Abstract
Background and Aims
The survival rate of patients with hepatocellular carcinoma is variable. The abnormal expression of RNA-binding proteins (RBPs) is closely related to the occurrence and development of malignant tumors. The primary aim of this study was to identify RBPs related to the prognosis of liver cancer and to construct a prognostic model of liver cancer.
Methods
We downloaded the hepatocellular carcinoma gene sequencing data from The Cancer Genome Atlas (cancergenome.nih.gov/) database, constructed a protein-protein interaction network, and used Cytoscape to realize the visualization. From among 325 abnormally expressed genes for RBPs, 9 (XPO5, enhancer of zeste 2 polycomb repressive complex 2 subunit [EZH2], CSTF2, BRCA1, RRP12, MRPL54, EIF2AK4, PPARGC1A, and SEPSECS) were selected for construction of the prognostic model. Then, we further verified the results through the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/) database and in vitro experiments.
Results
A prognostic model was constructed, which determined that the survival time of patients in the high-risk group was significantly shorter than that of the low-risk group (p<0.01). Univariate and multivariate Cox regression analysis suggested that the risk score was an independent prognostic factor (p<0.01). We also constructed a nomogram based on the risk score, survival time, and survival status. At the same time, we verified the high expression and cancer-promoting effects of EZH2 in tumors.
Conclusions
Survival, receiver operating characteristic curve and independent prognostic analyses demonstrated that we constructed a good prognostic model, which might be useful for estimating the survival of patients with hepatocellular carcinoma.
Keywords: Hepatocellular carcinoma, RNA binding protein, Prognostic model, Nomogram
Introduction
Worldwide, liver cancer is the fourth leading cause of cancer-related deaths and has the sixth highest incidence.1 It is estimated that 840,000 new cases of liver cancer are diagnosed and at least 780,000 people die of liver cancer every year, with China accounting for 47% of the total number of liver cancer cases as well as the related mortality.2,3 Hepatocellular carcinoma (HCC) is the predominant type of primary liver cancer, accounting for approximately 90% of all liver cancer cases.4 Although great progress has been made in the diagnosis and treatment of liver cancer, the 5-year survival rate of patients with advanced liver cancer is still less than 20%.5 In China, liver cancer ranks third in cancer-related mortality due to the large number of liver cancer patients, delayed diagnosis, and limited treatment options.6 Therefore, it is important to study the molecular mechanism of tumorigenesis and development, to find new targets of drug therapy, to identify new tumor markers, and to achieve an earlier diagnosis of liver cancer.
RNA-binding proteins (RBPs) are a group of proteins associated with RNA regulation and metabolism. Their main role is to mediate the maturation, transport, localization and translation of RNA, and their abnormal expression can cause a variety of diseases. At present, there are approximately 1,542 known human RBPs, accounting for approximately 7.5% of all protein-coding genes. It is now clear that RBPs are dysregulated in different types of cancer, affecting the expression and function of oncoproteins and tumor suppressors. For example, IGF-II mRNA-binding proteins (IMPs) are involved in the progression of tumors and the establishment and maintenance of tumor cell hierarchies.7 Therefore, studying the complex interaction network between RBPs and their cancer-related RNA targets will help in understanding the molecular mechanisms of RBPs in cancer progression, and may also enable the discovery of new cancer treatment targets. Although RBPs are known to be involved in the occurrence and development of a variety of tumors, we still know very little about the molecular mechanism of RBPs in tumor progression.
Because there are few studies on the role of RBPs in the occurrence and development of liver cancer, we designed this study to screen-out RBPs related to the prognosis of patients. We aimed to construct a predictive model that would be able to provide some help to clinical work and direction for future research, including of molecular mechanisms and the identification of molecular targets for treatment.
Methods
Clinical data and RNA sequencing data of the patients
We downloaded the RNA high-throughput sequencing data from The Cancer Genome Atlas (cancergenome.nih.gov/; TCGA) database, including 374 tumor tissue samples and 50 normal liver tissue samples (www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga). We also downloaded the clinical data of 377 HCC patients from the TCGA. After excluding six patients with incomplete data, a total of three hundred and seventy-one patients were finally included in the study, with each having data on follow-up time, survival status, disease stage, etc. The RNA high-throughput sequencing data included the expression data of 60,483 genes, and we extracted the expression levels of 1,473 RBPs from such. As this research did not involve human participants, no research ethics review was necessary.
Identification of the differentially-expressed RBPs (DERs) in HCC patients
We used R software to analyze expression of the extracted 1,473 RBPs. A total of 325 RBPs showed expression differences between the tumor tissues and normal tissues, including 203 up-regulated and 122 down-regulated DERs. The threshold for the DERs was set as |log fold-change (FC)|>1 and false discovery rate (FDR) <0.05.
Enrichment analysis and protein-protein interaction (PPI) network of the DERs
We divided the DERs into an up-regulated group and a down-regulated group, and then performed gene ontology (GO) function analysis on the two groups of RBPs for three GO domains: molecular functions (MF), biological processes (BP) and cellular components (CC). Next, we performed Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis on the two groups of patients. We used the clusterProfiler package in R software for the GO function enrichment analysis and the KEGG pathway enrichment analysis. Then, we uploaded these DERs to the online tool STRING (https://string-db.org) to construct the PPI network. We deleted the disconnected nodes, and the remaining RBPs were used for the next analysis. To further study the role of the DERs in HCC, we used Cytoscape software to create a PPI network that incorporated the nodes from the STRING database. At the same time, we also used the MCODE tool in the Cytoscape software to create a PPI subnet. Then, enrichment analysis and coexpression analysis were performed on the sub-network.
Screening of prognostic RBPs, construction and testing of the prognostic models
We used univariate Cox regression analysis to screen out 29 RBPs related to the patients’ outcomes, from among all of the DERs. The threshold of the Cox regression analysis was set to 0.001. All HCC patients were divided into a training group and a test group. Then, in the training group, a multivariate Cox regression analysis was performed to screen out nine RBPs. A prognostic model was constructed based on the relationship between the expression levels of these nine RBPs and the patients’ outcomes in the training group. The risk score of the HCC patients in the test group was calculated according to the prognostic model, and the patients in the test group were divided into high-risk and low-risk groups according to the risk scores. Then, we used survival curves, receiver operating characteristic (ROC) curves, risk curves and independent prognostic analysis to test the predictive power of the prognostic model. In addition, we still used the data GSE76427 in the Gene Expression Omnibus (GEO) database to verify our model externally.
Construction of the nomogram
The coefficients obtained by the Cox regression model were used to construct the nomogram of overall survival. To construct the nomogram, we first determined a scale axis of 0–100 to represent the score (in order to make the expression of the RBPs correspond to the scores on the scale axis) and we calculated the score of each RBP. We added the scores of each RBP to obtain the total score. Based on the correspondence between the total score axis and the survival rate, the 1-, 2-, 3- and 5-year survival rates of the patients could be predicted.
Cell culture, RNA isolation and quantitative real-time PCR (qRT-PCR) analysis
Because the risk ratio of enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2) is greater and the coefficient in the risk score calculation formula is greater, we choose EZH2 for further verification. A total of 20 HCC patient samples and paired non-tumor liver tissue samples were collected from the patients hospitalized at the Zhongnan Hospital, Wuhan University (Hubei Province, China). All patients provided written informed consent to the use of tissues for scientific research in the Department of Hepatobiliary and Pancreatic Surgery. All cell lines were purchased from the Chinese Type Culture Collection. All the cell lines were maintained in Dulbecco’s modified Eagle’s medium/high glucose (GE, USA) containing 10% fetal bovine serum (Gibco, USA). RNA was extracted using TRIzol reagent (TaKaRa, Japan) according to the manufacturer instructions. cDNAs were generated by the reverse transcription synthesis kit (TaKaRa) and the SYBR Green PCR Kit (TaKaRa) was used for qRT-PCR analysis. The primers used for EZH2 were 5′-GAGTTGGTGAATGCCCTTGGT-3′ and 5′-CATCTCGGTGATCCTCCAGATC-3′.
Gene knock-down
For generation of EZH2 knock-down cells, small interfering (si)RNA transfections were performed using the siRNA transfection reagent. The following siRNA sequences were used: EZH2-siRNA, CGGCUUCCCAAUAACAGUATT, UACUGUUAUUGGGAAGCCGTT.
Cell proliferation, migration, invasion, apoptosis, and cell cycle analyses
A CCK8 Kit (Dojindo, China) was used to measure cell viability. Transwell assay was used to assess cell migration. Cell migration was assessed by in vitro scratch wound assay. A BioCoat Matrigel Invasion Chamber (BD Biosciences, USA) was used to assess cell invasion; the number of cells migrating and invading was counted in three random areas. Apoptosis was assayed using the Annexin V-FITC Apoptosis Detection Kit (Invitrogen, USA), according to the manufacturer’s instructions, and the percentage of apoptotic cells was verified by flow cytometry (Beckman-Coulter, USA). To detect the cell cycle, 48 hours after transfection, the cells were stained with PI (propidium iodide) and assessed.
Statistical analysis
The datasets generated and analyzed during the current study are available in TCGA (https://cancergenome.nih.gov/) and GEO (www.ncbi.nlm.nih.gov/geo). Difference analysis and regression analysis of the data were processed in the R3.6.3 software (https://www.r-project.org). Statistical analysis of clinical data was performed using SPSS v.23.0 (IBM Corp., USA). Variables were compared by t-test or chi-square test.
Results
DERs in HCC patients
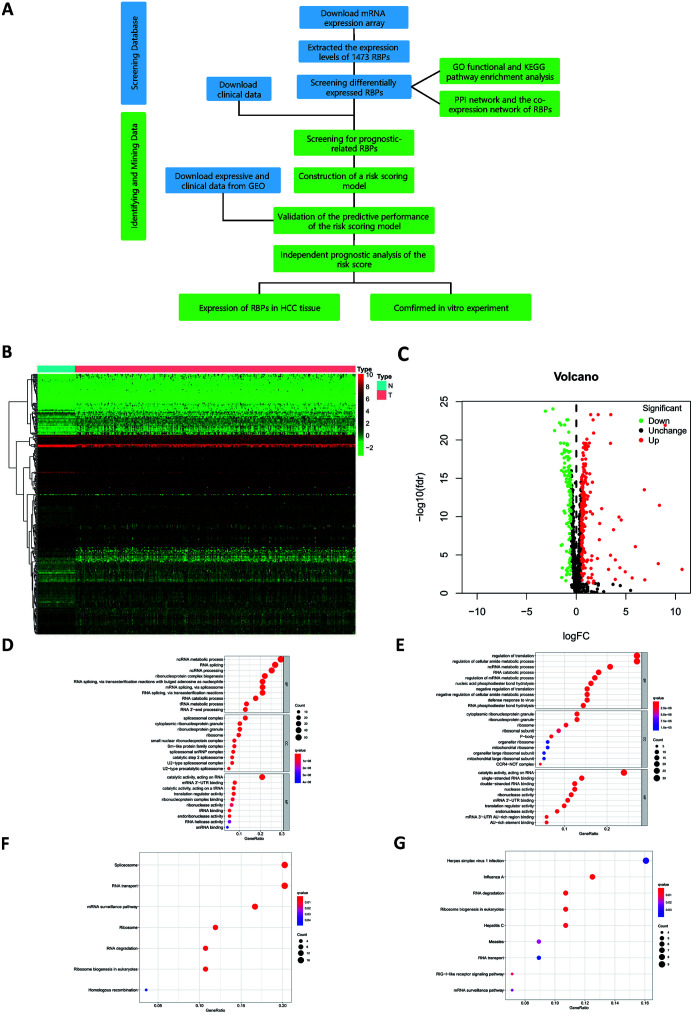
A flowchart showing the steps of data processing and integrative data analysis is provided in Figure 1A. The original data downloaded from TCGA included the RNA sequencing data of 50 normal tissues and 374 liver cancer tissue samples. The sequencing data itself comprised 60,483 sets of RNA expression data, including that of 1,473 RBPs. After data processing, it was found that 325 RBPs had expression differences (Fig. 1B), including 203 up-regulated and 122 down-regulated RBPs (Fig. 1C). The results of the DERs are presented in the form of heat maps and volcano maps.
Fig. 1. Flow chart of heat map, volcano plot, GO enrichment analysis and KEGG pathway enrichment of DERs.
(A) Flow chart. (B) Heat map of DERs. (C) Volcano plot of DERs. (D) GO enrichment analysis of up-regulated RBPs. (E) GO enrichment analysis of down-regulated RBPs. (F) KEGG pathway enrichment of up-regulated RBPs. (G) KEGG pathway enrichment of down-regulated RBPs.
GO functional and KEGG pathway enrichment analysis
The GO function enrichment analysis was performed on the obtained up-regulated RBPs and down-regulated RBPs. The GO terms included BP, MF, and CC. For BP, the up-regulated RBPs were mainly enriched in non-coding RNA metabolic process, RNA splicing, and non-coding RNA processing. For CC, the up-regulated RBPs were mainly enriched in spliceosomal complex, cytoplasmic ribonucleoprotein granules, and ribonucleoprotein granules. For MF, the RBPs were mainly enriched in catalytic activity and acting on RNA (Fig. 1D). Down-regulated RBPs were mainly enriched in regulation of translation, regulation of cellular amide metabolic processes, cytoplasmic ribonucleoprotein granules, ribonucleoprotein granules, and catalytic activity (Fig. 1E). KEGG pathway enrichment analysis of up-regulated RBPs showed that they were mainly enriched in spliceosome, RNA transport, and mRNA surveillance pathways (Fig. 1F). Down-regulated RBPs were enriched in herpes simplex virus 1 infection, influenza A, and RNA degradation (Fig. 1G). The results of the enrichment analysis are also displayed in the form of tables (Table 1 and Table 2). The results in the chart are sorted by p-values, and the top 15 results of the GO enrichment analysis are shown.
Table 1. GO functional enrichment analyses.
| ID | Description | GO term | p | q |
|---|---|---|---|---|
| Up-regulated RBPs | ||||
| GO:0034660 | ncRNA metabolic process | BP | 3.17E-43 | 4.16E-40 |
| GO:0034470 | ncRNA processing | BP | 7.52E-38 | 4.93E-35 |
| GO:0008380 | RNA splicing | BP | 2.34E-37 | 1.02E-34 |
| GO:0140098 | catalytic activity, acting on RNA | MF | 3.85E-29 | 6.17E-27 |
| GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile | BP | 2.08E-28 | 5.45E-26 |
| GO:0000398 | mRNA splicing, via spliceosome | BP | 2.08E-28 | 5.45E-26 |
| GO:0000375 | RNA splicing, via transesterification reactions | BP | 2.81E-28 | 6.14E-26 |
| GO:0022613 | ribonucleoprotein complex biogenesis | BP | 4.21E-27 | 7.87E-25 |
| GO:0031123 | RNA 3′-end processing | BP | 2.65E-22 | 4.34E-20 |
| GO:0005681 | spliceosomal complex | CC | 2.01E-21 | 2.70E-19 |
| GO:0006399 | tRNA metabolic process | BP | 2.53E-21 | 3.68E-19 |
| GO:0006401 | RNA catabolic process | BP | 3.97E-21 | 5.21E-19 |
| GO:0008033 | tRNA processing | BP | 4.46E-21 | 5.31E-19 |
| GO:0006402 | mRNA catabolic process | BP | 3.91E-18 | 4.27E-16 |
| GO:1903311 | regulation of mRNA metabolic process | BP | 1.97E-17 | 1.99E-15 |
| Down-regulated RBPs | ||||
| GO:0006417 | regulation of translation | BP | 3.73E-24 | 5.23E-21 |
| GO:0140098 | catalytic activity, acting on RNA | MF | 2.76E-22 | 4.31E-20 |
| GO:0034248 | regulation of cellular amide metabolic process | BP | 1.08E-22 | 7.55E-20 |
| GO:0003727 | single-stranded RNA binding | MF | 8.03E-20 | 6.26E-18 |
| GO:0003725 | double-stranded RNA binding | MF | 2.21E-18 | 1.15E-16 |
| GO:0090501 | RNA phosphodiester bond hydrolysis | BP | 8.66E-16 | 4.04E-13 |
| GO:0004540 | ribonuclease activity | MF | 3.98E-14 | 1.41E-12 |
| GO:0003730 | mRNA 3′-UTR binding | MF | 4.52E-14 | 1.41E-12 |
| GO:0034660 | ncRNA metabolic process | BP | 5.83E-15 | 1.79E-12 |
| GO:0017148 | negative regulation of translation | BP | 6.39E-15 | 1.79E-12 |
| GO:0034249 | negative regulation of cellular amide metabolic process | BP | 2.34E-14 | 5.47E-12 |
| GO:0051607 | defense response to virus | BP | 5.50E-14 | 1.01E-11 |
| GO:1903311 | regulation of mRNA metabolic process | BP | 5.75E-14 | 1.01E-11 |
| GO:0006401 | RNA catabolic process | BP | 2.03E-13 | 2.98E-11 |
| GO:0090305 | nucleic acid phosphodiester bond hydrolysis | BP | 2.13E-13 | 2.98E-11 |
nc, non-coding.
Table 2. KEGG enrichment analysis.
| ID | Description | p | q |
|---|---|---|---|
| Up-regulated RBPs | |||
| hsa03040 | spliceosome | 1.24E-13 | 4.85E-12 |
| hsa03015 | mRNA surveillance pathway | 3.33E-13 | 6.49E-12 |
| hsa03013 | RNA transport | 2.86E-12 | 3.71E-11 |
| hsa03018 | RNA degradation | 1.08E-07 | 1.05E-06 |
| hsa03008 | ribosome biogenesis in eukaryotes | 2.18E-06 | 1.70E-05 |
| hsa03010 | ribosome | 5.08E-06 | 3.30E-05 |
| hsa03440 | homologous recombination | 0.008791 | 0.048915 |
| Down-regulated RBPs | |||
| hsa03018 | RNA degradation | 1.62E-05 | 0.000718 |
| hsa03008 | ribosome biogenesis in eukaryotes | 0.000117 | 0.002460 |
| hsa05164 | influenza A | 0.000167 | 0.002460 |
| hsa05160 | hepatitis C | 0.000703 | 0.007773 |
| hsa04622 | RIG-I-like receptor signaling pathway | 0.001372 | 0.012129 |
| hsa05162 | measles | 0.002696 | 0.019863 |
| hsa03015 | mRNA surveillance pathway | 0.003590 | 0.022672 |
| hsa05168 | herpes simplex virus 1 infection | 0.006320 | 0.034924 |
| hsa03013 | RNA transport | 0.008050 | 0.039545 |
PPI network and the coexpression network of RBPs
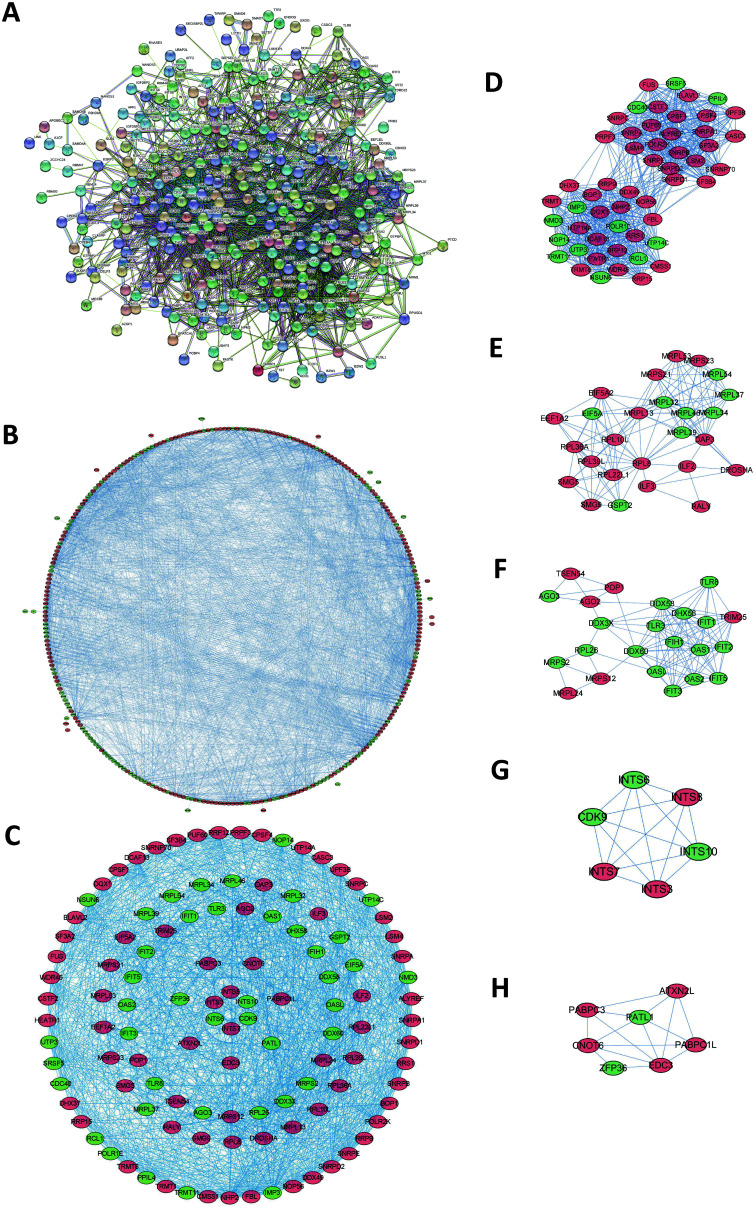
To study the interactions among the 325 RBPs, we used STRING to construct a PPI network. The connections between the molecules represent the possible interactions between two protein molecules, and the different colors of the lines represent different levels of evidence. After deleting 21 disconnected RBPs, there were a total of 304 nodes and 2,794 connections in the PPI diagram (Fig. 2A). Then, we used Cytoscape software to create a coexpression network of interactions among all nodes in the PPI. Among them, the block of proliferation 1 (i.e. BOP1) had the most interactive RBPs, and it had confirmed or potential interactions with 50 RBPs (Fig. 2B). We applied the MCODE tool in Cytoscape to construct a sub-network of the coexpression network (Fig. 2D–H), and then we selected the first five sub-networks according to their association score to identify the first important module, including 115 nodes and 1,295 edges (Fig. 2C).
Fig. 2. Interaction network of DERs.
(A) PPI network. (B) Network visualized using Cytoscape. (C) Network of important modules. (D-H) Important subnetworks. The light blue and purple lines, respectively, indicate known interactions from curated databases and experimentally determinations. Green, red and dark blue represent predicted interactions, including gene neighborhood, gene fusions and gene co-occurrences in Fig. 2A. Red circles: up-regulated RBPs; Green circles: down-regulated RBPs in Fig. 2B-H.
Screening of prognosis-related RBPs and construction of a risk scoring model
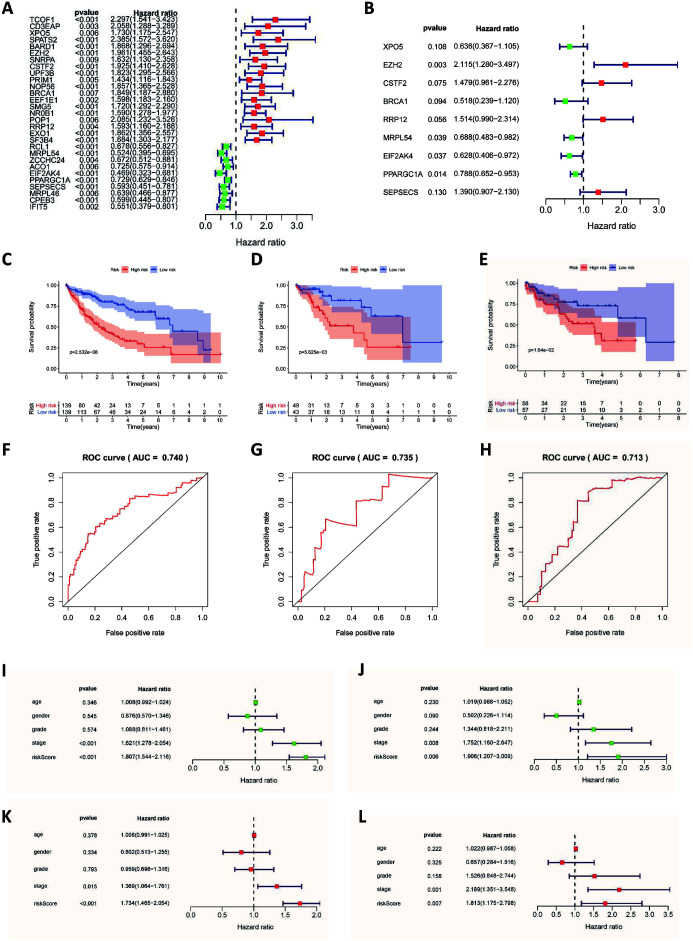
A total of 305 RBPs were included in the PPI network. To screen-out the RBPs related to prognosis, we used univariate Cox regression analysis to screen-out a total of 29 RBPs, and we calculated their hazard ratios (Fig. 3A). Among them, 19 RBPs had a hazard ratio greater than 1, which means they had a negative impact on patient prognosis, and 10 RBPs had a positive impact on the prognosis. We then divided the patients into a training group and a test group at a ratio of 7:3, and we performed multivariate Cox regression analysis on the expression of the selected RBPs of patients in the training group (Fig. 3B). Then, we built a prediction model based on the relationship between the expression of RBPs in the training group of patients and the patient’s survival and survival status. The formula for calculating the patient risk score was as follows:
| Risk score= (−0.4520×ExpXPO5) + (0.7493×ExpEZH2) + (0.3913×ExpCSTF2) + (−0.6586×ExpBRCA1) + (0.4145×ExpRRP12) + (−0.3734×ExpMRPL54) + (−0.4652× ExpEIF2AK4) + (−0.2380×ExpPPARGC1A) + (0.3293×ExpSEPSECS) |
Fig. 3. Results of the Cox regression analysis, survival and ROC curves.
(A) Univariate Cox regression analysis of DERs. (B) Multivariate Cox regression of DERs. (C) Survival curve of the training group. (D) Survival curve of the test group. (E) Survival curve of the GEO group. (F) ROC curve of the training group. (G) ROC curve of the test group. (H) ROC curve of the GEO group. (I) Univariate Cox regression analysis in the training group. (J) Univariate Cox regression analysis in the test group. (K) Multivariate Cox regression analysis in the training group. (L) Multivariate Cox regression analysis in the test group. Red and blue areas: 95% confidence interval.
To test the effectiveness of the predictive model, we used the survival curve method to evaluate the prognostic model. In the training group, patients with high-risk scores had a worse prognosis than patients with low-risk scores (p<0.01; Fig. 3C). In the test group, patients were divided into a high-scoring group and a low-scoring group according to the risk score model, and the survival rates of the two groups were also significantly different (p<0.01; Fig. 3D). In addition, we adopted the ROC test method and calculated the area under the ROC curve (AUC). The AUC of the training group was 0.735 and the AUC of the test group was 0.740, indicating that the risk scoring model we constructed has good diagnostic performance (Fig. 3F, G). Patients in the GEO cohort were also divided into high-risk groups and low-risk groups based on this model. There were also significant differences in survival rates between the two groups (p<0.05), and the AUC of the GEO cohort was 0.740 (Fig. 3E, H). This shows that the risk scoring formula we established can accurately divide patients into a high-risk group with a poor prognosis and a low-risk group with a good prognosis.
Independent prognostic analysis of the risk score
To evaluate whether the risk score is an independent prognosis-related factor, we conducted an independent prognostic analysis of risk scores for patients in the training group and test group. The prognostic analysis used univariate and multivariate Cox regressions. The univariate prognostic analysis of the training group showed that the clinical stage and risk score of the disease were independent factors affecting the prognosis (p<0.001; Fig. 3I). The multivariate prognostic analysis showed that the risk score (p<0.001) and clinical stage (p<0.05) were independent factors affecting the prognosis, and the risk score had a higher hazard ratio (Fig. 3J). In the test group of patients, both univariate and multivariate prognostic analysis showed that the risk score was an independent factor affecting the prognosis (p<0.01). This showed that the risk scoring model we built has good predictive ability.
Validation of the predictive performance of the risk scoring model
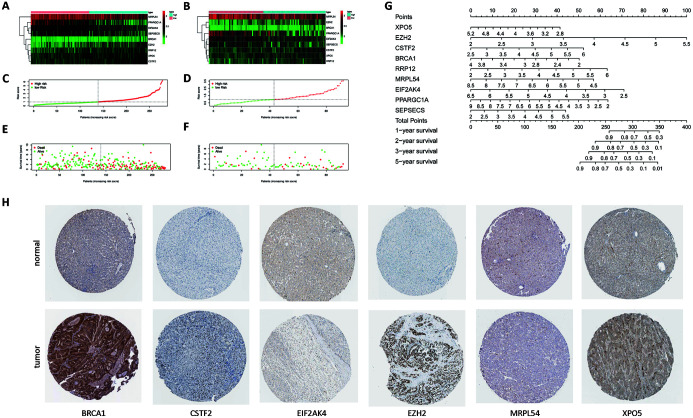
The expression levels of the nine RBPs in the training group and the test group were significantly different between the high-risk group and the low-risk group (Fig. 4A, B). Fig. 4C and D show the distribution of the patients’ risk scores. Fig. 4E and F show the relationship between the patient’s survival status, survival time, and risk score. The red dots represent high-risk patients, and the green dots represent low-risk patients. It can be seen that the higher the risk score, the higher the proportion of patients whose follow-up outcome is death and the shorter the follow-up time.
Fig. 4. Heat map and distribution of risk scores and survival status: nomogram and immunohistochemistry results.
(A) Heat map of RBPs in the training group. (B) Heat map of RBPs in the test group. (C) Distribution of risk scores in the training group. (D) Distribution of risk scores in the test group. (E) Distribution of survival status of patients in the training group. (F) Distribution of survival status of patients in the test group. (G) Nomogram for predicting 1-, 2-, 3- and 5-year overall survival of patients with HCC. (H) Immunohistochemistry results.
Clinical features of the high-risk group and low-risk group
We obtained the clinical characteristics of the two groups of patients, and performed a statistical analysis of the surgical methods, alpha-fetoprotein values, degree of liver cirrhosis, and other adjuvant treatments. The above-mentioned and other features of the two groups of patients found no significant difference, indicating risk score is an independent predictor of prognosis (Table 3).
Table 3. Clinical features of the high-risk group and low-risk group.
| Feature | Variables | High-risk group | Low-risk group | t/χ2 | p |
|---|---|---|---|---|---|
| Sex | |||||
| Male | 135 | 124 | 0.040 | 0.442 | |
| Female | 53 | 58 | |||
| Stage | |||||
| i | 76 | 95 | -0.052 | 0.321 | |
| ii | 50 | 35 | |||
| iii | 50 | 35 | |||
| iv | 1 | 4 | |||
| Unknown | 11 | 13 | |||
| AFP | 11,005.88±40,623.11 | 16,927.47±172,443.99 | -0.391 | 0.696 | |
| Fibrosis | |||||
| None | 22 | 32 | -0.074 | 0.155 | |
| Portal fibrosis | 18 | 13 | |||
| Fibrous septa | 12 | 16 | |||
| Nodular formation | 5 | 4 | |||
| Established cirrhosis | 33 | 36 | |||
| Unknown | 98 | 81 | |||
| Hepatitis | |||||
| HBV | 94 | 89 | -0.031 | 0.550 | |
| HCV | 14 | 18 | |||
| HBV+HCV | 34 | 44 | |||
| No hepatitis | 46 | 31 | |||
| Radiation | |||||
| Without | 115 | 124 | -0.074 | 0.157 | |
| With | 2 | 2 | |||
| Unknown | 71 | 56 | |||
| Surgical method | |||||
| Lobectomy | 77 | 64 | 0.042 | 0.421 | |
| Single segmentectomy | 42 | 45 | |||
| Multiple segmentectomy | 43 | 44 | |||
| Extended Lobectomy | 10 | 15 | |||
| Other | 16 | 14 | |||
| Ablation | |||||
| Without | 114 | 118 | 3.696 | 0.158 | |
| With | 4 | 9 | |||
| Unknown | 70 | 55 | |||
| Vascular invasion | |||||
| Without | 109 | 110 | 2.079 | 0.354 | |
| With | 50 | 53 | |||
| Unknown | 29 | 19 |
AFP, alpha-fetoprotein; HBV, hepatitis B virus; HCV, hepatitis C virus.
Nomogram construction
To better establish the relationship among RBPs’ expression, risk score and patient survival, we developed a nomogram. According to the nomogram, the expression levels of 9 RBPs can be converted into corresponding scores, and then the scores can be added to obtain the total risk score of the patient. The risk score corresponds to the estimated survival rate, including 1-, 2-, 3- and 5-year survival rates. According to the nomogram, the prognostic model can be applied in the clinic, and the long-term survival rate of a single patient can be predicted based on the expression of RBPs (Fig. 4G).
Expression of RBPs in HCC tissue
To further determine the expression of RBPs in liver cancer tissues for constructing this prognostic model, we used the immunohistochemical staining results in the database to show that BRCA1, CSTF2, EZH2 and XPO5 are highly expressed in liver cancer tissues. EIF2AK4 and MRPL54 are expressed at low levels in liver cancer tissues (Fig. 4H).
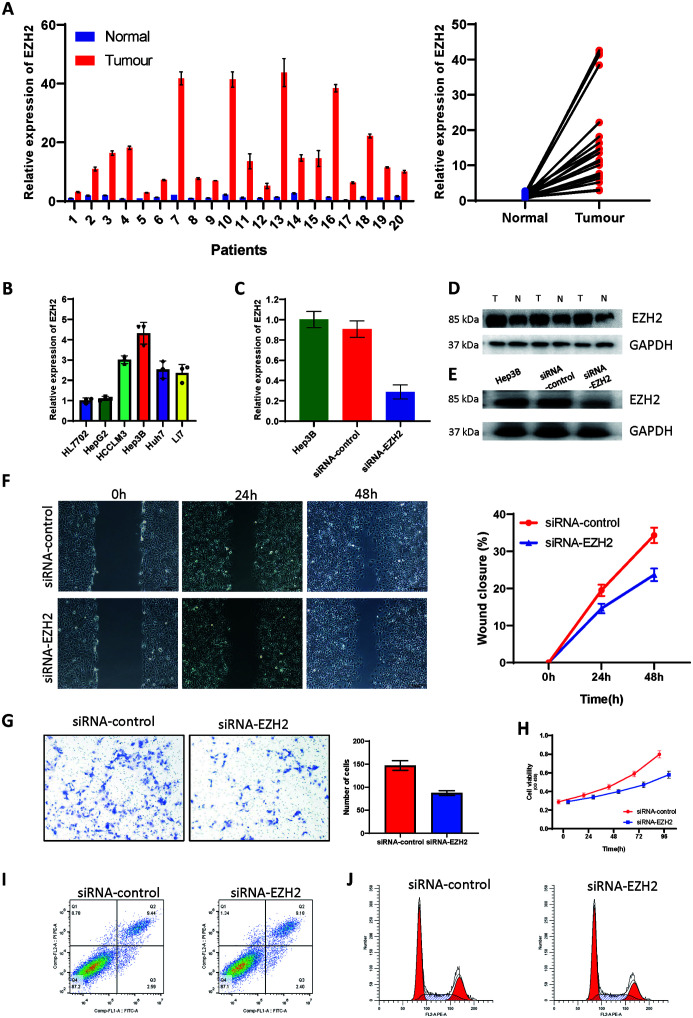
Verification of expression of EZH2 in tissues
The primers for EZH2 mRNA were designed, and the expression of 20 pairs of HCC and adjacent tissues was further verified by qRT-PCR. EZH2 showed a significant increase in liver cancer (Fig. 5A). In addition to the mRNA level, at the protein level, we also verified the high expression of EZH2 in tumors by western blotting (Fig. 5D). In addition, we also tested the data of EZH2 expression in normal liver cell lines and various HCC cell lines (Fig. 5B). Among them, the Hep3B cell line showed the highest expression of EZH2. We chose the Hep3B cell line for subsequent in vitro experiments.
Fig. 5. Expression of EZH2 and its cancer-promoting effect.
(A and D) EZH2 in tumor and paracarcinoma tissue for HCC patients. (B) EZH2 in different cell lines. (C and E) Regulatory effect of si-EZH2 transfection on the level of EZH2 in HCC cell lines. (F-H) Knock-down of EZH2 significantly inhibited invasion (F and G) and migration (H) of HCC cells. (H) Effect of EZH2 silencing on viability of HCC cell lines. (I) Knock-down of EZH2 had no effect on apoptosis of HCC cells. (J) Effects of EZH2 on the cell cycle.
Effect of EZH2 on the malignant behaviors of liver cancer cells
After transfection with siRNA, the mRNA and protein levels of EZH2 decreased significantly (Fig. 5C, E). We used the scratch test and the Transwell assay to determine whether reducing EZH2 affects the invasion and migration of HCC cells. Compared with the control group (siRNA-control), the migration ability of the HCC cells at the edge of the scratch in the siRNA-EZH2 group was significantly reduced (p<0.05; Fig. 5F). In addition, the number of HCC cells in the si-EZH2 group that passed through the Transwell chamber was decreased significantly (p<0.05; Fig. 5G). CCK8 experiment demonstrated that knock-down of EZH2 reduced the proliferation ability of cells (Fig. 5H). However, flow cytometry did not find a significant effect of EZH2 on cell apoptosis and cell cycle (Fig. 5I, J).
Discussion
RBPs are involved in almost all steps of RNA post-transcription regulation, regulating RNA splicing, polyadenylation, stability, localization, translation, and degradation.8,9 Studies have shown that the abnormal expression of certain RBPs is related to the HCC transcriptome and tumorigenicity and is related to the poor prognosis of liver cancer patients.10,11 However, due to the large number of RBPs, their diverse functions and complex mechanisms, there are still many RBPs whose mechanism of action has not been studied in depth.
To promote in-depth research of RBPs in HCC, we designed this study to screen-out the key RBPs that play a role in HCC. At the same time, we also developed a prognostic model of HCC patients based on the expression of RBPs.
To further study the interactions among the RBPs, we created a PPI network based on previous research, co-expression relationships, bioinformatics predictions, and gene-adjacent relationships. To study the relationships among the RBPs more intuitively, Cytoscape software was used to realize the visualization of the PPI network. In the graph created, we can see that some RBPs have a correlation with many RBPs, so we think these RBPs should have more biological functions and greater research value. In the network, BOP1 interacts with 50 RBPs. According to the results of enrichment analysis, it can be inferred that the research directions of RBPs mainly include ribonucleoprotein complex biogenesis, RNA splicing, non-coding RNA metabolic process, mRNA surveillance pathway, RNA transport, and others.
There are many studies on the mechanism of EZH2 in liver cancer. EZH2 is related to the prognosis of patients and can promote HCC progression by regulating the miR-22/galectin-9 axis or the expression of PD-L1 in hepatocellular carcinoma.12 PPARGC1A can interact with MiR-93-5p to promote the proliferation of liver cancer cells, and it can also interact with MiR-30b-5p to regulate the lipid metabolism of liver cancer cells.13,14 In addition, other mechanisms of PPARGC1A are also worthy of further study. There are few studies on EIF2AK4 and MRPL54 in HCC, but because these two RBPs are related to the prognosis of liver cancer in the results of the univariate and multivariate Cox regression analyses, they have great research value. EIF2AK4 and MRPL54 can be studied in terms of binding to RNA to affect the metabolism of RNA or to affect the variable shearing of RNA.
The nomogram makes the prognostic model used to predict the survival rate of patients at 1, 2, 3, and 5 years more intuitive and more convenient for clinical application. The cost of obtaining the expression level of nine RBPs is relatively low, and the survival rate calculated based on their expression level can help with clinical decision-making and selecting treatment options. For example, studies have shown that transarterial chemoembolization therapy for patients with poorly differentiated liver cancer and venous tumor thrombi after liver cancer resection can help prolong the survival of patients. However, for patients with early liver cancer and moderately differentiated liver cancer, whether to give interventional therapy is still controversial. According to our research, the treatment plan can be determined based on the risk score. If the risk score is high, it indicates a poor prognosis. It is thus recommended to give postoperative interventional chemotherapy, targeted therapy, and other treatment options.
In addition to the above-mentioned advantages, there are some shortcomings of this study. First, due to the need to construct the formula, not all RBPs included in the formula are prognosis-related RBPs in the multivariate Cox regression analysis, and some of the prognosis-related RBPs were not included in the prediction model. Second, in this study, the interaction and coexpression relationships among the RBPs were analyzed by an interaction network, but there is a lack of further research on the functions of these RBPs. In future research, the interactions among RBPs and mRNA or non-coding RNA need to be further studied, which can guide the functional research of these RBPs more effectively. Third, all data used in this research originated from public databases. In future studies, it will be more credible to collect single-center or multi-center clinical samples to test the predictive performance of the predictive model.
In short, we screened-out RBPs that are abnormally expressed in liver cancer and performed enrichment analysis and constructed a coexpression network. Some RBPs that play a role in the progression of liver cancer were identified, and some RBPs that need further research were highlighted. A prognostic model of liver cancer constructed based on the abnormal expression of RBPs has not been reported before. Our analysis results can provide certain guidance for studying the roles of RBPs in liver cancer. However, its actual predictive performance still needs to be verified with large clinical samples in the future. The constructed prediction model can be applied to clinical prognostication, and can also provide guidance for clinical work, drug treatment target selection and molecular marker research of liver cancer.
Acknowledgments
The authors would like to thank the medical research center, Zhongnan Hospital of Wuhan University, for providing equipment.
Abbreviations
- AUC
area under the curve
- BP
biological processes
- CC
cellular components
- DERs
differentially-expressed RBPs
- EZH2
enhancer of zeste 2 polycomb repressive complex 2 subunit
- FC
fold-change
- FDR
false discovery rate
- GEO
Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/)
- GO
gene ontology
- HCC
hepatocellular carcinoma
- ICC
intrahepatic cholangiocarcinoma
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MF
molecular functions
- PI
propidium Iodide
- PPI
protein-protein interactions network
- qRT-PCR
quantitative real-time PCR
- RBPs
RNA-binding proteins
- ROC
receiver operating characteristic
- si
small interfering
- TCGA
The Cancer Genome Atlas (cancergenome.nih.gov/)
Data sharing statement
All data are available upon request.
References
- 1.Villanueva A. Hepatocellular carcinoma. N Engl J Med. 2019;380(15):1450–1462. doi: 10.1056/NEJMra1713263. [DOI] [PubMed] [Google Scholar]
- 2.Zheng R, Qu C, Zhang S, Zeng H, Sun K, Gu X, et al. Liver cancer incidence and mortality in China: Temporal trends and projections to 2030. Chin J Cancer Res. 2018;30(6):571–579. doi: 10.21147/j.issn.1000-9604.2018.06.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petrick JL, Florio AA, Znaor A, Ruggieri D, Laversanne M, Alvarez CS, et al. International trends in hepatocellular carcinoma incidence, 1978-2012. Int J Cancer. 2020;147(2):317–330. doi: 10.1002/ijc.32723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Llovet JM, Zucman-Rossi J, Pikarsky E, Sangro B, Schwartz M, Sherman M, et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016;2:16018. doi: 10.1038/nrdp.2016.18. [DOI] [PubMed] [Google Scholar]
- 5.Kudo M. Systemic therapy for hepatocellular carcinoma: 2017 update. Oncology. 2017;93(Suppl 1):135–146. doi: 10.1159/000481244. [DOI] [PubMed] [Google Scholar]
- 6.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 7.Degrauwe N, Suvà ML, Janiszewska M, Riggi N, Stamenkovic I. IMPs: an RNA-binding protein family that provides a link between stem cell maintenance in normal development and cancer. Genes Dev. 2016;30(22):2459–2474. doi: 10.1101/gad.287540.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitchell SF, Parker R. Principles and properties of eukaryotic mRNPs. Mol Cell. 2014;54(4):547–558. doi: 10.1016/j.molcel.2014.04.033. [DOI] [PubMed] [Google Scholar]
- 9.Wang ZL, Li B, Luo YX, Lin Q, Liu SR, Zhang XQ, et al. Comprehensive genomic characterization of RNA-binding proteins across human cancers. Cell Rep. 2018;22(1):286–298. doi: 10.1016/j.celrep.2017.12.035. [DOI] [PubMed] [Google Scholar]
- 10.Dang H, Takai A, Forgues M, Pomyen Y, Mou H, Xue W, et al. Oncogenic activation of the RNA binding protein NELFE and MYC signaling in hepatocellular carcinoma. Cancer Cell. 2017;32(1):101–114.e8. doi: 10.1016/j.ccell.2017.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong W, Dai ZH, Liu FC, Guo XG, Ge CM, Ding J, et al. The RNA-binding protein RBM3 promotes cell proliferation in hepatocellular carcinoma by regulating circular RNA SCD-circRNA 2 production. EBioMedicine. 2019;45:155–167. doi: 10.1016/j.ebiom.2019.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao G, Jin LL, Liu CQ, Wang YC, Meng YM, Zhou ZG, et al. EZH2 negatively regulates PD-L1 expression in hepatocellular carcinoma. J Immunother Cancer. 2019;7(1):300. doi: 10.1186/s40425-019-0784-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, Zhao Q, Hu B. Community-based prevention and control of COVID-19: Experience from China. Am J Infect Control. 2020;48(6):716–717. doi: 10.1016/j.ajic.2020.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Li J, Kuang D, Wang X, Zhu Y, Xu S, et al. miR-148b-3p functions as a tumor suppressor in GISTs by directly targeting KIT. Cell Commun Signal. 2018;16(1):16. doi: 10.1186/s12964-018-0228-z. [DOI] [PMC free article] [PubMed] [Google Scholar]