Abstract
Gastric cancer (GC) continues to be the most common gastrointestinal malignancy in China, and tumor metastases are a major reason for poor prognosis. Circular RNAs (circRNAs) are an intriguing type of noncoding RNAs with important regulatory roles. However, the roles of circRNAs in GC metastasis have not been fully elucidated. Here, we reported that circ-transportin 3 (TNPO3) was significantly downregulated in 103 pairs of GC tissues compared with matched noncancerous tissues. The level of circ-TNPO3 expression correlated with differentiation of GC, and plasma circ-TNPO3 could serve as a potential diagnostic biomarker. Functionally, circ-TNPO3 inhibited proliferation and migration of GC in vitro and in vivo. We further verified that circ-TNPO3 competitively interacted with insulin-like growth factor 2 binding protein 3 (IGF2BP3) protein; thus, the role of IGF2BP3 in stabilizing MYC mRNA was weakened, which inhibited the expression of MYC and its target SNAIL. Taken together, circ-TNPO3 acts as a protein decoy for IGF2BP3 to regulate the MYC-SNAIL axis, thereby suppressing the proliferation and metastasis of GC. Therefore, circ-TNPO3 has the potential to serve as a therapeutic target for GC.
Keywords: circ-TNPO3, gastric cancer, IGF2BP3, MYC, SNAIL, protein decoy
Graphical abstract
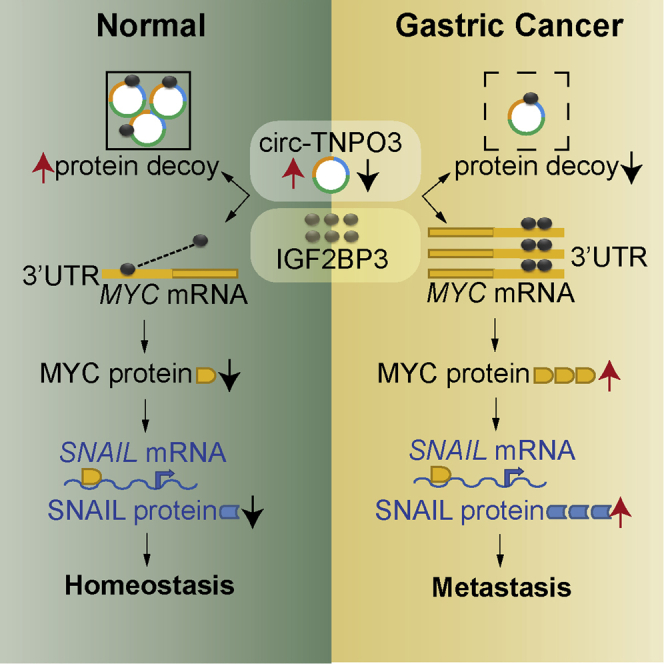
circ-TNPO3 acts as a protein decoy for IGF2BP3 to inhibit the expression of MYC and SNAIL. In GC, the downregulated circ-TNPO3 weakens its interaction with IGF2BP3 and promotes the expression of MYC and its target gene SNAIL, thereby resulting in GC metastasis.
Introduction
Gastric cancer (GC) continues to be one of the most commonly diagnosed and life-threatening malignant tumors worldwide. The incidence and mortality of GC occupy the second place of all cancers in China.1 Although early diagnosis and treatment methods of GC have improved greatly in recent years, patients still have a poor prognosis, predominantly due to the frequent presence of advanced-stage disease with lymphatic or distant metastases.2 Thus, discovery of novel therapeutic targets is urgently needed to improve the treatment and prognosis of GC patients.
Circular RNAs (circRNAs) are produced from pre-mRNA back splicing. They are evolutionarily conserved; they have tissue-specific expression and are more stable than linear mRNAs.3, 4, 5 Emerging evidence has demonstrated that circRNAs are abnormally expressed in multiple cancers and could act as vital regulators in carcinogenesis.6 Functionally, a number of studies have shown that circRNAs can regulate biological behaviors of cancer cells by serving as competitive endogenous RNAs (ceRNAs), microRNA (miRNA) sponges, protein baits or antagonists, RNA splicing regulators, parental genes transcriptional regulators, or even protein translation templates.7, 8, 9, 10
In GC, some differentially expressed circRNAs have been found and verified to play multiple roles in cells chemoresistance, proliferation, migration, and metastasis.7 However, most circRNAs are still largely uncharacterized, and the exact mechanisms of circRNAs’ involvement in GC development remain to be further studied.
RNA-binding protein IGF2BP3 (insulin-like growth factor 2 binding protein 3, also known as IMP3) has multiple post-transcriptional roles. In GC, IGF2BP3 promotes cancer cells proliferation and invasion, and it functions as an oncogene.11 MYC, a well-known oncogenic transcription factor, is frequently upregulated and contributes to tumorigenesis of GC.12,13 circ-Amotl1 can promote tumor development through inducing MYC nuclear translocation and promoting MYC stability.14 SNAIL is one of the most important epithelial-mesenchymal transition (EMT)-related transcription factors. It functions as a transcriptional repressor of E-cadherin and initiates the EMT process. In our previous studies, SNAIL has been shown to act as a promoter of GC progression.15,16
Here, based on the results of circRNA arrays, we identified that has_circ_0001741 (termed as circ-TNPO3) deriving from the linear gene TNPO3 (transportin 3) was poorly expressed in GC. We found that the expression of circ-TNPO3 reversely correlated with the degree of GC differentiation. circ-TNPO3 inhibited the tumorigenesis and metastasis of GC cells by interacting with RNA binding protein IGF2BP3 and inhibiting MYC and SNAIL signaling. These results indicate that circ-TNPO3 is a novel diagnostic and therapeutic target for GC.
Results
Expression profile of circRNAs in GC tissues and the validation, expression analysis, and characteristics of circ-TNPO3
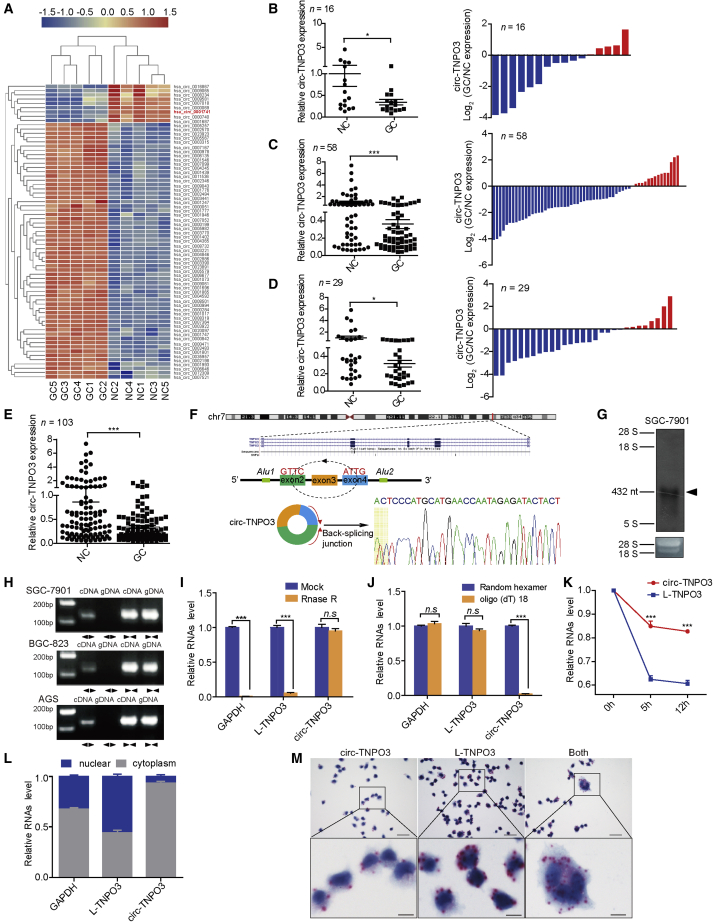
To characterize the circRNA expression profile in GC tissues, we performed circRNA microarray to compare five pairs of GC tissues and matched noncancerous (NC) tissues. We identified a circRNA signature in GC tissues with 184 upregulated and 113 downregulated circRNAs (|fold changes| > 1.5 and p < 0.05; Figure S1A; Table S1). We then further selected candidate circRNAs that satisfied the following criteria: (1) relatively high expression level of circRNAs in different cell lines or tissues (comparing and synthesizing our data with the data from Circbase) and (2) for functional research, inclusion of circRNAs with 300–6,000 bp in length. As a result, 60 upregulated and nine downregulated circRNAs fulfilled the abovementioned criteria and were included for further validation (Figure 1A; Table S2). Considering that, for some circRNAs with highly homologous sequences, it was hard to design appropriate divergent primers, 30 circRNAs were finally selected for additional verification. We performed qRT-PCR in an independent training phase consisting of 16 pairs of GC and matched NC tissues. We found that the level of circ-TNPO3 (hsa_circ_0001741) was lower in GC tissues (|fold change| = 0.34; p < 0.05; Figure 1B), whereas there was no difference in the expression of other circRNAs (Figure S1B). As shown in Figure 1B, the decrease in circ-TNPO3 was found in 11 of 16 GC tissue samples (68.8%) compared with the corresponding NC tissue samples.
Figure 1.
The level and characteristics of circ-TNPO3 in GC tissues or cells
(A) A heatmap shows the differential circRNA profiles in human GC tissues. (B–D) (Left) Levels of circ-TNPO3 in paired samples of GC from cohorts 1–3 are shown. (Right) Histograms indicate upregulation (red) or downregulation (blue) of circ-TNPO3 in GC tissues. (E) circ-TNPO3 expression in all 103 paired samples of GC is shown. (F) The genomic loci of circ-TNPO3 in TNPO3 gene are shown. Sanger sequencing of PCR product is shown. Arrows represent divergent primers of circ-TNPO3. (G) Northern blot shows the endogenous existence of circ-TNPO3 in SGC-7901 cells. (H) Existence of circ-TNPO3 in cDNA or genomic DNA (gDNA) of GC cells is shown. (I and J) Expression of circ-TNPO3, L-TNPO3, and GAPDH in total RNA treatment with RNase R (I) or reverse transcribed by random hexamer or oligo (dT)18 primer (J) is shown. (K) Stabilities of circ-TNPO3 and L-TNPO3 in SGC-7901 cells treated with actinomycin D are shown. (L) Expression of circ-TNPO3 and L-TNPO3 in nuclear and cytoplasmic fractions of SGC-7901 cells is shown. (M) Probes targeting circTNPO3 (left), L-TNPO3 (middle), or same sequences of the two genes (right) showed the location of circ-TNPO3, L-TNPO3, or total circ-TNPO3 + L-TNPO3 in SGC-7901 cells. The red dots represent positive signals; scale bars, upper 60 μm, lower 10 μm. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Next, we evaluated the expression of circ-TNPO3 in an independent validation set (n = 58). Consistent with the training phase, the expression of circ-TNPO3 obviously decreased in 75.9% of the GC tissues compared with NC tissues (|fold change| = 0.36; p < 0.001; Figure 1C). To exclude the differences caused by sample sources, another cohort, including 29 pairs of GC tissues and NC tissues from different areas and hospitals, was enrolled. As shown in Figure 1D, the consistent downregulation of the circ-TNPO3 in GC tissues was validated (|fold change| = 0.31; p < 0.05). In summary, through qRT-PCR of a total of 103 pairs of GC and NC tissues, we confirmed that circ-TNPO3 was poorly expressed in GC (Figure 1E).
circ-TNPO3 consists of three exons (432 nt) derived from TNPO3 gene. We confirmed the back-spliced junction of circ-TNPO3 by qRT-PCR with divergent primers consecutively with Sanger sequencing (Figure 1F). Existence of circ-TNPO3 was observed at 432 nt in northern blot assay using probe against the back-splicing junction site (Figure 1G). Only divergent primers, but not convergent primers, were able to amplify circ-TNPO3 in cDNA, but not in genomic DNA (gDNA) of SGC-7901, BGC-823, and AGS cell lines (Figure 1H). RNase R assay showed that circ-TNPO3 was resistant to RNase R, and linear TNPO3 (L-TNPO3) was susceptible to RNase R digestion, confirming that circ-TNPO3 was circular (Figure 1I). Random hexamer primers, rather than oligo (dT)18 primers, amplified intact circ-TNPO3 (Figure 1J). Furthermore, we used actinomycin D to inhibit transcription and then analyzed the levels of circ-TNPO3 and L-TNPO3. As shown in Figure 1K, circ-TNPO3 was more stable than L-TNPO3. Next, qRT-PCR of circ-TNPO3 in nuclear and cytoplasmic fractions (Figure 1L) and in situ hybridization (ISH) against circ-TNPO3 in GC cells (Figure 1M) or gastric tissues (Figure S2) demonstrated that circ-TNPO3 was predominately localized in the cytoplasm. Collectively, the above data show that circ-TNPO3 is a circular and stable transcript that is significantly downregulated in GC.
Level of circ-TNPO3 significantly correlates with differentiation of GC, and circ-TNPO3 is a potential diagnostic biomarker of GC
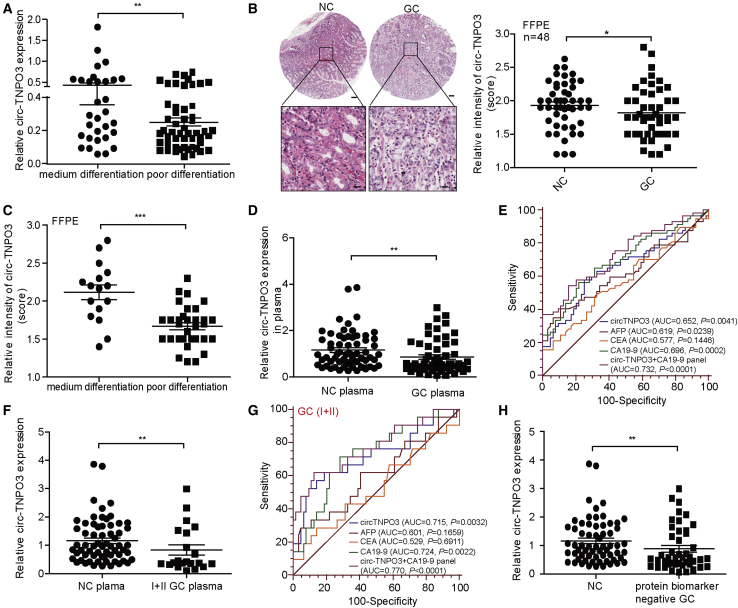
We also analyzed whether the decrease in circ-TNPO3 correlated with clinical characteristics. The level of circ-TNPO3 in patients with poorly differentiated GC was significantly lower than that in patients with medium (moderate or well) differentiated GC (Figure 2A). Nevertheless, there was no correlation between circ-TNPO3 expression and tumor stages or lymph node metastasis of GC (Figures S3A and S3B). To determine whether circ-TNPO3 level correlated with the prognosis of GC, an independent cohort of 48 pairs of formalin-fixed and paraffin-embedded (FFPE) GC and NC tissues was included. circ-TNPO3 expression was determined using ISH analysis on tissue array slides. As shown in Figure 2B, circ-TNPO3 was obviously downregulated in the GC group, which was consistent with the qRT-PCR results. In addition, circ-TNPO3 expression in FFPE tissues was related to the degree of GC differentiation (Figure 2C). However, we found no correlation between circ-TNPO3 expression and GC stage, metastasis, or overall survival (Figures S3C–S3E).
Figure 2.
circ-TNPO3 is associated with GC differentiation and acts as potential plasma biomarker of GC
(A) circ-TNPO3 levels in GC tissues with medium differentiation (n = 40) or poor differentiation (n = 63). (B) ISH analysis of circ-TNPO3 expression in 48 paired GC FFPE tissues and matched NC tissues is shown. The corresponding statistical plots are presented in the right panel; scale bars, upper 100 μm, lower 20 μm. (C) ISH analysis of circ-TNPO3 expression in GC FFPE samples with medium differentiation (n = 16) and poor differentiation (n = 32) is shown. (D) circ-TNPO3 level in plasma of patients with GC (n = 57) and in that of healthy controls (n = 64) is shown. (E) AUCs of plasma circ-TNPO3 and traditional biomarkers in distinguishing GC patients from healthy controls are shown. (F) The expression of plasma circ-TNPO3 in GC patients with early stages (n = 21) and in healthy controls (n = 64) is shown. (G) The AUCs of plasma circ-TNPO3 and traditional biomarkers in distinguishing early stages GC patients from healthy controls are shown. (H) The expression of circ-TNPO3 in plasma of patients with CEA-negative or CA19-9-negative GC (n = 45) and healthy controls (n = 64) is shown. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Considering that circulating circRNAs have potential to be diagnostic biomarkers for cancers, we compared the circ-TNPO3 level in plasma from patients with GC and healthy controls. Interestingly, circ-TNPO3 was significantly downregulated in plasma of GC patients (n = 57) compared with that from healthy controls (n = 64; Figure 2D). ROC analysis showed that circ-TNPO3 produced an area under the curve (AUC) of 0.65, which was better than traditional protein biomarkers, such as alpha fetal protein (AFP) (AUC = 0.62) and carcinoembryonic antigen (CEA) (AUC = 0.58). The plasma panel combining circ-TNPO3 and CA19-9 showed better diagnostic value (AUC = 0.73; Figure 2E). More importantly, we found that circ-TNPO3 was significantly downregulated in plasma of patients with early-stage GC (TNM stages I and II; n = 21) compared with healthy controls (Figure 2F). However, there were no significant differences in AFP or CEA levels between the early-stage GC and healthy controls. Similarly, combining circ-TNPO3 and CA19-9 produced better diagnostic value for early-stage GC (Figure 2G).
The traditional protein biomarkers often fail to diagnose GC due to its limited sensitivity. Then, we explored the potential of circ-TNPO3 as a diagnostic biomarker of GC with negative CEA or CA19-9 expression. We detected the circ-TNPO3 levels in 45 GC patients with normal CEA or CA19-9 levels; circ-TNPO3 significantly downregulated in these GC patients (Figure 2H), with an AUC of 0.76 (95% confidence interval [CI], 0.660–0.843; p < 0.0001). Overall, these data demonstrated that circ-TNPO3 significantly correlated with GC differentiation and could be a potential diagnostic biomarker of GC.
circ-TNPO3 inhibits migration and proliferation of GC cells in vitro
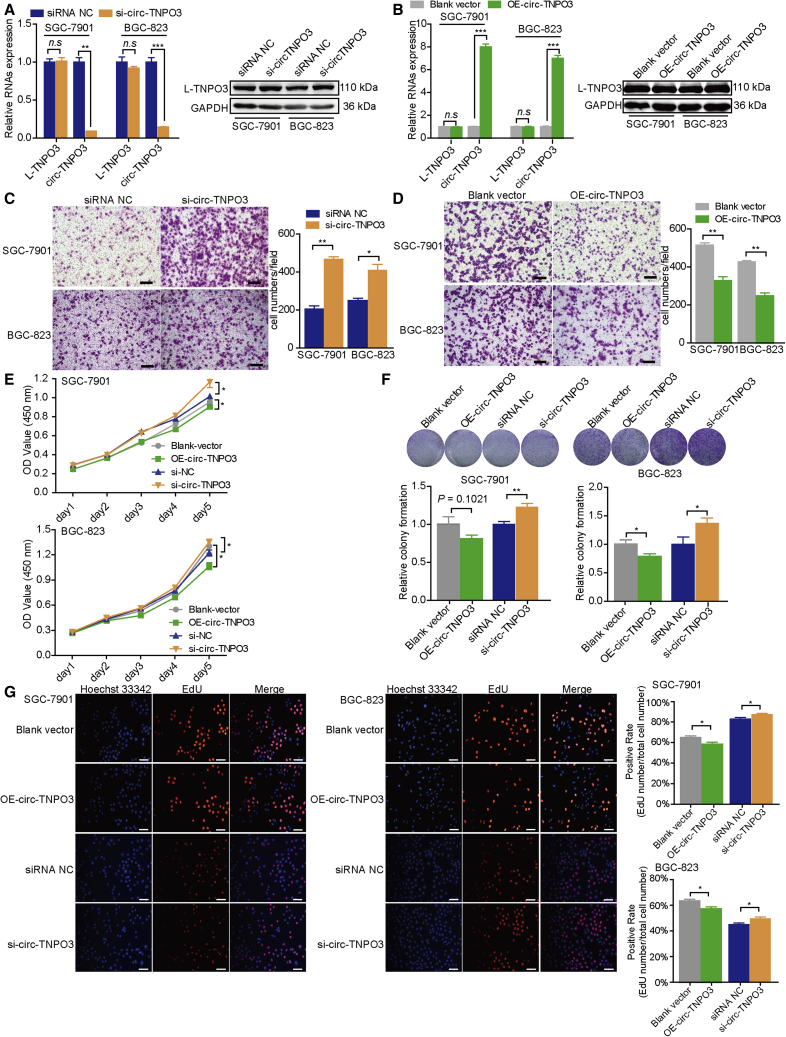
We then investigated the biological roles of circ-TNPO3 in GC. We first constructed a small interfering RNA (siRNA) targeting the back-splicing region of circ-TNPO3 and overexpression plasmid of circ-TNPO3. Then, we successfully manipulated the expression of circ-TNPO3 in SGC-7901 and BGC-823 cells without affecting the mRNA or protein levels of L-TNPO3 (Figures 3A and 3B). Then, we found that the inhibition of circ-TNPO3 significantly promoted although the overexpression of circ-TNPO3 suppressed migration abilities of GC cells (Figures 3C and 3D). Additionally, CCK8, colony formation, and EdU assays demonstrated that overexpression of circ-TNPO3 inhibited the proliferation of GC cells to some extent and inhibition of circ-TNPO3 moderately promoted the proliferation of GC cells (Figures 3E–3G). Meanwhile, circ-TNPO3 had no effect on cells apoptosis (Figure S4). These findings indicate that circ-TNPO3 can inhibit GC cells migration and proliferation in vitro.
Figure 3.
circ-TNPO3 inhibits GC cells migration and proliferation in vitro
(A and B) mRNA levels of circ-TNPO3, L-TNPO3 (A), and protein levels of L-TNPO3 (B) in GC cell lines. (C and D) Migration abilities of GC cells transfected with circ-TNPO3 siRNAs (C) or overexpression vectors (D) are shown; scale bar, 50 μm. (E–G) Proliferation abilities of GC cells transfected with circ-TNPO3 siRNAs or overexpression vectors were tested by CCK8 assays (E), colony formation (F), and Edu assay (G); scale bar, 50 μm. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
circ-TNPO3 inhibits migration and proliferation of GC cells in vivo
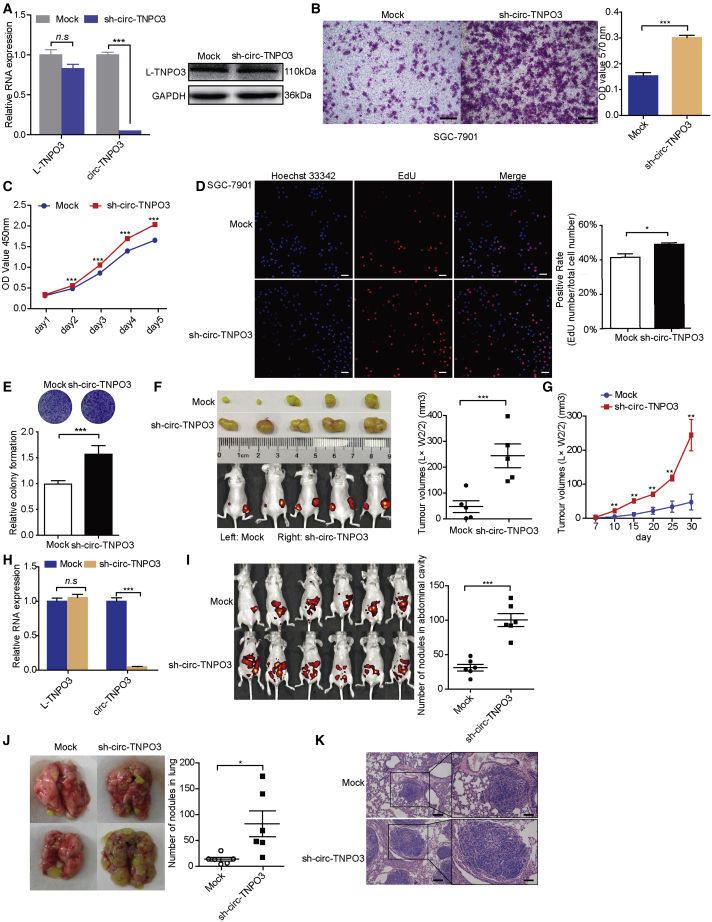
To further investigate biological roles of circ-TNPO3 in vivo, lentivirus-based circ-TNPO3 stable knockdown SGC-7901 cell line was constructed without affecting the level of L-TNPO3 (Figure 4A). sh-circ-TNPO3-SGC-7901 cells also showed increased migration ability (Figure 4B) and proliferation ability (Figures 4C–4E) in vitro, and the effect of stable transfection on cell proliferation was stronger than that of transient transfection. Next, multiple animal models were utilized to evaluate the function of circ-TNPO3 in vivo. In the tumor xenograft model, tumor growth and volume of subcutaneous xenografts significantly increased after the inhibition of circ-TNPO3 (Figures 4F and 4G). We verified the knockdown of circ-TNPO3 in xenografts derived from the sh-circ-TNPO3 group (Figure 4H). In the abdominal dissemination assays, the inhibition of circ-TNPO3 obviously elicited increased abdominal tumor nodules compared with the controls (Figure 4I). In the distal pulmonary metastasis models, nude mice that had received an injection of sh-circ-TNPO3-SGC-7901 cells displayed more lung metastatic colonies (Figures 4J and 4K). These results suggest that circ-TNPO3 exerts tumor suppressor role in vivo and inhibits GC growth and metastasis.
Figure 4.
circ-TNPO3 inhibits GC cells metastasis and proliferation in vivo
(A) mRNA levels (left) and protein levels (right) of circ-TNPO3 or L-TNPO3. (B) The migration ability of SGC-7901 cells is shown; scale bar, 50 μm. (C–E) Proliferation of SGC-7901 cells was measured by CCK8 assays (C), Edu assays (D; scale bar, 50 μm), and colony formation assays (E) at the indicated time. (F) Representative images of xenograft GC tumors (left) and the weight of xenografts were evaluated at the end point time (right). (G) The growth curves of xenograft tumors in vivo (n = 5) are shown. (H) Expression of circ-TNPO3 in tumor nodule tissues derived from tumor-bearing mice is shown. (I) Representative images of the GC abdominal spread model (left) are shown. Evaluation of the number of tumor nodules in mice abdominal cavities (right). (J and K) Representative images of lungs (J, left), quantification of lung metastatic colonization (J, right), and HE staining (K) of lungs biopsy from mice with injection of SGC-7901 cells via tail vein (n = 6) are shown; scale bars, 100 μm (left) and 60 μm (right). ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
circ-TNPO3 interacts with IGF2BP3 protein in GC cells
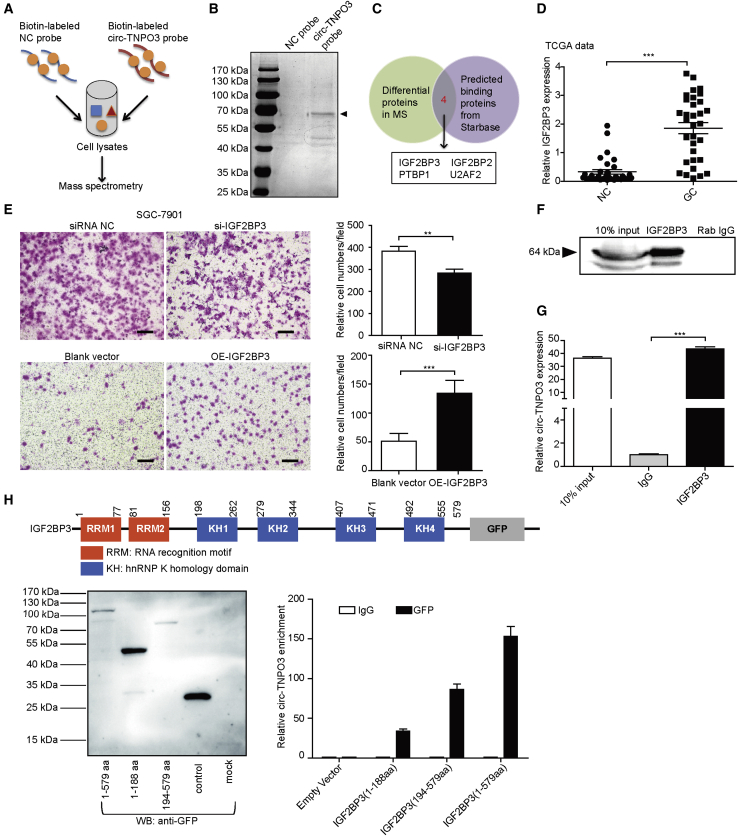
To explore the molecular mechanism responsible for the biological role of circ-TNPO3, we first investigated whether circ-TNPO3 was able to act as miRNAs sponge. Though several miRNAs were predicted to bind circ-TNPO3, few of them contain more than two binding sites. We selected 14 candidate miRNAs through overlapping the prediction results by miRanda, Targetscan, and miRNAs expression analysis of GC from the TCGA database (Figure S5A). However, no candidate miRNA binding to circ-TNPO3 was identified by RNA pull-down or luciferase reporter assays (Figures S5B–S5D). In addition, circRNADb database analysis showed that circ-TNPO3 had no open reading frame (ORF), and the possibility of encoding protein was relatively low (Figure S6).
Given that circRNAs have been demonstrated to bind with protein and serve as protein sponge or scaffold, we performed circ-TNPO3 RNA pull-down assay consecutively with mass spectrometry (MS) analysis; we identified 250 candidate proteins pulled down by circ-TNPO3 (Figures 5A and 5B; Table S3). KEGG analysis showed that these proteins were mainly related to ribosome, spliceosome, glycolysis, and RNA degradation (Figure S7). Overlapping of the pulled down proteins and the predicted circ-TNPO3 binding proteins from Starbase (Table S4) found four potential circ-TNPO3-interacting proteins: IGF2BP3; IGF2BP2; PTBP1; and U2AF2 (Figure 5C). Subsequently, we focused on IGF2BP3 for further validation because it had the highest score among all the proteins in the MS analysis. It has been demonstrated that IGF2BP3 is upregulated in various tumor types, including GC.11 In the current study, we confirmed that IGF2BP3 level was upregulated in GC based on the TCGA data (Figure 5D); moreover, we validated that silencing of IGF2BP3 inhibited although overexpression of IGF2BP3 promoted the migration of GC cells (Figures 5E and S8). Then, the interaction between circ-TNPO3 and IGF2BP3 was verified by RNA immunoprecipitation (RIP) analysis in SGC-7901 cells (Figures 5F and 5G). Given that IGF2BP3 contains two RNA recognition motifs (RRMs) and four KH domains, which are essential for its binding and function, we next constructed GFP-tagged full-length and truncated IGF2BP3 expression vectors and obtained the expected recombinant proteins by the confirmation with anti-GFP antibody (Figure 5H, left). RIP assays for full-length and truncated IGF2BP3 showed that the deletion of KH domains significantly abolished the enrichment of circ-TNPO3, indicating that circ-TNPO3 mainly binds to KH regions of IGF2BP3 (194–579 amino acids [aas]; Figure 5H, right). Furthermore, we confirmed that circ-TNPO3 and IGF2BP3 had no influence on the mutual expression (Figure S9). These results suggest that circ-TNPO3 can interact with IGF2BP3 protein.
Figure 5.
circ-TNPO3 directly binds to IGF2BP3 protein
(A) Schematic illustration of the process of exploring the circ-TNPO3-interacting proteins. (B) Coomassie bright blue staining of the differential proteins pulled down by circ-TNPO3 is shown. (C) Venn diagram shows the candidate proteins that may bind to circ-TNPO3. (D) IGF2BP3 expression in paired GC and NC tissues in the TCGA database (n = 33) is shown. (E) Cell migration abilities after transfection with si-IGF2BP3 (upper panel) or IGF2BP3 overexpression vector (lower panel) are shown; scale bar, 50 μm. (F) IGF2BP3 protein was pulled down via RIP assay. (G) Enrichment of circ-TNPO3 pulled down by IGF2BP3 protein is shown. (H) Schematic of GFP-tagged IGF2BP3 with its binding domains (upper panel) is shown. The full-length or truncated IGF2BP3 proteins pulled down by anti-GFP (left panel) are shown. RIP analysis of circ-TNPO3 enrichment pulled down by GFP in cells transfected with full-length or truncated IGF2BP3 constructs (right panel) is shown. ∗∗p < 0.01 and ∗∗∗p < 0.001.
circ-TNPO3 can function as a protein decoy for IGF2BP3 to regulate the expression of MYC
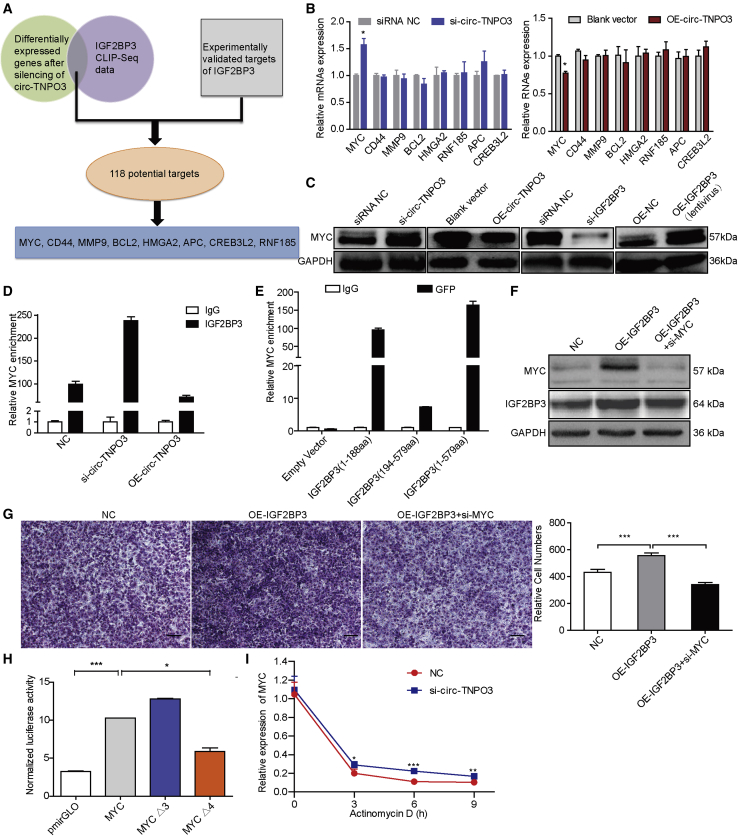
To explore the molecular mechanism by which circ-TNPO3 and IGF2BP3 regulate GC, we performed RNA sequencing (RNA-seq) of SGC-7901 cells with circ-TNPO3 inhibition. Subsequently, we found 205 upregulated and 204 downregulated genes (|fold change| > 2.0; p < 0.05) after the silencing of circ-TNPO3 (Figure S10A; Table S5). Additionally, GO analysis showed that these differentially expressed genes participated in cell migration signaling pathway, including extracellular matrix structural constituent, protein kinase B signaling, and cell-cell adhesion (Figure S10B). These results enabled us to focus on the role of circ-TNPO3 in regulating tumor metastasis.
Overlapping of the above differentially expressed genes with IGF2BP3 CLIP-seq data from the CLIPdb database and then combining them with experimentally validated published targets of IGF2BP family, we discovered 118 potential target genes of IGF2BP3 (Table S6). Based on the expression level of these candidate genes and their relation to cancer metastasis, we focused on MYC, RNF185, APC, CREB3L2, CD44, MMP9, BCL2, and HMGA2 for further validation (Figure 6A). Inhibiting circ-TNPO3 significantly increased although overexpressing circ-TNPO3 significantly decreased the mRNA and protein levels of MYC in SGC-7901 cells; however, circ-TNPO3 had no effect on the mRNA levels of other candidates (Figures 6B and 6C). We further illustrated that MYC level decreased in GC cells transfected with si-IGF2BP3 and increased upon enhancing IGF2BP3 level (Figure 6C). Then, RIP assays validated the interaction between IGF2BP3 protein and MYC mRNA (Figure 6D). The cells transfected with si-circ-TNPO3 enriched more MYC mRNA binding to IGF2BP3 although overexpression of circ-TNPO3 reduced MYC mRNA enriched by IGF2BP3 (Figure 6D). RIP assays for GFP-tagged full-length and truncated IGF2BP3 showed that the two RRMs (1–188 aas) were responsible for the interaction between IGF2BP3 and MYC mRNA (Figure 6E). In GC cells, inhibition of MYC impaired the ability of IGF2BP3 to promote cell migration (Figures 6F and 6G). To identify whether IGF2BP3 regulates MYC through binding to its 3′ UTR, we constructed luciferase reporter vectors containing the MYC 3′ UTR or deletion mutants. Increased luciferase activity was observed in HEK293 cells co-transfected with MYC 3′ UTR reporter and IGF2BP3; in contrast, decreased luciferase activity was observed after deleting the IGF2BP3 binding sites on MYC 3′ UTR (MYCΔ4; Figure 6H). In addition, the inhibition of circ-TNPO3 increased the stability of MYC mRNA (Figure 6I). In summary, we can conclude that MYC is a direct target of IGF2BP3 through its 3′ UTR, and circ-TNPO3 may function as a decoy or sponge for IGF2BP3 protein to regulate the expression of MYC.
Figure 6.
circ-TNPO3 functions as an IGF2BP3 protein sponge to regulate the expression of MYC
(A) A flow diagram shows the potential targets downstream of the circ-TNPO3-IGF2BP3 axis. (B) The relative mRNA levels of the candidate target genes in cells transfected with si-circ-TNPO3 (left) or OE-circ-TNPO3 vector (right) are shown. (C) The protein levels of MYC in SGC-7901 cells with inhibition or overexpression of circ-TNPO3 or IGF2BP3 are shown. (D) The enrichment of MYC mRNA pulled down by IGF2BP3 in SGC-7901 cells transfected with si-circ-TNPO3, OE-circ-TNPO3 vectors, or controls is shown. (E) RIP analysis for MYC enrichment in SGC-7901 cells after transfection with GFP-tagged-full-length or truncated IGF2BP3 constructs is shown. (F) Protein levels of MYC and IGF2BP3 in GC cells transfected with OE-IGF2BP3 or co-transfected with si-MYC are shown. (G) Migration abilities of GC cells transfected with OE-IGF2BP3 or co-transfected with si-MYC are shown; scale bar, 50 μm. (H) Luciferase assays performed with full-length MYC 3′ UTR or deletion mutants (MYC Δ3 or Δ4) are shown. (I) Stability of MYC in SGC-7901 cells with circ-TNPO3 knockdown or treated with actinomycin D at the indicated time points is shown. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
circ-TNPO3 regulates the migration of GC in an IGF2BP3- and SNAIL- dependent manner
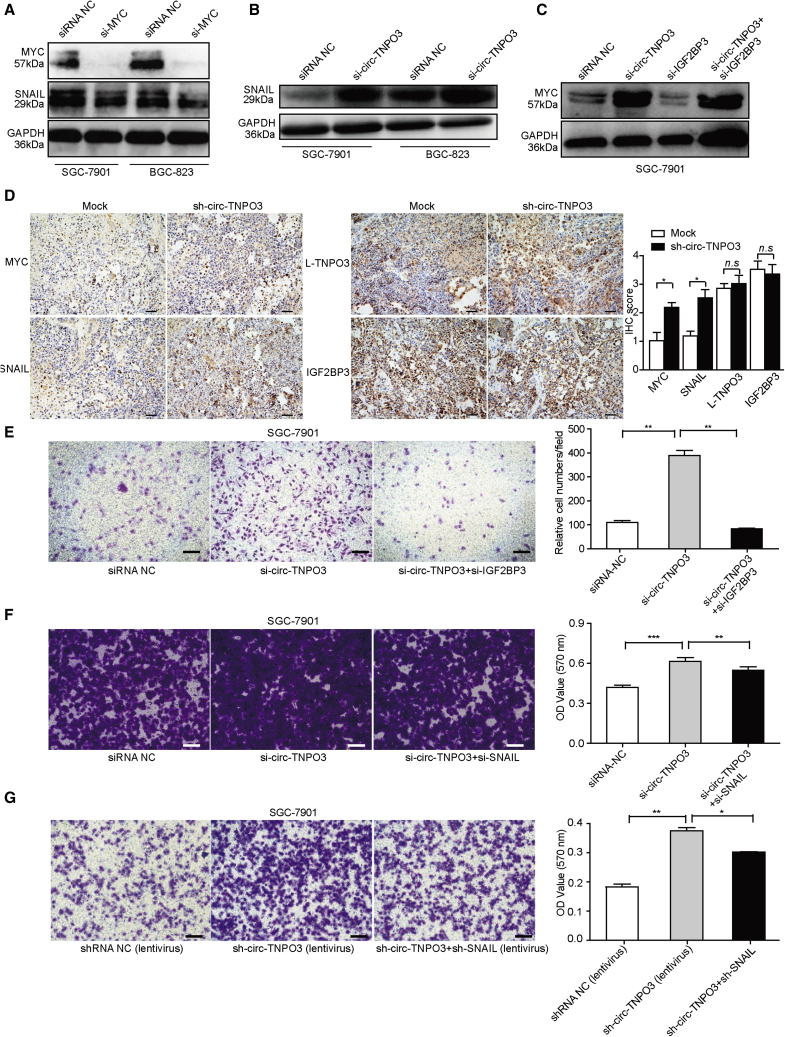
Given that MYC oncogene is highly amplified in many human cancers, including GC, and contributes to the genesis and progression of cancers, circ-TNPO3 may play its role through MYC and its downstream pathway. Combining the target genes of MYC and differently expressed genes after the silencing of circ-TNPO3, SNAIL attracted our attention. We validated that SNAIL played the key role in GC migration (Figure S11). It has been reported that MYC could bind to the promoter of SNAIL and participate in SNAIL activation.17 Consistent with a previous study, we found that the inhibition of MYC significantly inhibited the expression of SNAIL (Figure 7A). We further showed that si-circ-TNPO3 was able to enhance the expression of SNAIL and MYC, and si-circ-TNPO3 was able to counteract the inhibitory effect of si-IGF2BP3 on the expression of MYC (Figures 7B and 7C). In the xenograft mouse model, knockdown of circ-TNPO3 also increased the expression of MYC and SNAIL, but it had no effect on the levels of L-TNPO3 and IGF2BP3 (Figure 7D). Then, we tried to dissect whether the function of circ-TNPO3 as GC suppressor was dependent on the IGF2BP3-MYC-SNAIL axis. Transwell assays indicated that si-IGF2BP3 was able to rescue the increased metastatic potential induced by si-circ-TNPO3 (Figure 7E). Similarly, SNAIL was shown to be crucial for the enhanced migration ability induced by si-circ-TNPO3 (Figure 7F). We further constructed SGC-7901 cells with stably knocked down circ-TNPO3 and SNAIL by using recombinant lentivirus, and we also observed the expected rescuing effect on the cancer cell migration of SNAIL in vitro (Figure 7G). These data suggest that circ-TNPO3 regulates the migration of GC in an IGF2BP3-MYC-SNAIL-axis-dependent manner. Taken together, we demonstrated that circ-TNPO3 acts as a protein decoy for IGF2BP3 to suppress the metastasis of GC.
Figure 7.
circ-TNPO3 regulates GC migration via the IGF2BP3/MYC/SNAIL axis
(A and B) SNAIL protein levels in GC cells transfected with si-MYC (A) or si-circ-TNPO3 (B). (C) MYC levels in cells transfected with si-circ-TNPO3, si-IGF2BP3, and si-circ-TNPO3 + si-IGF2BP3 are shown. (D) IHC staining of MYC, SNAIL, IGF2BP3, and L-TNPO3 in the tumor xenograft tissues is shown; scale bar, 50 μm. (E) Migration abilities of SGC-7901 cells transfected with si-circ-TNPO3 alone and co-transfected with si-IGF2BP3 are shown. (F and G) Migration abilities of SGC-7901 cells transfected with si-circ-TNPO3/sh-circ-TNPO3 alone and co-transfected with si-SNAIL/sh-SNAIL are shown; scale bar, 50 μm. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Discussion
Emerging studies have demonstrated that widely dysregulated circRNAs exert important biological functions during tumor formation and development in multiple cancers.18,19 In particular, a group of circRNAs has been reported to regulate GC cell proliferation, apoptosis, migration, and invasion.20 For example, Zhang et al.21 discovered that circ-NRIP1 sponged miR-149-5p to affect the expression level of AKT1 and acted as a tumor promotor in GC. Chen et al.22 revealed that circ-AGO2 drove cancer progression by interacting with Hu-antigen R (HuR) protein and by derepressing downstream AGO2-miRNA complexes. Here, we identified that circ-TNPO3 was downregulated in GC tissues. We demonstrated that circ-TNPO3 expression correlated with GC differentiation, and circ-TNPO3 was capable of inhibiting proliferation and migration of GC both in vitro and in vivo, indicating its tumor suppressor role in GC. Notably, we observed that the effect of stable transfection cell strain on cell proliferation was stronger than that of transient transfection cell strains. The possible reasons for the difference may be: (1) the effect of circ-TNPO3 on GC cells proliferation is a time-dependent process. Stably transfected cell lines maintain low levels of circ-TNPO3 for an extended period so that the accumulation effect on proliferation is observed. (2) In vivo animal model provides an adequate context to foster tumor growth and is more like the clinical situation. Nevertheless, the exact mechanism behind the different effection on GC cells proliferation between stably and transiently transfected cell strains remains to be explored.
Currently, the mechanisms and biological functions of circRNAs are still unclear. In general, circRNAs have been shown to play important roles by acting as miRNA sponges, protein scaffolds, or by being translated themselves. The function of circRNAs as miRNA sponges has most often been reported. However, this action model is becoming controversial because very small number of circRNAs contain multiple binding sites for a single miRNA.23,24 A typical example is ciRS-7, which contains 73 conserved binding sites for miR-7.25 Alternatively, circRNAs can interact with RBP as protein sponges or scaffolds. circ-Mbl can strongly and specifically bind to MBL protein, and the modulation of MBL levels can affect circ-Mbl biosynthesis, suggesting that circ-Mbl can have a role in gene regulation by competing with linear splicing.26 Another example is circ-PABPN1. It acts as a protein sponge of HuR, leading to suppression of HuR binding to PABPN1 and weakening PABPN1 translation.27 Recently, there has been increasing evidence for the translation of circRNAs, such as circ-ZNF609, circ-Mbl, circ-FBXW7, and circ-SHPRH.9,28, 29, 30, 31 However, the functional relevance of most circRNA-derived peptides remains to be studied.
In our study, we focused on the interaction between circ-TNPO3 and cancer-associated RBPs. Using RNA pull-down, MS, and RIP assays, we demonstrated that circ-TNPO3 directly interacted with IGF2BP3 protein. However, in ovarian cancer, circ-TNPO3 was recently reported to act as a sponge for miR-1299, suggesting that circ-TNPO3 may play a versatile role in different tumors.32 IGF2BP3 and two other structurally and functionally related proteins, IGF2BP1 and IGF2BP2, belong to the IGF2BPs family. IGF2BPs have a role in RNA stability, trafficking, localization, and translation.33 IGF2BP3 has attracted particular interest because its expression is low in most adult tissues but is strongly expressed during embryogenesis and in diverse tumors. A recent study has documented that IGF2BP3 can interact with another circRNA, CDR1as, and mediate melanoma invasion and metastasis.34 Furthermore, accumulating evidence has demonstrated that IGF2BP3 can regulate the development and metastasis of many tumor types, so it represents a promising biomarker of tumors.35 For example, IGF2BP3 activates TAZ to facilitate alternative Wnt signaling through stabilizing WNT5B in breast cancer.36 Huang et al.37 revealed that IGF2BP3 played an oncogenic role in cancers through post-transcriptional regulation of MYC in m6A-dependent manner. In addition, IGF2BP3 dysfunction was found in other tumor types, such as pancreatic ductal adenocarcinoma,38 Ewing sarcoma,39 lung cancer,40 colon cancer,41 and renal cell carcinoma.42 In GC, upregulated IGF2BP3 expression was associated with poor survival.11 Wang et al.43 confirmed that IGF2BP3 was able to directly bind to HDGF mRNA, enhance HDGF mRNA stability, and promote GC angiogenesis. In the current study, we consistently showed that IGF2BP3 was upregulated in GC and was capable of promoting the migration of GC cells.
In mammals, the canonical structure of the three IGF2BP proteins is highly similar. IGF2BP3 contains two RRMs in their N-terminal regions and four KH domains in their C-terminal regions. We further showed that circ-TNPO3 mainly binds to KH regions of IGF2BP3 (194–579 aas). Although the exact mechanism of IGF2BP3 in modulating fate of mRNAs remains unknown, a series of target mRNAs was identified based on the CLIP data.35,44,45 Here, we identified that the downstream target of circ-TNPO3-IGF2BP3 axis was MYC. circ-TNPO3 was able to inhibit the expression of MYC through binding of IGF2BP3 in GC cells. Moreover, we found that circ-TNPO3 suppressed the expression of SNAIL through the IGF2BP3-MYC axis. The function of circ-TNPO3 as a GC suppressor gene was dependent on binding to IGF2BP3 and regulating downstream MYC-SNAIL axis.
MYC is an important proto-oncogene, and the alterations of MYC are commonly found on the path to cancer.46 For GC, copy number gains in MYC have been associated with poor survival. MYC may be a useful marker for clinical stratification and prognosis.47,48 SNAIL is a zinc-finger transcription factor, which is essential for the EMT process. Many studies have revealed that SNAIL is upregulated in GC and that it fosters tumor progression and prognosis.49,50 In our previous study, we have showed that SNAIL played key roles in GC migration and was modulated by several miRNAs, including miR-22 and miR-491-5p.15,16 This study provides a new avenue to regulate SNAIL pathway through the circ-TNPO3-IGF2BP3-MYC axis.
In addition, there is increasing evidence that circRNAs are emerging cancer biomarkers owing to their stable structure.7,51 In the current study, circ-TNPO3 was significantly downregulated in plasma of GC patients, especially in those with early stages of GC (TNM stages I and II), which cannot be discriminated by AFP or CEA. The plasma panel containing circ-TNPO3 and CA19-9 showed a better diagnostic value and has the potential to be a novel biomarker of GC.
In summary, for the first time, we identified that circ-TNPO3 functioned as a protein decoy to suppress the proliferation and migration of GC. We demonstrated that circ-TNPO3 competitively interacted with IGF2BP3 protein, resulting in the weakening of the role of IGF2BP3 in stabilizing MYC mRNA, which in turn inhibited the expression of MYC and SNAIL. In GC, the downregulated circ-TNPO3 weakened its interaction with IGF2BP3 and promoted the expression of MYC and its target gene SNAIL, thereby resulting in GC metastasis. The present study extends our knowledge about circRNA-RBP interaction and their regulatory role in GC progression and suggests that circ-TNPO3 has potential to be a diagnostic and therapeutic target in patients with GC.
Materials and methods
Human specimens
GC and paired noncancerous (at least 5 cm away from tumor edge) tissue samples were obtained from Southwest Hospital (58 pairs) and Xin Qiao Hospital (16 pairs) of Army Medical University. A total of 29 pairs of GC tissues were purchased from Super Chip (Shanghai, China). GC plasma samples (n = 57) were collected from Southwest Hospital, and samples of healthy controls matched for age and gender (n = 64) were collected from volunteers participating in the physical examination. These patients were all confirmed by histopathological analysis and did not undergo chemotherapy or radiotherapy at the time when tissue or blood samples were collected. Detailed patients’ information is documented in Tables S7 and S8. All the samples were collected with the consent of patients. This study was approved by the Ethics Review Board of Army Medical University.
Cell culture
GC cell lines used in this study were maintained in our research center. HEK293T cells were purchased from Han Biomedical (Shanghai, China). The cells were cultured in Dulbecco’s modified Eagle medium (Gibco) or RPMI 1640 medium (Gibco), supplemented with 10% heat-inactivated fetal bovine serum (FBS) at 37°C with 5% carbon dioxide.
Expression profile analysis of circRNAs
circRNA expression profile of five pairs of human GC tissues and their matched noncancerous tissues were analyzed using Arraystar Human circRNAs arrays, which contained 5,396 probes targeting the splicing sites of human circRNAs. The microarray experiments were performed by KangChen Bio-tech (Shanghai, China).
RNA extraction and qRT-PCR
The TRIzol (Invitrogen, Carlsbad, CA) method, PARIS Kit (Invitrogen, Carlsbad, CA), and miRNeasy Serum/Plasma Kit (QIAGEN, Germany) were used to isolate total RNAs, nuclear and cytoplasmic RNA, and plasma RNAs, respectively. The PrimeScript RT Master Mix (Takara, Dalian, China) and SYBR (Toyobo, Shanghai, China) were used to perform reverse transcription and real-time PCR analyses of mRNA and circRNAs. These data were normalized to β-actin. For the plasma samples, synthetic cel-miR-39 was used as a spiked-in standard and was tested by TaqMan MicroRNA Assays (Invitrogen, Carlsbad, CA, USA). The results were calculated in accordance with the 2−ΔΔCT method. The primer information is listed in Table S9.
RNase R and actinomycin D treatment
5 μg of total RNA from GC cells was treated with RNase R (3 U/μg, Epicenter Technologies) for 15 min at 37°C. After incubation, RNAs were purified by the RNeasy MinElute cleaning kit (QIAGEN, Hilden, Germany). To detect RNA stability, actinomycin D or DMSO (1 μg/mL) was added to the cell culture medium for the indicated time, consecutively with RNA extraction, reverse transcription, and qRT-PCR analyses.
Vector constructions
To construct circ-TNPO3 overexpression plasmid, the sequence of exon 2–4 of human TNPO3 mRNA was synthesized and cloned into pCD2.1-ciR vector (Geenseed Biotech, Guangzhou, China). To construct short hairpin RNAs (shRNAs) targeting circ-TNPO3 or SNAIL, the shRNA sequences targeting the back-splicing junction of circ-TNPO3 were ligated into the HBLV-GFP-Puro lentiviral vector (Han Biomedical, Shanghai, China), and the shRNA sequences targeting mRNA of SNAIL were ligated into the HBLV-mcherry-BSD lentiviral vector. Then, HBLV-GFP-circ-TNPO3 shRNA and HBLV-mcherry-SNAIL shRNA were used to generate stable GC cell lines. The sequences of shRNAs are shown in Table S10. IGF2BP3 full-length sequence, IGF2BP3-FL (1–579 aas), and IGF2BP3 truncated mutant fragments—IGF2BP3-N-terminal (NT, 1–188 aas) and IGF2BP3-C-terminal (CT, 194–579 aas)—were synthesized and cloned into pEGFP-N1 vector at the Xho I and Age I sites (Sangon Biotech, Shanghai, China). All the vectors were identified correctly by sequencing.
Transfection and construction of stable cell lines
siRNAs targeting circ-TNPO3, IGF2BP3, MYC, and SNAIL and corresponding negative controls were obtained from RiboBio (Guangzhou, China) or GenePharma (Shanghai, China). The information of siRNAs is shown in Table S10. Lipofectamine 2000 (Invitrogen, Carlsbad, CA) was used in transfection assays in line with the manufacturer’s instructions.
To establish stable transfectants, lentivirus-encoding circ-TNPO3 shRNA or SNAIL shRNA-infected SGC-7901 cells at MOI 1:100 were followed by selection with puromycin (1 μg/mL) or blasticidin S (10 μg/mL), respectively, for 7–10 days (after three passages). The transfection efficiencies were verified by qRT-PCR or western blot. The cells were then ready for subsequent assays.
Cell migration assay
GC cells migration ability was performed as described previously.15 Briefly, a total of 1 × 105 cells were seeded into the chambers with 5-μm or 8-μm pores (Corning). Culture medium containing 10% FBS or not was filled in the lower or upper chambers, respectively. The migrated cells were stained with crystal violet and quantified at the indicated time.
Cell proliferation assays
Cell proliferation abilities were detected using CCK8 assay, colony formation assay, and Edu assay. The detailed methods are shown in supplemental information.
RNA-seq
Gene expression profiles of GC cells with circ-TNPO3 knockdown or controls were analyzed using Illumina NovaSeq 6000 (Illumina, USA). Library construction and computational analyses were fulfilled by Shanghai Sinomics Corporation (Shanghai, China).
Animal experiments
The animal studies were approved by the Institutional Animal Care and Use Committee of Army Medical University and performed as described before.15 For xenograft incubation and in vivo metastasis assays, including peritoneal dissemination and distal pulmonary metastasis, stably transfected SGC-7901 cells with circ-TNPO3 knockdown or mock cells were injected into 6- to 8-week-old female BALB/c nude mice (Beijing HFK Bioscience, China) subcutaneously via abdominal cavities or via tail vein, respectively. The mice were sacrificed at the indicated time, and the tumor volume and metastatic tumor nodules were determined.
Northern blot
A northern blot experiment was performed in accordance with the instructions of the Digoxigenin (DIG) Northern Starter Kit (Roche). A specific probe was designed against the back-splicing junction of circ-TNPO3 with the length from 200 to 400 bp, and the T7 RNA polymerase promoter sequence was added to 5′ end of the probe. The labeled RNA probe reacted with digoxigenin-11-UTP using T7 RNA polymerase and labeling mixture. Then, 10- to 15-μg denatured RNA sample was used for running the formaldehyde gel and RNA was transferred to a nylon membrane. RNA was fixed to the membrane by UV-crosslink. The suitable treated DIG-labeled RNA probe was used for hybridization with 5-mL DIG Easy Hyb at 68°C overnight. After hybridization, the membrane was washed, blocked, and incubated with antibody and then immunological detection was carried out. The probe information is listed in Table S11.
RNA pull-down assays
The cells were lysed in 0.5% IGEPAL (Sigma) lysis buffer, including 50-mM Tris-HCl (pH 7.5), 10 U/μL Superase, 150-mM NaCl, protease and phosphatase inhibitor cocktail, and 5-mM MgCl2 (Invitrogen). The supernatant was hybridized with biotin-labeled RNA probes (GenePharma, Shanghai, China) at 4°C for 4 h, followed by adding prewashed and blocked streptavidin beads (Invitrogen); after incubation at 4°C for another 2 h and centrifugation, the beads were washed. The pull-down proteins and RNAs were extracted and analyzed by mass spectrometry and qRT-PCR, respectively. The sequences of biotin-labeled RNA probes are shown in Table S11.
Mass spectrometry
The protein sample pulled down by biotin-labeled circ-TNPO3 probe or control probe was resuspended in Nano-RPLC buffer. The analysis was then performed on a ChromXP C18 column (75 μm × 15 cm; C18; 3 μm; 120 Å). Data acquisition was performed with a Triple TOF 5600 System (AB SCIEX, USA).
RIP
RIP assays were performed with the MagnaRIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA). Briefly, approximately 1 × 107 cells were lysed in 50 μL of RIP Lysis Buffer and consecutively incubated with beads coated with 5 μg of control immunoglobulin G (IgG), α-IGF2BP3 (Millipore, Bedford, MA), or α-GFP (Roche). The mixture was rotated and incubated overnight at 4°C. Then, immunoprecipitated RNAs were extracted after treatment with proteinase K and expressions of specific genes were analyzed by qRT-PCR.
ISH
The locations of circ-TNPO3 and L-TNPO3 in GC cell lines or tissues were detected with custom BaseScope probes for each gene (Advanced Cell Diagnostics). The expression level of circ-TNPO3 in FFPE tissues was evaluated by using specific digoxin-labeled circ-TNPO3 probe (Exiqon) on tissue arrays containing the paired GC samples (Superchip Biotech, Shanghai). The scores of circ-TNPO3 levels (0–3) were assessed by evaluating the intensity of circ-TNPO3 staining in the cytoplasm of the cells. The information on these probes is shown in Table S11. The detailed methods are shown in supplemental information.
Luciferase assays
To validate the interaction between IGF2BP3 and MYC 3′ UTR, the 3′ UTR of human MYC mRNA was cloned into the pmirGlo vector. Based on the iCLIP data, IGF2BP3 binding sites for MYC 3′ UTRs were deleted and were named as MYCΔ3 and MYCΔ4. Sequences of MYC 3′ UTR, MYCΔ3 and MYCΔ were listed in Table S11. HEK293T cells were co-transfected with IGF2BP3 overexpression vector (200 ng/well), pRL-TK (10 ng/well), and luciferase reporter vectors (200 ng/well) for 48 h. Luciferase activities were measured on GLOMAX 20/20 luminometer (Promega, Madison, MI, USA) using a Dual Luciferase Reporter Assay (Promega, Madison, MI, USA). The detailed methods are shown in supplemental information.
Western blot assay
Antibodies against rabbit polyclonal IGF2BP3 (1:1,000), rabbit polyclonal SNAIL (1:500), rabbit polyclonal MYC (1:1,000), and TNPO3 (1:1,000) were obtained from Abcam (Cambridge, UK); internal reference monoclonal GAPDH antibody (1:5,000) was obtained from Cell Signaling Technology. Anti-rabbit or anti-mouse IgG conjugated with horseradish peroxidase (HRP) was used as secondary antibody (1:10,000; Zhong Shan, Shanghai, China). Signals were detected using the SuperSignal West Dura Extended Duration Substrate Kit (Thermo Scientific Pierce, Rockford, IL) and analyzed using the ImageJ software.
Statistical analysis
The results are presented as mean ± SEM. Statistical differences were calculated in the GraphPad Prism 5 software using Student’s t test, chi-square test, and Mann-Whitney U test, as appropriate. Paired t tests were used to analyze circ-TNPO3 levels in the paired GC and adjacent nontumor tissues samples. Sensitivity and specificity of plasma biomarkers were assessed using ROC curve and AUC. p < 0.05 indicated statistical significance.
Acknowledgments
This study was supported by National Natural Science Foundation of China (81872392 and 82073254), Scientific Research Foundation for Introduced High-Level Personnel in Chongqing Medical University (41021300160256), Chongqing Talents–Exceptional Young Talents Project (CQYC202005044), Science and technology research project of Chongqing Municipal Education Commission (KJZD-K202100405), National Program on Key Basic Research Project (973 Program) (2009CB522606), and National Science and Technology Major Projects for “Major New Drugs Innovation and Development” (2008ZX09101-033). We thank Prof. Huang Shenglin and Yong Lin for the experimental support on the work.
Author contributions
B.X., Q.Z., and T.Y. designed the study. B.X., T.Y., L.R., W.L., P.Y., J.L., H.M., D.C., Q.M., X.P., X.W., J.W., H. Zeng, W.Z., D.L., and P.L. performed the experiments. B.X., T.Y., L.R., H. Zhao, and Q.Z. participated in data analyses and drafting of the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.08.029.
Contributor Information
Quanming Zou, Email: qmzoutmmu@yeah.net.
Bin Xiao, Email: binxiaocqmu@cqmu.edu.cn.
Supplemental information
References
- 1.Chen W., Zheng R., Baade P.D., Zhang S., Zeng H., Bray F., Jemal A., Yu X.Q., He J. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Allemani C., Weir H.K., Carreira H., Harewood R., Spika D., Wang X.S., Bannon F., Ahn J.V., Johnson C.J., Bonaventure A., CONCORD Working Group Global surveillance of cancer survival 1995-2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2) Lancet. 2015;385:977–1010. doi: 10.1016/S0140-6736(14)62038-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salzman J., Chen R.E., Olsen M.N., Wang P.L., Brown P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 6.Zhou R., Wu Y., Wang W., Su W., Liu Y., Wang Y., Fan C., Li X., Li G., Li Y. Circular RNAs (circRNAs) in cancer. Cancer Lett. 2018;425:134–142. doi: 10.1016/j.canlet.2018.03.035. [DOI] [PubMed] [Google Scholar]
- 7.Shan C., Zhang Y., Hao X., Gao J., Chen X., Wang K. Biogenesis, functions and clinical significance of circRNAs in gastric cancer. Mol. Cancer. 2019;18:136. doi: 10.1186/s12943-019-1069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhong Y., Du Y., Yang X., Mo Y., Fan C., Xiong F., Ren D., Ye X., Li C., Wang Y. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol. Cancer. 2018;17:79. doi: 10.1186/s12943-018-0827-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pamudurti N.R., Bartok O., Jens M., Ashwal-Fluss R., Stottmeister C., Ruhe L., Hanan M., Wyler E., Perez-Hernandez D., Ramberger E. Translation of circRNAs. Mol. Cell. 2017;66:9–21.e7. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du W.W., Yang W., Liu E., Yang Z., Dhaliwal P., Yang B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou Y., Huang T., Siu H.L., Wong C.C., Dong Y., Wu F., Zhang B., Wu W.K., Cheng A.S., Yu J. IGF2BP3 functions as a potential oncogene and is a crucial target of miR-34a in gastric carcinogenesis. Mol. Cancer. 2017;16:77. doi: 10.1186/s12943-017-0647-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi W., Kim J., Park J., Lee D.H., Hwang D., Kim J.H., Ashktorab H., Smoot D., Kim S.Y., Choi C. YAP/TAZ initiates gastric tumorigenesis via upregulation of MYC. Cancer Res. 2018;78:3306–3320. doi: 10.1158/0008-5472.CAN-17-3487. [DOI] [PubMed] [Google Scholar]
- 13.Yang F., Xue X., Zheng L., Bi J., Zhou Y., Zhi K., Gu Y., Fang G. Long non-coding RNA GHET1 promotes gastric carcinoma cell proliferation by increasing c-Myc mRNA stability. FEBS J. 2014;281:802–813. doi: 10.1111/febs.12625. [DOI] [PubMed] [Google Scholar]
- 14.Yang Q., Du W.W., Wu N., Yang W., Awan F.M., Fang L., Ma J., Li X., Zeng Y., Yang Z. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ. 2017;24:1609–1620. doi: 10.1038/cdd.2017.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu T., Wang L.N., Li W., Zuo Q.F., Li M.M., Zou Q.M., Xiao B. Downregulation of miR-491-5p promotes gastric cancer metastasis by regulating SNAIL and FGFR4. Cancer Sci. 2018;109:1393–1403. doi: 10.1111/cas.13583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zuo Q.F., Cao L.Y., Yu T., Gong L., Wang L.N., Zhao Y.L., Xiao B., Zou Q.M. MicroRNA-22 inhibits tumor growth and metastasis in gastric cancer by directly targeting MMP14 and Snail. Cell Death Dis. 2015;6:e2000. doi: 10.1038/cddis.2015.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gebauer F., Schwarzl T., Valcárcel J., Hentze M.W. RNA-binding proteins in human genetic disease. Nat. Rev. Genet. 2021;22:185–198. doi: 10.1038/s41576-020-00302-y. [DOI] [PubMed] [Google Scholar]
- 18.Kristensen L.S., Hansen T.B., Venø M.T., Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37:555–565. doi: 10.1038/onc.2017.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kristensen L.S., Andersen M.S., Stagsted L.V.W., Ebbesen K.K., Hansen T.B., Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019;20:675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 20.Li R., Jiang J., Shi H., Qian H., Zhang X., Xu W. CircRNA: a rising star in gastric cancer. Cell. Mol. Life Sci. 2020;77:1661–1680. doi: 10.1007/s00018-019-03345-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X., Wang S., Wang H., Cao J., Huang X., Chen Z., Xu P., Sun G., Xu J., Lv J., Xu Z. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol. Cancer. 2019;18:20. doi: 10.1186/s12943-018-0935-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Y., Yang F., Fang E., Xiao W., Mei H., Li H., Li D., Song H., Wang J., Hong M. Circular RNA circAGO2 drives cancer progression through facilitating HuR-repressed functions of AGO2-miRNA complexes. Cell Death Differ. 2019;26:1346–1364. doi: 10.1038/s41418-018-0220-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo J.U., Agarwal V., Guo H., Bartel D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomson D.W., Dinger M.E. Endogenous microRNA sponges: evidence and controversy. Nat. Rev. Genet. 2016;17:272–283. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 25.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 26.Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 27.Abdelmohsen K., Panda A.C., Munk R., Grammatikakis I., Dudekula D.B., De S., Kim J., Noh J.H., Kim K.M., Martindale J.L., Gorospe M. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by circPABPN1. RNA Biol. 2017;14:361–369. doi: 10.1080/15476286.2017.1279788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O., Fatica A., Santini T., Andronache A., Wade M. circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell. 2017;66:22–37.e9. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Y., Gao X., Zhang M., Yan S., Sun C., Xiao F., Huang N., Yang X., Zhao K., Zhou H. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J. Natl. Cancer Inst. 2018;110:304–315. doi: 10.1093/jnci/djx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Y., Fan X., Mao M., Song X., Wu P., Zhang Y., Jin Y., Yang Y., Chen L.L., Wang Y. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27:626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang M., Huang N., Yang X., Luo J., Yan S., Xiao F., Chen W., Gao X., Zhao K., Zhou H. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37:1805–1814. doi: 10.1038/s41388-017-0019-9. [DOI] [PubMed] [Google Scholar]
- 32.Xia B., Zhao Z., Wu Y., Wang Y., Zhao Y., Wang J. Circular RNA circTNPO3 regulates paclitaxel resistance of ovarian cancer cells by miR-1299/NEK2 signaling pathway. Mol. Ther. Nucleic Acids. 2020;21:780–791. doi: 10.1016/j.omtn.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Degrauwe N., Suvà M.L., Janiszewska M., Riggi N., Stamenkovic I. IMPs: an RNA-binding protein family that provides a link between stem cell maintenance in normal development and cancer. Genes Dev. 2016;30:2459–2474. doi: 10.1101/gad.287540.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanniford D., Ulloa-Morales A., Karz A., Berzoti-Coelho M.G., Moubarak R.S., Sánchez-Sendra B., Kloetgen A., Davalos V., Imig J., Wu P. Epigenetic silencing of CDR1as drives IGF2BP3-mediated melanoma invasion and metastasis. Cancer Cell. 2020;37:55–70.e15. doi: 10.1016/j.ccell.2019.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lederer M., Bley N., Schleifer C., Hüttelmaier S. The role of the oncofetal IGF2 mRNA-binding protein 3 (IGF2BP3) in cancer. Semin. Cancer Biol. 2014;29:3–12. doi: 10.1016/j.semcancer.2014.07.006. [DOI] [PubMed] [Google Scholar]
- 36.Samanta S., Guru S., Elaimy A.L., Amante J.J., Ou J., Yu J., Zhu L.J., Mercurio A.M. IMP3 stabilization of WNT5B mRNA facilitates TAZ activation in breast cancer. Cell Rep. 2018;23:2559–2567. doi: 10.1016/j.celrep.2018.04.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang H., Weng H., Sun W., Qin X., Shi H., Wu H., Zhao B.S., Mesquita A., Liu C., Yuan C.L. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2018;20:285–295. doi: 10.1038/s41556-018-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ennajdaoui H., Howard J.M., Sterne-Weiler T., Jahanbani F., Coyne D.J., Uren P.J., Dargyte M., Katzman S., Draper J.M., Wallace A. IGF2BP3 modulates the interaction of invasion-associated transcripts with RISC. Cell Rep. 2016;15:1876–1883. doi: 10.1016/j.celrep.2016.04.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mancarella C., Pasello M., Ventura S., Grilli A., Calzolari L., Toracchio L., Lollini P.L., Donati D.M., Picci P., Ferrari S., Scotlandi K. Insulin-like growth factor 2 mRNA-binding protein 3 is a novel post-transcriptional regulator of Ewing sarcoma malignancy. Clin. Cancer Res. 2018;24:3704–3716. doi: 10.1158/1078-0432.CCR-17-2602. [DOI] [PubMed] [Google Scholar]
- 40.Sun X., Wei P., Shen C., Yang Y., Wang Y., Li Y., Du X. Prognostic value of the IASLC/ATS/ERS classification and IMP3 expression in lung adenocarcinoma of Chinese cases. Am. J. Cancer Res. 2015;5:2266–2276. [PMC free article] [PubMed] [Google Scholar]
- 41.Lochhead P., Imamura Y., Morikawa T., Kuchiba A., Yamauchi M., Liao X., Qian Z.R., Nishihara R., Wu K., Meyerhardt J.A. Insulin-like growth factor 2 messenger RNA binding protein 3 (IGF2BP3) is a marker of unfavourable prognosis in colorectal cancer. Eur. J. Cancer. 2012;48:3405–3413. doi: 10.1016/j.ejca.2012.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tschirdewahn S., Panic A., Püllen L., Harke N.N., Hadaschik B., Riesz P., Horváth A., Szalontai J., Nyirády P., Baba H.A. Circulating and tissue IMP3 levels are correlated with poor survival in renal cell carcinoma. Int. J. Cancer. 2019;145:531–539. doi: 10.1002/ijc.32124. [DOI] [PubMed] [Google Scholar]
- 43.Wang Q., Chen C., Ding Q., Zhao Y., Wang Z., Chen J., Jiang Z., Zhang Y., Xu G., Zhang J. METTL3-mediated m6A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut. 2020;69:1193–1205. doi: 10.1136/gutjnl-2019-319639. [DOI] [PubMed] [Google Scholar]
- 44.Jønson L., Christiansen J., Hansen T.V.O., Vikeså J., Yamamoto Y., Nielsen F.C. IMP3 RNP safe houses prevent miRNA-directed HMGA2 mRNA decay in cancer and development. Cell Rep. 2014;7:539–551. doi: 10.1016/j.celrep.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 45.Samanta S., Sharma V.M., Khan A., Mercurio A.M. Regulation of IMP3 by EGFR signaling and repression by ERβ: implications for triple-negative breast cancer. Oncogene. 2012;31:4689–4697. doi: 10.1038/onc.2011.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dang C.V. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X., Liu Y., Shao D., Qian Z., Dong Z., Sun Y., Xing X., Cheng X., Du H., Hu Y. Recurrent amplification of MYC and TNFRSF11B in 8q24 is associated with poor survival in patients with gastric cancer. Gastric Cancer. 2016;19:116–127. doi: 10.1007/s10120-015-0467-2. [DOI] [PubMed] [Google Scholar]
- 48.de Souza C.R., Leal M.F., Calcagno D.Q., Costa Sozinho E.K., Borges Bdo.N., Montenegro R.C., Dos Santos A.K., Dos Santos S.E., Ribeiro H.F., Assumpção P.P. MYC deregulation in gastric cancer and its clinicopathological implications. PLoS ONE. 2013;8:e64420. doi: 10.1371/journal.pone.0064420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He H., Chen W., Wang X., Wang C., Liu F., Shen Z., Xu J., Gu J., Sun Y. Snail is an independent prognostic predictor for progression and patient survival of gastric cancer. Cancer Sci. 2012;103:1296–1303. doi: 10.1111/j.1349-7006.2012.02295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee D.G., Kim H.S., Lee Y.S., Kim S., Cha S.Y., Ota I., Kim N.H., Cha Y.H., Yang D.H., Lee Y. Helicobacter pylori CagA promotes Snail-mediated epithelial-mesenchymal transition by reducing GSK-3 activity. Nat. Commun. 2014;5:4423. doi: 10.1038/ncomms5423. [DOI] [PubMed] [Google Scholar]
- 51.Zhang Y., Liang W., Zhang P., Chen J., Qian H., Zhang X., Xu W. Circular RNAs: emerging cancer biomarkers and targets. J. Exp. Clin. Cancer Res. 2017;36:152. doi: 10.1186/s13046-017-0624-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.