Abstract
Coding variants in apolipoprotein L1 (APOL1), termed G1 and G2, can explain most excess kidney disease risk in African Americans; however, the molecular pathways of APOL1-induced kidney dysfunction remain poorly understood. Here, we report that expression of G2 APOL1 in the podocytes of Nphs1rtTA/TRE-G2APOL1 (G2APOL1) mice leads to early activation of the cytosolic nucleotide sensor, stimulator of interferon genes (STING), and the NLR family pyrin domain–containing 3 (NLRP3) inflammasome. STING and NLRP3 expression was increased in podocytes from patients with high-risk APOL1 genotypes, and expression of APOL1 correlated with caspase-1 and gasdermin D (GSDMD) levels. To demonstrate the role of NLRP3 and STING in APOL1-associated kidney disease, we generated transgenic mice with the G2 APOL1 risk variant and genetic deletion of Nlrp3 (G2APOL1/Nlrp3 KO), Gsdmd (G2APOL1/Gsdmd KO), and STING (G2APOL1/STING KO). Knockout mice displayed marked reduction in albuminuria, azotemia, and kidney fibrosis compared with G2APOL1 mice. To evaluate the therapeutic potential of targeting NLRP3, GSDMD, and STING, we treated mice with MCC950, disulfiram, and C176, potent and selective inhibitors of NLRP3, GSDMD, and STING, respectively. G2APOL1 mice treated with MCC950, disulfiram, and C176 showed lower albuminuria and improved kidney function even when inhibitor treatment was initiated after the development of albuminuria.
Keywords: Cell Biology, Nephrology
Keywords: Chronic kidney disease
Introduction
African Americans represent 13% of the US population, and yet 35% of patients on chronic dialysis are African American (1, 2). This excess chronic kidney disease (CKD) risk can mostly be explained by G1 and G2 coding variants in apolipoprotein L1 (APOL1; refs. 3–5). APOL1 is a recently evolved member of the APOL gene family only found in humans and certain higher-order primates. The APOL1 G1 and G2 variants emerged as a result of a positive selection, as having a single copy of G1 or G2 risk variants protects from African trypanosomiasis; however, carrying 2 copies of G1 or G2 alleles (high-risk genotype) predisposes to kidney disease development (6).
To establish the causal link between risk variants and kidney disease, our laboratory previously generated cell-type-specific, doxycycline-inducible mouse models carrying G0 (G0APOL1, reference allele), or the G1 allele (G1APOL1) and/or G2APOL1 (7). We observed that mice with podocyte-specific expression of G1APOL1 and G2APOL1 manifested marked albuminuria, podocyte injury, and glomerulosclerosis, eventually leading to azotemia and chronic kidney failure, features similar to patients with the high-risk APOL1 genotype. The phenotype development was strongly associated with the risk genotypes, and mice carrying G0APOL1 showed only minor renal alterations.
APOL1-associated glomerular disease development is strongly linked to podocyte-specific expression (7), even though a large amount of APOL1 is present in plasma (8), as it is part of the innate immune system. Multiple cell types, including endothelial cells, vascular smooth muscle cells, and hepatocytes express APOL1 (9). Transgenic expression of APOL1 risk variants in kidney tubule epithelial or liver cells did not lead to observable phenotype development, indicating a critical cell-type dependence of the kidney phenotype (10). Human kidney transplant studies also support a strong correlation between kidney expression of APOL1 risk genotypes and kidney disease development (11–15).
Increased levels of APOL1 risk variants is an important trigger for APOL1-associated kidney disease development. In cultured cells, the APOL1-induced cytotoxicity is dose dependent (7, 16). In animal models, a strong correlation between phenotype and the degree of APOL1 RNA and protein expression was observed (7, 17). Increased APOL1 expression is considered to be the second hit triggering phenotype development in patients. Interferon (IFN) is the best-characterized regulator of APOL1 expression (18). Exogenous IFN administration to subjects with APOL1 high-risk genotype promotes rapid development of albuminuria and glomerulosclerosis (19). IFN administration increased APOL1 expression and induced proteinuria in a recently developed humanized-mouse-gene knockin model (20). Furthermore, APOL1 antisense oligonucleotides that reduce RNA stability and hence lower APOL1 protein expression ameliorated the renal phenotype in a humanized mouse model (17). These agents are currently being tested in trials for APOL1 nephropathy.
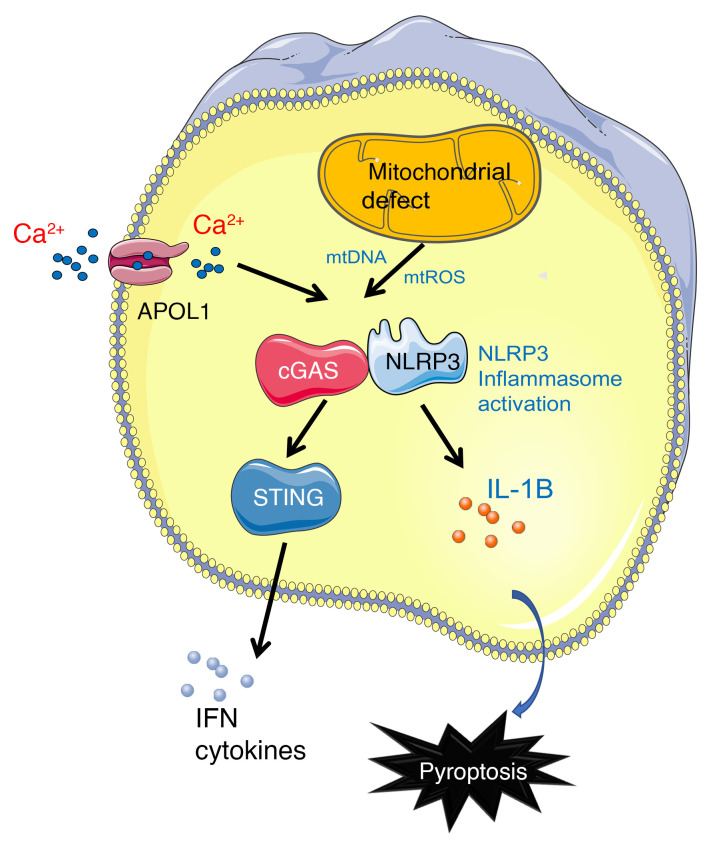
The mechanism of APOL1 risk variant–induced podocyte toxicity has been the subject of debate. APOL1 acts as an ion-conducting channel (or pore) in trypanosomes (21–24), and a similar ion-conducting activity might contribute to its podocyte-specific cytotoxicity as well (25). Other injury pathways have also been proposed, including increased K+ efflux causing the activation of stress-activated protein kinases, impaired endoplasmic reticulum trafficking, oligomerization resulting in mitochondrial pore opening, lysosomal leakage, disrupted endosomal trafficking, altered autophagy, and increased inflammatory stress (7, 16, 20, 26, 27, 28–31).
In the present study, we took a comprehensive approach to understand APOL1 risk variant–induced kidney disease development, using cultured human podocytes, cytotoxicity screening, podocyte-specific APOL1-transgenic mice, and patient samples. Our data indicate that APOL1 risk variant–induced cytotoxicity involves the cytosolic nucleotide sensor STING (the stimulator of interferon genes), as well as inflammasomes that contain Nod-like receptor protein 3 (NLRP3) and gasdermin D (GSDMD). Our results raise the possibility that targeting STING, NLRP3, and GSDMD is a potential therapeutic approach to treat APOL1-associated kidney disease.
Results
Podocyte APOL1 risk variant activates NLRP3 inflammasome signaling.
Mice expressing podocyte-specific doxycycline-inducible APOL1 risk variants manifest with podocyte loss leading to glomerulosclerosis (7), features characteristic of human APOL1-associated kidney disease. To understand the mechanism of podocyte injury, we compared mice expressing podocyte-specific APOL1 reference allele (G0) (Nphs1rtTA-TRE-G0APOL1-GFP; G0APOL1) and G2APOL1 on a doxycycline-containing diet for 3 weeks (Figure 1A). G0APOL1 mice showed no detectable albuminuria, while G2APOL1 mice developed marked albuminuria (Figure 1B) and glomerulosclerosis (Figure 1C). Both groups showed similar APOL1 protein expression levels when analyzed by Western blotting (Figure 1D).
Figure 1. Podocyte-specific APOL1 risk allele expression induced activation of cell death and signaling pathways in mouse kidneys.
(A) Experimental design: Nphs1-rtTA/TREG0APOL1-GFP (G0APOL1) and Npsh1-rtTA/TREG2APOL1-GFP (G2APOL1) mice were generated and placed on a doxycycline-containing diet for 3 weeks to induce kidney disease. (B) Albumin/creatinine ratio (ACR) of urine samples of G0APOL1 and G2APOL1 mice (n = 3). ***P < 0.001 vs. G0APOL1. (C) Representative PAS-stained sections of G0APOL1 and G2APOL1 mice. Scale bars: 30 μm (top) and 10 μm (bottom). (D) Representative Western blots of whole-kidney lysates of G0APOL1 and G2APOL1 mice. Immunoblots indicate markers of inflammasome activation (NLRP3, caspase-1, cleaved caspase-1 [cl-Casp1]), apoptosis (cleaved caspase-9 and cleaved caspase-3), and pyroptosis (caspase-11, gasdermin D [GSDMD], and cleaved N-terminal GSDMD [N-GSDMD]). GAPDH was used as a loading control. (E) Immunoblots indicate markers of STING activation (STING, phosphorylated STING, TBK1, phosphorylated TBK1, IRF3, phosphorylated IRF3). (F) Relative transcript levels of Ifit1, Ifitm1, Mx2, Isg15, and Stat1 in whole-kidney lysates of G20POL1 (n = 6) and G2APOL1 (n = 6) mice. *P < 0.05, ***P < 0.001 vs. G0APOL1. Significance was determined by Student’s 2-tailed t test (B) or 1-way ANOVA and SNK post hoc test (F). Data are expressed as the mean ± SEM.
RNA sequencing of whole-kidney lysates of WT, G0APOL1-expressing, and risk allele G1APOL1 and G2APOL1–expressing mice indicated higher expression of genes encoding cytokines (Tnfa, Il6, and Il18), markers of inflammasome and pyroptosis (Il1b, Casp1, Nlrp3, and Gsdmd), and cytosolic nucleotide sensors (Mb21d1/cGAS, Tmem173/Sting1, Irf7, and Tbk1) in mice expressing the APOL1 risk alleles when compared with WT and the reference G0 allele (ref. 7 and Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI136329DS1). Quantitative reverse transcription PCR (qRT-PCR) analysis further confirmed gene expression changes (Supplemental Figure 1B).
G2APOL1 mice showed a considerably higher expression of proteins associated with inflammasome activation, such as NLRP3, cleaved caspase-1 (10 kDa), caspase-11, and GSDMD (Figure 1D and Supplemental Figure 1C), confirming the activation of the inflammasome. In whole-kidney lysates of G2APOL1 mice, we also observed increased expression of markers of apoptosis, such as cleaved caspase-9 and cleaved caspase-3. Immunohistochemical analysis showed that immune and renal tubular epithelial cells expressed cleaved caspase-3 in G2APOL1 mice (Supplemental Figure 2A). We observed higher expression of intracellular nucleotide sensing pathways, including phosphorylated STING, TBK1, and IRF3 proteins in G2APOL1 mice (Figure 1E and Supplemental Figure 1C). Furthermore, transcriptional targets of STING, TBK, and IRF, including levels of IFN-stimulated genes (ISGs; including Ifit1, Ifitm1, Mx2, Isg15, and Stat1), were higher in mice with podocyte-specific risk variant expression (Figure 1F).
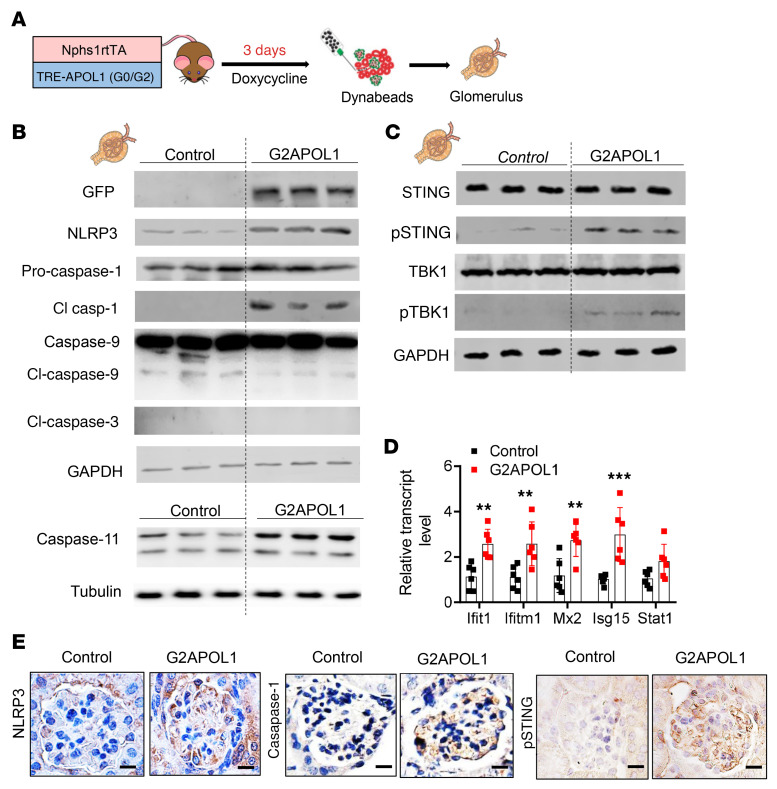
To understand early changes that are specific to glomeruli and podocytes, we analyzed G2APOL1 mice 3 days after initiation of doxycycline-containing food (Figure 2A). These animals developed detectable proteinuria but did not develop glomerulosclerosis (data not shown). We observed an increase in transgene expression, as visualized by green fluorescent protein (GFP) expression, in G2APOL1 mice compared with controls (Figure 2B). In isolated glomeruli, we failed to detect markers of apoptosis such as cleaved caspase-3 or caspase-9 (Figure 2B). On the other hand, we observed a marked increase in the expression of inflammasome proteins such as NLRP3 and downstream effectors, such as caspase-1 (pro-caspase-1 and 10 kDa cleaved caspase-1) and caspase-11 (Figure 2B). Protein expression levels of cytosolic nucleotide sensing pathways, including phosphorylated STING and phosphorylated TBK1 were higher in glomeruli of G2APOL1 mice (Figure 2C and Supplemental Figure 2B). In keeping with these findings, mRNA levels of ISGs were considerably higher in G2APOL1 mice (Figure 2D).
Figure 2. STING and inflammasome activation in glomeruli of G2APOL1 mice.
(A) Experimental design: Control and G2APOL1 mice were generated and placed on doxycycline diet for 3 days to induce APOL1 expression. Glomeruli were isolated using Dynabeads. (B) Representative Western blots from glomerular lysates of control or G2APOL1 mice. Immunoblots indicate markers of inflammasome activation (NLRP3, caspase-1, cleaved caspase-1, caspase-11) and apoptosis (cleaved caspase-9, cleaved caspase-3). GAPDH was used as a loading control. (C) Immunoblots indicate markers of STING activation (STING, phosphorylated STING, TBK1, and phosphorylated TBK1). (D) Relative transcript levels of Ifit1, Ifitm1, Mx2, Isg15, and Stat1 in kidneys of control (n = 6) and G2APOL1 (n = 6) mice. **P < 0.01, ***P < 0.001 vs. control. (E) Immunostaining analysis of NLRP3, caspase-1, and phosphorylated STING (pSTING) in control and G2APOL1 mice. Scale bars: 10 μm. Full images of NLRP3 and caspase-1 staining are shown in Supplemental Figure 6. Significance was determined by 1-way ANOVA and SNK post hoc test. Data are expressed as the mean ± SEM.
To examine the cell-type-specific expression of inflammasome markers, we performed immunostaining in kidneys of control and G2APOL1 mice. We observed a clear increase in NLRP3 and caspase-1 expression in G2APOL1 mice. In addition, phosphorylated-STING levels were also markedly higher in podocytes of G2APOL1 mice (Figure 2E).
In summary, our results indicate that the expression of podocyte-specific APOL1 risk variants in mice leads to early activation of the nucleotide sensing pathway STING and NLRP3 inflammasomes in podocytes.
APOL1-induced cytotoxicity in high-risk human podocytes is STING and NLRP3 dependent.
As mouse podocytes do not endogenously express APOL1, we next analyzed low-risk (G0/G0) and high-risk (G1/G2) genotype human podocyte-like epithelial cells (HUPECs) (7, 32). In patients, disease development is strongly linked to APOL1 variant level, which is controlled by IFN (1, 7, 18). IFN-γ treatment led to a marked increase in APOL1 expression in low- and high-risk genotype cells (Figure 3, A and B). The expression of the cytosolic nucleotide sensor STING and inflammasome proteins NLRP3 and caspase-1 (Figure 3, B and C, and Supplemental Figure 3, A–F) was higher in high-risk genotype cells compared with G0/G0 podocytes.
Figure 3. APOL1-induced cytotoxicity of high-risk human podocytes is inflammasome and STING dependent.
(A) Experimental design: Low- (G0/G0) and high-risk (G1/G2) HUPECs treated with IFN-γ. (B) Representative Western blots and (C) densitometric quantification of APOL1, NLRP3, cleaved caspase-1, STING, phosphorylated STING, and actin of G0/G0 and G1/G2 cells treated with vehicle or 2 ng/mL IFN-γ for 24 hours (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001. (D) Cytotoxicity, measured by LDH release, was normalized to calcein absorbance as an indicator of live cell count. G0/G0 and G1/G2 cells were treated for 24 hours with vehicle (n = 5), 0.2 ng/mL IFN-γ (n = 6), or 2 ng/mL IFN-γ (n = 6). ***P < 0.001 vs. control. (E) Cytotoxicity in G1/G2 HUPECs. G1/G2 HUPECs were treated with 2 ng/mL IFN-γ for 24 hours in the presence of inhibitors of NLRP3 (MCC950 [MCC]), caspase-1 (Ac-YVAD-CHO [Cho] and VX765 [VX]), caspase-9 (LEHD [Leh]), necroptosis (NEC1s [Nec]), ferroptosis (Liproxstatin [Lip]), p38 MAPK (SB 203580 [p38]), autophagy (choloroquine [CQ]), and inducers of autophagy (STF66247 [STF] and rapamycin [Rapa]). n = 3. ###P < 0.001 vs. control; **P < 0.01, ***P < 0.001 vs. IFN-γ. (F) Change in intracellular Ca2+ (measured by FURA-2 AM fluorescence) presented as percentage change from baseline. Cells were treated with 0.2 ng/mL, 2 ng/mL, or 20 ng/mL IFN-γ for 8 hours (n = 3). *P < 0.05 vs. control-treated cells. (G) Relative calcineurin activity of G0/G0 and G1/G2 HUPECs treated with sham or the indicated concentrations of IFN-γ. **P < 0.01, ***P < 0.001 vs. control (n = 6); ###P < 0.001 vs. indicated group. (H) Cytotoxicity of G1/G2 HUPECs. G1/G2 cells were treated for 24 hours with 2 ng/mL or 20 ng/mL IFN-γ with or without pretreatment with 0.5 μM BAPTA in Ca2+-free HBSS for 2 hours (n = 6). ***P < 0.001 vs. control-treated cells; red-colored, *P < 0.05 vs. indicated group. Significance was determined by 1-way ANOVA and SNK post hoc test. Data are expressed as the mean ± SEM.
High-risk genotype cells displayed a dose-dependent increase in cytotoxicity, evidenced by lactate dehydrogenase (LDH) release (Figure 3D). As observed previously (33), at higher doses of IFN-γ, even low-risk genotype cells showed some cytotoxicity (data not shown). To understand the mechanism of APOL1-induced cytotoxicity, we performed a cytotoxicity screen by targeting a variety of cell death and signaling pathways (Figure 3E and Supplemental Figure 3G). The NLRP3 inhibitor MCC950, cGAS inhibitor RU.521, STING inhibitor C176, and caspase-1 inhibitors Ac-YVAD-CHO and VX765 each significantly reduced IFN-γ–induced cytotoxicity in APOL1 high-risk-genotype cells. We did not observe a reduction in cytotoxicity following treatment with NEC1s, an inhibitor of necroptosis; Z-LEHD-FMK, a caspase-9 inhibitor; or liproxstatin-1, an inhibitor of ferroptosis (Figure 3E). Inhibition of autophagy with chloroquine prominently increased APOL1-induced cytotoxicity, while inducers of autophagy such as rapamycin and STF66247 lowered APOL1-induced cytotoxicity. SB 203580, an inhibitor of p38 MAPK, also lowered cytotoxicity in high-risk podocytes (Figure 3E). These studies highlighted the role of autophagy and the inflammasome pathway in APOL1-induced cytotoxicity.
APOL1 has been reported to function as a cation channel (16, 21, 22, 34) with potential calcium conductance (35); therefore, we next analyzed intracellular calcium levels. IFN-γ treatment of G0/G0 and G1/G2 human urinary podocytes (HUPECs) led to a dose-dependent increase in intracellular calcium. The increase in intracellular calcium was greater in G1/G2 podocytes (Figure 3F). The IFN-γ–treated high-risk genotype cells also displayed elevated calcineurin activity (Figure 3G). The rise in intracellular calcium contributed to cytotoxicity, as chelation of Ca2+ by BAPTA (Supplemental Figure 4A) lowered LDH release (Figure 3H). On the other hand, treatment of cultured podocytes with MCC950, Ac-YVAD-CHO, RU.521, or C176 did not alter intracellular calcium levels, indicating that CASP1, GSDMD, and STING activation is either downstream or independent of intracellular calcium changes (Supplemental Figure 4A). Increased cytosolic calcium levels contributed to caspase-1 activation, as IFN-γ treatment led to a marked increase in cleaved caspase-1 levels and chelation of intracellular calcium lowered caspase-1 levels, indicating that intracellular calcium changes were upstream of inflammasome activation (Supplemental Figure 4B). Moreover, treatment of APOL1 high-risk podocytes with a cGAS inhibitor (Ru.521) or STING inhibitor (C176) prevented the activation of NLRP3 and STING induced by IFN-γ (Supplemental Figure 4C), suggesting that NLRP3 is downstream of cGAS/STING. The increase in cytotoxicity, intracellular calcium, and calcineurin activity was confirmed using an independent line of high-risk-genotype HUPECs (Supplemental Figure 4, D–F).
Although the final mechanism of APOL1-induced cytotoxicity was linked to its channel activity, most studies agree that differences in intracellular trafficking and defects in autophagy and mitochondrial function contribute to risk variant–associated cell injury. Indeed, reanalysis of our prior RNA-sequencing data from WT, G0APOL1, and risk-allele mice confirmed the marked alterations in mitochondrial transcript and protein expression (Supplemental Figure 5, A and B). Interestingly, IFN-γ treatment resulted in dose-dependent alterations in maximal oxygen consumption capacity in high-risk APOL1 cells (Supplemental Figure 5, C and D). At the same time, we observed an increase in mitochondrial depolarization and increase in reactive oxygen species (ROS) generation (Supplemental Figure 5, E–H). The defect in mitochondrial function was associated with increased cytosolic accumulation of mitochondrial DNA (Supplemental Figure 5I). Inhibiting mitochondrial ROS production with mitoTEMPO or depletion of cytosolic mitochondrial DNA with ethidium bromide prevented NLRP3 and STING activation (Supplemental Figure 5J).
In summary, we found that IFN-γ treatment was associated with dose-dependent cytotoxicity in high-risk APOL1 podocytes. Expression of risk-variant APOL1 was associated with increased calcium flux and inflammasome activation and pyroptosis, altered mitophagy, cytosolic leakage of mitochondrial DNA, and activation of the cytosolic nucleotide sensor STING.
Genetic deletion of Nlrp3 in G2APOL1-transgenic mice markedly improved kidney function.
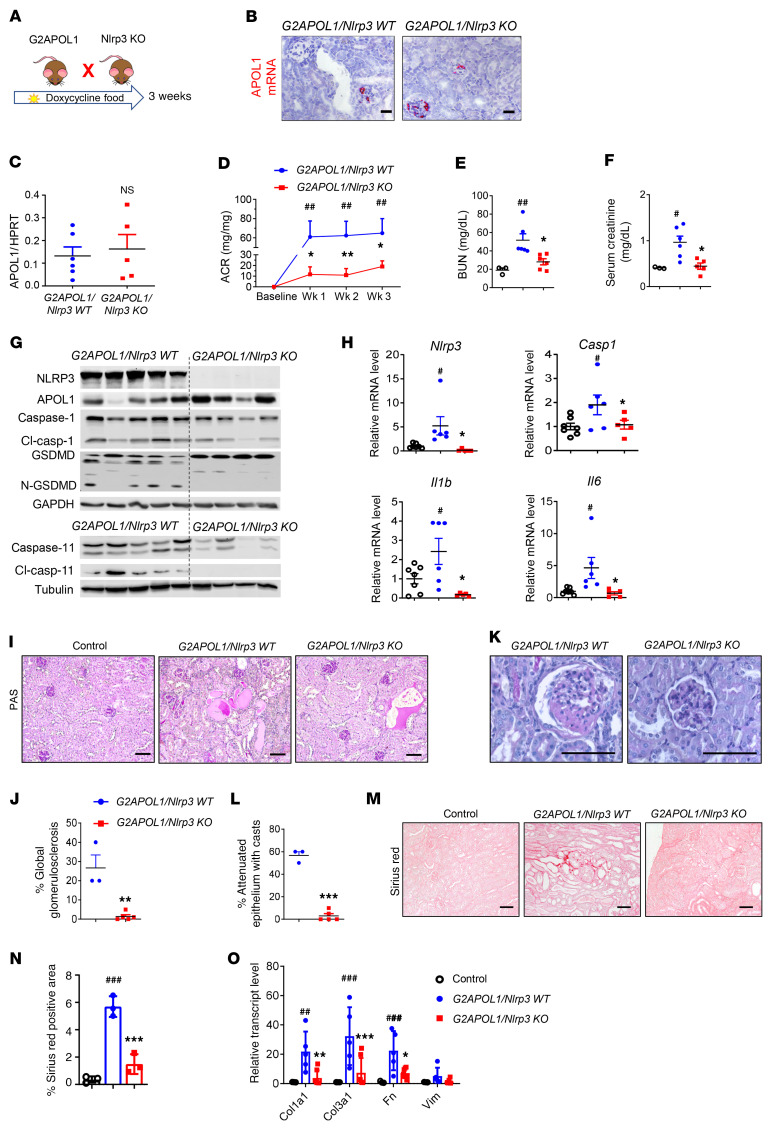
To examine the functional role of the NLRP3 inflammasome pathway in APOL1-associated kidney disease, we crossed global Nlrp3-knockout (Nlrp3-KO; ref. 36) and G2APOL1 mice to generate littermate G2APOL1/Nlrp3-WT and G2APOL1/Nlrp3-KO mice (Figure 4A). At 4 weeks of age, G2APOL1/Nlrp3-WT and G2APOL1/Nlrp3-KO mice were placed on doxycycline-containing diet for 3 weeks. Podocyte-specific APOL1 expression following doxycycline administration was confirmed by in situ hybridization (Figure 4B). Both groups showed similar levels of podocyte APOL1 expression by qRT-PCR (Figure 4C). G2APOL1/Nlrp3-WT mice developed proteinuria within 1 week of doxycycline administration, as evidenced by a high mean urinary albumin to creatinine ratio (ACR), and the proteinuria continued as the animals were kept on the doxycycline diet (Figure 4D). Mice with genetic deletion of Nlrp3 (i.e., G2APOL1/Nlrp3-KO) manifested with 70% lower urine ACR values.
Figure 4. Genetic deletion of Nlrp3 in G2APOL1-transgenic mice markedly improves kidney function.
(A) Experimental design for the generation of G2APOL1/Nlpr3-KO mice. (B) Representative images of APOL1 in situ hybridization. (C) Relative APOL1 transcript levels in kidneys of G2APOL1/Nlrp3-WT (n = 6) and G2APOL1/Nlrp3-KO (n = 5) mice. (D) ACR of G2APOL1/Nlpr3-KO (n = 6) and G2APOL1/Nlrp3-KO mice (n = 5) at baseline, 1, 2, and 3 weeks on doxycycline diet. ##P < 0.01 vs. baseline; *P < 0.05, **P < 0.01 vs. G2APOL1/Nlrp3-KO mice at the same time points. (E) BUN and (F) serum creatinine levels of control (n = 3), G2APOL1/Nlrp3-WT (n = 6), and G2APOL1/Nlrp3-KO mice (n = 5). *P < 0.05 vs. G2APOL1/Nlrp3-WT; #P < 0.05, ##P < 0.01 vs. control. (G) Western blots of whole-kidney lysates. (H) Relative transcript levels of Nlrp3, Casp1, Il1b, and Il6 in control (n = 7), G2APOL1/ Nlrp3-WT (n = 6), and G2APOL1/Nlrp3-KO (n = 5) mice. #P < 0.05 vs. control; *P < 0.05 vs. G2APOL1/Nlrp3-WT. (I) PAS-stained kidney sections. (J) Semiquantitative analysis of percentage of globally sclerotic glomeruli in G2APOL1/Nlrp3-WT (n = 3) and G2APOL1/NLRP3-KO (n = 5) mice. **P < 0.01 vs. G2APOL1/NLRP3-WT. (K) Representative images of glomeruli. (L) Percentage of attenuated epithelium with casts in G2APOL1/Nlrp3-WT (n = 3) and G2APOL1/NLRP3-KO (n = 5) mice. ***P < 0.001 vs. G2APOL1/NLRP3-WT. (M) Sirius red–stained kidney sections. (N) Quantification of Sirius red–positive area. ###P < 0.001 vs. control; ***P < 0.001 vs. G2APOL1/NLRP3-WT. (O) Relative mRNA levels of Col1a1, Col3a1, Fn1, and Vim in the kidneys of control (n = 3), G2APOL1/NLRP3-WT (n = 5), and G2APOL1/NLRP3-KO (n = 7) mice. ##P < 0.01, ###P < 0.001 vs. control; **P < 0.01, ***P < 0.001 vs. G2APOL1/NLRP3-WT. Scale bars: 30 μm. Significance was determined by 1-way ANOVA and SNK post hoc test. Data are expressed as the mean ± SEM.
Kidney function analysis, including measurements of serum blood urea nitrogen (BUN) and serum creatinine, showed that G2APOL1/Nlrp3-WT mice had significant azotemia compared with control (WT) mice (Figure 4, E and F). G2APOL1/Nlrp3-KO mice had markedly lower serum BUN and creatinine levels, indicating preservation of renal function and milder APOL1-associated kidney disease. We confirmed the genetic deletion of Nlrp3 by kidney tissue Western blotting and immunohistochemical analysis (Figure 4G and Supplemental Figure 6A). G2APOL1/Nlrp3-KO mice displayed markedly lower levels of cleaved caspase-1, cleaved GSDMD, and caspase-11 protein compared with G2APOL1/Nlrp3-WT mice (Figure 4G). In keeping with these findings, mRNA levels of caspase-1 (Casp1) and downstream inflammasome effectors IL-1β (Il1b) and IL-6 (Il6) were considerably lower in kidneys of G2APOL1/Nlrp3-KO mice (Figure 4H and Supplemental Figure 6B).
We next examined renal structural changes in G2APOL1/Nlrp3-WT and G2APOL1/Nlrp3-KO mice. Kidney sections stained with periodic acid–Schiff (PAS) indicated less global and segmental glomerulosclerosis in G2APOL1 mice in the absence of NLRP3 (Figure 4, I–K). Tubule damage was lower, including epithelial injury, brush border damage, and luminal cast number in Nlrp3-KO mice (Figure 4L). Sirius red staining, an indicator of kidney fibrosis, showed a marked reduction in collagen accumulation in G2APOL1/Nlrp3-KO mice (Figure 4, M and N). Expression of fibrosis markers such as collagen I α1 chain (Col1a1), collagen III α1 chain (Col3a1), fibronectin 1 (Fn1), and vimentin (Vim) in G2APOL1/Nlrp3-KO mice (Figure 4O) was markedly decreased.
Gasdermin D, encoded by GSDMD, plays a key role in inflammasome activation–induced pyroptotic cell death. To further confirm the role of the inflammasome pathway in APOL1-associated kidney disease, we crossed G2APOL1 mice with global Gsdmd-knockout (Gsdmd-KO) mice to generate G2APOL1/Gsdmd-WT and G2APOL1/Gsdmd-KO littermates (Supplemental Figure 7A). We confirmed the genetic deletion of Gsdmd by transcript and protein expression (Supplemental Figure 7, B and C). Genetic deletion of Gsdmd was associated with markedly lower albuminuria, serum BUN, and creatinine compared with podocyte-specific G2APOL1 mice (Supplemental Figure 7, D–F), and resulted in less glomerulosclerosis and tubulointerstitial fibrosis, as evidenced by immunohistochemistry and gene expression analysis (Supplemental Figure 7, G–I).
In summary, genetic deletion of the inflammasome component Nlrp3 and downstream target Gsdmd led to a marked improvement in kidney function and structure, and ameliorated inflammatory changes in APOL1-associated kidney disease models.
Genetic deletion of STING protects from APOL1-induced podocyte damage.
To understand the role of the cytosolic nucleotide sensors in APOL1 risk variant–induced kidney damage, we crossed podocyte-specific G2APOL1 mice with STING-KO mice to generate littermate G2APOL1/STING-WT and G2APOL1/STING-KO mice (Figure 5A). We confirmed the genetic deletion of STING by transcript and protein expression (Figure 5, B and C). STING-KO mice had improved kidney function indicated by lower albuminuria, serum BUN, and creatinine compared with G2APOL1 mice (Figure 5, D–I).
Figure 5. Genetic deletion of STING in G2APOL1-transgenic mice markedly improves kidney function.
(A) Experimental design for the generation of Nphs1rtTA/TREG2APOL1/STING-KO (G2APOL1/STING-KO) mice. (B) Western blots of whole-kidney lysates from G2APOL1/STING-WT and G2APOL1/STING-KO mice showing levels of STING and GAPDH. (C) Relative STING transcript levels in kidneys of G2APOL1/STING-WT (n = 5) and 2APOL1/STING-KO mice (n = 5). ***P < 0.01 vs. G2APOL1/STING-WT. (D) Albuminuria (ACR) of G2APOL1/STING-WT (n = 5) and G2APOL1/STING-KO mice (n = 5) at baseline, 1, 2, and 3 weeks on doxycycline diet. *P < 0.05, **P < 0.01, ***P < 0.001, comparing G2APOL1/STING-WT mice at the same time points. (E) Serum creatinine levels in control (n = 5), G2APOL1/STING-WT (n = 5), and G2APOL1/STING-KO mice (n = 5). ***P < 0.001 vs. control; #P < 0.05 vs. indicated group. (F) Serum urea nitrogen (BUN) levels in control, G2APOL1/ STING-WT, and G2APOL1/STING-KO mice. ***P < 0.001 vs. control; #P < 0.05 vs. indicated group. (G)PAS-stained and Sirius red–stained kidney sections of control, G2APOL1/STING-WT, and G2APOL1/STING-KO mice. Scale bars: 30 μm. (H) Quantification of globally sclerotic glomeruli and Sirius red–positive area of control, G2APOL1/STING-WT, and G2APOL1/STING-KO mice. n = 6 mice per group. ***P < 0.001 vs. control; ##P < 0.01, ###P < 0.001 vs. indicated group. (I) Relative mRNA levels of profibrotic genes Col1a1 (collagen type I α1 chain), Col3a1 (collagen type III α1 chain), Fn1 (fibronectin 1), and Vim (vimentin); and markers of inflammation Ccl2 (chemokine ligand 2), Tnfa (TNF-α), and Cxcl2 (CXC ligand 2) were evaluated in the kidneys of control, G2APOL1/STING-WT, and G2APOL1/STING-KO mice. n = 6 mice. ***P < 0.001 vs. control; #P < 0.05, ###P < 0.001 vs. indicated group. Significance was determined by 1-way ANOVA and SNK post hoc test. Data are expressed as the mean ± SEM.
PAS-stained kidney sections indicated reduced glomerular injury in STING-KO mice. Sirius red staining, as an indicator of kidney fibrosis, showed a marked reduction in collagen accumulation in G2APOL1/STING-KO mice (Figure 5G). Expression of profibrotic genes such as Col1a1, Col3a1, Fn1, and Vim (Figure 5I) was markedly decreased in G2APOL1/STING-KO mice. In summary, genetic deletion of the cytosolic nucleotide sensor STING resulted in marked functional and structural improvement in G2APOL1 mice.
Pharmacological inhibition of STING and GSDMD ameliorates kidney disease in G2APOL1 mice.
To evaluate the therapeutic potential of targeting cytosolic nucleotide sensing pathways such as STING, we treated mice with a small molecular STING inhibitor, C176 (37). Mice were treated with 750 nmol C176 before transgene induction and animals were euthanized 10 days following transgene induction (Figure 6A).
Figure 6. Pharmacological inhibition of GSDMD and STING improves kidney disease in G2APOL1 mice.
(A) Experimental design: G2APOL1 mice were placed on doxycycline diet and treated with GSDMD inhibitor (disulfiram) or STING inhibitor (C176) or sham for 10 days. (B) Relative APOL1 transcript levels in whole-kidney tissue of G2APOL1 sham (only doxycycline diet; n = 6), disulfiram (n = 6), and C176 (n = 6). (C) Albuminuria (ACR) levels of control (n = 6), G2APOL1 (n = 6), disulfiram-treated (n = 6), and C176-treated (n = 6) G2APOL1 mice. ***P < 0.001 vs. control; #P < 0.05 vs. G2APOL1. (D) Serum creatinine levels of control (n = 6), G2APOL1 (n = 6), disulfiram-treated (n = 6), and C176-treated (n = 6) G2APOL1 mice. ***P < 0.001 vs. control; #P < 0.05, ##P < 0.01 vs. G2APOL1. (E) BUN levels of control (n = 6), G2APOL1 (n = 6), disulfiram-treated (n = 6), and C176-treated (n = 6) G2APOL1 mice. ***P < 0.001 vs. control; #P < 0.05, ##P < 0.01 vs. G2APOL1. (F) PAS-stained and Sirius red–stained kidney sections of control, G2APOL1, disulfiram-, and C176-treated G2APOL1 mice. Scale bars: 30 μm. (G) Quantification of Sirius red–positive area of control, G2APOL1, disulfiram-, and C176-treated G2APOL1 mice. n = 6 mice per group. ***P < 0.001 vs. control; ###P < 0.001 vs. indicated group. (H) Relative mRNA levels of profibrotic genes Col1a1, Col3a1, Fn1, and Vim; and markers of inflammation Ccl2, Tnfa, and Cxcl2 were evaluated in the kidneys of control, G2APOL1, disulfiram-, and C176-treated G2APOL1 mice. n = 6 mice. **P < 0.01, ***P < 0.001 vs. control; #P < 0.05, ##P < 0.05, ###P < 0.001 vs. indicated group. Significance was determined by 1-way ANOVA and SNK post hoc test. Data are expressed as the mean ± SEM.
Mice receiving STING inhibitor showed protection from kidney disease, as demonstrated by functional changes such as markedly lower albuminuria, serum creatinine, and BUN (Figure 6, C–E). Animals also showed protection from histological damage, including substantially less glomerulosclerosis and tubulointerstitial fibrosis (Figure 6, F and G). Similarly, expression of fibrosis-associated genes such as Co1a1, Col 3a1, Fn1, and Vim was lower in inhibitor-treated mice. Expression of inflammation-associated genes (Ccl2, Tnfa, and Cxcl2) was lower following C176 treatment (Figure 6H). Similarly, beneficial results were obtained when mice were treated with the GSDMD inhibitor disulfiram initiated prior to G2APOL1 expression (Figure 6, A–H).
In summary, STING and GSDMD inhibitor treatment efficiently protected G2APOL1 mice from disease development.
Pharmacological inhibition of NLRP3 ameliorates kidney disease in G2APOL1 mice even when started after the initiation of proteinuria.
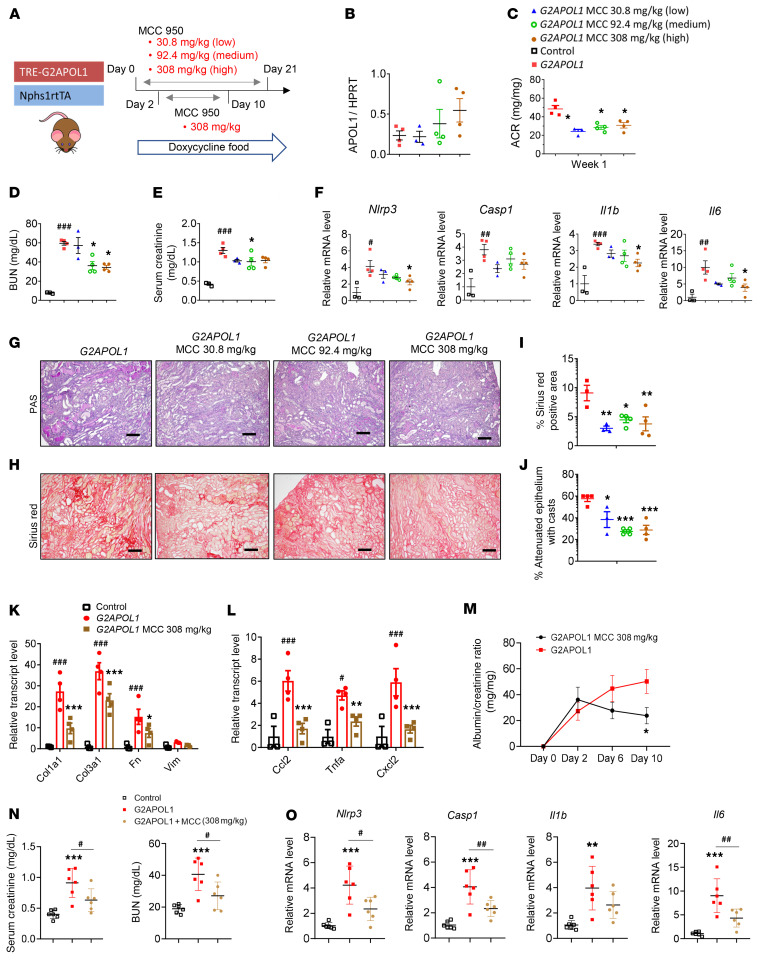
To understand the therapeutic potential of targeting NLRP3 for APOL1-associated kidney disease, we examined the effect of a potent, small molecular NLRP3 inhibitor, MCC950, also known as CRID3 or CP-456,773 (ref. 38 and Figure 7A). G2APOL1 mice were placed on doxycycline diet alone (sham) or doxycycline diet containing ± 30.8 mg/kg (low dose), 92.4 mg/kg (medium dose), or 308 mg/kg (high dose) MCC950 diet for 3 weeks. The inhibitor dose was determined by a pilot study based on food consumption to achieve therapeutic drug levels. APOL1 mRNA expression following doxycycline administration was confirmed by qRT-PCR; MCC950-treated animals exhibited slightly higher APOL1 expression, which did not reach statistical significance (Figure 7B).
Figure 7. Pharmacological inhibition of NLRP3 reduces inflammasome signaling and improves kidney disease in G2APOL1 mice.
(A) Experimental design: G2APOL1 mice were placed on doxycycline diet with or without 30.8 mg/kg (low), 92.4 mg/kg (medium), or 308 mg/kg (high) MCC950 for 3 weeks. (B) Relative APOL1 transcript levels in whole kidney, n = 4. (C) ACR at 1 week on doxycycline, n = 4. *P < 0.05 vs. G2APOL1 sham. (D) BUN, (E) serum creatinine, and (F) relative transcript levels of Nlrp3, Casp1, Il1b, and Il6 in control (n = 3), G2APOL1 (n = 4), low (n = 4), medium (n = 4), and high dose MCC950 (n = 4). #P < 0.05, ##P < 0.01, ###P < 0.001 vs. control; *P < 0.05 vs. G2APOL1. (G) Representative PAS-stained and (H) Sirius red–stained kidney section. Scale bars: 30 μm. (I) Quantification of Sirius red–positive area, n = 4 mice. *P < 0.05, **P < 0.01 vs. G2APOL1. (J) Percentage of attenuated epithelium with casts in G2APOL1 (n = 4), low (n = 4), medium (n = 4), and high dose MCC950 (n = 4). *P < 0.05, ***P < 0.001 vs. G2APOL1. (K) Relative mRNA levels of Col3a1, Fn1, and Vim, and (L) Ccl2, Tnfa, and Cxcl2 were evaluated in the kidneys of control (n = 3), G2APOL1 (n = 4), medium (n = 4), and high dose MCC950 (n = 4). ###P < 0.001 vs. control; **P < 0.01, ***P < 0.001 vs. G2APOL1 sham. (M) ACR in G2APOL1 mice treated with high dose MCC950 at baseline, 1, 2, 6, and 10 days on doxycycline diet (n = 6). *P < 0.05 vs. G2APOL1. (N) Serum creatinine levels and (O) relative transcript levels of Nlrp3, Casp1, Il1b, and Il6 in control, G2APOL1, and high dose MCC950 (n = 6). **P < 0.01, ***P < 0.001 vs. control; #P < 0.05, ##P < 0.01 vs. indicated group. Significance was determined by 1-way ANOVA and SNK post hoc test. Data are expressed as the mean ± SEM.
One week after the initiation of doxycycline treatment, G2APOL1 mice developed significant proteinuria (Figure 7C). Medium and high doses of MCC950 diet reduced urine ACR by approximately 36% (P = 0.036). Kidney function parameters, BUN and serum creatinine (Figure 7, D and E), indicated improvement in kidney function of MCC950-treated mice. The plasma level of MCC950 was within the therapeutic range (ref. 39 and Supplemental Figure 8A). BUN inversely correlated with plasma MCC950 concentration (Supplemental Figure 8B). Expression of the pyroptosis machinery–related genes Nlp3, Casp1, Il1b, and Il6 was lower in kidneys of MCC950-treated mice (Figure 7F).
We next evaluated renal structural damage in sham and MCC950-treated G2APOL1 mice using PAS- and Sirius red–stained kidney sections (Figure 7G). Markers of tubular damage, such as kidney tubule epithelial casts, were lower in MCC950-treated mice (Figure 7, I and J). The expression of profibrotic genes (Col1a1 and Col3a1) was noticeably lower in MCC950-treated G2APOL1 mice (Figure 7K). Similarly, macrophage and inflammatory markers (Ccl2, Tnfa, and Cxcl2) were lower in MCC950-treated mice (Figure 7L).
We next repeated the experiment by initiating the MCC950 treatment after the mice developed albuminuria; we posit that this would be a clinically more meaningful scenario. Two days after starting mice on the doxycycline-containing diet, animals developed proteinuria: approximately 35 mg/mg (albumin/creatinine). Mice were then started on MCC950-containing food and were compared to mice that remained on the regular doxycycline-containing diet for 10 days (Figure 7A). Albuminuria was markedly lower in mice treated with MCC950 compared with controls. BUN and creatinine were also lower in MCC950-treated mice (Figure 7M). Expression of genes associated with fibrosis and inflammation was reduced in mice receiving MCC950 (Figure 7O). Overall, our results indicated that pharmacological NLRP3 inhibition via MCC950 improved kidney function and structural parameters in G2APOL1 mice, even after albuminuria development, indicating its therapeutic potential.
Increase in podocyte NLRP3 and inflammasome expression in APOL1 high-risk individuals.
To determine whether inflammasome signaling plays a role in APOL1-associated kidney disease development in patients, we performed an unbiased RNA-sequencing analysis of microdissected glomeruli isolated from 427 human kidney samples (Figure 8A). The demographics of these patients are shown in Supplemental Table 1. It has been difficult to compare low- and high-risk APOL1 genotype patient samples due to their relative low sample numbers (40). In addition, high-risk-genotype patients usually present with more severe disease, confounding the results (7, 40). Therefore, we opted to identify genes whose expression correlates with glomerular APOL1 levels (Figure 8A), as APOL1 expression plays a key role in disease severity.
Figure 8. Increase in podocyte NLRP3 and inflammasome expression in APOL1 high-risk patients.
(A) We analyzed RNA-sequencing data from 427 microdissected human kidney glomeruli and correlated transcript level with APOL1 expression. (B) Relative APOL1 expression (y axis) and eGFR (x axis) in glomeruli isolated from 427 individual kidney samples (Pearson’s correlation). (C–I) Relative transcript expression (y axis) of IFI16 (C), STAT1 (D), CASP1 (E), GSDMD (F), STING (G), cGAS (H), and TBK1 (I) and APOL1 expression (x axis) in glomeruli isolated from 427 individual kidney samples (Pearson’s correlation and P values shown). (J) Representative immunofluorescence staining of NLRP3 (red) and podocalyxin (green), and DAPI staining (blue) in kidney samples of APOL1 high-risk control and CKD patients. Scale bars: 30 μm. (K) Representative images of NLRP3 in situ hybridization in kidney of APOL1 high-risk control and CKD patients. The red arrows indicate the expression of APOL1 mRNA. Scale bars: 30 μm. Student’s t test based on Pearson’s correlation coefficient was used to calculate the statistical significance of the association.
As reported by previous studies, APOL1 expression in glomeruli inversely correlated with estimated glomerular filtration rate (eGFR), such that expression was higher in human kidney tissue from individuals with lower eGFR (Figure 8B). Consistent with the in vitro podocyte data, expression of STAT1, the key transcription factor in the IFN-γ signaling pathway, strongly and positively correlated with APOL1 amount, indicating that STAT1 (and IFN signaling) is a key regulator of APOL1 in human kidney samples.
We observed that expression of genes in the inflammasome pathway, including GSDMD, CASP1 (caspase-1), CASP4, and CASP5 (Figure 8, C–F; CASP4 and CASP5 not shown), and genes in the cytosolic nucleotide sensing pathway, including STING, cGAS, and TBK1 (Figure 8, G–I), strongly correlated with APOL1 expression, suggesting a strong relationship between APOL1 expression and inflammasome/STING signaling in human kidney glomeruli. APOL1 protein expression showed good correlation with APOL1 mRNA expression in human kidney samples (Supplemental Figure 9A).
Furthermore, immunostaining studies of APOL1 high-risk-genotype kidneys of patients with normal and reduced eGFR showed a marked increase in NLRP3 expression in kidneys of high-risk CKD patients (Figure 8J). Dual immunofluorescence analysis to localize NLRP3 and kidney cell markers, include staining for podocalyxin (to mark podocytes), CD31 (for endothelial cells), Lotus tetragonolobus lectin (for proximal tubule cells), and CD11b (for myeloid cells) confirmed that the increase in NLRP3 expression is mainly located in podocytes of diseased kidney samples of patients (Figure 8J and Supplemental Figure 9B). Podocyte expression of NLPR3 mRNA in high-risk CKD patients was further validated by in situ hybridization (Figure 8K). In summary, we found a strong correlation between APOL1 and inflammasome/STING signaling in patient samples, indicating the clinical relevance of our observations.
Discussion
APOL1 genetic variants could explain almost all excess renal disease among African Americans (3, 41). Furthermore, the effect size (~2- to 80-fold) of APOL1 risk variants on kidney disease development is higher than any other observed complex trait variants. APOL1 variants have been associated with different forms of kidney disease, including HIV-associated nephropathy, focal segmental glomerulosclerosis, sickle cell nephropathy, lupus nephritis, and hypertensive kidney disease (42–45). Given the substantial impact of APOL1 variants in kidney disease development, the identification of new therapeutic targets is of key importance.
In addition to genetic variants, environmental factors are critically important for disease development (46). HIV-1 and SARS-Cov-2 infections, IFN treatment, inflammation, and elevated soluble urokinase-type plasminogen activator receptor (suPAR) levels (47) have been shown to strongly predict kidney disease development in individuals with a high-risk APOL1 genotype. All these factors are associated with elevated APOL1 levels, supporting the working hypothesis that APOL1 dose is the trigger or second hit to disease development. Recently, antisense oligonucleotide–based lowering of APOL1 expression has shown promise in a mouse model (17), confirming the crucial role of APOL1 dose in podocytes in disease development. Taking advantage of this information, we believe that the urinary podocyte system could be a valuable new screening tool (7, 32). This system tests the risk-variant effect on a susceptible, naturally occurring genetic background. APOL1 is a recently evolved gene with many coding variations (in addition to G1 and G2) that could also be important for cytotoxicity. It also does not require artificial genetic overexpression. Furthermore, it recapitulated the dose-dependent cytotoxicity induced by the risk variant. IFN administration was associated with dose-dependent necrosis, which was more severe in high-risk podocytes. Consistent with previous reports, we observed that at higher levels, the APOL1 reference allele was associated with cytotoxicity (33).
Here, using mouse models, patient samples, and in vitro–cultured cells, we defined mechanisms mediating APOL1-induced cytotoxicity, and provide avenues for potential therapeutic targeting. We observed the activation of a variety of key proteins in the cytosolic nucleotide sensing pathway (STING) and inflammasome (NLRP3, caspase-1, and GSDMD) when we analyzed glomeruli only 3 days after doxycycline-induced expression of risk-variant APOL1. Activation of the cytosolic nucleotide sensors and inflammasome was also observed in cultured podocytes and in cell lines with transient overexpression of risk-variant APOL1. The consistent activation and involvement of the NLRP3 inflammasome suggests that the cytotoxicity was mediated by pyroptosis, a form of regulated necrosis (48). Functional studies including cytotoxicity experiments confirmed the key role of NLRP3 and caspase-1, consistent with our in vivo findings.
STING and the NLRP3 inflammasome have been shown to play a role in tubule cells of the kidney in unilateral ureteral obstruction (49), adenine-induced tubulointerstitial nephritis (50), and remnant kidney (5/6 nephrectomy; ref. 51) models. In addition, NLRP3 inflammasome activation has been reported in lupus-prone mice and patients with lupus (52), indicating that the NLRP3 inflammasome might be a common downstream factor in a variety of injury models. Multiple pathways contribute to the activation of STING and NLRP3. Recent reports indicate that APOL1 functions as a cation channel in podocytes; however, no major difference in ion channel activity of risk and reference APOL1 was observed. Difference in APOL1 activity was mostly explained by intracellular trafficking, which plays a key role in channel activation (16, 21, 34). Consistently, we observed an increase in intracellular calcium levels in IFN-γ–treated podocytes. Intracellular calcium seems to play a critical upstream role in NLRP3 activation, as the chelation of intracellular calcium protects from cytotoxicity and NLRP3 activation (53). Most importantly, we observed a defect in intracellular trafficking, including autophagy which led to a defect in mitophagy (7). Defective mitophagy resulted in accumulation of defective mitochondria, including the release of mitochondrial DNA into the cytosol. Cytosolic mitochondrial DNA is a strong activator of cytosolic nucleotide sensors such as STING. Downstream of STING is TBK, IRF3, IRF7, and proinflammatory cytokine release. Future studies shall determine the exact mechanism of NLRP3 activation in podocytes.
Our studies using mice with genetic deletion of Nlrp3 and a potent small molecular inhibitor (38) of NLRP3 indicate the key role of the inflammasome in APOL1-mediated kidney disease development. The effect of genetic knockdown was more robust than the effect of the small molecular inhibitor. This might be due to the timing of administration, tissue penetration, or inhibitor dose, as shown by measuring free circulating inhibitor concentration. Nevertheless, administration of MCC950 even after the development of the albuminuria led to reduction in albuminuria, as well as improvements in functional parameters (creatinine and BUN) and structural changes (fibrosis and glomerulosclerosis). Inhibitors of STING and GSDMD show similar therapeutic promise, and their dosing should be tested in future pharmacologic studies.
Although the crucial role of APOL1 in the development of kidney disease has been extensively studied during the past decade, therapeutic development for APOL1-associated kidney disease has been difficult. Our results could have a broad therapeutic implication. At present, all clinical studies target APOL1 expression or APOL1 channel function. Given the key role of APOL1 in protecting against trypanosomiasis, these drugs could have very significant side effects in areas where trypanosomiasis is endemic. Our studies highlight an alternative mode for APOL1 targeting by blocking NLRP3, GSDMD, or STING. These are highly targetable pathways, as selective STING, caspase-1 (54), caspase-11, and GSDMD inhibitors have been developed. Antibodies that neutralize the downstream cytokine IL-1β, such as anakinra (55) and canakinumab (56), have already been approved for the treatment of various autoimmune conditions. In addition, an APOL1 pore blocker or antisense oligonucleotide will interfere with early disease. Disease-driving pathways might be different at later stages. Our work suggests that NLRP3 and STING will intervene at later stages of disease progression. Targeting both early and late pathways could offer important combination therapeutic opportunities. Future clinical studies will determine the effect of NLRP3, STING, and GSDMD in kidney disease development, but nevertheless our studies highlight an immediately translatable strategy.
An important limitation of our study is that the APOL1 gene is missing from the mouse genome, making it is unlikely that a single animal model will reflect the full spectrum of cytotoxic pathways and renal phenotypes seen in humans. Here, we compared humanized mice expressing a reference allele to those expressing a risk variant. Our previous studies highlighted that the model recapitulates kidney function changes (albuminuria, eGFR), histological changes (global and segmental sclerosis and fibrosis), and gene expression changes observed in patients. In addition, while the disease course appears rapid in mice, recent reports indicate a similar disease course in high-risk-APOL1 individuals following SARS-CoV-2 infection (57) or high-risk individuals following IFN injection (18).
In summary, our data demonstrate the activation of STING and the NLRP3 inflammasome in APOL1-mediated kidney disease. These studies suggest a key role of NLRP3 in mediating podocyte cytotoxicity by pyroptosis and the potential therapeutic targeting of this pathway in the treatment of APOL1-associated kidney disease.
Methods
Antibodies and reagents.
Primary antibodies against the following proteins were used: APOL1 (Proteintech, 66124-1-Ig; Western blot [WB] 1:1000); mouse caspase-1 (Santa Cruz Biotechnology, sc56036; WB 1:500; immunohistochemistry [IHC] 1:250); mouse caspase-1 (AdipoGen, AG-20B-0042-C100; WB 1:500, used only in Figure 1); human cleaved caspase-1 (Cell Signaling Technology [CST], 4199; WB 1:1000); cleaved caspase-3 (CST, 9664; WB 1:500; IHC 1:250); caspase-9 (Abcam, 25758; WB 1:1000); caspase-11 (Abcam, 180673; WB 1:1000); GAPDH (CST, 2118; WB 1:5000); GSDMD (Abcam, 219800; WB 1:1000), GSDMD (Sigma-Aldrich, G7422; WB 1:1000); GFP (Clontech, 632380; WB 1:5000); LC3II (CST, 2775; WB 1:1000), NLRP3 (CST, 15101; WB 1:500); NLRP3 (Novus, NBP2-12446; IHC 1:40, immunofluorescence [IF] 1:100), NLRP3 (AdipoGen, AG-20B-0014C; WB 1:1000); podocalyxin (R&D Systems, MAP1658; IF 1:100), tubulin (Sigma-Aldrich, T6199; WB 1:1000); STING (CST, 13647S; WB 1:1000); phosphorylated STING (CST, 19781S; WB 1:1000); TBK1 (CST, 3504S; WB 1:1000); phosphorylated TBK1 (CST, 5483; WB 1:1000); IRF3 (CST, 11904T; WB 1:1000); phosphorylated IRF3 (CST, 37829; WB 1:1000); CD31 (Abcam, 24590; IF 1:50), and CD11b (CST, 49420; IF 1:50). The following secondary antibodies were used for IF analysis: donkey anti–rabbit IgG (H+L), Alexa Fluor 555 (Invitrogen, A31572; IF 1:500) and chicken anti–mouse IgG (H+L), Alexa Fluor 488 (Invitrogen, 21200; IF 1:1000). The following reagents were used: Lotus tetragonolobus lectin, Fluorescein (Vector, FL-1321; IF 1:800); FBS (Atlanta Biologicals); DMEM 1× (Cellgro); G418 50 mg/mL (UPenn Cell Center Services); MEM Eagle (Sigma-Aldrich); penicillin-streptomycin (Cellgro); insulin-transferrin-selenium (ITS) (Cellgro); RPMI 1640 (Cellgro); and Tet system–approved FBS (Clontech). The following inhibitors were used: C176 (MedChemExpress, 1 μM); RU.521 (Invivogen, 4 μM); chloroquine (Sigma-Aldrich, 25 μM); 7-Cl-O-Nec1 (Abcam, 25 μM); Liproxstatin-1 (Tocris, 0.5 μM); MCC950 (Invivogen, 50 μM); rapamycin (LC Laboratories, 10 ng/mL); SB203580 (Tocris, 10 μM); STF62247 (Cayman Chemicals, 0.5 μM); VX765 (Toronto Research Chemicals, 10 μM); Z-LEHD-FMK (R&D Systems, 10 μM); and ZVAD-FMK (Promega, 10 μM).
Mouse studies.
TRE-APOL1 mice were generated as previously described (7). Briefly, APOL1 (G0/G2) cDNAs were cloned into the pBI-EGFP vector containing a tetracycline response element (TRE). The transgenic construct was injected into FVB/N oocytes.
B6.129S6-Nlrp3tm1Bhk/J (Nlrp3 KO; ref. 36) (stock no. 21302), B6(Cg)-Sting1tm1.2Camb/J (STING KO) (stock no. 025805), and C57BL/6N-Gsdmdem4Fcw/J (Gsdmd KO) (stock no. 032410) mice were obtained from The Jackson Laboratory (JAX), backcrossed with APOL1 FvB mice for 5 generations, and genotyped according to JAX genotyping protocols. Nlrp3-KO, STING-KO, and Gsdmd-KO mice appear normal and are viable and fertile. Nlrp3-KO, STING-KO, and Gsdmd-KO mice were crossed with Nphs1-rtTA/TREG0APOL1 (7) to generate littermate Nlrp3-KO Nphs1rtTA/TREG2APOL1 and Nlrp3-WT Nphs1rtTA/TREG2APOL1 mice; littermate STING-KO Nphs1rtTA/TREG2APOL1 and STING-KO Nphs1rtTA/TREG2APOL1 mice; and littermate Gsdmd-KO Nphs1rtTA/TREG2APOL1 and Gsdmd-KO Nphs1rtTA/TREG2APOL1 mice. At 4 weeks of age, APOL1 transgene expression was induced with doxycycline-containing food (200 mg/kg, Bio-Serv). Littermate mice were used for all experiments.
For NLRP3 inhibitor studies, Nphs1-rtTA/TREG2APOL1 (7) mice were divided into 4 groups. At 7 weeks of age, one group of mice received doxycycline diet only, while the other 3 groups received 30.8 mg/kg, 92.4 mg/kg, and 308 mg/kg MCC950 mixed with the doxycycline diet. MCC950 inhibitor was provided by Boehringer Ingelheim Pharmaceuticals, Inc. All groups of mice were provided with food and water ad libitum and monitored daily for 3 weeks.
Mice were injected intraperitoneally with disulfiram (50 mg/kg, Cayman) or C176 (750 nmol per mouse, Bio Vision) or DMSO dissolved in 85 μL corn oil daily. All mice were provided with food and water ad libitum and monitored for 10 days.
Mouse phenotype analysis.
Urine albumin and creatinine levels were determined using mouse albumin–specific ELISA (Bethyl Laboratories) and creatinine reagent set (Pointe Scientific), per the manufacturers’ protocols. BUN was determined by the TRACE DMA Urea kit (Thermo Electron Corporation).
Podocyte cell culture and transfection.
Transformed low-risk (G0/G0) and high-risk (G1/G2) HUPECs were generated as previously described (7, 32). HUPECs were cultured in RPMI 1640 supplemented with 10% FBS, 1% ITS, and 1% penicillin-streptomycin at 33°C.
Cytotoxicity assays.
HUPECs were plated in 96-well plates and treated with 2 ng/mL recombinant human IFN-γ (Peprotech) with or without inhibitors for 24 hours. The supernatant was used to quantify LDH release as a measure of cytotoxicity with a CytoTox 96 Non-Radioactive Cytotoxicity Assay (Promega), as described previously (7). Cells were treated with a solution containing 3 μM calcein (Invitrogen) and 3 μM propidium iodide (PI) (Invitrogen) in PBS for 30 minutes at 37°C. The plate was read at 485 nm/535 nm for calcein, and then at 530 nm/620 nm for PI.
Calcium measurements.
HUPECs (G0/G0 or G1/G2) were plated in 96-well plates and prepared for Ca2+ measurement using a modified protocol from PromoCell (PK-CA707-50029). Upon reaching 70% confluence, cells were serum starved overnight in RPMI and 0.2% FBS with 1% penicillin-streptomycin. Cells were loaded with 2.5 μM FURA-2 AM diluted in Hank’s balanced salt solution with HEPES (HBSS-HEPES) (Gibco) for 30 minutes at 33°C. Cells were washed with HBSS-HEPES and incubated in the same buffer for 45 minutes at 33°C. HUPECs were then treated with vehicle or 0.2 ng/mL, 2 ng/mL, or 20 ng/mL recombinant human IFN-γ in HBSS-HEPES (6 wells per treatment). At 8 hours following IFN-γ treatment, one set of cells was treated with 20 μM ionomycin for 30 seconds as a positive control. The FURA-2 fluorescence was measured at 37°C with excitation at 340 and 380 nm and emission at 500 nm. The F340/F380 ratio was recorded as a measure of intracellular calcium.
Calcineurin phosphatase activity assay.
Calcineurin activity in cultured HUPECs was analyzed with the Biomol Calcineurin Cellular Activity Assay kit (Abcam, ab139464) according to the manufacturer’s instructions.
Mitochondrial membrane potential measurement.
Mitochondrial membrane potential (Δψm) was measured according to the manufacturer’s instructions (Invitrogen, T3168). Briefly, cells were washed twice with PBS and then incubated with 5 μg/mL JC-1 for 10 minutes at 37°C and 5% CO2. After washing with PBS twice, the cells were analyzed for mitochondrial membrane potential by measuring the green/red ratio in an inverted fluorescence microscope (Nikon).
Oxygen consumption measurements.
For real-time measurement of the extracellular acidification rate (ECAR) and oxygen consumption rate (OCR), HUPECs were analyzed using an XF-96 Extracellular Flux Analyzer (Seahorse Bioscience). In brief, HUPECs were plated in XF-96 cell culture plates (7 × 104 cells/well in 70 μL) and either left unstimulated or stimulated with IFN-γ. At the indicated time points, HUPECs were washed and analyzed in XF Running Buffer per the manufacturer’s instructions to obtain real-time measurements of OCR and ECAR. Where indicated, ECAR and/or OCR were analyzed in response to 1 μM oligomycin, 1.5 μM fluoro-carbonyl cyanide phenylhydrazone (FCCP), and 100 nM rotenone plus 1μM antimycin A.
Subcellular fractionation.
Quantification of mitochondrial DNA in the cytosolic fraction was adapted from Lauren et al. (58) as follows: Briefly, cells were lysed by mild digitonin buffer containing 150 mM NaCl, 50 mM HEPES pH 7.4, 25 μg/mL digitonin (EMD Chemicals), protease and phosphatase inhibitors, and incubated on ice for 10 minutes followed by centrifugation at 2000g and 4°C for 10 minutes. Supernatants were centrifuged 3 times at 17,000g and 4°C for 10 minutes to yield the cytosolic fraction free of nuclear and mitochondrial contamination.
The pellet from the first spin was washed and resuspended in NP-40 buffer containing 150 mM NaCl, 50 mM HEPES pH 7.4, 1% NP-40, protease and phosphatase inhibitors, and incubated on ice for 30 minutes followed by centrifugation at 7000g and 4°C for 10 minutes to yield the crude mitochondrial fraction (supernatant) and nuclear fraction (pellet). DNA was then isolated from these pure cytosolic and nuclear fractions using a DNeasy Blood and Tissue kit (QIAGEN). qRT-PCR was performed on the cytosolic and nuclear fractions using nuclear DNA primer RPL13A and mitochondrial DNA primer mtCO1. The Ct values for RPL13A obtained from the respective nuclear fraction served as normalization controls.
Protein extraction and Western blotting.
All solutions, tubes, and centrifuges were maintained at 4°C. RIPA buffer (Cell Signaling Technology, 9806) with 1% SDS and protease cocktail (Complete Mini, Roche) was used to extract total protein lysates from tissues or cells according to the manufacturer’s instructions. Protein concentrations were measured using a BCA protein assay (Pierce, 23225). Whole-cell lysates (75 μg of protein) were heated (95°C) for 10 minutes in Laemmli sample buffer (Bio-Rad). Proteins were then separated by polyacrylamide gel electrophoresis in acrylamide gels (8%–15%) and transferred using a Bio-Rad Western system to nitrocellulose (Bio-Rad) membranes. Transferred blots were blocked for 1 hour in 5% nonfat milk in Tris-buffered saline. Membranes were incubated with specific primary antibodies at 4°C overnight, followed by incubation with a horseradish peroxidase–conjugated anti-mouse antibody (1:5000) or anti-rabbit antibody (1:5000) at 25°C for 1 hour. Resulting immunoblots were visualized using ECL Western Blotting Substrate (Pierce) in a LI-COR chemiluminescence imager (LI-COR), according to the manufacturers’ instructions.
Glomerular isolation.
Glomerular isolation was performed as described previously (59). Briefly, mice were euthanized and perfused through the heart with 150 μL of Dynabeads (Thermo Fisher Scientific, M-450) diluted in 30 mL of HBSS (Thermo Fisher Scientific). The kidneys were removed, minced, and digested in collagenase (1 mg/mL collagenase A, 100 U/mL deoxyribonuclease I in HBSS) at 37°C for 30 minutes with gentle agitation. The reaction was stopped by adding 10% FBS to the mixture. The collagenase-digested tissue was passed through a 100-μm cell strainer using a flattened pestle, and the cell strainer was then washed with 5 mL of HBSS. The cell suspension was then centrifuged at 1000 rpm for 5 minutes, and the cell pellet was resuspended in 2 mL of HBSS. Glomeruli containing Dynabeads were collected using a magnetic particle concentrator (Olympus) and washed at least 3 times with HBSS. The resulting glomeruli were lysed with RIPA/SDS lysis buffer, and the protein concentration was estimated as described above. Twenty micrograms of protein per sample was prepared for Western blotting by heating in Laemmli buffer.
Histological analysis.
For histological analysis, kidneys were fixed in 4% paraformaldehyde overnight, dehydrated, embedded into paraffin blocks, and sectioned onto glass slides. Sections were stained with PAS for histological analysis. Sirius red staining was performed to determine the degree of fibrosis. Immunohistochemical and immunofluorescence analyses were performed to visualize the expression of proteins in the kidney. Briefly, paraffin-embedded sections were deparaffinized, rehydrated, and incubated with indicated primary antibodies. For immunohistochemical analysis, samples were visualized using diaminobenzidine (DAB) substrate, and further counterstained with hematoxylin. For immunofluorescence, slides were incubated with fluorescently conjugated secondary antibodies, counterstained, and mounted with DAPI (nuclear stain). For visualizing mRNA expression in tissues using in situ hybridization, paraffin-embedded tissue samples were processed using an RNAscope 2.5 HD Duplex Detection Kit (Advanced Cell Diagnostics, 322436). The Hs-APOL1 (Advanced Cell Diagnostics, 459791-C) probe was used for the APOL1 RNAscope assay.
RNA extraction and qRT-PCR.
RNA was isolated from mouse kidney using TRIzol reagent (Invitrogen), as described previously (37). RNA was transcribed into cDNA using a cDNA Archival Kit (Life Technologies). qRT-PCR reactions were carried out with cDNA in a 384-well plate with 2× SYBR Green dye (Applied Biosystems) and gene-specific primer pairs (Supplemental Table 2) using the ViiA 7 System (Life Technologies). The data were normalized and analyzed using the ΔΔCt method with HPRT as the reference gene. The primers used are listed in Supplemental Table 2.
Mouse and human kidney RNA sequencing data.
Gene expression changes were reanalyzed from previously published mouse kidney RNA sequencing (7) and microdissected human kidney RNA sequencing data (37).
Plasma MCC950 measurements.
Plasma samples were subjected to plasma protein precipitation with acetonitrile/methanol (1:1) in the 96-well plate format and analyzed by liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS). The AB Sciex 6500+ triple quadrupole mass spectrometer equipped with a RapidFire sample injector was operated in the positive ion mode. The lower limit of quantification was 25 nmol/L.
Statistics.
Data are expressed as mean ± SEM. Unless noted otherwise, statistical significance was determined by 1-way analysis of variance (ANOVA) and Student-Newman-Keuls (SNK) post hoc test for multiple comparisons. Differences between 2 groups were analyzed using Student’s 2-tailed t test with unequal variance. A P value of less than 0.05 was considered significant. We did not perform analyses to predetermine sample sizes. Rather, the sample sizes were chosen empirically based on our previous experience in the calculation of experimental variability. The numbers of animals used are specified in corresponding figure legends. All experiments were performed with at least 3 biological replicates.
Study approval.
Human kidney samples were procured with approval from the Institutional Review Board (IRB) of the University of Pennsylvania.
For mouse studies, the protocol for all mice used in this study was approved by the Institutional Animal Care and Use Committee at the University of Pennsylvania.
Author contributions
J Wu and AR analyzed and interpreted data, designed figures, and wrote the manuscript. J Wu, NJC, XS, MJS, and ZM analyzed data and designed figures. PB, DL, J Wahba, and JBK collected data. JJK, SSP, and CMBK formulated the MCC950 drug dose and analyzed plasma. MBP performed pathological analysis and scoring. J Wu, AR, AL, and KS designed the study, analyzed and interpreted data, supervised the study, and wrote the manuscript.
Supplementary Material
Acknowledgments
The work was supported by NIH grant R01 DK105821 (to KS), the National Natural Science Foundation of China (grant 81870487 to J Wu), and the NIH Intramural Research Program (DK043308 to JBK). Support was also received from Boehringer Ingelheim.
Version 1. 10/15/2021
Electronic publication
Footnotes
Conflict of interest: CMBK, JJK, and SSP are full-time employees of Boehringer Ingelheim Pharmaceuticals, Inc. This work was partially supported by Boehringer Ingelheim Pharmaceuticals, Inc. (MCC inhibitor studies). KS is on the advisory board of Jnana Therapeutics.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(20):e136329.https://doi.org/10.1172/JCI136329.
Contributor Information
Junnan Wu, Email: junnan.wu@pennmedicine.upenn.edu.
Archana Raman, Email: archanaraman2889@gmail.com.
Nathan J. Coffey, Email: Nathan.Coffey@pennmedicine.upenn.edu.
Xin Sheng, Email: xin.sheng@pennmedicine.upenn.edu.
Joseph Wahba, Email: jwahba@sas.upenn.edu.
Matthew J. Seasock, Email: Matthew.Seasock@bcm.edu.
Ziyuan Ma, Email: ziyuan.ma@pennmedicine.upenn.edu.
Pazit Beckerman, Email: pazit.beckerman@gmail.com.
Matthew B. Palmer, Email: Matthew.Palmer@pennmedicine.upenn.edu.
Jeffrey B. Kopp, Email: jeffreyk@intra.niddk.nih.gov.
Jay J. Kuo, Email: jakekuo23@gmail.com.
Steven S. Pullen, Email: steven.pullen@boehringer-ingelheim.com.
Carine M. Boustany-Kari, Email: carine.boustany@boehringer-ingelheim.com.
Andreas Linkermann, Email: andreas.linkermann@uniklinikum-dresden.de.
Katalin Susztak, Email: ksusztak@pennmedicine.upenn.edu.
References
- 1.Beckerman P, Susztak K. APOL1: the balance imposed by infection, selection, and kidney disease. Trends Mol Med. 2018;24(8):682–695. doi: 10.1016/j.molmed.2018.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Norton JM, et al. Social determinants of racial disparities in CKD. J Am Soc Nephrol. 2016;27(9):2576–2595. doi: 10.1681/ASN.2016010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Genovese G, et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010;329(5993):841–845. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bajaj A, et al. Phenome-wide association analysis suggests the APOL1 linked disease spectrum primarily drives kidney-specific pathways. Kidney Int. 2020;97(5):1032–1041. doi: 10.1016/j.kint.2020.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freedman BI, et al. APOL1 at 10 years: progress and next steps. Kidney Int. 2021;99(6):1296–1302. doi: 10.1016/j.kint.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kruzel-Davila E, et al. APOL1 nephropathy: from gene to mechanisms of kidney injury. Nephrol Dial Transplant. 2016;31(3):349–358. doi: 10.1093/ndt/gfu391. [DOI] [PubMed] [Google Scholar]
- 7.Beckerman P, et al. Transgenic expression of human APOL1 risk variants in podocytes induces kidney disease in mice. Nat Med. 2017;23(4):429–438. doi: 10.1038/nm.4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weckerle A, et al. Characterization of circulating APOL1 protein complexes in African Americans. J Lipid Res. 2016;57(1):120–130. doi: 10.1194/jlr.M063453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shukha K, et al. Most ApoL1 is secreted by the liver. J Am Soc Nephrol. 2017;28(4):1079–1083. doi: 10.1681/ASN.2016040441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Molina-Portela MP, et al. Distinct roles of apolipoprotein components within the trypanosome lytic factor complex revealed in a novel transgenic mouse model. J Exp Med. 2008;205(8):1721–1728. doi: 10.1084/jem.20071463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reeves-Daniel AM, et al. The APOL1 gene and allograft survival after kidney transplantation. Am J Transplant. 2011;11(5):1025–1030. doi: 10.1111/j.1600-6143.2011.03513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freedman BI, et al. Apolipoprotein L1 gene variants in deceased organ donors are associated with renal allograft failure. Am J Transplant. 2015;15(6):1615–1622. doi: 10.1111/ajt.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freedman BI, et al. APOL1 genotype and kidney transplantation outcomes from deceased African American donors. Transplantation. 2016;100(1):194–202. doi: 10.1097/TP.0000000000000969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee BT, et al. The APOL1 genotype of African American kidney transplant recipients does not impact 5-year allograft survival. Am J Transplant. 2012;12(7):1924–1928. doi: 10.1111/j.1600-6143.2012.04033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riella LV, Sheridan AM. Testing for high-risk APOL1 alleles in potential living kidney donors. Am J Kidney Dis. 2015;66(3):396–401. doi: 10.1053/j.ajkd.2015.04.046. [DOI] [PubMed] [Google Scholar]
- 16.Olabisi OA, et al. APOL1 kidney disease risk variants cause cytotoxicity by depleting cellular potassium and inducing stress-activated protein kinases. Proc Natl Acad Sci U S A. 2016;113(4):830–837. doi: 10.1073/pnas.1522913113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aghajan M, et al. Antisense oligonucleotide treatment ameliorates IFN-γ-induced proteinuria in APOL1-transgenic mice. JCI Insight. 2019;4(12):e126124. doi: 10.1172/jci.insight.126124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nichols B, et al. Innate immunity pathways regulate the nephropathy gene apolipoprotein L1. Kidney Int. 2015;87(2):332–342. doi: 10.1038/ki.2014.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Markowitz GS, et al. Treatment with IFN-{alpha}, -{beta}, or -{gamma} is associated with collapsing focal segmental glomerulosclerosis. Clin J Am Soc Nephrol. 2010;5(4):607–615. doi: 10.2215/CJN.07311009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okamoto K, et al. APOL1 risk allele RNA contributes to renal toxicity by activating protein kinase R. Commun Biol. 2018;1:188. doi: 10.1038/s42003-018-0188-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomson R, Finkelstein A. Human trypanolytic factor APOL1 forms pH-gated cation-selective channels in planar lipid bilayers: relevance to trypanosome lysis. Proc Natl Acad Sci U S A. 2015;112(9):2894–2899. doi: 10.1073/pnas.1421953112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greene AS, Hajduk SL. Trypanosome lytic factor-1 initiates oxidation-stimulated osmotic lysis of trypanosoma brucei brucei. J Biol Chem. 2016;291(6):3063–3075. doi: 10.1074/jbc.M115.680371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Molina-Portela Mdel P, et al. Trypanosome lytic factor, a subclass of high-density lipoprotein, forms cation-selective pores in membranes. Mol Biochem Parasitol. 2005;144(2):218–226. doi: 10.1016/j.molbiopara.2005.08.018. [DOI] [PubMed] [Google Scholar]
- 24.Perez-Morga D, et al. Apolipoprotein L-I promotes trypanosome lysis by forming pores in lysosomal membranes. Science. 2005;309(5733):469–472. doi: 10.1126/science.1114566. [DOI] [PubMed] [Google Scholar]
- 25.Jha A, et al. Alterations in plasma membrane ion channel structures stimulate NLRP3 inflammasomes activation in APOL1 risk milieu. FEBS J. 2020;287(10):2000–2022. doi: 10.1111/febs.15133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chun J, et al. Recruitment of APOL1 kidney disease risk variants to lipid droplets attenuates cell toxicity. Proc Natl Acad Sci U S A. 2019;116(9):3712–3721. doi: 10.1073/pnas.1820414116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shah SS, et al. APOL1 kidney risk variants induce cell death via mitochondrial translocation and opening of the mitochondrial permeability transition pore. J Am Soc Nephrol. 2019;30(12):2355–2368. doi: 10.1681/ASN.2019020114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhaorigetu S, et al. ApoL1, a BH3-only lipid-binding protein, induces autophagic cell death. Autophagy. 2008;4(8):1079–1082. doi: 10.4161/auto.7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lan X, et al. APOL1 risk variants enhance podocyte necrosis through compromising lysosomal membrane permeability. Am J Physiol Renal Physiol. 2014;307(3):F326–F336. doi: 10.1152/ajprenal.00647.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kruzel-Davila E, et al. APOL1-mediated cell injury involves disruption of conserved trafficking processes. J Am Soc Nephrol. 2017;28(4):1117–1130. doi: 10.1681/ASN.2016050546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wakashin H, et al. APOL1 renal risk variants exacerbate podocyte injury by increasing inflammatory stress. BMC Nephrol. 2020;21(1):371. doi: 10.1186/s12882-020-01995-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakairi T, et al. Conditionally immortalized human podocyte cell lines established from urine. Am J Physiol Renal Physiol. 2010;298(3):F557–F567. doi: 10.1152/ajprenal.00509.2009. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.O’Toole JF, et al. ApoL1 overexpression drives variant-independent cytotoxicity. J Am Soc Nephrol. 2018;29(3):869–879. doi: 10.1681/ASN.2016121322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bruno J, et al. Apolipoprotein L1 confers pH-switchable ion permeability to phospholipid vesicles. J Biol Chem. 2017;292(44):18344–18353. doi: 10.1074/jbc.M117.813444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giovinazzo JA, et al. Apolipoprotein L-1 renal risk variants form active channels at the plasma membrane driving cytotoxicity. Elife. 2020;9:e51185. doi: 10.7554/eLife.51185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kovarova M, et al. NLRP1-dependent pyroptosis leads to acute lung injury and morbidity in mice. J Immunol. 2012;189(4):2006–2016. doi: 10.4049/jimmunol.1201065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chung KW, et al. Mitochondrial damage and activation of the STING pathway lead to renal inflammation and fibrosis. Cell Metab. 2019;30(4):784–799. doi: 10.1016/j.cmet.2019.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coll RC, et al. A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases. Nat Med. 2015;21(3):248–255. doi: 10.1038/nm.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Primiano MJ, et al. Efficacy and pharmacology of the NLRP3 inflammasome inhibitor CP-456,773 (CRID3) in murine models of dermal and pulmonary inflammation. J Immunol. 2016;197(6):2421–2433. doi: 10.4049/jimmunol.1600035. [DOI] [PubMed] [Google Scholar]
- 40.Sampson MG, et al. Integrative genomics identifies novel associations with APOL1 risk genotypes in Black NEPTUNE subjects. J Am Soc Nephrol. 2016;27(3):814–823. doi: 10.1681/ASN.2014111131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Limou S, et al. Sequencing rare and common APOL1 coding variants to determine kidney disease risk. Kidney Int. 2015;88(4):754–763. doi: 10.1038/ki.2015.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Freedman BI, et al. End-stage renal disease in African Americans with lupus nephritis is associated with APOL1. Arthritis Rheumatol. 2014;66(2):390–396. doi: 10.1002/art.38220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kopp JB, et al. Clinical features and histology of apolipoprotein L1-associated nephropathy in the FSGS Clinical Trial. J Am Soc Nephrol. 2015;26(6):1443–1448. doi: 10.1681/ASN.2013111242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lipkowitz MS, et al. Apolipoprotein L1 gene variants associate with hypertension-attributed nephropathy and the rate of kidney function decline in African Americans. Kidney Int. 2013;83(1):114–120. doi: 10.1038/ki.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Papeta N, et al. APOL1 variants increase risk for FSGS and HIVAN but not IgA nephropathy. J Am Soc Nephrol. 2011;22(11):1991–1996. doi: 10.1681/ASN.2011040434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Langefeld CD, et al. Genome-wide association studies suggest that APOL1-environment interactions more likely trigger kidney disease in African Americans with nondiabetic nephropathy than strong APOL1-second gene interactions. Kidney Int. 2018;94(3):599–607. doi: 10.1016/j.kint.2018.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hayek SS, et al. A tripartite complex of suPAR, APOL1 risk variants and αvβ3 integrin on podocytes mediates chronic kidney disease. Nat Med. 2017;23(8):945–953. doi: 10.1038/nm.4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sarhan M, et al. Origin and consequences of necroinflammation. Physiol Rev. 2018;98(2):727–780. doi: 10.1152/physrev.00041.2016. [DOI] [PubMed] [Google Scholar]
- 49.Vilaysane A, et al. The NLRP3 inflammasome promotes renal inflammation and contributes to CKD. J Am Soc Nephrol. 2010;21(10):1732–1744. doi: 10.1681/ASN.2010020143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Correa-Costa M, et al. Pivotal role of Toll-like receptors 2 and 4, its adaptor molecule MyD88, and inflammasome complex in experimental tubule-interstitial nephritis. PLoS One. 2011;6(12):e29004. doi: 10.1371/journal.pone.0029004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gong W, et al. NLRP3 deletion protects against renal fibrosis and attenuates mitochondrial abnormality in mouse with 5/6 nephrectomy. Am J Physiol Renal Physiol. 2016;310(10):F1081–F1088. doi: 10.1152/ajprenal.00534.2015. [DOI] [PubMed] [Google Scholar]
- 52.Fu R, et al. Podocyte activation of NLRP3 inflammasomes contributes to the development of proteinuria in lupus nephritis. Arthritis Rheumatol. 2017;69(8):1636–1646. doi: 10.1002/art.40155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murakami T, et al. Critical role for calcium mobilization in activation of the NLRP3 inflammasome. Proc Natl Acad Sci U S A. 2012;109(28):11282–11287. doi: 10.1073/pnas.1117765109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee H, et al. Caspase inhibitors: a review of recently patented compounds (2013-2015) Expert Opin Ther Pat. 2018;28(1):47–59. doi: 10.1080/13543776.2017.1378426. [DOI] [PubMed] [Google Scholar]
- 55.Mertens M, Singh JA. Anakinra for rheumatoid arthritis: a systematic review. J Rheumatol. 2009;36(6):1118–1125. doi: 10.3899/jrheum.090074. [DOI] [PubMed] [Google Scholar]
- 56.Ridker PM, et al. Antiinflammatory therapy with canakinumab for atherosclerotic disease. N Engl J Med. 2017;377(12):1119–1131. doi: 10.1056/NEJMoa1707914. [DOI] [PubMed] [Google Scholar]
- 57.May RM, et al. A multi-center retrospective cohort study defines the spectrum of kidney pathology in Coronavirus 2019 Disease (COVID-19) Kidney Int. doi: 10.1016/j.kint.2021.07.015. [published online August 2, 2021]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aarreberg LD, et al. Interleukin-1β induces mtDNA release to activate innate immune signaling via cGAS-STING. Mol Cell. 2019;74(4):801–815. doi: 10.1016/j.molcel.2019.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takemoto M, et al. A new method for large scale isolation of kidney glomeruli from mice. Am J Pathol. 2002;161(3):799–805. doi: 10.1016/S0002-9440(10)64239-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Harrow J, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22(9):1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Robinson MD, et al. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.