Abstract
Group A human rotavirus G genotypes were determined by means of reverse transcription-PCR in 170 stool specimens from children with acute diarrhea admitted to a Paris children’s hospital during a 1-year survey (1997 to 1998). The isolates all belonged to types G1 to G4, with type G4 predominating (60%).
Group A rotavirus is the main cause of acute gastroenteritis in children worldwide. The virus possesses a genome of 11 double-stranded RNA segments, each encoding one viral protein. The G and P serotypes of group A rotavirus are specified by two outer capsid proteins, respectively designated VP7 (encoded by genome segment 7, 8, or 9, depending on the strain) and VP4 (gene 4 product). Fourteen rotavirus G serotypes have been described, and 10 have been recovered from humans (4). Epidemiological studies based on G (VP7) serotyping or genotyping methods have indicated that serotypes G1 to G4 are the most widespread, and that type G1 is the most prevalent (3, 6, 7, 10, 13, 24, 27). Serotypes G1 to 4 are targeted by a rhesus rotavirus (RRV) tetravalent vaccine recently licensed by the U.S. Food and Drug Administration (11, 15). The vaccine provides moderate protection (∼50%) against rotavirus gastroenteritis of all severities and good to excellent protection (∼80 to 100%) against severe disease (2, 9, 17, 20, 25, 26). The perspective of using such a vaccine in Europe is attractive, but a precise knowledge of circulating G serotypes is crucial before and after vaccine introduction. Unusual G serotypes can be common in some parts of the world, especially developing countries, e.g., G9 in India (18) and G5 and G10 in Brazil (12, 21). Moreover, a recent study showed that G9 was the third most prevalent type in the United States, with an unusually high detection rate of 7.2% (19). To our knowledge, no data on G types circulating in France are available. We therefore determined the frequency and temporal distribution of human rotavirus (HRV) G types among children admitted to a Paris children’s hospital during a 1-year survey. Type G4 predominated during this period.
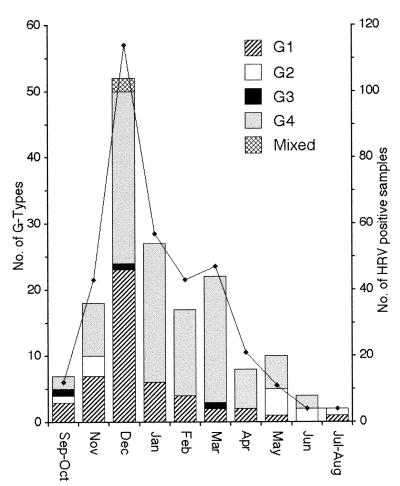
Between September 1997 and August 1998, 356 fecal samples from children under 5 years of age with acute diarrhea admitted to Trousseau Pediatric Hospital, Paris, France, were found positive for rotavirus infection by enzyme immunoassay (Abbott Diagnostic, Rungis, France) or electron microscopy. As usually reported in industrialized countries (4), a seasonal pattern of infection was observed, with the epidemic peak occurring in December (Fig. 1). At least 40% of each month’s positive samples were selected for further G-type characterization, in order to obtain a selection representative of the epidemic distribution. A total of 170 rotavirus-positive samples were selected; 138 were obtained from children hospitalized for community-acquired severe acute diarrhea (mean age, 10.6 months; range, 0.1 to 48 months), and 32 were from children who developed hospital-acquired diarrheal illness more than 3 days after admission (mean age, 6.3 months; range, 0.3 to 52 months).
FIG. 1.
Monthly distribution of rotavirus G types from September 1997 to August 1998. The solid line indicates the total number of rotavirus-positive samples. The G types of at least 40% of the rotavirus-positive samples obtained each month were determined.
G types were identified by the reverse transcription-PCR assay (RT-PCR) described by Gouvea et al. (8) with a few modifications. Stool suspensions (∼10% [wt/vol] in 9‰ NaCl) were clarified by low-speed centrifugation, and viral RNA was extracted from 200 μl of supernatant by using the QIAamp blood kit (Qiagen, Courtaboeuf, France) according to the manufacturer’s recommendations. Five microliters of the RNA extract was reverse-transcribed into gene 9 (VP7) full-length cDNA with the generic primers Beg9 and End9 (8) in a 50-μl reaction mixture containing 20 μM EDTA, 10 mM dithiothreitol, a 0.5 mM concentration of each deoxynucleoside triphosphate, a 0.1 μM concentration of each primer, 10 U of RNase inhibitor (Life Technologies, Cergy, France), 200 U of SuperScript II (Life Technologies), and 1× SuperScript buffer. After 45 min of incubation at 45°C the reaction was stopped by adding 1 μl of 0.5 M EDTA and 150 μl of water. Five microliters of cDNA was amplified with a mixture of G1 to G4 type-specific sense primers aBT1, aCT2, aET3, and aDT4 (8) and a generic antisense primer, EndA, whose sequence is conserved among G types (nucleotides 922 to 944: 5′-ATAGTATAAAATACTTGCCACCA-3′). The 50-μl reaction mixture consisted of 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 2 mM MgCl2, a 0.2 mM concentration of each deoxynucleoside triphosphate, a 0.25 μM concentration of each primer, and 1.25 U of AmpliTaq DNA polymerase (Perkin-Elmer, Villebon, France). Amplification was performed in a Perkin-Elmer thermocycler (model 9700), under PCR conditions of 35 cycles at 94°C for 30 s, 50°C for 1 min, and 72°C for 30 s. PCR products were analyzed by electrophoresis on 1.5% agarose gels. In this system, the sizes of the type-specific PCR products were 630 bp (G1), 533 bp (G2), 255 bp (G3), and 464 bp (G4). Rotaviruses of known G serotypes (strains Wa [G1], DS-1 [G2], SA11 and RRV [G3], and MtB2 [G4]) were used as controls in each experiment. Negative controls consisted of rotavirus-negative stool samples. By using this rapid extraction method and a one-step PCR assay, 128 samples (75%) could be G typed. All but three of the negative specimens were rescued after further RNA extraction with RNA-PLUS (Bioprobe Systems, Montreuil, France) followed by RT-PCR either under the same conditions or in a two-step technique. In the latter case, gene 9 cDNA was first amplified for 25 cycles (94°C for 30 s, 55°C for 1 min, and 72°C for 30 s) with the generic primers EndA and BegA (nucleotides 50 to 71: 5′-TGTATGGTATTGAATATACCAC-3′) complementary to conserved sequences located respectively in the 3′ and 5′ regions of gene 9. The PCR product (2 μl) was then amplified with type-specific primers as described above.
The G type was determined for 167 of the 170 rotavirus-positive specimens (98%), all of which belonged to conventional G types 1 to 4 (Table 1 and Fig. 1). Surprisingly, rotavirus type G4 was the most prevalent during the survey (n = 102; 60%). Type G4 was highly predominant during and after January 1998, when it accounted for 66 of the 90 typed cases (73%). Type G1 was detected at a lower frequency (n = 49; 29%) and was mainly found at the beginning of the epidemic, when it accounted for 43% of cases. Type G2 was infrequent (n = 11; 6.5%) and was only isolated at the beginning and end of the epidemic. G2 strains had short electropherotypes when analyzed by polyacrylamide gel electrophoresis (PAGE) (data not shown). Type G3 was detected sporadically, in only three of the specimens (1.8%). Two mixed G-type infections were detected (1.2%), and both were dual infections with G1 plus G4. These mixed infections occurred during the epidemic peak in December, when the prevalence of G1 strains was similar to that of G4 strains. The G-type distribution was not age dependent and was not associated with the hospital- or community-acquired nature of the infection (Table 1). This indicated that nosocomial rotavirus infection was probably due to continuous introduction of community strains, as previously observed (5, 23). Only 3 of the 170 rotavirus-positive samples could not be typed with the G1 to G4 primers. These samples were negative when analyzed by PAGE and were also negative when tested with the G8 (aAT8) and G9 (aFT9) type-specific primers described by Gouvea et al. (8). In addition, the generic primers repeatedly failed to amplify the full-length gene 9 in any of these samples. We considered that these strains were untypeable (because of RT-PCR failure due to a low amount of viral RNA or to the presence of strong inhibitors) rather than belonging to unconventional G types. Thus, no rotavirus strains of types other than G1 to G4 were identified in this survey, and their frequency could not have exceeded 1.8%.
TABLE 1.
Distribution of HRV G types during the 1997 to 1998 epidemic in Paris, France
| HRV strain category | No. of strains tested | No. (%) of strains belonging to type:
|
No. of nontypeable strains | ||||
|---|---|---|---|---|---|---|---|
| G1 | G2 | G3 | G4 | Mixeda | |||
| Community acquired | 138 | 38 (27.5) | 10 (7.2) | 3 (2.2) | 83 (60.1) | 1 (0.7) | 3b |
| Hospital acquired | 32 | 11 (34.4) | 1 (3.1) | 0 | 19 (59.4) | 1 (3.1) | 0 |
| Total | 170 | 49 (28.8) | 11 (6.5) | 3 (1.8) | 102 (60) | 2 (1.2) | 3 (1.8) |
Mixed rotavirus infections were dual infections with G1 plus G4.
All the nontypeable strains were negative when analyzed by PAGE and were also negative when tested with G8 and G9 type-specific primers.
This 1-year survey in a Paris children’s hospital provides the first data on G types circulating in France shortly before the proposed introduction of a rotavirus vaccine. We showed that all the rotavirus strains analyzed belonged to the most common types, G1 to G4, a situation encountered in the majority of industrialized countries (3, 6, 13, 24, 27), including European states (7, 16). We only detected three type G3 strains, in keeping with previous reports that type G3 infection is relatively uncommon in Europe (1, 7, 14, 16); alternatively, type G3 strains may produce a less severe illness, as our survey focused on children who were sufficiently ill to be admitted to a hospital. One interesting finding was the high prevalence of rotavirus type G4. Type G1 is usually the predominant strain worldwide (4, 6), and few epidemics associated with a predominant G4 strain have been recorded (1, 3, 14, 16). Whether or not the predominance of type G4 in our survey was restricted to the Paris area or to the 1997 to 1998 epidemic remains to be established. Additional long-term surveys conducted as part of a strain surveillance system will be needed to monitor the strains circulating in France, as data from a particular site or epidemic cannot be used as an indicator of what is happening throughout the country. In addition, such epidemics associated with a predominant G4 strain raise concerns over vaccine efficacy; indeed, the candidate rotavirus vaccine, RRV tetravalent vaccine, was mainly evaluated in trials in which serotype G1 (2, 9, 17, 20) or G3 (22) predominated.
Acknowledgments
This work was supported in part by Assistance Publique-Hôpitaux de Paris grant CRC 97135 and by the MESRT grant Programme de Recherches Fondamentales en Microbiologie, Maladies infectieuses et Parasitologie.
We thank J. Cohen and E. Kohli for providing the rotavirus control strains.
REFERENCES
- 1.Arista S, Vizzi E, Ferraro D, Cascio A, Di Stefano R. Distribution of VP7 serotypes and VP4 genotypes among rotavirus strains recovered from Italian children with diarrhea. Arch Virol. 1997;142:2065–2071. doi: 10.1007/s007050050224. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein D I, Glass R I, Rodgers G, Davidson B L, Sack D A. Evaluation of rhesus rotavirus monovalent and tetravalent reassortant vaccines in US children. US Rotavirus Vaccine Efficacy Group. JAMA. 1995;273:1191–1196. [PubMed] [Google Scholar]
- 3.Bishop R F, Unicomb L E, Barnes G L. Epidemiology of rotavirus serotypes in Melbourne, Australia, from 1973 to 1989. J Clin Microbiol. 1991;29:862–868. doi: 10.1128/jcm.29.5.862-868.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Estes M. Rotaviruses and their replication. In: Fields B, Knipe D, Howley P, et al., editors. Fields virology. New York, N.Y: Raven Press; 1996. pp. 1625–1655. [Google Scholar]
- 5.Gaggero A, Avendano L F, Fernandez J, Spencer E. Nosocomial transmission of rotavirus from patients admitted with diarrhea. J Clin Microbiol. 1992;30:3294–3297. doi: 10.1128/jcm.30.12.3294-3297.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gentsch J R, Woods P A, Ramachandran M, Das B K, Leite J P, Alfieri A, Kumar R, Bhan M K, Glass R I. Review of G and P typing results from a global collection of rotavirus strains: implications for vaccine development. J Infect Dis. 1996;174(Suppl. 1):S30–S36. doi: 10.1093/infdis/174.supplement_1.s30. [DOI] [PubMed] [Google Scholar]
- 7.Gerna G, Sarasini A, Arista S, di-Matteo A, Giovannelli L, Parea M, Halonen P. Prevalence of human rotavirus serotypes in some European countries 1981–1988. Scand J Infect Dis. 1990;22:5–10. doi: 10.3109/00365549009023112. [DOI] [PubMed] [Google Scholar]
- 8.Gouvea V, Glass R I, Woods P, Taniguchi K, Clark H F, Forrester B, Fang Z Y. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J Clin Microbiol. 1990;28:276–282. doi: 10.1128/jcm.28.2.276-282.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joensuu J, Koskenniemi E, Pang X L, Vesikari T. Randomised placebo-controlled trial of rhesus-human reassortant rotavirus vaccine for prevention of severe rotavirus gastroenteritis. Lancet. 1997;350:1205–1209. doi: 10.1016/S0140-6736(97)05118-0. [DOI] [PubMed] [Google Scholar]
- 10.Kaga E, Iizuka M, Nakagomi T, Nakagomi O. The distribution of G (VP7) and P (VP4) serotypes among human rotaviruses recovered from Japanese children with diarrhea. Microbiol Immunol. 1994;38:317–320. doi: 10.1111/j.1348-0421.1994.tb01784.x. [DOI] [PubMed] [Google Scholar]
- 11.Kapikian A Z, Hoshino Y, Chanock R M, Perez-Schael I. Efficacy of a quadrivalent rhesus rotavirus-based human rotavirus vaccine aimed at preventing severe rotavirus diarrhea in infants and young children. J Infect Dis. 1996;174(Suppl. 1):S65–S72. doi: 10.1093/infdis/174.supplement_1.s65. [DOI] [PubMed] [Google Scholar]
- 12.Leite J, Alfieri A A, Woods P A, Glass R I, Gentsch J R. Rotavirus G and P types circulating in Brazil: characterization by RT-PCR, probe hybridization, and sequence analysis. Arch Virol. 1996;141:2365–2374. doi: 10.1007/BF01718637. [DOI] [PubMed] [Google Scholar]
- 13.Matson D O, Estes M K, Burns J W, Greenberg H B, Taniguchi K, Urasawa S. Serotype variation of human group A rotaviruses in two regions of the USA. J Infect Dis. 1990;162:605–614. doi: 10.1093/infdis/162.3.605. [DOI] [PubMed] [Google Scholar]
- 14.Maunula L, van Bonsdorff C H. Rotavirus serotypes and electropherotypes in Finland from 1986 to 1990. Arch Virol. 1995;140:877–890. doi: 10.1007/BF01314964. [DOI] [PubMed] [Google Scholar]
- 15.Midthun K, Kapikian A Z. Rotavirus vaccines: an overview. Clin Microbiol Rev. 1996;9:423–434. doi: 10.1128/cmr.9.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noel J S, Beards G M, Cubitt W D. Epidemiological survey of human rotavirus serotypes and electropherotypes in young children admitted to two children’s hospitals in northeast London from 1984 to 1990. J Clin Microbiol. 1991;29:2213–2219. doi: 10.1128/jcm.29.10.2213-2219.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perez-Schael I, Guntinas M J, Perez M, Pagone V, Rojas A M, Gonzalez R, Cunto W, Hoshino Y, Kapikian A Z. Efficacy of the rhesus rotavirus-based quadrivalent vaccine in infants and young children in Venezuela. N Engl J Med. 1997;337:1181–1187. doi: 10.1056/NEJM199710233371701. [DOI] [PubMed] [Google Scholar]
- 18.Ramachandran M, Das B K, Vij A, Kumar R, Bhambal S S, Kesari N, Rawat H, Bahl L, Thakur S, Woods P A, Glass R I, Bhan M K, Gentsch J R. Unusual diversity of human rotavirus G and P genotypes in India. J Clin Microbiol. 1996;34:436–439. doi: 10.1128/jcm.34.2.436-439.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramachandran M, Gentsch J R, Parashar U D, Jin S, Woods P A, Holmes J L, Kirkwood C D, Bishop R F, Greenberg H B, Urasawa S, Gerna G, Coulson B S, Taniguchi K, Bresee J S, Glass R I. Detection and characterization of novel rotavirus strains in the United States. J Clin Microbiol. 1998;36:3223–3229. doi: 10.1128/jcm.36.11.3223-3229.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rennels M B, Glass R I, Dennehy P H, Bernstein D I, Pichichero M E, Zito E T, Mack M E, Davidson B L, Kapikian A Z. Safety and efficacy of high-dose rhesus-human reassortant rotavirus vaccines—report of the National Multicenter Trial. United States Rotavirus Vaccine Efficacy Group. Pediatrics. 1996;97:7–13. [PubMed] [Google Scholar]
- 21.Santos N, Lima R C, Pereira C F, Gouvea V. Detection of rotavirus types G8 and G10 among Brazilian children with diarrhea. J Clin Microbiol. 1998;36:2727–2729. doi: 10.1128/jcm.36.9.2727-2729.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Santosham M, Moulton L H, Reid R, Croll J, Weatherholt R, Ward R, Forro J, Zito E, Mack M, Brenneman G, Davidson B L. Efficacy and safety of high-dose rhesus-human reassortant rotavirus vaccine in Native American populations. J Pediatr. 1997;131:632–638. doi: 10.1016/s0022-3476(97)70076-3. [DOI] [PubMed] [Google Scholar]
- 23.Steele A D, Mnisi Y N, Williams M M, Bos P, Aspinall S. Electrophoretic typing of nosocomial rotavirus infection in a general paediatric unit showing the continual introduction of community strains. J Med Virol. 1993;40:126–132. doi: 10.1002/jmv.1890400209. [DOI] [PubMed] [Google Scholar]
- 24.Urasawa S, Urasawa T, Taniguchi K, Wakasugi F, Kobayashi N, Chiba S, Sakurada N, Morita M, Morita O, Tokieda M, Kawamoto H, Minekawa Y, Obseto M. Survey of human rotavirus serotypes in different locales in Japan by enzyme-linked immunosorbent assay with monoclonal antibodies. J Infect Dis. 1989;160:44–51. doi: 10.1093/infdis/160.1.44. [DOI] [PubMed] [Google Scholar]
- 25.Vesikari T. Rotavirus vaccines against diarrhoeal disease. Lancet. 1997;350:1538–1541. doi: 10.1016/S0140-6736(97)03254-6. [DOI] [PubMed] [Google Scholar]
- 26.Vesikari T, Joensuu J. Review of rotavirus vaccine trials in Finland. J Infect Dis. 1996;174(Suppl. 1):S81–S87. doi: 10.1093/infdis/174.supplement_1.s81. [DOI] [PubMed] [Google Scholar]
- 27.Woods P A, Gentsch J, Gouvea V, Mata L, Simhon A, Santosham M, Bai Z-S, Urasawa S, Glass R I. Distribution of serotypes of human rotavirus in different populations. J Clin Microbiol. 1992;30:781–785. doi: 10.1128/jcm.30.4.781-785.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]