Abstract
A novel colorimetric assay was developed and validated for accurate quantitation of human immunodeficiency virus (HIV) DNA in peripheral blood mononuclear cells (PBMCs). We tested 318 sequential samples from 56 subjects, 53 of whom were undergoing dual or triple therapy. Patients were considered responders when viremia levels were below 5,000 HIV RNA copies/ml. The mean DNA copy numbers for untreated and responder subjects were similar (72 and 75, respectively), while it was 4.54-fold higher for nonresponders (339). This report provides strong evidence that HIV DNA levels in PBMCs correlate with therapeutic efficacy and suggests that DNA quantitation is a useful tool to monitor the decay of the HIV reservoir toward disease remission, especially when viremia is undetectable.
Recent discoveries of the efficacy of highly active antiretroviral therapy (HAART) have provided important information on the dynamics of human immunodeficiency virus (HIV) replication during treatment and have also changed the management of HIV in infected individuals (12, 14, 18). The low or undetectable virus levels in plasma that were observed following the initiation of HAART (10, 15) and maintained throughout treatment are an indication of effective suppression of HIV replication. HIV DNA has been found in HAART-treated individuals without detectable plasma viremia (3, 19), showing the existence of latent reservoirs (4, 7). HIV infection can be reactivated from the latently infected resting T-cell population (20). Measurement of cell-associated viral DNA should provide greater understanding of the dynamics of HIV infection and could complement the information provided by plasma RNA when monitoring responsiveness to HAART in infected individuals.
In this study, we quantified HIV DNA in peripheral blood mononuclear cells (PMBCs) using a quantitative colorimetric assay. This novel method uses (i) lysates containing total DNA obtained from 106 PBMCs, (ii) single HIV DNA amplification with a biotinylated antisense primer, (iii) liquid hybridization of the biotinylated amplicons with a fluorescein-containing probe, (iv) hybrid capture into streptavidin-coated microplate wells, (v) single colorimetric detection with a monospecific antifluorescein antibody conjugated with horseradish peroxidase, and (vi) accurate quantitation performed by specific application of Quanti-Kin software, based on the color kinetics of an external reference standard curve with a dynamic range of 4 log units (8).
PCR procedures were performed with primer SK145 and the biotinylated primer SK431. Seventy-five microliters of a PCR mixture containing 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 200 μM deoxynucleoside triphosphates, a 0.15 μM concentration of each primer (TIB Molbiol, Genoa, Italy), and 2.5 U of Taq DNA polymerase was dispensed into microtubes placed on ice. A 25-μl aliquot of cell lysate (corresponding to 105 PBMCs) was added to the PCR mixture. Samples were placed in a thermal cycler (GeneAmp 9600; Perkin-Elmer, Monza, Italy) once the temperature of the cycler reached 80°C and were held at 95°C for 1 min before being subjected to 35 cycles of DNA amplification with the following thermal parameters: denaturation (10 s at 95°C), primer annealing (10 s at 60°C), and DNA extension (10 s at 72°C) for 5 cycles, followed by denaturation (10 s at 92°C), primer annealing (10 s at 55°C), and DNA extension (10 s at 72°C) for 30 cycles. Five microliters of the biotinylated amplification product was added to 120 μl of hybridization buffer (5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]) containing 2 pmol of the SK102 probe, which was synthesized with the insertion of a fluorescein molecule. The mixture was heated at 95°C for 5 min to denature the DNA duplex and then held at 57°C for 10 min to allow hybridization. Forty-five microliters of the hybridized product was transferred to a streptavidin-coated microplate well (Labsystems Oy, Helsinki, Finland) for hybrid capture. Following incubation for 1 h at 37°C, unbound components were removed by extensive washing. One hundred microliters of horseradish peroxidase-conjugated antifluorescein antibodies (Boehringer Mannheim, Milan, Italy), 100 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 3% fetal calf serum were added to the microwells. The microplate was incubated for 30 min at room temperature on a microplate shaker and, after a final wash to remove free conjugate, 100 μl of tetramethylbenzidine solution (Celbio, Milan, Italy) was added to each well. Color development was measured every 20 s for a period of 30 min at 650 nm with an automated microplate reader. The reaction was stopped by the addition of 0.2 M H2SO4, and the end point absorbance was read at 450 nm with a reference filter of 690 nm. All of the readings were evaluated with the Quanti-Kin program, which selects the best interpolation method for each reading and for each interval between the five reference standards (8). Quantitation of as few as 50 HIV DNA copies/106 PBMCs was ensured by the inclusion of an external reference standard curve of 5, 25, 100, 1,000, and 10,000 copies of the HIV genome (HIVZ6 plasmid; Perkin-Elmer) per reaction prepared in seronegative cell lysates (105 PBMCs/25 μl).
Amplification of the HLA DQ-α gene was performed with the HLA-specific primers GH26 and GH27 on HIV DNA-negative samples in order to assess the quality of the cell lysate (16).
The analysis of HIV DNA serial dilutions made in lysates of PBMCs from HIV-negative donors determined that the lower limit for quantitation was 5 copies per reaction. Assay specificity was determined by comparing the optical density (OD) values obtained from 20 negative controls with those of six 5-copy reference standards. The OD of the negative samples (mean ± standard deviation) was 0.095 ± 0.039, 5.4 times lower than the 5-copy reference standards’ OD (0.511 ± 0.197). There was no overlapping of the distributions of OD values obtained from the negative samples and the 5-copy reference standards, with a confidence interval of 99.95% (data not shown).
An external curve of 5, 25, 100, 1,000, and 10,000 HIV DNA copies was amplified by PCR and detected colorimetrically. Color production was linear for the low reference standards, which were quantified by the more sensitive end point reading. Exponential slopes reaching plateaus were seen for the high reference standards, and these were quantified by using the kinetic readings. Thus, a dynamic range from 5 to 10,000 copies per reaction was obtained.
Intra-assay variability was determined by the analysis of four replicates of each reference standard run together in the same assay. An external reference standard curve was used to quantify the replicate reference standards (Table 1). The coefficient of variation (CV) obtained for each reference standard concentration ranged from 6 to 37%, with a mean of 25%. To assess interassay variability, a reference standard curve was quantified in four separate and consecutive assays (Table 1). The CV obtained for each reference standard ranged from 18 to 53%, with a mean of 32%.
TABLE 1.
Interassay and intra-assay variability
| Type of comparison | No. of input copies | No. of HIV DNA copies/reaction in calculated standard replicate
|
Mean no. of HIV DNA copies/reaction | CV (%) | Maximum Z score | |||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |||||
| Intra-assay | 5 | 5 | 6 | 5 | 5 | 5 | 6 | 0.23 |
| 25 | 10 | 19 | 17 | 18 | 16 | 26 | 1.34 | |
| 100 | 97 | 122 | 44 | 89 | 88 | 37 | 2.07 | |
| 1,000 | 2,579 | 2,670 | 1,619 | 1,475 | 2,085 | 30 | 1.21 | |
| 10,000 | 10,000 | 7,782 | 4,943 | 7,661 | 7,596 | 27 | 1.44 | |
| Interassay | 5 | 3 | 2 | 7 | 7 | 5 | 53 | 2.56 |
| 25 | 61 | 46 | 34 | 41 | 46 | 25 | 1.20 | |
| 100 | 118 | 71 | 80 | 70 | 85 | 27 | 1.07 | |
| 1,000 | 1,344 | 1,773 | 2,136 | 849 | 1,525 | 36 | 1.89 | |
| 10,000 | 7,112 | 6,878 | 10,000 | 7,507 | 7,874 | 18 | 0.77 | |
The reproducibility of these estimations was further evaluated by using the Z score, which measures the significance of the differences in sets of quantitative values. The Z score was calculated for each set of reference standard replicates by using the following formula: Z = log10 (y1/y2)/(0.15 · 21/2), where y1 was the highest value and y2 was the lowest value obtained in each set. Theoretically, all concentrations should be identical (null hypothesis); therefore, the Z score is normally distributed with a mean of zero and a variance of one (17). However, Yen-Lieberman et al. (21) have shown that the multiple fluctuations affecting PCR quantification make differences irrelevant as long as the Z score remains lower than 1.96. If a Z score greater than 1.96 is obtained, a false difference is detected for that set of values. Two false differences were detected for these results, one at 100 copies (Z = 2.07) for the intra-assay estimation and the other at 5 copies (Z = 2.56) for the interassay estimation. In both cases a false difference was found in 5% of the total estimations (n = 20), indicating an overall good reproducibility.
Intra-assay variability was further evaluated by quantifying 24 clinical samples in duplicate in the same assay. The CVs obtained for the duplicate samples ranged from 5 to 51%, with a mean of 20%; 18 samples (75%) had CVs lower than 30% (data not shown). We quantified 17 clinical samples in duplicate in two different assays to assess interassay variability. The CVs obtained for the duplicate samples ranged from 3 to 64%, with a mean of 26%; 11 (65%) samples had CVs lower than 30%. These results demonstrate good reproducibility in clinical samples.
The objectives of this study were to determine the number range of HIV DNA-containing cells, its variation in sequential samples, and its correlation with therapy efficacy. A total of 318 blood samples were collected during a 3-year study period (1996 to 1998) from 53 HIV-infected individuals being treated with antiretroviral drugs and from 3 untreated HIV-infected individuals. Plasma RNA levels were determined by nucleic acid sequence-based amplification technology as previously described (1). PBMCs were isolated, counted, and aliquoted from whole blood treated with acid citrate dextrose by density gradient separation on Lympholyte-H medium (Cedarlane Laboratories, Hornby, Canada). Total cellular DNA was obtained from the PBMCs by cell lysis and stored at −20°C, as previously described (9).
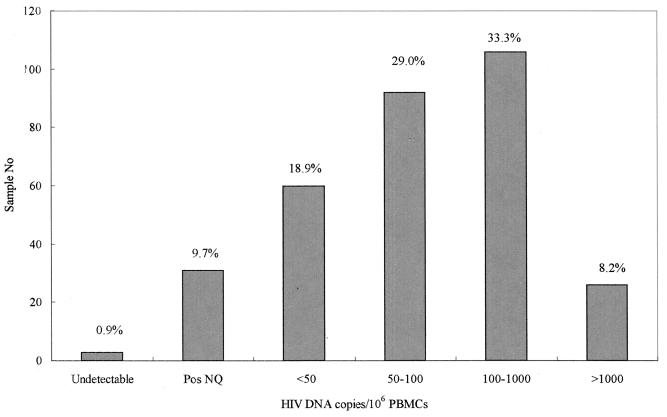
HIV DNA was detected in 315 (99.1%) of the 318 samples from HIV-infected individuals and quantified in 284 (89.3%) samples in a range of 50 to 8,900 copies per 106 PBMCs. As shown in Fig. 1, 92 (29%) samples had between 50 and 100 copies, 106 (33.3%) had between 100 and 1,000 copies, and 26 (8.2%) had more than 1,000 copies. Less than 50 copies per 106 PBMCs were detected in 60 (18.9%) samples by the standard protocol. Low DNA levels were detected in 31 (9.7%) samples by an extended amplification protocol with 45 cycles of PCR. HIV DNA was repeatedly not detected in three (0.9%) samples from three subjects whose other samples all contained fewer than 50 copies. These data show that the HIV DNA assay has an adequate range to quantify HIV DNA levels in clinical samples.
FIG. 1.
Distribution of HIV DNA in 318 clinical PBMC samples. Pos NQ, positive nonquantifiable.
The analysis of HIV DNA in sequential samples, collected from these 56 patients during a 14- to 24-month follow-up, showed a distinct distribution into two groups. In the first group, low DNA levels (<500 copies/106 PBMCs) were detected, with a linear trend throughout follow-up. High DNA levels (>500 copies/106 PBMCs) were observed in the second group, with wide fluctuations between samples.
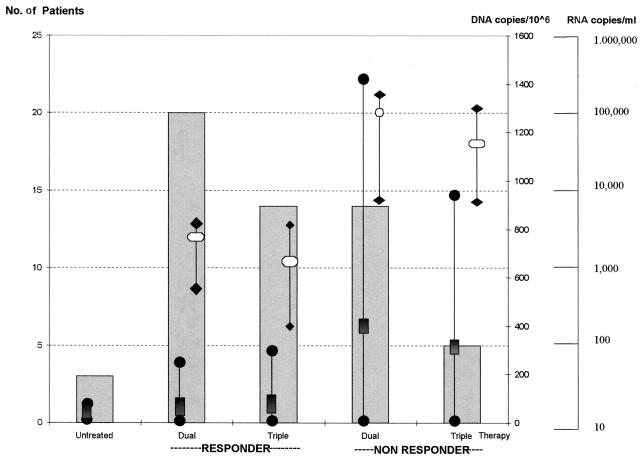
To further investigate this observation, we analyzed the relationship between HIV DNA levels and the efficacy of antiretroviral therapy in the last-tested PBMC sample from each of the 56 subjects. Three subjects were not receiving antiretroviral therapy at the time of the study, 34 were receiving combinations of two of the five available reverse transcriptase inhibitors, and the remaining 19 were treated with three drugs, including one of the four available protease inhibitors. Subjects were considered responders when viremia levels were below 5,000 copies of HIV RNA per ml of plasma. When these levels of virological markers were not observed, the patient was classified as a nonresponder. Twenty dual-therapy responder patients had a mean RNA copy number of 3,492 per ml of plasma, and 14 triple-therapy responders had a mean plasma RNA copy number of 1,242, while the RNA mean values were 55,916 and 34,880 in the two-drug and three-drug nonresponder patients, respectively.
As shown in Fig. 2, the mean HIV DNA copy numbers for untreated and responder subjects were similar (72 and 75, respectively), while it was 4.54-fold higher (339) for the nonresponders. Moreover, the mean DNA level was higher for dual-therapy nonresponders than for triple-therapy nonresponders, 390 and 288, respectively. These data clearly suggest that HIV DNA levels correlate with therapy efficacy.
FIG. 2.
HIV DNA levels and therapy efficacy. The relationship between DNA levels and therapy efficacy was evaluated with the last-tested samples from each patient.  , number of tested patients;
, number of tested patients;  and •|•, mean and range of DNA copy number; □ and ♦|♦, mean and range of RNA copy number.
and •|•, mean and range of DNA copy number; □ and ♦|♦, mean and range of RNA copy number.
The use of a potent combination of antiretroviral drugs has dramatically reduced HIV replication and viremia in and improved the quality of life of infected patients. For the first time, HIV disease progression can now be halted, and many patients are experiencing disease remission. As scientific debates about new strategies to eradicate HIV infection evolve, the scenario of HIV patient management continues to change and there is an urgent need for new virological markers, especially when conventional markers are negative (undetectable viremia) or of uncertain biological significance (low viremia).
This paper demonstrates that HIV DNA levels in PBMCs correlate with antiretroviral therapy efficacy. Convincing evidence has been provided from the longitudinal analysis of a large number of samples (318 samples, 4 to 10 samples per patient, 14 to 24 months of follow-up) collected from a cohort of 53 infected subjects being treated with a combination of two or three antiretroviral drugs and 3 infected, untreated subjects.
High DNA levels were observed in sequential samples from individuals who did not respond to therapy. In the presence of inefficient therapy, active viral replication results in elevated viremia (up to 5 × 105 RNA copies/ml) due to de novo infection of naïve cells and an increase in the number of cells containing viral DNA (5, 20). Individuals responding to dual or triple therapy had constantly low DNA levels, which should be composed mainly of proviral DNA integrated in latently infected CD4+ cells (4).
Earlier studies have reported discordant results for the quantitation of HIV proviral DNA and its relationship with antiretroviral therapy and disease progression. However, most of these studies monitored viral DNA during monotherapy clinical trials on a limited number of subjects and used short study periods (6, 11, 13).
Our results were obtained by an innovative and simple assay which has been validated for accurate quantitation of HIV DNA in PBMCs and is suitable for monitoring HIV DNA-containing cells in infected patients receiving HAART. The PBMC lysates used contained the total PBMC DNA population; therefore, both unintegrated and integrated HIV DNA forms were measured by the assay described. The HIV DNA population present in samples from responder subjects should be composed of mainly integrated proviral DNA, while the DNA in samples from nonresponders should contain a high percentage of unintegrated DNA, which accumulates during the initial phases of infection as a direct product of active viral replication, indicating therapy inefficacy (2).
It is widely accepted that the treatment time required for HIV eradication, if this is indeed possible, is considerably longer than previously suggested (15). At present there is a strong need to develop new strategies focused on eradicating the minute population of long-living chronically infected cells, which constitutes a reservoir of inducible infectious HIV. Quantitation of total HIV DNA could be useful in long-term longitudinal studies of patients receiving HAART.
Acknowledgments
This work was supported by grants from the Italian Research Program on AIDS (30A.0.71 and 9403-58) and from the School of Medicine, University of Genoa. A.A.G. and M.G. are fellows of the ISS Italian Research program on AIDS, and J.L.M. is a fellow of ANLAIDS.
We thank F. Indiveri and F. Puppo for continuous support and helpful discussions and Eddie Thüroff and Olfert Land for their expert advice.
REFERENCES
- 1.Bettini P, Boeri E, Lillo F, Abecasis C, Farma E, Crociati E, Uberti-Foppa C, Varnier O E. HIV-1 RNA quantitation by one-tube quantitative NASBA in HIV-infected patients. AIDS. 1996;10:1735–1737. [PubMed] [Google Scholar]
- 2.Cara A, Reitz M S., Jr New insights on the role of extrachromosomal forms of retroviral DNA. Leukemia. 1997;11:1395–1399. doi: 10.1038/sj.leu.2400776. [DOI] [PubMed] [Google Scholar]
- 3.Cavert W, Notermans D W, Staskus K, Wietgrefe S W, Zupancic M, Gebhard K, Henry K, Zhang Z Q, Mills R, McDade H, Goudsmit J, Danner S A, Haase A T. Kinetics of response in lymphoid tissues to antiretroviral therapy of HIV infections. Science. 1997;276:960–964. doi: 10.1126/science.276.5314.960. [DOI] [PubMed] [Google Scholar]
- 4.Chun T W, Carruth L, Finzi D, Shen K, DiGiuseppe J A, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn T C, Kuo Y H, Brookmeyer R, Zeiger M A, Barditch-Crovo T, Siliciano R F. Quantification of latent tissue reservoirs and total body viral load in HIV infection. Nature. 1997;387:183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 5.Chun T W, Stuyver L, Mizell S B, Ehler L A, Mican J A M, Baseler M, Lloyd A L, Nowak M A, Fauci A S. Presence of an inducible HIV latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci USA. 1997;94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donovan R M, Dickover R E, Goldstein E, Huth R G, Carlson J R. HIV proviral copy number in blood mononuclear cells from AIDS patients on zidovudine therapy. J Acquired Immune Defic Syndr. 1991;4:766–769. [PubMed] [Google Scholar]
- 7.Finzi D, Hermankova M, Pierson T, Carruth L M, Buck C, Chaisson R E, Quinn T C, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho D D, Richman D D, Siliciano R F. Identification of a reservoir for HIV in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 8.Giacomini M, McDermott J L, Giri A A, Martini I, Lillo F B, Varnier O E. A novel and innovative quantitative kinetic software for virological colorimetric assays. J Virol Methods. 1998;73:201–209. doi: 10.1016/s0166-0934(98)00059-7. [DOI] [PubMed] [Google Scholar]
- 9.Giri A A, Lillo F B, McDermott J L, Januzzi C, Risso S, Forni G, Concedi D R, Varnier O E. Detection of HIV sequences in children using radioactive and colorimetric polymerase chain reactions. J Med Virol. 1994;42:414–419. doi: 10.1002/jmv.1890420415. [DOI] [PubMed] [Google Scholar]
- 10.Gulick R M, Mellors J W, Havlir D, Eron J J, Gonzalez C, McMahon D, Richman D D, Valentine F T, Jonas L, Meibohm A, Emini E A, Chodakewitz J A. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N Engl J Med. 1997;337:734–739. doi: 10.1056/NEJM199709113371102. [DOI] [PubMed] [Google Scholar]
- 11.Ho D D, Moudgil T, Alam M. Quantitation of human immunodeficiency virus type 1 in the blood of infected persons. N Engl J Med. 1989;321:1621–1625. doi: 10.1056/NEJM198912143212401. [DOI] [PubMed] [Google Scholar]
- 12.Ho D D, Neumann A U, Perelson A S, Chen W, Leonard J M, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 13.Oka S, Urayama K, Hirabayashi Y, Kimura S, Mitamura K, Shimada K. Human immunodeficiency virus DNA copies as a virologic marker in a clinical trial with beta-interferon. J Acquired Immune Defic Syndr. 1992;5:707–711. [PubMed] [Google Scholar]
- 14.Perelson A S, Neumann A U, Markowitz M, Leonard J M, Ho D D. HIV dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 15.Perelson A S, Essunger P, Cao Y, Vesanen M, Hurley A, Saksela K, Markowitz M, Ho D D. Decay characteristics of HIV infected compartments during combination therapy. Nature. 1997;387:188–191. doi: 10.1038/387188a0. [DOI] [PubMed] [Google Scholar]
- 16.Scharf S J, Horn G T, Erlich H A. Direct cloning and sequence analysis of enzymatically amplified genomic sequences. Science. 1986;233:1076–1078. doi: 10.1126/science.3461561. [DOI] [PubMed] [Google Scholar]
- 17.Snedecor G W, Cochran W G. Statistical methods. Ames: Iowa State University Press; 1967. [Google Scholar]
- 18.Wei X, Ghosh S K, Taylor M E, Johnson V A, Emini E A, Deutsch P, Lifson J D, Bonhoeffer S, Nowak M A, Hahn B H, Saag M S, Show G M. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 19.Wong J K, Günthard H F, Havlir D V, Zhang Z Q, Haase A T, Ignacio C C, Kwok S, Emini E, Richman D D. Reduction of HIV in blood and lymph nodes following potent antiretroviral therapy and the virologic correlates of treatment failure. Proc Natl Acad Sci USA. 1997;94:12574–12579. doi: 10.1073/pnas.94.23.12574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong J K, Hezareh M, Günthard H F, Havlir D V, Ignacio C C, Spina C A, Richman D D. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278:1291–1294. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 21.Yen-Lieberman B, Brambilla D, Jackson B, Bremer J, Coombs R, Cronin M, Herman S, Katzenstein D, Leung S, Lin H J, Palumbo P, Rasheed S, Todd J, Vahey M, Reichelderfer P. Evaluation of a quality assurance program for quantitation of human immunodeficiency virus type 1 RNA in plasma by the AIDS Clinical Trials Group virology laboratories. J Clin Microbiol. 1996;34:2695–2701. doi: 10.1128/jcm.34.11.2695-2701.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]