE (Protein Interaction Network Extractor) for visualization of pathways and functional categories significantly enriched within each of the 3 protein sets (C1, C2, and C3) revealed by hierarchical clustering analysis in panel C. The x axis denotes the negative decimal logarithm of the FDR of enrichment (term P value corrected with Bonferroni’s test). Size of the node denotes number of proteins within each term.
Protein Interaction Network Extractor (PINE; ref. 22) analysis of differentially expressed proteins between the groups revealed an enrichment of protein networks involved in multiple inflammatory processes and pathways, including neutrophil degranulation, platelet activation, complement and coagulation cascades, phagocytosis, angiogenesis, acute-phase responses, oxidative stress, metabolism, and cell migration and adhesion (Supplemental Figure 2D).
Hierarchical clustering analysis led to the identification of 3 main clusters of disease samples, sample clusters S1, S2, and S3 (Figure 1C). A large set of proteins (protein cluster 1, C1) were upregulated in both KD and MIS-C, most strikingly in sample clusters S1 and S2, compared with healthy controls (Figure 1C). Functional annotation analysis revealed that this cluster was enriched with proteins involved in leukocyte-mediated immunity, neutrophil-mediated immunity, humoral immune responses, and extracellular matrix (Figure 1D and Supplemental Figure 3A). Functional annotation analyses revealed that a second set of proteins (C2) enriched in sample cluster S2 included proteins associated with platelet activation and aggregation, myofibrils, and smooth muscle cell contraction (Figure 1D and Supplemental Figure 3B). Proteins in C3 were upregulated exclusively in sample cluster S1, mostly severe MIS-C patients, and included heavy and light chain immunoglobins (Igs), as well as components of the classical complement cascade, C1qA, C1qB, and C1qC (Figure 1D and Supplemental Figure 3C).
Proteomic characterization reveals biomarkers that differentiate severe MIS-C from mild disease and from KD.
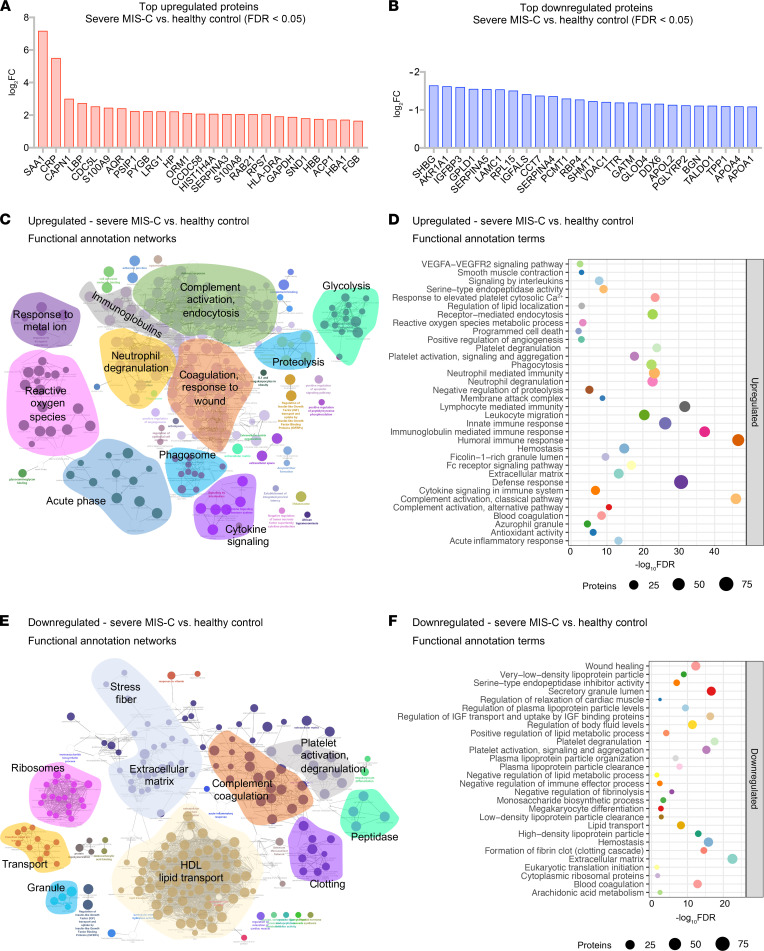
We next sought to identify proteins and associated pathways upregulated or downregulated in severe MIS-C and mild MIS-C patients compared with healthy controls (Figure 2 and Supplemental Figure 4). We identified 244 proteins increased and 135 proteins decreased in quantity in severe MIS-C compared with healthy controls (top 25 modulated proteins; Figure 2, A and B). Network and pathway analysis of significantly increased (Figure 2, C and D) or decreased (Figure 2, E and F) proteins revealed that humoral immune responses and complement pathways were highly enriched in severe MIS-C, including various Igs and C1q proteins, C1qA, C1qB, and C1qC (Supplemental Figure 5, A and B). The expression of proteins associated with platelet activation and coagulation pathways, including von Willebrand factor (VWF), F5, F9, F11, fibrinogen α and β chains (FGA and FGB), and SERPINF2, was also increased in severe MIS-C compared with healthy controls (Supplemental Figure 5, A and B). Fc receptor signaling, neutrophil-mediated responses, and phagocytosis pathways were also enriched in severe MIS-C. These include FCGR3A (CD16a), IgGFc-binding protein (FCGBP), calprotectin (S100A8 and S100A9), tissue inhibitor of metalloproteinases 1 (TIMP1), SERPINA1, SERPINA3, and acute-phase-reactant leucine-rich α2-glycoprotein 1 (LRG1) (Supplemental Figure 6, A and B, and Supplemental Figure 2C). Proteins involved in VEGF signaling and smooth muscle cell contraction were increased in severe MIS-C, including ICAM1, tropomyosin 4 (TPM4), p21-activated kinase (PAK2), FGA, FGB, and filamin B (FLNB) (Supplemental Figure 6C and Supplemental Figure 5B). As these proteins are highly expressed in vascular smooth muscle cells, their presence in the serum may reflect release from damaged blood vessels. Overall, the proteins and pathways increased in severe MIS-C indicate Ig-mediated inflammatory responses and endothelial dysfunction.
Figure 2. Characterization of severe MIS-C.
Protein expression was compared between severe MIS-C (n = 20) and healthy controls (n = 20). Proteins were considered significantly changed when FDR was less than 0.05, as determined by mapDIA statistical software for protein differential expression using MS/MS fragment-level quantitative data. (A) Top proteins enhanced in severe MIS-C, ranked by fold change. (B) Top proteins reduced in severe MIS-C, ranked by fold change. (C) ClueGO Ontology analysis via PINE visualized a network of pathways and functional annotation terms enriched in a set of proteins significantly increased in severe MIS-C population when compared with healthy controls. (D) Selected pathways and functional annotation terms from PINE analysis of proteins increased in severe MIS-C when compared with healthy controls. (E) Network plots visualized via PINE analysis of proteins reduced in severe MIS-C group when compared with healthy controls. (F) Selected pathways and functional annotation terms from protein functional enrichment analysis of proteins reduced in severe MIS-C group when compared with healthy controls.
Networks of proteins reduced in severe MIS-C revealed pathways involved in regulation of lipid transport, lipid metabolic processes, and lipoprotein clearance, including APOA1, APOA2, APOA4, APOC1, and APOM (Figure 2, E and F, and Supplemental Figure 6D). Some components of clotting and coagulation pathways were also downregulated in severe MIS-C compared with healthy controls (Figure 2, E and F, and Supplemental Figure 5A). Furthermore, there was downregulation of proteins involved in the regulation of body fluids and relaxation of cardiac muscle, including CAMK2D, which aligns with the increase in proBNP and the cardiovascular manifestations and shock observed in MIS-C (Figure 2, E and F, and Supplemental Figure 6E).
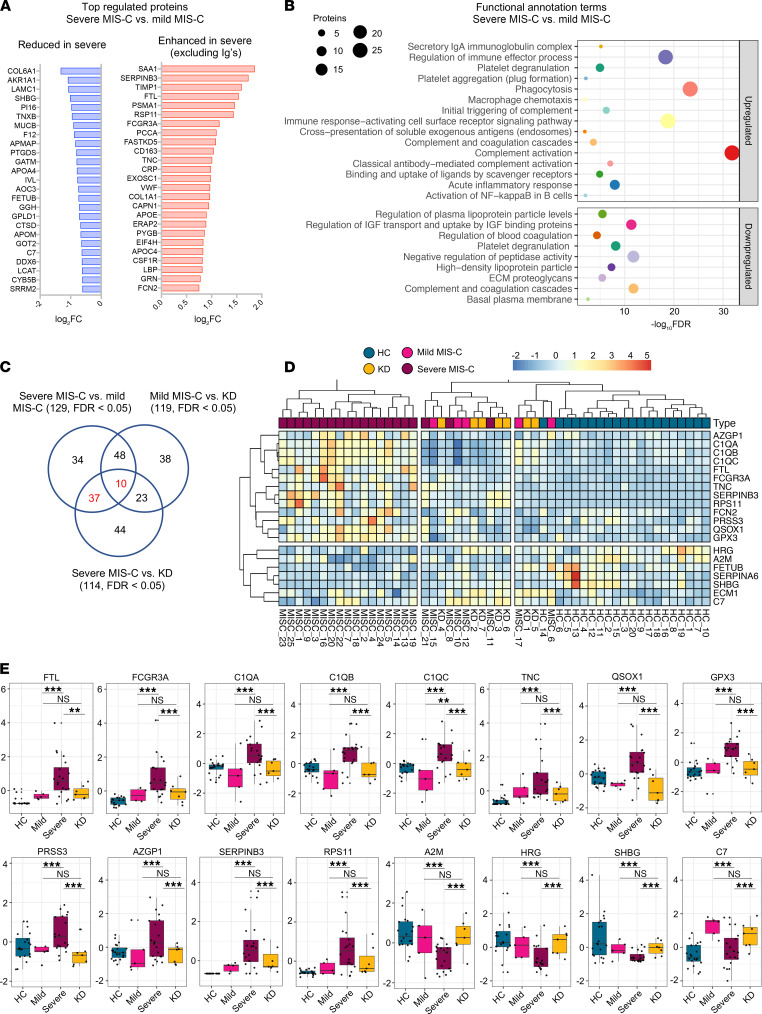
To determine factors contributing to severe disease, we compared severe MIS-C with mild MIS-C (Figure 3). We found that the expression of 75 proteins was significantly enhanced in severe MIS-C, while 61 proteins were significantly reduced when compared with mild MIS-C. Selected proteins differentially expressed between MIS-C severity groups are presented in Figure 3A. Compared with mild MIS-C, severe MIS-C patients had increased levels of proteins involved in pathways that included proteolysis, classical complement cascade, coagulation, acute-phase response, and inflammation (Figure 3, A and B, and Supplemental Figure 7). These included CRP, S100A9, SAA1, SAA2, STAT3, FCGR3A, LBP, CD163, ORM1, SERPINA1, SERPINA3, TIMP1, TLN-1, VWF, various Igs, and components of the C1 complex of the complement system (C1qA, C1qB, and C1qC) (Supplemental Figures 2C, 5B, 6B, and 7). In contrast, severe MIS-C patients had reduced expression of proteins in pathways, including negative regulators of peptidase activity, extracellular matrix proteoglycans, complement and coagulation cascades, and high-density lipid protein remodeling (Supplemental Figure 8, A and B).
Figure 3. Proteins distinguishing severe MIS-C from mild disease and KD.
Protein differential expression analysis was performed between severe MIS-C (n = 20) and mild MIS-C (n = 5) groups. Proteins were considered significantly changed when FDR was less than 0.05 as calculated by mapDIA statistical software. (A) Bar graphs show top increased and top decreased proteins in severe MIS-C when compared with mild, ranked by fold change and excluding Igs. (B) Selected pathways and functional annotation terms from protein functional enrichment analysis facilitated by PINE software using proteins increased (top panel) and decreased (bottom panel) in severe MIS-C compared with mild MIS-C. (C) Venn diagram of proteins differentially regulated between severe MIS-C, mild MIS-C, and KD. (D) Heatmap of selected proteins distinguishing severe MIS-C from mild MIS-C and KD. (E) Box-and-whisker plots of selected proteins found increased in severe MIS-C compared with mild MIS-C and KD. For improved visualization purposes, box-and-whisker plots show scaled protein expression values. Scaling was performed by mean centering and division by SD of each protein variable. For box-and-whisker plots, the bounds of the boxes represent IQR (Q1 to Q3) and the whiskers represent the nonoutlier minimum and maximum values, 1.5 × IQR. The median values are marked with a horizontal line in the boxes, and outliers are marked with black centered points outside the whiskers. Statistical analysis was calculated by mapDIA statistical software for protein differential expression using MS/MS fragment-level quantitative data. **P < 0.01, ***P < 0.001. NS, not significant.
Next, we aimed to determine which proteins distinguished severe MIS-C from mild MIS-C and KD (Figure 3, C–E), excluding clotting factors. Among proteins of interest, ferritin light chain (FTL) was highly expressed in severe MIS-C, as were proteins involved in Ig-mediated immune activation, including FCGR3A and components of the classical complement cascade, C1qA, C1qB, and C1qC (Figure 3E). Proteins that have been associated with heart failure were also identified as enhanced in severe MIS-C, including tenascin C (TNC; ref. 23) and QSOX1 (24) (Figure 3, D and E). Proteins with reduced expression in severe MIS-C were also identified and included histidine-rich glycoprotein (HRG), sex hormone–binding globulin (SHBG), and complement component 7 (C7) (Figure 3, D and E). Overall, the proteomic profiles of MIS-C and KD were similar, indicating shared pathogenic pathways. However, distinguishing proteins indicate MIS-C may be mediated more so by immune complexes, and have greater heart muscle involvement than KD. These proteins have potential to act as biomarkers to distinguish severe MIS-C from mild MIS-C or KD.
RNA-seq reveals a subgroup of hyperinflammatory MIS-C patients with enhanced myeloid responses, TRBV11-2 expansion, and SARS-CoV-2–specific antibodies.
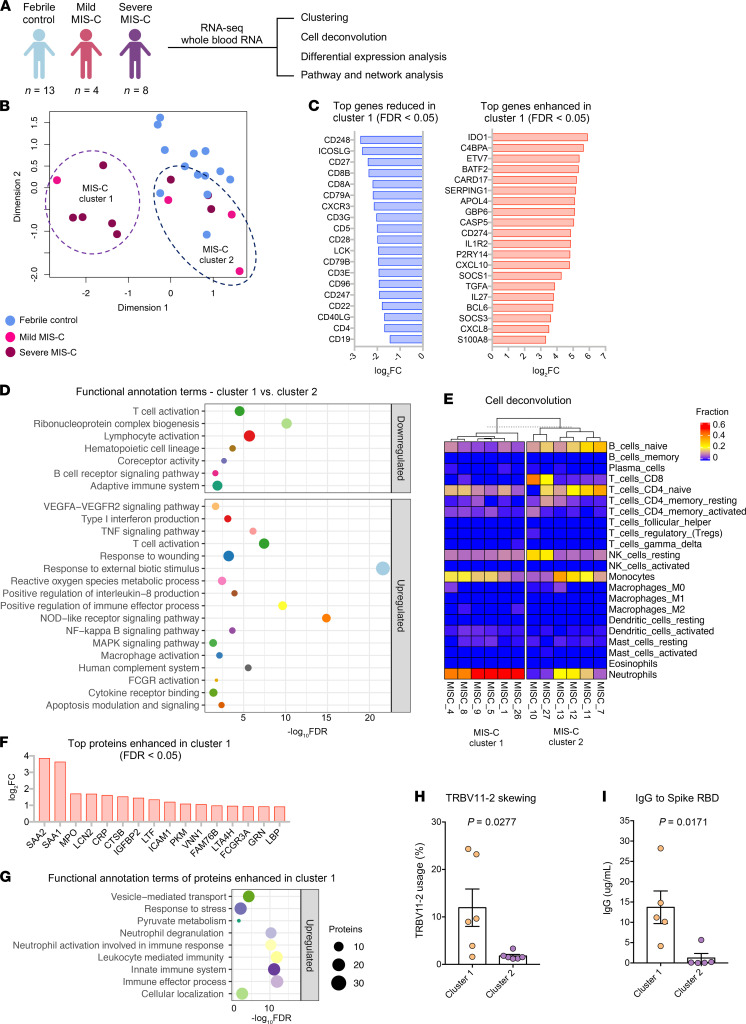
We performed RNA-seq analysis using RNA isolated from whole blood of febrile controls (n = 13), mild MIS-C (n = 4), and severe MIS-C patients (n = 8; Figure 4A). Hierarchical clustering and PCA demonstrated 2 subsets of MIS-C patients (Figure 4B and Supplemental Figure 9A). The first subset of MIS-C patients clustered separately (cluster 1) from febrile controls, while the other overlapped with febrile controls (cluster 2; Figure 4B). Cluster 1 consisted predominantly of severe MIS-C patients (5 severe and 1 mild), while cluster 2 contained an equal number of severe and mild MIS-C patients (3 severe and 3 mild; Figure 4B). Analyses revealed a large set of genes differentially expressed between the 2 MIS-C clusters (2895 genes upregulated and 2921 genes downregulated in cluster 1, FDR < 0.05, fold change [FC] > 2). The top 20 genes up- and downregulated in cluster 1 are presented in Figure 4C. Functional annotation analysis revealed that genes with increased expression in cluster 1 were involved in macrophage activation, neutrophil chemotaxis, innate signaling pathways, T cell activation, cytokine signaling, complement pathways, response to wounding, and apoptosis (Figure 4D and Supplemental Figure 9B). Cell deconvolution analysis identified increased relative abundance of neutrophils in cluster 1 MIS-C samples (Figure 4E). Genes with reduced expression in cluster 1 were involved in adaptive immune responses, as well as ribonucleoprotein complexes and RNA processing (Figure 4, C and D, and Supplemental Figure 9C). In line with these findings, cell deconvolution analysis revealed a reduction in adaptive immune cells in cluster 1, most strikingly a reduction in naive B cells, which may reflect lymphopenia that is observed in MIS-C (Figure 4E).
Figure 4. RNA-seq analysis of MIS-C.
RNA-seq was performed using whole-blood RNA isolated from febrile controls (n = 13), mild MIS-C (n = 4), and severe MIS-C (n = 8) patients. (A) Experimental design of RNA-seq analysis and patient groups. (B) PCA of RNA-seq profiles. (C) Genes up- or downregulated in cluster 1 vs. cluster 2 MIS-C patients (FDR < 0.05). (D) Selected pathways and functional annotation terms from gene functional enrichment analysis performed with PINE software using significantly up- and downregulated (FDR < 0.01, log2[FC] > 1.5 and < –1.25) genes in cluster 1 vs. cluster 2 MIS-C patients. (E) Cell deconvolution analysis of RNA-seq data by CIBERSORT. (F) Top proteins increased in cluster 1 vs. cluster 2, based on proteomics data. (G) Enriched pathways and functional annotation terms based on protein expression changes significantly (FDR < 0.05) upregulated in cluster 1 with respect to cluster 2. (H) TRBV11-2 expansion of RNA-seq samples (17). (I) IgG titers against Spike protein receptor binding domain (RBD). Data are presented as mean ± SEM. Statistical significance was determined by Mann-Whitney test (H and I).
We next compared the proteomes of cluster 1 with cluster 2 patients (Figure 4, F and G, and Supplemental Figure 9, D and E). Cluster 1, which was primarily severe MIS-C cases, was characterized by significantly enhanced expression of inflammatory markers, including CRP, SAA1, and SAA2, as well as proteins associated with neutrophil activation, including myeloperoxidase (MPO), lipocalin 2 (LCN2), cathepsin B (CATB), ICAM1, granulin (GRN), and LBP (Figure 4F). In line with this, pathway analysis of protein expression in cluster 1 identified an enrichment of neutrophil-mediated responses (Figure 4G). Analysis of proteins and pathways downregulated in cluster 1 identified lipoprotein-particle proteins, including APOA1 and APOA4 (Supplemental Figure 9, D and E), which were also observed as downregulated in severe MIS-C compared with mild MIS-C or healthy controls (Supplemental Figure 6D). Interestingly, we found a reduction in complement and coagulation cascade protein pathways in cluster 1 compared with cluster 2 (Supplemental Figure 9, D and E). Since complement pathways were identified as increased in cluster 1 by transcriptomics (Figure 4D), we analyzed the correlation between the direction of protein expression and gene expression changes when comparing cluster 1 with cluster 2 (Supplemental Figure 10A). We did not find a significant correlation between protein and gene expression changes, likely because for a subset, the gene expression and protein expression changes occurred in opposite directions (increased by gene expression yet decreased by protein expression in cluster 1). Functional annotation analysis showed that these genes/proteins, including C5, C3, C4BP, HP, F12, F5, and PF4, were enriched in complement and coagulation cascades (Supplemental Figure 10B). The increased gene expression but decreased protein expression of this subset may indicate excessive activation and consumption of these molecules. Interestingly, a reduction in C3 protein has also been observed in COVID-19 nonsurvivors compared with survivors (25).
We previously identified TRBV11-2 skewing in MIS-C patients, which correlated with disease severity and cytokine storm (17). As this study utilizes the same patient samples, we compared TRBV11-2 usage between the 2 MIS-C clusters identified by RNA-seq analysis (Figure 4B). The patients with TRBV11-2 expansion were restricted to cluster 1, which contained primarily severe MIS-C patients (Figure 4H). We also examined titers of antibodies against Spike protein between the 2 groups and found that cluster 1 MIS-C patients had higher levels of anti-Spike IgG antibodies than patients in cluster 2 (Figure 4I). This is similar to observations in adult COVID-19 patients, in which increased antibody titers against SARS-CoV-2 are associated with disease severity (26). Overall, these data indicate that the patients in MIS-C cluster 1 exhibited increased inflammatory makers, increased neutrophil responses, reduced lymphocytes, increased SARS-CoV-2 antibodies, and TRBV11-2 T cell expansion.
MIS-C autoantibodies are targeted to a diverse set of intracellular autoantigens and are enhanced in MIS-C cluster 1 patients.
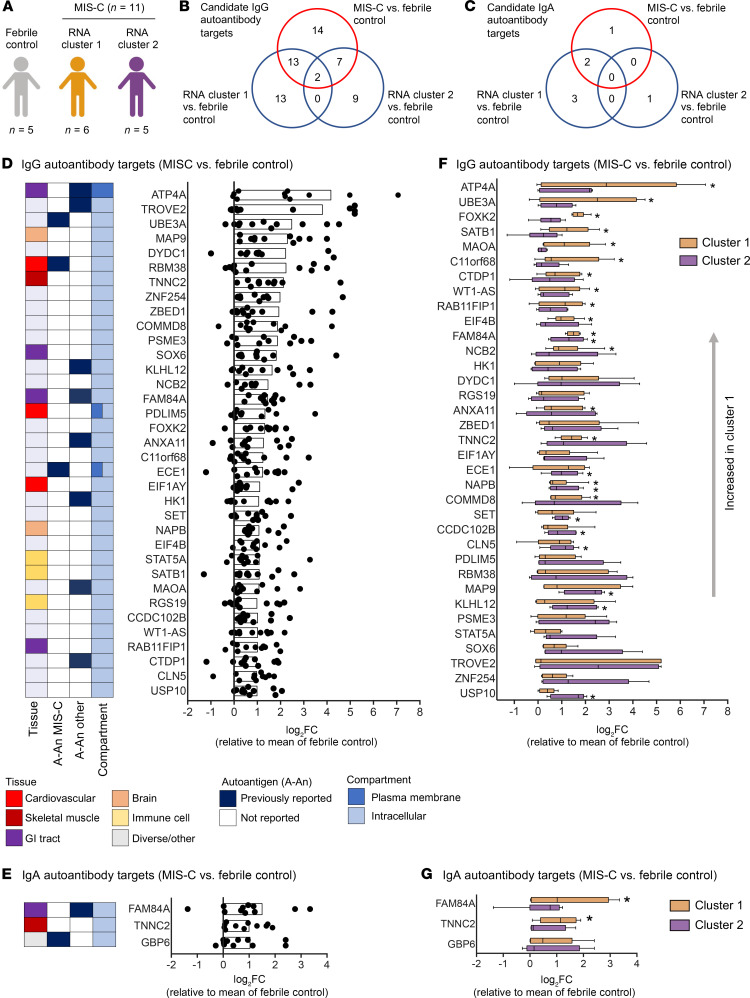
We next sought to characterize the levels of autoantibodies in our patient cohort and determine how these relate to the hyperinflammatory cluster 1 identified by RNA-seq analysis. Autoantibody analysis was performed using the HuProt array (CDI Labs) with serum from febrile controls (n = 5) and MIS-C patients (n = 11: 3 mild and 8 severe). The MIS-C patient group included 6 samples identified by RNA-seq as belonging to cluster 1, and 5 MIS-C samples from cluster 2 (Figure 5A). Candidate autoantibody targets were identified (P < 0.05, FC > 2) based on differential expression analysis of MIS-C samples, or RNA clusters, compared with febrile controls (Figure 5, B and C).
Figure 5. Autoantibody analysis of MIS-C.
(A) Autoantibody analysis was performed on serum from febrile controls (n = 5) and MIS-C patients (n = 11) using HuProt array. MIS-C samples correspond to RNA cluster 1 (n = 6) and RNA cluster 2 (n = 5) identified in Figure 4. (B) Venn diagram of candidate IgG autoantibody targets in MIS-C and RNA clusters (P < 0.05, FC > 2). (C) Venn diagram of candidate IgA autoantibody targets in MIS-C and RNA clusters (P < 0.05, FC > 2). (D) IgG autoantibody targets identified in MIS-C (n = 11) compared with febrile controls (n = 5). The bar represents log2(FC). Each symbol represents 1 MIS-C patient presented as log2(FC) above the mean of febrile controls. (E) IgA autoantibody targets identified in MIS-C (n = 11) compared with febrile controls (n = 5). The bar represents log2(FC). Each symbol represents 1 MIS-C patient presented as log2(FC) above the mean of febrile controls. (F) IgG autoantibody targets separated based on RNA cluster 1 (n = 6) and RNA cluster 2 (n = 5). Data are presented as log2(FC) above the mean of febrile controls. (G) IgA autoantibody targets separated based on RNA cluster 1 (n = 6) and RNA cluster 2 (n = 5). Data are presented as log2(FC) above the mean of febrile controls. For box-and-whisker plots, the bounds of the boxes represent the interquartile range (IQR, Q1 to Q3) and the whiskers represent the minimum and maximum values. The median values are marked with a horizontal line within the box. *FDR < 0.05 compared with febrile controls.
While the majority of IgG autoantibodies that significantly increased in MIS-C compared with febrile controls were targeted to ubiquitously expressed antigens, we identified a number of tissue-specific antigens from the GI tract and cardiovascular, skeletal muscle, and brain tissues, reflecting the systemic nature of MIS-C and the involvement of specific organs in clinical presentation of disease (Figure 5D). GI tract autoantigens included ATPase H+/K+-transporting α subunit (ATP4A), SRY-box 6 (SOX6), family with sequence similarity 84 member A (FAM84A), and RAB11 family interacting protein 1 (RAB11FIP1) (Figure 5D). Cardiovascular autoantigens included PDZ and LIM domain 5 (PDLIM5) and eukaryotic translation initiation factor 1A, Y linked (EIF1AY) (Figure 5D). Skeletal muscle autoantigens included RNA-binding motif protein 38 (RBM38) and skeletal troponin C2 (TNNC2) (Figure 5D). Brain autoantigens included microtubule-associated protein 9 (MAP9) and NSF attachment protein β (NAPB). Interestingly, several antigens highly expressed in neutrophils were identified, including endothelin-converting enzyme 1 (ECE1), SOX6, and RAB11FIP1 (Figure 5D). Autoantibodies were predominantly targeted to intracellular antigens, suggesting they may result from a secondary immune response to cell damage. We identified 3 IgA autoantibodies that were significantly increased in MIS-C compared with febrile controls, namely FAM84A, which is highly expressed in GI tissues, TNNC2, which, as mentioned above is highly expressed in skeletal muscle, and guanylate-binding protein family member 6 (GBP6) (Figure 5E). FAM84A and TNNC2 were significantly increased in RNA cluster 1, but not RNA cluster 2, compared with febrile controls.
We next examined how the 2 RNA clusters differed in autoantibody responses (Figure 5, F and G). Overall, patients in RNA cluster 1 had greater autoantibody responses than those in RNA cluster 2 (Figure 5, B and C), with the largest differences identified in IgG autoantibodies against ATP4A, UBE3A, FOXK2, SATB1, and MAOA (Figure 5F), and in IgA autoantibodies against FAM84A (Figure 5G). Overall, our data suggest systemic tissue damage and cell death may contribute to excessive antigenic drive against a diverse set of tissue-specific and ubiquitously expressed antigens. The enhanced levels of autoantibodies in RNA cluster 1 link autoantibody development to hyperinflammation, myeloid cell activation, lymphopenia, increased SARS-CoV-2 antibodies, and TRBV11-2 T cell expansion.
Patients belonging to RNA cluster 1 show BCR repertoires with highly connected networks of CDR3 sequences.
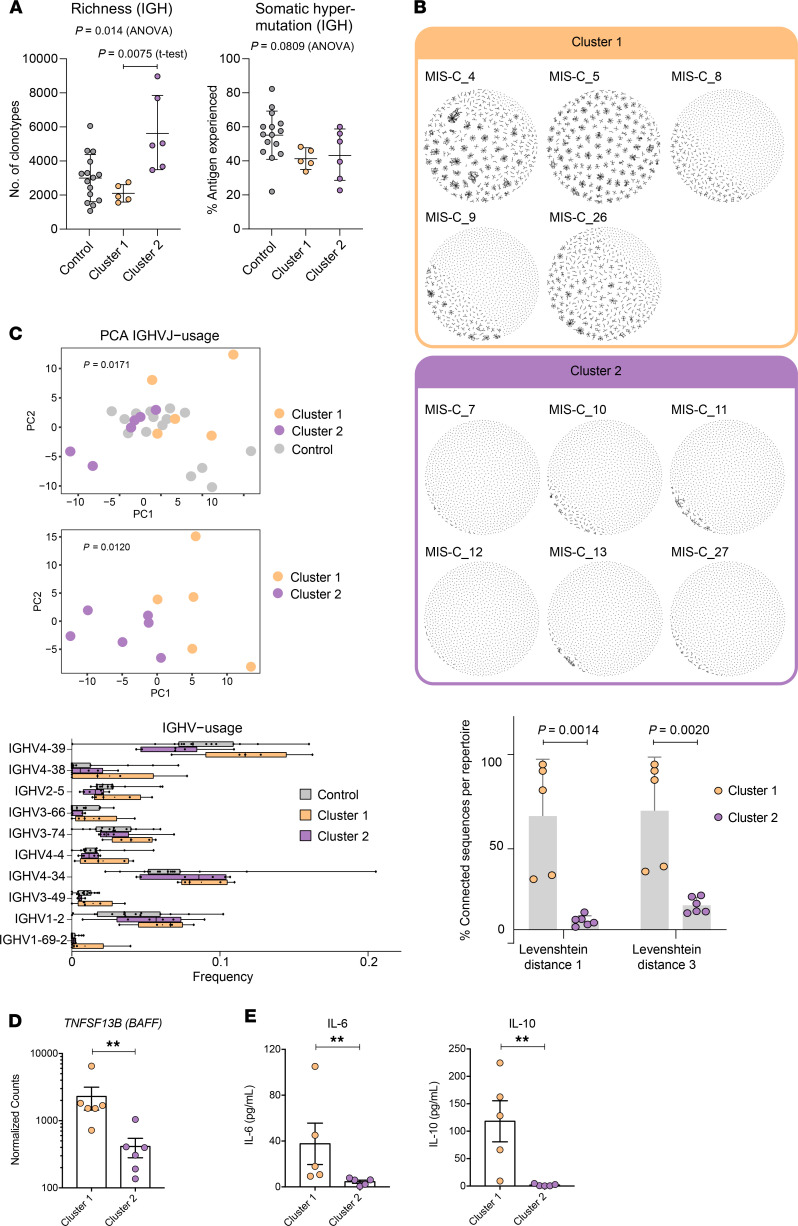
To further study B cell repertoire metrics and antigenic selection in our cohort, we performed BCR-seq on extracted RNA from blood samples of patients with mild (n = 4) or severe (n = 8) MIS-C, and age-matched febrile control patients (n = 15). We found a trend toward higher richness and a lower fraction of antigen-experienced B cells with somatic hypermutation in MIS-C patients than in febrile control patients. However, repertoire richness was distributed quite heterogeneously across patients, and the high richness pattern appeared to apply more to the small group of individuals with mild MIS-C without reaching statistical significance due to the small group size (Supplemental Figure 11). These distinct immune metrics were observable in all Ig chains — heavy chain (IGH) as well as κ (IGK) and λ (IGL) light chains — arguing in favor of the specificity of this finding. These findings were also consistent with the previously reported increased richness in T cell receptor repertoires in patients with mild MIS-C (17). Since our RNA-seq analysis revealed 2 clusters of MIS-C patients, with cluster 1 correlated with high levels of SARS-CoV-2 antibodies and autoantibodies as well as TRBV11-2 T cell expansion, we subdivided our MIS-C cohort into these clusters for the following analyses of BCR repertoires. Our aim was to determine imprints of (auto)antigenic selection or other B cell repertoire features that discriminate cluster 1 from cluster 2. MIS-C patients from cluster 1 showed lower B cell richness than febrile control patients and cluster 2 (Figure 6A), consistent with the contracted B cell compartment suggested by the transcriptome analysis of this cluster. Interestingly, although no differences in the level of somatic hypermutation were detectable between MIS-C patients of RNA cluster 1 and 2, the BCRs of all cluster 1 patients converged toward networks of highly similar CDR3 amino acid sequences (Figure 6B). The degree of connected sequences was significantly enriched in cluster 1 repertoires and comprised up to 99% of BCR clones when Levenshtein distances of 1 and 3 were used for network construction (Figure 6B). Thus, MIS-C patients in cluster 1 showed strong imprints of antigenic selection in their B cell repertoires.
Figure 6. B cell repertoire metrics, connectivity characteristics, and skewing of IGHV-J usage of MIS-C patients in RNA clusters 1 and 2.
(A) Richness and somatic hypermutation of productive IGH repertoires of MIS-C patients of RNA cluster 1 (n = 5) and RNA cluster 2 (n = 6) compared with age-matched febrile control patients (n = 15). Bars indicate mean ± SD. Statistical analysis: ordinary 1-way ANOVA for global analysis and unpaired Student’s t test for paired comparison. (B) Petri dish plots of IGH repertoire networks of MIS-C patients of RNA cluster 1 and 2. A sample of 1000 unique CDR3 amino acid clones per repertoire were subjected to imNet network analysis (75). Petri dish plots are shown for Levenshtein distance 1. Percentages of connected sequences of MIS-C patients of RNA cluster 1 and 2 obtained from networks with Levenshtein distance 1 and 3 are shown as bar plots. Bars indicate mean ± SD. Statistical analysis: unpaired Student’s t test. (C) PCA of differential IGHV-J gene usage in MIS-C patients of RNA cluster 1 (n = 5) versus cluster 2 (n = 6) versus age-matched febrile controls (n = 15). Statistical analysis: Pillai-Bartlett test of MANOVA of all principal components. Frequencies per repertoire of the 10 most skewed IGHV genes in MIS-C and febrile control patients are shown as box-and-whisker plots. The boxes extend from the 25th to 75th percentiles, whiskers from minimum to maximum, and the line within the box indicates the median. (D) BAFF expression in MIS-C cluster 1 and cluster 2, using the RNA-seq data in Figure 5. Data are presented as mean ± SEM. (E) IL-6 and IL-10 levels in serum of MIS-C cluster 1 and cluster 2 patients, using cytokine data from Supplemental Figure 1. Data are presented as mean ± SEM. Statistical analysis: Mann-Whitney test (D and E). **P < 0.01.
Higher frequency of autoantibody-associated IGHV genes IGHV4-34 and IGHV4-39 in MIS-C.
The higher autoantibody levels in RNA cluster 1 patients prompted us to investigate whether specific IGHV sequences known to be involved in the formation of autoantibodies are overrepresented in this cluster. To globally investigate IGHV-J gene usage in RNA cluster 1 versus cluster 2, we studied the repertoires by PCA. This revealed a significant skewing of IGHV-J gene usage between cluster 1 and cluster 2 (Figure 6C). Among the genes preferentially used in B cell repertoires of patients from RNA cluster 1 was IGHV4-39, which has been previously reported to be used by autoreactive lymphocytes in multiple sclerosis (27, 28). Moreover, IGHV4-34, a gene extensively studied for its usage in autoreactive B cells (29), was preferentially used in B cells from MIS-C patients in general, with a numerically higher expansion in cluster 1 repertoires. Furthermore, IGHV1-69, which is preferentially used in autoreactive B cells (30, 31), was also overrepresented in cluster 1 (Figure 6C).
We next asked which factors drive B lineage repertoires in MIS-C patients toward autoreactivity. The majority of patients from RNA cluster 1, where imprints of antigenic selection beyond SARS-CoV-2 reactivity were most obvious, showed superantigenic T cell interactions, which could be one driver promoting autoreactive B lymphocytes. The RNA transcriptomics data pointed to increased BAFF expression in RNA cluster 1, and cytokine analysis pointed to increased IL-6 and IL-10 levels in the serum (Figure 6, D and E). These results indicated that B cell dysregulation in MIS-C patients from RNA cluster 1 may not only be driven by superantigenic T cell interactions, but also by soluble factors derived from the pronounced myeloid/innate cell compartment in these patients.
Discussion
MIS-C is a hyperinflammatory disease with multi-organ involvement that is triggered by SARS-CoV-2 infection. Here, we sought to further understand the mechanisms driving MIS-C through a combination of discovery proteomics, transcriptomics, BCR repertoire analysis, and autoantibody arrays. Overall, our data point to an autoimmune phenotype in MIS-C characterized by dysregulated B cell responses, autoantibody production, and complement- and myeloid cell–mediated inflammation.
Our data highlight a central role for neutrophils and complement pathways in MIS-C, both of which are implicated in multiple systemic, autoimmune, and vasculitic diseases including anti-neutrophil cytoplasmic antibody–associated (ANCA-associated) vasculitis, Behcet’s disease, systemic lupus erythematosus (SLE), and KD (32–34). Similar mechanisms of neutrophil activation via FcγR and complement pathways as well as neutrophil-mediated pathogenesis through NETosis, cytokine and chemokine expression, and tissue damage via ROS and proteases are likely to occur in MIS-C (32, 34). Cytotoxic complement pathways, activated by antigen-autoantibody immune complexes as well as by alternative pathways may also cause direct cytotoxic tissue injury leading to multi-organ tissue damage (35, 36). Notably, IVIG is proposed to function through neutralization of FcγR proteins as well as inhibition of complement pathways (37), which may explain the success of IVIG in treating MIS-C (3, 6–8).
We found enhanced levels of multiple components of the complement pathway in MIS-C, although notably there were differences in complement components between mild and severe forms of the disease. Severe MIS-C patients expressed high levels of classical complement activation components C1qA, C1qB, and C1qC. The enhanced levels of C1q proteins and Igs in severe MIS-C may reflect an abundance of autoantibody immune complexes and an important role for immune complex–mediated pathology in MIS-C. The classical complement pathway through C1q plays a role in driving a number of autoimmune diseases via immune complex interactions (35). However, the role of C1q proteins in autoimmune disease is complex, as C1q deficiency can also enhance susceptibility to SLE, likely by reducing clearance of apoptotic cells (38). Interestingly, we found that severe MIS-C patients had reduced levels of C6 and C7, components of the membrane attack complex, compared with mild MIS-C patients. Furthermore, we found a reduction in complement C3 and C5 protein levels in serum of the hyperinflammatory RNA cluster 1 despite enhanced gene expression. These expression patterns may reflect excessive activation and consumption of complement components and a failure to clear autoantibody immune complexes.
We identified enhanced autoantibody levels in MIS-C patients, consistent with 2 previous reports (14, 16). Three autoantigens identified in our array were previously identified by Gruber et al. (14) as associated with MIS-C (UBE3A, ECE1, and RBM38). Another 8 IgG autoantigen candidates identified in our cohort have been previously reported as autoantigens in other diseases, including ATP4A (type I diabetes and corpus atrophic gastritis; refs. 39, 40), TROVE2 (Ro60, SSA2; systemic lupus erythematosus and Sjogren’s syndrome; ref. 41), KLHL12 (primary biliary cirrhosis and Sjogren’s syndrome; refs. 42, 43), FAM84A (inflammatory bowel disease; ref. 44), ANXA11 (prostate cancer; ref. 45), HK1 (primary biliary cirrhosis; ref. 42), MAOA (Alzheimer’s disease; ref. 46), and CTDP1 (Behcet’s disease; ref. 47). While we did not confirm the findings of Consiglio et al. (16), who identified autoantibodies targeting endoglin, this may be due to differences in assays and cohorts used. Most autoantigens identified in our array were intracellular and have diverse cell expression profiles, indicating that systemic tissue damage may drive autoantibody production in MIS-C. However, we also observed tissue-specific antigens from organs such as the gut, heart, endothelium, and skeletal muscle, reflecting the severe symptoms associated with these organs. Increased autoantibodies are common in various autoimmune and inflammatory diseases, and in response to some viral infections (48, 49). A number of factors can lead to autoantibody production, including abnormal survival of autoreactive lymphocytes, breakdown in B and T cell activation threshold, and excessive antigenic drive as a result of autoantigen release from damaged tissues and dying cells (49). Our data point to a combination of these events in MIS-C.
MIS-C patients belonging to RNA cluster 1 represented an interesting subset. These patients had high levels of inflammatory markers, activated myeloid function, and some showed superantigen-like T cell expansion. Moreover, this subset exhibited high levels of SARS-CoV-2 antibodies as well as autoantibodies. When looking in depth into the B cell immune repertoires of these patients, we found prominent B cell networks indicative of pronounced ongoing antigenic B cell selection. Immunogenetic repertoire biases with enrichments of genes previously described to be used in autoreactive lymphocytes (27–31) reflected the plasma autoantibody profiles of these patients. Although the promotion of autoreactive B cells could well be driven by superantigen-like T cell expansions in this cluster, it may also be favored by cytokines that were enriched in the cluster. One such candidate is BAFF, which may contribute to human autoimmune diseases not only by breaking tolerance during B cell development, but also by enhancing plasmablast survival. BAFF was strongly increased in patients from cluster 1, aligning with the evidence that these patients have activated neutrophil and innate cell compartments, which may give rise to elevated BAFF levels. Indeed, neutrophils can modulate B cell function through BAFF expression (50, 51) and promote autoimmune B cell responses (52). Positive feedback mechanisms between neutrophils/monocytes and B cell responses are likely important mechanisms driving MIS-C pathogenesis.
The association of TRBV11-2 T cell expansion with the hyperinflammatory RNA cluster 1 highlights the potential for TRBV11-2 expansion to act as a biomarker for severe disease. TRBV11-2 T cell activation and cytokine production may participate in the promotion of autoimmune responses through a number of mechanisms. Given the finding that HLA class I is associated with TRBV11-2 expansion (17), engagement of cytotoxic T cells with HLA class I molecules on endothelial cells may drive tissue damage and autoantigen release. Cytokine storm mediated by activated T cells may also contribute to tissue damage and modulation of immune cell functions that promote autoreactivity. IL-6, IL-10, and TNF-α expression correlate with TRBV11-2 expansion (17), all of which can drive B cell proliferation. However, whether TRBV11-2 T cell activation is the precipitating event that leads to development of MIS-C, or is an additional mechanism exacerbating the inflammatory autoimmune responses remains to be determined.
Gut dysfunction plays a role in driving autoimmunity, including autoreactive T cell and antibody responses (53). Severe GI symptoms are a common feature of MIS-C (5). We identified autoantibodies targeting gut-specific antigens in MIS-C patients that may drive some of these symptoms and lead to gut dysfunction. We identified increased serum levels of LBP and CD14 in MIS-C, which are indirect markers of gut dysfunction and are induced by gut microbial translocation (54, 55). Furthermore, a recent study identified persistent SARS-CoV-2 RNA in the stool of MIS-C patients, weeks after initial infection, and found sustained levels of shed Spike subunit 1 (S1) in the blood of MIS-C patients that correlated with high antibody titers against Spike and TRBV11-2 skewing (56). These data suggest that leakage of Spike and other gut antigens including LPS may occur across the gut barrier to promote a systemic immune response. This may include the TRBV11-2 expansion observed in MIS-C through a superantigen-like motif at the C-terminus of the Spike S1 subunit (17, 57).
Many immunological features of severe MIS-C are highly similar to those of severe COVID-19 in adults, including lymphopenia, CDR3-independent T cell expansion, reduced BCR diversity, high levels of autoantibodies, neutrophilia, and complement activation. Our data further demonstrate similarities between severe MIS-C and severe COVID-19 proteomic profiles, with common upregulated pathways including neutrophil degranulation, platelet degranulation, coagulation cascades, macrophage activation, and acute-phase response (58, 59). Furthermore, downregulation of proteins involved in plasma lipid regulation is evident for both diseases, including downregulation of protein constituents of HDL (58). Moreover, the hyperinflammatory cluster of MIS-C patients we identified (RNA cluster 1) shares many similarities with severe COVID-19 patients, including enhanced monocyte and neutrophil responses, increased SARS-CoV-2 antibody titers, high levels of autoantibodies, and an association with CDR3-independent TCR Vβ expansion. These similarities indicate that common mechanisms may be driving both diseases.
Overall, our study furthers our understanding of the immunopathogenesis of MIS-C, particularly the autoimmune aspects of disease, and highlights pathogenic pathways that may act as targets for more directed therapeutic interventions. We identify BCR profiles that are associated with hyperinflammation and autoimmune responses. We confirm our previous finding that TRBV11-2 T cell expansion is linked to a more severe disease phenotype and demonstrate a link between TRBV11-2 expansion and a cluster of hyperinflammatory MIS-C patients with autoimmune signatures. Furthermore, we identify serum proteins with potential to act as biomarkers to predict severity of MIS-C.
Limitations of this study.
Our study had several limitations due to sample availability. For proteomics, we could only obtain serum from control and MIS-C patients, and plasma from KD patients. Therefore, differences in coagulation pathways between MIS-C and KD could not be compared, and we removed clotting factors for any comparisons involving KD. Given that MIS-C is a new and rare disease, our RNA-seq, BCR-seq, and autoantibody analyses were limited by sample size. Validation in a larger cohort of patients is needed in future studies.
Methods
Study design
This is a descriptive study that aimed to characterize and identify pathogenic pathways, proteins, and signatures associated with MIS-C by multi-omics analysis, including proteomics, RNA-seq, autoantibody arrays, and BCR-seq. Samples consisted of mild MIS-C (n = 7), severe MIS-C (n = 20), KD (n = 7), healthy control (n = 20), and febrile control (n = 15) patients. MIS-C severity classification was based on admission to pediatric ICU requiring vasopressor use (severe) or no ICU admission (mild). Ninety-two percent of MIS-C serum samples and 100% of KD plasma samples were collected prior to IVIG administration. Patient data are presented in Table 1. The samples from the KD group were collected prior to the COVID-19 pandemic. As MIS-C is a new and rare syndrome, sample size was based on availability.
Case definitions
MIS-C was defined as per the Centers for Disease Control and Prevention (CDC) definition, which includes an individual age of less than 21 years, a persistent fever of greater than 38°C for more than 24 hours, laboratory evidence of inflammation, 2 or more organs involved, no plausible alternative diagnosis, and positivity or recent SARS-CoV-2 infection demonstrated by either real-time quantitative PCR (RT-qPCR) on nasopharyngeal swab, serology, or antigen testing, or exposure to an individual with COVID-19 within 4 weeks prior to the onset of symptoms (60). The American Heart Association definition was used for KD diagnosis (20).
Proteomics
Detailed proteomics methods can be found in the supplemental material and are outlined in McArdle et al. (21).
Depletion.
Plasma/serum samples were either analyzed as naive samples or were depleted of the 14 most abundant plasma proteins albumin, IgA, IgE, IgG, and IgM (κ and λ light chains), α-1-acid glycoprotein, α-1-antitrypsin, α-2-macroglobulin, apolipoprotein A1, fibrinogen, haptoglobin, and transferrin using High Select Top 14 Abundant Protein Depletion Resin composed of anti-camel antibodies (Thermo Fisher Scientific).
Digestion.
Plasma/serum and depleted plasma/serum trypsin digestion and desalting were performed using an automated workstation (Beckman i7), which is programed to perform reactions at a controlled temperature with uniform mixing, as previously described (61) using a standardized sample processing workflow (21).
LC-MS/MS.
Data-independent analysis (DIA) was performed on an Orbitrap Exploris 480 instrument (Thermo Fisher Scientific) interfaced with a flex source coupled to an Ultimate 3000 ultra-high-pressure chromatography system with mobile phase A (0.1% formic acid in water) and mobile phase B (0.1 % formic acid in acetonitrile). Peptides were separated on a linear gradient on a C18 column (15 cm, 3 μm) over the course of 60 minutes at a flow rate of 9.5 μL/min. Fragmented ions were detected across 50 DIA nonoverlapping precursor windows of 12-Da in size.
Bioinformatic data analysis.
DIA MS raw files were converted into mzML and the raw intensity data for peptide fragments was extracted from DIA files using the open source OpenSWATH workflow (62, 63), where experimental MS spectra were matched against a human twin population plasma peptide assay library (64). The total ion current–normalized transition-level data were scored (65), aligned (66), and processed using mapDIA software (67) to perform pairwise comparisons between groups at the peptide and protein level.
Clustering analysis and network analysis.
PCA was performed using the Factoextra package in R. Hierarchical clustering was performed using the pheatmap package in R. Protein network analysis was performed with PINE (22). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (68) partner repository with the data set identifier PXD025462. Data can be accessed at ProteomeXchange (69) (http://www.proteomexchange.org).
Cytokine array
Cytokine analysis of serum or plasma was performed using the V-PLEX Proinflammatory Panel 1 Human Kit (MSD). No detection of cytokine was assigned a value of 0.
RNA-seq
RNA-seq was performed using RNA isolated from whole blood. Library construction was performed using the SMARTer Stranded Total RNA-Seq Kit v3 – Pico Input Mammalian kit (Takara Bio USA, Inc.). Total RNA samples were assessed for concentration using a Qubit fluorometer (Thermo Fisher Scientific), and for quality using the 2100 Bioanalyzer (Agilent Technologies). Up to 10 ng of total RNA per sample was used for fragmentation, followed by reverse transcription. Indexing PCR added Illumina sequencing adapters with barcodes. Next, ribosomal cDNA was depleted, and the resulting library was PCR amplified. Sequencing was performed on the NovaSeq 6000 (Illumina) using 2 × 50 bp sequencing. Normalization and analysis of gene expression data was performed in R using edgeR and Limma-voom. Genes were considered differentially expressed with an adjusted P value of less than 0.05 and FC of greater than 2. Differentially expressed genes were analyzed using PINE (22). Cell deconvolution analysis was performed with CIBERSORT (70) (cell-type identification by estimating relative subsets of RNA transcripts) using the cell signature file LM22. The RNA-seq data have been deposited in the NCBI’s Gene Expression Omnibus (71) under accession number GSE179992.
SARS-CoV-2 Spike RBD antibody assay
SARS-CoV-2 serological Simoa assays for IgG against the Spike protein receptor binding domain (RBD) were prepared and performed as previously described (15, 72).
Autoantibody analysis
Autoantibody analysis was performed by CDI Labs using native HuProt arrays for IgG and IgA profiling of serum samples. Briefly, the arrays were blocked and probed with the samples at a 1:1000 dilution and incubated at room temperature for 1 hour. Then the arrays were washed and probed with Alexa Fluor 647–anti–human IgG (Fc) and Cy3–anti–human IgA secondary antibodies for signal detection. All samples were analyzed on the same day to avoid batch variations. Utilizing CDI Labs software, quantile normalization of the raw signal intensities (F635 median for IgG; F532 median for IgA) was performed on all arrays. A t test was used to compare the different groups and candidates were identified using the following criteria: FC of average signal intensity greater than 2 between the 2 groups, P value less than 0.05, and signal intensity of the candidates at least 5 SDs above the mean signal intensity in one group. Data are represented as log2(FC) relative to the mean of febrile controls. Tissue expression and cellular compartment analysis was performed with Human Protein Atlas (73) (http://www.proteinatlas.org).
BCR-seq
Library construction was performed using the SMARTer Human BCR IgG IgM H/K/L Profiling Sequencing Kit (Takara Bio Inc.). Total RNA samples were assessed for concentration using a Qubit fluorometer (Thermo Fisher Scientific), and for quality using the 2100 Bioanalyzer (Agilent Technologies). Up to 50 ng of total RNA per sample was used for reverse transcription, followed by 4 separate PCR amplification reactions for IgG, IgM, IgK, and IgL. A second round of PCR amplified the entire BCR variable region and a small portion of the constant region. After size selection, quantification and fragment analysis of the individual libraries were performed. Individual chains were then pooled and sequenced on the MiSeq (Illumina) using 2 × 300 bp sequencing. Fastq raw data have been deposited in the European Nucleotide Archive (ENA) under accession number PRJEB44566.
BCR analysis
Raw fastq files were converted to fasta sequences using seqtk tool (https://github.com/lh3/seqtk). Fasta sequences were submitted to IMGT/HighV-QUEST (74) for annotation of Ig rearrangements. All analyses and data plotting were performed using R version 3.5.1. Repertoires for heavy (IGH), κ (IGK), and λ light chains (IGL) were extracted and normalized to the lowest read count per chain by subsampling of 20,685, 137,544, or 126,509 productive IGH, IGK, and IGL reads, respectively. Richness was calculated as the number of unique CDR3 nucleotide sequences per repertoire. The percentage of antigen-experienced cells refers to cells that underwent somatic hypermutation. We calculated the percentage of sequence identity to germline alleles by dividing the number of identical nucleotides within the V gene by the length of the V gene per sequence. Sequences with less than 98% sequence identity to germline were counted as antigen-experienced cells. Dot plots for richness and somatic hypermutation were generated with Prism 8.0.2 (GraphPad Software). The networks of connected sequences were constructed for a subset of the 1000 most frequent different CDR3 amino acid sequences per repertoire using Imnet (75). Networks were constructed for Levenshtein distances 1 and 3. Petri dish plots and percentage of connected sequences were obtained from Imnet output files via R package igraph (76) (https://igraph.org). PCA and V gene usage box plots were visualized with R package ggplot2.
Statistics
For direct comparison of laboratory data between groups, Mann-Whitney t test was used. For multiple comparisons of cytokine data between groups, Kruskal-Wallis 1-way ANOVA with Dunn’s multiple comparison test was used. For proteomics data, differences between groups were identified by pair-wise comparisons with multiple testing correction and proteins were considered significantly different with an FDR of less than 0.05. For RNA-seq data, differences between clusters were identified by t test with Benjamini-Hochberg multiple testing correction and genes were considered significantly different with an FDR of less than 0.05. For autoantibody array, t test was used to compare the different groups and candidates were identified using the following criteria: FC of average signal intensity greater than 2 between the 2 groups, P less than 0.05, and signal intensity of the candidates at least 5 SDs above the mean signal intensity in 1 group. For BCR data, ordinary 1-way ANOVA was used for global analysis, unpaired Student’s t test was used for paired comparison, and Pillai-Bartlett test of MANOVA of principal components was used. Data were considered statistically different with a P value of less than 0.05.
Study approval
Biobanked, deidentified, patient blood remnant samples as well as control samples were obtained from Massachusetts General Hospital (Boston), Cedars Sinai Medical Center (Los Angeles), and Loma Linda University Hospital (Loma Linda) under ethics committee approval and informed consent.
Author contributions
RAP, AB, LP, MNR, JEVE, MB, and MA conceptualized the study. RAP, AB, AM, LP, and GA carried out the investigation. LMY, HKC, ML, and AF provided resources. RAP, AB, LP, MNR, JEVE, MB, and MA analyzed data. RAP, AB, LP, MNR, MB, and MA wrote the manuscript.
Supplementary Material
Acknowledgments
We thank the Applied Genomics, Computation & Translational core at Cedars Sinai and acknowledge BioRender. Funding was provided by a Cedars-Sinai Precision Health award (to RAP); NIH awards R01 AI072726 (to MA), 3RO1AI072726-10S1 (to MA), and R01HL11136 (to JEVE); the Erika Glazer Covid Fund (to JEVE); and the Advanced Clinical Biosystems Research Institute (to JEVE).
Version 1. 08/26/2021
In-Press Preview
Version 2. 10/15/2021
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(20):e151520.https://doi.org/10.1172/JCI151520.
Contributor Information
Rebecca A. Porritt, Email: Rebecca.Porritt@csmc.edu.
Aleksandra Binek, Email: Aleksandra.Binek@cshs.org.
Lisa Paschold, Email: lisa.paschold@uk-halle.de.
Magali Noval Rivas, Email: magali.novalrivas@csmc.edu.
Lael M. Yonker, Email: LYONKER@mgh.harvard.edu.
Galit Alter, Email: GALTER@mgh.harvard.edu.
Harsha K. Chandnani, Email: HChandnani@llu.edu.
Merrick Lopez, Email: MLopez@llu.edu.
Alessio Fasano, Email: afasano@mgh.harvard.edu.
Jennifer E. Van Eyk, Email: Jennifer.VanEyk@cshs.org.
Mascha Binder, Email: mascha.binder@uk-halle.de.
Moshe Arditi, Email: moshe.arditi@cshs.org.
References
- 1.Stokes EK, et al. Coronavirus Disease 2019 case surveillance - United States, January 22-May 30, 2020. MMWR Morb Mortal Wkly Rep. 2020;69(24):759–765. doi: 10.15585/mmwr.mm6924e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Götzinger F, et al. COVID-19 in children and adolescents in Europe: a multinational, multicentre cohort study. Lancet Child Adolesc Health. 2020;4(9):653–661. doi: 10.1016/S2352-4642(20)30177-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verdoni L, et al. An outbreak of severe Kawasaki-like disease at the Italian epicentre of the SARS-CoV-2 epidemic: an observational cohort study. Lancet. 2020;395(10239):1771–1778. doi: 10.1016/S0140-6736(20)31103-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toubiana J, et al. Kawasaki-like multisystem inflammatory syndrome in children during the covid-19 pandemic in Paris, France: prospective observational study. BMJ. 2020;369:m2094. doi: 10.1136/bmj.m2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belay ED, et al. Trends in geographic and temporal distribution of US children with multisystem inflammatory syndrome during the COVID-19 pandemic. JAMA Pediatr. 2021;175(8):837–845. doi: 10.1001/jamapediatrics.2021.0630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riphagen S, et al. Hyperinflammatory shock in children during COVID-19 pandemic. Lancet. 2020;395(10237):1607–1608. doi: 10.1016/S0140-6736(20)31094-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whittaker E, et al. Clinical characteristics of 58 children with a pediatric inflammatory multisystem syndrome temporally associated with SARS-CoV-2. JAMA. 2020;324(3):259–269. doi: 10.1001/jama.2020.10369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheung EW, et al. Multisystem inflammatory syndrome related to COVID-19 in previously healthy children and adolescents in New York City. JAMA. 2020;324(3):294–296. doi: 10.1001/jama.2020.10374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belhadjer Z, et al. Acute heart failure in multisystem inflammatory syndrome in children in the context of global SARS-CoV-2 pandemic. Circulation. 2020;142(5):429–436. doi: 10.1161/CIRCULATIONAHA.120.048360. [DOI] [PubMed] [Google Scholar]
- 10.McMurray JC, et al. Multisystem inflammatory syndrome in children (MIS-C), a post-viral myocarditis and systemic vasculitis-a critical review of its pathogenesis and treatment. Front Pediatr. 2020;8:626182. doi: 10.3389/fped.2020.626182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noval Rivas M, et al. COVID-19-associated multisystem inflammatory syndrome in children (MIS-C): A novel disease that mimics toxic shock syndrome-the superantigen hypothesis. J Allergy Clin Immunol. 2021;147(1):57–59. doi: 10.1016/j.jaci.2020.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pierce CA, et al. Immune responses to SARS-CoV-2 infection in hospitalized pediatric and adult patients. Sci Transl Med. 2020;12(564):eabd5487. doi: 10.1126/scitranslmed.abd5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vella LA, et al. Deep immune profiling of MIS-C demonstrates marked but transient immune activation compared to adult and pediatric COVID-19. Sci Immunol. 2021;6(57):eabf7570. doi: 10.1126/sciimmunol.abf7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gruber CN, et al. Mapping systemic inflammation and antibody responses in multisystem inflammatory syndrome in children (MIS-C) Cell. 2020;183(4):982–995. doi: 10.1016/j.cell.2020.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carter MJ, et al. Peripheral immunophenotypes in children with multisystem inflammatory syndrome associated with SARS-CoV-2 infection. Nat Med. 2020;26(11):1701–1707. doi: 10.1038/s41591-020-1054-6. [DOI] [PubMed] [Google Scholar]
- 16.Consiglio CR, et al. The immunology of multisystem inflammatory syndrome in children with COVID-19. Cell. 2020;183(4):968–981. doi: 10.1016/j.cell.2020.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Porritt RA, et al. HLA class I-associated expansion of TRBV11-2 T cells in multisystem inflammatory syndrome in children. J Clin Invest. 2021;131(10):146614. doi: 10.1172/JCI146614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramaswamy A, et al. Immune dysregulation and autoreactivity correlate with disease severity in SARS-CoV-2-associated multisystem inflammatory syndrome in children. Immunity. 2021;54(5):1083–1095. doi: 10.1016/j.immuni.2021.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartsch YC, et al. Humoral signatures of protective and pathological SARS-CoV-2 infection in children. Nat Med. 2021;27(3):454–462. doi: 10.1038/s41591-021-01263-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCrindle BW, et al. Diagnosis, treatment, and long-term management of kawasaki disease: a scientific statement for health professionals from the American Heart Association. Circulation. 2017;135(17):e927–e999. doi: 10.1161/CIR.0000000000000484. [DOI] [PubMed] [Google Scholar]
- 21. doi: 10.1101/2021.03.26.437268. Ardle AM, et al. Standardized workflow for precise mid- and high-throughput proteomics of blood biofluids [preprint]. Posted on bioRxiv March 26, 2021. [DOI] [PMC free article] [PubMed]
- 22.Sundararaman N, et al. PINE: an automation tool to extract and visualize protein-centric functional networks. J Am Soc Mass Spectrom. 2020;31(7):1410–1421. doi: 10.1021/jasms.0c00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Terasaki F, et al. Higher serum tenascin-C levels reflect the severity of heart failure, left ventricular dysfunction and remodeling in patients with dilated cardiomyopathy. Circ J. 2007;71(3):327–330. doi: 10.1253/circj.71.327. [DOI] [PubMed] [Google Scholar]
- 24.Vanhaverbeke M, et al. Peripheral blood RNA levels of QSOX1 and PLBD1 are new independent predictors of left ventricular dysfunction after acute myocardial infarction. Circ Genom Precis Med. 2019;12(12):e002656. doi: 10.1161/CIRCGEN.119.002656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fang S, et al. Decreased complement C3 levels are associated with poor prognosis in patients with COVID-19: A retrospective cohort study. Int Immunopharmacol. 2020;89(pt A):107070. doi: 10.1016/j.intimp.2020.107070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garcia-Beltran WF, et al. COVID-19-neutralizing antibodies predict disease severity and survival. Cell. 2021;184(2):476–488. doi: 10.1016/j.cell.2020.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.von Budingen HC, et al. B cell exchange across the blood-brain barrier in multiple sclerosis. J Clin Invest. 2012;122(12):4533–4543. doi: 10.1172/JCI63842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baranzini SE, et al. B cell repertoire diversity and clonal expansion in multiple sclerosis brain lesions. J Immunol. 1999;163(9):5133–5144. [PubMed] [Google Scholar]
- 29.Potter KN, et al. Evidence for involvement of a hydrophobic patch in framework region 1 of human V4-34-encoded Igs in recognition of the red blood cell I antigen. J Immunol. 2002;169(7):3777–3782. doi: 10.4049/jimmunol.169.7.3777. [DOI] [PubMed] [Google Scholar]
- 30.Avnir Y, et al. Structural determination of the broadly reactive anti-IGHV1-69 anti-idiotypic antibody G6 and its idiotope. Cell Rep. 2017;21(11):3243–3255. doi: 10.1016/j.celrep.2017.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hwang KK, et al. IGHV1-69 B cell chronic lymphocytic leukemia antibodies cross-react with HIV-1 and hepatitis C virus antigens as well as intestinal commensal bacteria. PLoS One. 2014;9(3):e90725. doi: 10.1371/journal.pone.0090725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thieblemont N, et al. Human neutrophils in auto-immunity. Semin Immunol. 2016;28(2):159–173. doi: 10.1016/j.smim.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 33.Biezeveld MH, et al. Sustained activation of neutrophils in the course of Kawasaki disease: an association with matrix metalloproteinases. Clin Exp Immunol. 2005;141(1):183–188. doi: 10.1111/j.1365-2249.2005.02829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gupta S, Kaplan MJ. The role of neutrophils and NETosis in autoimmune and renal diseases. Nat Rev Nephrol. 2016;12(7):402–413. doi: 10.1038/nrneph.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen M, et al. The complement system in systemic autoimmune disease. J Autoimmun. 2010;34(3):J276–J286. doi: 10.1016/j.jaut.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 36.Walport MJ. Complement. Second of two parts. N Engl J Med. 2001;344(15):1140–1144. doi: 10.1056/NEJM200104123441506. [DOI] [PubMed] [Google Scholar]
- 37.Galeotti C, et al. IVIG-mediated effector functions in autoimmune and inflammatory diseases. Int Immunol. 2017;29(11):491–498. doi: 10.1093/intimm/dxx039. [DOI] [PubMed] [Google Scholar]
- 38.Botto M, Walport MJ. C1q, autoimmunity and apoptosis. Immunobiology. 2002;205(4-5):395–406. doi: 10.1078/0171-2985-00141. [DOI] [PubMed] [Google Scholar]
- 39.Lahner E, et al. Autoantibodies toward ATP4A and ATP4B subunits of gastric proton pump H+,K+-ATPase are reliable serological pre-endoscopic markers of corpus atrophic gastritis. Clin Transl Gastroenterol. 2020;11(10):e00240. doi: 10.14309/ctg.0000000000000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wenzlau JM, et al. ATPase4A autoreactivity and its association with autoimmune phenotypes in the type 1 diabetes genetics consortium study. Diabetes Care. 2015;38 Suppl 2:S29–S36. doi: 10.2337/dcs15-2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen X, Wolin SL. The Ro 60 kDa autoantigen: insights into cellular function and role in autoimmunity. J Mol Med (Berl) 2004;82(4):232–239. doi: 10.1007/s00109-004-0529-0. [DOI] [PubMed] [Google Scholar]
- 42.Norman GL, et al. Anti-kelch-like 12 and anti-hexokinase 1: novel autoantibodies in primary biliary cirrhosis. Liver Int. 2015;35(2):642–651. doi: 10.1111/liv.12690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uchida K, et al. Identification of specific autoantigens in Sjögren’s syndrome by SEREX. Immunology. 2005;116(1):53–63. doi: 10.1111/j.1365-2567.2005.02197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vermeulen N, et al. Identification of a novel autoantigen in inflammatory bowel disease by protein microarray. Inflamm Bowel Dis. 2011;17(6):1291–1300. doi: 10.1002/ibd.21508. [DOI] [PubMed] [Google Scholar]
- 45.Ummanni R, et al. Prostate cancer-associated autoantibodies in serum against tumor-associated antigens as potential new biomarkers. J Proteomics. 2015;119:218–229. doi: 10.1016/j.jprot.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 46.de Oliveira-Junior LC, et al. Epitope fingerprinting for recognition of the polyclonal serum autoantibodies of Alzheimer’s disease. Biomed Res Int. 2015;2015:267989. doi: 10.1155/2015/267989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu CJ, et al. Identification of novel biomarkers for behcet disease diagnosis using human proteome microarray approach. Mol Cell Proteomics. 2017;16(2):147–156. doi: 10.1074/mcp.M116.061002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suurmond J, Diamond B. Autoantibodies in systemic autoimmune diseases: specificity and pathogenicity. J Clin Invest. 2015;125(6):2194–2202. doi: 10.1172/JCI78084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Elkon K, Casali P. Nature and functions of autoantibodies. Nat Clin Pract Rheumatol. 2008;4(9):491–498. doi: 10.1038/ncprheum0895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scapini P, et al. Regulation of B-cell-activating factor (BAFF)/B lymphocyte stimulator (BLyS) expression in human neutrophils. Immunol Lett. 2008;116(1):1–6. doi: 10.1016/j.imlet.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 51.Scapini P, et al. G-CSF-stimulated neutrophils are a prominent source of functional BLyS. J Exp Med. 2003;197(3):297–302. doi: 10.1084/jem.20021343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Costa S, et al. Recent advances on the crosstalk between neutrophils and B or T lymphocytes. Immunology. 2019;156(1):23–32. doi: 10.1111/imm.13005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ruff WE, Kriegel MA. Autoimmune host-microbiota interactions at barrier sites and beyond. Trends Mol Med. 2015;21(4):233–244. doi: 10.1016/j.molmed.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stehle JR, et al. Lipopolysaccharide-binding protein, a surrogate marker of microbial translocation, is associated with physical function in healthy older adults. J Gerontol A Biol Sci Med Sci. 2012;67(11):1212–1218. doi: 10.1093/gerona/gls178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Younas M, et al. Microbial translocation is linked to a specific immune activation profile in HIV-1-infected adults with suppressed viremia. Front Immunol. 2019;10:2185. doi: 10.3389/fimmu.2019.02185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yonker LM, et al. Multisystem inflammatory syndrome in children is driven by zonulin-dependent loss of gut mucosal barrier. J Clin Invest. 2021;131(14):149633. doi: 10.1172/JCI149633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cheng MH, et al. Superantigenic character of an insert unique to SARS-CoV-2 spike supported by skewed TCR repertoire in patients with hyperinflammation. Proc Natl Acad Sci U S A. 2020;117(41):25254–25262. doi: 10.1073/pnas.2010722117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Overmyer KA, et al. Large-scale multi-omic analysis of COVID-19 severity. Cell Syst. 2021;12(1):23–40. doi: 10.1016/j.cels.2020.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shen B, et al. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell. 2020;182(1):59–72. doi: 10.1016/j.cell.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. CDC. Information for Healthcare Providers about Multisystem Inflammatory Syndrome in Children (MIS-C). https://www.cdc.gov/mis-c/hcp/ Updated May 20, 2021. Accessed August 26, 2021.
- 61.Fu Q, et al. A plasma sample preparation for mass spectrometry using an automated workstation. J Vis Exp. 2020;(158)) doi: 10.3791/59842. [DOI] [PubMed] [Google Scholar]
- 62.Rost HL, et al. Automated SWATH data analysis using targeted extraction of ion chromatograms. Methods Mol Biol. 2017;1550:289–307. doi: 10.1007/978-1-4939-6747-6_20. [DOI] [PubMed] [Google Scholar]
- 63.Rost HL, et al. OpenSWATH enables automated, targeted analysis of data-independent acquisition MS data. Nat Biotechnol. 2014;32(3):219–223. doi: 10.1038/nbt.2841. [DOI] [PubMed] [Google Scholar]
- 64.Liu Y, et al. Quantitative variability of 342 plasma proteins in a human twin population. Mol Syst Biol. 2015;11(1):786. doi: 10.15252/msb.20145728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reiter L, et al. mProphet: automated data processing and statistical validation for large-scale SRM experiments. Nat Methods. 2011;8(5):430–435. doi: 10.1038/nmeth.1584. [DOI] [PubMed] [Google Scholar]
- 66.Rost HL, et al. TRIC: an automated alignment strategy for reproducible protein quantification in targeted proteomics. Nat Methods. 2016;13(9):777–783. doi: 10.1038/nmeth.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Teo G, et al. mapDIA: preprocessing and statistical analysis of quantitative proteomics data from data independent acquisition mass spectrometry. J Proteomics. 2015;129:108–120. doi: 10.1016/j.jprot.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Perez-Riverol Y, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47(D1):D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Deutsch EW, et al The ProteomeXchange consortium in 2020: enabling ‘big data’ approaches in proteomics. Nucleic Acids Res. 2020;48(D1):D1145–D1152. doi: 10.1093/nar/gkz984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Newman AM, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Edgar R, et al. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Norman M, et al. Ultrasensitive high-resolution profiling of early seroconversion in patients with COVID-19. Nat Biomed Eng. 2020;4(12):1180–1187. doi: 10.1038/s41551-020-00611-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Uhlen M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 74.Li S, et al. IMGT/HighV QUEST paradigm for T cell receptor IMGT clonotype diversity and next generation repertoire immunoprofiling. Nat Commun. 2013;4:2333. doi: 10.1038/ncomms3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Miho E, et al. Large-scale network analysis reveals the sequence space architecture of antibody repertoires. Nat Commun. 2019;10(1):1321. doi: 10.1038/s41467-019-09278-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Csardi G, Nepusz T. The igraph software package for complex network research. Int J Complex Syst. 2006;1695(5):1–9. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.