Abstract
Nuclear composition determines nuclear function. The early embryos of many species begin life with large pools of maternally provided components that become rapidly imported into an increasing number of nuclei as the cells undergo repeated cleavage divisions. Because early cell cycles are too fast for nuclei to achieve steady-state nucleocytoplasmic partitioning, the composition of cleavage stage nuclei is likely dominated by nuclear import. The end of the rapid cleavage stage and onset of major zygotic transcription, known as the mid-blastula transition (MBT), is controlled by the ratio of nuclei/cytoplasm, indicating that changes in nuclear composition likely mediate MBT timing. Here, we explore how different nuclear import regimes can affect protein accumulation in the nucleus in the early Drosophila embryo. We find that nuclear import differs dramatically for a general nuclear cargo (NLS (nuclear localization signal)-mRFP) and a proposed MBT regulator (histone H3). We show that nuclear import rates of NLS-mRFP in a given nucleus remain relatively unchanged throughout the cleavage cycles, whereas those of H3 halve with each cycle. We model these two distinct modes of nuclear import as “nucleus-limited” and “import-limited” and examine how the two different modes can contribute to different protein accumulation dynamics. Finally, we incorporate these distinct modes of nuclear import into a model for cell-cycle regulation at the MBT and find that the import-limited H3 dynamics contribute to increased robustness and allow for stepwise cell-cycle slowing at the MBT.
Significance
Nuclear import dynamics are likely to contribute significantly to cell-cycle regulation at the mid-blastula transition (MBT). We measure the nuclear import behavior of a general nuclear marker NLS-mRFP and histone H3 in living Drosophila embryos and find that they represent distinct nuclear import behaviors. We then model different regimes of nuclear import and explore how these modes may contribute to cell-cycle behavior near the MBT. We find that the presumably unusual import behavior of histone H3 contributes to the observed robust, stepwise cell-cycle slowing at the MBT.
Introduction
The concentrations of nuclear components determine their abilities to regulate cellular functioning. The state of the nucleus changes dramatically throughout early development as cells differentiate into specific cell fates. One dramatic example of such a state change is the mid-blastula transition (MBT), in which cells switch from a fast-dividing, low transcribing regime to a slow-dividing, highly transcribing state (1,2). This transition is controlled by the ratio of nuclei/cytoplasm (N/C ratio) (3, 4, 5, 6, 7). Initially, the embryo comes loaded with a vast pool of maternally provided components that are almost entirely cytoplasmic because of the low nuclear content at fertilization. A subset of these components are then gradually partitioned into an exponentially increasing number of nuclei because the number of nuclei doubles with each cell cycle, but the total amount of embryonic protein remains approximately constant (8,9). Alterations to the N/C ratio result in corresponding advancement or delay of the MBT (3, 4, 5, 6, 7). A longstanding hypothesis has been that titration of some critical components into the nuclei is responsible for timing the onset of the MBT (3,4). Several candidate titrated components have been identified, including histones, replication factors, and deoxyribonucleotides, all of which are required in the nucleus (10, 11, 12, 13, 14, 15, 16, 17, 18, 19).
If the titration of nuclear proteins is responsible for timing the MBT as proposed, then the dynamics of translocation of such factors into the nucleus would be of vital importance in regulating nuclear state as cells undergo the transition. Indeed, alteration of active nuclear transport and nuclear size have been shown to result in changes in MBT behavior akin to those resulting from manipulating the N/C ratio (20, 21, 22). Active transport of molecules across the nuclear membrane is mediated by karyopherins, such as importins, that recognize the nuclear localization signals (NLSs) of cargos in the cytoplasm and facilitate their passage across nuclear pore complexes (NPCs) (23). In the pre-MBT cell cycles, entry into mitosis and the accompanying nuclear envelope breakdown (NEB) occur before the nuclei have stopped growing, indicating that bulk nuclear import does not reach steady state by the end of a given cycle (24, 25, 26). In this regime, the dynamics of nuclear import likely dominate the transient composition of the nucleus within a given cell cycle and may thereby affect activities of downstream pathways, including the cell cycle and transcription. However, the effect of nuclear import dynamics on MBT control has been little investigated.
Here, we have sought to model how nuclear import dynamics can contribute to cell-cycle slowing at the MBT in the context of early Drosophila embryogenesis. The early Drosophila embryo undergoes 13 rounds of rapid syncytial divisions before pausing the cell cycle and undergoing the MBT at nuclear cycle 14 (NC14) (Fig. 1 A). We focused on NC11 through NC13, during which cell-cycle durations gradually lengthen from ∼11 min at NC11 to ∼14 min at NC12 and ∼21 min at NC13 (Fig. 1 B). We observe distinct dynamics of nuclear import depending on cargo proteins and model import regimes that explain these observed behaviors. Furthermore, we investigate how nuclear import dynamics of histone H3, which we have recently characterized as a cell-cycle regulator (27), contribute to the cell-cycle dynamics in NC11–13. Our results show that nuclear import dynamics play a key role in regulating the early embryonic cell cycle.
Figure 1.
Distinct dynamics of NLS-mRFP and histone H3 nuclear import. (A) The cell cycle in the early Drosophila embryo. The early Drosophila embryo undergoes 13 rounds of rapid and synchronous divisions before pausing the cell cycle at nuclear cycle (NC) 14 and undergoing the mid-blastula transition (MBT). (B) Cell-cycle durations gradually slow from NC11 to NC13. (C) Nuclear growth dynamics in NC11–13 (25). (D and E) Total nuclear intensities of NLS-mRFP and H3-Dendra2 during interphase in NC11–13. H3-Dendra2 data were reported previously (25). Gray boxes represent mitosis. These two proteins represent two distinct modes of nuclear import with each cell cycle. (F) Nuclear import rates of NLS-mRFP and H3-Dendra2 as measured by initial slopes of nuclear intensities in (D) and (E). NLS-mRFP import is relatively constant as per nucleus basis (total import doubles with the number of nuclei), whereas H3-Dendra2 halves with each cell cycle (total import capacity remains constant embryo wide). Data represent the mean ± standard deviation (SD).
Materials and methods
Drosophila stocks and crosses
NLS-mRFP (Bloomington Drosophila Stock Center (BDSC) #31418), His2Av-mRFP (BDSC #23650), and yw (BDSC #1495) lines were obtained from the Bloomington Drosophila Stock Center. The nos-PBac line was a gift from Robert Marmion and Stanislav Shvartsman. All fly stocks were maintained at room temperature of ∼22°C on a standard cornmeal or standard molasses media.
Generation of an endogenously tagged Chk1-sfGFP line
For CRISPR-Cas9 editing of the endogenous chk1/grp gene, a single CRISPR target site was selected using Target Finder (28) near the stop codon, and the designed guide RNA (gRNA) was subcloned into the pU6-BbsI-chiRNA vector (a gift from Melissa Harrison and Kate O’Connor-Giles and Jill Wildonger, Addgene plasmid #45946; http://www.addgene.org/45946/; Research Resource Identifiers (RRID): Addgene_45946; Watertown, MA). Approximately 1 kb of homology arms with a synonymous mutation at the PAM site were subcloned into the pScarlessHD-sfGFP-DsRed plasmid (a gift from Kate O’Connor-Giles, Addgene plasmid #80811; http://www.addgene.org/80811/; RRID: Addgene_80811) (GenScript, Piscataway, NJ). gRNA and homology arm plasmids were co-injected into nos-Cas9 embryos (TH00787.N) and DsRed+ progeny were screened (BestGene, Chino Hills, CA). The DsRed marker was removed through a cross to nos-PBac flies. Expression of Chk1-sfGFP was confirmed by Western blotting with anti-GFP antibody (1:2000, ab290; Abcam, Cambridge, UK).
Microscopy
Embryos were collected on yeasted apple juice agar plates for 2 h at 25°C and dechorionated with 4% sodium hypochlorite for 2 min followed by a wash in deionized water. Dechorionated embryos were mounted in deionized water on a glass-bottom microwell dish (MatTek, Ashland, MA), and fluorescent images were acquired by a Nikon A1R laser scanning confocal microscope (Nikon, Tokyo, Japan) with a 20× 0.75 NA objective at room temperature of ∼22°C. Time-lapse videos were obtained at a time resolution of either 30 s (Chk1-sfGFP) or 45 s (NLS-mRFP).
Image analysis and quantification
Nuclear regions were segmented from either NLS-mRFP or His2Av-mRFP images using ilastik (29), and nuclear intensities of NLS-mRFP or Chk1-sfGFP were quantified. Nuclear volumes and nuclear H3-Dendra2 data were derived from our previous work (25).
Numerical simulation, modeling, and parameters
The ordinary differential equations were numerically computed using the scipy.integrate.ode (30). For modeling, we first considered two simple nuclear import regimes, in which either embryo-wide import or per-nuclear import was constant regardless of the increasing number of nuclei during embryogenesis, to recapitulate different behaviors of NLS-mRFP and histone H3 nuclear import. To investigate how nuclear import dynamics affect downstream pathway activities, we constructed an integrative model for H3 nuclear import and H3 control of the early embryonic cell cycle by extending previous models (25,27,31, 32, 33, 34). Model details are described in the main text and Supporting materials and methods. The values of parameters used for simulation are based on previous works (25,27,34) with slight modifications to closely match experimental measurements and are summarized in Table S1.
Results
Distinct dynamics of nuclear import during early embryogenesis
In the early embryo, nuclei display remarkably rapid growth (Fig. 1 C) indicative of large-scale translocation of proteins from cytoplasmic pools into the nucleus. The early Drosophila embryo develops as a syncytium in which nuclei share a common cytoplasmic pool until NC14. Because of the increasing number of nuclei, demand for nuclear import increases with each cycle relative to a fixed cytoplasm. To analyze dynamics of nuclear import in vivo, we visualized an NLS-mRFP marker as a proxy for general nuclear import in NC11–13. To accurately measure the nuclear import of a single protein within these rapidly growing nuclei, we report total integrated nuclear intensities rather than average intensities (i.e., concentrations; Fig. S1). We observed total nuclear NLS-mRFP increased approximately linearly over time with little indication of slowing over the course of the cell cycle, suggesting that the kinetics of nuclear import are saturated and NLS-mRFP does not approach steady state in the early cycles (Fig. 1 D). In addition, the initial slope of NLS-mRFP accumulation was similar for the final three cell cycles before the MBT (Fig. 1 F). This indicates that embryo-wide NLS-mRFP import keeps pace with the increase in the number of nuclei and per-nuclear import is independent of the number of nuclei.
The dynamics of NLS-mRFP nuclear import stand in marked contrast to histone H3, which we have previously characterized using H3-Dendra2 (25). Nuclear import rates of H3 as measured by initial slopes of total nuclear intensities halve with each cycle (Fig. 1, E and F). This indicates that total embryo-wide H3 import is approximately constant and is divided between an increasingly large number of nuclei with each division. Moreover, total final nuclear protein amounts decrease from NC11 to NC13, indicating that H3 pools are exhausted near the MBT. H3 also differs from NLS-mRFP in that a fraction of H3 protein remains stably associated with DNA through mitosis resulting in a portion of the H3 pool that is independent of nuclear import. However, the higher H3-Dendra2 concentrations in early cycles than in late cycles are the result of a large pool of non-DNA-bound H3 in early nuclei (25). These observations indicate that H3 and NLS-mRFP must depend on different nuclear import regimes to result in such different import dynamics.
Given the large pool of cytoplasm in the early embryo, it has typically been assumed that nuclear import rates are limited by the availability of nuclei to uptake proteins (35, 36, 37). For example, the number of NPCs may be limiting for bulk nuclear import. In this regime, which we refer to as “nucleus-limited,” rates of nuclear import in each nucleus (per-nuclear import) are unaffected by the increase in the number of nuclei: import rate = r(c), where the import rate of a given cargo is a function of its cytoplasmic concentrations. Therefore, the initial slope of nuclear import in each nucleus remains the same, but cytoplasmic consumption doubles as the number of nuclei doubles with each cycle (Fig. 2 A). This model is consistent with the measured behavior of NLS-mRFP. However, H3 nuclear import does not fall into this category. H3 nuclear import rates become diluted as the number of nuclei increase and can be phenomenologically described as import rate = r′(c)/Nn, where Nn is the number of nuclei. Here, the initial slope of per-nuclear import halves and cytoplasmic consumption remains unchanged with each cycle. We refer to this regime as “import-limited” because embryo-wide import does not keep pace with the increase in the demand, and therefore, import capacity is distributed over the increasing number of nuclei (Fig. 2 B).
Figure 2.
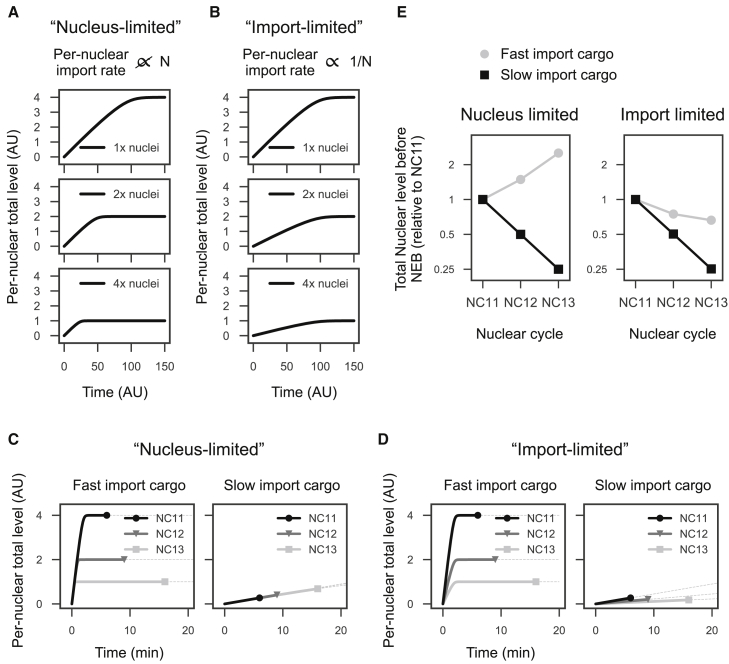
Dynamics of nuclear accumulation in response to doubling the number of nuclei in the “nucleus-limited” and “import-limited” models. (A) In the nucleus-limited regime, nuclear import in an individual nucleus (per-nuclear import) is unaffected by an increase in the number of nuclei. The equations for the model are dn/dt = Vc/(K + c) and dc/dt = −NnVc/(K + c), where n represents levels of a cargo in a given nucleus, c indicates cytoplasmic pools, Nn is the number of nuclei in the embryo (750, 1500, and 3000 in NC11, NC12, and NC13, respectively), n0 = 0, c0 = 3000, V = 0.05, and K = 300. (B) In the import-limited regime, per-nuclear import halves as the number of nuclei doubles with each cycle. The equations for the model are dn/dt = Vc/(K + c)N and dc/dt = −Vc/(K + c), where n0 = 0, c0 = 3000, V = 37.5, and K = 300. (C and D) Dynamics of nuclear import in NC11–13 with respect to different timescales of nuclear import. When the timescale of nuclear import is sufficiently faster than cell-cycle durations, per-nuclear levels halve with each cell cycle regardless of the method of nuclear import. When nuclear import is slow compared to cell-cycle times, the nucleus-limited mechanism results in more accumulation in late cycles because of longer cell-cycle durations, whereas the import-limited mechanism still reduces nuclear accumulation in late cell cycles. The equations for (C) are dn/dt = Vc/(K + c) and dc/dt = −NVc/(K + c), where n0 = 0, c0 = 3000, V = 2.5 (left) or 0.05 (right), and K = 300. The equations for (D) are dn/dt = Vc/(K + c) and dc/dt = −NVc/(K + c), where n0 = 0, c0 = 3000, V = 1875 (left) or 37.5 (right), and K = 300. (E) Levels of nuclear accumulation before NEB. The two import modes can result in distinct dynamics of nuclear accumulation depending on the timescale of nuclear import and cell-cycle durations.
To understand how the two import mechanisms might affect nuclear import dynamics differently in the early embryo, we calculated levels of nuclear accumulation in NC11, NC12, and NC13 under both nuclei-limited and import-limited regimes. To model nuclear import, we used the Michaelis-Menten equation Vc/(K + c) with a relatively small value of K compared to the cytoplasmic pool. This well recapitulates the linear accumulation of nuclear proteins in vivo (Fig. 1). An important consideration for how finite cytoplasmic pools of a given cargo will accumulate in the nucleus is how the timescale of nuclear import compares to the length of the cell cycle. When nuclear import is sufficiently faster than the timescale of cell cycles, cytoplasmic pools are rapidly exhausted, and the final amount of the cargo in a given nucleus halves as the number of nuclei doubles with each cycle regardless of the methods of nuclear import (Fig. 2, C–E). By contrast, when the timescale of nuclear import is slower than cell-cycle durations, the nucleus-limited regime results in an increased level of nuclear accumulation from NC11 to NC12 and from NC12 to NC13 (Fig. 2, C–E). This is because cell-cycle durations lengthen during this period, allowing more time for protein accumulation (Fig. 2 C, right). However, in the import-limited regime, nuclear protein accumulation is reduced with each cycle (Fig. 2 D, right) because the increased cell-cycle duration cannot overcome the decreased slope of accumulation. Together, the nucleus-limited regime can both increase and decrease nuclear accumulation in an ∼10-fold dynamic range from NC11 to NC13 depending on import parameters, whereas the import-limited regime robustly reduces nuclear accumulation with each cycle.
Investigating the role of nuclear import dynamics in early cell cycles
Thus far, we have shown that diverse behaviors of protein nuclear accumulation can arise purely from differences in import regimes and without the need for cargo-specific active regulation (e.g., modifications to specific cargo). However, how these different import regimes might affect downstream cellular behaviors remains unclear. Given the proposed role for nuclear titration of histone H3 in cell-cycle regulation at the MBT (13,15), we sought to tackle this question by creating an integrative model for H3 nuclear import and H3 control of embryonic cell-cycle progression (Fig. 3 A).
Figure 3.
(A) Schematic of the model for import-limited H3 nuclear import (WT) and cell-cycle regulation. Rates of H3 nuclear import are expressed by the Michaelis-Menten equation and become diluted over the increasing number of nuclei (N). Cell-cycle progression is driven by the cyclin-Cdk1 cell-cycle oscillator. H3 regulates the cell cycle through inhibition of the checkpoint kinase, Chk1, that interferes with cell-cycle progression. See Supporting materials and methods for full details of the model. (B) Chk1 concentrations in the nucleus remain unchanged from NC11 through NC13. The endogenous chk1/grp locus was tagged with sfGFP using CRISPR-Cas9 genome editing. Average Chk1-sfGFP intensities in NC11–13 are shown. Data represent the mean ± SD. (C) Simulation of H3 nuclear import, nuclear growth, and Cyc-Cdk1 oscillations in the WT embryo, consistent with previous observations of H3 nuclear import and nuclear growth (Figs. 1, C and E, and S4 (25), as well as the dynamics of cell-cycle slowing in NC11–13 (Fig. S5).
Cell-cycle progression in the early embryo is driven by the cyclin-Cdk1 (Cyc-Cdk1) system. Based on previous studies (31,32), the core Cyc-Cdk1 system was modeled with two ordinary differential equations:
and
where [A] and [I] represent concentrations of active and inactive forms of cyclin-Cdk1 complex, respectively. The active form is produced at a rate of ks, and , , and are ultrasensitive functions of [A], reflecting Wee1-dependent double-negative feedback, Cdc25-dependent positive feedback, and APC/C-dependent ultrasensitive negative feedback, respectively (Fig. 3 A). It is well established that the checkpoint kinase, Chk1, regulates the Cyc-Cdk1 double-negative feedback and positive feedback regulation and slows cell-cycle progression as the embryo approaches the MBT (Fig. 3 A; (38, 39, 40, 41)). Together, and were modeled by and , where represents Chk1 activity (27,33,34). Measured values of nuclear concentrations of Chk1 remained almost unchanged from NC11 through NC13 (Figs. 3 B and S2), but Chk1 activity must increase to slow the cell cycle at the MBT. We have recently shown that H3 acts as a competitive inhibitor of Chk1 and that the decrease in H3 concentrations is crucial for cell-cycle slowing (27). However, it is unclear how dynamics of H3 nuclear import contributes to the H3 regulation of cell cycle.
We have previously shown that the per-nuclear import rates of H3 in interphase are well recapitulated by what we referred to as the import-limited regime:
where Hc represents total cytoplasmic H3 and Nn is the number of nuclei in the early embryo (25). Again, note that Nn doubles as the embryo undergoes divisions, and therefore, per-nuclear import rates halve with each cell cycle. We also tested whether the nucleus-limited regime could explain the dynamics of H3 nuclear import, but no parameter was found to better recapitulate the experimental data (Fig. S3; Supporting materials and methods). Total H3 in the embryo—Htotal = Hc + Nn × Hn, where Hn indicates total nuclear H3 in a given nucleus—was approximated to be constant during the cell cycle (25). The value of KH has been estimated to be significantly small compared to Htotal, indicating that import rates are saturated until Hc is almost completely exhausted (25). This is required to recapitulate the biphasic dynamics of H3 nuclear accumulation at NC13 (Fig. 1 E). Dynamics of nuclear growth, Cyc-Cdk1-dependent NEB, and loss of non-DNA-bound H3 in mitosis were also included in this model to calculate nuclear H3 concentrations, [Hn] (Fig. S4). Finally, we modeled Chk1 inhibition by H3 as (27). See Supporting materials and methods for full details of the model.
Simulated time courses of total nuclear H3, nuclear volumes, and Cyc-Cdk1 activity are shown in Fig. 3 C. Nuclear H3 accumulates almost linearly until either NEB (NC11 and NC12) or cytoplasmic H3 pools are fully exhausted (NC13), recapitulating previous observations of H3 nuclear import (Fig. 1 E). Note that total nuclear H3 and nuclear volumes after NEB are twice as large as their initial values at the beginning of each cell cycle, reflecting replication of the genome and H3 inclusion into chromatin. Cell-cycle durations, as defined by the period of the Cyc-Cdk1 oscillation, lengthened with each cell cycle, consistent with previous experimental data (Fig. S5; (15,27,39,42)). Together, our model quantitatively recapitulates the dynamics of H3 nuclear import and its downstream activity, cell-cycle progression, during early embryogenesis.
Nuclear import results in differential sensitivity to H3 pool size in each cell cycle
We have previously examined how histone abundance controls embryonic cell-cycle dynamics using histone depleted embryos and observed that 60% of histone depletion results in dramatic cell-cycle lengthening in NC13 but has little effect on early cycles through NC11 (15). To ask whether histone import could explain the distinct sensitivities of different cell cycles to changes in histone pool size, we simulated histone depletion and overexpression (Fig. 4 A). We found that even 60% of H3 depletion is still sufficient to saturate nuclear import at NC11 and that the dynamics of H3 accumulation in NC11 are almost the same between wild-type (WT) and H3 depletion. However, H3-depleted embryos started running out of cytoplasmic H3 pools in NC12 and NC13, resulting in reductions in nuclear H3 and prolonged cell-cycle durations, consistent with previous experimental observations. In contrast, we found that histone overexpression does not dramatically alter dynamics of H3 nuclear import and cell-cycle durations in NC11 and NC12 and only slightly affects NC13 durations. These simulation results may explain why histone depletion experiments frequently show more robust cell-cycle phenotypes than histone overexpression, although we note that there are also technical difficulties in substantially increasing histone levels in vivo because of the large size of the endogenous histone pool (13,15).
Figure 4.
(A) Simulation of H3 depletion (40% of WT, red lines) and H3 overexpression (200% of WT, blue lines). Simulated dynamics of nuclear H3 and Cyc-Cdk1 are shown along with WT (gray dashed lines). Histone reduction barely affects NC11 durations, slightly lengthens NC12 durations, and dramatically prolongs NC13 durations. This behavior is consistent with experimental observations of histone-depleted embryos (15). By contrast, cell-cycle durations were mostly unaffected by histone overexpression. (B) Simulated cell-cycle durations are plotted as a function of total H3 in the embryo. Note that depletion of more than 60% H3 results in loss of Cyc-Cdk1 oscillations in NC13, which can be interpreted as premature cell-cycle arrest (15). Simulated embryos show greater sensitivity to histone reduction than overexpression under this import regime. To see this figure in color, go online.
To better understand the relationship between total histone levels and cell-cycle dynamics, we simulated cell-cycle durations with respect to various sizes of total H3 pools (Htotal). We found that cell-cycle durations are robust until the pool size of total H3 falls below a “threshold” (Fig. 4 B). This threshold corresponds to the level at which cytoplasmic H3 pools start running out during interphase and is different for different cell cycles because demand for histones increases with each cycle. Above the threshold, dynamics of nuclear H3 levels are the same within the timescale of early embryonic cell cycles. Thus, nuclear import sets the lower limit of cell-cycle durations. In the WT condition, the pool size of total H3 is far from the NC11 and NC12 thresholds and close to the NC13 threshold, allowing robust and rapid cell cycles in NC11 and NC12 as well as flexibility of the NC13 cell cycle that might be beneficial to be prepared for large-scale cell-cycle remodeling at NC14.
Dilution of H3 nuclear import contributes to both stepwise and robust cell-cycle slowing
Next, we sought to understand the importance of the presumably unusual import-limited regime of H3 nuclear import for cell-cycle control. We analyzed how cell-cycle dynamics would change with each cell cycle if H3 nuclear import were described by the nucleus-limited regime, as was the case for NLS-mRFP (Fig. 5 A). In this “mutant” import model, increasing the number of nuclei with each cycle does not affect import rates from NC11 through NC13 (Figs. 5 B and S6 A). Note that in Fig. 5 B, we used the parameter value by which the import rate at NC11 is the same as the import-limited NC11. Therefore, import rates in NC12 and NC13 were increased by twofold and fourfold compared to those in the “WT” import-limited model, respectively. In this regime, the dynamics of H3 nuclear import in NC11 and NC12 were barely distinguishable because the pool size of cytoplasmic H3 is too large to become limiting in NC11 and NC12. Consistent with the H3 dynamics, we found that the nuclei-limited regime does not recapitulate the stepwise increase in cell-cycle durations from NC11 to NC12, and from NC12 to NC13 (Fig. 5 C). Rather, NC12 is equivalent in length to NC11. Additionally, although this regime resulted in a similar extent of cell-cycle slowing at NC13 to experimental observations, NC13 durations become more sensitive to fluctuations in the total H3 pool size, losing robustness. In the above analysis, we chose the parameter value such that the import rate at NC11 in the mutant import model corresponds to that of NC11 in the WT model, but the same conclusions hold when we matched the import rate at either NC12 or NC13 to that of the WT import model (Figs. S6 and S7). Together, these results indicate that dilution of H3 nuclear import due to the import-limited regime is critical for robust gradual cell-cycle slowing during early embryogenesis.
Figure 5.
(A) Schematic of the model for nucleus-limited H3 nuclear import and cell-cycle regulation. Unlike the import-limited model in Fig. 3 A that is consistent with experimental data, we assumed that the import rate of H3 does not depend on the number of nuclei, as was the case for NLS-mRFP (Fig. 1D). (B) Simulated dynamics of H3 nuclear import with the parameter value by which rate of nuclear import at NC11 is the same as the import-limited model. Because there is no dilution over nuclei, import rates in NC12 and NC13 are increased by twofold and fourfold compared with those in the original import-limited model (Fig. 3), respectively. (C) Simulated cell-cycle durations as a function of total H3 in the embryo with the parameter value used in Fig. 5B. Note that data for NC11 correspond to data for NC11 in Fig. 4B.
Discussion
In the early embryos that rapidly alternate between interphase and mitosis, nucleocytoplasmic partitioning is far from steady state for most proteins, and the kinetics of nuclear import determine transient nuclear composition. Here, we have investigated the role of nuclear import dynamics in the context of early embryogenesis of Drosophila. We considered two regimes in which either nuclear capacity or import capacity is limiting and showed that these mechanisms well recapitulate distinct dynamics of NLS-mRFP, which accumulates more in late cycles, and histone H3-Dendra2, which becomes diluted with each cycle. Importantly, these distinct dynamics can arise from only a difference in the nuclear import regime and the timescale of import and do not require cargo-specific active regulation. Using an integrative model for H3 nuclear import and cell-cycle control, we showed that dilution of H3 nuclear import contributes to the stepwise regulation of cell-cycle durations as well as its robustness. Together, this study defines the critical role of nuclear import dynamics in early embryogenesis.
In the pre-MBT cell cycles, nuclei proceed to mitosis before nuclear growth reaches steady state, suggesting that the timescale of bulk nuclear import is slower than the timescale of the cell cycle. For relatively slow import cargos, effects of the different import regimes on the dynamics of nuclear accumulation becomes pronounced (Fig. 2). However, some nuclear proteins are imported much more rapidly than bulk nuclear proteins. For example, the transcription factor, Bicoid, reaches steady state within minutes into a given nuclear cycle (36,43). Another example of rapid nuclear import is the pioneer factor, Zelda (44), which binds to thousands of sites in the genome as early as at NC8 (45,46). For transcription factors such as Bicoid and Zelda, the speed of import may be more important than the specific import regime, as steady-state concentrations are likely to be more robust in patterning the correct spatial and temporal expression of target genes. It will be of interest to find whether and how dynamics of nuclear import behavior may link to the function of the cargo.
The specific molecular underpinnings of the observed different behaviors between nucleus-limited cargos (e.g., NLS-mRFP) and import-limited cargos (e.g., H3) require further study. One strong candidate for the nucleus-limited regime is the number of NPCs that act as the gates for nuclear transport. NPCs are preassembled in the endoplasmic reticulum membrane sheets called annulate lamellae and are supplied to the nuclear envelope, maintaining NPC concentrations on the nuclear membrane even in the rapidly dividing embryo (47). Therefore, the number of NPCs likely remains relatively fixed for a given nucleus and increases embryo wide as more nuclei are made. For the import-limited regime, any component that defines import capacity for the cargo could become limiting, for example, karyopherins (Kaps). Eukaryote genomes encode tens of Kaps: at least 14 in yeast, 16 in Drosophila, and 20 in humans (48,49). Different Kaps prefer specific NLSs, although many cargos can be recognized by multiple Kaps with different binding affinities (50). Expression levels of different Kaps differ by at least eightfold in yeast, and both Kap concentrations and binding affinities of Kap-cargo pairs affect import rates (51). It may be that differences in a cargo dependency on import pathways contribute to distinct regimes of nuclear import behavior. Understanding the relationship between these quantitative parameters and the behaviors of nuclear import dynamics is of high importance for a full quantitative understanding of nuclear state changes during early development.
Author contributions
Conceptualization, Y.S. and A.A.A.; investigation, Y.S.; writing—original draft, Y.S. and A.A.A.; writing—review and editing, Y.S. and A.A.A.
Acknowledgments
We thank Thao Nguyen and Martin Wühr for discussion and comments; Melissa Harrison, Kate O’Connor-Giles, and Jill Wildonger for plasmids; Robert Marmion, Stanislav Shvartsman, and the Bloomington Drosophila Stock Center (National Institutes of Health P40OD018537) for fly stocks. We also thank Gary Laevsky and the Confocal Imaging Facility at Princeton University, Gordon Gray and the Drosophila Media Core Facility at Princeton University, and Ann Lavanway, Britton Johnson, and Drosophila Media Core Facility at Dartmouth College. Y.S. was supported by Japan Society for the Promotion of Science Overseas Research Fellowships, the Postdoctoral Fellowship from the Uehara Memorial Foundation, and the Osamu Hayaishi Memorial Scholarship for Study Abroad.
Editor: Stanislav Shvartsman.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2021.05.005.
Contributor Information
Yuki Shindo, Email: yuki.shindo@dartmouth.edu.
Amanda A. Amodeo, Email: amanda.a.amodeo@dartmouth.edu.
Supporting citations
Reference (52) can be found in the Supporting material.
Supporting material
References
- 1.Blythe S.A., Wieschaus E.F. Coordinating cell cycle remodeling with transcriptional activation at the Drosophila MBT. Curr. Top. Dev. Biol. 2015;113:113–148. doi: 10.1016/bs.ctdb.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 2.Yuan K., Seller C.A., O’Farrell P.H. Timing the Drosophila mid-blastula transition: a cell cycle-centered view. Trends Genet. 2016;32:496–507. doi: 10.1016/j.tig.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Newport J., Kirschner M. A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell. 1982;30:675–686. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- 4.Newport J., Kirschner M. A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell. 1982;30:687–696. doi: 10.1016/0092-8674(82)90273-2. [DOI] [PubMed] [Google Scholar]
- 5.Edgar B.A., Kiehle C.P., Schubiger G. Cell cycle control by the nucleo-cytoplasmic ratio in early Drosophila development. Cell. 1986;44:365–372. doi: 10.1016/0092-8674(86)90771-3. [DOI] [PubMed] [Google Scholar]
- 6.Kane D.A., Kimmel C.B. The zebrafish midblastula transition. Development. 1993;119:447–456. doi: 10.1242/dev.119.2.447. [DOI] [PubMed] [Google Scholar]
- 7.Chen H., Einstein L.C., Good M.C. Spatiotemporal patterning of zygotic genome activation in a model vertebrate embryo. Dev. Cell. 2019;49:852–866.e7. doi: 10.1016/j.devcel.2019.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casas-Vila N., Bluhm A., Butter F. The developmental proteome of Drosophila melanogaster. Genome Res. 2017;27:1273–1285. doi: 10.1101/gr.213694.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peshkin L., Wühr M., Kirschner M.W. On the relationship of protein and mRNA dynamics in vertebrate embryonic development. Dev. Cell. 2015;35:383–394. doi: 10.1016/j.devcel.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Almouzni G., Wolffe A.P. Replication-coupled chromatin assembly is required for the repression of basal transcription in vivo. Genes Dev. 1993;7:2033–2047. doi: 10.1101/gad.7.10.2033. [DOI] [PubMed] [Google Scholar]
- 11.Prioleau M.N., Huet J., Méchali M. Competition between chromatin and transcription complex assembly regulates gene expression during early development. Cell. 1994;77:439–449. doi: 10.1016/0092-8674(94)90158-9. [DOI] [PubMed] [Google Scholar]
- 12.Almouzni G., Wolffe A.P. Constraints on transcriptional activator function contribute to transcriptional quiescence during early Xenopus embryogenesis. EMBO J. 1995;14:1752–1765. doi: 10.1002/j.1460-2075.1995.tb07164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amodeo A.A., Jukam D., Skotheim J.M. Histone titration against the genome sets the DNA-to-cytoplasm threshold for the Xenopus midblastula transition. Proc. Natl. Acad. Sci. USA. 2015;112:E1086–E1095. doi: 10.1073/pnas.1413990112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joseph S.R., Pálfy M., Vastenhouw N.L. Competition between histone and transcription factor binding regulates the onset of transcription in zebrafish embryos. eLife. 2017;6:e23326. doi: 10.7554/eLife.23326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chari S., Wilky H., Amodeo A.A. Histone concentration regulates the cell cycle and transcription in early development. Development. 2019;146:dev177402. doi: 10.1242/dev.177402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collart C., Allen G.E., Zegerman P. Titration of four replication factors is essential for the Xenopus laevis midblastula transition. Science. 2013;341:893–896. doi: 10.1126/science.1241530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vastag L., Jorgensen P., Kirschner M.W. Remodeling of the metabolome during early frog development. PLoS One. 2011;6:e16881. doi: 10.1371/journal.pone.0016881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Djabrayan N.J.-V., Smits C.M., Shvartsman S.Y. Metabolic regulation of developmental cell cycles and zygotic transcription. Curr. Biol. 2019;29:1193–1198.e5. doi: 10.1016/j.cub.2019.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu B., Winkler F., Großhans J. A link between deoxyribonucleotide metabolites and embryonic cell-cycle control. Curr. Biol. 2019;29:1187–1192.e3. doi: 10.1016/j.cub.2019.02.021. [DOI] [PubMed] [Google Scholar]
- 20.Levy D.L., Heald R. Nuclear size is regulated by importin α and Ntf2 in Xenopus. Cell. 2010;143:288–298. doi: 10.1016/j.cell.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jevtić P., Levy D.L. Nuclear size scaling during Xenopus early development contributes to midblastula transition timing. Curr. Biol. 2015;25:45–52. doi: 10.1016/j.cub.2014.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jevtić P., Levy D.L. Both nuclear size and DNA amount contribute to midblastula transition timing in Xenopus laevis. Sci. Rep. 2017;7:7908. doi: 10.1038/s41598-017-08243-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wente S.R., Rout M.P. The nuclear pore complex and nuclear transport. Cold Spring Harb. Perspect. Biol. 2010;2:a000562. doi: 10.1101/cshperspect.a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerhart J.C. In: Biological Regulation and Development. Goldberger R.F., editor. Volume 2. Springer; 1980. Mechanisms regulating pattern formation in the amphibian egg and early embryo; pp. 133–316. [Google Scholar]
- 25.Shindo Y., Amodeo A.A. Dynamics of free and chromatin-bound histone H3 during early embryogenesis. Curr. Biol. 2019;29:359–366.e4. doi: 10.1016/j.cub.2018.12.020. [DOI] [PubMed] [Google Scholar]
- 26.Mukherjee R.N., Sallé J., Levy D.L. The perinuclear ER scales nuclear size independently of cell size in early embryos. Dev. Cell. 2020;54:395–409.e7. doi: 10.1016/j.devcel.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shindo Y., Amodeo A.A. Excess histone H3 is a competitive Chk1 inhibitor that controls cell-cycle remodeling in the early Drosophila embryo. Curr. Biol. 2021 doi: 10.1016/j.cub.2021.03.035. S0960-9822(21)00374-2. Published online April 6, 2021. [DOI] [PubMed] [Google Scholar]
- 28.Gratz S.J., Ukken F.P., O’Connor-Giles K.M. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics. 2014;196:961–971. doi: 10.1534/genetics.113.160713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berg S., Kutra D., Kreshuk A. ilastik: interactive machine learning for (bio)image analysis. Nat. Methods. 2019;16:1226–1232. doi: 10.1038/s41592-019-0582-9. [DOI] [PubMed] [Google Scholar]
- 30.Virtanen P., Gommers R., van Mulbregt P., SciPy 1.0 Contributors SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods. 2020;17:261–272. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Novak B., Tyson J.J. Modeling the cell division cycle: M-phase trigger, oscillations, and size control. J. Theor. Biol. 1993;165:101–134. [Google Scholar]
- 32.Yang Q., Ferrell J.E., Jr. The Cdk1-APC/C cell cycle oscillator circuit functions as a time-delayed, ultrasensitive switch. Nat. Cell Biol. 2013;15:519–525. doi: 10.1038/ncb2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deneke V.E., Melbinger A., Di Talia S. Waves of Cdk1 activity in S phase synchronize the cell cycle in Drosophila embryos. Dev. Cell. 2016;38:399–412. doi: 10.1016/j.devcel.2016.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vergassola M., Deneke V.E., Di Talia S. Mitotic waves in the early embryogenesis of Drosophila: bistability traded for speed. Proc. Natl. Acad. Sci. USA. 2018;115:E2165–E2174. doi: 10.1073/pnas.1714873115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Calzone L., Thieffry D., Novak B. Dynamical modeling of syncytial mitotic cycles in Drosophila embryos. Mol. Syst. Biol. 2007;3:131. doi: 10.1038/msb4100171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grimm O., Wieschaus E. The Bicoid gradient is shaped independently of nuclei. Development. 2010;137:2857–2862. doi: 10.1242/dev.052589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grimm O., Sanchez Zini V., Wieschaus E. Torso RTK controls Capicua degradation by changing its subcellular localization. Development. 2012;139:3962–3968. doi: 10.1242/dev.084327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fogarty P., Campbell S.D., Sullivan W. The Drosophila grapes gene is related to checkpoint gene chk1/rad27 and is required for late syncytial division fidelity. Curr. Biol. 1997;7:418–426. doi: 10.1016/s0960-9822(06)00189-8. [DOI] [PubMed] [Google Scholar]
- 39.Sibon O.C., Stevenson V.A., Theurkauf W.E. DNA-replication checkpoint control at the Drosophila midblastula transition. Nature. 1997;388:93–97. doi: 10.1038/40439. [DOI] [PubMed] [Google Scholar]
- 40.Farrell J.A., O’Farrell P.H. Mechanism and regulation of Cdc25/Twine protein destruction in embryonic cell-cycle remodeling. Curr. Biol. 2013;23:118–126. doi: 10.1016/j.cub.2012.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Di Talia S., She R., Wieschaus E.F. Posttranslational control of Cdc25 degradation terminates Drosophila’s early cell-cycle program. Curr. Biol. 2013;23:127–132. doi: 10.1016/j.cub.2012.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takada S., Kwak S., Theurkauf W.E. grp (chk1) replication-checkpoint mutations and DNA damage trigger a Chk2-dependent block at the Drosophila midblastula transition. Development. 2007;134:1737–1744. doi: 10.1242/dev.02831. [DOI] [PubMed] [Google Scholar]
- 43.Gregor T., Wieschaus E.F., Tank D.W. Stability and nuclear dynamics of the bicoid morphogen gradient. Cell. 2007;130:141–152. doi: 10.1016/j.cell.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dufourt J., Trullo A., Lagha M. Temporal control of gene expression by the pioneer factor Zelda through transient interactions in hubs. Nat. Commun. 2018;9:5194. doi: 10.1038/s41467-018-07613-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liang H.-L., Nien C.-Y., Rushlow C. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature. 2008;456:400–403. doi: 10.1038/nature07388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harrison M.M., Li X.Y., Eisen M.B. Zelda binding in the early Drosophila melanogaster embryo marks regions subsequently activated at the maternal-to-zygotic transition. PLoS Genet. 2011;7:e1002266. doi: 10.1371/journal.pgen.1002266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hampoelz B., Mackmull M.T., Beck M. Pre-assembled nuclear pores insert into the nuclear envelope during early development. Cell. 2016;166:664–678. doi: 10.1016/j.cell.2016.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mosammaparast N., Pemberton L.F. Karyopherins: from nuclear-transport mediators to nuclear-function regulators. Trends Cell Biol. 2004;14:547–556. doi: 10.1016/j.tcb.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 49.Quan Y., Ji Z.-L., Tao T. Evolutionary and transcriptional analysis of karyopherin β superfamily proteins. Mol. Cell. Proteomics. 2008;7:1254–1269. doi: 10.1074/mcp.M700511-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chook Y.M., Süel K.E. Nuclear import by karyopherin-βs: recognition and inhibition. Biochim. Biophys. Acta. 2011;1813:1593–1606. doi: 10.1016/j.bbamcr.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Timney B.L., Tetenbaum-Novatt J., Rout M.P. Simple kinetic relationships and nonspecific competition govern nuclear import rates in vivo. J. Cell Biol. 2006;175:579–593. doi: 10.1083/jcb.200608141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Royou A., McCusker D., Sullivan W. Grapes(Chk1) prevents nuclear CDK1 activation by delaying cyclin B nuclear accumulation. J. Cell Biol. 2008;183:63–75. doi: 10.1083/jcb.200801153. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.