Abstract
Computational models of the brain and its biomechanical response to skull accelerations are important tools for understanding and predicting traumatic brain injuries (TBIs). However, most models have been developed using experimental data collected on animal models and cadaveric specimens, both of which differ from the living human brain. Here we describe efforts to noninvasively measure the biomechanical response of the human brain with MRI—at non-injurious strain levels—and generate data that can be used to develop, calibrate, and evaluate computational brain biomechanics models. Specifically, this paper reports on a project supported by the National Institute of Neurological Disorders and Stroke to comprehensively image brain anatomy and geometry, mechanical properties, and brain deformations that arise from impulsive and harmonic skull loadings. The outcome of this work will be a publicly available dataset (http://www.nitrc.org/projects/bbir) that includes measurements on both males and females across an age range from adolescence to older adulthood. This article describes the rationale and approach for this study, the data available, and how these data may be used to develop new computational models and augment existing approaches; it will serve as a reference to researchers interested in using these data.
Keywords: traumatic brain injury, computational models, magnetic resonance imaging, deformation, strain
1. Introduction
1.1. Scope and Purpose
Traumatic brain injury (TBI), including mild TBI and concussion, is widespread65,95 and can have lasting and severe consequences10,63. Large, fast deformations of brain tissue caused by high skull accelerations undoubtedly contribute to TBI pathology43,105,110, and repeated, smaller brain deformations caused by sub-concussive accelerations likely underlie features of chronic traumatic encephalopathy (CTE)10,110. However, the relationships between skull acceleration, brain deformation, pathophysiology, and brain function are not fully understood. These biomechanical relationships are critical to unraveling the mechanisms of TBI and CTE and ultimately improving approaches to prevention and treatment. For instance, knowing how much acceleration and deformation leads to injury and what regions of the brain are likely to undergo injurious deformations can inform helmet design91. However, it is ethically impossible to answer these questions by experimental studies that risk injury to human subjects.
Computer simulation of human skull and brain mechanics offers an appealing and potentially powerful approach to provide both qualitative and quantitative insights into TBI51. A computational model can simulate the brain’s response to skull accelerations of arbitrary magnitude and direction, providing predictions of 3D displacement vector fields and strain tensor fields with high spatial and temporal resolution throughout the entire brain. However, it is crucial to remember that the predictions of computational models are not measurements. The accuracy and utility of these predictions depends on many factors, including the assumptions and approximations used to build the model38,69, but most critically on the data used in model development. To create an accurate and useful model, parameters such as the mechanical properties of brain tissue and its connections to the skull must be obtained from experimental measurements76. Similarly, the accuracy of predicted deformations can only be truly assessed by quantitative comparison to corresponding measurements of deformation (strain).
Given the present inability to capture experimental data on the living human brain at high (injurious) strains and strain rates, researchers have instead employed alternative methods to study skull-brain mechanics using animal models21,109 and cadaveric specimens1,2,45,46. Unfortunately, both the animal and cadaver brains differ from the intact, living human brain, thus limiting the utility of these data alone for developing models of TBI. Instead, we propose that noninvasive MRI measurements of deformation of the living human brain can be used to augment the development, calibration, and evaluation of computational models of brain biomechanics (Figure 1).
Figure 1.
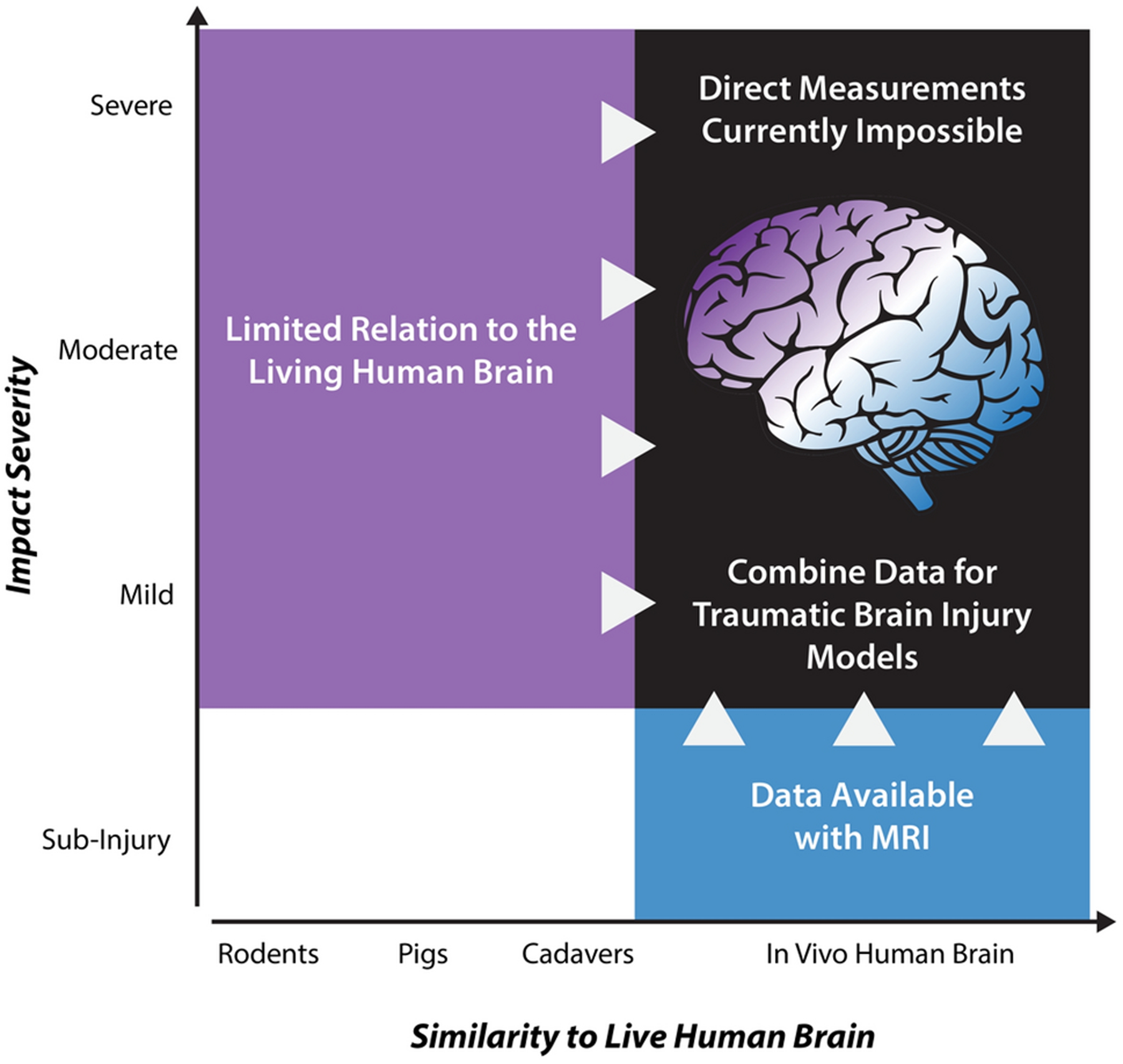
Computational models of traumatic brain injury (TBI) require experimental measurements of brain deformation in response to skull accelerations. Most models are built from data obtained in model systems (cells, animals, or cadaveric specimens) at high strains and strain rates relevant to TBI. However, none of these systems completely recapitulates the behavior of the living human brain. We propose to augment these data with noninvasive MRI measures of brain deformation at safe strains and strain rates to develop, calibrate, and evaluate TBI models.
To that end, we report new data on the mechanical behavior of the human brain, obtained using MRI techniques in human volunteers during sub-injury levels of skull acceleration, and made publicly available with support from the National Institute of Neurological Disorders and Stroke (NINDS) of the National Institutes of Health (NIH). We aim to provide comprehensive measurements of 3D deformation of the human brain through (i) measurements of motion in response to impulsive or harmonic skull motion, (ii) material properties, (iii) anatomy, and (iv) microstructure of the human brain. These measurements capture key features of the response of the intact, living human brain to skull motion and complement data available from cadaveric and animal studies. The major outcome of the project will be a publicly available dataset (http://www.nitrc.org/projects/bbir) accompanied by tools for using these data to parameterize and evaluate computational models of brain biomechanics under sub-injurious loading conditions.
1.2. Computational TBI Models
Computational models of the brain have become a valuable tool in biomechanics research to investigate TBI mechanisms, predict injury risk, and develop safety gear51,55,111. Models can illuminate the biomechanics of the brain during an impact by applying potentially injurious kinematic conditions that are impossible to achieve with human subjects, or even cadaveric specimens. These models have been used to create injury risk curves through proposed summary metrics of deformation, such as maximum principal strain (MPS), maximum axonal strain (MAS), and the cumulative strain damage measurement (CSDM)30,60,111,122. These risk curves, in turn, have informed injury mitigation approaches and consumer safety standards. The convenience and wide use of brain computational models emphasize the critical need for assessment of their biofidelity.
To draw physiologically relevant conclusions about TBI risk and mitigation, a model’s biomechanical response should ideally resemble that of a live human brain. However, few computational brain models have been evaluated by comparison to experimental measurements of living human brain deformation81,124. Instead, modelers have largely relied on cadaveric or animal studies. Many brain models have been evaluated using a single set of data from cadaveric brain motion under blunt impact45,46; more recently, measurements of high-rate, rotational cadaveric brain motion have been obtained using sonomicrometry1,2. Some models have also leveraged data from animal studies44,111, reconstructed impacts23,102,116, and on-field measurements in contact sports30,31, though none of these data directly provide the necessary measurements of brain deformation to properly develop or calibrate a model for the in vivo human brain.
The evaluation and interpretation of computational brain models is confounded by a number of factors. Model predictions depend on: (i) physical parameters like material properties, applied loading, and boundary and interface conditions; (ii) numerical parameters like mesh element type and size, time step, and temporal integration scheme; and (iii) geometry and anatomy, which may differ between the model and an individual subject38,69. Model outputs, usually displacement or strain time-history throughout the brain, are also reduced to a single global strain measure (e.g., MPS) thus ignoring the richness of spatial and temporal variations which could be used to evaluate a given model.
Detailed anatomy, microstructure, and strain measures from individual subjects can be used to create subject-specific models that account for variations in head and brain morphometry. Incorporating subject-specific anatomy has been motivated by studies showing variability in predicted brain deformation due to variations in geometry66, axonal tract alignment36, and impact kinematics117. While many models have recreated patient-specific impact scenarios, only a few studies have evaluated models using individual subject anatomy31,55,66,68,80 and strain data124. Additionally, subject-specific material properties, such as those derived from magnetic resonance elastography (MRE), have the potential to improve models though this has been explored only recently37,70.
1.3. Imaging Brain Biomechanics with MRI
MRI is well-suited to elucidate the mechanics of the live, human brain. Structural MRI offers excellent contrast in soft tissues, which enables high spatial resolution of subject-specific brain anatomy that can be segmented and classified using automated algorithms22,27,52. Diffusion tensor imaging (DTI) provides information about the orientation of white matter fibers9,82. Most importantly, there are several well-developed MRI acquisition techniques that are sensitive to motion and can be used to image how the brain moves in different conditions88,106,112. These methods offer opportunities for examining brain motion in ways that illuminate traumatic events, but at safe displacement and strain levels, using both harmonic and impulsive loading of the skull.
Magnetic resonance elastography (MRE) is a phase-contrast MRI technique that is sensitive to harmonic motion at scales of hundreds of nanometers to microns71,85. In MRE, vibration by an external driver leads to shear waves in soft tissue and the speed of shear waves is used to estimate tissue mechanical properties. MRE has been used to study the effects of neurodegeneration on brain viscoelasticity48,83 (the diseased brain is significantly softer than the healthy brain) as well to study normal development58,78,119 and aging4,49,100. Notable differences in tissue properties have been observed across the lifespan and between individuals47,50. Outcomes from MRE include whole-brain maps with spatially resolved tissue stiffness and viscosity, which are valuable data to incorporate into subject-specific TBI models. Additionally, the computed displacement fields illustrate the brain response to harmonic loading, including how waves propagate through the brain20,107 and generate different regional strain patterns86. Recent efforts have also sought to measure relative motion between the skull and brain to characterize their coupling6,7,120.
The brain’s response to impulsive loading of the skull (as in blunt impact) can be imaged at sub-injury levels using tagged MRI, an imaging method originally developed to noninvasively measure contractile function of the heart5,121. In tagged MRI, the longitudinal magnetization in the tissue is modulated to create temporary patterns of contrast that move with the tissue and can be analyzed to create maps of deformation and strain67,87,118. Advances in imaging and analysis techniques have established the ability to track 3D strains over time throughout the brain41. In the current method, repeated impulsive loading is applied to the skull through custom devices that restrict head motion to generate a repeatable accelerative loading on the order of 2–3 g11,25 (20–30 m/s2) or 150–350 rad/s2 17,61. These loading conditions are comparable to or smaller than accelerations due to other non-injurious, everyday activities29. This loading has been applied in several directions, including neck rotation17,41,61,62,98 (rotation in the axial plane), neck extension11,41,61 (translation and rotation in the sagittal plane), and neck flexion25. Diverse loading scenarios can uncover potential similarities and differences in brain response from impacts in different locations and directions.
Importantly, computational models that predict injury based on strain should be evaluated by comparison to experimental strain measurements124. The full-field measurements of dynamic, 3D strain throughout the brain in vivo at sub-injury levels provided by MRI-based methods are well-suited to this purpose, and complement estimates of strain from relatively sparse marker sets in the cadaveric brain125,126 at higher loading levels.
1.4. Study Design & Overview
The goal of this paper is to provide an overview and guide to new data describing the biomechanics of the living human brain. These new imaging and brain deformation datasets will help address gaps in the development and evaluation of computational TBI models. These data reflect spatially-resolved, three-dimensional motion from the whole brain in living humans. They will enable model construction and parameterization with anatomy (from MRI), microstructure (from DTI), and material properties (from MRE), and evaluation by comparison to subject-specific brain deformation (from tagged MRI). Data sets from multiple subjects will be collected and released to enable characterization of variability in the biomechanical response.
Data are being collected at three different sites: Washington University in St. Louis, University of Delaware, and the Clinical Center at the National Institutes of Health. The scan protocols differ between sites and focus on different aspects of brain biomechanics. Procedures include tagged MRI to measure brain response to impulsive loading, MRE to measure brain response to harmonic motion at multiple frequencies, and high-resolution MRE to map brain material properties. In the following section, we describe the measurements that will be shared with the community, and the methods used to obtain these data. We then discuss considerations and challenges in developing, calibrating, and evaluating TBI models using the full field data from this study.
2. MRI-Based Measurements and Analysis
Participants in this study will complete a scanning session that includes one of tagged MRI of impulsive loading, MRE of harmonic loading, or high-resolution MRE for material property mapping, depending on the site where data is collected, along with associated anatomical images. Each site will acquire data on approximately 100 participants including both males and females across a large age range (14–80 years old, split into four groups) to capture relevant age and sex differences. The complete dataset will include approximately 300 participants with data to be uploaded as it is collected.
2.1. Brain Response to Impulsive Loading Measured by Tagged MRI
Data on how the brain deforms in response to impulsive loading are acquired at the Clinical Center of the National Institutes of Health (NIH). These studies use tagged MRI to generate measurements of 3D deformation throughout the entire brain in response to repeated skull loading at sub-injury acceleration levels41,61.
We use the harmonic phase finite element method (HARP-FE) to compute 3D displacements and strain fields41 from multi-slice tagged MRI acquired during impulsive loading of the skull. We have previously demonstrated that this approach provides accurate and reliable 3D measurements of brain deformation41,61. The motions of interest are neck rotation (rotation within the axial plane of the head) and neck extension (rotation within the sagittal plane of the head). MRI-compatible devices constrain motion of the volunteer’s head to generate repeatable, mild head accelerations of approximately 150–350 rad/s2 and angular velocities at impact of roughly 1.5–3.5 rad/s, which are well below injury-level conditions. An angular position sensor provides measurements of angular position, velocity, and acceleration of the head cradle during impact. Brain motion is imaged with a multi-slice acquisition that incorporates tags along three orthogonal directions to provide full brain coverage at 18 ms temporal resolution, using fewer than 150 repetitions61. All tagged MRI data at NIH is acquired on a Siemens 3T Biograph scanner with flexible array and spine receive coils used together to accommodate the head support device.
Following MRI acquisition, 3D motion tracking is performed using the HARP-FE method, which uses HARP images as a forcing function for a finite element mesh41. HARP-FE generates dense, 3D measurements of Lagrangian displacement that are then projected from the finite element mesh back to the image space for the calculation of Lagrangian strains61. The outcomes of the tagged MRI experiments are full vector, time resolved measures of strain at each point in the brain for the entire duration of sampling around the impact (Figure 2). From these data many summary outcomes describing strain response can be computed, and anatomical and diffusion-weighted images can be combined with these measurements to estimate axonal strain and to map deformation of brain structures and vasculature61. In a previous study with the same acquisition and impact device, measured MPS values (95th percentile) in the brain ranged from 0.019 to 0.05361.
Figure 2.

Deformation (strain) in the brain of a human volunteer, measured with tagged MRI during its response to impulsive angular acceleration of the head. All components of the strain tensor (εij) are captured throughout the entire brain and across time. These strain fields can be used to calculate summary strain metrics, such as maximum principal strain (MPS).
2.2. Brain Response to Harmonic Skull Motion Measured by MR Elastography
Data on the brain response to harmonic motion are acquired at Washington University in St. Louis (WUSTL) using MRE over a wide range of frequencies. MRE offers a robust technique to capture full vector, 3D displacement fields throughout the entire brain48. The human brain appears to exhibit preferred modes of oscillation which may be selectively excited at different frequencies24,64, and thus these studies complement tagged MRI studies, in which broadband excitation is provided by impulsive loading. We additionally seek to capture both skull motion and brain motion6,7,120 to illuminate features of the skull-brain interface relevant to TBI.
Harmonic skull displacement is generated by the Resoundant pneumatic actuator system (Resoundant, Rochester, MN) with one of two passive drivers in contact with the head: a soft pillow driver for occipital actuation84, as is commonly used in brain MRE, or a flexible silicone bottle for lateral actuation, developed more recently for multi-excitation MRE experiments3,107. The WUSTL MRE protocol includes five separate scans with either occipital or lateral actuation at 20, 30, 50, 70, and 90 Hz. MRE displacement data is acquired with an echoplanar imaging (EPI) sequence for robust, whole-brain displacement data across a range of frequencies. Motion-encoding gradients (MEGs) applied in the sequence are tailored for frequency to balance motion sensitivity with echo time through fractional encoding97 or additional gradients. At each frequency, we acquire MRE displacement data over the whole brain (240×240×132 mm3) with 3 mm isotropic voxel resolution and 4 time points per period. Immediately following the whole brain acquisition, we use the same EPI sequence to acquire MRE displacement data with reduced MEG strength on six slices (3 mm isotropic voxels, 18 mm slice spacing) and 8 time points per period. This data captures rigid-body motion with minimal or no phase wrapping and allows us to reliably temporally unwrap the whole brain phase data6. Acquisition time for both scans at each frequency range from 5 minutes (20 Hz) to 3 minutes (90 Hz). The remainder of the protocol includes anatomical and diffusion-weighted scans, as described below. All imaging data at WUSTL are acquired on a Siemens 3T Prisma scanner with a 20-channel head/neck coil.
MRE images are masked to isolate voxels in the brain and the skull and scalp. From the reduced MEG strength displacement data, we estimate the rigid-body motion of the brain and scalp7, which we then use to temporally unwrap the whole-brain phase field to obtain accurate full-field displacement data (3D displacement vector at each voxel)6. Displacement data are processed to obtain rigid-body motion and dynamic deformation, including a full 3D strain tensor in each brain voxel at each frequency. These field variables are used to calculate scalar and vector measures that characterize the frequency response of the brain to applied harmonic motion86 (Figure 3), such as wave propagation direction20, octahedral shear strain75, and axonal strain by incorporating fiber direction from DTI data.
Figure 3.

Illustration of brain response to applied harmonic motion from MRE: Total displacement (u) and dynamic deformation (“wave displacement”, , obtained by subtracting rigid-body motion from u) of the brain with (A) occipital actuation and (B) lateral actuation at 50 Hz. Note the difference in scale for total displacement between actuation types. (C) Median octahedral shear strain (OSS) in the brain as a function of actuation frequency. Error bars show standard deviation of median values between subjects (N=4, occipital; N=7, lateral). Normalized OSS distributions, calculated as OSS divided by median OSS in the brain, for (D) occipital actuation and (E) lateral actuation.
2.3. Brain Material Properties Measured by MR Elastography
Measurements of the mechanical properties of the human brain are performed at the University of Delaware (UD). At UD, we use a high-resolution MRE sequence to capture whole-brain displacement data and an advanced inversion algorithm to generate spatially resolved maps of complex shear modulus, stiffness, and damping ratio (Figure 4). We have previously demonstrated that these methods result in property maps with improved resolution and reliability57, which can be used for subject-specific modeling with heterogeneous properties. As brain tissue mechanical properties are frequency-dependent19,99,113, capturing this behavior is critical for accurate and appropriate modeling, and thus MRE exams are performed at multiple frequencies.
Figure 4.

Illustration of mechanical property map outcomes from high-resolution MRE: (A) Corresponding anatomical image; (B) shear storage modulus, G’; (C) shear loss modulus, G”; (D) shear stiffness, μ; and (E) damping ratio, ξ. Property maps are created for each of three frequencies separately, 30, 50, and 70 Hz (only 50 Hz shown).
MRE displacement data are acquired with our 3D multiband, multishot spiral sequence56. This sequence allows for high spatial resolution through data sampling designed to maximize signal-to-noise ratio efficiency and minimize artifacts, with further correction for field inhomogeneities and motion-induced phase errors during image reconstruction in PowerGrid16. The MRE protocol includes three separate scans at 30, 50, and 70 Hz, with 1.5 mm isotropic imaging resolution and whole-brain coverage (240×240×120 mm3). To reduce scan time, we also implemented the OSCILLATE encoding scheme77. OSCILLATE exploits spatiotemporal correlations in MRE to reconstruct a set of images with reduced rank from undersampled data. This acceleration results in acquisition times of approximately 5 minutes per frequency. MRE scans encode displacements generated by the Resoundant pneumatic actuator system with a soft pillow driver. The remainder of the protocol includes reduced MEG scans to measure rigid body motion, as described above, and anatomical and diffusion-weighted scans, as described below. All imaging data at UD are being acquired on a Siemens 3T Prisma scanner with a 64-channel head/neck coil.
From each imaged displacement field, we estimate brain tissue material properties using the nonlinear inversion algorithm (NLI)74. NLI is a finite element-based optimization approach to the MRE inverse problem that models tissue as a heterogeneous, viscoelastic solid. NLI estimates the complex shear modulus G* = G’+iG”, with the storage modulus G’ describing elastic behavior of tissue and the loss modulus G” describing the viscous behavior. Additionally, we calculate the shear stiffness μ = 2|G*|2/(|G*|+G’), which generally describes the resistance of a viscoelastic material to harmonic forcing and is related to the square of the wave speed71, as well as the damping ratio ξ = G”/2G’, which describes the relative viscous-to-elastic behavior of the material72. These parameters are all determined at a single frequency and thus are reported for each of 30, 50, and 70 Hz; however, opportunity exists to model the frequency-dependence of the shear modulus directly in the NLI formulation113.
2.4. Brain Geometry and Microstructure from Anatomical and Diffusion-Weighted MRI
In addition to probing the mechanical behavior of the brain, MRI is used to characterize its anatomical structure, thus enabling construction of detailed brain models. MRI sequences can be tailored to map soft tissue structure, axonal orientation, and vasculature. T1- and T2-weighted images are acquired at approximately 1 mm isotropic spatial resolution as part of the protocol at each site. T1-weighted imaging provides exquisite contrast between gray and white matter, while T2-weighted imaging offers superior definition of the subarachnoid space, a key component in defining accurate boundary conditions in computational brain injury models. Diffusion tensor imaging (DTI) data are acquired using at least 30 non-collinear gradient directions at approximately 2 mm isotropic spatial resolution to characterize white matter fiber tracts. Finally, both susceptibility-weighted imaging (SWI) and time-of-flight MR angiography (MRA) are acquired in a subset of participants. These two acquisitions offer complementary depictions of cerebral vasculature, with MRA more revealing of arteries and SWI more sensitive to veins. Because of its ability to achieve submillimeter spatial resolution within a reasonable acquisition time103, SWI is also useful in finding thin dural membranes such as the falx cerebri and tentorium cerebelli.
Images are processed to provide voxel-wise label maps of various anatomical structures as well as to measure diffusion properties in white matter (Figure 5). To label cortical and subcortical gray matter structures, T1- and T2-weighted images will be skull-stripped96 and processed with a multi-atlas segmentation and cortical reconstruction algorithm52. From these same images and the segmentation labels, the subarachnoid space can be automatically defined, as preliminarily described by Glaister, et al40. Both the falx and tentorium will be reconstructed using a fast-marching, multi-atlas-based segmentation based on T1-weighted and SWI images39. Diffusion-weighted images will be processed using the TORTOISE software package53,90, leading to estimates of the diffusion tensor, fractional anisotropy, and mean diffusivity at every voxel, with major white matter fiber tracts labeled automatically12. Tools for automatically defining vasculature from MRA images are currently in development, with initial results described by Bilgel, et al14. This processing pipeline is executed on each dataset collected according to the exact protocol from each site, with original and processed images shared along with biomechanics imaging data.
Figure 5.

Images acquired to extract brain geometry and microstructure features relevant to computational brain models. Imaging data includes T1- and T2-weighted imaging, diffusion tensor imaging, susceptibility-weighted imaging, and time-of-flight MR angiography. From these data we extract labels of anatomical regions, white matter tract orientation, location of the falx and tentorium dural membranes, and the location of vasculature and meninges. Imaging data and extracted features are made available along with corresponding biomechanical imaging data.
2.5. Data Sharing
To facilitate the dissemination of the data and promote the sharing of results we have created a website hosted on the Neuroimaging Tools and Resources Collaboratory (NITRC) titled “Brain Biomechanics Imaging Resources” (http://www.nitrc.org/projects/bbir). The site lists the available datasets to download and Matlab software to facilitate initial exploration of the data. There are two corpuses of data: tagged MRI and MRE. The tagged MRI data to be shared consists of: 1) estimated displacement fields; 2) calculated strain fields; 3) head kinematics (angular positions, velocity, acceleration); and 4) corresponding anatomical MR images. The MRE data to be shared consists of: 1) raw displacement fields; 2) calculated strain fields; 3) estimated shear modulus maps; 4) head acceleration (if recorded); and 5) corresponding anatomical MR images. MRE and tagged MRI data will be shared in their original native image space, and the processed anatomical data will be registered to a standardized space (MNI-15228) and resampled at 0.8 mm isotropic digital resolution prior to sharing. For both the tagged MRI and MRE subjects, demographic and anthropometric information—e.g., age, sex, height, and weight—is provided. The data are currently distributed in the Neuroimaging Informatics Technology Initiative (NIfTI) file format. NIfTI is an open file format that provides a standardized way to disseminate the data without custom software libraries. All datasets are released under GNU general Public License v3.0 and with the acceptance of the NITRC copyright and license terms. All data collected in this study is collected following procedures approved by the appropriate Institutional Review Board, and all participants provide written, informed consent that included the public sharing of de-identified data.
Data acquisition is ongoing, so the number of subjects and available data will increase. Additionally, as processing pipelines enter stable phases, additional processed results will be made available on the data repository site. Furthermore, we anticipate that as these pipelines undergo revisions, there may be multiple updates to the available processed data.
3. Using Imaging Data to Improve Computational Models of Brain Biomechanics
The data described in this work and made publicly available through this project can contribute to the development, calibration, and evaluation (or validation) of new brain biomechanics models, as illustrated by Figure 6, which, in turn, can be used to make engineering predictions about brain deformation in TBI. Even with the availability of such a comprehensive dataset, building an ideal computational model is challenging and requires the development of tools to appropriately extract information relevant to brain injury, as well as the careful consideration of prediction uncertainty.
Figure 6.

Illustration of an example workflow for developing a computational brain biomechanics model from MRI data. Shown schematically from left to right, model development requires: (i) parameterization with anatomy, microstructure, material properties, boundary conditions, and input kinematics; (ii) calibration by comparison to a subset of measurements; and (iii) evaluation by comparison to different measurements.
3.1. Perspectives on the Next Generation of Computational Models
We envision that the next generation of computational models of brain biomechanics will comprise a large ensemble of subject-specific models, spanning a wide range of ages and both sexes, as well as population-average models for groups of different age and sex; these models would be developed, calibrated, and quantitatively evaluated by comparison to a wide range of high-quality experimental data.
Creating models based on subject-specific imaging data can overcome a significant source of uncertainty in model development and model performance: the anthropometric, biomechanical, and physiological differences between subjects which may affect the brain’s mechanical response. A model of a single subject also provides greater insights into model biofidelity, as well as an individualized prediction of TBI risk. An ideal scenario for model evaluation would be to build subject-specific computational models using geometry from anatomical images of each subject, and then to compare the output of these models with experimental brain deformation data for that subject. Each model would be calibrated using data from the individual subject under certain loading conditions, before predicting subject brain deformation under other loading conditions. To this end, a major contribution of this study is to provide datasets for creating and evaluating these models. We note that not every subject will complete every biomechanics imaging scan, and thus only a subset of subject-specific models will be built using all classes of available data. Specifically, we expect that fewer than 5 subjects will undergo both tagged MRI studies, which provide response to impact and thus data for model calibration, and MRE studies, which provide subject-specific material properties. Most subject-specific models will need to be built using individual anatomy and group-average material properties from 12 subjects per group (see section 3.3).
As an alternative to subject-specific models, group-average models that represent groups of similar age and gender (e.g. 14–17 year old males, 55–80 year old females, etc.) can be developed to investigate and predict differences in TBI risk. In this case, imaging data from the subjects within a particular group must be co-registered to some common space, which presents several challenges (see section 3.2). However, this approach offers opportunities for understanding the range of expected observations in a group and providing confidence intervals on predictions. This insight can similarly be achieved through an ensemble of subject-specific models that represent the range of anatomy and physiology in the general population and in which the accuracy of each model can be quantified. Such an ensemble of models would provide insight into the range of outcomes of a typical impact, and could identify common features of brain mechanics that might inform strategies for preventing or treating TBI. Both approaches (group-average models and ensembles of subject-specific models) require many experimental data sets across the population, which is a major contribution of this study.
3.2. Opportunities and Challenges with Incorporating Imaging Data in Models
While the imaging data collected in the study offer a wealth of information to modelers, there are multiple challenges when using imaging data in creating models. Many models use discrete regions based on anatomical image segmentations of white and gray matter, ventricles, and the falx and tentorium; however, such segmentations often impart sharp boundaries in the model or have to be smoothed to generate a realistic surface. Additionally, many regions important for brain biomechanics such as the cortical cerebrospinal fluid, meninges, and skull are not easily distinguished and require refined algorithms to segment automatically. The issue of discrete region segmentation is particularly important in building group-averaged models as the imaging data from multiple subjects must be co-registered to create necessary inputs. Registration strategies should be considered carefully, and ideally the error and uncertainty involved in these steps should be quantified.
The MRE data collected in this study offers an opportunity to also incorporate spatial heterogeneity in mechanical properties through detailed maps of shear modulus, which exhibit considerable variability between regions and across subjects47. However, it is important to note that while MRE measurements can provide linear viscoelastic material properties that govern very small shear deformations to the brain (on the order of ~10−4–10−3 strain)86, realistic head injury scenarios involve much larger strain magnitudes (~10−1–100 strain)54,101,123. Thus, MRE data do not capture stress-strain nonlinearity or features of finite strain behavior such as strain stiffening and compression-tension asymmetry. In contrast, many 3D brain simulations have implemented visco-hyperelastic material models15,38,69,79,94, with quasi-linear viscoelasticity using a Prony-series formulation of the relaxation modulus to describe behavior at dynamic strain rates31,123. Care and insight are required in using MRE data to parameterize the material models used for studying brain injury. Possible strategies include combining the spatial heterogeneity and magnitudes of shear moduli from MRE at small strains (<0.01 strain), with data from tagged MRI (0.05–0.10 strain), and data from large-strain responses in ex vivo tissue. Some researchers have already begun incorporating the MRE data in their models37,70, yet the optimal approach to this problem remains an open challenge.
Quantifying the uncertainty and accuracy of models also requires consideration. Each choice in creating a computational model from experimental data involves approximations or assumptions that impart uncertainty to the model. Quantifying this net uncertainty and its effect on the model predictions is desirable for a fair and rigorous comparison of model predictions to experimental observations (which have their own uncertainties). A typical uncertainty quantification (UQ) framework for computational models comprises108: (i) modeling the system, (ii) defining probabilistic models of input parameters to quantify the sources of uncertainty, and (iii) propagating the uncertainties from the input parameters through the computational model.
Monte Carlo (MC) simulations are a highly versatile and popular approach for propagating uncertainties through computational models, though computationally expensive13. For complex systems such as 3D models of brain biomechanics, surrogate models (also called metamodels) offer an efficient and powerful tool for UQ, as they approximate the behavior of a computational model with negligible computational cost per run. In addition to UQ, these models can also be used to conduct sensitivity analyses to identify the key parameters that have the strongest influence on the model predictions. Such information is helpful for both modelers (as it identifies “important” and “unimportant” input parameters) and experimentalists (as it identifies the measurements in which uncertainty should be minimized). Specific objectives in this area include developing agreement on metrics relevant to brain mechanics and TBI, and identifying metadata that captures uncertainties in both experiments and simulations.
A particular challenge in evaluating the accuracy of predicted 3D displacement or strain fields from computational models is defining objective metrics that capture the most important features of the prediction. Some simulation studies focus on prediction of injury from global, scalar quantities such as the maximum principal strain or the fraction of elements exceeding a certain threshold30,89; if these global scalar quantities are used for model evaluation, it discounts much of the time-resolved, tensorial field data available. Metrics proposed for model evaluation include the Correlation Score (CS)59, Correlation and Analysis (CORA)33 metric, ISO score8, and other generic error metrics such as root mean square error (RMSE) or sum of squared error (SSE). Such scores are helpful in summarizing the relationships between dynamic 3D data sets, typically using one number that indicates a benchmark level of accuracy. However, there does not yet appear to be consensus on how these scores can be used for calibration or evaluation124. In part this is because of the tensorial nature of the key fields: deformation and strain, along with their temporal variations. To that end, there have been efforts to recalibrate35 or design anew other metrics that measure spatiotemporal similarity of a tensor field42. Given the evolution of imaging methods that provide the full strain tensor as a function of position (even if at lower spatial or temporal resolution), we advocate the comparison of deformation (strain) predictions from a simulation with all of the available experimental strain data from corresponding loading conditions, including their variations across time. Continuing investigations into evaluation metrics and their interpretation, supplemented with the increased availability of brain deformation data, will allow for objective and unbiased evaluation of the next generation of computational brain models.
This project will provide data on brain anatomy and material properties from individuals grouped by age and sex, with the goal of allowing for the development of group-average models. The target for this project is to have such data from at least 12 individuals within each group; this number is a feasible contribution within the project period, but it is not known a priori whether it is a sufficient number for robust statistical characterization of the population at large. Challenges remain for the modeling community in how to combine these anatomical and physical data sets into useful models. For example, it is not obvious what is the best way to construct a mechanically-representative, group-average geometry from a set of anatomical images. The variability in anatomy and properties within each group, however, might inform confidence intervals for model parameters and predictions.
3.3. Limitations
As noted above, although all subjects in these studies will undergo multiple types of imaging scans, only a small subset of the subjects (< 5 subjects) will undergo every type of scan; thus, only a few of the subject-specific models can incorporate all classes of imaging data (subject specific anatomy, microstructure, and material properties). These few models will be valuable to compare with models that are subject-specific in terms of anatomy and microstructure, but with group-averaged material properties.
The sub-injury levels of skull acceleration in these studies mean that observations apply only indirectly to injurious events, thus a computational model that is calibrated solely using in vivo tagged MRI data for predicting injury may have significant uncertainty in predicting the response to higher accelerations. Existing computational models of the human brain have been evaluated using both high-rate cadaveric impact data46 and in vivo head rotation data62. While the former is generally associated with large peak acceleration amplitudes (≿ 8000 rad/s2) and small impulse durations (~3–5 ms), the latter consists of much smaller peak accelerations (< 500 rad/s2) and longer impulse durations (~40 ms). Importantly, many of the real-life concussive and subconcussive head impacts involve peak accelerations and loading durations that lie between these two extremes. This highlights the importance of considering both high-amplitude cadaveric and low-amplitude in vivo (tagged MRI) datasets for model evaluation123. Similarly, a model built and calibrated using only cadaveric data may be poor at predicting the response of the brain in vivo. Opportunities exist to bridge this gap by using both experimental imaging data collected in the living brain, and data from the cadaveric brain collected under higher accelerations and shorter loading durations. Complementary animal studies, for example in the pig, might be used to compare the response of in vivo and ex vivo brain behavior, although comparisons must account for the differences in size and anatomy.
MRE data are obtained under small strain conditions and thus do not directly reflect large-strain behavior important for modeling injury. Additionally, in MRE the assumption is typically made that brain tissue is isotropic, to simplify the property estimation problem. This assumption ignores the mechanical anisotropy imparted by the organization of axons in white matter tracts25,26,92,115. Anisotropy of white matter can be incorporated in the constitutive model18,31,34 or modeled explicitly,32,117 including the spatially varying direction of fiber alignment and the degree of dispersion both of which can be measured using DTI. Currently, assumptions about mechanical anisotropy are supported only by very limited data. While the degree of mechanical anisotropy in white matter is still a topic of active research15, anisotropic MRE of the brain is currently under development by our group and others73,93,114, with preliminary estimates of shear and tensile anisotropy of at least 10–30% in the fiber direction in white matter tracts104,107.
Practical considerations like MRI scan time and scanner coil size limit the type, duration, and frequency of loading for data collected in this project. All these aspects remain open areas of development for our group. In future work we anticipate broadening the types of motion and range of impact duration in tagged MRI experiments, and the harmonic excitation frequencies used in the MRE experiments.
4. Outlook
Through the data collected, processed, and disseminated in this project, we aim to provide new resources for developing computational brain models. Using MRI techniques, full field measurements of displacement, strain, and mechanical properties throughout the brain, acquired at sub-injury levels of skull acceleration, are now available in male and female subjects of ages from adolescence through older adulthood, with more data to be released as it is collected over the next several years. These data complement traditional biomechanics measurements from animal studies and cadaveric specimens, and they provide new avenues for parameterizing and evaluating models of brain biomechanics. With the range of participants involved, we expect models will be able to account for differences between individuals and investigate the potential effects of age and sex on the biomechanical response of the brain and risk of injury.
We look forward to working with the modeling community to incorporate these data, along with other brain biomechanics measurements from animal models and cadaveric specimens, into the next generation of brain biomechanics models. We believe this approach will be synergistic, enhancing the utility of both (i) the current, MRI-derived data from human volunteers, and (ii) complementary cadaveric data from other studies, for modeling the response of the intact, living human brain to injury-level accelerations. We hope these studies stimulate future interactions with and among those who develop and use computational models of brain biomechanics.
Acknowledgments
We acknowledge support from the National Institutes of Health grants U01NS112120 and R01/R56NS055951, the intramural research program in the Clinical Center of the National Institutes of Health, and the Department of Defense in the Center for Neuroscience and Regenerative Medicine.
Footnotes
Publisher's Disclaimer: The Version of Record of this article, as published and maintained by the publisher, is available online at: https://doi.org/10.1007/s10439-021-02820-0. The Version of Record is the version of the article after copy-editing and typesetting, and connected to open research data, open protocols, and open code where available. Any supplementary information can be found on the journal website, connected to the Version of Record.
Conflict of Interest Statement
Dr. Prince and Dr. Johnson have intellectual property rights related to some of the imaging methods described in this work.
Data Availability Statement
All data described in this work is available at http://www.nitrc.org/projects/bbir.
References
- 1.Alshareef A, Giudice JS, Forman J, Salzar RS, and Panzer MB. A novel method for quantifying human in situ whole brain deformation under rotational loading using sonomicrometry. J. Neurotrauma 35:780–789, 2018. [DOI] [PubMed] [Google Scholar]
- 2.Alshareef A, Giudice JS, Forman J, Shedd DF, Reynier KA, Wu T, Sochor S, Sochor MR, Salzar RS, and Panzer MB. Biomechanics of the Human Brain during Dynamic Rotation of the Head. J. Neurotrauma 37:1546–1555, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson AT, Van Houten EEW, McGarry MDJ, Paulsen KD, Holtrop JL, Sutton BP, Georgiadis JG, and Johnson CL. Observation of direction-dependent mechanical properties in the human brain with multi-excitation MR elastography. J. Mech. Behav. Biomed. Mater 59:538–546, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arani A, Murphy MC, Glaser KJ, Manduca A, Lake DS, Kruse SA, Jack CR, Ehman RL, and Huston J. Measuring the effects of aging and sex on regional brain stiffness with MR elastography in healthy older adults. Neuroimage 111:59–64, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Axel L, and Dougherty L. MR Imaging of Motion with Spatial Modulation of Magnetization. Radiology 171:841–845, 1989. [DOI] [PubMed] [Google Scholar]
- 6.Badachhape AA, Okamoto RJ, Durham RS, Efron BD, Nadell SJ, Johnson CL, and Bayly PV. The Relationship of Three-Dimensional Human Skull Motion to Brain Tissue Deformation in Magnetic Resonance Elastography Studies. J. Biomech. Eng 139:1–12, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Badachhape AA, Okamoto RJ, Johnson CL, and Bayly PV. Relationships between scalp, brain, and skull motion estimated using magnetic resonance elastography. J. Biomech 73:40–49, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barbat S, Fu Y, Zhan Z, and Gehre C. Objective rating metric for dynamic systems. Enhanc. Saf. Veh, 2013. [Google Scholar]
- 9.Basser PJ, and Pierpaoli C. Microstructural and physiological features of tissues elucidated by quantitative-diffusion-tensor MRI. 1996. J. Magn. Reson 111:209–219, 1996. [DOI] [PubMed] [Google Scholar]
- 10.Baugh CM, Stamm JM, Riley DO, Gavett BE, Shenton ME, Lin A, Nowinski CJ, Cantu RC, McKee AC, and Stern RA. Chronic traumatic encephalopathy: Neurodegeneration following repetitive concussive and subconcussive brain trauma. Brain Imaging Behav. 6:244–254, 2012. [DOI] [PubMed] [Google Scholar]
- 11.Bayly PV, Cohen TS, Leister EP, Ajo D, Leuthardt EC, and Genin GM. Deformation of the Human Brain Induced by Mild Acceleration. J. Neurotrauma 22:845–856, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bazin PL, Ye C, Bogovic JA, Shiee N, Reich DS, Prince JL, and Pham DL. Direct segmentation of the major white matter tracts in diffusion tensor images. Neuroimage 58:458–468, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berthaume MA, Dechow PC, Iriarte-Diaz J, Ross CF, Strait DS, Wang Q, and Grosse IR. Probabilistic finite element analysis of a craniofacial finite element model. J. Theor. Biol 300:242–253, 2012. [DOI] [PubMed] [Google Scholar]
- 14.Bilgel M, Roy S, Carass A, Nyquist PA, and Prince JL. Automated anatomical labeling of the cerebral arteries using belief propagation. Proc SPIE 8669:866918, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Budday S, Ovaert TC, Holzapfel GA, Steinmann P, and Kuhl E. Fifty Shades of Brain: A Review on the Mechanical Testing and Modeling of Brain Tissue. Arch. Comput. Methods Eng 27:1187–1230, 2020. [Google Scholar]
- 16.Cerjanic A, Holtrop JL, Ngo G-C, Leback B, Arnold G, Van Moer M, LaBelle G, Fessler JA, and Sutton BP. PowerGrid: A open source library for accelerated iterative magnetic resonance image reconstruction. Proc. Intl. Soc. Mag. Res. Med 525, 2016. [Google Scholar]
- 17.Chan DD, Knutsen AK, Lu YC, Yang SH, Magrath E, Wang WT, Bayly PV, Butman JA, and Pham DL. Statistical Characterization of Human Brain Deformation during Mild Angular Acceleration Measured in Vivo by Tagged Magnetic Resonance Imaging. J. Biomech. Eng 140:101005, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chatelin S, Deck C, and Willinger R. An anisotropic viscous hyperelastic constitutive law for brain material finite-element modeling. J. Biorheol 27:26–37, 2013. [Google Scholar]
- 19.Clayton EH, Garbow JR, and Bayly PV. Frequency-dependent viscoelastic parameters of mouse brain tissue estimated by MR elastography. Phys. Med. Biol 56:2391–2406, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clayton EH, Genin GM, and Bayly PV. Transmission, attenuation and reflection of shear waves in the human brain. J. R. Soc. Interface 9:2899–2910, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coats B, Binenbaum G, Smith C, Peiffer RL, Christian CW, Duhaime AC, and Margulies SS. Cyclic head rotations produce modest brain injury in infant piglets. J. Neurotrauma 34:235–247, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dale AM, Fischl B, and Sereno MI. Cortical Surface-Based Analysis. I. Segmentation and surface reconstruction. Neuroimage 9:179–194, 1999. [DOI] [PubMed] [Google Scholar]
- 23.Elkin BS, Gabler LF, Panzer MB, and Siegmund GP. Brain tissue strains vary with head impact location: A possible explanation for increased concussion risk in struck versus striking football players. Clin. Biomech 64:49–57, 2019. [DOI] [PubMed] [Google Scholar]
- 24.Escarcega JD, Knutsen AK, Okamoto RJ, Pham DL, and Bayly PV. Natural oscillatory modes of 3D deformation of the human brain in vivo. J. Biomech 119:110259, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng Y, Abney TM, Okamoto RJ, Pless RB, Genin GM, and Bayly PV. Relative brain displacement and deformation during constrained mild frontal head impact. J. R. Soc. Interface 7:1677–1688, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng Y, Okamoto RJ, Namani R, Genin GM, and Bayly PV. Measurements of mechanical anisotropy in brain tissue and implications for transversely isotropic material models of white matter. J. Mech. Behav. Biomed. Mater 23:117–132, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fischl B, and Dale AM. Measuring the thickness of the human cerebral cortex. Proc. Natl. Acad. Sci 97:11050–11055, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fonov VS, Evans AC, McKinstry RC, Almli CR, and Collins DL. Unbiased nonlinear average age-appropriate brain templates from birth to adulthood. Neuroimage 47:S102, 2009. [Google Scholar]
- 29.Funk JR, Cormier JM, Bain CE, Guzman H, Bonugli E, and Manoogian SJ. Head and Neck loading in everyday and vigorous activities. Ann. Biomed. Eng 39:766–776, 2011. [DOI] [PubMed] [Google Scholar]
- 30.Gabler LF, Crandall JR, and Panzer MB. Assessment of Kinematic Brain Injury Metrics for Predicting Strain Responses in Diverse Automotive Impact Conditions. Ann. Biomed. Eng 44:3705–3718, 2016. [DOI] [PubMed] [Google Scholar]
- 31.Ganpule S, Daphalapurkar NP, Ramesh KT, Knutsen AK, Pham DL, Bayly PV, and Prince JL. A Three-Dimensional Computational Human Head Model That Captures Live Human Brain Dynamics. J. Neurotrauma 34:2154–2166, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garimella HT, and Kraft RH. Modeling the mechanics of axonal fiber tracts using the embedded finite element method. Int. j. numer. method. biomed. eng 33:26–35, 2017. [DOI] [PubMed] [Google Scholar]
- 33.Gehre C, Gades H, and Wernicke P. Objective Rating of Signals Using Test and Simulation Responses. Proc. Int. Tech. Conf. Enhanc. Saf. Veh 2009:, 2009. [Google Scholar]
- 34.Giordano C, and Kleiven S. Evaluation of Axonal Strain as a Predictor for Mild Traumatic Brain Injuries Using Finite Element Modeling. Stapp Car Crash J. 58:29–61, 2014. [DOI] [PubMed] [Google Scholar]
- 35.Giordano C, and Kleiven S. Development of an Unbiased Validation Protocol to Assess the Biofidelity of Finite Element Head Models used in Prediction of Traumatic Brain Injury. Stapp Car Crash J. 60:363–471, 2016. [DOI] [PubMed] [Google Scholar]
- 36.Giordano C, Zappalà S, and Kleiven S. Anisotropic finite element models for brain injury prediction: the sensitivity of axonal strain to white matter tract inter-subject variability. Biomech. Model. Mechanobiol 16:1269–1293, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giudice JS, Alshareef A, Wu T, Knutsen AK, V Hiscox L, Johnson CL, and Panzer MB. Calibration of a Heterogeneous Brain Model Using a Subject-Specic Inverse Finite Element Approach. Front. Bioeng. Biotechnol, 2021.doi: 10.3389/fbioe.2021.664268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giudice JS, Zeng W, Wu T, Alshareef A, Shedd DF, and Panzer MB. An Analytical Review of the Numerical Methods used for Finite Element Modeling of Traumatic Brain Injury. Ann. Biomed. Eng 47:1855–1872, 2019. [DOI] [PubMed] [Google Scholar]
- 39.Glaister J, Carass A, Pham DL, Butman JA, and Prince JL. Falx cerebri segmentation via multi-atlas boundary fusion. Med Image Comput Comput Assist Interv 10433:92–99, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Glaister J, Shao M, Li X, Carass A, Roy S, Blitz AM, Prince JL, and Ellingsen L. Deformable model reconstruction of the subarachnoid space. Proc SPIE 10574:1057431, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gomez AD, Knutsen AK, Xing F, Lu YC, Chan D, Pham DL, Bayly P, and Prince JL. 3-D Measurements of Acceleration-Induced Brain Deformation via Harmonic Phase Analysis and Finite-Element Models. IEEE Trans. Biomed. Eng 66:1456–1467, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gomez AD, Stone ML, Bayly PV, and Prince JL. Quantifying tensor field similarity with global distributions and optimal transport. Lect. Notes Comput. Sci 11071 LNCS:428–436, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Graham DI, Adams JH, Nicoll JAR, Maxwell WL, and Gennarelli TA. The Nature, Distribution and Causes of Traumatic Brain Injury. Brain Pathol. 5:397–406, 1995. [DOI] [PubMed] [Google Scholar]
- 44.Hajiaghamemar M, Seidi M, and Margulies SS. Head Rotational Kinematics, Tissue Deformations, and Their Relationships to the Acute Traumatic Axonal Injury. J. Biomech. Eng 142:031006, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hardy WN, Foster CD, Mason MJ, Yang KH, King AI, and Tashman S. Investigation of Head Injury Mechanisms Using Neutral Density Technology and High-Speed Biplanar X-ray. Stapp Car Crash J 45:337–368, 2001. [DOI] [PubMed] [Google Scholar]
- 46.Hardy WN, Mason MJ, Foster CD, Shah CS, Kopacz JM, Yang KH, King AI, Bishop J, Bey M, Anderst W, and Tashman S. A Study of the Response of the Human Cadaver Head to Impact. Stapp Car Crash J 51:17–80, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hiscox LV, McGarry MDJ, Schwarb H, Van Houten EEW, Pohlig RT, Roberts N, Huesmann GR, Burzynska AZ, Sutton BP, Hillman CH, Kramer AF, Cohen NJ, Barbey AK, Paulsen KD, and Johnson CL. Standard-space atlas of the viscoelastic properties of the human brain. Hum. Brain Mapp 41:5282–5300, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hiscox LV, Johnson CL, Barnhill E, McGarry MDJ, Huston J, Van Beek EJR, Starr JM, and Roberts N. Magnetic resonance elastography (MRE) of the human brain: Technique, findings and clinical applications. Phys. Med. Biol 61:R401–R437, 2016. [DOI] [PubMed] [Google Scholar]
- 49.Hiscox LV, Johnson CL, McGarry MDJ, Perrins M, Littlejohn A, van Beek EJR, Roberts N, and Starr JM. High-resolution magnetic resonance elastography reveals differences in subcortical gray matter viscoelasticity between young and healthy older adults. Neurobiol. Aging 65:158–167, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hiscox LV, Schwarb H, McGarry MDJ, and Johnson CL. Aging brain mechanics: Progress and promise of magnetic resonance elastography. Neuroimage 232:117889, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Horstemeyer MF, Panzer MB, and Prabhu RK. State-of-the-Art Modeling and Simulation of the Brain’s Response to Mechanical Loads. Ann. Biomed. Eng 47:1829–1831, 2019. [DOI] [PubMed] [Google Scholar]
- 52.Huo Y, Plassard AJ, Carass A, Resnick SM, Pham DL, Prince JL, and Landman BA. Consistent cortical reconstruction and multi-atlas brain segmentation. Neuroimage 138:197–210, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Irfanoglu MO, Modi P, Nayak A, Hutchinson EB, Sarlls J, and Pierpaoli C. DRBUDDI (Diffeomorphic Registration for Blip-Up blip-Down Diffusion Imaging) method for correcting echo planar imaging distortions. Neuroimage 106:284–299, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ji S, and Zhao W. A Pre-computed Brain Response Atlas for Instantaneous Strain Estimation in Contact Sports. Ann. Biomed. Eng 43:1877–1895, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ji S, Zhao W, Ford JC, Beckwith JG, Bolander RP, Greenwald RM, Flashman LA, Paulsen KD, and McAllister TW. Group-wise evaluation and comparison of white matter fiber strain and maximum principal strain in sports-related concussion. J. Neurotrauma 32:441–454, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnson CL, Holtrop JL, Anderson AT, and Sutton BP. Brain MR elastography with multiband excitation and nonlinear motion-induced phase error correction. Proc. Intl. Soc. Mag. Res. Med 1951, 2016. [Google Scholar]
- 57.Johnson CL, Schwarb H, McGarry MDJ, Anderson AT, Huesmann GR, Sutton BP, and Cohen NJ. Viscoelasticity of subcortical gray matter structures. Hum. Brain Mapp 37:4221–4233, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnson CL, and Telzer EH. Magnetic resonance elastography for examining developmental changes in the mechanical properties of the brain. Dev. Cogn. Neurosci 33:176–181, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kimpara H, Nakahira Y, Iwamoto M, Miki K, Ichihara K, ichi Kawano S, and Taguchi T. Investigation of anteroposterior head-neck responses during severe frontal impacts using a brain-spinal cord complex FE model. Stapp Car Crash J. 50:509–544, 2006. [DOI] [PubMed] [Google Scholar]
- 60.Kleiven S Predictors for Traumatic Brain Injuries Evaluated through Accident Reconstructions. Stapp Car Crash J. 51:81–114, 2007. [DOI] [PubMed] [Google Scholar]
- 61.Knutsen AK, Gomez AD, Gangolli M, Wang W-T, Chan D, Lu Y-C, Christoforou E, Prince JL, Bayly PV, Butman JA, and Pham DL. In vivo estimates of axonal stretch and 3D brain deformation during mild head impact. Brain Multiphysics 1:100015, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Knutsen AK, Magrath E, McEntee JE, Xing F, Prince JL, Bayly PV, Butman JA, and Pham DL. Improved measurement of brain deformation during mild head acceleration using a novel tagged MRI sequence. J. Biomech 47:3475–3481, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Koerte IK, Lin AP, Willems A, Muehlmann M, Hufschmidt J, Coleman MJ, Green I, Liao H, Tate DF, Wilde EA, Pasternak O, Bouix S, Rathi Y, Bigler ED, Stern RA, and Shenton ME. A review of neuroimaging findings in repetitive brain trauma. Brain Pathol. 25:318–349, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Laksari K, Kurt M, Babaee H, Kleiven S, and Camarillo D. Mechanistic Insights into Human Brain Impact Dynamics through Modal Analysis. Phys. Rev. Lett 120:138101, 2018. [DOI] [PubMed] [Google Scholar]
- 65.Langlois JA, Rutland-Brown W, and Wald MM. The epidemiology and impact of traumatic brain injury: A brief overview. J. Head Trauma Rehabil 21:375–378, 2006. [DOI] [PubMed] [Google Scholar]
- 66.Li X, Zhou Z, and Kleiven S. An anatomically detailed and personalizable head injury model: Significance of brain and white matter tract morphological variability on strain. Biomech. Model. Mechanobiol 20:403–431, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu X, and Prince JL. Shortest path refinement for motion estimation from tagged MR images. IEEE Trans. Med. Imaging 29:1560–1572, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lu YC, Daphalapurkar NP, Knutsen AK, Glaister J, Pham DL, Butman JA, Prince JL, Bayly PV, and Ramesh KT. A 3D Computational Head Model Under Dynamic Head Rotation and Head Extension Validated Using Live Human Brain Data, Including the Falx and the Tentorium. Ann. Biomed. Eng 47:1923–1940, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Madhukar A, and Ostoja-Starzewski M. Finite Element Methods in Human Head Impact Simulations: A Review. Ann. Biomed. Eng 47:1832–1854, 2019. [DOI] [PubMed] [Google Scholar]
- 70.Madhukar A, and Ostoja-Starzewski M. Modeling and Simulation of Head Trauma Utilizing White Matter Properties from Magnetic Resonance Elastography. Modelling 1:225–241, 2020. [Google Scholar]
- 71.Manduca A, Oliphant TE, Dresner MA, Mahowald JL, Kruse SA, Amromin E, Felmlee JP, Greenleaf JF, and Ehman RL. Magnetic Resonance Elastography: Non-Invasive Mapping of Tissue Elasticity. Med. Image Anal 5:237–254, 2001. [DOI] [PubMed] [Google Scholar]
- 72.McGarry MDJ, and Van Houten EEW. Use of a Rayleigh damping model in elastography. Med. Biol. Eng. Comput 46:759–766, 2008. [DOI] [PubMed] [Google Scholar]
- 73.McGarry MDJ, Van Houten EEW, Guertler C, Okamoto RJ, Smith DR, Sowinski D, Johnson CL, Bayly PV, Weaver JB, and Paulsen KD. A heterogenous, time harmonic, nearly incompressible transverse isotropic finite element brain simulation platform for MR elastography. Phys. Med. Biol 66:055029, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McGarry MDJ, Van Houten EEW, Johnson CL, Georgiadis JG, Sutton BP, Weaver JB, and Paulsen KD. Multiresolution MR elastography using nonlinear inversion. Med. Phys 39:6388–6396, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McGarry MDJ, Van Houten EEW, Perrĩez PR, Pattison AJ, Weaver JB, and Paulsen KD. An octahedral shear strain-based measure of SNR for 3D MR elastography. Phys. Med. Biol 56:N153–N164, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McGill K, Teixeira-Dias F, and Callanan A. A review of validation methods for the intracranial response of FEHM to blunt impacts. Appl. Sci 10:1–34, 2020. [Google Scholar]
- 77.McIlvain G, Cerjanic A, Christodoulou AG, Mcgarry MDJ, and Johnson CL. OSCILLATE: A Low-Rank Approach for Accelerated Magnetic Resonance Elastography. Proc. Intl. Soc. Mag. Res. Med 169, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McIlvain G, Schwarb H, Cohen NJ, Telzer EH, and Johnson CL. Mechanical properties of the in vivo adolescent human brain. Dev. Cogn. Neurosci 34:27–33, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mihai LA, Budday S, Holzapfel GA, Kuhl E, and Goriely A. A family of hyperelastic models for human brain tissue. J. Mech. Phys. Solids 106:60–79, 2017. [Google Scholar]
- 80.Miller LE, Urban JE, and Stitzel JD. Development and validation of an atlas-based finite element brain model. Biomech. Model. Mechanobiol 15:1201–1214, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Miller LE, Urban JE, and Stitzel JD. Validation performance comparison for finite element models of the human brain. Comput. Methods Biomech. Biomed. Engin 20:1273–1288, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mori S, and Zhang J. Principles of Diffusion Tensor Imaging and Its Applications to Basic Neuroscience Research. Neuron 51:527–539, 2006. [DOI] [PubMed] [Google Scholar]
- 83.Murphy MC, Huston J, and Ehman RL. MR elastography of the brain and its application in neurological diseases. Neuroimage 187:176–183, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Murphy MC, Huston J, Jack CR, Glaser KJ, Manduca A, Felmlee JP, and Ehman RL. Decreased brain stiffness in Alzheimer’s disease determined by magnetic resonance elastography. J. Magn. Reson. Imaging 34:494–498, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Muthupillai R, Lomas DJ, Rossman PJ, Greenleaf JF, Manduca A, and Ehman RL. Magnetic resonance elastography by direct visualization of propagating acoustic strain waves. Science (80-.) 269:1854–1857, 1995. [DOI] [PubMed] [Google Scholar]
- 86.Okamoto RJ, Romano AJ, Johnson CL, and V Bayly P. Insights Into Traumatic Brain Injury From MRI of Harmonic Brain Motion. J. Exp. Neurosci 13:117906951984044, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Osman NF, Kerwin WS, McVeigh ER, and Prince JL. Cardiac motion tracking using CINE harmonic phase (HARP) magnetic resonance imaging. Magn. Reson. Med 42:1048–1060, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pahlavian SH, Oshinski J, Zhong X, Loth F, and Amini R. Regional Quantification of Brain Tissue Strain Using Displacement-Encoding with Stimulated Echoes Magnetic Resonance Imaging. J. Biomech. Eng 140:1–13, 2018. [DOI] [PubMed] [Google Scholar]
- 89.Panzer MB, Myers BS, Capehart BP, and Bass CR. Development of a finite element model for blast brain injury and the effects of CSF cavitation. Ann. Biomed. Eng 40:1530–1544, 2012. [DOI] [PubMed] [Google Scholar]
- 90.Pierpaoli C, Walker L, Irfanoglu MO, Barnett A, Basser P, Chang L-C, Koay CG, Pajevic S, Sarlls J, and Wu M. TORTOISE: an integrated software package for processing of diffusion MRI data. Proc. Intl. Soc. Mag. Res. Med 1597, 2010. [Google Scholar]
- 91.Post A, Oeur A, Hoshizaki B, and Gilchrist MD. An examination of American football helmets using brain deformation metrics associated with concussion. Mater. Des 45:653–662, 2013. [Google Scholar]
- 92.Prange MT, and Margulies SS. Regional, directional, and age-dependent properties of the brain undergoing large deformation. J. Biomech. Eng 124:244–252, 2002. [DOI] [PubMed] [Google Scholar]
- 93.Romano A, Scheel M, Hirsch S, Braun J, and Sack I. In vivo waveguide elastography of white matter tracts in the human brain. Magn. Reson. Med 68:1410–1422, 2012. [DOI] [PubMed] [Google Scholar]
- 94.De Rooij R, and Kuhl E. Constitutive Modeling of Brain Tissue: Current Perspectives. Appl. Mech. Rev 68:010801, 2016. [Google Scholar]
- 95.Roozenbeek B, Maas AIR, and Menon DK. Changing patterns in the epidemiology of traumatic brain injury. Nat. Rev. Neurol 9:231–236, 2013. [DOI] [PubMed] [Google Scholar]
- 96.Roy S, Butman JA, and Pham DL. Robust skull stripping using multiple MR image contrasts insensitive to pathology. Neuroimage 146:132–147, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rump J, Klatt D, Braun J, Warmuth C, and Sack I. Fractional encoding of harmonic motions in MR elastography. Magn. Reson. Med 57:388–395, 2007. [DOI] [PubMed] [Google Scholar]
- 98.Sabet AA, Christoforou E, Zatlin B, Genin GM, and Bayly PV. Deformation of the human brain induced by mild angular head acceleration. J. Biomech 41:307–315, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sack I, Jöhrens K, Wuerfel J, and Braun J. Structure-sensitive elastography: on the viscoelastic powerlaw behavior of in vivo human tissue in health and disease. Soft Matter 9:5672–5680, 2013. [Google Scholar]
- 100.Sack I, Streitberger KJ, Krefting D, Paul F, and Braun J. The influence of physiological aging and atrophy on brain viscoelastic properties in humans. PLoS One 6:e23451, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sahoo D, Deck C, and Willinger R. Brain injury tolerance limit based on computation of axonal strain. Accid. Anal. Prev 92:53–70, 2016. [DOI] [PubMed] [Google Scholar]
- 102.Sanchez EJ, Gabler LF, Good AB, Funk JR, Crandall JR, and Panzer MB. A reanalysis of football impact reconstructions for head kinematics and finite element modeling. Clin. Biomech 64:82–89, 2019. [DOI] [PubMed] [Google Scholar]
- 103.Sati P, Thomasson DM, Li N, Pham DL, Biassou NM, Reich DS, and Butman JA. Rapid, high-resolution, whole-brain, susceptibility-based MRI of multiple sclerosis. Mult. Scler. J 20:1464–1470, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schmidt JL, Tweten DJ, Badachhape AA, Reiter AJ, Okamoto RJ, Garbow JR, and Bayly PV. Measurement of anisotropic mechanical properties in porcine brain white matter ex vivo using magnetic resonance elastography. J. Mech. Behav. Biomed. Mater 79:30–37, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shaw NA The neurophysiology of concussion. Prog. Neurobiol 67:281–344, 2002. [DOI] [PubMed] [Google Scholar]
- 106.Sloots JJ, Biessels GJ, and Zwanenburg JJM. Cardiac and respiration-induced brain deformations in humans quantified with high-field MRI. Neuroimage 210:116581, 2020. [DOI] [PubMed] [Google Scholar]
- 107.Smith DR, Guertler CA, Okamoto RJ, Romano AJ, Bayly PV, and Johnson CL. Multi-excitation magnetic resonance elastography of the brain: Wave propagation in anisotropic white matter. J. Biomech. Eng 142:071005, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sudret B, Marelli S, and Wiart J. Surrogate models for uncertainty quantification: An overview. Eur. Conf. Antennas Propag 793–797, 2017. [Google Scholar]
- 109.Sullivan S, Eucker SA, Gabrieli D, Bradfield C, Coats B, Maltese MR, Lee J, Smith C, and Margulies SS. White matter tract-oriented deformation predicts traumatic axonal brain injury and reveals rotational direction-specific vulnerabilities. Biomech. Model. Mechanobiol 14:877–896, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tagge CA et al. Concussion, microvascular injury, and early tauopathy in young athletes after impact head injury and an impact concussion mouse model. Brain 141:422–458, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Takhounts EG, Craig MJ, Moorhouse K, McFadden J, and Hasija V. Development of Brain Injury Criteria (BrIC). Stapp Car Crash J. 57:243–266, 2013. [DOI] [PubMed] [Google Scholar]
- 112.Terem I, Ni WW, Goubran M, Rahimi MS, Zaharchuk G, Yeom KW, Moseley ME, Kurt M, and Holdsworth SJ. Revealing sub-voxel motions of brain tissue using phase-based amplified MRI (aMRI). Magn. Reson. Med 80:2549–2559, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Testu J, McGarry MDJ, Dittmann F, Weaver JB, Paulsen KD, Sack I, and Van Houten EEW. Viscoelastic power law parameters of in vivo human brain estimated by MR elastography. J. Mech. Behav. Biomed. Mater 74:333–341, 2017. [DOI] [PubMed] [Google Scholar]
- 114.Tweten DJ, Okamoto RJ, Schmidt JL, Garbow JR, and Bayly PV. Estimation of material parameters from slow and fast shear waves in an incompressible, transversely isotropic material. J. Biomech 48:4002–4009, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Velardi F, Fraternali F, and Angelillo M. Anisotropic constitutive equations and experimental tensile behavior of brain tissue. Biomech. Model. Mechanobiol 5:53–61, 2006. [DOI] [PubMed] [Google Scholar]
- 116.Viano DC, Casson IR, Pellman EJ, Zhang L, King AI, and Yang KH. Concussion in professional football: Brain responses by finite element analysis: Part 9. Neurosurgery 57:891–915, 2005. [DOI] [PubMed] [Google Scholar]
- 117.Wu T, Alshareef A, Giudice JS, and Panzer MB. Explicit Modeling of White Matter Axonal Fiber Tracts in a Finite Element Brain Model. Ann. Biomed. Eng 47:1908–1922, 2019. [DOI] [PubMed] [Google Scholar]
- 118.Xing F, Woo J, Gomez AD, Pham DL, Bayly PV, Stone M, and Prince JL. Phase Vector Incompressible Registration Algorithm for Motion Estimation from Tagged Magnetic Resonance Images. IEEE Trans. Med. Imaging 36:2116–2128, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yeung J, Jugé L, Hatt A, and Bilston LE. Paediatric brain tissue properties measured with magnetic resonance elastography. Biomech. Model. Mechanobiol 18:1497–1505, 2019. [DOI] [PubMed] [Google Scholar]
- 120.Yin Z, Sui Y, Trzasko JD, Rossman PJ, Manduca A, Ehman RL, and Huston J. In vivo characterization of 3D skull and brain motion during dynamic head vibration using magnetic resonance elastography. Magn. Reson. Med 80:2573–2585, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zerhouni EA, Parish DM, Rogers WJ, Yang A, and Shapiro EP. Human heart: Tagging with MR imaging - A new method for noninvasive assessment of myocardial motion. Radiology 169:59–63, 1988. [DOI] [PubMed] [Google Scholar]
- 122.Zhang L, Yang KH, and King AI. A Proposed Injury Threshold for Mild Traumatic Brain Injury. J. Biomech. Eng 126:226–236, 2004. [DOI] [PubMed] [Google Scholar]
- 123.Zhao W, Choate B, and Ji S. Material properties of the brain in injury-relevant conditions – Experiments and computational modeling. J. Mech. Behav. Biomed. Mater 80:222–234, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhao W, and Ji S. Displacement- and Strain-Based Discrimination of Head Injury Models across a Wide Range of Blunt Conditions. Ann. Biomed. Eng 48:1661–1677, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhao W, Wu Z, and Ji S. Displacement Error Propagation From Embedded Markers to Brain Strain. J. Biomech. Eng, 2021.doi: 10.1115/1.4051050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhou Z, Li X, Kleiven S, and Hardy WN. Brain Strain from Motion of Sparse Markers. Stapp Car Crash J. 63:1–26, 2019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data described in this work is available at http://www.nitrc.org/projects/bbir.


