Abstract
PICALM and CLU genes have been linked to alterations in brain biochemical processes that may have an impact on Alzheimer’s disease (AD) development and neurophysiological dynamics. The aim of this study is to analyze the relationship between the electroencephalographic (EEG) activity and the PICALM and CLU alleles described as conferring risk or protective effects on AD patients and healthy controls. For this purpose, EEG activity was acquired from: 18 AD patients and 12 controls carrying risk alleles of both PICALM and CLU genes, and 35 AD patients and 12 controls carrying both protective alleles. Relative power (RP) in the conventional EEG frequency bands (delta, theta, alpha, beta, and gamma) was computed to quantify the brain activity at source level. In addition, spatial entropy (SE) was calculated in each band to characterize the regional distribution of the RP values throughout the brain. Statistically significant differences in global RP and SE at beta band (p-values < 0.05, Mann–Whitney U-test) were found between genotypes in the AD group. Furthermore, RP showed statistically significant differences in 58 cortical regions out of the 68 analyzed in AD. No statistically significant differences were found in the control group at any frequency band. Our results suggest that PICALM and CLU AD-inducing genotypes are involved in physiological processes related to disruption in beta power, which may be associated with physiological disturbances such as alterations in beta-amyloid and neurotransmitter metabolism.
Subject terms: Genetics, Neuroscience, Neurology, Risk factors, Mathematics and computing
Introduction
Alzheimer’s disease (AD) is the most prevalent cause of dementia worldwide, with an estimate of over 50 million affected people in 20191. This number is expected to nearly double every 20 years, leading to 152 million by 20501. AD symptomatology is defined by memory loss and cognitive and behavioral dysfunctions. Age and genetics are the most prominent risk factors for developing AD2. Among the genetic factors attributed to the disease, amyloid precursor protein (APP), presenilin-1 (PSEN1), and presenilin-2 (PSEN2) mutations have been found to be correlated with early-onset AD3–5. On the other hand, late-onset AD is known for being strongly associated with the presence of apolipoprotein E (ApoE) 4 allele2. Previous research has reported other genetic aspects associated with diverse physiological mechanisms that could contribute to the development of the disease. For instance, alterations in genes regulating autophagy6, cell adhesion7, endocytosis8, or apoptosis9 also have an impact on AD progression. One of the most insidious causes of neural disruption is chronic inflammation due to extracellular accumulation of insoluble beta-amyloid peptide (A)10. Aggregates of A come in the form of neuritic plaques, which have been considered the neuropathological hallmark of AD2,11. In fact, confirmation of histologic presence of A is established as one of the main AD biomarkers according to the Aging and Alzheimer’s Association (NIA-AA)12. Although the role of A as the origin of the inflammation remains unclear, its relationship with localized neurophysiological disturbances in brain tissue implies to be one of the key aspects in the AD pathogenesis13. In this way, evidence of genes participating in A-related pathways has been reported. Among them, phosphatidylinositol binding clathrin assembly protein (PICALM) and clusterin (CLU) genes have been found to be involved in A deposition and clearance14–17.
Other widely studied phenomena associated with neurodegeneration are changes in the behavior of neurotransmitter neuronal populations. Particularly, AD is characterized by significant alterations in neurotransmitter balance. Acetylcholine and glutamate deficits have been previously associated to AD progression, since they may play key roles in cognitive impairment18. Acetylcholine and glutamate neurotransmitters are intimately involved in memory and learning processes, in which they exhibit functional synergy in neuronal firing of pyramidal neurons18. However, many other neurotransmitter systems, such as GABAergic, serotonergic, and possibly dopaminergic neurons, have been associated with AD abnormalities19. The importance of these aspects in the understanding of dementia are apparent, as new methods of AD diagnosis have been developed measuring these biochemical alterations20, along with therapy strategies targeting these deficits modulating neurotransmitter expression via drug administration21. Neurotransmission functionality has also been related to genetic features. For instance, PICALM and CLU have been suggested to be implicated in several neurotransmission mechanisms22–24. These relationships imply that alterations in the genetic expression could manifest in synaptic disturbances. In addition to the evidence linking PICALM and CLU to neurochemical processes, these genes have also been associated with AD through genome-wide association studies25,26. Single nucleotide polymorphisms (SNP) rs3851179 and rs11136000 showed correlation with reduced risk of late-onset AD25,26. Afterwards, the association between the aforementioned SNPs and the risk of late-onset AD was confirmed27.
Another interesting approach to investigate the neurodegeneration associated to AD is the examination of alterations in functional brain activity. For this purpose, the analysis of electroencephalographic (EEG) activity has been extensively used. EEG can keep track of the rapid and transient nature of neuroelectric dynamics due to its high temporal resolution. This technique comes with advantages like low cost, portability, and non-invasiveness. The usefulness of EEG signal analyses has been demonstrated in the study of brain activity disruptions caused by AD28,29. However, EEG measurements are acquired on the scalp surface, being unable to illustrate the activity and location of individual neuronal oscillators and thus identify aberrant neural behavior in specific brain areas. To overcome this limitation, source localization procedures have been developed. Standardized low resolution electromagnetic tomography (sLORETA) is based on the assumption that neighboring neurons are strongly correlated, yielding zero localization error in the simulations30. Brain activity alterations in AD have been broadly studied analyzing source-level signals acquired by sLORETA. For instance, cortical sources of resting-state EEG were assessed in mild AD patients, obtaining a power increase of widespread delta sources and a power decrease of posterior alpha sources31. Furthermore, functional connectivity was investigated including deeper regions of the brain; the results revealed that AD networks were associated with lower global information processing and higher local information processing32. Previous research indicated a correlation between source-level EEG alterations and presence of PICALM and CLU risk alleles. Ponomareva et al.33 investigated the EEG from healthy subjects and obtained higher power in alpha-3 band (10.9–12.9 Hz) in the CLU CC allele group, in addition to higher beta power and lower alpha inter-hemispheric connectivity in the PICALM GG allele group34,35. These insights reveal an association between abnormal power fluctuations and certain SNPs in PICALM and CLU, which could be linked to dysfunctions caused by higher A accumulations or disrupted neurotransmission mechanisms, even in pre-clinical stages of the disease33–35. However, to the best of our knowledge, no assessment of multiple EEG sources under the effects of both PICALM and CLU risk alleles in AD patients has been conducted before. Thus, the novelty of this study is the investigation of the relationship between source-level EEG and AD-inducing genotypes associated with shared aspects of neurochemical metabolism.
As suggested by the aforementioned evidence, the relationship between AD-inducing variants of PICALM and CLU and neuronal dysfunctions are mainly twofold. First, the association with impaired performance of A clearance, and thus to greater inflammatory responses that could affect neurodegeneration. Second, the implications in primary neurotransmitter balance, which could lead to abnormal synaptic behavior. Therefore, we hypothesize that the presence of these alleles (rs3851179 and rs11136000 SNPs) may have an impact on brain activity. Consequently, this study is aimed at analyzing source-level EEG in AD patients and healthy controls with risk and protective alleles of PICALM and CLU in order to detect which alterations of brain electrical dynamics could be potentially connected to localized A presence and neurotransmission deficits.
Methods
Subjects
Fifty-three AD patients (16 males and 37 females) and 24 healthy controls (13 males and 11 females) were enrolled in this study. AD patients met the diagnosis criteria according to the NIA-AA36. Additional inclusion and exclusion conditions were applied to participate in the study. Inclusion requisites were ages older than 65 and presence of risk or protector alleles for both rs3851179 and rs11136000 simultaneously. Exclusion conditions included clinical history of neoplasia (active or under treatment), recent surgery, hypercatabolic states, or vascular pathology. Also, cases with atypical signs in the evolution of the disease in AD patients or chronic alcoholism were disregarded. PICALM risk and protective genotypes were established as the presence of GG and AG+AA genotypes, respectively. In the case of CLU, risk and protective alleles were CC and CT+TT. These decisions were made consistently with previous research assessing the impact of PICALM and CLU alleles that confer risk or protection against AD33,34. Evaluation of cognitive state was performed for each subject using Mini-Mental State Examination (MMSE)37. Informed consent was acquired from each subject, relative or legal representative of each subject, in line with the recommendations of the Code of Ethics of the World Medical Association. The study was in compliance with the Declaration of Helsinki. The study protocol was approved by The Ethics Committee at the Porto University (Porto, Portugal. Report n 38/CEUP/2018). Demographic data is presented in Table 1. Statistical analyses were performed to identify potential confounding factors using test ( = 0.05). Subjects were matched by sex ((1, N = 24) = 1.51, p-value = 0.219; (1, N = 53) = 1.23, p-value = 0.266), and ApoE 4 presence ((1, N = 22) = 0.386, p-value = 0.534; (1, N = 49) = 1.34, p-value = 0.249). Regarding AD subgroups, patients were matched by severity ((2, N = 53) = 2.65, p-value = 0.265). Also, no statistically significant differences between genetic groups were found considering the age or MMSE score of the subjects (p-values > 0.05, Mann–Whitney U-test). AD severity was adjusted to Reisberg scale criteria38 according to 3 severity stages: mild AD (), moderate AD (), and severe AD (). All the comparisons showed no evidence of statistical dependency of any factor towards presence of risk or protective alleles.
Table 1.
Demographic data.
| Genotype | Group | N | Age (mean ± SD) (years) | Sex (M:F) | MMSE (mean ± SD) |
|---|---|---|---|---|---|
| Risk alleles | Healthy controls | 12 | 80.33 ± 6.64 | 8:4 | 29.33 ± 0.65 |
| patients | 6 | 80.00 ± 4.38 | 2:4 | 21.00 ± 1.26 | |
| patients | 8 | 84.25 ± 3.24 | 2:6 | 12.50 ± 1.60 | |
| patients | 4 | 76.50 ± 7.94 | 0:4 | 2.25 ± 4.50 | |
| Protective alleles | Healthy controls | 12 | 81.41 ± 7.75 | 5:7 | 28.59 ± 1.08 |
| patients | 17 | 79.29 ± 8.42 | 9:8 | 23.06 ± 2.75 | |
| patients | 8 | 83.13 ± 6.94 | 0:8 | 14.13 ± 2.30 | |
| patients | 10 | 80.70 ± 5.08 | 3:7 | 4.40 ± 4.27 |
SD, standard deviation; M, male; F, female; , mild AD; , moderate AD; , severe AD; MMSE, Mini-Mental State Examination score.
Genetic analysis
A sample of saliva from each subject was collected by means of a DNA Genotek Oragene DNA kit (OG-500). The biological samples were genotyped using Thermo Fisher Scientific Axiom Spain Biobank Arrays at the Spanish National Center for Genotyping (CeGEN, Santiago de Compostela, Spain), and SNP calling was performed with Affymetrix Power Tools.
Variant calling quality control (QC) was implemented in the genotyping. This protocol was followed by both individual and marker analyses, according to Affymetrix best practices guide. Criteria of variant calling QC invalidates subjects with dish QC or QC call rates below the defined thresholds. Also, subjects with heterozygosity rates greater than the defined acceptance threshold were not considered for further analysis. PICALM and CLU risk variants (rs3851179 and rs11136000) qualified as valid in the QC analysis.
EEG recordings and pre-processing
For each subject, five minutes of resting-state EEG data were recorded with a 19-channel Nihon Kohden Neurofax JE-921A EEG System at electrodes F3, F4, F7, F8, Fp1, Fp2, T3, T4, T5, T6, C3, C4, P3, P4, O1, O2, Fz, Cz, and Pz of the international 10–20 system. The sampling frequency was set to 500 Hz. During acquisition, common average reference was applied. Participants were asked to stay relaxed in a noise-free environment along the procedure. The state of vigilance was controlled by the researchers to prevent drowsiness.
EEG data were stored as ASCII files in a personal computer. Signals were preprocessed according to previous studies39,40: (i) mean removal; (ii) 50 Hz notch filter; (iii) Hamming-window bandpass filter between 1 and 70 Hz; (iv) independent component analysis (ICA) to remove components associated with other biosignals; (v) segmentation into epochs of 5 s duration; and (vi) visual rejection of epochs containing artifacts. The average number of artifact-free epochs selected per subject was 39.95 ± 12.74 (mean ± SD).
After EEG acquisition, source-level activity was estimated using sLORETA source localization algorithm30. sLORETA computes 3D linear solutions for the EEG inverse problem, allowing a noise-normalized estimation of the distribution of electrical activity within a brain model. An identity matrix was used as noise covariance, as no noise recordings were available. A total of 68 brain regions of interest (ROI) were defined according to the Desikan-Killiany atlas, a gyrus-based classification of regions corresponding to their dominant function41. Digital pre-processing and analysis of the signals were carried out with MATLAB (R2018a version, Mathworks, Natick, MA) and Brainstorm toolbox (documented and freely available for download online under the GNU general public license, http://neuroimage.usc.edu/brainstorm42).
EEG processing
Relative power (RP) was calculated for each artifact-free epoch for the 68 ROIs proposed in the Desikan-Killiany atlas41. RP is a commonly used metric to estimate the relative contribution of power of each frequency band to the global power spectrum of a signal43. RP has relatively low inter-subject variability and provides power estimations that do not depend on the filtering and amplification characteristics of the recording device43. RP equation is described below:
| 1 |
where PSD is the normalized power spectral density, associated with each frequency component f. RP was computed in the traditional frequency bands: delta (, 1–4 Hz), theta (, 4–8 Hz), alpha (, 8–13 Hz), beta (, 13–30 Hz), and gamma (, 30–70 Hz). Once RP values were obtained for each artifact-free epoch, they were averaged for each ROI, obtaining an array of 68 values per subject and frequency band.
In order to assess the heterogeneity of the distribution of RP values, spatial entropy (SE) was calculated. This parameter is based on the Shannon definition of entropy. Homogeneous distributions result in low values of SE, and vice versa. SE was calculated from the RP values from the 68 ROIs for each frequency band in each artifact-free epoch. Then, the averaged SE along epochs was calculated. SE is defined as follows:
| 2 |
where n is each bin in the probability density function (PDF), and N is the total number of bins. The probability density function was estimated by means of the normalized histogram of RP values at each frequency band. The Freedman-Diaconis rule was used to set the number of bins of the histogram44.
Statistical analysis
Initially, we carried out an exploratory analysis to evaluate normality and homoscedasticity of RP distributions at each frequency band. For this purpose, Kolmogorov–Smirnov and Levene tests were conducted. Nor RP neither SE values met parametric assumptions; therefore, statistical differences between risk and protective alleles subgroups were calculated with Mann–Whitney U-tests ( = 0.05). In addition, a false discovering rate (FDR) controlling procedure was used to deal with multiple comparisons assessing differences for each ROI45.
Results and discussion
Potential biochemical implications of PICALM and CLU risk variants and their relationship with RP alterations
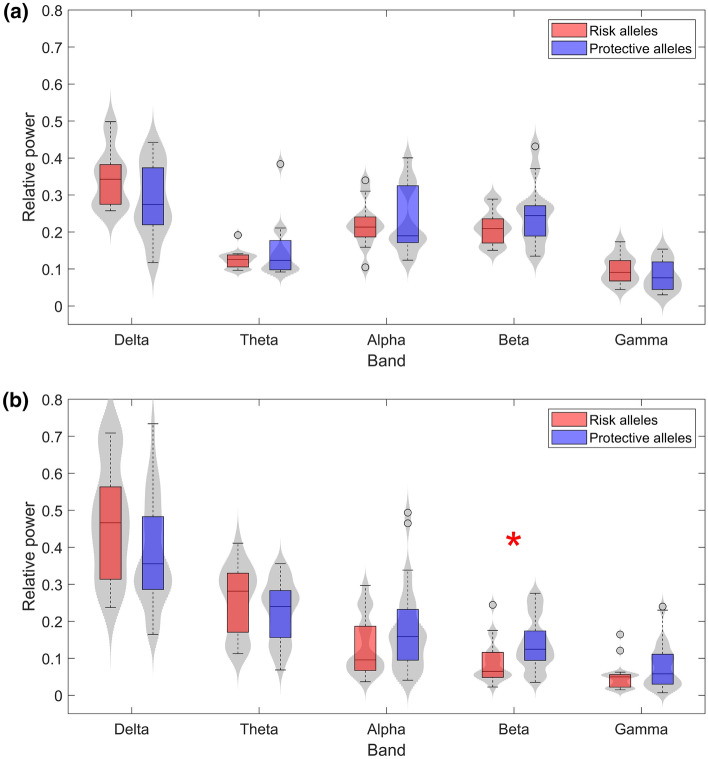
RP in each ROI was calculated for 53 AD patients (18 with risk alleles of both PICALM and CLU genes and 35 with the protective variants) and 24 healthy controls (12 with risk alleles and 12 with protective alleles) in the traditional frequency bands. Figure 1 displays the distribution of the grand-average RP values of all ROIs in both control (1.a) and AD patients (1.b). In AD, the risk group showed higher RP values in lower frequency bands (delta and theta) and lower RP values in higher frequency bands (alpha, beta, and gamma). Only significant statistical differentiation between risk and protective genetic variants was found in beta frequency band (U-value 342, p-value 0.007, Mann–Whitney U-test). No other band showed statistically significant differences, although delta band was close to the limit of statistical significance (U-value 584, p-value = 0.067, Mann–Whitney U-test). In general, signs indicating slowing of the EEG have been commonly identified as a quantitative manifestation of neurodegeneration46. Interestingly, the observed disturbances in each single frequency band are in line with the typical spectral alterations found in dementia46, which suggests that risk genotypes may be associated with worse cognitive status. Given that statistical analyses indicated no evidence of significant confounding factors, alterations of EEG global spectral features are influenced most likely by the genetic AD-inducing effects of the proposed SNPs, and not by an AD severity bias.
Figure 1.
Grand-averaged RP values across ROIs for (a) controls and (b) AD patients in each frequency band. Subjects with risk alleles for both PICALM and CLU are represented in red, whereas subjects with both protective alleles are displayed in blue. Statistically significant differences (p-value < 0.05, Mann–Whitney U-test) are highlighted with a red asterisk.
On the other hand, the control group exhibited slightly different trends in alpha and gamma. Beta band kept showing higher RP values in the protective genotype, however, significant differences were not obtained. This finding may be due to the opposite influence of PICALM alleles in healthy subjects, which has been previously reported34. This new insight suggests that the effect of PICALM gene could differ depending on the cognitive status of the subject or, at least, on the severity of neurodegeneration. Beta band has been previously associated with not only sensory-motor processing47, but also with cognitive processes like spatial and temporal orienting of attention48 or working memory49. Previous evidence reported that excess of amplitude in beta is associated with stress, anxiety, overthinking, and overstimulation50. On the other hand, the hallmark of neurophysiological rhythms alterations in AD is a general slowing of the EEG46, in which beta activity is significantly reduced51,52. These points altogether lead to consider that certain ranges of EEG beta power could be associated with better cognitive performance. Since PICALM GG genotype carriers have been previously suggested to be prone to stress reactions34, we could hypothesize that this version of PICALM, in general, tends to destabilize neural dynamics under abnormal conditions. In other words, the protective allele of PICALM may be associated with more stable ranges of beta activity, closer to healthier brain functioning. This reasoning could also be applied to other frequency bands. For instance, alpha band exhibited a change of tendency in control RP values in comparison with AD. Although no statistically significant differences were observed, this finding may be another case of different effects of the genotype on the EEG dynamics depending on the status of neurodegeneration. A previous study reported an association between CLU and RP in upper alpha band for healthy cognitive subjects33, which presented higher values for the risk allele. Alpha oscillations reflect neural processing related with essential cognitive functions, such as memory, intelligence, and attention53,54. However, excessive values of alpha power have been associated with cholinergic network alterations that could affect synaptic efficacy and reduce cortical connectivity55. Thus, the tendency change observed in our results may be due to significant disturbances on physiological mechanisms involving neurotransmitter populations. In this way, CLU protective allele could be implicated in cholinergic endeavors closer to healthy conditions in any cognitive status.
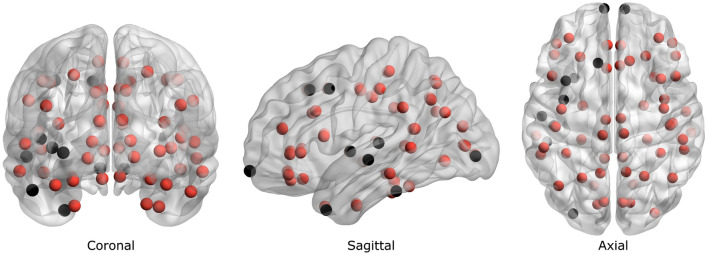
Given that PICALM and CLU genes seem to be related with A clearance processes, we suggest that malfunction of these mechanisms may have some impact in establishing physiological conditions for greater A deposition in the brain, and hence, amplify the detrimental effects of dementia. In this study, statistically significant differences in RP values at beta band were reported in 58 out of the 68 ROIs in AD (FDR-corrected p-values < 0.05, Mann–Whitney U-test), and no differences in any ROI at all in any frequency band for controls. The obtained differences in AD were represented in a 3D brain model (BrainNet Viewer, v1.6356) in Fig. 2, which exhibited beta RP disruptions throughout the brain. According to previous research investigating histological alterations in AD, no spatial preference was observed in the deposition of neuritic plaques in affected brains57. In fact, the distribution of neuritic plaques presented large between-subject variation, being opposite to what was found regarding with neurofibrillary tangles57. This study assumes that PICALM and CLU risk variants may stimulate A deposition hindering A clearance mechanisms, since these genes may be involved in these processes14–17. Given that A is broadly distributed across the brain in AD, and EEG disturbances in patients with risk alleles of both PICALM and CLU are also obtained in most ROIs, it can be proposed that the reported EEG abnormalities may be associated with increased accumulation of A. Noteworthy, all the regions that did not show evidence of significant differences were found in the left hemisphere, except a single ROI located in the right frontopolar lobe. This observation should be cautiously interpreted, since p-values obtained in the spatial analysis were close to the significance threshold ( = 0.05). This finding may not be due to the distribution of A, but to the functional asymmetry of the brain. In fact, left hemisphere and beta band activity have been reported to be preferentially engaged in time perception58–60, a cognitive process that could be heavily present during resting-state, while the patient is waiting till the end of the EEG acquisition. Slight variations in beta activity caused by functional specialization of the brain may be the reason why some ROIs are not expressed as significantly different by the statistical analyses.
Figure 2.
Brain model representing localizations of each ROI according to the Desikan-Killiany atlas. Red balls represent ROIs that showed statistically significant differences in beta RP values between AD patients with risk and protective PICALM and CLU alleles (FDR-corrected p-value < 0.05, Mann–Whitney U-test).
Apart from its inflammatory effect10, A may have a significant impact on neurophysiology on additional scales. For instance, the implications of greater presence of A in synaptic functionality have been previously examined. Several studies connecting dopamine metabolism with A deposition were reported, obtaining a decreased release of this neurotransmitter in A-infused rat brains61,62. Thus, we suggest that PICALM and CLU risk variants (and hence greater accumulations of A in brain tissue) may be closely related to EEG alterations in beta band caused by dopaminergic deficits. Also, the relation between A-mediated inflammation and neurotransmitter disturbances has been assessed, obtaining significant interactions with cholinergic, serotonergic, and dopaminergic neuronal populations63. However, confirming direct causality between AD-inducing genotypes and neurotransmission alterations caused by A-related pathological conditions, such as chronic inflammation, requires further investigation.
Alterations on RP distributions and their potential relationship with AD as a disconnection syndrome
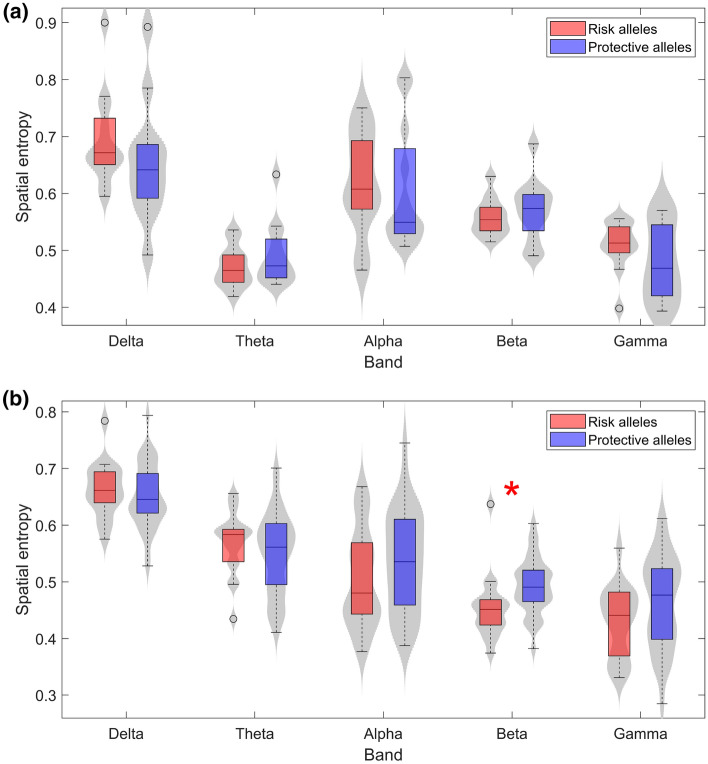
The homogeneity of the RP values throughout the brain was assessed by applying the spatial entropy to RP values. Figure 3 displays the distribution of the SE values in each frequency band in controls (Fig. 3a) and AD patients (Fig. 3b). In AD, the analysis exposed higher values of entropy in subjects with protective alleles in alpha, beta, and gamma bands, and lower values in delta and theta bands. Furthermore, beta band revealed statistically significant differences between subjects with risk and protective alleles in the AD group (U-value 321, p-value 0.002, Mann–Whitney U-test). As specialized cortical areas working interdependently were previously identified64, entropy alterations could be an effect of impaired communication between different neural populations. The synchronous activation of these neural networks to perform complex processes may result in electrical activity of diverse power and frequencies. Additionally, it has been previously proposed that higher frequencies build local patches of synchrony, while slower rhythms are more present in long-distance interactions65,66. Previous research investigated these inter-neural synergies by means of EEG coherence analyses while assessing intelligence performance67. In this study67, lower coherence results were obtained in higher IQ subjects, which is suggested to be associated with increased neural efficiency and brain complexity. Since coherence is defined by the spectral correlation between two time series or, in other words, how likely a series could be predicted by another one, low coherence values are expected to be related with high SE values and vice versa. These assumptions are consistent with a model that relates lower coherences to increased spatial differentiation and hence, to higher processing efficiency68,69. As higher frequencies may be more sensitive to changes in the spatial organization of specialized neural populations, it seems reasonable that RPs in higher frequency bands could be more affected by physiological disturbances that alter connectivity, albeit in a subtle way, since they work locally, influencing specific and smaller neighboring neural populations. On the other hand, Fig. 3a shows the SE values obtained for the control group. The significant differences found in beta for AD patients were not obtained. Furthermore, SE trends in controls changed from AD similarly to what was observed in RP. This finding could indicate that, analogously with RP, SE values in certain intervals may be associated with optimal brain functionality. Also, protective alleles of PICALM and CLU could participate in maintaining SE in those ranges. To conclude, our SE results in beta band suggest a possible association between AD patients with PICALM and CLU risk alleles and a worse cognitive functioning.
Figure 3.
Spatial entropy of RP values for (a) controls and (b) AD patients in each frequency band. Subjects with risk alleles for both PICALM and CLU are represented in red, whereas subjects with both protective alleles are displayed in blue. Statistically significant differences (p-value < 0.05, Mann–Whitney U-test) are highlighted with a red asterisk.
Impact of PICALM and CLU risk alleles on the EEG spectral properties and neurotransmitter metabolism along the AD continuum
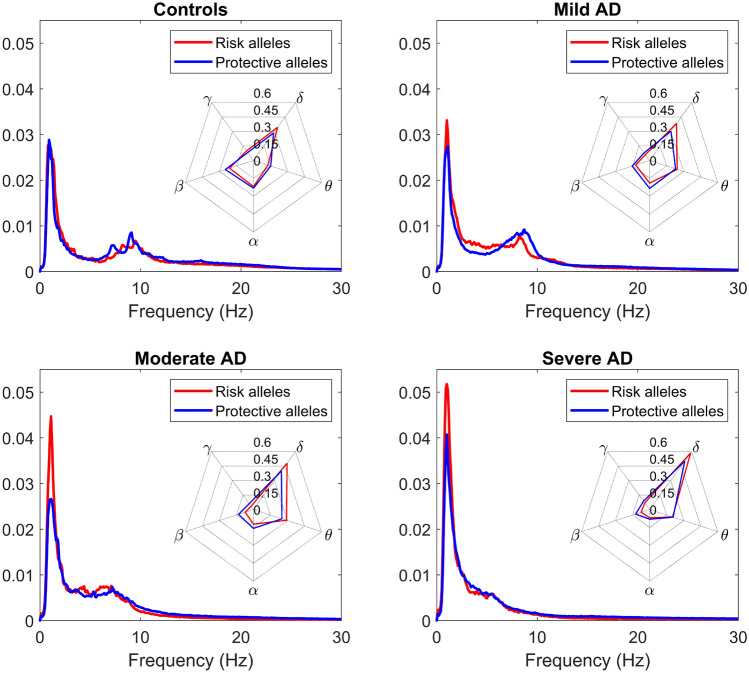
Finally, PSDs were inspected along the AD continuum. AD patients were organized corresponding to the stage of the disease: mild, moderate, or severe, according to the Reisberg scale38. PSDs were displayed for controls and AD patients at each stage of the disease (Fig. 4), which showed a general slowing of the EEG in the risk group compared to the protective one. Slowdowns in the EEG have been previously associated with disruption of information processing and cognitive dysfunction46. Hence, our results suggest that patients with both PICALM and CLU risk alleles have a deterioration status equivalent to more severe stages of neurodegeneration. Indeed, the normalized PSD from mild AD subjects with the risk alleles is apparently more similar to the PSD from moderate AD subjects with protective alleles than to their protective counterpart. Furthermore, PSDs showed greater differences between genotypes in the mild AD group than in the severe AD group; the EEG slowdown is more evident between patients subgroups (subgroup with risk alleles and subgroup with carriers of protective alleles) than between patients subgroups. This observation suggests that the effects of PICALM and CLU risk alleles may be more notorious in earlier stages of the disease. In fact, cytopathology in cortical cholinergic pathways has been found to be an early event in the AD continuum70. As the genes under study may be involved on neurotransmitter disturbances, the presence of risk alleles could lead to a greater impact in early AD. On the other hand, given the correlation between EEG spectral alterations throughout the brain and A presence, and the common implication of these genes in A modulation14–17, we suggest that increased A accumulation may be related to the disruption of spectral dynamics. In addition, the presence of AD-inducing PICALM and CLU variants seems to have a stronger effect on beta power.
Figure 4.
Normalized PSDs for controls and AD patients at different stages of the disease. The grand-averaged spectra are displayed from subjects with risk and protective alleles. Spider plots presenting RP values at each frequency band are shown for the four groups (Controls, Mild AD, Moderate AD, and Severe AD).
Beta frequency band also showed consistently lower RP values for risk-genotype subjects along the AD continuum. Besides, differences in beta band are preserved as severity increases, while they decrease in theta, alpha, and gamma. This observation suggests the persistent effect that AD-inducing genotypes cause to neural dynamics along the disease progression. This issue may be due to augmented expression of the implicated genes in the last stages of the AD. In this line, previous research observed plasma clusterin levels correlating positively with deterioration status71,72. In addition, the brain dopaminergic system was found to be affected by AD, decreasing the ability of dopamine reuptake, and correlating with the severity of the extrapyramidal symptoms73. The significant differences observed in beta band may be a manifestation of other neurotransmitter abnormalities. In a previous study, a loss of beta synchronization was found in the early stages of AD development, but not in other frequency bands74. Connectivity metrics allow evaluating how local specialized neural networks work together to integrate information. Beta-specialized neuronal populations may become isolated due to alterations in anatomical pathways and, therefore, a decrease in RP at beta band may be a reasonable effect of this aspect. However, if AD is identified as a disconnection syndrome from the perspective of neuronal loss exclusively, alterations in a single frequency band would be difficult to explain74. For this reason, other factors that contribute to loss of synchrony, such as cholinergic disturbances, may be relevant. In fact, blockade of the cholinergic system has previously been associated with reduced coherence of the EEG at rest75. Given the association of PICALM and CLU with different neurotransmitter metabolisms22–24, carriers of AD-inducing alleles may be expressing these physiological anomalies in the form of RP changes at beta band. To sum up, disruptions in several neurotransmitter systems may play a crucial role in the EEG alterations due to PICALM and CLU risk genotypes.
Conclusion
In the present study, an association between AD-inducing PICALM and CLU alleles and disturbances in the EEG was revealed. Despite our outcomes being relevant, some methodological issues should be taken into account. First, although our results indicate a potential relation between A accumulation or neurotransmitter deficits and alterations in beta EEG activity, these associations are merely speculative. Unfortunately, we have no access to medical imaging data to validate causal relationships between our results and alterations in neurochemical metabolism. For this reason, this study is not so much about ascertaining the associations between alleles and EEG alterations, but rather to exhibit possible physiological causes related to these alleles that may be associated to neurodynamic disturbances. Hence, there is the possibility that other underlying factors may be causing these changes as a hidden side effect of the PICALM and CLU alleles. In order to clarify the main objective of this article, the associations reported here are a summary of all the potential biochemical causes involving PICALM and CLU after an extensive investigation of the literature. A main part of the study is the elucidation of how these aspects are potentially related to beta power in brain activity. However, finding causality between the proposed physiological elements and EEG perturbations will be planned in subsequent research projects, in which we intend to have medical imaging available. Obtaining information on the hallmark brain changes of AD will allow verifying the results showed in this study.
In addition, due to the highly restrictive genetic conditions to participate in the study (AD patients and controls with a pair of either risk or protective variants), the number of selected participants became relatively small. In order to increase robustness of additional investigation on this topic, larger databases should be considered. Finally, in line with the former point, our database was quite unbalanced towards subjects with protective genetic variants. This issue is partially due to the categorization of 2 of the 3 possible genotypes as protective (e.g. CLU TT and TC genotypes as protective and CC ones conferring risk). This criterion to classify individuals in each category resulted in unbalanced groups and may introduce some bias to our calculations that should be addressed enrolling new participants in future studies.
Our results showed that the brain activity from AD patients with risk alleles is characterized by a statistically significant decrease of RP and SE in beta band, in comparison with subjects with two protective alleles. Since these genetic elements are associated with alterations in biochemical pathways in neurophysiological patterns, the changes obtained in beta activity may have some relation with greater accumulations of A or neurotransmission abnormalities. However, further research is needed to improve our understanding of AD at the molecular level.
Acknowledgements
We deeply thank all participants, families and institutions involved, namely: Asociación de Familiares de Enfermos de Alzheimer de Ávila, Ávila; Associação de Pensionistas e Reformados de Viana do Castelo, Viana do Castelo; Casa do Povo de Alvito S.Pedro, Barcelos; Santa Casa da Misericórdia de Vila Nova de Gaia; Obra Social Nossa Senhora da Boa Viagem, Porto; Gero Vida, Villaralbo (Zamora); Asociación de Familiares de Alzheimer de León; Residencia San Raimundo en Coreses; Centro de Dia S. João de Deus, da Santa Casa da Misericórdia do Porto; Lar Santa Rita, da Santa Casa da Misericórdia de Caminha; Centro Social e Cultural de Vila Praia de Âncora; Lar Casa de Magalhães; Armonía Centro de Día, Zamora. Also, we want to express our gratitude to Patricia Sousa from ‘Associação Portuguesa de Familiares e Amigos dos Doentes de Alzheimer’ and Carmen Pita from ‘Asociación de Familiares y Amigos de Enfermos de Alzheimer y Otras Demencias de Zamora’. They contributed with their psychological and caregiver skills to ease the stress of the patients.
Author contributions
A.M.-C., C.G. and J.P. collected the EEG signals. A.L. and N.P. processed the genetic data. V.R.-G. preprocessed the signals, A.M.-C. processed the signals, analyzed the data, and wrote the manuscript. A.M.-C. and R.H. designed the study and interpreted the results. C.G., J.P., and V.G.-d.P. interpreted the results. All authors have read, revised, and approved the final manuscript.
Funding
This research was supported by ‘Ministerio de Ciencia, Innovación y Universidades - Agencia Estatal de Investigación’ and ‘European Regional Development Fund’ (FEDER) under project PGC2018-098214-A-I00, by ‘European Commission’ and ‘FEDER’ under projects ‘Análisis y correlación entre el genoma completo y la actividad cerebral para la ayuda en el diagnóstico de la enfermedad de Alzheimer’ and ‘Análisis y correlación entre la epigenética y la actividad cerebral para evaluar el riesgo de migraña crónica y episódica en mujeres’ (‘Cooperation Programme Interreg V-A Spain-Portugal POCTEP 2014–2020’), by ‘CIBER de Bioingeniería, Biomateriales y Nanomedicina (CIBER-BBN)’ through ‘Instituto de Salud Carlos III’ co-funded with FEDER funds, and by Portuguese funds through FCT-Fundação para a Ciência e a Tecnologia/Ministério da Ciência, Tecnologia e Inovação and the projects ‘Institute for Research and Innovation in Health Sciences’ (POCI-01-0145-FEDER-007274) and ‘Center of Mathematics of the University of Porto’ (UID/MAT/00144/2013). V.R.-G. was in receipt of a PIF-UVa grant from the University of Valladolid. A.L. and N.P. are funded by FCT: IF/01262/2014, and through the Decreto-Lei n 57/2016 de 29 de Agosto, respectively. The genotyping service was carried out at CEGEN-PRB3-ISCIII; it is supported by grant PT17/0019, of the PE I+D+i 2013-2016, funded by ISCIII and ERDF.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Alzheimer’s disease international. In World Alzheimer Report 2019: Attitudes to dementia Alzheimer’s Disease International (2019). https://www.alz.co.uk/research/world-report-2019.
- 2.Alzheimer’s Association 2019 Alzheimer’s disease facts and figures. Alzheimers. Dement. 2019;15:321–387. doi: 10.1016/j.jalz.2019.01.010. [DOI] [Google Scholar]
- 3.Goate A, et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature. 1991;349:704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- 4.Sherrington R, et al. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer’s disease. Nature. 1995;375:754–760. doi: 10.1038/375754a0. [DOI] [PubMed] [Google Scholar]
- 5.Levy-Lahad E, et al. Candidate gene for the chromosome 1 familial Alzheimer’s disease locus. Science. 1995;269:973–977. doi: 10.1126/science.7638622. [DOI] [PubMed] [Google Scholar]
- 6.Nixon RA, Yang DS. Autophagy failure in Alzheimer’s disease-locating the primary defect. Neurobiol. Dis. 2011;43:38–45. doi: 10.1016/j.nbd.2011.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bao X, et al. Cell adhesion molecule pathway genes are regulated by cis-regulatory SNPs and show significantly altered expression in Alzheimer’s disease brains. Neurobiol. Aging. 2015;36(2904):e1–2904.e7. doi: 10.1016/j.neurobiolaging.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 8.Kimura N, Yanagisawa K. Traffic jam hypothesis: Relationship between endocytic dysfunction and Alzheimer’s disease. Neurochem. Int. 2018;119:35–41. doi: 10.1016/j.neuint.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 9.Gu C, et al. Long noncoding RNA EBF3-AS promotes neuron apoptosis in Alzheimer’s disease. DNA Cell Biol. 2018;37:220–226. doi: 10.1089/dna.2017.4012. [DOI] [PubMed] [Google Scholar]
- 10.Akiyama H, et al. Inflammation and Alzheimer’s disease. Neurobiol. Aging. 2000;21:383–421. doi: 10.1016/S0197-4580(00)00124-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Selkoe DJ. Cell biology of the amyloid -protein precursor and the mechanism of Alzheimer’s disease. Annu. Rev. Cell Biol. 1994;10:373–403. doi: 10.1146/annurev.cb.10.110194.002105. [DOI] [PubMed] [Google Scholar]
- 12.Jack CR, et al. NIA-AA research famework: Toward a biological definition of Alzheimer’s disease. Alzheimers. Dement. 2018;14:535–562. doi: 10.1016/j.jalz.2018.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meraz-Ríos MA, Toral-Rios D, Franco-Bocanegra D, Villeda-Hernández J, Campos-Peña V. Inflammatory process in Alzheimer’s disease. Front. Integr. Neurosci. 2013;7:59. doi: 10.3389/fnint.2013.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu W, Tan L, Yu JT. The role of PICALM in Alzheimer’s disease. Mol. Neurobiol. 2015;52:399–413. doi: 10.1007/s12035-014-8878-3. [DOI] [PubMed] [Google Scholar]
- 15.Zhao Z, et al. Central role for PICALM in amyloid- blood–brain barrier transcytosis and clearance. Nat. Neurosci. 2015;18:978–987. doi: 10.1038/nn.4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeMattos RB, et al. Clusterin promotes amyloid plaque formation and is critical for neuritic toxicity in a mouse model of Alzheimer’s disease. Proc. Natl. Acad. Sci. USA. 2002;99:10843–10848. doi: 10.1073/pnas.162228299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zlokovic BV, et al. Brain Uptake of circulating apolipoproteins J and E complexed to Alzheimer’s amyloid . Biochem. Biophys. Res. Commun. 1994;205:1431–1437. doi: 10.1006/bbrc.1994.2825. [DOI] [PubMed] [Google Scholar]
- 18.Francis PT. The interplay of neurotransmitters in Alzheimer’s disease. CNS Spectr. 2005;10:6–9. doi: 10.1017/S1092852900014164. [DOI] [PubMed] [Google Scholar]
- 19.Reinikainen KJ, Soininen H, Riekkinen PJ. Neurotransmitter changes in Alzheimer’s disease: Implications to diagnostics and therapy. J. Neurosci. Res. 1990;27:576–586. doi: 10.1002/jnr.490270419. [DOI] [PubMed] [Google Scholar]
- 20.Sangubotla R, Kim J. Recent trends in analytical approaches for detecting neurotransmitters in Alzheimer’s disease. Trends Analyt. Chem. 2018;105:240–250. doi: 10.1016/j.trac.2018.05.014. [DOI] [Google Scholar]
- 21.Prakash A, Kalra J, Mani V, Ramasamy K, Majeed ABA. Pharmacological approaches for Alzheimer’s disease: Neurotransmitter as drug targets. Expert Rev. Neurother. 2014;15:53–71. doi: 10.1586/14737175.2015.988709. [DOI] [PubMed] [Google Scholar]
- 22.Harel A, Wu F, Mattson MP, Morris CM, Yao PJ. Evidence for CALM in directing VAMP2 trafficking. Traffic. 2008;9:417–429. doi: 10.1111/j.1600-0854.2007.00694.x. [DOI] [PubMed] [Google Scholar]
- 23.Harel A, Mattson MP, Yao PJ. CALM, a clathrin assembly protein, influences cell surface GluR2 abundance. Neuromol. Med. 2011;13:88–90. doi: 10.1007/s12017-010-8142-6. [DOI] [PubMed] [Google Scholar]
- 24.Chen F, et al. Clusterin secreted from astrocyte promotes excitatory synaptic transmission and ameliorates Alzheimer’s disease neuropathology. Mol. Neurodegener. 2021;16:5. doi: 10.1186/s13024-021-00426-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harold D, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat. Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lambert JC, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat. Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 27.Carrasquillo MM, et al. Replication of CLU, CR1, and PICALM associations with Alzheimer disease. Arch. Neurol. 2010;67:961–964. doi: 10.1001/archneurol.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horvath A, et al. EEG and ERP biomarkers of Alzheimer’s disease: A critical review. Front. Biosci. 2018;23:183–220. doi: 10.2741/4587. [DOI] [PubMed] [Google Scholar]
- 29.Vecchio F, et al. Resting state cortical EEG rhythms in Alzheimer’s disease: Toward EEG markers for clinical applications: A review. Suppl. Clin. Neurophysiol. 2013;62:223–236. doi: 10.1016/B978-0-7020-5307-8.00015-6. [DOI] [PubMed] [Google Scholar]
- 30.Pascual-Marqui, R. D. Standardized low resolution brain electromagnetic tomography (sLORETA): technical details (Tech, Rep, 2002). [PubMed]
- 31.Babiloni C, et al. Cortical sources of resting state EEG rhythms are sensitive to the progression of early stage Alzheimer’s disease. J. Alzheimer’s Dis. 2013;34:1015–1035. doi: 10.3233/JAD-121750. [DOI] [PubMed] [Google Scholar]
- 32.Kabbara A, et al. Reduced integration and improved segregation of functional brain networks in Alzheimer’s disease. J. Neural Eng. 2018;15:026023. doi: 10.1088/1741-2552/aaaa76. [DOI] [PubMed] [Google Scholar]
- 33.Ponomareva NV, et al. Age-dependent effect of Alzheimer’s risk variant of CLU on EEG alpha rhythm in non-demented adults. Front. Aging Neurosci. 2013;5:86. doi: 10.3389/fnagi.2013.00086/abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ponomareva NV, et al. Quantitative EEG during normal aging: Association with the Alzheimer’s disease genetic risk variant in PICALM gene. Neurobiol. Aging. 2017;51(177):e1-177.e8. doi: 10.1016/j.neurobiolaging.2016.12.010. [DOI] [PubMed] [Google Scholar]
- 35.Ponomareva NV, et al. Genetic association between Alzheimer’s disease risk variant of the PICALM gene and EEG functional connectivity in non-demented adults. Front. Neurosci. 2020;14:324. doi: 10.3389/fnins.2020.00324/full. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McKhann GM, et al. The diagnosis of dementia due to Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers. Dement. 2011;7:263–269. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Folstein MF, Folstein SE, McHugh PR. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J. Psychiatr. Res. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- 38.Reisberg B. Dementia: A systematic approach to identifying reversible causes. Geriatrics. 1986;41:30–46. [PubMed] [Google Scholar]
- 39.Maturana-Candelas A, Gómez C, Poza J, Pinto N, Hornero R. EEG characterization of the Alzheimer’s disease continuum by means of multiscale entropies. Entropy. 2019;21:544. doi: 10.3390/e21060544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruiz-Gómez SJ, et al. Measuring alterations of spontaneous EEG neural coupling in Alzheimer’s disease and mild cognitive impairment by means of cross-entropy metrics. Front. Neuroinform. 2018;12:76. doi: 10.3389/fninf.2018.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Desikan RS, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. NeuroImage. 2006;31:968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 42.Tadel F, Baillet S, Mosher JC, Pantazis D, Leahy RM. Brainstorm: A user-friendly application for MEG/EEG analysis. Comput. Intell. Neurosci. 2011;2011:13. doi: 10.1155/2011/879716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bachiller A, et al. Decreased spectral entropy modulation in patients with schizophrenia during a P300 task. Eur. Arch. Psychiatry Clin. Neurosci. 2014;264:533–543. doi: 10.1007/s00406-014-0488-6. [DOI] [PubMed] [Google Scholar]
- 44.Freedman D, Diaconis P. On the histogram as a density estimator: L2 theory. Probab. Theory Relat. Fields. 1981;57:453–476. [Google Scholar]
- 45.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995;57:289–300. [Google Scholar]
- 46.Jeong J. EEG dynamics in patients with Alzheimer’s disease. Clin. Neurophysiol. 2004;115:1490–1505. doi: 10.1016/j.clinph.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 47.Hari R. Human cortical oscillations: A neuromagnetic view through the skull. Trends Neurosci. 1997;20:44–49. doi: 10.1016/S0166-2236(96)10065-5. [DOI] [PubMed] [Google Scholar]
- 48.van Ede F, de Lange F, Jensen O, Maris E. Orienting attention to an upcoming tactile event involves a spatially and temporally specific modulation of sensorimotor alpha- and beta-band oscillations. J. Neurosci. 2011;31:2016–2024. doi: 10.1523/JNEUROSCI.5630-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hsu C-C, Cheng C-W, Chiu Y-S. Analyze the beta waves of electroencephalogram signals from young musicians and non-musicians in major scale working memory task. Neurosci. Lett. 2017;640:42–46. doi: 10.1016/j.neulet.2017.01.022. [DOI] [PubMed] [Google Scholar]
- 50.Herrmann CS, Strüber D, Helfrich RF, Engel AK. EEG oscillations: From correlation to causality. Int. J. Psychophysiol. 2016;103:12–21. doi: 10.1016/j.ijpsycho.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 51.Coben LA, Danziger WL, Berg L. Frequency analysis of the resting awake EEG in mild senile dementia of Alzheimer type. Electroencephalogr. Clin. Neurophysiol. 1983;55:372–380. doi: 10.1016/0013-4694(83)90124-4. [DOI] [PubMed] [Google Scholar]
- 52.Coben LA, Danziger W, Storandt M. A longitudinal EEG study of mild senile dementia of Alzheimer type: Changes at 1 year and at 2.5 years. Electroencephalogr. Clin. Neurophysiol. 1985;61:101–112. doi: 10.1016/0013-4694(85)91048-X. [DOI] [PubMed] [Google Scholar]
- 53.Cooper NR, Burgess AP, Croft RJ, Gruzelier JH. Investigating evoked and induced electroencephalogram activity in task-related alpha power increases during an internally directed attention task. NeuroReport. 2006;17:205–208. doi: 10.1097/01.wnr.0000198433.29389.54. [DOI] [PubMed] [Google Scholar]
- 54.Klimesch W. Alpha-band oscillations, attention, and controlled access to stored information. Trends Cogn. Sci. 2012;16:606–617. doi: 10.1016/j.tics.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steriade M. Grouping of brain rhythms in corticothalamic systems. Neuroscience. 2006;137:1087–1106. doi: 10.1016/j.neuroscience.2005.10.029. [DOI] [PubMed] [Google Scholar]
- 56.Xia M, Wang J, He Y. BrainNet viewer: A network visualization tool for human brain connectomics. PLoS ONE. 2013;8:68910. doi: 10.1371/journal.pone.0068910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 58.Nicholls ME, Schier M, Stough CK, Box A. Psychophysical and electrophysiologic support for a left hemisphere temporal processing advantage. Neuropsychiatry Neuropsychol. Behav. Neurol. 1999;12:11–6. [PubMed] [Google Scholar]
- 59.Wiener M, Kanai R. Frequency tuning for temporal perception and prediction. Curr. Opin. Behav. Sci. 2016;8:1–6. doi: 10.1016/j.cobeha.2016.01.001. [DOI] [Google Scholar]
- 60.Ghaderi AH, et al. Time estimation and beta segregation: An EEG study and graph theoretical approach. PLoS ONE. 2018;13:e0195380. doi: 10.1371/journal.pone.0195380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Itoh A, et al. Dysfunction of cholinergic and dopaminergic neuronal systems in -amyloid protein-infused rats. J. Neurochem. 2002;66:1113–1117. doi: 10.1046/j.1471-4159.1996.66031113.x. [DOI] [PubMed] [Google Scholar]
- 62.Trabace L, et al. Soluble amyloid beta1-42 reduces dopamine levels in rat prefrontal cortex: Relationship to nitric oxide. Neuroscience. 2007;147:652–663. doi: 10.1016/j.neuroscience.2007.04.056. [DOI] [PubMed] [Google Scholar]
- 63.Hochstrasser T, Hohsfield LA, Sperner-Unterweger B, Humpel C. -Amyloid induced effects on cholinergic, serotonergic, and dopaminergic neurons is differentially counteracted by anti-inflammatory drugs. J. Neurosci. Res. 2012;91:83–94. doi: 10.1002/jnr.23126. [DOI] [PubMed] [Google Scholar]
- 64.Bressler SL. Large-scale cortical networks and cognition. Brain Res. Rev. 1995;20:288–304. doi: 10.1016/0165-0173(94)00016-I. [DOI] [PubMed] [Google Scholar]
- 65.Von Stein A, Sarnthein J. Different frequencies for different scales of cortical integration: From local gamma to long range alpha/theta synchronization. Int. J. Psychophysiol. 2000;38:301–313. doi: 10.1016/S0167-8760(00)00172-0. [DOI] [PubMed] [Google Scholar]
- 66.Varela F, Lachaux JP, Rodriguez E, Martinerie J. The brainweb: Phase synchronization and large-scale integration. Nat. Rev. Neurosci. 2001;2:229–239. doi: 10.1038/35067550. [DOI] [PubMed] [Google Scholar]
- 67.Thatcher RW, North D, Biver C. EEG and intelligence: Relations between EEG coherence, EEG phase delay and power. Clin. Neurophysiol. 2005;116:2129–2141. doi: 10.1016/j.clinph.2005.04.026. [DOI] [PubMed] [Google Scholar]
- 68.Silberstein RB, Song J, Nunez PL, Park W. Dynamic sculpting of brain functional connectivity is correlated with performance. Brain Topogr. 2004;16:249–254. doi: 10.1023/B:BRAT.0000032860.04812.b1. [DOI] [PubMed] [Google Scholar]
- 69.Thatcher RW, Krause PJ, Hrybyk M. Cortico-cortical associations and EEG coherence: A two-compartmental model. Electroencephalogr. Clin. Neurophysiol. 1986;64:123–143. doi: 10.1016/0013-4694(86)90107-0. [DOI] [PubMed] [Google Scholar]
- 70.Mesulam M, Shaw P, Mash D, Weintraub S. Cholinergic nucleus basalis tauopathy emerges early in the aging-MCI-AD continuum. Ann. Neurol. 2004;55:815–828. doi: 10.1002/ana.20100. [DOI] [PubMed] [Google Scholar]
- 71.Schrijvers EM, Koudstaal PJ, Hofman A, Breteler MM. Plasma clusterin and the risk of Alzheimer disease. JAMA. 2011;305:1322–1326. doi: 10.1001/jama.2011.381. [DOI] [PubMed] [Google Scholar]
- 72.Thambisetty M, et al. Association of plasma clusterin concentration with severity, pathology, and progression in Alzheimer disease. Arch. Gen. Psychiatry. 2010;67:739–748. doi: 10.1001/archgenpsychiatry.2010.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rinne JO, Sahlberg N, Ruottinen H, Någren K, Lehikoinen P. Striatal uptake of the dopamine reuptake ligand [11C]-CFT is reduced in Alzheimer’s disease assessed by positron emission tomography. Neurology. 1998;50:152–156. doi: 10.1212/WNL.50.1.152. [DOI] [PubMed] [Google Scholar]
- 74.Stam CJ, Van Der Made Y, Pijnenburg YAL, Scheltens P. EEG synchronization in mild cognitive impairment and Alzheimer’s disease. Acta Neurol. Scand. 2003;108:90–96. doi: 10.1034/j.1600-0404.2003.02067.x. [DOI] [PubMed] [Google Scholar]
- 75.Kikuchi M, Wada Y, Koshino Y, Nanbu Y, Hashimoto T. Effects of scopolamine on interhemispheric EEG coherence in healthy subjects: Analysis during rest and photic stimulation. Clin. EEG Electroencephalogr. 2000;31:109–115. doi: 10.1177/155005940003100210. [DOI] [PubMed] [Google Scholar]