Abstract
Urinary extracellular vesicles (uEV) are a topical source of non‐invasive biomarkers for health and diseases of the urogenital system. However, several challenges have become evident in the standardization of uEV pipelines from collection of urine to biomarker analysis. Here, we studied the effect of pre‐analytical variables and developed means of quality control for uEV isolates to be used in transcriptomic biomarker research. We included urine samples from healthy controls and individuals with type 1 or type 2 diabetes and normo‐, micro‐ or macroalbuminuria and isolated uEV by ultracentrifugation. We studied the effect of storage temperature (‐20°C vs. ‐80°C), time (up to 4 years) and storage format (urine or isolated uEV) on quality of uEV by nanoparticle tracking analysis, electron microscopy, Western blotting and qPCR. Urinary EV RNA was compared in terms of quantity, quality, and by mRNA or miRNA sequencing. To study the stability of miRNA levels in samples isolated by different methods, we created and tested a list of miRNAs commonly enriched in uEV isolates. uEV and their transcriptome were preserved in urine or as isolated uEV even after long‐term storage at ‐80°C. However, storage at ‐20°C degraded particularly the GC‐rich part of the transcriptome and EV protein markers. Transcriptome was preserved in RNA samples extracted with and without DNAse, but read distributions still showed some differences in e.g. intergenic and intronic reads. MiRNAs commonly enriched in uEV isolates were stable and concordant between different EV isolation methods. Analysis of never frozen uEV helped to identify surface characteristics of particles by EM. In addition to uEV, qPCR assays demonstrated that uEV isolates commonly contained polyoma viruses. Based on our results, we present recommendations how to store and handle uEV isolates for transcriptomics studies that may help to expedite standardization of the EV biomarker field.
Keywords: biomarkers, DNAse, microRNA, mRNA, storage temperature, storage time, transcriptomics, urinary exosomes, urinary extracellular vesicles, virus
1. INTRODUCTION
Extracellular vesicles (EV) are heterogeneous membrane‐enclosed sacculi secreted by cells. EV cargo is comprised of proteins, several RNA species, DNA, metabolites and lipids (Malkin & Bratman, 2020; Raposo & Stoorvogel, 2013), which are loaded to EV during their biogenesis (Merchant et al., 2017). As EV cargo can capture the physiological or pathophysiological state of their parental cells, EV are a promising source of biomarkers for diseases of diverse aetiology (Dickhout & Koenen, 2018; Guha et al., 2019; Pang et al., 2020; Wang et al., 2021). In addition, the bilipid‐layer encapsulated cargo is protected, for example, from proteases and nucleases (Kim et al., 2017). As RNA is prone to degradation outside the cell, protected RNA inside vesicles has raised the interest in transcriptomic biomarker research (Pös et al., 2018).
EV have been described in diverse biofluids e.g. cerebrospinal fluid, plasma and urine (Yuana et al., 2013). Urine is a valuable biofluid because it can be obtained non‐invasively and it is rich in EV derived from the urinary system cells. Thus, urine is considered as an attractive source of biomarkers for the genitourinary track (Erdbrugger & Le, 2016; Oliveira et al., 2020; Svenningsen et al., 2020) or even for neurological disorders (Wang et al., 2019).
Studies that intend to discover reliable candidate biomarkers should include a relatively large number of samples (Yekula et al., 2020). A sufficient sample size can compensate factors such as inter‐individual variability and technical variability related to EV isolation and downstream analytics (Oeyen et al., 2019). However, collecting large sets of suitable samples from healthy controls and donors in different disease stages can take years, especially when potential confounding factors such as sex, age and medication are considered. Alternatively, samples already collected and stored with various protocols for other than uEV studies have been repurposed for uEV biomarker studies. Generally, lack of standardization in biofluid collection, pre‐processing, storage and coding as well as lack of identified housekeeping genes for EV studies (Lehmann et al., 2012; Théry et al., 2018; Witwer et al., 2013) hinder multi‐centre collaborative efforts and comparison between published studies.
Some efforts have been made to assess the effect of pre‐analytical variables such as urine storage time and temperature on particle concentration, uEV markers, electron microscopy, and RNA yield. However, most of the studies have focused on short storage times of up to 10 days at different temperatures (Armstrong et al., 2018; Oosthuyzen et al., 2013; Zhou et al., 2006) or up to 1 year at ‐80°C (Yuana et al., 2015; Zhou et al., 2006). Of note, while there is a handful of investigations comparing EV isolation methods using messenger RNA sequencing (mRNA‐seq) (Barreiro et al., 2020) and micro RNA sequencing (miRNA‐seq) (Barreiro et al., 2020; Cheng et al., 2014; Mussack et al., 2019; Park et al., 2020; Srinivasan et al., 2019), comparison of other pre‐analytical variables with sequencing as an end‐point are largely lacking (Miranda et al., 2014). With few exceptions (Armstrong et al., 2018; Barreiro et al., 2020; Park et al., 2020), studies have so far included only healthy control samples, which could bias results interpretation since diseases that influence urine composition could also alter composition and stability of uEV isolates. This is especially true in diabetic kidney disease, where the kidney filtration barrier is damaged causing albuminuria (Thomas et al., 2015) i.e. leakage of proteins into the urine.
Here, we studied the effect of pre‐analytical variables on the quality of the uEV isolates from > 100 urine samples of healthy and diabetic individuals. Based on results, we developed a quality control and recommendations to be used in transcriptomic biomarker research of uEV.
2. MATERIALS AND METHODS
Experimental workflows used in this study are presented in Figure 1.
FIGURE 1.
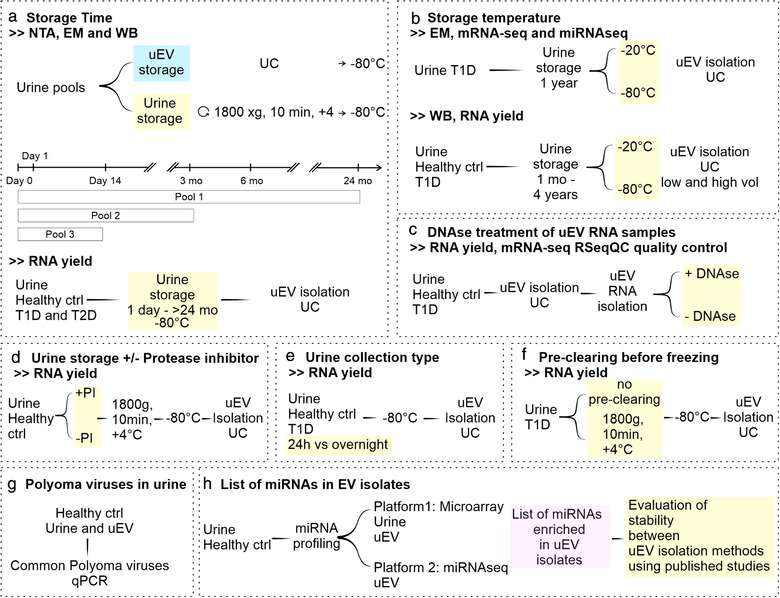
Experimental workflows employed in this study to evaluate the effects of pre‐analytical variables on uEV quality and transcriptomics: a. Effect of storage time on the uEV particle number, size distribution, morphology, protein markers and RNA yield. b. Effect of storage temperature on the uEV morphology, protein markers, RNA yield, miRNA‐seq and mRNA‐seq. c. Effect of DNAse treatment on uEV RNA yield and quality of mRNA‐seq alignment as evaluated with RSeqQC. d. Effect of urine storage with or without protease inhibitors on uEV RNA yield. e. Effect of urine collection type (24 h vs. overnight) on uEV RNA yield. f. Effect of pre‐clearing urine before freezing on uEV RNA yield. g. Co‐isolation of common polyoma viruses present in urine and uEV isolates. h. Analysis of miRNAs enriched in the uEV isolates and testing the list to evaluate stability of the miRNA between isolation methods using published datasets. Healthy control individuals (Healthy ctrl), messenger RNA sequencing (mRNA‐seq), micro RNA sequencing (miRNA‐seq), months (mo), individuals with type 1 diabetes (T1D) or type 2 diabetes (T2D), ultracentrifugation (UC), urinary extracellular vesicles (uEV), volume (vol)
2.1. Ethical permissions
All experiments were performed in accordance with the Declaration of Helsinki and the study protocols were approved by the Ethical Committees of Helsinki and Uusimaa Hospital District, Turku University Hospital or Vaasa Central Hospital. Urine samples and clinical data were obtained from donors participating in the nation‐wide prospective FinnDiane Study (Finnish Diabetic Nephropathy Study) targeting risk factors for diabetic nephropathy in type 1 diabetes (T1D) (FinnDiane 163/E5/04, 199/E5/05, 491/E5/2006; 238/13/03/00/15), and DIREVA study (Diabetes Registry in Vaasa), inviting all diabetic individuals living in the Vaasa Central Hospital District to participate since 2007 ((48/1801/2014; 116/1805/2016), or from participants in the Helsinki Urological Biobank and SalWe “Get It Done” research project addressing the use of EV for Personalized diagnostics and Care (Dnro 263/13/03/02/2011379/13/03/02/2012 and Dnro § 212).
2.2. Urine collection and storage
We collected urine samples from donors including healthy volunteers and individuals with type 1 or type 2 diabetic kidney disease. Individuals with T1D or type 2 diabetes (T2D) from FinnDiane and DIREVA cohorts were classified into normo‐, micro‐ and macroalbuminuria groups. Classification in Finndiane cohort was based on urinary albumin excretion rate (AER) on 24 h urine collection (two out of three consecutive urine collections). Individuals were considered macroalbuminuric with AER > 300 mg/24 h, microalbuminuric with AER 30–300 mg/24 h, and normoalbuminuric with AER < 30 mg/24 h. In DIREVA cohort, macroalbuminuria was defined based on AER (on overnight urine collection) and/or albumin to creatinine ratio (ACR) (spot urine albumin/creatinine ratio). Individuals were considered macroalbuminuric with AER > 200 μg/min or ACR > 35 mg/mmol, microalbuminuric with AER 20–200 μg/min and ACR > 3.5 mg/mmol at the last clinical visit and normoalbuminuric with AER < 20 μg/min and ACR < 3.5 (mg/mmol) at all of the clinical visits. All details concerning pre‐analytical variables, demographic data and albuminuria status are presented in the Supplementary Table 1.
For long‐, mid‐ and short‐term storage experiments I, II, and III, we collected pools of the first or second morning urine samples (Pool 1–3). For studies measuring RNA yield as a function of storage time, we collected individual 24 h (donors collected full void urine during 24 h) or overnight (full first void urine) urine samples, from healthy controls and donors with T1D or T2D and normo‐, micro‐ or macroalbuminuria. For storage temperature studies, we collected individual morning (midstream first morning urine void) urine or 24 h samples, from healthy controls or donors with T1D and normoalbuminuria or macroalbuminuria. After collection, the samples were directly divided into aliquots and stored in ‐20°C and ‐80°C until EV preparation.
To assess the effect of protease inhibitors on RNA yield, we collected individual or pooled overnight or first morning urine samples from healthy controls. Samples were stored at ‐80°C with protease inhibitors (+PI) or without (‐PI) for short‐ and long‐ term. Sample pairs used for assessing RNA yield from different urine collection types consisted of 24 h urine and overnight urine samples provided by the same donor on the same day. We included samples from healthy controls and individuals with T1D and micro‐ or macroalbuminuria. For assessing the effect of pre‐clearing, 40 ml aliquots of urine from individuals with T1D and micro‐ or macroalbuminuria were taken from the 24 h collections and processed with or without centrifugation at 1800 g, 10 min at +4°C before freezing at ‐80°C. Samples used to derive lists of miRNA enriched in the uEV isolates were pooled (pool 1, n = 11, microarray) or individual (n = 10, Qiagen set) first or second morning urine samples from < 45 years old healthy male controls.
Urines used for studying RNA yield as a function of storage time, storage temperature, type of urine collection, protease inhibitor addition and pre‐clearing step before freezing were supplemented with Calbiochem EDTA‐free Protease Inhibitor Cocktail Set III (Merck Millipore, Burlington, MA) at a dilution of 1/1000‐1/2000 before storage at ‐80°C.
All samples except for 24 h collections, urine collection format and storage temperature study were centrifuged at 1800 g for 10 min at +4°C before EV isolation or freezing the urine supernatant in aliquots at ‐80°C.
2.3. EV isolation and storage
EV were isolated from 7.8‐50 ml urine (Supplementary Table 1). Isolation from > 10 ml urine volumes was done by ultracentrifugation (UC) as previously described (Barreiro et al., 2020; Puhka et al., 2017). Urine samples were melted at +37°C in a water bath, vortexed for 90 s and centrifuged at 8000 g at +4°C (breaking) for 15 min using a fixed angle AG‐6512C rotor (Kubota Corp.). Supernatants were filtered using Whatman 1.2 μm cellulose acetate syringe filters (GE Healthcare, Buckinghamshire, UK). Thereafter, 30 ml of processed urine was centrifuged at 4°C for 90 min at 100,000 g (maximum breaking) in a SW28 rotor, k‐factor 254.5 (Beckman Coulter, Inc., Brea, CA). Supernatants were discarded and the pellets were washed in 20 ml of PBS repeating the ultracentrifugation configuration. Then, supernatants were removed and uEV samples were suspended in filtered PBS (0.22 μm PES filter; Jet Bio‐Filtration, Guangzhou, China) and stored in protein or DNA LoBind tubes (Eppendorf, Hamburg, Germany) at ‐80°C.
To isolate uEV from low volumes of urine (< 10 ml), a modification of the protocol was applied. Samples were melted and pre‐processed as above and then ultracentrifuged using the low volumes at 4°C for 2 h 30 min at 61,909 g (maximum breaking) in a 70.1 ti rotor, k‐factor 198 (Beckman Coulter, Inc., Brea, CA). Supernatants were discarded and the pellets were suspended in filtered PBS and stored in protein or DNA LoBind tubes at ‐80°C.
For storage experiments, where storage in urine or as isolated uEV was compared, we isolated the uEV directly from the fresh urine pool and from frozen replicate urine samples at time points ≥day 14. The uEV isolated from three replicate frozen urine aliquots were directly analysed at each time point i.e. they were not refrozen. Fresh uEV (day 0) were divided in aliquots of 3 replicate uEV isolates per time point, and either directly analysed (d0), stored at ‐80°C o/n and then analysed (d1), or stored at ‐80°C until analysis at the later time points.
2.4. Particle concentration and particle size distribution of uEV isolates
The particle concentration and particle size distribution were analysed from three replicate uEV isolates of the same urine pool using nanoparticle tracking analysis (NTA) instrument LM14C equipped with a violet (405 nm, 70 mW) laser (Malvern Instruments Ltd., Malvern, UK) and an sCMOS camera (Hamamatsu Photonics K.K., Hamamatsu, Japan) at controlled temperature of 22.0°C and camera level 14 using Nanosight software 3.0 (Malvern Instruments Ltd.). EV samples were diluted 1:25000 with filtered (0.2 μm) Hepes buffered saline (10 mM Hepes, 0.14 M NaCl) resulting in 15–50 particles/frame, and three videos of 90 s were recorded from each sample, mixing the sample manually between measurements. Data were analysed with NanoSight NTA 3.0 software using a detection threshold 5 and a gain of 10. Each technical replicate was measured 3 times.
2.5. Electron microscopy (EM)
Immunostaining was done as previously described (Puhka et al., 2017). Briefly, after loading to 200 mesh copper grids, samples were fixed in 2% PFA (Electron Microscopy Sciences). Thereafter, grids were incubated with anti‐CD59 clone MEM‐43 (1:100 dilution, Thermo Scientific, Waltham, MA). Then, samples were incubated with 10 nm gold‐conjugated anti‐mouse secondary antibody (1:80 dilution, BBI Solutions, Cardiff, UK) and washed.
Negative staining was performed as previously described (Puhka et al., 2017). Briefly, 5 μl of the uEV isolates (equivalent to 0.5‐1.5 ml of urine) was loaded on Formvar/Carbon 200 mesh TH, Copper grids (Ted Pella Inc., Redding, CA). Thereafter, grids were fixed with 2% PFA (Electron Microscopy Sciences, Hatfield, PA) and stained with 2% neutral uranyl acetate and embedded in methyl cellulose uranyl acetate mixture (1,8/0,4%). Images were captured using Jeol JEM‐1400 (Jeol Ltd, Tokyo, Japan) operating at 80 kV, equipped with a Gatan Orius SC 1000B CCD‐camera (Gatan Inc., Pleasanton, CA) and using an image size of 4008 × 2672 pixels.
2.6. Western blotting
For storage time and temperature studies, uEV isolates derived from equal volumes of urine were denatured in Laemmli buffer and, loaded to MINI‐Protean TGX 4%–20% gradient gels or 4%–20% Mini‐PROTEAN TGX Stain‐Free Protein Gels (Bio‐Rad laboratories Inc., Hercules, CA) to perform SDS‐PAGE and Western blotting as previously described (Barreiro et al., 2020; Puhka et al., 2017) . Proteins were stained either with QC colloidal Coomassie Stain or visualized in stain‐free gels. Membranes were immunostained for CD9, TSG101, Tamm‐Horsfall protein (THP), Podocalyxin (PDX), calnexin, GM130, CD59, CD63 and/or HSP70. Details on primary and secondary antibodies used in this study are provided in the Supplementary Table 2. Secondary antibody was detected using Super Signal West Femto Maximum Sensitivity kit (Thermo Fisher Scientific) or Pierce™ ECL Plus Western Blotting Substrate (Thermo Fisher Scientific).
Urine pellet was obtained by centrifuging 40 ml of fresh urine at 1800 g, for 10 min at +4°C. Pellet was resuspended in 200 μl of 0.2 um filtered PBS and protein concentration was measured using Micro BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA).
HeLA cells were incubated with RIPA lysis (Cold Spring Harbor Press, 2017) buffer for 30 min on ice after which they were vortexed. Homogenate was centrifuged for 10 min, 10000 RPM at +4°C. Supernatant was recovered and proteins were measured using DC Protein Assay Kit II (Bio‐Rad laboratories Inc.).
2.7. Polyoma virus detection
Urine and uEV isolates were analysed at the Helsinki university hospital laboratory (HUSlab). Test denomination for polyomaviruses was U –PoVNh, which includes testing for common BK‐polyomavirus and John Cunningham (JC)‐polyomavirus (U ‐BKVNh ja U –JCVNh, respectively). Viruses were detected by real‐time polymerase chain reaction (PCR). The test is quantitative for urine and the limit of quantification of the test is 3000 copies/ml.
2.8. RNA isolations for pre‐analytical variable studies
For storage time and storage temperature experiments, the total RNA isolation method was chosen depending on the starting volume of urine in order to obtain a suitable concentration of RNA compatible with sequencing applications: i) uEV isolates derived from urine samples with > 10 ml starting volume, total RNA was isolated using miRNeasy mini kit, (Qiagen, Hilden, Germany) procedure following manufacturer's protocol for Purification of Total RNA, Including Small RNAs, from Animal Cells. RNA was eluted in 30 μl of RNase‐free water. Depending on the experiment, samples were processed with DNAse treatment (+DNAse) or without (‐DNAse), and ii) uEV isolates derived from urine samples with < 10 ml starting volume, total RNA was isolated using miRNeasy micro kit, (Qiagen) procedure following manufacturer's protocol for Purification of Total RNA, Including Small RNAs, from Animal Cells. RNA was eluted in 14 μl of RNase free water.
For +/‐ DNAse experiments, total RNA was isolated from pairs of samples using miRNeasy micro kit (Qiagen) or miRNeasy mini kit (Qiagen) or Nucleospin miRNA kit (Macherey‐Nagel GmbH & Co. KG, Duren, Germany) with or without DNAse treatment according to manufacturer's instructions. Different elution volumes were used, but the elution volume was conserved between sample pairs. Please refer to the Supplementary Table 1 for detailed information on RNA isolation methods and elution volumes for each sub‐study.
RNAs were analysed using Agilent RNA 6000 Pico kit (Agilent Technologies, Santa Clara, CA) or Qubit RNA or dsDNA HS Assay Kits (Thermo Fisher Scientific).
2.9. mRNA‐seq and data analysis for storage temperature and DNAse treatment comparisons
Messenger RNA sequencing libraries were prepared from 1 ng of DNAse‐treated total RNA using SMART‐Seq v4 Ultra Low Input RNA Kit for Sequencing (Takara BIo Inc., Mountain View, CA) and Nextera XT kit (Ilumina Inc., San Diego, CA) according to the manufacturer's protocol. Libraries were sequenced on the Illumina Hiseq 2000 platform (Illumina Inc.) as 100 bp cycle paired end reads. RNAs isolated without DNAse treatment from the same uEV isolates (19 out of the 20 samples stored at ‐80°C) were prepared in the same way for mRNA‐seq and sequenced on the Illumina NextSeq 500 platform (Illumina Inc.) as 150 bp cycle paired end reads.
For data analysis, mRNA‐seq reads were mapped to the GRCh38.95 reference genome using STAR v2.5.3a (Dobin et al., 2013). RNA sequencing metrics were assessed using RseQC v2.6.4 (L. Wang et al., 2012) and mapped reads were counted using HTseq v0.6.0 (Anders et al., 2015). STAR, RseQC and HTseq were run using chipster v4 (https://chipster.csc.fi/) (Kallio et al., 2011) . Differential expression analysis was performed using edgeR (Robinson et al., 2010) in R v4.0.3 (Team, 2020). Genes with a false discovery rate (FDR) < 0.05 and a log2 fold change ≥ 0.75 or ≤ ‐0.75 were considered differentially expressed. Differentially expressed genes were subjected to gene ontology enrichment analysis (http://geneontology.org/) (Ashburner et al., 2000; Mi et al., 2019; “The Gene Ontology Consortium", 2021) (accession date 03.12.2020). Transcript length and gene GC‐content data was obtained by using BioMart (Smedley et al., 2009). Kidney‐enriched genes list was obtained from the human protein atlas (Uhlen et al., 2015) (http://www.proteinatlas.org). P‐body enriched transcripts were accessed from Hubstenberger et al., 2017.
2.10. miRNA‐seq and data analysis for storage temperature comparison
miRNA sequencing libraries were prepared from 10 ng of total RNA using TailorMix miRNA sample prep v2 (SeqMatic, Union City, CA) according to the manufacturer's protocol. Libraries were sequenced on the Illumina Hiseq 2000 platform (Illumina Inc.) as 100 bp cycle paired end reads.
For data analysis, sequences were aligned to known miRNAs using miRExpress (W. C. Wang et al., 2009). Differential expression analysis was performed using edgeR in R v4.0.3. Genes with an FDR lower than 0.05 and a log2 Fold change ≥ 0.75 or ≤ ‐0.75 were considered differentially expressed. The presence or absence of miRNAs in EV, reported by other studies, were assessed using Vesiclepedia (http://microvesicles.org/) (accession date 12.01.2021).
2.11. miRNA‐seq and microarray analysis of urine and uEV for deriving the list of miRNAs enriched in uEV isolates
For miRNA analysis, we used a sequencing platform at Qiagen and Toray's 3D Gene microarray platform from TATAA. In Qiagen, spike‐ins were added prior to RNA isolation from EVs using Qiagen's miRNeasy Serum/Plasma kit according to the manufacturers’ instructions. Libraries were prepared with QIAseq miRNA Library Prep kit including the unique molecular index tagging and sequenced with NextSeq500 (Illumina) using 75 nt read length. For data analysis, reads were mapped to miRBase and to the host genome using Bowtie2 (2.2.2). Data were normalized with weighted trimmed mean of M‐values (TMM) method.
For microarray, RNA was extracted from 10 and 30 ml of urine using Urine Cell‐ Free Circulating RNA purification Maxi kit (Norgen Biocorp) and from uEV with miRNeasy Mini Kit (Qiagen) and the Toray extraction reagent (TRT‐XE111) according to manufacturers’ instructions. Samples were concentrated down to 8 μl in the Biovac 060, iLMVAC and run‐on capillary electrophoresis on the Fragment Analyzer, Advanced Analytical with a High Sensitivity RNA kit (DNF472‐0500) to determine the presence of small RNAs. The maximum input volume of 2.2 μl of each sample was then mixed with miRNA spike (Cat No TRT‐XR304, Toray) and further labelled, hybridized and washed according to the instruction manual from Toray (H‐M‐R miRNA protocol 4‐Plex V3).
The intensity of each miRNA was analysed with the 3D‐Gene Scanner 3000 (Toray) with auto gain, auto focus and auto analysis settings, all according to manufacturer's instructions. Quality control was performed on all samples based on the QC report from the instrument. Background correction and global normalization of data was done according to Toray instructions.
2.12. Deriving the list of miRNAs enriched in uEV isolates and assessment of their expression in published datasets
To build a list of miRNAs typically found in uEV isolates, we used microarray to analyse RNA from urine samples and uEV isolated by UC from the same samples. The signals from both were arranged in decreasing order and cross‐compared to find miRNAs highly expressed in urine only (normalized signal > 100), in EV only or higher in EV than urine. Then, these lists were compared to the UC derived uEV‐miRNA sequencing results, organized in decreasing order with a minimum of > 1000 normalized counts on average. Micro‐RNAs present in the uEV enrichment lists from both platforms, and missing from the urine enrichment list, were assigned as enriched in the uEV isolates. Finally, this list was compared to entries in Vesiclepedia (accession date 10.07.2019).
We analysed the expression levels of the listed miRNAs enriched in the uEV isolates using previously published data. Data provided as Reads Per Million Scaled miRNA Reads (RPMSmiR) was log2 transformed. (Srinivasan et al., 2019). Data provided as raw count table (Barreiro et al., 2020) was TMM normalized and log2CPM transformed using edgeR. Raw (trimmed) sequencing data (Mussack et al., 2019) was aligned using Chimira (Vitsios & Enright, 2015), TMM normalized and log2CPM transformed using edgeR.
2.13. Statistics
OriginPro 2020 (OriginLab Corporation, Northampton, MA, USA) was used for the statistical analysis of NTA data, the significant difference was assessed using ANOVA and Bonferroni comparison of means, considering p‐values < 0.05 significant. IBM SPSS statistics V24 was used for statistical analysis (IBM Corporation, Armonk, NY). RNA yield (log10 transformed) mean differences for more than two groups were tested using ANOVA and Bonferroni post hoc tests. Mean differences for two groups were tested using Student's' t‐Test for Independent Samples or paired samples depending on the experimental design. GC proportion values were arcsin transformed (transformed x = 2*ARCSIN(SQRT(x)). Values are represented as mean ± SEM. P‐values < 0.05 were considered statistically significant. ggplot2 (Wickham, 2016) and pheatmap (Kolde, 2019) were used for data visualization.
2.14. Data availability
The raw sequencing data analysed in this study is not publicly available due to local regulations. We provide raw mRNA‐seq count data in the Supplementary Table 10 and raw miRNA‐seq count data in the Supplementary Table 11. Datasets used to derive the list of miRNAs enriched in uEV isolates are available in the Supplementary Table 12 (microarray raw data, background subtracted data, and global normalization) and Supplementary Table 13 (miRNA‐seq raw counts). Raw mRNA‐seq count data from uEV RNA samples isolated without DNAse treatment is available in Supplementary Table 14. Raw sequencing data is available to qualified investigators upon reasonable request.
3. RESULTS
3.1. Preservation of uEV during cryostorage – comparison of storing either urine or isolated uEV in PBS
3.1.1. Particle size and concentration of uEV remained unchanged during the storage of 24 months at –80°C regardless of the storage medium
To assess the overall preservation of uEV at –80°C, we conducted studies storing urine and isolated uEV in PBS for up to 24 months (Table 1, see also Supplementary Figure 1). No significant alterations in the particle concentrations of uEV isolates were observed and the particle size, assessed using mean size, modal size, median size, and 10th – 90th percentile range, remained stable regardless of the storage time or medium. To assess shorter term stability of uEV more carefully, a separate 3‐month study was conducted. Although a statistically significant decrease in the particle concentration was detected after 14 days of storage in both storage modalities, this was not observed in the 3‐months’ time point and the size distribution remained similar (Table 2, see also Supplementary Figure 2). In both studies the variation in measurements between time points was comparable to the variation between technical replicate measurements in individual time points confirming the overall stability of uEV irrespective of the storage medium.
TABLE 1.
Particle concentration and size distribution of uEV stored (‐80°C) up to 24 months in PBS or urine
| uEV storage modality, time | Mean concentration (particles/ml) | Mean diameter (nm) | Modal diameter (nm) | Median diameter (nm) | 10th percentile ‐ 90th percentile (nm) |
|---|---|---|---|---|---|
| Fresh | 1,13E13 ± 8,69E11 | 145,3 ± 13,7 | 101,5 ± 5,5 | 115,4 ± 7,1 | 72,2 ± 9,4 ‐ 253,7 ± 38,9 |
| PBS, 3 mo | 1,81E13 ± 1,74E12 | 129,6 ± 12,3 | 87,7 ± 5,9 | 102,9 ± 13,7 | 66,9 ± 9,5 ‐ 213,3 ± 19,9 |
| Urine, 3 mo | 1,30E13 ± 4,88E12 | 139,5 ± 12,0 | 91,6 ± 8,5 | 107,7 ± 7,5 | 68,9 ± 5,7 ‐ 260,9 ± 34,4 |
| PBS, 6 mo | 1,31E13 ± 5,41E12 | 128,8 ± 4,2 | 87,8 ± 2,5 | 107,6 ± 4,9 | 70,2 ± 2,6 ‐ 206,8 ± 12,5 |
| Urine, 6 mo | 1,48E13 ± 4,53E12 | 141,3 ± 15,9 | 90,1 ± 11,1 | 106,2 ± 12,2 | 67,4 ± 5,4 ‐ 254,7 ± 40,4 |
| PBS, 24 mo | 9,00E12 ± 8,97E11 | 148,7 ± 7,6 | 106,7 ± 3,0 | 123,3 ± 4,1 | 82,7 ± 1,2 ‐ 236,8 ± 12,0 |
| Urine, 24 mo | 8,97E12 ± 2,43E12 | 164,4 ± 7,7 | 100,7 ± 2,0 | 131,0 ± 13,5 | 83,3 ± 1,9 ‐ 289,4 ± 15,8 |
| uEV stored in PBS | 1,29E13 ± 4,28E12 | 138,1 ± 12,8 | 95,9 ± 9,5 | 112,3 ± 10,8 | 73,0 ± 8,5 ‐ 227,7 ± 28,0 |
| uEV stored in urine | 1,20E13 ± 3,78E12 | 147,6 ± 15,0 | 95,9 ± 8,4 | 115,1 ± 13,6 | 73,0 ± 8,4 ‐ 264,7 ± 32,6 |
Values represent mean ± SD, n = 3 technical replicates, bolded values represent mean values of given storage modality across all time points ± SD, n = 12. Months (mo), phosphate buffered saline (PBS), urinary extracellular vesicles (uEV)
TABLE 2.
Particle concentration and size distribution of uEV stored (‐80°C) up to 90 days in PBS or urine
| uEV storage modality, time | Mean concentration (particles/ml) | Mean diameter (nm) | Modal diameter (nm) | Median diameter (nm) | 10th percentile ‐ 90th percentile (nm) |
|---|---|---|---|---|---|
| Fresh | 4,8E12 ± 1,1E12 | 163,9 ± 7,4 | 124,3 ± 24,3 | 143,2 ± 18,6 | 82,1 ± 6,9 ‐ 254,5 ± 14,3 |
| PBS, d1 | 3,2E12 ± 2,9E11 | 170,6 ± 6,3 | 127,4 ± 40,7 | 163,6 ± 20,1 | 87,0 ± 6,6 ‐ 262,5 ± 15,7 |
| PBS, d14 | 1,4E12 ± 7,3E11 ** | 140,5 ± 11,1 | 117,4 ± 21,6 | 124,5 ± 9,0 | 73,3 ± 8,0 ‐ 213,0 ± 31,1 |
| Urine d14 | 2,0E12 ± 1,6E12 * | 124,5 ± 28,7 | 105,6 ± 14,7 | 104,8 ± 17,9 | 70,4 ± 13,1 ‐ 187,3 ± 63,9 |
| PBS, d90 | 3,4E12 ± 1,9E11 | 158,7 ± 9,3 | 111,1 ± 2,8 | 129,9 ± 8,1 | 86,4 ± 2,9 ‐ 254,4 ± 11,1 |
| Urine, d90 | 2,2E12 ± 1,4E11 | 168,6 ± 7,7 | 113,3 ± 5,9 | 141,5 ± 10,3 | 90,8 ± 4,6 ‐ 271,5 ± 9,9 |
| uEV stored in PBS | 3,20E12 ± 1,38E12 | 158,4 ± 13,8 | 120,1 ± 23,2 | 140,3 ± 20,3 | 82,2 ± 7,9 ‐ 246,1 ± 26,3 |
| uEV stored in urine | 3,01E12 ± 1,67E12 | 152,3 ± 26,0 | 114,4 ± 16,6 | 129,9 ± 23,4 | 81,1 ± 11,8 ‐ 237,8 ± 50,8 |
Values represent mean ± SD, n = 3 technical replicates, bolded values represent mean values of given storage modality across all time points ± SD, n = 12 for PBS stored uEV, n = 9 for uEV stored in urine. * = P < 0.05, ** = P < 0.01 compared to freshly isolated uEV using Bonferroni comparison of means. Days (d), phosphate buffered saline (PBS), urinary extracellular vesicles (uEV).
3.1.2. uEV morphology and protein markers were conserved for up to 24 months at ‐80°C in both storage media
We used transmission electron microscopy to evaluate the morphology of uEV from the fresh and –80°C frozen samples stored for up to 24 months in urine or as isolated uEV in PBS. We observed uEV of typical morphology and variable sizes in all samples (Figure 2).
FIGURE 2.
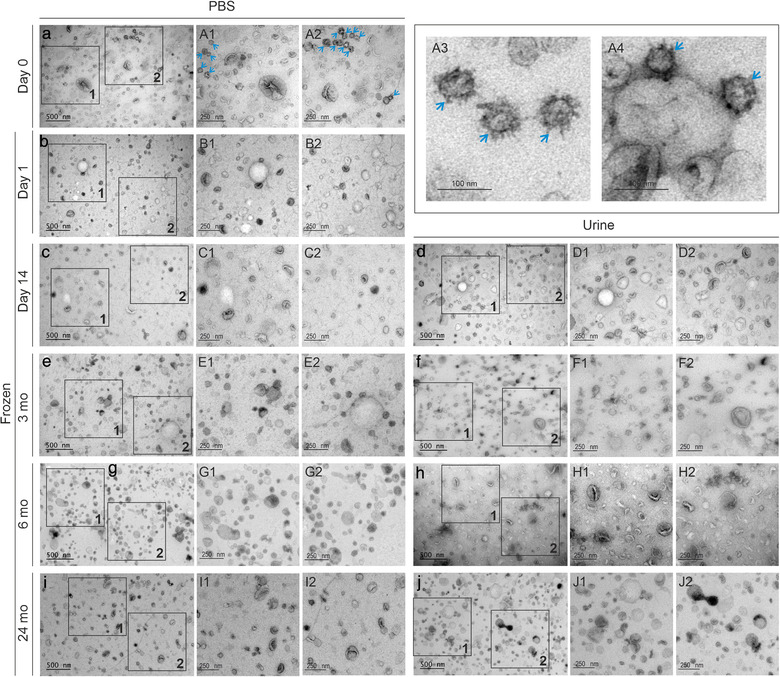
Effect of storage time (‐80°C) and storage medium on uEV morphology. Transmission electron micrographs of fresh uEV (day 0) or ‐80°C stored uEV divided based on storage medium i.e. uEV were isolated from urine stored at ‐80°C or uEV were isolated from fresh urine and then stored in PBS at ‐80°C for 1 day, 14 days and 3, 6, or 24 months. uEV derived from equal volumes of urine were loaded to grids from each time point. Blue arrowheads: particles with spikes visible in fresh samples. Months (mo), phosphate buffered saline (PBS), urinary extracellular vesicles (uEV)
SDS‐PAGE of stain‐free and Coomassie blue stained gels showed similar protein profiles of uEV isolates across all storage time points and storage medium (Figure 3a). Similarly, Western blotting showed the presence of EV markers TSG101, CD9 and HSP70 in all samples (Figure 3b). Gels for SDS‐PAGE and Western blotting were loaded with uEV derived from equal urine volumes.
FIGURE 3.

Effect of storage time (‐80°C) and storage medium on uEV protein profile and markers. Data is from fresh uEV (day 0) or ‐80°C stored uEV divided based on storage medium i.e. uEV were isolated from urine stored at ‐80°C or uEV were isolated from fresh urine and then stored in PBS at ‐80°C for 1 day, 14 days and 3, 6, or 24 months. uEV derived from equal volumes of urine were loaded from each time point. At each time point, medium was run as three or two technical replicates. a. SDS‐PAGE protein profiles of uEV detected in stain free‐gels (day 0 and 1) or in coomassie blue ‐stained gels (day 14 and 3, 6, or 24 months). b. Western blotting of uEV enriched markers. Immunodetection of EV markers TSG101, HSP70 and CD9 is shown. Months (mo), phosphate buffered saline (PBS), urinary extracellular vesicles (uEV)
3.1.3. Fresh uEV isolates presented particles with surface spikes – detection of commonly coisolated polyoma viruses
Although the overall morphology of uEV appeared rather stable during cryostorage, we observed particles with surface spikes (Figure 2a1, a2, a3, a4) and average size of 56.3 ± 0.1 nm (142 particles measured from negative staining EM images from 3 unique samples) in some of the samples and with higher frequency or better visibility in the fresh, unfrozen samples (4 out of 9 fresh individual or pooled samples). Spiked particles were further explored by staining 5 fresh and frozen (‐80°C) sample pairs. Again, we found spiked particles in the fresh uEV isolates, but not in the frozen counterpart (Figure 4a and c) in 3 out of 5 pairs. To explore whether the spiked particles were positive for common EV markers, we performed immunostaining against CD59. CD59 was selected, because it is a common uEV surface marker, and the staining is specific with minimal background (Puhka et al., 2017). The CD59 immunostaining was observed in uEV but no gold particles labelled the spiked particles (Figure 4c1 and c2).
FIGURE 4.

Effect of freezing (‐80°C) on particle surface spikes. Transmission electron micrographs of fresh and frozen (‐80°C) uEV from the same sample. Vesicles from equal volume of urine were loaded onto grids. a and b. Negative staining. c and d. Immunodetection of CD59. Blue arrows: vesicles with spikes, back arrows: gold particles indicating presence of CD59. Urinary extracellular vesicles (uEV)
Having observed spiked particles of uniform size in the fresh uEV isolates, we wanted to explore, if urinary viruses co‐isolated with uEV, and used qPCR to detect common urinary polyoma viruses. BK‐polyomavirus was detected in two uEV isolates from individual donors and in the uEV isolate from pooled urine (pool 1), but it was not detected in the pooled urine. JC‐polyoma virus was detected in both the pooled urine and uEV isolate from this pooled urine (Supplementary Table 3).
These results show that storing urine or uEV in PBS at –80°C for up to 24 months preserves particle size, particle concentration, uEV morphology and protein markers. However, particle surface characteristics are better observed in never frozen samples. Polyoma viruses co‐isolate with uEV and should be considered as common contaminants in uEV isolates.
3.2. Effect of storage time, temperature and sample handling on uEV
3.2.1. Urine storage temperature affected more uEV‐enriched protein markers than uEV morphology
Electron microscopy showed that uEV of typical morphology could be isolated from urines stored for 1 year at ‐20°C or ‐80°C including donors with T1D and micro‐ or macroalbuminuria (Figure 5a). To study the presence of EV markers in uEV derived from urine stored up to 24 months at ‐20°C or ‐80°C, we isolated vesicles from low (7.8) and large (30 ml) volumes of urine from healthy controls and donors with T1D and normo‐ or macroalbuminuria. By Western blotting of uEV from low volumes of urine, we found that samples stored at ‐20°C had less TSG101, CD9 and CD63, especially in normo‐ and macroalbuminuria samples which were stored for more than 4 months (Figure 5b). All EV markers were still detected in the uEV isolated from healthy controls urine stored at ‐20°C for 1.5 months (Figure 5b) with similar levels as in‐ 80°C. Tamm‐Horsfall protein (THP) was detected in all samples, while Golgi marker GM130 was negative for all samples. In uEV from large volumes of urine, the results were similar. EV markers TSG101, CD9 and podocalyxin gave fainter signals for the samples stored for more than 4 months, THP was detected in all uEV isolates at variable levels, GM130 was negative and the endoplasmic reticulum marker calnexin only faintly detected in the uEV isolates in comparison to the urine pellet and HeLa cells lysate which exhibited strong staining (Supplementary Figure 3).
FIGURE 5.
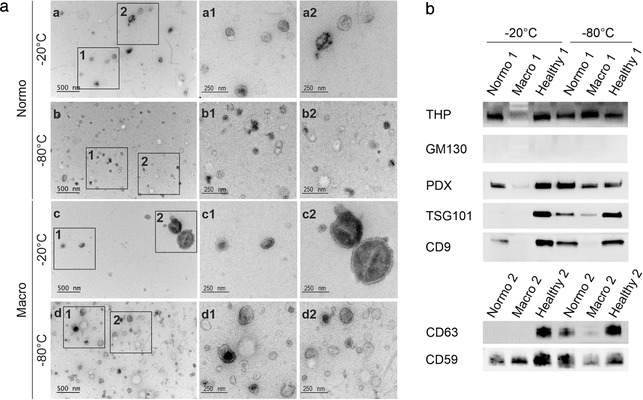
Effect of urine storage temperature (‐20°C vs. ‐80°C) on uEV morphology and protein markers. a. Transmission electron micrographs of frozen uEV from donors with type 1 diabetes and normo‐ or macroalbuminuria. Urine samples were stored at ‐20°C or ‐80°C for 1 year before uEV isolation. b. Western blotting of uEV isolated from urine stored at ‐20°C or ‐80°C for 1.5 months (healthy) or longer, up to 4 years (normo and macro). Here, uEV were isolated from low urine volume, 7.8 ml (uEV isolated from high urine volumes in Supplementary Figure 3). uEV derived from equal volumes of urine were loaded for each sample. Immunodetection of EV markers (PDX, TSG101, CD9, CD63 and CD59), a common protein that co‐isolates with uEV (THP) and a Golgi marker (GM130). Healthy control (Healthy), macroalbuminuria (macro), normoalbuminuria (normo), podocalyxin (PDX), Tamm–Horsfall protein (THP), urinary extracellular vesicles (uEV)
3.2.2. Urine storage at ‐20°C and DNAse treatment decreased RNA yield from uEV, whereas storage at ‐80°C did not
We explored the uEV RNA yields focusing on the effect of different pre‐analytical variables: urine storage time and temperature, addition of protease inhibitors, collection type, pre‐clearing and DNAse treatment including samples from donors with varying albuminuria status (Supplementary Table 1). To study the effect of storage time on RNA yield, uEV were isolated from 107 urine samples stored for 1 day to up to 4 years at ‐80°C. Samples were obtained from healthy controls and individuals with T1D or T2D and normo‐, micro‐ or macroalbuminuria and divided into storage time groups. Statistical testing with ANOVA did not show significant differences between the time groups (average RNA yields ranging between 517–900 pg/ml of urine) (Figure 6a, Supplementary Table 4). Since 8.4% of the urine samples were donated by women and it has been reported that uEV from women may carry more RNA than uEV from men (Ben‐Dov et al., 2016), we repeated the analysis excluding women. However, we obtained similar results: the time groups did not differ (Supplementary Table 4).
FIGURE 6.

Effect of storage time, storage temperature and DNAse treatment on RNA or DNA yield from uEV. Total RNA yield (expressed as pg/ml of urine) was quantified using pico 6000 RNA chip in Agilent 2100 Bioanalyzer. DNA content was quantified using Qubit dsDNA HS Assay kit. a. RNA yield and urine storage time. uEV were isolated from urines stored for up to 4 years at ‐80°C including samples from healthy controls and donors with T1D or T2D and normo‐ micro‐ or macroalbuminuria. Differences in the RNA yield between time points were tested using One‐way ANOVA and Bonferroni Post‐Hoc test. b. RNA yield and urine storage temperature (‐20°C, ‐80°C). uEV were isolated from healthy controls and donors with T1D and normo‐ or macroalbuminuria including urines stored for up to 4 years. T‐test for paired samples was used to assess the difference between groups. c and d. DNAse treatment. RNA yield (C) and DNA concentration (D) of uEV‐RNA treated with and without DNAse during RNA isolation. uEV were isolated from healthy control samples. T‐test for paired samples was used to assess difference between groups. Months (mo), type 1 diabetes (T1D), type 2 diabetes (T2D), urinary extracellular vesicles (uEV), with DNAse treatment (+DNAse), without DNAse treatment (‐DNAse)
We further investigated storage time –induced differences by dividing the 107 samples into two groups: non‐albuminuria (healthy controls + T1D or T2D and normoalbuminuria) and albuminuria (T1D or T2D and micro‐ or macroalbuminuria). Again, no statistically significant differences were found between the time groups for non‐albuminuria (average RNA yield range 408–886 pg/ml), or albuminuria (561‐962 pg/ml) (Supplementary Figure 4A and B, Supplementary Table 4).
To assess the effect of storage temperature on uEV RNA yield, urine samples from healthy controls and individuals with T1D and normo‐ or macroalbuminuria were divided into aliquots and stored either at ‐20°C or ‐80°C for 1.5 months to 4 years. We found that the mean RNA yield from the ‐20°C stored samples was significantly reduced compared to the ‐80°C stored samples (T‐test for paired samples; P < 0.01, Figure 6b, Supplementary Figure 5, Supplementary Table 4). We repeated the analysis by dividing the above samples in non‐albuminuria (healthy controls + T1D and normoalbuminuria) and albuminuria (T1D and macroalbuminuria) groups. In both groups, the mean uEV RNA yield was higher from the urines stored at ‐80°C compared to urines stored at ‐20°C, but the difference was statistically significant only for the non‐albuminuria group (t‐test for paired samples; P < 0.05, Supplementary Figure 6A and B, Supplementary Table 4). To assess the effect of short‐term storage (1.5 months) separately, we compared uEV RNA yields between the storage temperatures and using low or high volumes of healthy control urines for UC –however, no significant changes were found (Supplementary Figure 6C and D).
The urine samples for the storage time and temperature studies had protease inhibitors. However, in many biobank collections they are omitted, thus we analysed the effect of protease inhibitors on uEV RNA yield from healthy control urine samples stored at ‐80°C for up to 3 years. First, we assessed the effect of protease inhibitors on RNA yield as a function of urine storage time within groups i.e. short‐term (1‐14 days) versus long‐term (> 24 months) storage in samples stored with protease inhibitors (+PI) or without (‐PI). Urine storage +/‐ PI appeared to preserve uEV RNA yield similarly over time: we did not find statistically significant differences in yield between the short and long term storage (Supplementary Figure 6E and F, Supplementary Table 4). Second, we analysed the effect of protease inhibitors on uEV RNA yield as a function of storage time between groups: urines stored +PI versus ‐PI for short‐ and long term. Again, we did not find statistically significant differences (Supplementary Figure 6G and H, Supplementary Table 4).
We next compared the uEV RNA yields from two main clinical urine collection types (24 h vs. overnight). We isolated the uEV from pairs of urine samples collected from individual donors on the same day and then stored for up to 20 months at ‐80°C. Donors included healthy controls and individuals with T1D and micro‐ or macro‐albuminuria. There were no statistically significant differences between the RNA yields from 24 h and overnight collections (Supplementary Figure 6I, Supplementary Table 4).
To study the effect of pre‐clearing urine before freezing, we included urine samples from individuals with T1D and micro‐ or macroalbuminuria stored for up to 2 months at ‐80°C. Again, we found no statistically significant differences in uEV RNA yields from the paired urines processed with or without the pre‐clearing step (Supplementary Figure 6J, Supplementary Table 4).
We finally evaluated the effect of in‐column DNAse treatment during uEV RNA‐isolation on RNA and DNA yield. Here, we used healthy control samples, different RNA isolation kits and uEV isolated by UC from different starting volumes of urine. However, between sample pairs, the processing was always done in parallel, and the only difference was the treatment with DNAse (+DNAse) and without DNAse (‐DNAse). We found that both RNA and DNA yield from samples ‐DNAse was 20% and 23% higher than from samples +DNAse, respectively (t‐test for paired samples; P < 0.05; Figure 6c and d, Supplementary Figure 5, Supplementary Table 4).
3.2.3. Urine storage temperature (‐20°C vs. ‐80°C) affected the mRNA transcriptome of uEV
To study the effect of urine storage temperature on the transcriptome of the uEV, we isolated uEV from 24 urine samples from T1D individuals with normo‐, micro‐ or macroalbuminuria stored for 1 year. Samples included 4 pairs of urine stored at ‐20°C or ‐80°C and 16 more samples stored at ‐80°C.
Quality of the mRNA sequence alignment, evaluated as mapped versus unmapped reads, appeared not to be affected by storage temperature (Figure 7a). Number of raw reads evaluated between sample pairs, was slightly lower for the ‐20°C samples (4.5E+06 ± 0.8 E+06 reads, n = 4) compared to the ‐80°C samples (5.3E+06 ± 0.4 E+06 reads, n = 4), but the difference was not statistically significant.
FIGURE 7.

Effect of urine storage temperature (‐20°C versus ‐80°C) on mRNA sequencing of uEV. Results depict 24 uEV‐mRNA samples including 4 pairs of urine samples stored in ‐20°C versus ‐80°C (pairs 1–4) and 16 more samples stored at –80°C. All samples were stored for 1 year at the indicated temperatures. Data was TMM normalized and analysed using edgeR. a. Alignment of raw sequencing reads to human genome using STAR. b. Sample to sample correlation heatmap (10136 genes, with > 1 raw count in at least 90% of the samples). c. Principal component analysis (10136 genes, with > 1 raw count in at least 90% of the samples). d. Heatmap of 22 kidney‐enriched genes (www.proteinatlas.org). e. Differential expression analysis of the ‐20°C versus ‐80°C pairs (4 pairs) (12642 genes, with > 5 raw counts in at least 50% of the samples). Volcano plot depicts differentially expressed genes. Adjusted p‐values lower than 0.05 and log2 fold change ≥ 0.75 or ≤ ‐0.75 are highlighted in red. Principal component analysis (PCA), urinary extracellular vesicle (uEV)
Genes with raw counts of > 1 in at least 90% of the samples were selected for an exploratory analysis of the data (10136 genes). Sample‐to‐sample correlation analysis (Figure 7b) as well as PCA (Figure 7c) showed two clear clusters corresponding to ‐20°C or ‐80°C storage. Considering the separation, we assessed whether kidney‐enriched genes were affected in the ‐20°C samples. Out of 53 genes classified as kidney‐enriched in the human protein atlas, 22 were found in our filtered dataset. Over 50% of these kidney‐enriched genes were less expressed in the ‐20°C samples compared to ‐80°C samples (Figure 7d).
To identify the differentially expressed genes between the ‐20°C and ‐80°C samples, only sample pairs and genes with > 5 raw counts in at least 50% of the samples (12642 genes) were included in the analysis. The analysis identified 137 upregulated and 1967 downregulated genes in the ‐20°C samples (Figure 7e, Supplementary Table 5). To characterize the genes that were up ‐and downregulated, transcript length and GC‐content information of the genes was retrieved using BioMart. While the transcript lengths did not differ, the GC‐content was higher in genes downregulated by urine storage at ‐20°C (Figure 8a and b). For further exploration of the GC‐contents, we analysed all genes with ≥5 raw counts. This confirmed that samples stored at ‐20°C contained a significantly higher proportion of genes with a low (0‐50%) GC‐content (81.4% ± 1.1%, n = 4) than the samples stored at ‐80°C (71.6% ± 0.1% n = 4) (t‐test for paired samples; P < 0.01; Figure 8c). Inversely, for the high %GC categories (51%‐60% and 61%–80%), samples stored at ‐80°C showed higher proportions (23.6% ± 0.1%, 4.7% ± 0.8%, respectively, n = 4) than the samples stored at ‐20°C (15.9% ± 0.9%, 2.6% ± 0.3%, respectively, n = 4) (t‐test for paired samples; P < 0.01; Figure 8c). As P‐bodies have been found to enrich AU‐rich transcripts (Courel et al., 2019), we next investigated whether the transcripts preserved at ‐20°C could be derived from P‐bodies. Here, we used mRNA‐seq data of 5202 transcripts shown to be enriched in sorted P‐bodies (log2FC > 1, FDR < 0.05, (Hubstenberger et al., 2017)) and compared them to the transcripts up‐ and downregulated by ‐20°C storage. Venn analysis showed that 67 out of the 137 (49%) upregulated mRNAs were also enriched in P‐bodies compared to only 202 out of 1967 (10%) of the downregulated (Figure 8d). Thus, the ‐20°C upregulated transcripts were relatively enriched in P‐body transcripts.
FIGURE 8.

Gene length, GC content and overlap with P‐body transcripts of up‐ and down‐regulated genes in uEV isolated from urines stored at ‐20°C versus ‐80°C. a. Transcript length (if several transcripts were retrieved, the average length was calculated). b. Gene %GC content. c. Distribution of gene %GC content in ‐20°C and ‐80°C samples. Genes with raw counts ≥ 5 were selected for the analysis. T‐test for four paired samples (Type 1 Diabetes) was used to assess differences between groups per percentage category. Urinary extracellular vesicles (uEV). d. Venn diagram of P‐body enriched transcripts and transcripts upregulated or downregulated in uEV by urine storage at ‐20°C versus ‐80°C
By gene set enrichment analyses (GO terms), we found that the downregulated genes in the ‐20°C group were mainly involved in biological processes associated to carbohydrate and lipid metabolism (Figure 9a, Supplementary Table 6). Molecular functions of downregulated genes in the ‐20°C samples were related to DNA, GTPase, and cadherin binding (Figure 9c, Supplementary Table 6), while most cellular components were associated to extracellular vesicle, extracellular exosome, extracellular organelle, cytosol nucleoplasm and vesicle (Figure 9e, Supplementary Table 6). For up‐regulated genes in the ‐20°C group, no enriched biological processes were found. Molecular functions of the up‐regulated genes were linked to RNA binding and nucleic acid binding (Figure 9b, Supplementary Table 6). Most cellular component ontologies in the upregulated genes from the ‐20°C samples were associated to nucleoid, mitochondrial nucleoid, spliceosome, mitochondrial inner membrane and organelle inner membrane (Figure 9d, Supplementary Table 6).
FIGURE 9.
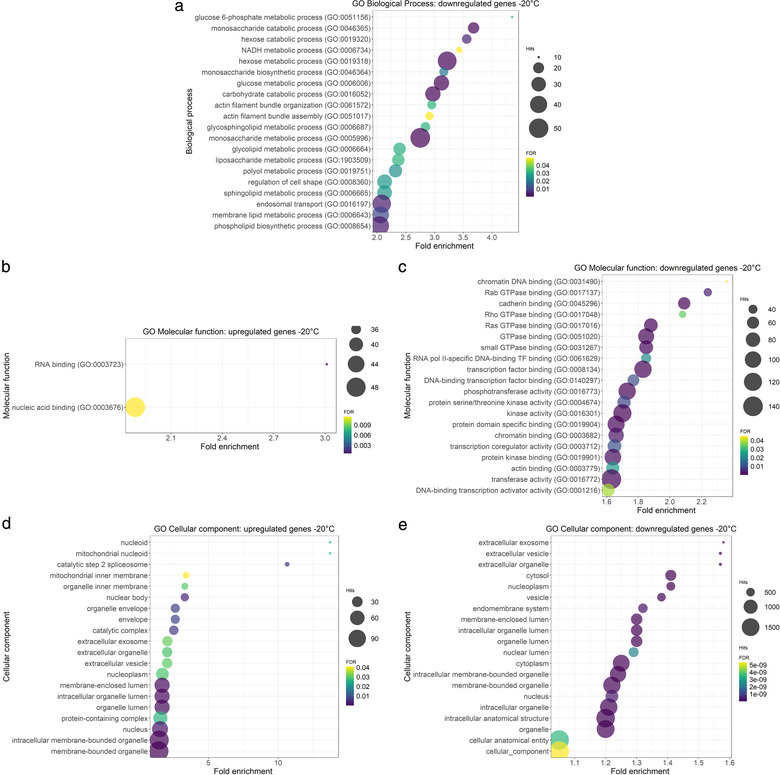
Gene ontology enrichment analysis of differentially expressed genes in uEV isolated from urine stored at ‐20°C versus ‐80°C. a. Top 20 GO biological process of downregulated genes in the ‐20°C group. No significant biological processes were found for upregulated genes. b. GO molecular function of upregulated genes in the ‐20°C group. c. Top 20 GO molecular function of downregulated genes in the ‐20°C group. d. Top 20 GO cellular component of upregulated genes in the ‐20°C group. e. Top 20 GO cellular component of downregulated genes in the ‐20°C group. Gene ontology (GO), Urinary extracellular vesicles (uEV)
3.2.4. DNAse treatment affected the read distributions from mRNA‐seq
We analysed the effect of in‐column DNAse treatment during uEV RNA‐isolation on mRNA‐seq results. To this end, we used sequencing data from 19 pairs of uEV isolates from which RNA was isolated +/‐ DNAse. uEV were isolated from urines stored at –80°C including urine donors with T1D and normo‐, micro and macroalbuminuria. RNA profiles obtained using Agilent RNA 6000 Pico Kit were similar between +/‐DNAse treatments (representative profiles in Supplementary Figure 5). Alignment of raw sequencing reads showed that > 90% of reads mapped uniquely to human genome in both groups (‐DNAse: 92.7% ± 1.5 %; +DNAse 90.6±1.6). Analysing the distribution of the mapped reads with RSeQC, we found that the RNA samples treated +DNAse had a higher average proportion of 3′ untranslated region exons (3′UTR Exons), while the samples ‐DNAse treatment had a higher average proportion of coding sequence exons (CDC Exons), 5′ untranslated region exons (5′UTR Exons), introns, and intergenic parts, transcription start and end sites (TSS or TES within 10 kb) (Supplementary Figure 7 and 8, Supplementary Table 7).
To estimate the number of genes captured in each group, we filtered the data to include genes with > 1 raw counts in at least 50% of the samples. The number of genes that passed the filter in the +DNAse group was slightly higher than for the ‐DNAse group (16131 and 14803 genes, respectively). We applied a similar filter for protein coding genes and found that their numbers were closely similar in both groups (+DNAse: 12950 genes, ‐DNAse: 12512 genes).
Exploratory analysis of the mRNA of log2CPM normalized counts by PCA showed a batch effect between groups (Supplementary Figure 9), and consequently, no further comparisons of datasets were performed.
3.2.5. Urine storage temperature (‐20°C vs. ‐80°C) affected the miRNA transcriptome of uEVs
To evaluate the effect of storage temperature on miRNA transcriptome of uEV, the same total RNA which was used for the mRNA‐seq was subjected to miRNA‐seq. Comparison of raw miRNA counts between the 4 sample pairs (‐20°C vs. ‐80°C) showed that the mean counts for the ‐20°C samples (4.57E + 04 ± 1.3 E + 04 reads, n = 4) were significantly lower than for the ‐80°C samples (8.7E + 05 ± 4.4 E + 05 reads, n = 4) (t‐test for paired samples; P < 0.05, Figure 10a). MicroRNAs with > 1 raw counts in at least one sample were selected for an exploratory analysis of the data (623 miRNAs). By sample‐to‐sample correlation analysis or PCA of the 4 pairs (‐20°C vs. ‐80°C) and 16 more samples (–80°C), we did not find a clear clustering of the ‐20°C and ‐80°C samples (Figure 10b and c). However, when only sample pairs were compared, a distance between the ‐20°C and ‐80°C groups was evident in the correlation heatmap and by PCA (Supplementary Figure 10). To estimate the number of miRNAs in the ‐20°C and ‐80°C samples, we filtered miRNAs with > 1 raw counts in all samples for each group. The number of miRNAs that passed the filter in the ‐80°C samples (162 miRNAs) was higher than in the ‐20°C samples (48 miRNAs). To evaluate the differentially expressed miRNAs between the ‐20°C and ‐80°C samples, only sample pairs were compared including miRNAs with raw counts of > 1 in at least 50% of the samples (183 miRNAs). We found 4 upregulated and 29 downregulated miRNAs in ‐20°C samples (Figure 10d, Supplementary Table 8).
FIGURE 10.

Effect of urine storage temperature (‐20°C vs. ‐80°C) on uEV miRNA sequencing. Results depict 24 uEV miRNA samples including 4 pairs of urine samples stored in ‐20°C versus ‐80°C (pairs 1–4) and 16 more samples stored at –80°C. All samples were stored for 1 year at the indicated temperatures. Data was TMM normalized and analysed using edgeR. a. miRNA raw sequencing reads. b. Sample‐to‐sample correlation heatmap (623 miRNAs, with > 1 raw count in at least one sample). c. Principal component analysis (623 miRNAs, with > 1 raw count in at least one sample). d. Differential expression analysis of ‐20°C versus ‐80°C pairs (4 pairs) (183 miRNAs, with > 1 raw count in at least 50% of the samples). Heatmap showing the 33 differentially expressed miRNAs (4 upregulated and 29 downregulated in ‐20°C samples). MicroRNAs with adjusted p‐values lower than 0.05 and a log2 fold change ≥ 0.75 or ≤ ‐0.75 were considered differentially expressed between groups. Principal component analysis (PCA), urinary extracellular vesicles (uEV)
In summary, storage of urine at –20°C compared to storage at –80°C affected the detection of uEV protein markers, RNA yield, mRNA and miRNA transcriptome. The type of urine collection, use of protease inhibitors and pre‐clearing the urine before freezing at ‐80°C had no effect on the RNA yield. DNAse treatment had a small effect on RNA yield and the mRNA‐seq results.
3.3. Development of an enrichment list to evaluate miRNA stability and concordance between different uEV isolation methods
Most of the current uEV biomarker research is focused on miRNA but progress is hampered by varied outputs from different uEV isolation methods and lack of housekeeping miRNAs. Thus, we decided to develop a miRNA enrichment list to identify stable miRNAs from different uEV isolation methods. First, we defined a list of miRNAs commonly enriched in the uEV isolates (enriched miRNAs) using miRNA expression data derived from one miRNA‐seq platform and one microarray platform, as well as data in Vesiclepedia (Pathan et al., 2019). The miRNA‐seq data was from uEV isolates and microarray data from uEV isolates and their paired urine samples ‐ all from healthy male controls. For miRNAs enriched in EV or urine, we ranked the results from both platforms from the highest to the lowest expression, after which the top expressed miRNAs were cross compared. From miRNAs in uEV, we selected those that were highly expressed in both platforms and absent or lower expressed in urine samples according to the microarray data. In addition, we cross‐compared that the selected miRNAs had entries in Vesiclepedia (Supplementary Table 9).
For piloting the stability of the 14 miRNAs in the enrichment list, we evaluated 3 published miRNA‐seq datasets comparing uEV isolation methods. Here we selected studies with more than 5000 raw miRNA counts in all samples and isolation methods.
Dataset from Barreiro et al consisted of miRNAs from uEV isolates obtained by 3 methods (hydrostatic filtration dialysis, UC and Norgen Biotek urine exosome purification and RNA Isolation midi kit). Urines were derived from 5 healthy male controls and 5 men with diabetic kidney disease manifesting with macroalbuminuria. The expression levels of enriched miRNAs in uEV isolates were high and did not differ between methods (Figure 11a).
FIGURE 11.

Comparison of miRNAs enriched in the uEV isolates from this study or in previous publications evaluating uEV isolation methods. The list of miRNA enriched in uEV isolates was build based on healthy male ‐derived data from two different miRNA platforms and Vesiclepedia. The plots show the miRNA expression levels from different miRNA‐seq studies including only men. a. Barreiro et al., 2020. uEV were isolated from healthy controls and individuals with type 1 diabetes and macroalbuminuria. Raw counts were TMM normalized using edgeR. b. Mussack et al., 2019. uEV were isolated from healthy controls. Trimmed sequences were aligned using Chimira and counts were TMM normalized using edgeR. c. Srinivasan et al., 2019. Data selected for our analysis is derived from uEV or exRNA isolated from pooled urines of healthy controls (technical replicates). RPMSmiR counts were log2 transformed. d. uEV isolated by UC from urine sample pairs (4 pairs) stored at ‐20°C and ‐80°C. Urine samples included were from individuals with type 1 diabetes and micro‐ or macroalbuminuria. Hsa‐miR‐10a‐5p was found to be upregulated in ‐20°C group (figure 9). Counts per million (CPM), exosome Isolation Kit Pan, human (Exosome isolation kit), false discovery rate (FDR), hydrostatic filtration dialysis (HFD), ultracentrifugation (UC), reads per million scaled miRNA (RPMSmiR), urinary extracellular vesicles (uEV), urine Exosome Purification and RNA Isolation Midi Kit (NG)
Dataset of Mussack et al included miRNAs from uEV isolates derived from six healthy men and isolation by 5 methods (Norgen Biotek urine exosome purification and RNA Isolation midi kit, Miltenyi Biotec exosome isolation kit pan, Qiagen exoRNeasy serum/plasma midi kit, miRCURY exosome isolation kit – cells, urine and CSF, and UC). Analysis of the enriched miRNAs in uEV isolates showed that for most of the isolation methods, these miRNAs were highly and stably expressed. Some of the miRNAs appeared to be less expressed in samples isolated by Norgen isolation kit (Figure 11b).
Dataset from Srinivasan et al comprised miRNAs from uEV isolates of 3 technical replicates of pooled healthy male urines. Authors filtered samples per library size. Four uEV isolation methods (System Biosciences ExoQuick, Qiagen ExoRNeasy Midi kit, UC, Millipore ultrafiltration) and a total RNA isolation method (Qiagen miRNeasy micro kit) passed author's filters and were included in our analysis. Similarly to other datasets, enriched miRNAs were highly and stably expressed in uEV isolates from most of the isolation methods. Only hsa‐miR‐23b‐3p appeared to be less expressed in the samples isolated by ExoRNeasy, UC and ultrafiltration (Figure 11c).
Finally, we studied our data from the uEV isolates from urines stored at ‐20°C and ‐80°C. The enriched miRNAs were equally expressed in both groups except for hsa‐miR‐10a‐5p, which was significantly higher in uEV isolates from ‐20°C than ‐80°C urines (Figure 11d).
Taken together, the results showed that the enriched miRNAs in our list were highly and stably expressed in diverse datasets which highlights their robustness.
4. DISCUSSION
Interest in uEV as a source for biomarkers has increased steadily in recent years. However, little is known about the effects of pre‐analytical variables on urine and uEV cargo stability in the long term. To contribute to the standardization efforts in the EV field including those of the International Society of Extracellular Vesicles and urine biobanking activities, we explored the effect of basic pre‐analytical variables on the quality of the uEV isolates for their application in transcriptomics research.
We first evaluated the effect of short‐ to long‐term storage, up to 24 months, of both urine and isolated uEV in PBS at ‐80°C. By NTA, we found that uEV were stable during the storage, and both storage modalities stored the assessed particles equally well in terms of particle concentration and size distribution (Tables 1 and 2). Oosthuyzen et al., 2013 reported a decrease in particle concentration (evaluated as area under the curve for particles sized 20–100 nm) already after 2 h of storage at ‐80°C, while Yuana et al., 2015 reported a modest increase of particle concentration (2‐fold increase) and diameter (17%) in samples stored at ‐80°C for 1 year. However, these studies measured the particles in cell‐free urine while we measured particles from uEV isolates obtained by UC. Other reports on short‐term storage of EV derived from cell cultures and plasma are in line with our findings (Sarker et al., 2014; Sokolova et al., 2011). As the current isolation and analysis workflows are prone to some variation, we suggest that changes in the particle concentrations should be > > 3x (and statistically significant) to be considered truly significant. Our data also showed preservation of uEV morphology, protein profiles, and EV markers at ‐80°C for at least 2 years either as urine or as isolated uEV in PBS (Figures 2 and 3). These findings agree with the previous reports on storage of urine or cell free urine at ‐80°C up to 1 year (Yuana et al., 2015; Zhou et al., 2006). The equal preservation of uEV in PBS and urine is relevant information for biobanks and other sample collections considering storing isolated uEV, which would consume a lot less cryostorage space than urine.
The uEV RNA yields from > 100 urine samples stored up to 4 years at ‐80°C appeared stable in various pre‐analytical study settings. As we included samples with different albuminuria status, this suggests that urine composition does not affect uEV RNA stability (Figures 6a and Supplementary Figure 4 and 6). Focusing on the pre‐analytical steps relevant to biobanks, we observed no effect of collection type or addition of (relatively expensive) protease inhibitors before freezing, or pre‐clearing of urine, which adds hands‐on time prior freezing. However, it should be noted that the effect of pre‐clearing depends e.g. on the collection properties‐ the amount of cells in urine ‐ and pre‐clearing is generally recommended for uEV studies (Erdbrügger et al., 2021) (Supplementary Figure 6).
Studying the morphology of uEV in fresh versus frozen samples by EM, we observed the presence of spiked particles in some of the fresh urine samples (Figure 4). These spiked particles were scarcely detected in frozen samples even after a short‐term storage at ‐80°C. A plausible explanation for this phenomenon is a change of the spike protein structure due to freezing as reported e.g. for SARS‐CoV2 under diverse storage conditions (Edwards et al., 2021). As the spiked particles had a homogenous size (56 nm) compatible with not only uEV, but also viruses, we addressed the question whether the most prevalent urinary polyomaviruses were commonly co‐isolating with uEV (Goetsch et al., 2018; Polo et al., 2004). JC‐ and BK‐polyomavirus infections are common and usually asymptomatic during childhood, but the virus persists in the kidney and urinary tract in adulthood and may be detectable in asymptomatic adults (Goetsch et al., 2018). By qPCR, we found both the JC‐ and BK‐polyomavirus in uEV isolates and JC‐polyomavirus also in urine. The discrepancy between the detection of BK‐polyomavirus in uEV isolates but not in urine could be due to an enrichment of viral particles by UC. In conclusion, the possible presence of viral particles in urine samples should be considered when isolating uEV, since they can contribute to the total number of particles in the isolates and introduce nucleic acid contamination, DNA in the case of polyomaviruses, to the samples. Our results suggest that never frozen urine is the best choice for quality control of uEV isolates by EM, as it allows the visualization of surface characteristics of particles.
Many large urine sample collections are stored at ‐20°C for years. This may be due to lack of cold storage resources or guidelines in urine storage for uEV studies. One of the aims of the present study was to explore how urine samples stored at ‐20°C differ from the ones stored at ‐80°C, and whether the ‐20°C samples can be used for uEV research. Even if we did not observe morphological changes in uEV by EM, storage at ‐20°C reduced uEV RNA and EV protein marker yields after 4 months (Figure 6b, Supplementary Figure 3, 6A‐D). As the uEV RNA and proteins were still preserved at the 1.5 month storage time point, our results agree with the previous reports of short‐term stability of EV proteins at ‐20°C including urine stored for 1 week and cell ‐derived EV stored for 10 days (Lee et al., 2016; Zhou et al., 2006). We thus hypothesize that already a mid‐term storage of urine at ‐20°C (2‐3 months) may affect the stability of some EV markers and RNA yield. In contrast to our data, no differences were observed in RNA yield of EV isolated from plasma stored up to 5 years at ‐20°C compared to fresh samples (Ge et al., 2014). Thus, results may vary between biofluids and EV (and RNA) stability during storage should be assessed separately in each case.
Storage at –20°C affected also the sequencing results. Although the number of raw reads in uEV mRNA‐seq from the urines stored at ‐20°C for 1 year was acceptable (above 4 million reads), they were lower than from the urines stored at ‐80°C. PCA and sample‐to‐sample correlation analysis showed sample separation by storage temperature, which was explained by the finding that 16% of the genes were differentially expressed, with a predominant downregulation, also among the kidney‐enriched genes, in the uEV of the ‐20°C group (Figure 7). As these results suggested degradation of a part of the uEV transcriptome, we assessed whether some specific types of transcripts, e.g. long, were degraded more than others. Interestingly, even if the transcript lengths of the up‐ and down‐ regulated genes did not differ, the ‐20°C down‐regulated genes had a higher GC‐content than the up‐regulated ones (Figure 8b and c). Conversely, ‐20°C samples were rich in genes with high AT‐content. Transcription of GC‐rich genes is more efficient than that of AT‐rich genes, and consequently, GC‐rich transcripts are more abundant (Kudla et al., 2006). High GC‐content of transcripts also contributes to their cytosolic localization (Mordstein et al., 2020). AU‐rich transcripts are less abundant and mostly located in cytoplasmic processing bodies (P‐bodies) (Courel et al., 2019) or in the nucleus (Mordstein et al., 2020). Transcripts located in the cytosol and P‐bodies can be exported to EV (Fabbiano et al., 2020). Thus, both GC‐ and AU‐rich transcripts can be found in uEV isolates inside EV or as part of cellular remnants. Therefore, a plausible explanation for our observations is that transcripts that suffer degradation at ‐20°C are the most abundant ones (high GC‐content). Then, during sequencing library preparation, there would be proportionally more AU‐rich transcripts in the ‐20°C samples available for amplification resulting in over amplification. This could mean that even when nominal levels of AU‐rich transcripts would not differ between ‐20°C and ‐80°C samples, such transcripts could become over‐represented in the ‐20°C libraries. However, this finding should be confirmed for example by using library preparation kits, which include unique molecular identifiers (UMIs). In addition, as almost half of the ‐20°C upregulated transcripts were also enriched in P‐bodies (Figure 8d), it is possible that AU‐rich transcripts enjoy a better protection from degradation at ‐20°C by e.g., some P‐body or EV‐components than the GC‐rich transcripts. Likewise, factors known to influence RNA degradation such as mRNA structure (Akiyama et al., 2016; Andrzejewska et al., 2020), poly‐A tail length contributing to protective structures at 3′ end (Akiyama et al., 2016), and preference of nucleases for certain sequences (Courel et al., 2019) should be considered. Gene ontology analysis revealed that the downregulated genes in the ‐20°C group were associated with biological functions of glucose and lipid metabolism and with the compartment of EV (Figure 9). Hence, uEV isolated from urine stored at ‐20°C are a poor source for studies of EV or sugar and lipid metabolism in e.g. diabetes or diabetic kidney disease. Based on the mRNA‐seq results, we would recommend to avoid ‐20°C stored samples, or, at least, avoid mixing mRNA‐seq data from samples stored at different temperatures. Many of the results from mRNA‐seq were replicated by miRNA‐seq, including differential expression of 18% of the miRNA with predominant downregulation in the ‐20°C stored samples (Figure 10 and Supplementary Figure 10). The ‐20°C stored samples gave significantly less miRNA raw reads and detected miRNAs (‐3 fold) than ‐80°C stored samples. Thus, the same recommendations apply to miRNA‐seq as presented for mRNA‐seq. Nevertheless, if analysis of –20°C stored samples or mixing of samples from different storage temperatures cannot be avoided in sequencing, the storage time of the compared samples should be considered and/or the analysis could be directed towards the most stable transcriptomes (Figure 8 and Figure 11).
It is commonly recommended to treat RNA samples to be sequenced with DNAse I to reduce the misinterpretation of DNA reads as RNA reads. In practice, the effect of DNAse treatment should be observed as a reduction of the fraction of intergenic reads (Griffith et al., 2015), if significant amounts of DNA are present in the sequenced samples. However, the amount of DNA is generally low in cell‐free samples, including urine (Oreskovic et al., 2019; Streleckiene et al., 2018). As the RNA content of EV samples is also low in comparison to cell or tissue RNA and the DNAse treatment reduces RNA yields (Norhazlin et al., 2015; Pang et al., 2020), the risk of losing the precious EV‐RNA due to the treatment is high. We thus regarded relevant to investigate the necessity of the DNAse treatment by analysing RNA yield and mRNA‐seq results from uEV. In agreement with previous studies, we observed a slightly reduced RNA and DNA yield in the +DNAse samples with all RNA isolation methods tested (Figure 6c and d). The mRNA‐seq read distributions had low, but consistent, differences between the +/‐DNAse groups. Intergenic regions were higher in the ‐DNAse samples compared to the +DNAse samples, but the proportion was less than 1% in both sample types. Coding exonic reads were higher in the ‐DNAse samples, but the percentage of this category was over 80% in both groups, which indicates a good library preparation (Supplementary Figure 7 and 8). Further, as there were no great differences in the number of all detected and protein coding genes, we conclude that RNA isolation both +/‐DNAse coupled to poly‐A approach for library preparation generated acceptable quality of sequencing. It is worth noting that this result is not likely to apply if random primers are used to generate sequencing libraries, because this approach yields a higher fraction of intergenic reads (Sultan et al., 2014; Zhao et al., 2014). However, as read distributions differed and PCA showed a batch effect between groups (Supplementary Figure 9), we would not recommend mixing samples extracted +/‐DNase in the same analysis. Either sample type can be analysed alone or a batch effect correction method tested.
EV isolation methods are based on variable physicochemical principles and therefore they may enrich different EV populations or other sample components (Merchant et al., 2017). Thus, when isolating uEV with different methods, miRNA‐seq results usually differ (Barreiro et al., 2020; Mussack et al., 2019; Park et al., 2020). As a result, the EV field has not yet been able to identify stable housekeeping miRNAs. We approached this problem by compiling a list of 14 miRNA commonly enriched in uEV isolates in a hope to identify stable miRNAs between EV isolation methods (Figure 11). We validated the list using miRNA‐seq datasets from three published studies, where authors compared three or more uEV isolation methods (Barreiro et al., 2020; Mussack et al., 2019 and Srinivasan et al., 2019); the miRNA expression levels were stable and highly concordant between the uEV methods, except for one isolation method and one miRNA in two of the studies. We also found a high concordance between the enriched miRNAs in uEV isolates from urines stored at –20°C and –80°C – surprising, given the lower number of reads from the ‐20°C samples. It is remarkable that the enriched miRNAs were stable and concordant in all the datasets despite that they were generated from different urine samples using various uEV and RNA isolation workflows, with different library preparation kits and different sequencing platforms. We thus conclude that our enrichment list is quite robust. We propose that the enrichment list could be tested for data normalization. It could also be further developed by incorporating data from women and e.g. groups of more variable miRNAs to better assess the concordance of uEV isolation methods for miRNA‐seq. We also want to point out that the robustness is a positive sign that a common stable transcriptome, “biofluid ‐specific housekeeping exRNA signature”, exists and seems feasible to be defined in future efforts of uEV method standardization including those of the extracellular RNA consortium or miREV (Hildebrandt et al., 2021).
We acknowledge that we only used UC for isolating the uEV. Many isolation methods are currently available which use other principles of EV isolation. For example, our results of long‐term storage stability of isolated uEV in PBS may not be reproduced if uEV were isolated using a kit with a different suspension buffer than PBS. However, since UC has been and is still the most used method to isolate EV (Royo et al., 2020), our results of urine storage are useful and may also serve as a guidance even when other isolation methods will be employed. For urine storage temperature experiments assessing EV protein marker stabilities, only a limited number of time points were tested. Thus, a study with higher number of samples and time points could allow determination of the exact time period (between 1.5 and 4 months), when uEV become unstable at ‐20°C. We did not include NTA analysis for the samples stored at different temperatures, although it could help to understand if the reduced yields of RNA and EV protein markers in the ‐20°C samples correlate with a reduction in the uEV numbers. Our study set‐up for protease inhibitors was better for comparing the RNA yields between the short and long storage time points within a group (either + or – PI) than for comparing between groups (+ vs. – PI). This was due to other differences between the groups than the inclusion or omission of PI, e.g., urine collection type. In the pre‐clearing test, the number of samples was low, which affects the statistical power. Thus, the protease inhibitors and pre‐clearing results should be confirmed using a higher number of paired samples. We used poly‐A library preparation to compare RNA‐seq between samples stored at ‐20°C and ‐80°C, but poly‐A approach is not optimal for degraded samples (Schuierer et al., 2017). As part of the RNA from the ‐20°C samples was degraded (apparent from the RNA yield and sequencing results and Bioanalyzer profiles, Supplementary Figure 5), a library preparation approach suitable for degraded RNA could be tested to improve sequencing results (Adiconis et al., 2013).
Finally, we have summarized our results and present recommendations on how to store and handle samples for transcriptomic studies of uEV in Figure 12. Our enrichment list comprising miRNAs commonly enriched in the uEV isolates can be used as a starting point to define the least variable uEV transcriptome independent from the used isolation and analytics workflows and for normalization attempts. We hope that the list and our findings and recommendations help to expedite standardization of the uEV biomarker field.
FIGURE 12.

Summary of results and recommendations. Never frozen refers to uEV isolated from fresh urine and kept at +4°C for a maximum of 1 day. Electron microscopy (EM), not applicable (NA), not determined (ND), not compared but data presented in the manuscript supports the statement (No difference)*, overnight (ON), principal component analysis (PCA), sequencing (seq), urinary extracellular vesicles (uEV), with DNAse treatment (+DNAse), with pre‐clearing before freezing (+pre‐clearing), with protease inhibitor (+PI), without DNAse treatment (‐DNAse), without pre‐clearing before freezing (‐pre‐clearing), without protease inhibitor (‐PI).
CONFLICT OF INTEREST
KB, OPD, SV, CF, PG, TT, HH, MY, PS, ES, TH, SL, AR, LG, MP, have no conflict of interest to declare.
Supporting information
Supporting information.
ACKNOWLEDGEMENTS
We thank Prof. Olli Kallioniemi for providing laboratory facilities, MD Olli Veikkola for technical assistance and Doc Pirkko Mattila for setting up EV sequencing technologies at FIMM, University of Helsinki. We also acknowledge the Sequencing Units of FIMM Tech Center supported by Biocenter Finland, and the Department of Clinical Sciences, Diabetes and Endocrinology, Lund University, Malmö, Sweden, for sequencing services, and Electron Microscopy Unit of the Institute of Biotechnology, University of Helsinki, for providing microscopy facilities. This project has received funding from the Innovative Medicines Initiative 2 Joint Undertaking under grant agreement no. 115974 (BEAt‐DKD). This Joint Undertaking receives support from the European Union's Horizon 2020 research and innovation program and EFPIA with JDRF. The research was also supported by the Academy of Finland (grants no. 263401, 267882, 312063 to L.G., and 317599 to O.P.D.). This project also received funding from SalWe Research Program Personalized Diagnostics and Care (GET IT DONE); Tekes – the Finnish Funding Agency for Technology and Innovation (Business Finland) grant 3986/31/2013.
Barreiro, K. , Dwivedi, O. P. , Valkonen, S. , Groop, P.‐H. , Tuomi, T. , Holthofer, H. , Rannikko, A. , Yliperttula, M. , Siljander, P. , Laitinen, S. , Serkkola, E. , af Hällström, T. , Forsblom, C. , Groop, L. , & Puhka, M. (2021). Urinary extracellular vesicles: Assessment of pre‐analytical variables and development of a quality control with focus on transcriptomic biomarker research. Journal of Extracellular Vesicles, 10, e12158. 10.1002/jev2.12158
REFERENCES
- Adiconis, X. , Borges‐Rivera, D. , Satija, R. , Deluca, D. S. , Busby, M. A. , Berlin, A. M. , Sivachenko, A. , Thompson, D. A. , Wysoker, A. , Fennell, T. , Gnirke, A. , Pochet, N. , Regev, A. , & Levin, J. Z. (2013). Comparative analysis of RNA sequencing methods for degraded or low‐input samples. Nature Methods, 10(7), 623–629. 10.1038/nmeth.2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama, B. M. , Eiler, D. , & Kieft, J. S. (2016). Structured RNAs that evade or confound exonucleases: Function follows form. Current Opinion in Structural Biology, 36, 40–47. 10.1016/j.sbi.2015.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders, S. , Pyl, P. T. , & Huber, W. (2015). HTSeq–a Python framework to work with high‐throughput sequencing data. Bioinformatics, 31(2), 166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrzejewska, A. , Zawadzka, M. , & Pachulska‐Wieczorek, K. (2020). On the way to understanding the interplay between the RNA structure and functions in cells: A genome‐wide perspective. International Journal of Molecular Sciences, 21(18), 6770. 10.3390/ijms21186770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong, D. A. , Dessaint, J. A. , Ringelberg, C. S. , Hazlett, H. F. , Howard, L. , Abdalla, M. A.K. , Barnaby, R. L. , Stanton, B. A. , Cervinski, M. A. , & Ashare, A. (2018). Pre‐analytical handling conditions and small RNA recovery from urine for miRNA profiling. The Journal of Molecular Diagnostics, 20(5), 565–571. 10.1016/j.jmoldx.2018.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner, M. , Ball, C. A. , Blake, J. A. , Botstein, D. , Butler, H. , Cherry, J. M , Davis, A. P. , Dolinski, K. , Dwight, S. S. , Eppig, J. T. , Harris, M. A. , Hill, D. P. , Issel‐Tarver, L. , Kasarskis, A. , Lewis, S. , Matese, J. C. , Richardson, J. E. , Ringwald, M. , Rubin, G. M. , & Sherlock, G. (2000). Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nature Genetics, 25(1), 25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreiro, K. , Dwivedi, O. P. , Leparc, G. , Rolser, M. , Delic, D. , Forsblom, C. , Groop, P. ‐. H. , Groop, L. , Huber, T. B. , Puhka, M. , & Holthofer, H. (2020). Comparison of urinary extracellular vesicle isolation methods for transcriptomic biomarker research in diabetic kidney disease. Journal of Extracellular Vesicles, 10(2), e12038. 10.1002/jev2.12038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben‐Dov, I. Z. , Whalen, V. M. , Goilav, B. , Max, K. E. A. , & Tuschl, T. (2016). Cell and microvesicle urine microRNA deep sequencing profiles from healthy individuals: Observations with potential impact on biomarker studies. Plos One, 11(1), e0147249. 10.1371/journal.pone.0147249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, L. , Sun, X. , Scicluna, B. J. , Coleman, B. M. , & Hill, A. F. (2014). Characterization and deep sequencing analysis of exosomal and non‐exosomal miRNA in human urine. Kidney International, 86(2), 433–444. 10.1038/ki.2013.502 [DOI] [PubMed] [Google Scholar]
- Cold Spring Harbor Press . (2017). RIPA lysis buffer .: Cold Spring Harb. Protoc. (pdb.rec101428).
- Courel, M. , Clément, Y. , Bossevain, C. , Foretek, D. , Vidal Cruchez, O. , Yi, Z. , Bénard, M. , Benassy, M.‐N. , Kress, M. , Vindry, C. , Ernoult‐Lange, M. , Antoniewski, C. , Morillon, A. , Brest, P. , Hubstenberger, A. , Roest Crollius, H. , Standart, N. , & Weil, D. (2019). GC content shapes mRNA storage and decay in human cells. Elife, 8. 10.7554/eLife.49708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickhout, A. , & Koenen, R. R. (2018). Extracellular vesicles as biomarkers in cardiovascular disease; chances and risks. Front Cardiovasc Med, 5, 113. 10.3389/fcvm.2018.00113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin, A. , Davis, C. A. , Schlesinger, F. , Drenkow, J. , Zaleski, C. , Jha, S. , Batut, P. , Chaisson, M. , & Gingeras, T. R. (2013). STAR: ultrafast universal RNA‐seq aligner. Bioinformatics, 29(1), 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, R. J. , Mansouri, K. , Stalls, V. , Manne, K. , Watts, B. , Parks, R. , Janowska, K. , Gobeil, S. M. C. , Kopp, M. , Li, D. , Lu, X. , Mu, Z. , Deyton, M. , Oguin, T. H. , Sprenz, J. , Williams, W. , Saunders, K. O. , Montefiori, D. , Sempowski, G. D., … Acharya, P. (2021). Cold sensitivity of the SARS‐CoV‐2 spike ectodomain. Nature Structural & Molecular Biology, 28(2), 128–131. 10.1038/s41594-020-00547-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdbrügger, U. , Le, T. H. (2016). Extracellular vesicles in renal diseases: More than novel biomarkers? Journal of the American Society of Nephrology, 27(1), 12–26. 10.1681/asn.2015010074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdbrügger, U. , Blijdorp, C. J. , Bijnsdorp, I. V. , Borràs, F. E. , Burger, D. , Bussolati, B. , Byrd, J. B. , Clayton, A. , Dear, J. W. , Falcón‐Pérez, J. M. , Grange, C. , Hill, A. F. , Holthöfer, H. , Hoorn, E. J. , Jenster, G. , Jimenez, C. R. , Junker, K. , Klein, J. , Knepper, M. A., … Martens‐Uzunova, E. S. (2021). Urinary extracellular vesicles: A position paper by the urine task force of the international society for extracellular vesicles. Journal of Extracellular Vesicles, 10(7), e12093. 10.1002/jev2.12093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbiano, F. , Corsi, J. , Gurrieri, E. , Trevisan, C. , Notarangelo, M. , & D'agostino, V. G. . (2020). RNA packaging into extracellular vesicles: An orchestra of RNA‐binding proteins? Journal of Extracellular Vesicles, 10(2), e12043. 10.1002/jev2.12043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, Q. , Zhou, Y. , Lu, J. , Bai, Y. , Xie, X. , & Lu, Z. (2014). miRNA in plasma exosome is stable under different storage conditions. Molecules (Basel, Switzerland), 19(2), 1568–1575. 10.3390/molecules19021568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium . (2021). The gene ontology resource: Enriching a GOld mine Nucleic Acids Research, 49(D1), D325–D334. 10.1093/nar/gkaa1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetsch, H. E. , Zhao, L. , Gnegy, M. , Imperiale, M. J. , Love, N. G. , & Wigginton, K. R. (2018). Fate of the urinary tract virus BK human polyomavirus in source‐separated urine. Applied and Environmental Microbiology, 84(7). 10.1128/aem.02374-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith, M. , Walker, J. R. , Spies, N. C. , Ainscough, B. J. , & Griffith, O. L. (2015). Informatics for RNA sequencing: A web resource for analysis on the cloud. Plos Computational Biology, 11(8), e1004393. 10.1371/journal.pcbi.1004393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha, D. , Lorenz, D. R. , Misra, V. , Chettimada, S. , Morgello, S. , & Gabuzda, D. (2019). Proteomic analysis of cerebrospinal fluid extracellular vesicles reveals synaptic injury, inflammation, and stress response markers in HIV patients with cognitive impairment. Journal of Neuroinflammation, 16(1), 254. 10.1186/s12974-019-1617-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt, A. , Kirchner, B. , Nolte‐'T Hoen, E. N.M. , & Pfaffl, M. W. (2021). miREV: An online database and tool to uncover potential reference rnas and biomarkers in small‐RNA sequencing data sets from extracellular vesicles enriched samples. Journal of Molecular Biology, 433(15), 167070. 10.1016/j.jmb.2021.167070 [DOI] [PubMed] [Google Scholar]
- Hubstenberger, A. , Courel, M. , Bénard, M. , Souquere, S. , Ernoult‐Lange, M. , Chouaib, R. , Yi, Z. , Morlot, J.‐B. , Munier, A. , Fradet, M. , Daunesse, M. , Bertrand, E. , Pierron, G. , Mozziconacci, J. , Kress, M. , & Weil, D. (2017). P‐body purification reveals the condensation of repressed mRNA regulons. Molecular Cell, 68(1), 144–157.e5.e145. 10.1016/j.molcel.2017.09.003 [DOI] [PubMed] [Google Scholar]
- Kallio, M. A. , Tuimala, J. T. , Hupponen, T. , Klemelä, P. , Gentile, M. , Scheinin, I. , Koski, M. , Käki, J. , & Korpelainen, E. I. (2011). Chipster: User‐friendly analysis software for microarray and other high‐throughput data. BMC Genomics [Electronic Resource], 12, 507. 10.1186/1471-2164-12-507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, K. M.i , Abdelmohsen, K. , Mustapic, M. , Kapogiannis, D. , & Gorospe, M. (2017). RNA in extracellular vesicles. Wiley interdisciplinary reviews. RNA, 8(4), e1413. 10.1002/wrna.1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolde, R. (2019). pheatmap: Pretty Heatmaps. R package version 1.0.12. https://CRAN.R‐project.org/package=pheatmap
- Kudla, G. , Lipinski, L. , Caffin, F. , Helwak, A. , & Zylicz, M. (2006). High guanine and cytosine content increases mRNA levels in mammalian cells. Plos Biology, 4(6), e180. 10.1371/journal.pbio.0040180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M. , Ban, J.‐J. , Im, W. , & Kim, M. (2016). Influence of storage condition on exosome recovery. Biotechnology and Bioprocess Engineering, 21(2), 299–304. 10.1007/s12257-015-0781-x [DOI] [Google Scholar]
- Lehmann, S. , Guadagni, F. , Moore, H. , Ashton, G. , Barnes, M. , Benson, E. , Clements, J. , Koppandi, I. , Coppola, D. , Demiroglu, S. Y. , Desouza, Y. , De Wilde, A. , Duker, J. , Eliason, J. , Glazer, B. , Harding, K. , Jeon, J. P. , Kessler, J. , Kokkat, T., … International Society for Biological and Environmental Repositories (ISBER) Working Group on Biospecimen Science (2012). Standard preanalytical coding for biospecimens: Review and implementation of the Sample PREanalytical Code (SPREC). Biopreserv Biobank, 10(4), 366–374. 10.1089/bio.2012.0012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkin, E. Z. , & Bratman, S. V. (2020). Bioactive DNA from extracellular vesicles and particles. Cell Death & Disease, 11(7), 584. 10.1038/s41419-020-02803-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant, M. L. , Rood, I. M. , Deegens, J. K. J. , & Klein, J. B. (2017). Isolation and characterization of urinary extracellular vesicles: Implications for biomarker discovery. Nature Reviews Nephrology, 13(12), 731–749. 10.1038/nrneph.2017.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi, H. , Muruganujan, A. , Ebert, D. , Huang, X. , & Thomas, P. D. (2019). PANTHER version 14: More genomes, a new PANTHER GO‐slim and improvements in enrichment analysis tools. Nucleic Acids Research, 47(D1), D419–D426. 10.1093/nar/gky1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda, K. C. , Bond, D. T. , Levin, J. Z. , Adiconis, X. , Sivachenko, A. , Russ, C. , Brown, D. , Nusbaum, C. , & Russo, L. M. (2014). Massively parallel sequencing of human urinary exosome/microvesicle RNA reveals a predominance of non‐coding RNA. Plos One, 9(5), e96094. 10.1371/journal.pone.0096094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordstein, C. , Savisaar, R. , Young, R. S. , Bazile, J. , Talmane, L. , Luft, J. , Liss, M. , Taylor, M. S. , Hurst, L. D. , & Kudla, G. (2020). Codon usage and splicing jointly influence mRNA localization. Cell Systems, 10(4), 351–362.e8.e358. 10.1016/j.cels.2020.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussack, V. , Wittmann, G. , & Pfaffl, M. W. (2019). Comparing small urinary extracellular vesicle purification methods with a view to RNA sequencing‐Enabling robust and non‐invasive biomarker research. Biomolecular Detection and Quantification, 17, 100089. 10.1016/j.bdq.2019.100089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norhazlin, J. , Nor‐Ashikin, M. N. K. , Hoh, B. P. , Sheikh Abdul Kadir, S. H. , Norita, S. , Mohd‐Fazirul, M. , Wan‐Hafizah, W. J. , Razif, D. , Rajikin, M. H. , & Abdullah, B. (2015). Effect of DNase treatment on RNA extraction from preimplantation murine embryos. Genetics and Molecular Research [Electronic Resource], 14(3), 10172–10184. 10.4238/2015.August.28.1 [DOI] [PubMed] [Google Scholar]
- Oeyen, E. , Willems, H. , ’T Kindt, R. , Sandra, K. , Boonen, K. , Hoekx, L. , De Wachter, S. , Ameye, F. , & Mertens, I. (2019). Determination of variability due to biological and technical variation in urinary extracellular vesicles as a crucial step in biomarker discovery studies. Journal of Extracellular Vesicles, 8(1), 1676035. 10.1080/20013078.2019.1676035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Oliveira, M. C. , Caires, H. R. , Oliveira, M. J. , Fraga, A. , Vasconcelos, M. H. , & Ribeiro, R. (2020). Urinary biomarkers in bladder cancer: Where do we stand and potential role of extracellular vesicles. Cancers (Basel), 12(6), 1400. 10.3390/cancers12061400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosthuyzen, W. , Sime, N. E. L. , Ivy, J. R. , Turtle, E. J. , Street, J. M. , Pound, J. , Bath, L. E. , Webb, D. J. , Gregory, C. D. , Bailey, M. A. , & Dear, J. W. (2013). Quantification of human urinary exosomes by nanoparticle tracking analysis. Journal of Physiology, 591(23), 5833–5842. 10.1113/jphysiol.2013.264069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oreskovic, A. , Brault, N. D. , Panpradist, N. , Lai, J. J. , Lutz, B. R. (2019). Analytical comparison of methods for extraction of short cell‐free DNA from urine. The Journal of Molecular Diagnostics, 21(6), 1067–1078. 10.1016/j.jmoldx.2019.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang, B. , Zhu, Y. , Ni, J. , Thompson, J. , Malouf, D. , Bucci, J. , Graham, P. , & Li, Y. (2020). Extracellular vesicles: The next generation of biomarkers for liquid biopsy‐based prostate cancer diagnosis. Theranostics, 10(5), 2309–2326. 10.7150/thno.39486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang, W.‐K. , Kang, S. , Ryu, D.‐Y. , Rahman, M. S. , Park, Y.‐J. , & Pang, M.‐G. (2020). Optimization of sperm RNA processing for developmental research. Scientific Reports, 10(1), 11606. 10.1038/s41598-020-68486-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, S. , Lee, K. , Park, I. B. , Kim, N. H. , Cho, S. , Rhee, W. J. , Oh, Y. , Choi, J. , Nam, S. , & Lee, D. H.o (2020). The profiles of microRNAs from urinary extracellular vesicles (EVs) prepared by various isolation methods and their correlation with serum EV microRNAs. Diabetes Research and Clinical Practice, 160, 108010. 10.1016/j.diabres.2020.108010 [DOI] [PubMed] [Google Scholar]
- Pathan, M. , Fonseka, P. , Chitti, S. V. , Kang, T. , Sanwlani, R. , Van Deun, J. , Hendrix, A. , & Mathivanan, S. (2019). Vesiclepedia 2019: A compendium of RNA, proteins, lipids and metabolites in extracellular vesicles. Nucleic Acids Research, 47(D1), D516–D519. 10.1093/nar/gky1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo, C. , Pérez, J. L. , Mielnichuck, A. , Fedele, C. G. , Niubo, J. , & Tenorio, A. (2004). Prevalence and patterns of polyomavirus urinary excretion in immunocompetent adults and children. Clinical Microbiology and Infection, 10(7), 640–644. 10.1111/j.1469-0691.2004.00882.x [DOI] [PubMed] [Google Scholar]
- Puhka, M. , Nordberg, M.‐E. , Valkonen, S. , Rannikko, A. , Kallioniemi, O. , Siljander, P. , & Af Hällström, T. M. (2017). KeepEX, a simple dilution protocol for improving extracellular vesicle yields from urine. European Journal of Pharmaceutical Sciences, 98, 30–39. 10.1016/j.ejps.2016.10.021 [DOI] [PubMed] [Google Scholar]
- Puhka, M. , Takatalo, M. , Nordberg, M.‐E. , Valkonen, S. , Nandania, J. , Aatonen, M. , Yliperttula, M. , Laitinen, S. , Velagapudi, V. , Mirtti, T. , Kallioniemi, O. , Rannikko, A. , Siljander, P. R. ‐. M. , & Af Hällström, T. M. (2017). Metabolomic profiling of extracellular vesicles and alternative normalization methods reveal enriched metabolites and strategies to study prostate cancer‐related changes. Theranostics, 7(16), 3824–3841. 10.7150/thno.19890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pös, O. , Biró, O. , Szemes, T. , & Nagy, B. (2018). Circulating cell‐free nucleic acids: Characteristics and applications. European Journal of Human Genetics, 26(7), 937–945. 10.1038/s41431-018-0132-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo, G. , & Stoorvogel, W. (2013). Extracellular vesicles: Exosomes, microvesicles, and friends. Journal of Cell Biology, 200(4), 373–383. 10.1083/jcb.201211138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, M. D. , Mccarthy, D. J. , & Smyth, G. K. (2010). edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26(1), 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royo, F. , Théry, C. , Falcón‐Pérez, J. M. , Nieuwland, R. , & Witwer, K. W. (2020). Methods for separation and characterization of extracellular vesicles: results of a worldwide survey performed by the ISEV rigor and standardization subcommittee. Cells, 9(9), 1955. 10.3390/cells9091955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarker, S. , Scholz‐Romero, K. , Perez, A. , Illanes, S. E. , Mitchell, M. D. , Rice, G. E. , & Salomon, C. (2014). Placenta‐derived exosomes continuously increase in maternal circulation over the first trimester of pregnancy. Journal of Translational Medicine, 12, 204. 10.1186/1479-5876-12-204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuierer, S. , Carbone, W. , Knehr, J. , Petitjean, V. , Fernandez, A. , Sultan, M. , & Roma, G. (2017). A comprehensive assessment of RNA‐seq protocols for degraded and low‐quantity samples. Bmc Genomics [Electronic Resource], 18(1), 442. 10.1186/s12864-017-3827-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedley, D. , Haider, S. , Ballester, B. , Holland, R. , London, D. , Thorisson, G. , & Kasprzyk, A. (2009). BioMart–biological queries made easy. Bmc Genomics [Electronic Resource], 10, 22. 10.1186/1471-2164-10-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolova, V. , Ludwig, A.‐K. , Hornung, S. , Rotan, O. , Horn, P. A. , Epple, M. , & Giebel, B. (2011). Characterisation of exosomes derived from human cells by nanoparticle tracking analysis and scanning electron microscopy. Colloids and Surfaces. B, Biointerfaces, 87(1), 146–150. 10.1016/j.colsurfb.2011.05.013 [DOI] [PubMed] [Google Scholar]
- Srinivasan, S. , Yeri, A. , Cheah, P. S. , Chung, A. , Danielson, K. , De Hoff, P. , Filant, J. , Laurent, C. D. , Laurent, L. D. , Magee, R. , Moeller, C. , Murthy, V. L. , Nejad, P. , Paul, A. , Rigoutsos, I. , Rodosthenous, R. , Shah, R. V. , Simonson, B. , To, C., … Laurent, L. C. (2019). Small RNA sequencing across diverse biofluids identifies optimal methods for exRNA isolation. Cell, 177(2), 446–462.e16.e416. 10.1016/j.cell.2019.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streleckiene, G. , Reid, H. M. , Arnold, N. , Bauerschlag, D. , Forster, M. (2018). Quantifying cell free DNA in urine: Comparison between commercial kits, impact of gender and inter‐individual variation. Biotechniques, 64(5), 225–230. 10.2144/btn-2018-0003 [DOI] [PubMed] [Google Scholar]
- Sultan, M. , Amstislavskiy, V. , Risch, T. , Schuette, M. , Dökel, S. , Ralser, M. , Balzereit, D. , Lehrach, H. , & Yaspo, M.‐L. (2014). Influence of RNA extraction methods and library selection schemes on RNA‐seq data. Bmc Genomics [Electronic Resource], 15(1), 675. 10.1186/1471-2164-15-675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenningsen, P. , Sabaratnam, R. , & Jensen, B. L. (2020). Urinary extracellular vesicles: Origin, role as intercellular messengers and biomarkers; efficient sorting and potential treatment options. Acta Physiologica, 228(1), e13346. 10.1111/apha.13346 [DOI] [PubMed] [Google Scholar]
- Team, R. C. (2020). R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. URL http://www.R‐project.org/ [Google Scholar]
- Théry, C. , Witwer, K. W. , Aikawa, E. , Alcaraz, M. J. , Anderson, J. D. , Andriantsitohaina, R. , Antoniou, A. , Arab, T. , Archer, F. , Atkin‐Smith, G. K. , Ayre, D. C. , Bach, J.‐M. , Bachurski, D. , Baharvand, H. , Balaj, L. , Baldacchino, S. , Bauer, N. N. , Baxter, A. A. , Bebawy, M., … Zuba‐Surma, E. K. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of Extracellular Vesicles, 7(1), 1535750. 10.1080/20013078.2018.1535750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, M. C. , Brownlee, M. , Susztak, K. , Sharma, K. , Jandeleit‐Dahm, K. A. M. , Zoungas, S. , Rossing, P. , Groop, P.‐H. , & Cooper, M. E. (2015). Diabetic kidney disease. Nature Reviews Disease Primers, 1, 15018. 10.1038/nrdp.2015.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen, M. , Fagerberg, L. , Hallstrom, B. M. , Lindskog, C. , Oksvold, P. , Mardinoglu, A. , Sivertsson, A. , Kampf, C. , Sjostedt, E. , Asplund, A. , Olsson, I. , Edlund, K. , Lundberg, E. , Navani, S. , Szigyarto, C. A.‐K. , Odeberg, J. , Djureinovic, D. , Takanen, J. O. , Hober, S., … Ponten, F. (2015). Proteomics. Tissue‐based map of the human proteome. Science, 347(6220), 1260419. 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- Wang, L. , Wang, S. , & Li, W. (2012). RSeQC: Quality control of RNA‐seq experiments. Bioinformatics, 28(16), 2184–2185. 10.1093/bioinformatics/bts356 [DOI] [PubMed] [Google Scholar]
- Wang, S. , Dong, Y. , Gong, A. , Kong, H. , Gao, J. , Hao, X. , Liu, Y. , Wang, Z. , Fan, Y. , Liu, C. , & Xu, W. (2021). Exosomal circRNAs as novel cancer biomarkers: Challenges and opportunities. International Journal of Biological Sciences, 17(2), 562–573. 10.7150/ijbs.48782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S. , Kojima, K. , Mobley, J. A. , & West, A. B. (2019). Proteomic analysis of urinary extracellular vesicles reveal biomarkers for neurologic disease. EBioMedicine, 45, 351–361. 10.1016/j.ebiom.2019.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W.‐C. , Lin, F.‐M. , Chang, W.‐C. , Lin, K.‐Y. , Huang, H.‐D. , & Lin, N.‐S. (2009). miRExpress: Analyzing high‐throughput sequencing data for profiling microRNA expression. BMC Bioinformatics, 10, 328. 10.1186/1471-2105-10-328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham, H. (2016). ggplot2: Elegant graphics for data analysis. Springer‐Verlag. [Google Scholar]
- Vitsios, D. M. , & Enright, A. J. (2015). Chimira: Analysis of small RNA sequencing data and microRNA modifications. Bioinformatics, 31(20), 3365–3367. 10.1093/bioinformatics/btv380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witwer, K. W. , Buzás, E. I. , Bemis, L. T. , Bora, A. , Lässer, C. , Lötvall, J. , Nolte‐‘T Hoen, E. N. , Piper, M. G. , Sivaraman, S. , Skog, J. , Théry, C. , Wauben, M. H. , & Hochberg, F. (2013). Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. Journal of Extracellular Vesicles, 2, 20360. 10.3402/jev.v2i0.20360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekula, A. , Muralidharan, K. , Kang, K. M. , Wang, L. , Balaj, L. , Carter, B. S. (2020). From laboratory to clinic: Translation of extracellular vesicle based cancer biomarkers. Methods (San Diego, Calif.), 177, 58–66. 10.1016/j.ymeth.2020.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuana, Y. , Böing, A. N. , Grootemaat, A. E. , Van Der Pol, E. , Hau, C. M. , Cizmar, P. , Buhr, E. , Sturk, A. , & Nieuwland, R. (2015). Handling and storage of human body fluids for analysis of extracellular vesicles. Journal of Extracellular Vesicles, 4, 29260. 10.3402/jev.v4.29260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuana, Y. , Sturk, A. , & Nieuwland, R. (2013). Extracellular vesicles in physiological and pathological conditions. Blood Reviews, 27(1), 31–39. 10.1016/j.blre.2012.12.002 [DOI] [PubMed] [Google Scholar]
- Zhao, W. , He, X. , Hoadley, K. A. , Parker, J. S. , Hayes, D. N. , & Perou, C. M. (2014). Comparison of RNA‐Seq by poly (A) capture, ribosomal RNA depletion, and DNA microarray for expression profiling. Bmc Genomics [Electronic Resource], 15(1), 419. 10.1186/1471-2164-15-419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, H. , Yuen, P. S. T. , Pisitkun, T. , Gonzales, P. A. , Yasuda, H. , Dear, J. W. , Gross, P. , Knepper, M. A. , & Star, R. A. (2006). Collection, storage, preservation, and normalization of human urinary exosomes for biomarker discovery. Kidney International, 69(8), 1471–1476. 10.1038/sj.ki.5000273 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Data Availability Statement
The raw sequencing data analysed in this study is not publicly available due to local regulations. We provide raw mRNA‐seq count data in the Supplementary Table 10 and raw miRNA‐seq count data in the Supplementary Table 11. Datasets used to derive the list of miRNAs enriched in uEV isolates are available in the Supplementary Table 12 (microarray raw data, background subtracted data, and global normalization) and Supplementary Table 13 (miRNA‐seq raw counts). Raw mRNA‐seq count data from uEV RNA samples isolated without DNAse treatment is available in Supplementary Table 14. Raw sequencing data is available to qualified investigators upon reasonable request.


