Abstract
An immunoglobulin M (IgM) enzyme-linked immunosorbent assay (ELISA) was developed by using a 24-amino-acid peptide of the 18-kDa Epstein-Barr virus (EBV) viral capsid antigen (VCAp18). IgM detection was increased by 23% by using this antigen. Detection of IgM antibodies to the EBV proteins in the new ELISA was 100% specific and 95% sensitive.
Epstein-Barr virus (EBV) is the causative agent of infectious mononucleosis (2). However, other pathogens, mainly cytomegalovirus but also adenovirus, rubella virus, hepatitis A virus, human immunodeficiency virus, and Toxoplasma gondii, or other as yet unknown agents can cause an infectious mononucleosis-like disease (8, 9). EBV-specific immunoglobulin M (IgM) detection is used for confirmation of a diagnosis of infectious mononucleosis or acute primary EBV infection but leads to false-positive and false-negative results for EBV-specific IgM for adults and for children, especially those under 12 years old (10, 11, 15). False-positive EBV-specific IgM reactivity has been attributed to the presence of cross-reactive cytomegalovirus and toxoplasma IgM antibodies or contaminating cellular proteins in EBV antigen preparation. The development of synthetic peptides used as solid-phase antigens for the serodiagnosis may overcome these problems, but these methods demonstrate low sensitivity. The combination of several synthetic epitopes of EBV proteins and, alternatively, the degeneration of a unique continuous epitope by constructing a convergent combinatorial peptide library, or mixotope (7), have been shown to improve the sensitivity of EBV-specific Ig (IgG, IgA, or IgM) enzyme-linked immunosorbent assay (ELISA) (14, 16). The objective of the present study was to develop and evaluate, for its utility in the diagnosis of primary EBV infection, an EBV-specific IgM ELISA based on a 24-amino-acid peptide of the 18-kDa EBV viral capsid antigen p18 (VCA p18) used alone or in association with its mixotope obtained by creating an artificial degeneration in the VCAp18 sequence.
An immunodominant epitope of the EBV protein VCA p18, the VCAp18(153-176) sequence (16) herein referred to as VCAp18 peptide, and its mixotope were synthesized by using the conventional solid-phase “Boc-benzyl” strategy in an automated peptide synthesizer (model 430A; Applied Biosystems, Inc.). Briefly, amino acids were introduced by HBTU/HOBt activation protocol with systematic double coupling on a Boc-Gln-Pam resin (Applied Biosystems). The VCAp18 peptide was purified to >90% by preparative reversed-phase high-performance liquid chromatography. Identity was confirmed by determination of amino acid composition and by mass spectrometry (apparatus from Bio Ion AB, Uppsala, Sweden). As indicated in Table 1, the mixotope, called MIXO(P,G) was designed from the sequence of VCAp18(153-176), in which the original amino acid was mixed during the synthesis with a second, selected from the replaceability matrix defined experimentally by Geysen et al. (6), excluding the glycine-rich sequence (GSGGGG) and the proline residues that were likely to diminish the immunoreactivity. We selected the second residue presenting the best score in this matrix. Due to the complexity of the mixotope construct, detailed characterization proved impossible. An aliquot of purified mixotope was submitted to total acid hydrolysis for analysis of amino acid composition. Results were as expected, confirming that no preferential amino acid incorporation had occurred.
TABLE 1.
Compositions of the peptide and mixotope used as solid-phase antigens in ELISAs of IgM against EBVa
| Antigen | Sequence |
|---|---|
| VCA p18 | A V D T G S G G G G Q P H D T A P R G A R K K Q |
| A V D T Q H D T A R G A R K K Q | |
| MIXO (P,G) | G S G G G G P P |
| G I E S N G E S G K S G K R R N |
Equimolar amounts of the appropriate subsets of amino acids (shown vertically for the mixotope) were introduced in each degenerate position during the synthesis. Theoretically, this produced sets of peptides containing 65,536 closely related combinatorial sequences.
Microtiter plates (96 wells) (Maxisorp; Nunc, Roskilde, Denmark) were coated overnight with 1 μg of VCAp18 peptide and, in some experiments, also with 10 μg of MIXO(P,G) antigen for 1 h at 37°C in coating buffer (0.05 M sodium carbonate–bicarbonate buffer [pH 9.6]). After a wash (0.05% Tween 20 in phosphate-buffered saline), the wells were blocked with 2% bovine serum albumin (37°C; 1 h) and filled with 200 μl of a patient’s serum (diluted 1 to 50) and diluted peroxidase-conjugated goat anti-human IgM or IgG antibody (Sanofi Diagnostics Pasteur, Marnes-la-Coquette, France), followed by a 1-h incubation at 37°C in a humidified atmosphere and three washes each time. Bound conjugated antibody was revealed by using peroxidase substrate (Sigma Chemical Co., St. Louis, Mo.), and color development was stopped after 30 min by addition of 2 N sulfuric acid. Absorbance at 492 nm (A492) was measured. Blank wells and negative and positive sera were included on each plate. Three standard deviations above the mean A492 of EBV-seronegative samples was used as a cutoff value in the ELISAs. Before incubation, sera tested for IgM analysis were treated with rheumatoid factor-absorbent serum (Behringwerke AG, Marburg, Germany) or with goat anti-human IgG serum (Sanofi Diagnostics Pasteur, Marnes-la-Coquette, France) according to each manufacturer’s instructions.
One hundred fourteen sequential patient sera submitted to the Clinical Laboratory of the Pasteur Institute, Lille, France, for diagnosis of primary EBV infection were used for this study. Two standard serological tests for diagnosing recent primary infection, (i) ELISA for detection of IgM to VCA-EA-EBNA (EBV-encoded nuclear antigen) and (ii) the VCA-specific IgG–EBNA antibody profile, were used as the reference assays for evaluating the new VCAp18 peptide and VCAp18-MIXO(P,G) IgM ELISAs. Primary infection was defined as the presence of IgM antibody to VCA-EA-EBNA or the presence of IgG antibody to EBV VCA in the absence of IgG antibody to EBNA, a marker of the latent phase of an EBV infection. Past infection was defined as the presence of IgG antibodies both to EBV VCA and to EBNA. Seronegative individuals were those who had no antibodies against either VCA-EA-EBNA, EBV VCA, or EBNA.
The correlations of the four assays with each other are shown in Table 2. The panel of sera giving only concordant results by both reference assays can be regarded as a serological “gold standard” for diagnosing primary EBV infection. In comparison to the reference tests indicating a recent infection, the sensitivity of the VCAp18-peptide IgM ELISA was 72% (P = 0.2) and that of the VCAp18-MIXO(P,G) IgM ELISA was 95% (P = 1.0); the specificities were 100 and 98%, respectively.
TABLE 2.
Evaluation of IgM ELISAs based on a 24-amino-acid VCAp18 peptide used alone or in association with its mixotope, MIXO(P,G), for the diagnosis of primary EBV infection and comparison with reference assays
| Result | No. of sera | Results of reference tests
|
Consensus interpretation of reference test results | No. (%) of sera positive by:
|
|||
|---|---|---|---|---|---|---|---|
| VCA-EA-EBNA-specific IgM ELISA | EBV VCA-specific IgG IF | EBNA-specific IgG ELISA | VCAp18 peptide-specific IgM ELISA | VCAp18-MIXO(P,G)-specific IgM ELISA | |||
| Concordant | 40 | Positive | Positive | Negative | Recent primary EBV infection | 29 (72) | 38 (95) |
| 46 | Negative | Positive | Positive | Past EBV infection | 0 (0) | 1 (2) | |
| 28 | Negative | Negative | Negative | No evidence of recent or past EBV infection | 0 (0) | 0 (0) | |
| Possibly discordant | 0 | Negative | Positive | Negative | Suggested recent infection | 0 | 0 |
| 0 | Positive | Positive | Positive | Suggested recent infection | 0 | 0 | |
Two sera from VCA-EA-EBNA IgM ELISA-positive sera from children younger than 4 years escaped VCAp18-MIXO(P,G) IgM detection by ELISA and are considered to show false-negative results. These results are not inconsistent with results obtained with the reference IgM ELISA, as a different set of EBV antigens was used. These sera were available for further analysis and were shown to have very low titers (1/10 and 1/40) of VCA IgM antibody as determined by indirect immunofluorescence test and no VCAp18-MIXO(P,G) IgG antibody. For this range of titers, some cross-reactivities with other VCA proteins have been observed for samples from patients with cytomegalovirus, hepatitis A virus, parvovirus, and leptospiral infections, as well as for samples containing rheumatoid factor (8, 9). The fact that our model peptide, VCAp18(153-176), appeared to have no sequence homology with other human herpesviruses (1, 3, 12) could explain the absence of reactivity of the VCAp18-MIXO(P,G) IgM and IgG ELISAs for these sera.
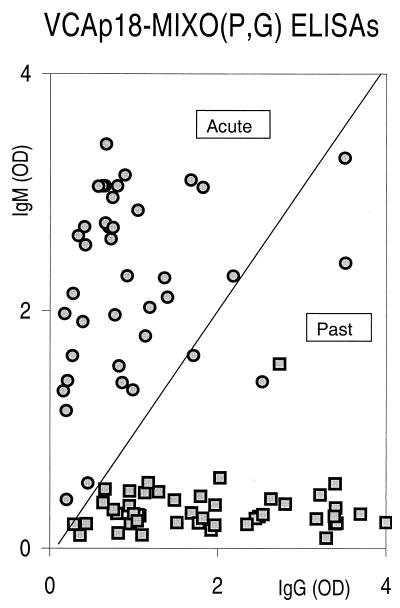
One patient with no evidence of recent EBV infection revealed by either of the reference assays had VCAp18 IgM detectable by ELISA and is considered to have shown a false-positive result. This patient has been shown to have high-affinity IgG antibody (an independent marker of past infection) and a high level of VCAp18 IgG antibody. It has been reported that false-positive serum could be the result of EBV reactivation due to cross-reaction with IgM against other viruses or to the reappearance of EBV-specific IgM due to polyclonal activation induced by pathogens that produce an infectious mononucleosis-like disease (8, 9). To test this hypothesis, we tentatively compared the relative VCAp18-MIXO(P,G)-specific IgM and IgG antibody levels obtained by ELISAs for the positive sera identified in the two reference tests. Figure 1 shows that all the sera from patients with no evidence of recent EBV infection revealed by either of the reference assays were classified as having past infection. The false-positive result for VCAp18-MIXO(P,G)-specific IgM could be effectively attributed to EBV reactivation and is interpreted in our VCAp18-MIXO(P,G)-specific IgM and IgG antibody profile as indicating a past EBV infection. It was evident that the specificity of the new ELISA for IgM increased from 98 to 100% when VCAp18-MIXO(P,G)-specific IgM and IgG profiling was used. In addition, only 2 (5%) of 40 sera identified as revealing recent infection by one of the reference assays were not found in the acute infection section of our representation and should be considered to show evidence of past infection in spite of their VCA IgG-EBNA antibody profile demonstrating acute infection. The possibility of false-positive or, for these two sera, false-negative results cannot be excluded when profiles of VCA IgG-EBNA antibodies are used for diagnosing recent primary EBV infection, as has been reported for children under 12 years old and for immunosuppressed patients, who are often unable to develop an EBNA-1 antibody response, making differentiation of acute and past infections difficult (4, 5, 10, 11, 13).
FIG. 1.
Comparison of IgM and IgG antibody levels obtained by VCAp18-MIXO(P,G) ELISAs for patients diagnosed as having primary (circles) or past (squares) EBV infection based on the results of the two reference tests (VCA-EA-EBNA-specific IgM ELISA and VCA IgG-EBNA antibody profiling). The diagonal line bisecting the figure is the limit of identity between IgM and IgG absorbance values. OD, optical density.
The preliminary evaluation of the VCAp18-MIXO(P,G) IgM ELISA suggests that it may provide a sensitive and very specific alternative for diagnosis of primary EBV infection by using peptides as solid-phase antigens and that larger “in-use” studies are warranted. Our study indicates that a convergent combinatorial peptide library, or mixotope, designed based on statistical data describing the replaceability of amino acids in linear epitopes, could be a useful adjunct to a nonvariable peptide antigen used in immunoenzymatic serodiagnosis.
Acknowledgments
This study was supported by grants from CNRS (UMR 8527 and 8525), the Institut Pasteur de Lille, and the Université de Lille II.
REFERENCES
- 1.Chee M S, Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Horsnell T, Hutchinson C A, Kousarides T, Martignetti J A, Preddie E, Satchwell S C, Tomlinson P, Weston K M, Barrel B G. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–170. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 2.Cheeseman S H. Infectious mononucleosis. Semin Hematol. 1988;25:261–268. [PubMed] [Google Scholar]
- 3.Davidson A J, Scott J E. The complete DNA sequence of varicella zoster virus. J Gen Virol. 1986;67:1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- 4.Field P R, Dwyer D E. Difficulties with the serologic diagnosis of infectious mononucleosis: a review of the RCPA quality assurance programs. Pathology. 1996;28:270–276. doi: 10.1080/00313029600169144. [DOI] [PubMed] [Google Scholar]
- 5.Fleisher G R, Collins M, Fager S. Limitations of available tests for diagnosis of infectious mononucleosis. J Clin Microbiol. 1983;17:619–624. doi: 10.1128/jcm.17.4.619-624.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geysen H M, Mason T J, Rodda S J. Cognitive features of continuous antigenic determinant. J Mol Recognit. 1988;1:32–41. doi: 10.1002/jmr.300010107. [DOI] [PubMed] [Google Scholar]
- 7.Gras-Masse H, Georges B, Estaquier J, Tranchand-Bunel D, Tartar A, Druilhe P, Auriault C. Convergent peptide libraries, or mixotopes, to elicit or to identify specific immune responses. Curr Opin Immunol. 1999;11:223–228. doi: 10.1016/s0952-7915(99)80038-7. [DOI] [PubMed] [Google Scholar]
- 8.Gray J J, Caldwell J, Valerio M. The rapid serological diagnosis of infectious mononucleosis. J Infect. 1992;25:39–46. doi: 10.1016/0163-4453(92)93465-3. [DOI] [PubMed] [Google Scholar]
- 9.Ho D W T, Field P R, Cunningham A L. Rapid diagnosis of acute Epstein-Barr virus infection by an indirect enzyme-linked immunosorbent assay for specific immunoglobulin M (IgM) antibody without rheumatoid factor and specific IgG interference. J Clin Microbiol. 1989;27:952–958. doi: 10.1128/jcm.27.5.952-958.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levine D, Tilton R C, Parry M F, Klenk R, Morelli A, Hofreuter N. False-positive EBNA IgM and IgG antibody tests for infectious mononucleosis in children. Pediatrics. 1994;94:892–894. [PubMed] [Google Scholar]
- 11.Linderholm M, Boman J, Juto P, Linde A. Comparative evaluation of nine kits for rapid diagnosis of infectious mononucleosis and Epstein-Barr virus-specific serology. J Clin Microbiol. 1994;32:259–261. doi: 10.1128/jcm.32.1.259-261.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rickinson A B, Kieff E. Epstein-Barr virus. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2397–2446. [Google Scholar]
- 13.Seigneurin J M, Lavoue M F, Genoulaz O, Bornkamm G W, Lenoir G M. Antibody response against the Epstein-Barr virus-coded nuclear antigen 2 (EBNA2) in different groups of individuals. Int J Cancer. 1987;40:349–353. doi: 10.1002/ijc.2910400311. [DOI] [PubMed] [Google Scholar]
- 14.Tranchand-Bunel D, Auriault C, Diesis E, Gras-Masse H. Detection of human antibodies using “convergent” combinatorial peptide libraries or “mixotopes” designed from non-variable antigen: application to the EBV viral capsid antigen p18. J Pept Res. 1998;52:495–508. doi: 10.1111/j.1399-3011.1998.tb01254.x. [DOI] [PubMed] [Google Scholar]
- 15.vanGrunsven W M J, Nabbe A, Middeldorp J M. Identification and molecular characterization of two diagnostically relevant marker proteins of the Epstein-Barr virus capsid antigen. J Med Virol. 1993;40:161–169. doi: 10.1002/jmv.1890400215. [DOI] [PubMed] [Google Scholar]
- 16.vanGrunsven W M J, Spaan W J M, Middeldorp J M. Localization and diagnostic application of immunodominant domains of the BFRF3-encoded Epstein-Barr virus capsid protein. J Infect Dis. 1994;170:13–19. doi: 10.1093/infdis/170.1.13. [DOI] [PubMed] [Google Scholar]