Abstract
Numerous therapeutic compounds have been isolated from naturally abundant organic resources, which may offer economical and sustainable sources of compounds with safe and efficacious biological activities. In the cosmetics industry, natural compounds with anti-aging activities are eagerly sought. Thus, we prepared various extracts from Rubus fraxinifolius leaves and used enzyme inhibition assays to isolate compounds with protective effects against skin aging. Two triterpenoids were isolated from Rubus fraxinifolius Poir. leaves. The structures were characterized by spectroscopic analyses (LC-ESI-MS, 1D/2D NMR) and comparison to reported data. Compound 1 and 2 were determined as 2,3-O-ethyleneglycol, 19-hydroxyurs-12-en-23,28-dioic acid and 2,3-O-propanediol,19-hydroxyurs-12-en-28-oic acid. Methanol extract and isolates were assessed for their inhibitory effects on elastase and tyrosinase. Compounds 1 and 2 inhibited elastase with IC50 122.199 µg/mL and 98.22 µg/mL, and also inhibited tyrosinase with IC50 207.79 µg/mL and 221.51 µg/mL, respectively. The molecular docking proved that both compounds have affinities toward the enzymes.
Subject terms: Chemical biology, Drug discovery, Plant sciences
Introduction
Although challenging to achieve, natural anti-aging substances that are safe for humans can be economically and sustainably harvested from abundant natural resources. About 700 species of Rubus (Rosaceae) are distributed globally, even though few species are found in the tropics. This genus containing nutrients (sugar, vitamins, etc.), secondary metabolites (triterpenoids, flavonoids, polyphenols, etc.) and have many anti-aging activities (antioxidant, ant elastase, ant tyrosinase, anti-collagenase, anti-UV, anti-inflammatory, wound healing, etc.)1,2. In addition, Rubus species also reported containing diverse triterpenes with various biological activities3–7.
In Indonesia, R. fraxinifolius Poir. (Rosaceae) is distributed in Java, Borneo, etc., and popularly known as ‘arben.’ Moreover, some local farmers harvest these plants, and the berry fruit is usually consumed either fresh or frozen. Our previous study demonstrated that R. fraxinifolius stem extract has the activity to inhibit elastase, tyrosinase, and as an antioxidant8. Some other investigations also showed that the leaf and fruit of R. fraxinifolius have a potent antioxidant activity9,10. However, the study of R. fraxinifolius has been little researched, and no studies of chemical components have been reported.
Elastase is a serine protease enzyme, which has a crucial role in skin wrinkling or sagging through the degradation of dermal elastic fiber (elastin) and causes loss of skin elasticity. One of these is skin fibroblast-derived elastase. Thus, as moderators of elastin fiber degradation that causes skin aging, elastase inhibitors have attracted attention as agents for cosmetic preparations11,12. Melanogenesis is mediated by tyrosinase and regulates melanin biosynthesis through a two-stage reaction. In the initial step, L-tyrosine is hydroxylated to L-3,4-dihydroxyphenylalanine (L-DOPA). In the second step, L-DOPA is oxidized to the corresponding O-quinone. Therefore, natural compounds with tyrosinase inhibitory activity are commonly used in cosmetics that inhibit hyperpigmentation with melanin and hence favor skin whitening13. The elastase and tyrosinase enzyme inhibitors could be developed as skin whitening, anti-aging, or anti-wrinkle agents to treat dermatological disorders14.
This study isolated ursane triterpenoids from R. fraxinifolius leaves and elucidated their structures in spectroscopic analyses using electrospray ionization mass spectroscopy (ESI-MS) and 1H NMR, 13C NMR, and 2D NMR (DEPT, HSQC, HMQC, and HMBC). Subsequently, we performed assays of elastase and tyrosinase inhibitory activities of crude extracts, fractions, and isolated compounds.
The activity of two selected compounds was also observed through in silico method in this research. A molecular docking approach was performed with DockThor15,16. The macromolecules used in this research were obtained from protein data bank (RCSB PDB, 2000) with identity 2y9x and 3hgp for tyrosinase and elastase, respectively17,18. The macromolecules were optimized in UCSF Chimera and the molecular docking results were observed with PyMOL19,20.
Results and discussion
The ethyl acetate and methanolic extracts of R. fraxinifolius leaves showed potential as elastase inhibitors with percent inhibitory > 40% in 100 µg/mL, whereas n-hexane extract had no activity (Fig. 1). Some Rubus also showed elastase inhibitory activity such R. sanctus (% inhibition 14.68–49.20 in 100 µg/mL), R. compactus, R. robustus, etc.2,21. Hence, we fractionated the active extracts using vacuum liquid chromatography/VLC and collected 11 fractions from each. Fractions from ethyl acetate extract showed weak elastase inhibitory activities (< 20% in 100 µg/mL), but some methanol fractions showed potential activity in these assays. We choose methanol fraction 8 (M8) for further isolation because it had the largest yield (57%) and also have elastase inhibitory activity (44.82%). M8 was further partitioned and purified over silica gel and through a Sephadex column, and two amorphous powders were produced.
Figure 1.
Elastase inhibition by R. fraxinifolius leaf extracts and fractions.
Isolates were identified and characterized using liquid chromatography-mass spectroscopy (LCMS), 1H and 13C NMR, DEPT, heteronuclear single quantum coherence (HSQC), heteronuclear multiple quantum correlation (HMQC), and heteronuclear multiple bond correlation (HMBC). Table 1, showed the NMR spectral data of compounds.
Table 1.
1H and 13C NMR spectra for compounds 1 and 2.
| Compound 1 (500 MHz, in CD3OD) |
Compound 2 (500 MHz, in CD3OD) |
Tormentic acid22 (400 MHz, in CD3OD) |
|||||
|---|---|---|---|---|---|---|---|
| δ 13C | δ 1H | δ 13C | δ 1H | δ 13C | δ 1H | ||
| 1 | CH2 | 46.39 | 1.92 | 39.16 | 1.7 | 48.0 | 2.04 |
| 2 | CH | 68.53 | 3.85m | 78.49 | 3.35m | 68.6 | 3.62 ddd |
| 3 | CH | 78.18 | 3.47 (d,9.5 Hz) | 86.09 | 3.03 (d,10 Hz) | 83.8 | 2.91(d,9.8 Hz) |
| 4 | C | 41.34 | – | 43.17 | – | 41.1 | – |
| 5 | CH | 46.93 | 1.72 | 57.15 | 0.917 | 55.4 | 1.04 |
| 6 | CH2 | 18.40 | 1.53; 1.28 | 19.34 | 1.59; 1.25 | 19.1 | 1.59 |
| 7 | CH2 | 32.6 | 1.61 (m) | 33.65 | 1.95; 0.903 | 33.5 | 1.37/1.64 |
| 8 | C | 39.69 | – | 44.22 | – | 40.6 | – |
| 9 | CH | 47.50 | 1.43 | 47.96 | 1.43 | 47.9 | 1.82 |
| 10 | C | 37.49 | – | 42.7 | – | 39.9 | – |
| 11 | CH2 | 23.65 | 2.04; 1.79 | 23.87 | 2.04 | 24.2 | 2.04 |
| 12 | CH | 127.23 | 5.3 (t,7) | 129.36 | 5.37 | 128.2 | 5.28 (t,3.2) |
| 13 | C | 139.49 | – | 140.24 | – | 139.5 | – |
| 14 | C | 48.39 | – | 42.84 | – | 42.3 | – |
| 15 | CH2 | 28.49 | 1.83; 1.13 | 29.66 | 1.0, 1.76 | 29.3 | 1.02, 1.85 |
| 16 | CH2 | 25.59 | 1.61;1.36 | 34.5 | 1.28; 1,51 | 26.8 | 1.75, 1.28 |
| 17 | C | 49.83 | – | 48.34 | – | 48.6 | – |
| 18 | CH | 54.20 | 2.6s | 55.15 | 2.42s | 54.4 | 2.50s |
| 19 | C | 72.70 | – | 73.68 | – | 72.6 | – |
| 20 | CH | 41.73 | 1.68 | 41.18 | 1.68 | 42.2 | 1.37 |
| 21 | CH2 | 26.28 | 1.53;1.28 | 27.38 | 1.53;1.28 | 26.1 | 1.54, 2.59 |
| 22 | CH2 | 38.07 | 1.77; 2.02 | 26.69 | 2.55; 1.51 | 37.8 | 1.75 |
| 23 | C | 178.96 | – | 23.86 | 1.22s | 29.5 | 0.9s |
| 24 | CH3 | 22.63 | 1.89s | 17.46 | 0.76s | 16.8 | 1.01s |
| 25 | CH3 | 17.54 | 1.02s | 17.46 | 0.97s | 17.1 | 1.08s |
| 26 | CH3 | 17.86 | 0.82s | 13.96 | 0.69s | 17.8 | 0.82s |
| 27 | CH3 | 24.98 | 1.32s | 24.86 | 1.30s | 24.6 | 1.37s |
| 28 | C | 183.70 | – | 182.23 | – | 182.7 | – |
| 29 | CH3 | 27.36 | 1.20s | 27.17 | 1.17s | 27.0 | 1.2s |
| 30 | CH3 | 16.82 | 0.92 (d,6) | 16.69 | 0.91 (d,6) | 17.5 | 0.93 (d 6) |
| 31 | CH2 | 61.27 | 3.62 (d,1.15); 4.05 (d,11.5) | 66.54 | 3.22 (d,1.15); 3.42 | – | – |
| 32 | CH2 | 63.20 | 3.52 (d,11.5); 4.06 (d,11.5) | 20.05 | 1.4; 1.6 | – | – |
| 33 | CH2 | – | – | 66.31 | 4.02 (d,11.5); 3.36 | – | – |
Data were measured in CD3OD (500 MHz).
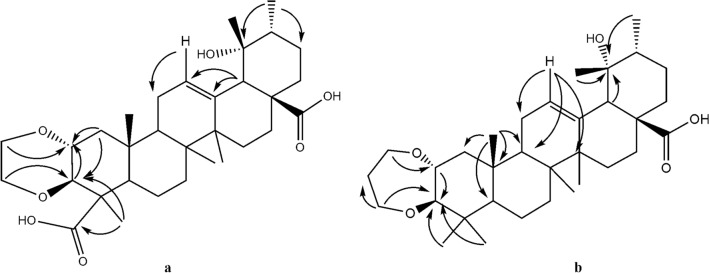
Compound 1: was isolated as an amorphous white powder. LCMS–ESI spectra for this compound displayed a molecular ion peak at m/z 543.32 [M-H]+, suggesting the molecular formula C32H48O7 with nine unsaturation equivalents. IR spectra revealed absorption maxima corresponding with hydroxyl (3.343.4 cm−1) and olefinic (1.684 cm−1) functional groups. Moreover, 1H NMR spectra of compound 1 revealed methine proton (CH) a singlet signal at δH 2.6, a characteristic signal for the H-18 of ursane-type triterpenes with 19-O-substitutions. The triterpene type 19α-hydroxy-ursane also has a typical proton signal in the area around δH 2.6 ppm. This deshielded signal can differ markedly from other ordinal methylene proton signals, with a characteristic shift due to an olefinic proton (H12) at δH 5.28 (t, J = 6 Hz). Complete and unequivocal 1H and 13C chemical shift assignments were assisted by HMQC (13C × 1H) and HMBC (13C × 1H) spectra. From the HMBC cross peak, the proton at δH 2.60 was confirmed to be H-16 (Fig. 2a). Other specific characters were found for the five methyl singlets at δH 1.89, 1.02, 0.82, 1.32, 1.20 (each, H24-27; 29), a methyl doublet at δH 0.92 (d, J = 6 Hz), and the olefinic proton signal at δH 5.30 (t, J = 7 Hz, H-12). The present 13C NMR spectra show 32 carbon resonances, and with DEPT and HMQC spectra, displayed two carbonyl carbons (δ C 183.70, C-28 and δC 178.96, C-23), an olefinic carbon at δC 127.23, C-12, an olefinic quaternary carbon (δC 139.49, C-13), an oxygen-bearing quaternary carbon (δC 72.7, C-19), ten aliphatic methylenes and six methyl carbons. The two olefinic carbons at δC 127.23 (C-12) and 139.49 (C-13) were characteristic of the ursane-triterpenoid type. Another signal indicated the presence of ethylene glycol (-OCH2-CH2O-) was showed at signals at δH 3.62 (d, J = 11.5 Hz), 4.05 (d, J = 11.5 Hz), 3.52 (d, J = 11.5 Hz) and 4.06 (d, J = 11.5 Hz) and supported with δC 61.27 (t) and 63.20 (t). It was located at C-2 and C-3 based on the presence of long-range correlation in the HMBC spectra. Hence, compound 1 was identified as 2,3-O-ethylene glycol, 19-hydroxyurs-12-en-23,28-dioic acid (Fig. 3a).
Figure 2.
Key HMBC correlations. a = compound 1; b = compound 2.
Figure 3.
Structures of compounds 1 and 2. a = 2,3-O-ethylene glycol, 19-hydroxyurs-12-en-23,28-dioic acid; b = 2,3-O-propanediol, 19-hydroxyurs-12-en-28-oic acid.
Compound 2: was obtained as a white amorphous powder. MS–ESI m/z 527.33 [M-H]+ (calcd. For 528.3815). In LCMS-ESI spectra, a molecular ion peak is present at m/z 527.33 [M-H]+, indicating the molecular formula C33H52O5, which required 8 degrees of unsaturation. The IR spectrum contained absorption maxima corresponding with hydroxyl (2,927.8 cm−1) and olefinic (1,686 cm−1) functional groups. Moreover, 1H NMR spectra for compound 2 showed a singlet at δH 2.42, which is characteristic of the H18 of a ursane-type triterpene with 19-O-substitution. Other specific characteristic spectra included the presence of six methyl singlets at δH 1.22, 0.76, 0.97, 0.69, 1.30 and 1.17 (each, H-24–28, H-30), a methyl doublet at 0.91 (d, J = 7 Hz), and the olefinic proton signal at δH 5.37 (d, J = 7 Hz, H-12). Corresponding 13C NMR and DEPT spectra show 33 carbon resonances, and with HSQC and HMQC spectra indicate the presence of a carbonyl carbon (δC 182.23, C-28), an olefinic carbon (δC 129.36, C-12), an olefinic quaternary carbon (δC 140.24, C-13), an oxygen-bearing quaternary carbon (δC 73.68, C-19), eleven aliphatic methylenes, and seven methyl carbons. These data indicated that compound 2 carries a ursane-triterpenoid skeleton. The other signals are indicated the presence of 1,3-propanediol functional group, at δH 3.22 (d, J = 11.5 Hz), 3.42 (d, J = 10.5 Hz), 1.38 (m), 1.51 (m) and 4.02 (d, J = 11.5 Hz) and 3.36 (t, J = 10.5 Hz). Its functional group has any long-range correlation with C2-C3. Based on these results, compound 2 was thus identified as 2,3-O-Propanediol, 19-hydroxyurs-12-en-28-oic acid (Figs. 2b and 3b). A similar compound is 2,3-O-Isopropylidene tormentic acid, isolated from Rubus xanthocarpus.7.
In Table 1, all 1H and 13C NMR spectra were compared with tormentic acid (TA/2α,3α, 19-trihydroxy-12-ursen-28-oic acid)22, which is a ursane triterpenoid that has been found in several Rubus species4,6,23–25. Tormentic acid had strong similarities with compounds 1 and 2, except that chemical shifts related to C-23 and C31-33 differed. This moiety was also confirmed in DEPT, HMQC, and HMBC experiments (Fig. 2).
The triterpenoid tormentic acid is widely distributed in natural plant foods. It has various bioactivities: hypoglycaemic effects, anti-inflammatory, and anti-atherogenic properties, reduced vascular smooth muscle cell proliferation, antiproliferative activities in renal, prostate, and melanoma cancer cell lines24,26. Therefore, because these compounds have the same skeleton, they may have possible appropriate potential activities.
Figure 4, represents the inhibitory elastase and tyrosinase activity of isolates. The data obtained from in vitro enzyme inhibition assays were expressed as the standard deviation (SD). Compounds 1 and 2 inhibited elastase with IC50 122.199 and 98.22 µg/mL, and also inhibited tyrosinase with IC50 207.79 and 221.51 µg/mL, respectively. Some pentacyclic triterpenoid (ursolic acid and oleanolic acid) reported having elastase inhibition activity27,28. The IC50 elastase inhibition value of compound 2 is lower than that of compound 1. Both compounds were less inhibition activity than the positive control oleanolic acid, which had an IC50 value of 90.39 µg/mL. In agreement, previous studies showed an IC50 for oleanolic acid of 76.5 μg/mL and an IC50 value of 31.0 μg/mL for ursolic acid28. Previously reported kinetic analyses of pentacyclic triterpenes showed that these compounds competitively and reversibly inhibit neutrophil elastase. In the same study, molecular docking experiments showed that the molecular scaffolding moiety 28-COOH and double bonds in pentacyclic triterpenes are essential for their inhibitory activities29.
Figure 4.
Anti elastase and anti-tyrosinase activities of extracts and isolates from R. fraxinifolius leaves.
One of the problems that arise with increasing age is hyperpigmentation. Hence, there is an ongoing search for skin-lightening agents or new depigmenting. Suppression of tyrosinase can act against melanogenesis30. As shown in Fig. 4, the methanol extract and compounds 1 and 2 demonstrated moderate activities as tyrosinase inhibitors.
In this research, molecular docking was also used to analyze the binding activities of selected compounds to tyrosinase and elastase as their targets. Crystal structures used were 2Y9X, a crystal structure of tyrosinase from Agaricus bisporus with inhibitor tropolone; and 3HGP, a crystal structure of porcine pancreatic elastase complexed with a potent peptidyl inhibitor FR130180. Both were selected because they were obtained from the same organism used for the in vitro assay of this research. The macromolecules are also bound to their respective inhibitors so that the active state of the enzyme conformations could be obtained. The cocrystals were used as the center of molecular docking target to narrow down the compound binding probabilities so that the scoring process becoming more efficient.
From the molecular docking, average binding affinities were obtained for both compounds as shown in Figs. 5 and 6. The affinity prediction is used to rank different ligands considering the top energy pose of each compound, a prediction of better energy pose is shown by lower binding affinities scores16. The redocking of designated inhibitor tropolone was done with average binding affinities − 7.41 kcal/mol. The average binding affinities of compounds 1 and 2 to tyrosinase were − 7.84 kcal/mol and − 8.37 kcal/mol, respectively (Table 2). Both compounds were predicted to have a better affinity than the inhibitor.
Figure 5.
Visualization of elastase binding site using PyMOL. Blue = cocrystal; green = compound 1; yellow = compound 2.
Figure 6.
Visualization of tyrosinase binding site using PyMOL. Magenta = cocrystal; green = compound 1; yellow = compound 2.
Table 2.
Binding score of compounds to tyrosinase (PDBID 2Y9X) and elastase (PDBID 3HGP), evaluated using DockThor.
| Compound | Average binding score to tyrosinase (kcal/mol) | Average binding score to elastase (kcal/mol) |
|---|---|---|
| Compound 1 | − 7.84 | − 7.58 |
| Compound 2 | − 8.37 | − 8.06 |
| 0TR (2Y9X cocrystal) | − 7.41 | |
| FRW (3HGP cocrystal) | − 7.84 |
Meanwhile, the average binding affinities of elastase redocking was − 7.84 kcal/mol. The average binding affinities of compounds 1 and 2 to elastase were − 7.58 kcal/mol and -8.06 kcal/mol, respectively. Even though the result showed that compound 1 has lower affinities than the cocrystal, it is slightly different (< 0.5 kcal/mol) compared to the inhibitor used hence the scores may overlap31. These scores showed that both compounds were predicted to have affinities toward the enzymes, which is in conjunction with the in vitro assay results.
Methods
General experimental procedures
1H and 13C NMR spectra were recorded at 500 MHz using a JEOL JNM-ECZ500R/S1 instrument. Infrared (IR) spectra were measured with an FTIR, IRPrestige-21, Shimadzu. Other analyses were performed using a Waters UPLC-MS XEVO G2-XS QTof instrument, a VersaMax Microplate reader and a BioTek ELX800 Microplate Reader, pre-coated aluminum sheets TLC-Silica gel 60 GF254 (Merck, Darmstadt, Germany), column chromatography (CC) was performed using Silica gel 60 (Merck, Darmstadt, Germany) with 70–230 mesh for open chromatography and 230–400 mesh for vacuum chromatography.
Plant materials
R. fraxinifolius leaves were collected from plantation area in mount Pangrango, West Java at the height of 4,343 ft, in December 2018. The specimen was identified by a botanist (Dr. Joni Setijo) at the Research Centre for Biology, Indonesian Institute of Sciences, Indonesia, with specimen number 033/IPH.1.01/If.07. The collection of plant material had received permission from farmer and complied with institutional regulations and guidelines.
Extraction and isolation
Air-dried powdered leaves (2300 g) were extracted using a Soxhlet apparatus with gradient solvent (n-hexane, EtOAc, and MeOH) to provide the respective extracts, then evaporated with a rotary evaporator and vacuum oven. The methanol extract (291 g) and the ethyl acetate extract (65 g) were adsorbed on the silica gel and performed vacuum liquid chromatography/VLC, eluting with a stepwise gradient of EtOAc: MeOH (from 1:0 to 0:1) to produce 11 fractions for each extract (Ea1–Ea11 and M1–M11 and). Similar fractions which had positive reactions with vanillin sulfuric reagents in TLC were combined. Fraction yield: Ea1-3 (2.27 g); Ea4-6 (8.47 g); Ea7-8 (12.9 g); Ea9-11 (22.3 g); and M1-3 (4.89 g); M4-5 (5.1 g); M6-7 (6.02 g); M8 (166.42 g); M9 (10.53 g); M10-11 (2.9 g). All fractions were submitted for inhibitor elastase activity, Fr. M8 gave the greatest yield and had potent activity. Fr. M8 successively further partitioning by CC over silica gel (eluent CH2Cl2/MeOH with a stepwise gradient) and purified using Sephadex LH-20 column (eluting with CHCl3–MeOH 100:10, v/v. Two fractions showed solid nature and were crystallized with chloroform and methanol to get two isolates: compound 1 (18 mg) and 2 (31 mg). The purity of all the isolates was evaluated by two-dimensional TLC and visualize the spot using 5% sulphuric acid in methanol, followed by heating the plates at 110 °C for 5 min.
The crystals were identified and characterized using liquid chromatography-mass spectroscopy (LCMS), 1H, and 13C distortions enhancement by polarization transfer (DEPT) NMR, heteronuclear single quantum coherence (HSQC), heteronuclear multiple quantum correlation (HMQC), and heteronuclear multiple bond correlation (HMBC).
3-O-ethylene glycol, 19-hydroxyurs-12-en-23,28-dioic acid
White amorphous powder; 1H NMR (500 MHz, in CD3OD), 13C NMR (500 MHz, in CD3OD) spectroscopic data see Table 1; MS–ESI m/z 543.32 [M-H]+ (calcd for C32H48O7).
3-O-Propanediol, 19-hydroxyurs-12-en-28-oic acid
White amorphous powder; 1H NMR (500 MHz, in CD3OD), 13C NMR (500 MHz, in CD3OD) spectroscopic data see Table 1; MS–ESI m/z 527.33 [M-H]+ (calcd for C33H52O5).
Elastase inhibition assay
Elastase inhibition assay was performed as described previously with some modifications32. Briefly, in Nunc-96 well microtiter plates, 20-µL aliquots of 0.8-units/mL PPE in Trizma® base buffer (pH 8.0) were mixed with 20-µL samples, and the mixtures were then diluted to 180 µL in Trizma® base buffer. Test extracts were preincubated with enzyme for 15 min, and 20-μL aliquots of the substrate N-succinyl-Ala-Ala-Ala-p-nitroanilide (A3PVN; 2.9 mM) were added and incubated for another 15 min. Positive control and blank wells contained oleanolic acid and water, respectively. Experiments were conducted in triplicate, and inhibition rates were determined according to absorbance at 401 nm using a VersaMax microplate reader.
Percentage inhibition was calculated using the following equation:
where E is the absorbance of the enzyme reaction, Eb is the absorbance of the enzyme blank, T is the absorbance of the test sample, and Tb is the absorbance of the test blank. IC50 values were determined from a linear graph of percent elastase inhibition against concentration (50, 75, 100,125,150 µg/mL).
Tyrosinase inhibition assay
Tyrosinase inhibition was determined using the DOPA-chrome formation method as described previously with slight modifications13. Briefly, in 96-well plates, 20 μL aliquots of DMSO (control) or test compounds at varying concentrations were mixed with 40 μL aliquots of 30 U/mL mushroom tyrosinase (Sigma Aldrich) and 100-μL of 0.1-M phosphate buffer (pH 6.8). They were preincubated for 10 min at room temperature. Reactions were initiated by adding 4 μL aliquots of 10 mM L-DOPA to each well and incubating at 37 °C for 20 min. Tyrosinase activity was then determined by measuring absorbance at 475 nm. Kojic acid was used as a positive control. Experiments were performed in triplicate. Percentage of tyrosinase inhibition was calculated using the following equation:
where E is the absorbance of the enzyme reaction, Eb is the absorbance of the enzyme blank, T is the absorbance of the test sample, and Tb is the absorbance of the test blank. IC50 values were determined from a linear graph of percent elastase inhibition against concentration (250, 125, 62.5, 31.25, 15.6 µg/mL).
Molecular docking
In this research, in silico pharmacological activity was predicted with molecular docking using DockThor. Targeted molecular docking was done by using a cocrystallized ligand as the center of the active site. Structure 2Y9X was used for tyrosinase molecular docking with tropolone, an inhibitor of mushroom tyrosinase, as its cocrystal. The center of the molecular docking site is defined at − 10.032; − 28.769 and − 43.467 as X, Y, Z dimensions respectively. Chain A was separated for use and the Holmium atom was removed from the structure using UCSF Chimera.
The structure of 3HGP, a porcine elastase was also used in this research. FR130180, as the cocrystal was used as the center of the molecular docking site. The coordinates were 12.453, 9.237, and 1.199 for X, Y, Z dimensions, respectively. The binding affinity was analyzed from the ten best conformers of each molecular docking calculation.
Supplementary Information
Acknowledgements
This research was funded by the “Publikasi Terindeks Internasional (PUTI) Doktor Universitas Indonesia 2020” No: NKB-525/UN2.RST/HKP.05.00/2020NKB-525/UN2.RST/HKP.05.00/2020.
Author contributions
B.E. and Y.D. conceived the experiments; Y.D., E.A.R., and R.R.S. conducted the experiments; Y.D., M.H. and F.C.S. analysed the results. All authors reviewed the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-99970-x.
References
- 1.Zeng HJ, Liu Z, Wang YP, Yang D, Yang R, Qu LB. Studies on the anti-aging activity of a glycoprotein isolated from Fupenzi (Rubus chingii Hu.) and its regulation on Klotho Gene Expression in Mice Kidney. Int. J. Biol. Macromol. 2018;119:470–476. doi: 10.1016/j.ijbiomac.2018.07.157. [DOI] [PubMed] [Google Scholar]
- 2.Bravo K, Alzate F, Osorio E. Fruits of selected wild and cultivated andean plants as sources of potential compounds with antioxidant and anti-aging activity. Ind. Crops Prod. 2016;85(February):341–352. doi: 10.1016/j.indcrop.2015.12.074. [DOI] [Google Scholar]
- 3.Ono M, Yasuda S, Nishi K, Yamamoto K, Fuchizaki S, Higuchi S, Komatsu H, Okawa M, Kinjo J, Yoshimitsu H, Nohara T. Two new triterpenoids from the seeds of blackberry (Rubus fructicosus) Nat. Prod. Res. 2016;30(8):904–911. doi: 10.1080/14786419.2015.1076820. [DOI] [PubMed] [Google Scholar]
- 4.Mei Q, Chen X, Xia X, Fang Z, Zhou H, Gao Y, Dai W, Jiang R. Isolation and chemotaxonomic significance of chemical constituents from Rubus parvifolius. Chin. Herb. Med. 2016;8(1):75–79. doi: 10.1016/S1674-6384(16)60011-4. [DOI] [Google Scholar]
- 5.Yu G, Luo Z, Wang W, Li Y, Zhou Y, Shi Y. Rubus chingii Hu: a review of the phytochemistry and pharmacology. Front. Pharmacol. 2019;10:799. doi: 10.3389/fphar.2019.00799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanegusuku M, Sbors D, Bastos E. Phytochemical and analgesic activity of extract, fractions and a 19-hydroxyursane-type triterpenoid obtained from Rubus rosaefolius (Rosaceae) Biol. Pharm. Bull. 2007;30(5):999–1002. doi: 10.1248/bpb.30.999. [DOI] [PubMed] [Google Scholar]
- 7.Li BZ, Wang BG, Jia ZJ. Pentacyclic triterpenoids from Rubus xanthocarpus. Phytochemistry. 1998;49(8):2477–2481. doi: 10.1016/S0031-9422(98)00161-7. [DOI] [Google Scholar]
- 8.Desmiaty Y, Saputri FC, Hanafi M, Prastiwi R, Elya B. Anti-elastase, anti-tyrosinase, and anti-oxidant of Rubus fraxinifolius stem methanolic extract. Pharmacogn. J. 2020;12(2):271–275. doi: 10.5530/pj.2020.12.42. [DOI] [Google Scholar]
- 9.Galvez MAC. Evaluation of DPPH free radical scavenging activity and phytochemical screening of selected folkloric medicinal plants in Tinoc, Ifugao, Cordillera Administrative Region, Philippines. Int. J. Sci. Res. Publ. 2014;5(12):440–445. [Google Scholar]
- 10.Bakar MFA, Ismail NA, Isha A, Ling ALM. Phytochemical composition and biological activities of selected wild berries (Rubus moluccanus L., R. fraxinifolius Poir, and R. alpestris Blume) Evidence Based Complement. Altern. Med. 2016;2016(November):1–10. doi: 10.1155/2016/2482930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imokawa G, Nakajima H, Ishida K. Biological mechanisms underlying the ultraviolet radiation-induced formation of skin wrinkling and sagging II: over-expression of Neprilysin plays an essential role. Int. J. Mol. Sci. 2015;16(4):7776–7795. doi: 10.3390/ijms16047776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pientaweeratch S, Panapisal V, Tansirikongkol A. Antioxidant, anti-collagenase and anti-elastase activities of Phyllanthus emblica, Manilkara zapota and Silymarin: an in vitro comparative study for anti-aging applications. Pharm. Biol. 2016;54(9):1865–1872. doi: 10.3109/13880209.2015.1133658. [DOI] [PubMed] [Google Scholar]
- 13.Tu PTB, Tawata S. Anti-oxidant, anti-aging, and anti-melanogenic properties of the essential oils from two varieties of alpinia zerumbet. Molecules. 2015;20(9):16723–16740. doi: 10.3390/molecules200916723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiocchio I, Mandrone M, Sanna C, Maxia A, Tacchini M, Poli F. Screening of a hundred plant extracts as tyrosinase and elastase inhibitors, two enzymatic targets of cosmetic interest. Ind. Crops Prod. 2018;122(February):498–505. doi: 10.1016/j.indcrop.2018.06.029. [DOI] [Google Scholar]
- 15.Guedes IA, Barreto AMS, Marinho D, Krempser E, Kuenemann MA, Sperandio O, Dardenne LE, Miteva MA. New machine learning and physics-based scoring functions for drug discovery. Sci. Rep. 2021;11(1):1–19. doi: 10.1038/s41598-021-82410-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santos KB, Guedes IA, Karl ALM, Dardenne LE. Highly flexible ligand docking: benchmarking of the dockthor program on the LEADS-PEP protein-peptide data set. J. Chem. Inf. Model. 2020;60(2):667–683. doi: 10.1021/ACS.JCIM.9B00905. [DOI] [PubMed] [Google Scholar]
- 17.Tamada, T.K.T. . K. R. and T. T. Structure of porcine pancreatic elastase complexed with a potent peptidyl inhibitor FR130180 determined by high resolution crystallography https://www.google.com/search?q=Tamada%2C+T.%2C+Kinoshita%2C+T.%2C+Kuroki%2C+R.+and+Tada%2C+T.%2C+2009.+Structure+of+porcine+pancreatic+elastase+complexed+with+a+potent+peptidyl+inhibitor+FR130180+determined+by+high+resolution+crystallography.&oq=Tamada%2C+T.%2C+Kinoshita%2C+T.%2C+Kuroki%2C+R.+and+Tada%2C+T.%2C+2009.+Structure+of+porcine+pancreatic+elastase+complexed+with+a+potent+peptidyl+inhibitor+FR130180+determined+by+high+resolution+crystallography.&aqs=chrome.0.69i59.1149j0j9&sourceid=chrome&ie=UTF-8. Accessed Aug 26, 2021.
- 18.Ismaya WT, Rozeboom HJ, Weijn A, Mes JJ, Fusetti F, Wichers HJ, Dijkstra BW. Crystal structure of agaricus bisporus mushroom tyrosinase: identity of the tetramer subunits and interaction with tropolone. Biochemistry. 2011;50(24):5477–5486. doi: 10.1021/BI200395T. [DOI] [PubMed] [Google Scholar]
- 19.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25(13):1605–1612. doi: 10.1002/JCC.20084. [DOI] [PubMed] [Google Scholar]
- 20.Schrödinger, LLC. 2010. The PyMOL Molecular Graphics System, Version 2.0 - Google Search https://www.google.com/search?q=Schrödinger%2C+LLC.+2010.+The+PyMOL+Molecular+Graphics+System%2C+Version+2.0&oq=Schrödinger%2C+LLC.+2010.+The+PyMOL+Molecular+Graphics+System%2C+Version+2.0&aqs=chrome..69i57.1037j0j4&sourceid=chrome&ie=UTF-8. Accessed Aug 26, 2021.
- 21.Akkol EK, Süntar I, Ilhan M, Aras E. In vitro enzyme inhibitory effects of Rubus Sanctus Schreber and its active metabolite as a function of wound healing activity. J. Herb. Med. 2015;5(4):207–210. doi: 10.1016/j.hermed.2015.09.002. [DOI] [Google Scholar]
- 22.Tanaka JCA, Vidotti GJ, Da Silva CC. A new tormentic acid derivative from luehea divaricata Mart. (Tiliaceae) J. Braz. Chem. Soc. 2003;14(3):475–478. doi: 10.1590/S0103-50532003000300024. [DOI] [Google Scholar]
- 23.Chen Z, Tong L, Feng Y, Wu J, Zhao X, Ruan H, Pi H, Zhang P. Ursane-type nortriterpenes with a five-membered a-ring from rubus innominatus. Phytochemistry. 2015;116(1):329–336. doi: 10.1016/j.phytochem.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 24.Petreanu M, Ferreira EK, Sagaz APM, Vendramini-Costa DB, Ruiz ALTG, De Carvalho JE, Campos A, Cechinel Filho V, Delle Monache F, Niero R. Uncommon trimethoxylated flavonol obtained from rubus rosaefolius leaves and its antiproliferative activity. Evidence-based Complement. Altern. Med. 2015;2015:6–11. doi: 10.1155/2015/341216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang H, Han H, Man WJ, Hou AJ, Guo XY, Xing XD, Yan ML, Yang L, Yang L. Ursane-type triterpenoids from the roots of rosa multiflora with their anti-inflammatory activity. J. Asian Nat. Prod. Res. 2020;22(2):131–137. doi: 10.1080/10286020.2018.1541135. [DOI] [PubMed] [Google Scholar]
- 26.Yang Y, Wang Y, Wang Y, Zhao M, Jia H, Li B, Xing D. Tormentic acid inhibits IL-1β-induced inflammatory response in human osteoarthritic chondrocytes. Inflammation. 2016;39(3):1151–1159. doi: 10.1007/s10753-016-0349-8. [DOI] [PubMed] [Google Scholar]
- 27.Nema NK, Maity N, Sarkar BK, Mukherjee PK. Matrix metalloproteinase, hyaluronidase and elastase inhibitory potential of standardized extract of centella asiatica. Pharm. Biol. 2013;51(9):1182–1187. doi: 10.3109/13880209.2013.782505. [DOI] [PubMed] [Google Scholar]
- 28.Lee KK, Cho JJ, Park EJ, Choi JD. Anti-elastase and anti-hyaluronidase of phenolic substance from areca catechu as a new anti-ageing agent. Int. J. Cosmet. Sci. 2001;23(6):341–346. doi: 10.1046/j.0412-5463.2001.00102.x. [DOI] [PubMed] [Google Scholar]
- 29.Feng L, Liu X, Zhu W, Guo F, YingchunWu S, Wang R, Chen K, Huang C, Li Y. Inhibition of human neutrophil elastase by pentacyclic triterpenes. PLoS ONE. 2013;8(12):e82794. doi: 10.1371/journal.pone.0082794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dzialo M, Mierziak J, Korzun U, Preisner M, Szopa J, Kulma A. The potential of plant phenolics in prevention and therapy of skin disorders. Int. J. Mol. Sci. 2016;17(2):1–42. doi: 10.3390/ijms17020160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castro-Alvarez A, Costa AM, Vilarrasa J. The Performance of Several Docking Programs at Reproducing Protein–Macrolide-Like Crystal Structures. Mol. 2017;22(1):136. doi: 10.3390/MOLECULES22010136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thring TSA, Hili P, Naughton DP. Anti-collagenase, anti-elastase and anti-oxidant activities of extracts from 21 plants. BMC Complement. Altern. Med. 2009;9(1):27. doi: 10.1186/1472-6882-9-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.