Abstract
Transcription factors are the major agents that read the regulatory sequence information in the genome to initiate changes in expression of specific genes, both in development and in physiological activation responses. Their actions depend on site-specific DNA binding and are largely guided by their individual DNA target sequence specificities. However, their action is far more conditional in a real developmental context than would be expected for simple reading of local genomic DNA sequence, which is common to all cells in the organism. They are constrained by slow-changing chromatin states and by interactions with other transcription factors, which affect their occupancy patterns of potential sites across the genome. These mechanisms lead to emergent discontinuities in function even for transcription factors with minimally changing expression. This is well revealed by diverse lineages of blood cells developing throughout life from hematopoietic stem cells, which use overlapping combinations of transcription factors to drive strongly divergent gene regulation programs. Here, using development of T lymphocytes from hematopoietic multipotent progenitor cells as a focus, recent evidence is reviewed on how binding specificity and dynamics, transcription factor cooperativity, and chromatin state changes impact the effective regulatory functions of key transcription factors including PU.1, Runx1, Notch-RBPJ, and Bcl11b.
Significance
Transcription factors “read” the genomic sequence to determine when and where gene expression should take place. They are vital for sequential developmental changes, and their collaboration in different combinations allows gene networks to operate that generate thousands of different cell types. However, their mechanisms of action on the genome in stem-cell-based mammalian systems are based on many additional factors in addition to their ability to bind specifically recognized sites in the DNA. Here, the early T-cell development system provides a specific vantage on how transcription factors interact with each other and both alter and respond to chromatin states to create developmental discontinuities. The different time constants of these effects have a great impact on prediction of transcription factor behavior.
Introduction
Transcription factors (TFs) are the key readers of genomic regulatory sequence, and they bring specificity to developmental gene regulation. Unlike other transcriptional regulators, they work 1) in trans, 2) coordinating activities of sites across the genome, and 3) in a sequence-specific way, discriminating one potential regulatory site from another. Combinatorial requirements for concerted TF actions underlie the logic operating at nodes of gene regulatory networks. Gene regulation mechanisms not only direct development but also mediate the acute physiological responses of mature differentiated cells to perturbation (1, 2, 3, 4). However, most physiological responses are temporary, allowing the cell to return to something very much like the initial ground state after the stimulus is removed. By contrast, development forecloses previous regulatory options and opens others at each step, causing a permanent change in the ground state. A central question is how different mechanisms interact to generate this irreversibility.
In an embryo, the overall complexity increase can itself be a source of irreversibility. Complex patterns of transcription factor expression in different cells of the embryo are generated by hierarchical gene regulatory network operation, and boundaries between different cell types are often direct outcomes of all-or-none differences between the expression of specific transcription factors on each side of the future boundary (reviewed in (5)). These expression differences are often triggered or reinforced by signaling between the neighboring cells as soon as they become distinct, with new signal-interaction interfaces appearing as cell division proceeds, thus relating gene network progression to the increasing anatomical complexity of the embryo. However, hematopoiesis, the best-studied example of development from stem cells, is different. In hematopoiesis, progressive changes in cell state also occur. However, because each hematopoietic cell’s fate is determined independent of others and maintained despite its migration through different microenvironments, these changes must be enforced at the single-cell level, at which there is no increase in complexity. The direction of developmental change must be based on cell-intrinsic, heritable mechanisms enabling each cell to keep track of where it is in a multistep developmental pathway. Some of these mechanisms are evident in sharp shifts in TF site-binding preferences across the genome at specific developmental state transitions. As described below, recent evidence shows that even the same TF expressed at virtually unchanging levels in one of these lineages can regulate completely different target genes before and after a state transition (6,7).
The stage discontinuities may help to preserve the directionality of this process. Thus, the question of how system irreversibility is achieved can be approached through the mechanisms constraining the scope of action of individual TFs in different developmental contexts. Major candidates for these mechanisms are ordered activation or deactivation of collaborating TF activities for joint target gene regulation, and/or long-term changes in chromatin accessibility, that transform the ground states for gene regulation.
This article will first review the modes of interaction between TF-based and chromatin states that can contribute to gene regulatory logic in the context of differentiating cells in long-lived, complex organisms. It will then draw upon highly detailed evidence from the early T-cell developmental pathway to consider how distinct biophysical mechanisms may respectively contribute to the regulatory logic, kinetics, and irreversibility of developmental progression. The results in this system show that we must account not only for different stable cell-type identities but also for the mechanisms that can transiently break this stability at key developmental transitions, to shift a progenitor to a new, restabilized identity in which many old components play new roles.
Binding, detection of binding, and functional association
Sequence-specific TFs are normally assumed to work through DNA binding and to bind to specific sites based on DNA sequence motif recognition (8, 9, 10, 11). However, it does not follow that all potential target sites are bound by an available TF at a given time or that their binding necessarily corresponds with function. This has become clear as in the past 15 years, ChIP-seq (12) and its derivatives, ChIP-exo (13) and CUT&RUN (14), have vastly simplified the job of determining where on the genome TFs are actually binding at a given moment in a particular cell type. They have technical limitations (cell numbers needed) and caveats for interpretation (cross-linking issues, possible open chromatin bias). Nevertheless, these methods have demonstrated two important features of the relationship of regulatory genome sequence to TF function, especially in mammalian cells: namely selective, conditional site-binding choice and rare functionality.
First, ChIP-seq data show that many TFs, at least in well-defined mammalian cell types, bind detectably to only a small subset of the potential target sequences encoded by the genome. On the order of 104 sites are typically found occupied by a given TF in a genome that should contain 105–106 equally good copies of their target motifs. This might be explained on the basis of differential affinity and TF expression levels (“fugacity,” (15)) (Fig. 1 A, top). However, occupancy patterns for the same TF can differ substantially in different cell types, and this is discussed extensively below.
Figure 1.
Modulators of transcription factor (TF) binding. (A) Affinity and accessibility: comparison of binding of a generic TF “factor A” (magenta triangles) of TF affinity for target sites in open and closed chromatin. Two conditions are shown: occupancy in the presence of high levels of factor A or in the presence of low factor A. Blue shading of sites indicates relative site affinities as shown in key. Left: in a cell with high factor A. Right: in a cell with low factor A. Top: all sites open. Bottom: with closed chromatin (horizontal gray) limiting access to a block of sites as indicated. Sizes of triangles approximate relative levels of occupancy. Bent arrows show potential impacts on target gene activation, assuming that transcription at each promoter depends on factor A binding to the neighboring site. Question mark: uncertain regulatory impact of low-level occupancy. (B) Combinatorial action via direct binding collaboration shows impact of binding of factor A to higher and lower affinity sites (as in A) with a second factor, B (yellow ovals). Left genomic region: hypothetical sites where genomic sequences allow independent binding of A. Right genomic region: different hypothetical sites where A site binding depends on interaction of A with another partner (sites with light blue outlines). Here, juxtaposed sites for A and B allow highly enhanced binding of A, but only if B is present. As shown in the diagram, the impact of factor B on binding of A is often greatest at sites where the affinity of A for the site is otherwise low (lighter blue). Question mark: uncertain ability of site affinity alone to overcome requirement for partner. (C) Combinatorial action via local chromatin opening shows impact on binding of factor A to closed sites when factor B acts as a pioneer. In this example, A is blocked by closed chromatin, but B is not. Top: comparison of open and closed binding site occupancies by A. Bottom: zoom-in to show effect of B on the closed sites specifically (assuming no change in the open sites). Note that advent and binding of factor B creates an island of sharply localized chromatin access for A, where binding of A is not necessarily based on affinity of A for the site itself. (D) Chromatin domain opening shows impact on factor A binding to multiple sites in an extended region (under “closed chromatin” bar at left) when factor B binding can catalyze broad opening of a large chromatin domain (e.g., (16)).
Second, at the same time, the sites that TFs bind to are far more numerous than are accounted for by the genetic loci that actually respond to the presence of those TFs in gain or loss of function perturbations. Typical disparities for the factors described below are ∼20,000 binding sites but only ∼500 responsive loci (activated and/or repressed). A particular hotspot for “false” positives can be promoters. Strong TF binding is often seen at promoters and apparent open chromatin regions around genes that are actively being expressed, whether these genes actually respond to those TFs or not. In descriptive whole genome surveys, such genes are often assumed to be TF targets, but when actual functional tests are done by perturbation of TF levels, most genes linked with TF binding are found not to be regulated by the TF in that cell context (for example, (17, 18, 19, 20, 21)). Although there are several possible explanations, the results emphasize that ChIP-seq evidence for TF binding on its own cannot be equated with function. It regularly occurs at a large excess of “nonfunctional” sites, demanding another explanation for what distinguishes the functional ones.
Combinatorial binding stabilization by direct TF-TF interaction
Genome-wide binding data have raised questions about the degree to which each TF’s recognition specificity contributes independently to its own binding profile. Because TFs recognize different DNA sequence motifs, in the early 2000s it was a natural assumption that combinations of TFs would only be found binding together at genomic sites where all of them independently recognized their cognate motifs; thus, the genome would use the full combinatorial complexity potential of TF specificities to encode the requirements for activity at a given enhancer site.
Different TFs with overlapping but nonidentical patterns of expression can indeed convey independent logical inputs for gene regulation if each depends on the other to stabilize its binding (Fig. 1 B). At sites where DNA sequence provides weak to moderate matches to each of the target motifs, the “AND” logic through which the two factors collaborate for gene regulation can be manifest directly through a structural requirement for simultaneous binding. Such AND logic, as partner factor expression changes, can clearly provide part of the explanation for stage specificity of TF binding (see below; Runx). Collaborative binding has also been argued to limit the potential of each individual TF to bind to developmentally irrelevant regulatory elements (22) in a given context.
Collaborative mechanisms can be more or less structurally specific. Some TFs have structural features that require them to be recruited to a given site by interaction with another specific TF or TFs (23). In such cases, the expression of one of these other factors in the cell can make an all-or-none difference for binding at a given site. The Ets1 transcription factor, for example, has an autoinhibitory domain that reduces its own binding to naked DNA. However, this inhibition can be removed allosterically by interaction with either Runx1 or Pax5. In B-lymphoid lineage cells, which uniquely express Pax5, this gives Ets1 access to Ets-Pax composite sites that Ets1 cannot bind in other cells (24). In T lineage cells, Ets1 collaborates with Runx1 to bind to composite Ets-Runx sites (25, 26, 27). In other examples, the interferon response factor IRF4 shows an ability to form highly specific collaborations with the Ets subfamily member PU.1 at composite Ets-IRF elements (28,29) (called “EICE”), whereas it forms alternative collaborations with basic leucine zipper (bZIP) AP-1/BATF family members at other composite sites (AICE) (30). The differential availability of these partners in different lineages and under different signaling conditions then effectively changes the affinity of the shared factor for different composite sites. Other well-known cases are those of PU.1 interacting selectively with the basic leucine zipper C/EBP family TFs in myeloid cells, although not in B cells (31). Natural single-nucleotide polymorphisms between different mouse strains show that the binding of either PU.1 or C/EBP to regions of PU.1 and C/EBP motifs in normal myeloid cells depends on the integrity of the motif for the other factor at that site as well as its own (32).
In initially studied examples, sites for functionally interacting TFs were closely juxtaposed in regulatory element DNA (33, 34, 35, 36, 37). This could suggest that the genome encodes stereospecific requirements for TF combinatorial action. However, evolutionary comparisons have shown that the binding site spacings and orders in a regulatory system can be highly variable, yet still keep the target gene similarly responsive to combined action by the same TFs (38,39). This makes it likely that TF-TF functional collaboration is also favored by mechanisms that do not require such structural precision.
Chromatin gating of TF access
The possibility of a TF’s engaging a given binding site may also be affected by accessibility constraints because of local chromatin states, which are a function of the lineage history of the cell type. Accurate modeling of the kinetics of TF actions in development depends on taking account of these mechanisms, even in fast-developing Drosophila embryos (40), and as described below, their impact is likely to be much stronger in slow mammalian systems. Hints of the power of chromatin accessibility to bias TF binding choices have come from ChIP-seq analyses of multiple, structurally disparate TFs within the same cell type. Although genomic sequence would predict different TFs to display completely different binding patterns, ∼10 years ago the earliest multi-TF ChIP-seq analyses in a highly-defined, hematopoietic precursor cell type showed a sobering picture of TF binding deployment (41). When 10 distinct TFs were interrogated, instead of finding 10 unique binding profiles with only a few, highly significant sites of overlap, the patterns of binding for many of the TFs had unexpectedly high similarities. Among the sites bound were sites that had previously been identified as functional enhancers, but a heptad of structurally different TFs binding together to the same sites was unexpectedly prevalent in the pattern (42).
Chromatin compaction can affect TF binding access (Fig. 1 A, bottom), and different cell types show dramatically distinct patterns of nucleosome packing in chromatin across the genome. These patterns have long been detected by nuclease accessibility (43, 44, 45) and have recently become much easier to detect by accessibility to transposase action (ATAC-seq) (46). Using genome-wide mapping of chromatin accessibility, it has commonly been found that the binding patterns of diverse TFs are concentrated at “open” sites within the given cell. The question is whether this association is a consequence of the TF action or a constraint upon it.
Those TFs that can bind their sites even when chromatin appears closed have special roles as “pioneer” factors (Fig. 1 C, role of “factor B” (47,48)). The initial displacement of a nucleosome by a stably bound TF can broadly enhance access for others as long as the site remains open (Fig. 1 C; (49)). In at least some cases, such as endodermal pioneering by FoxA1 and lymphomyeloid cell pioneering by PU.1 (see below), the role of a pioneer TF indeed correlates with establishment of a new, heritable developmental ground state.
The physical accessibility detected by DNase or transposase is only one parameter of chromatin configuration that shows cell-type-specific differences without being encoded by DNA sequences per se, and it is possible that some of these other chromatin features also affect TF access. Locally, nucleosomes can be modified with “active” marks such as H3K4 di- and tri-methylation and acetylation of H3K9 and H3K27 or “repressive” marks such as trimethylation of H3K9 or H3K27. Locally, also, methylation of DNA may interfere with a TF’s recognition of its target sites. At a larger scale, chromosomal loci can be sequestered in different nuclear regions in different cell types (50), in “active” or “inactive” compartments (51,52), enclosed in different topologically associated domains, and engaged in different local CTCF-anchored loops generated by cohesin-dependent loop extrusion (53,54). All of these chromatin differences that distinguish cell types are regularly correlated with differential gene expression in the steady state. Furthermore, acute gene induction responses, e.g., in immune cells undergoing stimulation by antigens or cytokines, nicely correlate a variety of rapid local chromatin changes with changes in gene expression, e.g., local increases in accessibility and deposition of active histone marks. Similarly, there are numerous larger-scale chromatin topological changes in mature T cells whenever they get activated by T-cell receptor signals (55, 56, 57).
An insight into the mechanisms used to couple TF action to large scale-chromatin change comes from the example of a well characterized TF that causes widely propagated chromatin reconfiguration at a key developmental transition in T-cell development (cf. Fig. 1 D). At the end of the pro-T-cell stages (see below), the E protein E2A acts abruptly to open the locus encoding the Rag1 and Rag2 transposases that assemble the T-cell antigen receptor genes for expression (16). E2A and its paralog HEB are known to be functionally crucial for the expression of the Rag1-Rag2 locus, working through a T-lineage-specific enhancer element (16,58). Mutational disruption of seven E2A binding sites clustered together within this ∼2 kb enhancer in vivo has recently confirmed that E2A binding at this relatively compact element is essential for impacts that propagate all across the ∼200 kb topologically associated domain containing the Rag1-Rag2 locus. Binding at this enhancer is indispensable to allow loading of cohesin, to flip interaction compartment status from inactive to active, and to maintain open chromatin structure over this wide region (16).
Chromatin and regulatory dynamics: time and reversibility
Such chromatin changes could be important to explain the processivity of development if they alter the playing field for future gene regulation in the cells, with different time constants than those of TF binding itself. Chromatin stage change is usually induced by action of TF combinations (Fig. 1, C and D; (59, 60, 61, 62, 63, 64, 65, 66)). In addition to the chromatin-opening effects of pioneer factors described above, chromosome looping and local depositions of active chromatin marks are rapidly increased by the activation of signal-dependent TFs like STAT and Notch/RBPJ (2,4,67).
In principle, the timescales of the effects of TF-TF collaboration on open sites and effects of TF-induced chromatin change could be different. TF-TF collaboration at accessible sites is immediate (Fig. 1 B), and at a given site, concerted TF binding per se is memoryless. In contrast, a new pattern of chromatin accessibility (or modification, or architecture; Fig. 1, C and D) could be inherited, perpetuated by default through cell division by reader-writer mechanisms that are sequence nonspecific. Such passive propagation of a chromatin state would represent a source of regulatory inertia within an established cell lineage, making regulatory baselines dependent on lineage history and resisting change until the statuses of specific loci are disrupted by specific TF combinations, either in normal development or in artificial reprogramming (65,68,69). However, the ease of altering many chromatin state features any time an immune cell is activated raises questions about which of these mechanisms really are most important to affect future gene regulation. Only a subset of these induction-dependent chromatin changes appears to cause lasting changes in regulatory thresholds (60,62).
Higher-order chromatin looping may also change during development or cell activation, but its own regulatory impacts are not fully clear. Promoter-enhancer interactions of course occur during transcriptional activity, but it is not clear how strongly larger-scale topology affects regulation. In immune cells, activation-induced changes in looping do not appear to alter the larger pattern of gene associations between active and inactive chromatin compartments (57) and may be largely reversible when the cells revert to resting states. Elegant experiments in which cohesin or CTCF was acutely degraded in tissue culture cells or in noncycling thymocytes (to filter out confounding population effects due to cohesin roles in mitosis) showed that although loss of these components rapidly dissociated loops, a large fraction of the cell-type-specific gene expression in the steady state was independent of the loop structure (70, 71, 72).
A better candidate for a long-term threshold-setter for gene regulatory baselines may be DNA CpG methylation, which has long been known to be propagated across cell cycles (73). Although not strictly correlated with transcription, it has been implicated in long-term shifts in gene expression defaults in systems from T helper cell lineage choice to B-cell development to reprogramming of differentiated cells to iPS cells (35,63,65,74, 75, 76, 77, 78). In a deliberately simplified system, among four different repressive mechanisms, DNA methylation gave the most durable response changes (79). Not simply a symptom of closed chromatin, target motif methylation also specifically disfavors recognition by TFs of multiple families (80). This mechanism is not used in classic developmental gene regulatory systems such as the Drosophila embryo, though, and it is challenging to monitor in a truly genome-wide manner with small cell numbers. These issues have limited the developmental contexts in which functional regulatory impacts of methylation have been studied closely to date. Still, demethylation of specific sites via TET-dependent oxidation appears to be highly correlated with developmental gene regulation in the systems described below (81, 82, 83, 84, 85), and some novel approaches to manipulate methylation in a targeted way offer huge future promise (86).
Thus, beyond showing that TFs can affect certain features of chromatin, there is an acute need for quantitative measurement of the strengths of these chromatin state effects and the kinetics and reversibilities of chromatin state changes to gauge the extent to which they really cause discontinuities in the normal “playing field” for developmental gene regulation. The relationships of both directly collaborative and chromatin state-based mechanisms to modulate TF regulatory function, in a system that could be exploited to yield such measurements, are discussed below.
Hematopoietic development as a distinct window on TF rules of action
Hematopoiesis is an informative, distinctive system in which to examine cause-and-effect relationships in transcriptional regulation. Hematopoietic development (after the first cohorts in the embryo) begins from “set-aside” cells that are apparently restricted to hematopoietic fates but maintain access to ∼12 different major fate options. As stem cells, they are mitotically quiescent for long periods of time, occasionally carrying out a self-renewal cell division, but are capable of great proliferative expansion if triggered. Individual hematopoietic stem cells become activated rarely and asynchronously through life and then generate large clones of rapidly dividing multipotent progenitors with various developmental repertoires, which go on to differentiate along different combinations of pathways. Thus, hematopoiesis is a developmental system that generates new differentiated cells continuously through the entire lifespan. Fig. 2 A presents a simplified roadmap of mammalian hematopoiesis and shows some of the key TFs that are essential for major differentiation branches. For example, development of red blood cells (erythrocytes) and platelets (from megakaryocytes) depends heavily on GATA1 and GATA2 dual-C4 zinc finger factors. “Myeloid” innate-immune and inflammatory cells, including monocytes, macrophages, dendritic cells, and neutrophils, depend on PU.1 (divergent Ets subfamily). Lymphoid cells of all types depend on multi-C2H2 zinc finger factor Ikaros, and B and T lymphocytes in particular depend on basic helix-loop-helix E proteins (87, 88, 89, 90, 91). (Additional lymphoid-essential factors of the Runx and Bcl11 families are described in detail below.) This system provides vivid examples to show that individual TF roles in the relevant gene regulatory networks are highly conditional, not simply based on binding to genomic motifs with the highest possible affinities.
Figure 2.
Hematopoiesis and gene regulatory network circuits for conditional TF action. (A) A simplified overview of hematopoiesis in young adult mice. Hematopoietic stem cells (HSCs) give rise to functionally heterogeneous multipotent progenitors (MPPs), which proliferate to generate mixed populations of partially lineage-restricted progenitors as shown, which in turn proliferate and then undergo stereotyped terminal differentiation into the mature cell types. Not all intermediates are shown. Importantly, single HSCs, MPPs, and partially restricted progenitor cells are individually capable of giving rise to diverse cell types in their clonal progeny. Lineage decisions in different pathways are made roughly in parallel. It is likely that some initial branches of differentiation are chosen probabilistically, and the choices that generate different types of partially restricted intermediates appear to be variable from one HSC or MPP clone to another (dashed arrows). Thus, strictly binary branches are not shown. However, differentiation appears to be unidirectional for all hematopoietic cell types, and the terminal programs that generate output cells are canonical and well-defined. The requirement for certain key TFs, shown in the figure, is shared among certain groups of lineage branches. Although the differentiated progeny types are very distinct, their shared regulatory requirements correspond well to the ability of these groups of fates to be generated from the same single-cell progenitors in clonal assays. (B) Classic mutual antagonism circuit for PU.1 and GATA1 in myeloid versus erythroid development. PU.1 and GATA1 are shown to repress each other’s expression at the level of transcriptional repression and also to interfere with each other’s transactivation functions via protein-level interference (dashed lines). The results are mutually exclusive activation of a “myeloid” program in cells dominated by PU.1 or an “erythroid” program in cells dominated by GATA1. Note that mutual repression at the transcriptional level depends not only on PU.1 and GATA1 but also on their respective partners, C/EBPα and FOG1 (product of Zfpm1). Shown is an updated model from references cited in the text. (C) Ability of PU.1 and GATA1 to collaborate for mast-cell development and gene regulation. Mast-cell development, an alternative to both erythroid and myeloid pathways, results from the moderate levels of expression of PU.1 and GATA1 combined with the absence of both C/EBPα and FOG1, as described in the text. Here, myeloid and erythroid roles of these factors are attenuated or absent (dotted lines), whereas the distinct mast-cell program genes are activated by combinatorial activity of PU.1 and GATA1 (or GATA2) binding (solid lines). Although myeloid and erythroid genes might still be engaged by PU.1 and GATA1, they are not efficiently activated in mast cells, possibly because of the lower levels of these regulators than in (B) and the lack of C/EBPα and FOG1.
In classic work, cell fate determination in mammalian hematopoiesis has been explained by simple gene regulatory network circuits (e.g., Fig. 2 B; (90,92,93)). Various lineages of descendants are distinguished by expression of TF that are expressed in apparently lineage-specific ways (Fig. 2 A) and show mutual antagonism in experimental tests (Fig. 2 B). Importantly, however, a regulated ability to switch network states is inherent to the function of the hematopoietic system. The long-term stability of the poised state in the stem cells and multipotent precursors is based on the active operation of “stem/progenitor” gene regulatory networks (94), which must regularly get overridden when the cells commit to differentiate. Despite the association of key TFs with different, mutually exclusive pathways of differentiation, the hematopoietic precursors start out with overlapping expression of these same TFs, which can play roles both in the “stem/progenitor” network and in individual lineage differentiation networks (87,89,95, 96, 97, 98, 99). Furthermore, the same TFs that can oppose each other in a mutual-exclusivity gene regulatory network circuit in one cell type can collaborate in another (100, 101, 102). In part, this is because the same TFs collaborate with different, independently regulated TFs when they participate in different developmental programs. Examples are GATA family factors and the Ets family factor PU.1 (92,103, 104, 105). GATA1 opposes PU.1 in erythrocyte versus monocyte/macrophage specification, mutually repressing each other and mutually inhibiting each other’s transcriptional regulatory activities (Fig. 2 B). However, these two factors collaborate in mast-cell development and function (106). The highly stable mast-cell state is apparently made possible by the absence of certain partners of PU.1 and GATA1, C/EBPα, and FOG1, respectively (Fig. 2 C; (93,102,107)). Thus, because of the availability of different partners, the logic relating a given TF’s action to a particular target gene’s expression is dependent on the cell lineage (101,108).
There is conditionality also in the dose dependence of factor action in hematopoiesis. PU.1 at high levels blocks B-cell development to favor macrophage development instead (109,110), whereas PU.1 at low levels is vital for most B-cell precursors to be produced at all (111,112). GATA3 is indispensable for T-cell generation at multiple successive steps, but modest overexpression of GATA3 is profoundly inhibitory to the T-cell pathway (113, 114, 115, 116, 117). Twofold differences count; many hematopoietic TFs show haploinsufficiency phenotypes, as reviewed elsewhere (87). A key question is whether this high dose sensitivity comes from highly cooperative DNA binding, from competitive TF-TF interactions, or from other mechanisms, described below.
Hematopoiesis thus offers a gallery of distinct contexts in which to examine closely why a given TF behaves differently in different contexts within any desired degree of developmental relatedness. Different roles can be related directly to different levels of expression, different binding partners, and/or different inherited chromatin states. Furthermore, when a precursor-product series has been defined, it is possible to test which modifiers of TF action are heritable from stage to stage and which act reversibly. Early T-cell development is a good case.
Cellular and chromatin contexts of early T-cell development
The steps through which the hematopoietic stem/progenitor gene regulatory network gets reconfigured into a committed cell-type gene regulatory network are most readily tracked in the T-lymphocyte developmental pathway. Multipotent precursors normally undergo this program in the thymus, but there are excellent in vitro differentiation systems that reproduce the T lineage commitment process with high efficiency, and fine-scale distinctions among the successive developmental stages have been resolved by numerous cellular markers. This has enabled the component steps in the process to be dissected at unusually high resolution, with clear definition of the transition from multipotentiality to commitment at the single-cell level. Main features of the pathway are briefly summarized here (for more detail, see (118, 119, 120, 121, 122)).
Upon arrival in the thymus, multipotent precursors are guided into the T-cell pathway by multiple days of exposure to strong Notch pathway signals (Fig. 3 A; (118,125)). Notch signaling simultaneously inhibits the cells from taking alternative, non-T developmental pathways and stepwise drives the T-cell specification program forward. In the process, over a dozen transcriptional regulators that the multipotent cells had been expressing upon entering the thymus are silenced, and new regulatory genes encoding signature TFs of the T-cell program are gradually upregulated. Whereas the cells entering the thymus are initially multipotent, through this process they intrinsically lose competence for alternative developmental fates, thus undergoing commitment. Soon after, they become eligible to rearrange their T-cell receptor (TCR) genes in the distinctive recombination and somatic mutation process that ends up giving each cell a unique immune-receptor specificity, important for its future immune-system roles (126,127). However, cell identity and commitment are established before TCR gene assembly, in the “pro-T cell” stages.
Figure 3.
Early T-cell development and binding behavior of pro-T-cell transcription factors. (A) The pro-T-cell developmental framework. Shown is the sequence of stages that each wave of multipotent precursors goes through in the thymus, dependent on Notch signaling, from thymic entry to first expression of T-cell receptor (TCR) genes. This progression in a young mouse takes ∼2 weeks for each cohort of cells, with one to two cell cycles per day, yielding ∼104 fold expansion. The stages shown are followed by a series of later intrathymic events in which cells undergo positive or negative selection on the basis of the TCR genes that they express, important for immune repertoire selection but outside the scope of the events described here. TSP, thymus-seeding progenitor; DN1 (ETP), first main intrathymic stage; ETP, early T-cell progenitor; DN2a, second stage, before commitment; DN2b, third stage, after commitment; DN3, fourth stage, initiation of first major phase of TCR locus gene rearrangements that assemble the TCR coding gene sequences to determine cell’s future immunological specificity. (B) Zoomed-in view of the commitment transition (stages DN2a to DN2b) with dynamics of expression of select key TFs across this interval. The figure contrasts strongly changing TFs PU.1 and Bcl11b with minimally changing TFs Ikaros, E2A, and Runx1+3. In addition to those shown, TCF1 and GATA3—T-cell specific factors first expressed in the earlier, DN1 (ETP) stage—increase slightly or remain constant across this interval (data not shown for simplicity). Note: vertical scale approximates a logarithmic expression scale. (C) Observed patterns of engagement of PU.1 (blue triangles) in pre- and postcommitment pro-T cells. PU.1 expression decreases substantially during commitment, thus demonstrating a natural titration experiment in vivo as pro-T cells developmentally progress. Some residual binding is seen after commitment because PU.1 protein is long-lived, therefore persisting somewhat longer than Spi1 (PU.1-encoding) RNA expression. This demonstrates the way PU.1 protein levels relate to occupancy thresholds for different affinity classes of sites (20). Affinities of binding to motifs at indicated classes of binding sites, based on log-odds position weight matrix motif scores validated by in vitro binding (123), are shown by blue shading, and closed chromatin (here defined by ATAC-seq criteria) is indicated as in Fig. 1. (D) Observed patterns of engagement of Bcl11b (orange triangles) in pro-T cells. Bcl11b is only expressed after commitment. Note that the most common Bcl11b binding sites, including sites that appear to mediate Bcl11b transcriptional function, do not match the motif identified for Bcl11b binding in vitro. Not shown: global analysis of three-dimensional genome organization suggests that Bcl11b binding is enriched near chromatin loop anchor sites (124), although it is also abundant elsewhere. (E) Observed patterns of engagement of Runx1 and/or Runx3 (purple triangles) in pre- and postcommitment pro-T cells. Observed Runx binding is poorly correlated with Runx-specific motif “quality” based on log-odds position weight matrix scores, and so only two levels of site quality are shown for simplicity. Note that regardless of presumed affinity, many Runx binding sites are stage specific and that blocks of neighboring sites (dashed-line brackets) may become bound or unbound in a stage-specific way.
This process is notably slow. In the young postnatal mouse thymus, precursors undergo at least 7–10 cell divisions under the influence of Notch signaling before crossing the transition linked to T-cell commitment (128, 129, 130). In young postnatal mice, individual stage transitions along the way each take ∼2 days (two to four cell cycles) apiece, not hours or minutes (128). Although these pro-T cells undoubtedly undergo stochastic transcriptional bursting at active loci like other cells, the slowness of the global state changes during their differentiation is an important indicator that the developmental impacts of these transcriptional bursts are mostly being buffered. The duration of each substage indicates regulatory metastability across multiple cell cycles. This, then, increases the distinctiveness of the specific transitions that occur when cells do make a sharp state change. It is the programmed alternation between metastable and transition phases that requires explanation.
At a molecular level, commitment is marked not only by TF expression changes but also by multiple coordinated chromatin changes. T-cell precursors before commitment have chromatin accessibility profiles, chromatin looping profiles, and chromatin A/B compartment profiles that strikingly resemble those of hematopoietic stem cells and multipotent progenitors (124,131). Then, after multiple cell cycles, these profiles change rather sharply to resemble typical T lineage profiles, and the watershed of the most pronounced changes is at the commitment transition itself (124). By following a few key TFs, the impacts of these changes can be seen and also how the activities of those TFs may contribute both to stability and to change.
Key transcription factors in early T-cell development
The TF-coding genes across the genome that change expression during commitment have been fully surveyed (131, 132, 133), and several with prominent roles have been subjected to years of genetic and molecular analysis. Seven TFs that contribute importantly to the T-cell specification pathway are the progenitor-specific factor PU.1 (an Ets family factor of a distinctive subgroup), the stably expressed factors E2A (a basic helix-loop-helix factor), Ikaros (a multi-C2H2 Zn finger factor), and Runx1 (a Runt domain factor), and specifically T-lineage-associated factors TCF1 (encoded by Tcf7; a high mobility group factor), GATA3 (a dual-C4 Zn finger factor), and Bcl11b (another multi-C2H2 Zn finger factor). The expression of these regulatory factors is highly stereotyped and has an important period of overlap during the lineage commitment decision (Fig. 3 B; (118,125)). In addition, Notch1 and initially also Notch2 with their TF partner RBPJ (also called CSL, CBF-1/Suppressor of Hairless/Lag-1) provide crucial inputs (6,134, 135, 136, 137, 138). The roles of several of these TFs correspond to their developmental expression patterns, with PU.1 governing genes that are expressed mostly in multipotent progenitors (20), TCF1 promoting a T-lineage-specific chromatin state (139), and TCF1 and Bcl11b active in T-lineage-specific gene expression (17,140, 141, 142). GATA3, Runx1, Notch-RBPJ, basic helix-loop-helix E proteins, and Ikaros work broadly throughout this process, with distinctive and indispensable T-cell roles (6,7,113,114,143, 144, 145). However, the functions of the stably expressed TFs are much more dynamic than their seemingly flat, unchanging expression patterns would suggest. Stably expressed factors in this system such as Runx1 and E2A and Notch signaling repeated across different stages can all exert notably stage-specific effects. This stage-specific action of continuously available factors means that these TFs offer important points of entry to the mechanisms creating developmental discontinuity.
Of these factors, the relationships of binding to function have been investigated most deeply for PU.1, Bcl11b, and Runx1 in the context of the T-cell commitment process. PU.1 is expressed robustly until commitment, when it is turned off. Bcl11b is silent in hematopoietic progenitors and nearly all non-T hematopoietic cells and turns on only at T lineage commitment, but then remains on permanently afterwards. Runx1 is highly expressed in T lineage cells, at similar levels before and after commitment, and is expressed more strongly in early T lineage cells than in any other major hematopoietic cell type (131). All of these factors are functionally important for T-cell development (7,146, 147, 148, 149, 150, 151, 152), but each of these factors behaves differently from the others as a molecular agent interacting with the chromatinized genome.
Binding and access rules: PU.1
PU.1 has been studied extensively in myeloid cells, dendritic cells, and B cells, with deep analyses of its binding requirements, pioneering activity, preferred TF activity partners, and dose-dependent functions (31,98,123,153, 154, 155, 156, 157, 158, 159, 160, 161). In early T lineage cells, its role is confined to the multipotent, precommitment stages. It appears to act primarily as a positive regulator of most of its direct targets (genes responding functionally when it binds de novo to linked sites) and as a marker for open chromatin sites during the stages when it is expressed (20), binding >30,000 sites in pro-T cells before commitment (132). As the cells undergo commitment, it is silenced, and then most T cells never express it again. However, it is functionally important for establishing the T-cell precursor pool (147). PU.1 can block Notch signaling if expressed too highly (108), which can result in transdifferentiation to a myeloid pathway (162,163), but at normal levels it promotes early T lineage proliferation and protects the cells from deviation to the NK cell pathway (147). Its target motif is consistently seen to be the most enriched motif among chromatin sites that are open specifically before commitment (7,124,131,139).
PU.1 is a biophysically “well-behaved” TF that shows motif-faithful in vivo binding behavior that is overall consistent with its in vitro DNA-binding activity (20,123). Even as its expression declines during pro-T-cell commitment, it maintains binding to a consistent pattern of sites, albeit with declining occupancy, and the sites occupied longest as PU.1 concentration drops are those with stronger matches to the PU.1 motif position weight matrix (PWM), as would be predicted by mass action (Fig. 3 C). It also acts as a pioneer factor in early T lineage cells, with the ability to locate and bind to many of its physiological target sites whether in open or certain kinds of closed chromatin, provided that the sites are high affinity (Fig. 3 C). This activity can be seen if exogenous PU.1 is added (back) to immature T lineage cells after they have undergone commitment and shut off their endogenous PU.1 and when the normal target sites of PU.1 have become closed (20). Such “restored” PU.1 appears to be capable of (re-)opening a large subset of these sites by nucleosome displacement (49,164) and recruiting histone acetyltransferases (20). PU.1’s pioneering roles in vivo are helped by its structural features enabling it to bind sites even in methylated DNA regions (165) and by its ability to recruit either DNA methyltransferases or TET enzymes for DNA demethylation to its binding sites in different contexts (83). The genes in pro-T cells that respond most sensitively to PU.1 do not necessarily bind PU.1 at their promoters, but rather at distal or intronic sites where chromatin opening also depends on PU.1 (20). Thus, PU.1 appears to work in this system as a positive regulator that functions in large part through opening of enhancer chromatin.
Genome-wide, its binding pattern is a major determinant of the overall pattern of open chromatin through the earliest stages of T-cell development, which is maintained as long as PU.1 is present (20,147). Nevertheless, its ability to engage closed chromatin sites as well depends not only on its DNA-binding domain but also on the integrity of its non-DNA-binding, protein-interaction domains (20), even when the sites have very high-quality binding motifs. These are unstructured domains (166) which are likely to promote noncovalent protein condensation and liquid-liquid phase separation, very similar to the domain in a B-cell pioneer factor, EBF1, also recently shown to be required for its chromatin-opening activity (167,168). This supports evidence from other systems that pioneering may often depend on protein-protein cooperation (169). In the context of non-T hematopoietic cell types, PU.1 associates strongly with interferon response factors 4 and 8, NF-κB, and LXR and has especially prominent partnerships involving C/EBP and AP-1 bZIP family factors in myeloid cells (28,29,31,32,157), all of which have motifs coenriched with PU.1 occupancy sites in myeloid cells. However, in pro-T cells, PU.1 interacts preferentially with Runx factors, and the most commonly coenriched motif at PU.1 sites in pro-T cells is a Runx motif (20,170).
In summary, PU.1 responds to lineage-specific availabilities of partner factors to establish moderately lineage-specific patterns of site choice, but many of its pro-T-cell functional target genes are in fact regulated similarly by it in other lineages as well (171). Rather than responding sensitively to regulatory discontinuities, its own developmental silencing creates regulatory discontinuities.
Binding and access rules: BCL11B
Bcl11b is a bifunctional TF, expressed in a reciprocal pattern to that of PU.1, encoded by a gene that only becomes transcriptionally active as pro-T cells undergo commitment (172). Bcl11b is important for commitment itself, as well as for later T-cell functional regulation, with a specific role in blocking aberrant activation and effector-differentiation processes of the T cells as well as licensing genes for expression that encode crucial T-cell signaling molecules (17,140,148, 149, 150,173). Like PU.1 in earlier stages, it binds ∼20,000–30,000 sites across the genome in newly committed pro-T cells and often engages sites that are open and associated with active enhancer histone modifications (17,124,140). The relationship between its binding and function is less clear cut than for PU.1, however. More Bcl11b-bound responsive targets appear to be negatively regulated by Bcl11b than positively regulated, and Bcl11b forms particularly common interactions with NuRD chromatin remodeling complexes, which are often associated with repression (Fig. 3 D). However, Bcl11b binding has also been shown to be highly associated with an increased frequency of chromatin looping in the Bcl11b-rich regions, and loop density usually correlates most with positive regulation (Fig. 3 D; (124)). Although Bcl11b deletion blocks T-cell developmental progression past commitment, acute deletion of Bcl11b in mature T cells measurably reduces the number of loops involving sites with at least one Bcl11b-associated anchor (124). Thus, Bcl11b appears to have a role in chromatin architecture, distinct from but complementary to that of PU.1.
The site choice biochemistry of Bcl11b in vivo poses a stark contrast to that of PU.1. In early T lineage cells, Bcl11b’s own DNA-binding specificity appears to be substantially masked by its interactions with other proteins. Several studies have identified GC-rich DNA sequence motifs that are bound efficiently by Bcl11b with purified DNA in vitro (174,175). However, these motifs are not well enriched in pro-T cells at the sites where Bcl11b is detected to bind in vivo or in the vicinity of genes that respond to Bcl11b functionally (17). Instead, in pro-T cells and later T cells alike, Bcl11b is found binding to sites most highly enriched for Runx and Ets family target motifs (17,124,140,176,177), and its occupancy is usually accompanied by detection of Runx1 protein at the same sites (Fig. 3 D; (7,17)). In many cases, Bcl11b could be binding DNA indirectly, possibly via Runx1. However, functionally responsive Bcl11b-regulated targets are enriched for linkage to the minority of Bcl11b occupancy sites where Runx1 recruitment or site choice depends on the presence of Bcl11b as a binding partner (17). Bcl11b function is thus connected with the subset of its sites where it is needed to nucleate assemblies of complexes with Runx1 and chromatin remodeling factors.
Bcl11b thus creates a novel regulatory state potential with its sudden advent during commitment. However, the effect of this new regulatory state is focused less on genes with Bcl11b-specific sites than on genes where Bcl11b action can alter the impact of other factors.
Binding and access rules: RUNX factors
Runx family factors are bifunctional and play diverse roles, critical for early hematopoiesis and indispensable for T-cell development (178,179). When Runx1 is first expressed in early hematopoiesis, it has the capacity to drive reorganization of the binding of other TFs to establish the hematopoietic progenitor state (180,181). In that context, it has provided a paradigm of how hit-and-run action can create lasting effects via chromatin alterations (182). Runx factors are also expressed very robustly in all stages of the development of precursors into committed pro-T cells, enabling these factors to work both as common partners for PU.1 at open chromatin sites before commitment and as common partners for Bcl11b after commitment. The expression of individual Runx family members shifts from the earliest stages through commitment, with Runx1 becoming the most prominent family member after commitment, but the changes in one factor are largely balanced by reciprocal changes in another Runx family member so that the total RNA, protein, and ChIP-seq-inferred binding activity levels appear almost unchanging (7).
These factors are of extreme interest despite their stable expression. First, Runx factors collectively are essential for cells to enter and progress through the T-cell program (151,152). Second, open chromatin sites in pro-T cells both before and after commitment are strikingly rich in Runx target motifs, whether the sites are opening or closing (7). Runx motifs are accordingly coenriched at functional binding sites of numerous other lymphoid TFs, especially both PU.1 and Bcl11b sites (17,20), as noted above in pro-T cells, and at sites for lineage-determining factors E2A and EBF1 in pro-B cells (183,184). Third, the genes that Runx factors regulate positively include many of the signature genes of the T-cell program, whereas the genes that they regulate negatively are involved in progenitor states and developmental alternatives (7). Thus, they are critically implicated at the core of the T-cell program.
Binding site choice for Runx factors is superficially conventional. Before, during, and after commitment, Runx factors are found binding to sites that are enriched for Runx motifs. Interestingly, their sites are almost equally enriched for motifs for Ets family TFs, PU.1 before commitment and Ets1/Erg type after commitment (7). However, in contrast to PU.1, the hierarchy of Runx1 binding strengths in vivo does not match the PWM log-odds scores of the individual sites (170). In further contrast to PU.1, which remains bound to a similar choice of sites even as its concentration drops, Runx binding site choices shift considerably within the T-cell lineage as the cells go through commitment, even though Runx activity levels stay the same (Fig. 3 E). In fact, most of the 25,000–30,000 nonpromoter sites that Runx factors bind before and after commitment are different, with ∼1/3 of the total sites abandoned and 1/3 of the sites newly gained as the cells progress through commitment (7). Thus, even at constant levels in the nucleus, Runx occupancy choices appear to be strongly affected by shifts in regulatory “context.”
Runx factors thus provide an ideal readout and probe for the mechanisms transforming the regulatory contexts in these two states, just a few days apart in cells of the same lineage. They feature strongly in mechanistic insights about system behavior in early T-cell development.
System behavior: stage-specific functions of non-stage-specific factors
Runx factors provide a glimpse of the mechanisms that may also account for stage-specific actions of other non-stage-specific factors. In early T-cell development, Runx factors have sharp, highly stage-specific impacts on gene expression (7). First, despite their constant levels, they preferentially activate or repress the minority of expressed genes that undergo great changes in expression during commitment, either positively or negatively (7). Even more surprisingly, their effects on the same target genes, whether positive or negative, can change dramatically before and after commitment. Many genes dependent on Runx (or repressed by Runx) before commitment are completely insensitive to Runx factors after commitment, and many genes regulated by Runx after commitment are completely insensitive to Runx factors before commitment. Thus, not only target choice but also the nature of impact is altered by the changes in regulatory state.
Besides Runx1, Notch signaling itself, E2A, and Ikaros also show evidence for highly stage-specific roles in gene expression. The range of actions of these factors could be guessed in some cases from the developmental patterns of expression of the genes they affect (16,134,143,144,185). However, the abruptness of such changes in function has been most fully demonstrated through recent Cas9-based experimental methods for carrying out the same knockout perturbation in pro-T cells at distinct, successive developmental stages (6,7). For the cleanest results, the Cas9-mediated gene disruption has also acutely knocked out redundant paralogs simultaneously to eliminate compensation (6,7). Although Notch is the most important environmental signal driving cells through pro-T developmental progression both before and after commitment, the transcriptional responses to signaling by the Notch pathway were found to be acutely stage dependent, as for Runx factors. Most genes significantly regulated by Notch signaling (and Notch-RBPJ activity) before commitment were not detectably regulated by Notch signaling after commitment at all and vice versa (6).
Such results of direct comparisons show that TF-target gene functional regulatory connections need to be analyzed in the relevant stage and cannot be assumed to be constant, even within the same cell lineage. Furthermore, expression of the regulating TF in these cases is a prerequisite but is not correlated with expression of the target gene or with the window of time in which the target is sensitive to the regulator.
Stage specificity via TF collaboration and via partner TF competition
In pro-T cells, Runx factors are highly subject to partner effects. Before commitment, PU.1 is its preferred partner, but the relationship is unequal, as already noted. The binding of PU.1 appears not to be Runx dependent, for acute Runx1 disruption does not appear to affect PU.1 binding patterns (20) However, the addition of PU.1 to a postcommitment pro-T cell (to drive development “backward”) markedly shifts the occupancy pattern of Runx1 to favor sites where it could co-bind with PU.1, abandoning many postcommitment specific sites (170). The PU.1/Runx1 co-bound sites often show very weak versions of a Runx PWM, even when they are occupied at the expense of seemingly better sites for Runx (“cofactor theft”) (Fig. 4 A; (170)). The poor quality of Runx motifs at these co-binding sites is a likely symptom of recruitment by protein-protein interactions rather than by pure DNA recognition and is also typical of many Runx sites in the unperturbed cells that naturally express PU.1 before commitment. Conversely, PU.1 co-binding sites are enriched among the sites that Runx abandons normally after commitment, when PU.1 is no longer present (7). Thus, PU.1 is one of the partners that strongly influences the target site choices that Runx makes.
Figure 4.
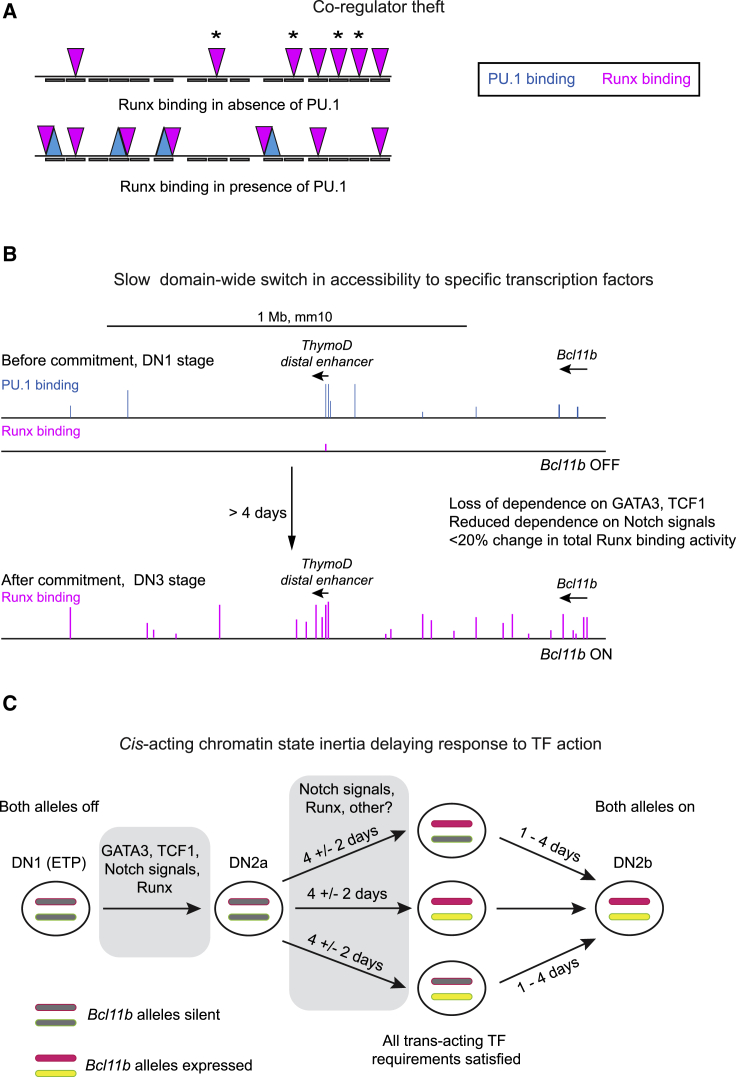
Discontinuity mechanisms modulating TF action. (A) Redeployment by “cofactor theft” (170). Schematically shown is the effect of PU.1 (blue, upward-pointing triangles) on binding choices of Runx1 (magenta, downward-pointing triangles) at sites in a hypothetical genomic region. Top: schematic of Runx binding without PU.1 in a postcommitment T-cell precursor. Asterisks: theft-sensitive sites. Bottom: Runx binding when PU.1 is introduced (cyan, upward-pointing triangles). (B) PU.1 and Runx binding to the extended Bcl11b regulatory region before and after commitment. Major peaks of binding are shown across ∼1.7 Mb of mouse chromosome 12, traced manually from data shown in (7). Arrows: direction and extent of transcription units (partial map). Note the absence of Runx binding signals (magenta) across this region before commitment, despite the equal presence of Runx factors before and after commitment and the readily detectable binding of PU.1 (cyan). PU.1 is not shown after commitment because its expression is strongly reduced. (C) Functional evidence for kinetic impact of closed chromatin at the Bcl11b regulatory region: stochastic, cis-acting kinetic drag breaks synchrony between alleles in same nucleus. Summary of evidence from (172,186) is given. Left to right: DN1 (ETP) stage to DN2b stage. Bcl11b transcriptional activation onset occurs at variable times after the cells reach DN2a stage and correlates functionally with commitment. Trans-acting factors: action of GATA3, TCF1, Notch signaling, and Runx1 + Runx3 in DN1 stage and the DN1 to DN2a transition are needed to prime locus for opening, although Runx factors bind inefficiently in DN1 stage (cf. B). For DN2a stage cells, Runx and Notch signaling still show regulatory impact, but GATA3 and TCF1 do not. Cis-acting mechanism: even when all trans-acting factor requirements are met, individual alleles of Bcl11b still have indeterminate activation timing within the same nucleus, as shown by tagging each allele with a different color fluorescent protein. Cells transitioning from Bcl11b-off to Bcl11b-on may activate “Red” allele first, “Yellow” allele first, or both alleles simultaneously. Activation of either allele shows the latest possible time when the cell has satisfied all trans-acting requirements for Bcl11b activation. However, the time between activation of the first and second alleles activated varies from no delay to a delay of >4 days (>4 cell cycles), in different individual clones of DN2a cells, showing that the activation process depends on a slow step that can persistently distinguish the two alleles of the same responding gene across multiple cell cycles. In the most extreme case, the interval between the last required time of action of positive regulator GATA3 and first detectable expression of the later-activated allele of Bcl11b can be ∼7 days. Kinetic relationships between expression of an allele and that allele’s increased accessibility to Runx binding are under investigation.
After commitment, Runx1 binds most frequently with Bcl11b, although this overlap may be somewhat exaggerated by the fixation conditions used to score both factors. The mutual dependences have not been examined in both directions, but only a small fraction of Runx binding sites appear to depend on the presence of Bcl11b. Interestingly, though, if Bcl11b is deleted, Runx1 binding then appears at numerous new sites, again of comparable or better enrichment for the Runx1 PWM (17). Thus, Bcl11b after commitment is also involved in redirection of some Runx binding behavior, though not as dominantly as PU.1 before commitment. Motif analysis at developmentally shifting sites (7) suggests possible sites of Runx1 recruitment by E proteins, as well.
The notable point is not just that one TF can strongly recruit another TF to its binding sites; it is rather that this occurs at the expense of a discrete subset of alternative occupancy sites. The result suggests that TF partners compete for a functionally limited pool of Runx1 (or other TFs subject to “theft” as well), with collateral impacts on many sites without their being bound by the perturbing partner TF at all (170). How often this site occupancy shift alters expression of target genes needs further investigation. Many genes that are functionally responsive targets of these TFs are surrounded with multiple sites of TF binding, spread over tens or hundreds of kb, only some of which may be subject to these redirection/cooperative recruitment effects, interspersed with others that remain unaffected (17,170). However, even if TF-TF collaboration may only affect an interspersed subset of binding sites for each partner, it may play a greater role in longer-scale collective effects on chromatin structure, discussed below.
Stage-specific functions: hit-and-run TF roles in gene regulation
Hit-and-run developmental regulation is a particularly important variant of stage-specific TF activity. If TF expression were always correlated with expression of the TF’s target genes, then lineages would require stable expression of a core of mutually sustaining, “identity-promoting” TFs. For example, macrophages (a major myeloid hematopoietic cell type) do appear to maintain high expression of PU.1 as a cell-type identifier, whatever their other functional specializations, and this plays both a gene network role and a biochemical role as a pioneer factor for macrophage-specific cis-regulatory elements (156). However, T lineage cells are unusually long-lived and become extremely diversified once mature, and some of the most important lineage-specific TFs activated during the T-cell specification program, including TCF1, GATA3, and E2A, can be downregulated in particular, specialized T-cell descendants (133). The crucial Notch/RBPJ complex itself, for example, ceases to be essential shortly after the expression of the first version of the T-cell receptor complex (187). In general, important T-cell identity target genes that initially depend upon these TFs for their activation during T-cell specification continue to be expressed in a sustained way throughout later mature T-cell activity states, even when these TFs are gone.
Specific genes regulated by Bcl11b, Notch-RBPJ, and Runx factors during commitment in a hit-and-run fashion show that the same target gene can switch from acute dependence on a regulator to near-complete independence within days. Genes of the Cd3 complex, which encode the signaling machinery for the T-cell receptor complex, are highly dependent on Bcl11b activity for their initial activation in pro-T cells but become independent of Bcl11b by later stages of T-cell development and in mature T cells (17,140,149,188,189). As already noted, Runx factor activities show numerous cases of hit-and-run function, not only in the T-cell lineage (7) but also in establishing the progenitor-cell phenotypes at the embryonic origins of definitive hematopoiesis (182). The gene Tcf7, encoding the essential TF TCF1, itself depends intensely on both Notch/RBPJ and Runx activity for its expression before commitment but becomes completely insensitive to changes in Notch and Runx activity after commitment (6,7). Finally, Bcl11b itself is also regulated by hit-and-run mechanisms. Despite a strong dependence on Notch signaling to support its eligibility to be activated, Bcl11b becomes completely Notch independent almost immediately once it is expressed (172). Also, it is only turned on if TCF1 and GATA3 are active during the earliest pro-T-cell stage, but it can still be turned on normally if either of these factors is deleted a few days later, yet also before Bcl11b turns on (172). Thus, the activating role of TCF1 and GATA3 is required but can be temporally uncoupled from Bcl11b transcriptional initiation in this system almost completely. This is not unique, as a strikingly similar mechanism has been reported in Caenorhabditis elegans neurogenesis (190).
These examples emphasize that in these slow-kinetic developmental systems, requirements for TF activity are highest to trigger changes in gene expression state, not necessarily to maintain them. This is a very different role than to sustain a given rate of RNA polymerase II recruitment and transcriptional bursting from a given promoter. Although in some cases, such hit-and-run behavior could be due to a TF role “relay race,” as suggested for some mature T-cell responses (60), it could also be mediated by durable chromatin state changes. This demands a focused examination of the mechanisms involved in creating these discontinuities in gene regulation.
Differential impacts of chromatin states on TF binding
As discussed earlier, many TFs cannot gain access to their binding sites in closed chromatin until other “pioneer” factors have opened the chromatin or at least have established a beachhead (40,48,191,192). For TFs to open closed chromatin, combinatorial action may be required (169,193). However, different TFs appear to be restricted by qualitatively different chromatin state constraints.
PU.1 binds to many target sites in closed as well as open chromatin (20,123), with half of its observed binding sites in pro-T cells appearing “closed” by ATAC-seq criteria (20). These “closed” binding sites provide evidence for the energetic cost that it pays for establishing occupancy in these more restricted regions (20,123). In pro-T cells, the PU.1 occupancy sites in closed regions have an extremely high enrichment of classic PU.1 target motifs and are skewed to higher-quality position weight matrix matches with those motifs than its binding sites in open regions (Fig. 3 C). As PU.1 levels in the cell decrease, the differences between open and closed sites widen by log-odds differences of 1.0–1.5 (20), corresponding to KD differences from ∼0.5 to 0.2 μM (123). This higher apparent affinity threshold, plus the requirement for non-DNA-binding domains that may enable PU.1 to interact with other proteins (20), suggest the minimal cost of overcoming the ATAC-closed barrier.
Polycomb repression complex 2 (PRC2)-modified chromatin presents a different kind of barrier. PU.1 binding appears to be virtually excluded from H3K27me3-modified chromatin sites, both for occupancy by endogenously expressed PU.1 at its natural levels and for binding by PU.1 experimentally reintroduced in later T-cell stages, after endogenous PU.1 is turned off (20,132). In contrast, GATA3, which is otherwise far less robust in its measured site-binding preferences (132,194), readily binds to certain sites even when they are within regions of strong H3K27me3 marking. Thus, Polycomb-“repressed” as opposed to simply “non-open” chromatin sites may provide a stronger barrier for PU.1 action than for GATA3 action.
Although a frequent partner of PU.1, Runx1 observes different chromatin state restrictions. A striking example is seen at a distal enhancer for the Bcl11b locus, where the chromatin is generally closed before commitment and open afterwards (Fig. 4 B). Runx1, like GATA3, TCF1, and Notch, is a functionally important positive regulator of Bcl11b expression, needed for Bcl11b activation (7,172). The distal Bcl11b enhancer harbors a cluster of PU.1 sites and Runx sites, which are strongly occupied by PU.1 before commitment and by Runx1 after commitment. However, whereas PU.1 can freely bind this enhancer in its “closed” state before commitment (132,195), Runx factor binding is harshly excluded (Fig. 4 B). In fact, ChIP-seq profiles show a stark inability of Runx factors to bind across almost 2 Mb around the distal enhancer, before commitment. Then, during commitment, Runx binding suddenly appears at >30 occupancy sites across this domain (Fig. 4 B; (7)). At the same time, the region undergoes a compartment flip, chromatin looping to the Bcl11b gene body greatly increases across the whole 2-Mb interval (124), and Bcl11b is turned on.
These distinctions show that “closed” chromatin is not a monolithic state but is perceived differently by different transcription factors. Thus, developmental changes in particular aspects of chromatin state can cause selective discontinuities in the access of particular TFs to the affected genomic loci.
Chromatin state inertia can exert substantial kinetic drag on TF response manifestation
As argued above, TFs can have a long-term, durable impact on gene expression if they trigger alterations in local chromatin state, for example, by flipping chromatin at a key regulatory locus from “repressed” to “active” and/or by altering DNA methylation. However, the requirement for an extensive chromatin state change can delay the transcriptional response substantially, indeed affecting the kinetics of developmental progression (130). Thus, despite the durability of the effect, the response may not appear with tight temporal coupling to the timing of action of the TFs themselves.
In this system, in fact, local chromatin state can provide an “inertial” drag that can delay gene expression responses to TFs for days, lagging through multiple cell cycles. A particularly clear example is provided by the kinetics of activation of the two alleles of Bcl11b itself (Fig. 4 C). The allelic comparison discriminates between effects mediated by trans-acting TFs and effects mediated by cis-acting chromatin states. Clearly neither allele would be expected to be active until requisite positive regulators were present and negative regulators were removed, but the activation of even one allele of Bcl11b would indicate the presence of a fully permissive quorum of trans-acting factors. Tagging of the two alleles of the Bcl11b gene with different fluorescent reporters (without affecting the coding function) showed that the two alleles can undergo activation noncoordinately at disparate, stochastically distributed times within the same cells (186). This was not due to maternal or paternal imprinting, because the alleles turned on in random order among different pro-T cells from the same mouse. In cells purified based on their activation of one allele and then followed over time, the other allele showed very variable activation timing from one clone to the next, with delays ranging up to 4 days (186). Because TFs rapidly bind and dissociate from DNA over timescales that are orders of magnitude shorter than this, there would be no basis for trans-acting TFs alone to activate one allele in a cell and its descendants multiple cell cycles before the other allele. Thus, a cis-acting mechanism must be part of the brake on activation, delaying it beyond any gene network delays in activating the initiating TFs.
In the case of Bcl11b, several cis-acting mechanisms could be involved. Polycomb complex 2-modified chromatin (H3K27me3) on some Bcl11b regulatory elements or DNA methylation could contribute to the initial deep silencing (195,196). In addition, Bcl11b is initially sequestered at the nuclear periphery from which it shifts to the nuclear interior around the time that it is activated (196,197). This relocalization is least partly under control of a cis-acting lncRNA called “ThymoD” that is crucial for Bcl11b activation (197) and must be transcribed from the vicinity of the key distal enhancer (186,195). It is unclear how general an example Bcl11b may be; it may be subject to special mechanisms making Bcl11b expression particularly hard to induce but easy to maintain. However, this identification of a durable cis-acting constraint depended on comparing activation of individual alleles within the same cells, which has not been attempted at many loci. Conceivably, such a mechanism could cause stochastic delays to activation timing of many other developmentally activated loci as well.
Concluding remarks
Transcription factors guide a rapidly unfolding cascade of transcriptional change, with remarkably coordinated kinetics, in embryonic life. In contrast, in the development of individual lymphocytes from hematopoietic stem cells in mammals, developmental progression is slow compared to cell cycle rate, fate choices have a strong stochastic component, timing is highly variable among cells even within the same clone, and cells establish their own differentiation programs independent of the choices made in parallel by other cells in the cohort. A question has been how such a “lone wolf” developmental mechanism can be made faithful at all, considering that it is based necessarily on TF-DNA binding, which is biochemically transient and reversible. This article suggests that it could draw some of its irreversibility from the same chromatin-associated mechanisms that provide “inertial drag” to make the progression so slow. Further, the suggestion is that chromatin state change dynamics, as well as system-level effects of TFs on the activities of their partner TFs, together provide a controllable switch between stages when the cells’ transcriptional identity is relatively stable and self-renewing and stages when cells are undergoing rapid change, which are followed by new stages of metastability that are functionally different from the preceding ones.
It is well understood that TFs interact with chromatin mechanisms, but most of the work on this area has focused on fully differentiated, immortalized cell lines on the one hand, where gene regulation is only measured in a “maintenance” mode, or else on the highly abnormal process of lineage erasure and reprogramming caused by forced ectopic expression of a full set of “Yamanaka factors.” The relationships of these identity-deconstruction processes to natural development have not been well established. Here, therefore, the progression of normal T-cell precursors from multipotency to T lineage commitment is proposed as an instructive model in which the constraints of TF activity by prior chromatin states can be measured and the impacts of TF activity to cause chromatin states to change can be illustrated, tracked, and dissected.
The work reviewed here identifies several features of these real developmental events that are important to consider in future modeling of TF action for developmental gene regulation. First, binding site choices are highly conditional, based on inherited chromatin state and current availabilities of collaborating factors, not simply predictable from genomically encoded site affinities and total TF pools in the nucleus. Second, partner interactions compete for functionally limited TF pool sizes, causing some indirect effects by depleting particular sites of occupancy to give preference to others. The rules that define such “theft”-vulnerable sites and their functional significance need to be defined. Third, TF regulatory action can be much more stage-specific than TF expression, breaking the promise of correlation between responsible TF and responding target genes. Finally, although some TF actions can rapidly cause changes in chromatin looping or histone modification, other developmental changes may require extended times to modify cis-acting chromatin states, which in some cases may completely uncouple the timing of TF action from response of its target. These features offer fascinating glimpses of new aspects of gene regulation that must be taken into account for full understanding of complex developmental systems.
Author contributions
E.V.R. wrote the article and made the figures.
Acknowledgments
The author thanks Barbara Wold, Constanze Bonifer, the late Eric H. Davidson, and past and current members of my group for illuminating, critical discussions on this subject.
Work on transcription factor functional genomics in the author’s lab has been supported by USPHS grants RC2CA148278, R01AI083514, R01AI095943, R01AI135200, R01HD076915, and R01HL119102. Past support from the A. B. Ruddock Professorship is also gratefully acknowledged. E.V.R. is a member of the Scientific Advisory Board for Century Therapeutics and has served as consultant or advisor to A2 Biotherapeutics and Kite Pharma.
Editor: Stanislav Y. Shvartsman.
References
- 1.Ramirez-Carrozzi V.R., Braas D., Smale S.T. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138:114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vahedi G., Takahashi H., O’Shea J.J. STATs shape the active enhancer landscape of T cell populations. Cell. 2012;151:981–993. doi: 10.1016/j.cell.2012.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spitz F., Furlong E.E.M. Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 2012;13:613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- 4.Li P., Leonard W.J. Chromatin accessibility and interactions in the transcriptional regulation of T cells. Front. Immunol. 2018;9:2738. doi: 10.3389/fimmu.2018.02738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peter I.S., Davidson E.H. Academic Press, Elsevier; San Diego, CA: 2015. Genomic Control Process: Development and Evolution. [Google Scholar]
- 6.Romero-Wolf M., Shin B., Hosokawa H. Notch2 complements Notch1 to mediate inductive signaling that initiates early T cell development. J. Cell Biol. 2020;219:e202005093. doi: 10.1083/jcb.202005093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shin B., Hosokawa H., Rothenberg E.V. Runx1 and Runx3 drive progenitor to T-lineage transcriptome conversion in mouse T cell commitment via dynamic genomic site switching. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2019655118. e2019655118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jolma A., Yan J., Taipale J. DNA-binding specificities of human transcription factors. Cell. 2013;152:327–339. doi: 10.1016/j.cell.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 9.Badis G., Berger M.F., Bulyk M.L. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gordân R., Hartemink A.J., Bulyk M.L. Distinguishing direct versus indirect transcription factor-DNA interactions. Genome Res. 2009;19:2090–2100. doi: 10.1101/gr.094144.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weirauch M.T., Yang A., Hughes T.R. Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 2014;158:1431–1443. doi: 10.1016/j.cell.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wold B., Myers R.M. Sequence census methods for functional genomics. Nat. Methods. 2008;5:19–21. doi: 10.1038/nmeth1157. [DOI] [PubMed] [Google Scholar]
- 13.Matteau D., Rodrigue S. Precise identification of DNA-binding proteins genomic location by exonuclease coupled chromatin immunoprecipitation (ChIP-exo) Methods Mol. Biol. 2015;1334:173–193. doi: 10.1007/978-1-4939-2877-4_11. [DOI] [PubMed] [Google Scholar]
- 14.Skene P.J., Henikoff J.G., Henikoff S. Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat. Protoc. 2018;13:1006–1019. doi: 10.1038/nprot.2018.015. [DOI] [PubMed] [Google Scholar]
- 15.Weinert F.M., Brewster R.C., Kegel W.K. Scaling of gene expression with transcription-factor fugacity. Phys. Rev. Lett. 2014;113:258101. doi: 10.1103/PhysRevLett.113.258101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyazaki K., Watanabe H., Miyazaki M. The transcription factor E2A activates multiple enhancers that drive Rag expression in developing T and B cells. Sci. Immunol. 2020;5:eabb1455. doi: 10.1126/sciimmunol.abb1455. [DOI] [PubMed] [Google Scholar]
- 17.Hosokawa H., Romero-Wolf M., Rothenberg E.V. Bcl11b sets pro-T cell fate by site-specific cofactor recruitment and by repressing Id2 and Zbtb16. Nat. Immunol. 2018;19:1427–1440. doi: 10.1038/s41590-018-0238-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Revilla-I-Domingo R., Bilic I., Busslinger M. The B-cell identity factor Pax5 regulates distinct transcriptional programmes in early and late B lymphopoiesis. EMBO J. 2012;31:3130–3146. doi: 10.1038/emboj.2012.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McManus S., Ebert A., Busslinger M. The transcription factor Pax5 regulates its target genes by recruiting chromatin-modifying proteins in committed B cells. EMBO J. 2011;30:2388–2404. doi: 10.1038/emboj.2011.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ungerbäck J., Hosokawa H., Rothenberg E.V. Pioneering, chromatin remodeling, and epigenetic constraint in early T-cell gene regulation by SPI1 (PU.1) Genome Res. 2018;28:1508–1519. doi: 10.1101/gr.231423.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biggin M.D. Animal transcription networks as highly connected, quantitative continua. Dev. Cell. 2011;21:611–626. doi: 10.1016/j.devcel.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 22.Luna-Zurita L., Stirnimann C.U., Bruneau B.G. Complex interdependence regulates heterotypic transcription factor distribution and coordinates cardiogenesis. Cell. 2016;164:999–1014. doi: 10.1016/j.cell.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogata K., Sato K., Tahirov T.H. Eukaryotic transcriptional regulatory complexes: cooperativity from near and afar. Curr. Opin. Struct. Biol. 2003;13:40–48. doi: 10.1016/s0959-440x(03)00012-5. [DOI] [PubMed] [Google Scholar]
- 24.Garvie C.W., Hagman J., Wolberger C. Structural studies of Ets-1/Pax5 complex formation on DNA. Mol. Cell. 2001;8:1267–1276. doi: 10.1016/s1097-2765(01)00410-5. [DOI] [PubMed] [Google Scholar]
- 25.Goetz T.L., Gu T.L., Graves B.J. Auto-inhibition of Ets-1 is counteracted by DNA binding cooperativity with core-binding factor α2. Mol. Cell. Biol. 2000;20:81–90. doi: 10.1128/mcb.20.1.81-90.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu T.L., Goetz T.L., Speck N.A. Auto-inhibition and partner proteins, core-binding factor β (CBFβ) and Ets-1, modulate DNA binding by CBFα2 (AML1) Mol. Cell. Biol. 2000;20:91–103. doi: 10.1128/mcb.20.1.91-103.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hollenhorst P.C., Chandler K.J., Graves B.J. DNA specificity determinants associate with distinct transcription factor functions. PLoS Genet. 2009;5:e1000778. doi: 10.1371/journal.pgen.1000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Escalante C.R., Brass A.L., Aggarwal A.K. Crystal structure of PU.1/IRF-4/DNA ternary complex. Mol. Cell. 2002;10:1097–1105. doi: 10.1016/s1097-2765(02)00703-7. [DOI] [PubMed] [Google Scholar]
- 29.Brass A.L., Zhu A.Q., Singh H. Assembly requirements of PU.1-Pip (IRF-4) activator complexes: inhibiting function in vivo using fused dimers. EMBO J. 1999;18:977–991. doi: 10.1093/emboj/18.4.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glasmacher E., Agrawal S., Singh H. A genomic regulatory element that directs assembly and function of immune-specific AP-1-IRF complexes. Science. 2012;338:975–980. doi: 10.1126/science.1228309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heinz S., Benner C., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heinz S., Romanoski C.E., Glass C.K. Effect of natural genetic variation on enhancer selection and function. Nature. 2013;503:487–492. doi: 10.1038/nature12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thanos D., Maniatis T. Virus induction of human IFN β gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 34.Giese K., Kingsley C., Grosschedl R. Assembly and function of a TCR α enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein-protein interactions. Genes Dev. 1995;9:995–1008. doi: 10.1101/gad.9.8.995. [DOI] [PubMed] [Google Scholar]
- 35.Maier H., Ostraat R., Hagman J. Early B cell factor cooperates with Runx1 and mediates epigenetic changes associated with mb-1 transcription. Nat. Immunol. 2004;5:1069–1077. doi: 10.1038/ni1119. [DOI] [PubMed] [Google Scholar]
- 36.Jain J., Loh C., Rao A. Transcriptional regulation of the IL-2 gene. Curr. Opin. Immunol. 1995;7:333–342. doi: 10.1016/0952-7915(95)80107-3. [DOI] [PubMed] [Google Scholar]
- 37.Rothenberg E.V., Ward S.B. A dynamic assembly of diverse transcription factors integrates activation and cell-type information for interleukin 2 gene regulation. Proc. Natl. Acad. Sci. USA. 1996;93:9358–9365. doi: 10.1073/pnas.93.18.9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hare E.E., Peterson B.K., Eisen M.B. Sepsid even-skipped enhancers are functionally conserved in Drosophila despite lack of sequence conservation. PLoS Genet. 2008;4:e1000106. doi: 10.1371/journal.pgen.1000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmidt D., Wilson M.D., Odom D.T. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science. 2010;328:1036–1040. doi: 10.1126/science.1186176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eck E., Liu J., Garcia H.G. Quantitative dissection of transcription in development yields evidence for transcription-factor-driven chromatin accessibility. eLife. 2020;9:e56429. doi: 10.7554/eLife.56429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson N.K., Foster S.D., Göttgens B. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7:532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 42.Beck D., Thoms J.A., Pimanda J.E. Genome-wide analysis of transcriptional regulators in human HSPCs reveals a densely interconnected network of coding and noncoding genes. Blood. 2013;122:e12–e22. doi: 10.1182/blood-2013-03-490425. [DOI] [PubMed] [Google Scholar]
- 43.Boyes J., Felsenfeld G. Tissue-specific factors additively increase the probability of the all-or-none formation of a hypersensitive site. EMBO J. 1996;15:2496–2507. [PMC free article] [PubMed] [Google Scholar]
- 44.Felsenfeld G. Chromatin as an essential part of the transcriptional mechanism. Nature. 1992;355:219–224. doi: 10.1038/355219a0. [DOI] [PubMed] [Google Scholar]
- 45.Thurman R.E., Rynes E., Stamatoyannopoulos J.A. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buenrostro J.D., Giresi P.G., Greenleaf W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iwafuchi-Doi M., Donahue G., Zaret K.S. The pioneer transcription factor FoxA maintains an accessible nucleosome configuration at enhancers for tissue-specific gene activation. Mol. Cell. 2016;62:79–91. doi: 10.1016/j.molcel.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zaret K.S., Carroll J.S. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Comoglio F., Simonatto M., Natoli G. Dissection of acute stimulus-inducible nucleosome remodeling in mammalian cells. Genes Dev. 2019;33:1159–1174. doi: 10.1101/gad.326348.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin Y.C., Benner C., Murre C. Global changes in the nuclear positioning of genes and intra- and interdomain genomic interactions that orchestrate B cell fate. Nat. Immunol. 2012;13:1196–1204. doi: 10.1038/ni.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lieberman-Aiden E., van Berkum N.L., Dekker J. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rao S.S., Huntley M.H., Aiden E.L. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vian L., Pękowska A., Casellas R. The energetics and physiological impact of cohesin extrusion. Cell. 2018;173:1165–1178.e20. doi: 10.1016/j.cell.2018.03.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hansen A.S., Cattoglio C., Tjian R. Recent evidence that TADs and chromatin loops are dynamic structures. Nucleus. 2018;9:20–32. doi: 10.1080/19491034.2017.1389365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Calderon D., Nguyen M.L.T., Pritchard J.K. Landscape of stimulation-responsive chromatin across diverse human immune cells. Nat. Genet. 2019;51:1494–1505. doi: 10.1038/s41588-019-0505-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Javierre B.M., Burren O.S., Fraser P., BLUEPRINT Consortium Lineage-specific genome architecture links enhancers and non-coding disease variants to target gene promoters. Cell. 2016;167:1369–1384.e19. doi: 10.1016/j.cell.2016.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bediaga N.G., Coughlan H.D., Harrison L.C. Multi-level remodelling of chromatin underlying activation of human T cells. Sci. Rep. 2021;11:528. doi: 10.1038/s41598-020-80165-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Naik A.K., Byrd A.T., Krangel M.S. Hierarchical assembly and disassembly of a transcriptionally active RAG locus in CD4+CD8+ thymocytes. J. Exp. Med. 2019;216:231–243. doi: 10.1084/jem.20181402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Voss T.C., Hager G.L. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat. Rev. Genet. 2014;15:69–81. doi: 10.1038/nrg3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bevington S.L., Keane P., Cockerill P.N. IL-2/IL-7-inducible factors pioneer the path to T cell differentiation in advance of lineage-defining factors. EMBO J. 2020;39:e105220. doi: 10.15252/embj.2020105220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bevington S.L., Cauchy P., Cockerill P.N. T cell receptor and cytokine signaling can function at different stages to establish and maintain transcriptional memory and enable T helper cell differentiation. Front. Immunol. 2017;8:204. doi: 10.3389/fimmu.2017.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van der Veeken J., Zhong Y., Rudensky A.Y. Natural genetic variation reveals key features of epigenetic and transcriptional memory in virus-specific CD8 T cells. Immunity. 2019;50:1202–1217.e7. doi: 10.1016/j.immuni.2019.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sardina J.L., Collombet S., Graf T. Transcription factors drive Tet2-mediated enhancer demethylation to reprogram cell fate. Cell Stem Cell. 2018;23:727–741.e9. doi: 10.1016/j.stem.2018.08.016. [DOI] [PubMed] [Google Scholar]
- 64.Stadhouders R., Vidal E., Graf T. Transcription factors orchestrate dynamic interplay between genome topology and gene regulation during cell reprogramming. Nat. Genet. 2018;50:238–249. doi: 10.1038/s41588-017-0030-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Apostolou E., Stadtfeld M. Cellular trajectories and molecular mechanisms of iPSC reprogramming. Curr. Opin. Genet. Dev. 2018;52:77–85. doi: 10.1016/j.gde.2018.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Koche R.P., Smith Z.D., Meissner A. Reprogramming factor expression initiates widespread targeted chromatin remodeling. Cell Stem Cell. 2011;8:96–105. doi: 10.1016/j.stem.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Petrovic J., Zhou Y., Faryabi R.B. Oncogenic Notch promotes long-range regulatory interactions within hyperconnected 3D cliques. Mol. Cell. 2019;73:1174–1190.e12. doi: 10.1016/j.molcel.2019.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Takahashi K., Yamanaka S. A decade of transcription factor-mediated reprogramming to pluripotency. Nat. Rev. Mol. Cell Biol. 2016;17:183–193. doi: 10.1038/nrm.2016.8. [DOI] [PubMed] [Google Scholar]
- 69.van Oevelen C., Kallin E.M., Graf T. Transcription factor-induced enhancer modulations during cell fate conversions. Curr. Opin. Genet. Dev. 2013;23:562–567. doi: 10.1016/j.gde.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 70.Rao S.S.P., Huang S.C., Aiden E.L. Cohesin loss eliminates all loop domains. Cell. 2017;171:305–320.e24. doi: 10.1016/j.cell.2017.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nora E.P., Goloborodko A., Bruneau B.G. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell. 2017;169:930–944.e22. doi: 10.1016/j.cell.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Seitan V.C., Faure A.J., Merkenschlager M. Cohesin-based chromatin interactions enable regulated gene expression within preexisting architectural compartments. Genome Res. 2013;23:2066–2077. doi: 10.1101/gr.161620.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee P.P., Fitzpatrick D.R., Wilson C.B. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15:763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- 74.Zviran A., Mor N., Hanna J.H. Deterministic somatic cell reprogramming involves continuous transcriptional changes governed by Myc and epigenetic-driven modules. Cell Stem Cell. 2019;24:328–341.e9. doi: 10.1016/j.stem.2018.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Durek P., Nordström K., Polansky J.K., DEEP Consortium Epigenomic profiling of human CD4+ T cells supports a linear differentiation model and highlights molecular regulators of memory development. Immunity. 2016;45:1148–1161. doi: 10.1016/j.immuni.2016.10.022. [DOI] [PubMed] [Google Scholar]
- 76.Vincenzetti L., Leoni C., Monticelli S. The contribution of active and passive mechanisms of 5mC and 5hmC removal in human T lymphocytes is differentiation- and activation-dependent. Eur. J. Immunol. 2019;49:611–625. doi: 10.1002/eji.201847967. [DOI] [PubMed] [Google Scholar]
- 77.Wilson C.B., Makar K.W., Fitzpatrick D.R. DNA methylation and the expanding epigenetics of T cell lineage commitment. Semin. Immunol. 2005;17:105–119. doi: 10.1016/j.smim.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 78.Avni O., Rao A. T cell differentiation: a mechanistic view. Curr. Opin. Immunol. 2000;12:654–659. doi: 10.1016/s0952-7915(00)00158-8. [DOI] [PubMed] [Google Scholar]
- 79.Bintu L., Yong J., Elowitz M.B. Dynamics of epigenetic regulation at the single-cell level. Science. 2016;351:720–724. doi: 10.1126/science.aab2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yin Y., Morgunova E., Taipale J. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science. 2017;356:eaaj2239. doi: 10.1126/science.aaj2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cieslak A., Charbonnier G., Asnafi V. Blueprint of human thymopoiesis reveals molecular mechanisms of stage-specific TCR enhancer activation. J. Exp. Med. 2020;217:e20192360. doi: 10.1084/jem.20192360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tsagaratou A., Äijö T., Rao A. Dissecting the dynamic changes of 5-hydroxymethylcytosine in T-cell development and differentiation. Proc. Natl. Acad. Sci. USA. 2014;111:E3306–E3315. doi: 10.1073/pnas.1412327111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.de la Rica L., Rodríguez-Ubreva J., Ballestar E. PU.1 target genes undergo Tet2-coupled demethylation and DNMT3b-mediated methylation in monocyte-to-osteoclast differentiation. Genome Biol. 2013;14:R99. doi: 10.1186/gb-2013-14-9-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ji H., Ehrlich L.I., Feinberg A.P. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature. 2010;467:338–342. doi: 10.1038/nature09367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jeong M., Sun D., Goodell M.A. Large conserved domains of low DNA methylation maintained by Dnmt3a. Nat. Genet. 2014;46:17–23. doi: 10.1038/ng.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kressler C., Gasparoni G., Polansky J.K. Targeted de-methylation of the FOXP3-TSDR is sufficient to induce physiological FOXP3 expression but not a functional Treg phenotype. Front. Immunol. 2021;11:609891. doi: 10.3389/fimmu.2020.609891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rothenberg E.V., Kueh H.Y., Zhang J.A. Hematopoiesis and T-cell specification as a model developmental system. Immunol. Rev. 2016;271:72–97. doi: 10.1111/imr.12417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Göttgens B. Regulatory network control of blood stem cells. Blood. 2015;125:2614–2620. doi: 10.1182/blood-2014-08-570226. [DOI] [PubMed] [Google Scholar]
- 89.Mercer E.M., Lin Y.C., Murre C. Factors and networks that underpin early hematopoiesis. Semin. Immunol. 2011;23:317–325. doi: 10.1016/j.smim.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Laslo P., Pongubala J.M., Singh H. Gene regulatory networks directing myeloid and lymphoid cell fates within the immune system. Semin. Immunol. 2008;20:228–235. doi: 10.1016/j.smim.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 91.Georgopoulos K. The making of a lymphocyte: the choice among disparate cell fates and the IKAROS enigma. Genes Dev. 2017;31:439–450. doi: 10.1101/gad.297002.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huang S., Guo Y.P., Enver T. Bifurcation dynamics in lineage-commitment in bipotent progenitor cells. Dev. Biol. 2007;305:695–713. doi: 10.1016/j.ydbio.2007.02.036. [DOI] [PubMed] [Google Scholar]
- 93.Chickarmane V., Enver T., Peterson C. Computational modeling of the hematopoietic erythroid-myeloid switch reveals insights into cooperativity, priming, and irreversibility. PLoS Comput. Biol. 2009;5:e1000268. doi: 10.1371/journal.pcbi.1000268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schütte J., Wang H., Göttgens B. An experimentally validated network of nine haematopoietic transcription factors reveals mechanisms of cell state stability. eLife. 2016;5:e11469. doi: 10.7554/eLife.11469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ciau-Uitz A., Patient R. Gene regulatory networks governing the generation and regeneration of blood. J. Comput. Biol. 2019;26:719–725. doi: 10.1089/cmb.2019.0114. [DOI] [PubMed] [Google Scholar]
- 96.Hoang T., Lambert J.A., Martin R. SCL/TAL1 in hematopoiesis and cellular reprogramming. Curr. Top. Dev. Biol. 2016;118:163–204. doi: 10.1016/bs.ctdb.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 97.DeVilbiss A.W., Sanalkumar R., Bresnick E.H. Hematopoietic transcriptional mechanisms: from locus-specific to genome-wide vantage points. Exp. Hematol. 2014;42:618–629. doi: 10.1016/j.exphem.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pang S.H.M., de Graaf C.A., Nutt S.L. PU.1 is required for the developmental progression of multipotent progenitors to common lymphoid progenitors. Front. Immunol. 2018;9:1264. doi: 10.3389/fimmu.2018.01264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Imperato M.R., Cauchy P., Bonifer C. The RUNX1-PU.1 axis in the control of hematopoiesis. Int. J. Hematol. 2015;101:319–329. doi: 10.1007/s12185-015-1762-8. [DOI] [PubMed] [Google Scholar]
- 100.Monteiro R., Pouget C., Patient R. The gata1/pu.1 lineage fate paradigm varies between blood populations and is modulated by tif1γ. EMBO J. 2011;30:1093–1103. doi: 10.1038/emboj.2011.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hosokawa H., Romero-Wolf M., Rothenberg E.V. Cell type-specific actions of Bcl11b in early T-lineage and group 2 innate lymphoid cells. J. Exp. Med. 2020;217:e20190972. doi: 10.1084/jem.20190972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cantor A.B., Iwasaki H., Orkin S.H. Antagonism of FOG-1 and GATA factors in fate choice for the mast cell lineage. J. Exp. Med. 2008;205:611–624. doi: 10.1084/jem.20070544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gillespie M.A., Palii C.G., Brand M. Absolute quantification of transcription factors reveals principles of gene regulation in erythropoiesis. Mol. Cell. 2020;78:960–974.e11. doi: 10.1016/j.molcel.2020.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Swiers G., Patient R., Loose M. Genetic regulatory networks programming hematopoietic stem cells and erythroid lineage specification. Dev. Biol. 2006;294:525–540. doi: 10.1016/j.ydbio.2006.02.051. [DOI] [PubMed] [Google Scholar]
- 105.Graf T., Zon L.I. In: Hematopoiesis: A Developmental Approach. Zon L.I., editor. Oxford University Press; 2001. Transcription factors that induce commitment of multipotent hematopoietic progenitors: lessons from the MEP system; pp. 355–362. [Google Scholar]
- 106.Walsh J.C., DeKoter R.P., Singh H. Cooperative and antagonistic interplay between PU.1 and GATA-2 in the specification of myeloid cell fates. Immunity. 2002;17:665–676. doi: 10.1016/s1074-7613(02)00452-1. [DOI] [PubMed] [Google Scholar]
- 107.Iwasaki H., Mizuno S., Akashi K. The order of expression of transcription factors directs hierarchical specification of hematopoietic lineages. Genes Dev. 2006;20:3010–3021. doi: 10.1101/gad.1493506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Del Real M.M., Rothenberg E.V. Architecture of a lymphomyeloid developmental switch controlled by PU.1, Notch and Gata3. Development. 2013;140:1207–1219. doi: 10.1242/dev.088559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.DeKoter R.P., Lee H.J., Singh H. PU.1 regulates expression of the interleukin-7 receptor in lymphoid progenitors. Immunity. 2002;16:297–309. doi: 10.1016/s1074-7613(02)00269-8. [DOI] [PubMed] [Google Scholar]
- 110.Kueh H.Y., Champhekar A., Rothenberg E.V. Positive feedback between PU.1 and the cell cycle controls myeloid differentiation. Science. 2013;341:670–673. doi: 10.1126/science.1240831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.DeKoter R.P., Singh H. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science. 2000;288:1439–1441. doi: 10.1126/science.288.5470.1439. [DOI] [PubMed] [Google Scholar]
- 112.Ye M., Ermakova O., Graf T. PU.1 is not strictly required for B cell development and its absence induces a B-2 to B-1 cell switch. J. Exp. Med. 2005;202:1411–1422. doi: 10.1084/jem.20051089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.García-Ojeda M.E., Klein Wolterink R.G., Di Santo J.P. GATA-3 promotes T-cell specification by repressing B-cell potential in pro-T cells in mice. Blood. 2013;121:1749–1759. doi: 10.1182/blood-2012-06-440065. [DOI] [PubMed] [Google Scholar]
- 114.Scripture-Adams D.D., Damle S.S., Rothenberg E.V. GATA-3 dose-dependent checkpoints in early T cell commitment. J. Immunol. 2014;193:3470–3491. doi: 10.4049/jimmunol.1301663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Taghon T., Yui M.A., Rothenberg E.V. Mast cell lineage diversion of T lineage precursors by the essential T cell transcription factor GATA-3. Nat. Immunol. 2007;8:845–855. doi: 10.1038/ni1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Xu W., Carr T., Kee B.L. E2A transcription factors limit expression of Gata3 to facilitate T lymphocyte lineage commitment. Blood. 2013;121:1534–1542. doi: 10.1182/blood-2012-08-449447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hosoya T., Maillard I., Engel J.D. From the cradle to the grave: activities of GATA-3 throughout T-cell development and differentiation. Immunol. Rev. 2010;238:110–125. doi: 10.1111/j.1600-065X.2010.00954.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hosokawa H., Rothenberg E.V. How transcription factors drive choice of the T cell fate. Nat. Rev. Immunol. 2021;21:162–176. doi: 10.1038/s41577-020-00426-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Seo W., Taniuchi I. Transcriptional regulation of early T-cell development in the thymus. Eur. J. Immunol. 2016;46:531–538. doi: 10.1002/eji.201545821. [DOI] [PubMed] [Google Scholar]
- 120.Thompson P.K., Zúñiga-Pflücker J.C. On becoming a T cell, a convergence of factors kick it up a Notch along the way. Semin. Immunol. 2011;23:350–359. doi: 10.1016/j.smim.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 121.De Obaldia M.E., Bhandoola A. Transcriptional regulation of innate and adaptive lymphocyte lineages. Annu. Rev. Immunol. 2015;33:607–642. doi: 10.1146/annurev-immunol-032414-112032. [DOI] [PubMed] [Google Scholar]
- 122.Rothenberg E.V. Programming for T-lymphocyte fates: modularity and mechanisms. Genes Dev. 2019;33:1117–1135. doi: 10.1101/gad.327163.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pham T.-H., Minderjahn J., Rehli M. Mechanisms of in vivo binding site selection of the hematopoietic master transcription factor PU.1. Nucleic Acids Res. 2013;41:6391–6402. doi: 10.1093/nar/gkt355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hu G., Cui K., Zhao K. Transformation of accessible chromatin and 3D nucleome underlies lineage commitment of early T cells. Immunity. 2018;48:227–242.e8. doi: 10.1016/j.immuni.2018.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yui M.A., Rothenberg E.V. Developmental gene networks: a triathlon on the course to T cell identity. Nat. Rev. Immunol. 2014;14:529–545. doi: 10.1038/nri3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Spicuglia S., Franchini D.M., Ferrier P. Regulation of V(D)J recombination. Curr. Opin. Immunol. 2006;18:158–163. doi: 10.1016/j.coi.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 127.Carico Z., Krangel M.S. Chromatin dynamics and the development of the TCRα and TCRδ repertoires. Adv. Immunol. 2015;128:307–361. doi: 10.1016/bs.ai.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 128.Manesso E., Chickarmane V., Peterson C. Computational modelling of T-cell formation kinetics: output regulated by initial proliferation-linked deferral of developmental competence. J. R. Soc. Interface. 2013;10:20120774. doi: 10.1098/rsif.2012.0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kawamoto H., Ohmura K., Katsura Y. Extensive proliferation of T cell lineage-restricted progenitors in the thymus: an essential process for clonal expression of diverse T cell receptor β chains. Eur. J. Immunol. 2003;33:606–615. doi: 10.1002/eji.200323461. [DOI] [PubMed] [Google Scholar]
- 130.Olariu V., Yui M.A., Peterson C. Multi-scale dynamical modeling of T cell development from an early thymic progenitor state to lineage commitment. Cell Rep. 2021;34:108622. doi: 10.1016/j.celrep.2020.108622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yoshida H., Lareau C.A., Benoist C., Immunological Genome Project The cis-regulatory atlas of the mouse immune system. Cell. 2019;176:897–912.e20. doi: 10.1016/j.cell.2018.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhang J.A., Mortazavi A., Rothenberg E.V. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell. 2012;149:467–482. doi: 10.1016/j.cell.2012.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mingueneau M., Kreslavsky T., Turley S., Immunological Genome Consortium The transcriptional landscape of αβ T cell differentiation. Nat. Immunol. 2013;14:619–632. doi: 10.1038/ni.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wang Q., Yan R., Chiang M.Y. Stage-specific roles for Zmiz1 in Notch-dependent steps of early T-cell development. Blood. 2018;132:1279–1292. doi: 10.1182/blood-2018-02-835850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Maillard I., Tu L., Pear W.S. The requirement for Notch signaling at the β-selection checkpoint in vivo is absolute and independent of the pre-T cell receptor. J. Exp. Med. 2006;203:2239–2245. doi: 10.1084/jem.20061020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chen E.L.Y., Thompson P.K., Zúñiga-Pflücker J.C. RBPJ-dependent Notch signaling initiates the T cell program in a subset of thymus-seeding progenitors. Nat. Immunol. 2019;20:1456–1468. doi: 10.1038/s41590-019-0518-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Tanigaki K., Tsuji M., Honjo T. Regulation of αβ/γδ T cell lineage commitment and peripheral T cell responses by Notch/RBP-J signaling. Immunity. 2004;20:611–622. doi: 10.1016/s1074-7613(04)00109-8. [DOI] [PubMed] [Google Scholar]
- 138.Radtke F., Wilson A., MacDonald H.R. Notch regulation of lymphocyte development and function. Nat. Immunol. 2004;5:247–253. doi: 10.1038/ni1045. [DOI] [PubMed] [Google Scholar]
- 139.Johnson J.L., Georgakilas G., Vahedi G. Lineage-determining transcription factor TCF-1 initiates the epigenetic identity of T cells. Immunity. 2018;48:243–257.e10. doi: 10.1016/j.immuni.2018.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Longabaugh W.J.R., Zeng W., Rothenberg E.V. Bcl11b and combinatorial resolution of cell fate in the T-cell gene regulatory network. Proc. Natl. Acad. Sci. USA. 2017;114:5800–5807. doi: 10.1073/pnas.1610617114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Weber B.N., Chi A.W., Bhandoola A. A critical role for TCF-1 in T-lineage specification and differentiation. Nature. 2011;476:63–68. doi: 10.1038/nature10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Germar K., Dose M., Gounari F. T-cell factor 1 is a gatekeeper for T-cell specification in response to Notch signaling. Proc. Natl. Acad. Sci. USA. 2011;108:20060–20065. doi: 10.1073/pnas.1110230108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Oravecz A., Apostolov A., Kastner P. Ikaros mediates gene silencing in T cells through Polycomb repressive complex 2. Nat. Commun. 2015;6:8823. doi: 10.1038/ncomms9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Arenzana T.L., Schjerven H., Smale S.T. Regulation of gene expression dynamics during developmental transitions by the Ikaros transcription factor. Genes Dev. 2015;29:1801–1816. doi: 10.1101/gad.266999.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Miyazaki M., Miyazaki K., Murre C. The E-Id protein axis specifies adaptive lymphoid cell identity and suppresses thymic innate lymphoid cell development. Immunity. 2017;46:818–834.e4. doi: 10.1016/j.immuni.2017.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Dakic A., Metcalf D., Nutt S.L. PU.1 regulates the commitment of adult hematopoietic progenitors and restricts granulopoiesis. J. Exp. Med. 2005;201:1487–1502. doi: 10.1084/jem.20050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Champhekar A., Damle S.S., Rothenberg E.V. Regulation of early T-lineage gene expression and developmental progression by the progenitor cell transcription factor PU.1. Genes Dev. 2015;29:832–848. doi: 10.1101/gad.259879.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ikawa T., Hirose S., Kawamoto H. An essential developmental checkpoint for production of the T cell lineage. Science. 2010;329:93–96. doi: 10.1126/science.1188995. [DOI] [PubMed] [Google Scholar]
- 149.Li P., Burke S., Liu P. Reprogramming of T cells to natural killer-like cells upon Bcl11b deletion. Science. 2010;329:85–89. doi: 10.1126/science.1188063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Li L., Leid M., Rothenberg E.V. An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science. 2010;329:89–93. doi: 10.1126/science.1188989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Talebian L., Li Z., Speck N.A. T-lymphoid, megakaryocyte, and granulocyte development are sensitive to decreases in CBFβ dosage. Blood. 2007;109:11–21. doi: 10.1182/blood-2006-05-021188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Guo Y., Maillard I., Speck N.A. Core binding factors are necessary for natural killer cell development and cooperate with Notch signaling during T-cell specification. Blood. 2008;112:480–492. doi: 10.1182/blood-2007-10-120261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Pham T.H., Benner C., Rehli M. Dynamic epigenetic enhancer signatures reveal key transcription factors associated with monocytic differentiation states. Blood. 2012;119:e161–e171. doi: 10.1182/blood-2012-01-402453. [DOI] [PubMed] [Google Scholar]
- 154.Kamath M.B., Houston I.B., DeKoter R.P. Dose-dependent repression of T-cell and natural killer cell genes by PU.1 enforces myeloid and B-cell identity. Leukemia. 2008;22:1214–1225. doi: 10.1038/leu.2008.67. [DOI] [PubMed] [Google Scholar]
- 155.Minderjahn J., Schmidt A., Rehli M. Mechanisms governing the pioneering and redistribution capabilities of the non-classical pioneer PU.1. Nat. Commun. 2020;11:402. doi: 10.1038/s41467-019-13960-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Glass C.K., Natoli G. Molecular control of activation and priming in macrophages. Nat. Immunol. 2016;17:26–33. doi: 10.1038/ni.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Natoli G., Ghisletti S., Barozzi I. The genomic landscapes of inflammation. Genes Dev. 2011;25:101–106. doi: 10.1101/gad.2018811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ghisletti S., Barozzi I., Natoli G. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32:317–328. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 159.Iwasaki H., Somoza C., Akashi K. Distinctive and indispensable roles of PU.1 in maintenance of hematopoietic stem cells and their differentiation. Blood. 2005;106:1590–1600. doi: 10.1182/blood-2005-03-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Leddin M., Perrod C., Rosenbauer F. Two distinct auto-regulatory loops operate at the PU.1 locus in B cells and myeloid cells. Blood. 2011;117:2827–2838. doi: 10.1182/blood-2010-08-302976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lin Q., Chauvistré H., Zenke M. Epigenetic program and transcription factor circuitry of dendritic cell development. Nucleic Acids Res. 2015;43:9680–9693. doi: 10.1093/nar/gkv1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Laiosa C.V., Stadtfeld M., Graf T. Reprogramming of committed T cell progenitors to macrophages and dendritic cells by C/EBP α and PU.1 transcription factors. Immunity. 2006;25:731–744. doi: 10.1016/j.immuni.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 163.Franco C.B., Scripture-Adams D.D., Rothenberg E.V. Notch/Delta signaling constrains reengineering of pro-T cells by PU.1. Proc. Natl. Acad. Sci. USA. 2006;103:11993–11998. doi: 10.1073/pnas.0601188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Barozzi I., Simonatto M., Natoli G. Coregulation of transcription factor binding and nucleosome occupancy through DNA features of mammalian enhancers. Mol. Cell. 2014;54:844–857. doi: 10.1016/j.molcel.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Stephens D.C., Poon G.M. Differential sensitivity to methylated DNA by ETS-family transcription factors is intrinsically encoded in their DNA-binding domains. Nucleic Acids Res. 2016;44:8671–8681. doi: 10.1093/nar/gkw528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Xhani S., Lee S., Poon G.M.K. Intrinsic disorder controls two functionally distinct dimers of the master transcription factor PU.1. Sci. Adv. 2020;6:eaay3178. doi: 10.1126/sciadv.aay3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wang Y., Zolotarev N., Grosschedl R. A prion-like domain in transcription factor EBF1 promotes phase separation and enables B cell programming of progenitor chromatin. Immunity. 2020;53:1151–1167.e6. doi: 10.1016/j.immuni.2020.10.009. [DOI] [PubMed] [Google Scholar]
- 168.Boller S., Ramamoorthy S., Grosschedl R. Pioneering activity of the C-terminal domain of EBF1 shapes the chromatin landscape for B cell programming. Immunity. 2016;44:527–541. doi: 10.1016/j.immuni.2016.02.021. [DOI] [PubMed] [Google Scholar]
- 169.Soufi A., Garcia M.F., Zaret K.S. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell. 2015;161:555–568. doi: 10.1016/j.cell.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Hosokawa H., Ungerbäck J., Rothenberg E.V. Transcription factor PU.1 represses and activates gene expression in early T cells by redirecting partner transcription factor binding. Immunity. 2018;48:1119–1134.e7. doi: 10.1016/j.immuni.2018.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Rothenberg E.V., Hosokawa H., Ungerbäck J. Mechanisms of action of hematopoietic transcription factor PU.1 in initiation of T-cell development. Front. Immunol. 2019;10:228. doi: 10.3389/fimmu.2019.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kueh H.Y., Yui M.A., Rothenberg E.V. Asynchronous combinatorial action of four regulatory factors activates Bcl11b for T cell commitment. Nat. Immunol. 2016;17:956–965. doi: 10.1038/ni.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Avram D., Califano D. The multifaceted roles of Bcl11b in thymic and peripheral T cells: impact on immune diseases. J. Immunol. 2014;193:2059–2065. doi: 10.4049/jimmunol.1400930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Avram D., Fields A., Leid M. COUP-TF (chicken ovalbumin upstream promoter transcription factor)-interacting protein 1 (CTIP1) is a sequence-specific DNA binding protein. Biochem. J. 2002;368:555–563. doi: 10.1042/BJ20020496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Liu N., Hargreaves V.V., Orkin S.H. Direct promoter repression by BCL11A controls the fetal to adult hemoglobin switch. Cell. 2018;173:430–442.e17. doi: 10.1016/j.cell.2018.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Drashansky T.T., Helm E., Avram D. Bcl11b prevents fatal autoimmunity by promoting Treg cell program and constraining innate lineages in Treg cells. Sci. Adv. 2019;5:eaaw0480. doi: 10.1126/sciadv.aaw0480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Lorentsen K.J., Cho J.J., Avram D. Bcl11b is essential for licensing Th2 differentiation during helminth infection and allergic asthma. Nat. Commun. 2018;9:1679. doi: 10.1038/s41467-018-04111-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Mevel R., Draper J.E., Lacaud G. RUNX transcription factors: orchestrators of development. Development. 2019;146:dev148296. doi: 10.1242/dev.148296. [DOI] [PubMed] [Google Scholar]
- 179.Ebihara T., Seo W., Taniuchi I. Roles of RUNX complexes in immune cell development. Adv. Exp. Med. Biol. 2017;962:395–413. doi: 10.1007/978-981-10-3233-2_24. [DOI] [PubMed] [Google Scholar]
- 180.Gilmour J., Assi S.A., Bonifer C. The co-operation of RUNX1 with LDB1, CDK9 and BRD4 drives transcription factor complex relocation during haematopoietic specification. Sci. Rep. 2018;8:10410. doi: 10.1038/s41598-018-28506-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Lichtinger M., Ingram R., Bonifer C. RUNX1 reshapes the epigenetic landscape at the onset of haematopoiesis. EMBO J. 2012;31:4318–4333. doi: 10.1038/emboj.2012.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Hoogenkamp M., Lichtinger M., Bonifer C. Early chromatin unfolding by RUNX1: a molecular explanation for differential requirements during specification versus maintenance of the hematopoietic gene expression program. Blood. 2009;114:299–309. doi: 10.1182/blood-2008-11-191890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Lukin K., Fields S., Hagman J. Compound haploinsufficiencies of Ebf1 and Runx1 genes impede B cell lineage progression. Proc. Natl. Acad. Sci. USA. 2010;107:7869–7874. doi: 10.1073/pnas.1003525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Lin Y.C., Jhunjhunwala S., Murre C. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat. Immunol. 2010;11:635–643. doi: 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wang H., Zang C., Aster J.C. NOTCH1-RBPJ complexes drive target gene expression through dynamic interactions with superenhancers. Proc. Natl. Acad. Sci. USA. 2014;111:705–710. doi: 10.1073/pnas.1315023111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Ng K.K.H., Yui M.A., Kueh H.Y. A stochastic epigenetic switch controls the dynamics of T-cell lineage commitment. eLife. 2018;7:e37851. doi: 10.7554/eLife.37851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Wolfer A., Bakker T., Radtke F. Inactivation of Notch 1 in immature thymocytes does not perturb CD4 or CD8T cell development. Nat. Immunol. 2001;2:235–241. doi: 10.1038/85294. [DOI] [PubMed] [Google Scholar]
- 188.Kastner P., Chan S., Leid M. Bcl11b represses a mature T-cell gene expression program in immature CD4+CD8+ thymocytes. Eur. J. Immunol. 2010;40:2143–2154. doi: 10.1002/eji.200940258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Albu D.I., Feng D., Avram D. BCL11B is required for positive selection and survival of double-positive thymocytes. J. Exp. Med. 2007;204:3003–3015. doi: 10.1084/jem.20070863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Charest J., Daniele T., Cochella L. Combinatorial action of temporally segregated transcription factors. Dev. Cell. 2020;55:483–499.e7. doi: 10.1016/j.devcel.2020.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Soufi A., Donahue G., Zaret K.S. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell. 2012;151:994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Xu Z., Chen H., Small S. Impacts of the ubiquitous factor Zelda on Bicoid-dependent DNA binding and transcription in Drosophila. Genes Dev. 2014;28:608–621. doi: 10.1101/gad.234534.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Chronis C., Fiziev P., Plath K. Cooperative binding of transcription factors orchestrates reprogramming. Cell. 2017;168:442–459.e20. doi: 10.1016/j.cell.2016.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Wei G., Abraham B.J., Zhao K. Genome-wide analyses of transcription factor GATA3-mediated gene regulation in distinct T cell types. Immunity. 2011;35:299–311. doi: 10.1016/j.immuni.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Li L., Zhang J.A., Rothenberg E.V. A far downstream enhancer for murine Bcl11b controls its T-cell specific expression. Blood. 2013;122:902–911. doi: 10.1182/blood-2012-08-447839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Pease N.A., Nguyen P.H.B., Kueh H.Y. Tunable, division-independent control of gene activation timing by a polycomb switch. Cell Rep. 2021;34:108888. doi: 10.1016/j.celrep.2021.108888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Isoda T., Moore A.J., Murre C. Non-coding transcription instructs chromatin folding and compartmentalization to dictate enhancer-promoter communication and T cell fate. Cell. 2017;171:103–119.e18. doi: 10.1016/j.cell.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]