Abstract
The potential clinical applications of the powerful in vitro-transcribed (IVT)-mRNAs, to restore defective protein functions, strongly depend on their successful intracellular delivery and transient translation through the development of safe and efficient delivery platforms. In this study, an innovative (international patent-pending) methodology was developed, combining the IVT-mRNAs with the protein transduction domain (PTD) technology, as an efficient delivery platform. Based on the PTD technology, which enables the intracellular delivery of various cargoes intracellularly, successful conjugation of a PTD to the IVT-mRNAs was achieved and evaluated by band-shift assay and NMR spectroscopy. In addition, the PTD-IVT-mRNAs were applied and evaluated in two protein-disease models, including the mitochondrial disorder fatal infantile cardioencephalomyopathy and cytochrome c oxidase (COX) deficiency (attributed to SCO2 gene mutations) and β-thalassemia. The PTD-IVT-mRNA of SCO2 was successfully transduced and translated to the corresponding Sco2 protein inside the primary fibroblasts of a SCO2/COX-deficient patient, whereas the PTD-IVT-mRNA of β-globin was transduced and translated in bone marrow cells, derived from three β-thalassemic patients. The transducibility and the structural stability of the PDT-IVT-mRNAs, in both cases, were confirmed at the RNA and protein levels. We propose that our novel delivery platform could be clinically applicable as a protein therapy for metabolic/genetic disorders.
Keywords: IVT-mRNA, protein transduction domain technology, PTD technology, delivery platform, protein therapy, SCO2, β-globin
Graphical abstract

The corresponding author and colleagues successfully developed a universal delivery platform of therapeutic in vitro-transcribed (IVT)-mRNAs conjugated to a selected PTD through an innovative patent-pending methodology. PTD-IVT-mRNAs exhibited significant stability, intracellular transduction, and protein expression efficiency and were applied in patients’ cells of two different monogenic/metabolic disorders, revealing powerful future perspectives.
Introduction
Although in vitro-transcribed (IVT)-mRNA is produced extracellularly, it retains the ability to mediate the translation of genetic information into proteins in recipient cells.1 The major advantage of IVT-mRNA technology is that transduction of mRNA molecules does not induce oncogenic mutations, unlike gene therapy strategies applied via retro-/lenti-viral vectors. In principle, IVT-mRNA-based therapeutic approaches are applicable to a wide range of acute and chronic diseases, including diseases that can be cured by protein replacement therapy (PRT), genome editing, cell fate reprogramming, and cancer immunotherapy.2 It is worth mentioning that IVT-mRNA-based vaccines have been brought into the limelight, due to the urgent need to vaccinate people worldwide against the COVID-19 pandemic, caused by the SARS-CoV-2 coronavirus. Of the various approaches that have been developed, BioNTech/Pfizer and Moderna IVT-mRNA vaccines were the first to be injected into human volunteers and received preliminary approval for emergency use. They offer significant advantages, such as faster and easier manufacturing and clinical development, compared to the more time-consuming traditional vaccines.
IVT-mRNA can be transduced into eukaryotic cells by a receptor-mediated mechanism, although at an extremely low uptake rate in most cell types.3 Moreover, the problem of IVT-mRNA susceptibility to enzymatic degradation, which affects its structural and functional stability in fluids with high RNase activity,3, 4, 5 is of great concern. However, it has been found that appropriate construct design of IVT-mRNA through chemical and structural modifications can potentially control the half-life and translation rate of the mRNA6 by increasing its translation efficiency. These modifications include, but are not limited to, the following: (1) addition of anti-reverse cap analogs (ARCAs);2,7 (2) involvement of modified nucleosides, such as 5mC (5-methylcytidine) and Ψ (pseudo-uridine);8 (3) polyadenylation at the 3′ end of the IVT-mRNA;2 (4) replacement of the Adenylate/Uridylate–rich elements (AU-rich elements, AREs; destabilizing elements), with more stable 5′ UTRs (untranslated regions), like the murine 5′ UTR of the β-globin gene, as well as the human 3′ UTR of the α-globin or β-globin genes;9 and (5) existence of a strong Kozak sequence to initiate translation at the 5′ UTR.
Intracellular delivery of IVT-mRNA can be mediated by several methods, including cell membrane disruption (by electroporation or gene gun), resulting in a fairly high percentage of dead cells,10 although there are ongoing efforts to improve safety and efficacy. In addition, IVT-mRNA delivery can be mediated via various nanoparticle (NP)-mediated delivery systems. The latter includes lipid-based and polymer-based systems with cationic surface charge, as the main feature for complexation with the negatively charged mRNA molecules. However, because surface charge, colloidal instability, poor stability in serum, and rapid clearance play important roles,11, 12, 13 there is an ongoing need for alternative IVT-mRNA delivery systems.2
In this study, we sought to design a novel conjugation reaction between the IVT-mRNAs and protein transduction domains (PTDs)14, 15, 16, 17 to achieve an alternative, effective, and potentially safer approach to delivering IVT-mRNAs into cultured human cell lines. PTD technology uses short peptides capable of penetrating almost all cellular membranes and intracellularly carrying a variety of “cargoes,” ranging from micromolecules and small interfering RNAs (siRNAs) to macromolecules (proteins, RNAs, plasmid DNA, NPs), with relatively high transduction efficiency as well as low cytotoxicity.
There are two strategies to generate a PTD/nucleic acid complex. The first is based on electrostatic self-assembly of non-covalent, stable bonds between the PTD and its cargo,18,19 whereas the second strategy involves the use of chemical ligands that covalently bind the PTD to the cargo, mainly through a cleavable disulfide,20 amide,21,22 thiazolidine, oxime, or hydrazine bond.23 The non-covalent strategy relies on the amphipathic properties of short peptides (e.g., MPG, Pep-1, or CADY), which can form complexes with cargoes, without the need for chemical modification or crosslinking. The covalent method, although it is limited from a chemical point of view because of the risk of altering the biological activity of the cargo, as well as the efficiency of the released cargo, does offer the advantage of less cargo loss. Alternatively, covalent binding offers the advantage of less “cargo loss” and protection of the cargo under adverse conditions during in vivo delivery, due to the existence of RNases.24
The developed PTD-IVT-mRNA platform system, described in this work, was evaluated as a potential therapeutic approach in two protein-defective and genetically aberrant disease models. The first model refers to a mitochondrial disorder specifically attributed to SCO2 mutations. Mutations in the human SCO2 gene lead to primary mitochondrial disorders, such as fatal infantile cardioencephalomyopathy and cytochrome c oxidase (COX) deficiency,25 as well as other neurodegenerative disorders, such as spinal muscular atrophy (SMA)26 and Leigh syndrome.27 Sco2 is a human mitochondrial inner-membrane protein25 involved in mitochondrial copper transfer,28 redox signaling,25,29, 30, 31, 32 and bioenergetics, via the p53 regulatory pathway.33 SCO2 has been listed among ∼2,000 genes essential for human cell viability.34 Treatment of mitochondrial protein deficiencies remains an unmet need, as there are no established therapeutic approaches, only supportive treatment with agents, such as vitamins (mainly B, but also C and E, as antioxidants) and other substances, such as co-enzyme Q10 (CoQ10), α-lipoic acid (α-ALA), L-carnitine, and creatine.35 In relation to mitochondrial disorders, due to SCO2 mutations, combinations of copper and bezafibrate36,37 or elesclomol (ES) have been proposed as a potential copper carrier.38 Our group has proposed PRT, via the production and successful delivery of the human recombinant full-length mitochondrial protein Sco2, fused with Transactivator of Transcription peptide (TAT peptide) via PTD technology, into the mitochondria of a SCO2/COX-deficient cell culture model, complementing COX deficiency in fibroblasts, derived from a SCO2-deficient patient.15,17 The second model, in which our IVT-mRNA delivery platform has been applied, is β-thalassemia, a hereditary anemia, caused by the lack or insufficient production of the β-hemoglobin (HBB) chain, due to a broad spectrum of mutations. With 270 million heterozygotes worldwide, β-thalassemia represents the most common monogenic disorder and is potentially amenable to gene therapy.39 However, the current management of β-thalassemia consists of prenatal diagnosis, transfusion therapy, and allogeneic bone marrow (BM) transplantation (BMT),40 the latter of which is the only potentially curative to date. In addition, a gene therapy product (Zynteglo), using a lentiviral vector carrying the human β-globin, has been developed and recently authorized by the US Food and Drug Administration (FDA) and European Medicines Agency (EMA) (with recommendation provided by the EMA Pharmacovigilance Risk Assessment Committee) for the treatment of transfusion-dependent β-thalassemia (TDT). This product is under post-marketing surveillance and additional commitments. Additionally, many other outstanding efforts are being conducted, such as gene therapy in hematopoietic stem cells (HSCs), mediated by retroviral or lentiviral vectors (as mentioned above) or genome editing. The latter technologies aim to correct β-thalassemia mutations via the following: (1) zinc-finger nucleases (ZFN)-induced homology-directed repair (HDR) or non-homologous end-joining (NHEJ) with one ongoing clinical trial; (2) meganucleases (MNs); (3) transcription activator-like effector nucleases (TALENs); and (4) the RNA-guided CRISPR-CRISPR-associated protein 9 (Cas9) system,39 with two ongoing clinical trials in transfusion-dependent β-thalassemic patients. However, despite the undeniable success of the approaches described above, in the case of thalassemias, these strategies pose major challenges curing patients with the β0/β0 genotype in terms of safety and control of transgene expression, raising the need to explore alternative genetic and cellular therapeutic approaches. In the context of PRT for β-thalassemia, our group recently has published the study of the production of the recombinant β-globin chain, fused to TAT, and its successful intracellular transduction in human chronic myelogenous leukemia K-562 cells, replacing the endogenous missing β-globin chain and interacting with endogenous α-globin chains to eventually form α2β2-tetramers.14
The central goal of the work presented here was to design a method to construct and develop a novel PTD-IVT-mRNA platform, using an established PTD, to effectively deliver IVT-mRNA into human fibroblasts, as well as hematopoietic cells, deficient in Sco2 and β-globin proteins, respectively. Our platform tackles problems of stability, transduction, and translation of the IVT-mRNAs and allows transient production of the proteins of interest via efficient translation of their corresponding transduced IVT-mRNAs. Altogether, this platform provides a suitable approach for efficient PRT, applicable to various monogenic/metabolic disorders.
Results
Construction of the template for in vitro transcription and production of IVT-mRNA
The final plasmid IVT templates pcDNA3-5′ UTR-SCO2-3′ UTR (Figure S1A) and pcDNA3-5′ UTR-β-globin-3′ UTR (Figure S2A) are shown in the DNA plasmid maps, and each successful cloning was confirmed by sequencing. For the construction of the final IVT reaction templates, several recombinant plasmid vectors were designed (Figures S3 and S4), constructed, transformed into the E. coli strain TOP10F′, and then isolated. Maps of all recombinant plasmid constructs produced are displayed via SnapGene software (from GSL Biotech; available at https://www.snapgene.com/). Each IVT plasmid template also contained the eukaryotic ribosomal binding sequence (RBS), known as the Kozak sequence, which is essential for initiating the translation of the IVT-mRNA strands by binding of the eukaryotic ribosome to the IVT-mRNA strands. This Kozak sequence used was present within the fused 5′ UTR of murine β-globin mRNA (Table S1). In addition, the IVT plasmid template contained the corresponding CDS of SCO2/β-globin, followed by the 3′ UTR of the human β-globin mRNA.
Each IVT-mRNA (5′ UTR-SCO2-3′ UTR, simplified as IVT-mRNA of SCO2/5′ UTR-β-globin-3′ UTR, simplified as IVT-mRNA of β-globin) was prepared by the T7 polymerase-based in vitro transcription method and purified by phenol-chloroform after linearization of the recombinant plasmid vector by digestion with the XbaI restriction enzyme. IVT-mRNA maps are also shown via SnapGene software (Figures S1B and S2B). Successful synthesis of capped IVT-mRNAs with poly(A) tailing was confirmed by denaturing 6% PAGE/8 M urea gel electrophoresis (Figure S5), as well as by cDNA synthesis and PCR with specific primers.
Conjugation of the PTD to the IVT-mRNA
According to our innovative design, a selected PTD (PFVYLI) was used to transduce the IVT-mRNA. To detect the conjugation of the PTD to each IVT-mRNA, a band-shift assay was performed. The PTD-IVT-mRNAs showed delayed transposition into the denaturing 6% PAGE/8 M urea gel compared to their corresponding naked IVT-mRNAs (without PTD), which were used as internal negative controls (Figure 1). In addition, a non-PTD peptide (WSYGLRPG) was used as a negative control peptide (in 2× concentration) to assess the relative penetration capacity of the PTD used in the PTD-IVT-mRNA platform. The conjugation product of the control peptide showed slower transposition into the denaturing 6% PAGE/8 M urea gel, as well, compared to the naked IVT-mRNA (Figure S6A).
Figure 1.
Band-shift assay of the PTD-IVT-mRNAs
Denaturing 6% PAGE/8 M urea gel electrophoresis of the PTD-IVT-mRNAs as compared with the corresponding naked IVT-mRNA ofSCO2(A) and IVT-mRNA of β-globin.(B). MR, RNA molecular weight marker.
Nuclear Magnetic Resonance (NMR) experiments (in DMSO-d6) were performed to record the 1D 1H spectra of the PTD and the PTD-IVT-mRNA. Figure 2A shows the amino group (NH) region of the 1H spectra of the PTD, which exhibits a low dispersion of the NH chemical shifts and is a typical feature of a short, mobile, and disordered (poly)peptide. Nevertheless, the NHs of the free PTD (most of them appearing as degenerate doublets in 7.6−8.1 ppm due to NH-Hα coupling; J = 8.80−9.00 Hz) shifted significantly in the PTD-IVT-mRNA reaction mixture (Figure 2B). In the PTD-IVT-mRNA, changes in the chemical and magnetic environment resulted in significant chemical shift differences in the PTD backbone NH peaks, appeared as doublets, and were distributed over the range of 7.5−8.8 ppm.
Figure 2.
NMR analysis of the PTD-IVT-mRNA of SCO2
(A) NH region of the 1H spectra of the free PTD in DMSO. (B) NH region of the 1H spectra of the PTD-IVT-mRNA in DMSO (plus D2O).
To provide further evidence for the successful conjugation of the PTD to IVT-mRNA, based on the PTD backbone amide NH groups, an attempt was made to investigate the labile nature of these protons. Therefore, 15% D2O in 500 μL of DMSO-d6 (plus 90 μL reaction mixture; see Materials and methods) was added to the NMR solution buffer, and additional 1H 1D NMR spectra were recorded with the same experimental parameters (Figure 2B). The proton resonances of the amino acid (aa) methyl groups were identified, and their intensities, which appeared in both NMR samples (without and with 15% D2O), were normalized. Then, the intensities of the backbone NH peaks were compared. It can be clearly seen (Figure 2B) that the intensity of the exchangeable NH peaks was significantly reduced by the addition of the D2O, as the result of the Hydrogen–deuterium (H/D) exchange.
The NMR analysis of the control peptide-IVT-mRNA of SCO2 also confirmed the successful conjugation, as chemical shift changes were observed in the 1H spectrum of the conjugation product, showing chemical shift difference with respect to the NH resonances of the free control peptide (especially for those in the regions of 7.0–7.3 ppm and 7.75–8.15 ppm) (Figure S6B). Therefore, the control peptide conjugated to the IVT-mRNA of SCO2 was also used in subsequent experiments.
Assessment of the structural stability of PTD-IVT-mRNA
Degradation of the PTD-IVT-mRNA of SCO2 was examined after treatment with fetal bovine serum (FBS), which contains nucleases, for 1 h. Gel electrophoresis showed significant degradation of the naked IVT-mRNA after 1 h exposure to 10% FBS. On the other hand, PTD-IVT-mRNA of SCO2 showed a statistically significant increased tolerance to nucleases under the same treatment conditions (∗p = 0.0430). Even after 1 h of exposure to 10% FBS, PTD-IVT-mRNA was still trapped in the loading wells, indicating that the IVT-mRNA was still conjugated to the PTD and protected (Figures 3A and 3B). Interestingly, the control peptide-IVT-mRNA of the SCO2 conjugation product appeared to be also resistant to nucleases found within 10% FBS, although to a much lesser degree than the PTD-IVT-mRNA (Figures S7A and S7B).
Figure 3.
Stability evaluation assays for the PTD-IVT-mRNA of SCO2
(A) 2% agarose gel electrophoresis of IVT-mRNA and PTD-IVT-mRNA, incubated at 37°C in 10% FBS for 1 h. (B) Bar graph of band intensity of the IVT-mRNA and PTD-IVT-mRNA (compared to the corresponding untreated IVT-mRNA [lane 2] and PTD-IVT-mRNA [lane 6], respectively) after FBS treatment for 1 h with ∗p = 0.0430. (C) 2% agarose gel electrophoresis of IVT-mRNA and PTD-IVT-mRNA, incubated at 37°C with RNase A. (D) Bar graph of band intensity of the IVT-mRNA and PTD-IVT-mRNA (compared to the corresponding untreated IVT-mRNA [lane 2] and PTD-IVT-mRNA [lane 6], respectively) after RNase treatment for 1 h with ∗∗p = 0.0050.
PTD-IVT-mRNA of SCO2 showed a statistically significant higher stability compared to the naked IVT-mRNA (∗∗p = 0.005) after incubation with RNase A for 1 h at 37°C. The naked IVT-mRNA of SCO2 showed a decrease in band intensity (fold difference [FD]: 6.7) compared to the untreated IVT-mRNA, indicating that only the 14.9% of the IVT-mRNA remained stable after RNase A treatment (Figures 3C and 3D). The PTD appears to protect the IVT-mRNA from RNase degradation, as band intensity was only reduced with an FD of 1.13 compared to untreated PTD-IVT-mRNA, as 72% of the IVT-mRNA remained stable after incubation with RNase (Figures 3C and 3D), solely due to the conjugation with the peptide (minus the IVT-mRNA portion). These results are from three independent experiments, each performed in triplicates. As shown in Figures S7C and S7D, the control peptide-IVT-mRNA of SCO2 remained stable after treatment with RNase A, as the band intensity was reduced with an FD of 1.40 only, compared to untreated control peptide-IVT-mRNA of SCO2, as 42% of the IVT-mRNA remained stable after incubation with RNase, solely due to the conjugation with the peptide (minus the IVT-mRNA portion).
Presence of proteinase K
The proteinase K assay was performed to investigate the nature of the bond formed between the PTD and the IVT-mRNA. After treatment for 1 h at 37°C, PTD-IVT-mRNA of SCO2 appeared to be resistant to digestion with proteinase K. The IVT-mRNA conjugated to PTD showed a slower transposition into the gel, having the same profile as the untreated PTD-IVT-mRNA (Figure S8). The failure of proteinase K to dissociate the bond formed during the conjugation of the PTD to the IVT-mRNA is probably because of the PTD aa sequence or the formation of a “protective” secondary structure of the PTD and the IVT-mRNA.
PTD conjugation allows efficient IVT-mRNA uptake and translation
PTD-IVT-mRNA of SCO2
To achieve efficient translation of the IVT-mRNA of the SCO2 into its corresponding gene product, it was first conjugated to the selected PTD to be effectively delivered into cells. Based on the IVT-mRNA amount (0.5 μg), PTD-IVT-mRNA of SCO2 was first incubated with K-562 cells (Figure 4) and then with primary SCO2-deficient fibroblasts (Figure 5).
Figure 4.
Kinetic analysis of SCO2 translation in K-562 cells after transduction with the PTD-IVT-mRNA of SCO2
(A) Flow cytometry results, corresponding to the proportion of Sco2+ cells after staining with anti-Sco2.IgG in untreated cells (0 h) and those transduced with PTD-IVT-mRNA of SCO2 from 30 min to 96 h. (B) Representative histograms from flow cytometry analysis showing Sco2 positivity in untreated cells (0 h) and those transduced with PTD-IVT-mRNA of SCO2 from 30 min to 96 h. ANOVA test: ∗∗p = 0.0055; t test: 0 h−30 min, ∗∗∗p = 0.0002; 0 h−24 h, p = 0.0710 not significant (ns); 0 h−48 h, ∗p = 0.01; 0 h−72 h, ∗p = 0.0170; 0 h−96 h, ∗p = 0.0147. (C) Increase (indicated as ↑) of fold difference (FD) in SCO2 translation in K-562 cells (∗p = 0.0227) after transduction with PTD-IVT-mRNA of SCO2 at 72 h compared to corresponding untreated cells. (D) 0.5% agarose gel electrophoresis of RT-PCR products with cDNA as a template, derived from total RNA extracts of the cells, untreated (0 h) and transduced with PTD-IVT-mRNA of SCO2 up to 96 h. MD, DNA molecular weight marker. (E) The levels of the IVT-mRNA of SCO2, verified by qPCR, using cDNA as a template, derived from total RNA extracts of K-562 cells, untreated (0 h) and transduced with PTD-IVT-mRNA of SCO2 up to 96 h. ANOVA test: ∗∗∗∗p ≤ 0.0001.
Figure 5.
Kinetic analysis of SCO2 translation in fibroblasts derived from a SCO2/COX-deficient patient after transduction with the PTD-IVT-mRNA of SCO2
(A) Flow cytometry results corresponding to the proportion of Sco2+ cells after staining with anti-Sco2.IgG in untreated cells (0 h) and those transduced with PTD-IVT-mRNA of SCO2 from 30 min to 96 h. (B) Representative histograms from flow cytometry analysis showing Sco2 positivity in untreated cells (0 h) and those transduced with PTD-IVT-mRNA of SCO2 from 30 min to 96 h. ANOVA test: ∗∗∗∗p ≤ 0.0001; t test: 0 h−30 min, ∗∗∗p = 0.0002; 0 h−1 h, ∗∗∗p = 0.0003; 0 h−4 h, ∗∗∗∗p ≤ 0.0001; 0 h−24 h, ∗∗∗p = 0.0007; 0 h−48 h, ∗∗∗p = 0.0002; 0 h−72 h, ∗∗∗p = 0.0004; 0 h−96 h, ∗∗p = 0.0040. (C) Increase (indicated as ↑) of FD in SCO2 translation in fibroblasts derived from a SCO2/COX-deficient patient (∗∗p = 0.0013) after transduction with PTD-IVT-mRNA of SCO2 at 72 h compared to corresponding untreated cells. (D) 0.5% agarose gel electrophoresis of PCR products with cDNA as a template, derived from total RNA extracts of the cells, untreated (0 h) and transduced with PTD-IVT-mRNA of SCO2 up to 96 h. (E) The levels of the IVT-mRNA of SCO2 verified by qPCR using cDNA as a template, derived from total RNA extracts of fibroblasts derived from a SCO2/COX-deficient patient, untreated (0 h) and transduced with PTD-IVT-mRNA of SCO2 up to 96 h. ANOVA test: ∗∗∗p = 0.0004.
Regarding the efficiency in protein level, treatment of K-562 cells with PTD-IVT-mRNA of SCO2 showed increased SCO2 translation from the first 30 min, which was increased over time and reached its maximum level at 96 h post-transduction (Figures 4A and 4B). The SCO2 translation was statistically significantly increased (FD: 1.53, ∗p = 0.0227) at 72 h post-transduction (Figure 4C). In K-562 cells transduced with PTD-IVT-mRNA of SCO2, corresponding to 0.5 μg, no growth inhibition was observed up to 96 h, as shown in Figure S9, nor was cell death (data not shown). The efficient delivery of PTD-IVT-mRNA of SCO2 was confirmed from the first 30 min up to 96 h (Figure 4D), where the IVT-mRNA was detected in K-562 cells (up to an FD of 9) compared to untreated cells and normalized with β-actin gene expression (Figure 4E).
In the case of primary SCO2/COX-deficient fibroblasts, transduction with PTD-IVT-mRNA of SCO2 showed an increase in SCO2 translation, which was observed after only 30 min of exposure and persisted until 96 h post-transduction (Figures 5A and 5B), without notably affecting cell viability (Figure S10). After transduction of 0.5 μg of the PTD-IVT-mRNA into primary SCO2-deficient fibroblasts for 72 h, flow cytometry analysis showed a statistically significant (FD: 7.45, ∗∗p = 0.0013) increase in SCO2 translation compared to the corresponding untreated SCO2-deficient fibroblasts (Figure 5C). The efficient delivery of PTD-IVT-mRNA of SCO2 was confirmed from the first 30 min up to 96 h (Figure 5D), where the IVT-mRNA was detected in SCO2/COX-deficient fibroblasts (up to an FD of 11.9), compared to untreated cells and normalized with β-actin gene expression (Figure 5E).
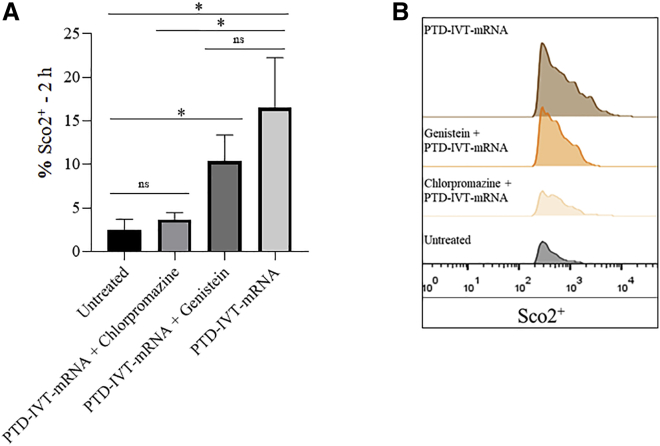
Regarding the uptake mechanism of PTD-IVT-mRNA of SCO2 in SCO2/COX-deficient fibroblasts, cells were preincubated with chlorpromazine and then treated with PTD-IVT-mRNA of SCO2. Compared with untreated cells, no significant differences in SCO2 translation were observed. However, compared to cells treated solely with the PTD-IVT-mRNA, the SCO2 translation was statistically significantly decreased (∗p = 0.0189). However, preincubation with genistein and then treatment with PTD-IVT-mRNA of SCO2 showed a significant increase (FD: 4, ∗p = 0.0136) in SCO2 translation as compared to untreated cells. There is a slight uptake inhibition in genistein-treated cells compared to solely PTD-IVT-mRNA-treated cells; however, this is not statistically significant (p = 0.1779) (Figures 6A and 6B). These data indicate that only in the presence of the active clathrin-dependent endocytosis inhibitor, chlorpromazine, the internalization of PTD-IVT-mRNA of SCO2 is blocked and not in the presence of the active caveolin-dependent endocytosis inhibitor, genistein. Taken together, PTD-IVT-mRNA of SCO2 uptake is mediated via clathrin-dependent endocytosis.
Figure 6.
Uptake mechanism of PTD-IVT-mRNA of SCO2 in SCO2/COX-deficient fibroblasts
(A) Effects of inhibitors of clathrin- and caveolin-mediated endocytosis on protein translation after transduction with PTD-IVT-mRNA of SCO2. The PTD-IVT-mRNA of SCO2 uptake was evaluated using flow cytometry. (B) Representative histograms from flow cytometry analysis, showing Sco2 positivity in untreated cells (0 h), in PTD-IVT-mRNA of SCO2-treated cells, with or without prior incubation, with inhibitors of clathrin- and caveolin-mediated endocytosis. t test: untreated−chlorpromazine, ns; untreated−genistein, ∗p = 0.0136; untreated−PTD-IVT-mRNA, ∗p = 0.0147; chlorpromazine−PTD-IVT-mRNA, ∗p = 0.0189; genistein−PTD-IVT-mRNA, ns.
During the kinetic analysis of SCO2 translation in fibroblasts derived from a SCO2/COX-deficient patient, the control peptide-IVT-mRNA of SCO2 showed no statistically significant increase in SCO2 translation compared to untreated fibroblasts (0 h) from 30 min to 96 h (Figures 7A and 7B), and western blot analysis revealed that Sco2 was successfully probed only in fibroblasts of the SCO2/COX patient treated with the PTD-IVT-mRNA of SCO2 (Figure 7C) and not in untreated or control peptide-IVT-mRNA of SCO2-treated cells. Furthermore, the IVT-mRNA of SCO2 was not detected into the primary SCO2-deficient fibroblasts treated with the control peptide-IVT-mRNA of the SCO2 conjugation product by qPCR and RT-PCR (Figures 7D and 7E) compared to PTD-IVT-mRNA-treated cells.
Figure 7.
Kinetic analysis of SCO2 translation in fibroblasts derived from a SCO2/COX-deficient patient
(A) Flow cytometry results, corresponding to the proportion of Sco2+ cells after staining with anti-Sco2.IgG in untreated cells (0 h), treated with control peptide-IVT-mRNA of SCO2, and treated with PTD-IVT-mRNA of SCO2 from 30 min to 96 h. (B) Representative histogram from flow cytometry analysis, showing Sco2 positivity. Control peptide-IVT-mRNA of SCO2, ANOVA test: ∗∗p = 0.0037; t test: 0 h−30 min, ns; 0 h−24 h, ns; 0 h−48 h, ∗p = 0.0125; 0 h−72 h, ns; 0 h−96 h, ∗p = 0.0249. PTD-IVT-mRNA of SCO2, ANOVA test: ∗∗∗∗p ≤ 0.0001; t test: 0 h−30 min, ∗∗∗∗p ≤ 0.0001; 0 h−24 h, ∗∗∗∗p ≤ 0.0001; 0 h−48 h, ∗∗∗∗p ≤ 0.0001; 0 h−72 h, ∗∗∗∗p ≤ 0.0001; 0 h−96 h, ∗∗∗p = 0.0001. (C) Representative western blot for Sco2, probing in fibroblasts of a SCO2/COX patient in (1) untreated, (2) treated with the control peptide-IVT-mRNA of SCO2, and (3) treated with the PTD-IVT-mRNA of SCO2. (D) The levels of the IVT-mRNA of SCO2, verified by qPCR, using cDNA as a template, derived from total RNA extracts of fibroblasts derived from a SCO2/COX-deficient patient, untreated (0 h); treated with the control peptide-IVT-mRNA of SCO2; and treated with PTD-IVT-mRNA of SCO2. One sample t-test: PTD-IVT-mRNA, ∗∗p = 0.0043. (E) 0.5% agarose gel electrophoresis of PCR products, with cDNA as a template, derived from total RNA extracts of the cells: (1) untreated, (2) treated with the IVT-mRNA of SCO2, (3) treated with the control peptide-IVT-mRNA of SCO2, and (4) treated with PTD-IVT-mRNA of SCO2. (F) Histochemical assessment of COX activity in primary fibroblasts, derived from the SCO2-deficient patient, untreated or treated for 72 h with control peptide-IVT-mRNA of SCO2 or PTD-IVT-mRNA of SCO2 (10× magnification).
Histochemical analysis of COX enzymatic activity was performed using primary fibroblasts, derived from the SCO2/COX-deficient patient,25 which were grown in a 96-well plate and incubated for several time intervals, up to 96 h, with the PTD-IVT-mRNA of SCO2 (0.1 μg/0.05 mL). Cells transduced with the PTD-IVT-mRNA of SCO2 showed increased staining for COX compared to untreated cells or cells exposed to control peptide-IVT-mRNA of SCO2 (Figure 7F). The PTD-IVT-mRNA-induced COX staining was enhanced in a time-dependent manner after the transduction process, indicating successful intracellular delivery of PTD-IVT-mRNA of SCO2, efficient SCO2 translation, and sufficient recovery of COX activity (Figure 8).
Figure 8.
Histochemical assessment of COX activity in SCO2-deficient primary fibroblasts after transduction with the PTD-IVT-mRNA of SCO2 (0.1 μg/0.05 mL)
Primary fibroblasts, derived from the SCO2-deficient patient M.C.,25 incubated for various time intervals (0 h−96 h) with the PTD-IVT-mRNA of SCO2, were histochemically stained to detect the recovery of COX activity (10× magnification).
PTD-IVT-mRNA of β-globin
Concerning the model system of β-thalassemia, the PTD-IVT-mRNA of β-globin was generated, and based on the IVT-mRNA used at different amounts (0.5−0.75 μg), the in vitro cellular translation kinetics of PTD-IVT-mRNA of β-globin was studied first in K-562 cells and later in β-thalassemia BM cells from three individual patients.
The transduced PTD-IVT-mRNA of β-globin in K-562 cells showed an increase in β-globin translation from the first 30 min of exposure to 96 h post-transduction (Figures 9A and 9B). At the 72-h post-transduction time-point with PTD-IVT-mRNA of β-globin, a statistically significant increase in translation of β-globin was detected (FD: 3.13, ∗∗∗p = 0.0007), compared to untreated cells (Figure 9C).
Figure 9.
Kinetic analysis of β-globin translation in K-562 cells after transduction of PTD-IVT-mRNA of β-globin at various time intervals from 0 h to 96 h
(A) Flow cytometry results, corresponding to the proportion of β-globin+ cells after staining with anti-hemoglobin β.IgG. (B) Representative histograms from flow cytometry analysis, showing β-globin positivity in untreated (0 h) and those transduced with PTD-IVT-mRNA β-globin K-562 cells, from 30 min to 96 h. ANOVA test: ∗∗∗∗p ≤ 0.0001; t test: 0 h−30 min, ∗∗p = 0.005; 0 h−24 h, ∗∗∗∗p ≤ 0.0001; 0 h−48 h, ∗∗p = 0.0033; 0 h−72 h, ∗∗∗∗p ≤ 0.0001; 0 h−96 h, ∗p = 0.0121]. (C) Increase (indicated as ↑) of FD in β-globin translation in K-562 cells after transduction with PTD-IVT-mRNA of β-globin at 72 h compared to untreated cells (∗∗∗p = 0.0007). (D) 0.5% agarose gel electrophoresis of PCR products with cDNA as a template, derived from total RNA extracts of K-562 cells, untreated (0 h) and transduced with PTD-IVT-mRNA of β-globin up to 96 h. (E) The levels of the IVT-mRNA of β-globin, verified by qPCR, using cDNA as a template, derived from total RNA extracts of K-562 cells, untreated (0 h) and transduced with PTD-IVT-mRNA of β-globin up to 96 h. ANOVA test: ∗∗∗∗p ≤ 0.0001.
Regarding the delivery efficiency of PTD-IVT-mRNA of β-globin into cells, it was assessed by RT-PCR and qPCR. Regarding RT-PCR, although a less-intense band at 631 bp was also detected in the untreated cells, the band intensity was much higher in K-562 cells treated with PTD-IVT-mRNA of β-globin, from 30 min up to 96 h, demonstrating the successful intracellular transduction (Figure 9D). Regarding qPCR, the IVT-mRNA of β-globin was detected in PTD-IVT-mRNA-treated K-562 cells (increased up to an FD of 5.75) compared to untreated cells and normalized with β-actin gene expression (Figure 9E).
BM-nucleated cells, derived from β-thalassemia patients (Figure 10A, 1−3, for patient 2 and Figure 10B, 1−3, for patient 3) and transduced with the PTD-IVT-mRNA of β-globin, also showed increased β-globin production from the first 30 min, which appeared to be stably increased, up to 96 h after transduction. In general, all three samples derived from the individual β-thalassemia patients, transduced with the PTD-IVT-mRNA of β-globin, showed a statistically significant increase (n = 3, FD: 1.58, ∗∗∗p = 0.0002) of β-globin translation. Notably, patient 3 showed a statistically significant increase (2×) in β-globin translation (FD: 2.05, ∗∗∗p = 0.003) at 72 h as compared to untreated cells (Figure 10C) and up to an FD of 3.73 (∗∗∗∗p ≤ 0.0001) at 96 h (Figure 10B).
Figure 10.
Kinetic analysis of β-globin translation in a β-thalassemic patient’s bone marrow (BM) cells
(A) Patient 2 (I.) and (B) patient 3 (N.) after transduction of PTD-IVT-mRNA of β-globin at various time intervals from 0 h to 96 h. [1] Flow cytometry results, corresponding to the proportion of β-globin+ cells after staining with anti-hemoglobin β.IgG. [2] Representative histograms from flow cytometry analysis, showing β-globin positivity in untreated (0 h) and those transduced with the PTD-IVT-mRNA of β-globin in a β-thalassemic patient’s BM cells from 30 min to 96 h. Patient 2, ANOVA test: ∗∗∗∗p ≤ 0.0001; t test: 0 h−30 min, ∗∗∗p = 0.0007; 0 h−24 h, ∗∗p = 0.0036; 0 h−48 h, ∗∗p = 0.0036; 0 h−72 h, ∗∗p = 0.0031; 0 h−96 h, ∗∗p = 0.0019. Patient 3, ANOVA test: ∗∗∗∗p ≤ 0.0001; t test: 0 h−30 min, p = 0.1482, ns; 0 h−24 h, ∗p = 0.0179; 0 h−48 h, ∗∗∗p = 0.0002; 0 h−72 h, ∗∗p = 0.0031; 0 h−96 h, ∗∗∗∗p ≤ 0.0001. [3] 0.5% agarose gel electrophoresis of PCR products with cDNA as a template, derived from total RNA extracts of a β-thalassemic patient’s BM cells, untreated (0 h) and transduced with the PTD-IVT-mRNA of β-globin up to 96 h. (C) Increase (indicated as ↑) of FD in β-globin translation in three independent samples, derived from three individual β-thalassemic patients after transduction with PTD-IVT-mRNA of β-globin at 72 h compared to untreated cells. Patient 1 (K.): ∗∗∗∗p ≤ 0.0001; patient 2 (I.): ∗∗p = 0.0027; and patient 3 (N.): ∗∗∗p = 0.0003.
Discussion
By conjugating a selected PTD (PFVYLI) to IVT-mRNA, this study developed a novel IVT-mRNA delivery platform that has potential clinical applications for protein therapy of monogenetic/metabolic diseases. Existing state-of-the-art IVT-mRNA delivery methods, such as those using lipid NPs, show high transfectability and high translational outcomes. Pfizer-BioNTech’s BNT162b2 vaccine and Moderna’s mRNA-1273, already marketed as safe and effective vaccines against SARS-CoV-2, both use lipid NPs as mRNA carriers.41,42 These vaccines result in robust immune responses with good tolerability after intramuscular injection.43 However, in the context of ongoing research into the ideal delivery system for IVT-mRNA,44 the use of our novel PTD IVT-mRNA, which appears to be a promising platform for IVT-mRNA delivery and its subsequent translation into a gene product, has been proposed as an alternative option.
Nowadays, systems based on peptide vectors for the delivery of nucleic acids are becoming more popular and widely used due to their flexibility. Peptides have low molecular weight; degradable aa sequences; and distinct biological properties (such as cell permeability efficiency and a targeting nuclear surface), which are their main advantageous properties compared to cationic lipid carriers.45 For over 20 years, PTDs have been used as an alternative to gene-therapy tools to increase the efficiency of transduction of proteins as well as other cargoes into target cells, with relatively low cytotoxicity observed. Several preclinical and clinical trials are currently underway, using PTDs as therapeutic carriers, such as PEP-010 (for metastatic solid tumor cancer, phase Ia/Ib; PEP-Therapy, Evry Cedex, France), ATP128 (for colorectal cancer, phase Ib; Boehringer-Ingelheim, Ingelheim am Rhein, Germany), AVB-620 (for breast cancer, phase I, phase II/III; Avelas Biosciences, Inc., La Jolla, CA, USA), XG-102 (for hearing loss, phase IIΙ; Auris Medical, Basel, Switzerland), and DTS-108 (for cancer, phase I; Diatos SA, Paris, France).46,47
The utility of our novel platform was validated in parallel in two individual monogenic diseases—mitochondrial disorder due to SCO2 mutations and β-thalassemia due to β-globin mutations—to establish its flexibility of use as well as its applicability for the potential delivery and translation of any IVT-mRNA of interest. Herein, we present the conjugation of a selected PTD to two individual IVT-mRNAs (SCO2 and β-globin) by an effective innovative delivery methodology (international patent pending) that forms a stable moiety, allowing sufficient intracellular transduction and translation of the IVT-mRNAs into the corresponding proteins, Sco2 and β-globin.
Briefly, we cloned the IVT plasmid templates containing each CDS, SCO2 and β-globin, with the addition of the 5′ and 3′ UTRs of the β-globin, which increases the stability of the IVT-mRNA. IVT-mRNA production was confirmed by both direct electrophoresis and cDNA synthesis, followed by PCR, providing information about the correct size and sequence integrity of the IVT-mRNA, respectively.
Subsequently, the IVT-mRNAs, both of SCO2 and β-globin, were conjugated to the selected PTD, and the conjugation was first confirmed by the specific band shift, as conjugation to the PTD showed an expected slower transposition of the IVT-mRNA into the polyacrylamide gel compared to the corresponding naked IVT-mRNA. In parallel, a control peptide (WSYGLRPG) was recruited (in 2× concentration compared to the concentration used for the PTD) to serve as a negative control in all subsequent experiments. The control peptide-IVT-mRNA of SCO2 also showed a—similar to the PTD—slower transposition profile into the polyacrylamide gel compared to the naked IVT-mRNA of SCO2.
Furthermore, NMR data analysis provided experimental evidence for the successful conjugation of the PTD-IVT-mRNA, as significant chemical shift differences were observed to the NH peaks of the PTD backbone compared to the NH peaks of the free PTD. These simple experiments provided unequivocal evidence for the nature of the PTD backbone amide peaks and enabled their identification in the reaction mixture of the successfully formed PTD-IVT-mRNA. Similarly, the NMR analysis of the control peptide-IVT-mRNA of SCO2 confirmed the conjugation of the control peptide with the IVT-mRNA, as changes were observed, showing chemical shift differences to the control peptide NH peaks.
Furthermore, the question of whether the IVT-mRNA can be protected from nuclease digestion was investigated, as this is one of the major obstacles in IVT-mRNA-based gene delivery. For this assay, the IVT-mRNA of SCO2 was selected, and the corresponding PTD-IVT-mRNA showed statistically significant stability in RNase-rich environments. After treatment with FBS, a significant portion PTD-IVT-mRNA of SCO2 remained intact, whereas the corresponding naked IVT-mRNA strands disappeared and were not detected, suggesting that PTD protects the IVT-mRNA from immediate nuclease digestion. The control peptide showed a lower ability of protection of the IVT-mRNA of SCO2. Similarly, direct incubation with RNase A showed statistically significant differences between the stability of the IVT-mRNA conjugated to PTD and the naked counterpart. Similarly, a high percentage of protection was observed during the RNase assay of the control peptide-IVT-mRNA of SCO2.
Intracellular delivery of the PTD-IVT-mRNA, of both SCO2 and β-globin, was first studied in K-562 cells, an easy-to-grow and manipulate cell line. In the study of the mitochondrial-defective disease, due to SCO2 mutations, the efficiency of the transduction and translation of the PTD-IVT-mRNA of SCO2 was more than encouraging. The IVT-mRNA of SCO2 was early and highly detected in PTD-IVT-mRNA-treated cells via qPCR analysis in K-562 cells and fibroblasts derived from a SCO2/COX-deficient patient. In the preliminary transduction and translation experiments of K-562 cells, the levels of Sco2 protein were progressively increased and reached the highest protein level 96 h after transduction. The levels of SCO2 translation, 72 h post-transduction, were higher (FD: 1.53) than the untreated K-562 cells. We further proceeded with PTD-IVT-mRNA of SCO2 transduction of fibroblasts derived from a SCO2/COX-deficient patient.15,25 Our delivery platform showed sufficient translational rates of Sco2 protein, from 30 min to 96 h post-transduction, compared to the kinetic analysis of SCO2 translation in cells treated with control peptide-IVT-mRNA of SCO2. Notably, PTD-IVT-mRNA of SCO2 increased SCO2 translation in cells from the SCO2/COX-deficient patient (up to an FD of 7.45), 72 h after transduction. We further investigated the cellular uptake mechanism of PTD IVT-mRNA from SCO2 in fibroblasts, derived from a SCO2/COX-deficient patient, using inhibitors of clathrin- or caveolin-mediated endocytic pathways. Thus, our data suggested that PTD-IVT-mRNA was mediated via a clathrin-dependent pathway.
Furthermore, phenotype complementation was achieved in the primary fibroblasts, derived from a SCO2/COX-deficient patient. The COX activity of these cells (compound heterozygous, with Q53X and E140K mutations) corresponded to 30% residual activity compared to control fibroblasts.25 The increased staining of the transduced patient’s fibroblasts with the PTD-IVT-mRNA of SCO2 (compared to untreated cells or control peptide-IVT-mRNA-treated cells) indicated its successful transduction, followed by translation of the IVT-mRNA of SCO2 to the Sco2 protein, confirming its continued sufficient functionality and restoration of the COX activity. Taking into consideration that even a 9% of reduced COX activity48 is related to a patient phenotype, the transduction efficiency of the PTD-IVT-mRNA of SCO2 and the folds of protein increase observed have the potential to lead to a sufficient phenotype complementation.
In the study of β-globin in K-562 cells, PTD-IVT-mRNA of β-globin entered the cells as early as the first 30 min, confirmed via qPCR analysis, and “triggered” increased translation of the β-globin protein, showing a peak at 72 h post-transduction (FD: 3.13) compared to untreated K-562 cells. The K-562 cell line is also used as a suitable model for β-globin thalassemia research, because of the lack of mature β-globin nascent chains.14 Interestingly, the failure to produce β-globin in K-562 cells could result from an acquired mutation or from an alteration in the environment of the regulatory factors of the β-globin gene, suggesting that the β-globin mRNA is efficiently transcribed, but its translation is blocked by trans-acting factors.49 This could explain a less-intense band at 631 bp, which was also detected in untreated K-562 cells after RNA extraction, cDNA synthesis, and PCR with 5′ UTR-β-globin-3′ UTR-specific primers. Moreover, the band intensity was much higher in K-562 cells transduced with PTD-IVT-mRNA of β-globin, demonstrating successful intracellular transduction. Our next step was to transduce PTD-IVT-mRNA of β-globin into cells from β-thalassemia patients’ BM. The PTD-IVT-mRNA of β-globin was immediately translated into the corresponding β-globin in the BM cells starting from the first 30 min after transduction and showed increased protein translation over a long period of time (up to 96 h studied). More specifically, our novel delivery platform showed an increase in β-globin translation (FD: 1.58) compared to untreated cells in the three individual β-thalassemia patient BM cells at 72 h post-transduction at concentrations that do not cause cytotoxic effects. At this point, it is worth mentioning that the heterogeneity of the cell population of BM cells may reflect the reason for the relatively lower FD of the increase in β-globin translation in the patients’ BM cells compared to the K-562 cells or to the fold of the increase in Sco2 protein in the SCO2/COX -deficient cells. Despite the heterogeneity in the BM population used, at these early stages of development of our novel PTD-IVT-mRNA platform, it was interesting to evaluate the applicability and transduction capacity of our group’s proposed PTD-IVT-mRNA of β-globin.
The anticipated discrepancy, in terms of protein translation profile, of each of the two IVT-mRNAs in the different cell types studied is quite expected, as cell dynamics and requirements differ for each IVT-mRNA to be translated. Moreover, many factors are involved in the cellular uptake and translational efficiency of the delivered IVT-mRNA, depending on both their physicochemical properties and the biomechanical properties of cell membranes.50 Moreover, the altered effect of the PTD-IVT-mRNA in the transduced cells is highly dependent on the needs of each cell type. For example, with respect to the IVT-mRNA of SCO2, it is worth noting that although K-562 endogenous SCO2 transcription and translation proceed normally after the transduction of the PTD-IVT-mRNA of SCO2, the SCO2 translation levels were statistically significantly increased (FD: 1.53 compared to untreated K-562 cells). In contrast, in the mitochondrial defective disease, due to mutations in SCO2 (which unfortunately leads to early death of affected patients in the first stages of their lives), SCO2 translation levels in the SCO2/COX-deficient patient’s cells were significantly increased (FD: 7.45 compared to untreated patient’s cells) after transduction of the PTD-IVT-mRNA of SCO2.
In summary, our work demonstrated that the selected PTD was successfully conjugated to various IVT-mRNAs by an innovative international patent-pending methodology; remained stable even in RNase-rich environments; and was intracellularly transduced into three types of human cells leading to an increase in the production of Sco2 and β-globin proteins in two different disease models of monogenic/metabolic disorders. In the case of COX deficiency mediated by SCO2 mutations, PTD-IVT-mRNA of SCO2 even reversed the monogenic disease phenotype in cell culture and led to the recovery of the suppressed COX activity. These results show that our PTD-IVT-mRNA delivery approach would be a promising platform for successful gene expression, able to be clinically exploited as a PRT for metabolic/genetic disorders.
Materials and methods
Oligonucleotides
All pairs of primers (forward [F]/reverse [R]) used in this study are listed below. Restriction enzyme sites, incorporated to enable cloning, are underlined.
For the SCO2 sequence:
F-H5SCO: 5′-CTA AAG CTT GGA AAC AAA GCA ATC TAT TCT GAT AGA CTC AGG AAG CAA AATG CTG ACT-3′ (67-mer)
R-NSCO: 5′-CCT AT GCG GCC GC TCA AGA CAG GAC ACT GCG GAA AGC CG-3′ (39-mer)
F-N-3′ UTR: 5′-AAG AT GCG GCC GC TAAGCTCGCTTTCTTGCTGTC-3′ (34-mer)
R-BGLO-3′ UTR-X: 5′-CCT AT CTC GAG GCA ATG AAA ATA AAT GTT TTT-3′ (32-mer)
FAMU2SCO: 5′-GAT AGA CTC AGG AAG CAA AAT GC-3′ (23-mer)
RAMU2SCO: 5′-GAC AAA AGC CAG GAC CTC AGA-3′ (21-mer)
For the β-globin sequence:
B-glo-F: 5′-ATGGTGCACCTGACTCCTGA-3′ (20-mer)
Bglo-3′ UTR-R: 5′-GCA ATG AAA ATA AAT GTT TTT-3′ (21-mer)
Bglo-H-5′ UTR-F: 5′-CTA AAG CTT GGA AAC AAA GCA ATC TAT TCT GAT AGA CTC AGG AAG CAA AAT GGT GCA CCT GAC T-3′ (67-mer)
R-BGLO-3′ UTR-X: 5′-CCT AT CTC GAG GCA ATG AAA ATA AAT GTT TTT-3′ (32-mer)
F-beta2: 5′-ATA GAC TCA GGA AGC AAA ATG GT-3′ (23-mer)
R-beta-2: 5′-GTC CAA GGG TAG ACC AG-3′ (20-mer)
For the β-actin sequence:
F-ACTB: 5′-TTG CTG ACA GGA TGC AGA AG-3′ (20-mer)
R-ACTB: 5′-TGA TCC ACA TCT GCT GGA AG-3′ (20-mer)
Construction of recombinant vectors as the in vitro transcription’s templates
The cloning of the SCO2 and β-globin coding sequences (CDSs), each one fused to the optimized 5′ UTR of the murine β-globin mRNA (upstream), including a strong Kozak sequence51 (Table S1), and the 3′ UTR of the human β-globin mRNA (downstream), was carried out individually (for each CDS) in the pcDNA3.1(+) vector mammalian expression vector (Invitrogen Life Technologies, USA). The pcDNA3.1(+) vector includes a multiple cloning site (MCS) for cloning the sequence to be transcribed, downstream of a T7 promoter and upstream of a unique restriction enzyme site to be used for linearization.
Total RNA was extracted from the peripheral blood of a healthy individual and the corresponding produced cDNA served as the template for the amplification of the Homo sapiens HBB complete CDS (GenBank: BC007075.1; 443 bp), including its 3′ UTR (135 bp), using the primer pair B-glo-F/Bglo-3′ UTR-R. The resulting PCR product (576 bp) was ligated to the pCRII-TOPO vector (TOPO TA Cloning Kit; Invitrogen Life Technologies, USA) to generate the pCRII-TOPO-β-globin-3′ UTR vector. From the produced plasmid, the 5′ UTR-β-globin-3′ UTR (631 bp) was amplified, using the primer Bglo-H-5′ UTR-F (including the 5′ UTR of the murine β-globin mRNA) and the primer R-BGLO-3′ UTR-X to add a restriction enzyme (XhoI) cut site. All PCR products, as well as the DNA plasmids, were electrophoresed in 0.5% agarose gels, and 100 bp or 1 kb DNA molecular weight markers (Nippon Genetics, Germany) were used (indicated as MD). TOPO TA cloning was followed, in order to produce the pCRII-TOPO-5′ UTR-β-globin-3′ UTR vector, which was further digested with the restriction enzymes HindIII and XhoI (Takara Bio, Japan), to subclone 5′ UTR-β-globin-3′ UTR into the corresponding cut sites of the pcDNA3 vector. The recombinant plasmid pcDNA3-5′ UTR-β-globin-3′ UTR was generated as the final IVT reaction template for 5′ UTR-β-globin-3′ UTR.
Using the plasmid pHSCO125 as a template, the primer pair F-H5SCO/R-NSCO was used to amplify the F1B1 SCO2F sequence (801 bp; the corresponding CDS of the SCO2, encoding the human full-length Sco2 protein), and then the SCO2 CDS was amplified in fusion to the 5′ UTR of the murine β-globin mRNA (which was included in the sequence of the forward primer, upstream of the CDS sequence). The Sco2 protein consists of 266 aa, including the first 41 aa, predicted to correspond to the N-terminal leader peptide or mitochondrial targeting signal (MTS) peptide (https://ihg.helmholtz-muenchen.de/ihg/mitoprot.html).15 The resulting PCR product (866 bp) was ligated to the pCRII-TOPO vector to generate the pCRII-TOPO-5′ UTR-SCO2 vector. For the amplification of the 3′ UTR of the human β-globin mRNA (159 bp), the primer pair F-N-3′ UTR/R-BGLO-3′ UTR-X was used, whereas the pcDNA3-5′ UTR-β-globin-3′ UTR vector served as a template, followed by the cloning of this sequence to the pCRII-TOPO vector. The pCRII-TOPO-3′ UTR vector was digested by the NotI and XhoI (Takara Bio, Japan) restriction enzymes to be subcloned to the corresponding cut sites of the pcDNA3 vector. The pCRII-TOPO-5′ UTR-SCO2 was then digested with HindIII and NotI (Takara Bio, Japan) restriction enzymes, and the 5′ UTR-SCO2 digested sequence was subcloned into the HindIII and NotI sites of pcDNA3-3′ UTR to generate the corresponding recombinant plasmid pcDNA3-5′ UTR-SCO2-3′ UTR, the final SCO2 IVT reaction template.
For each cloning procedure, freshly prepared, competent E. coli strain TOP10F′ was transformed with the recombinant prokaryotic plasmids. Plasmid DNA isolation was carried out (Nucleospin Plasmid Kit; Macherey-Nagel, Germany), and the clones containing the correct construct inserts were selected via restriction fragment-length polymorphism (RFLP) analysis and verified by automatic sequence analysis (CeMIA SA, Greece).
Preparation and purification of IVT-mRNA
The templates of the IVT reaction—(1) the pcDNA3-5′ UTR-SCO2-3′ UTR, encoding the human full-length Sco2 protein, and (2) the pcDNA3-5′ UTR-β-globin-3′ UTR, encoding the human β-globin, both under the control of a T7 promoter—were propagated in E. coli strain TOP10F′ and purified using standard phenol-chloroform extraction and the ethanol precipitation method. The plasmids were linearized with the XbaI (Takara Bio, Japan) restriction enzyme and purified using a phenol-chloroform extraction method and ethanol precipitation as well, and the digested plasmids were used (one at a time) as a DNA template for the in vitro transcription.
Capped and poly(A)-tailed IVT-mRNA transcripts were produced using the HiScribe T7 ARCA mRNA Kit (with tailing; New England Biolabs [NEB], USA). Each IVT reaction was assembled at 37°C for 2 h using nuclease-free water, the reaction’s buffer/T7 RNA polymerase mixture, 2.5 μg of the plasmid DNA template, and the four ribonucleoside tri-phosphate (rNTP) solution (also containing the ARCAs). DNase I digestion was performed for 15 min at 37°C. Then, adenine residues were added to the 3′ end during the poly(A) tailing reaction and catalyzed by the E. coli poly(A) polymerase (PAP) for 30 min at 37°C; IVT-mRNA purification was performed by LiCl precipitation; and the IVT-mRNA pellet was dissolved in RNase-free water and stored at −20°C.
Each IVT-mRNA prepared was then quantified by spectrophotometric analysis at 260 nm, assessed by a UV spectrophotometer (Nanodrop; Thermo Fisher Scientific, USA). Furthermore, the IVT-mRNAs were analyzed by denaturing 6% PAGE/8 M urea gel electrophoresis at 10 mA/40 V for about 1 h and stained with 0.5 μg/mL ethidium bromide in Tris-borate-EDTA (TBE; 1×) solution for 15 min to confirm its full-length size, using a low-range RNA molecular weight marker (Thermo Fisher Scientific, USA), indicated as MR. Finally, the integrity of the sequence of the IVT-mRNA was assessed by cDNA synthesis, followed by PCR with the use of the SCO2- or β-globin-specific primers.
Conjugation of the PTD to the IVT-mRNA of SCO2/β-globin
The selected PTD is a 6-aa peptide, the PFVYLI, with a well-known protein transduction/penetrating capacity of DNA NPs, larger proteins, siRNAs, etc. The peptide PFVYLI is a shorter version of the peptide C105Y, a synthetic cell-penetrating peptide based on the aa sequence corresponding to residues 359−374 of α1-antitrypsin.52, 53, 54, 55 PFVYLI was synthesized upon order (Thermo Fisher Scientific, USA, and GeneCust, Belgium). PFVYLI is referred to as “PTD.”
The methodology for the preparation of PTD-IVT-mRNAs is described in the international patent application filed on November 11, 2020, under number (no.) PCT/GR2020/000059, which was published on May 20, 2021, under no. WO2021/094792A1, claiming priority of the Greek patent application no. GR20190100504, filed on November 11, 2019, supported by the applicant’s Aristotle University of Thessaloniki, Thessaloniki, Greece, and the University of Thessaly, Volos, Greece, entitled “Method for the development of a delivery platform to produce deliverable PTD-IVT-mRNA therapeutics.”
In brief, the selected PTD—the PFVYLI peptide—is exploited as a neutral surface charge carrier for any therapeutic IVT-mRNA of interest through the covalent chemical reaction of the IVT-mRNA molecule with puromycin, which was conjugated via an amide bond to the PTD. Upon forming the puromycin-PTD product, it was subsequently phosphorylated and then ligated to the therapeutic IVT-mRNA.
The successful conjugation of the known PTD to the IVT-mRNA of either SCO2 or β-globin was evaluated by a band-shift assay compared to the naked IVT-mRNA. Electrophoresis was performed on denaturing 6% PAGE/8 M urea gel, which was visualized and imaged by ethidium bromide staining.
In parallel, the same procedures (novel conjugation reaction, band-shift assay, and NMR analysis in comparison with the naked IVT-mRNA) were conducted with a non-PTD peptide, as a peptide-negative control conjugated with the IVT-mRNA of SCO2 in order to evaluate the penetrating capacity of the selected PTD. The selected control peptide is predicted to be a non-PTD with the sequence WSYGLRPG56 of a preprotein, which is proteolytically processed to generate a peptide that is a member of the gonadotropin-releasing hormone (GnRH) family of proteins. In order to investigate the optimum conditions and achieve the highest reaction yield possible, the novel conjugation reaction was also conducted with two different amounts of this non-PTD control peptide (1× and 2×; data not shown).
NMR analysis of the PTD-IVT-mRNA
NMR experiments, aimed for the verification of the successful PTD conjugation to the IVT-mRNA, were conducted with the PTD-IVT-mRNA of SCO2 (as well as with the control peptide-IVT-mRNA of SCO2) and were recorded at 298 K on a Bruker AVANCE III high-definition four-channel 700 MHz NMR spectrometer equipped with a cryogenically cooled 5 mm 1H/13C/15N/D Z-gradient probe (TCI; Bruker). A typical 1H 1D zgesgp pulse sequence was used for the acquisition of 1H-1D with D1 = 1 s, and D1 = 2 s. All experiments were performed at 25°C. Data were processed with Bruker’s TopSpin 3.3.
Assessment of structural stability of the conjugated PTD-IVT-mRNA
To assess the structural stability of the produced PTD-IVT-mRNA of SCO2 (for these assays, the IVT-mRNA of SCO2 was selected) to remain structurally intact under the cell culture conditions employed in vitro and in vivo, it was incubated as the following: (1) in serum (10% FBS) and (2) in the direct presence of exogenous RNase A (DNase free; AppliChem, Germany) at 37°C and for different time intervals. Thus:
-
(1)
0.35 μg of the IVT-mRNA of SCO2, naked or conjugated to PTD, was incubated in 10% FBS for 5 min or 1 h at 37°C and inactivated by heating for 20 min at 70°C,57 and
-
(2)
0.8 μg of the IVT-mRNA of naked or conjugated to PTD was incubated with 1 ng/mL of RNase A for 1 h at 37°C and inactivated by heating for 20 min at 70°C.
In parallel, the stability of the control peptide-IVT-mRNA of SCO2 was assessed after treatment with 10% FBS and RNase A at the same conditions as the PTD-IVT-mRNA.
Assessment of structural stability was performed via gel electrophoresis, carried out on a 2% agarose gel at 95 V for 60 min, and stained in ethidium bromide to examine the degradation of the naked IVT-mRNA and the PTD-IVT-mRNA of SCO2.
-
(3)
For further investigation of the nature of the type of the conjugation of the PTD to the IVT-mRNA of SCO2, a proteinase K assay was performed. 0.8 μg of the IVT-mRNA of naked or conjugated PTD was incubated at 37°C for 1 h in the absence or presence of 50 μg/mL proteinase K (Finnzymes Oy, Finland) in a total volume of 20 μL. The digestion was stopped by heating at 70°C for 10 min. Samples were then analyzed by agarose gel electrophoresis at 95 V for 60 min and staining in ethidium bromide.
Cell cultures
Human-cultured K-56249 cells were employed as a suitable model system to assess both the transduction as well as expression efficiency of our IVT-mRNA and conjugated PTD-delivery platform in preliminary experiments. K-562 cells were seeded in suspension culture in Roswell Park Memorial Institute (RPMI)-1640 medium (Gibco, Thermo Fisher Scientific, USA), supplemented with 10% v/v FBS and 1% Gibco Antibiotic-Antimycotic (amphotericin B [25 μg/mL], streptomycin [100 μg/mL], and penicillin [100 U/mL]), and maintained in exponential growth at 37°C in a 5% CO2 humidified atmosphere.
Primary fibroblasts, derived from a SCO2-deficient patient (M.C.) with compound heterozygous (Q53X and E140K) mutations,25 were grown as monolayer culture in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Thermo Fisher Scientific, USA); supplemented with 15% v/v FBS, 1% Gibco Antibiotic-Antimycotic, and 1% L-glutamine (GlutaMAX supplement; Gibco, Thermo Fisher Scientific, USA); and maintained in exponential growth at 37°C in a 5% CO2 humidified atmosphere. The cells were suspended by trypsinization with Trypsin-EDTA solution (0.1%; Gibco, Thermo Fisher Scientific, USA). Cells used corresponded to low passage number (less than 9).
BM cells were collected from three β-thalassemic patients (patient 1 [K.], patient 2 [I.], and patient 3 [N.]), and all subjects gave written, informed consent. Venous blood (10 mL) was drawn from all subjects and collected into sterile tubes containing unfractionated conventional heparin as an anticoagulant. BM-derived mononuclear cells (BMDMCs) were isolated from blood samples by centrifugation with Ficoll-Hypaque (Sigma-Aldrich, USA)58 and were cultured under the same conditions, described above for K-562 cells.
Cell number was assessed, as previously described, by using a Neubauer hematocytometer under a light microscope, whereas cell death was assessed by using the trypan blue (Sigma-Aldrich, USA) exclusion assay.59
Intracellular transduction of the PTD-IVT-mRNA
All intracellular transduction experiments were conducted with a specific PTD-IVT-mRNA volume each time, which solely depended on the amount of the respective IVT-mRNA, permitting the treatment with 0.5 μg IVT-mRNA.
In the case of SCO2, the patient’s fibroblast cells were seeded in a 24-well plate at the density of 3−5 × 105 cells/mL (in 500 μL of complete medium) per well, 1 day before transduction. After 24 h, complete medium was replaced by 250 μL of Opti-MEM I Reduced Serum Medium (Gibco, Thermo Fisher Scientific, USA). All cell treatments were harvested after 96 h, whereas 0.5 μg of PTD-IVT-mRNA of SCO2 was added (in triplicates concerning each time point [1 h−96 h]) with comparisons made to untreated cells. Cell numbers were assessed by using a Neubauer hematocytometer.
In the case of the suspension cell cultures concerning the transduction experiments, the cells were seeded in a 24-well culture dish on the same day of transduction, and cells were suspended in 250 μL of Opti-MEM. Concerning the PTD-IVT-mRNA of SCO2, it was added in the amount of 0.5 μg in K-562 for each time point (1 h−96 h). The PTD-IVT-mRNA of β-globin was added in the amounts of 0.5 and 0.75 μg to K-562 cells and BM cells, derived from β-thalassemic patients, respectively. 2 h post-transduction, 500 μL of complete medium was added. Then, the cells were incubated at 37°C and 5% CO2 until their analysis (0−96 h). Cell numbers were assessed, by using a Neubauer hematocytometer, for each individual time-point.
Cellular uptake mechanism
To uncover the mechanism via which the PTD-IVT-mRNA, used in our novel delivery platform, is taken up via the cell membrane, the amount of PTD-IVT-mRNA of SCO2 transduced was evaluated in the presence and absence of chlorpromazine and genistein, known inhibitors of clathrin- or caveolin-mediated endocytosis, respectively. Primary fibroblasts, derived from a SCO2-deficient patient, were preincubated for 30 min at 37°C in the presence of 50 μM chlorpromazine (Sigma-Aldrich, USA) or 25 nM genistein (Sigma-Aldrich, USA),60 followed by media replacement with fresh Opti-MEM and a 2-h-treatment with 0.5 μg of PTD-IVT-mRNA of SCO2.
Delivery efficiency analysis
The successful intracellular delivery of the IVT-mRNA, through its conjugation to the PTD, was evaluated via total RNA extraction from transduced cells, using Tri-Sure, as described above. Two-step RT-PCR was performed, with the SuperScript First-Strand System for RT-PCR (cDNA synthesis; Invitrogen Life Technologies, USA) and the DreamTaq DNA Polymerase (Thermo Fisher Scientific, USA) for the PCR reaction using specific primers.
For the IVT-mRNA intracellular detection via qPCR, the primers FAMU2SCO/BAMU2SCO (for SCO2) and F-beta2/R-beta2 (for β-globin) were designed, as previously described. The delivery levels of IVT-mRNA were quantified by two-step qPCR analysis with the KAPA SYBR FAST qPCR Kit (KK4602; Kapa Biosystems), according to the manufacturer’s instructions, and all reactions were run in duplicate. In this study, β-actin mRNA was chosen as the endogenous control. The relative levels of the IVT-mRNAs were calculated with the 2[-Delta Delta C(t)] (2−ΔΔCt) method,61 and the differences in IVT-mRNA intracellular amounts between the treated and control cells were expressed as FD changes.
Translation efficiency analysis
To quantify the percentage of Sco2-positive or β-globin-positive cells, untreated and transduced cells were harvested, washed with PBS (1×), blocked with 1% FBS, fixed (using BD Cytofix/Cytoperm Fixation/Permeabilization Kit; BD Biosciences), and intracellularly stained with anti-Sco2.immunoglobulin G (IgG)15 or HBB antibody (37-8) (Santa Cruz Biotechnology, USA) as the primary antibody and the Goat Anti-Rabbit IgG (H+L) Secondary Antibody, FITC (fluorescein isothiocyanate) Conjugate (Thermo Fisher Scientific, USA), or anti-mouse IgG kappa binding protein (m-IgGκ BP), FITC, as the secondary antibody (dilution 1:200 with BD Perm/Wash buffer 1×), respectively. The samples were analyzed by the CyFlow Cube 8 flow cytometer (Sysmex Partec, Germany). Dead cells were excluded using FVS660 dye (Fixable Viability Stain 660; BD Biosciences, USA), and then living cells were gated based on their large forward-scatter (FSC) and large side-scatter (SSC) profiles to avoid cell debris. As a negative control, cells were stained only with the secondary antibody. Analysis of raw data was followed by using FlowJo software (BD Biosciences). Data were determined as the mean of three independent experiments. 72 h post-transduction was selected as a representative time-point in all cell types and in all PTD-IVT-mRNA experiments to illustrate the PTD-IVT-mRNA’s transduction efficiency. The increase of the corresponding protein’s translation, assessed via flow cytometry, was demonstrated in FD of increase compared to the mean of the proportion of the positive cells in untreated cells. The untreated cell proportion of positive cells was normalized (valued as 1) and illustrated as a dashed line in FD bar graphs.
SDS-PAGE and western blot analysis were carried out as previously described,14,15,62 and isolated mitochondrial proteins were blotted with either the purified polyclonal antiserum raised against recombinant human Sco2 protein (anti-Sco2.IgG)64 (1:700) or the mouse monoclonal anti-β-actin.IgG (1:2,000) (Santa Cruz Biotechnology). Finally, membranes were incubated with alkaline phosphatase-conjugated goat anti-rabbit IgG-Alkaline phosphatase (AP) (Sigma-Aldrich) (1:2,500) or goat anti-mouse.IgG-AP (1:1,000) (Santa Cruz Biotechnology) as secondary antibodies. Proteins were visualized by using 5-bromo-4-chloro-3-indolyl phosphate (BCIP)/nitro blue tetrazolium (NBT) (Biotium) substrates for alkaline phosphatase.
Histochemical staining for COX activity
A SCO2/COX-deficient patient’s fibroblasts, grown in 96-well plates, were either untreated or incubated for 0−96 h with the PTD-IVT-mRNA of SCO2, corresponding to 0.1 μg of IVT-mRNA, in 0.05 mL of Opti-MEM and 0.1 mL of complete medium, 2 h post-transduction. Then, the cells were washed with PBS (1×) and air dried for 45 min at room temperature. Afterward, the cells were pre-incubated with 200 μL of a solution containing 1 mM CoCl2, 50 μL DMSO in 50 mM Tris-HCl (pH 7.6), and 10% w/v sucrose for 15 min at room temperature. After rinsing once with 0.1 × PBS:10% w/v sucrose (1:1), the cells were incubated for 3 h at 37°C in a 5% CO2-humidified atmosphere with 200 μL of substrate solution (10 mg of cytochrome c, 10 mg of 3,3′-diaminobenzidine, 2 mg of catalase, and 25 μL of DMSO in 10 mL of 0.1 × PBS, pH 7.6, containing 10% w/v sucrose). In the following step, cells (still attached on a 96-well plate) were incubated for 5 min in 0.1 × PBS:10% w/v sucrose (1:1) for another 5 min in PBS (1×) and for 5 min in double-distilled water (ddH2O).15 After air-drying, the plate was examined under an inverted microscope, and photos were taken. Data were determined as the mean of three independent experiments. Primary fibroblasts were also exposed for 72 h to respective amounts of the IVT-mRNA of SCO2 or to control peptide-IVT-mRNA of SCO2 serving as negative control experiments (10× magnification).
Statistical analysis
Statistical analysis was performed using GraphPad Prism version 8.0 software. All data are presented as the mean ± standard error of the mean (SEM). For comparisons among different groups, an ANOVA test was executed. For comparisons of differences between two groups, a t test was performed. p < 0.05 was considered to be significant: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.01, and ∗∗∗∗p < 0.0001.
Acknowledgments
All experiments and manuscript preparation were conducted at Aristotle University of Thessaloniki, Thessaloniki, Macedonia, Greece (except NMR analysis, conducted at the University of Patras, Patras, Greece). We would like to thank Prof. E. A. Schon (Columbia University Medical Center, New York, NY, USA) for kindly donating the SCO2 patient’s primary fibroblast cells to Assoc. Prof. L.C.P.15,25 We would also like to thank Dr. Gavriilides George for his valuable and important assistance in conducting flow cytometry experiments. A.N.M. has been financially supported by the General Secretariat for Research and Technology (GSRT) and the Hellenic Foundation for Research and Innovation (HFRI) (scholarship code: 1533).
Author contributions
A.N.M., as co-inventor of the patent-pending methodology to produce PTD-IVT-mRNAs, performed all of the experimental work and the data analysis and wrote the manuscript. I.S.P. designed, in collaboration with L.C.P., as co-inventor, the patent-pending methodology to produce PTD-IVT-mRNAs and had a crucial role in the successful development of the PTD-IVT-mRNAs and in revising of the paper. G.S. performed the NMR analysis. E.V., as a hematologist, was responsible for the β-thalassemic patients. A.S.T. had an all-around consulting role in the therapeutic approach of a mitochondrial disorder, due to SCO2 mutations, as well as of β-thalassemia, via the PTD technology and contributed to the writing of the paper. I.S.V., as co-inventor of the patent-pending methodology to produce PTD-IVT-mRNAs, had a crucial consulting role in the experimental work and revision of the paper. L.C.P. was the main inventor of the patent-pending methodology to produce PTD-IVT-mRNAs, designed the experimental plan, supervised the experiments and analysis of data, contributed to the writing of the manuscript, and provided the final approval for the manuscript.
Declaration of interests
Authors L.C.P., I.S.P., I.S.V., and A.N.M. are all patent applicants (supported by the Aristotle University of Thessaloniki and the University of Thessaly) in the international patent application filed on November 11, 2020, under no. PCT/GR2020/000059, which was published on May 20, 2021, under no. WO2021/094792A1, claiming priority of the Greek patent application no. GR20190100504, filed on November 11, 2019, supported by the Applicants Aristotle University of Thessaloniki, Thessaloniki, Greece, and the University of Thessaly, Volos, Greece, entitled “Method for the development of a delivery platform to produce deliverable PTD-IVT-mRNA therapeutics.”
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.09.008.
Supplemental information
References
- 1.Zhang X., Men K., Zhang Y., Zhang R., Yang L., Duan X. Local and systemic delivery of mRNA encoding survivin-T34A by lipoplex for efficient colon cancer gene therapy. Int. J. Nanomedicine. 2019;14:2733–2751. doi: 10.2147/IJN.S198747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miliotou A.N., Papadopoulou L.C. In Vitro Transcribed-mRNA CAR Therapy Development. Methods Mol. Biol. 2020;2086:87–117. doi: 10.1007/978-1-0716-0146-4_7. [DOI] [PubMed] [Google Scholar]
- 3.Weissman D. mRNA transcript therapy. Expert Rev. Vaccines. 2015;14:265–281. doi: 10.1586/14760584.2015.973859. [DOI] [PubMed] [Google Scholar]
- 4.Houseley J., Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 5.Tsui N.B., Ng E.K., Lo Y.M. Stability of endogenous and added RNA in blood specimens, serum, and plasma. Clin. Chem. 2002;48:1647–1653. [PubMed] [Google Scholar]
- 6.Sergeeva O.V., Koteliansky V.E., Zatsepin T.S. mRNA-Based Therapeutics - Advances and Perspectives. Biochemistry (Mosc.) 2016;81:709–722. doi: 10.1134/S0006297916070075. [DOI] [PubMed] [Google Scholar]
- 7.Grudzien-Nogalska E., Stepinski J., Jemielity J., Zuberek J., Stolarski R., Rhoads R.E., Darzynkiewicz E. Synthesis of anti-reverse cap analogs (ARCAs) and their applications in mRNA translation and stability. Methods Enzymol. 2007;431:203–227. doi: 10.1016/S0076-6879(07)31011-2. [DOI] [PubMed] [Google Scholar]
- 8.Andries O., Mc Cafferty S., De Smedt S.C., Weiss R., Sanders N.N., Kitada T. N(1)-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J. Control. Release. 2015;217:337–344. doi: 10.1016/j.jconrel.2015.08.051. [DOI] [PubMed] [Google Scholar]
- 9.Ross J., Sullivan T.D. Half-lives of beta and gamma globin messenger RNAs and of protein synthetic capacity in cultured human reticulocytes. Blood. 1985;66:1149–1154. [PubMed] [Google Scholar]
- 10.Broderick K.E., Humeau L.M. Electroporation-enhanced delivery of nucleic acid vaccines. Expert Rev. Vaccines. 2015;14:195–204. doi: 10.1586/14760584.2015.990890. [DOI] [PubMed] [Google Scholar]
- 11.Kauffman K.J., Webber M.J., Anderson D.G. Materials for non-viral intracellular delivery of messenger RNA therapeutics. J. Control. Release. 2016;240:227–234. doi: 10.1016/j.jconrel.2015.12.032. [DOI] [PubMed] [Google Scholar]
- 12.Yin H., Kanasty R.L., Eltoukhy A.A., Vegas A.J., Dorkin J.R., Anderson D.G. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014;15:541–555. doi: 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- 13.Tros de Ilarduya C., Sun Y., Düzgüneş N. Gene delivery by lipoplexes and polyplexes. Eur. J. Pharm. Sci. 2010;40:159–170. doi: 10.1016/j.ejps.2010.03.019. [DOI] [PubMed] [Google Scholar]
- 14.Papadopoulou L.C., Ingendoh-Tsakmakidis A., Mpoutoureli C.N., Tzikalou L.D., Spyridou E.D., Gavriilidis G.I., Kaiafas G.C., Ntaska A.T., Vlachaki E., Panayotou G. Production and Transduction of a Human Recombinant β-Globin Chain into Proerythroid K-562 Cells To Replace Missing Endogenous β-Globin. Mol. Pharm. 2018;15:5665–5677. doi: 10.1021/acs.molpharmaceut.8b00857. [DOI] [PubMed] [Google Scholar]
- 15.Foltopoulou P.F., Tsiftsoglou A.S., Bonovolias I.D., Ingendoh A.T., Papadopoulou L.C. Intracellular delivery of full length recombinant human mitochondrial L-Sco2 protein into the mitochondria of permanent cell lines and SCO2 deficient patient’s primary cells. Biochim. Biophys. Acta. 2010;1802:497–508. doi: 10.1016/j.bbadis.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 16.Papadopoulou L.C., Tsiftsoglou A.S. The potential role of cell penetrating peptides in the intracellular delivery of proteins for therapy of erythroid related disorders. Pharmaceuticals (Basel) 2013;6:32–53. doi: 10.3390/ph6010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Papadopoulou L.C., Tsiftsoglou A.S. Transduction of human recombinant proteins into mitochondria as a protein therapeutic approach for mitochondrial disorders. Pharm. Res. 2011;28:2639–2656. doi: 10.1007/s11095-011-0546-y. [DOI] [PubMed] [Google Scholar]
- 18.Bell G.D., Yang Y., Leung E., Krissansen G.W. mRNA transfection by a Xentry-protamine cell-penetrating peptide is enhanced by TLR antagonist E6446. PLoS ONE. 2018;13:e0201464. doi: 10.1371/journal.pone.0201464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van den Brand D., Gorris M.A.J., van Asbeck A.H., Palmen E., Ebisch I., Dolstra H., Hällbrink M., Massuger L.F.A.G., Brock R. Peptide-mediated delivery of therapeutic mRNA in ovarian cancer. Eur. J. Pharm. Biopharm. 2019;141:180–190. doi: 10.1016/j.ejpb.2019.05.014. [DOI] [PubMed] [Google Scholar]
- 20.Aviñó A., Grijalvo S., Pérez-Rentero S., Garibotti A., Terrazas M., Eritja R. Synthesis of oligonucleotide-peptide conjugates for biomedical and technological applications. Methods Mol. Biol. 2011;751:223–238. doi: 10.1007/978-1-61779-151-2_14. [DOI] [PubMed] [Google Scholar]
- 21.Haralambidis J., Duncan L., Angus K., Tregear G.W. The synthesis of polyamide-oligonucleotide conjugate molecules. Nucleic Acids Res. 1990;18:493–499. doi: 10.1093/nar/18.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kachalova A., Zubin E., Stetsenko D., Gait M., Oretskaya T. Oligonucleotides with 2′-O-carboxymethyl group: synthesis and 2′-conjugation via amide bond formation on solid phase. Org. Biomol. Chem. 2004;2:2793–2797. doi: 10.1039/B409496D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zatsepin T.S., Stetsenko D.A., Arzumanov A.A., Romanova E.A., Gait M.J., Oretskaya T.S. Synthesis of peptide-oligonucleotide conjugates with single and multiple peptides attached to 2′-aldehydes through thiazolidine, oxime, and hydrazine linkages. Bioconjug. Chem. 2002;13:822–830. doi: 10.1021/bc020016+. [DOI] [PubMed] [Google Scholar]
- 24.Meade B.R., Dowdy S.F. Exogenous siRNA delivery using peptide transduction domains/cell penetrating peptides. Adv. Drug Deliv. Rev. 2007;59:134–140. doi: 10.1016/j.addr.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 25.Papadopoulou L.C., Sue C.M., Davidson M.M., Tanji K., Nishino I., Sadlock J.E., Krishna S., Walker W., Selby J., Glerum D.M. Fatal infantile cardioencephalomyopathy with COX deficiency and mutations in SCO2, a COX assembly gene. Nat. Genet. 1999;23:333–337. doi: 10.1038/15513. [DOI] [PubMed] [Google Scholar]
- 26.Tarnopolsky M.A., Bourgeois J.M., Fu M.H., Kataeva G., Shah J., Simon D.K., Mahoney D., Johns D., MacKay N., Robinson B.H. Novel SCO2 mutation (G1521A) presenting as a spinal muscular atrophy type I phenotype. Am. J. Med. Genet. A. 2004;125A:310–314. doi: 10.1002/ajmg.a.20466. [DOI] [PubMed] [Google Scholar]
- 27.Szymanska-Debinska T., Karkucinska-Wieckowska A., Piekutowska-Abramczuk D., Jurkiewicz E., Iwanicka-Pronicka K., Rokicki D., Pronicki M. Leigh disease due to SCO2 mutations revealed at extended autopsy. J. Clin. Pathol. 2015;68:397–399. doi: 10.1136/jclinpath-2014-202606. [DOI] [PubMed] [Google Scholar]
- 28.DiMauro S., Hirano M., Schon E.A. Approaches to the treatment of mitochondrial diseases. Muscle Nerve. 2006;34:265–283. doi: 10.1002/mus.20598. [DOI] [PubMed] [Google Scholar]
- 29.Ekim Kocabey A., Kost L., Gehlhar M., Rödel G., Gey U. Mitochondrial Sco proteins are involved in oxidative stress defense. Redox Biol. 2019;21:101079. doi: 10.1016/j.redox.2018.101079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams J.C., Sue C., Banting G.S., Yang H., Glerum D.M., Hendrickson W.A., Schon E.A. Crystal structure of human SCO1: implications for redox signaling by a mitochondrial cytochrome c oxidase “assembly” protein. J. Biol. Chem. 2005;280:15202–15211. doi: 10.1074/jbc.M410705200. [DOI] [PubMed] [Google Scholar]
- 31.Timón-Gómez A., Nývltová E., Abriata L.A., Vila A.J., Hosler J., Barrientos A. Mitochondrial cytochrome c oxidase biogenesis: Recent developments. Semin. Cell Dev. Biol. 2018;76:163–178. doi: 10.1016/j.semcdb.2017.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pacheu-Grau D., Wasilewski M., Oeljeklaus S., Gibhardt C.S., Aich A., Chudenkova M., Dennerlein S., Deckers M., Bogeski I., Warscheid B. COA6 Facilitates Cytochrome c Oxidase Biogenesis as Thiol-reductase for Copper Metallochaperones in Mitochondria. J. Mol. Biol. 2020;432:2067–2079. doi: 10.1016/j.jmb.2020.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matoba S., Kang J.G., Patino W.D., Wragg A., Boehm M., Gavrilova O., Hurley P.J., Bunz F., Hwang P.M. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 34.Wang T., Birsoy K., Hughes N.W., Krupczak K.M., Post Y., Wei J.J., Lander E.S., Sabatini D.M. Identification and characterization of essential genes in the human genome. Science. 2015;350:1096–1101. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuszak A.J., Espey M.G., Falk M.J., Holmbeck M.A., Manfredi G., Shadel G.S., Vernon H.J., Zolkipli-Cunningham Z. Nutritional Interventions for Mitochondrial OXPHOS Deficiencies: Mechanisms and Model Systems. Annu. Rev. Pathol. 2018;13:163–191. doi: 10.1146/annurev-pathol-020117-043644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Casarin A., Giorgi G., Pertegato V., Siviero R., Cerqua C., Doimo M., Basso G., Sacconi S., Cassina M., Rizzuto R. Copper and bezafibrate cooperate to rescue cytochrome c oxidase deficiency in cells of patients with SCO2 mutations. Orphanet J. Rare Dis. 2012;7:21. doi: 10.1186/1750-1172-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.El-Hattab A.W., Zarante A.M., Almannai M., Scaglia F. Therapies for mitochondrial diseases and current clinical trials. Mol. Genet. Metab. 2017;122:1–9. doi: 10.1016/j.ymgme.2017.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soma S., Latimer A.J., Chun H., Vicary A.C., Timbalia S.A., Boulet A., Rahn J.J., Chan S.S.L., Leary S.C., Kim B.E. Elesclomol restores mitochondrial function in genetic models of copper deficiency. Proc. Natl. Acad. Sci. USA. 2018;115:8161–8166. doi: 10.1073/pnas.1806296115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karponi G., Zogas N. Gene Therapy For Beta-Thalassemia: Updated Perspectives. Appl. Clin. Genet. 2019;12:167–180. doi: 10.2147/TACG.S178546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giardini C., Lucarelli G. Bone marrow transplantation in the treatment of thalassemia. Curr. Opin. Hematol. 1994;1:170–176. [PubMed] [Google Scholar]
- 41.Corbett K.S., Flynn B., Foulds K.E., Francica J.R., Boyoglu-Barnum S., Werner A.P., Flach B., O’Connell S., Bock K.W., Minai M. Evaluation of the mRNA-1273 Vaccine against SARS-CoV-2 in Nonhuman Primates. N. Engl. J. Med. 2020;383:1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., C4591001 Clinical Trial Group Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hassett K.J., Benenato K.E., Jacquinet E., Lee A., Woods A., Yuzhakov O., Himansu S., Deterling J., Geilich B.M., Ketova T. Optimization of Lipid Nanoparticles for Intramuscular Administration of mRNA Vaccines. Mol. Ther. Nucleic Acids. 2019;15:1–11. doi: 10.1016/j.omtn.2019.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xue H.Y., Guo P., Wen W.C., Wong H.L. Lipid-Based Nanocarriers for RNA Delivery. Curr. Pharm. Des. 2015;21:3140–3147. doi: 10.2174/1381612821666150531164540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ahmed M. Peptides, polypeptides and peptide-polymer hybrids as nucleic acid carriers. Biomater. Sci. 2017;5:2188–2211. doi: 10.1039/c7bm00584a. [DOI] [PubMed] [Google Scholar]
- 46.Xie J., Bi Y., Zhang H., Dong S., Teng L., Lee R.J., Yang Z. Cell-Penetrating Peptides in Diagnosis and Treatment of Human Diseases: From Preclinical Research to Clinical Application. Front. Pharmacol. 2020;11:697. doi: 10.3389/fphar.2020.00697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matijass M., Neundorf I. Cell-penetrating peptides as part of therapeutics used in cancer research. Med. Drug Discov. 2021;10:100092. [Google Scholar]
- 48.Glerum D.M., Yanamura W., Capaldi R.A., Robinson B.H. Characterization of cytochrome-c oxidase mutants in human fibroblasts. FEBS Lett. 1988;236:100–104. doi: 10.1016/0014-5793(88)80293-x. [DOI] [PubMed] [Google Scholar]
- 49.Fordis C.M., Anagnou N.P., Dean A., Nienhuis A.W., Schechter A.N. A beta-globin gene, inactive in the K562 leukemic cell, functions normally in a heterologous expression system. Proc. Natl. Acad. Sci. USA. 1984;81:4485–4489. doi: 10.1073/pnas.81.14.4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bhosle S.M., Loomis K.H., Kirschman J.L., Blanchard E.L., Vanover D.A., Zurla C., Habrant D., Edwards D., Baumhof P., Pitard B., Santangelo P.J. Unifying in vitro and in vivo IVT mRNA expression discrepancies in skeletal muscle via mechanotransduction. Biomaterials. 2018;159:189–203. doi: 10.1016/j.biomaterials.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 51.Koblas T., Leontovyc I., Loukotova S., Kosinova L., Saudek F. Reprogramming of Pancreatic Exocrine Cells AR42J Into Insulin-producing Cells Using mRNAs for Pdx1, Ngn3, and MafA Transcription Factors. Mol. Ther. Nucleic Acids. 2016;5:e320. doi: 10.1038/mtna.2016.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rhee M., Davis P. Mechanism of uptake of C105Y, a novel cell-penetrating peptide. J. Biol. Chem. 2006;281:1233–1240. doi: 10.1074/jbc.M509813200. [DOI] [PubMed] [Google Scholar]
- 53.Jones S., Lukanowska M., Suhorutsenko J., Oxenham S., Barratt C., Publicover S., Copolovici D.M., Langel Ü., Howl J. Intracellular translocation and differential accumulation of cell-penetrating peptides in bovine spermatozoa: evaluation of efficient delivery vectors that do not compromise human sperm motility. Hum. Reprod. 2013;28:1874–1889. doi: 10.1093/humrep/det064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barkalina N., Jones C., Townley H., Coward K. Functionalization of mesoporous silica nanoparticles with a cell-penetrating peptide to target mammalian sperm in vitro. Nanomedicine (Lond.) 2015;10:1539–1553. doi: 10.2217/nnm.14.235. [DOI] [PubMed] [Google Scholar]
- 55.Fang B., Guo H.Y., Zhang M., Jiang L., Ren F.Z. The six amino acid antimicrobial peptide bLFcin6 penetrates cells and delivers siRNA. FEBS J. 2013;280:1007–1017. doi: 10.1111/febs.12093. [DOI] [PubMed] [Google Scholar]
- 56.Hällbrink M., Kilk K., Elmquist A., Lundberg P., Lindgren M., Jiang Y., Pooga M., Soomets U., Langel Ü. Prediction of Cell-Penetrating Peptides. Int. J. Pept. Res. Ther. 2005;11:249–259. [Google Scholar]
- 57.Kim H., Park Y., Lee J.B. Self-assembled Messenger RNA Nanoparticles (mRNA-NPs) for Efficient Gene Expression. Sci. Rep. 2015;5:12737. doi: 10.1038/srep12737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vlachaki E., Ioannidou-Papagiannaki E., Tziomalos K., Haralambidou-Vranitsa S., Perifanis V., Klonizakis I., Athanassiou-Metaxa M. Peripheral blood haematopoietic progenitor cells in patients with beta thalassaemia major receiving desferrioxamine or deferiprone as chelation therapy. Eur. J. Haematol. 2007;78:48–51. doi: 10.1111/j.1600-0609.2006.00773.x. [DOI] [PubMed] [Google Scholar]
- 59.Tsiftsoglou A.S., Hwang K.M., Agrawal K.C., Sartorelli A.C. Strand-scission of Sarcoma 180 tumor cell DNA induced by 1-formylisoquinoline thiosemicarbazone. Biochem. Pharmacol. 1975;24:1631–1633. doi: 10.1016/0006-2952(75)90091-x. [DOI] [PubMed] [Google Scholar]
- 60.Takahashi S., Tada R., Negishi Y., Aramaki Y. Mechanisms of Enhanced Antigen Delivery to Murine Dendritic Cells by the Cationic Liposomes. Open J. Immunol. 2017;7:85–101. [Google Scholar]
- 61.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 62.Kaiafas G.C., Papagiannopoulou D., Miliotou Α.N., Tsingotjidou A.S., Chalkidou P.C., Tsika A.C., Spyroulias G.A., Tsiftsoglou A.S., Papadopoulou L.C. In vivo biodistribution study of TAT-L-Sco2 fusion protein, developed as protein therapeutic for mitochondrial disorders attributed to SCO2 mutations. Mol. Genet. Metab. Rep. 2020;25:100683. doi: 10.1016/j.ymgmr.2020.100683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Foltopoulou P.F., Zachariadis G.A., Politou A.S., Tsiftsoglou A.S., Papadopoulou L.C. Human recombinant mutated forms of the mitochondrial COX assembly Sco2 protein differ from wild-type in physical state and copper binding capacity. Mol. Genet. Metab. 2004;81:225–236. doi: 10.1016/j.ymgme.2003.11.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.