Abstract
Distinct patterns of actomyosin contractility are often associated with particular epithelial tissue shape changes during development. For example, a planar-polarized pattern of myosin II localization regulated by Rho1 signaling during Drosophila body axis elongation is thought to drive cell behaviors that contribute to convergent extension. However, it is not well understood how specific aspects of a myosin pattern influence the multiple cell behaviors, including cell intercalation, cell shape changes, and apical cell area fluctuations, that simultaneously occur during morphogenesis. Here, we developed two optogenetic tools, optoGEF and optoGAP, to activate or deactivate Rho1 signaling, respectively. We used these tools to manipulate myosin patterns at the apical side of the germband epithelium during Drosophila axis elongation and analyzed the effects on contractile cell behaviors. We show that uniform activation or inactivation of Rho1 signaling across the apical surface of the germband is sufficient to disrupt the planar-polarized pattern of myosin at cell junctions on the timescale of 3–5 min, leading to distinct changes in junctional and medial myosin patterns in optoGEF and optoGAP embryos. These two perturbations to Rho1 activity both disrupt axis elongation and cell intercalation but have distinct effects on cell area fluctuations and cell packings that are linked with changes in the medial and junctional myosin pools. These studies demonstrate that acute optogenetic perturbations to Rho1 activity are sufficient to rapidly override the endogenous planar-polarized myosin pattern in the germband during axis elongation. Moreover, our results reveal that the levels of Rho1 activity and the balance between medial and junctional myosin play key roles not only in organizing the cell rearrangements that are known to directly contribute to axis elongation but also in regulating cell area fluctuations and cell packings, which have been proposed to be important factors influencing the mechanics of tissue deformation and flow.
Significance
Tissues are shaped by forces produced by dynamic patterns of actomyosin contractility. However, the mechanisms underlying these myosin patterns and their translation into cell behavior and tissue-level movements are not understood. Here, we show that optogenetic tools designed to control upstream regulators of myosin II can be used to rapidly manipulate myosin patterns and analyze the downstream effects on cell behaviors. Combining optogenetics with live imaging in the fruit fly embryo, we show that acute optogenetic perturbations are sufficient to rapidly override the existing myosin pattern and alter cell movements and shapes during body axis elongation, resulting in abnormalities in embryo structure. These results link myosin patterns to cell behaviors, providing new insights into the mechanisms that generate functional tissues.
Introduction
During development, tissues undergo dramatic changes in shape that are largely driven by patterns of contractile forces generated by the cellular actomyosin cytoskeleton (1, 2, 3, 4). Patterns of myosin II localization and activity are responsible for producing spatially and temporally regulated cell behaviors that physically sculpt tissues and organs. In epithelial tissues, for example, planar-polarized patterns of myosin localization at cell junctions, as well as polarized flows of apical actomyosin, are often associated with cell rearrangements that narrow and elongate tissues, whereas pulsed, radial patterns of myosin at the apical surface of cells are often associated with apical constriction during tissue invagination (4,5). Such myosin localization patterns are conserved, and significant insight into the roles of specific myosin patterns has been inferred from correlating myosin patterns with cell behaviors and tissue movements. However, gaining insight into how myosin localization and dynamics control distinct aspects of cell behavior requires experiments in which the myosin localization pattern is perturbed and the resulting cell behaviors are analyzed.
Various methods, such as drug inhibition, genetic mutations, and protein knockdown or overexpression, have been used to perturb myosin II and its regulators to study their functions during morphogenesis. These approaches can have significant limitations, depending on the process of interest. For example, during embryonic body axis elongation in Drosophila, drug inhibitors are typically injected into the embryo, limiting spatial control and potentially causing destructive effects at the injection site. Traditional molecular genetics perturbations cannot always be easily or flexibly targeted at specific time points or to specific groups of cells at this stage of development, limiting spatial and temporal control. This makes it difficult to separate the effects of the perturbation on the tissue of interest from the effects in neighboring regions of the embryo or prior events during development. Thus, a major obstacle to understanding how patterns of actomyosin localization influence cell behaviors during tissue morphogenesis has been the lack of tools for flexible and precise manipulation of patterns of actomyosin contractility in vivo (6). Optogenetic technologies have emerged as attractive approaches for flexible and noninvasive manipulation of protein activities and cell behaviors (6, 7, 8, 9, 10). In particular, new optogenetic tools designed to manipulate actomyosin contractility with high spatiotemporal precision can now be used for studying the spatial and temporal requirements for myosin II in dynamic cell and tissue behaviors (11, 12, 13, 14, 15, 16, 17, 18, 19, 20). For example, optogenetic tools designed to disrupt the actin cytoskeleton (12) or activate actomyosin contractility (16,20) have been used to investigate mechanisms of tissue bending in Drosophila.
Convergent extension of the head-to-tail axis of the D. melanogaster embryo is a powerful system for studying the mechanisms by which dynamic patterns of actomyosin contractility influence cell behaviors and tissue-level morphogenesis. During body axis elongation, the anterior-posterior (AP) axis of the embryo extends by more than twofold. The majority of extension occurs during the first 30 min and is driven by cell intercalation (oriented cell rearrangement) (21, 22, 23, 24, 25) and cell shape changes (24,26, 27, 28) in the germband epithelium. Just before the onset of body axis elongation, F-actin and myosin II become asymmetrically localized near adherens junctions at cell interfaces between anterior and posterior cell neighbors (“AP” or “vertical” edges), generating a planar-polarized pattern of myosin II localization (22,24,29, 30, 31) and promoting cell intercalation that contributes to tissue elongation (22,23,32, 33, 34, 35). In addition to junctional actomyosin, a dynamic actomyosin meshwork in the medial-apical domain of germband cells exhibits polarized flows that are thought to contribute to pulsatile fluctuations in cell junction lengths and in apical cell areas and to cell intercalation (34, 35, 36, 37, 38, 39). Recent theoretical and experimental studies point toward important roles both for active fluctuations in cell shape and junctions (40,41) and for details of cellular packings within tissues (42, 43, 44, 45) in the physics of cell rearrangements and the ability of a tissue to remodel and flow (i.e., tissue fluidity). It remains unclear exactly how cell intercalation, cell shape changes, and pulsatile cell area fluctuations are organized by the myosin localization pattern and together contribute to tissue shape change.
The patterns of myosin II localization during Drosophila axis elongation are regulated by the Rho/Rho-kinase signaling pathway (30,35,38,46, 47, 48, 49, 50). In general, patterns of Rho activity are directed by Rho-specific guanine nucleotide exchange factors (RhoGEFs), which promote the active state of Rho, and by Rho GTPase-activating proteins (RhoGAPs), which promote the inactive state of Rho (51, 52, 53, 54). During axis elongation, the active state of the RhoGTPase Drosophila Rho1 is promoted by the exchange factors RhoGEF2 and Cysts/Dp114RhoGEF of the RhoGEF family (38,46, 47, 48, 49, 50). Although RhoGAP activity is required for proper Rho signaling in some contexts (54, 55, 56), RhoGAPs essential for Drosophila axis elongation have not yet been described. Drosophila RhoGAP71E has been shown to play a role, along with RhoGEF2, in organizing radial patterns of myosin activity that drive apical constriction during invagination of the presumptive mesoderm (56).
Because Rho activity controlled by RhoGEFs and RhoGAPs is thought to direct actomyosin contractility patterns in many contexts, it is an attractive target for optogenetic manipulation. Indeed, domains of RhoGEFs have been used successfully in optogenetic tools to activate actomyosin contractility and induce tissue folding in the early Drosophila embryo during cellularization and ventral furrow formation stages (16, 17, 18,20), just before the strong planar-polarized accumulation of myosin at adherens junctions present in the germband during axis elongation. To our knowledge, optogenetic tools based on RhoGAPs to inactivate Rho1 signaling and reduce actomyosin contractility have not yet been described. It remains unclear whether optogenetic technologies based on RhoGEFs and RhoGAPs will be sufficient to override and manipulate strong endogenous planar-polarized myosin patterns present in various tissues and stages of development, for example during Drosophila axis elongation.
Here, we develop optogenetic tools to activate (optoGEF) or deactivate (optoGAP) Rho1 signaling. We use optogenetic activation and deactivation of Rho1 signaling to perturb the myosin pattern at the apical surface of cells in the germband epithelium during Drosophila body axis elongation and analyze the effects on contractile cell behaviors within the tissue. We find that uniform photoactivation of optoGEF or optoGAP in the germband is sufficient to rapidly override the endogenous myosin pattern on the timescale of 3–5 min, abolishing myosin planar polarity and reducing cell intercalation and convergent extension. However, these two perturbations have distinct effects on junctional and medial myosin localization, apical cell area fluctuations, and cell packings within the germband. Activation of Rho1 signaling in optoGEF embryos increases myosin accumulation in the medial-apical domain of germband cells, leading to increased amplitudes of apical cell area fluctuations. This enhanced contractility is associated with heterogeneous reductions in apical cell areas across the tissue, disrupting cellular packings within the germband. Conversely, inactivation of Rho1 signaling in optoGAP embryos decreases both medial and junctional myosin accumulation, which is associated with a reduction in cell area fluctuations. These results demonstrate that the level of Rho1 activity and the balance between junctional and medial myosin regulate apical cell area fluctuations and cellular packings in the germband, which have been proposed to influence the biophysics of cell rearrangements.
Materials and methods
Cloning
To generate CRY2-RhoGEF2, the full-length RhoGEF2 cDNA of D. melanogaster was amplified by PCR from plasmid UAS-akkordion dNhe (plasmid number 41976; Addgene, Watertown, NY) (57), and the cDNA of CRY2PHR was PCR amplified from the pCRY2PHR-mCherryN1 plasmid from the Tucker Lab (plasmid number 26866; Addgene, Watertown, NY) (7). DNA fragments were linked using the NEBuilder HiFi DNA Assembly Kit (New England BioLabs, Ipswich, MA). RhoGAP71E was PCR amplified from the exons corresponding to RhoGAP71E protein from pACMAN CH321-72J07 (BACPAC Resources, Children’s Hospital Oakland Research Institute, Oakland, CA), and DNA fragments were assembled using a combination of restriction-ligation and DNA fragment assembly to obtain the full-length RhoGAP71E cDNA. To produce an N-terminal fluorescently tagged version of constructs, the sequence encoding mCherry from the pCRY2PHR-mCherryN1 plasmid from the Tucker Lab (plasmid number 26866; Addgene, Watertown, NY) (7) was included during DNA fragment assembly. In the constructs, a (GA)5 linker was introduced between mCherry and CRYPHR and a GG(SG)4 linker was introduced between CRY2 and RhoGEF2 or RhoGAP71E. Constructs were cloned into the pENTR/D-Topo vector (Life Technologies, Carlsbad, CA) and recombined into the UASp-attB destination vector (gift of F. Wirtz-Peitz) using the Gateway cloning system (Life Technologies, Carlsbad, CA) for subsequent expression using the Gal4-UAS system (58). The resulting plasmids were purified, sequenced, and used to generate transgenic flies (BestGene, Chino Hills, CA). To ensure comparable expression levels, all transgenes corresponding to CRY2 constructs were inserted into the attP2 site on chromosome III.
Fly stocks
Stocks and crosses were maintained at 23°C, and experiments were performed at room temperature (~21°C). The stocks w∗;; w+, UASp>mCherry-CRY2PHR-RhoGEF2 and w∗;; w+, UASp>mCherry-CRY2PHR-RhoGAP71E were generated in this study, crossed with w∗; P[w+, UASp>CIBN-pmGFP]/Cyo; Sb/TM3,Ser (gift of Stefano De Renzis, European Molecular Biology Laboratory, Heidelberg, Germany) and subsequently expressed using the maternal α-tubulin matα-tub15 or matα-tub67 Gal4-VP16 drivers (mat67, mat15; a gift of D. St Johnston). To visualize myosin II, mat15 and sqh>sqh-mCherry (BDSC 59024; donated by Beth Stronach, University of Pittsburgh, Pittsburgh, PA) were recombined on chromosome III and used alone or in combination with mat67. Crosses and embryos were kept protected from light. Fly sorting was performed in the dark on a stereomicroscope equipped with a Red 25 Wratten Filter (Kodak, Rochester, NY). Embryos studied were progeny of females of the following genotypes:
-
1)
UASp>CIBN-pmGFP/mat67 (II); sqh>sqh-mCherry/+ (III)
-
2)
UASp>CIBN-pmGFP/+ (II); UASp>mCherry-CRY2PHR-RhoGEF2/mat15 (III)
-
3)
UASp>CIBN-pmGFP/+ (II); UASp>CRY2PHR-RhoGEF2/sqh>sqh-mCherry, mat15 (III)
-
4)
UASp>CIBN-pmGFP/mat67 (II); UASp>mCherry-CRY2PHR-RhoGAP71E/mat15 (III)
-
5)
UASp>CIBN-pmGFP/mat67 (II); UASp>CRY2PHR-RhoGAP71E/sqh>sqh-mCherry, mat15 (III)
Fly stocks used in this study were as follows:
-
1)
w∗;; w+, UASp>mCherry-CRY2PHR-RhoGEF2/TM3,Sb (this study)
-
2)
w∗;; w+, UASp>CRY2PHR-RhoGEF2/TM3,Sb (this study)
-
3)
w∗;; w+, UASp>mCherry-CRY2PHR-RhoGAP71E/TM3,Sb (this study)
-
4)
w∗;; w+, UASp>CRY2PHR-RhoGAP71E/TM3,Sb (this study)
-
5)
w∗; w+, UASp>CIBN-pmGFP/CyO; Sb/TM3,Ser (Stefano De Renzis, European Molecular Biology Laboratory, Heidelberg, Germany)
-
6)
w∗;; sqh>sqh-mCherry (Beth Stronach, University of Pittsburgh, Pittsburgh, PA)
-
7)
w∗; w+, UASp>CIBN-pmGFP, mat67; sqh>sqh-mCherry
-
8)
w∗;; sqh>sqh-mCherry, mat15
-
9)
w∗; mat67; sqh>sqh-mCherry, mat15
Assessing axis elongation phenotypes and embryo viability
Samples for imaging were prepared in a dark room illuminated with red light. Embryos were collected under red light, maintained in darkness for 4 h after being laid, briefly exposed to red light to manually score body axis elongation phenotypes on a stereomicroscope, and returned to darkness. After 1 day, embryo viability was assessed from the embryo hatching rate.
Live imaging
Samples for imaging were prepared in a dark room illuminated with red light. Embryos in early stage 6 were selected under Halocarbon oil 27 (Sigma-Aldrich, St. Louis, MO), dechorionated with 50% bleach for 2 min, washed with water, and mounted using a 50:50 mixture of Halocarbon oil 27:700 on a custom-made imaging chamber between an oxygen-permeable membrane (YSI, Yellow Springs, OH) and a glass coverslip. Embryos were positioned ventrolaterally for observation of the germband and were imaged on a Zeiss LSM 880 confocal microscope equipped with a diode laser for 561 nm excitation and an argon laser for 488 nm excitation, a standard LSM confocal detector, and an Airyscan detector with FAST module (Carl Zeiss, Oberkochen, Germany). Embryos were photoactivated with 488 nm light. Imaging was performed with a C-Apo 40×/1.2 NA water immersion objective or a Plan-Apo 63×/1.40 NA oil immersion objective (Carl Zeiss). Images were acquired using the standard LSM mode, unless otherwise noted. For myosin imaging, the Airyscan FAST module was utilized. The light for bright field illumination was filtered through a Red 25 Kodak Wratten Filter to prevent unwanted photoactivation during embryo selection. Imaging conditions were identical for embryos within a given group for each set of experiments. Image acquisition was performed with ZEN Black software (Carl Zeiss, Oberkochen Germany). Images of the germband were acquired as 10 μm z-stacks with a 1 μm z-step (40× objective, LSM mode) or a 0.7 μm z-step (63× objective, Airyscan FAST), beginning at the apical side of the tissue. Z-stacks were acquired every 15 s. Photoactivation was achieved by scanning the same ventrolateral region of the germband with blue laser light (λ = 488 nm) that was being imaged with blue and/or green light (λ = 488 nm, λ = 561 nm) at every imaging z-plane and time step. To visualize myosin before photoactivation, mCherry-tagged myosin regulatory light chain was imaged with 561 nm light for 2.5 min. For quantification of protein localization patterns at different positions along the z-axis, a 10 or 15 μm z-stack (beginning at the apical side) with a 0.7 or 0.5 μm z-step was taken before and after photoactivation; photoactivation was achieved by scanning the blue laser (λ = 488 nm) over a 10 μm z-stack with 1 μm z-steps every 15 s for 2.5 min (40× objective, LSM mode).
Image analysis and quantification
Tissue elongation during body axis elongation was calculated from confocal time-lapse videos using the particle image velocimetry software PIVlab version 1.41 (59) in MATLAB (The MathWorks, Natick, MA), as previously described (30,59). For myosin analysis, still frames shown and confocal videos analyzed correspond to the maximal intensity projections of 5 μm at the apical side of the tissue, starting apically just underneath the vitelline membrane, unless otherwise noted. Images were processed and analyzed using the ImageJ distribution Fiji (60,61). Unless otherwise noted, protein fluorescence intensities are reported relative to the values at t = 0 in each video. The mean junctional myosin intensity at each cell edge was quantified manually using 0.5-μm-wide lines (excluding vertex regions) in a 40 μm square region, and the intensity and orientation of each edge was obtained. Medial-apical myosin intensity was quantified manually as the mean intensity in a polygon manually drawn just inside the cell membrane outline. At least 10 cells and 40 junctions were analyzed per embryo. Planar polarity was obtained as the ratio of the mean intensity of AP (“vertical,” 90 ± 30°) edges to the mean intensity of DV (“horizontal,” 0 ± 30°) edges for each embryo. Cell outlines were visualized using CIBN-pmGFP for manually obtaining cell areas. At least six cells per embryo and three to four embryos per condition were analyzed. For quantifying protein localization patterns at different z-positions along the apical to basal axis, 0.5-μm-wide lines (excluding vertex regions) were used to measure intensity at cell edges and polygons drawn just inside the cell outline were used to quantify medial and/or cytoplasmic intensity. For automated image segmentation analysis, the SEGGA tissue segmentation and analysis software was used, cell outlines were visualized with CIBN-pmGFP, and three to five embryos per genotype were analyzed (Figs. 1 C and 3, B and D) (24).
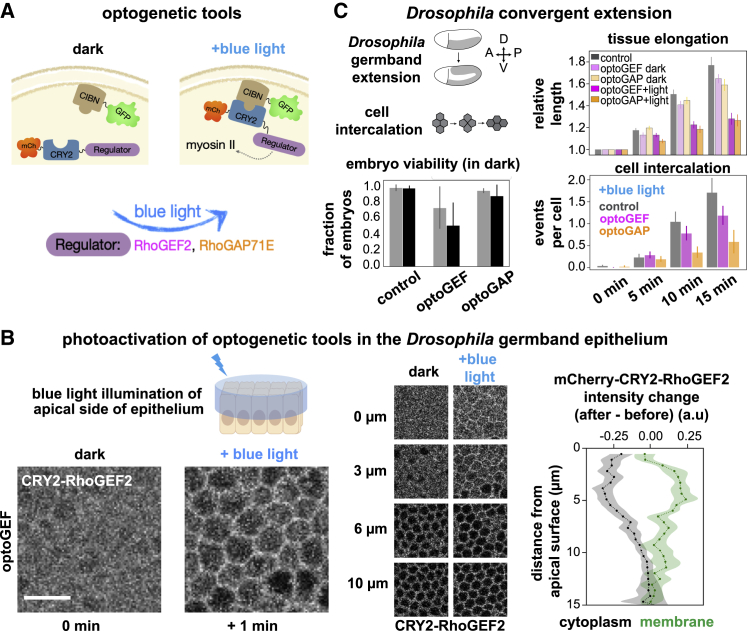
Figure 1.
Optogenetic activation (optoGEF) or inactivation (optoGAP) of Rho1 signaling disrupts convergent extension during Drosophila axis elongation. (A) Schematic of optogenetic tools. CIBN is tagged with GFP and targeted to the cell membrane. The light-sensitive PHR domain of CRY2 is tagged with mCherry and fused to upstream myosin II regulators of the Rho/Rho-kinase pathway. Blue-light illumination induces dimerization of CRY2 and CIBN, leading to accumulation of the regulator at the cell membrane. The full-length Drosophila RhoGEF2 (optoGEF) or RhoGAP71E (optoGAP) proteins were used to create the optogenetic tools. (B) Blue-light illumination of the apical surface of the germband epithelium in the Drosophila embryo to activate optogenetic tools during axis elongation. (Left) Before blue-light exposure, the mCherry-CRY2 regulator component of the tool is localized in the cytoplasm of germband cells (optoGEF shown here). After 1 min of blue-light exposure, the mCherry-CRY2 regulator component of the tool accumulates at the cell membrane and is depleted from the cytoplasm. Apical view shows projection of z-slices from 2 to 8 μm. Scale bar, 10 μm. (Center) Localization of the mCherry-CRY2 regulator component of the tool (optoGEF shown here) before and after blue-light activation at different z-positions along the apical to basal axis of the epithelium. Medial-apical (0 μm), junctional (3 μm), and lateral (6, 10 μm) are shown. (Right) Quantification of the change in intensity of mCherry-CRY2-RhoGEF2 at the cell membrane in optoGEF embryos after 1 min of blue-light activation. The greatest change is observed from 1 to 6 μm below the apical side of cells. Mean ± SE between embryos is shown; n = 4 embryos with six cells per embryo. (C) (Top left) Schematics of convergent extension and cell intercalation during Drosophila body axis elongation. (Bottom left) Effects of expression of the optogenetic tools (in the absence of activating blue light) on the viability of embryos are shown. Fraction of control, optoGEF, and optoGAP embryos that successfully completed axis elongation (gray) and hatch into larvae (black) are shown. Mean ± SD between replicate experiments is shown; n = 163–227 embryos per group. (Top right) When the apical surface of the germband is illuminated with blue light during axis elongation, tissue elongation is reduced in optoGEF and optoGAP embryos compared with controls. Mean ± SE between embryos is shown; n = 3–8 embryos per group. (Bottom right) Cell intercalation is reduced in optoGEF and optoGAP embryos compared with controls. Mean ± SE between embryos is shown; n = 3–5 embryos per group.
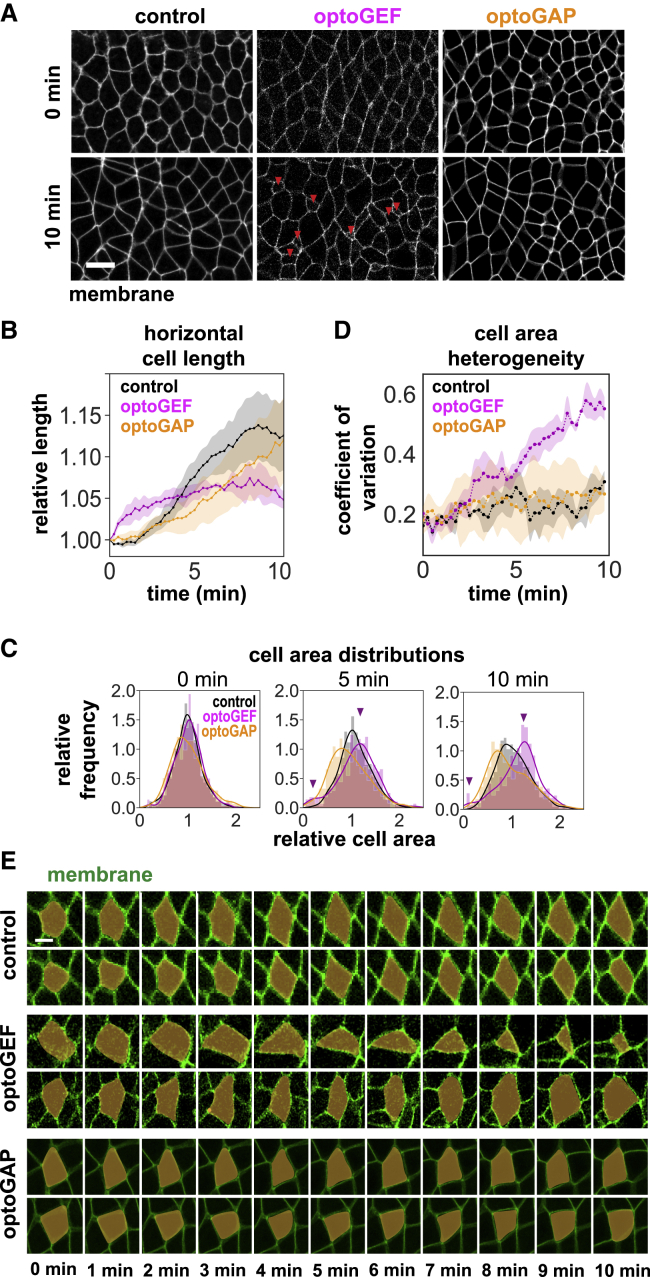
Figure 3.
Perturbing Rho1 activity across the germband using optoGEF or optoGAP alters apical cell areas. (A) Still images from confocal videos of apical cell shapes in the germband during axis elongation. Cells with small apical areas are observed in optoGEF embryos (red arrows). Anterior is left. Ventral is down. Scale bar, 10 μm. (B) Mean cell length along the AP axis (horizontal length) normalized to the value at t = 0, a metric for changes in cell shape that contribute to tissue elongation. Mean ± SE between embryos is shown; n = 3–5 embryos per condition. (C) Histograms of apical cell areas at 0, 5, and 10 min, normalized to the mean value at t = 0 in each embryo; n = 3–7 embryos per condition. (D) The coefficient of variation of apical cell area as a function of time in control, optoGEF, and optoGAP embryos. Cell area heterogeneity increases in optoGEF embryos compared with control or optoGAP embryos. Mean ± SE between embryos is shown; n = 3–5 embryos per genotype. (E) Still images from time-lapse videos of apical cell areas from two characteristic individual cells each in control, optoGEF, or optoGAP embryos. In control and optoGAP embryos, cells tend to maintain apical area over time. In optoGEF embryos, some cells shrink apical area progressively, whereas other cells increase apical area over time. Cell membrane is labeled with CIBN-pmGFP. Scale bar, 5 μm.
Cell behavior analysis
Horizontal cell length, coefficient of variation of apical cell area, cell rearrangements, and tissue elongation were calculated from cell segmentation analysis using the SEGGA software (24), and three to five embryos were analyzed per genotype. For horizontal cell length, the horizontal span of an ellipse fitted to each cell was calculated, and the mean value across the tissue is shown (24). Analyses of single-cell areas, medial myosin, and junctional myosin were performed manually. 8–12 cells per embryo and three to four embryos per genotype were analyzed. For Figs. 5 and 6, raw cell area and myosin intensity data were treated with a Savitzky-Golay filter (filter window: 9, order: 5). Myosin intensities were normalized to the value at t = 0. Changes of cell area and myosin correspond to the differences between 15 s time steps.
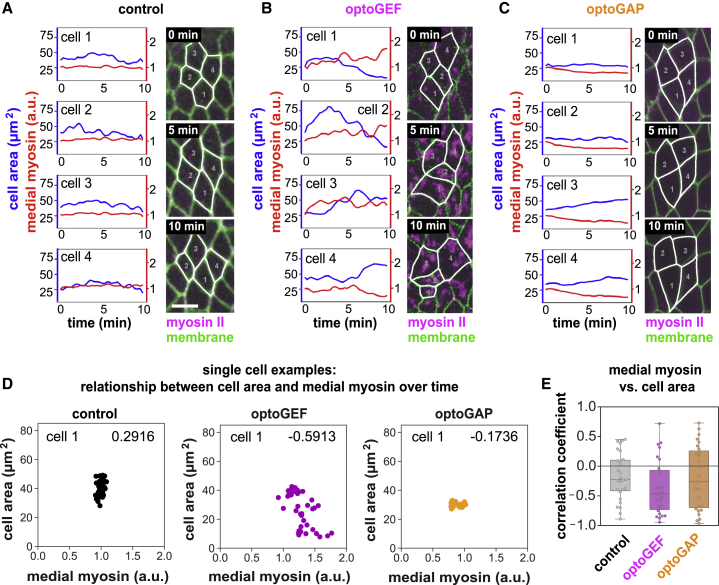
Figure 5.
Changes in apical cell area over time are associated with medial myosin patterns in single cells during convergent extension. (A–C) Examples of apical cell area and medial myosin intensity in individual cells over time in control (A), optoGEF (B), and optoGAP (C) embryos. Cell area (blue) and medial myosin intensity (red) in four neighboring cells are shown. Myosin intensities are relative to the value at t = 0 for each cell. Stills from confocal videos showing cells at 0, 5, and 10 min. Myosin II labeled with mCherry-tagged myosin regulatory light chain is shown in magenta. Cell membrane labeled with CIBN-pmGFP is shown in green. Overlaid cell outlines are shown in white. Scale bar, 10 μm. (D) Relationship between apical cell area and medial myosin intensity over time for example individual cells. Each data point represents the cell values at a single time point. See also Fig. S2. (E) Correlation coefficient between medial myosin intensity and apical cell area over 10 min for individual cells. Each point represents the correlation for data points from a single cell over time; n = 28 cells from three to four embryos per genotype.
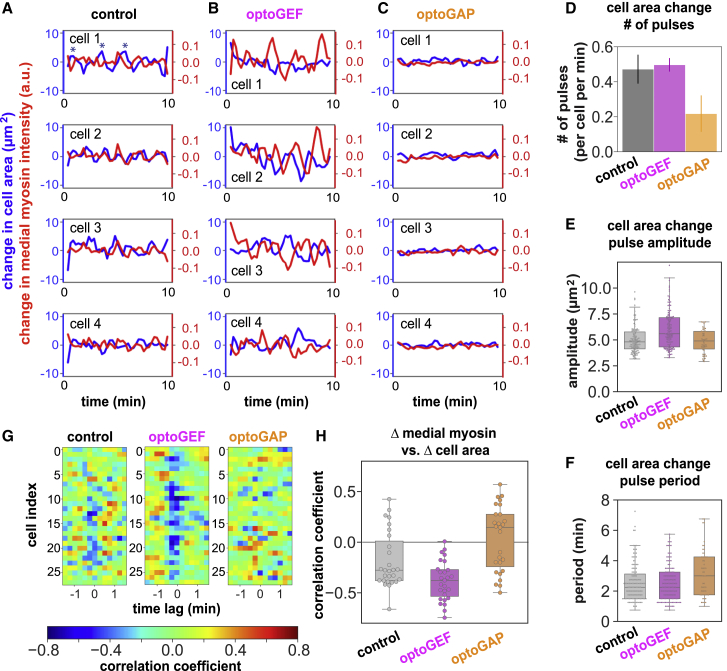
Figure 6.
Apical cell area fluctuations are increased in optoGEF and decreased in optoGAP embryos. (A–C) Changes in apical cell area (blue) and changes in normalized medial myosin intensity (red) between 15 s time points in four typical cells in control (A), optoGEF (B), and optoGAP (C) embryos photoactivated with blue light. Example pulses are indicated by asterisks (∗). (D) Number of pulses per cell per time. The error in area associated with cell segmentation was 3 μm2, so pulses less than 3 μm2 in amplitude were not included in analysis. (E) Amplitude of area change pulses is increased in optoGEF and not significantly changed in optoGAP compared with control embryos (optoGEF, p = 0.06; optoGAP, p = 0.37). Each point corresponds to one cell area pulse; n = 12 cells per embryo for three to four embryos per genotype. (F) Time period between successive cell area pulses. (G and H) Cross-correlation between changes in medial myosin and changes in apical cell area. (G) Correlation coefficients at different time lags for individual cells. (H) Correlation coefficients for control, optoGEF, and optoGAP cells (optoGEF, p = 0.03; optoGAP, p = 0.03 compared with control embryos); the correlation coefficient for each cell was taken as the maximal absolute value at the 0 or −15 s time lags. n = 28 cells from three to four embryos per genotype.
Statistical analysis
Mean values and mean ± SE were determined as indicated in each figure legend. The nonparametric Kruskal-Wallis test with post hoc Dunn test was used to determine p-values, unless otherwise noted.
Results
Optogenetic activation or inactivation of Rho1 signaling disrupts convergent extension in Drosophila
To dissect how patterns of actomyosin contractility influence cell behaviors that drive convergent extension, we used an optogenetic system to rapidly manipulate Rho1 signaling with high spatiotemporal precision at the apical surface of the epithelium of the developing Drosophila embryo. During convergent extension, myosin II in the germband epithelium is present in a planar-polarized pattern at apical cell junctions (22,23,29) as well as in meshworks in the medial-apical domain of cells (34,37) and is activated by the Rho1/Rho-kinase pathway (35,38,46, 47, 48, 49, 50). To manipulate patterns of Rho1 activity, we generated tools for blue-light-gated recruitment of Rho1 regulators to the cell membrane using the CRY2/CIB1 heterodimerization system (7,11,12,16) (Fig. 1 A).
The first tool, optoGEF, was designed to control the cell membrane localization of Drosophila RhoGEF2, which promotes myosin activity in the germband and the presumptive mesoderm (38,48,56,62, 63, 64, 65, 66). We generated embryos that coexpress CRY2-RhoGEF2 (the full-length RhoGEF2 fused to the light-sensitive CRY2 PHR domain, generated in this study) and CIBN-pmGFP (the N-terminal domain of the CRY2 binding partner CIB1 tagged with a cell membrane anchor and GFP) (7,12). Blue-light-induced accumulation of CRY2-RhoGEF2 at the cell membrane is predicted to increase Rho1 activity, leading to increased actomyosin contractility. The second tool, optoGAP, was designed to control the cell membrane localization of Drosophila RhoGAP71E, which plays a role in organizing radial patterns of myosin activity in presumptive mesoderm cells (56). The optoGAP tool (the full-length RhoGAP71E fused to the light-sensitive CRY2 PHR domain, generated in this study) is predicted to promote the inactive state of Rho1, leading to decreased actomyosin contractility.
To test the light-gated behavior of optoGEF and optoGAP during Drosophila axis elongation, we expressed the components of the tools in the early Drosophila embryo using the Gal4-UAS system. In the dark, the CRY2 component of the optogenetic tools localized to the cytoplasm (Fig. 1 B). We then photoactivated the apical surface of the ventrolateral region of the germband epithelium with 488 nm laser light, scanned over the same volume used for imaging (Fig. 1 B). Blue-light exposure led to rapid accumulation of the CRY2 component of the tools at the cell membrane and depletion from the cytoplasm (Fig. 1 B). The maximal membrane recruitment and cytoplasmic depletion occurred from 1 to 6 μm below the apical cell surface (Fig. 1 B), overlapping with the positions of myosin II at adherens junctions and in the medial-apical actomyosin meshwork, both of which contribute to contractile cell behaviors during convergent extension.
First, we analyzed the effects of the optogenetic tools on embryonic development in the absence of activating blue light. Embryos expressing the optoGAP tool displayed a slight reduction in embryonic viability, assessed by quantifying the fraction of embryos that go on to hatch into larvae, compared with control embryos, whereas optoGEF embryos showed a greater reduction in viability (Fig. 1 C). We observed that some optoGEF embryos, those with very high expression levels of the tool components, displayed defects in egg shape, appearing more rounded and smaller than wild-type controls, or failed to cellularize. These results are consistent with the possibility that expression of the optogenetic tools at high levels may cause defects earlier in development, potentially due to overexpression effects of RhoGEF2 or RhoGAP71E as part of the transgenic optogenetic tools. We therefore restricted our attention to embryos expressing low to moderate levels of optoGEF or optoGAP in this study.
Next, we analyzed how photoactivation with 488 nm blue-light influenced axis elongation in optoGEF and optoGAP embryos. Experiments were performed such that photoactivation of the germband was initiated at the onset of axis elongation, defined as t = 0, which reduced potential effects on other developmental events in the embryo. Under continuous, uniform photoactivation achieved by scanning 488 nm laser light over the apical surface of the germband during imaging, embryos expressing optoGEF elongated 1.28 ± 0.04-fold, and embryos expressing optoGAP elongated 1.27 ± 0.06-fold during the first 15 min of axis elongation, significantly less than control embryos expressing only the CIBN-pmGFP component of the optogenetic system (1.77 ± 0.08-fold) (p = 0.018 and p = 0.019, respectively) or dark control optoGEF and optoGAP embryos not exposed to blue light (p = 0.065 and p = 0.066, respectively) (Fig. 1 C). Consistent with the reduced tissue-level elongation, the number of oriented cell rearrangements per cell was reduced to 1.2 ± 0.2 in optoGEF embryos (p = 0.28) and 0.67 ± 0.22 in optoGAP embryos (p = 0.03), compared with 1.7 ± 0.3 rearrangements per cell in control embryos (Fig. 1 C; t = 15 min). These results indicate that rapid optogenetic recruitment of Rho1 regulators to the apical surface of the germband epithelium is sufficient to disrupt axis elongation, providing an opportunity to dissect how Rho1 activity and the patterns of actomyosin contractility it directs influence contractile cell behaviors during convergent extension.
Optogenetic control of Rho1 activity allows rapid manipulation of myosin patterns in the germband during axis elongation
Patterns of myosin localization and activity and the resulting contractile forces generated drive a wide array of cell behaviors that contribute to epithelial morphogenesis. For example, polarized actomyosin contractility, involving both the junctional and medial-apical pools of myosin, is required for cell intercalation during axis elongation (22,23,27,29,34,48). Whereas significant insight has been gained by correlating myosin patterns with cell and tissue behaviors during development, understanding how myosin patterns influence cell behaviors requires a direct perturbation approach. Drug injections do not allow fine spatial control and can be destructive to the embryo, and traditional genetic manipulations cannot always be easily or flexibly targeted to specific groups of cells or developmental events. To test whether optogenetic manipulation of Rho1 activity in the germband might overcome the limitations of traditional perturbation approaches, we analyzed myosin localization patterns in optoGEF and optoGAP embryos.
To analyze how recruitment of Rho1 regulators at the apical surface of the tissue influences the spatial and temporal dynamics of myosin, we performed time-lapse confocal imaging in optoGEF and optoGAP embryos expressing an mCherry-tagged myosin regulatory light chain (sqh) transgene. In the absence of activating blue light, myosin II was present in a planar-polarized pattern at cell junctions and in the medial-apical cortex in optoGEF and optoGAP embryos, similar to control embryos (Fig. 2, A–C). We next tested how blue-light illumination, beginning at the onset of axis elongation (t = 0) and continuing throughout the process, affected myosin localization. In control embryos expressing only the CIBN-pmGFP component of the system, myosin II was present at cell junctions and the medial-apical cortex of germband cells (Fig. 2, D–D’’). Just before the onset of axis elongation and before blue-light exposure, myosin began to accumulate more strongly at junctions between anterior and posterior cell neighbors (AP edges) than at junctions between dorsal and ventral cell neighbors (DV edges) (Fig. 2 D). During axis elongation and blue-light exposure, myosin continued to accumulate at AP edges, resulting in a planar-polarized pattern (Fig. 2, D’ and D’’), consistent with previous studies of myosin planar polarity in wild-type embryos (22, 23, 24,29, 30, 31, 32, 33).
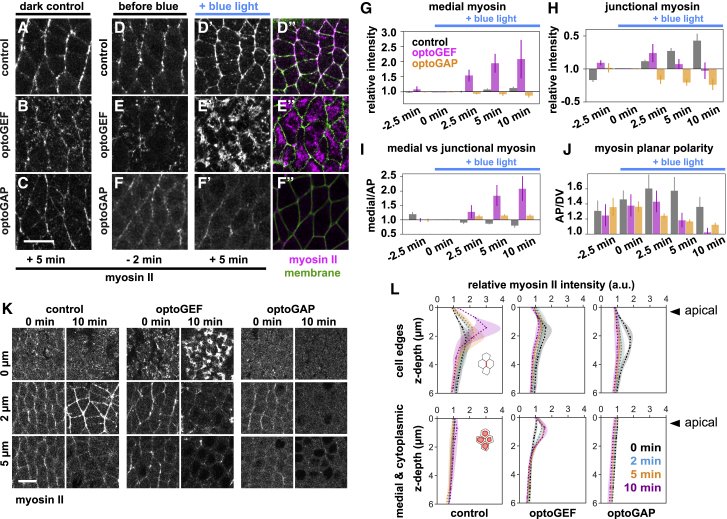
Figure 2.
Optogenetic manipulation of Rho1 signaling with optoGEF or optoGAP alters junctional and medial myosin II localization patterns and disrupts myosin planar polarity during axis elongation. (A–F) Stills from videos of epithelial germband cells during Drosophila body axis elongation showing myosin II localization at the apical side of cells. Maximal intensity projection of z-slices in the most apical 5 μm is shown. Myosin II (myosin) was visualized using an mCherry-tagged myosin regulatory light chain (sqh) transgene. (A–C) Dark control embryos without blue-light activation (imaging only with 561 nm light). Myosin II is present in a planar-polarized pattern, showing increased accumulation at “vertical” AP cell interfaces. (D–F’’) Embryos with 488 nm blue-light activation (imaging with 488 and 561 nm light). (D–F) Myosin localization 2 min before blue-light exposure. (D’–F’ and D’’–F’’) After 5 min of activating blue-light exposure at the apical surface of the germband, optoGEF embryos showed an overall increase in cortical myosin levels (E’ and E’’), and optoGAP embryos showed an overall decrease in cortical myosin levels (F’ and F’’) compared with control embryos (D’ and D’’). (D’’–F’’) myosin II (magenta), cell membrane (green). Anterior is to the left. Ventral is down. Scale bar, 10 μm. (G) Mean myosin intensity in the medial-apical domain over time relative to the value at t = 0. (H) Mean myosin intensity at vertical AP cell junctions over time relative to the value at t = 0. (I) Ratio of mean myosin intensities in the medial-apical domain compared with vertical AP junctions. Medial myosin accumulation relative to at AP junctions was increased in optoGEF compared with control or optoGAP embryos (2.09 ± 0.43 in optoGEF and 1.16 ± 0.05 in optoGAP, compared with 0.79 ± 0.07 in control embryos at t = 10 min; p = 0.01 and p = 0.26, respectively). (J) Ratio of mean myosin intensities at vertical AP junctions compared with horizontal DV junctions. Myosin planar polarity was disrupted in optoGEF and optoGAP embryos (1.04 ± 0.06 in optoGEF and 1.13 ± 0.02 in optoGAP, compared with 1.41 ± 0.12 in control embryos at t = 10 min; p = 0.04 and p = 0.1, respectively). (I and J) Ratios of mean medial to mean AP intensities or ratios of mean AP to mean DV intensities for each embryo at each time point were calculated. (G–J) Mean ± SE between embryos is shown; n = 3–4 embryos per condition. (K) Myosin localization at different z-planes along the apical-basal axis at 0 and 10 min of blue-light illumination, beginning from the onset of axis elongation. The most apical side of cells corresponds to z = 0 μm. Scale bar, 10 μm. (L) Myosin intensity profiles along the apical-basal axis at AP cell edges (top), which represent contacts between anterior and posterior cell neighbors, and in the medial and cytoplasmic domain (bottom) at 0, 2, 5, and 10 min of blue-light illumination, beginning from the onset of axis elongation. Mean ± SE between embryos is shown; n = 3 embryos per condition with 10 cells per embryo.
In contrast, myosin localization patterns changed dramatically in cells of optoGEF embryos immediately after blue-light exposure, indicating that these changes in myosin localization were induced by cell membrane accumulation of the Rho1 regulator RhoGEF2. In optoGEF embryos, myosin was initially localized at cell junctions before blue-light exposure (Fig. 2, E, G, and H). This myosin pattern was similar to that of control embryos (Fig. 2, D, G, and H), but with slightly reduced junctional levels. After blue-light exposure starting at t = 0 to recruit RhoGEF2 to the cell membrane, myosin rapidly accumulated at the medial-apical side of cells. The rapid increase in medial myosin in optoGEF embryos was accompanied by a decrease in junctional myosin levels (Fig. 2, E’, E’’, G, and H), producing a strong increase in the ratio of medial to junctional myosin and a decrease in myosin planar polarity within 3–5 min of blue-light illumination. At t = 10 min, the ratio of medial to junctional myosin increased to a value of 2.09 ± 0.43 (compared with 0.79 ± 0.07 in controls, p = 0.01) (Fig. 2 I) and myosin planar polarity decreased to a value of 1.04 ± 0.06 (compared with 1.41 ± 0.12 in controls, p = 0.04) (Fig. 2 J). The resulting myosin pattern is reminiscent of the radial pattern in apically constricting cells in the presumptive mesoderm of Drosophila embryos (67, 68, 69). These results demonstrate that acute activation of optoGEF across the apical side of the germband rapidly disrupts the planar-polarized pattern of junctional myosin localization organized by endogenous Rho1 signaling.
In optoGAP embryos, cortical myosin localization decreased after blue-light exposure, indicating that these changes in myosin localization were induced by cell membrane accumulation of the Rho1 regulator RhoGAP71E. In these embryos, we observed gradual decreases in both medial and junctional myosin (Fig. 2, F’, F’’, G, and H). This decrease in the cortical localization of myosin is consistent with the role of RhoGAPs in promoting the inactive state of Rho1. The changes in cortical myosin localization in optoGAP embryos were associated with a decrease in myosin planar polarity that was observable within 3 min and continuously decreased thereafter (1.13 ± 0.02 compared with 1.41 ± 0.12 in control embryos at t = 10 min, p = 0.1) (Fig. 2 J). These results demonstrate that acute activation of optoGAP across the apical side of the germband disrupts the levels of myosin at the cortex, including the planar-polarized junctional accumulation.
To test how the optogenetic perturbations influence myosin localization along the apical and lateral sides of cells, we analyzed myosin localization patterns at different z-positions in the confocal time-lapse videos. In control embryos, myosin became strongly enriched at cell contacts between anterior and posterior cell neighbors from 1.5 to 3 μm below the apical side of cells (Fig. 2, K and L), consistent with localization at adherens junctions. Myosin was also present in the medial-apical domain at the very apical side of the cells from 0 to 1 μm and was not detectable at high levels along the lateral sides of cells below junctions (Fig. 2, K and L). In optoGEF embryos, myosin became strongly enriched at the medial-apical side of cells and showed decreased junctional enrichment compared with controls, again with little detectable lateral accumulation (Fig. 2, K and L). In contrast, myosin localization was reduced both at cell edges and in the medial-apical domain of optoGAP embryos (Fig. 2, K and L).
Taken together, these findings demonstrate that activation of either optoGEF or optoGAP at the apical side of the germband, beginning at the onset of axis elongation, is sufficient to override the planar-polarized myosin patterns that are organized by endogenous Rho1 signaling in the germband, converting the planar-polarized junctional myosin pattern into a more medial-apical radial pattern in optoGEF or strongly reducing overall cortical myosin levels in optoGAP, over the timescale of a few minutes. In both optoGEF and optoGAP embryos, myosin planar polarity was significantly reduced, consistent with the strong reductions in cell rearrangements and tissue elongation in these embryos (Fig. 1 C). Such rapid, local, and noninvasive optogenetic perturbations provide a unique opportunity to study how myosin patterns affect contractile cell behavior during convergent extension.
Optogenetic manipulation of Rho1 activity is associated with changes in apical cell area
Actomyosin activity within cells is thought to drive a wide variety of cell behaviors. It is known that the planar-polarized myosin pattern in the germband is required for oriented cell rearrangements that drive axis elongation, and our results of reduced myosin planar polarity, cell rearrangement, and tissue elongation in optoGEF and optoGAP embryos (see Figs. 1 C and 2 J) are consistent with this. Yet, it remains less clear how myosin patterns in the germband influence other aspects of contractile cell behaviors, such as cell shape and apical cell area. To address this, we analyzed how optogenetically perturbed myosin patterns in the germband of optoGEF and optoGAP embryos influence cell shape and apical cell area.
First, we analyzed cell stretching along the AP axis as a metric for cell shape changes that contribute to tissue-level elongation (24,26,28). In control embryos, the mean horizontal length of cells along the AP axis increased by 12 ± 4% at t = 10 min relative to t = 0 min (Fig. 3, A and B), similar to previous studies of wild-type embryos (24,27,28,45). Because cell stretching along the AP axis is thought to be associated with external forces from posterior midgut invagination and is also thought to depend on cell rearrangements in the germband that can relax this stretch (26,28,45,70), we hypothesized that the reduced cell rearrangements in optoGEF and optoGAP embryos might contribute to increased stretching. We found that the effects of optogenetic perturbations on cell length changes were not statistically significant in photoactivated optoGEF (p = 0.67) or optoGAP (p = 0.87) compared with control embryos (Fig. 3, A and B). These results suggest that, although changes in myosin patterns have the potential to impact cell stretching during axis elongation, the effects in these studies were not strong.
Next, we analyzed cell areas in the germband tissue because medial-apical actomyosin contractility plays key roles in apical constriction in a number of contexts. For example, during ventral furrow invagination, a group of cells exhibiting strong medial-apical myosin contractility constrict their apical sides, leading to apical cell area reduction and tissue invagination (68). In prior studies by other groups, apical constriction and invagination have been reproduced via local optogenetic activation of Rho1 in small groups of cells in the early Drosophila embryo (16,20). Naively, one might expect similar behavior here, with reduced apical cell area in the photoactivated germband of optoGEF embryos. However, when we activated Rho1 across the ventrolateral region of the germband in optoGEF embryos, we did not observe uniform apical cell area reductions across the tissue and, instead, observed the presence of a small fraction of cells with aberrantly small apical cell areas (Fig. 3 A, red arrows).
To quantify this heterogeneity, we measured the apical areas of cells within the tissue at different time points during axis elongation. At the beginning of axis elongation (t = 0), control embryos display a normal distribution of cell areas (Fig. 3 C). Over time, the distribution widens as cells undergo cell rearrangements during axis elongation. In contrast, although optoGEF embryos start out with a cell area distribution similar to controls at t = 0, we observe the appearance of a population of cells with drastic reductions in area, whereas other cells display a modest increase in area at 5 and 10 min of blue-light illumination (Fig. 3 C). In optoGAP embryos, we observed a cell area distribution in which some cells exhibit increased area relative to the mean during axis elongation (Fig. 3 C).
To directly compare cell area heterogeneity across these different perturbations, we quantified the coefficient of variation in cell area and plotted this as a function of time (Fig. 3 D). In control embryos, the variation in area among cells started out at a low level of 0.16 at t = 0 and increased by 1.9 ± 0.16-fold during the first 10 min of axis elongation. Cell areas in optoGAP embryos displayed a similar coefficient of variation over time as control embryos (1.5 ± 0.07-fold change from t = 0 to 10 min, p = 0.27). In contrast, cells in optoGEF embryos showed a greater increase in cell area variation starting around 3 min after tool activation, reaching a 2.81 ± 0.42-fold increase at t = 10 min (p = 0.03).
Next, we wanted to investigate the origins of the heterogeneous cell area reductions in optoGEF embryos. In one model, the cell population with aberrantly small areas in optoGEF embryos might represent cells that are progressively shrinking or ratcheting down their apical area over time. Alternatively, the small cell population might reflect cells that are fluctuating or pulsing but not progressively changing apical area and just happen to have a small area at the time point captured. To distinguish between these possibilities, we observed individual cells over time in control, optoGEF, and optoGAP embryos (Fig. 3 E). In control embryos, the apical areas of individual cells were maintained over time during axis elongation (Fig. 3 E). Similar behavior was observed in photoactivated optoGAP embryos (Fig. 3 E). In contrast, we observed distinct cell behaviors in photoactivated optoGEF embryos. One group of cells tended to progressively shrink apical areas over time, whereas another group tended to increase areas over time. In other words, the cells with aberrantly small areas in optoGEF embryos did not just transiently display small apical areas but were instead progressively reducing their apical areas over time. These observations point toward the conclusion that the heterogeneity in cell areas in optoGEF embryos arises because of progressive apical constriction of some cells and expansion of other cells in the population. Taken together, these results demonstrate that rapid optogenetic perturbation of Rho1 activity alters the apical areas of epithelial cells within the germband.
Cell area heterogeneity is linked to tissue-level patterns of medial and junctional myosin
To gain insight into these heterogeneities in contractile cell behaviors and link them to tissue-level myosin patterns, we quantified the correlations between cell area, junctional myosin, and medial myosin over time in control, optoGEF, and optoGAP embryos (Fig. 4). We reasoned that one potential mechanism for heterogeneity in cell area might be the overall balance between junctional and medial myosin, which might help to stabilize apical cell area. Alternatively, cell area heterogeneity might be linked to heterogeneity in myosin localization between cells within the tissue.
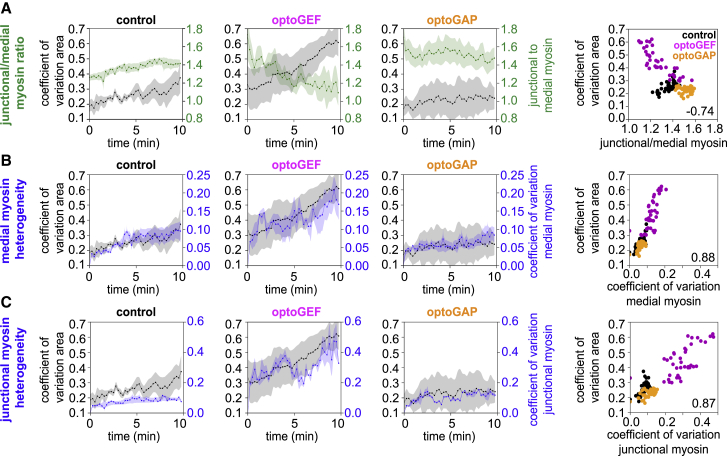
Figure 4.
Heterogeneity in apical cell area is associated with tissue-level myosin patterns during convergent extension. Relationships between the coefficient of variation in apical cell area and various metrics of tissue-level myosin patterns for germband cells in control, optoGEF, and optoGAP embryos during 10 min of blue-light illumination of the apical surface of the tissue during axis elongation. (A) (Left) Cell area coefficient of variation (black) and ratio of junctional to medial myosin intensities (green) over time. (Right) Cell area coefficient of variation versus the ratio of junctional to medial myosin intensities for control, optoGEF, and optoGAP embryos is shown. Each data point represents the mean between embryos at an individual time point. Value in bottom right corner of plot indicates correlation coefficient. (B) (Left) Cell area coefficient of variation (black) and medial myosin intensity coefficient of variation (blue) over time. (Right) Cell area coefficient of variation versus medial myosin coefficient of variation is shown. (C) (Left) Cell area coefficient of variation (black) and junctional myosin intensity coefficient of variation (blue) over time. (Right) Cell area coefficient of variation versus junctional myosin coefficient of variation is shown. Mean ± SE between embryos is shown; n = 12 cells per embryo from three to four embryos per genotype. See also Fig. S1.
First, we tested how the cell area coefficient of variation was correlated with overall levels of junctional and medial myosin. We find that the increase in cell area heterogeneity in optoGEF embryos correlates with the decrease in junctional myosin and increase in medial myosin after optogenetic perturbation (Figs. 4 A and S1). In contrast, control and optoGAP embryos maintain low area heterogeneity, which correlates with maintaining high levels of junctional relative to medial myosin, even when overall levels of myosin are decreased in optoGAP embryos (Figs. 4 A and S1). Across all groups, cell area heterogeneity was inversely correlated with the ratio of junctional to medial myosin intensity (correlation coefficient = −0.74 for data from all groups). These results are consistent with a role for high levels of junctional myosin relative to medial myosin in cell area maintenance.
Next, we tested if the heterogeneity in cell area was associated with heterogeneity in myosin localization patterns between cells. We quantified how the cell area coefficient of variation was correlated with the coefficient of variation of junctional or of medial myosin levels between cells. Interestingly, cell area heterogeneity was correlated with both medial myosin heterogeneity across the tissue (correlation coefficient = 0.88 for data from all groups) and junctional myosin heterogeneity across the tissue (correlation coefficient = 0.87) in control, optoGEF, and optoGAP embryos (Fig. 4, B and C). These results are consistent with the idea that the observed cell area heterogeneity in optoGEF embryos is linked with the more heterogeneous accumulation of myosin in cells across the germband.
Taken together, these results suggest that maintaining a balance between junctional and medial myosin contributes to apical cell area maintenance. In addition, the uniformity of myosin levels between cells in the tissue also might contribute to cell area maintenance, with increased heterogeneity in myosin levels between cells correlated with heterogeneity in cell areas. Thus, optogenetic manipulation to activate Rho1 signaling across the germband, which is associated with increased medial and decreased junctional myosin, leads to heterogeneous changes in myosin levels and heterogeneous decreases in cell area. In contrast, inactivating Rho1 signaling across the germband, which is associated with homogeneous decreases in medial and junctional myosin, leads to less prominent and less heterogeneous cell area changes.
Apical cell area changes are linked with medial myosin changes in single cells over time
To gain insight into these contractile cell behaviors at the single-cell level, we tracked and quantified cell area and medial myosin in small groups of individual cells within the germband of photoactivated control, optoGEF, and optoGAP embryos (Fig. 5).
In control embryos, cells maintained relatively constant apical areas, with some fluctuations over time during axis elongation (Figs. 5, A and D and S2). This was associated with relatively constant medial myosin levels over this time period. Similar behavior was observed in photoactivated optoGAP embryos, although the area fluctuations appeared less prominent, and medial myosin levels decreased over time (Figs. 5, C and D and S2). In contrast, in photoactivated optoGEF embryos, some cells in a group progressively decreased in area over time, whereas neighboring cells progressively increased their apical area, and the pulsed cell area fluctuations were more prominent (Figs. 5, B and D and S2). This was associated with more dramatic changes in medial myosin levels within cells. On average, this resulted in a high variation between cells in the optoGEF group compared with controls at the 10 min time point.
Next, we tested whether apical area is correlated with medial myosin levels in individual cells over the course of the 10 min experiment. Although there was significant cell-to-cell variability, on average, cell area was negatively correlated with medial myosin levels in optoGEF embryos (p = 0.006 compared with null hypothesis) (Figs. 5, D and E and S2). The correlation was not statistically significant in control or optoGAP embryos. Taken together, these results demonstrate that optogenetic perturbation of Rho1 signaling leads to changes in medial myosin levels that are linked with apical cell area changes over the 10 minute timescale.
Pulsatile cell area fluctuations are tuned by optogenetic perturbation of Rho1 activity
To further investigate how optogenetic Rho1 perturbations influence contractile cell behaviors, we quantified the effects on short timescale pulsatile cell area fluctuations. Although short timescale fluctuations have been reported in the germband during axis elongation (34,37,71), their role in the process remains unclear. We wondered whether the short timescale fluctuations in cell area that we observed in control and optoGEF embryos were correlated with myosin dynamics.
To test this, we first measured changes in cell area and in medial myosin between successive 15 s time points in confocal time-lapse videos (Fig. 6, A–C). In control embryos, the peaks in cell area change pulses had an amplitude of 5.0 ± 0.1 μm2 and period of 2.57 ± 0.11 min (Fig. 6, A, D, E, and F), comparable with prior reports (37,71). The amplitude of changes in apical cell area in optoGEF embryos was increased slightly to 6.0 ± 0.2 μm2 (p = 0.06). In contrast, large pulses were nearly absent in optoGAP embryos (Fig. 6, C–E). The time period between pulses was not significantly altered in optoGEF (2.51 ± 0.1 min) or optoGAP (2.77 ± 0.2 min) compared with control embryos (Fig. 6 F).
Next, we tested whether the cell area fluctuations were correlated with the medial myosin fluctuations. We found a modest temporal correlation in control embryos over the 10 min period analyzed at the beginning of axis elongation (median correlation coefficient = −0.28, p = 0.02 compared with null hypothesis) (Fig. 6, G and H). This is somewhat weaker than correlations between cell area and medial myosin changes observed at later stages of axis elongation, when medial myosin accumulation was found to precede cell area changes by several seconds (37,71); this difference is potentially explained by the higher levels of medial myosin at later stages of axis elongation. The correlations between cell area changes and medial myosin fluctuations were increased in optoGEF embryos (median correlation coefficient = −0.37, p = 0.03) and decreased in optoGAP (median correlation coefficient = 0.125, p = 0.03) compared with controls (Fig. 6, G and H). This is consistent with trends observed in previous studies in wild-type embryos later in axis elongation (37,71) and in optogenetically induced actomyosin contractility mimicking furrow formation (16) that report correlations between medial myosin accumulation and cell area changes. Taken together, these results demonstrate that cell area fluctuations are tuned by Rho1 activity in the germband, with pulse amplitudes increased in optoGEF compared with control embryos.
Discussion
Patterns of actomyosin contractility influence multiple aspects of cell behavior within tissues that contribute to tissue-level morphogenesis. Here, we used optogenetic tools to activate or deactivate Rho1 signaling across the apical side of the germband epithelium during Drosophila axis elongation to manipulate myosin patterns and analyze the effects on cell behaviors within the tissue. We demonstrate the ability to rapidly override the endogenous planar-polarized myosin pattern with both an optoGEF and an optoGAP tool. In optoGEF embryos, the planar-polarized pattern of myosin localization at cell junctions is transformed into a radial pattern at the medial-apical cortex of cells after recruitment of RhoGEF2 to the apical cell membrane. In optoGAP embryos, recruitment of RhoGAP71E results in a decrease in myosin accumulation at the cell cortex in both the junctional and medial-apical domains, abolishing the endogenous myosin pattern. Both optogenetic perturbations resulted in a loss of myosin planar polarity and strong reductions in cell intercalation and tissue elongation. However, optoGEF and optoGAP had distinct effects on cell shapes and pulsatile cell area fluctuations that can contribute to tissue behavior during convergent extension. Whereas depletion of cortical myosin in optoGAP embryos was associated with a reduction in apical cell area fluctuations, medial myosin accumulation in optoGEF embryos led to enhanced apical area fluctuations and heterogeneous decreases in apical cell area.
For this study, we developed two CRY2/CIBN-based optogenetic tools to manipulate actomyosin in Drosophila: optoGEF to activate and optoGAP to deactivate Rho1 signaling. To our knowledge, a RhoGAP-based optogenetic tool to manipulate actomyosin contractility in Drosophila has not been previously described. Related optogenetic approaches based on RhoGEFs using the DHPH catalytic domains of RhoGEF2 in a CRY2/CIBN system (16, 17, 18) or the catalytic DH domain of the LARG GEF guanine nucleotide exchange factor in a LOV-domain-based system (20) have previously been used to manipulate Rho1 activity in the Drosophila embryo. The optoGEF tool described here is distinct from these prior tools in that it includes the full RhoGEF2 protein, instead of only the catalytic domains, and so has the potential to recapitulate additional aspects of RhoGEF2 function. Prior studies using RhoGEF-based optogenetic tools focused on manipulating Rho1 activity and actomyosin contractility during cellularization and ventral furrow formation (16, 17, 18,20), revealing the potential to reconstitute aspects of morphogenesis via ectopic, local Rho1 activation in the epithelium. In contrast, in this study, we perturb an existing myosin localization and activity pattern in the epithelium at a stage of development when myosin is strongly planar polarized, and we analyze how these perturbations influence cell behaviors during axis elongation. Consistent with prior observations (16,20), we find that optogenetic activation of Rho1 on the apical side of the Drosophila epithelium leads to rapid accumulation of myosin in the medial-apical domain of cells. We observe pulsatile apical cell area fluctuations in optoGEF embryos, consistent with pulsatile cell behaviors observed in optogenetic studies using the RhoGEF2 DHPH catalytic domains in the early embryo (16). The more heterogenous cell area reductions and absence of tissue folding that we observe in the germband of optoGEF embryos compared with prior optogenetic studies conducted in other stages and regions of the embryo may reflect a number of factors: differing gene expression patterns at these developmental stages, existence of anisotropies associated with planar polarity in the germband, size of the optogenetic activation region, mechanical properties of the germband tissue, mechanical or geometrical constraints, or differences in the optogenetic tools themselves.
The effects of optoGEF and optoGAP perturbations on myosin planar polarity in the germband are rapid. During the first 2.5 min, myosin planar polarity reduction occurs faster in optoGAP than in optoGEF embryos, but optoGEF embryos show a greater final reduction in planar polarity than optoGAP. These distinct dynamics may reflect the molecular mechanisms and timescales for Rho1 activation versus inactivation, Rho-kinase activation versus inactivation, and myosin regulatory light chain phosphorylation versus dephosphorylation. In addition, Rho1 and RhoGEF2 are known to play roles in regulating the actin cytoskeleton (72), which could contribute to the observed changes in myosin localization patterns and cell behaviors. Interestingly, whereas myosin planar polarity was more strongly reduced in optoGEF compared with optoGAP embryos, we observed higher rates of cell rearrangements in the optoGEF case, reflecting that other aspects of the myosin localization pattern may influence contractile cell behaviors that contribute to axis elongation. One possibility is that the enhanced medial myosin and pulsatile apical cell area fluctuations in optoGEF embryos may promote cell rearrangement and intercalation (discussed below).
The medial-apical myosin pattern in optoGEF embryos was similar to that observed in the germband of embryos overexpressing RhoGEF2, expressing constitutively active concertina, or treated to depolymerize microtubules (48,50,66). Interestingly, the medial myosin pattern in optoGEF embryos was distinct from the more junctional myosin pattern in embryos expressing a phosphomimetic myosin regulatory light chain locked in to the “on” state (30), possibly suggesting additional effects of the optoGEF tool on the actomyosin meshwork beyond increasing myosin activity. Alternatively, the difference in myosin localization between optoGEF and phosphomimetic myosin embryos might reflect depletion of myosin from junctions due to a limited myosin pool or changes in the balance between membrane localization of RhoGEF2 and Cysts/Dp114RhoGEF, which are thought to have distinct roles in regulating medial and junctional myosin in the germband (49,50). The condensed cluster of medial myosin observed in optoGEF embryos is reminiscent of the myosin in apically constricting cells under isotropic tension in the invaginating posterior midgut of wild-type embryos and in the presumptive mesoderm of embryos with disrupted tension anisotropy (67,73), which raises the possibility that the optogenetically induced change in myosin pattern alters the tension distribution in the tissue. Furthermore, myosin recruitment to the cell cortex can be mechanosensitive (33), raising the possibility of complex effects of the optogenetic perturbations both within and outside of the germband. Future studies to quantify the mechanical tension in the tissue will be needed to directly link changes in myosin localization with changes in mechanical tension in the germband.
During axis elongation, accumulation of actomyosin at the medial-apical domain of germband cells is associated with fluctuations in apical cell area in wild-type embryos (37,71). We found that the optogenetic perturbations tuned these pulsatile cell area changes. In optoGEF embryos, the accumulation of myosin in the medial-apical domain and formation of a radial myosin pattern was associated with increased magnitude of area fluctuations, enhanced net area changes, and high cell area heterogeneity compared with control embryos. The increase in cell area heterogeneity in the germband contrasts with the more uniform cell area decreases after optogenetic activation of Rho1 in localized regions of tissue in the early embryo (16,20). One possible explanation is that a tissue-wide pattern of medial myosin accumulation has different effects on apical cell area than a local pattern, potentially due to the size of the contractile region and the geometrical and mechanical constraints in the embryo. For example, if all cells in a large region of the germband tissue try to contract at the same time, perhaps the tissue-scale deformation cannot be accommodated by surrounding tissue, leading to some activated cells contracting and others expanding. Alternatively, the heterogeneity in cell areas may be influenced by junctional and medial myosin levels and the heterogeneity in myosin levels between cells in the tissue. This may reflect cell-to-cell variability in expression of the optogenetic tools or changes to the actin cytoskeleton or cell-cell adhesions that contribute to apical cell area maintenance. Aspects of the endogenous planar polarity pattern in the tissue may contribute to this phenotype. Future studies that locally activate Rho1 signaling in small groups of cells in the germband will be essential to understanding the effects of activation region size in a planar-polarized tissue.
Whereas a population of cells in optoGEF embryos displayed progressive cell area decreases, other cells displayed a combination of transient periods of area reduction and expansion with no stabilization of cell area changes, similar to in bcd nos tsl mutant embryos that have high levels of medial relative to junctional myosin (37). The absence of a net reduction in apical cell area over time, despite increased medial myosin, might be caused by the concomitant reduction of junctional myosin in these embryos, which could disrupt tension stabilization among groups of cells. Consistent with this notion, progressive apical constriction was observed in ventrolateral germband cells in JAK/STAT mutant embryos, which have high levels of medial-apical myosin but normal levels of junctional myosin (36). A role for junctional myosin in stabilization of cell area was also evident in optoGAP embryos, in which the reduction in junctional and medial myosin generated a subtle area relaxation in a considerable fraction of cells. In addition, the observed changes in myosin localization in optoGEF and optoGAP embryos could be coupled with changes in the actin cytoskeleton or cell adhesions, which might contribute to these phenotypes. Future studies analyzing the effects of these optogenetic perturbations on other cytoskeletal and adhesive proteins will be needed to distinguish between these possibilities.
Interestingly, the amplitudes of apical cell area fluctuations during germband extension in optoGEF and optoGAP embryos, in which myosin planar polarity is disrupted, correlate with the rates of cell rearrangement in the germband. As myosin planar polarity is thought to generate the anisotropic internal tensions that drive cell rearrangement, a complete lack of myosin planar polarity would be predicted to block cell rearrangements in the germband. Although myosin planar polarity is nearly abolished in both optoGEF and optoGAP embryos, cell rearrangements are less severely impacted in optoGEF embryos. One explanation is that small remaining differences in myosin localization and activity at junctions is not detectable in imaging but still sufficient to drive rearrangements. Alternatively, mechanisms that are independent of junctional myosin planar polarity might be contributing to cell rearrangement in this context (74). One potential mechanism is that active myosin-associated cell area fluctuations might help promote cell rearrangements. Consistent with this notion, recent theoretical and experimental work highlights that cells must overcome a physical energy barrier associated with cell shape changes to proceed through a cell rearrangement (43, 44, 45). Active fluctuations in tension and shape can help to overcome these energy barriers and promote the ability of a tissue to remodel and flow (tissue fluidity). Indeed, recent experimental studies point toward exactly such a role for active fluctuations in promoting fluidity within tissues (40,41). Future studies of the biophysics of cell rearrangements in the germband will be needed to explore further how active fluctuations might contribute to oriented cell rearrangements in both wild-type and mutant embryos.
In this study, optogenetic perturbations to Rho1 activity in the germband epithelium allowed us to link cellular junctional and medial myosin patterns to distinct aspects of cell behavior that contribute to convergent extension. Indeed, the changes in myosin patterns and resulting cell behaviors in response to tuning Rho1 signaling with optoGEF or optoGAP are consistent with prior studies demonstrating that quantitative levels of G protein activity can tune distinct morphogenetic behaviors (48). However, the rapid changes in the myosin pattern in the germband of optoGEF embryos (from the endogenous planar-polarized pattern to one reminiscent of the radial patterns present in invaginating cells of the presumptive mesoderm) were not sufficient to completely transform cell behaviors to recapitulate coordinated apical constriction and tissue invagination. This points to the existence of additional mechanisms beyond the myosin pattern that are required to ectopically recapitulate morphogenetic movements at other stages of development. Going forward, it will be interesting to further explore how internal biochemical regulation couples with mechanical cues and constraints to give rise to the dynamic myosin patterns that orchestrate diverse cell behaviors and morphogenetic events. In addition, it will be important to investigate how anisotropic patterns of junctional actomyosin couple with active fluctuations driven by the apical actomyosin cytoskeleton to promote cell rearrangements and epithelial tissue fluidity during development. The optogenetic tools and approaches we developed here will be valuable in these investigations.
Author contributions
R.M.H.-P. designed research, performed research, analyzed data, and wrote the manuscript. C.C. analyzed data. C.A. analyzed data. A.L. analyzed data. K.E.K. designed research, analyzed data, and wrote the manuscript.
Acknowledgments
We thank Stefano De Renzis for the UASp>CIBN::pmGFP fly stock, Jennifer Zallen and Frederik Wirtz-Peitz for the UASp-attB destination vectors, the Bloomington Drosophila Stock Center for fly stocks, BACPAC Genomics at Children’s Hospital Oakland Research Institute (now BACPAC Genomics) for BAC clones, and members of the Kasza Lab for helpful discussion and comments on the manuscript.
This work was supported by a National Science Foundation Civil, Mechanical, and Manufacturing Innovation Grant 1751841 (to K.E.K.). K.E.K. holds a Career Award at the Scientific Interface from the Burroughs Wellcome Fund, a Clare Boothe Luce Professorship, and a Packard Fellowship.
Editor: Jeremiah Zartman.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2021.06.041.
Supporting material
References
- 1.Lecuit T., Lenne P.-F. Cell surface mechanics and the control of cell shape, tissue patterns and morphogenesis. Nat. Rev. Mol. Cell Biol. 2007;8:633–644. doi: 10.1038/nrm2222. [DOI] [PubMed] [Google Scholar]
- 2.Kasza K.E., Zallen J.A. Dynamics and regulation of contractile actin-myosin networks in morphogenesis. Curr. Opin. Cell Biol. 2011;23:30–38. doi: 10.1016/j.ceb.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heisenberg C.P., Bellaïche Y. Forces in tissue morphogenesis and patterning. Cell. 2013;153:948–962. doi: 10.1016/j.cell.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 4.Herrera-Perez R.M., Kasza K.E. Biophysical control of the cell rearrangements and cell shape changes that build epithelial tissues. Curr. Opin. Genet. Dev. 2018;51:88–95. doi: 10.1016/j.gde.2018.07.005. [DOI] [PubMed] [Google Scholar]
- 5.Martin A.C. Pulsation and stabilization: contractile forces that underlie morphogenesis. Dev. Biol. 2010;341:114–125. doi: 10.1016/j.ydbio.2009.10.031. [DOI] [PubMed] [Google Scholar]
- 6.Herrera-Perez R.M., Kasza K.E. Manipulating the patterns of mechanical forces that shape multicellular tissues. Physiology (Bethesda) 2019;34:381–391. doi: 10.1152/physiol.00018.2019. [DOI] [PubMed] [Google Scholar]
- 7.Kennedy M.J., Hughes R.M., Tucker C.L. Rapid blue-light-mediated induction of protein interactions in living cells. Nat. Methods. 2010;7:973–975. doi: 10.1038/nmeth.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guglielmi G., Falk H.J., De Renzis S. Optogenetic control of protein function: from intracellular processes to tissue morphogenesis. Trends Cell Biol. 2016;26:864–874. doi: 10.1016/j.tcb.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Q., Tucker C.L. Engineering genetically-encoded tools for optogenetic control of protein activity. Curr. Opin. Chem. Biol. 2017;40:17–23. doi: 10.1016/j.cbpa.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson H.E., Toettcher J.E. Illuminating developmental biology with cellular optogenetics. Curr. Opin. Biotechnol. 2018;52:42–48. doi: 10.1016/j.copbio.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Idevall-Hagren O., Dickson E.J., De Camilli P. Optogenetic control of phosphoinositide metabolism. Proc. Natl. Acad. Sci. USA. 2012;109:E2316–E2323. doi: 10.1073/pnas.1211305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guglielmi G., Barry J.D., De Renzis S. An optogenetic method to modulate cell contractility during tissue morphogenesis. Dev. Cell. 2015;35:646–660. doi: 10.1016/j.devcel.2015.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wagner E., Glotzer M. Local RhoA activation induces cytokinetic furrows independent of spindle position and cell cycle stage. J. Cell Biol. 2016;213:641–649. doi: 10.1083/jcb.201603025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oakes P.W., Wagner E., Gardel M.L. Optogenetic control of RhoA reveals zyxin-mediated elasticity of stress fibres. Nat. Commun. 2017;8:15817. doi: 10.1038/ncomms15817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Valon L., Marín-Llauradó A., Trepat X. Optogenetic control of cellular forces and mechanotransduction. Nat. Commun. 2017;8:14396. doi: 10.1038/ncomms14396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Izquierdo E., Quinkler T., De Renzis S. Guided morphogenesis through optogenetic activation of Rho signalling during early Drosophila embryogenesis. Nat. Commun. 2018;9:2366. doi: 10.1038/s41467-018-04754-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krueger D., Tardivo P., De Renzis S. Downregulation of basal myosin-II is required for cell shape changes and tissue invagination. EMBO J. 2018;37:e100170. doi: 10.15252/embj.2018100170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krueger D., Quinkler T., De Renzis S. Cross-linker-mediated regulation of actin network organization controls tissue morphogenesis. J. Cell Biol. 2019;218:2743–2761. doi: 10.1083/jcb.201811127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cavanaugh K.E., Staddon M.F., Gardel M.L. RhoA mediates epithelial cell shape changes via mechanosensitive endocytosis. Dev. Cell. 2020;52:152–166.e5. doi: 10.1016/j.devcel.2019.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rich A., Fehon R.G., Glotzer M. Rho1 activation recapitulates early gastrulation events in the ventral, but not dorsal, epithelium of Drosophila embryos. eLife. 2020;9:e56893. doi: 10.7554/eLife.56893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Irvine K.D., Wieschaus E. Cell intercalation during Drosophila germband extension and its regulation by pair-rule segmentation genes. Development. 1994;120:827–841. doi: 10.1242/dev.120.4.827. [DOI] [PubMed] [Google Scholar]
- 22.Bertet C., Sulak L., Lecuit T. Myosin-dependent junction remodelling controls planar cell intercalation and axis elongation. Nature. 2004;429:667–671. doi: 10.1038/nature02590. [DOI] [PubMed] [Google Scholar]
- 23.Blankenship J.T., Backovic S.T., Zallen J.A. Multicellular rosette formation links planar cell polarity to tissue morphogenesis. Dev. Cell. 2006;11:459–470. doi: 10.1016/j.devcel.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 24.Farrell D.L., Weitz O., Zallen J.A. SEGGA: a toolset for rapid automated analysis of epithelial cell polarity and dynamics. Development. 2017;144:1725–1734. doi: 10.1242/dev.146837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kasza K.E., Supriyatno S., Zallen J.A. Cellular defects resulting from disease-related myosin II mutations in Drosophila. Proc. Natl. Acad. Sci. USA. 2019;116:22205–22211. doi: 10.1073/pnas.1909227116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Butler L.C., Blanchard G.B., Sanson B. Cell shape changes indicate a role for extrinsic tensile forces in Drosophila germ-band extension. Nat. Cell Biol. 2009;11:859–864. doi: 10.1038/ncb1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collinet C., Rauzi M., Lecuit T. Local and tissue-scale forces drive oriented junction growth during tissue extension. Nat. Cell Biol. 2015;17:1247–1258. doi: 10.1038/ncb3226. [DOI] [PubMed] [Google Scholar]
- 28.Lye C.M., Blanchard G.B., Sanson B. Mechanical coupling between endoderm invagination and axis extension in Drosophila. PLoS Biol. 2015;13:e1002292. doi: 10.1371/journal.pbio.1002292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zallen J.A., Wieschaus E. Patterned gene expression directs bipolar planar polarity in Drosophila. Dev. Cell. 2004;6:343–355. doi: 10.1016/s1534-5807(04)00060-7. [DOI] [PubMed] [Google Scholar]
- 30.Kasza K.E., Farrell D.L., Zallen J.A. Spatiotemporal control of epithelial remodeling by regulated myosin phosphorylation. Proc. Natl. Acad. Sci. USA. 2014;111:11732–11737. doi: 10.1073/pnas.1400520111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tetley R.J., Blanchard G.B., Sanson B. Unipolar distributions of junctional Myosin II identify cell stripe boundaries that drive cell intercalation throughout Drosophila axis extension. eLife. 2016;5:e12094. doi: 10.7554/eLife.12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rauzi M., Verant P., Lenne P.-F. Nature and anisotropy of cortical forces orienting Drosophila tissue morphogenesis. Nat. Cell Biol. 2008;10:1401–1410. doi: 10.1038/ncb1798. [DOI] [PubMed] [Google Scholar]
- 33.Fernandez-Gonzalez R., Simoes S., Zallen J.A. Myosin II dynamics are regulated by tension in intercalating cells. Dev. Cell. 2009;17:736–743. doi: 10.1016/j.devcel.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rauzi M., Lenne P.-F., Lecuit T. Planar polarized actomyosin contractile flows control epithelial junction remodelling. Nature. 2010;468:1110–1114. doi: 10.1038/nature09566. [DOI] [PubMed] [Google Scholar]
- 35.Munjal A., Philippe J.M., Lecuit T. A self-organized biomechanical network drives shape changes during tissue morphogenesis. Nature. 2015;524:351–355. doi: 10.1038/nature14603. [DOI] [PubMed] [Google Scholar]
- 36.Bertet C., Rauzi M., Lecuit T. Repression of Wasp by JAK/STAT signalling inhibits medial actomyosin network assembly and apical cell constriction in intercalating epithelial cells. Development. 2009;136:4199–4212. doi: 10.1242/dev.040402. [DOI] [PubMed] [Google Scholar]
- 37.Fernandez-Gonzalez R., Zallen J.A. Oscillatory behaviors and hierarchical assembly of contractile structures in intercalating cells. Phys. Biol. 2011;8:045005. doi: 10.1088/1478-3975/8/4/045005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levayer R., Pelissier-Monier A., Lecuit T. Spatial regulation of Dia and Myosin-II by RhoGEF2 controls initiation of E-cadherin endocytosis during epithelial morphogenesis. Nat. Cell Biol. 2011;13:529–540. doi: 10.1038/ncb2224. [DOI] [PubMed] [Google Scholar]
- 39.Yu J.C., Fernandez-Gonzalez R. Local mechanical forces promote polarized junctional assembly and axis elongation in Drosophila. eLife. 2016;5:1–15. doi: 10.7554/eLife.10757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krajnc M. Solid-fluid transition and cell sorting in epithelia with junctional tension fluctuations. Soft Matter. 2020;16:3209–3215. doi: 10.1039/c9sm02310k. [DOI] [PubMed] [Google Scholar]
- 41.Kim S., Pochitaloff M., Campàs O. Embryonic tissues as active foams. Nat. Phys. 2021;17:859–866. doi: 10.1038/s41567-021-01215-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Farhadifar R., Röper J.C., Jülicher F. The influence of cell mechanics, cell-cell interactions, and proliferation on epithelial packing. Curr. Biol. 2007;17:2095–2104. doi: 10.1016/j.cub.2007.11.049. [DOI] [PubMed] [Google Scholar]
- 43.Bi D., Yang X., Manning M.L. Motility-driven glass and jamming transitions in biological tissues. Phys. Rev. X. 2016;6:21011. doi: 10.1103/PhysRevX.6.021011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park J.-A., Kim J.H., Fredberg J.J. Unjamming and cell shape in the asthmatic airway epithelium. Nat. Mater. 2015;14:1040–1048. doi: 10.1038/nmat4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang X., Merkel M., Kasza K.E. Anisotropy links cell shapes to tissue flow during convergent extension. Proc. Natl. Acad. Sci. USA. 2020;117:13541–13551. doi: 10.1073/pnas.1916418117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simões S., Blankenship J.T., Zallen J.A. Rho-kinase directs Bazooka/Par-3 planar polarity during Drosophila axis elongation. Dev. Cell. 2010;19:377–388. doi: 10.1016/j.devcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simões S., Mainieri A., Zallen J.A. Rho GTPase and Shroom direct planar polarized actomyosin contractility during convergent extension. J. Cell Biol. 2014;204:575–589. doi: 10.1083/jcb.201307070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kerridge S., Munjal A., Lecuit T. Modular activation of Rho1 by GPCR signalling imparts polarized myosin II activation during morphogenesis. Nat. Cell Biol. 2016;18:261–270. doi: 10.1038/ncb3302. [DOI] [PubMed] [Google Scholar]
- 49.Silver J.T., Wirtz-Peitz F., Tepass U. Apical polarity proteins recruit the RhoGEF Cysts to promote junctional myosin assembly. J. Cell Biol. 2019;218:3397–3414. doi: 10.1083/jcb.201807106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garcia De Las Bayonas A., Philippe J.-M., Lecuit T. Distinct RhoGEFs activate apical and junctional contractility under control of G proteins during epithelial morphogenesis. Curr. Biol. 2019;29:3370–3385.e7. doi: 10.1016/j.cub.2019.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jaffe A.B., Hall A. Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 52.Amano M., Nakayama M., Kaibuchi K. Rho-kinase/ROCK: a key regulator of the cytoskeleton and cell polarity. Cytoskeleton (Hoboken) 2010;67:545–554. doi: 10.1002/cm.20472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cherfils J., Zeghouf M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol. Rev. 2013;93:269–309. doi: 10.1152/physrev.00003.2012. [DOI] [PubMed] [Google Scholar]
- 54.Denk-Lobnig M., Martin A.C. Modular regulation of Rho family GTPases in development. Small GTPases. 2019;10:122–129. doi: 10.1080/21541248.2017.1294234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Simões S., Denholm B., Jacinto A. Compartmentalisation of Rho regulators directs cell invagination during tissue morphogenesis. Development. 2006;133:4257–4267. doi: 10.1242/dev.02588. [DOI] [PubMed] [Google Scholar]
- 56.Mason F.M., Xie S., Martin A.C. RhoA GTPase inhibition organizes contraction during epithelial morphogenesis. J. Cell Biol. 2016;214:603–617. doi: 10.1083/jcb.201603077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Häcker U., Perrimon N. DRhoGEF2 encodes a member of the Dbl family of oncogenes and controls cell shape changes during gastrulation in Drosophila. Genes Dev. 1998;12:274–284. doi: 10.1101/gad.12.2.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brand A.H., Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 59.Thielicke W., Stamhuis E.J. PIVlab – towards user-friendly, affordable and accurate digital particle image velocimetry in MATLAB. J. Open Res. Softw. 2014;2:e30. [Google Scholar]
- 60.Schindelin J., Arganda-Carreras I., Cardona A. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rueden C.T., Schindelin J., Eliceiri K.W. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics. 2017;18:529. doi: 10.1186/s12859-017-1934-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barrett K., Leptin M., Settleman J. The Rho GTPase and a putative RhoGEF mediate a signaling pathway for the cell shape changes in Drosophila gastrulation. Cell. 1997;91:905–915. doi: 10.1016/s0092-8674(00)80482-1. [DOI] [PubMed] [Google Scholar]
- 63.Nikolaidou K.K., Barrett K. A Rho GTPase signaling pathway is used reiteratively in epithelial folding and potentially selects the outcome of Rho activation. Curr. Biol. 2004;14:1822–1826. doi: 10.1016/j.cub.2004.09.080. [DOI] [PubMed] [Google Scholar]
- 64.Dawes-Hoang R.E., Parmar K.M., Wieschaus E.F. Folded gastrulation, cell shape change and the control of myosin localization. Development. 2005;132:4165–4178. doi: 10.1242/dev.01938. [DOI] [PubMed] [Google Scholar]
- 65.Fox D.T., Peifer M. Abelson kinase (Abl) and RhoGEF2 regulate actin organization during cell constriction in Drosophila. Development. 2007;134:567–578. doi: 10.1242/dev.02748. [DOI] [PubMed] [Google Scholar]
- 66.Kölsch V., Seher T., Leptin M. Control of Drosophila gastrulation by apical localization of adherens junctions and RhoGEF2. Science. 2007;315:384–386. doi: 10.1126/science.1134833. [DOI] [PubMed] [Google Scholar]
- 67.Chanet S., Miller C.J., Martin A.C. Actomyosin meshwork mechanosensing enables tissue shape to orient cell force. Nat. Commun. 2017;8:15014. doi: 10.1038/ncomms15014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martin A.C., Kaschube M., Wieschaus E.F. Pulsed contractions of an actin-myosin network drive apical constriction. Nature. 2009;457:495–499. doi: 10.1038/nature07522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mason F.M., Tworoger M., Martin A.C. Apical domain polarization localizes actin-myosin activity to drive ratchet-like apical constriction. Nat. Cell Biol. 2013;15:926–936. doi: 10.1038/ncb2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rauzi M., Krzic U., Leptin M. Embryo-scale tissue mechanics during Drosophila gastrulation movements. Nat. Commun. 2015;6:8677. doi: 10.1038/ncomms9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sawyer J.K., Choi W., Peifer M. A contractile actomyosin network linked to adherens junctions by Canoe/afadin helps drive convergent extension. Mol. Biol. Cell. 2011;22:2491–2508. doi: 10.1091/mbc.E11-05-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grosshans J., Wenzl C., Müller H.-A. RhoGEF2 and the formin Dia control the formation of the furrow canal by directed actin assembly during Drosophila cellularisation. Development. 2005;132:1009–1020. doi: 10.1242/dev.01669. [DOI] [PubMed] [Google Scholar]
- 73.Martin A.C., Gelbart M., Wieschaus E.F. Integration of contractile forces during tissue invagination. J. Cell Biol. 2010;188:735–749. doi: 10.1083/jcb.200910099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sun Z., Amourda C., Toyama Y. Basolateral protrusion and apical contraction cooperatively drive Drosophila germ-band extension. Nat. Cell Biol. 2017;19:375–383. doi: 10.1038/ncb3497. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.