Abstract
Background
GLS4 is a first-in-class hepatitis B virus (HBV) capsid assembly modulator (class I) that can inhibit HBV replication by interfering with the assembly and disassembly of HBV nucleocapsid. Here, we evaluated its antiviral activity, pharmacokinetics, and tolerability in a double-blind, randomized, parallel, entecavir-controlled study.
Methods
Twenty-four patients with chronic HBV were randomized to receive a 28-day course of GLS4 (120 or 240 mg) and ritonavir (100 mg) combination (cohorts A and B, respectively) or entecavir treatment (cohort C) at a 1:1:1 ratio. Patients were followed up for 40 days in a phase 1b study.
Results
The GLS4/ritonavir combination was a tolerated combination for the treatment of chronic HBV infection. A total of 2, 3, and 3 subjects presented with alanine aminotransferase flare in cohorts A, B, and C, respectively. This contributed to the withdrawal of 1, 2, and 1 patient from cohorts A, B, and C, respectively. The mean Ctrough of GLS4 was 205–218 ng/mL, which was approximately 3.7–3.9 times the 90% effective concentration (55.8 ng/mL), with a lower accumulation (accumulation rate, 1.1–2.0). In cohorts A, B, and C, the mean declines in HBV DNA after 28 days of treatment were −1.42, −2.13, and −3.5 log10 IU/mL; in hepatitis B surface antigen were −0.06, −0.14, and −0.33 log10 IU/mL; in pregenomic RNA were −0.75, −1.78, and −0.96 log10 copies/mL; and in hepatitis B core antigen were −0.23, −0.5, and −0.44 log10 U/mL, respectively.
Conclusions
Treatment with 120 mg GLS4 was tolerated and had antiviral activity in patients with chronic HBV infection.
Clinical Trials Registration
Chinese Clinical Trial Registry; CTR20160068. http://www.chinadrugtrials.org.cn.
Keywords: hepatitis B treatment, capsid assembly modulator, clinical trial, hepatitis B virus, response
GLS4 is a first-in-class hepatitis B virus (HBV) capsid assembly modulator (class I). Here, we confirm that GLS4 (120 mg) is tolerated by patients and can inhibit HBV DNA, HBsAg, and HBeAg, as well as HBV pgRNA and HBcrAg.
Chronic viral hepatitis due to hepatitis B virus (HBV) remains a leading cause of premature mortality worldwide, with an estimated 0.6–1 million deaths/year [1, 2]. Currently, treatment of chronic HBV infection is mainly based on the use of interferon and nucleos(t)ide analogues (NUCs). Nevertheless, the emergence of drug resistance and viral relapse and side effects pose a major concern [3].
HBV capsid protein plays an important role in viral DNA synthesis from pregenomic RNA (pgRNA) [4, 5]. The encapsidation of HBV pgRNA is an evolutionarily conserved step [6, 7]. Therefore, developing pharmacological agents that target the HBV capsid may be efficient for various HBV genotypes [8].
GLS4 is a novel HBV class I capsid assembly modulator (CpAM), which induces the formation of aberrant nucleocapsid structures (Supplementary Material) [9, 10]. GLS4 is derived from phenylpropenamide and has better anti-HBV activity in vitro than Bay 41–4109 (a phenylpropenamide derivative) [11]. GLS4 was shown to be well tolerated and metabolized by cytochrome P450 3A4 (CYP3A4) metabolic enzymes in a preclinical study in dogs [12]. To increase the exposure level of GLS4, ritonavir was selected to inhibit CYP3A4 metabolic enzymes. Following GLS4 (120 or 240 mg) in combination with 100 mg ritonavir treatment, the steady-state minimum concentration (Cmin) was increased to the 90% effective concentration (EC90) (mean steady-state Cmin was 187.9 ng/mL and 300.7 ng/mL, ~3.37- and 5.38-fold that of the EC90 value in vitro), and the treatment was shown to be tolerated by healthy subjects (unpublished observations).
In this study, we examined the tolerance, pharmacokinetics (PK), and efficacy of GLS4 (120 or 240 mg) in combination with 100 mg ritonavir for treating chronic HBV infection compared with entecavir in a phase 1b trial.
METHODS
Study Design and Participants
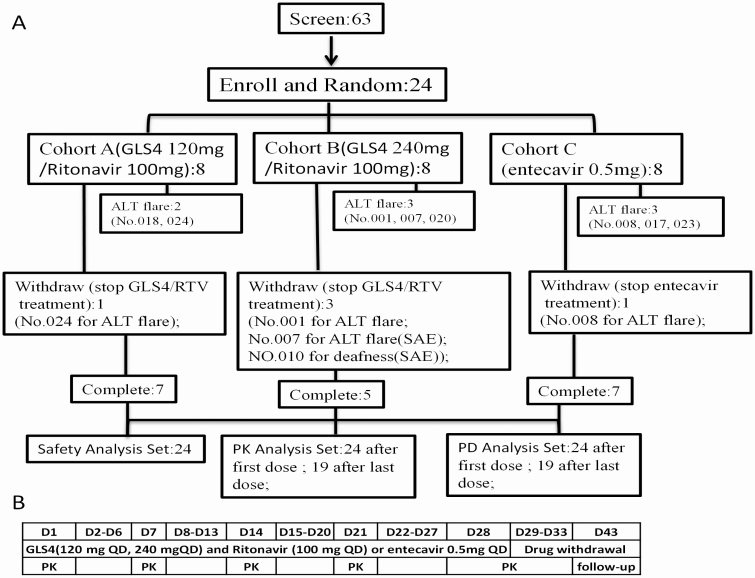
This was a randomized, open-label, active-controlled, phase 1b study (Chinese Clinical Trial Registry, registration no. CTR20160068). Patients received a combination of GLS4 and ritonavir or entecavir. A total of 24 eligible patients with chronic HBV infection were enrolled between April 2016 and July 2016. They were randomized 1:1:1 to receive a combination of GLS4 120 mg/day and ritonavir 100 mg/day or GLS4 240 mg/day and ritonavir 100 mg/day (cohorts A or B, respectively) or entecavir 0.5 mg/day (cohort C) for 28 days (Figure 1). Each group included 8 subjects to avoid exposing patients to toxic or ineffective dosages, according to the guidance on the clinical PK of novel candidate compounds.
Figure 1.
A, Study flow chart. B, The trial duration and test schedules in the study. Abbreviations: ALT, alanine aminotransferase; D, day; PD, pharmacodynamics; PK, pharmacokinetics; QD, once a day; RTV, ritonavir; SAE, serious adverse events.
The main inclusion criteria were as follows: patients with a negative blood human chorionic gonadotropin; presence of chronic HBV infection (hepatitis B surface antigen [HBsAg] positive [+], hepatitis B e antigen positive [HBeAg+] and hepatitis B core antibody [HBcAb] immunoglobulin M [IgM] negative [−]); HBV DNA ≥1 × 105 IU/mL; serum alanine aminotransferase (ALT) levels within 2–10 times the upper limit of normal (ULN); a FibroScan score of 17.5 or less within 6 months or liver tissue showing no cirrhosis within 12 months prior to enrollment; treatment-naive patients or patients who stopped interferon or NUCs treatment at least 6 months prior to enrollment; and patients who did not receive any antiviral therapy including Chinese herbal medicine, immunomodulators, thymosin, or other immune-stimulating factors within 6 months before enrollment. The main exclusion criteria included a serum total bilirubin of more than 2 times the ULN; coinfection with human immunodeficiency virus or hepatitis C virus or syphilis; subjects with concurrent severe chronic medical conditions or with malignancy; and subjects with liver cirrhosis (FibroScan score >17.5 within 6 months and clinical judgement [liver and spleen ultrasound and platelet count or liver tissue showing signs of cirrhosis]) were excluded (Supplementary Material).
Procedures
Patients were required to visit the facility for follow-up at days 7, 14, 21, 27, and 40; and early termination (Supplementary Material). FibroScan was performed during screening. Alcohol consumption and smoking were not allowed 7 days before the initial dosing until the end of the study.
This study was conducted at the Phase I Clinical Research Center, The First Hospital of Jilin University. The study protocol and informed consent forms adhered to the principles of the Declaration of Helsinki and were approved by an independent ethics committee or institutional review board. All subjects provided written informed consent before participating in any study-related procedure.
Treatment with silibinin and glutathione was done according to the physician’s discretion to protect and repair the hepatocyte membrane and reduce the hepatic cell damage in subjects with ALT flares. Therefore, several subjects were treated with silibinin and glutathione, while others were not.
Outcomes
The primary objective of this study was to evaluate the safety/tolerability of GLS4 after administration for 28 days. Secondary objectives included the PK and antiviral activities of GLS4.
Assessments
Safety and tolerability evaluations were based on adverse events (AEs), clinical laboratory tests, vital signs, electrocardiograms (ECGs), and physical examination. The Common Terminology Criteria for Adverse Events (CTCAE) 4.03 were used to grade AEs and laboratory and ECG abnormalities; PK assessments were obtained with the noncompartmental model. Antiviral activity assessments were based on observed antiviral activity, and the primary endpoint was the change in serum HBV DNA within the 28 days of treatment. Other endpoints included the change in the serum HBsAg, HBeAg, pgRNA and hepatitis B core-related antigen (HBcrAg). These factors were detected by quantitative polymerase chain reaction (PCR), Abbott Architect assays, and the Lumipulse G HBcrAg assay. The virus resistance profile was detected by PCR or DNA sequencing in all subjects as previously described [1, 2]. Detection methods are detailed in the Supplementary Material.
Statistical Analysis
Plasma PK parameters, including the maximum observed plasma concentration (Cmax), time to maximum observed plasma concentration (Tmax), area under the concentration-time curve from the dosing start point to the last time point with measurable plasma concentration (AUC0–t) prior to next dose, AUC from time of dosing extrapolated to infinity (AUC0–∞), as well as the terminal elimination half-life of the drug in plasma (t½), clearance (CL/F), volume (Vz/F), accumulation rate, and degree of fluctuation, were estimated by the noncompartmental PK approach using WinNonlin software. Variables were also analyzed by the Student’s t test or Kruskal–Wallis test, regression analysis, or correlation analysis using SAS software (SAS Institute).
RESULTS
Among the 63 recruited patients, 24 fulfilled the inclusion and exclusion criteria and all of them were NUCs naive. A total of 20 of 24 (83%) patients were males and 4 of 24 (17%) patients were females. Five patients withdrew from the study without completing the antiviral treatment and PK analysis. Specifically, 4 patients withdrew due to ALT flares (cohort A, patient 024; cohort B, patients 001 and 007 [serious AEs, SAEs]; and cohort C, patient 008) and 1 patient withdrew from cohort B (010; SAE) due to deafness (Figure 1). The demographic characteristics and clinical features were matched among the different cohorts (Table 1). However, HBsAg and HBeAg levels were lower in cohort C (entecavir treatment) compared with cohorts A and B. All enrolled patients were HBeAg-positive and of Han Chinese ethnicity.
Table 1.
Baseline Demographic Characteristics and Clinical Features
| Baseline Parameter | Cohort A (n = 8) | Cohort B (n = 8) | Cohort C (n = 8) |
|---|---|---|---|
| Gender (male/female), n/n | 6/2 | 7/1 | 4/4 |
| Ethnicity (Han/other), n/n | 6/2 | 7/1 | 7/1 |
| Age, years | 33.6 ± 9.05 | 36.4 ± 10.56 | 31.5 ± 4.28 |
| Smoking (yes/no), n | 3/5 | 4/4 | 0/8 |
| Drinking (yes/no), n | 0/8 | 2/6 | 0/8 |
| BMI, kg/m2 | 24.30 ± 3.42 | 23.85 ± 3.49 | 23.54 ± 2.07 |
| HBV DNA, log10 IU/mL | 8.24 ± 0.89 | 8.274 ± 0.89 | 8.044 ± 0.49 |
| HBsAg, log10 IU/mL | 4.51 ± 0.67 | 4.21 ± 0.52 | 3.77 ± 0.52 |
| HBeAg, log10 IU/mL | 3.09 ± 0.38 | 2.96 ± 0.53 | 2.50 ± 1.15 |
| HBV pgRNA, log10 copies/mL | 7.50 ± 1.00 | 7.50 ± 1.00 | 7.33 ± 0.61 |
| ALT, U/L | 144 ± 97 | 160 ± 87 | 162 ± 107 |
| FibroScan score | 8.44 ± 4.01 | 9.01 ± 2.40 | 8.15 ± 2.20 |
| AFP,a ng/mL | 4.14(3.07,6.42) | 5.01(2.98,7.03) | 11.61(2.93,23.51) |
| Genotype | B = 1, C = 6, other = 1 | B = 0, C = 5, other = 3 | B = 0, C = 7, other = 1 |
Data are presented as means ± SDs unless otherwise indicated.
Abbreviations: AFP, alpha fetoprotein; ALT, alanine aminotransferase; BMI, body mass index; HBeAg, hepatitis B e antigen; HBsAg, hepatitis B surface antigen; HBV, hepatitis B virus; pgRNA, pregenomic RNA; Q1, quartile1; Q3, quartile3.
aValues are medians (Q1, Q3).
Tolerability
Among the enrolled patients, 18 of 24 (75%) subjects experienced an adverse reaction (5, 6, and 7 patients in cohorts A, B, and C, respectively), with adverse reaction frequencies of 15, 29, and 22 in cohorts A, B, and C, respectively. The most common adverse reactions were ALT elevation, aspartate aminotransferase elevation, and γ-glutamyl transferase elevation (Supplementary Table 1). Most adverse reactions were mild or moderate in intensity and did not require treatment. Eight subjects had ALT flares, and 4 discontinued GLS4/ritonavir or entecavir treatment, withdrew from the study, and needed silibinin and glutathione treatment. In addition, another subject experienced ALT flares and required silibinin and glutathione treatment without discontinuing the antiviral treatment (Figure 1, Supplementary Table 2).
Patient 007 experienced ALT flares (maximum ALT, 511 IU/mL, grade 4 according to the CTCAE criteria) after 8 days of GLS4 treatment, with liver injury (SAE), and this patient recovered after receiving silibinin and glutathione for 31 days. In addition, patients 001, 008, and 024 experienced ALT flares (grade 3–4), which led to their withdrawal from the study at 7 to 8 days. Those patients recovered after taking silibinin and glutathione for 9–35 days. Other subjects with ALT flares were not treated and they spontaneously recovered, except for patient 023 who received treatment for 15 days. Silibinin and glutathione treatment was effective in reducing ALT flares, as demonstrated by the decline in ALT level (Supplementary Figure 1, Supplementary Table 2).
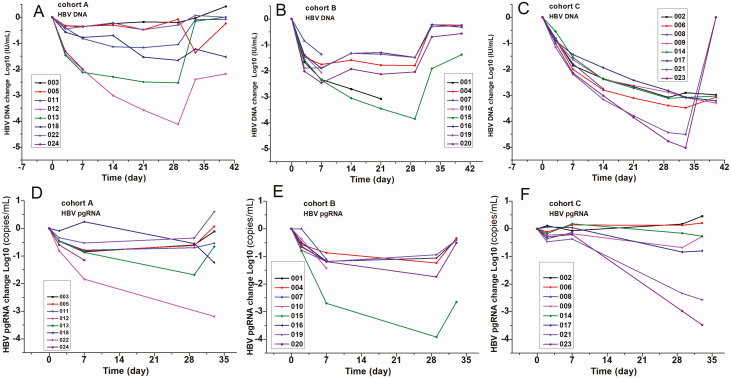
Patient 010 presented with deafness (SAE) in the right ear, which occurred after 8 days of GLS4 treatment. However, it was not related to treatment as judged by an otolaryngologist because early signs of deafness were present before starting the treatment. This patient withdrew from the study at 8 days, and his laboratory results were normal or similar to the level at initial screening. Furthermore, the screening FibroScan scores ranged from 4.3 to 11.7, except for patient 012 (cohort A) who had a score of 16.9. Interestingly, this patient did not show signs of ALT flare and GLS4 treatment resulted in antiviral activity (Figure 2A).
Figure 2.
Changes in therapeutic effect factors at different time points following treatment in each cohort. Patients in cohort A were treated with a combination of GLS4 120 mg/day and ritonavir 100 mg/day; cohort B, GLS4 240 mg/day and ritonavir 100 mg/day; and cohort C, entecavir 0.5 mg/day for 28 days. A–C, Change in HBV DNA among patients in cohorts A, B, and C, respectively. D–F, Change in the HBV pgRNA value of cohorts A–C, respectively. Abbreviations: HBV, hepatitis B virus; pgRNA, pregenomic RNA.
Antiviral Activity
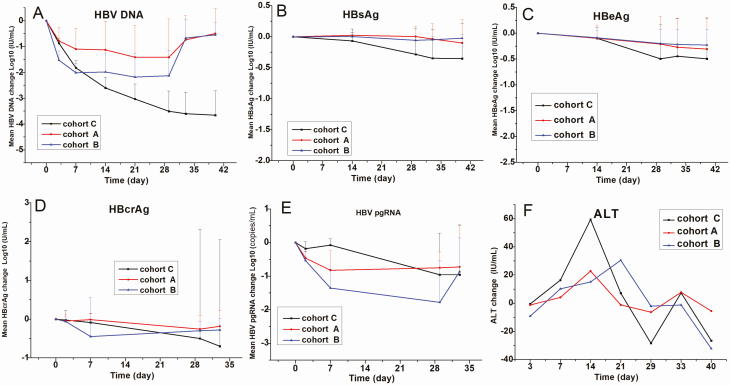
The mean declines in level of HBV DNA were −1.42, −2.13, and −3.5 log10 IU/mL in cohorts A, B, and C, respectively (Figures 2 and 3). On day 40, virological relapse was more frequently observed in cohorts A and B. It is worth noting that patients 003, 005, and 022 in cohort A had a poor response to GLS4 treatment (decline in HBV DNA levels of <0.5 log10 IU/mL). Following the 28 days of treatment, the mean declines in HBsAg were −0.06, −0.14, and −0.33 log10 IU/mL, and the mean declines in HBeAg were −0.25, −0.30, and −0.43 log10 IU/mL in cohorts A, B, and C, respectively. None of the treated patients had HBsAg clearance, HBeAg seroconversion, or HBsAg seroconversion (Figure 3, Supplementary Figure 2).
Figure 3.
The mean change of therapeutic effect factors at different time points following GLS4 and ritonavir combination or entecavir treatment. Patients in cohort A were treated with a combination of GLS4 120 mg/day and ritonavir 100 mg/day; cohort B, GLS4 240 mg/day and ritonavir 100 mg/day; and cohort C, entecavir 0.5 mg/day for 28 days. A–F show the means changes in mean HBV DNA, HBsAg, HBeAg, HBcrAg, HBV pgRNA, and ALT values, respectively. Abbreviations: ALT, alanine aminotransferase; HBeAg, hepatitis B e antigen; HBsAg, hepatitis B surface antigen; HBcrAg, hepatitis B core-related antigen; HBV, hepatitis B virus; pgRNA, pregenomic RNA.
The mean declines in pgRNA were −0.75, −1.78, and −0.96 log10 copies/mL in cohorts A, B, and C, respectively. HBV pgRNA significantly declined in cohort B compared with cohorts A and C (Supplementary Material). None of the enrolled patients had undetectable HBV pgRNA by the end of treatment, and pgRNA levels returned to baseline levels in most patients. After the 28 days of treatment, the mean declines in HBcrAg were −0.23, −0.50, and −0.44 log10 U/mL in cohorts A, B, and C, respectively. Patient 007 had a clear decline in HBcrAg in cohort B (Figures 2 and 3, Supplementary Figure 2).
Compared with baseline levels, the mean change in ALT levels demonstrated a decreasing trend among all patients (Figure 3F). It is noteworthy that GLS4 resistance was not observed among patients in cohorts A and B, and GLS4 was effective even in those who were resistant to lamivudine or adefovir dipivoxil treatment (cohort A: patient 013; cohort B: patient 020). Interestingly, all subjects with ALT flares had lower HBV DNA and HBV pgRNA, and a total of 6 patients (018, 001, 007, 008, 017, and 023) had decreased HBsAg and HBeAg after treatment (Figure 2, Supplementary Figure 1).
Pharmacokinetics of GLS4
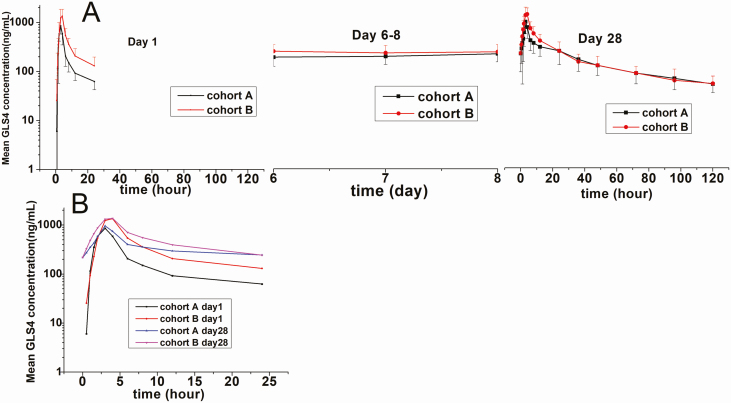
The GLS4 concentration-time profiles of the different cohorts are shown in Figure 4, and their PK parameters are shown in Table 2. Following the first and last dose of GLS4/ritonavir on days 1 and 28, respectively, GLS4 concentrations increased after drug administration, reaching a maximum level at approximately 3–3.5 hours. GLS4 exposure also increased with dose and time. The end-stage elimination of GLS4 was a 2-phase process. The half-life (t1/2) of GLS4 was longer after multiple doses (Supplementary Material). GLS4 concentration reached a steady state approximately 6 days after administration. The mean trough concentration (Ctrough) of GLS4 was 205–218 ng/mL (cohorts A and B). GLS4 was metabolized into GLS4-M1 to GLS4-M4 by oxidation, and these metabolites did not demonstrate anti-HBV activity. The accumulation rates (AUC0–24 of day 28 vs AUC0–24 of day 1 or Cmax of day 28 vs Cmax of day 1) were similar between cohorts A and B: 1.6–2.0 with AUC0–24 and 1.1–1.2 with Cmax. The AUC, t1/2, and Vz/F of GLS4 increased, while CL/F decreased, and Cmax and Tmax did not change between the first and the last dose. The range of GLS4 exposures (Cmax and AUC) demonstrated a saturation trend (regression coefficient, 0.23 to 0.91).
Figure 4.
The plasma concentration-time profiles in cohorts A and B following the first and last treatment dose. Patients in cohort A were treated with combination of GLS4 120 mg/day and ritonavir 100 mg/day, and patients in cohort B were treated with GLS4 240 mg/day and ritonavir 100 mg/day for 28 days. A, Mean log (±SD) GLS4 plasma concentration-time profiles. B, Mean log GLS4 plasma concentration-time profiles at 0–24 hours.
Table 2.
Comparison of Pharmacokinetic Parameters of GLS4 Between the First and Last Dose in Each Treatment Cohort
| Study Day | ||||||||
|---|---|---|---|---|---|---|---|---|
| Cohort A | Cohort B | |||||||
| Parameter | Day 1 (24 Hours) | Day 28 (24 Hours) | Day 28 (120 Hours) | P | Day 1 (24 Hours) | Day 28 (24 Hours) | Day 28 (120 Hours) | P |
| AUC0–∞, ng × hours/mL | … | … | 24 240 (39) | … | … | 22 585 (60) | ||
| AUC0–t, ng × hours/mL | 4268 (27) | 7871(56) | 17 161 (49) | <.01 | 8055 (34) | 11 289(29) | 20 166 (37) | <.01 |
| Cmax, ng/mL | 885 (27) | 865 (66) | 865 (66) | .9 | 1447 (39) | 1461 (24) | 1461 (24) | .98 |
| Median Tmax (minimum–maximum), hours | 3 (1.5,4) | 3 (0.5, 4) | 3 (0.5, 4) | .625 | 3.5 (2, 4) | 3 (2, 4) | 3 (2, 4) | .89 |
| CL/F, mL/min | 22 134 (27) | 6727(76) | 5523 (45) | <.01 | 24 094 (42) | 14 461(49) | 9538 (38) | .01 |
| Vz/F, mL | 408 677 (37) | 294 145(51) | 499 250 (60) | >.05 | 355 766 (31) | 310 831(32) | 873 579 (58) | <.01 |
| t1/2, hours | 13.1 (3.4) | 32.1(11.7) | 64.2 (14.8) | <.01 | 10.6 (3.3) | 16.3(7.6) | 67.3 (24.2) | <.01 |
| AUC0–24, ng × hours/mL | 4268 (27) | 7871 (56) | 7871 (56) | >.05 | 8055 (34) | 11 289 (29) | 11 289 (29) | >.05 |
| Df, % | … | … | 204 (36) | … | … | 265 (33) | ||
| Ctrough, ng/mL | … | 218(59) | … | … | 205(63) | … | ||
| Accumulation rate AUC0–24 | … | … | 2.0(45) | … | … | 1.6(7) | ||
| Accumulation rate Cmax | … | … | 1.1(53) | … | … | 1.2(31) | ||
Data are presented as means (CV). The time range for PK parameter calculation were 24, 24 and 120 hours at day1 (24 hours), day 28 (24 hours), and day 28 (120 hours), respectively.
Abbreviations: AUC0–t, area under the concentration-time curve from time of dosing to the last time point with measurable plasma concentration prior to next dose; AUC0–∞, AUC from time of dosing extrapolated to infinity; CL/F, clearance; Cmax, maximum observed plasma concentration; Ctrough, trough concentration; CV, coefficient of variation; Df, degree of fluctuation; Tmax, time to maximum observed plasma concentration; t1/2, terminal elimination half-life of the drug in plasma; Vz/F, volume.
Relationship Between Maximum ALT Level With Baseline Factors and GLS4 Exposure Level
A strong relationship was observed between maximum ALT levels and baseline ALT levels during treatment. A positive correlation was also observed between baseline ALT levels and the intensity of AEs. In particular, a baseline ALT level greater than 3 times the ULN was associated with the patient being prone to ALT flares, whereas a baseline ALT level of more than 5 times the ULN was associated with the patient being prone to treatment discontinuation (patients 001, 007, and 024) (Supplementary Figures 3 and 4). There was a minor association between baseline HBV DNA and GLS4 exposure with maximum ALT levels (Supplementary Figure 3).
DISCUSSION
In this study, we examined the clinical tolerability, efficacy, and PK of GLS4 in patients with chronic HBV infection. Interestingly, our results demonstrated a decline in serum HBV DNA and HBV pgRNA in patients of cohorts A and B. Compared with cohort B, patients in cohort A were less susceptible to ALT flares (2/8 vs 3/8), withdrawal from the study due to ALT flares (1/8 vs 2/8), frequency of adverse reactions (15 vs 29), frequency of grade 3–4 adverse reactions (2 vs 7), and presence of SAEs (0 vs 1). Therefore, it is plausible that the drug combination in cohort A was safer than that in cohort B, and thus tolerance in cohort A was acceptable.
A total of 5 patients were treated with silibinin and glutathione treatment following ALT flares. One subject in the control group (cohort C) was given silibinin and glutathione treatment starting on day 14 of the study, for a total of 15 days. This had some confounding effects on the study, such as tolerance, but this subject was diagnosed with ALT flare. The remaining 4 patients were treated with silibinin and glutathione after discontinuation of the clinical study drug, and were also diagnosed with ALT flares. Therefore, the confounding effect on tolerance was relatively small. In terms of efficacy, silibinin and glutathione have no antiviral effects and only protect and repair the hepatocyte membranes [13, 14]. Therefore, it is plausible to state that silibinin and glutathione treatment had little confounding effect on efficacy.
In this study, subjects with high baseline ALT levels were prone to ALT flares. However, ALT flares were also correlated with the decline in HBV DNA and viral antigens, indicating the occurrence of therapeutic flares [15]. Therefore, higher ALT level at baseline is a double-edged sword as it indicates immune system activation and liver cell damage [15]. Following treatment, the rise in ALT level can possibly be attributed to a further activation of the immune system [16]. In our previous study, we observed that adefovir dipivoxil treatment resulted in an increase in T-helper cell 1 (Th1)/Th2 cytokines producing T cells and serum cytokine levels and a decline in HBV DNA level [17]. Those findings (decreased viral antigens and HBV DNA level observed in some patients) were observed in cohort C following treatment, which also supports the above-mentioned analysis (Supplementary Material). Further, due to the absence of ALT flares in healthy subjects, ALT flares may be immune mediated or attributed to the induction of death in infected hepatocytes [15].
Our patient cohort had higher mean baseline ALT levels (144–162 vs 27–45 IU/mL) than the baseline ALT levels observed in class II heteroaryldihydropyrimidines (capsid assembly modulators [CpAMs]) phase I clinical studies such as ABI-H0731, without ALT flare [16, 18]. Subjects with higher baseline ALT levels were prone to have higher maximum ALT levels. Therefore, appropriate ALT levels such as those less than 5 times the ULN can be recommended as inclusion criteria for GLS4-based therapy. The small sample size (8 subjects/cohort) and inconsistent ALT baseline of our patients hindered the computation of relationship analysis between GLS4 exposure and maximum ALT level. Therefore, future studies are needed to fully address this issue.
Finally, the ALT flare was neither accompanied by an increase in bilirubin, international normalized ratio, or prothrombin time nor a decrease in serum albumin; thus, the risk of developing severe liver injury is low, albeit this requires carefully designed and larger cohort studies. It is worth mentioning that we reported SAEs in the initial phase of the study when ALT was increased by 10 times even if those patients recovered after silibinin treatment or spontaneously. Meanwhile, future immune-function evaluation will be needed to analyze the causes of the elevated transaminase.
GLS4 could trigger aberrant HBV core particle assembly in vitro, thereby inhibiting the accumulation of covalent close circle DNA (cccDNA) and core gene expression [12, 19]. The primary efficacy analysis of serum HBV DNA levels revealed a consistent antiviral effect in the GLS4-treated cohort. Following GLS4 treatment, the mean change in HBV DNA was approximately −1.42 to −2.13 log10 IU/mL, which was higher than that observed after class II CpAMs NVR 3–778 treatment (−1.43 log10 IU/mL). The mean declines (log10 IU/mL) in HBsAg and HBeAg levels were also superior than those observed after NVR 3–778 treatment (HBsAg: −0.14 vs −0.02; HBeAg: −0.43 vs −0.09, respectively) [9]. These results indicate that class I CpAMs GLS4 can efficiently inhibit HBV DNA, HBsAg, and HBeAg levels.
Serum HBV pgRNA and HBcrAg levels reflect the status of viral replication and cccDNA inside hepatocytes [20]. Previous preclinical research revealed that capsid inhibitors have multifaceted antiviral mechanisms [11]. CpAMs was found to induce HBV core protein aggregation and degradation, which can induce endoplasmic reticulum stress and cell death. In addition, the destruction of capsid protein increases the viral nucleic acid exposure, thereby stimulating the innate immune response through a pattern-recognition receptor [21–23]. GLS4 belongs to CpAMs. Moreover, the HBcrAg level could also reflect HBV core protein level. The decreased HBcrAg level observed in this study supports the degradation of core protein, thereby resulting in a stronger HBV-specific immune response in the infected hepatocytes [24]. Meanwhile, GLS4 prevents capsid assembly, depletes cccDNA levels, inhibits the production of HBV pgRNA-containing particles, and prevents viral replication [20, 25–27]. On the other hand, entecavir inhibits the polymerase-mediated reverse transcription of encapsidated HBV pgRNA to DNA, but does not prevent continued formation and secretion of pgRNA-containing particles [9]. Therefore, theoretically, entecavir should not inhibit pgRNA levels within the 28 days of treatment.
Interestingly, the correlation between HBV DNA and pgRNA responses was lower in cohort C (regression coefficient = 0.39) than in cohorts A and B (regression coefficients = 0.62 and 0.76, respectively). These results likely reflect the different antiviral mechanisms of HBV nucleo(s)tides and class I CpAMs.
In patients enrolled in cohorts A and B, our results demonstrated that the efficient absorption of GLS4 (Tmax ~3 h) with slow plasma elimination (t1/2 ~64.2–67.3 hours when combined with ritonavir vs 1.09–15.8 hours of GLS4 monotherapy in healthy subjects; data not shown). GLS4 accumulation was lower and its mean Ctrough (steady-state concentration: 205–218 ng/mL) after dosing reached 3.7–3.9 times the EC90 (55.8 ng/mL). Therefore, the addition of ritonavir increased the antiviral effect of GLS4 by increasing its steady-state concentration beyond that of EC90 [11].
This study provides the first clinical evidence that the inhibition of viral production can be achieved in patients with HBV by the novel class I CpAMs, GLS4 [9]. Long-term entecavir therapy is associated with a decreased incidence of liver decompensation and hepatocellular carcinoma [28]. Nevertheless, long-term oral antiviral therapy is associated with several disadvantages and virologic relapse is frequently reported after stopping the treatment [28]. Thus, it seems plausible that combination therapies consisting of a capsid inhibitor and another target drug may be more efficient in treating patients with HBV [29]. Indeed, the combination of anti-HBV drugs of different mechanisms (such as NUCs) to treat HBV could overcome the possibility of resistance to HBV monotherapy, which may lead to a functional cure.
With regard to limitations, the lack of immune inflammatory factor analysis can be considered as a major study limitation. This will be addressed in a future multicenter cohort study with longer follow-up period.
In conclusion, administration of 120 mg GLS4 for 28 days resulted in the reduction in serum HBV DNA and HBV pgRNA levels in the treated patients without major AEs, especially in patients with lower baseline ALT level (<5 times the ULN). Further, HBsAg, HBeAg, and HBcrAg reductions were also observed in some patients. Therefore, 120 mg GLS4/100 ritonavir may be a potential drug treatment for chronic HBV infection.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Author contributions. Y. D. and J. N. contributed to the conception and design of the study, data acquisition, and data analysis and interpretation. H. C., X. L., X. Z., and M. W. contributed to the data acquisition, data analysis, and interpretation. H. Z., F. W., Y. C., Y. Z., H. C., X. L., M. W., C. L., and J. L. contributed to the data analysis and interpretation. All authors made critical revisions to the draft versions of the manuscript and approved the final manuscript.
Disclaimer. The funding sources did not have any influence on study design, data collection, analysis and interpretation of the data, writing of the report, or the decision to submit for publication.
Financial support. This work was supported by the National Major Scientific and Technological Special Project for Significant New Drug Development during the Thirteenth Five-Year Plan Period of China (project numbers 2017ZX09304004, 2017ZX09101001-002-004), the National Natural Science Foundation of China (project number 81602897), and the program for the JLU Science and Technology Innovative Research Team (number 2017TD-08).
Potential conflicts of interest. The authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1. Stanaway JD, Flaxman AD, Naghavi M, et al. . The global burden of viral hepatitis from 1990 to 2013: findings from the Global Burden of Disease Study 2013. Lancet 2016; 388:1081–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schweitzer A, Horn J, Mikolajczyk RT, Krause G, Ott JJ. Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013. Lancet 2015; 386:1546–55. [DOI] [PubMed] [Google Scholar]
- 3. Mu D, Yuan FC, Chen Y, et al. . Baseline value of intrahepatic HBV DNA over cccDNA predicts patient’s response to interferon therapy. Sci Rep 2017; 7:5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zlotnick A, Stray SJ. How does your virus grow? Understanding and interfering with virus assembly. Trends Biotechnol 2003; 21:536–42. [DOI] [PubMed] [Google Scholar]
- 5. Le Pogam S, Yuan TT, Sahu GK, Chatterjee S, Shih C. Low-level secretion of human hepatitis B virus virions caused by two independent, naturally occurring mutations (P5T and L60V) in the capsid protein. J Virol 2000; 74:9099–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Steven AC, Conway JF, Cheng N, et al. . Structure, assembly, and antigenicity of hepatitis B virus capsid proteins. Adv Virus Res 2005; 64:125–64. [DOI] [PubMed] [Google Scholar]
- 7. Chain BM, Myers R. Variability and conservation in hepatitis B virus core protein. BMC Microbiol 2005; 5:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Choi IG, Yu YG. Interaction and assembly of HBV structural proteins: novel target sites of anti-HBV agents. Infect Disord Drug Targets 2007; 7:251–6. [DOI] [PubMed] [Google Scholar]
- 9. Yuen MF, Gane EJ, Kim DJ, et al. . Antiviral activity, safety, and pharmacokinetics of capsid assembly modulator NVR 3-778 in patients with chronic HBV infection. Gastroenterology 2019; 156:1392–403.e7. [DOI] [PubMed] [Google Scholar]
- 10. Fanning GC, Zoulim F, Hou J, Bertoletti A. Therapeutic strategies for hepatitis B virus infection: towards a cure. Nat Rev Drug Discov 2019; 18:827–44. [DOI] [PubMed] [Google Scholar]
- 11. Ren Q, Liu X, Luo Z, et al. . Discovery of hepatitis B virus capsid assembly inhibitors leading to a heteroaryldihydropyrimidine based clinical candidate (GLS4). Bioorg Med Chem 2017; 25:1042–56. [DOI] [PubMed] [Google Scholar]
- 12. Zhou X, Gao ZW, Meng J, Chen XY, Zhong DF. Effects of ketoconazole and rifampicin on the pharmacokinetics of GLS4, a novel anti-hepatitis B virus compound, in dogs. Acta Pharmacol Sin 2013; 34:1420–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hou JL, Zhao W, Lee C, et al. . Outcomes of long-term treatment of chronic HBV infection with entecavir or other agents from a randomized trial in 24 countries. Clin Gastroenterol Hepatol 2020; 18:457–67, e21. [DOI] [PubMed] [Google Scholar]
- 14. Soriano V, Barreiro P, Benitez L, Peña JM, de Mendoza C. New antivirals for the treatment of chronic hepatitis B. Expert Opin Investig Drugs 2017; 26:843–51. [DOI] [PubMed] [Google Scholar]
- 15. Deres K, Schröder CH, Paessens A, et al. . Inhibition of hepatitis B virus replication by drug-induced depletion of nucleocapsids. Science 2003; 299:893–6. [DOI] [PubMed] [Google Scholar]
- 16. Boni C, Vecchi A, Rossi M, et al. . TLR7 agonist increases responses of hepatitis B virus-specific T cells and natural killer cells in patients with chronic hepatitis B treated with nucleos(t)ide analogues. Gastroenterology 2018; 154:1764–77.e7. [DOI] [PubMed] [Google Scholar]
- 17. Jiang Y, Ma Z, Xin G, et al. . Th1 and Th2 immune response in chronic hepatitis B patients during a long-term treatment with adefovir dipivoxil. Mediators Inflamm 2010; 2010:143026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yuen Man-Fung AK, Gane Edward J. Final results of a phase 1b 28-day study of ABI-H0731, a novel core inhibitor, in non-cirrhotic viremic subjects with chronic hepatitis B. Hepatology. 2018; 68:46A–7A. [Google Scholar]
- 19. Billioud G, Pichoud C, Puerstinger G, Neyts J, Zoulim F. The main hepatitis B virus (HBV) mutants resistant to nucleoside analogs are susceptible in vitro to non-nucleoside inhibitors of HBV replication. Antiviral Res 2011; 92:271–6. [DOI] [PubMed] [Google Scholar]
- 20. Lam AM, Ren S, Espiritu C, et al. . Hepatitis B virus capsid assembly modulators, but not nucleoside analogs, inhibit the production of extracellular pregenomic RNA and spliced RNA variants. Antimicrob Agents Chemother. 2017; 61:e00680-–17.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang XY, Wei ZM, Wu GY, et al. . In vitro inhibition of HBV replication by a novel compound, GLS4, and its efficacy against adefovir-dipivoxil-resistant HBV mutations. Antivir Ther 2012; 17:793–803. [DOI] [PubMed] [Google Scholar]
- 22. Schlee M, Hartmann G. Discriminating self from non-self in nucleic acid sensing. Nat Rev Immunol 2016; 16:566–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Venkatakrishnan B, Katen SP, Francis S, Chirapu S, Finn MG, Zlotnick A. Hepatitis B virus capsids have diverse structural responses to small-molecule ligands bound to the heteroaryldihydropyrimidine pocket. J Virol 2016; 90:3994–4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mak LY, Seto WK, Fung J, Yuen MF. Novel developments of hepatitis B: treatment goals, agents and monitoring tools. Expert Rev Clin Pharmacol 2019; 12:109–20. [DOI] [PubMed] [Google Scholar]
- 25. Berke JM, Dehertogh P, Vergauwen K, et al. . Capsid assembly modulators have a dual mechanism of action in primary human hepatocytes infected with hepatitis B virus. Antimicrob Agents Chemother 2019; 64:e01686–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jansen L, Kootstra NA, van Dort KA, Takkenberg RB, Reesink HW, Zaaijer HL. Hepatitis B virus pregenomic RNA is present in virions in plasma and is associated with a response to pegylated interferon alfa-2a and nucleos(t)ide analogues. J Infect Dis 2016; 213:224–32. [DOI] [PubMed] [Google Scholar]
- 27. Wang J, Sheng Q, Ding Y, et al. . HBV RNA virion-like particles produced under nucleos(t)ide analogues treatment are mainly replication-deficient. J Hepatol 2018; 68:847–9. [DOI] [PubMed] [Google Scholar]
- 28. Klumpp K, Lam AM, Lukacs C, et al. . High-resolution crystal structure of a hepatitis B virus replication inhibitor bound to the viral core protein. Proc Natl Acad Sci USA 2015; 112:15196–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gane EJ. Future anti-HBV strategies. Liver Int 2017; 37(Suppl 1):40–4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.