Abstract
Objectives.
Although T cells have been implicated in the pathogenesis of systemic sclerosis (SSc), a comprehensive study of T-cell mediated immune responses in the affected skin of patients with progressive SSc is lacking. Droplet-based single-cell transcriptome analysis of SSc skin biopsies opens avenues for dissecting patient-specific T-cell heterogeneity, providing a basis for identifying novel gene expression related to functional pathways associated with severity of SSc skin disease.
Methods.
Single-cell RNA-sequencing was performed by Droplet-based sequencing (10X Genomics), focusing on 3,729 CD3+ lymphocytes (867 cells from normal and 2,862 cells from SSc skin samples) from skin biopsies of 27 patients with active SSc and 10 healthy donors. Confocal immunofluorescence microscopy of progressive SSc skin samples validated transcriptional results and visualized spatial localizations of T-cell subsets.
Results.
We identified several subsets of recirculating and tissue-resident T cells in healthy and SSc skin that were associated with distinct signaling pathways. While most clusters shared a common gene expression signature between patients and controls, we identified a unique cluster of recirculating CXCL13+ T cells in SSc skin which expressed a T helper follicular-like gene expression signature and that appears to be poised to promote B-cell responses within the inflamed skin of patients.
Conclusions.
Current available therapies to reverse or even slow progression of SSc lead to broad killing of immune cells and consequent toxicities, including death. Identifying the precise immune mechanism(s) driving SSc pathogenesis could lead to innovative therapies that selectively target the aberrant immune response, resulting in better efficacy and less toxicity.
Keywords: systemic sclerosis, T lymphocytes, skin, single-cell transcriptome
INTRODUCTION
Systemic sclerosis (SSc) is a disabling and often fatal autoimmune disease characterized by vasculopathy, inflammation, and extensive cutaneous and visceral fibrosis.[1] SSc has the highest fatality rate among connective tissue diseases and few therapies are available that reverse or even slow progression. Several T-lymphocyte subsets and the cytokines they produce have been implicated in the inflammatory and fibrotic processes of SSc.[2, 3] Particularly, we and others identified increased numbers of pro-fibrotic T-cell subsets including Th2 and Tc2 cells producing IL-4 and IL-13[4, 5] as well as IL-21-producing T follicular helper (TFH)-like cells[6] in the blood and in the sclerotic skin of patients with active SSc. Moreover, skin infiltrating Th17 and Th22 cells producing IL-17-family cytokines were shown to drive inflammatory responses involving fibroblasts and endothelial cells,[3] whereas IL-9 produced by Th9 cells was shown to induce NETosis, expansion of mast cells, and increased production of SSc‐related autoantibodies by B lymphocytes.[7] Deregulation of T regulatory (Treg) cell function was also associated with altered immune homeostasis and fibrosis in SSc.[8, 9] Finally, cytotoxic CD8+ and CD4+ T cells[4, 10] in the lesional skin of patients with early active SSc were also detected. These cells express markers of cytotoxicity and are likely involved in tissue damage and in perpetuating autoimmune responses.[11] However, a comprehensive study of resident and recirculating T-cell subpopulations in SSc skin disease remains elusive.
Recent advances in single-cell transcriptome technology, including droplet-based single-cell RNA sequencing (scRNAseq),[12] profile gene expression across thousands of individual cells from a large heterogeneous population such as a patient biopsy. This high-resolution analysis of cellular heterogeneity reveals individual cell functions in the context of their microenvironment and provides striking insights into the complex cellular composition of normal and diseased tissue. Here we report scRNAseq analysis of T cells from the affected skin of patients with progressive SSc. This analysis provides an unprecedented view of lymphocyte heterogeneity within the skin-microenvironment of individual SSc skin sample and offers important implications for personalized disease management.
METHODS
Full-thickness 3mm skin biopsies were obtained from 32 patients with confirmed diagnosis of active diffuse cutaneous SSc (dcSSc)[13, 14] at the Scleroderma Clinic of UPMC and at the Scleroderma Program of the University of Michigan Medical School. Disease subtype and internal organ involvement were assessed according to established criteria.[13–15] Research protocols involving human subjects were approved by the Institutional Review Boards of the Universities of Pittsburgh and Michigan. All participants gave written informed consent in accordance with the Declaration of Helsinki. Twenty-seven patient samples were used for scRNA-seq and 5 for immunofluorescence (IF) microscopy. Experimental procedures for scRNAseq followed established techniques using the Chromium Single Cell 3’ Library V2 kit (10x Genomics).[16] Multicolor antibody staining using tyramide signal amplification (ThermoFisher) was performed on formalin-fixed, paraffin-embedded skin samples as previously described.[17] See Supplementary Methods for details. All scRNA-seq data have been deposited in the Gene Expression Omnibus: GSE138669.
RESULTS
Single-cell transcriptome profiles reveal T lymphocyte heterogeneity in dcSSc skin lesions
ScRNAseq was used to profile the transcriptome of T lymphocytes obtained from the enzymatically digested skin of 27 active dcSSc and 10 HC samples (Table S1 and Figure S1). Histological evaluation of SSc skin lesions shows increased collagen deposition in the dermis, thickening of the collagen fiber bundles and subintimal thickening of blood vessels, as well as perivascular mononuclear cell infiltration compared to control skin (Figure 1A). In total, we analyzed 3,729 CD3+ cells: 867 from HC and 2,862 from SSc skin (Figure 1B). Comparison of the transcriptional profiles of each lymphocyte subset from the patient and control skin samples identified 9 clusters (Figures 1C and Table S2). In parallel to PCA, Harmony[18] was used to demonstrate that the samples did not suffer from any batch effects (Figure S2). By comparing gene expression from each cell in the cluster to that of all other cells in the dataset, we determined the differential expression (DE) of genes in each cluster. The cut-off for significance was p<0.05 and we required that gene expression be from at least 25% of cells in the cluster. The heatmap in Figure 1D reports examples of the most highly significant DE genes for each cluster. Conventional CD4+ T cells comprised three clusters (#0, 1, and 4) distinguished from each other by only subtle transcriptome differences. Cluster #2 identifies CD8+ T cells, while cluster #3 identifies Tregs. Cytokine IL-26 is the signature gene of cluster #5, CXCL13 of cluster #7, and TRDC of cluster #8. Cells in cluster #6 express proliferation genes. We next employed Ingenuity Pathway Analysis[19] to identify activation of key molecular pathways in each T-cell cluster (Figure 1E). Examples of pathways activated in each lymphocyte cluster included IL-7 and PI3K/AKT signaling (cluster #0); glucocorticoid receptor and TNFR2 signaling (cluster #1); Th1, Th2 activation and T-cell exhaustion signaling (cluster #2); and ICOS-ICOSL and PD1-PDL1 signaling (cluster #3). Clusters #4 ,6, and 7 up-regulated expression of oxidative phosphorylation and mitochondrial pathways. The ERK/MAPK signaling pathway was also upregulated by cluster #7, while clusters #5 and 8 up-regulated the HMGB1 signaling pathway. Within each cluster, no consistent changes were observed in the proportion of T cells between SSc and control samples, apart from cluster #6 where we found a statistically significant increase of SSc T cells and in cluster #7 that was comprised uniquely of cells from SSc samples (Figure 1F). Figure 1G demonstrates the SSc-exclusive expression of signature genes (CXCL13, CD200, and MS4A6A) from cluster #7. Thus, scRNAseq analysis provides an unprecedented view of T lymphocyte heterogeneity in the skin lesions of patients with active dcSSc and identifies a unique subset found only in patient samples.
Figure 1. Transcriptional profiles of T lymphocytes from SSc and HC skin samples.
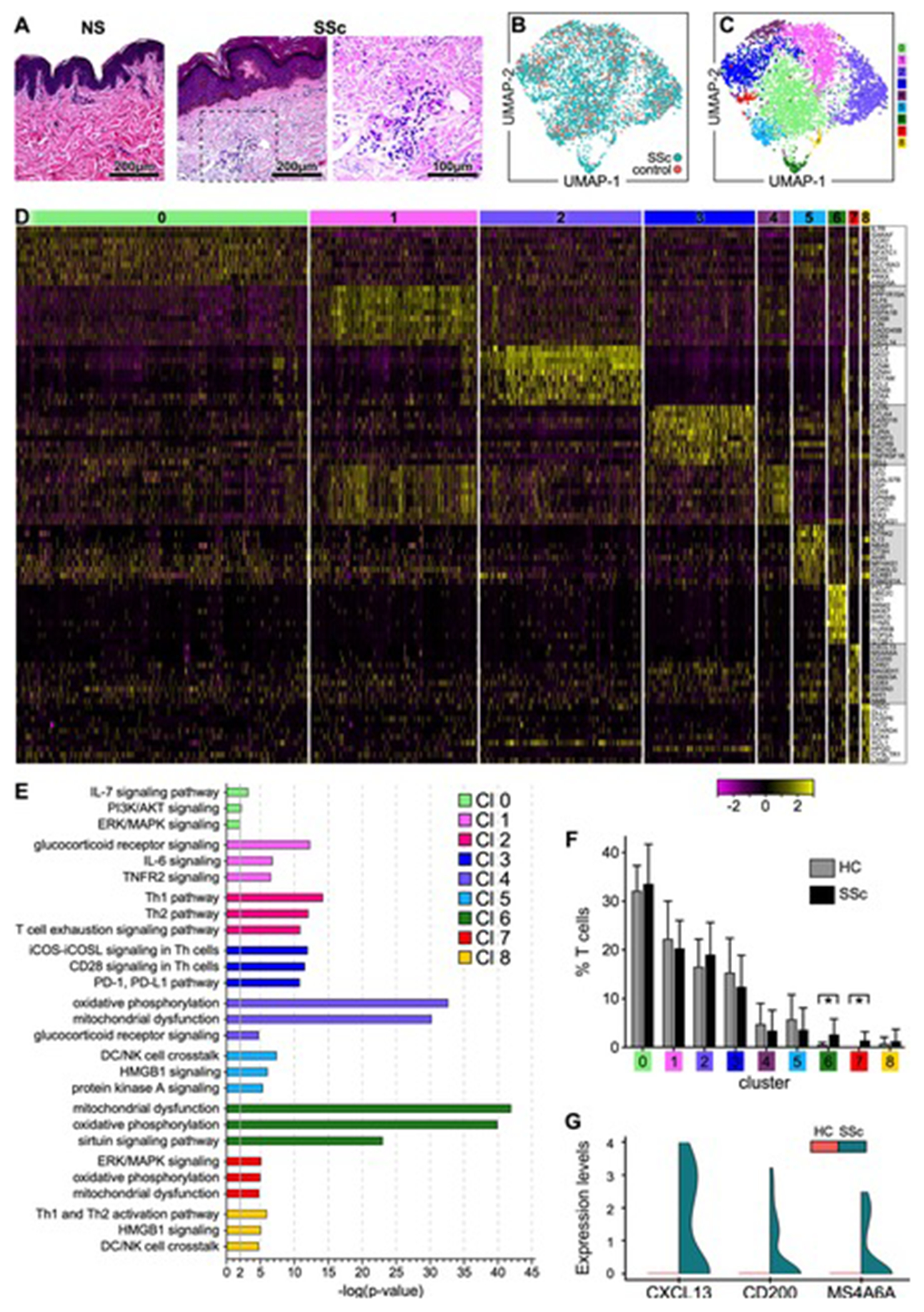
(A) Hematoxylin and eosin staining (H&E) of skin biopsies from representative healthy control (HC) and SSc skin samples analyzed by scRNA-seq (200X, 400X). (B) Transcriptomes of 3,729 CD3+ cells (867 cells from HC and 2,862 cells from SSc skin samples) (C) revealed 9 discrete Louvain clusters using Seurat.[58] (D) Heatmap showing examples of the most highly significant differentially expressed genes (n=10) for each cluster from (C). Cluster numbers are indicated at the top. Each column represents a cell. (E) Pathway analysis by Ingenuity (Qiagen). Highly significant examples of distinct pathways activated in each cluster are shown. In IPA, the pathways were compared using a P-value cut-off of 0.05. (F) Proportion of T cells from each patient or control sample within each cluster. Statistics by Mann-Whitney test. (G) Violin plot showing the expression levels of CXCL13, CD200, and MS4A6A by cells in cluster #7 comparing SSc to HC samples. SSc and HC UMAP: Uniform Manifold Approximation and Projection for dimension reduction.
Cell state transition of resident and recirculating T cells in healthy and SSc skin
The tissue distribution of skin-resident and -recirculating T cells was established by determining expression of CD69, ITGAE, CCR7, and SELL.[20] The normalized single-cell expression showed that CD69 was expressed by a large number of cells in most T-cell clusters, exhibiting modestly upregulated expression in SSc T cells, whereas ITGAE was detected in fewer cells, primarily from cluster #6 (Figure 2A–B). CCR7 and SELL expression also appeared to be confined to specific clusters (Figure 2A–B). Specifically, CCR7 was mainly expressed by cells in clusters #0, 6, and 7, with a higher number of CCR7+ cells in SSc versus HC skin samples (Figure 2B). SELL was primarily expressed by SSc T cells in clusters #0, 3, and 6. Although we cannot exclude presence of cells in transitional states within each cluster, our findings indicated that T cells in clusters #1, 2, 3, 4, 5, and 8 exhibited predominantly a skin-resident phenotype (TRM, CD69+ITGAE+/−). These included CD4+, CD8+ and Treg populations. In contrast, most SSc T cells in clusters #0 and 7 expressed markers of recirculating memory migratory cells (TMM, CCR7+SELL−), the latter representing the population most highly upregulated in SSc skin. Finally, proliferating T cells in cluster #6, also mainly present in SSc skin, represented a mix of TRM and TMM cells.
Figure 2. Identification of resident and recirculating T cells in healthy and SSc skin.
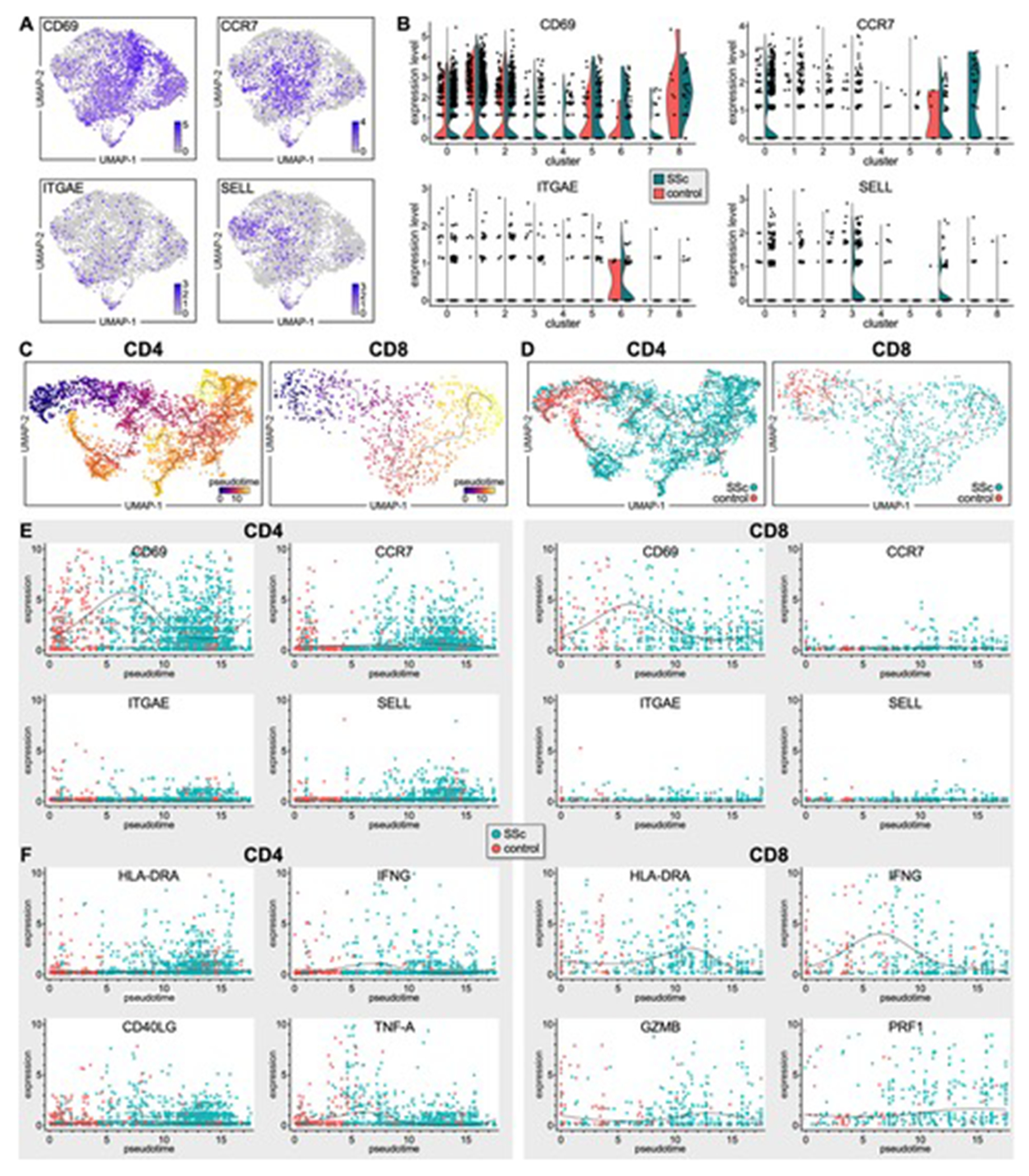
(A) Transcriptomes of CD69+, CCR7+, ITGAE+, SELL+ T lymphocytes from patient and HC skin samples. (B) Violin plots showing expression levels of CD69, CCR7, ITGAE, SELL by SSc and HC T lymphocytes from each T-cell cluster (Figure 1C). (C-D) Single-cell pseudotime trajectories of CD4+ and CD8+ T cells estimated using Monocle 3. A continuous value from 0 to 20 was assigned to each cell as a pseudotime. Expression dynamics of skin-residency (E) or T-cell activation (F) markers along the pseudotime of SSc and HC T cells by scatter plots with regression curves.
We performed a trajectory analysis using Monocle 3[21, 22] to examine the connection between skin-residency and kinetics of gene expression during CD4+ and CD8+ T-cell differentiation in SSc skin. This algorithm utilizes a concept referred to as pseudotime, in theory examining the change in phenotype of cell types over time. However, here it examines changes in gene expression, relating each cell to other cells and ordering the cells showing the closest transcriptomes adjacent to each other. It may but does not necessarily indicate a descendant-progenitor relationship. All T cells from SSc and healthy skin were placed on these trajectories based on changes in their transcriptome (Figure 2C). Strikingly, both CD4+ and CD8+ T cells from HC samples were distributed to early pseudotimes, whereas SSc T cells were enriched in late pseudotimes, showing a clear temporal separation (Figure 2D). We then examined the transition of average expression values along pseudotime for a panel of marker genes associated with T-cell residency. We found that HC CD4+ and CD8+ T cells exhibited a TRM phenotype as these cells highly expressed CD69 and expressed only low levels of CCR7, ITGAE and SELL. This phenotype was further confirmed by the increased expression of CXCR6 and little to no expression of KLF2, S1PR1, and SELPLG (gene for CLA) by these cells (Figures 2E and S3).[20, 23–25] In contrast, CD4+ and CD8+ T cells from SSc skin were clearly shifted towards more differentiated states compared to controls and corresponded to several TRM subsets (CD69+ITGAE+/−CXCR6+) from earlier to late pseudotime as well as of some recirculating T cells expressing CCR7, SELL, KLF2, S1PR1, and SELPLG, particularly among SSc CD4+ cells at the latest pseudotimes (Figures 2E and S3A). Interestingly, SSc T cells at the latest pseudotimes expressed high levels of activation and effector markers (Figure 2F and S3B) including HLA-DRA, IFNG, CRTAM (CD4+ and CD8+ T cells), CD40L and TNF-A (CD4+ cells), as well as GZMB and PRF1 (CD8+ T cells). Finally, most SSc CD4+ and CD8+ T cells from early to late pseudotime expressed CD27 and CD28 implying that SSc T cells exhibit an activated effector-memory phenotype.
Single-cell RNAseq identifies a novel CXCL13+ T-cell subset in dcSSc skin lesions
Chemokine CXCL13 was selectively expressed by most CD4+ T cells from the SSc-specific cluster #7 (Figure 3A). Moreover, no other cell-type from patient or HC skin samples, apart from a negligible number of SSc CD8+ T cells, expressed CXCL13 (Figures 3A and S4A–B). These transcriptional results were validated by multicolor immunofluorescence microscopy showing several CD3+CXCL13+ cells within perivascular inflammatory infiltrates in the lesional skin of patients with active SSc but not in HC skin samples (Figure 3B). Consistently, most CD3+CXCL13+ are CD4+ (Figure 3C). Flow cytometry to identify circulating CD4+CXCL13+ T cells in the blood of healthy and dcSSc samples showed no difference in the frequency of this cell-type in patients and controls (Figure S4C–E). Although we detected CD3+CXCL13+ cells in the skin of 44% of the patients tested, we found no statistically significant association between the frequency of CD3+CXCL13+ cells and the skin score of patients or their autoantibody specificity (Figure 3D). In contrast, we observed a significant correlation with presence of interstitial lung disease (ILD) and a trend for increased numbers of CD3+ CXCL13+ cells at the earliest disease stage. The frequency of CD3+CXCL13+ cells did not correlate with the age or sex of patients. Finally, we observed that patients treated with immunosuppressants exhibited a statistically significant lower frequency of CD3+CXCL13+ cells compared to untreated patients (Figure 3D).
Figure 3. Characterization of the SSc-specific CXCL13+ T-cell subset in the lesional skin of patients with active dcSSc.
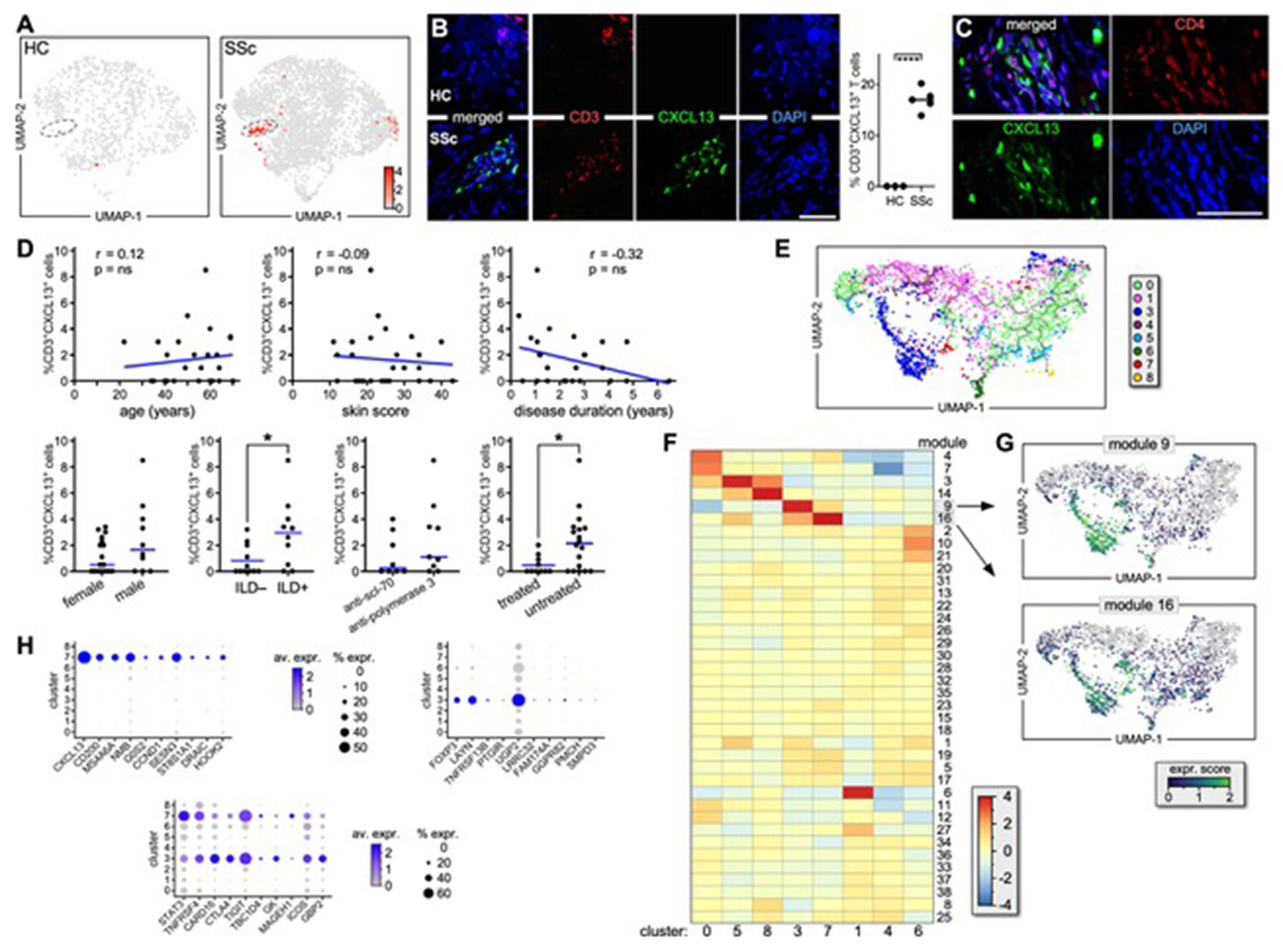
(A) CXCL13 expression by T lymphocytes from HC (left) and SSc (right) skin samples. (B) Immunofluorescence microscopy shows co-expression of CXCL13 and CD3 in skin samples from active dcSSc (n=5) and HC (n=3). A representative example and the proportion of CD3+CXCL13+ cells are shown. Results are expressed as a percentage of positive cells out of the entire infiltrate following quantification of 8 high-power fields/sample (HPF; magnification X600). Error bars are mean ± SD. Statistics by t-Student’s test. (C) Immunofluorescence microscopy shows co-expression of CXCL13 and CD4 in dcSSc skin samples (n=5). A representative experiment is shown at 1000X. (D) Correlation between the frequency of CD3+CXCL13+ cells by scRNAseq and the clinical parameters of patients. (E) Single-cell trajectory of SSc and HC CD4+ T-cell clusters. (F) Heatmap shows gene expression of T-cell clusters based on Louvain-community analysis[26] (G) Expression transition of genes from modules #16 and 9 along the pseudotime. (H) Dot plots show the proportion of cells and the scaled average gene expression of genes uniquely expressed by CD3+CXCL13+ cells or Tregs or commonly expressed.
To understand the CD4+CXCL13+ T cells in the context of SSc CD4+ T-cell differentiation, we constructed a single-cell trajectory for the CD4+ T-cell clusters identified in Figure 1C. We observed that CD4+CXCL13+ T cells were located in close proximity to SSc Tregs, suggesting a developmental connection between these two subsets (Figure 3E). Contrary to HCs, SSc Tregs up-regulated expression of ICOS, FOXP3, IL2RA, and PPP1CB, while down-regulating expression of CTLA4 (Figure S4F). To reveal gene expression similarities between clusters along the pseudotime, we grouped expressed genes into modules based on Louvain-community analysis (Figure 4G).[26] A module heatmap indicates that modules #6 and 9 exhibit gene expression similarities between Tregs (cluster #3) and CD3+CXCL13+ cells (cluster #7; Figure 3F). Indeed, although CD3+CXCL13+ cells and Tregs present distinct transcriptional profiles, they also exhibit common expression of genes not expressed significantly by other T cell populations (Figure 3H and Table S3) such as STAT3, TNFRSF4, CTLA4, TIGIT, ICOS. Genes uniquely expressed by Tregs include FOXP3, LAYYN, TNFRSF13B, UGP2, PTGIR; while examples of uniquely expressed gene by CD3+CXCL13+ cells include CD200, MS4A6A, DRAIC, CCND1, G0S2, NMB. Although SSc CD3+CXCL13+ cells express several genes also found in TFH cells,[27] they lack expression of the canonical TFH genes CXCR5 and BCL6 (Figure 4A and S4F–G). By immunofluorescence microscopy we visualized CD3+CXCL13+ cells co-expressing ICOS, TIGIT, CTLA4 as well as producing IL-21 and IFNγ (Figure 4B–E) in dcSSc skin lesions. Significantly, we also showed that CD3+CXCL13+ cells are adjacent to CD20+ B cells within inflammatory infiltrates of dcSSc skin (Figure 4F). Notably, microarray data from multiple early dcSSc patient and control skin samples demonstrated the presence of up-regulation of CXCL13 (Figure S5, red arrow) in a subset of patients, which was associated with a T- and B-cell gene expression signature (e.g.: CD3, CD247, CD8A, GZMM, GZMH, IL15RA, CCR7 and CR2, CD19, CD22, MS4A1, respectively). Thus, chemokine CXCL13 identifies a TFH-like subset in the lesional skin of dcSSc patients with active skin disease.
Figure 4. SSc CD3+CXCL13+ T cells are a TFH-like subset in inflamed SSc skin.
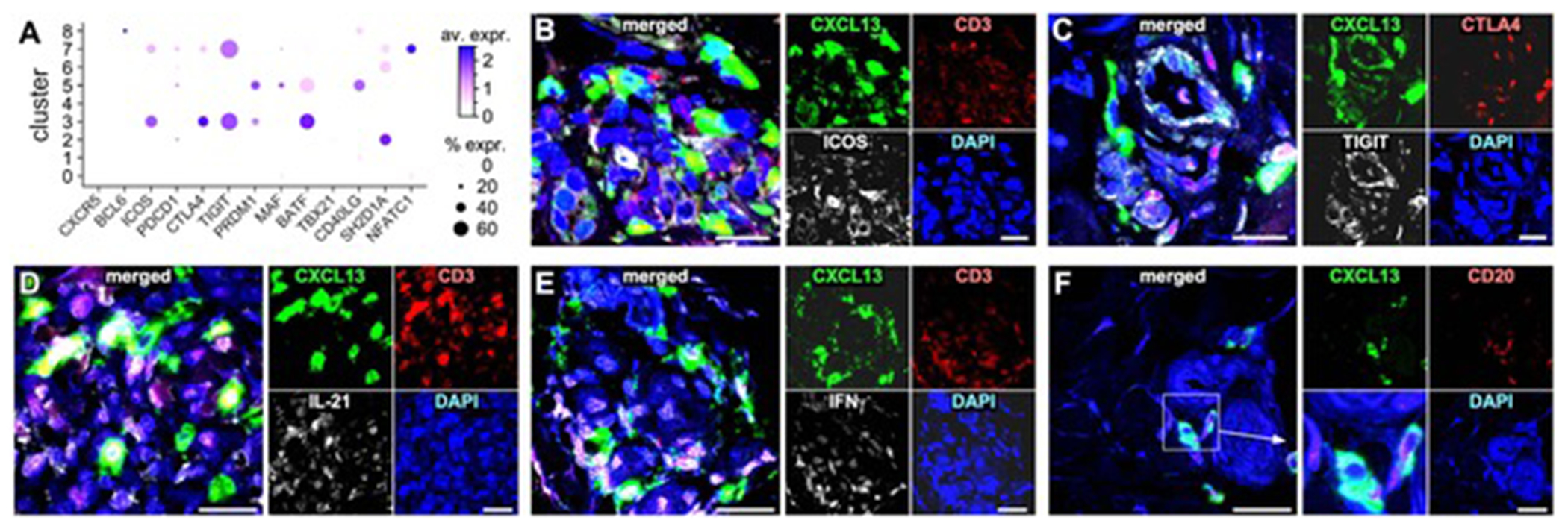
(A) Dot-plot showing the proportion of cells and the scaled average gene expression of selected TFH genes [27] by T-cell clusters identified in Figure 1C. Multicolor immunofluorescence microscopy visualizes co-expression of CXCL13, CD3, ICOS (B), CXCL13, TIGIT, CTLA4 (C), CXCL13, CD3, IL-21 (D), CXCL13, CD3, IFNγ (E), and CXCL13, CD3, CD20 (F) in active dcSSc skin samples (n=5). Representative examples are shown at 1000X, inset in (F) is zoomed-in by 3X. DAPI stains nuclei.
DISCUSSION
Abnormal T-cell responses are central in the development of SSc skin disease. Here we employed scRNAseq to comprehensively profile the transcriptomes of recirculating and resident T cells in the skin lesions of a large cohort of patients with dcSSc. Our analysis provides an unprecedented view of T-cell heterogeneity in SSc skin and identifies a distinct subset that expressed factors enabling B-cell help and likely promoting B-cell responses and possibly autoantibodies production within pathological inflamed skin. Although the role of autoantibodies in SSc in uncertain, they have been implicated in innate immune responses.[28] Furthermore, the perivascular location of T cell infiltration in SSc skin suggests that these cells may play a direct role in vascular damage the other major driver of SSc clinical disease. Thus, a better understanding of T cell responses in SSc skin disease could lead to more targeted T cell therapy in SSc, an urgent need in view of recent clinical trials, indicating that myeloablation is effective in treating the disease but with significant risks. The specific immune cell targets leading to improved outcomes remain uncertain and targeting T cell activation more generally has not provided a clear clinical benefit.[29] Our observations that only a very specific population of T cells is expanded in SSc skin suggests that these cells are playing a critical role in disease progression and that targeting this select T cell population might interrupt the disease process.
Recirculating and resident memory T cells play essential roles in localized immune responses in human skin and both populations have been implicated in a variety of inflammatory and autoimmune conditions.[20, 23, 30–32] Here we show that while most CD4+ and CD8+ T cells in HC skin are TRM, T cells in SSc lesions are characterized by a combination of skin-resident and recirculating T cells (~ 30%), particularly within the CD4+ T-cell compartment. These cells present a TMM [20] phenotype as they lack CD69 expression but co-express CCR7 and skin homing receptors (CLA/CCR4). Lack of SELL expression by TMM prevents these cells from recirculating via the lymphatics. Rather, SSc TMM cells likely recirculate between blood and skin, in agreement with our previous studies showing skin-tropic T cells in the blood of SSc patients.[4, 5] However, a small fraction of SSc TMM cells co-express CCR7 and SELL, and may recirculate between the blood, skin, and non-cutaneous distal lymph nodes. We observed that some TMM cells also co-express CD69, suggesting they may be in a transitional state. While it has been proposed that TMM represent the direct precursors of TRM cells, [20] pathogenic TMM cells might contribute directly to the development of SSc skin disease. As cutaneous TRM cells cannot leave the skin and recirculate nor differentiate into other memory T cell subsets they are believed to proliferate in situ at a low level.[33] Importantly, we also identified a cluster of proliferating TRM cells (#6) containing a higher number of cells deriving from SSc compared to HC samples. Resident and circulating SSc T cells exhibited a core gene expression signature including various genes involved in adaptive immune responses as well as oxidative phosphorylation and mitochondrial metabolic reprograming (clusters #4, 6, 7), likely reflecting chronic antigenic stimulation, as described in other autoimmune skin diseases.[34, 35] In agreement with our previous studies, demonstrating T-cell plasticity in blood and skin-resident SSc T cells compared to their HC counterparts,[4] we show here that some SSc TRM subsets (clusters #5, 8, 2) up-regulate Th1 and Th2 activation pathways and cytokine dysregulation. As previously shown,[36] these T-cell clusters also up-regulate expression of GATA-3, the master regulator of type-2 cytokines while maintaining adequate levels of TBX21. Interestingly, a recent phase II proof-of-concept study using romilkimab, a novel humanized antibody that neutralizes IL-4 and IL-13, in early dcSSc patients induced a significant reduction in modified Rodnan skin score in patients compared to placebo.[37] Cells from patient-samples also differentially express pro-inflammatory cytokines such IL-32, IL-26, IL-16, which are up-regulated in the serum and in the lesional skin of SSc patients.[38–40] These cytokines have been implicated in the pathogenesis of other inflammatory skin diseases[41–44] by promoting secretion of other inflammatory cytokines (TNFa, IL6) and chemokines (IL8, CXCL2) by macrophages and by acting as chemoattractants and modulators of T-cell activation. Single-cell trajectory analysis indicates that SSc CD4+ and CD8+ TRM cells are distributed throughout pseudotime, while SSc TMM are mostly found in late pseudotime, indicating distinct gene expression. Strikingly, both CD4+ and CD8+ TRM cells from HC samples were mostly distributed to early pseudotime whereas SSc T cells were enriched in late pseudotime and were characterized by up-regulation of genes associated with an activated effector-memory state. One of the trajectory branches showed enriched expression of primary markers of Tregs (FOXP3, IL2RA) and demonstrate enrichment of ICOS+ Tregs[45] in SSc samples. Several studies indicate that ICOS+ Tregs exhibit strong suppressive potential via up-regulation of CTLA-4, GITR (TNFRSF18), LAG3, and TIGIT as well as of IL-10 and TGFb.[45, 46] While we observed that most of these molecules were up-regulated in SSc compared to HC Tregs, CTLA-4, a major regulator of Treg suppressive function,[47] was down-regulated in SSc Tregs, implying a potential functional defect. Additionally, SSc Tregs up-regulate expression of PPP1CB, which was shown to make functionally defective Tregs in rheumatoid arthritis (RA) by dephosphorylating FOXP3 at the Ser 418 residue,[48] underlying another potential dysfunction in SSc Tregs.
We identified a subset of CD4+ T cells (cluster #7) in SSc skin that express a core gene signature including CXCL13, CD200, MS4A6A, ICOS, PD1, CTLA4, TIGIT and produce high levels of IL21 and IFNγ. A T-cell subset with a similar phenotype (CD4+CXCL13+CXCR5−) was recently described in the synovium and peripheral blood of RA patients as well as in the tumor microenvironment of breast cancer (BC).[49–51] This subset, named TFHX13 for the high-level expression of the signature chemokine CXCL13, phenotypically resembled TFH cells and was linked with long-term survival and an increase in tumor-infiltrating lymphocytes in BC whereas was associated with progressive disease in RA.[50, 51] Both studies showed that infiltrating TFHX13 cells induce B cell responses by promoting tissue-localized T–B-cell interactions and contribute to the development of tertiary lymphoid structures (TLOs). CXCL13 is a potent B-cell chemoattractant and a key factor for TLO formation.[52] Significantly, we showed by microarray analysis of cutaneous biopsies from patients with early dcSSc up-regulation of CXCL13 in a distinct subset of patients, which was associated with a T-cell and a B-cell gene expression signature. Our data indicate that CD3+CXCL13+ cells colocalize with B cells in inflamed dcSSc skin, suggesting in situ interactions as seen also in RA synovial tissues. Moreover, other studies observed upregulation of CXCL13 in the serum of SSc patients that correlates with disease severity,[53] as well as in idiopathic pulmonary fibrosis, where CXCL13 represents a prognostic marker.[54] Intriguingly, our data indicate a significant correlation between increased frequencies of skin recirculating CD3+CXCL13+ cells and ILD in the patients tested, warranting further investigation on the role this cell type in ILD pathogenesis. However, the frequency of CD3+CXCL13+ cells did not correlate with the skin score. We have known for several years that bulk SSc skin gene expression of T cell markers does not generally correlate well with the skin score.[55] The perivascular location of the CXCL13 T cells and lymphoid aggregates in the skin suggests that CXCL13 T cells are more important in the vascular disease in SSc than the fibrotic disease manifestations. These findings, as well as our observation that the frequency of CD3+CXCL13+ cells was significantly lower in patients treated with immunosuppressants, suggests that a comprehensive study based on a larger patient cohort with complete clinical details is necessary for understanding the pathogenic mechanisms at the clinical level.
Taylor et al, recently identified a similar subset of TFH-like cells (CD4+ICOShighPD-1highCXCR5+BCL6+) in SSc skin that contributed to dermal fibrosis via IL-21 production and that correlated with disease scores.[6] Moreover, Ricard et al, observed that circulating TFH cells (CD4+CXCR5+PD1+BCL6+) correlate with disease severity in SSc and induce B-cell differentiation into plasmablasts secreting Ig via IL-21 production.[56] In comparison, we show that SSc CD3+CXCL13+ cells express lower levels of ICOS and PD1, lack expression of the canonical TFH markers BCL6 and CXCR5, and express a TMM phenotype, suggesting a potentially distinct migratory capacity within inflamed skin. While BCL6 was long considered critical for IL-21 production, recent studies indicate that MAF may be involved instead[27] and indeed we found up-regulation of MAF in SSc CD3+CXCL13+ cells. Our data suggest that CD3+CXCL13+ cells are developmentally connected with ICOS+ Tregs. Significantly, studies in SLE show that high ICOS+ Tregs frequencies correlate positively with disease activity scores and serum auto-antibody titer,[57] and suggest that ICOS+ Tregs might represent the precursors of inflammatory cells. Thus, SSc CD3+CXCL13+ T cells may represent a distinct TFH subset that differentiates from conversion of ICOS+ Tregs and which is uniquely poised to promote B-cell responses and antibody production within pathologically inflamed non-lymphoid SSc skin lesions.
In conclusion, single-cell transcriptome profiling provides novel insights into SSc pathogenesis by revealing specific landscapes of T lymphocyte subsets. A better understanding of the immunological mechanisms underlying disease processes will lead to novel and targeted therapeutic approaches in SSc, thus realizing the goal of precision medicine.
Supplementary Material
KEY MESSAGES.
What is already known about this subject?
Current available therapies to reverse or even slow progression of systemic sclerosis (SSc), such as myeloablation, lead to broad killing of immune cells and consequent toxicities, including death.
Although T cells have been implicated in the pathogenesis of SSc, a comprehensive study of T-cell mediated immune responses in the affected skin of patients with progressive SSc is lacking.
What does this study add?
Our analysis provides an unprecedented view of T-cell heterogeneity in SSc skin.
We identified a distinct CXCL13+ T-cell subset that expresses factors enabling B-cell help and likely promoting B-cell responses and possibly autoantibodies production within pathological inflamed skin.
How might this impact on clinical practice or future developments?
These results provide a better understanding of T cell responses in SSc skin disease, allowing development of more targeted T cell therapy in SSc that results in better efficacy and less toxicity, thus realizing the goal of precision medicine.
Acknowledgements:
This work was supported by National Institutes of Health grant P50-AR060780 (PI: R. L.) and by the Pittsburgh Autoimmunity Center of Excellence in Rheumatology (PACER) to P.F.
Footnotes
Disclosure of Conflict of Interest: The authors declare no competing financial interests.
REFERENCES
- 1.Denton CP, Khanna D. Systemic sclerosis. Lancet. 2017. October 7; 390(10103):1685–1699. [DOI] [PubMed] [Google Scholar]
- 2.Fuschiotti P T cells and cytokines in systemic sclerosis. Current opinion in rheumatology. 2018. November; 30(6):594–599. [DOI] [PubMed] [Google Scholar]
- 3.Chizzolini C, Boin F. The role of the acquired immune response in systemic sclerosis. Seminars in immunopathology. 2015. September; 37(5):519–528. [DOI] [PubMed] [Google Scholar]
- 4.Li G, Larregina AT, Domsic RT, Stolz DB, Medsger TA Jr., Lafyatis R, et al. Skin-Resident Effector Memory CD8(+)CD28(−) T Cells Exhibit a Profibrotic Phenotype in Patients with Systemic Sclerosis. J Invest Dermatol. 2017. May; 137(5):1042–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuschiotti P, Larregina AT, Ho J, Feghali-Bostwick C, Medsger TA, Jr. Interleukin-13-producing CD8+ T cells mediate dermal fibrosis in patients with systemic sclerosis. Arthritis Rheum. 2013. January; 65(1):236–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor DK, Mittereder N, Kuta E, Delaney T, Burwell T, Dacosta K, et al. T follicular helper-like cells contribute to skin fibrosis. Sci Transl Med. 2018. March 7; 10(431). [DOI] [PubMed] [Google Scholar]
- 7.Guggino G, Lo Pizzo M, Di Liberto D, Rizzo A, Cipriani P, Ruscitti P, et al. Interleukin-9 over-expression and T helper 9 polarization in systemic sclerosis patients. Clinical and experimental immunology. 2017. November; 190(2):208–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mathian A, Parizot C, Dorgham K, Trad S, Arnaud L, Larsen M, et al. Activated and resting regulatory T cell exhaustion concurs with high levels of interleukin-22 expression in systemic sclerosis lesions. Ann Rheum Dis. 2012. July; 71(7):1227–1234. [DOI] [PubMed] [Google Scholar]
- 9.MacDonald KG, Dawson NA, Huang Q, Dunne JV, Levings MK, Broady R. Regulatory T cells produce profibrotic cytokines in the skin of patients with systemic sclerosis. J Allergy Clin Immunol. 2015. April; 135(4):946–e949. [DOI] [PubMed] [Google Scholar]
- 10.Maehara T, Kaneko N, Perugino CA, Mattoo H, Kers J, Allard-Chamard H, et al. Cytotoxic CD4+ T lymphocytes may induce endothelial cell apoptosis in systemic sclerosis. J Clin Invest. 2020. May 1; 130(5):2451–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuschiotti P T cells in SSc skin lesions: knowing your enemy. Nature reviews Rheumatology. 2020. May; 16(5):253–254. [DOI] [PubMed] [Google Scholar]
- 12.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015. May 21; 161(5):1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.LeRoy EC, Black C, Fleischmajer R, Jablonska S, Krieg T, Medsger TA Jr., et al. Scleroderma (systemic sclerosis): classification, subsets and pathogenesis. J Rheumatol. 1988. February; 15(2):202–205. [PubMed] [Google Scholar]
- 14.Steen VD, Medsger TA Jr. Severe organ involvement in systemic sclerosis with diffuse scleroderma. Arthritis Rheum. 2000. November; 43(11):2437–2444. [DOI] [PubMed] [Google Scholar]
- 15.Medsger TA Jr., Bombardieri S, Czirjak L, Scorza R, Della Rossa A, Bencivelli W Assessment of disease severity and prognosis. Clin Exp Rheumatol. 2003; 21(3 Suppl 29):S42–46. [PubMed] [Google Scholar]
- 16.Gaydosik AM, Tabib T, Geskin LJ, Bayan CA, Conway JF, Lafyatis R, et al. Single-Cell Lymphocyte Heterogeneity in Advanced Cutaneous T-cell Lymphoma Skin Tumors. Clin Cancer Res. 2019. July 15; 25(14):4443–4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaydosik AM, Queen DS, Trager MH, Akilov OE, Geskin L, Fuschiotti P. Genome-wide transcriptome analysis of the STAT6-regulated genes in advanced-stage cutaneous T-cell lymphoma. Blood. 2020. May 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nature methods. 2019. December; 16(12):1289–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kramer A, Green J, Pollard J Jr., Tugendreich S Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014. February 15; 30(4):523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watanabe R, Gehad A, Yang C, Scott LL, Teague JE, Schlapbach C, et al. Human skin is protected by four functionally and phenotypically discrete populations of resident and recirculating memory T cells. Sci Transl Med 2015. March 18; 7(279):279ra239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019. February; 566(7745):496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nature biotechnology. 2014. April; 32(4):381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szabo PA, Levitin HM, Miron M, Snyder ME, Senda T, Yuan J, et al. Single-cell transcriptomics of human T cells reveals tissue and activation signatures in health and disease. Nat Commun. 2019. October 17; 10(1):4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Milner JJ, Toma C, Yu B, Zhang K, Omilusik K, Phan AT, et al. Runx3 programs CD8(+) T cell residency in non-lymphoid tissues and tumours. Nature. 2017. December 14; 552(7684):253–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skon CN, Lee JY, Anderson KG, Masopust D, Hogquist KA, Jameson SC. Transcriptional downregulation of S1pr1 is required for the establishment of resident memory CD8+ T cells. Nat Immunol. 2013. December; 14(12):1285–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng C, Liu S, Zhang H, Guan R, Li D, Zhou F, et al. Dimension Reduction and Clustering Models for Single-Cell RNA Sequencing Data: A Comparative Study. Int J Mol Sci. 2020. March 22; 21(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jogdand GM, Mohanty S, Devadas S. Regulators of Tfh Cell Differentiation. Frontiers in immunology. 2016; 7:520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim D, Peck A, Santer D, Patole P, Schwartz SM, Molitor JA, et al. Induction of interferon-alpha by scleroderma sera containing autoantibodies to topoisomerase I: association of higher interferon-alpha activity with lung fibrosis. Arthritis Rheum. 2008. July; 58(7):2163–2173. [DOI] [PubMed] [Google Scholar]
- 29.Khanna D, Spino C, Johnson S, Chung L, Whitfield ML, Denton CP, et al. Abatacept in Early Diffuse Cutaneous Systemic Sclerosis: Results of a Phase II Investigator-Initiated, Multicenter, Double-Blind, Randomized, Placebo-Controlled Trial. Arthritis & rheumatology. 2020. January; 72(1):125–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ho AW, Kupper TS. T cells and the skin: from protective immunity to inflammatory skin disorders. Nat Rev Immunol. 2019. August; 19(8):490–502. [DOI] [PubMed] [Google Scholar]
- 31.Szabo PA, Miron M, Farber DL. Location, location, location: Tissue resident memory T cells in mice and humans. Sci Immunol. 2019. April 5; 4(34). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schenkel JM, Masopust D. Tissue-resident memory T cells. Immunity. 2014. December 18; 41(6):886–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park SL, Zaid A, Hor JL, Christo SN, Prier JE, Davies B, et al. Local proliferation maintains a stable pool of tissue-resident memory T cells after antiviral recall responses. Nat Immunol. 2018. February; 19(2):183–191. [DOI] [PubMed] [Google Scholar]
- 34.Geltink RIK, Kyle RL, Pearce EL. Unraveling the Complex Interplay Between T Cell Metabolism and Function. Annual review of immunology. 2018. April 26; 36:461–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Z, Fujii H, Mohan SV, Goronzy JJ, Weyand CM. Phosphofructokinase deficiency impairs ATP generation, autophagy, and redox balance in rheumatoid arthritis T cells. J Exp Med. 2013. September 23; 210(10):2119–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cascio S, Medsger TA Jr., Hawse WF, Watkins SC, Milcarek C, Moreland LW, et al. 14-3-3z sequesters cytosolic T-bet, upregulating IL-13 levels in TC2 and CD8(+) lymphocytes from patients with scleroderma. J Allergy Clin Immunol. 2018. July; 142(1):109–119 e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Allanore Y, Wung P, Soubrane C, Esperet C, Marrache F, Bejuit R, et al. A randomised, double-blind, placebo-controlled, 24-week, phase II, proof-of-concept study of romilkimab (SAR156597) in early diffuse cutaneous systemic sclerosis. Ann Rheum Dis. 2020. December; 79(12):1600–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Di Benedetto P, Guggino G, Manzi G, Ruscitti P, Berardicurti O, Panzera N, et al. Interleukin-32 in systemic sclerosis, a potential new biomarker for pulmonary arterial hypertension. Arthritis Res Ther. 2020. June 1; 22(1):127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou Y, Hou W, Xu K, Han D, Jiang C, Mou K, et al. The elevated expression of Th17-related cytokines and receptors is associated with skin lesion severity in early systemic sclerosis. Hum Immunol. 2015. January; 76(1):22–29. [DOI] [PubMed] [Google Scholar]
- 40.Kawabata K, Makino T, Makino K, Kajihara I, Fukushima S, Ihn H. IL-16 expression is increased in the skin and sera of patients with systemic sclerosis. Rheumatology. 2020. March 1; 59(3):519–523. [DOI] [PubMed] [Google Scholar]
- 41.Itoh T, Hatano R, Komiya E, Otsuka H, Narita Y, Aune TM, et al. Biological Effects of IL-26 on T Cell-Mediated Skin Inflammation, Including Psoriasis. J Invest Dermatol. 2019. April; 139(4):878–889. [DOI] [PubMed] [Google Scholar]
- 42.Kamijo H, Miyagaki T, Hayashi Y, Akatsuka T, Watanabe-Otobe S, Oka T, et al. Increased IL-26 Expression Promotes T Helper Type 17- and T Helper Type 2-Associated Cytokine Production by Keratinocytes in Atopic Dermatitis. J Invest Dermatol. 2020. March; 140(3):636–644 e632. [DOI] [PubMed] [Google Scholar]
- 43.Meyer N, Zimmermann M, Burgler S, Bassin C, Woehrl S, Moritz K, et al. IL-32 is expressed by human primary keratinocytes and modulates keratinocyte apoptosis in atopic dermatitis. J Allergy Clin Immunol. 2010. April; 125(4):858–865 e810. [DOI] [PubMed] [Google Scholar]
- 44.Kempuraj D, Conti P, Vasiadi M, Alysandratos KD, Tagen M, Kalogeromitros D, et al. IL-32 is increased along with tryptase in lesional psoriatic skin and is up-regulated by substance P in human mast cells. Eur J Dermatol. 2010. Nov-Dec; 20(6):865–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li DY, Xiong XZ. ICOS(+) Tregs: A Functional Subset of Tregs in Immune Diseases. Frontiers in immunology. 2020; 11:2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miragaia RJ, Gomes T, Chomka A, Jardine L, Riedel A, Hegazy AN, et al. Single-Cell Transcriptomics of Regulatory T Cells Reveals Trajectories of Tissue Adaptation. Immunity. 2019. February 19; 50(2):493–504 e497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rudd CE. CTLA-4 co-receptor impacts on the function of Treg and CD8+ T-cell subsets. Eur J Immunol. 2009. March; 39(3):687–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nie H, Zheng Y, Li R, Guo TB, He D, Fang L, et al. Phosphorylation of FOXP3 controls regulatory T cell function and is inhibited by TNF-alpha in rheumatoid arthritis. Nat Med. 2013. March; 19(3):322–328. [DOI] [PubMed] [Google Scholar]
- 49.Gu-Trantien C, Willard-Gallo K. PD-1(hi)CXCR5(−)CD4(+) TFH Cells Play Defense in Cancer and Offense in Arthritis. Trends Immunol. 2017. December; 38(12):875–878. [DOI] [PubMed] [Google Scholar]
- 50.Gu-Trantien C, Migliori E, Buisseret L, de Wind A, Brohee S, Garaud S, et al. CXCL13-producing TFH cells link immune suppression and adaptive memory in human breast cancer. JCI Insight. 2017. June 2; 2(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rao DA, Gurish MF, Marshall JL, Slowikowski K, Fonseka CY, Liu Y, et al. Pathologically expanded peripheral T helper cell subset drives B cells in rheumatoid arthritis. Nature. 2017. February 1; 542(7639):110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ansel KM, Ngo VN, Hyman PL, Luther SA, Forster R, Sedgwick JD, et al. A chemokine-driven positive feedback loop organizes lymphoid follicles. Nature. 2000. July 20; 406(6793):309–314. [DOI] [PubMed] [Google Scholar]
- 53.Wutte N, Kovacs G, Berghold A, Reiter H, Aberer W, Aberer E. CXCL13 and B-cell activating factor as putative biomarkers in systemic sclerosis. Br J Dermatol. 2013. September; 169(3):723–725. [DOI] [PubMed] [Google Scholar]
- 54.Vuga LJ, Tedrow JR, Pandit KV, Tan J, Kass DJ, Xue J, et al. C-X-C motif chemokine 13 (CXCL13) is a prognostic biomarker of idiopathic pulmonary fibrosis. American journal of respiratory and critical care medicine. 2014. April 15; 189(8):966–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Milano A, Pendergrass SA, Sargent JL, George LK, McCalmont TH, Connolly MK, et al. Molecular subsets in the gene expression signatures of scleroderma skin. PLoS One. 2008; 3(7):e2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ricard L, Jachiet V, Malard F, Ye Y, Stocker N, Riviere S, et al. Circulating follicular helper T cells are increased in systemic sclerosis and promote plasmablast differentiation through the IL-21 pathway which can be inhibited by ruxolitinib. Ann Rheum Dis. 2019. April; 78(4):539–550. [DOI] [PubMed] [Google Scholar]
- 57.Liu Y, Zhu T, Cai G, Qin Y, Wang W, Tang G, et al. Elevated circulating CD4+ ICOS+ Foxp3+ T cells contribute to overproduction of IL-10 and are correlated with disease severity in patients with systemic lupus erythematosus. Lupus. 2011. May; 20(6):620–627. [DOI] [PubMed] [Google Scholar]
- 58.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nature biotechnology. 2015. May; 33(5):495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


