Abstract
The metabolism of polyphenolic polymers is essential to the development and response to environmental changes of organisms from all kingdoms of life, but shows particular diversity in plants. In contrast to other biopolymers, whose polymerisation is catalysed by homologous gene families, polyphenolic metabolism depends on phenoloxidases, a group of heterogeneous oxidases that share little beyond the eponymous common substrate. In this review, we provide an overview of the differences and similarities between phenoloxidases in their protein structure, reaction mechanism, substrate specificity, and functional roles. Using the example of laccases (LACs), we also performed a meta-analysis of enzyme kinetics, a comprehensive phylogenetic analysis and machine-learning based protein structure modelling to link functions, evolution, and structures in this group of phenoloxidases. With these approaches, we generated a framework to explain the reported functional differences between paralogs, while also hinting at the likely diversity of yet undescribed LAC functions. Altogether, this review provides a basis to better understand the functional overlaps and specificities between and within the three major families of phenoloxidases, their evolutionary trajectories, and their importance for plant primary and secondary metabolism.
Keywords: lignin, polyphenolic polymers, laccase, polyphenol oxidase, peroxidase, bayesian phylogeny, protein modelling
Introduction
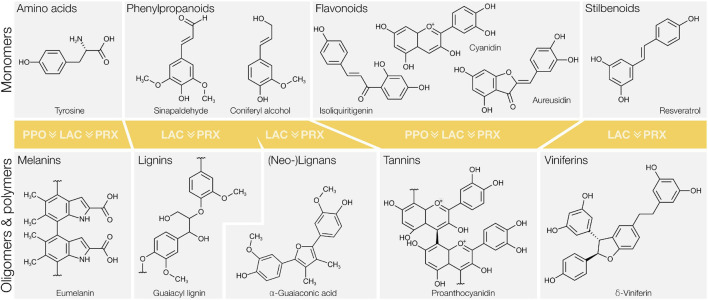
Phenolic compounds form a large and heterogeneous group of primary and secondary metabolites that contain at least one hydroxylated aromatic ring. Phenolics provide solutions to many of the difficulties posed by terrestrial habitats, and their chemical diversification is closely associated with the transition to life on land (Stafford, 2000). Phenolic pigments, like melanins and flavonoids, are antioxidants that protect all major prokaryotic and eukaryotic taxa against UV radiation and reactive oxygen species and function as visual signals to pollinators or seed dispersers in plants (Cheynier et al., 2013; Carletti et al., 2014). Lignin and other structural phenolic polymers accumulate in cuticle, seed coat, and vascular system to enable plant vertical growth, resistance to desiccation and herbivores, as well as long distance water transport (Barros et al., 2015). Smaller phenolics such as salicylic acid, tannins, (neo)lignans or phytoalexins act as chemical or olfactory signals to coordinate responses to environmental factors and biotic interactions (Treutter, 2006).
The majority of known phenolic metabolites derive from the shikimate pathway present in plants, prokaryotes, fungi and some protists. It produces simple phenolic and aromatic amino acids. In plants, phenylalanine and tyrosine establish the starting point of the C6C3 phenylpropanoid pathway. This pathway forms a metabolic crossroad with multiple branching points leading to the formation of different complex phenolics (Barros et al., 2015; Tohge et al., 2017). Once synthesised and transported to specific cellular compartments, many C6C3 phenylpropanoid monomers undergo oxidative cross-coupling to form oligo- or polymers (Figure 1). These polymerisation reactions are catalysed by peroxidases (PRXs), polyphenol oxidases (PPOs), and laccases (LACs), a heterogeneous group of enzymes often called phenoloxidases. Phenolic polymerisation occurs constitutively during development and homeostasis but can also be triggered by wounding or defence pathways (Pourcel et al., 2007; Chong et al., 2009; Barros et al., 2015). The most abundant phenolic polymer in the biosphere is lignin, present in vascular plants (Barros et al., 2015) and red algae (Martone et al., 2009). Lignin derives from the oxidative polymerisation of phenylpropanoids secreted to the cell wall, and forms complex structures specific to distinct cell types and cell wall layers (Terashima and Fukushima, 1988). Other polyphenolics have more defined and repetitive structures than lignin. This includes cross-linked phloroglucinol monomers forming phlorotannins in brown algae (Berglin et al., 2004; Meslet-Cladiere et al., 2013) and oxidised tyrosine forming melanins in the cuticle of insects and mammalian melanosomes (Mason, 1947). Beside developmental processes, some polyphenolics are formed specifically as a wound response. In these cases, the phenolic monomers are spatially separated from the phenoloxidase(s) in different subcellular sites, enabling contact only if the tissue is ruptured. A readily observable example of this mechanism is the O2 dependent browning of cut fruits, which results from the polymerisation of flavonoids and aromatic amino acids into melanins (Figure 1). Stilbenoids are also known to undergo oxidative coupling in response to biotic and abiotic stresses, forming phenolic oligomers called viniferins (Figure 1; Pezet et al., 2003). The oxidising capacity of phenoloxidases derives from the reduction of either molecular oxygen or peroxides. These enzymes thereby fulfil two functions that were crucial for plant adaptation to life on land: they remove excess oxygen species to detoxify their high atmospheric concentrations (Decker and Terwilliger, 2000), and catalyse the formation of various polyphenolic compounds enabling plants to adapt and thrive to changing environmental conditions. Phenoloxidases are therefore essential not only to better understand fundamental physiological processes, but also regarding their potential uses to modify plant biomass and/or adjust abiotic and biotic responses. Such engineered plants, like non-browning apples with a silenced PPO, are readily commercialised. In the present article, we will review the three families of unrelated enzymes that compose the functional group of phenoloxidases: PRXs (Welinder, 1992), PPOs (Sánchez-Ferrer et al., 1995), and LACs (McCaig et al., 2005). To further elucidate the diversity within each type of phenoloxidases, we performed deeper analyses using the example of LACs which have been functionally demonstrated to oxidise different phenolic compounds. We generated a comprehensive phylogeny of plant LACs to estimate their evolutionary emergence and subsequent diversification. We also used machine learning based predictive three-dimensional (3D) protein modelling of LAC paralogs to start bridging the gap between sequence information and putative biological functions.
FIGURE 1.
Main groups of phenolic compounds in monomeric state and after oxidative coupling catalysed by phenoloxidases. Note that peroxidases (PRX) and laccases (LAC) can oxidise most types of phenolics in contrast to polyphenol oxidases (PPO). Note also the historical substrate (α-guaiaconic acid) used for the discovery of phenoloxidases, which turns blue after enzymatic oxidation by forming quinones.
Historical Perspective
The term phenoloxidase is used today to encompass three main families of unrelated oxidising enzymes: PRXs, PPOs, and LACs. However, the definition of phenoloxidase has evolved with time, technologies, and model organisms. In plant and fungal organisms, phenoloxidases refer to LACs, sometimes including PPOs and even PRXs (Ander and Eriksson, 1976; Liu et al., 1994; Ranocha et al., 1999; Barros et al., 2015; Kües, 2015). In animals, phenoloxidases usually refer to PPOs, sometimes including LACs but not PRXs (Hattori et al., 2005; Terwilliger and Ryan, 2006; Luna-Acosta et al., 2011; Rao et al., 2014). These differences derive from the original definition of the term, based a specific enzymatic activity before the advent of DNA sequencing and protein phylogenetics. Two centuries ago, first Planche (1820) and then Schönbein (1856) became intrigued with boletes, whose fruiting bodies rapidly turn blue when damaged and exposed to air. They moreover observed that many plant and fungal tissues were able to turn guaiacum (α-guaiaconic acid, a C6C3 phenolic lignan extracted from the resin of Guaiacum sp.; Figure 1) from colourless to blue, and that this capacity was abolished after boiling. Schönbein (1856) also observed that the alcoholic extracts of fungi were only able to produce the blue colour in the presence of either “activated oxygen” from pressed mushroom juice, or peroxides, thereby describing PRX activity for the first time. Later on, an enzyme from Rhus vernicifera was shown to harden the tree’s sap into lacquer (Yoshida, 1883) and named laccase. Shortly after, LAC activity was shown to turn guaiacum blue (Bertrand, 1894), using molecular oxygen as a co-substrate (Kastle and Loevenhart, 1901). The discovery of PPOs was made from observing that certain fungal species turned not blue but red, and then black after cutting. These fungi could oxidise tyrosine in the presence of O2, marking the first description of the tyrosinase activity of PPOs (Bourquelot and Bertrand, 1896). Already then, it was observed that LACs were far more thermostable than PPOs, a criterion then used to distinguish between the two phenoloxidases (Bourquelot and Bertrand, 1896). The term oxydase was introduced by Bertrand (1896) as a general term for these water-soluble oxidising enzymes using O2, replacing the previous term of oxidising ferments coined by Traube (1877). As these oxidases were all phenoloxidases, the two terms were used synonymously at the time (Kastle, 1910; Onslow, 1920; Szent-Györgyi, 1930). In 1903, the “activated oxygen” initially described by Schönbein was identified as hydrogen peroxide (Bach and Chodat, 1903). This result led the same authors to postulate that all phenoloxidases were two-component systems comprising an H2O2 generating oxygenase and a phenol oxidising PRX (Chodat and Bach, 1903; Onslow, 1920). However, Szent-Györgyi (1925) rebutted this two-component model and showed that the blueing of guaiacum by a potato oxydase was independent from peroxide and PRX activity. Szent-Györgyi (1925) moreover demonstrated that the blueing reaction was indirect and depended on the oxidation of an intermediate catechol, which then oxidised the guaiacum itself. This represented the first description of indirect phenoloxidase activity via redox shuttles that are now known as mediators. Three decades later, the phenoldehydrogenase enzyme that Freudenberg et al. (1952) had associated to lignification was shown to be a LAC (Higuchi, 1958), leading to the synonymous use of LAC and phenoloxidase by plant scientists. Altogether, the term phenoloxidase evolved through time depending on both individual author and scientific field. Nowadays, phenoloxidases describe structurally heterogeneous and phylogenetically unrelated enzymes including LACs, PPOs, and PRXs, grouped together only by their common capacity to oxidise directly and/or indirectly substrates presenting a phenolic ring.
Peroxidases
Distribution of Peroxidases Among Kingdoms and Species
Every organism in the biosphere contains PRXs (EC 1.11.1.X) which oxidise their substrate using the reduction of H2O2 or organic peroxides (Shigeto and Tsutsumi, 2016). The substrates, co-substrates, active centres, protein structures, and reaction mechanisms of the different PRX families and superfamilies are so diverse and different that the relevance of the classification of all PRXs into one EC 1.11.1 has been previously questioned (Hofrichter et al., 2010). Even when focusing on PRXs that primarily oxidise phenolic substrates, there are fundamental differences between plant class III PRXs, fungal class II PRXs such as lignin PRXs (LiPs), manganese PRXs (MnPs), and versatile PRXs (VPs), as well as bacterial dye decolourising PRXs (DyPs). Within these groups, however, PRXs are more conserved. Class III PRXs have a minimum of 25% protein sequence identity between plant species (Table 1 and Figure 2). Compared to LACs and PPOs, class III PRXs show the steepest rise in number of paralogs with increasing genome size, suggesting that repeated gene duplication events occurred throughout evolution (Figure 3A). In extant angiosperms, Arabidopsis thaliana has 73 paralogs, and Eucalyptus grandis has almost 200. Despite some computational predictions of alternative splicing of class III PRX genes, there is no experimental evidence defining either their existence or importance. Class III PRXs are exclusive to streptophytes (Nishiyama et al., 2018; Mbadinga Mbadinga et al., 2020), suggesting that phenol oxidising PRXs appeared after the transition of plants to terrestrial habitat but prior to the appearance of vascular tissues.
TABLE 1.
Overview of the three groups of phenoloxidases in plants.
| Class III PRXs | PPOs | LACs | ||
| Distribution | Chlorophytes | – | ? | ? |
| Charophytes | + | + | – | |
| Bryophytes | + | + | + | |
| Lycophytes | + | + | + | |
| Polypodiophytes | + | + | + | |
| Gymnosperms | + | + | + | |
| Angiosperms | + | + | + | |
| Protein sequence identity | > 25% | > 35% | > 35% | |
| Number of paralogs | 20–200 | 0–15 | 1–80 | |
| Structure | Type | Monomeric | Homomeric dimers and oligomers | Monomers, homo-/ heteromeric dimers and oligomers |
| Glycosylation | Universal | Some | Universal | |
| Peptide signal | Many | Some | Many | |
| Transit peptide | – | Many | – | |
| Shielding domain | – | Universal | ? | |
| Cofactors | 1 Fe (heme), 2 Ca | 2 Cu | 4 Cu | |
| Active protein size (kDa) | 30–45 | 35–60 | 55–70 | |
| Co-substrate | H2O2 | O2 | O2 | |
| Main subcellular localisation | Cell Wall | Plastid | Cell Wall | |
| Main phenolic metabolism | Lignin | Melanin | Lignin |
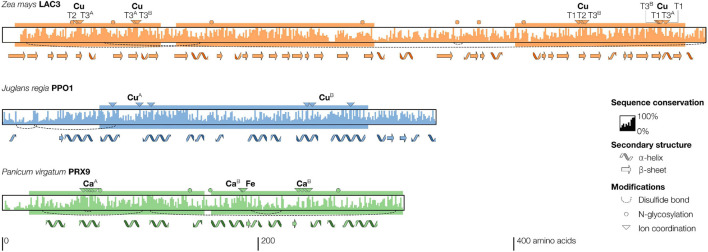
FIGURE 2.
Structural features and sequence conservation in mature protein chains of phenoloxidases. Conserved domains (plastocyanin, tyrosinase, and calcium-binding for LAC3, PPO1, and PRX9, respectively) are shown as solid rectangles. Secondary structures, positions of ion coordinating residues, N-glycosylation sites and disulfide bonds are indicated according to the respective published crystal structures. The bar coded sequence conservation is calculated across all paralogs from Populus trichocarpa, Brachypodium distachyon, Physcomitrium patens, and Selaginella moellendorffii. We have adopted the recent revision of the nomenclature of Physcomitrella patens to Physcomitrium patens (see Rensing et al., 2020).
FIGURE 3.
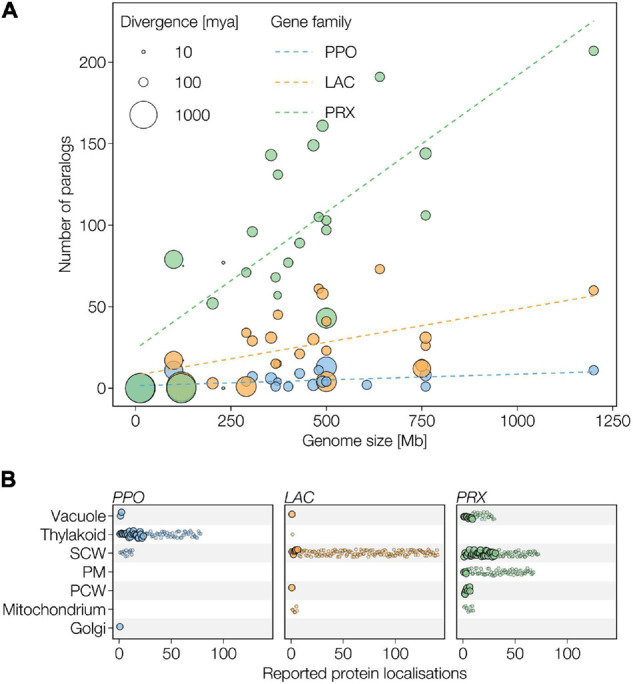
Differences in evolutionary duplication and sub-cellular localisation of the different type of plant phenoloxidases. (A) Evolution of plant phenoloxidase multigenic families. Number of genes encoding for PPOs, LACs, and class III PRXs are plotted against the genome size of the respective species. Circle size indicates evolutionary divergence from A. thaliana in million years ago. Fitted lines represent the trends of multigenic family sizes against total genome size. (B) Predicted and experimentally confirmed subcellular localisations of PPO, LAC, and class III PRX paralogs in plants. Large dots represent experimentally verified localisations, small dots are predictions. PCW, primary cell wall; PM, plasma membrane; SCW, secondary cell wall. All paralogs predicted to the apoplast are placed at SCW.
Expression and Localisation of Peroxidases
Class III PRXs are expressed in all plant organs and tissues, during various developmental stages and stress responses, mirroring the many functions fulfilled by these enzymes (Welinder et al., 2002; Cosio and Dunand, 2009; Wang et al., 2015b). Most PRXs have an N-terminal peptide signal targeting them via the secretory pathway toward membrane structures, vacuole, cell wall, and/or apoplast (Figure 3B). Some PRXs even exhibit specific cell wall layer localisations. Zinnia violacea ZPO-C is exclusively localised in the secondary cell walls of tracheary elements (Sato et al., 2006). Arabidopsis AtPRX64 is present only in the middle lamella and cell corners of interfascicular fibers (Chou et al., 2018) but restricted to the casparian strip in endodermal cells (Lee et al., 2013). Other PRX paralogs have been predicted to be targeted to the mitochondria or bound to membranes (Lüthje and Martinez-Cortes, 2018). These membrane-bound forms have been confirmed biochemically although it remains unclear on which side of the membrane these PRXs are located (Mika and Lüthje, 2003; Mika et al., 2010). Overall, class III PRXs appeared to be mostly associated with cell wall, membrane-bound and vacuolar phenolic metabolism.
Peroxidase Protein Structure
Plant class III PRXs are heme-dependent PRXs whose activity relies on two calcium ions and a heme centred on an iron atom (Fe) coordinated within a protoporphyrin IX (Figure 2). In contrast to fungal class II PRXs, the heme in class III PRXs is non-covalently linked between histidine residues (Moural et al., 2017). Class III PRXs are formed by two domains, called proximal and distal, each binding one calcium ion (Figure 2), which are hypothesised to originate from an ancestral internal gene duplication event (Passardi et al., 2007). Class III PRXs do not appear to require proteolytic activation. Both class II and III PRXs contain highly conserved disulphide bridges that are required for heme coordination and enzyme activity (Ogawa et al., 1979; Howes et al., 2001). Class III and II PRXs are generally monomeric (Janusz et al., 2013; Bernardes et al., 2015) whereas bacterial DyPs form dimers and oligomers (Colpa et al., 2014). Class III PRXs are heavily glycosylated, which is important for their stability and activity (Lige et al., 2001; Hofrichter et al., 2010; Palm et al., 2014) although the glycosylation sites are not conserved (Figure 2).
Reaction Mechanism
Class III PRXs possess two distinct reaction mechanisms: a peroxidative cycle that uses H2O2 or other peroxides to oxidise their substrate (Figure 4), and a hydroxylic cycle that converts H2O2 into other types of reactive oxygen species (Liszkay et al., 2003). In its peroxidative cycle, PRXs are the most potent oxidants of all phenoloxidases with redox potentials (E°) sometimes exceeding 1 V. This enables PRXs to oxidise substrates unusable by other phenoloxidases (Welinder et al., 2002; Hofrichter et al., 2010). The optimal pH of phenol-oxidising PRXs usually ranges from neutral to basic, with the exception of DyPs which function best in acidic conditions (Colpa et al., 2014). Mechanistically, PRX activity depends on their H2O2 mediated two-electron oxidation into an intermediate state, named compound I, in which the heme Fe(III) is oxidised into Fe(IV) and a radical free electron is present on the key residues of the active site. Compound I can then oxidise one substrate molecule with the radical electron, and subsequently a second substrate molecule via the reduction of Fe(IV) back to Fe(III).
FIGURE 4.
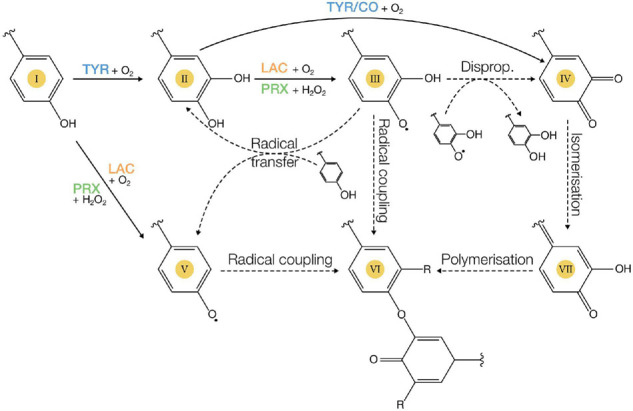
Classical reactions catalysed by PPOs (CO, catechol oxidase; TYR, tyrosinase), LACs, and class III PRXs. Enzymatic and non-enzymatic reactions are indicated with solid and dashed arrows respectively on the example of simple mono- and diphenolic molecules. Monophenol hydroxylation (I → II) is generally considered to be exclusive to TYRs. The one electron oxidation of diphenols (II) by LACs or PRXs leads to a semiquinone radical (III). This can then couple with another semiquinone to form a dimer or polymer (VI), transfer its radical to another compound (III + I → II + V), or disproportionate with another semiquinone to form a quinone (2 III → IV + II). Quinones (IV) can isomerise to quinone methides (VII), which undergo non-enzymatic coupling reactions to form dimers (VI). Lastly, LACs and PRXs can also oxidise monophenols (I) into phenoxy radicals (V).
To oxidise substrates that do not fit into their substrate binding pocket, PRXs exploit two alternative mechanisms of substrate oxidation: indirect oxidation via mediators, and long-range electron transfer from the enzyme core to its periphery. Mediators are small molecules that act as transiently oxidised intermediates, freely diffusing and transferring their radical charge onto other molecules that are either too large or inaccessible to PRXs. LiPs (Harvey et al., 1986), MnPs (Wariishi et al., 1991), and VPs (Gómez-Toribio et al., 2001) use mediators during the oxidative depolymerisation of lignin. The intervention of mediators has also been suggested during the oxidative polymerisation of lignin in plants (Önnerud et al., 2002; Ralph et al., 2004). Long-range electron transfer functions by relocating the site of substrate oxidation from the heme group in the core of the protein to exposed amino acids at the surface of PRXs, enabling the oxidation of large substrates such as lignin polymers. Such long-range electron transfer is used by VPs (Ruiz-Dueñas et al., 2008), LiPs (Miki et al., 2011), and DyPs (Strittmatter et al., 2013) during the oxidative degradation of lignin. Similarly, this mechanism has also been suggested to occur during the oxidative extension of lignin polymers by plant class III PRXs (Shigeto et al., 2014). Despite this flexibility in oxidative mechanism, paralogs of class III PRXs in plants exhibit different in vitro affinities toward artificial substrates (Shigeto et al., 2014) and monomeric model compounds similar to precursors of syringyl (S) and guaiacyl (G) residues of lignin (Shigeto and Tsutsumi, 2016; Shigeto et al., 2017). Altogether, the exact biological substrate(s), the site(s), and mechanism(s) of oxidation remain uncertain for most PRX paralogs.
Functional Roles of Peroxidases
Class III plant PRXs have been associated to multiple processes during development and stress responses (Cosio and Dunand, 2009). One of the main proposed roles of PRXs is during lignin formation to oxidise secreted lignin phenolic monomers in specific cell wall layers of distinct cell types (Herrero et al., 2013; Shigeto et al., 2013). PRXs are the main phenoloxidase responsible of the lignification of the casparian strip in endodermal cells of A. thaliana (Lee et al., 2013; Rojas-Murcia et al., 2020). Ectopic lignin formation in the cell walls of flax bast fibres (Chantreau et al., 2014) and in the extracellular medium of Norway spruce cell cultures (Laitinen et al., 2017) also depend on PRXs. Loss-of-function mutations of class III PRXs as well as their ectopic over-expression have varying effects on lignin amount and residue composition (Table 2) mirroring their diverse in vitro affinities (Shigeto and Tsutsumi, 2016). Beside lignification, class III PRXs are also associated with the cross-linking of extensins in cell walls (Jacobowitz et al., 2019), the vacuolar degradation of anthocyanin in Brunfelsia (Zipor et al., 2015), auxin homeostasis (Cosio et al., 2009), as well as the partial cell wall degradation of seed coats (Kunieda et al., 2013). Using their hydroxylic cycle, class III PRXs are moreover involved in oxidative burst responses (Choi et al., 2007; Daudi et al., 2012) and cell wall extension during cell elongation and lateral root formation (Mei et al., 2009; Müller et al., 2009). In contrast to plant PRXs, fungal MnPs and LiPs as well as bacterial DyPs are exclusively implicated in the breakdown of lignin and other polyphenolic compounds (Hammel and Cullen, 2008). Altogether, we are beginning to outline the overall diversity of class III PRXs but the specific biological functions and redundancies between its many paralogs remain unclear.
TABLE 2.
Reported impact of phenoloxidase knock-out (KO), knock-down (KD), and over-expression (OE) on lignin amount and composition.
| Gene family | Gene/Target | Species | Lignin impact | Type | References |
| Class III PRX | AtPRX2/25/71 | Arabidopsis thaliana | – S-units | KO | Shigeto et al., 2013 |
| AtPRX3/9/39/64/72 | Arabidopsis thaliana | Unlignified casparian strip | KD/KO | Rojas-Murcia et al., 2020 | |
| AtPRX4 | Arabidopsis thaliana | – S-units | KO | Fernández-Pérez et al., 2015b | |
| AtPRX17 | Arabidopsis thaliana | – Lignin | KO | Cosio et al., 2017 | |
| AtPRX17 | Arabidopsis thaliana | + Lignin | OE | Cosio et al., 2017 | |
| AtPRX52 | Arabidopsis thaliana | – S-units | KO | Fernández-Pérez et al., 2015a | |
| AtPRX64 | Arabidopsis thaliana | Delayed casparian strip | KD | Lee et al., 2013 | |
| AtPRX72 | Arabidopsis thaliana | – Lignin | KO | Herrero et al., 2013 | |
| CsPRX25 | Citrus sinensis | + Lignin | OE | Li et al., 2020 | |
| OsPRX38 | Arabidopsis thaliana | + Lignin | OE | Kidwai et al., 2019 | |
| PtrPO21 | Populus trichocarpa | – Lignin | KD | Lin et al., 2016 | |
| ZePRX | Nicotiana tabacum | + S-units | OE | García-Ulloa et al., 2020 | |
| LAC | AtLAC17 | Arabidopsis thaliana | – G-units | KD | Cesarino et al., 2013 |
| AtLAC2 | Arabidopsis thaliana | + Root lignin | KO | Khandal et al., 2020 | |
| AtLAC2 | Arabidopsis thaliana | – Root lignin | OE | Khandal et al., 2020 | |
| AtLAC4 | Arabidopsis thaliana | – G-units | KD | Zhao et al., 2013 | |
| AtLAC4 | Arabidopsis thaliana | + Lignin | OE | Wang et al., 2014 | |
| AtLAC4/17 | Arabidopsis thaliana | – G-units | KD/KO | Berthet et al., 2011 | |
| AtLAC4/17/11 | Arabidopsis thaliana | Unlignified vasculature | KD/KO | Zhao et al., 2013 | |
| BdLAC5 | Brachypodium dystachion | – G-units | KD | Wang et al., 2015a | |
| BdLAC5/8 | Brachypodium dystachion | – Lignin and G-units | KD | Le Bris et al., 2019 | |
| ChLAC8 | Arabidopsis thaliana | + C-units (exogeneous) | OE | Wang X. et al., 2020 | |
| ChLAC8 | Cleome hassleriana | – C-units | KD | Wang X. et al., 2020 | |
| GhLAC1 | Gossypium hirsutum | + Lignin | OE | Hu et al., 2017 | |
| miR397: 15 laccases | Oryza sativa | – Lignin | OE | Swetha et al., 2018 | |
| miR397a: 12 laccases | Populus trichocarpa | – Lignin and G-units | OE | Lu et al., 2013 | |
| miR397b: AtLAC2,4,17 | Arabidopsis thaliana | – Shoot lignin and G-units | OE | Wang et al., 2014 | |
| miR397b: AtLAC2,4,17 | Arabidopsis thaliana | + Root lignin | OE | Khandal et al., 2020 | |
| miR397b: AtLAC2,4,17 | Arabidopsis thaliana | – Root lignin | KD | Khandal et al., 2020 | |
| miR528: ZmLAC3, 5 | Zea mays | + Stem lignin | KD | Sun et al., 2018 | |
| miR857: AtLAC7 | Arabidopsis thaliana | – Lignin and S/G | OE | Zhao et al., 2015 | |
| miR857: AtLAC7 | Arabidopsis thaliana | + Lignin and S/G | KD | Zhao et al., 2015 | |
| MsLAC1 | Arabidopsis thaliana | + Lignin and G-units | OE | He et al., 2019 | |
| PtLAC2 | Populus trichocarpa | – G-units | KD | Bryan et al., 2016 | |
| ZmLAC3 | Zea mays | + Lignin | OE | Sun et al., 2018 |
G, guaiacyl units of lignin; S, syringyl units of lignin; C, caffeyl units of lignin; +, increase; –, decrease.
Polyphenol Oxidases
Distribution of Polyphenol Oxidases Among Kingdoms and Species
Polyphenol oxidases (EC 1.10.3.1, 1.14.18.1) are copper containing enzymes that are almost universally present in plants, fungi and animals (Sánchez-Ferrer et al., 1995), common in bacteria (Claus and Decker, 2006), and have more recently been found in some archaea (Kim et al., 2016). They usually form small gene families that rarely exceed 10 paralogs (Figure 3A; Esposito et al., 2012; Tran et al., 2012; Martínez-García et al., 2016). A systematic genome analysis found no PPO orthologs in green algea (Tran et al., 2012). However, isolated reports of PPO activity in chlorophytes (Tocher et al., 1966) and charophytes (Holst and Yopp, 1976) together with putative PPO sequences in the genome of Chara braunii (Nishiyama et al., 2018) suggest an evolutionary origin before the emergence of terrestrial plants (Table 1). During the course of plant evolution, PPOs are unique among phenoloxidases in showing no significant increases in paralog numbers with increasing genome size (Figure 3A) and have even been lost completely in the genus Arabidopsis.
Expression and Localisation of Polyphenol Oxidases
Plant PPO genes are generally up-regulated in response to biotic and abiotic stresses. In tomato, different stresses and stress-associated compounds affect PPOs expression in different tissues: jasmonate up-regulated PPO expression in young leaves, ethylene in older leaves and salicylic acid in whole shoots (Thipyapong and Steffens, 1997). In pineapple, two PPO genes are expressed constitutively in whole plants, and are drastically up-regulated in fruits submitted to cold stress (Stewart et al., 2001). The promoter of one PPO associated to the biosynthesis of the anthocyanin betalain in red swiss chard is developmentally controlled in roots and petioles even when introduced heterelogously in A. thaliana (Yu et al., 2015). In plants, ∼75% of PPOs possess a plastid transit peptide and are predicted to accumulate in the thylakoid lumen using the twin arginine-dependent translocation pathway. Only a few PPOs have signal peptides and are predicted to the secretory pathway (Tran et al., 2012; Figure 3B). These non-plastidial localisation of PPO in plants were confirmed for the aureusidine synthase in Antirrhinum majus (Ono et al., 2006) and PPO13 in Populus trichocarpa (Tran and Constabel, 2011) in the vacuolar lumen. Additionally, another PPO was shown to localise in the golgi-network in Annona cherimola (Olmedo et al., 2018; Figure 3B). Across kingdoms, PPO localisation is more diverse: animal and fungal PPOs are located in the cytosol and associated to clotting after wounding in insects (Schmid et al., 2019) or secreted to the apoplast to form fungal cell walls or insect cuticles (Barrett, 1986; Mayer, 2006). In contrast, mammalian PPOs are bound to membranes of specialised melanosomes (Wang and Hebert, 2006). Based on these differences in localisation between kingdoms and species, PPOs are likely involved in specialised phenolic metabolism.
Structure of Polyphenol Oxidases
Polyphenol oxidases generally form homodimers or -oligomers in plants (Dirks-Hofmeister et al., 2012; Molitor et al., 2016), and homo- and hetero-oligomers in mammals (Wang and Hebert, 2006), arthropods (Li et al., 2009), molluscs (Jaenicke and Decker, 2003), and bacteria (Kong et al., 2000). Although N-glycosylation is common in animal PPOs (Wang and Hebert, 2006), they are rarely glycosylated in plants (Table 1). Aureusidine synthase is the only reported glycosylated PPO (Nakayama et al., 2000) although putative glycosylation sites have been predicted for the A. cherimola PPO (Olmedo et al., 2018). A common feature of most PPOs is the need for catalytic activation. In plants, PPOs are translated as latent pro-PPOs composed of the N-terminal plastidial transit peptide, the catalytic domain housing two copper atoms, followed by a disordered linker and a C-terminal shielding domain (Marusek et al., 2006). Fungal PPOs have a similar structure but lack the transit peptide (Marusek et al., 2006). In arthropods, the shielding domain is instead N-terminal (Li et al., 2009) although some paralogs in Drosophila lack this shielding domain (Chen et al., 2012). Mammalian PPOs contain a C-terminal transmembrane domain, but no shielding domain (Wang and Hebert, 2006), and bacterial PPOs exist in a wide variety of structures (Faccio et al., 2012). The shielding domain, when present, contains a placeholder residue that makes the site of substrate oxidation inaccessible in pro-PPOs. Highly specific serine proteases activate arthropod PPOs by cleaving off the N-terminal shielding domain (Li et al., 2009). In plants or fungi, no PPO activating protease has been identified, but a similar specific proteolytic activation is hypothesised for the aurone synthase of Coreopsis grandiflora (Molitor et al., 2016). Alternatively, both plant and insect pro-PPOs have been shown to be activated by low pH (∼3.5) or detergents instead of proteolytic cleavage (Bidla et al., 2007; Leufken et al., 2015). In plants, these treatments lead to a conformational change of the shielding domain due to the disordered nature of its linker (Leufken et al., 2015). Some bacterial PPOs alternatively recruit the placeholder residue from an associated caddie protein (Decker et al., 2007). PPOs containing a shielding domain are relatively conserved in size between species and range between 40 and 70 kDa (Mayer, 2006; Li et al., 2009), whereas PPOs without a shielding domain range from only 15 kDa in bacteria (Faccio et al., 2012) to above 70 kDa in mammals (Wang and Hebert, 2006). Within kingdoms, PPO protein sequence identity ranges from 30 to 50%, but decreases to 5% between kingdoms as only the copper and oxygen binding motifs are conserved (Figure 2). Although PPOs are very heterogeneous in structure between kingdoms, their conserved activation mechanism suggests that this post-translational regulation plays a pivotal role in their physiological functions.
Reaction Mechanism
The enzymatic activity of PPOs depends on a dinuclear type 3 copper pair which is coordinated by 6 histidine residues (Figure 2; Bijelic et al., 2015). The E° of this copper pair is estimated at ∼260 mV (Ghosh and Mukherjee, 1998), making PPOs the least potent oxidisers among phenoloxidases. PPOs best function between pH 5 and 6.5 at temperatures of 20–40°C (Queiroz et al., 2008). PPOs can catalyse two distinct reactions using O2: (i) the ortho-hydroxylation of monophenols, like tyrosine and tyramine, into ortho-diphenols (monophenolase activity) and (ii) the oxidation of ortho-diphenols or catechols into ortho-quinones (diphenolase or catecholase activity) (Figure 4; Solomon et al., 1996). These different activities establish the distinctive criterion separating PPOs into tyrosinases (TYR, EC 1.14.18.1, monophenol/o-diphenol:O2 oxido-reductases) capable of catalysing both reactions, and catechol oxidases (CO, EC 1.10.3.1, o-diphenol:O2 oxido-reductases) only possessing the diphenolase activity (Figure 4; Solomon et al., 1996). The structural reason behind this biochemical distinction is still unclear as no fundamental differences were identified in either the protein structure, localisation, or expression between TYRs and COs (Bijelic et al., 2015; Solem et al., 2016). An asparagine-glutamate couple stabilising one water molecule in the active site appears to be key for the electron abstraction of monophenolic substrates. Site-directed mutagenesis to introduce an asparagine residue into Vitis vinifera CO enabled a novel monophenolase activity toward tyrosine (Solem et al., 2016). However, several known TYRs lack this asparagine residue, suggesting other explanations for the CO to TYR specificity (Pretzler and Rompel, 2017). Alternatively, the monophenolase activity has been proposed to depend on whether the substrate can be stabilised at the active site (Bijelic et al., 2015; Molitor et al., 2016). Indeed, a leucine residue gating the entry to the active site was shown to stabilise classic TYR substrates in enzymes classified as TYRs (Goldfeder et al., 2014; Bijelic et al., 2015). In COs, this leucine is replaced by an arginine (Goldfeder et al., 2014; Bijelic et al., 2015). Again, however, the universality of this rule is questioned by some TYRs containing a supposedly destabilising arginine at this position (Pretzler and Rompel, 2017). Beside the absence of a clear structural determinant, the biochemical distinction between TYRs and COs based on their ability to oxidise classical TYR substrates like tyrosine and tyramine has also been questioned (Molitor et al., 2016). The C. grandiflora aurone synthase lacks activity toward these substrates and is accordingly classified as a CO (Molitor et al., 2015). The enzyme does however exhibit monophenolase activity toward its physiological substrate the chalcone isoliquiritigenin (Figure 1; Molitor et al., 2016). The oxidation of tyrosine or tyramine therefore does not seem to enable a relevant mechanistic distinction between PPOs but rather detects differences in substrate specificities. Consequently, many enzymes categorised as COs may biologically function as TYRs (monophenolase activity) on their physiological substrates.
Biological Function(s) of Polyphenol Oxidases in Plants
Despite their structural heterogeneity, most PPOs in animal and fungal species exclusively initiate the reaction cascade leading to complex phenolic polymers such as melanin (Figure 1). In plants, PPOs primary respond to wounding, which ruptures the compartmentalisation separating PPOs in plastids from their substrates stored in vacuoles. The expression of PPOs is up-regulated by major defence pathways (Constabel and Ryan, 1998) and their functional loss increases disease susceptibility (Thipyapong et al., 2004). For the post-harvest conservation of fresh plant produces, silencing of PPOs in potato (Chi et al., 2014; González et al., 2020), rice (Yu et al., 2008), and apple (Waltz, 2015) almost completely abolishes the browning of tubers, seeds, and fruits. PPOs have also been associated in the wounding independent biosynthesis of anthocyanin (Gao et al., 2009; Nakatsuka et al., 2013), aurones (Nakayama et al., 2000; Kaintz et al., 2014), and lignans (Cho et al., 2003). While these examples demonstrate the versatility of PPOs, the exact substrates of most of these enzymes and whether they act as TYRs or COs are unclear (Sullivan, 2015; Boeckx et al., 2017). However, the fact that PPOs were not duplicated and even lost in Arabidopsis suggests that they are implicated in non-essential pathways, or that their loss has been compensated by other phenoloxidases with greater E°.
Laccases
Distribution of Laccases Among Kingdoms and Species
Laccases (EC 1.10.3.2, p-diphenol oxygen oxidoreductases) are members of the multi-copper-oxidase family, together with ascorbate oxidases and ferroxidases, which all share a copper-mediated reaction but oxidise distinct substrates (Kües and Ruhl, 2011; Reiss et al., 2013). LACs are present in all plants (Weng and Chapple, 2010), widely distributed in fungi (Baldrian, 2006), and have also been found in bacteria (Santhanam et al., 2011), archaea (Uthandi et al., 2010), arthropods (Barrett, 1986; Hattori et al., 2005), and molluscs (Luna-Acosta et al., 2011) but not in mammals. In plants, the number of LAC paralog genes ranges from 1 in Marchantia polymorpha to more than 50 in P. trichocarpa and E. grandis (Figures 3A, 5). LACs in other kingdoms are however present as single genes or form small multigenic families. LACs share around 40% protein sequence identity within kingdoms (Figure 2) but conservation between kingdoms is limited to residues around the active site (∼10–30% total sequence identity). The conservation of LAC genes in plants as well as the increases of paralog numbers with increasing genome size (Table 1 and Figure 3A) suggest both critical roles in the plant life cycle and repeated events of sub- and/or neo-functionalisation during plant speciation. There are conflicting reports on exact appearance of LACs in plants. Green unicellular algae, such as Volvox carteri and Chlamydomonas reinhardtii, were suggested to have genes encoding for LACs (Weng and Chapple, 2010; Zhao et al., 2013) although no LAC enzymatic activities had been detected in these species (Otto et al., 2015). To address this open question, we generated a comprehensive phylogeny of all LACs from 10 taxonomically diverse species with published reference genomes (Figure 5). In contrast to previous phylogenies (McCaig et al., 2005; Turlapati et al., 2011; Zhao et al., 2013; Wang X. et al., 2020; Yonekura-Sakakibara et al., 2020), we used only full-length sequences (to avoid partial homology due to incomplete sequences) and included ascorbate oxidases as an outgroup to distinguish between the two families of multicopper oxidases. We moreover chose a bayesian approach to provide probabilities (i.e., statistical support) for each computed branch (Supplementary Figure 2). This new phylogenetic analysis first enabled us to determine that the LAC-like sequences present in the genome of unicellular green algae are more likely ascorbate oxidases than bona fide LACs (Figure 5). We determined that the most basal bona fide LACs are from M. polymorpha and Physcomitrium patens, which together with sequences from Azolla filiculoides form the paraphyletic group of basal plant LACs (Figure 5). Our analysis therefore suggests that ancestral bona fide LACs originated in multicellular green algae or early land plants. The remaining LACs formed eight well supported (posterior probabilities >0.9 except for clade II at 0.62; Supplementary Figure 2) monophyletic clades named in order of divergence from I to VIII. After the appearance of basal LACs, multiple waves of gene duplication events occurred with the sequential emergence of vascular plants, spermatophytes and angiosperms, leading to repeated opportunities for sub- and/or neo-functionalisations (Figure 5). These duplication events predominantly affected clades IV–VIII, which contained the majority of LACs from gymnosperms and angiosperms but no lycophyte, fern, or moss sequences. This imbalance suggests that the emergent functional diversity of LAC paralogs is specifically associated with the evolution of spermatophytes.
FIGURE 5.
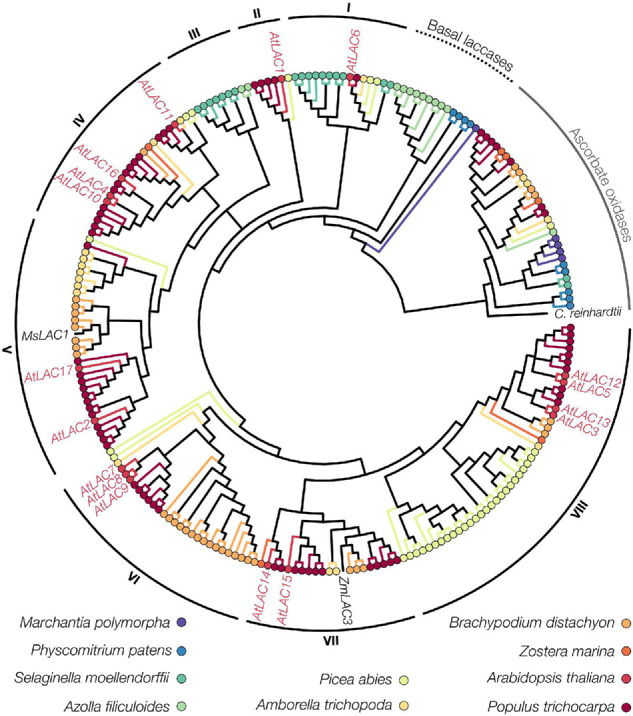
Phylogenetic analysis of LAC homologs. Bayesian phylogeny of high-confidence LAC homologs from 10 species and ascorbate oxidases as the outgroup. The A. thaliana paralogs can be grouped into eight clades which broadly correspond to previous results (Turlapati et al., 2011; Zhao et al., 2013), except for composition and position of clades I and II, and the occurrence of clade III, which have not been described before. Note that the only C. reinhardtii sequence is closer related to ascorbate oxidases than LACs, while no sequence from V. carteri or C. braunii passed the motif-based sequence filtering.
Localisation and Expression of Laccases
At the whole plant level, LACs are mainly expressed in the different lignified tissues. In A. thaliana, AtLAC4, 5, 10, 12, and 17 are expressed in vascular bundles (Turlapati et al., 2011) and co-regulated with secondary cell wall formation in tracheary elements (Derbyshire et al., 2015) whereas AtLAC1, 3, 5, and 13 are expressed in endodermal cells (Rojas-Murcia et al., 2020). AtLAC5 (Yonekura-Sakakibara et al., 2020) and AtLAC15 (Turlapati et al., 2011) which catalyse the formation of neolignans and proanthocyanidins respectively are strongly expressed in seed coats. Pollen grains, which are sterile in loss-of-function mutants affecting phenylpropanoid biosynthesis (Rohde et al., 2004; Schilmiller et al., 2009; Weng et al., 2010), exclusively express AtLAC8 (Turlapati et al., 2011). The unlignified phloem and cortex express AtLAC8 and AtLAC9, respectively (Turlapati et al., 2011) which both undergo alternative splicing (Zhang et al., 2010). Overall, different LAC paralogs are specifically expressed in different lignified and unlignified cell types, thereby suggesting neo-functionalisation in which LAC paralogs do not all function redundantly.
The majority of LACs present an N-terminal signal peptide targeting them to the secretory pathway (Figure 3B). LACs generally accumulate in the cell walls of plants (McCaig et al., 2005; Chou et al., 2018), in the extracellular space of fungi and archaea (Baldrian, 2006; Uthandi et al., 2010), or in the saliva, digestive apparatus, and/or exoskeletal cuticle for insects (Dittmer et al., 2004; Arakane et al., 2005; Hattori et al., 2005). In contrast, LACs in bacteria are often intracellular or periplasmic (Rosconi et al., 2005; Santhanam et al., 2011). Secreted LACs in plants are not free in the apoplast but ionically or covalently bound to the cell wall (Bao et al., 1993; Liu et al., 1994; Ranocha et al., 1999). Moreover, different plant LACs localise in specific cell wall layers. In A. thaliana, AtLAC4 fluorescent fusions are immobilised to the secondary cell wall of interfascicular fibers (Chou et al., 2018) whereas immunolocalisation of AtLAC4 and AtLAC17 show more accumulation in the S3 layer of these secondary walls (Berthet et al., 2011). Other LAC paralogs such as AtLAC1, 3, 5, and 13 also specifically accumulate in the casparian strip of endodermal cells (Rojas-Murcia et al., 2020). In Chamaecyparis obtusa, CoLAC1 and CoLAC3 were respectively localised in the inner and outer S2 layers of tracheid compression wood (Hiraide et al., 2021). Beside cell wall localisation, LACs can be targeted to vacuoles in litchi (Fang et al., 2015), to the cytoplasm in hairy roots of Brassica juncea (Telke et al., 2011), but are also predicted to mitochondria in Pinus taeda, Oryza sativa, and Gossypium spp. (Figure 3) and peroxisome in Lolium perenne (Gavnholt and Larsen, 2002). Overall, however, the majority of LACs in plants are targeted to the cell walls (Figure 3B).
Structure of Laccases
Laccases are active as monomers but also as homomeric and heteromeric oligomers in plants (Jaiswal et al., 2014, 2015), algae (Otto and Schlosser, 2014), fungi (Perry et al., 1993; Ng and Wang, 2004; Junghanns et al., 2009), and bacteria (Diamantidis et al., 2000; Rosconi et al., 2005). Although glycosylation is universally predicted for eukaryotic LACs, glycosylation sites are not conserved (Figure 2). Partial or complete deglycosylation of fungal LACs does not significantly alter their enzymatic activity, but increases their susceptibility to proteolysis (Yoshitake et al., 1993; Vite-Vallejo et al., 2009). However, heterologous expression in Pichia pastoris of fungal LAC mutated in single glycosylation sites resulted in LACs with more than 50% reduced activity (Maestre-Reyna et al., 2015). This observation suggested potential roles of glycosylation sites for specific LAC paralogs. Each LAC monomer contains three distinct cupredoxin-like domains (Figure 2), housing the catalytic copper atoms. These domains are characterised by several tightly packed anti-parallel β-sheets known as a greek-key motif, which forms the hydrophobic core of the enzyme (Figure 2; Hakulinen and Rouvinen, 2015). An intriguing exception to the three-domain structure are bacterial two-domain or small LACs, which only contain two cupredoxin-like domains and are obligate homotrimers to be active, with the third copper binding site formed at the interface between the interacting monomers (Endo et al., 2002; Skálová et al., 2009). Beside bacterial small LACs, the size of LAC monomers is conserved across kingdoms at 55–70 kDa without the glycan moieties. Some LACs from ascomycetes (Hakulinen et al., 2002) and basidiomycetes (Bleve et al., 2013) are encoded as pro-proteins with a C-terminal blocker tail which needs to be proteolytically removed to activate LACs (Bulter et al., 2003; Kiiskinen and Saloheimo, 2004; Bleve et al., 2013). In contrast to PPOs, this tail is only 10–15 amino acids long and specifically blocks the O2 reduction site. Among the plant LACs analysed in Figure 5, we found potentially analogous C-terminal blocker tails in AtLAC8, 9, three predicted P. patens LACs and in several Brachypodium distachyon LACs (Supplementary Figure 1). Altogether, our understanding of LAC activity in plants and their regulation via proteolysis, complex formation, and/or allosteric interactions still remains incomplete.
Laccase Reaction Mechanism
Laccase activity relies on four copper atoms for substrate oxidation and for O2 reduction. Two of these copper atoms form a binuclear T3 copper centre which is similar but not identical to the one found in PPOs (Jones and Solomon, 2015). LACs possess in addition a type 1 copper atom (T1) and a type 2 copper atom (T2) (Table 1 and Figure 2). Because the S–Cu bond between T1 copper and a coordinating cysteine residue leads to strong absorption at ∼600 nm (Rodgers et al., 2010), LACs are also called blue-copper oxidases or enzymes (Hoegger et al., 2006). LACs possess one site for the one-electron substrate oxidation at the T1 copper (Solomon et al., 1996) and another for O2 reduction close to the trinuclear copper cluster (1 Cu in T2 + 2 Cu in T3) resulting in an overall E° for LACs of 0.4–0.8 V (Xu, 1997; Xu et al., 1998, 1999; Durão et al., 2006). The E° of T1, controlling the speed of electron abstraction from the substrate, represents the main limiting factor for both reaction speed and substrate specificity (Xu et al., 1996; Tadesse et al., 2008). The most influential residue on LAC E° is the axial residue at the T1 copper, which can either be coordinating (Met) or non-coordinating (Leu, Ile, and Phe) (Xu et al., 1999; Durão et al., 2006). The axial residue is responsible for roughly half (∼200 mV) of the observed natural variation in LAC E°, which is complemented by several second coordination sphere effects (Hadt et al., 2012). When the axial residue is methionine, it reduces the E° by coordinating the T1 together with the two histidines and one cysteine that are universally conserved, stabilising the oxidised intermediate form of the LAC (Ghosh et al., 2009). These low E° LACs are found primarily in bryophytes, insects, and bacteria. Previous reports using primary structure sequence alignment concluded that plant LACs also presented an axial methionine (Jones and Solomon, 2015; Mate and Alcalde, 2015). This is not however a general feature, and our systematic analysis of plant LACs revealed that 143 out of 194 LACs presented a non-coordinating leucine in the axial position of the T1 centre. Overall, paralogs with an axial leucine are likely to have high E° and are potentially involved in phenylpropanoid metabolism such as lignification. In contrast, LACs with an axial methionine and accordingly lower E°, such as ADE/LAC and AtLAC15, have been implicated in the oxidation of other phenolic substrates such as flavonoids.
Laccase Substrate Specificity
Laccases can oxidise various o- and p-mono- and diphenols, but also accept a broad range of other small phenolic and non-phenolic substrates such as phenolic heterocycles (phenothiazine), amines (aniline, diaminofluorene) and amides (syringamide) (Jeon and Chang, 2013; Reiss et al., 2013). Unlike other phenoloxidases, LACs are highly stable in time and temperature (Bourquelot and Bertrand, 1896; Hildén et al., 2009) and generally exhibit high optimal reaction temperatures (Figure 6A). The optimal pH of LACs is substrate specific, due to pH-dependent changes of substrate E°, easing the oxidation of phenolic substrates at higher pH compared to non-phenolic substrates which are pH independent (Rodgers et al., 2010). Because increasing pH concomitantly increases inhibition of the T2/T3 centre by OH–, the LAC activity profiles toward phenolic substrates are generally biphasic (Xu, 1997, 2001). Fungal LACs have however been reported to be more sensitive to these pH changes than the plant LAC from R. vernicifera (Nakamura, 1958). At lower pH, fungal LACs use a conserved aspartate residue around position 206 (Asp206) to deprotonate phenolic substrates (Madzak et al., 2006; Tadesse et al., 2008). Replacement of the Asp206 with an Asn leads to an increase of the optimal pH for phenolic substrates by almost two units but also significantly decreases its oxidation efficiency (Madzak et al., 2006; Mate et al., 2013). Primary sequence alignments show that this Asp is replaced with an Asn in most plant and bacterial LACs (Madzak et al., 2006). Both the presence of Asn and higher theoretical isoelectric points (Figure 6B) suggested that bacterial (Rosado et al., 2012; Martins et al., 2015) and plant LACs (Dwivedi et al., 2011) best operate in neutral to basic pH, in contrast to the acidic pH optimum for fungal LACs (Baldrian, 2006). To evaluate this assumption, we performed a meta-analysis of published enzymatic activity on both phenolic (SGZ and DMP) and non-phenolic synthetic substrates (ABTS). The comparison of enzymatic parameters between kingdoms is complicated as only a handful of plant LACs have been isolated and characterised (Bao et al., 1993; Ranocha et al., 1999; Telke et al., 2011; Jaiswal et al., 2014, 2015; Fang et al., 2015; Koutaniemi et al., 2015). Moreover, heterologous expression of plant LACs in bacteria or P. pastoris is possible (Wang X. et al., 2020) but often problematic. Heterologous expression has resulted in inactive enzymes (Sato et al., 2001) or enzymes displaying unexpected in vitro substrate preferences differing from whole plant functional studies (He et al., 2019). Overall, LAC activity for these different substrates was similar between kingdoms and showed a large variability within kingdoms (Figure 6C). Only bacterial LACs with phenolic substrates (SGZ and DMP) followed the assumption of higher pH optima (Figure 6D). In contrast, plant LACs presented an optimal pH similar to fungal LACs and the overall LAC activity independently of the kingdom depended more on the structure of the substrate used than the pH (Figures 6D,E). This observation implies that LACs can oxidise different substrates at different pH depending on their chemical structure. In addition, LAC activity can also be indirect, using small redox-shuttle mediators, to oxidise substrates that either have prohibitively high E° or do not fit their binding pockets. Altogether, the high E° and the capacity for indirect oxidation potentially enables LACs to oxidise a wide range of substrates.
FIGURE 6.
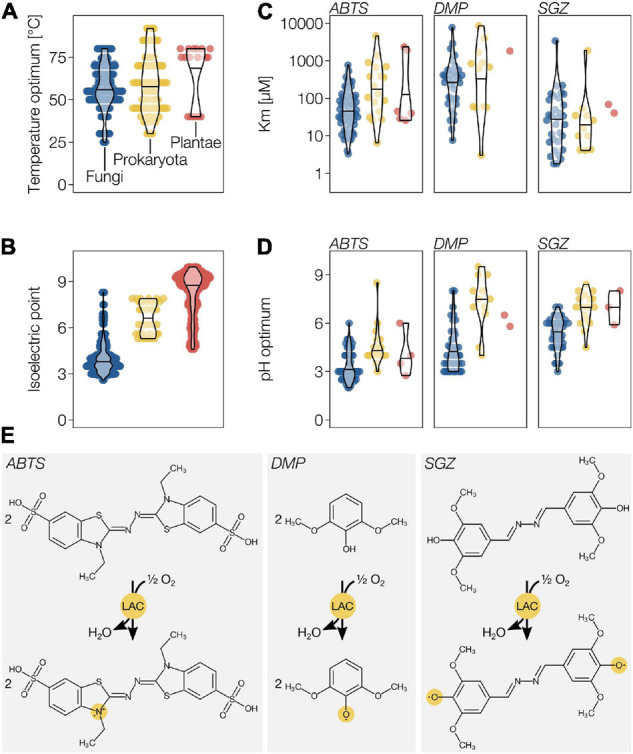
Bibliometric analysis of LAC enzymatic parameters. (A) Temperature optimum of LACs activity from fungi, prokaryota and plants. (B) Isoelectric points of LACs from the three kingdoms. Note that most of the isoelectric point data represent calculated values rather than experimental ones. (C) Km values of LACs from the three kingdoms for the classical non-phenolic substrate 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid) (ABTS) and two common phenolic substrates (DMP, 2,6-dimethoxyphenol; SGZ, syringaldazine). (D) Optimal pH for the oxidation of ABTS, DMP and SGZ. (E) Schematic representation of LAC mediated oxidation of ABTS, DMP and SGZ.
Roles of Laccases
Laccases from all kingdoms are primarily involved in the metabolism of phenolic polymers. In plants, LACs oxidise lignin monomers to form lignin (Freudenberg et al., 1952). In wood-rotting fungi and bacteria, LACs have the opposite function of breaking down lignin (Ander and Eriksson, 1976; Bourbonnais and Paice, 1990; Majumdar et al., 2014). Other fungal, bacterial, and insect LACs are involved in the formation of polyphenolic pigments such as melanin, thus acting directly downstream of PPOs (Clutterbuck, 1972; Martins et al., 2002; Arakane et al., 2005). LACs produced by phloem sucking insects have been suggested to polymerise and inactivate defence-associated plant phenolics (Hattori et al., 2005). In plants, the functional importance of LACs in lignin biosynthesis was shown by genetic modulation studies in Arabidopsis (Berthet et al., 2011; Zhao et al., 2013, 2015; Schuetz et al., 2014; Wang et al., 2014), Brachypodium (Wang et al., 2015a), and Populus (Ranocha et al., 2002; Lu et al., 2013) (Table 2). Synergistic action of several LAC paralogs is necessary to control lignin amount and composition. In contrast to the Arabidopsis lac11 single mutant with no visible defects and the lac4/17 double mutant with only minor growth alterations in continuous light conditions (Berthet et al., 2011), the lac4/17/11 triple mutant is dwarfed, completely sterile and forms no lignin in its vascular tissues (Zhao et al., 2013). Beside lignification, specific LAC paralogs oxidise other phenylpropanoids to form stereo-specific (neo)lignans together with dirigent proteins in the Arabidopsis seed coat (Yonekura-Sakakibara et al., 2020). AtLAC15 and litchi ADE/LAC oxidise flavonoids, showing potential overlap in function with PPOs (Pourcel et al., 2005; Fang et al., 2015). Similar to lignin metabolism in which different LACs either polymerise or break down the polymer, specific LAC paralogs are associated to either the anabolic or catabolic oxidation of flavonoids: AtLAC15 polymerises flavonoids into proanthocyanidin (Pourcel et al., 2005), whereas litchi ADE/LAC degrades anthocyanins (Fang et al., 2015). Altogether, their importance for vascular cell wall lignification makes LACs essential for plant growth, while other paralogs play additional roles in diverse aspects of other phenolic metabolism. However, the molecular mechanisms underlying their synergistic functions, distinct substrate specificity and anabolic/catabolic activities is still unclear.
Modelling the Structural Differences Between Laccase Paralogs
To evaluate how the overall protein structure and its substrate binding pocket topology related to the different roles/activity of specific LAC paralogs in plants, we built 3D protein homology models. Using the recently published AlphaFold 2 algorithm (Jumper et al., 2021), we computed 3D models for all 17 A. thaliana LAC paralogs as well as five paralogs from other plant species previously functionally characterised (Figure 7). The AlphaFold 2 models were consistently of considerably better quality (as estimated by discrete optimised protein energy, or DOPE; Shen and Sali, 2006) than those computed using traditional single template modelling based on the crystal structure of the only crystallised plant laccase ZmLAC3 (Xie et al., 2020; Supplementary Figure 3). The high quality of these models allowed us to precisely measure the substrate binding pocket volume, compactness or pocket shape (the pocket volume relative to the protein surface forming the pocket), mouth area (the steric limitation of the entrance to reach the binding pocket), and depth (distance to the protein surface of the two histidines—451 and 519 in ZmLAC3—coordinating the T1 copper) for each paralog.
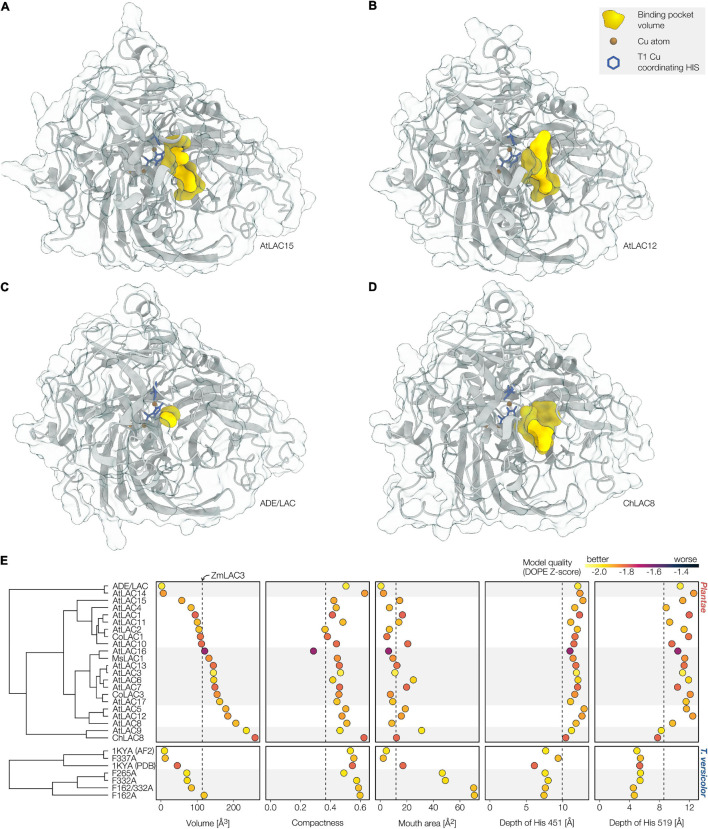
FIGURE 7.
Structural analysis of modelled LAC binding pockets. (A–D) AlphaFold2 structural predictions of AtLAC15, AtLAC12, ADE/LAC, and ChLAC8. The binding pocket volume detected with CASTp is shown in yellow, the copper atoms in brown and the histidines coordinating the T1 copper in blue. (E) Hierarchical clustering of LACs based on the topology of their substrate binding pockets. Pockets were characterised using binding pocket volume, mouth area (the surface area of the yellow pocket volume that is not obscured behind the semi-transparent protein surface) and compactness (pocket volume relative to pocket forming protein surface area), as well as the distance from the protein surface of the two T1 coordinating histidines at the bottom of the pocket (451 and 519 in ZmLAC3). The results for the binding pocket from the crystal structure of ZmLAC3 are indicated by a dashed line. The quality of each individual model is colour coded, where values below –1.2 indicate native-like models. Trametes versicolor LacIIIb (PDB identifier 1KYA) was included for validation, showing expected increases in binding pocket size and mouth area in the targeted mutagenesis LacIIIb versions F265A, F332A, F162A, and F162A/F332A but not in F337A. PDB, crystal structure from the protein database; AF2, AlphaFold 2 model.
To validate the reliability of our modelling approach for such precise measurements, we generated 3D models for the wild-type and multiple point-mutants of the fungal LAC IIIb from Trametes versicolor. The structure of the wild-type enzyme had been solved by X-ray crystallography (Bertrand et al., 2002; PDB: 1KYA) and revealed that the binding pocket is gated by multiple phenylalanines. Galli et al. (2011) generated multiple Phe to Ala point-mutants in these residues and showed that this enabled bulkier substrates to be oxidised more efficiently. This observation suggested that the Phe to Ala replacements increased the size of the binding pocket and/or of the binding pocket mouth (Galli et al., 2011). Indeed, our modelling analysis showed that replacements F162A, F265A, F332A, and F162A/F332A increased entrance area, confirming the structural consequences of these mutations (Figure 7E). In contrast, the replacement of F337, which is involved in electron transport but not pocket formation (Galli et al., 2011), had no effect on binding pocket topology (Figure 7E). Having validated our modelling approach, we used it to characterise the binding pockets of the multiple plant LAC paralogs. The plant LAC binding pockets were delimited by regions that were highly variable in both sequence and structure, exhibiting no conserved gating residues (Figure 7). However, in paralogs with larger binding pockets, bulky residues such as Phe, Tyr, and Pro (Pro265 and Phe352 in ChLAC8, Pro276 and Phe362 in AtLAC12; Figures 7B,D and Supplementary Video 1) fulfilled a structural role similar to the ones of the Phe gating the entrance of the T. versicolor LAC (Figure 7E). These residues delineated a binding pocket mouth relatively far away from the T1 copper-coordinating histidine (roughly 11Å in ChLAC8 and AtLAC12), likely restricting the access to specific substrates that can fully enter the binding pocket to reach the active site. In contrast, in paralogs with smaller binding pockets, these bulky residues are replaced with smaller ones and/or oriented away from the binding pocket (Glu161 and Asn438 in ADE/LAC, Ala159 and Ile268 in AtLAC15, Figures 7A,C and Supplementary Video 1). This placed the entrance of the pocket closer to the active site (∼6Å in AtLAC15, ∼7Å in ADE/LAC), facilitating access to the active site. Altogether the different combinations of binding pocket size, mouth area, and pocket shape suggest that the different modelled LAC paralogs are likely adapted to specifically oxidise different substrates. LACs with smaller and more exposed pockets could oxidise single groups/tails/sidechains of bulkier substrates, whereas LACs with larger binding pockets would require smaller or more specific substrates to enter the pocket.
When considering the substrate stabilisation and its deprotonation, previous assumptions based on 2D sequence alignments predicted higher optimal reaction pH for plant LACs. The analyses of the 3D models of plant LACs showed that, similar to the structure of ZmLAC3 (Xie et al., 2020), the residue analogous to the fungal Asp206 in plants is in position 449 (Glu449) and filled by a Glu in 157 paralogs or by an Asp in 21 paralogs of the 194 plant LACs analysed (Supplementary Movie 1). Both Glu and Asp residues in this position facilitate phenolic deprotonation similarly to the Asp206 of fungal LACs (Madzak et al., 2006). Among the 3D-modelled paralogs, the prediction for a higher pH optimum only holds for ADE/LAC (with a glutamine), AtLAC14 (with an asparagine) and AtLAC15 (with a glycine). In contrast to previous prediction, our analysis suggested that both the oxidative capacity and pH optimum of plant LACs are generally similar to their fungal homologs except for a few paralogs with higher pH optimum. Our analysis further corroborated the empirical measurements (Figure 6) showing similar pH optima between purified plant and fungal LACs. These results highlight the universal importance of key conserved residues for deprotonating phenolic substrates.
Hierarchical clustering of all the different LAC paralogs based on binding pocket topology resulted in five clusters which showed considerable overlap with previously published functional similarities (Figure 7E). The cluster with LAC paralogs known to oxidise flavonoids had the smallest binding pockets with moderate (AtLAC15) to minimal (ADE/LAC) compactness and pocket mouth areas (Figure 7E). Another cluster grouped paralogs pivotal for vascular lignification (AtLAC4 and AtLAC11) as well as CoLAC1 shown to preferentially oxidise lignin hydroxyphenyl (H) residue precursors of lignin (Hiraide et al., 2021). This group presented intermediate sized binding pockets of generally low compactness gated by mostly small mouth area (Figure 7E). In contrast, LAC paralogs shown to alter lignin G residue accumulation in loss/gain-of-function experiments (AtLAC17, MsLAC1, and CoLAC3; Table 2) were grouped by intermediate sized binding pockets of intermediate compactness gated by variable sized mouth area (Figure 7E). AtLAC5, 8, and 12 formed a cluster with the larger pockets of intermediate compactness and gating, whereas AtLAC9 and ChLAC8 constituted the group with the largest pocket and moderate to high compactness and mouth areas (Figure 7E). In line with the observations previously made on the crystal structure of ZmLAC3 (Xie et al., 2020), all analysed plant LAC protein structures exhibited much deeper binding pockets of lower compactness than fungal LACs (Figure 7E). This observation suggested that plant LACs might be less efficient in the oxidation of bulky substrates such as large lignin polymers. Lastly, our results showed that little correlation linked LAC function/activity to their phylogenetic relationship. The clustering according to binding pocket topology differed drastically from that based on sequence homology (Supplementary Figure 4) but better reflected LAC function/activity. This approach might thus be the more reliable approach to predict functional similarities in LACs.
Common Features of Phenoloxidases
Critical Comparison of Phenoloxidases
The biological requirement for so many different and diverse phenoloxidases in plants remains unclear. However, their increasing paralog numbers suggest pivotal roles in plant development and/or stress response, especially for PRXs and LACs. The extreme diversity of phenoloxidases and their functional roles can partly be explained by differences in localisation and activation. Their regulation can be separated into constitutive or inducible phenoloxidases which will act at specific subcellular sites in distinct cell types during development and/or stress response. The distinction between constitutive and inducible phenoloxidases, generally defined at the transcriptional level, provides long-term and short-term responses respectively. When considering lignin formation for example, the function(s) of phenoloxidases will either be constitutive during growth (formation of vascular tissues—Zhao et al., 2013), inducible for growth under constraints (altered by gravity in reaction wood—Hiraide et al., 2021) or induced during biotic stress response (bacterial infection in leaves—Lee et al., 2019). We can subcategorise constitutive phenoloxidases into “in action” or “in waiting,” as phenoloxidases can be regulated by proteolytic activation and/or substrate availability. An example of phenoloxidases “in waiting” are PPOs in apple fruits, which only become active when the tissue is ruptured. Phenoloxidases that are constitutively “in action” include cell wall resident LACs in the vasculature, which continuously lignify the cell wall long after the cell itself has died (Pesquet et al., 2013, 2019; Ménard et al., 2021). Another aspect behind the diversity of phenoloxidases is their capacity to synergistically act in the same reaction cascade by sequential action or complex formation (Barros et al., 2015). Sequential action of different groups of phenoloxidases occurs in melanin formation, where initial oxidation of amino acids by PPOs is followed by the polymerisation of the intermediates by LACs. On the other hand, the functional roles of potential heteromeric protein complexes, especially in LACs, are still completely unclear. Altogether, the various complementary modes of action of phenoloxidases call for future extensive functional studies to investigate the genetic and physical interactions of phenoloxidases at the cellular and subcellular levels.
Direct and Indirect Oxidation Mechanisms
The identity of the biological substrates oxidised by plant phenoloxidases and the factors determining the direction of the oxidative reaction (polymerising or depolymerising) in the metabolism of phenolic polymers remain open questions. Most if not all phenoloxidases can use indirect reaction via radical redox shuttle mediators. In lignolytic fungal PRXs, MnPs activity is mediated by the oxidation of Mn2+ to Mn3+ to cleave lignin (Wariishi et al., 1991), whereas LiPs use a veratryl alcohol mediator (Harvey et al., 1986; Akamatsu et al., 1990). VPs are called versatile for their capacity to oxidise substrates both directly and through Mn2+ mediators (Gómez-Toribio et al., 2001). The presence and identity of mediators has also been suggested to determine the direction of the oxidative reaction (Jeon and Chang, 2013; Hilgers et al., 2018). Some fungal LAC paralogs that polymerise phenolic moieties into lignin-like structures in the absence of mediators will instead break-down polymers in the presence of mediators (Bourbonnais et al., 1995; Shleev et al., 2006; Maijala et al., 2012; Munk et al., 2015). The mediators involved in lignin depolymerisation in vivo are still unknown and candidates include (i) small lignin-related monomeric phenolics such as vanillin, ferulic acid or syringylic compounds (Lahtinen et al., 2009; Cañas and Camarero, 2010), (ii) Mn2+ (Schlosser and Höffer, 2002), and/or (iii) secreted hydroquinones (Wei et al., 2010). The presence of these mediators however cannot be the only factor determining the direction of the oxidative reaction because many predicted mediators are present during plant cell wall lignification and even incorporated into lignin (Barros et al., 2015). In fact, easily oxidised compounds such as coniferyl alcohol, p-coumarate (Takahama et al., 1996; Ralph et al., 2004) or Mn2+/Mn3+ (Önnerud et al., 2002) can be used as intermediate to transfer the radical charge to growing lignin polymers, oligomers and/or bulky monomers. Altogether, it appears that both substrate specificity and the direction of oxidising reaction are defined by a combination of protein structure, binding pocket anatomy, mediator availability, and other not yet determined reaction conditions or interactions.
Limitation of Phenoloxidase Activity by Co-substrate Availability
Every phenoloxidase requires a specific co-substrate to oxidise phenolic compounds, H2O2 for PRX and O2 for PPOs and LACs. Local control of O2 and H2O2 concentrations therefore represents an essential aspect regulating the in situ activity of phenoloxidases. Although present in high concentrations in the atmosphere, O2 concentration in plant tissues generally decreases with increasing distance from the epidermis (Spicer and Holbrook, 2005) and lignified tissues such as wood mostly remain in a state of hypoxia (Sorz and Hietz, 2006; Gansert and Blossfeld, 2008). To increase aeration, O2 not only diffuses inward from the air through the bark (Sorz and Hietz, 2006), but is also transported throughout the plant by the xylem sap (Gansert, 2003). However, even in aqueous solutions in equilibrium with the atmosphere, the dissolved O2 concentration is only roughly equivalent to the fungal LAC Km toward O2 (Xu, 2001; Zumarraga et al., 2008). This suggests that in conditions of phenolic substrate excess, LAC activity in planta is limited by O2 just like LAC activity in vitro in aqueous solutions (Ortner et al., 2015). To fuel PRX activity, H2O2 production directly depends on the activity of plasma membrane localised NAPDH oxidases, also called respiratory burst oxidase homolog (RBOH), which release superoxide O2⋅– that is then dismutated by superoxide dismutase (SOD) to form H2O2 (Podgórska et al., 2017). Both the dismutation reaction by SOD to form H2O2 and its breaking down by catalase release O2, and both SOD and catalase activity have been detected in the cell wall (Podgórska et al., 2017). Interestingly, H2O2 production in plants is enhanced in condition of hypoxia (Vergara et al., 2012). Generation and transport of reactive oxygen species, and the associated O2 produced by their dismutation and breakdown, might therefore be an underestimated regulator of not only PRX, but also LAC activity.
Impact of pH on Phenoloxidase Activity and Phenolic Compound Oxidation
Our metadata analysis revealed differences between optimal pH and substrate type for phenoloxidases (Figure 6D), suggesting that local pH represents an essential factor which controls the activity of phenoloxidases. Local pH also directly affects the E° of phenolic substrates and facilitates their oxidation at higher pH. Some phenolic compounds, such as L-DOPA or pyrogallol, even auto-oxidise and polymerise non-enzymatically at neutral and higher pH (Gao et al., 1998; Eslami et al., 2012). This potential regulation of phenoloxidase activity and phenol oxidation by pH is of particular interest when considering that tracheary elements, the water conducting cells of vascular plants, accumulate their lignin post-mortem (Pesquet et al., 2013, 2019; Barros et al., 2015; Ménard et al., 2021) once their cell wall is exposed to xylem sap. Available data shows that the pH of the xylem sap is consistently 1 to 2 units higher than that of the cell wall in living cells (Table 3). Additionally, xylem sap pH is highly regulated with developmental state in each organ, time of the day and season (Alves et al., 2004; Aubrey et al., 2011) as well as in response to environmental stress conditions such as water availability (Wilkinson and Davies, 1997; Gloser et al., 2016; Pagliarani et al., 2019). The tight regulation of pH at the level of every cell, if not in every cell wall layer, undergoing phenolic oxidation might represent an additional mechanism to control phenoloxidase activity in development and stress response.
TABLE 3.
Xylem sap and cell wall pH in different plant species.
| Species | Xylem sap pH | Cell wall pH | References |
| Acer pseudoplatanus | 6.9 | 6.2 | Essiamah, 1980; Taylor and Davies, 1985 |
| Arabidopsis thaliana | 6 | 5.5 | Bibikova et al., 1998; Martinière et al., 2018 |
| Betula pendula | 7.5 | 5.5 | Taylor and Davies, 1985; Sauter and Ambrosius, 1986 |
| Brassica napus | 7.4 | 6 | Husted and Schjoerring, 1995; Gloser et al., 2016 |
| Helianthus annuus | 7.2 | 5.3 | Jia and Davies, 2007; Gloser et al., 2016 |
| Phaseolus coccineus | 6.6 | 5.9 | Starrach and Mayer, 1989; Gloser et al., 2016 |
| Pisum sativum | 6.2 | 6 | Jacobs and Ray, 1976; Gloser et al., 2016 |
Conclusion
Phenoloxidases include multiple unrelated and very diverse enzymes responsible of oxidising phenolics. From a mechanistic perspective, phenoloxidases could show relatively little substrate specificity due to indirect oxidation mechanisms using mediators and long-range electron transfer. LACs and class III PRXs have been suggested to act redundantly in the oxidative polymerisation of the earth’s most abundant phenolic polymer, lignin (Boerjan et al., 2003; Ralph et al., 2004). This assumption, based on the low substrate specificity of these different phenoloxidases when oxidising small phenolics in vitro, is effectively supported by the multitude of “non-canonical” constituents incorporated in lignin such as flavonoids (Lan et al., 2015) and hydroxystilbenes (del Río et al., 2017). However, these observations rarely differentiate between the cell walls of different cell types, as well as between their different cell wall layers, which exhibit drastically distinct monomeric composition, amount and structure of lignin (Terashima and Fukushima, 1988; Terashima et al., 2012; Blaschek et al., 2020a,b; Mottiar et al., 2020; Yamamoto et al., 2020). As cell wall lignification is a cell-cell cooperative process (Pesquet et al., 2013; Smith et al., 2013) mediated by the release of mobile lignin monomers in the apoplast, lignin formation in the specific cell wall layers of each cell type will require a directing force to control their distinct amount and composition, such as using different combinations of phenoloxidases. Whether the potential non-redundant roles of phenoloxidases are due to intrinsic differences in monomer specificity, sequential action, or distinct requirements in the catalytic environment still remains unclear. In addition, the phenoloxidases glycosylation state, nature of mediators, cell wall micro-environments, and protein interactions have all been shown to affect activity, specificity, and even reaction direction (anabolic vs. catabolic). Altogether, we are only beginning to understand the diverse roles played by phenoloxidases. Further research, focusing on comprehensive in situ functional characterisation of these phenoloxidases, will be necessary to clarify their precise roles and regulation.
Methods
Evolution of Phenoloxidase Gene Families
The numbers of paralogs (Table 1 and Figure 3) are taken from the bibliography or, in the case of PRXs, from PeroxiBase (Savelli et al., 2019). The time since divergence from A. thaliana for each species was taken from the timetree project (Kumar et al., 2017).
Structure and Sequence Conservation in Phenoloxidases
One plant phenoloxidase with resolved crystal structure was chosen per group to visualise secondary structure and coordinating residues (Figure 2). Sequence conservation was estimated based on a multiple sequence alignment of all full-length paralogs from P. patens (formerly named Physcomitrella patens), Selaginella moellendorffii, B. distachyon, and P. trichocarpa.
Laccase Phylogeny
Laccase sequences were identified by protein blast against all 17 A. thaliana LACs in P. trichocarpa, Zostera marina, B. distachyon, Amborella trichocarpa, S. moellendorffii, P. patens, M. polymorpha, C. braunii, V. carteri, C. reinhardtii (NCBI), Picea abies1 (Sundell et al., 2015), and A. filiculoides2 (Li et al., 2018). Sequences that were duplicates, incomplete, or missing core copper binding motifs (McCaig et al., 2005) were removed, and Signal peptides and extensive gaps were trimmed. The non-LAC sequences that remained after this filtering (exclusively ascorbate oxidases) were included as an outgroup. An appropriate amino acid replacement model (WAG with empirical frequencies and a proportion of invariant sites) was selected with ModelTest-NG v0.1.5 (Darriba et al., 2020). MrBayes v3.2.2 (Ronquist et al., 2012) was run on CIPRES3 (disabled BEAGLE) for one million generations to compute the phylogenetic tree (for the log-likelihood plot of chain convergence; see Supplementary Figure 2). The tree was visualised in R v4.0.4 using the “treeio” v1.14.3 (Wang L.-G. et al., 2020) and “ggtree” v2.4.1 (Yu et al., 2017) packages.
Laccase Homology Modelling
Laccase homology models were built using AlphaFold 2 with amber relaxation (Jumper et al., 2021) based on MMseqs2 multiple sequence alignments (Mirdita et al., 2019). Signal peptides of the modelled sequences were removed using SignalP v4.1 (Petersen et al., 2011). The single template models in Supplementary Figure 4 were built using Modeller v10.1 (Webb and Sali, 2016), based on the crystal structure of the maize laccase ZmLAC3 (PDB: 6klg; Xie et al., 2020), including the 4 copper ions as rigid bodies. A total of 30 single-template models were built per paralog (5 individual models with 5 loop-refinement iterations each). Model quality was assessed using modeller’s normalised DOPE score (Shen and Sali, 2006). For each model, the distances of the T1 copper coordinating histidines from the protein surface were estimated using DEPTH v2.0.0 (Tan et al., 2013). Binding pockets were characterised using CASTp (Tian et al., 2018) with a probe radius of 1.4 Å. The correct binding pocket for each paralog was identified as the pocket formed by the highest number of residues aligning to the pocket-forming residues of ZmLAC3. Pocket compactness was calculated as , where V is the volume of the binding pocket and A is the protein surface area forming the pocket. Modelled protein structures were visualised in UCSF ChimeraX v1.2.5 (Pettersen et al., 2021). The models were clustered based on the medians for each parameter using average linkage clustering in R v4.0.4. The correlations between the parameters used for clustering were moderate at most (Supplementary Figure 5). To compare the structure-based clustering with sequence homology, a bayesian phylogenetic tree was generated from the modelled sequences using the same approach as described in the previous paragraph. The two dendrograms were then compared using the “dendextend” package v1.14.0 (Galili, 2015) in R v4.0.4.
Author Contributions
LB compiled and analysed the data. EP ensured financial support. LB and EP wrote the manuscript. Both authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank William Gardi and Ulrich Theopold for critical comments and Laura Schat for her help in troubleshooting and interpreting the phylogenetic analyses. We also thank Bio4Energy (a strategic research environment appointed by the Swedish government) and the Department of Ecology, Environment and Plant Sciences (DEEP) and the Bolin Centre for Climate Research of Stockholm University (SU).
Footnotes
Funding
This work was supported by Vetenskapsrådet (VR) research grants 2010-4620 and 2016-04727 (EP), the Stiftelsen för Strategisk Forskning ValueTree (EP), and the Bolin Centre for Climate Research RA3, RA4 and RA5 “seed money” (EP).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.754601/full#supplementary-material
Animations of the predicted protein structures for AtLAC12, AtLAC15, ADE/LAC, and ChLAC8. The protein surface is coloured by its distance to the T1 copper, the binding pocket volume is shown in yellow. Residues shaping the binding pocket mouth are shown in pink and labelled. The two histidines coordinating the T1 copper are shown in blue.
References
- Akamatsu Y., Ma D. B., Higuchi T., Shimada M. (1990). A novel enzymatic decarboxylation of oxalic acid by the lignin peroxidase system of white-rot fungus Phanerochaete chrysosporium. FEBS Lett. 269 261–263. [DOI] [PubMed] [Google Scholar]
- Alves G., Ameglio T., Guilliot A., Fleurat-Lessard P., Lacointe A., Sakr S., et al. (2004). Winter variation in xylem sap pH of walnut trees: involvement of plasma membrane H+-ATPase of vessel-associated cells. Tree Physiol. 24 99–105. 10.1093/treephys/24.1.99 [DOI] [PubMed] [Google Scholar]
- Ander P., Eriksson K.-E. L. (1976). The importance of phenol oxidase activity in lignin degradation by the white-rot fungus Sporotrichum pulverulentum. Arch. Microbiol. 109 1–8. 10.1007/BF00425105 [DOI] [Google Scholar]
- Arakane Y., Muthukrishnan S., Beeman R. W., Kanost M. R., Kramer K. J. (2005). Laccase 2 is the phenoloxidase gene required for beetle cuticle tanning. Proc. Natl. Acad. Sci. U.S.A. 102 11337–11342. 10.1073/pnas.0504982102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubrey D. P., Boyles J. G., Krysinsky L. S., Teskey R. O. (2011). Spatial and temporal patterns of xylem sap pH derived from stems and twigs of Populus deltoides L. Environ. Exp. Bot. 71 376–381. 10.1016/j.envexpbot.2011.02.006 [DOI] [Google Scholar]
- Bach A., Chodat R. (1903). Untersuchungen über die Rolle der Peroxyde in der Chemie der lebenden Zelle. IV. Ueber peroxydase. Berichte Dtsch. Chem. Ges. 36 600–605. [Google Scholar]
- Baldrian P. (2006). Fungal laccases–occurrence and properties. FEMS Microbiol. Rev. 30 215–242. 10.1111/j.1574-4976.2005.00010.x [DOI] [PubMed] [Google Scholar]
- Bao W., O’Malley D. M., Whetten R., Sederoff R. R. (1993). A laccase associated with lignification in loblolly pine xylem. Science 260 672–674. 10.1126/science.260.5108.672 [DOI] [PubMed] [Google Scholar]
- Barrett F. M. (1986). Characterization of phenoloxidase from larval cuticle of Sarcophaga bullata and a comparison with cuticular enzymes from other species. Can. J. Zool. 65 1158–1166. 10.1139/z87-181 33356898 [DOI] [Google Scholar]
- Barros J., Serk H., Granlund I., Pesquet E. (2015). The cell biology of lignification in higher plants. Ann. Bot. 115 1053–1074. 10.1093/aob/mcv046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglin M., Delage L., Potin P., Vilter H., Elwing H. (2004). Enzymatic cross-linking of a phenolic polymer extracted from the marine alga Fucus serratus. Biomacromolecules 5 2376–2383. 10.1021/bm0496864 [DOI] [PubMed] [Google Scholar]
- Bernardes A., Textor L. C., Santos J. C., Cuadrado N. H., Kostetsky E. Y., Roig M. G., et al. (2015). Crystal structure analysis of peroxidase from the palm tree Chamaerops excelsa. Biochimie 111 58–69. 10.1016/j.biochi.2015.01.014 [DOI] [PubMed] [Google Scholar]
- Berthet S., Demont-Caulet N., Pollet B., Bidzinski P., Cezard L., Le Bris P., et al. (2011). Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell 23 1124–1137. 10.1105/tpc.110.082792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand G. (1894). Sur le Latex de l’Arbre à Laque et sur une Nouvelle diastase contenue dans ce latex. Compt. Rendus Soc. Biol. 46 478–480. [Google Scholar]
- Bertrand G. (1896). Sur les rapports qui existent entre la constitution chimique des composés organiques et leur oxydabilité sous l’influence de la laccase. Bull. Soc. Chim. Paris Ser. 3 791–793. [Google Scholar]
- Bertrand T., Jolivalt C., Briozzo P., Caminade E., Joly N., Madzak C., et al. (2002). Crystal structure of a four-copper laccase complexed with an arylamine:? insights into substrate recognition and correlation with kinetics. Biochemistry 41 7325–7333. 10.1021/bi0201318 [DOI] [PubMed] [Google Scholar]
- Bibikova T. N., Jacob T., Dahse I., Gilroy S. (1998). Localized changes in apoplastic and cytoplasmic pH are associated with root hair development in Arabidopsis thaliana. Development 125 2925–2934. 10.1242/dev.125.15.2925 [DOI] [PubMed] [Google Scholar]
- Bidla G., Dushay M. S., Theopold U. (2007). Crystal cell rupture after injury in Drosophila requires the JNK pathway, small GTPases and the TNF homolog Eiger. J. Cell Sci. 120 1209–1215. 10.1242/jcs.03420 [DOI] [PubMed] [Google Scholar]
- Bijelic A., Pretzler M., Molitor C., Zekiri F., Rompel A. (2015). The structure of a plant tyrosinase from Walnut leaves reveals the importance of “substrate-guiding residues” for enzymatic specificity. Angew. Chem. Int. Ed. 54 14677–14680. 10.1002/anie.201506994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaschek L., Champagne A., Dimotakis C., Nuoendagula, Decou R., Hishiyama S., et al. (2020a). Cellular and genetic regulation of coniferaldehyde incorporation in lignin of herbaceous and woody plants by quantitative wiesner staining. Front. Plant Sci. 11:109. 10.3389/fpls.2020.00109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaschek L., Nuoendagula, Bacsik Z., Kajita S., Pesquet E. (2020b). Determining the genetic regulation and coordination of lignification in stem tissues of Arabidopsis using semiquantitative Raman microspectroscopy. ACS Sustain. Chem. Eng. 8 4900–4909. 10.1021/acssuschemeng.0c00194 [DOI] [Google Scholar]
- Bleve G., Lezzi C., Spagnolo S., Tasco G., Tufariello M., Casadio R., et al. (2013). Role of the C-terminus of Pleurotus eryngii Ery4 laccase in determining enzyme structure, catalytic properties and stability. Protein Eng. Des. Sel. 26 1–13. 10.1093/protein/gzs056 [DOI] [PubMed] [Google Scholar]
- Boeckx T., Winters A., Webb K. J., Kingston-Smith A. H. (2017). Detection of potential chloroplastic substrates for polyphenol oxidase suggests a role in undamaged leaves. Front. Plant Sci. 8:237. 10.3389/fpls.2017.00237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W., Ralph J., Baucher M. (2003). Lignin biosynthesis. Annu. Rev. Plant Biol. 54 519–546. 10.1146/annurev.arplant.54.031902.134938 [DOI] [PubMed] [Google Scholar]
- Bourbonnais R., Paice M. G. (1990). Oxidation of non-phenolic substrates. An expanded role for laccase in lignin biodegradation. FEBS Lett. 267 99–102. 10.1016/0014-5793(90)80298-W [DOI] [PubMed] [Google Scholar]
- Bourbonnais R., Paice M. G., Reid I. D., Lanthier P., Yaguchi M. (1995). Lignin oxidation by laccase isozymes from Trametes versicolor and role of the mediator 2,2’-azinobis(3-ethylbenzthiazoline-6-sulfonate) in kraft lignin depolymerization. Appl. Environ. Microbiol. 61 1876–1880. 10.1128/aem.61.5.1876-1880.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourquelot E., Bertrand G. (1896). Les ferments oxydants dans les Champignons. Bull. Soc. Mycol. France 12 18–26. [Google Scholar]
- Bryan A. C., Jawdy S., Gunter L., Gjersing E., Sykes R., Hinchee M. A. W., et al. (2016). Knockdown of a laccase in Populus deltoides confers altered cell wall chemistry and increased sugar release. Plant Biotechnol. J. 14 2010–2020. 10.1111/pbi.12560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulter T., Alcalde M., Sieber V., Meinhold P., Schlachtbauer C., Arnold F. H. (2003). Functional expression of a fungal laccase in Saccharomyces cerevisiae by directed evolution. Appl. Environ. Microbiol. 69 987–995. 10.1128/AEM.69.2.987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cañas A. I., Camarero S. (2010). Laccases and their natural mediators: biotechnological tools for sustainable eco-friendly processes. Biotechnol. Adv. 28 694–705. 10.1016/j.biotechadv.2010.05.002 [DOI] [PubMed] [Google Scholar]
- Carletti G., Nervo G., Cattivelli L. (2014). Flavonoids and melanins: a common strategy across two kingdoms. Int. J. Biol. Sci. 10 1159–1170. 10.7150/ijbs.9672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesarino I., Araújo P., Sampaio Mayer J. L., Vicentini R., Berthet S., Demedts B., et al. (2013). Expression of SofLAC, a new laccase in sugarcane, restores lignin content but not S:G ratio of Arabidopsis lac17 mutant. J. Exp. Bot. 64 1769–1781. 10.1093/jxb/ert045 [DOI] [PubMed] [Google Scholar]
- Chantreau M., Portelette A., Dauwe R., Kiyoto S., Crônier D., Morreel K., et al. (2014). Ectopic lignification in the flax lignified bast fiber1 mutant stem is associated with tissue-specific modifications in gene expression and cell wall composition. Plant Cell Online 26 4462–4482. 10.1105/tpc.114.130443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Liu F., Yang B., Lu A., Wang S., Wang J., et al. (2012). Specific amino acids affecting Drosophila melanogaster prophenoloxidase activity in vitro. Dev. Comp. Immunol. 38 88–97. 10.1016/j.dci.2012.04.007 [DOI] [PubMed] [Google Scholar]
- Cheynier V., Comte G., Davies K. M., Lattanzio V., Martens S. (2013). Plant phenolics: recent advances on their biosynthesis, genetics, and ecophysiology. Plant Physiol. Biochem. 72 1–20. 10.1016/j.plaphy.2013.05.009 [DOI] [PubMed] [Google Scholar]
- Chi M., Bhagwat B., Lane W. D., Tang G., Su Y., Sun R., et al. (2014). Reduced polyphenol oxidase gene expression and enzymatic browning in potato (Solanum tuberosum L.) with artificial microRNAs. BMC Plant Biol. 14:62. 10.1186/1471-2229-14-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho M.-H., Moinuddin S. G. A., Helms G. L., Hishiyama S., Eichinger D., Davin L. B., et al. (2003). (+)-Larreatricin hydroxylase, an enantio-specific polyphenol oxidase from the creosote bush (Larrea tridentata). Proc. Natl. Acad. Sci. U.S.A. 100 10641–10646. 10.1073/pnas.1934562100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodat R., Bach A. (1903). Untersuchungen über die Rolle der peroxyde in der chemie der lebenden Zelle. V. Zerlegung der sogenannten oxydasen in oxygenasen und peroxydasen. Berichte Dtsch. Chem. Ges. 36 606–609. [Google Scholar]
- Choi H. W., Kim Y. J., Lee S. C., Hong J. K., Hwang B. K. (2007). Hydrogen peroxide generation by the pepper extracellular peroxidase CaPO2 activates local and systemic cell death and defense response to bacterial pathogens. Plant Physiol. 145 890–904. 10.1104/pp.107.103325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J., Poutaraud A., Hugueney P. (2009). Metabolism and roles of stilbenes in plants. Plant Sci. 177 143–155. 10.1016/j.plantsci.2009.05.012 [DOI] [Google Scholar]
- Chou E. Y., Schuetz M., Hoffmann N., Watanabe Y., Sibout R., Samuels A. L. (2018). Distribution, mobility, and anchoring of lignin-related oxidative enzymes in Arabidopsis secondary cell walls. J. Exp. Bot. 69 1849–1859. 10.1093/jxb/ery067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claus H., Decker H. (2006). Bacterial tyrosinases. Syst. Appl. Microbiol. 29 3–14. 10.1016/j.syapm.2005.07.012 [DOI] [PubMed] [Google Scholar]
- Clutterbuck A. J. (1972). Absence of laccase from yellow-spored mutants of Aspergillus nidulans. J. Gen. Microbiol. 70 423–435. 10.1099/00221287-70-3-423 [DOI] [PubMed] [Google Scholar]
- Colpa D. I., Fraaije M. W., Van Bloois E. (2014). DyP-type peroxidases: a promising and versatile class of enzymes. J. Ind. Microbiol. Biotechnol. 41 1–7. 10.1007/s10295-013-1371-6 [DOI] [PubMed] [Google Scholar]
- Constabel C. P., Ryan C. A. (1998). A survey of wound- and methyl jasmonate-induced leaf polyphenol oxidase in crop plants. Phytochemistry 47 507–511. 10.1016/S0031-9422(97)00539-6 [DOI] [Google Scholar]
- Cosio C., Dunand C. (2009). Specific functions of individual class III peroxidase genes. J. Exp. Bot. 60 391–408. 10.1093/jxb/ern318 [DOI] [PubMed] [Google Scholar]
- Cosio C., Ranocha P., Francoz E., Burlat V., Zheng Y., Perry S. E., et al. (2017). The class III peroxidase PRX17 is a direct target of the MADS-box transcription factor AGAMOUS-LIKE15 (AGL15) and participates in lignified tissue formation. New Phytol. 213 250–263. 10.1111/nph.14127 [DOI] [PubMed] [Google Scholar]
- Cosio C., Vuillemin L., De Meyer M., Kevers C., Penel C., Dunand C. (2009). An anionic class III peroxidase from zucchini may regulate hypocotyl elongation through its auxin oxidase activity. Planta 229 823–836. 10.1007/s00425-008-0876-0 [DOI] [PubMed] [Google Scholar]
- Darriba D., Posada D., Kozlov A. M., Stamatakis A., Morel B., Flouri T. (2020). ModelTest-NG: a new and scalable tool for the selection of DNA and protein evolutionary models. Mol. Biol. Evol. 37 291–294. 10.1093/molbev/msz189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daudi A., Cheng Z., O’Brien J. A., Mammarella N., Khan S., Ausubel F. M., et al. (2012). The apoplastic oxidative burst peroxidase in Arabidopsis is a major component of pattern-triggered immunity. Plant Cell 24 275–287. 10.1105/tpc.111.093039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker H., Terwilliger N. (2000). Cops and robbers: putative evolution of copper oxygen-binding proteins. J. Exp. Biol. 203 1777–1782. 10.1242/jeb.203.12.1777 [DOI] [PubMed] [Google Scholar]
- Decker H., Schweikardt T., Nillius D., Salzbrunn U., Jaenicke E., Tuczek F. (2007). Similar enzyme activation and catalysis in hemocyanins and tyrosinases. Gene 398 183–191. 10.1016/j.gene.2007.02.051 [DOI] [PubMed] [Google Scholar]
- del Río J. C., Rencoret J., Gutiérrez A., Kim H., Ralph J. (2017). Hydroxystilbenes are monomers in palm fruit endocarp lignins. Plant Physiol. 174 2072–2082. 10.1104/pp.17.00362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire P., Ménard D., Green P., Saalbach G., Buschmann H., Lloyd C. W., et al. (2015). Proteomic analysis of microtubule interacting proteins over the course of xylem tracheary element formation in Arabidopsis. Plant Cell 27 2709–2726. 10.1105/tpc.15.00314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamantidis G., Effosse A., Potier P., Bally R. (2000). Purification and characterization of the first bacterial laccase in the rhizospheric bacterium Azospirillum lipoferum. Soil Biol. Biochem. 32 919–927. 10.1016/S0038-0717(99)00221-7 [DOI] [Google Scholar]
- Dirks-Hofmeister M. E., Inlow J. K., Moerschbacher B. M. (2012). Site-directed mutagenesis of a tetrameric dandelion polyphenol oxidase (PPO-6) reveals the site of subunit interaction. Plant Mol. Biol. 80 203–217. 10.1007/s11103-012-9943-9 [DOI] [PubMed] [Google Scholar]
- Dittmer N. T., Suderman R. J., Jiang H., Zhu Y. C., Gorman M. J., Kramer K. J., et al. (2004). Characterization of cDNAs encoding putative laccase-like multicopper oxidases and developmental expression in the tobacco hornworm, Manduca sexta, and the malaria mosquito, Anopheles gambiae. Insect Biochem. Mol. Biol. 34 29–41. 10.1016/j.ibmb.2003.08.003 [DOI] [PubMed] [Google Scholar]
- Durão P., Bento I., Fernandes A. T., Melo E. P., Lindley P. F., Martins L. O. (2006). Perturbations of the T1 copper site in the CotA laccase from Bacillus subtilis: structural, biochemical, enzymatic and stability studies. J. Biol. Inorg. Chem. 11 514–526. 10.1007/s00775-006-0102-0 [DOI] [PubMed] [Google Scholar]
- Dwivedi U. N., Singh P., Pandey V. P., Kumar A. (2011). Structure-function relationship among bacterial, fungal and plant laccases. J. Mol. Catal. B Enzym. 68 117–128. 10.1016/j.molcatb.2010.11.002 [DOI] [Google Scholar]
- Endo K., Hosono K., Beppu T., Ueda K. (2002). A novel extracytoplasmic phenol oxidase of streptomyces: its possible involvement in the onset of morphogenesis. Microbiology 148 1767–1776. 10.1099/00221287-148-6-1767 [DOI] [PubMed] [Google Scholar]
- Eslami M., Zare H. R., Namazian M. (2012). Thermodynamic parameters of electrochemical oxidation of L - DOPA: experimental and theoretical studies. J. Phys. Chem. B 116 12552–12557. 10.1021/jp3054229 [DOI] [PubMed] [Google Scholar]
- Esposito R., D’Aniello S., Squarzoni P., Pezzotti M. R., Ristoratore F., Spagnuolo A. (2012). New insights into the evolution of metazoan tyrosinase gene family. PLoS One 7:e35731. 10.1371/journal.pone.0035731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essiamah S. K. (1980). Spring sap of trees. Berichte Dtsch. Bot. Ges. 93 257–267. 10.1111/j.1438-8677.1980.tb03337.x [DOI] [Google Scholar]
- Faccio G., Kruus K., Saloheimo M., Thöny-Meyer L. (2012). Bacterial tyrosinases and their applications. Process Biochem. 47 1749–1760. 10.1016/j.procbio.2012.08.018 [DOI] [Google Scholar]
- Fang F., Zhang X., Luo H., Zhou J., Gong Y., Li W., et al. (2015). An intracellular laccase is responsible for the epicatechin mediated anthocyanin degradation in litchi fruit pericarp. Plant Physiol. 169 2391–2408. 10.1104/pp.15.00359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Pérez F., Vivar T., Pomar F., Pedreño M. A., Novo-Uzal E. (2015b). Peroxidase 4 is involved in syringyl lignin formation in Arabidopsis thaliana. J. Plant Physiol. 175 86–94. 10.1016/j.jplph.2014.11.006 [DOI] [PubMed] [Google Scholar]
- Fernández-Pérez F., Pomar F., Pedreño M. A., Novo-Uzal E. (2015a). The suppression of AtPrx52 affects fibers but not xylem lignification in Arabidopsis by altering the proportion of syringyl units. Physiol. Plant. 154 395–406. 10.1111/ppl.12310 [DOI] [PubMed] [Google Scholar]
- Freudenberg K., Reznik H., Boesenberg H., Rasenack D. (1952). Das an der Verholzung beteiligte Fermentsystem. Chem. Ber. 85 641–647. [Google Scholar]
- Galili T. (2015). dendextend: an R package for visualizing, adjusting and comparing trees of hierarchical clustering. Bioinformatics 31 3718–3720. 10.1093/bioinformatics/btv428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli C., Gentili P., Jolivalt C., Madzak C., Vadalà R. (2011). How is the reactivity of laccase affected by single-point mutations? Engineering laccase for improved activity towards sterically demanding substrates. Appl. Microbiol. Biotechnol. 91 123–131. 10.1007/s00253-011-3240-4 [DOI] [PubMed] [Google Scholar]
- Gansert D. (2003). Xylem sap flow as a major pathway for oxygen supply to the sapwood of birch (Betula pubescens Ehr.). Plant Cell Environ. 26 1803–1814. 10.1046/j.1365-3040.2003.01097.x [DOI] [Google Scholar]
- Gansert D., Blossfeld S. (2008). “The application of novel optical sensors (optodes) in experimental plant ecology,” in Progress in Botany, eds Lüttge U., Beyschlag W., Murata J. (Berlin: Springer; ), 333–358. 10.1007/978-3-540-72954-9_14 [DOI] [Google Scholar]
- Gao R., Yuan Z., Zhao Z., Gao X. (1998). Mechanism of pyrogallol autoxidation and determination of superoxide dismutase enzyme activity. Bioelectrochem. Bioenerg. 45 41–45. 10.1016/S0302-4598(98)00072-5 [DOI] [Google Scholar]
- Gao Z. J., Han X. H., Xiao X. G. (2009). Purification and characterisation of polyphenol oxidase from red Swiss chard (Beta vulgaris subspecies cicla) leaves. Food Chem. 117 342–348. 10.1016/j.foodchem.2009.04.013 [DOI] [Google Scholar]
- García-Ulloa A., Sanjurjo L., Cimini S., Encina A., Martínez-Rubio R., Bouza R., et al. (2020). Overexpression of ZePrx in Nicotiana tabacum affects lignin biosynthesis without altering redox homeostasis. Front. Plant Sci. 11:900. 10.3389/fpls.2020.00900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavnholt B., Larsen K. (2002). Molecular biology of plant laccases in relation to lignin formation. Physiol. Plant. 116 273–280. 10.1034/j.1399-3054.2002.1160301.x 11841302 [DOI] [Google Scholar]
- Ghosh D., Mukherjee R. (1998). Modeling tyrosinase monooxygenase activity. spectroscopic and magnetic investigations of products due to reactions between copper(I) complexes of xylyl-based dinucleating ligands and dioxygen: aromatic ring hydroxylation and irreversible oxidation product. Inorg. Chem. 37 6597–6605. 10.1021/ic9713689 [DOI] [PubMed] [Google Scholar]
- Ghosh S., Xie X., Dey A., Sun Y., Scholes C. P., Solomon E. I. (2009). Thermodynamic equilibrium between blue and green copper sites and the role of the protein in controlling function. Proc. Natl. Acad. Sci. U.S.A. 106 4969–4974. 10.1073/pnas.0900995106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloser V., Korovetska H., Martín-Vertedor A. I., Hájíèková M., Prokop Z., Wilkinson S., et al. (2016). The dynamics of xylem sap pH under drought: a universal response in herbs? Plant Soil 409 259–272. 10.1007/s11104-016-2962-6 [DOI] [Google Scholar]
- Goldfeder M., Kanteev M., Isaschar-Ovdat S., Adir N., Fishman A. (2014). Determination of tyrosinase substrate-binding modes reveals mechanistic differences between type-3 copper proteins. Nat. Commun. 5:4505. 10.1038/ncomms5505 [DOI] [PubMed] [Google Scholar]
- Gómez-Toribio V., Martínez A. T., Martínez M. J., Guillén F. (2001). Oxidation of hydroquinones by the versatile ligninolytic peroxidase from Pleurotus eryngii: H2O2 generation and the influence of Mn2+. Eur. J. Biochem. 268 4787–4793. 10.1046/j.1432-1327.2001.02405.x [DOI] [PubMed] [Google Scholar]
- González M. N., Massa G. A., Andersson M., Turesson H., Olsson N., Fält A.-S., et al. (2020). Reduced enzymatic browning in potato tubers by specific editing of a polyphenol oxidase gene via ribonucleoprotein complexes delivery of the CRISPR/Cas9 system. Front. Plant Sci. 10:1649. 10.3389/fpls.2019.01649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadt R. G., Sun N., Marshall N. M., Hodgson K. O., Hedman B., Lu Y., et al. (2012). Spectroscopic and DFT studies of second-sphere variants of the type 1 copper site in azurin: covalent and nonlocal electrostatic contributions to reduction potentials. J. Am. Chem. Soc. 134 16701–16716. 10.1021/ja306438n [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakulinen N., Rouvinen J. (2015). Three-dimensional structures of laccases. Cell. Mol. Life Sci. 72 857–868. 10.1007/s00018-014-1827-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakulinen N., Kiiskinen L. L., Kruus K., Saloheimo M., Paanen A., Koivula A., et al. (2002). Crystal structure of a laccase from melanocarpus albomyces with an intact trinuclear copper site. Nat. Struct. Biol. 9 601–605. 10.1038/nsb823 [DOI] [PubMed] [Google Scholar]
- Hammel K. E., Cullen D. (2008). Role of fungal peroxidases in biological ligninolysis. Curr. Opin. Plant Biol. 11 349–355. 10.1016/j.pbi.2008.02.003 [DOI] [PubMed] [Google Scholar]
- Harvey P. J., Schoemaker H. E., Palmer J. M. (1986). Veratryl alcohol as a mediator and the role of radical cations in lignin biodegradation by Phanerochaete chrysosporium. FEBS Lett. 195 242–246. 10.1016/0014-5793(86)80168-5 [DOI] [Google Scholar]
- Hattori M., Konishi H., Tamura Y., Konno K., Sogawa K. (2005). Laccase-type phenoloxidase in salivary glands and watery saliva of the green rice leafhopper, Nephotettix cincticeps. J. Insect Physiol. 51 1359–1365. 10.1016/j.jinsphys.2005.08.010 [DOI] [PubMed] [Google Scholar]
- He F., Machemer-Noonan K., Golfier P., Unda F., Dechert J., Zhang W., et al. (2019). The in vivo impact of MsLAC1, a Miscanthus laccase isoform, on lignification and lignin composition contrasts with its in vitro substrate preference. BMC Plant Biol. 19:552. 10.1186/s12870-019-2174-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero J., Fernández-Pérez F., Yebra T., Novo-Uzal E., Pomar F., Pedreño M. Á, et al. (2013). Bioinformatic and functional characterization of the basic peroxidase 72 from Arabidopsis thaliana involved in lignin biosynthesis. Planta 237 1599–1612. 10.1007/s00425-013-1865-5 [DOI] [PubMed] [Google Scholar]
- Higuchi T. (1958). Further studies on phenol oxidase related to the lignin biosynthesis. J. Biochem. 45 515–528. [Google Scholar]
- Hildén K., Hakala T. K., Lundell T. (2009). Thermotolerant and thermostable laccases. Biotechnol. Lett. 31 1117–1128. 10.1007/s10529-009-9998-0 [DOI] [PubMed] [Google Scholar]
- Hilgers R., Vincken J.-P., Gruppen H., Kabel M. A. (2018). Laccase/mediator systems: their reactivity toward phenolic lignin structures. ACS Sustain. Chem. Eng. 6 2037–2046. 10.1021/acssuschemeng.7b03451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraide H., Tobimatsu Y., Yoshinaga A., Lam P. Y., Kobayashi M., Matsushita Y., et al. (2021). Localised laccase activity modulates distribution of lignin polymers in gymnosperm compression wood. New Phytol. 230 2186–2199. 10.1111/nph.17264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoegger P. J., Kilaru S., James T. Y., Thacker J. R., Kües U. (2006). Phylogenetic comparison and classification of laccase and related multicopper oxidase protein sequences. FEBS J. 273 2308–2326. 10.1111/j.1742-4658.2006.05247.x [DOI] [PubMed] [Google Scholar]
- Hofrichter M., Ullrich R., Pecyna M. J., Liers C., Lundell T. (2010). New and classic families of secreted fungal heme peroxidases. Appl. Microbiol. Biotechnol. 87 871–897. 10.1007/s00253-010-2633-0 [DOI] [PubMed] [Google Scholar]
- Holst R. W., Yopp J. H. (1976). An algal polyphenol oxidase: characterization of the o-diphenol-oxidase from the charophyte, Nitella mirabilis. Phycologia 15 119–124. 10.2216/i0031-8884-15-2-119.1 [DOI] [Google Scholar]
- Howes B. D. D., Feis A., Raimondi L., Indiani C., Smulevich G. (2001). The critical role of the proximal calcium ion in the structural properties of horseradish peroxidase. J. Biol. Chem. 276 40704–40711. 10.1074/jbc.M107489200 [DOI] [PubMed] [Google Scholar]
- Hu Q., Min L., Yang X., Jin S., Zhang L., Li Y., et al. (2017). Laccase GhLac1 modulates broad-spectrum biotic stress tolerance via DAMP-triggered immunity. Plant Physiol. 176 1808–1823. 10.1104/pp.17.01628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husted S., Schjoerring J. K. (1995). Apoplastic pH and ammonium concentration in leaves of Brassica napus L. Plant Physiol. 109 1453–1460. 10.1104/pp.109.4.1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobowitz J. R., Doyle W. C., Weng J.-K. (2019). PRX9 and PRX40 are extensin peroxidases essential for maintaining tapetum and microspore cell wall integrity during arabidopsis anther development. Plant Cell 31 848–861. 10.1105/tpc.18.00907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs M., Ray P. M. (1976). Rapid auxin-induced decrease in free space pH and its relationship to auxin-induced growth in maize and pea 1. Plant Physiol. 58 203–209. 10.1104/pp.58.2.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenicke E., Decker H. (2003). Tyrosinases from crustaceans form hexamers. Biochem. J. 371 515–523. 10.1042/BJ20021058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal N., Pandey V. P., Dwivedi U. N. (2014). Purification of a thermostable laccase from Leucaena leucocephala using a copper alginate entrapment approach and the application of the laccase in dye decolorization. Process Biochem. 49 1196–1204. 10.1016/j.procbio.2014.04.002 [DOI] [Google Scholar]
- Jaiswal N., Pandey V. P., Dwivedi U. N. (2015). Purification of a thermostable alkaline laccase from papaya (Carica papaya) using affinity chromatography. Int. J. Biol. Macromol. 72 326–332. 10.1016/j.ijbiomac.2014.08.032 [DOI] [PubMed] [Google Scholar]
- Janusz G., Kucharzyk K. H., Pawlik A., Staszczak M., Paszczynski A. J. (2013). Fungal laccase, manganese peroxidase and lignin peroxidase: gene expression and regulation. Enzyme Microb. Technol. 52 1–12. 10.1016/j.enzmictec.2012.10.003 [DOI] [PubMed] [Google Scholar]
- Jeon J. R., Chang Y. S. (2013). Laccase-mediated oxidation of small organics: bifunctional roles for versatile applications. Trends Biotechnol. 31 335–341. 10.1016/j.tibtech.2013.04.002 [DOI] [PubMed] [Google Scholar]
- Jia W., Davies W. J. (2007). Modification of leaf apoplastic ph in relation to stomatal sensitivity to root-sourced abscisic acid signals. Plant Physiol. 143 68–77. 10.1104/pp.106.089110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S. M., Solomon E. I. (2015). Electron transfer and reaction mechanism of laccases. Cell. Mol. Life Sci. 72 869–883. 10.1007/s00018-014-1826-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596 583–589. 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junghanns C., Pecyna M. J., Böhm D., Jehmlich N., Martin C., Von Bergen M., et al. (2009). Biochemical and molecular genetic characterisation of a novel laccase produced by the aquatic ascomycete Phoma sp. UHH 5-1-03. Appl. Microbiol. Biotechnol. 84 1095–1105. 10.1007/s00253-009-2028-2 [DOI] [PubMed] [Google Scholar]
- Kaintz C., Molitor C., Thill J., Kampatsikas I., Michael C., Halbwirth H., et al. (2014). Cloning and functional expression in E. coli of a polyphenol oxidase transcript from Coreopsis grandiflora involved in aurone formation. FEBS Lett. 588 3417–3426. 10.1016/j.febslet.2014.07.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastle J. H. (1910). The oxidases. Treas. Dep. Public Health Mar. Hosp. Serv. U. S. Lab. Bull. 59 7–161. [Google Scholar]
- Kastle J. H., Loevenhart H. S. (1901). On the nature of certain of the oxidizing ferments. Am. Chem. J. 26 539–566. [Google Scholar]
- Khandal H., Singh A. P., Chattopadhyay D. (2020). The microRNA397b-LACCASE2 module regulates root lignification under water and phosphate deficiency. Plant Physiol. 182 1387–1403. 10.1104/pp.19.00921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwai M., Dhar Y. V., Gautam N., Tiwari M., Ahmad I. Z., Asif M. H., et al. (2019). Oryza sativa class III peroxidase (OsPRX38) overexpression in Arabidopsis thaliana reduces arsenic accumulation due to apoplastic lignification. J. Hazard. Mater. 362 383–393. 10.1016/j.jhazmat.2018.09.029 [DOI] [PubMed] [Google Scholar]
- Kiiskinen L., Saloheimo M. (2004). Molecular cloning and expression in Saccharomyces cerevisiae of a laccase gene from the ascomycete melanocarpus albomyces. Appl. Environ. Microbiol. 70 137–144. 10.1128/AEM.70.1.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Yeon Y. J., Choi Y. R., Song W., Pack S. P., Choi Y. S. (2016). A cold-adapted tyrosinase with an abnormally high monophenolase/diphenolase activity ratio originating from the marine archaeon Candidatus Nitrosopumilus koreensis. Biotechnol. Lett. 38 1535–1542. 10.1007/s10529-016-2125-0 [DOI] [PubMed] [Google Scholar]
- Kong K.-H., Hong M.-P., Choi S.-S., Kim Y.-T., Cho S.-H. (2000). Purification and characterization of a highly stable tyrosinase from Thermomicrobium roseum. Biotechnol. Appl. Biochem. 31 113–118. 10.1042/BA19990096 [DOI] [PubMed] [Google Scholar]
- Koutaniemi S., Malmberg H. A., Simola L. K., Teeri T. H. (2015). Norway spruce (Picea abies) laccases: characterization of a laccase in a lignin-forming tissue culture. J. Integr. Plant Biol. 57 341–348. 10.1111/jipb.12333 [DOI] [PubMed] [Google Scholar]
- Kües U. (2015). Fungal enzymes for environmental management. Curr. Opin. Biotechnol. 33 268–278. 10.1016/j.copbio.2015.03.006 [DOI] [PubMed] [Google Scholar]
- Kües U., Ruhl M. (2011). Multiple multi-copper oxidase gene families in basidiomycetes – what for? Curr. Genomics 12 72–94. 10.2174/138920211795564377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Suleski M., Hedges S. B. (2017). TimeTree: a resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 34 1812–1819. 10.1093/molbev/msx116 [DOI] [PubMed] [Google Scholar]
- Kunieda T., Shimada T., Kondo M., Nishimura M., Nishitani K., Hara-Nishimura I. (2013). Spatiotemporal secretion of peroxidase36 is required for seed coat mucilage extrusion in Arabidopsis. Plant Cell 25 1355–1367. 10.1105/tpc.113.110072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahtinen M., Kruus K., Boer H., Kemell M., Andberg M., Viikari L., et al. (2009). The effect of lignin model compound structure on the rate of oxidation catalyzed by two different fungal laccases. J. Mol. Catal. B Enzym. 57 204–210. 10.1016/j.molcatb.2008.09.004 [DOI] [Google Scholar]
- Laitinen T., Morreel K., Delhomme N., Gauthier A., Schiffthaler B., Nickolov K., et al. (2017). A key role for apoplastic H 2 O 2 in Norway spruce phenolic metabolism. Plant Physiol. 174 1449–1475. 10.1104/pp.17.00085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan W., Lu F., Regner M., Zhu Y., Rencoret J., Ralph S. A., et al. (2015). Tricin, a flavonoid monomer in monocot lignification. Plant Physiol. 167 1284–1295. 10.1104/pp.114.253757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bris P., Wang Y., Barbereau C., Antelme S., Cézard L., Legée F., et al. (2019). Inactivation of LACCASE8 and LACCASE5 genes in Brachypodium distachyon leads to severe decrease in lignin content and high increase in saccharification yield without impacting plant integrity. Biotechnol. Biofuels 12:181. 10.1186/s13068-019-1525-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. H., Jeon H. S., Kim S. H., Chung J. H., Roppolo D., Lee H. J., et al. (2019). Lignin-based barrier restricts pathogens to the infection site and confers resistance in plants. EMBO J. 38:e101948. 10.15252/embj.2019101948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Rubio M. C., Alassimone J., Geldner N. (2013). A mechanism for localized lignin deposition in the endodermis. Cell 153 402–412. 10.1016/j.cell.2013.02.045 [DOI] [PubMed] [Google Scholar]
- Leufken C. M., Moerschbacher B. M., Dirks-Hofmeister M. E. (2015). Dandelion PPO-1/PPO-2 domain-swaps: the C-terminal domain modulates the pH optimum and the linker affects SDS-mediated activation and stability. Biochim. Biophys. Acta 1854 178–186. 10.1016/j.bbapap.2014.11.007 [DOI] [PubMed] [Google Scholar]
- Li F.-W., Brouwer P., Carretero-Paulet L., Cheng S., de Vries J., Delaux P.-M., et al. (2018). Fern genomes elucidate land plant evolution and cyanobacterial symbioses. Nat. Plants 4 460–472. 10.1038/s41477-018-0188-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Qin X., Qi J., Dou W., Dunand C., Chen S., et al. (2020). CsPrx25, a class III peroxidase in Citrus sinensis, confers resistance to citrus bacterial canker through the maintenance of ROS homeostasis and cell wall lignification. Hortic. Res. 7 1–11. 10.1038/s41438-020-00415-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wang Y., Jiang H., Deng J. (2009). Crystal structure of Manduca sexta prophenoloxidase provides insights into the mechanism of type 3 copper enzymes. Proc. Natl. Acad. Sci. U.S.A. 106 17002–17006. 10.1073/pnas.0906095106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lige B., Ma S., Van Huystee R. B. (2001). The effects of the site-directed removal of N-glycosylation from cationic peanut peroxidase on its function. Arch. Biochem. Biophys. 386 17–24. 10.1006/abbi.2000.2187 [DOI] [PubMed] [Google Scholar]
- Lin C.-Y., Li Q., Tunlaya-Anukit S., Shi R., Sun Y.-H., Wang J. P., et al. (2016). A cell wall-bound anionic peroxidase, PtrPO21, is involved in lignin polymerization in Populus trichocarpa. Tree Genet. Genomes 12:22. 10.1007/s11295-016-0978-y [DOI] [Google Scholar]
- Liszkay A., Kenk B., Schopfer P. (2003). Evidence for the involvement of cell wall peroxidase in the generation of hydroxyl radicals mediating extension growth. Planta 217 658–667. 10.1007/s00425-003-1028-1 [DOI] [PubMed] [Google Scholar]
- Liu L., Dean J. F. D., Friedman W. E., Eriksson K.-E. L. (1994). A laccase-like phenoloxidase is correlated with lignin biosynthesis in Zinnia elegans stem tissues. Plant J. 6 213–224. 10.1046/j.1365-313X.1994.6020213.x [DOI] [Google Scholar]
- Lu S., Li Q., Wei H., Chang M.-J., Tunlaya-Anukit S., Kim H., et al. (2013). Ptr-miR397a is a negative regulator of laccase genes affecting lignin content in Populus trichocarpa. Proc. Natl. Acad. Sci. U.S.A. 110 10848–10853. 10.1073/pnas.1308936110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna-Acosta A., Saulnier D., Pommier M., Haffner P., de Decker S., Renault T., et al. (2011). First evidence of a potential antibacterial activity involving a laccase-type enzyme of the phenoloxidase system in Pacific oyster Crassostrea gigas haemocytes. Fish Shellfish Immunol. 31 795–800. 10.1016/j.fsi.2011.07.016 [DOI] [PubMed] [Google Scholar]
- Lüthje S., Martinez-Cortes T. (2018). Membrane-bound class III peroxidases: unexpected enzymes with exciting functions. Int. J. Mol. Sci. 19 10–12. 10.3390/ijms19102876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madzak C., Mimmi M. C., Caminade E., Brault A., Baumberger S., Briozzo P., et al. (2006). Shifting the optimal pH of activity for a laccase from the fungus Trametes versicolor by structure-based mutagenesis. Protein Eng. Des. Sel. 19 77–84. 10.1093/protein/gzj004 [DOI] [PubMed] [Google Scholar]
- Maestre-Reyna M., Liu W. C., Jeng W. Y., Lee C. C., Hsu C. A., Wen T. N., et al. (2015). Structural and functional roles of glycosylation in fungal laccase from lentinus sp. PLoS One 10:e0120601. 10.1371/journal.pone.0120601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maijala P., Mattinen M.-L., Nousiainen P., Kontro J., Asikkala J., Sipilä J., et al. (2012). Action of fungal laccases on lignin model compounds in organic solvents. J. Mol. Catal. B Enzym. 76 59–67. 10.1016/j.molcatb.2011.12.009 [DOI] [Google Scholar]
- Majumdar S., Lukk T., Solbiati J. O., Bauer S., Nair S. K., Cronan J. E., et al. (2014). Roles of small laccases from streptomyces in lignin degradation. Biochemistry 53 4047–4058. 10.1021/bi500285t [DOI] [PubMed] [Google Scholar]
- Martínez-García P. J., Crepeau M. W., Puiu D., Gonzalez-Ibeas D., Whalen J., Stevens K. A., et al. (2016). The walnut (Juglans regia) genome sequence reveals diversity in genes coding for the biosynthesis of non-structural polyphenols. Plant J. Cell Mol. Biol. 87 507–532. 10.1111/tpj.13207 [DOI] [PubMed] [Google Scholar]
- Martinière A., Gibrat R., Sentenac H., Dumont X., Gaillard I., Paris N. (2018). Uncovering pH at both sides of the root plasma membrane interface using noninvasive imaging. Proc. Natl. Acad. Sci. U.S.A. 115 6488–6493. 10.1073/pnas.1721769115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins L. O., Durão P., Brissos V., Lindley P. F. (2015). Laccases of prokaryotic origin: enzymes at the interface of protein science and protein technology. Cell. Mol. Life Sci. 72 911–922. 10.1007/s00018-014-1822-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins L. O., Soares C. M., Pereira M. M., Teixeira M., Costa T., Jones G. H., et al. (2002). Molecular and biochemical characterization of a highly stable bacterial laccase that occurs as a structural component of the Bacillus subtilis endospore coat. J. Biol. Chem. 277 18849–18859. 10.1074/jbc.M200827200 [DOI] [PubMed] [Google Scholar]
- Martone P. T., Estevez J. M., Lu F., Ruel K., Denny M. W., Somerville C., et al. (2009). Discovery of lignin in seaweed reveals convergent evolution of cell-wall architecture. Curr. Biol. 19 169–175. 10.1016/j.cub.2008.12.031 [DOI] [PubMed] [Google Scholar]
- Marusek C. M., Trobaugh N. M., Flurkey W. H., Inlow J. K. (2006). Comparative analysis of polyphenol oxidase from plant and fungal species. J. Inorg. Biochem. 100 108–123. 10.1016/j.jinorgbio.2005.10.008 [DOI] [PubMed] [Google Scholar]
- Mason H. S. (1947). The chemistry of melanin. J. Biol. Chem. 168 433–438. 10.1016/S0021-9258(17)30903-1 [DOI] [PubMed] [Google Scholar]
- Mate D. M., Alcalde M. (2015). Laccase engineering: from rational design to directed evolution. Biotechnol. Adv. 33 25–40. 10.1016/j.biotechadv.2014.12.007 [DOI] [PubMed] [Google Scholar]
- Mate D. M., Gonzalez-Perez D., Falk M., Kittl R., Pita M., De Lacey A. L., et al. (2013). Blood tolerant laccase by directed evolution. Chem. Biol. 20 223–231. 10.1016/j.chembiol.2013.01.001 [DOI] [PubMed] [Google Scholar]
- Mayer A. M. (2006). Polyphenol oxidases in plants and fungi: going places? A review. Phytochemistry 67 2318–2331. 10.1016/j.phytochem.2006.08.006 [DOI] [PubMed] [Google Scholar]
- Mbadinga Mbadinga D. L., Li Q., Ranocha P., Martinez Y., Dunand C. (2020). Global analysis of non-animal peroxidases provides insights into the evolution of this gene family in the green lineage. J. Exp. Bot. 71 3350–3360. 10.1093/jxb/eraa141 [DOI] [PubMed] [Google Scholar]
- McCaig B. C., Meagher R. B., Dean J. F. D. (2005). Gene structure and molecular analysis of the laccase-like multicopper oxidase (LMCO) gene family in Arabidopsis thaliana. Planta 221 619–636. 10.1007/s00425-004-1472-6 [DOI] [PubMed] [Google Scholar]
- Mei W., Qin Y., Song W., Li J., Zhu Y. (2009). Cotton GhPOX1 encoding plant class III peroxidase may be responsible for the high level of reactive oxygen species production that is related to cotton fiber elongation. J. Genet. Genomics 36 141–150. 10.1016/S1673-8527(08)60101-0 [DOI] [PubMed] [Google Scholar]
- Ménard D., Blaschek L., Kriechbaum K., Lee C. C., Zhu C., Nuoendagula, et al. (2021). Specific and dynamic lignification at the cell-type level controls plant physiology and adaptability. bioRxiv [Preprint]. 10.1101/2021.06.12.447240 [DOI] [Google Scholar]
- Meslet-Cladiere L., Delage L., Leroux C. J.-J., Goulitquer S., Leblanc C., Creis E., et al. (2013). Structure/function analysis of a type III polyketide synthase in the brown alga Ectocarpus siliculosus reveals a biochemical pathway in phlorotannin monomer biosynthesis. Plant Cell 25 3089–3103. 10.1105/tpc.113.111336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mika A., Lüthje S. (2003). Properties of guaiacol peroxidase activities isolated from corn root plasma membranes. Plant Physiol. 132 1489–1498. 10.1104/pp.103.020396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mika A., Boenisch M. J., Hopff D., Lüthje S. (2010). Membrane-bound guaiacol peroxidases from maize (Zea mays L.) roots are regulated by methyl jasmonate, salicylic acid, and pathogen elicitors. J. Exp. Bot. 61 831–841. 10.1093/jxb/erp353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki Y., Calviño F. R., Pogni R., Giansanti S., Ruiz-Dueñas F. J., Martínez M. J., et al. (2011). Crystallographic, kinetic, and spectroscopic study of the first ligninolytic peroxidase presenting a catalytic tyrosine. J. Biol. Chem. 286 15525–15534. 10.1074/jbc.M111.220996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirdita M., Steinegger M., Söding J. (2019). MMseqs2 desktop and local web server app for fast, interactive sequence searches. Bioinformatics 35 2856–2858. 10.1093/bioinformatics/bty1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molitor C., Mauracher S. G., Rompel A. (2016). Aurone synthase is a catechol oxidase with hydroxylase activity and provides insights into the mechanism of plant polyphenol oxidases. Proc. Natl. Acad. Sci. U.S.A. 113 E1806–E1815. 10.1073/pnas.1523575113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molitor C., Mauracher S. G., Pargan S., Mayer R. L., Halbwirth H., Rompel A. (2015). Latent and active aurone synthase from petals of C. grandiflora: a polyphenol oxidase with unique characteristics. Planta 242 519–537. 10.1007/s00425-015-2261-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottiar Y., Gierlinger N., Jeremic D., Master E. R., Mansfield S. D. (2020). Atypical lignification in eastern leatherwood (Dirca palustris). New Phytol. 226 704–713. 10.1111/nph.16394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moural T. W., Lewis K. M., Barnaba C., Zhu F., Palmer N. A., Sarath G., et al. (2017). Characterization of class III peroxidases from switchgrass. Plant Physiol. 173 417–433. 10.1104/pp.16.01426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller K., Linkies A., Vreeburg R. A. M., Fry S. C., Krieger-Liszkay A., Leubner-Metzger G. (2009). In vivo cell wall loosening by hydroxyl radicals during cress seed germination and elongation growth. Plant Physiol. 150 1855–1865. 10.1104/pp.109.139204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munk L., Sitarz A. K., Kalyani D. C., Mikkelsen J. D., Meyer A. S. (2015). Can laccases catalyze bond cleavage in lignin? Biotechnol. Adv. 33 13–24. 10.1016/j.biotechadv.2014.12.008 [DOI] [PubMed] [Google Scholar]
- Nakamura T. (1958). Purification and physico-chemical properties of laccase. Biochim. Biophys. Acta 30 44–52. 10.1016/0006-3002(58)90239-7 [DOI] [PubMed] [Google Scholar]
- Nakatsuka T., Yamada E., Takahashi H., Imamura T., Suzuki M., Ozeki Y., et al. (2013). Genetic engineering of yellow betalain pigments beyond the species barrier. Sci. Rep. 3:1970. 10.1038/srep01970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T., Yonekura-Sakakibara K., Sato T., Kikuchi S., Fukui Y., Fukuchi-Mizutani M., et al. (2000). Aureusidin synthase: a polyphenol oxidase homolog responsible for flower coloration. Science 290 1163–1166. 10.1126/science.290.5494.1163 [DOI] [PubMed] [Google Scholar]
- Ng T. B., Wang H. X. (2004). A homodimeric laccase with unique characteristics from the yellow mushroom Cantharellus cibarius. Biochem. Biophys. Res. Commun. 313 37–41. 10.1016/j.bbrc.2003.11.087 [DOI] [PubMed] [Google Scholar]
- Nishiyama T., Sakayama H., de Vries J., Buschmann H., Saint-Marcoux D., Ullrich K. K., et al. (2018). The chara genome: secondary complexity and implications for plant terrestrialization. Cell 174 448–464.e24. 10.1016/j.cell.2018.06.033 [DOI] [PubMed] [Google Scholar]
- Ogawa S., Shiro Y., Morishima I. (1979). Calcium binding by horseradish peroxidase c and the heme environmental structure. Biochem. Biophys. Res. Commun. 90 674–678. 10.1016/0006-291X(79)91288-9 [DOI] [PubMed] [Google Scholar]
- Olmedo P., Moreno A. A., Sanhueza D., Balic I., Silva-Sanzana C., Zepeda B., et al. (2018). A catechol oxidase AcPPO from cherimoya (Annona cherimola Mill.) is localized to the Golgi apparatus. Plant Sci. 266 46–54. 10.1016/j.plantsci.2017.10.012 [DOI] [PubMed] [Google Scholar]
- Önnerud H., Zhang L., Gellerstedt G., Henriksson G. (2002). Polymerization of monolignols by redox shuttle – mediated enzymatic oxidation?: a new model in lignin biosynthesis I. Plant Cell 14 1953–1962. 10.1105/tpc.001487.wood [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono E., Hatayama M., Isono Y., Sato T., Watanabe R., Yonekura-Sakakibara K., et al. (2006). Localization of a flavonoid biosynthetic polyphenol oxidase in vacuoles. Plant J. 45 133–143. 10.1111/j.1365-313X.2005.02625.x [DOI] [PubMed] [Google Scholar]
- Onslow M. W. (1920). Oxidising enzymes. II. The nature of the enzymes associated with certain direct oxidising systems in plants. Biochem. J. 14 535–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortner A., Huber D., Haske-Cornelius O., Weber H. K., Hofer K., Bauer W., et al. (2015). Laccase mediated oxidation of industrial lignins: is oxygen limiting? Process Biochem. 50 1277–1283. 10.1016/j.procbio.2015.05.003 [DOI] [Google Scholar]
- Otto B., Schlosser D. (2014). First laccase in green algae: purification and characterization of an extracellular phenol oxidase from Tetracystis aeria. Planta 240 1225–1236. 10.1007/s00425-014-2144-9 [DOI] [PubMed] [Google Scholar]
- Otto B., Beuchel C., Liers C., Reisser W., Harms H., Schlosser D. (2015). Laccase-like enzyme activities from chlorophycean green algae with potential for bioconversion of phenolic pollutants. FEMS Microbiol. Lett. 362 1–8. 10.1093/femsle/fnv072 [DOI] [PubMed] [Google Scholar]
- Pagliarani C., Casolo V., Beiragi M. A., Cavalletto S., Siciliano I., Schubert A., et al. (2019). Priming xylem for stress recovery depends on coordinated activity of sugar metabolic pathways and changes in xylem sap pH. Plant Cell Environ. 42 1775–1787. 10.1111/pce.13533 [DOI] [PubMed] [Google Scholar]
- Palm G. J., Sharma A., Kumari M., Panjikar S., Albrecht D., Jagannadham M. V., et al. (2014). Post-translational modification and extended glycosylation pattern of a plant latex peroxidase of native source characterized by X-ray crystallography. FEBS J. 281 4319–4333. 10.1111/febs.12900 [DOI] [PubMed] [Google Scholar]
- Passardi F., Bakalovic N., Teixeira F. K., Margis-Pinheiro M., Penel C., Dunand C. (2007). Prokaryotic origins of the non-animal peroxidase superfamily and organelle-mediated transmission to eukaryotes. Genomics 89 567–579. 10.1016/j.ygeno.2007.01.006 [DOI] [PubMed] [Google Scholar]
- Perry C. R., Matcham S. E., Wood D. A., Thurston C. F. (1993). The structure of laccase protein and its synthesis by the commercial mushroom Agaricus bisporus. J. Gen. Microbiol. 139 171–178. 10.1099/00221287-139-1-171 [DOI] [PubMed] [Google Scholar]
- Pesquet E., Wagner A., Grabber J. H. (2019). Cell culture systems: invaluable tools to investigate lignin formation and cell wall properties. Curr. Opin. Biotechnol. 56 215–222. 10.1016/j.copbio.2019.02.001 [DOI] [PubMed] [Google Scholar]
- Pesquet E., Zhang B., Gorzsás A., Puhakainen T., Serk H., Escamez S., et al. (2013). Non-cell-autonomous postmortem lignification of tracheary elements in Zinnia elegans. Plant Cell 25 1314–1328. 10.1105/tpc.113.110593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen T. N., Brunak S., von Heijne G., Nielsen H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8 785–786. 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- Pettersen E. F., Goddard T. D., Huang C. C., Meng E. C., Couch G. S., Croll T. I., et al. (2021). UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30 70–82. 10.1002/pro.3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezet R., Perret C., Jean-Denis J. B., Tabacchi R., Gindro K., Viret O. (2003). δ-viniferin, a resveratrol dehydrodimer: one of the major stilbenes synthesized by stressed grapevine leaves. J. Agric. Food Chem. 51 5488–5492. 10.1021/jf030227o [DOI] [PubMed] [Google Scholar]
- Planche L. A. (1820). Experiénces sur les substances qui développent la couleur bleue dans la résine de guïac. J. Pharm. Chim. 6 16–25. [Google Scholar]
- Podgórska A., Burian M., Szal B. (2017). Extra-cellular but extra-ordinarily important for cells: apoplastic reactive oxygen species metabolism. Front. Plant Sci. 8:1353. 10.3389/fpls.2017.01353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourcel L., Routaboul J. M., Cheynier V., Lepiniec L., Debeaujon I. (2007). Flavonoid oxidation in plants: from biochemical properties to physiological functions. Trends Plant Sci. 12 29–36. 10.1016/j.tplants.2006.11.006 [DOI] [PubMed] [Google Scholar]
- Pourcel L., Routaboul J. M., Kerhoas L., Caboche M., Lepiniec L., Debeaujon I. (2005). Transparent Testa10 encodes a laccase-like enzyme involved in oxidative polymerization of flavonoids in Arabidopsis seed coat. Plant Cell 17 2966–2980. 10.1105/tpc.105.035154.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pretzler M., Rompel A. (2017). What causes the different functionality in type-III-copper enzymes? A state of the art perspective. Inorg. Chim. Acta 481 25–31. 10.1016/j.ica.2017.04.041 [DOI] [Google Scholar]
- Queiroz C., Lopes M. L. M., Fialho E., Valente-Mesquita V. L. (2008). Polyphenol oxidase: characteristics and mechanisms of browning control. Food Rev. Int. 24 361–375. 10.1080/87559120802089332 [DOI] [Google Scholar]
- Ralph J., Bunzel M., Marita J. M., Hatfield R. D., Lu F., Kim H., et al. (2004). Peroxidase-dependent cross-linking reactions of p-hydroxycinnamates in plant cell walls. Phytochem. Rev. 3 79–96. 10.1023/B:PHYT.0000047811.13837.fb [DOI] [Google Scholar]
- Ranocha P., Chabannes M., Chamayou S., Jauneau A., Boudet A., Goffner D. (2002). Laccase down-regulation causes alterations in phenolic metabolism and cell wall structure in poplar 1. Plant Physiol. 129 145–155. 10.1104/pp.010988.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranocha P., McDougall G., Hawkins S., Sterjiades R., Borderies G., Stewart D., et al. (1999). Biochemical characterization, molecular cloning and expression of laccases – a divergent gene family – in poplar. Eur. J. Biochem. 259 485–495. 10.1046/j.1432-1327.1999.00061.x [DOI] [PubMed] [Google Scholar]
- Rao M. A., Scelza R., Acevedo F., Diez M. C., Gianfreda L. (2014). Enzymes as useful tools for environmental purposes. Chemosphere 107 145–162. 10.1016/j.chemosphere.2013.12.059 [DOI] [PubMed] [Google Scholar]
- Reiss R., Ihssen J., Richter M., Eichhorn E., Schilling B., Thöny-Meyer L. (2013). Laccase versus laccase-like multi-copper oxidase: a comparative study of similar enzymes with diverse substrate spectra. PLoS One 8:e65633. 10.1371/journal.pone.0065633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing S. A., Goffinet B., Meyberg R., Wu S.-Z., Bezanilla M. (2020). The moss Physcomitrium (Physcomitrella) patens: a model organism for non-seed plants. Plant Cell 32 1361–1376. 10.1105/tpc.19.00828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers C. J., Blanford C. F., Giddens S. R., Skamnioti P., Armstrong F. A., Gurr S. J. (2010). Designer laccases: a vogue for high-potential fungal enzymes? Trends Biotechnol. 28 63–72. 10.1016/j.tibtech.2009.11.001 [DOI] [PubMed] [Google Scholar]
- Rohde A., Morreel K., Ralph J., Goeminne G., Hostyn V., De Rycke R., et al. (2004). Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far-reaching consequences on phenylpropanoid, amino acid, and carbohydrate metabolism. Plant Cell 16 2749–2771. 10.1105/tpc.104.023705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas-Murcia N., Hématy K., Lee Y., Emonet A., Ursache R., Fujita S., et al. (2020). High-order mutants reveal an essential requirement for peroxidases but not laccases in Casparian strip lignification. Proc. Natl. Acad. Sci. U.S.A. 117 29166–29177. 10.1073/pnas.2012728117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F., Teslenko M., van der Mark P., Ayres D. L., Darling A., Höhna S., et al. (2012). MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado T., Bernardo P., Koci K., Coelho A. V., Robalo M. P., Martins L. O. (2012). Methyl syringate: an efficient phenolic mediator for bacterial and fungal laccases. Bioresour. Technol. 124 371–378. 10.1016/j.biortech.2012.08.023 [DOI] [PubMed] [Google Scholar]
- Rosconi F., Fraguas L. F., Martínez-Drets G., Castro-Sowinski S. (2005). Purification and characterization of a periplasmic laccase produced by Sinorhizobium meliloti. Enzyme Microb. Technol. 36 800–807. 10.1016/j.enzmictec.2005.01.003 [DOI] [Google Scholar]
- Ruiz-Dueñas F. J., Morales M., Mate M. J., Romero A., Martínez M. J., Smith A. T., et al. (2008). Site-directed mutagenesis of the catalytic tryptophan environment in Pleurotus eryngii versatile peroxidase. Biochemistry 47 1685–1695. 10.1021/bi7020298 [DOI] [PubMed] [Google Scholar]
- Sánchez-Ferrer Á, Neptuno Rodríguez-López J., García-Cánovas F., García-Carmona F. (1995). Tyrosinase: a comprehensive review of its mechanism. Biochim. Biophys. Acta 1247 1–11. 10.1016/0167-4838(94)00204-T [DOI] [PubMed] [Google Scholar]
- Santhanam N., Vivanco J. M., Decker S. R., Reardon K. F. (2011). Expression of industrially relevant laccases: prokaryotic style. Trends Biotechnol. 29 480–489. 10.1016/j.tibtech.2011.04.005 [DOI] [PubMed] [Google Scholar]
- Sato Y., Demura T., Yamawaki K., Inoue Y., Sato S., Sugiyama M., et al. (2006). Isolation and characterization of a novel peroxidase gene ZPO-C whose expression and function are closely associated with lignification during tracheary element differentiation. Plant Cell Physiol. 47 493–503. 10.1093/pcp/pcj016 [DOI] [PubMed] [Google Scholar]
- Sato Y., Wuli B., Sederoff R., Whetten R. (2001). Molecular cloning and expression of eight laccase cDNAs in loblolly pine (Pinus taeda). J. Plant Res. 114 147–155. 10.1007/PL00013978 [DOI] [Google Scholar]
- Sauter J. J., Ambrosius T. (1986). Changes in the partitioning of carbohydrates in the wood during bud break in Betula pendula Roth. J. Plant Physiol. 124 31–43. 10.1016/S0176-1617(86)80175-4 [DOI] [Google Scholar]
- Savelli B., Li Q., Webber M., Jemmat A. M., Robitaille A., Zamocky M., et al. (2019). RedoxiBase: a database for ROS homeostasis regulated proteins. Redox Biol. 26:101247. 10.1016/j.redox.2019.101247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller A. L., Stout J., Weng J. K., Humphreys J., Ruegger M. O., Chapple C. (2009). Mutations in the cinnamate 4-hydroxylase gene impact metabolism, growth and development in Arabidopsis. Plant J. 60 771–782. 10.1111/j.1365-313X.2009.03996.x [DOI] [PubMed] [Google Scholar]
- Schlosser D., Höffer C. (2002). Laccase-catalyzed oxidation of Mn 2+ in the presence of natural Mn 3+ chelators as a novel source of extracellular H 2 O 2 production and its impact on manganese peroxidase. Appl. Environ. Microbiol. 68 3514–3521. 10.1128/AEM.68.7.3514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M. R., Dziedziech A., Arefin B., Kienzle T., Wang Z., Akhter M., et al. (2019). Insect hemolymph coagulation: kinetics of classically and non-classically secreted clotting factors. Insect Biochem. Mol. Biol. 109 63–71. 10.1016/j.ibmb.2019.04.007 [DOI] [PubMed] [Google Scholar]
- Schönbein C. F. (1856). XVI. On ozone and ozonic actions in mushrooms. Lond. Edinb. Dublin Philos. Mag. J. Sci. 11 137–141. 10.1080/14786445608642035 [DOI] [Google Scholar]
- Schuetz M., Benske A., Smith R. A., Watanabe Y., Tobimatsu Y., Ralph J., et al. (2014). Laccases direct lignification in the discrete secondary cell wall domains of protoxylem. Plant Physiol. 166 798–807. 10.1104/pp.114.245597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M., Sali A. (2006). Statistical potential for assessment and prediction of protein structures. Protein Sci. Publ. Protein Soc. 15 2507–2524. 10.1110/ps.062416606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeto J., Tsutsumi Y. (2016). Diverse functions and reactions of class III peroxidases. New Phytol. 209 1395–1402. 10.1111/nph.13738 [DOI] [PubMed] [Google Scholar]
- Shigeto J., Honjo H., Fujita K., Tsutsumi Y. (2017). Generation of lignin polymer models via dehydrogenative polymerization of coniferyl alcohol and syringyl alcohol via several plant peroxidases involved in lignification and analysis of the resulting DHPs by MALDI-TOF analysis. Holzforschung 72 267–274. 10.1515/hf-2017-0125 [DOI] [Google Scholar]
- Shigeto J., Kiyonaga Y., Fujita K., Kondo R., Tsutsumi Y. (2013). Putative cationic cell-wall-bound peroxidase homologues in arabidopsis, AtPrx2, AtPrx25, and AtPrx71, are involved in lignification. J. Agric. Food Chem. 61 3781–3788. 10.1021/jf400426g [DOI] [PubMed] [Google Scholar]
- Shigeto J., Nagano M., Fujita K., Tsutsumi Y. (2014). Catalytic profile of Arabidopsis peroxidases, AtPrx-2, 25 and 71, contributing to stem lignification. PLoS One 9:e105332. 10.1371/journal.pone.0105332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shleev S., Persson P., Shumakovich G., Mazhugo Y., Yaropolov A., Ruzgas T., et al. (2006). Interaction of fungal laccases and laccase-mediator systems with lignin. Enzyme Microb. Technol. 39 841–847. 10.1016/j.enzmictec.2006.01.010 [DOI] [Google Scholar]
- Skálová T., Dohnálek J., Østergaard L. H., Østergaard P. R., Kolenko P., Dušková J., et al. (2009). The structure of the small laccase from Streptomyces coelicolor reveals a link between laccases and nitrite reductases. J. Mol. Biol. 385 1165–1178. 10.1016/j.jmb.2008.11.024 [DOI] [PubMed] [Google Scholar]
- Smith R. A., Schuetz M., Roach M., Mansfield S. D., Ellis B., Samuels L. (2013). Neighboring parenchyma cells contribute to Arabidopsis xylem lignification, while lignification of interfascicular fibers is cell autonomous. Plant Cell 25 3988–3999. 10.1105/tpc.113.117176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solem E., Tuczek F., Decker H. (2016). Tyrosinase versus catechol oxidase: one asparagine makes the difference. Angew. Chem. Int. Ed. 55 2884–2888. 10.1002/anie.201508534 [DOI] [PubMed] [Google Scholar]
- Solomon E. I., Sundaram U. M., Machonkin T. E. (1996). Multicopper oxidases and oxygenases. Chem. Rev. 96 2563–2606. 10.1021/cr950046o [DOI] [PubMed] [Google Scholar]
- Sorz J., Hietz P. (2006). Gas diffusion through wood: implications for oxygen supply. Trees 20 34–41. 10.1007/s00468-005-0010-x [DOI] [Google Scholar]
- Spicer R., Holbrook N. M. (2005). Within-stem oxygen concentration and sap flow in four temperate tree species: does long-lived xylem parenchyma experience hypoxia? Plant Cell Environ. 28 192–201. 10.1111/j.1365-3040.2004.01262.x [DOI] [Google Scholar]
- Stafford H. A. (2000). “The evolution of phenolics in plants,” in Evolution of Metabolic Pathways, eds J. T. Romeo, R. Ibrahim, L. Varin, and V. De Luca (Amsterdam: Elsevier Science Ltd; ), 25–54. 10.1016/S0079-9920(00)80003-9 [DOI] [Google Scholar]
- Starrach N., Mayer W.-E. (1989). Changes of the apoplastic pH and K+ concentration in the phaseolus pulvinus in situ in relation to rhythmic leaf movements. J. Exp. Bot. 40 865–873. 10.1093/jxb/40.8.865 12432039 [DOI] [Google Scholar]
- Stewart R. J., Sawyer B. J. B., Bucheli C. S., Robinson S. P. (2001). Polyphenol oxidase is induced by chilling and wounding in pineapple. Funct. Plant Biol. 28 181–191. 10.1071/pp00094 [DOI] [Google Scholar]
- Strittmatter E., Wachter S., Liers C., Ullrich R., Hofrichter M., Plattner D. A., et al. (2013). Radical formation on a conserved tyrosine residue is crucial for DyP activity. Arch. Biochem. Biophys. 537 161–167. 10.1016/j.abb.2013.07.007 [DOI] [PubMed] [Google Scholar]
- Sullivan M. L. (2015). Beyond brown: polyphenol oxidases as enzymes of plant specialized metabolism. Front. Plant Sci. 5:783. 10.3389/fpls.2014.00783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Liu X., Yang J., Liu W., Du Q., Wang H., et al. (2018). MicroRNA528 affects lodging resistance of maize by regulating lignin biosynthesis under nitrogen-luxury conditions. Mol. Plant 11 806–814. 10.1016/j.molp.2018.03.013 [DOI] [PubMed] [Google Scholar]
- Sundell D., Mannapperuma C., Netotea S., Delhomme N., Lin Y.-C., Sjödin A., et al. (2015). The plant genome integrative explorer resource: plantgenIE.org. New Phytol. 208 1149–1156. 10.1111/nph.13557 [DOI] [PubMed] [Google Scholar]
- Swetha C., Basu D., Pachamuthu K., Tirumalai V., Nair A., Prasad M., et al. (2018). Major domestication-related phenotypes in indica rice are due to loss of miRNA-mediated laccase silencing. Plant Cell 30 2649–2662. 10.1105/tpc.18.00472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szent-Györgyi A. (1925). Zellatmung. IV. Mitteilung: Über den oxydationsmechanismus der Kartoffeln. Biochem. Z. 162 399–412. [Google Scholar]
- Szent-Györgyi A. (1930). On the mechanism of biological oxidation and the function of the suprarenal gland. Science 72 125–126. [DOI] [PubMed] [Google Scholar]
- Tadesse M. A., D’Annibale A., Galli C., Gentili P., Sergi F. (2008). An assessment of the relative contributions of redox and steric issues to laccase specificity towards putative substrates. Org. Biomol. Chem. 6:868. 10.1039/b716002j [DOI] [PubMed] [Google Scholar]
- Takahama U., Oniki T., Shimokawa H. (1996). A possible mechanism for the oxidation of sinapyl alcohol by peroxidase-dependent reactions in the apoplast: enhancement of the oxidation by hydroxycinnamic acids and components of the apoplast. Plant Cell Physiol. 37 499–504. 10.1093/oxfordjournals.pcp.a028972 [DOI] [Google Scholar]
- Tan K. P., Nguyen T. B., Patel S., Varadarajan R., Madhusudhan M. S. (2013). Depth: a web server to compute depth, cavity sizes, detect potential small-molecule ligand-binding cavities and predict the pKa of ionizable residues in proteins. Nucleic Acids Res. 41 W314–W321. 10.1093/nar/gkt503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor G., Davies W. J. (1985). The control of leaf growth of betula and acer by photoenvironment. New Phytol. 101 259–268. 10.1111/j.1469-8137.1985.tb02833.x [DOI] [Google Scholar]
- Telke A. A., Kagalkar A. N., Jagtap U. B., Desai N. S., Bapat V. A., Govindwar S. P. (2011). Biochemical characterization of laccase from hairy root culture of Brassica juncea L. and role of redox mediators to enhance its potential for the decolorization of textile dyes. Planta 234 1137–1149. 10.1007/s00425-011-1469-x [DOI] [PubMed] [Google Scholar]
- Terashima N., Fukushima K. (1988). Heterogeneity in formation of lignin-XI: an autoradiographic study of the heterogeneous formation and structure of pine lignin. Wood Sci. Technol. 22 259–270. 10.1007/BF00386021 [DOI] [Google Scholar]
- Terashima N., Yoshida M., Hafrén J., Fukushima K., Westermark U. (2012). Proposed supramolecular structure of lignin in softwood tracheid compound middle lamella regions. Holzforschung 66 907–915. 10.1515/hf-2012-0021 [DOI] [Google Scholar]
- Terwilliger N. B., Ryan M. C. (2006). Functional and phylogenetic analyses of phenoloxidases from brachyuran (Cancer magister) and branchiopod (Artemia franciscana, Triopslongicaudatus) crustaceans. Biol. Bull. 210 38–50. 10.2307/4134535 [DOI] [PubMed] [Google Scholar]
- Thipyapong P., Steffens J. C. (1997). Tomato polyphenol oxidase (differential response of the polyphenol oxidase F promoter to injuries and wound signals). Plant Physiol. 115 409–418. 10.1104/pp.115.2.409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thipyapong P., Hunt M. D., Steffens J. C. (2004). Antisense downregulation of polyphenol oxidase results in enhanced disease susceptibility. Planta 220 105–117. 10.1007/s00425-004-1330-6 [DOI] [PubMed] [Google Scholar]
- Tian W., Chen C., Lei X., Zhao J., Liang J. (2018). CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res. 46 W363–W367. 10.1093/nar/gky473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tocher R. D., Tocher R. D., Meeuse B. J. D. (1966). Enzymes of marine algae: i. Studies on phenolase in the green alga, Monostroma fuscum. Can. J. Bot. 44 551–561. 10.1139/b66-067 33356898 [DOI] [Google Scholar]
- Tohge T., De Souza L. P., Fernie A. R. (2017). Current understanding of the pathways of flavonoid biosynthesis in model and crop plants. J. Exp. Bot. 68 4013–4028. 10.1093/jxb/erx177 [DOI] [PubMed] [Google Scholar]
- Tran L. T., Constabel C. P. (2011). The polyphenol oxidase gene family in poplar: phylogeny, differential expression and identification of a novel, vacuolar isoform. Planta 234 799–813. 10.1007/s00425-011-1441-9 [DOI] [PubMed] [Google Scholar]
- Tran L. T., Taylor J. S., Constabel C. P. (2012). The polyphenol oxidase gene family in land plants: lineage-specific duplication and expansion. BMC Genomics 13:395. 10.1186/1471-2164-13-395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traube M. (1877). Die chemische theorie der Fermentwirkungen und der Chemismus der respiration. Antwort auf die Aeusserungen des Hrn. Hoppe-Seyler. Berichte Dtsch. Chem. Ges. 10 1984–1992. [Google Scholar]
- Treutter D. (2006). Significance of flavonoids in plant resistance: a review. Environ. Chem. Lett. 4 147–157. 10.1007/s10311-006-0068-8 [DOI] [Google Scholar]
- Turlapati P. V., Kim K. W., Davin L. B., Lewis N. G. (2011). The laccase multigene family in Arabidopsis thaliana: towards addressing the mystery of their gene function(s). Planta 233 439–470. 10.1007/s00425-010-1298-3 [DOI] [PubMed] [Google Scholar]
- Uthandi S., Saad B., Humbard M. A., Maupin-Furlow J. A. (2010). LccA, an archaeal laccase secreted as a highly stable glycoprotein into the extracellular medium by Haloferax volcanii. Appl. Environ. Microbiol. 76 733–743. 10.1128/AEM.01757-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergara R., Parada F., Rubio S., Pérez F. J. (2012). Hypoxia induces H2O2 production and activates antioxidant defence system in grapevine buds through mediation of H2O2 and ethylene. J. Exp. Bot. 63 4123–4131. 10.1093/jxb/ers094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vite-Vallejo O., Palomares L. A., Dantán-González E., Ayala-Castro H. G., Martínez-Anaya C., Valderrama B., et al. (2009). The role of N-glycosylation on the enzymatic activity of a Pycnoporus sanguineus laccase. Enzyme Microb. Technol. 45 233–239. 10.1016/j.enzmictec.2009.05.007 [DOI] [Google Scholar]
- Waltz E. (2015). Nonbrowning GM apple cleared for market. Nat. Biotechnol. 33 326–327. 10.1038/nbt0415-326c [DOI] [PubMed] [Google Scholar]
- Wang C. Y., Zhang S., Yu Y., Luo Y. C., Liu Q., Ju C., et al. (2014). MiR397b regulates both lignin content and seed number in Arabidopsis via modulating a laccase involved in lignin biosynthesis. Plant Biotechnol. J. 12 1132–1142. 10.1111/pbi.12222 [DOI] [PubMed] [Google Scholar]
- Wang L.-G., Lam T. T.-Y., Xu S., Dai Z., Zhou L., Feng T., et al. (2020). Treeio: an R package for phylogenetic tree input and output with richly annotated and associated data. Mol. Biol. Evol. 37 599–603. 10.1093/molbev/msz240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Hebert D. N. (2006). Tyrosinase maturation through the mammalian secretory pathway: bringing color to life. Pigment Cell Res. 19 3–18. 10.1111/j.1600-0749.2005.00288.x [DOI] [PubMed] [Google Scholar]
- Wang X., Zhuo C., Xiao X., Wang X., Docampo-Palacios M., Chen F., et al. (2020). Substrate specificity of LACCASE8 facilitates polymerization of caffeyl alcohol for C-lignin biosynthesis in the seed coat of Cleome hassleriana. Plant Cell 32 3825–3845. 10.1105/tpc.20.00598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wang Q., Zhao Y., Han G., Zhu S. (2015b). Systematic analysis of maize class III peroxidase gene family reveals a conserved subfamily involved in abiotic stress response. Gene 566 95–108. 10.1016/j.gene.2015.04.041 [DOI] [PubMed] [Google Scholar]
- Wang Y., Bouchabke-Coussa O., Lebris P., Antelme S., Soulhat C., Gineau E., et al. (2015a). LACCASE5 is required for lignification of the Brachypodium distachyon culm. Plant Physiol. 168 192–204. 10.1104/pp.114.255489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wariishi H., Valli K., Gold M. H. (1991). In vitro depolymerisation of lignin by manganese peroxidase of Phanerochaete chrysosporium. Biochem. Biophys. Res. Commun. 176 269–275. 10.1016/0006-291X(91)90919-X [DOI] [PubMed] [Google Scholar]
- Webb B., Sali A. (2016). Comparative protein structure modeling using modeller. Curr. Protoc. Bioinformatics 54 5.6.1–5.6.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei D., Houtman C. J., Kapich A. N., Hunt C. G., Cullen D., Hammel K. E. (2010). Laccase and its role in production of extracellular reactive oxygen species during wood decay by the brown rot basidiomycete postia placenta. Appl. Environ. Microbiol. 76 2091–2097. 10.1128/AEM.02929-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welinder K. G. (1992). Superfamily of plant, fungal and bacterial peroxidases. Curr. Opin. Struct. Biol. 2 388–393. 10.1016/0959-440X(92)90230-5 [DOI] [Google Scholar]
- Welinder K. G., Justesen A. F., Kjærsgård I. V. H., Jensen R. B., Rasmussen S. K., Jespersen H. M., et al. (2002). Structural diversity and transcription of class III peroxidases from Arabidopsis thaliana. Eur. J. Biochem. 269 6063–6081. 10.1046/j.1432-1033.2002.03311.x [DOI] [PubMed] [Google Scholar]
- Weng J.-K., Chapple C. (2010). The origin and evolution of lignin biosynthesis. New Phytol. 187 273–285. 10.1111/j.1469-8137.2010.03327.x [DOI] [PubMed] [Google Scholar]
- Weng J.-K., Mo H., Chapple C. (2010). Over-expression of F5H in COMT-deficient Arabidopsis leads to enrichment of an unusual lignin and disruption of pollen wall formation. Plant J. 64 898–911. 10.1111/j.1365-313X.2010.04391.x [DOI] [PubMed] [Google Scholar]
- Wilkinson S., Davies W. J. (1997). Xylem sap pH increase: a drought signal received at the apoplastic face of the guard cell that involves the suppression of saturable abscisic acid uptake by the epidermal symplast. Plant Physiol. 113 559–573. 10.1104/pp.113.2.559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T., Liu Z., Wang G. (2020). Structural basis for monolignol oxidation by a maize laccase. Nat. Plants 6 231–237. 10.1038/s41477-020-0595-5 [DOI] [PubMed] [Google Scholar]
- Xu F. (1997). Effects of redox potential and hydroxide inhibition on the pH activity profile of fungal laccases. J. Biol. Chem. 272 924–928. 10.1074/jbc.272.2.924 [DOI] [PubMed] [Google Scholar]
- Xu F. (2001). Dioxygen reactivity of laccase. Appl. Biochem. Biotechnol. 95:125. [DOI] [PubMed] [Google Scholar]
- Xu F., Berka R. M., Wahleithner J. A., Nelson B. A., Shuster J. R., Brown S. H., et al. (1998). Site-directed mutations in fungal laccase: effect on redox potential, activity and pH profile. Biochem. J. 334 63–70. 10.1042/bj3340063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F., Palmer A. E., Yaver D. S., Berka R. M., Gambretta G. A., Brown S. H., et al. (1999). Targeted mutations in a Trametes villosa laccase. Biochemistry 274 12372–12375. 10.1074/jbc.274.18.12372 [DOI] [PubMed] [Google Scholar]
- Xu F., Shin W., Brown S. H., Wahleithner J. A., Sundaram U. M., Solomon E. I. (1996). A study of a series of recombinant fungal laccases and bilirubin oxidase that exhibit significant differences in redox potential, substrate specificity, and stability. Biochim. Biophys. Acta 1292 303–311. 10.1016/0167-4838(95)00210-3 [DOI] [PubMed] [Google Scholar]
- Yamamoto M., Blaschek L., Subbotina E., Kajita S., Pesquet E. (2020). Importance of lignin coniferaldehyde residues for plant properties and sustainable uses. ChemSusChem 13 4400–4408. 10.1002/cssc.202001242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K., Yamamura M., Matsuda F., Ono E., Nakabayashi R., Sugawara S., et al. (2020). Seed-coat protective neolignans are produced by the dirigent protein AtDP1 and the laccase AtLAC5 in Arabidopsis. Plant Cell 33 129–152. 10.1093/plcell/koaa014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H. (1883). Chemistry of lacquer (Urushi). Part I. J. Chem. Soc. Trans. 43 472–486. 10.1039/ct8834300472 [DOI] [Google Scholar]
- Yoshitake A., Katayama Y., Nakamura M., Iimura Y., Kawai S., Morohoshi N. (1993). N-linked carbohydrate chains protect laccase III from proteolysis in Coriolus versicolor. J. Gen. Microbiol. 139 179–185. 10.1099/00221287-139-1-179 [DOI] [Google Scholar]
- Yu G., Smith D. K., Zhu H., Guan Y., Lam T. T.-Y. (2017). ggtree: an r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 8 28–36. 10.1111/2041-210X.12628 [DOI] [Google Scholar]
- Yu Y., Tang T., Qian Q., Wang Y., Yan M., Zeng D., et al. (2008). Independent losses of function in a polyphenol oxidase in rice: differentiation in grain discoloration between subspecies and the role of positive selection under domestication. Plant Cell 20 2946–2959. 10.1105/tpc.108.060426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z.-H., Han Y.-N., Xiao X.-G. (2015). A PPO promoter from betalain-producing red swiss chard, directs petiole- and root-preferential expression of foreign gene in anthocyanins-producing plants. Int. J. Mol. Sci. 16 27032–27043. 10.3390/ijms161126011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P. G., Huang S. Z., Pin A.-L., Adams K. L. (2010). Extensive divergence in alternative splicing patterns after gene and genome duplication during the evolutionary history of Arabidopsis. Mol. Biol. Evol. 27 1686–1697. 10.1093/molbev/msq054 [DOI] [PubMed] [Google Scholar]
- Zhao Q., Nakashima J., Chen F., Yin Y., Fu C., Yun J., et al. (2013). LACCASE is necessary and nonredundant with PEROXIDASE for lignin polymerization during vascular development in Arabidopsis. Plant Cell 25 3976–3987. 10.1105/tpc.113.117770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Lin S., Qiu Z., Cao D., Wen J., Deng X., et al. (2015). MicroRNA857 is involved in the regulation of secondary growth of vascular tissues in Arabidopsis. Plant Physiol. 169 2539–2552. 10.1104/pp.15.01011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipor G., Duarte P., Carqueijeiro I., Shahar L., Ovadia R., Teper-Bamnolker P., et al. (2015). In planta anthocyanin degradation by a vacuolar class III peroxidase in Brunfelsia calycina flowers. New Phytol. 205 653–665. 10.1111/nph.13038 [DOI] [PubMed] [Google Scholar]
- Zumarraga M., Dominguez C., Camarero S., Shleev S., Polaina J., Martinez-Arias A., et al. (2008). Combinatorial saturation mutagenesis of the myceliophthora thermophila laccase T2 mutant: the connection between the C-terminal plug and the conserved VSG tripeptide. Comb. Chem. High Throughput Screen 11 807–816. 10.2174/138620708786734235 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Animations of the predicted protein structures for AtLAC12, AtLAC15, ADE/LAC, and ChLAC8. The protein surface is coloured by its distance to the T1 copper, the binding pocket volume is shown in yellow. Residues shaping the binding pocket mouth are shown in pink and labelled. The two histidines coordinating the T1 copper are shown in blue.