Abstract
Neoantigens are now recognized drivers of the antitumor immune response. Recurrent neoantigens, shared among groups of patients, have thus become increasingly coveted therapeutic targets. Here, we report on the data-driven identification of a robustly presented, immunogenic neoantigen that is derived from the combination of HLA-A*01:01 and RAS.Q61K. Analysis of large patient cohorts indicated that this combination applies to 3% of patients with melanoma. Using HLA peptidomics, we were able to demonstrate robust endogenous presentation of the neoantigen in 10 tumor samples. We detected specific reactivity to the mutated peptide within tumor-infiltrating lymphocytes (TILs) from 2 unrelated patients, thus confirming its natural immunogenicity. We further investigated the neoantigen-specific clones and their T cell receptors (TCRs) via a combination of TCR sequencing, TCR overexpression, functional assays, and single-cell transcriptomics. Our analysis revealed a diverse repertoire of neoantigen-specific clones with both intra- and interpatient TCR similarities. Moreover, 1 dominant clone proved to cross-react with the highly prevalent RAS.Q61R variant. Transcriptome analysis revealed a high association of TCR clones with specific T cell phenotypes in response to cognate melanoma, with neoantigen-specific cells showing an activated and dysfunctional phenotype. Identification of recurrent neoantigens and their reactive TCRs can promote “off-the-shelf” precision immunotherapies, alleviating limitations of personalized treatments.
Keywords: Genetics, Oncology
Keywords: Antigen, MHC class 1, Melanoma
Introduction
Immunotherapy sparked new hope for oncology in recent years, due to its remarkable ability to induce long-term tumor regression of metastatic cancer. This feature is shared across immunotherapeutic modalities, including both checkpoint blockade and adoptive cell transfer (ACT) of tumor-infiltrating lymphocytes (TILs). It is believed that the final common pathway of these 2 treatments is the specific recognition of tumor antigens by cytotoxic T lymphocytes (1–3). In-depth dissection of immunotherapy success stories revealed a center-stage role for mutation-derived antigens, designated as neoantigens, in mediating the antitumor immune response (4–11).
Neoantigens are cell-surface peptide–HLA complexes in which the peptide component, i.e., the neopeptide, is the altered degradation product of a mutated protein. Restricted in expression to the diseased tissue, and uncurbed by immune tolerance, neoantigens may elicit specific antitumor reactivity upon T cell receptor (TCR) engagement and are therefore ideal therapeutic targets.
The grand majority of neoantigens identified from treated patients derive from private, nonrecurring, mutations and thus, although effective, cannot be generalized beyond the individual patient (12, 13). Hotspot neoantigens, i.e., neoantigens that appear in a large group of patients with cancer, are highly sought after for 2 main reasons. First, hotspot neoantigens may pave the way toward “off-the-shelf” cellular treatments, vaccines, and patient screening strategies (14). Tumor cells expressing validated mutation/HLA combinations should be amenable to immunotherapy. Even in the absence of a priori immune recognition, predetermined TCRs, from other patients or even healthy donors, can be used to redirect autologous T cells against neglected hotspot neoantigens (15). Second, hotspot neoantigens are potentially superior to private neoantigens as treatment targets. This is because immunotherapy directed at subclonal mutations of heterogeneous tumors might give way to immune escape, whereas hotspot neoantigens, which are typically derived from clonal oncogenic mutations, are expected to present more homogenously within tumors.
Few potentially recurrent neoantigens have been uncovered over the years, stemming from major oncogenes, such as BRAF, NRAS, and p53 (16–19). The clinical relevance of such neoantigens, however, was directly demonstrated only of late, through successful ACT treatment of a patient with metastatic colon cancer, using autologous TILs directed at a newly discovered HLA-C*08:02/KRAS.G12D recurrent neoantigen (7, 20). These recent successes propelled the reemergence of endeavors to discover hotspot neoantigens. A p53-centered screen revealed native TIL reactivity toward derived neoantigens in 8% of screened patients, with recurring neoantigens identified in several cases (21, 22). Other efforts focused on the identification of T cells from the peripheral blood of patients or healthy donors that target specific hotspots, thereby expanding the repertoire of known KRAS and other oncogene-derived neoantigens for both HLA class I and HLA class II (23, 24).
Some of the above-mentioned recurrent neoantigens were uncovered by serendipity. In other studies, efforts were focused on hotspot mutations and/or highly prevalent HLA alleles. Truly recurrent neoantigens, however, may only form at the productive intersection of recurrent oncogenic mutations and common HLA alleles. As early events in neoplasm development, recurrent oncogenic mutations are potentially vulnerable to the effects of immunoediting (25). Recent bioinformatic analysis found patients with cancer to be less likely to have oncogenic mutations that are predicted to bind their HLA alleles (26), whereas others did not find evidence for such skewing (27). To properly select for high-yield candidates, HLA allele/mutation co-occurrences should best be evaluated within the potentially skewed cancer patient population. With patients’ sequencing data becoming increasingly available, such analyses are now feasible. Moreover, HLA-binding algorithms, which are well trained on prevalent HLA alleles, may help point to the most promising HLA-mutation combinations.
To date, neoantigen discovery efforts are almost exclusively T cell centric. In these methods, candidate neopeptides are artificially expressed in antigen-presenting cells (APCs), either as pulsed synthetic peptides or via minigene overexpression. APCs are then coincubated with T cells, most commonly TILs, and their response profile interpreted for indirect identification of neoantigens. Further characterization and validation rely heavily on binding predictions, such that identified neoantigens are restricted to those that are both predicted to bind and are immunogenic in the tested patient (15, 20). Moreover, presented neoantigens that do not induce reactivity in the tested individual may be overlooked (12). HLA peptidomics uses immunoaffinity purification of peptide-HLA complexes followed by liquid chromatography tandem mass spectrometry (LC/MS-MS) to characterize the actively presented antigen repertoire. Contrary to the methods described above, it enables the direct identification of neoantigens from tumor samples (28). In our laboratory, we used HLA peptidomics successfully to identify neoantigens from tumor cells (12, 13).
The causal role of RAS proteins in cancer has long been recognized, with activating mutations appearing in one-third of all human cancers (29). RAS-derived recurrent neoantigens are thus of great interest (7). The 3 main isoforms, KRAS, NRAS, and HRAS, share identical, 86-amino-acid-long N-termini. Within this identical stretch, 3 mutational hotspots were recognized at positions 12, 13, and 61. KRAS is the most highly mutated RAS isoform across cancers (85% of RAS mutations). However, in melanoma, the most successful immunotherapy target to date, NRAS mutations dominate. Specifically, NRAS.61 is the second most highly mutated position in melanoma, appearing in as many as 20% of patients. NRAS-mutant melanoma is associated with poorer outcomes, compared with non–NRAS-mutant melanoma. Multiple attempts to develop RAS-targeted therapy have yet to yield effective approved treatments. In the void of successful NRAS-targeted agents, immunotherapy occupies a key role in current therapeutic regimens aimed at NRAS-mutant melanoma.
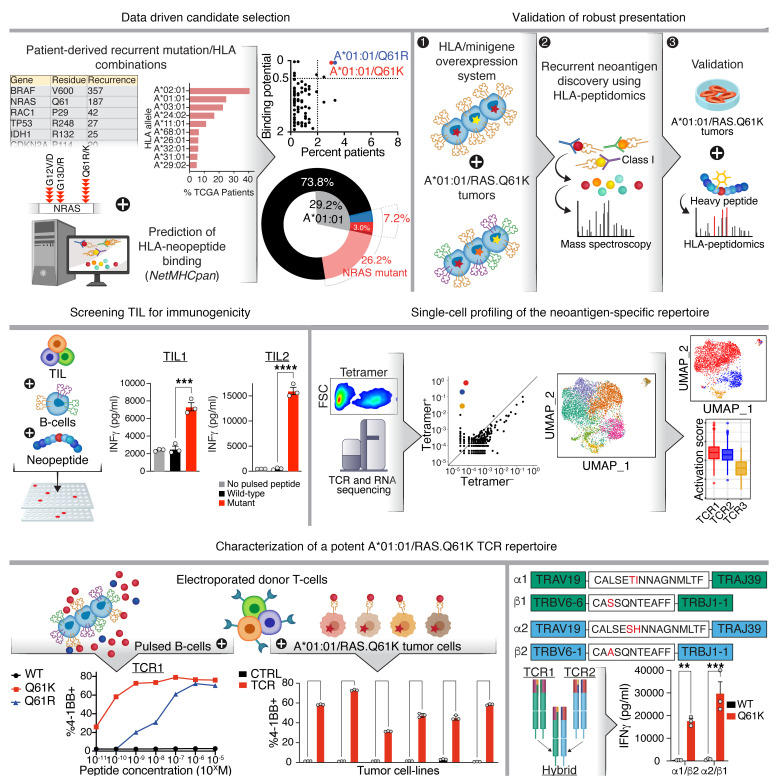
Here, we used a data-driven pipeline for the discovery of recurrent neoantigens, with combined analysis of presentation and immunogenicity (Figure 1). We report on the discovery of a robustly presented and immunogenic RAS.61-derived hotspot neoantigen present in a substantial group of cancer patients. We further identified the neoantigen-specific TCRs within TILs from 2 unrelated patients and transcriptionally characterized the neoantigen-specific T cell response at the single-cell level.
Figure 1. Recurrent neoantigen discovery pipeline.
Data-driven analysis of cancer patient cohorts was used to select high-recurrence RAS.Q61/HLA allele combinations that were predicted to yield neoantigens, with HLA-A*01:01/RAS.Q61 heading the list. Robust neoantigen presentation was corroborated and quantified via HLA peptidomics using a panel of tumor samples with endogenous expression. Specific reactivity was identified within TILs from 2 unrelated patients, thus validating the immunogenicity of the neoantigen. Tetramer-specific cells were TCR- and RNA-sequenced at the single-cell level, allowing transcriptional profiling of neoantigen-specific clones. A repertoire of sensitive and specific TCRs were characterized, with striking intra- and interpatient TCR sequence similarity between clones. Parts of Figure 1 were created using BioRender.com.
Results
Data-driven selection of neoantigen candidates.
NRAS.Q61 is the second most highly mutated protein position in melanoma, but not all of its derived neoantigens will be equally recurrent. To qualify as an “interesting” hotspot neoantigen, its combined HLA allele/mutation frequency should be high in patients with cancer (26). Here, we applied a data-driven approach to neoantigen discovery by analyzing patients’ genetic data to focus on high-potential candidates. Considering only those HLA alleles that co-occurred with RAS.Q61 mutations in the analyzed data sets, we used NetMHCpan to predict binding, 8- to 14-amino-acid-long, RAS.Q61-derived neopeptides (30, 31). Using these data, we assigned each HLA-mutation combination 2 scores: (a) its frequency in the cohort, and (b) its best binding prediction score (i.e., best percentage rank [referred to hereafter as “%Rank”] enumerated over the predicted binding neopeptides). We then applied stepwise filtration and sorting, setting a frequency threshold and then ordering the remaining candidates according to their estimated binding potential.
Because the 3 RAS isoforms, i.e., NRAS, KRAS, and HRAS, have an identical N-terminus sequence, we included them all in our analyses, along with the 4 most prevalent RAS.Q61 amino acid substitutions: arginine, lysine, leucine, and histidine.
Using the above-described scheme, we analyzed 3 melanoma cohorts: The Cancer Genome Atlas (TCGA), aggregating both mutation and HLA-I annotations for 363 melanoma cases, the Hartwig database, containing data on 221 such patients, and the previously published MELA-AU data set for 69 patients (32–35).
Ninety-five (26%), 60 (27%), and 15 (22%) individuals within these cohorts have RAS.Q61-mutated tumors, respectively (Figure 2, D–F, and Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI129466DS1). As expected, we found that the most frequent amino acid substitutions at position 61 were arginine (46%–60%) and lysine (35%–47%); and NRAS was the most abundantly mutated RAS isoform at position 61 (96%–100% of RAS.61 mutations). Comparison of HLA allele frequencies between TCGA patients with RAS.Q61-mutant and those with WT RAS.Q61 melanoma did not reveal significant prevalence skews (Fisher’s exact test with FDR correction, Supplemental Figure 1 and Supplemental Table 1).
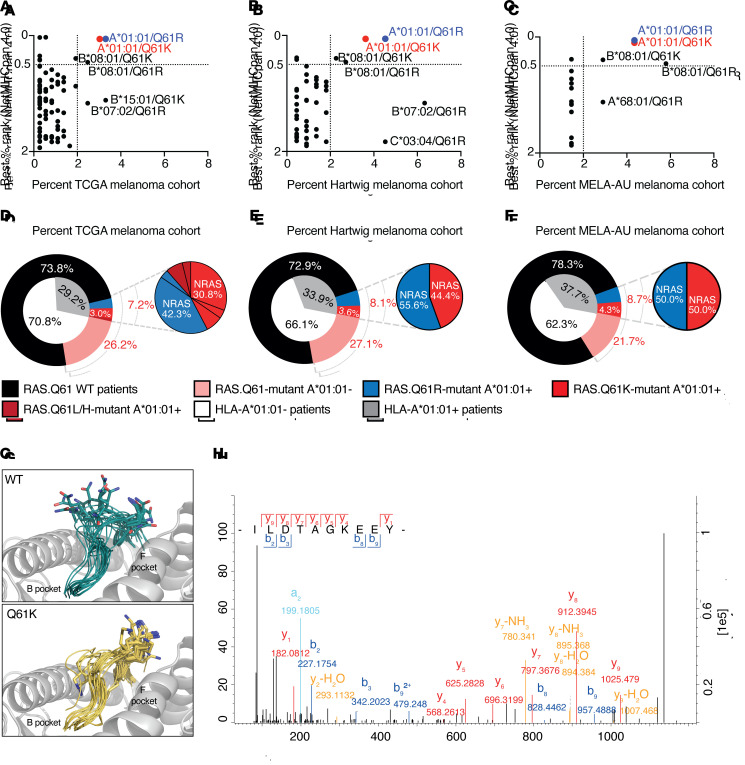
Figure 2. From data-driven candidate selection to verification of presentation.
(A–C) Percentage of patients with HLA/RAS.Q61 combination in cohorts of patients with melanoma (x axis) and NetMHCpan 4.0 binding predictions Best%Rank (y axis). %Rank≤0.5 were considered strong binders; 0.5<%Rank≤2 were considered weak binders. (A) TCGA, (B) Hartwig, (C) MELA-AU melanoma cohorts. (D–F) Frequency of A*01:01/RAS.Q61-mutant combinations in melanoma cohorts. (D) TCGA, (E) Hartwig, (F) MELA-AU melanoma cohorts. (G) Predicted structures of A*01:01 in complex with RAS peptides. Gray indicates HLA; turquoise indicates ILDTAGQEEY (WT); yellow indicates ILDTAGKEEY (mutant, RAS.Q61K). The peptide backbones are represented as ribbons, with P7 residue (position 61) side-chain atoms shown. Hydrogens were omitted for clarity. (H) Tandem mass spectra of ILDTAGKEEY as it was identified in HLA peptidomics of the melanoma cell line 17T, endogenously expressing HLA-A*01:01/NRAS.Q61K. Method: Fourier transform mass spectrometry (FTMS); higher-energy collisional dissociation (HCD) score: 91.55; m/z: 569.79. Peptide identification was further validated in 2 separate targeted MS repeats.
Plotting HLA/mutation combinations by their scores was demonstrative of how the most promising candidates were prioritized (Figure 2, A–C). Across the 3 data sets, HLA-A*01:01 stood out for contributing the top 2 candidates: HLA-A*01:01/RAS.Q61R and HLA-A*01:01/RAS.Q61K. We therefore selected HLA-A*01:01 for further analysis.
Consistent with its high abundance in the general population, 26%–38% of patients in the analyzed cancer cohorts were found to carry HLA-A*01:01 (35). Importantly, the allele’s frequency did not diminish when restricted to the RAS.Q61-mutant melanoma population: 27%–40% of mutants possessed it. HLA-A*01:01 co-appeared with RAS.Q61 mutations in 7%–8.7% of patients with melanoma. Specifically, 3.3%–4.5% and 3%–4.3% of patients had the combinations HLA-A*01:01/RAS.Q61R and HLA-A*01:01/RAS.Q61K, respectively, in our analysis.
NetMHCpan 4.0 predicted 32 RAS.Q61-derived neopeptides for A*01:01, including 9 predicted strong binders (Supplemental Table 1). Importantly, the same “canonical peptide,” ILDTAGXEEY, was the top binding prediction for both RAS.Q61R and RAS.Q61K (predicted binding affinities of 202.4 nM and 218.5 nM, respectively), while its less prevalent variants (RAS.Q61L and RAS.Q61H) were also predicted to bind the A*01:01 allele (58.2 nM and 101.8 nM, respectively).
Of note, the pan-cancer analysis recapitulated our melanoma-based candidate prioritization (Supplemental Figure 1). Still frequent, RAS.Q61 mutations appeared in 3.3%–5.9% of the pan-cancer cases (226 of 6824 and 123 of 2091 individuals from TCGA and Hartwig data sets, respectively). The relevance of an HLA-A*01:01/RAS.Q61 neoantigen would therefore not be restricted to melanoma.
Direct identification of HLA-A*01:01/RAS.Q61–derived neoantigens using a monoallelic overexpression system.
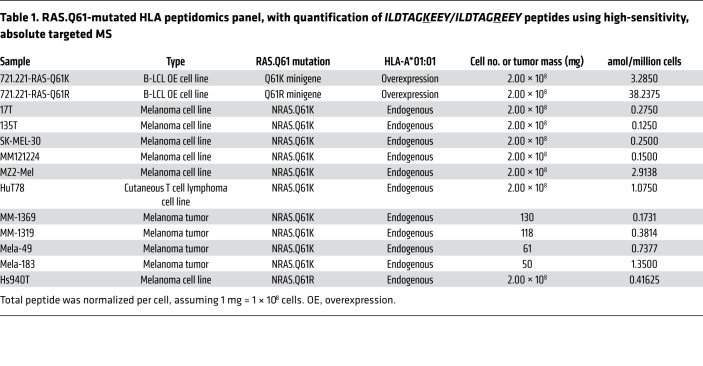
Unlike for RAS.Q61R, no neoantigen had been previously reported for the highly recurrent HLA-A*01:01/RAS.Q61K combination (16, 36). Focusing on RAS.Q61K, we therefore set out to query the HLA-A*01:01–bound neoantigen landscape in an unbiased manner. To this end, we established a 721.221-based HLA-A*01:01 monoallelic cell line that co-overexpressed a 25 mer RAS.Q61K minigene and subjected it to HLA peptidomics. This overexpression setup had the advantages of availability and neoantigen amplification, while still maintaining native antigen processing and presentation. Mass spectrometry (MS) results were analyzed using MaxQuant (37) and queried against the human proteome data set (Uniprot), to which we manually added the minigene sequence. We detected a total of 2403 peptides for 721.221A*01:01;mRAS.Q61K (see “721-OE-HLA-I peptidomics” in Supplemental Table 2). Although NetMHCpan predicted 7 different HLA-A*01:01–binding neopeptides for RAS.Q61K (see “input 27 mers” in Supplemental Table 1), a single decapeptide, ILDTAGKEEY, was identified. We further validated this finding by comparing the identification spectra with that of a synthetic peptide (Supplemental Figure 2). Interestingly, ILDTAGKEEY and the closely related ILDTAGREEY were the best-scoring neopeptide predictions for RAS.Q61K and RAS.Q61R, respectively, in the context of HLA-A*01:01. We quantified neopeptide presentation in our overexpression system, as well as validated the presentation of RAS.Q61R-derived ILDTAGREEY using high-sensitivity absolute targeted MS. ILDTAGKEEY and ILDTAGREEY were detected at 3.2850 and 38.2375 amol per 1 × 106 cells for 721.221A*01:01;mRAS.Q61K and 721.221A*01:01;mRAS.Q61R, respectively.
ILDTAGKEEY is robustly presented on HLA-A*01:01/RAS.Q61K–expressing tumors.
To further validate our findings in the malignant context and under endogenous mutation/HLA expression, we subjected the 17T tumor–derived cell line to HLA peptidomics. As previous whole-exome efforts in our laboratory (38) identified 17T melanoma as NRAS.Q61K mutant, and HLA typing granted it HLA-A*01:01+, we used MS in discovery mode to unbiasedly identify neoantigens, regardless of prediction or prior identifications. The database for MS consisted of all normal sequences in the human proteome as well as of mutated protein sequences as defined by 17T whole-exome sequencing. Indeed, among 2356 peptides, a single neopeptide was detected — the decapeptide ILDTAGKEEY (Figure 2H and see “17T HLA-I peptidomics” in Supplemental Table 2).
To investigate the robustness of presentation, we curated a panel of 4 snap-frozen tumors and 5 additional tumor-derived cell lines, all endogenously expressing the HLA-A*01:01/NRAS.Q61K combination (Table 1). We included Hs940T, an HLA-A*01:01/NRAS.Q61R melanoma cell line, in our analysis to also test for endogenous presentation of ILDTAGREEY. All cell lines were validated as expressing mutant NRAS transcripts (Supplemental Figures 3 and 4).
Table 1. RAS.Q61-mutated HLA peptidomics panel, with quantification of ILDTAGKEEY/ILDTAGREEY peptides using high-sensitivity, absolute targeted MS.
HLA peptidomics using high-sensitivity absolute targeted MS identified the ILDTAGKEEY neopeptide in all NRAS.Q61K-mutant samples at quantities ranging from 0.1250 to 2.9138 amol per 1 × 106 cells, assuming 1 × 108 cells per 1 mg tumor tissue (Supplemental Figures 5–16 and ref. 39). As expected, we identified ILDTAGREEY for Hs940T (Supplemental Figure 17). Importantly, the HuT78 cell line from our panel was derived from a T cell lymphoma, signifying the cross-cancer-type relevance of the identified neoantigens.
We conclude that ILDTAGKEEY is a robust, naturally processed, RAS.Q61K-derived neopeptide that is presented in the context of HLA allele A*01:01.
Mutant lysine of HLA-A*01:01/ILDTAGKEEY is free to interact with TCRs.
WT ILDTAGQEEY was shown to strongly bind HLA-A*01:01 (40), prompting us to gauge the availability of mutant lysine to interact with TCRs. We threaded the side chains of ILDTAGKEEY and ILDTAGQEEY onto the backbone of a previously resolved HLA-A*01:01/ALK decapeptide crystal structure (Protein Data Bank [PDB]: 6at9) (41) and sampled conformations by 5 independent molecular dynamics (MD) trajectories per complex as well as by docking using freely accessible web server interfaces for FlexPepDock (42), ClusPro (43), and DINC (44). Hydrogen bond analysis indicated that mutant and WT variants interacted with HLA-A*01:01 in a similar manner. Hydrogen bonds were observed between the HLA receptor for both the WT and mutant variants (Supplemental Figure 18A) at position 3 (P3) and the C-terminus (PΩ) as expected (45). In contrast, P7 (residue 61) formed fewer hydrogen bonds with the HLA receptor. While P7’s backbone remained within the peptide-binding groove throughout the simulation, the side chain was solvent-exposed and directed toward the TCR-facing surface (Figure 2G). This structural modeling suggests that residue 61 faces outward of the HLA pocket and is available to interact with TCRs, possibly supporting differential immunogenicity toward the mutant versus WT peptides.
HLA-A*01:01/ILDTAGKEEY is immunogenic.
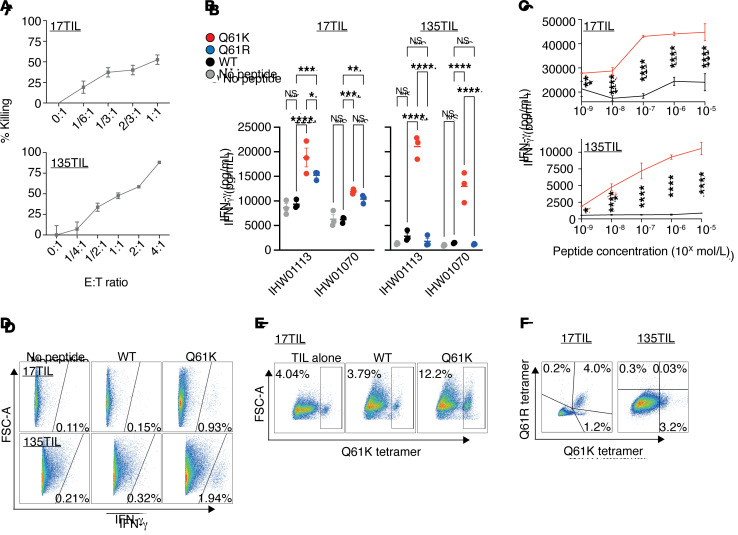
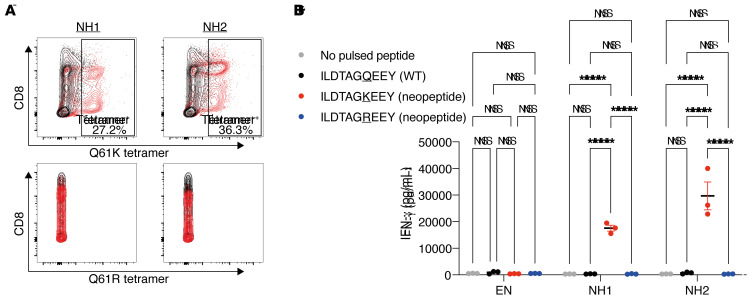
We used the TIL products 17TIL/135TIL, relating to melanoma cell lines 17T and 135T of our panel, respectively, to evaluate the immunogenicity of the identified RAS.Q61K neoantigen. These TIL products have a killing capacity toward cognate melanoma, as demonstrated in GFP-based killing assays (Figure 3A). To distinguish neoantigen-specific from bulk reactivity, we quantified IFN-γ release by the TIL products in response to peptide-pulsed, HLA-A*01:01+–presenting cells. B lymphoblastoid cell lines (B-LCLs) (46) were pulsed with either a DMSO vehicle or WT (ILDTAGQEEY) or mutant (ILDTAGKEEY or ILDTAGREEY) synthetic peptides, followed by overnight coincubation with either 17TIL or 135TIL, and then IFN-γ ELISA of the coincubation medium (Supplemental Table 3). As depicted in Figure 3B, RAS.Q61K stimulated IFN-γ release from both 17TIL and 135TIL, whereas the WT variant did not elicit a significant increase over a nonpulsed B cell control. Remarkably, although 17T is not a RAS.Q61R mutant, 17TIL exhibited cross-reactivity with RAS.Q61R. Peptide titration assays confirmed dose-dependent reactivity with HLA-A*01:01/ILDTAGKEEY, for both 17TIL and 135TIL (Figure 3C).
Figure 3. A*01:01/ILDTAGKEEY is immunogenic.
(A) Bulk TILs were able to kill cognate melanoma in a fluorescent cell in vitro–killing assay. The fluorescently labeled melanoma cell line 17T was coincubated with IHW01161 and the cell line 135T was coincubated with IHW01070. Error bars represent the SEM of biological triplicates. (B and C) Bulk TILs were specifically reactive toward the neoantigen. TILs were coincubated with minimal epitope–pulsed A*01:01+ B-LCLs. (B) IFN-γ ELISA results. Error bars represent the SEM of biological triplicates; 2-way ANOVA with Tukey’s correction for multiple comparisons. (C) IFN-γ ELISA peptide titration assay with 17TIL (IHW0116) and 135TIL (IHW01070). Plots show the mean with SD of biological triplicates; 2-way ANOVA with Sidak’s correction for multiple comparisons. (D–F) Delineation of the neoantigen-specific subpopulations. (D) IFN-γ secretion assay of the melanoma cells 17TIL (coincubatad with peptide-pulsed IHW01161 cells) and 135TIL (coincubated with peptide-pulsed IHW01070 cells). P < 0.00001, by χ2 for both. (E) Differential expansion of 17TILs in response to coincubation with B-LCL IHW01161 and either the WT or mutant peptide. P < 0.00001, by χ2 test. (F) CD8+ gated TILs double-stained with the Q61K and Q61R tetramers. FSC-A, forward scatter area. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Flow cytometric analysis revealed a majority of CD4+ cells for 17TIL (58%–80%) and CD8+ for 135TIL (90%) (Supplemental Figure 19). A flow cytometry–based IFN-γ secretion assay and tetramer staining delineated the neoantigen-specific subpopulations (Supplemental Figure 20). We observed neoantigen-specific IFN-γ secretion in 0.93% and 1.94% of CD4– TILs for 17TIL and 135TIL, respectively, upon incubation with peptide-pulsed B-LCLs (P < 0.00001 for both, χ2 test, 10 μg/mL peptide, Figure 3D). With HLA-A*01:01/ILDTAGKEEY-tetramer staining, 5.2% and 3.2% of CD8+ cells bound RAS.Q61K, for 17TIL and 135TIL, respectively (1.46% and 2.8% of total TILs, respectively; Figure 3F). A proliferation assay, performed for 17TIL, demonstrated neoantigen-dependent expansion (P < 0.00001, χ2; Figure 3E). Moreover, costaining TILs with HLA-A*01:01/ILDTAGREEY-tetramer corroborated the cross-reactivity of 17TIL with this variant and confirmed that the majority (76%) of HLA-A*01:01/ILDTAGKEEY–binding cells cross-react with the arginine-substituted peptide (Figure 3F). CD8– subpopulations did not stain with the tetramer, as expected (Supplemental Figure 19).
We thus demonstrate specific TIL reactivity toward HLA-A*01:01/ILDTAGKEEY in 2 unrelated patients, 1 of whom showed cross-reactivity toward HLA-A*01:01/ILDTAGREEY. We conclude that both of these neoantigens are immunogenic.
Identification of neoantigen-binding TCR chains.
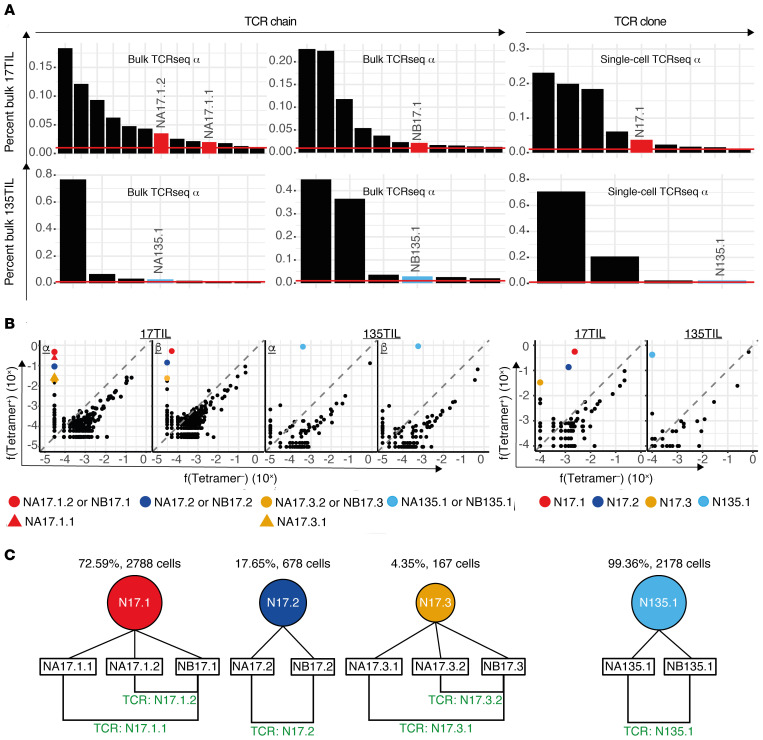
TCRs that target HLA-A*01:01/ILDTAGKEEY have translational value; ACT of engineered T cells using these TCRs is expected to apply to 3% of patients with melanoma, and to 2.9:1000 patients pan-cancer. We performed RNA-based TCR sequencing on tetramer-sorted TILs to identify relevant α and β chains. For 17TIL and 135TIL each, 3 cell populations were individually sequenced: bulk CD4– and its A*01:01/ILDTAGKEEY-tetramer+ and tetramer– subpopulations. We contrasted the tetramer+ and tetramer– repertoires in order to extract tetramer-enriched (i.e., tetramer-specific) chains, while sequencing of the bulk cell population allowed us to place these chains in context. Our sorting and sequencing experiments (3 biological replicates) were highly consistent (Supplemental Figure 21). Although each sequencing experiment revealed hundreds to thousands of distinct, productive TCR chains (Supplemental Table 4), both 17TIL and 135TIL clearly exhibited oligoclonal distributions (Figure 4). With restriction to amino acid sequences that reflected 1% or more of the transcripts, 11 β (13 α) chains dominated the CD4– repertoire of 17TIL, with cumulative coverage of 76.5% (69.6%) of the transcripts. Similarly, for 135TIL, 6 β (7 α) chains accounted for 92.5% (93.6%) of the CD4– repertoire. The tetramer+ and tetramer– subpopulations were likewise characterized by oligoclonal TCR distributions (Supplemental Figure 22). When we further focused on TCR chains with at least 100-fold tetramer+/tetramer– transcript frequency enrichment, 4 β and 6 α chains emerged as neoantigen specific for 17TIL (Figure 4), Table 2, and Supplemental Table 4). The cumulative frequencies for these 4 β chains were 69.6%, 3%, and 0.005% in the tetramer+, bulk CD4–, and tetramer– populations, respectively. Likewise, cumulative frequencies for the 6 α chains were 83.8%, 6.8%, and 0% in the tetramer+, bulk CD4–, and tetramer– populations, respectively. For 135TIL, a single αβ pair met the above criteria (Figure 4), with frequencies of 89.9% and 85.2% within tetramer+ 135TIL, respectively. These chains made up only 2.9% and 2.8% of the bulk CD4– α and β 135TIL repertoires, respectively. Interestingly, the α and β frequency distributions were consistent enough to enable an educated guess regarding chain pairing on the basis of bulk TCR sequencing (Figure 4).
Figure 4. Combined bulk and single-cell TCR sequencing identifies a repertoire of neoantigen-specific TCRs.
(A) 17TIL and 135TIL had oligoclonal distributions. Frequency distributions for bulk 17TIL (top) and 135TIL (bottom) are shown. Left panel: TCR α and β chain frequencies from bulk TCR sequencing of CD4– bulk populations. Right panel: Single-cell TCR sequencing of CD8+ bulk populations. Only chains/clones with a frequency of at least 1% are depicted. A representative result of 3 replicates is shown for the bulk TCR sequencing. (B) Tetramer+ versus tetramer– frequencies (f). Only TCR chains/clones that were enriched in the tetramer+ population (i.e., tetramer+/tetramer– >> 1) were considered tetramer specific and potentially neoantigen specific. Left panel: Bulk TCR sequencing α and β chains (a representative result of 3 replicates is shown). Right panel: Single-cell TCR sequencing. For validation, we focused on the TCRs that were greater than 100-fold tetramer+-enriched and made up at least 1% of the tetramer+ repertoire (see colored TCR chains/clones). (C) Nomenclature for the leading neoantigen-specific clones in 17TIL and 135TIL and their derived TCRs.
Table 2. List of tetramer-specific TCRs analyzed.
Single-cell characterization of the TCR repertoire.
To further investigate these TCR repertoires, and to uncover tetramer-binding αβ pairings, we performed single-cell TCR sequencing. After overnight coincubation of TILs and cognate melanoma cells (i.e., 17TIL with 17T, 135TIL with 135T), we sorted the samples into populations of interest (Supplemental Figure 23); we then proceeded to perform dual RNA-Seq and TCR single-cell sequencing using the 10x Genomics platform. As with our bulk sequencing experiment, we focused on 3 TIL populations: bulk CD8+ and its HLA-A*01:01/ILDTAGKEEY-tetramer+ and tetramer– subpopulations. We again used tetramer+/tetramer– frequency enrichment to hone in on tetramer-specific clones.
As expected, our bulk and single-cell sequencing data were highly consistent (Supplemental Figures 24–27). We identified 343 and 44 CD8+ clones for 17TIL and 135TIL, respectively, with a small subset of clones dominating the CD8+ repertoire (Figure 4). Using the same filtration criteria of 100-fold or greater enrichment and clonal expansion of at least 1% of tetramer+ cells, we identified 3 leading tetramer-specific clones for 17TIL and a single clone for 135TIL (Figure 4, Table 2, and Supplemental Figure 4). These tetramer-specific clones made up merely 5.1% and 2.2% of the CD8+ cells for 17TIL and 135TIL, respectively, with the dominant clone accounting for more than 70% of the tetramer-enriched subset. Importantly, our candidate chain pairings, deduced from bulk TCR sequencing, were confirmed by the single-cell data.
Clone and TCR chain nomenclature.
Tetramer-specific clones are hereafter defined as single-cell clones that were at least 100-fold tetramer+/tetramer– frequency enriched in both the single-cell and bulk TCR-sequencing experiments. Specifically, a clone was considered tetramer enriched according to bulk TCR sequencing if all of its TCR chains were tetramer enriched in at least 1 of the 3 bulk sequencing replicates.
This filtration retained a total of 15 tetramer-specific clones, 6 of which had more than 5 cells in the tetramer+ subpopulation (Supplemental Table 4). Therefore, we annotated these clones as N{TIL: 17/135}.{clone number} in descending order of frequency.
For functional validation, we focused, as above, on clones consisting of at least 1% of the tetramer-specific repertoire: these were the top 3 clones in 17TIL — N17.1, N17.2, and N17.3 — and a single dominant tetramer-specific clone for 135TIL, N135.1 (Figure 4). N17.2, N17.4, and N135.1 are all αβ cells, whereas N17.3 is ααβ. Conventional analysis pipelines deemed N17.1 an αβ clone. However, as will be further elucidated below, this clone has in fact a second α chain, rearranged with the δ gene TRDV1 (Supplemental Figure 28).
The respective TCR chains of the above-mentioned clones are designated as follows: N{chain type: A/B}{TIL: 17/135}.{clone number}.{chain number [where applicable]} (Figure 4). The TCR products of αβ clones will bear the same name as the clone (i.e., TCR N17.2 is the combination of NA17.2 and NB17.2), whereas for ααβ cells, the TCR name will be determined by the differentiating α chain (i.e., TCR N17.1.2 is the combination of NA17.1.2 and NB17.1).
Intra- and interpatient sequence convergence of neoantigen-specific TCRs.
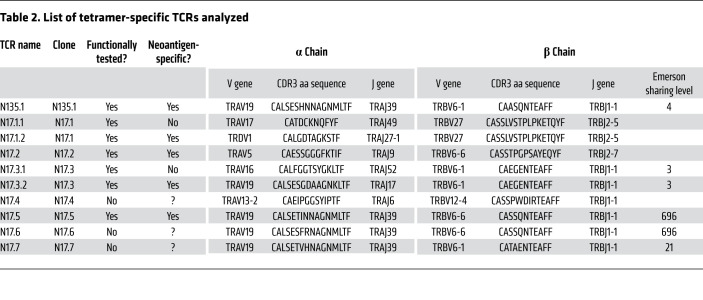
The amino acid sequences of annotated tetramer-specific TCRs (Table 2) revealed a cross-patient TCR resemblance between N17.3, N17.5, and N135.1: the β chains had 11-amino-acid-long CDR3 regions, with a Hamming distance of 1–3 between them, and V/J genes of the same family (TRBV6-1 or TRBV6-6, TRBJ1-1). The α chains were also similar, with Hamming distance of 2–4 between their 16-amino-acid-long CDR3, a shared V gene (TRAV19), and the same J gene (TRAJ39) in 2 of the chains. With restriction to a Hamming distance of 3 or less and the same V/J gene families, we found, strikingly, 2 lower-frequency α chains and 1 β chain resembling NA135.1 and NB135.1, respectively. Moreover, our single-cell experiment confirmed the pairing of these chains into 2 additional clones resembling N135.1. Using the same naming conventions as before, we denoted these clones N17.6 and N17.7 (Figure 5, Table 2, and Supplemental Table 4), with N17.5 and N17.6 sharing the exact same β chain sequence. Although their frequencies within the repertoire were lower, these additional clones were tetramer specific (Figure 5A).
Figure 5. N135.1 TCR similarity cluster within 17TIL converges on both α and β chains.
Inspection of the 17TIL repertoire revealed a cluster of 4 TCRs that were similar to N135.1: N17.3.2, N17.5, N17.6, and N17.7. The top most frequent TCRs on this list, N17.3.2 and N17.5, were validated as potent and neoantigen specific (see Figures 6 and 7). (A) Both α and β chains for the similarity cluster were enriched in the tetramer+ subpopulation. α/β Pairings were confirmed by single-cell TCR sequencing. Representative data (replicate 1 of 3) from bulk TCR sequencing. (B) Sequence comparison of the TCRs’ variable regions. Note the Hamming distance of up to 4 amino acids and the similarity of the V/J genes. (C) TCRs were plotted according to the Pgen of their α (x axis) and β chains (y axis). NH1 (NRAS hybrid TCR no. 1) combines the most probable α/β within the similarity cluster; it was generated through chain swapping between N135.1 and N17.5. NH2 is the counterpart to NH1, mixing NA17.5 with NB135.1. (D) Illustration of the chain swapping generating NH1, combining NA135.1 with NB17.5.
All in all, we uncovered a convergent class of 5 tetramer-specific clones. Two of these, N17.3 and N135.1, were already selected for functional validation on the basis of their frequency. We added the most frequent among the similarity-selected TCRs, N17.5 (consisting of 7 cells in our single-cell data), to our TCR validation pipeline.
Validation and characterization of the neoantigen-binding TCR repertoire.
Having identified multiple tetramer-specific T cell clones, we aimed to validate their individual sensitivity and specificity toward the HLA-A*01:01/ILDTAGKEEY neoantigen.
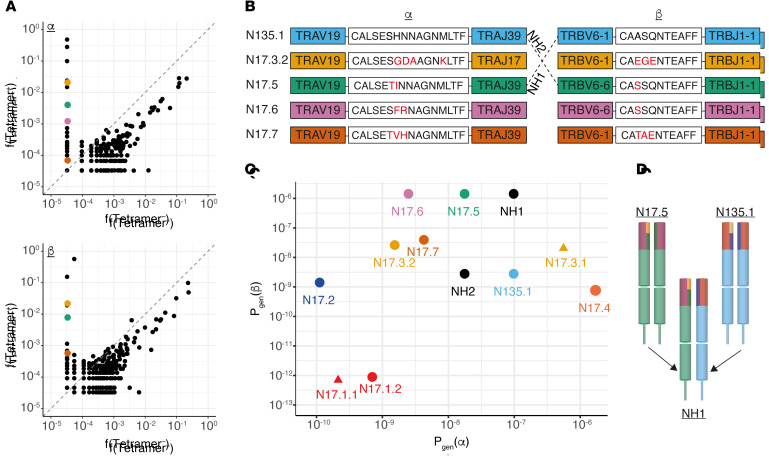
To this end, we induced overexpression of αβ combinations of interest in healthy donor peripheral T cells. As shown in Figure 6A, tetramer-binding experiments confirmed neoantigen binding for N17.2, N17.5, and N135.1 and discerned the relevant αβ combination for ααβ clone N17.3 (NA17.3.2/NB17.3). Surprisingly, the αβ pair pertaining to the most prominent of 17TIL’s tetramer-specific clones, N17.1, did not validate, even though it was efficiently expressed (Supplemental Figure 29). Clonal expression of the δ-gene TRDV1 in this clone, as observed in single-cell RNA-Seq, led us to reinspect the TCR-sequencing data and uncover a second α chain for N17.1, NA17.1.2, resulting from an uncommon event of TRDV1/TRAJ27 recombination (ref. 47 and Supplemental Figure 28). NA17.1.2 has been identified in frequency equivalent to that of NA17.1.1 in bulk TCR sequencing (40.4%–54.9% and 25.6%–41.3%, respectively) and detected in 99.1% (2905 of 2930) of N17.1 cells in our single-cell analysis. Reanalysis of our TCR-sequencing data revealed 2 additional TRDV1 α chains for 17TIL and 5 such chains in 135TIL. No other TRDV genes were implicated in α chain rearrangements. As expected, the NA17.1.2/NB17.1 chain combination, denoted by N17.1.2, validated on tetramer staining (Figure 6A).
Figure 6. A repertoire of potent neoantigen-specific TCRs.
Healthy donor T cells were electroporated with in vitro–transcribed TCR α and β chains. (A) Flow cytometric plots of A*01:01/ILDTAGKEEY-tetramer–stained cells. Negative controls were cells electroporated without mRNA (EN, electroporated nothing). (B and C) Electroporated T cells were coincubated overnight with peptide-pulsed IHW01161 cells. (B) IFN-γ ELISA with 10 μM peptide. ****P < 0.0001, by 1-way ANOVA with Tukey’s correction for multiple comparisons. Error bars represent the SEM of biological triplicates. (C) 4-1BB peptide titration assay. Plots show the mean with SD of biological triplicates. ****P < 0.0001, by 2-way ANOVA with Sidak’s correction for multiple comparisons.
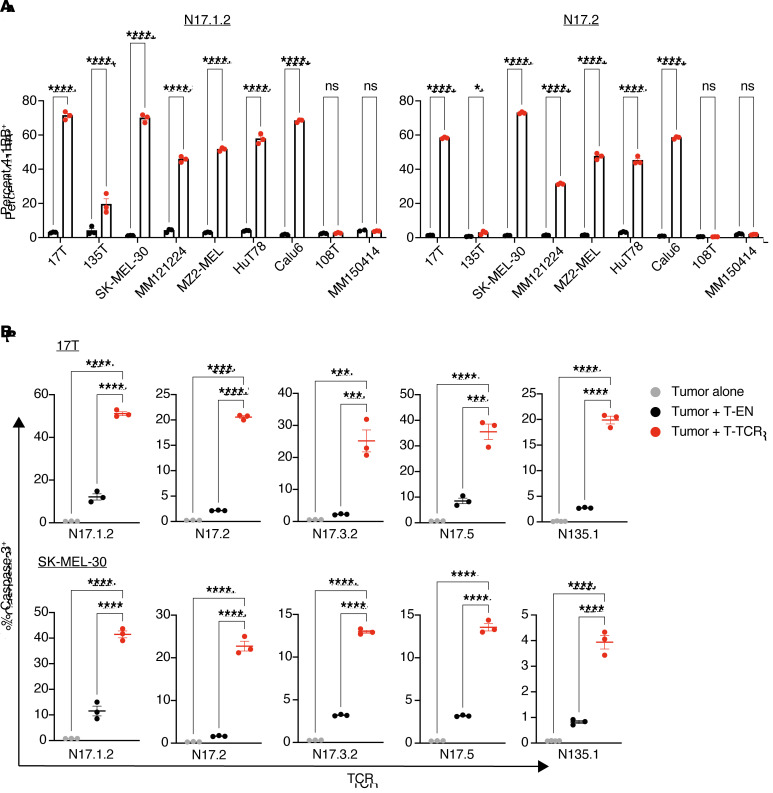
We proceeded to coincubate the different TCR-engineered T cells with HLA-A*01:01+ B-LCLs pulsed with either the ILDTAGKEEY neopeptide or its WT counterpart at different concentrations. All 5 tested TCRs proved to be neoantigen specific, as evidenced by differential reactivity toward the mutated peptide at supraphysiologic pulsed-peptide concentrations (Figure 6B). Peptide titration experiments further elucidated that these TCRs potently recognized the ILDTAGKEEY neopeptide at concentrations ranging from 0.01 nM to 10 nM (Figure 6C and Supplemental Figures 30–39). Interestingly, we observed an inverse relationship between TCR frequency and sensitivity within 17TIL (Supplemental Figure 40). To further evaluate the reactivity and cytotoxic capacity of the TCRs toward endogenously expressed neoantigen, we coincubated TCR-engineered T cells with HLA-A*01:01+NRAS.Q61K+ tumor cell lines. Testing the 2 most potent TCRs, N17.1.2 and N17.2, in a reactivity assay against a panel of tumor cell lines recapitulated our immunopeptidomics results (Figure 7A and Supplemental Figures 41–58). Moreover, we observed significant 4-1BB upregulation upon coincubation with the HLA-A*01:01+KRAS.Q61K+ lung adenocarcinoma cell line Calu6, attesting to the cross-isoform, cross-cancer-type relevance of the HLA-A*01:01/ILDTAGKEEY neoantigen, as predicted. All 5 tested TCRs had cytotoxic capacity toward neoantigen-expressing melanoma, as demonstrated in a cleaved caspase-3 assay (Figure 7B and Supplemental Figures 59–67).
Figure 7. Neoantigen-specific TCRs convey reactivity and cytotoxic capacity toward tumor cells that endogenously express the neoantigen.
Healthy donor T cells were electroporated with in vitro–transcribed TCR α and β chains and coincubated with tumor-derived cell lines. Donor cells electroporated without mRNA were used as T cell negative controls. (A) Percentage of T cells expressing the activation marker 4-1BB after an overnight 1:1 coincubation; 17T, 135T, SK-MEL-30, MM121224 and MZ2-MEL are all melanoma cell lines endogenously expressing HLA-A*01:01/NRAS.Q61K. HuT78 is a T cell lymphoma cell line that endogenously expresses HLA-A*01:01/NRAS.Q61K. Calu6 is a lung adenocarcinoma cell line that endogenously expresses HLA-A*01:01/KRAS.Q61K. Negative control cell lines: 108T (A*01:01+/NRAS WT); MM150414 (A*01:01–/NRAS.Q61K). See Methods for further information on the cell lines. *P < 0.05 and ****P < 0.0001, by 2-way ANOVA with Sidak’s correction for multiple comparisons. (B) Cleaved caspase-3 killing assay: percentage of 17T (top) and SK-MEL-30 (bottom) tumor cells expressing cleaved caspase-3 after a 3-hour coincubation at a 3:1 effector/target ratio. ***P < 0.001 and ****P < 0.0001, by 1-way ANOVA with Tukey’s correction for multiple comparisons. Error bars represent the SEM of biological triplicates throughout.
Since the majority of tetramer-binding 17TIL also cross-binds to the HLA-A*01:01/ILDTAGREEY-tetramer, we speculated that it is N17.1.2 that possesses both reactivities. Remarkably, indeed, N17.1.2 proved to recognize the ILDTAGREEY neopeptide at concentration as low as 1 nM (Figure 6C). We confirmed that the remaining 4 TCRs did not recognize the ILDTAGREEY neopeptide.
TCR generation considerations for the neoantigen-specific similarity cluster.
As mentioned above, similarity cluster TCRs N17.5 and N17.6 share an identical β chain. Interestingly, the CDR3 sequence of chain NB17.5, CASSQNTEAFF, is in fact “public,” as it is shared among 696 (of 786) samples in the Emerson data set (48). Of note, although in the Emerson data set this same CDR3 sequence can be attained with 39 different TRBV genes (i.e., high convergent recombination), in our data it solely recombined from TRBV6-6. Taken together with TRBV6 conservation across all similarity cluster TCRs, a hypothesis emerged that TRBV6 has functional significance outside of the CDR3 region for this convergent neoantigen-specific class. Thirty percent of HLA-annotated CASSQNTEAFF+ samples in the Emerson data set possess HLA allele A*01:01 (189 vs. 440). Recombination with TRBV6-6 (or TRBV6) was not statistically increased for this subset of samples (Fisher’s exact test, P = 0.45). Although similar to NB17.5, all other β chains of the convergent class have low Emerson sharing levels (Table 2).
It is instructive to plot the generation probabilities (denoted herein as Pgen) of the convergent class clones (Figure 5C). Our 2 most potent TCRs, N17.1.2 and N17.2, exhibited low Pgen for both their α and β chains. N17.5 and N17.6 excelled in their Pgen(β), as expected for a public clonotype. N135.1 had the highest Pgen(α) of the set.
From an engineering point of view, it is interesting to investigate whether some of the α/β chains are interchangeable. As depicted in Figure 5, B–D, NH1 (NA135.1/NB17.5) is a hypothetical clone that maximizes Pgen for both α and β within the similarity cluster. We tested NH1 for neoantigen reactivity, and it validated with both tetramer staining and neoantigen-specific IFN-γ release (Figure 8). The reverse combination, NA17.5/NB135.1, validated as well, whereas transposing the chains of N17.3.2 with either N17.5 or N135.1 failed to maintain neoantigen specificity (Supplemental Figure 68).
Figure 8. α/β Chain-swapping within the TCR similarity cluster perserves neoantigen specificity.
Donor T cells were electroporated with in vitro–transcribed TCR α and β chains to express NH1 and NH2. Donor cells electroporated without mRNA (EN) were used as T cell negative controls. (A) Flow cytometric analysis of anti-CD8– and tetramer-stained cells. (B) IFN-γ ELISA after overnight 1:1 coincubation with IHW01161-presenting cells, pulsed with the WT or mutant peptides. IHW01161 without pulsed peptide (DMSO only) served as a negative control. Error bars represent the SEM of biological triplicates. ****P < 0.0001, by 2-way ANOVA with Tukey’s correction for multiple comparisons.
Single-cell profiling of CD8+ 17TIL and 135TIL.
We next sought to investigate the transcriptional profile of neoantigen-specific clones in response to cognate melanoma and compare their cell states with other CD8+ TIL populations. Briefly, bulk 17TIL and 135TIL were coincubated with the cognate melanoma cell line overnight at a 1:1 ratio, sorted into bulk CD8+, CD8+ tetramer+, and CD8+ tetramer– T cell populations, and profiled using paired single-cell RNA-Seq and TCR sequencing on the 10x Genomics platform.
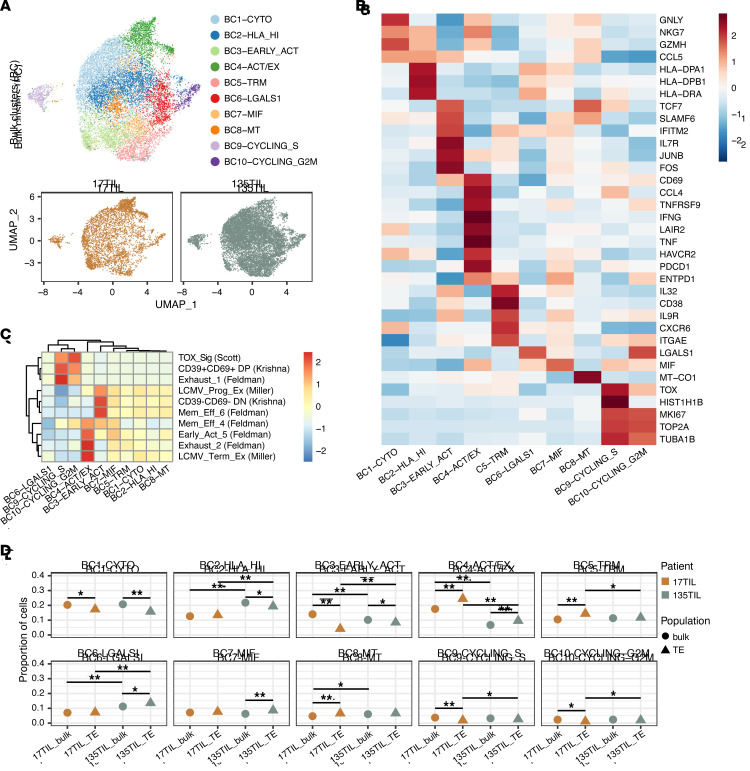
After quality filtering based on the TCR chains captured, the number of UMIs, and the percentage of mitochondrial reads per cell (see “single-cell analysis cell-filtering steps” in Supplemental Table 5), we obtained 14,012 cells from the sorted bulk CD8+ cell populations (3815 cells, 17TIL bulk; 10,197 cells, 135TIL bulk). To generate an overview of the overall CD8+ TIL landscape, we integrated these bulk populations and performed unsupervised clustering, which revealed 10 phenotypic clusters (bulk clusters, BC1-10, Figure 9A), including a cytotoxic cluster (3275 cells) expressing high levels of GNLY, NKG7, and GZMH (Figure 9B and Supplemental Figure 69). We also identified a cluster of 1708 cells that expressed markers of an early activation state, such as IL7R, FOS, TCF7, and SLAMF6, and showed similarities to progenitor and stem-like gene signatures from previous studies (BC3, Figure 9C), including a CD39–CD69– double-negative signature that was associated with an effective clinical response in the settings of adoptive cell transfer and immune checkpoint blockade (49). BC4 differentially expressed a mixture of activation markers, including IFNG, TNFRSF9 (4-1BB), TNF (TNF-α), ENTPD1 (CD39), CD69, and inhibitory checkpoint genes, such as HAVCR2 (TIM-3), PDCD1 (PD-1), TIGIT, and LAG3 (Figure 9B, Supplemental Figure 69, and Supplemental Table 5), and displayed transcriptional overlap with established exhaustion signatures (Figure 9C). Additionally, the bulk CD8+ T cell landscape contained a cluster of tissue-resident memory-like cells that expressed ITGAE, CXCR6, and ENTPD1 (CD39), as well as 2 clusters of proliferating cells that expressed high levels of G2M/S cell-cycle–related genes including MKI67, TOP2A, and TUBA1B. Indicative of shared cell states and effective data integration, clustering did not segregate 17TIL and 135TIL into separate clusters (Figure 9A). Nonetheless, 17TIL had an increased proportion of early-activated (BC3) and activated/exhausted (BC4) cells (Figure 9D and Supplemental Figure 70D), suggesting that there were also patient-specific phenotypic differences between the bulk CD8+ T cell populations.
Figure 9. Unsupervised clustering of bulk TILs.
Tetramer-enriched clones showed increased exhaustion and decreased early-activation proportions compared with bulk TILs. (A) UMAP of cells from bulk-sorted populations colored by phenotype cluster (top). UMAP of cells from each patient sample (bottom). (B) Heatmap depicting gene expression of select cluster markers and canonical T cell subset marker genes across the phenotypic clusters identified. (C) Gene expression module scoring of clusters with previously published CD8+ T cell gene sets (select signatures from ref. 43). (D) Comparison of phenotypic proportions among 17TIL and 135TIL bulk and tetramer+ populations. *P < 0.05 and **P < 0.001, by χ2 test.
In order to focus our analysis on true tetramer-enriched clones, we filtered the 17TIL and 135TIL CD8+ tetramer+ populations, as described above, to only include tetramer+/tetramer– frequency-enriched clones with at least 5 cells (i.e., N17.1-5, N135.1), in addition to clones identified through TCR similarity (N17.6, N17.7). This yielded 3679 and 2178 cells derived from 7 clones and a single clone in the tetramer+ populations of 17TIL and 135TIL, respectively (see “single-cell analysis cell-filtering steps” in Supplemental Table 5). These tetramer-enriched clones were also present within the bulk population, albeit at much lower frequencies, with the exception of N17.5, N17.6, and N17.7, which were not found in the bulk sample (Supplemental Figure 70, E and F).
To compare the transcriptional states of these tetramer-enriched clones with the broader CD8+ landscape, we integrated the filtered 17TIL and 135TIL tetramer+ samples with the bulk CD8+ samples, using the bulk populations as a reference (see Methods). Unsupervised clustering of the tetramer-enriched and bulk TILs generated a phenotypic clustering profile highly similar to that of bulk CD8+ populations alone (Supplemental Figure 70, A and B, and Figure 9A). Thus, BC cluster annotations were maintained for the integrated bulk and tetramer-enriched analysis. By comparing phenotypic proportions between tetramer-enriched and bulk populations, we observed that both 17TIL and 135TIL tetramer-enriched populations were significantly enriched for activated/exhausted phenotypes (Figure 9D and Supplemental Figure 70, C and D). Furthermore, the 17TIL tetramer-enriched population was also more prevalent in the tissue-resident memory cluster than in the 17TIL bulk CD8+ population. Conversely, the tetramer-enriched cell populations were depleted for early-activated and cytotoxic phenotypes in comparison with their bulk counterparts. However, all of the well-represented neoantigen-specific clones (N135.1, N17.1, N17.2, N17.3, and N17.4) contained cells in the early-activated cluster (Supplemental Figure 70D), which is related to the CD39–CD69– stem-like progenitor state (49). Interestingly, the dominant and sensitive N17.1 clone contained the smallest proportion of these early-activated cells, with only 1.3% of cells within the clone belonging to BC3 (Supplemental Figure 70D). Taken together, these results suggest that, although neoantigen-specific clones are generally present in a more mature activated and/or exhausted state in comparison with other CD8+ clones, the degree to which this is true varies between neoantigen-specific clones and may reflect their expansion and functional capabilities.
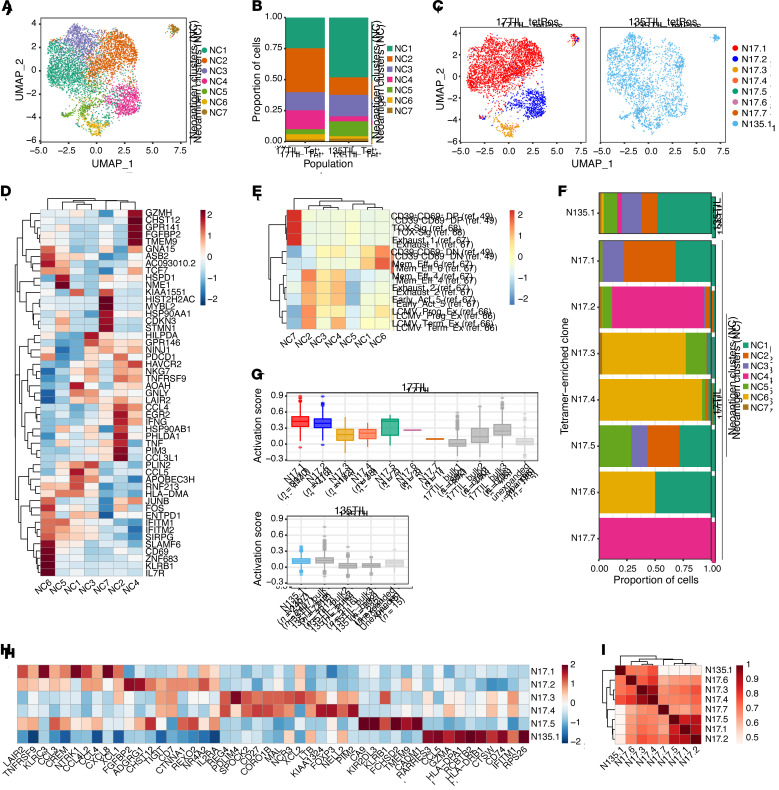
To further characterize and compare the phenotypic states of individual neoantigen-specific clones, we analyzed the tetramer-enriched cell populations alone. Unsupervised clustering of 17TIL and 135TIL tetramer-enriched cell populations revealed 7 clusters (neoantigen clusters, NC1-7) that encompassed clones from both patients and did not correspond directly to the phenotypic clusters derived from the bulk sample integration (Figure 10, A and B, and Supplemental Figure 71). NC2 differentially expressed markers of activation including IFNG, TNFRSF9 (4-1BB), and TNF, as well as cytotoxicity-related genes such as NKG7 (Figure 10D and Supplemental Figure 72). NC1 and NC3 also expressed the cytotoxicity genes NKG7 and GNLY, although their cellular states were less well defined. While NC4 exhibited phenotypic similarities to the activated effector-like NC2 (i.e., expression of NKG7, TNFRSF9, and IFNG), it expressed higher levels of certain inhibitory receptors such as HAVCR2 (TIM-3) and TIGIT, suggesting that cells in this cluster may exist in a more exhausted state, which was corroborated by a modest association with established exhaustion signatures (Figure 10E). NC6 consisted of cells expressing a mixture of the early-activation genes TCF7, SLAMF6, and FOS and the memory marker KLRB1 (Figure 10D and Supplemental Figure 72), and displayed the highest correspondence to stem-like and memory signatures described in the literature (Figure 10E).
Figure 10. Contrasting tetramer-specific clones reveals transcriptional clonal segregation.
(A) UMAP of cells from tetramer+ populations colored by phenotype cluster. (B) Distribution of cells across clusters within each patient sample. Tet+, tetramer+. (C) UMAP of cells from each patient sample colored by tetramer-enriched clone identity. tetPos, tetramer+. (D) Heatmap depicting gene expression of select cluster markers and canonical T cell subset marker genes across the phenotypic clusters identified. (E) Gene expression module scoring of tetramer+ clusters with previously published CD8+ T cell gene sets (select signatures from ref. 49 and refs. 66–68). (F) Distribution of cells across phenotype clusters within each tetramer-enriched clone. (G) Comparison of T cell activation scores (from ref. 50) between each tetramer-enriched clone, other highly expanded clones in the bulk population, and all other unexpanded clones. The number of cells in each clone or group is indicated. (H) Gene signatures of each tetramer-enriched clone (for clones with >5 cells). (I) Spearman correlation of tetramer-enriched clones based on clone-specific gene signatures as defined in H.
Mapping TCR clonal identities onto this phenotypic clustering revealed a strong transcriptional association among cells within the same clone, despite the exclusion of TCR genes from the dimensionality reduction and clustering process (see Methods). Concretely, cells from N17.1 predominantly occupied NC1-3, while N17.2 was largely restricted to NC4 (Figure 10, C and F). The less frequent (N17.3, N17.4) and sensitive (N17.3) well-represented clones were mainly composed of cells in the early-activated NC6. This observation that different clones against a single neoantigen assumed distinct clonal states may suggest that differential T cell states are induced by the unique characteristics of the specific TCR-neoantigen interaction. By contrast, N135.1 was distributed among all clusters (Figure 10, C and F). Since different tumors may differentially present the same neoantigen, as we have shown for 17T and 135T, it is difficult to conclude whether the wide range of phenotypes observed within N135.1 is TCR intrinsic, neoantigen dependent, or both. It is interesting to note that N17.5, which resembles N135.1 in both sequence and function, also exhibited a dispersed phenotypic state across several clusters (Figure 10F), although its underrepresentation and small clone size (7 cells) preclude any definitive conclusion (Supplemental Figure 70E).
To compare the extent of activation, we scored neoantigen-specific clones, the 3 most highly expanded bulk clones, and the aggregation of all unexpanded clones (bulk clones presenting with 1 cell) on the basis of an established T cell activation gene signature (ref. 50 and Figure 10G). Counterintuitively, 17TIL neoantigen–specific clones exhibited high activation scores when compared with the top expanded bulk clones despite being orders of magnitude less frequent within the bulk TIL population (Supplemental Figure 70, E and F). Consistent with the sensitivity hierarchy delineated by our peptide titration experiments (Figure 6), cells from N17.1 were present in the most activated state on average, followed closely by N17.2, whereas N17.3 and N17.4 displayed comparable levels of lower activation states (Figure 10G). While N17.5 contains cells with high activation scores, its low clonal frequency and large range of activation scores within the clone precludes any significant interpretation of its activation pattern. The unexpanded 17TIL clones collectively displayed significantly lower activation scores than did either the neoantigen-specific clones or the top expanded bulk clones. Although there was less variability in activation scores for clones from 135TIL (Figure 10G), we also observed a similar pattern in which the neoantigen-specific clone N135.1 had a significantly higher activation score than the second and third most-expanded bulk clones despite comprising only 2.2% of the bulk 135TIL population (P < 0.001, when using the activation signature, as in ref. 50, as well as individual activation markers IFN-γ, 4-1BB, TNF-α, and CD69; Wilcoxon test with Bonferroni’s correction for multiple comparisons; Supplemental Figure 70F). These trends were also seen when analyzing the expression levels of individual activation markers, such as IFNG and TNFRSF9 (4-1BB) (Supplemental Figure 73). It is important to note that 17TIL and 135TIL were both expanded in vitro using the rapid expansion protocol (REP), which sometimes skews native clonal frequencies (51). Moreover, it is possible that the 17T and 135T cell lines do not present certain antigens that were dominant in the original tumors and may be recognized by the most expanded TIL clones.
To generate clone-specific gene signatures, we performed differential gene expression analysis between the tetramer-enriched clones from 17TIL and 135TIL with a clone size of greater than 5 cells (Figure 10H). N17.1 displayed high expression levels of LAIR2, TNFRSF9 (4-1BB), and several chemokine molecules, including CCL3, CCL4, CXCL8, and CCL4L2. These genes were also expressed by N17.2 and N17.5, the latter of which additionally expresses high levels of KLRB1 and the killer cell immunoglobulin-like receptor KIR2DL3. The gene signatures of N17.3 and N17.4 were highly concordant, with both clones overexpressing REG4, CD27, and SPOCK2, whereas N135.1 exhibited a distinct gene signature consisting of RARRES3, CCL5, and GZMA. Clustering of the neoantigen-specific clones based on these clone-specific gene signatures revealed that N135.1 was indeed transcriptionally less similar to the other 17TIL-derived clones (Figure 10I). Among the 17TIL neoantigen–specific clones, we observed 2 clusters of high correlation, with N17.1, N17.2, and N17.5 in 1 cluster and N17.3 and N17.4 in the other, which recapitulated their shared gene signature and activation score patterns (Figure 10, G and H). Notably, we did not observe a tendency for clones with similar TCR sequences to cluster together according to their phenotypic profile.
Discussion
The identification of recurrent neoantigens presented by tumors and recognized by T cells has been the main challenge in applying robust and effective cancer immunotherapies in the clinic. Identification of neoantigens has primarily been carried out using approaches based on the overexpression of neopeptides of interest in APCs and the assessment of their reactivity using HLA-matched TILs. This allowed the identification of numerous immune-reactive neopeptides. However, this approach has 2 main drawbacks: (a) since neopeptide identification is based on T cell reactivity, there is no certainty that the neoantigen is actually presented on the patient’s tumor tissue; and (b) most identified neoantigens are private, limiting their robust clinical application.
Alongside the few recurrent neoantigens discovered by serendipity (7, 20, 52), recent efforts used the above-mentioned methods to directly look for such antigens. These studies usually selected samples either on the basis of specific recurrent mutations (21, 23, 24), or according to the HLA allele frequency in the cell population (24). However, the population-wide applicability of a recurrent neoantigen depends on the productive intersection of both. A highly recurrent mutation may produce an infrequent neoantigen if combined with a rare HLA allele and vice versa.
Given these limitations, we developed a new approach that increases the likelihood of identifying both presented and recurrent neopeptides. To the best of our knowledge, this is the first study to use such an approach, in which our data-driven analysis combined the use of a recurrent mutation database with knowledge of the HLA allotypes of the patients who harbored the recurrent mutation. Such an analysis allowed the accurate and rational selection of candidates to be assessed for recurrent neopeptide presentation.
Moreover, we directly demonstrated neoantigen presentation. To query a candidate’s neopeptide space in an unbiased manner, we induced co-overexpression of the relevant minigene and HLA combination and evaluated it directly using HLA peptidomics. This not only pushed neopeptide presentation to its maximum, but also obviated the need for hard-to-come-by tumor samples in the early screening phase. Once a specific neopeptide was validated, we proceed to test for its robust presentation within the malignant context. To increase our detection capabilities, we perform HLA peptidomics in a targeted fashion using heavy synthetic peptides that were spiked into the HLA peptidomics sample. This not only increased the sensitivity of the process but also allowed an approximate assessment of the concentration of the endogenous peptide of interest.
As a melanoma-oriented laboratory, and in light of the therapeutic benefit immunotherapy has brought specifically to patients with melanoma, we decided to direct our proof-of-concept efforts for the pipeline described above to mutations that are highly recurrent in melanoma.NRAS-mutant melanoma is notorious for the bad prognosis it confers. Unlike for BRAF-mutant melanoma, there are currently no available targeted agents to treat this subtype. Although patients with NRAS mutations are more reliant on immunotherapy for their treatment, current data are conflicting regarding the benefit they receive from checkpoint blockade (53, 54). We therefore further focused on RAS.Q61, the second most highly mutated position in melanoma, for the study at hand. The discovery pipeline described is of course readily generalizable to any recurrent mutations, and we are currently using it to uncover additional recurrent neoantigens.
Applying this approach to RAS.Q61 yielded several important findings. First, we identified a NRAS.Q61K-derived neopeptide that is recurrently presented by HLA-A*01:01, demonstrating the strength of our approach. Second, we showed that the NRAS.Q61K-derived neopeptide was robustly presented on malignant cells. Third, we demonstrated the immunogenicity of our identified neoantigen using TILs from 2 unrelated patients. Fourth, TCR sequencing of tetramer+ TILs uncovered functional neoantigen-specific TCRs that may prove therapeutically relevant beyond the individual patient.
Importantly, our results extend beyond the NRAS.Q61K mutation in melanoma. The ILDTAGKEEY sequence is shared across RAS variants, thus, we expected KRAS- and HRAS-mutant tumors to also present the neoantigen. Indeed, our findings in HLA peptidomics and reactivity assays corroborated that the HLA-A*01:01/ILDTAGKEEY neoantigen is relevant across tumor types and is presented on a KRAS-mutant tumor.
The main RAS.Q61 variants are all predicted to be strong binders of HLA-A*01:01. Indeed, T cell reactivity toward HLA-A*01:01/ILDTAGREEY, the RAS.Q61R variant, was previously identified in TILs from a patient with melanoma (16). The cumulative frequency of the HLA/mutation combinations A*01:01/NRAS.Q61K and A*01:01/NRAS.Q61R amounts to 7% of patients with melanoma. Importantly, a dominant, TRDV1 recombined TCR we uncovered cross-reacted with both the RAS.Q61K and RAS.Q61R variants of our identified neoantigen, thereby more than doubling the therapeutic potential of our findings. Finally, although our study focused specifically on the HLA allotype A*01:01, our RAS.Q61K peptide was predicted to bind other HLA allotypes, albeit weakly. Future studies will determine whether these neopeptides also bind these other predicted allotypes.
Our neopeptide-HLA 3D structural model supports potential binding of both WT and mutant RAS.Q61 peptides to HLA-A*01:01. Indeed, although not identified in our data set, there is MS evidence that the WT ILDTAGQEEY peptide is processed and presented in HLA-A*01:01–expressing cell lines (45). However, our neopeptide-HLA structural model also predicted that the mutated amino acid is free to engage with the TCR. Thus, although both the WT and mutant peptides may be presented, differential reactivity toward the WT and mutant peptides is expected. This was indeed corroborated by our reactivity assays, which showed dose-dependent TIL reactivity specifically toward the mutant, but not the WT NRAS peptide.
Bulk TCR sequencing of tetramer-enriched TILs provided us with TCR α and β sequences that were highly likely to participate in neoantigen-specific receptors. Similar α and β chain frequency distributions hinted at specific chain pairings that were corroborated by single-cell TCR sequencing. Moreover, all of these TCRs were validated as potent and neoantigen specific.
While historically more emphasis was put on the TCR β chain, as demonstrated in the 2 neoantigen-specific ααβ clones we analyzed, the α chain plays a crucial role in determining antigen recognition.
The neoantigen-specific repertoire we uncovered exhibited remarkable intra- and interpatient TCR sequence similarities. These similarities spanned both the α and β chains of their respective TCRs. For their HLA-C*08:02/RAS.G12D recurrent neoantigen, Tran et al., too, reported on an identical TCR appearing in 2 unrelated patients (7, 20). These statistically improbable phenomena are reminiscent of “public” convergent sequences appearing in certain viral and autoimmune contexts and deserve further exploration that is beyond the scope of the current work.
Our single-cell analyses of neoantigen-specific clones recapitulated the activation we observed in functional assays. As a whole, these cells showed increased exhaustion (BC4) and decreased early-activation (BC3) proportions compared with bulk TILs, in line with continuous antigen exposure.
Although in vitro expansion might have skewed the original clone frequencies, exposure to cognate melanoma spotlighted neoantigen-specific clones over even the most expanded bulk TIL clones. When compared side by side, the larger neoantigen-specific clones trended toward better activation scores. While the proportion of early-activated cells tended to decrease with increased TCR sensitivity, all well-represented neoantigen-specific clones retained some cells in BC3 upon exposure to melanoma. This is important, as BC3 resembles the recently described stem-like CD39–CD69– signature, which has been shown to be imperative for ACT success (49).
Contrasting neoantigen-specific cells strikingly resulted in clonal segregation for 17TIL, highlighting the idea that “clonal states” are driven by the unique TCR-tumor interaction. Although beyond the scope of the current work, studying this relationship between TCR and neoantigen-induced T cell states may inform the rational design of adoptive T cell therapy, especially as TCR libraries like the one we present here for the A*01:01/ILDTAGKEEY neoantigen become available.
Unlike for 17TIL, N135.1 distributes over the entire uniform manifold approximation and projection (UMAP). Moreover, although clone N135.1 is similar in its TCR sequence and functional profile to some neoantigen-specific clones in 17TIL, gene signature–based clustering revealed it to be transcriptionally distinct from these clones. This may be due to the tumor-end of the equation, as we have shown that 17T and 135T differentially presented the neoantigen. Gene signature–based clustering did put together the most highly activated (N17.1, N17.2, N17.5) versus early-activated (N17.3, N17.4) clones.
Interestingly, LAIR2 was differentially expressed in the highly activated neoantigen-specific clones. The role of LAIR2 in the tumor microenvironment is yet to be fully elucidated. As a secreted antagonist of LAIR1, LAIR2 has been shown to induce an immune-excitatory net effect and has been linked to autoimmunity (55–57). Recently, LAIR2 expression has been suggested to promote T cell exhaustion by activation of CD8+ T cells and to associate with poor prognosis in non–small-cell lung cancer (58). Future work may aim to validate LAIR2 secretion in neoantigen-specific CD8+ cells and explore its immunomodulatory role in the cancer microenvironment.
Our new pipeline enabled us to identify a RAS.Q61K-derived neopeptide that was recurrently presented by HLA-A*01:01. As NRAS.Q61K is harbored by 3% of patients with melanoma (2.9:1000 patients pan-cancer) and the HLA-A*01:01 allotype is found in 25% of the population, this discovery is relevant for a high fraction of the cancer patient population. These findings strongly suggest that our data-driven analysis in cancer patient cohorts, in combination with binding predictions, increases the chances to successfully discover the most promising candidate neopeptides. Indeed, pan-melanoma as well as pan-cancer applications of this pipeline are highly feasible, as indicated in Figure 1.
Although it has been suggested that recurrent driver neoantigens may be immune edited (26), and only a handful of recurrent neoantigens have been discovered to date (7, 16–21, 23, 24), the insight gathered through our analysis strengthens the notion that these neoantigens may actually be more abundant than previously suggested. As the delineation of validated HLA neoantigen pairs derived from recurrent “hotspot” mutations together with their paired TCRs could serve as predefined “off-the-shelf” immunotherapy targets relevant for numerous patients, systematic application of our approach is of high relevance for personalized treatments.
Methods
Further information can be found in the Supplemental Methods.
Data-driven analysis for recurrent neoantigen candidate prioritization
HLA/mutation co-occurrence analysis using patient data.
TCGA data on provisional cohorts were downloaded via cBioportal on October 10, 2018, accumulating to a total of 8038 patients (33, 59, 60). Previously published high-resolution HLA-I typing data for 9176 TCGA patients were obtained courtesy of Hannah Carter (UCSD, San Diego, California, USA) (26). Intersection of these 2 sources yielded a mutation- and HLA-annotated cohort of 6840 patients in total, 368 of whom had melanoma. Mutation and HLA data for 11,033 patients pan-cancer, including 221 patients with melanoma, were downloaded from the Hartwig Medical Foundation database on January 17, 2020, and were processed as previously described (34, 61). HLA-typed samples were annotated on the basis of their RAS.Q61 status. Canonical transcript identifiers for NRAS (ENST00000369535), KRAS (ENST00000256078), and HRAS (ENST00000451590) were obtained from Ensembl, version 101, on August 31, 2020, and their protein-coding sequence coordinates were obtained from Gencode, version 35, on August 30, 2020.
The previously published MELA-AU data set consisting of data on 69 patients with melanoma was obtained from David Adams of the Wellcome Sanger Institute (Hinxton, Cambridge, United Kingdom) (35).
For each of these 3 data sets, we computed pan-cancer and melanoma lists of all HLA-I alleles co-appearing with RAS.Q61 mutations. For each HLA/mutation combination, we calculated its frequency in melanoma and pan-cancer (Supplemental Table 1). For TCGA data set, HLA-I frequencies were compared between the RAS.Q61-mutated and general populations (Fisher’s exact test with FDR correction).
NetMHCpan predictions.
We used netMHCpan 4.0 to predict RAS.Q61-derived neopeptides (30). Using the protein sequence of NRAS, we extracted the 27-amino-acid-long stretch flanking position 61. This sequence is conserved among RAS variants; therefore, it is representative of NRAS, KRAS, and HRAS. Four mutated variants were constructed by substituting glutamine 61 with arginine, lysine, leucine, or histidine (see “input 27 mers” in Supplemental Table 1). All 5 versions were wrapped into the FASTA format and served as input for netMHCpan, along with our compiled list of HLA alleles co-appearing with RAS.Q61 mutations in the analyzed patient databases (Supplemental Table 1). The algorithm was set to scan for binding peptides of 8–14 amino acids in length, with peptides ranked at 0.5≤%Rank≤2 considered weak binders and those with %Rank≤0.5 considered strong binders. The output was filtered to retain only peptides spanning position 61. A “best %Rank” score was assigned to each HLA/mutation combination by enumerating over the predicted neopeptides.
Tumor tissue, cell lines, TILs, and PBMCs
Metastatic melanoma cell lines 17T, 135T, 108T and their cognate TIL products were established as previously described (62). Whole-exome sequencing was performed for 17T as part of a previous sequencing effort in our laboratory (63). Established TILs were expanded via the REP, as described previously (64).
The commercial melanoma cell line SK-MEL-30 (ACC-151) was purchased from DSMZ (https://www.dsmz.de). Additional tumor cell lines HuT78, Hs940T and Calu6, as well as HB95 (W6/32) and HB145 (IVA12) hybridoma cells, were purchased from American Type Culture Collection (ATCC). The cell lines MM121224 and MM150414 were provided by Mitch Levesque (University of Zurich Hospital, Zurich, Switzerland). The cell line MZ2-MEL, established by Thierry Boon and colleagues (65), was obtained courtesy of Ugur Sahin (Translational Oncology at the University Medical Center of the Johannes Gutenberg University, Mainz, Germany). The EBV-transformed B-LCLs IHW01161, IHW01113, IHW01070, and 721.221 (IHW00001) were purchased from the Fred Hutch International Histocompatibility Working Group (IHWG) Cell and DNA Bank (https://www.fredhutch.org/en.html). NRAS.Q61K-mutant malignant melanoma snap-frozen tumors were provided in-house (Mela049, Mela183, MM1369, and MM1319).
Healthy donor leukocyte preparations from blood donations intended for research were purchased from MDA (Israel’s national blood service). PBMCs were extracted from these using Ficoll-Paque PLUS (GE Healthcare) via density-gradient centrifugation, cryopreserved in aliquots in FCS with 10% DMSO, and thawed as needed per experiments.
The hybridoma cells HB95 and HB145 were used to purify pan–HLA-I and pan–HLA-II antibodies for the preparation of the HLA-affinity columns. All cell lines were tested regularly (EZ-PCR Mycoplasma Kit, Biological Industries) and were found negative for mycoplasma contamination.
Melanoma cell lines were cultured in complete RPMI medium consisting of RPMI supplemented with 10% FCS, 2 mM l-glutamine, 2.5% HEPES, 100 IU/mL penicillin, and 0.1 mg/mL streptomycin. B-LCLs were cultured in B cell medium consisting of complete RPMI medium supplemented with 1 mM sodium pyruvate. TILs and PBMCs were cultured in T cell medium consisting of RPMI supplemented with 10% heat-inactivated human serum AB, 2 mM l-glutamine, 100 U/mL penicillin, 0.1 mg/mL streptomycin, and 5.5 × 10–5 M β-mercaptoethanol. TILs were thawed and recovered in T cell medium containing 6000 IU/mL IL-2 (Proleukin) for 3 days prior to use in downstream assays. PBMCs were thawed and selected for T cells in T cell medium containing 300 IU/mL IL-2 and 50 ng/mL OKT3 anti-CD3 antibody (317304, BioLegend) 7 days prior to experiments. PBMCs were subcultured in T cell medium supplemented with 300 IU/mL IL-2 on day 3 or 4 after thawing. T cell medium was used for coincubations in the caspase-3 killing and neoantigen-induced expansion assays. Complete RPMI medium was used for overnight TIL/B-LCL and PBMC/B-LCL coincubations.
Statistics.
Statistical analysis was executed using GraphPad Prism (version 9.0.2), base R (version 3.5.0), and Real Statistics Resource Pack software for Excel (release 7.7.1). Comparison of HLA allele frequencies between the pan-cancer, melanoma, and RAS.Q61-mutated TCGA populations was conducted using Fisher’s exact test with FDR correction. IFN-γ ELISA, 4-1BB–based reactivity, and cleaved caspase-3 assays were analyzed using 1-way ANOVA with Tukey’s multiple-comparison test. Where multiple assays were conducted concurrently and could therefore be reliably compared, a 2-way ANOVA was used. Peptide titration assays were analyzed by 2-way ANOVA with Sidak’s multiple-comparison test. Comparison of single-cell phenotypic proportions was performed using the χ2 test. Differences in activation scores between TCR clones were analyzed using a Wilcoxon rank-sum test with Bonferroni’s multiple hypothesis testing correction. Corrected P values of 0.05 less were considered significant.
Study approval.
The metastatic melanoma cell lines 17T, 135T, 108T, and their cognate TIL products were established as previously described (33), with informed patient consent under a protocol approved by the NIH’s IRB Ethics Committee.
Author contributions
AP and YS conceived of and designed the study. AP, EG, NF, JAP, and ATS developed the study methodology. AP, MA, JAP, SRZ, EG, TD, EB, SK, PG, YL, and GB conducted experiments and acquired data. AP, EG, NF, JAP, ATS, CB, TD, MYN, CAP, NLB, BD, EF, and RL analyzed and interpreted the data (e.g., statistical analysis, biostatistics, and computational analysis). AP, EG, NF, and YS wrote, reviewed, and/or revised the manuscript. All authors contributed to the final version of the manuscript. MPL, ML, AA, SAR, MYN, DJA, JSW, PS, RAS, GBJ, and CJC provided administrative, technical, or material support. YS and NF supervised the study.
Supplementary Material
Acknowledgments
We acknowledge the NIH Tetramer Facility for their help in producing the tetramers. This publication and the underlying research were partly facilitated by TCGA Research Network, the Hartwig Medical Foundation, and the Center for Personalized Cancer Treatment (CPCT), which generated, analyzed, and made available data for this purpose (32, 33). We are grateful to Astar Shamul and Aviad Pato for teaching us the TCR cloning and electroporation procedures and to Zelig Eshhar (The Weizmann Institute of Science, Rehovot, Israel and Tel-Aviv Sourasky Medical Center, Tel-Aviv, Israel) and Ugur Sahin (Translational Oncology at the University Medical Center of the Johannes Gutenberg University, Mainz, Germany) for providing the pGEM-4Z-EGFP-A64 plasmid and the MZ2-mel cell line, respectively. This work was supported by the Intramural Research Programs of the NCI and by a National Health and Medical Research Council of Australia (NHMRC) Program Grant (APP1093017, to RAS). YS is supported by the European Union’s Horizon 2020 Research and Innovation Program (no. 770854); ERC-2017-CoG (no. 754282); MRA (no. 622106); The Rising Tide Foundation; the Garvan Institute; the Graf Family Foundation; the Green Family Charitable Foundation; and the Knell Family and Hamburger Family. MYN and YS are supported by grants from the Israel Science Foundation (nos. 494/16, ISF-NSFC 2463/16, and 696/17, respectively). ATS is supported by NIH grant K08CA230188; the Parker Institute for Cancer Immunotherapy; a Technology Impact Award from the Cancer Research Institute; and a Career Award for Medical Scientists from the Burroughs Wellcome Fund. JAP is supported by NIH T32AI007290. RAS is supported by an NHMRC Practitioner Fellowship (APP1141295). Support from colleagues at the Melanoma Institute Australia and the Royal Prince Alfred Hospital and investigators of the Australian Melanoma Genome Project is gratefully acknowledged.
Version 1. 10/15/2021
Electronic publication
Footnotes
Conflict of interest: RAS has received fees for professional services from Evaxion, Provectus Biopharmaceuticals Australia, Qbiotics, Novartis, Merck Sharp & Dohme, NeraCare, AMGEN Inc., Bristol-Myers Squibb, Myriad Genetics, and GlaxoSmithKline. YS, AP, EG, MA, SRZ, and NF are inventors on a patent titled “Identification of recurrent mutated neopeptides” (WO2020/234875) and “TCRS directed at RAS-derived recurrent neoantigens” (Israeli patent application no. 282814).
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(20):e129466.https://doi.org/10.1172/JCI129466.
Contributor Information
Aviyah Peri, Email: aviyah.peri@weizmann.ac.il.
Erez Greenstein, Email: erez.greenstein@weizmann.ac.il.
Michal Alon, Email: michal.alon@weizmann.ac.il.
Joy A. Pai, Email: joypai@stanford.edu.
Tamir Dingjan, Email: tamir.dingjan@weizmann.ac.il.
Shlomit Reich-Zeliger, Email: Shlomit.Reich-zeliger@weizmann.ac.il.
Eilon Barnea, Email: eilonb@technion.ac.il.
Chaya Barbolin, Email: chaya.barbolin@weizmann.ac.il.
Ronen Levy, Email: ronen.levy@weizmann.ac.il.
Claudia Arnedo-Pac, Email: claudia.arnedo@irbbarcelona.org.
Shelly Kalaora, Email: shelly.kalaora@weizmann.ac.il.
Bareket Dassa, Email: bareket.dassa@weizmann.ac.il.
Ester Feldmesser, Email: Ester.Feldmesser@weizmann.ac.il.
Ping Shang, Email: Ping.Shang@melanoma.org.au.
Polina Greenberg, Email: polina.greenberg@weizmann.ac.il.
Yishai Levin, Email: Yishai.Levin@weizmann.ac.il.
Gil Benedek, Email: GILB@HADASSAH.ORG.IL.
Mitchell P. Levesque, Email: Mitchell.Levesque@usz.ch.
David J. Adams, Email: david.adams@sanger.ac.uk.
Michal Lotem, Email: mlotem@hadassah.org.il.
Richard A. Scolyer, Email: richard.scolyer@sydney.edu.au.
Arie Admon, Email: admon@tx.technion.ac.il.
Cyrille J. Cohen, Email: cohency@biu.ac.il.
Nuria Lopez-Bigas, Email: nuria.lopez@irbbarcelona.org.
Ansuman T. Satpathy, Email: satpathy@stanford.edu.
Nir Friedman, Email: Nir.Friedman@weizmann.ac.il.
Yardena Samuels, Email: Yardena.samuels@weizmann.ac.il.
References
- 1.Tran E, et al. “Final common pathway” of human cancer immunotherapy: targeting random somatic mutations. Nat Immunol. 2017;18(3):255–262. doi: 10.1038/ni.3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gubin MM, et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 2014;515(7528):577–581. doi: 10.1038/nature13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tumeh PC, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515(7528):568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robbins PF, et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med. 2013;19(6):747–752. doi: 10.1038/nm.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu YC, et al. Efficient identification of mutated cancer antigens recognized by T cells associated with durable tumor regressions. Clin Cancer Res. 2014;20(13):3401–3410. doi: 10.1158/1078-0432.CCR-14-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tran E, et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science. 2014;344(6184):641–645. doi: 10.1126/science.1251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tran E, et al. T-cell transfer therapy targeting mutant KRAS in cancer. N Engl J Med. 2016;375(23):2255–2262. doi: 10.1056/NEJMoa1609279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosenberg SA. Raising the bar: the curative potential of human cancer immunotherapy. Sci Transl Med. 2012;4(127):127ps8. doi: 10.1126/scitranslmed.3003634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ott PA, et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature. 2017;547(7662):217–221. doi: 10.1038/nature22991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sahin U, et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature. 2017;547(7662):222–226. doi: 10.1038/nature23003. [DOI] [PubMed] [Google Scholar]
- 11.Carreno BM, et al. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science. 2015;348(6236):803–808. doi: 10.1126/science.aaa3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalaora S, et al. Use of HLA peptidomics and whole exome sequencing to identify human immunogenic neo-antigens. Oncotarget. 2016;7(5):5110–5117. doi: 10.18632/oncotarget.6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalaora S, et al. Combined analysis of antigen presentation and T-cell recognition reveals restricted immune responses in melanoma. Cancer Discov. 2018;8(11):1366–1375. doi: 10.1158/2159-8290.CD-17-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carbone DP, et al. Immunization with mutant p53- and K-ras-derived peptides in cancer patients: immune response and clinical outcome. J Clin Oncol. 2005;23(22):5099–5107. doi: 10.1200/JCO.2005.03.158. [DOI] [PubMed] [Google Scholar]
- 15.Strønen E, et al. Targeting of cancer neoantigens with donor-derived T cell receptor repertoires. Science. 2016;352(6291):1337–1341. doi: 10.1126/science.aaf2288. [DOI] [PubMed] [Google Scholar]
- 16.Linard B, et al. A ras-mutated peptide targeted by CTL infiltrating a human melanoma lesion. J Immunol. 2002;168(9):4802–4808. doi: 10.4049/jimmunol.168.9.4802. [DOI] [PubMed] [Google Scholar]
- 17.Iii TGD, et al. T cell epitopes encompassing the mutational hot spot position 61 of p21 ras. Promiscuity in ras peptide binding to HLA. Eur J Immunol. 1994;24(2):410–414. doi: 10.1002/eji.1830240221. [DOI] [PubMed] [Google Scholar]
- 18.Sharkey MS, et al. CD4(+) T-cell recognition of mutated B-RAF in melanoma patients harboring the V599E mutation. Cancer Res. 2004;64(5):1595–1599. doi: 10.1158/0008-5472.can-03-3231. [DOI] [PubMed] [Google Scholar]
- 19.Shamalov K, et al. The mutational status of p53 can influence its recognition by human T-cells. Oncoimmunology. 2017;6(4):e1285990. doi: 10.1080/2162402X.2017.1285990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tran E, et al. Immunogenicity of somatic mutations in human gastrointestinal cancers. Science. 2015;350(6266):1387–1390. doi: 10.1126/science.aad1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malekzadeh P, et al. Neoantigen screening identifies broad TP53 mutant immunogenicity in patients with epithelial cancers. J Clin Invest. 2019;129(3):1109–1114. doi: 10.1172/JCI123791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deniger DC, et al. T-cell responses to TP53 “Hotspot” mutations and unique neoantigens expressed by human ovarian cancers. Clin Cancer Res. 2018;24(22):5562–5573. doi: 10.1158/1078-0432.CCR-18-0573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cafri G, et al. Memory T cells targeting oncogenic mutations detected in peripheral blood of epithelial cancer patients. Nat Commun. 2019;10(1):449. doi: 10.1038/s41467-019-08304-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iiizumi S, et al. Identification of novel HLA class II-restricted neoantigens derived from driver mutations. Cancers (Basel) 2019;11(2):266. doi: 10.3390/cancers11020266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499(7457):214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.et al. MHC-I genotype restricts the oncogenic mutational landscape. Cell. 2017;171(6):1272–1283. doi: 10.1016/j.cell.2017.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van den Eynden J, et al. Lack of detectable neoantigen depletion signals in the untreated cancer genome. Nat Genet. 2019;51(12):1741–1748. doi: 10.1038/s41588-019-0532-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yadav M, et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature. 2014;515(7528):572–576. doi: 10.1038/nature14001. [DOI] [PubMed] [Google Scholar]
- 29.Simanshu DK, et al. RAS proteins and their regulators in human disease. Cell. 2017;170(1):17–33. doi: 10.1016/j.cell.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jurtz V, et al. NetMHCpan-4.0: improved peptide-MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data. J Immunol. 2017;199(9):3360–3368. doi: 10.4049/jimmunol.1700893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fritsch EF, et al. Cancer immunology miniatures HLA-binding properties of tumor neoepitopes in humans. Cancer Immunol Res. 2014;2(6):522–529. doi: 10.1158/2326-6066.CIR-13-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang J, et al. International Cancer Genome Consortium Data Portal – a one-stop shop for cancer genomics data. Database (Oxford) 2011;2011:bar026. doi: 10.1093/database/bar026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bailey MH, et al. Comprehensive characterization of cancer driver genes and mutations. Cell. 2018;173(2):371–385. doi: 10.1016/j.cell.2018.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Priestley P, et al. Pan-cancer whole-genome analyses of metastatic solid tumours. Nature. 2019;575(7781):210–216. doi: 10.1038/s41586-019-1689-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hayward NK, et al. Whole-genome landscapes of major melanoma subtypes. Nature. 2017;545(7653):175–180. doi: 10.1038/nature22071. [DOI] [PubMed] [Google Scholar]
- 36.Wang Q, et al. Cancer immunology miniatures direct detection and quantification of neoantigens. Cancer Immunol Res. 2019;7(11):1748–1754. doi: 10.1158/2326-6066.CIR-19-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 38.Arafeh R, et al. Recurrent inactivating RASA2 mutations in melanoma. Nat Genet. 2015;47(12):1408–1410. doi: 10.1038/ng.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Monte U Del Does the cell number 10 9 still really fit one gram of tumor tissue? Cell Cycle. 2009;8(3):505–506. doi: 10.4161/cc.8.3.7608. [DOI] [PubMed] [Google Scholar]
- 40.Giam K, et al. A comprehensive analysis of peptides presented by HLA-A1. Tissue Antigens. 2015;85(6):492–496. doi: 10.1111/tan.12565. [DOI] [PubMed] [Google Scholar]
- 41.Toor JS, et al. A Recurrent Mutation in Anaplastic Lymphoma Kinase with Distinct Neoepitope Conformations. Front Immunol. 2018;9:99. doi: 10.3389/fimmu.2018.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.London N, et al. Rosetta FlexPepDock web server — high resolution modeling of peptide-protein interactions. Nucleic Acids Res. 2011;39(Web Server issue):W249–253. doi: 10.1093/nar/gkr431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vajda S, et al. New additions to the ClusPro server motivated by CAPRI. Proteins. 2017;85(3):435–444. doi: 10.1002/prot.25219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Antunes DA, et al. DINC 2.0: a new protein–peptide focking eebserver using an incremental approach. Cancer Res. 2017;77(21):e55–e57. doi: 10.1158/0008-5472.CAN-17-0511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Greiff V, et al. Bioinformatic and statistical analysis of adaptive immune repertoires. Trends Immunol. 2015;36(11):738–749. doi: 10.1016/j.it.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 46. Fred Hutch. Fred Hutchinson Cancer Research Center. https://www.fredhutch.org/en.html Accessed September 3, 2021.
- 47.Liu P, et al. Characterization of human αβTCR repertoire and discovery of D-D fusion in TCRβ chains. Protein Cell. 2014;5(8):603–615. doi: 10.1007/s13238-014-0060-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emerson RO, et al. Immunosequencing identifies signatures of cytomegalovirus exposure history and HLA-mediated effects on the T cell repertoire. Nat Genet. 2017;49(5):659–665. doi: 10.1038/ng.3822. [DOI] [PubMed] [Google Scholar]
- 49.Krishna S, et al. Stem-like CD8 T cells mediate response of adoptive cell immunotherapy against human cancer. Science. 2020;370(6522):1328–1334. doi: 10.1126/science.abb9847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Szabo PA, et al. Single-cell transcriptomics of human T cells reveals tissue and activation signatures in health and disease. Nat Commun. 2019;10(1):1–16. doi: 10.1038/s41467-019-12464-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Poschke IC, et al. The outcome of ex vivo TIL expansion is highly influenced by spatial heterogeneity of the tumor T-cell repertoire and differences in intrinsic in vitro growth capacity between T-cell clones. Clin Cancer Res. 2020;26(16):4289–4301. doi: 10.1158/1078-0432.CCR-19-3845. [DOI] [PubMed] [Google Scholar]
- 52.Sim MJW, et al. High-affinity oligoclonal TCRs define effective adoptive T cell therapy targeting mutant KRAS-G12D. Proc Natl Acad Sci U S A. 2020;117(23):12826–12835. doi: 10.1073/pnas.1921964117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guida M, et al. No impact of NRAS mutation on features of primary and metastatic melanoma or on outcomes of checkpoint inhibitor immunotherapy: An italian melanoma intergroup (IMI) study. Cancers (Basel) 2021;13(3):1–15. doi: 10.3390/cancers13030475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnson DB, et al. Impact of NRAS mutations for patients with advanced melanoma treated with immune therapies. Cancer Immunol Res. 2015;3(3):288–295. doi: 10.1158/2326-6066.CIR-14-0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lebbink RJ, et al. The soluble leukocyte-associated Ig-like receptor (LAIR)-2 antagonizes the collagen/LAIR-1 inhibitory immune interaction. J Immunol. 2008;180(3):1662–1669. doi: 10.4049/jimmunol.180.3.1662. [DOI] [PubMed] [Google Scholar]
- 56.Simone R, et al. Serum LAIR-2 is increased in autoimmune thyroid diseases. PLoS One. 2013;8(5):e63282. doi: 10.1371/journal.pone.0063282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nordkamp MJMO, et al. Enhanced secretion of leukocyte-associated immunoglobulin-like receptor 2 (LAIR-2) and soluble LAIR-1 in rheumatoid arthritis: LAIR-2 is a more efficient antagonist of the LAIR-1-collagen inhibitory interaction than is soluble LAIR-1. Arthritis Rheum. 2011;63(12):3749–3757. doi: 10.1002/art.30612. [DOI] [PubMed] [Google Scholar]
- 58.Ly D, et al. P1.04-05 elucidating the role of leukocyte-associated immunoglobin-like receptor 2 (LAIR2) in lung cancer development. J Thorac Oncol. 2018;13(10):S526–S527. doi: 10.1016/j.jtho.2018.08.720. [DOI] [Google Scholar]
- 59.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data: Figure 1. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martínez-Jiménez F, et al. A compendium of mutational cancer driver genes. Nat Rev Cancer. 2020;20(10):555–572. doi: 10.1038/s41568-020-0290-x. [DOI] [PubMed] [Google Scholar]
- 62.Lu YC, et al. Mutated PPP1R3B is recognized by T cells used to treat a melanoma patient who experienced a durable complete tumor regression. J Immunol. 2013;190(12):6034–6042. doi: 10.4049/jimmunol.1202830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wei X, et al. Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat Genet. 2011;43(5):442–446. doi: 10.1038/ng.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Klapper JA, et al. Single-pass, closed-system rapid expansion of lymphocyte cultures for adoptive cell therapy. J Immunol Methods. 2009;345(1–2):90–99. doi: 10.1016/j.jim.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hérin M, et al. Production of stable cytolytic T-cell clones directed against autologous human melanoma. Int J Cancer. 1987;39(3):390–396. doi: 10.1002/ijc.2910390320. [DOI] [PubMed] [Google Scholar]
- 66.Miller BC, et al. Subsets of exhausted CD8+ T cells differentially mediate tumor control and respond to checkpoint blockade. Nat Immunol. 2019;20(3):326–336. doi: 10.1038/s41590-019-0312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sade-Feldman M, et al. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell. 2019;176(1–2):404. doi: 10.1016/j.cell.2018.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Scott AC, et al. TOX is a critical regulator of tumour-specific T cell differentiation. Nature. 2019;571(7764):270–274. doi: 10.1038/s41586-019-1324-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.