Abstract
B cells within germinal centers (GCs) enter cycles of antibody affinity maturation or exit the GC as memory cells or plasma cells. Here we examined the contribution of interleukin (IL)-4 on B cell fate decisions in the GC. Single cell RNA-sequencing identified a subset of light zone GC B cells expressing high IL4 receptor-a (IL4Ra) and CD23 and lacking a Myc-associated signature. These cells could differentiate into pre-memory cells. B cell-specific deletion of IL4Ra or STAT6 favored the pre-memory cell trajectory, and provision of exogenous IL4 in a wild-type context reduced pre-memory cell frequencies. IL4 acted during antigen-specific interactions but also influenced bystander cells. Deletion of IL4Ra from follicular dendritic cells (FDCs) increased IL4 availability in the GC, impaired selection of affinity-matured B cells and reduced memory cell generation. We propose that GC FDCs establish a niche that limits bystander IL4 activity, focusing IL4 action on B cells undergoing selection and enhancing memory cell differentiation.
Graphiccal Abstract
eTOC blurb
B cells within germinal centers (GCs) enter cycles of antibody affinity maturation or exit the GC as memory cells or plasma cells. Duan, Liu et al. examine the contribution of interleukin (IL)-4 on B cell fate decisions in the GC and reveal that IL4Ra+ GC follicular dendritic cells limit bystander IL-4 activity, focusing IL4 action on B cells undergoing selection and enhancing memory cell differentiation.
Introduction
Germinal centers (GCs) are important sites of affinity-based B cell selection (Bannard and Cyster, 2017; MacLennan, 1994; Victora and Nussenzweig, 2012). GCs are divided into a dark zone (DZ) and a light zone (LZ) and GC B cells move cyclically between these compartments. The DZ is composed of proliferating GC B cells that highly express activation induced-cytidine deaminase (AID) and undergo Ig gene somatic hypermutation. The LZ contains GC B cells and a dense network of follicular dendritic cells (FDCs) that play a crucial role in displaying opsonized antigens to the B cells. The LZ is also the main site for GC B cell interaction with T follicular helper (Tfh) cells. Pre-memory cells are found within the LZ, marked by reduced expression of sphingosine-1-phosphate receptor 2 (S1PR2) and the transcription factor Bcl6 and increased expression of several surface markers including CCR6 and CD38 (Laidlaw et al., 2017; Suan et al., 2017; Wang et al., 2017). Pre-plasma cells can be found amongst both LZ and DZ GC cells (Ise et al., 2018; Krautler et al., 2017; Radtke and Bannard, 2018; Zhang et al., 2018).
GC responses are T cell-dependent and positive selection of GC B cells requires receipt of CD40L signals from T follicular helper (Tfh) cells. Tfh cells also are an important source of several cytokines, including interleukin (IL)-4. Mice lacking IL4 or IL4Ra, or lacking STAT6 and thus having defective IL4Ra signaling, exhibit strong defects in isotype switching to IgG1 and IgE but variable effects on the GC response, with some studies reporting no effect and others showing a marked defect in the magnitude of the response or a reduction in affinity maturation (Andoh et al., 2000; Chevrier et al., 2017; Cunningham et al., 2004; Gaya et al., 2018; Gonzalez et al., 2018; Reinhardt et al., 2009; Reiter and Pfeffer, 2002; Turqueti-Neves et al., 2014; Vajdy et al., 1995). Deficiency in IL4 (and IL13) or in STAT6 can lead to a reduction in the GC LZ relative to the DZ compartment (Gonzalez et al., 2018; Turqueti-Neves et al., 2014).
The magnitude of IL4 production by Tfh cells is thought to be lower than by Th2-type effector cells and this has been suggested to allow more restricted delivery of IL4 to cognate partner B cells (Crotty, 2014). However, T cells release IL4 in a non-directed manner (‘IL-4 broadcasting’), in contrast to the polarized secretion of IL-2 (Huse et al., 2006). In lymph nodes (LNs) responding to a helminth infection, a strong Th2-inducing condition, IL-4 is available non-cognately across the entire LN (Perona-Wright et al., 2010; Turqueti-Neves et al., 2014). These findings raise the question of whether there are mechanisms that cooperate with low IL-4 production to favor the cognate actions of IL-4 during B cell selection in GCs.
In an earlier scRNA-seq analyses of stromal cell subsets in lymphoid tissue (Rodda et al., 2018), we found that IL4Ra is expressed in FDCs, raising questions as to whether the IL-4 receptor acts intrinsically in LZ cells and whether FDC IL4Ra could affect cell decision-making during this GC stage. We therefore performed experiments to better define how IL-4 acts within the GC. Using droplet-based scRNA-seq, we identified a LZ GC B cell cluster marked by high IL4Ra and CD23 expression and lacking a Myc gene-expression signature. RNA-based cell-fate trajectory and flow cytometry analysis demonstrated that this CD23+ cluster contained pre-memory cells and had the potential to generate pre-plasma cells. scV(D)J-seq revealed that pre-memory cells harbor fewer affinity-improving mutations than pre-plasma cells. IL4Ra and STAT6 functioned intrinsically to promote the CD23+ LZ state and restrained pre-memory B cell generation. This trajectory was dependent on IL-4 availability: Loss of IL4Ra from FDCs was associated with increased IL-4 availability, less stringent affinity-based selection and reduced memory cell generation. We propose that FDCs provide a niche that limits bystander IL-4 activity, focusing IL-4 action on B cells undergoing selection and enhancing memory B cell generation.
Results
Identification of an IL4Ra+ CD23+ LZ GC B cell subset by scRNA-seq
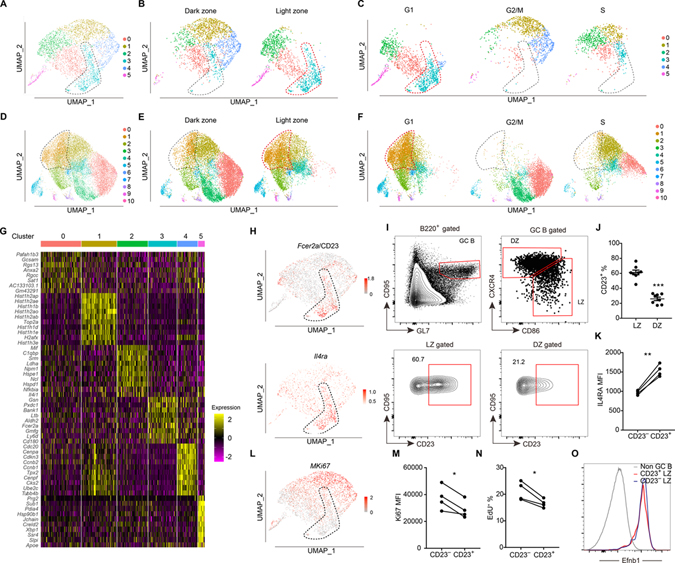
To obtain insight into the IL4-responsive B cells within GCs, we performed scRNA-seq on 3542 GC B cells at day 7 after immunization with 4-hydroxy-3-nitrophenyl (NP)-haptenated antigen (NP-CGG). Clustering analysis led us to identify a subset (cluster 3) of GC B cells that mostly had a LZ gene expression signature and was at the G1 phase of cell cycle (Fig. 1A–C). A lower depth sequencing analysis of 23091 cells at day 14 after immunization revealed a greater amount of GC cell heterogeneity but identified an analogous cluster (cluster 1, Fig. 1D–F and Fig. S1A). Expression of mRNA for Il4ra was enriched in a predominantly LZ cluster and other distinguishing genes included CD23 (Fcer2a), lymphotoxin-b (Ltb), Bank1 and Gelsolin (Gsn) (Fig. 1G, H, and Table S1). Each of these genes is IL4 inducible in activated B cells (Defrance et al., 1987; Paterson et al., 1996) and CD23 is also inducible in transitional B cells (Granato et al., 2014). For simplicity, we refer to day 7 cluster 3 and day 14 cluster 1 as the ‘CD23+ cluster’, though it should be recognized that smaller frequencies of Fcer2a high cells were present in some other clusters.
Figure 1. Identification of a CD23+ LZ GC B cell subset by scRNA-seq.
(A-C) UMAP plots of sorted B220+ CD38− GL7hi IgD− GC B cells at day 7 post-immunization (A), split between LZ and DZ (B) and split among different cell cycle phases (C). (D-F) UMAP plots of sorted B220+ CD95hi GL7hi IgD− GC B cells at day 14 post-immunization (D), split between LZ and DZ (E) and split among different cell cycle phases (F). Each point is a single cell colored by cluster assignment. (G) Top 10 marker genes distinguishing the different clusters of GC cells detected at day 7. (H) Expression of Fcer2a (CD23) and Il4ra in the day 7 dataset projected onto UMAP plots. Color scaled for each gene with log normalized expression level. (I, J) Representative FACS profiles (I) and frequencies of CD23+ subset (J) in LZ (CXCR4lo CD86hi) and DZ (CXCR4hi CD86lo) GC B cells 12–14 days after immunization. Each symbol represents one mouse. Data are pooled from two experiments. (K) Geometric mean fluorescence intensity (MFI) of IL4Ra in CD23− and CD23+ LZ GC B cells at day 13 following NP-KLH immunization. (L) Expression of Mki67 from day 7 GC B cell dataset projected onto UMAP plots. (M) MFI of Ki67 in CD23− and CD23+ LZ GC B cells at day 13 following NP-KLH immunization. One of two experiments with similar result is shown. (N) Frequencies of EdU+ cells in CD23+ and CD23− LZ GC B cells after 1 hour EdU labeling. In K, M, N each symbol represents one mouse, each line represents the same mouse, and one of two experiments with similar results is shown. (O) Representative histograms of Efnb1 (Ephrin B1) in non-GC B cells (B220+ CD95lo GL7lo), CD23+ and CD23− LZ GC B cells 12–14 days after immunization. Data represent three experiments, with at least 3 mice per experiment. See also Figure S1 and Table S1.
In this and all subsequent figures, data are presented as mean ± SEM; n.s., not significant, *P < 0.05, **P < 0.01, ***P < 0.001.
Flow cytometric analysis showed that LZ GC B cells could be divided into CD23+ and CD23− subsets, while DZ cells were largely CD23− (Fig. 1I, J). In accord with the higher Il4ra mRNA, the CD23+ subset had higher IL4Ra protein expression (Fig. 1K). In agreement with the G1 cell cycle status designation of the majority of the CD23+ cluster, expression of the cell cycle associated marker Ki67 was low in most of these cells (Fig. 1L). Flow cytometric analysis showed lower expression of Ki67 protein in CD23+ than CD23− LZ cells (Fig. 1M). One-hour EdU (5-ethynyl-2’-deoxyuridine) labeling confirmed that fewer CD23+ than CD23− LZ cells were in cell cycle (Fig. 1N). CD23 may also be highly expressed by pre-GC B cells. However, pre-GC B cells are CD38+ and undergo rapid proliferation and show high Ki67 expression (Ise et al., 2018; Schwickert et al., 2009; Wang et al., 2017). The low CD38, low cell cycle gene expression and low EdU labeling indicated that CD23+ LZ GC B cells were not pre-GC B cells. CD23+ LZ cells were also confirmed to express the definitive GC marker, EprhinB1 (Laidlaw et al., 2017; Wang et al., 2017) (Fig. 1O). Combined, these data indicate that CD23+ LZ cells have properties that distinguish them from other mature GC B cells.
CD23+ LZ B cells can differentiate into pre-memory and pre-plasma cells
To better understand the difference between clusters of GC B cells, we calculated the score of different gene signatures using VISION (DeTomaso et al., 2019). GC B cells that have recently received strong positive selection signals from Tfh cells are known to be enriched for Myc expression and function (Calado et al., 2012; Dominguez-Sola et al., 2012; Ersching et al., 2017; Finkin et al., 2019), and both Myc and Myc target genes were more enriched in LZ cells outside of the CD23+ cluster, particularly in cluster 2 (Fig. S1B, C). In accord with having received T cell help, the populations enriched for Myc+ cells showed a high score for a CD40 signaling gene signature (Fig. S1D, E) and for hallmark mTORC1 signaling (Fig. S1F). Additionally, hexose kinase-2 (Hk2), an enzyme important for glycolysis, was expressed by the Myc+ population and these cells were enriched for gluconeogenesis pathway activity (Fig. S1G). In contrast to the populations enriched for Myc+ cells, CD23+ LZ cells showed enrichment for a BCR signaling signature (Fig. S1H). These data suggest that this population may have encountered antigen, but not recently had strong cognate interactions with helper T cells. However, there was heterogeneity as can be seen in the upper part of day 7 cluster 3 in the UMAP plot where there were some Myc target gene-expressing cells and some S-phase cells. Thus, while the cells cluster due to their overall gene expression signature, they were not all in a single state, likely indicating the cluster has multiple outputs.
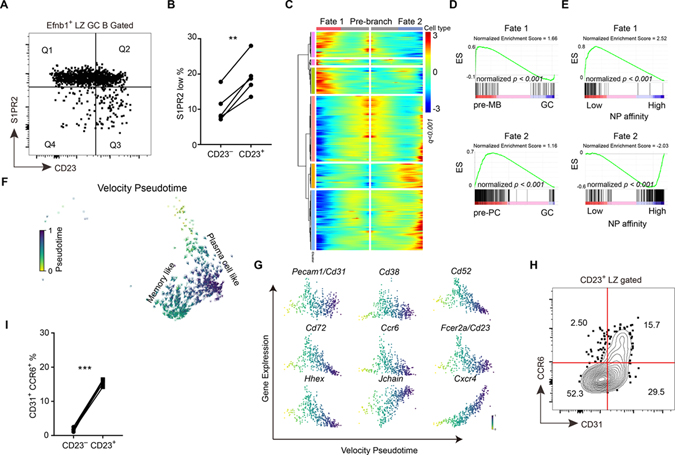
Previous work has shown that pre-memory and pre-plasma B cells in the GC begin to downregulate the GC marker, S1pr2 (detected using a Venus reporter) (Laidlaw et al., 2017). Flow cytometric analysis showed that the S1pr2-Venus low cells were enriched in the CD23+ compartment (Fig. 2A, B). Using VISION analysis, the gene expression signature of EphrinB1+ S1pr2low post GC B cells was enriched in the CD23+ cluster (Fig. S2A, B). The Myc+ cluster also included some cells with post GC properties (Fig. S2A), consistent with some Myc+ cells being pre-plasma cells (Ise et al., 2018). Cluster 5 at day 7 and clusters 8 and 9 at day 14 corresponded to contaminating plasmablasts or plasma cells and cluster 6 at day 14 to memory B cells (purity was lower at day 14 because CD38 was not included in the stain).
Figure 2. CD23+ LZ B cells can differentiate into pre-memory cells.
(A, B) S1pr2Venus/+ reporter mice were immunized with NP-haptenated antigen. (A) Representative FACS profile displaying gates between CD23 and S1PR2 in EFNB1+ LZ GC B cells. (B) Percentages of S1PR2lo cells in CD23+ or CD23− amongst Efnb1+ LZ GC B cells. ** P<0.01. (C) Heatmap of differentially expressed genes between cell fate 1 and cell fate 2 within cluster 3 of day 7 LZ GC B cell dataset by RNA trajectory pseudotime analysis. Colored bars on side indicate groups of similarly expressed genes (all genes were listed in Table S3). (D) GSEA analysis of differentially expressed genes from the two cell fates compared to pre-memory B cell (Wang et al., 2017) and pre-plasma cell (Ise et al., 2018) RNA sequencing datasets. Enrichment profiles for cell fate 1 compared with pre-memory (upper) and cell fate 2 compared with pre-plasma cell (lower). (E) GSEA of differentially expressed genes from different cell fates compared to RNA sequencing dataset of different NP affinity GC B cells (Shinnakasu et al., 2016). Enrichment profiles for cell fate 1 compared with low-affinity (upper) and cell fate 2 compared with high-affinity (lower). ES represents enrichment score. (F) RNA velocity pseudotime analysis for CD23+ LZ GC B cells are embedded in a UMAP plot. Color represents the velocity pseudotime. (G) Expression level of genes of interest along the velocity pseudotime. Color represents the velocity pseudotime. Determined using scVelo. (H) Representative FACS profile displaying gates between CD31 and CCR6 in CD23+ LZ GC B cells 12–14 days after immunization. Data are representative of two experiments, with at least 3 mice per experiment. (I) Percentages of CD31+CCR6+ cells in CD23− and CD23+ LZ GC B cells on day 13 after immunization. In B and I each symbol represents one mouse, and each line represents the same mouse. *** P<0.001. See also Figure S2 and Table S3.
Applying trajectory inference pseudotime analysis (using Monocle 2 (Qiu et al., 2017)) to the day 7 (Fig. 2C–E) and day 14 (Fig. S2C–E) CD23+ cluster LZ cells, we found that these cells could differentiate via two major branches (Fig. 2C, S2C). We compared the differentially expressed genes between these two branches to published pre-memory B cell, pre-plasma cell, and different NP affinity GC B cell RNA sequencing datasets (Ise et al., 2018; Shinnakasu et al., 2016; Wang et al., 2017). The highly expressed genes in branch 1 were enriched in the pre-memory B cell signature and also in an NP low-affinity GC B cell signature (Fig. 2D, E, S2D, E). The genes highly expressed in branch 2 were enriched in the pre-plasma cell and NP high-affinity GC B cell signatures (Fig. 2D, E, S2D, E). Comparing to pre-memory B cells, the cells in branch 2 were more like GC B cells (Fig. S2F,G). Besides the gene similarity-based pseudotime analysis, we also carried out RNA velocity-based pseudotime analysis (Bergen et al., 2020). Consistent with the previous analysis, CD23+ LZ GC B cell were found to differentiate in two directions (Fig. 2F). The markers of pre-memory B cells which were identified by Monocle 2 were also increased in their expression in one direction (Fig. 2G). These cells correspond to the pre-memory cells identified by Monocle 2. By pseudotime analysis, Ccr6, Cd38, Cd52, Fcer2a (Cd23), Cd72, Pecam1 (Cd31) and Hhex were upregulated during memory B cell differentiation (Fig. 2G). CCR6 and CD38 have been used to identify memory B cell precursors (Laidlaw et al., 2017; Suan et al., 2017) and Hhex was recently identified as a key transcription factor for memory B cell formation (Laidlaw et al., 2020). Using antibodies to CCR6 and CD31, a CCR6+CD31+ pre-memory subset could be identified amongst CD23+ LZ GC B cells (Fig. 2H) while there were few such cells amongst CD23− LZ GC B cells (Fig. 2I). A similar fraction of CD23+CD31+ GC cells stained for the pre-memory marker CD38 (Fig. S2H). The pre-memory gene set was also enriched for CD72 and CD31 and CD72 tended to be co-expressed (Fig. S2I). IL4Ra expression was also highest on the CD23+CD31+ cells (Fig. S2J). In the other branch, CD23+ LZ GC B cells were predicted to differentiate into CD23− pre-plasma cell-like cells (Fig. 2G). Consistent with this prediction, Blimp1-GFP positive cells were enriched in the CD23− GC B cells (Fig. S2K, L). By RNA velocity pseudotime analysis, the expression of J chain and CXCR4 were predicted to be upregulated in the pre-plasma cell direction (Fig. 2G). CXCR4 upregulation at this early stage is consistent with plasma cells leaving the GC through the DZ (Krautler et al., 2017; Radtke and Bannard, 2018; Zhang et al., 2018). However, the lack of robust pre-plasma cell surface markers and the late appearance of the Blimp1-GFP reporter during differentiation limited our ability to flow cytometrically quantitate the representation of pre-plasma cells amongst CD23+ cells compared to other GC subsets from where they may also emerge, and we therefore focused our analysis on memory B cell generation.
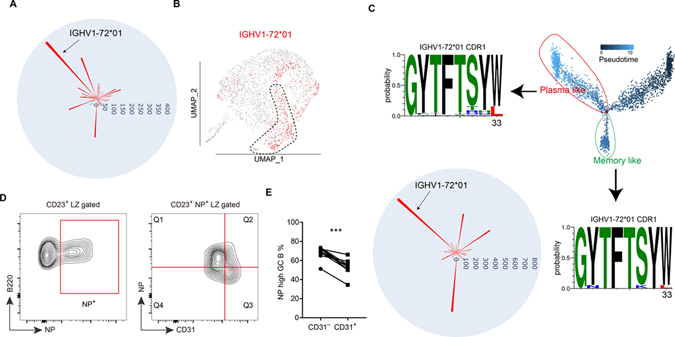
Single cell VDJ-seq reveals selected VH gene enrichment in CD23+ LZ cells
To track BCR heavy chain gene selection and affinity maturation, we performed scVDJ-seq at day 7 and 14 post NP-haptenated antigen immunization. At the early period after NP immunization, several VH genes are used but starting from day 6 IGHV1-72*01 (Vh186.2) begins to dominate (Jacob et al., 1993). By scVDJ-seq analysis, IGHV1-72*01 was the most used heavy chain gene at day 7 (Fig. 3A). After mapping the heavy chain information to the single cell transcriptome, cells in the CD23+ cluster were enriched for usage of IGHV1-72*01 (Fig. 3B and Fig. S2M). Interestingly, the next two most frequent NP-binding associated VHs, IGHV1-64*01 and IGHV1-53*01 (Jacob et al., 1993), were under-represented in the CD23+ cluster (Fig. S2M) indicating that some feature of IGHV1-72*01 was driving representation in the CD23+ cluster, possibly antigen-binding affinity. Although few mutations of IGHV1-72*01 were detected at day 7, the high NP affinity W33L mutation of IGHV1-72*01 could be detected at day 14 (Fig. 3C). By this time point IGHV1-72*01 was no longer enriched in the CD23+ cluster, and cells carrying the W33L mutation did not show a strong bias for any cluster (Fig. S2N, O). By pseudotime analysis of the day 14 data, similar to day 7, we could divide the CD23+ cluster into pre-memory like cells and pre-plasma like cells (Fig. 3C, S2C, D). We next analyzed the scVDJ-seq data in these two subclusters. The W33L mutation was readily detected in the pre-plasma like cells while this affinity-improving mutation was rare in pre-memory B cells (Fig. 3C). GSEA also showed that day 14 pre-memory B cells more highly expressed genes enriched in NP low-affinity GC B cells (Fig. S2E). Consistent with the sequencing data, 14 days after NP-KLH immunization, CD23+ CD31+ LZ B cells showed lower NP binding than CD23+ CD31− LZ B cells (Fig. 3D, E), despite the CD31+ cells having slightly higher total surface BCR levels as determined by CD79b (Igβ) staining (Fig. S2P, Q). Lower NP-binding in flow cytometry assays was previously found to be reflective of lower affinity for NP (Shinnakasu et al., 2016). These data demonstrate the ability of scRNA-seq to identify and link the early selection and differentiation events of NP-binding GC B cells.
Figure 3. Single cell VDJ-seq analysis reveals selected heavy chain gene enrichment in CD23+ LZ cells.
(A) Circular bar plot shows the cell number of each heavy chain gene detected by scVDJ-seq of day 7 GC B cells. (B) UMAP plot of the distribution of NP specific heavy chain (IGHV1-72*01) in different clusters of day 7 GC B cells. (C) Sequence logos of IGHV1-72*01 CDR1 of plasma like cells and memory like B cells divided by pseudotime analysis. Circular bar plot shows the cell number of each heavy chain gene detected by the scVDJ-seq of day 14 GC B cells. (D, E) Representative FACS profiles (D) and percentages of NPhi subset in CD31− cells and CD31+ cells amongst NP+ CD23+ LZ cells 12–14 days after immunization. Each symbol represents one mouse, and each line represents the same mouse. Data are pooled from two experiments. *** P<0.001. See also Figure S2.
IL4Ra signaling restrains memory B cell development at the CD23+ LZ stage
IL4Ra activation leads to phosphorylation and activation of the transcription factor STAT6 (McCormick and Heller, 2015). Intracellular staining showed pSTAT6 levels were higher in CD23+ than CD23− LZ GC B cells (Fig. S2R). Moreover, VISION analysis showed a concentration of IL4 pathway activity in the CD23+ cluster (Fig. S2S). Thus, while this cluster has low Myc and mTOR target gene activity suggesting the cells are not receiving the strongest levels of T cell help, the IL4 pathway activity suggests they are exposed to some helper signals. Lower T cell help could occur due to presentation of lower amounts of MHCII-peptide complexes leading to less strong triggering of the T cell. Additionally, lower T cell help could reflect less cumulative contact time with helper T cells. A further possibility, suggested by data below, is that some B cells could be receiving IL4 in a non-cognate manner, as bystander cells.
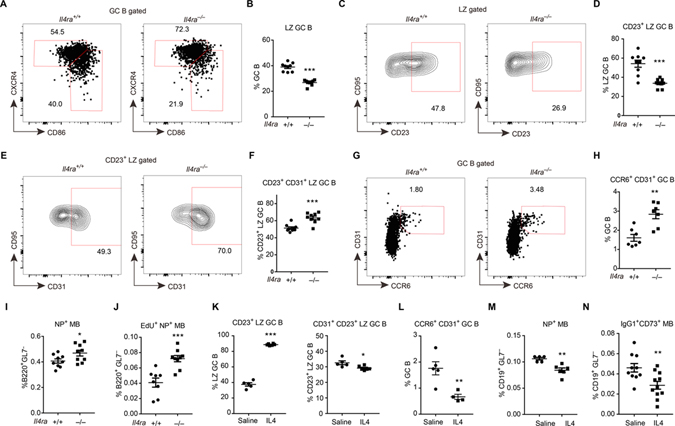
To study the role of IL4Ra signaling in GC LZ cells, Il4ra−/− mice were immunized with NP-KLH and analyzed at day 12–14. IL4Ra-deficient mice mounted a GC response of normal magnitude, but there was a 40% reduction in the frequency of LZ cells (Fig. 4A, B, S3A, B). In accord with the IL4Ra expression data, IL4Ra-deficiency caused a preferential depletion of the CD23+ LZ subset (Fig. 4C, D). Within the CD23+ LZ compartment, however, there was an increased frequency of CD31+ cells (Fig. 4E, F). In agreement with the CD23+CD31+ cells being enriched for pre-memory B cells, Il4ra−/− mice had an increased frequency of CD31+CCR6+ GC B cells (Fig. 4G, H). Il4ra−/− B cells were more competent in memory formation, as there were more B220+GL7−NP+ memory cells (Fig. 4I and S3C, D). To further test if these memory cells were GC-derived, we injected EdU to label GC B cells at day 11 post immunization and detected newly generated (EdU labeled) memory B cells three days later. Memory B cell differentiation was significantly increased for Il4ra−/− B cells, confirming a role for IL4Ra in restraining memory cell generation (Fig. 4J). Measurement of isotype switched cells is also often used to track memory cell generation, but this could not be used for Il4ra−/− cells because of the strong defect in IgG1 isotype switching (Fig. S3E). In addition to the effects on memory B cell generation, there was also a small increase in the abundance of IgD−CD138+ plasma cells in Il4ra−/− mice (Fig. S3F, G).
Figure 4. IL4Ra signaling restrains memory B cell development at the CD23+ LZ stage.
(A-I) Immunized Il4ra+/+ and Il4ra−/− mice were analyzed on day 13. (A, B) Representative FACS profiles (A) and frequencies of CXCR4lo CD86hi LZ cells (B) in GC B cells. (C, D) Representative FACS profiles (C) and frequencies of CD23+ cells (D) in LZ GC B cells. (E, F) Representative FACS profiles (E) and frequencies of CD31+ cells (F) in CD23+ LZ GC B cells. (G, H) Representative FACS profiles (G) and frequencies of CD31+ CCR6+ cells (H) in GC B cells. (I) Frequencies NP+ memory B cells in B220+ GL7− non-GC B cells. Data are pooled from two experiments. (J) Immunized Il4ra+/+ and Il4ra−/− mice were treated with EdU on day 11 post immunization and were analyzed on day 14. Frequencies of NP+ EdU+ memory B cells in B220+ GL7− non-GC B cells. Data are pooled from two experiments. (K-N) Immunized mice received IL4-αIL4 complex or saline on day 11, 12, 13 post immunization and were analyzed on day 14. (K, L) Frequencies of CD23+ cells in LZ GC B cells (K left), CD31+ cells in CD23+ LZ GC B cells (K right), and CD31+CCR6+ cells in GC B cells (L). (M, N) Frequencies of NP+ memory B cells (M) and IgG1+ CD73+ memory B cells (N) in CD19+ GL7− non-GC B cells. Each symbol represents one mouse. (K-M) One of three experiments with similar result is shown. (N) Data are pooled from three experiments. See also Figure S3.
To investigate if IL4 was sufficient to restrain memory B cell differentiation in vivo, we treated mice with IL4-αIL4 complexes (Finkelman et al., 1993; Ricardo-Gonzalez et al., 2010) for 3 days starting at day 11. The frequency of GC B cells was not changed by the treatment (Fig. S3H), but the frequency of IgG1+ GC B cells was increased (Fig. S3I). The frequency of LZ cells and CD23+ LZ cells was markedly increased (Fig. 4K, S3J). The percentages of CD23+CD31+ LZ B cells (Fig. 4K) and CCR6+CD31+ pre-memory cells (Fig. 4L) were significantly reduced. Similar reductions were noted for NP+ memory B cells (Fig. 4M). CD73 is a marker of many though not all memory B cells and it can be differentially induced depending on immunization conditions, with some studies indicating it is preferentially expressed on memory cells generated in a Bcl6-dependent manner (Anderson et al., 2007; Bemark et al., 2016; He et al., 2017; Kaji et al., 2012; Onodera et al., 2012; Taylor et al., 2012; Tomayko et al., 2010). We also gated IgG1+CD73+ memory cells and found that their frequency amongst CD19+GL7− cells was reduced (Fig. 4N, S3K) even though the overall amount of B cell IgG1 switching was increased (Fig. S3I). The frequency of plasma cell was also reduced following IL4-αIL4 complex treatment (Fig. S3L). These data suggest that IL4 is sufficient to restrain GC B cell differentiation from a CD23+ LZ state to the memory state and that it may also restrain plasma cell generation. In keeping with these findings, previous in vitro work has shown that IL4 pathway activity can restrict plasma cell differentiation (Ettinger et al., 2005; Jenks et al., 2018; Pignarre et al., 2021).
Memory B cell differentiation can be affected by BCR affinity and/or Ig class switch (Gitlin et al., 2016; Viant et al., 2020). To investigate whether the repression effect of IL4 on memory cell generation was caused by affinity and/or class switch modulation, we treated AID-deficient mice with IL4-αIL4 complexes for three days starting at day 11. Similar to the wild type mice, IL4 treatment caused a reduction in differentiation of memory B cells (Fig. S3M–P). These data suggest that IL4 restrains memory B cell differentiation independently of effects on BCR affinity or class switch.
In agreement with a repressive action of IL4 on memory B cell development, an analysis of published B cell RNA sequencing data (Mokada-Gopal et al., 2017) revealed that in vitro incubation of follicular B cells with IL4 inhibits the expression of many markers expressed by memory B cells such as Cd31, Cd38, Cd72, Ccr6 and Cr2 and the memory B cell differentiation related transcription factors Hhex, Bach2, and Zbtb32 while promoting expression of Fcer2a and Il4ra (Fig. S3Q). This regulation of gene expression could be released by deletion of Stat6 (Fig. S3Q).
IL4Ra acts intrinsically in GC B cells
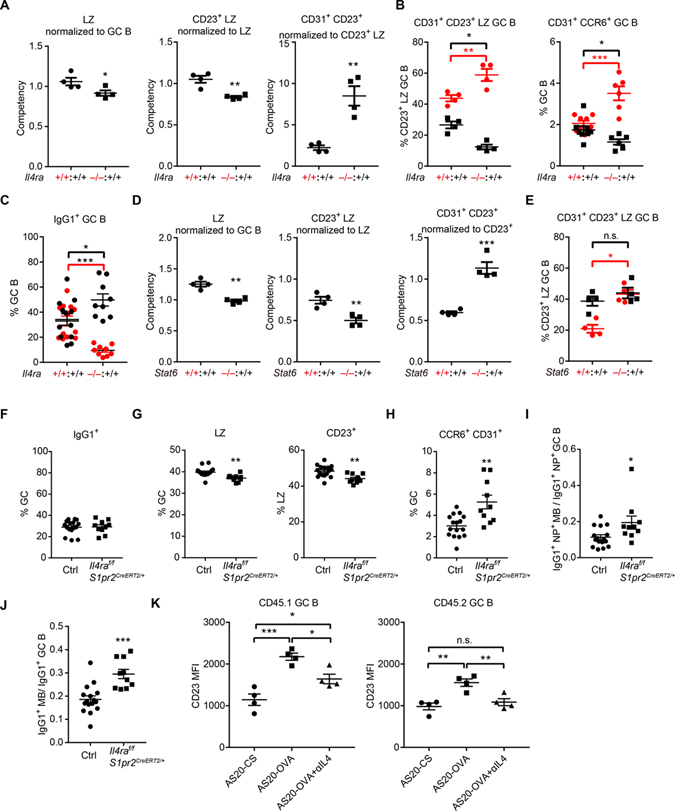
To determine whether the increased predisposition of GC B cells to a memory cell fate in the absence of IL4Ra was due to a cell intrinsic role of the receptor, we generated WT CD45.1 and WT or Il4ra−/− CD45.2 mixed BM chimeras. The influence of IL4Ra-deficiency on B cell representation in a particular stage of the GC response was determined by comparing the ratio of CD45.2+ (WT or Il4ra−/− depending on the chimera) to CD45.1+ (WT) B cells in the target population (e.g. LZ) with the corresponding ratio in the parent population for that subset (total GC in the LZ case) to obtain a Competency measurement (Fig. S4A–C). Thus, if Il4ra−/− cells were present at a ratio of 3:1 in the target population but had been 2:1 in the parent population, then the competency measurement would be 1.5. This method allows standardization across different sets of mice that had different levels of hematopoietic chimerism. The chimeric mice showed a notable under-representation of Il4ra−/− B cells in the GC (Fig. S4C, D), likely due to lower competitiveness of these cells for expansion in the pre-GC compartment. We also noted a reduction in the frequency of total GC cells in the WT:Il4ra−/− mixed chimeras compared to the control mixed chimeras (Fig. S4E). These findings were unexpected given the unaltered GC response magnitude in the full Il4ra−/− mice (Fig. S3B) and suggest complex interactions between WT and Il4ra−/− cells that we dissect further in later experiments.
In the mixed chimeras, IL4Ra-deficient B cells showed a disadvantage in total LZ and CD23+ LZ (Fig. 5A) but showed an advantage in the CD23+CD31+ and CD31+CCR6+ pre-memory compartment (Fig. 5A, B, S4F). Assessment of the effect of IL4Ra-deficiency on the memory compartment in the mixed chimeras was problematic because of the low representation of Il4ra−/− cells in GCs and our lack of a definitive marker for GC-derived memory cells. Interestingly, when we examined the behavior of CD45.1 WT cells in GCs containing about 50% Il4ra−/− CD45.2 cells, we noted that there were reductions in the frequencies of CD23+CD31+ and CD31+CCR6+ WT cells compared to their representation in GCs composed of only WT cells (Fig. 5B, black symbols; CD45.2 WT or Il4ra−/− cells shown in red symbols). A reduction in WT CD23+CD31+ and CD31+CCR6+ cells was also seen after IL4 injection (Fig. 4K, L) and could be occurring in the WT:Il4ra−/− mixed chimeras due to reduced IL4 consumption by Il4ra−/− GC cells leading to increased IL4 availability for WT cells. In agreement with the idea of a general increase in splenic IL4 availability, the WT cells in WT:Il4ra−/− mixed chimeras showed an increased extent of switching to the IL4-dependent IgG1 isotype compared to cells in WT:WT mixed chimeras (Fig. 5C).
Figure 5. IL4RA and STAT6 are intrinsically required for restraining memory B cell development.
(A-C) Mixed BM chimeras were made with CD45.1 wild-type (Il4ra+/+) and CD45.2 Il4ra+/+ or Il4ra−/− BM cells. (A) Competency values of LZ (B220+ CD95hi GL7hi CXCR4lo CD86hi) (left), CD23+ LZ (middle), CD31+ CD23+ LZ (right) GC compartment at day 13 following immunization. (B) Frequencies of CD31+ cells in CD23+ LZ GC B cells (left) and CCR6+ CD31+ cells in GC B cells (right) in different compartments (black CD45.1+ and red CD45.2+) of mixed chimeras. (C) Frequencies of IgG1+ cells in GC B cells in different compartments (black CD45.1+ and red CD45.2+) of mixed chimeras. Gating strategy and equation for calculating differentiation ratios shown in Figure S4A–C. (A-B) One of three experiments with similar results is shown. (C) Data are pooled from three experiments. (D, E) Mixed (50:50) BM chimeras were made with CD45.1 wild-type (Stat6+/+) and CD45.2 Stat6+/+ or Stat6−/− BM cells. (D) Competency values of LZ (left), CD23+ LZ (middle), CD31+ CD23+ LZ (right) GC compartment at day 13 following immunization. (E) Frequencies of CD31+ cells in CD23+ LZ GC B cells in different compartments (black CD45.1+ and red CD45.2+) of mixed chimeras. One of two experiments with similar results is shown. (F-J) Control and S1pr2CreERT2/+ IL4raf/f mice were immunized with NP-KLH, treated with tamoxifen to induce Cre expression, and analyzed on day 13. (F) Frequencies of IgG1+ cells in GC B cells. (G, H) Frequencies of LZ cells in GC B cells (G left), CD23+ cells in LZ GC B cells (G right), CD31+CCR6+ cells in GC B cells (H). (I) Ratio of NP+ IgG1+ memory B cells to NP+IgG1+ GC B cells. (J) Ratio of CD38+IgG1+ memory B cells to CD38−IgG1+ GC B cells. Data are pooled from three experiments. (K) Geometric mean fluorescence intensity (MFI) of CD23 in CD45.1+ (left) and CD45.2+ MD4 GC B cells with indicated treatment. One of two experiments with similar results is shown. In all plots each symbol represents one mouse. In K, one-way ANOVA with Sidak multiple comparisons test was used. See also Figure S5 and S6.
An analogous assessment of WT:Stat6−/− mixed BM chimaeras showed a similar advantage of Stat6−/− cells in the CD23+CD31+ LZ subset (Fig. 5D). Importantly, however, these mixed chimeras did not show the disadvantage of CD23+CD31+ WT cells competing with the Stat6−/− cells (Fig. 5E). Stat6-deficiency did not alter IL4Ra surface levels on GC B cells (Fig. S4G) and there was not an increase in IgG1+ WT GC B cells in the WT:Stat6−/− mixed chimeras (Fig. S4H). Therefore, we speculate that in contrast to the situation in Il4ra−/− mixed BM chimeras, the IL4 available in the lymphoid tissue microenvironment to WT cells is unlikely to be changed in Stat6−/− mixed BM chimeras.
Besides Stat6, IL4 could also signal via IRS1/2 (Wills-Karp and Finkelman, 2008). To further investigate the Stat6 dependence of IL4 signaling in the GC we treated Stat6-deficient mice with IL4 complexes for 3 days starting 11 days after immunization. Although IL4 could still slightly increase the percentage of LZ cells and the expression of CD23, the frequency of CCR6+CD31+ GC B cells and memory B cell was not altered (Fig. S4I, J) indicating that IL4 signaling via STAT6 is needed for effects on memory cell generation.
Our studies in WT:Il4ra−/− mixed BM chimeras could not exclude the possibility that the influence of IL4Ra-deficiency on memory cell generation was due to effects occurring prior to GC formation. We took two approaches to address this issue. First, we reconstituted mice with Cγ1Cre/+ Cas9f/+ BM that had been transduced with sgRNA against IL4Ra (using a vector containing a BFP reporter) (Fig. S5A). This approach achieved knockdown of IL4Ra in many BFP+ GC B cells and was associated with a reduction in the LZ compartment and an increase in the frequency of CCR6+CD31+ pre-memory cells amongst BFP+ GC cells (Fig. S5B). The frequency of IgG1+ cells amongst BFP+ GC cells was unchanged compared to BFP− GC cells (Fig. S5C). Comparing the frequency of IgG1+CD38+ memory B cells with IgG1+ GC B cells or the frequency of NP+CD38+ memory B cells with NP+ GC B cells showed that the conditional loss of IL4Ra was associated with an increase in memory cell generation (Fig. S5D, E). However, Cγ1Cre is expressed in some activated B cells as well as in GC B cells and thus does not strictly limit the IL4Ra knockdown to the GC stage. In our second approach, we generated S1pr2CreERT2/+Il4raf/f mice and treated them early after immunization with tamoxifen to induce selective ablation of IL4Ra in GC B cells. This approach caused a reduction in IL4Ra in approximately 50% of GC B cells without altering expression in follicular B cells (Fig. S5F). Interestingly, as in the WT:Il4ra−/− BM chimeras (Fig. S4E) but not full Il4ra−/− mice (Fig. S3B), these mice showed a reduction in the magnitude of the overall GC response (Fig. S5G). The conditional IL4Ra-deficient mice showed unaltered frequencies of IgG1+ cells amongst GC cells (Fig. 5F). The LZ and CD23+ LZ was reduced while the CCR6+CD31+ pre-memory cells were increased (Fig. 5G, H). Referencing to IgG1+NP+ GC cells to control for the smaller GC size, there was an increased proportion of IgG1+NP+CD38+ memory B cells; this increase was also seen for total IgG1+ memory B cells (Fig. 5I, J). Taken together, these data indicate that intrinsic IL4Ra signaling within GC B cells restrains the extent of pre-memory and memory B cell generation.
Another notable feature of the CD23+ GC B cells was their higher expression of Ltα and Ltβ than most other GC B cells (Fig. 1G, S5H). Flow cytometric analysis with LTβR-Ig confirmed that CD23+ LZ GC B cell had higher LTα1β2 expression than total GC B cells (Fig. S5I, J). Moreover, IL4Ra and Stat6 were intrinsically required for normal LTα1β2 expression in GC B cells (Fig. S5K–N). These findings are in accord with the activity of IL4 in promoting LTα1β2 expression in B and T lymphocytes (Cortes-Selva et al., 2019; Dubey et al., 2017; Luther et al., 2002). GC FDCs are strongly dependent on LTα1β2 for their maintenance (Fu and Chaplin, 1999; Haberman et al., 2019; Mackay and Browning, 1998). Therefore, the extent of IL4 signaling in CD23+ GC B cells might influence properties of the GC FDC network.
Non-cognate IL4 actions in the GC
Findings in the WT:Il4ra−/− mixed BM chimeras provided evidence for bystander actions of IL4 in the GC. As a further test for possible non-cognate actions of IL4 in the GC, we asked whether augmenting the delivery of T cell help to a subset of GC B cells would lead to evidence of increased IL4 signaling in bystander GC B cells. Following a similar strategy to the DEC205-OVA method for antigen loading of B cells independently from the BCR (Victora et al., 2010), we coupled OVA to the AS20 (A20–1.7) antibody that is specific for CD45.1. Adoptive transfers were performed with 10:90 mixtures of CD45.1 MD4 and CD45.2 MD4 hen egg lysozyme (HEL) specific B cells together with ovalbumin (OVA) specific OT2 T cells into CD45.2 recipients. After immunizing with HEL-OVA to establish a GC response, the mice were treated with AS20-OVA (Fig. S5O). The treatment led to expansion of the OVA loaded (CD45.1) MD4 B cells in the LZ at 12 hrs and in the DZ at 72 hrs (Fig. S5P, Q). These findings are comparable to those observed with the DEC205-OVA based GC B cell antigen loading system (Victora et al., 2010) and they establish a versatile method for differential antigen loading in vivo using widely available CD45 congenic mice. Using this cell transfer and antigen-loading approach, we tested for cognate and non-cognate influences of T cells on CD23 expression after 12 hrs. In these experiments we transferred 80:20 mixtures of CD45.1 MD4 and CD45.2 MD4 cells. The antigen-loaded CD45.1 MD4 GC B cells showed a strong increase in CD23, consistent with cognate receipt of CD40 and IL4 signals; importantly, the bystander CD45.2 GC B cells also showed a measurable increase in CD23 (Fig. 5K). The bystander increase in CD23 was fully blocked by treatment with anti-IL4 (Fig. 5K). These data indicate that under conditions where many GC B cells are interacting strongly with Tfh cells, bystander GC B cells can be exposed to IL4.
Searching for an explanation for the reduced GC magnitude in WT:Il4ra−/− BM chimeras and tamoxifen treated S1pr2CreERT2/+Il4raf/f mice (that were also chimeric in their IL4Ra loss) versus the intact GC response in Il4ra−/− mice, we considered the possibility that in the mixed setting the chronically elevated available IL4 was having a repressive influence on the WT GC cells. In accord with this notion, when mice were treated with IL4 complexes daily for 3 days, their GC B cells showed lower Bcl6 protein levels (Fig. S5R); by contrast, Bcl6 was increased in follicular B cells, in keeping with in vitro findings for activated B cells (Chevrier et al., 2017; Haniuda et al., 2020; Robinson et al., 2019). Moreover, by the fourth day of IL4 complex treatment, there was a reduction in the magnitude of the GC response (Fig. S5S). In vitro studies have shown that under some conditions IL4 can suppress B cell proliferation or promote apoptosis (Jelinek and Lipsky, 1988; Pignarre et al., 2021). Together, these findings further highlight the importance of appropriately controlled IL4 signaling during the GC response.
IL4Ra expression by FDCs limits IL4 availability to GC B cells
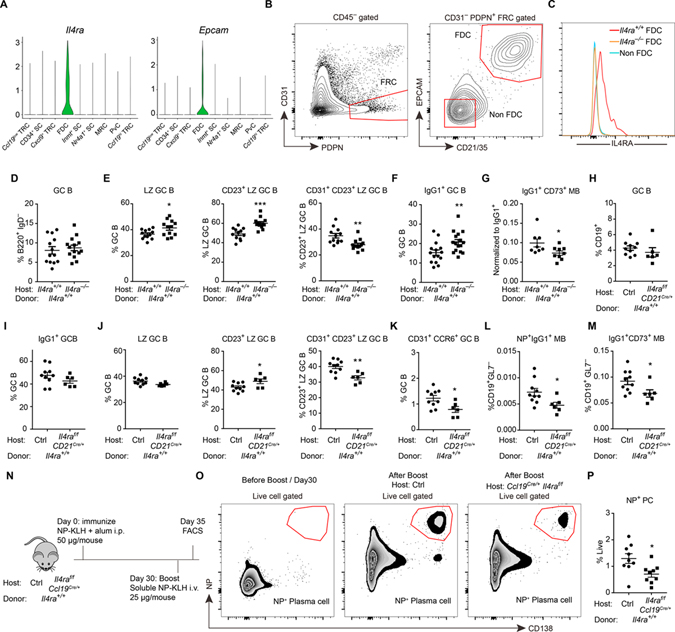
Single cell RNA-seq data of lymph node (LN) and spleen stromal cells (Cheng et al., 2019; Rodda et al., 2018) showed IL4Ra was expressed by FDCs but not other types of fibroblastic reticular cells (Fig. 6A, S6A). During this analysis we also found that epithelial cell adhesion molecule (Epcam) is specifically expressed by FDCs (Fig. 6A, S6A). Co-staining Epcam with the FDC markers Cr2 and Tmem119 in tissue sections confirmed the selective expression of this adhesion molecule by FDCs (Fig. S6B, C). Combining Epcam with CR2 (CD21) staining in flow cytometry allowed for improved identification of FDCs (Fig. 6B). Consistent with the scRNA-seq data, ILR4a was selectively expressed on FDCs by flow cytometry (Fig. 6C).
Figure 6. IL4RA expression by FDCs limits IL4 availability to GC B cells.
(A) Violin plots of Il4ra and Epcam expression in different stromal cell subsets. (B, C) Representative FACS profiles (B) and histograms (C) of IL4RA in non-FDCs (CD45− PDPN+ CD31− CD21/35− EPCAM−) and FDCs (CD45− PDPN+ CD31− CD21/35+ EPCAM+) isolated from mesenteric LNs in mice with indicated genotype. Data represent two experiments, with at least three mice per experiment. (D-G) Reverse chimaeras were made by reconstituting irradiated Il4ra+/+ and Il4ra−/− mice with WT BM. (D-F) Frequencies of CD95hi GL7hi GC B cells in B220+ IgD− B cells (D), CXCR4lo CD86hi LZ cells in GC B cells (E left), CD23+ cells in LZ B cells (E middle), CD31+ cells in CD23+ LZ B cells (E right) and IgG1+ cells in GC B cells (F) 13 days after immunization. (G) Ratio of CD73+ IgG1+ memory B cells to IgG1+ cells. Data are pooled from three experiments. (H-M) Reverse chimeras were made by reconstituting irradiated control and Cd21Cre/+ Il4raf/f mice with wild-type (Il4ra+/+) BM. (H-K) Frequencies of CD95hi GL7hi GC B cells in CD19+ B cells (H), IgG1+ cells in GC B cells (I), LZ cells in GC B cells (J left), CD23+ cells in LZ B cells (J middle), CD31+ cells in CD23+ LZ B cells (J right) and CD31+ CCR6+ cells in GC B cells (K) 13 days after immunization. (L, M) Frequencies of NP+IgG1+ (L) and CD73+ IgG1+ (M) memory cells in non-GC B cells. (N-P) Scheme for testing memory response in Ccl19Cre/+ Il4raf/f mice that had been reconstituted with wild-type BM (N), flow cytometry plots from the indicated mice (O), and summary data (P) showing frequency of CD138+NP+ plasma cells in the spleen of boosted control or Ccl19Cre/+ Il4raf/f chimeric mice. Data are pooled from two of three experiments. In D-M and P, each symbol represents one mouse. See also Figure S6.
In an effort to determine whether the IL4Ra expressed by FDCs impacts the GC response, we first generated reverse BM chimeric mice that lacked IL4Ra on all radiation-resistant cells but had intact expression on B cells, T cells and other hematopoietic cells. Immunized Il4ra−/− reverse BM chimeras revealed a normal frequency of GC B cells (Fig. 6D) and of FDCs (Fig. S6D) but had an increased frequency of LZ GC B cells (Fig. 6E). Among the LZ cells, there was a higher percentage of CD23+ cells and a lower percentage of CD31+ cells in the CD23+ LZ compartment (Fig. 6E). The fraction of GC cells that switched to IgG1 was increased (Fig. 6F). In these experiments a lower valency form of the immunogen (NP14-CGG) was used than in other experiments (where NP28-CGG, NP28-KLH or NP32-KLH was used) and this was associated with a weaker NP-specific response that precluded quantitating NP+ memory B cell cells. However, compared to the overall frequency of IgG1+ cells, there was a reduction in IgG1+CD73+ memory cells (Fig. 6G). Plasma cell frequencies were also down (Fig. S6E). To determine whether signaling via the IL4 receptor was important in stromal cells, we also studied BM chimeras lacking the cytokine cγ chain that is needed for IL4R signaling (McCormick and Heller, 2015) and was also expressed by FDCs (Fig. S6F). In these animals, there was no significant difference in GC, LZ, memory or plasma cell compartments (Fig. S6G–L). IL4 can also signal via the IL4Ra/IL13Ra complex (McCormick and Heller, 2015). We detected minimal IL13Ra expression by FDCs (Fig. S6M) making it unlikely that this complex is involved in FDC responses to IL4 though our data do not fully exclude this possibility.
Given the widespread expression of IL4Ra by tissue cell types, it was important to perform more restricted deletion of IL4Ra. We took advantage of Ccl19Cre/+ to restrict IL4Ra deletion to lymphoid stromal cells, including FDCs (Cheng et al., 2019) and of CD21Cre/+ to restrict ablation to FDCs amongst stromal cells (Kraus et al., 2004; Wang et al., 2014). In the latter animals, ablation also occurs in B cells, but expression here was rescued by studying irradiated mice that have been reconstituted with WT BM. Unexpectedly, we found that Ccl19Cre/+ led to deletion of IL4Ra in a fraction of B cells and we therefore also reconstituted these mice with WT BM prior to performing the immunizations. Staining of spleen sections for CD35 (CR1) showed the GC FDC networks were comparable between control and Ccl19Cre/+ Il4raf/f chimeras (Fig. S6N). In contrast to the full Il4ra−/− BM chimeras, IgG1+ GC cell frequencies were not increased in either the CD21Cre/+ Il4raf/f chimeras (Fig. 6H, I) or the Ccl19Cre/+ Il4raf/f chimeras (Fig. S6O, P). Importantly, however, in both the CD21Cre/+ Il4raf/f chimeras (Fig. 6J, K) and the Ccl19Cre/+ Il4raf/f chimeras (Fig. S6Q, R), there was an increase in the CD23+ LZ, a decrease in CD31+CD23+ cells within the CD23+ LZ compartment and a decrease in CD31+CCR6+ pre-memory cells. There was also a decrease in NP+ and CD73+ IgG1+ memory B cells (Fig. 6L, M, S6S). When considered together with the above findings for IL4 treated mice and mice lacking IL4Ra on some GC B cells (Fig. 4K–N and Fig. 5B), it can be suggested that IL4 abundance in the GC is increased when stromal cells lack IL4Ra. To account for the differential effects on IgG1 switching we speculate that in mice lacking IL4Ra on all non-hematopoietic cells, IL4 availability is widely increased, including in pre-GC niches where isotype switching occurs, as well as within GCs; when IL4Ra loss is restricted to FDCs, IL4 availability may only be increased in the GC, a microenvironment where little if any isotype switching occurs (Roco et al., 2019; Sundling et al., 2021).
To further test the impact of stromal IL4Ra deficiency on the memory B cell compartment, immunized Ccl19Cre/+ Il4raf/f chimeras were boosted at 4 weeks and their plasma cell response was analyzed 5 days later. The stromal IL4Ra-deficient mice showed a significant reduction in generation of secondary NP+ plasma cells compared to WT controls (Fig. 6N–P). These data are consistent with reduced memory cell generation when FDCs are unable to restrict IL4 availability.
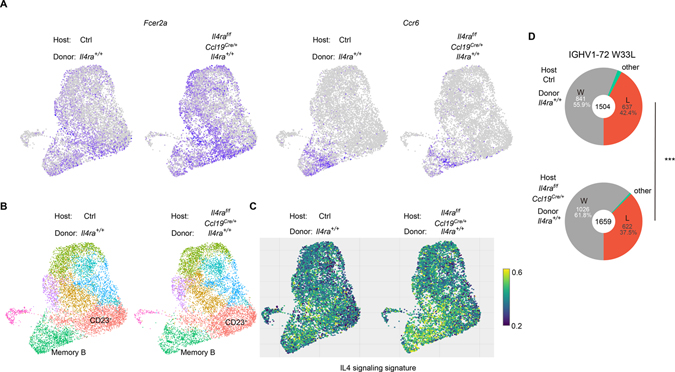
As a further approach to test for FDC IL4Ra-mediated restraint of IL4 signaling in GC B cells we performed scRNA-seq and scVDJ-seq on GC cells from WT and Ccl19Cre/+ Il4raf/f chimeras. Consistent with the flow cytometric findings, the expression of Fcer2a was increased and the expression of Ccr6 was decreased in the GC B cells from stromal IL4Ra-deficient mice (Fig. 7A). Clustering analysis also revealed a decrease in memory B cells (16.22% in WT and 12.92% in Ccl19Cre/+ Il4raf/f) and plasma cells (3.58% in WT and 1.86% in Ccl19Cre/+ Il4raf/f) (Fig. 7B and S7A). We also calculated the signature score of IL4 pathway activity by VISION, revealing an increase of IL4 signaling in the stromal IL4Ra-deficient chimeras (Fig. 7C).
Figure 7. Single cell RNA-seq analysis of stromal cell IL4Ra deficient mice.
(A-D) Reverse chimaeras were made by reconstituting irradiated Il4ra+/+ and Ccl19Cre/+ Il4raf/f mice with WT BM. Single cell RNA-seq and scVDJ-seq was performed on B220+ CD95hi GL7hi IgD− cells sorted at 14 post NP-KLH immunization. Cells from 4 mice were pooled together in each condition. (A) UMAP plots of the expression of Fcer2a and Ccr6. Color scaled for each gene with log normalized expression level. (B) UMAP plots of different clusters. (C) Visualize signature scores of IL4 signaling in GC B cells. Color scaled for signature score. (D) Pie charts displaying the frequencies of W33L mutations in IGHV1-72*01 sequences. Chi-square test. See also Figure S7.
Analysis of the scVDJ-seq data showed that there was no difference in the usage of IGHV1-72 heavy chain (Fig. S7B) or in the class switch of IGHV1-72 (Fig. S7C) between WT and Ccl19Cre/+ Il4raf/f chimeras. Importantly, however, there was a decrease in the frequency of W33L high affinity cells in the Ccl19Cre/+ Il4raf/f chimeras (Fig. 7D), suggesting a reduction in selection stringency when mice lack IL4Ra on FDCs. This was also suggested by a 30% enrichment for the affinity-reducing D50G mutation (Dal Porto et al., 1998) in the Ccl19Cre/+ Il4raf/f chimeras (0.937% in WT and 1.25% in Ccl19Cre/+ Il4raf/f). Finally, we also used these data to further test the relationship between affinity and memory and again found that the frequency of W33L mutations was lower in the memory B cell than in the GC B cell and plasma cell compartments in both WT and Ccl19Cre/+ Il4raf/f chimeras (Fig. S7D). Although Ig λ+ cells are dominant in the NP GC response (Jack et al., 1977) their frequency in memory B cells was lower than in GC B cells and plasma cells (Fig. S7E).
Discussion
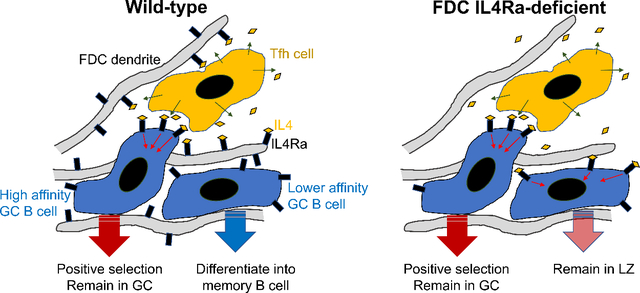
The experiments in this study were motivated by the finding from stromal cell scRNA-seq analysis that IL4Ra is expressed by FDCs (Rodda et al., 2018). Investigating IL4 actions in the GC, we characterized an IL4-responsive CD23+ LZ population of GC B cells that was enriched in pre-memory B cells and cells with features of pre-plasma cells. IL4Ra-STAT6 signaling was shown to disfavor pre-memory and memory cell development. Several lines of evidence indicated that IL4 could act on bystander B cells within the GC. Using conditional gene deficient mice, we found that FDC IL4Ra expression contributed to restraining IL4 availability in the GC. We propose that FDC IL4Ra expression helps prevent non-cognate actions of IL4 in the GC and thereby promotes antibody affinity maturation and GC B cell differentiation. While IL4 broadcasting by effector Th2 cells may be beneficial for type 2 immune responses, such as during worm expulsion, non-cognate actions of IL4 may be detrimental during selection events in GCs. We suggest that the GC evolved as a specialized microenvironment that enhances the ability of IL4 to act in a targeted manner.
Our findings extend recent work establishing that scRNA-seq can be used to identify pre-memory and pre-plasma cells within the GC (Holmes et al., 2020; Laidlaw et al., 2020) and provide evidence that it can be used to identify cells at an even earlier stage that are projected to take on one or other of these fates. Signaling pathway network analysis (Chen et al., 2012) of the day 7 scRNA-seq data showed that the CD23+ cluster was enriched for RelA activity (unpubl. obs.), in line with genetic studies showing that RelA was dispensable for GC maintenance but important for differentiation to post-GC states (Heise et al., 2014). The significance of the enriched BCR gene expression signature in the CD23+ LZ cells is not yet clear although some studies have suggested that BCR signaling is important for GC B cell differentiation into plasma cells (Krautler et al., 2017; Li et al., 2018).
Recent reports have provided evidence that memory B cells are often of lower affinity than GC B cells that are sampled from the animal at the same time (Shinnakasu et al., 2016; Suan et al., 2017; Sundling et al., 2021), though this has not been seen in every case (Wang et al., 2017). A limitation in some of these studies has been the possibility that the memory B cells being sequenced were generated at an earlier stage in the response than the GC cells, a time when less affinity enhancing mutations had accrued. However, in some studies the lower antigen binding ability was noted for pre-memory cells compared to contemporaneous GC cells (Shinnakasu et al., 2016; Suan et al., 2017). Our use of scVDJ-seq establishes a further approach to examine this issue and provides evidence supporting the conclusion that memory B cells and plasma cells are strongly selected for antigen binding, but cells taking on the memory fate are less strongly affinity matured than those taking on the plasma cell fate.
Cells expressing low levels of the plasma cell transcription factor Blimp1 have been identified amongst both LZ and DZ phenotype cells, with greater abundance in the DZ (Krautler et al., 2017; Radtke and Bannard, 2018). Our RNA trajectory analysis suggests some cells can take on early plasma cell gene expression properties while in a CD23+ LZ state, including beginning to upregulate CXCR4, a key marker of the DZ state. These cells may therefore give rise to DZ pre-plasma cells. These observations do not exclude the possibility of pre-plasma cells also emerging separately through a pathway in the DZ. Moreover, even when cells reach a Blimp1-low state, their fate can be unstable with some cells returning to a GC state (Radtke and Bannard, 2018).
CD23 expression by LZ cells has been previously noted, with expression being higher on pre-memory cells than most other GC cells, but the significance of this expression was not addressed (Gonzalez et al., 2018; Suan et al., 2017). The ability of IL4 to cause induction of CD23+ LZ cells, the detection of higher pStat6 in CD23+ than CD23− LZ cells, and the evidence of IL4 signaling pathway activity in the CD23+ cell cluster provides evidence that CD23 marks IL4-responding cells within the GC. CD23 can also be induced by CD40 signaling (Paterson et al., 1996; Siepmann et al., 1996) and we observed small numbers of cells in the Myc+ cluster that had high Fcer2a transcript levels. The basis for their being a separate population of CD23hi Myclo/− cells is not yet clear but they might reflect cells that have not yet received sufficient cognate T cell help to turn on Myc. We also suggest that some of the CD23+ cells exhibiting IL4 pathway activity in the absence of Myc expression may be cells receiving IL4 in a bystander manner.
The role of IL4 in GC responses has been examined in multiple studies but with variable conclusions. The findings presented here agree with observations indicating that IL4 is not universally required for mounting a GC response of normal magnitude. However, the data do add to evidence that IL4 can augment the size of the LZ compartment (Gonzalez et al., 2018; Turqueti-Neves et al., 2014). We propose that IL4 augments LZ cell accumulation at least in part by restraining effector cell output from the CD23+ LZ compartment. IL4 acts to intrinsically restrain CD23+ LZ B cell differentiation into memory B cells. When IL4Ra is absent, the cells appear to transit more readily to a CD23+CD31+ pre-memory state, likely contributing to an overall reduction in the LZ and an enlargement of the memory compartment. We noted similar trends for IL4Ra in restraining plasma cell generation and although this was less completely studied due to marker limitations, the finding is consistent with several in vitro studies (Ettinger et al., 2005; Jenks et al., 2018; Pignarre et al., 2021). Importantly, while it has been possible to track effects of IL4 on memory cell output, we have not been able to assess to what extent CD23+ LZ cells continue on to other stages of the GC. It is likely that some of the cells receive sufficient cumulative positive selection signals to undergo cyclic-reentry to the DZ.
A key aspect of this study is the finding that FDC IL4Ra helps restrict IL4 availability in the GC. Past work has highlighted that Tfh cells produce low amounts of IL4 compared to Th2 cells and this was suggested to be important for limiting IL4 delivery to cognate partner B cells (Crotty, 2014). However, IL4 was found to be broadcast from activated T cells in vitro and not released selectively at the synapse (Huse et al., 2006) and in accord with this observation, our data indicate that when the IL4Ra abundance in GCs is reduced, non-cognate actions of IL4 become notable. Moreover, our work adds support to the notion that the amounts of IL4 acting in the GC need to be finely tuned since both too little and too much can be associated with LZ defects, less affinity maturation and reduced GC responses; too much can also be associated with reduced memory and plasma cell generation. Our findings suggest that high IL4Ra expression on FDCs is part of the specialization of the GC microenvironment, helping establish a niche that limits non-cognate actions of IL4. The mechanism of FDC IL4Ra action in restraining IL4 availability is not yet clear, though IL4Ra has a well-established ability to mediate internalization of IL4 (Friedrich et al., 1999). It is also possible that soluble IL4Ra is generated though whether this has positive or negative effects on IL4 availability may be context dependent (Chilton and Fernandez-Botran, 1993; Khodoun et al., 2007). We speculate that in GCs containing some high affinity B cells that trigger strong activation of Tfh cells, this leads to transient production of sufficient amounts of IL4 to overcome the depletion mechanisms of the GC niche and promote ongoing participation of bystander lower affinity GC cells, helping sustain the diversity of individual GC responses.
Limitations
All the experiments were performed with intraperitoneal immunizations and analysis of GC responses in the spleen. It is not yet known if FDC IL4Ra will have equivalent functions in lymph nodes or across other types of GC responses. A technical limitation was our inability to perform ex vivo pSTAT6 staining in stromal IL4Ra-deficient mice due to poor assay sensitivity. The lack of markers to track emergence of pre-plasma cells was a limitation in assessing the accuracy of the trajectory analysis. We note that our study has not fully elucidated the effects of IL4 signaling in GC B cells, such as determining target genes engaged by Stat6 or the fate of IL4-stimulated cells that stay within the GC. Finally, we acknowledge that the clustering analysis has limitations in the precision with which it represents GC cells that are in a single state.
STAR Methods
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Jason Cyster (Jason.Cyster@ucsf.edu).
Materials availability
AS20-OVA generated in this study is available from the Lead Contact with a completed Materials Transfer Agreement.
Data and code availability
Single-cell RNA-seq data have been deposited at GEO(GSE180920).
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
B6 (Jax 000664) and Boy/J (Jax 002014) were originally purchased from the Jackson Laboratory. B6 (NCI 556) and B6-Ly5 (NCI 564) mice were purchased from National Cancer Institute at age 6–8 weeks. Il4ra−/− mice (Jax 003514) and Stat6−/− mice on a B6 background were provided by Drs. Mark Ansel and Richard Locksley, respectively. Some Il4ra−/− mice developed severe dermatological lesions and poor health and were excluded from the study. IL-2Rγ-chain (Cγ) deficient mice (Jax 003174) were purchased form Jackson Laboratory. S1pr2Venus/+ were generated as previously described (Moriyama et al., 2014). Aicdatm1Hon (Muramatsu et al., 2000) and Blimp1Gfp/+ (Prdm1Gfp/+) mice (Kallies et al., 2004) were provided by Chris Allen. Il4ra floxed mice (Herbert et al., 2004) were provided by Mark Ansel. Ccl19Cre/+ mice (Chai et al., 2013) and CD21Cre/+ mice (Kraus et al., 2004) were from an internal colony. Cas9f/f (Rosa26-LSL-Cas9, 026175) and Cγ1Cre/+ (010611) mice were from Jax. S1pr2CreERT2/+ mice were provided by T. Okada at the RIKEN Center for Integrative Medical Sciences (Shinnakasu et al., 2016). All mice were maintained under specific-pathogen free conditions and used according to institutional and governmental guidelines for animal welfare.
METHOD DETAILS
Immunization and treatment
For various in vivo experiments, mice were given 50 μg NP-haptenated antigen (Biosearch Technology) plus 1 μg LPS in 50 μl aluminium hydroxide gel (Invivogen) by i.p. injection. NP28-CGG, NP28-KLH or NP32-KLH was used in all experiments except in the Il4ra−/−, Cγ−/− and Ccl19-Cre Il4raf/f mice that were reconstituted with WT BM; in these chimeric mice NP14-CGG was used. To boost the immunization, 25 μg soluble NP-haptenated antigen was given by i.v. injection. To induce Cre expression, 20mg/ml corn oil dissolved tamoxifen (Sigma–Aldrich) was injected at 2 mg per 20g mouse i.p. at days 4, 6 and 8 post immunization. Normal chow was replaced with TAM diet (Envigo) on the day after the final tamoxifen dose.
Mixed or reverse bone marrow chimeras
For mixed chimeras, CD45.1 B6 mice were lethally irradiated by X-ray and transferred intravenously with BM cells mixed at a 1:1 ratio for WT CD45.1:WT CD45.2 control chimeras and a 2:3 ratio for WT CD45.1:Il4ra−/− CD45.2 chimeras. Typically we observed enhanced reconstitution of the follicular B cell compartment by Il4ra−/− BM such that the latter chimeras had on average 80% Il4ra−/− follicular B cells. For reverse chimeras, CD45.2 Il4ra+/+ and Il4ra−/− mice were lethally irradiated and transferred intravenously with CD45.1 B6 BM cells. Chimeras were immunized 6–8 weeks post reconstitution.
Retroviral constructs and transductions
Il4ra sgRNA sequences were selected using Benchling’s CRISPR Guide tool. The following Il4ra sgRNA sequences were used: sgRNA 1 (5′-CAAAATCAGACAGCCCACAG-3′); sgRNA 2 (5′-GATGTAGTCAGAGAAGCAGG-3′). The tRNA-gRNA-tRNA pMIA plasmid was used as a template for generating the tRNA-gRNA arrays, and these arrays were cloned into a pMCP-BFP vector by golden gate cloning. The pMCP-BFP sgRNA vector was generated by digesting the pMCPA vector (Kotov et al., 2019) with NcoI and NotI to remove Ametrine and replace it with tagBFP by In-Fusion Cloning. Retrovirus was generated by transfecting the Plat-E packaging cell line with 7.5 μg plasmid DNA and 16 μl Lipofectamine 2000 (Thermo Fisher Scientific). Cγ1Cre/+ Cas9f/+ mice were treated with 3 mg 5-fluorouracil (Sigma–Aldrich) by i.v. injection. Bone marrow was collected after 4–5 days and cultured in complete DMEM containing 10% fetal bovine serum, antibiotics (penicillin (50 IU/ml) and streptomycin (50 mg/ml); Cellgro) and 10 mM HEPES (pH 7.2; Cellgro), supplemented with IL-3, IL-6 and stem cell factor. Bone marrow cells were spin-infected twice at days 1 and 2 and then transferred into irradiated recipients.
EdU labeling and analysis
For 1h labeling, following immunization, mice were given intravenously 0.5 mg EdU 1h before being sacrificed. For long term labeling, immunized mice were intravenously given 0.5 mg EdU every 4 hours for 3 times 11 days post immunization and analyzed 3 days after EdU labeling. Cells were stained for incorporated EdU with the Click-iT EdU Alexa Fluor 647 Flow Cytometry Assay Kit (Invitrogen), according to the manufacturer’s instructions.
IL4 injection and analysis
2 μg mouse IL4 (PeproTech) and 10 μg anti-IL4 (clone 11B11, Bio X cell) were incubated at RT for 15min to make IL4-αIL4 complex. Immunized mice were intravenously given 2 μg complex once per day at day 11, 12, 13 post immunization, and analyzed at day 14.
AS20-OVA stimulation
The expressed Hc and Lc antibody genes in the CD45.1 specific AS20-1.17 hybridoma were sequenced. The Hc VDJ region was cloned into a vector (pBR322) containing the IgG1 constant regions coupled at the C-terminus to chicken OVA or a peptide (aa 57–346) from the malaria circumsporozoite (CS) protein as a control (vectors kindly provided by Michel Nussenzweig); the light chain VJ region was cloned into a vector containing the kappa C1 constant region (Boscardin et al., 2006). The antibody conjugates were expressed in HEK293 cells and purified using protein G. Naïve OT-II T cells were intravenously injected into B6 mice in combination with CD45.1+ and CD45.2+ MD4 B cells. Each recipient mouse was given 30 μg HEL-OVA antigen plus 1 μg LPS in alum (Thermo Scientific). 4–5 days post immunization, recipients were intravenously given 30 μg AS20-CS or AS20-OVA alone or with 100 μg anti-IL4 antibody (11B11) and analyzed 12 hours later.
Flow cytometry
Single-cell suspension of splenic cells were washed, blocked with 2.4G2 antibody (BioXcell), and stained with antibodies of indicated specificities in MACS buffer. Staining reagents include: BV786 anti-CD19 (1D3), PE-Cy7 anti-CD38 (90), Alexa Fluor 647 anti-CCR6 (140706), BV786 anti-B220 (RA3-6B2), BV421 anti-CD138 (281–2), BV711 anti-CD138 (281–2), PE-Cy7 anti-CD95 (Jo2), eFlour450 anti-GL7 (Ly-77), FITC anti-GL7 (Ly-77), PE-Cy7 anti-CD31 (MEC13.3), APC anti-CD23 (B3B4), PerCP/Cy5.5 anti-IgD (11–26c.2a), APC anti-Ki67 (16A8), BV605 anti-CD45.2 (104), BV605 anti-CD45.1 (A20) and BV605 anti-CD86 (GL-1) from Biolegend; biotinylated anti-CXCR4 (2B11), PE anti-CD31 (MEC 13.3), PE anti-IL4Ra (mIL4R-M1), Biotin anti-IL4Ra (mIL4R-M1), FITC anti-CD79b (HM79b), purified anti-CD72 b and c Alloantigens (JY/93), streptavidin-BV711 and streptavidin-BV605 from BD Biosciences; biotinylated anti-Ephrin-B1 polyclonal antibody from R&D System; NP-PE and NIP-BSA-biotin were used to stain for NP-binding cells (Biosearch Technology). Dead cell exclusion was based on Fixable Viability Dye eFluor 780 staining (eBioscience) and non-singlet events were excluded with FSC-W/FSC-H characteristics. For intracellular staining, cells were stained using Cytoperm/Cytofix kit (BD Biosciences) or Foxp3/Transcription Factor staining Buffer set (eBioscience) according to the manufacturer’s protocol. All data were collected on an LSR II cytometer (BD) and analyzed with FlowJo software (TreeStar).
Immunofluorescent staining
Mesenteric lymph nodes were fixed in 4% paraformaldehyde in PBS for 4 hours on ice and incubated in 30% sucrose overnight, followed by embedding in tissue freezing medium (OCT). Cryosections of 10 μm were stained with goat anti-IgD, rabbit anti-Tmem119, APC-conjugated anti-CD21/35, biotin-conjugated EpCAM, streptavidin-PE (Biolegend) and AMCA-conjugated donkey anti-goat IgG (H+L) (Jackson Immunoresearch). Images were captured with a Zeiss AxioOberver Z1 inverted microscope.
Single cell sequencing and analysis
Two sets of 7-day and 14-day wild-type GC B cells were sorted from mice immunized as above. Single cells were captured using the 10X Chromium (10X Genomics), and libraries was prepared according the manufacturer’s instructions (Chromium™ Single Cell 5’ Library & Gel Bead Kit, Chromium Single Cell 5′ Library Construction Kit, and Chromium Single Cell V(D)J Enrichment Kit, 10x Genomics). Libraries were run on the HiSeq4000 for Illumina sequencing. Sequencing data were processed with the 10X Cell Ranger (10X Genomics). Seurat (v3) was used for the further analysis. For day 7, sample 1 mean reads per cell was 314,827 and sample 2 was 565,017. Median genes per cell of sample 1 was 4213 and sample 2 was 4118. For day 14, sample 1 mean reads per cell was 82,769 and sample 2 was 96,660. Median genes per cell of sample 1 was 1590 and for sample 2 was 1456. Cells expressing less than 200 genes were filtered out. Cells with the percentage of mitochondrial genes per cell less than 5% were used for the day 7 dataset; due to the lower read depth in the day 14 dataset, cells with the percentage of mitochondrial genes per cell 20% were used. Cells with the frequency of ribosome genes per cell more than 30% were filtered out. 3542 (1012+2530) cells from the day 7 group and 19757 (9109+10648) cells from the day 14 group remained for further analyzed. We used “LogNormalize” (scale factor: 10,000) to normalize gene expression in each cell. The influence of batch effects and percentage of mitochondrial genes in each group/condition were regressed out using Seurat. The top 2000 variable genes of each group were selected for the PCA analysis. According to the PCElbowPlot, we chose the first 30 principal components (PCs) for the UMAP dimension reduction and clustering (resolution is 0.4). The cell markers of different clusters were identified using the FindAllMarkers function. Cell cycle score and classification were calculated by CellCycleScoring function based on the expression of G2/M and S phase markers (Kowalczyk et al., 2015). We used the published list of genes (Milpied et al., 2018; Victora et al., 2012) to calculate the DZ and LZ scores with the AddModuleScore function from the Seurat package. A post GC B cell RNA sequencing dataset (Laidlaw et al., 2017) was used to calculated the post GC B cell gene set signature with DESeq2. Log2 fold change more than 0.3 and adjusted p value less than 0.001 genes were used as the post GC B cell gene set signature (Table S2). Gene set signature score were calculated with VISION (DeTomaso et al., 2019) on the Seurat object.
The pseudo time analysis on CD23+ LZ GC B cell were carried out with Monocle v2 (Qiu et al., 2017). Top 1000 marker genes for each cluster were used for unsupervised ordering of the cells and the DDRTree algorithm implemented in Monocle2 was applied for trajectory reconstruction. Branch specific genes were calculated by the BEAM algorithm. Genes with q value less than 0.001 were plotted and used for GSEA analysis. To determine the fate of each branch, we compared the branch specific genes with the q value less than 0.001 to the published RNA sequencing datasets (Ise et al., 2018; Laidlaw et al., 2017; Shinnakasu et al., 2016) using GSEA..
For RNA velocity analysis, the spliced reads and unspliced reads were recounted by the Velocyto package (La Manno et al., 2018). We calculated the RNA velocity values for each gene each cell and embedded the RNA velocity vector to low-dimension space following the scVelo pipeline. The RNA velocity graph based pseudo time was calculated by Velocity pseudotime function from scVelo (Bergen et al., 2020).
BCR sequence reads were processed using Cell Ranger V3. AssignGenes and MakeDb python scripts were used to process 10X V(D)J data and generate a Change-O format file. ParseDb python script was used to parse the output file into heavy chain Change-O file. Only the record from which the barcode was detected and that passed the quality control for 5’ gene expression analysis was kept and merged to the Seurat object as meta information. CD23+ LZ GC B cells were grouped into pre-memory B cell and pre-plasma cell according the pseudotime analysis, and the cell expressing IGHV1-72*01 were kept. The CDR1 sequence of each cell in each group was used to generate the sequence logo by the WebLogo V3 (Crooks et al., 2004).
To determine the difference between control and stromal cell IL4ra-deficient mice, we sorted GC B cells from four mice and pooled them together in each condition. For the control sample, mean reads per cell was 25534, median genes per cell was 1727. For the stromal cell IL4Ra deficient sample, mean reads per cell was 28062, median genes per cell was 1791. Naïve B cells, non-B cells and low-quality cells were removed first. Cells expressing less than 200 genes were filtered out. Cells with a percentage of mitochondrial genes more than 10% were removed. 8038 cells were left in the control group and 7314 cells were left in the stromal cell IL4Ra-deficient group. Count data was normalized to log scale. The top 2000 variable genes of each group were selected. Two datasets were integrated with Seurat by using CCA based method. Clustering was carried out with this integrated dataset. For the BCR heavy chain analysis, the same preprocessing was carried out as described above. If the cell barcode was detected by more than one sequence only one was kept. Only functional sequences were used for analysis. 7791 heavy chain sequences were left in the control group and 8419 heavy chain sequences were left in the in the stromal cell IL4Ra-deficient group. BCR sequences in which the barcode was also detected and kept in the gene expression dataset were kept and mapped into Seurat object as meta information.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistics and graphing were done with Prism (GraphPad). Two-tailed Student’s t-tests were used to compare endpoint means of different groups unless otherwise indicated.
Supplementary Material
Markers of different clusters of Day7 GC B cells, Related to Figure 1
Post GC B cell signature, Related to Figure S2
BCR heavy chain information of GC B cells, Related to Figure 3, 7, S2, S7, and STAR Methods
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-CD19 BV785 (clone 1D3) | BD Bioscience | Cat#563333; RRID: AB_2738141 |
| anti-B220 BV785 (clone RA3–6B2) | BioLegend | Cat#103246; RRID: AB_2563256 |
| anti-IgD FITC (clone 11–26c.2a) | BioLegend | Cat#405704; RRID: AB_394859 |
| anti-IgD Alexa Fluor 647 (clone 11–26c.2a) | BioLegend | Cat#405708; RRID: AB_893528 |
| anti-IgD Pacific Blue (clone 11–26c.2a) | BioLegend | Cat#405712; RRID: AB_1937244 |
| anti-IgD BV605 | BD Bioscience | Cat#563003; RRID: AB_2737944 |
| anti-IgD PerCP/Cy5.5 | BioLegend | Cat#405710; RRID: AB_1575113 |
| anti-GL-7 (Ly-77) Pacific Blue (clone GL7) | BioLegend | Cat#144614; RRID: AB_2563292 |
| anti-GL-7 (Ly-77) FITC (clone GL7) | BioLegend | Cat#144604; RRID: AB_2561697 |
| anti-CD95 PE-CY7 (clone Jo2) | BD Bioscience | Cat#557653; RRID: AB_396768 |
| anti-CD45.1 PERCP-CY5.5 (clone A20) | Tonbo | Cat#65–0453-U100 |
| anti-CD45.1 BV605 (clone A20) | BioLegend | Cat#110738; RRID: AB_2562565 |
| anti-CD45.2 BV605 (clone 104) | BioLegend | Cat#109841; RRID: AB_2563485 |
| anti-Ephrin-B1 Biotin (polyclonal) | R&D | Cat#BAF473; RRID: AB_2293418 |
| anti-CXCR4 Biotin (clone 2B11) | BD Bioscience | Cat#551968; RRID: AB_394307 |
| anti-CD86 BV605 | Biolegend | Cat#105037; RRID: AB_11204429 |
| anti-IgG1 FITC (clone RMG1–1) | BioLegend | Cat#406606; RRID: AB_493293 |
| anti-IgG1 APC | BioLegend | Cat#406610; RRID: AB_10696420 |
| anti-CD138 BV421 (clone 281–2) | BioLegend | Cat#142508; RRID: AB_11203544 |
| anti-CD138 BV711 (clone 281–2) | BD Bioscience | Cat#563193; RRID: AB_2631190 |
| anti-CD98 BV711 | BD Bioscience | Cat#745466; RRID: AB_2743009 |
| anti-CD73 PERCP-CY5.5 (clone TY/11.8) | BioLegend | Cat#127214; RRID: AB_11219403 |
| anti-gp38 APC (clone 8.1.1) | BioLegend | Cat#127410; RRID: AB_10613649 |
| anti-CD31 PE (clone MEC 13.3) | BD Biosciences | Cat#553373; RRID: AB_394819 |
| anti-CD31 PE-Cy7 (clone MEC 13.3) | BioLegend | Cat#102417; RRID: AB_830756 |
| anti-Epcam BV711 | BioLegend | Cat#118233; RRID: AB_2632775 |
| anti-CD21/35 pacific Blue | BioLegend | Cat#123414; RRID: AB_2085158 |
| Streptavidin BV711 | BD Bioscience | Cat#563262; RRID: AB_2869478 |
| anti-CD79b FITC | BD Bioscience | Cat#555303; RRID: AB_395717 |
| anti-CD23 Alexa Fluor 488 | BioLegend | Cat#101609; RRID: AB_493362 |
| anti-CD23 Alexa Fluor 647 | BioLegend | Cat#101612; RRID: AB_2103038 |
| anti-CD72 | BD Bioscience | Cat#550268; RRID: AB_393565 |
| anti-IL4ra PE (Clone mIL4R-M1) | BD Bioscience | Cat#552509; RRID: AB_394407 |
| anti-IL4 (clone 11B11) | Bio X cell | Cat#BE0045; RRID: AB_1107707 |
| Fixable Viability Dye eFluor780 | eBioscience | Cat#65–0865-14 |
| anti-CD38 PE-Cy7 | BioLegend | Cat#102718; RRID: AB_2275531 |
| anti-CD38 FITC | BioLegend | Cat# 102705; RRID: AB_312926 |
| anti-CD196 Alexa Fluor 647 | BD Bioscience | Cat#557976; RRID: AB_2228793 |
| anti-Ki67 Alexa Fluor 647 | BD Bioscience | Cat#558615; RRID: AB_647130 |
| anti-Bcl-6 Alexa Fluor 647 | BD Bioscience | Cat#561525; RRID: AB_10898007 |
| anti-pStat6 (Tyr641) (46H1L12) | Thermo Fisher Scientific | Cat# 700247; RRID: AB_2532305 |
| Rabbit anti-Tmem119 | B. Barres | Bennett et al., 2016 |
| AS20/anti-CD45.1-OVA | This paper | NA |
| Bacterial and virus strains | ||
| Biological samples | ||
| Chemicals, peptides, and recombinant proteins | ||
| Lipopolysaccharides (Escherichia coli, serotype O111:B4) | Sigma | Cat# L2630–25MG |
| NP-CGG | Biosearch Technologies | Cat#N-5055C-5 |
| NP-KLH | Biosearch Technologies |
Cat#N-5060 |
| NP-PE | Biosearch Technologies | Cat#N-5070–1 |
| Collagenase IV | Worthington Biochemical | Cat#LS004188 |
| DNase I, bovine pancreas (single cell stromal cell analysis) | Sigma-Aldrich | Cat#DN25 |
| EdU (5-ethynyl-2’-deoxyuridine) | Setareh Biotech | Cat#7180 |
| mLTbR-huIgG1 (LTbR-Fc) | J. Browning | N/A |
| Aluminium hydroxide gel | Invivogen | Cat#vac-alu-250 |
| Murine recombinant IL-4 | PeproTech | Cat#214–14 |
| Critical commercial assays | ||
| Chromium Single Cell 5′ Library & Gel Bead Kit | 10x Genomics | Cat#1000014 |
| Chromium i7 Multiplex Kit | 10x Genomics | Cat#120262 |
| Chromium Single Cell A Chip Kit | 10x Genomics | Cat#1000151 |
| Chromium Single Cell 5′ Library Construction Kit | 10x Genomics | Cat#1000020 |
| Chromium Single Cell V(D)J Enrichment Kit, Mouse B Cell | 10x Genomics | Cat#1000072 |
| Click-iT Plus EdU Alexa Fluor 647 Flow Cytometry Assay Kit 100 Tests | Invitrogen | Cat#C10635 |
| Lipofectamine 2000 Transfection Reagent | Thermo Fisher Scientific | Cat# 11668500 |
| Deposited data | ||
| 10x single cell RNA-seq data | This paper | GSE180920 |
| BCR heavy chain sequencing data | This paper | Table S4 |
| IL4 incubation RNA-seq data | Mokada-Gopal et al., 2017 | GSE84075 |
| Spleen stromal cell single cell RNA-seq data | Cheng et al., 2019 | E-MTAB-7703, E-MTAB-7094 |
| NP-specific high- and low-affinity GC B cell RNA-seq data | Shinnakasu et al., 2016 | GSE73729 |
| Pre plasma cell RNA-seq data | Ise et al., 2018 | GSE109732 |
| Post GC B cell RNA-seq data | Laidlaw et al., 2017 | GSE89897 |
| Pre memory B cell RNA-seq data | Wang et al., 2017 | GSE85018 |
| Experimental models: Cell lines | ||
| Platinum-E (Plat-E) Retroviral Packaging Cell Line | Gift from S. Schwab | N/A |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | JAX: 000664 |
| Mouse: B6.SJL-Ptprca Pepcb/BoyJ | The Jackson Laboratory | JAX: 002014 |
| Mouse: Rosa26-LSL-Cas9 | The Jackson Laboratory | JAX: 024857 |
| Mouse: Il4ra−/− | The Jackson Laboratory | JAX:003514 |
| Mouse: Cγ1Cre/+ | The Jackson Laboratory | JAX: 010611 |
| Mouse: B6.129S4-Il2rgtm1Wjl/J | The Jackson Laboratory | JAX:003174 |
| Mouse: Aicdatm1Hon | Muramatsu et al., 2000 | MGI: 2156156 |
| Mouse: Blimp1Gfp/+ | Kallies et al., 2004 | MGI: 3510704 |
| Mouse: Il4raf/f | Herbert et al., 2004 | N/A |
| Mouse: Ccl19Cre/+ | Chai et al., 2013 | MGI: 5526991 |
| Mouse: CD21Cre/+ | Kraus et al., 2004 | MGI: 3047571 |
| Mouse: S1pr2CreERT2/+ | Shinnakasu et al., 2016 | MGI: 6435090 |
| Mouse: Stat6−/− | Takeda et al., 1996 | MGI: 1888385 |
| Mouse: S1pr2Venus/+ | Moriyama et al., 2014 | N/A |
| Oligonucleotides | ||
| Il4ra sgRNA 1 (5’-CAAAATCAGACAGCCCACAG-3’) | This paper | N/A |
| Il4ra sgRNA 2 (5’-GATGTAGTCAGAGAAGCAGG-3’) | This paper | N/A |
| Recombinant DNA | ||
| pMCP-BFP | Kotov et al., 2019 | N/A |
| Software and algorithms | ||
| GraphPad Prism | GraphPad Software |
http://www.graphpad.com/scientific-software/prism/ RRID: SCR_002798 |
| Fiji | NIH |
http://fiji.sc/ RRID: SCR_002285 |
| R v3.6.2 | R Foundation for Statistical Computing | https://www.r-project.org/ |
| Cell Ranger v3.1.0 | 10x Genomics |
https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/what-is-cell-ranger RRID: SCR_017344 |
| Seurat v3.1.1 | Satija Lab |
http://satijalab.org/seurat/ RRID: SCR_007322 |
| Scanpy v1.4.6 | Theis Lab |
https://icb-scanpy.readthedocs-hosted.com/en/stable/ RRID: SCR_018139 |
| ScVelo v0.1.25 | Theis Lab |
https://scvelo.readthedocs.io/index.html RRID: SCR_018168 |
| Velocyto v0.17 | velocyto-team |
http://velocyto.org/ RRID: SCR_018167 |
| LoomR v0.2.1.9 | Satija Lab | https://github.com/mojaveazure/loomR |
| loompy v3.0 | Linnarsson Lab |
http://loompy.org/ RRID: SCR_016666 |
| Monocle 2 | Trapnell lab |
http://cole-trapnell-lab.github.io/monocle-release/docs/ RRID: SCR_016339 |
| VISION v1.1.0 | Yosef Lab | https://yoseflab.github.io/VISION/news/index.html |
| IgBLAST V1.14.0 | NCBI |
https://ncbi.github.io/igblast/rel/Release-notes.html RRID: SCR_002873 |
| Immcantation | Kleinstein Lab | https://immcantation.readthedocs.io/en/stable/index.html |
| Plotly Python Open Source Graphing Library v4.5.3 | Plotly |
https://github.com/plotly/plotly.py/ RRID: SCR_013991 |
| RStudio | RStudio |
https://www.rstudio.com/products/rstudio/ RRID: SCR_000432 |
| GSEA v4 for Linux | GSEA team |
https://www.gsea-msigdb.org/gsea/index.jsp RRID: SCR_003199 |
| Weblogo 3 | Crooks et al., 2004 |
http://weblogo.threeplusone.com/ RRID: SCR_010236 |
| Flowjo | Treestar |
https://www.flowjo.com/ RRID: SCR_008520 |
| Other | ||
Highlights.
scRNA-seq identified an IL4Ra+ CD23+ LZ GC B cell subset
IL4 restrained CD23+ GC B cell differentiation into memory cells
IL4 exerted non-cognate actions within the GC
FDC IL4Ra restricted GC IL4 availability and augmented memory B cell generation
Acknowledgements
We thank Chris Allen, Mark Ansel, Marlys Fassett, Rich Locksley, Burkhard Ludewig and Takaharu Okada for mice. This work was supported by grants from the NIH (R01AI045073). D.L. and H.C. were supported by the Irvington Cancer Research Institute. J.G.C. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Declaration of Interests
J.G.C. is an SAB member of Be Biopharma and MiroBio Ltd.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- Anderson SM, Tomayko MM, Ahuja A, Haberman AM, and Shlomchik MJ (2007). New markers for murine memory B cells that define mutated and unmutated subsets. J. Exp. Med. 204, 2103–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andoh A, Masuda A, Yamakawa M, Kumazawa Y, and Kasajima T (2000). Absence of interleukin-4 enhances germinal center reaction in secondary immune response. Immunol. Lett. 73, 35–41. [DOI] [PubMed] [Google Scholar]
- Bannard O, and Cyster JG (2017). Germinal centers: programmed for affinity maturation and antibody diversification. Curr. Opin. Immunol. 45, 21–30. [DOI] [PubMed] [Google Scholar]
- Bemark M, Hazanov H, Stromberg A, Komban R, Holmqvist J, Koster S, Mattsson J, Sikora P, Mehr R, and Lycke NY (2016). Limited clonal relatedness between gut IgA plasma cells and memory B cells after oral immunization. Nature communications 7, 12698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergen V, Lange M, Peidli S, Wolf A, and Theis FJ (2020). Generalizing RNA velocity to transient cell states through dynamical modeling. bioRxiv 820936; doi: 10.1101/820936 [DOI] [PubMed] [Google Scholar]
- Boscardin SB, Hafalla JC, Masilamani RF, Kamphorst AO, Zebroski HA, Rai U, Morrot A, Zavala F, Steinman RM, Nussenzweig RS, and Nussenzweig MC (2006). Antigen targeting to dendritic cells elicits long-lived T cell help for antibody responses. J. Exp. Med. 203, 599–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calado DP, Sasaki Y, Godinho SA, Pellerin A, Kochert K, Sleckman BP, de Alboran IM, Janz M, Rodig S, and Rajewsky K (2012). The cell-cycle regulator c-Myc is essential for the formation and maintenance of germinal centers. Nat. Immunol. 13, 1092–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Q, Onder L, Scandella E, Gil-Cruz C, Perez-Shibayama C, Cupovic J, Danuser R, Sparwasser T, Luther SA, Thiel V, et al. (2013). Maturation of lymph node fibroblastic reticular cells from myofibroblastic precursors is critical for antiviral immunity. Immunity 38, 1013–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, Xu H, Gordonov S, Lim MP, Perkins MH, and Ma’ayan A (2012). Expression2Kinases: mRNA profiling linked to multiple upstream regulatory layers. Bioinformatics 28, 105–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HW, Onder L, Novkovic M, Soneson C, Lutge M, Pikor N, Scandella E, Robinson MD, Miyazaki JI, Tersteegen A, et al. (2019). Origin and differentiation trajectories of fibroblastic reticular cells in the splenic white pulp. Nature communications 10, 1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevrier S, Kratina T, Emslie D, Tarlinton DM, and Corcoran LM (2017). IL4 and IL21 cooperate to induce the high Bcl6 protein level required for germinal center formation. Immunol. Cell Biol. 95, 925–932. [DOI] [PubMed] [Google Scholar]
- Chilton PM, and Fernandez-Botran R (1993). Production of soluble IL-4 receptors by murine spleen cells is regulated by T cell activation and IL-4. J. Immunol. 151, 5907–5917. [PubMed] [Google Scholar]
- Cortes-Selva D, Ready A, Gibbs L, Rajwa B, and Fairfax KC (2019). IL-4 promotes stromal cell expansion and is critical for development of a type-2, but not a type 1 immune response. Eur. J. Immunol. 49, 428–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, and Brenner SE (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S (2014). T follicular helper cell differentiation, function, and roles in disease. Immunity 41, 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham AF, Serre K, Toellner KM, Khan M, Alexander J, Brombacher F, and MacLennan IC (2004). Pinpointing IL-4-independent acquisition and IL-4-influenced maintenance of Th2 activity by CD4 T cells. Eur. J. Immunol. 34, 686–694. [DOI] [PubMed] [Google Scholar]
- Dal Porto JM, Haberman AM, Shlomchik MJ, and Kelsoe G (1998). Antigen drives very low affinity B cells to become plasmacytes and enter germinal centers. J. Immunol. 161, 5373–5381. [PubMed] [Google Scholar]
- Defrance T, Aubry JP, Rousset F, Vanbervliet B, Bonnefoy JY, Arai N, Takebe Y, Yokota T, Lee F, Arai K, and et al. (1987). Human recombinant interleukin 4 induces Fc epsilon receptors (CD23) on normal human B lymphocytes. J. Exp. Med. 165, 1459–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeTomaso D, Jones MG, Subramaniam M, Ashuach T, Ye CJ, and Yosef N (2019). Functional interpretation of single cell similarity maps. Nature communications 10, 4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Sola D, Victora GD, Ying CY, Phan RT, Saito M, Nussenzweig MC, and Dalla-Favera R (2012). The proto-oncogene MYC is required for selection in the germinal center and cyclic reentry. Nat. Immunol. 13, 1083–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey LK, Karempudi P, Luther SA, Ludewig B, and Harris NL (2017). Interactions between fibroblastic reticular cells and B cells promote mesenteric lymph node lymphangiogenesis. Nature communications 8, 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ersching J, Efeyan A, Mesin L, Jacobsen JT, Pasqual G, Grabiner BC, Dominguez-Sola D, Sabatini DM, and Victora GD (2017). Germinal Center Selection and Affinity Maturation Require Dynamic Regulation of mTORC1 Kinase. Immunity 46, 1045–1058 e1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettinger R, Sims GP, Fairhurst AM, Robbins R, da Silva YS, Spolski R, Leonard WJ, and Lipsky PE (2005). IL-21 induces differentiation of human naive and memory B cells into antibody-secreting plasma cells. J. Immunol. 175, 7867–7879. [DOI] [PubMed] [Google Scholar]
- Finkelman FD, Madden KB, Morris SC, Holmes JM, Boiani N, Katona IM, and Maliszewski CR (1993). Anti-cytokine antibodies as carrier proteins. Prolongation of in vivo effects of exogenous cytokines by injection of cytokine-anti-cytokine antibody complexes. J. Immunol. 151, 1235–1244. [PubMed] [Google Scholar]
- Finkin S, Hartweger H, Oliveira TY, Kara EE, and Nussenzweig MC (2019). Protein Amounts of the MYC Transcription Factor Determine Germinal Center B Cell Division Capacity. Immunity 51, 324–336 e325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich K, Kammer W, Erhardt I, Brandlein S, Arnold S, and Sebald W (1999). The two subunits of the interleukin-4 receptor mediate independent and distinct patterns of ligand endocytosis. Eur. J. Biochem. 265, 457–465. [DOI] [PubMed] [Google Scholar]
- Fu Y-X, and Chaplin DD (1999). Development and maturation of secondary lymphoid tissues. Ann. Rev. Immunol. 17, 399–433. [DOI] [PubMed] [Google Scholar]
- Gaya M, Barral P, Burbage M, Aggarwal S, Montaner B, Warren Navia A, Aid M, Tsui C, Maldonado P, Nair U, et al. (2018). Initiation of Antiviral B Cell Immunity Relies on Innate Signals from Spatially Positioned NKT Cells. Cell 172, 517–533 e520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitlin AD, von Boehmer L, Gazumyan A, Shulman Z, Oliveira TY, and Nussenzweig MC (2016). Independent Roles of Switching and Hypermutation in the Development and Persistence of B Lymphocyte Memory. Immunity 44, 769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez DG, Cote CM, Patel JR, Smith CB, Zhang Y, Nickerson KM, Zhang T, Kerfoot SM, and Haberman AM (2018). Nonredundant Roles of IL-21 and IL-4 in the Phased Initiation of Germinal Center B Cells and Subsequent Self-Renewal Transitions. J. Immunol. 201, 3569–3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granato A, Hayashi EA, Baptista BJ, Bellio M, and Nobrega A (2014). IL-4 regulates Bim expression and promotes B cell maturation in synergy with BAFF conferring resistance to cell death at negative selection checkpoints. J. Immunol. 192, 5761–5775. [DOI] [PubMed] [Google Scholar]
- Haberman AM, Gonzalez DG, Wong P, Zhang TT, and Kerfoot SM (2019). Germinal center B cell initiation, GC maturation, and the coevolution of its stromal cell niches. Immunol. Rev. 288, 10–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haniuda K, Fukao S, and Kitamura D (2020). Metabolic Reprogramming Induces Germinal Center B Cell Differentiation through Bcl6 Locus Remodeling. Cell Rep 33, 108333. [DOI] [PubMed] [Google Scholar]
- He JS, Subramaniam S, Narang V, Srinivasan K, Saunders SP, Carbajo D, Wen-Shan T, Hidayah Hamadee N, Lum J, Lee A, et al. (2017). IgG1 memory B cells keep the memory of IgE responses. Nature communications 8, 641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heise N, De Silva NS, Silva K, Carette A, Simonetti G, Pasparakis M, and Klein U (2014). Germinal center B cell maintenance and differentiation are controlled by distinct NF-kappaB transcription factor subunits. J. Exp. Med. 211, 2103–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert DR, Holscher C, Mohrs M, Arendse B, Schwegmann A, Radwanska M, Leeto M, Kirsch R, Hall P, Mossmann H, et al. (2004). Alternative macrophage activation is essential for survival during schistosomiasis and downmodulates T helper 1 responses and immunopathology. Immunity 20, 623–635. [DOI] [PubMed] [Google Scholar]
- Holmes AB, Corinaldesi C, Shen Q, Kumar R, Compagno N, Wang Z, Nitzan M, Grunstein E, Pasqualucci L, Dalla-Favera R, and Basso K (2020). Single-cell analysis of germinal-center B cells informs on lymphoma cell of origin and outcome. J. Exp. Med. 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse M, Lillemeier BF, Kuhns MS, Chen DS, and Davis MM (2006). T cells use two directionally distinct pathways for cytokine secretion. Nat. Immunol. 7, 247–255. [DOI] [PubMed] [Google Scholar]
- Ise W, Fujii K, Shiroguchi K, Ito A, Kometani K, Takeda K, Kawakami E, Yamashita K, Suzuki K, Okada T, and Kurosaki T (2018). T Follicular Helper Cell-Germinal Center B Cell Interaction Strength Regulates Entry into Plasma Cell or Recycling Germinal Center Cell Fate. Immunity 48, 702–715 e704. [DOI] [PubMed] [Google Scholar]
- Jack RS, Imanishi-Kari T, and Rajewsky K (1977). Idiotypic analysis of the response of C57BL/6 mice to the (4-hydroxy-3-nitrophenyl)acetyl group. Eur. J. Immunol. 7, 559–565. [DOI] [PubMed] [Google Scholar]
- Jacob J, Przylepa J, Miller C, and Kelsoe G (1993). In situ studies of the primary immune response to (4-hydroxy-3-nitrophenyl)acetyl. III. The kinetics of V region mutation and selection in germinal center B cells. J. Exp. Med. 178, 1293–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinek DF, and Lipsky PE (1988). Inhibitory influence of IL-4 on human B cell responsiveness. J. Immunol. 141, 164–173. [PubMed] [Google Scholar]
- Jenks SA, Cashman KS, Zumaquero E, Marigorta UM, Patel AV, Wang X, Tomar D, Woodruff MC, Simon Z, Bugrovsky R, et al. (2018). Distinct Effector B Cells Induced by Unregulated Toll-like Receptor 7 Contribute to Pathogenic Responses in Systemic Lupus Erythematosus. Immunity 49, 725–739 e726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji T, Ishige A, Hikida M, Taka J, Hijikata A, Kubo M, Nagashima T, Takahashi Y, Kurosaki T, Okada M, et al. (2012). Distinct cellular pathways select germline-encoded and somatically mutated antibodies into immunological memory. J. Exp. Med. 209, 2079–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallies A, Hasbold J, Tarlinton DM, Dietrich W, Corcoran LM, Hodgkin PD, and Nutt SL (2004). Plasma cell ontogeny defined by quantitative changes in blimp-1 expression. J. Exp. Med. 200, 967–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodoun M, Lewis CC, Yang JQ, Orekov T, Potter C, Wynn T, Mentink-Kane M, Hershey GK, Wills-Karp M, and Finkelman FD (2007). Differences in expression, affinity, and function of soluble (s)IL-4Ralpha and sIL-13Ralpha2 suggest opposite effects on allergic responses. J. Immunol. 179, 6429–6438. [DOI] [PubMed] [Google Scholar]
- Kotov DI, Mitchell JS, Pengo T, Ruedl C, Way SS, Langlois RA, Fife BT, and Jenkins MK (2019). TCR Affinity Biases Th Cell Differentiation by Regulating CD25, Eef1e1, and Gbp2. J. Immunol. 202, 2535–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk MS, Tirosh I, Heckl D, Rao TN, Dixit A, Haas BJ, Schneider RK, Wagers AJ, Ebert BL, and Regev A (2015). Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res. 25, 1860–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus M, Alimzhanov MB, Rajewsky N, and Rajewsky K (2004). Survival of resting mature B lymphocytes depends on BCR signaling via the Igalpha/beta heterodimer. Cell 117, 787–800. [DOI] [PubMed] [Google Scholar]
- Krautler NJ, Suan D, Butt D, Bourne K, Hermes JR, Chan TD, Sundling C, Kaplan W, Schofield P, Jackson J, et al. (2017). Differentiation of germinal center B cells into plasma cells is initiated by high-affinity antigen and completed by Tfh cells. J. Exp. Med. 214, 1259–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lonnerberg P, Furlan A, et al. (2018). RNA velocity of single cells. Nature 560, 494–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laidlaw BJ, Duan L, Xu Y, Vazquez SE, and Cyster JG (2020). The transcription factor Hhex cooperates with the corepressor Tle3 to promote memory B cell development. Nat. Immunol. 21, 1082–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laidlaw BJ, Schmidt TH, Green JA, Allen CD, Okada T, and Cyster JG (2017). The Eph-related tyrosine kinase ligand Ephrin-B1 marks germinal center and memory precursor B cells. J. Exp. Med. 214, 639–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Gadzinsky A, Gong L, Tong H, Calderon V, Li Y, Kitamura D, Klein U, Langdon WY, Hou F, et al. (2018). Cbl Ubiquitin Ligases Control B Cell Exit from the Germinal-Center Reaction. Immunity 48, 530–541 e536. [DOI] [PubMed] [Google Scholar]
- Luther SA, Bidgol A, Hargreaves DC, Schmidt A, Xu Y, Paniyadi J, Matloubian M, and Cyster JG (2002). Differing Activities of Homeostatic Chemokines CCL19, CCL21, and CXCL12 in Lymphocyte and Dendritic Cell Recruitment and Lymphoid Neogenesis. J. Immunol. 169, 424–433. [DOI] [PubMed] [Google Scholar]
- Mackay F, and Browning JL (1998). Turning off follicular dendritic cells. Nature 395, 26–27. [DOI] [PubMed] [Google Scholar]
- MacLennan ICM (1994). Germinal Centers. Annu. Rev. Immunol. 12, 117–139. [DOI] [PubMed] [Google Scholar]
- McCormick SM, and Heller NM (2015). Commentary: IL-4 and IL-13 receptors and signaling. Cytokine 75, 38–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milpied P, Cervera-Marzal I, Mollichella ML, Tesson B, Brisou G, Traverse-Glehen A, Salles G, Spinelli L, and Nadel B (2018). Human germinal center transcriptional programs are de-synchronized in B cell lymphoma. Nat. Immunol. 19, 1013–1024. [DOI] [PubMed] [Google Scholar]
- Mokada-Gopal L, Boeser A, Lehmann CHK, Drepper F, Dudziak D, Warscheid B, and Voehringer D (2017). Identification of Novel STAT6-Regulated Proteins in Mouse B Cells by Comparative Transcriptome and Proteome Analysis. J. Immunol. 198, 3737–3745. [DOI] [PubMed] [Google Scholar]
- Moriyama S, Takahashi N, Green JA, Hori S, Kubo M, Cyster JG, and Okada T (2014). Sphingosine-1-phosphate receptor 2 is critical for follicular helper T cell retention in germinal centers. J. Exp. Med. 211, 1297–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, and Honjo T (2000). Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102, 553–563. [DOI] [PubMed] [Google Scholar]
- Onodera T, Takahashi Y, Yokoi Y, Ato M, Kodama Y, Hachimura S, Kurosaki T, and Kobayashi K (2012). Memory B cells in the lung participate in protective humoral immune responses to pulmonary influenza virus reinfection. Proc. Natl. Acad. Sci. U. S. A. 109, 2485–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson RL, Lack G, Domenico JM, Delespesse G, Leung DY, Finkel TH, and Gelfand EW (1996). Triggering through CD40 promotes interleukin-4-induced CD23 production and enhanced soluble CD23 release in atopic disease. Eur. J. Immunol. 26, 1979–1984. [DOI] [PubMed] [Google Scholar]
- Perona-Wright G, Mohrs K, and Mohrs M (2010). Sustained signaling by canonical helper T cell cytokines throughout the reactive lymph node. Nat. Immunol. 11, 520–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignarre A, Chatonnet F, Caron G, Haas M, Desmots F, and Fest T (2021). Plasmablasts derive from CD23-activated B cells after the extinction of IL-4/STAT6 signaling and IRF4 induction. Blood 137, 1166–1180. [DOI] [PubMed] [Google Scholar]
- Qiu X, Hill A, Packer J, Lin D, Ma YA, and Trapnell C (2017). Single-cell mRNA quantification and differential analysis with Census. Nat. Methods 14, 309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radtke D, and Bannard O (2018). Expression of the Plasma Cell Transcriptional Regulator Blimp-1 by Dark Zone Germinal Center B Cells During Periods of Proliferation. Front Immunol 9, 3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt RL, Liang HE, and Locksley RM (2009). Cytokine-secreting follicular T cells shape the antibody repertoire. Nat. Immunol. 10, 385–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter R, and Pfeffer K (2002). Impaired germinal centre formation and humoral immune response in the absence of CD28 and interleukin-4. Immunology 106, 222–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricardo-Gonzalez RR, Red Eagle A, Odegaard JI, Jouihan H, Morel CR, Heredia JE, Mukundan L, Wu D, Locksley RM, and Chawla A (2010). IL-4/STAT6 immune axis regulates peripheral nutrient metabolism and insulin sensitivity. Proc. Natl. Acad. Sci. U. S. A. 107, 22617–22622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MJ, Pitt C, Brodie EJ, Valk AM, O’Donnell K, Nitschke L, Jones S, and Tarlinton DM (2019). BAFF, IL-4 and IL-21 separably program germinal center-like phenotype acquisition, BCL6 expression, proliferation and survival of CD40L-activated B cells in vitro. Immunol. Cell Biol. 97, 826–839. [DOI] [PubMed] [Google Scholar]
- Roco JA, Mesin L, Binder SC, Nefzger C, Gonzalez-Figueroa P, Canete PF, Ellyard J, Shen Q, Robert PA, Cappello J, et al. (2019). Class-Switch Recombination Occurs Infrequently in Germinal Centers. Immunity 51, 337–350 e337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodda LB, Lu E, Bennett ML, Sokol CL, Wang X, Luther SA, Barres BA, Luster AD, Ye CJ, and Cyster JG (2018). Single-Cell RNA Sequencing of Lymph Node Stromal Cells Reveals Niche-Associated Heterogeneity. Immunity 48, 1014–1028 e1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwickert TA, Alabyev B, Manser T, and Nussenzweig MC (2009). Germinal center reutilization by newly activated B cells. J. Exp. Med. 206, 2907–2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinnakasu R, Inoue T, Kometani K, Moriyama S, Adachi Y, Nakayama M, Takahashi Y, Fukuyama H, Okada T, and Kurosaki T (2016). Regulated selection of germinal-center cells into the memory B cell compartment. Nat. Immunol. 17, 861–869. [DOI] [PubMed] [Google Scholar]
- Siepmann K, Wohlleben G, and Gray D (1996). CD40-mediated regulation of interleukin-4 signaling pathways in B lymphocytes. Eur. J. Immunol. 26, 1544–1552. [DOI] [PubMed] [Google Scholar]
- Suan D, Krautler NJ, Maag JLV, Butt D, Bourne K, Hermes JR, Avery DT, Young C, Statham A, Elliott M, et al. (2017). CCR6 Defines Memory B Cell Precursors in Mouse and Human Germinal Centers, Revealing Light-Zone Location and Predominant Low Antigen Affinity. Immunity 47, 1142–1153 e1144. [DOI] [PubMed] [Google Scholar]
- Sundling C, Lau AWY, Bourne K, Young C, Laurianto C, Hermes JR, Menzies RJ, Butt D, Krautler NJ, Zahra D, et al. (2021). Positive selection of IgG(+) over IgM(+) B cells in the germinal center reaction. Immunity. [DOI] [PubMed] [Google Scholar]
- Takeda K, Tanaka T, Shi W, Matsumoto M, Minami M, Kashiwamura S, Nakanishi K, Yoshida N, Kishimoto T, and Akira S (1996). Essential role of Stat6 in IL-4 signalling. Nature 380, 627–630. [DOI] [PubMed] [Google Scholar]
- Taylor JJ, Pape KA, and Jenkins MK (2012). A germinal center-independent pathway generates unswitched memory B cells early in the primary response. J. Exp. Med. 209, 597–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomayko MM, Steinel NC, Anderson SM, and Shlomchik MJ (2010). Cutting edge: Hierarchy of maturity of murine memory B cell subsets. J. Immunol. 185, 7146–7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turqueti-Neves A, Otte M, da Costa OP, Hopken UE, Lipp M, Buch T, and Voehringer D (2014). B-cell-intrinsic STAT6 signaling controls germinal center formation. Eur. J. Immunol. [DOI] [PubMed] [Google Scholar]
- Vajdy M, Kosco-Vilbois MH, Kopf M, Kohler G, and Lycke N (1995). Impaired mucosal immune responses in interleukin 4-targeted mice. J. Exp. Med. 181, 41–53. [DOI] [PubMed] [Google Scholar]
- Viant C, Weymar GHJ, Escolano A, Chen S, Hartweger H, Cipolla M, Gazumyan A, and Nussenzweig MC (2020). Antibody Affinity Shapes the Choice between Memory and Germinal Center B Cell Fates. Cell 183, 1298–1311 e1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora GD, Dominguez-Sola D, Holmes AB, Deroubaix S, Dalla-Favera R, and Nussenzweig MC (2012). Identification of human germinal center light and dark zone cells and their relationship to human B-cell lymphomas. Blood 120, 2240–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora GD, and Nussenzweig MC (2012). Germinal centers. Annu. Rev. Immunol. 30, 429–457. [DOI] [PubMed] [Google Scholar]
- Victora GD, Schwickert TA, Fooksman DR, Kamphorst AO, Meyer-Hermann M, Dustin ML, and Nussenzweig MC (2010). Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell 143, 592–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Rodda LB, Bannard O, and Cyster JG (2014). Integrin-mediated interactions between B cells and follicular dendritic cells influence germinal center B cell fitness. J. Immunol. 192, 4601–4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Shi J, Yan J, Xiao Z, Hou X, Lu P, Hou S, Mao T, Liu W, Ma Y, et al. (2017). Germinal-center development of memory B cells driven by IL-9 from follicular helper T cells. Nat. Immunol. 18, 921–930. [DOI] [PubMed] [Google Scholar]
- Wills-Karp M, and Finkelman FD (2008). Untangling the complex web of IL-4- and IL-13-mediated signaling pathways. Sci Signal 1, pe55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Tech L, George LA, Acs A, Durrett RE, Hess H, Walker LSK, Tarlinton DM, Fletcher AL, Hauser AE, and Toellner KM (2018). Plasma cell output from germinal centers is regulated by signals from Tfh and stromal cells. J. Exp. Med. 215, 1227–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Markers of different clusters of Day7 GC B cells, Related to Figure 1
Post GC B cell signature, Related to Figure S2
BCR heavy chain information of GC B cells, Related to Figure 3, 7, S2, S7, and STAR Methods
Data Availability Statement
Single-cell RNA-seq data have been deposited at GEO(GSE180920).
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.