Abstract
Primordial germ cells (PGCs) are common ancestors of all germline cells. However, mechanistic understanding of how PGC specification occurs is limited. Here, we identified transcription factor CP2-like 1 (Tfcp2l1), an important pluripotency factor, as a pivotal factor for PGC-like cell (PGCLC) specification. High-throughput sequencing and quantitative real-time PCR analysis showed that Tfcp2l1 expression is gradually increased during mouse and human epiblast differentiation into PGCLCs in vivo and in vitro. Consequently, overexpression of Tfcp2l1 can enhance the specification efficiency even without inductive cytokines in mouse epiblast-like cells derived from embryonic stem cells, while knockdown of Tfcp2l1 significantly inhibits PGCLC generation. Mechanistic studies revealed that Tfcp2l1 exerts its function partially through the direct induction of PR domain zinc finger protein 14, a key PGC marker, as downregulation of the PR domain zinc finger protein 14 transcript can impair the ability of Tfcp2l1 to direct PGCLC commitment. Importantly, we finally demonstrated that the crucial role of the human homolog Tfcp2l1 in promoting PGCLC specification is conserved in human pluripotent stem cells. Together, our data uncover a novel function of Tfcp2l1 in PGCLC fate determination and facilitate a better understanding of germ cell development.
Keywords: embryonic stem cells, primordial germ cell–like cells, TFCP2l1, Prdm14
Abbreviations: Blimp1, B lymphocyte–induced maturation protein-1; BMP, bone morphogenetic protein; ChIP, chromatin immunoprecipitation; DOX, doxycycline; EGF, epidermal growth factor; EpiLCs, epiblast-like cells; EpiSCs, epiblast stem cells; ESCs, embryonic stem cells; FACS, fluorescence-activated cell sorting; i-Tfcp2l1, inducible Tfcp2l1; iMeLCs, incipient mesoderm-like cells; iPSCs, induced pluripotent stem cells; KSR, KnockOut Serum Replacement; LIF, leukemia inhibitory factor; Oct4, octamer-binding transcription factor 4; PB, PiggyBac; PGCLCs, PGC-like cells; PGCs, primordial germ cells; Prdm14, PR domain zinc finger protein 14; qRT-PCR, quantitative real-time PCR; ROCK, Rho-associated protein kinase; SCF, stem cell factor; Tfap2c, transcription factor AP-2 gamma; Tfcp2l1, transcription factor CP2-like 1; Wnt, wingless/integrated
In mammals, all gametes, including sperm and oocytes, originate from primordial germ cells (PGCs), which were first discovered at the posterior end of the primitive streak in the extraembryonic mesoderm at embryonic day 7.25 in mice (1). Deciphering the mechanisms involved in PGC specification and development is important for understanding the associated diseases and infertility. To date, gene KO studies using KO mice have identified several essential inductive signals for the fate of PGCs, such as the bone morphogenetic protein (BMP) and wingless/integrated (Wnt) pathways (2, 3, 4), and many downstream core transcription factors, among which PR domain zinc finger protein 14 (Prdm14), B lymphocyte–induced maturation protein-1 (Blimp1, also named Prdm1), and transcription factor AP-2 gamma (Tfap2c, also named Ap2-γ) are the most critical in mice (5, 6, 7). BMP4 secreted from extraembryonic ectoderm is sufficient to trigger Blimp1 and Prdm14 expression in the epiblast and to induce the formation of PGC-like cells (PGCLCs) (2, 4). Meanwhile, Wnt3 is able to enable epiblast cells to respond to BMP4 to form PGCs via Brachyury (4), which functions downstream of Wnt3 by directly inducing Prdm1 synergistically with BMP4 (8). In fact, the specification of PGCs is a very complicated process, and the molecular mechanisms are poorly understood, especially for human PGCs because of technical and ethical obstacles to obtaining such cells from early embryos.
Identification and functional assessment of PGC commitment–associated pathways and factors requires an in vitro model, for which the most appropriate approach is the induction of PGCLCs from pluripotent stem cells in vitro, including embryonic stem cells (ESCs), derived from the inner cell mass of the blastocyst and induced pluripotent stem cells (iPSCs), reprogrammed from somatic cells (9, 10, 11, 12, 13). The initial work was tested in mouse ESCs and iPSCs (14). In this procedure, mouse ESCs and iPSCs were first converted into epiblast-like cells (EpiLCs) through exposure to activin A and basic fibroblast growth factor for 2 days, and then, these cells were further differentiated into PGCLCs in response to cytokines for 4 to 6 days, principally BMP4 and leukemia inhibitory factor (LIF) (14). These mouse PGCLCs exhibited analogous transcriptomic and epigenetic profiles compared with those of E9.5 migratory mouse PGCs in vivo (14). With this PGCLC specification system, Prdm14 alone or in combination with Blimp1 and Tfap2c is sufficient to efficiently direct EpiLCs, but not ESCs, into a PGCLC state even in the absence of cytokines (15). Notably, human pluripotent stem cells differ from mouse ESCs and iPSCs (9, 10, 11) but are similar to mouse epiblast stem cells (EpiSCs) (16, 17). Unlike the formation process of mouse PGCLCs, human iPSCs first have to transform into incipient mesoderm-like cells (iMeLCs) stimulated by activin A, Rho-associated protein kinase (ROCK) inhibitor, and Wnt signaling agonist for 2 days (18). After this treatment, iMeLCs robustly generated human PGCLCs corresponding to week 7 human PGCs in vivo when cultured under BMP4, LIF, stem cell factor (SCF), and epidermal growth factor (EGF) conditions for 4 days (18). Together, based on stem cells as a model, more regulators important for PGCLC development need to be explored.
Many stem cell pluripotency factors have been demonstrated to be essential for the survival and formation of PGCLCs, such as octamer-binding transcription factor 4 (Oct4) (19) and Nanog (20). More recently, we identified transcription factor CP2-like 1 (Tfcp2l1) as a key marker of pluripotency and a critical gene for the maintenance of the undifferentiated state of mouse and human ESCs (21, 22, 23), whereas its function is still unclear during PGCLC commitment. For the first time, this project found that Tfcp2l1 transcripts were gradually increased when epiblast cells became PGCs in vivo and in vitro. Therefore, overexpression of Tfcp2l1 was able to facilitate the differentiation of mouse and human pluripotent stem cells into PGCLCs. These results provide a new perspective for people to understand the regulatory network of PGCLC specification.
Results
The transcription of Tfcp2l1 is increased during mouse PGC formation in vivo and in vitro
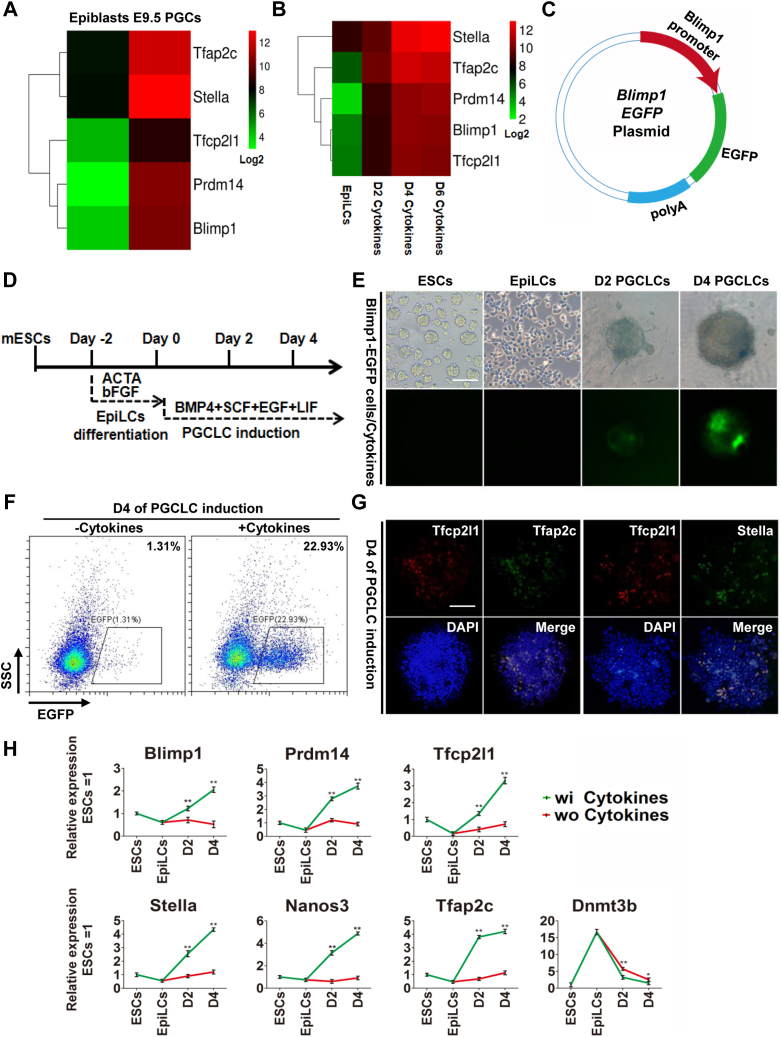
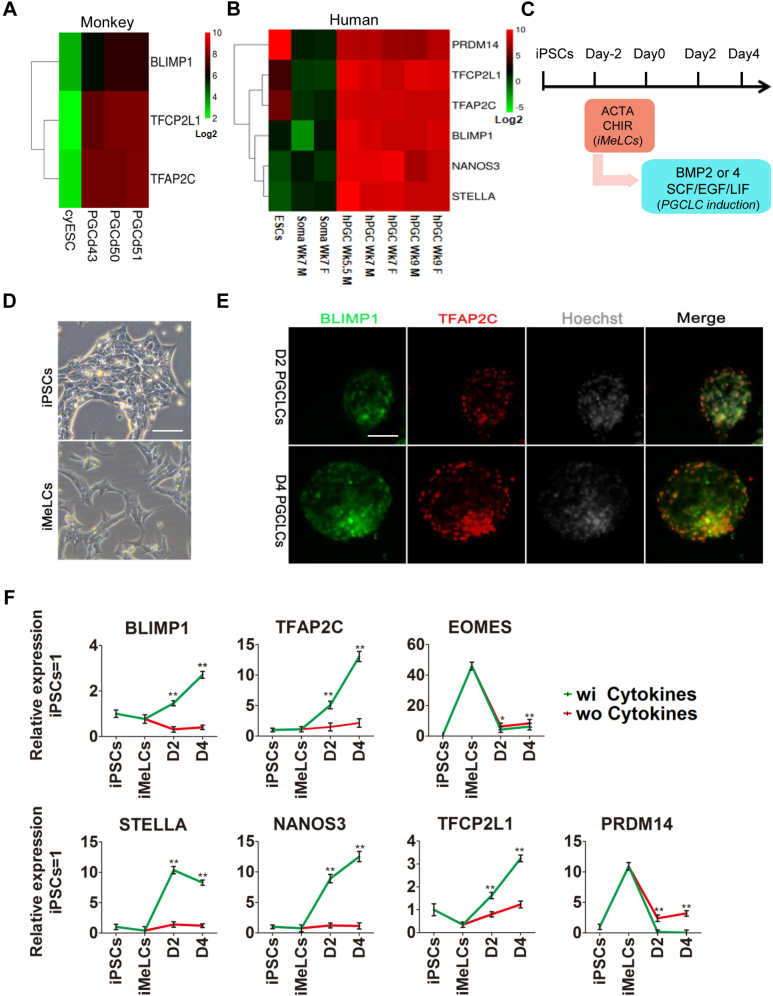
To compare the expression pattern of Tfcp2l1 during PGC specification in vivo, we first analyzed the transcript data in mouse epiblasts and E9.5 PGCs (GEO: GSE46855) (14). The results showed that E9.5 PGCs exhibited higher transcript levels of Tfcp2l1 and the PGC markers Tfap2c, Prdm14, Blimp1, and Stella than epiblasts (Fig. 1A). In addition, E11.5 and E12.5 male and female PGCs, testis, liver, and heart also expressed higher levels of Tfcp2l1 than R1 mouse ESCs and other adult tissues, such as the brain, spleen, and thymus (Fig. S1, A–C) (24).
Figure 1.
Tfcp2l1 is upregulated in PGCs.A, heatmap showing the expression of Tfcp2l1 and PGC markers in mouse epiblasts and E9.5 PGCs. B, heatmap revealing the levels of Tfcp2l1 and PGC markers during the conversion of EpiLC into PGCLCs. C, the construction of the Blimp1-EGFP report plasmid. D, schematic diagram of mouse ESC differentiation into PGCLCs. E, green fluorescence changes during the process of mouse PGCLC induction in Blimp1-EGFP cells. The scale bar represents 100 μm. F, FACS analysis of Blimp1-EGFP cells on day 4 embryoids after mouse PGCLC induction. The percentage of EGFP expression has been indicated. G, immunofluorescence analysis of Tfcp2l1 (red), Tfap2c (green), and Stella (green) in day 4 mouse PGCLC embryoids. The scale bar represents 100 μm. H, qRT-PCR analysis of the expression levels of Tfcp2l1 and several core genes of mouse PGCLCs treated with (wi) or without (wo) cytokines, including BMP4, LIF, SCF, and EGF. Data are the mean ± SD (N = 3 biological replicates). ∗p < 0.05, ∗∗p < 0.01 versus wo cytokines. D2, day 2; D4, day 4; EpiLCs, epiblast-like cells; LIF, leukemia inhibitory factor; PGCLCs, PGC-like cells; PGCs, primordial germ cells; SCF, stem cell factor; Tfcp2l1, transcription factor CP2-like 1.
Next, we analyzed a previous high-throughput sequencing result of PGCLC specification and found that the expression of Tfcp2l1 was also gradually upregulated during EpiLCs differentiation into PGCLCs (GEO: GSE46855) (14) (Fig. 1B). To validate these data in vitro, we established a mouse ESC cell line transfected with a construct in which the expression of the EGFP gene was driven by a DNA sequence representing the mouse Blimp1 reporter (Fig. 1C). When ESCs differentiated into PGCLCs by the addition of BMP4, LIF, SCF, and EGF, the intensity of the Blimp1-EGFP reporter increased progressively until day 4, as analyzed by the fluorescence microscopy and fluorescence-activated cell sorting (FACS) (Fig. 1, D–F). Immunofluorescence staining showed that most of the Tfcp2l1-positive cells coexpressed the PGC marker genes Tfap2c and Stella (Fig. 1G). In addition, quantitative real-time PCR (qRT-PCR) detection revealed that Tfcp2l1 had an expression pattern similar to those of the PGC markers Prdm14, Blimp1, Tfap2c, Nanos3, and Stella (Fig. 1H). They were decreased significantly when mouse ESCs differentiated into EpiLCs but gradually increased when PGCLCs emerged (Fig. 1H). In contrast, the epigenetic modifier Dnmt3b showed opposite expression profile (Fig. 1H). Collectively, these data indicate that Tfcp2l1 is highly expressed in PGCLCs during early embryonic development.
Tfcp2l1 enhances the generation of PGCLCs from mouse ESCs
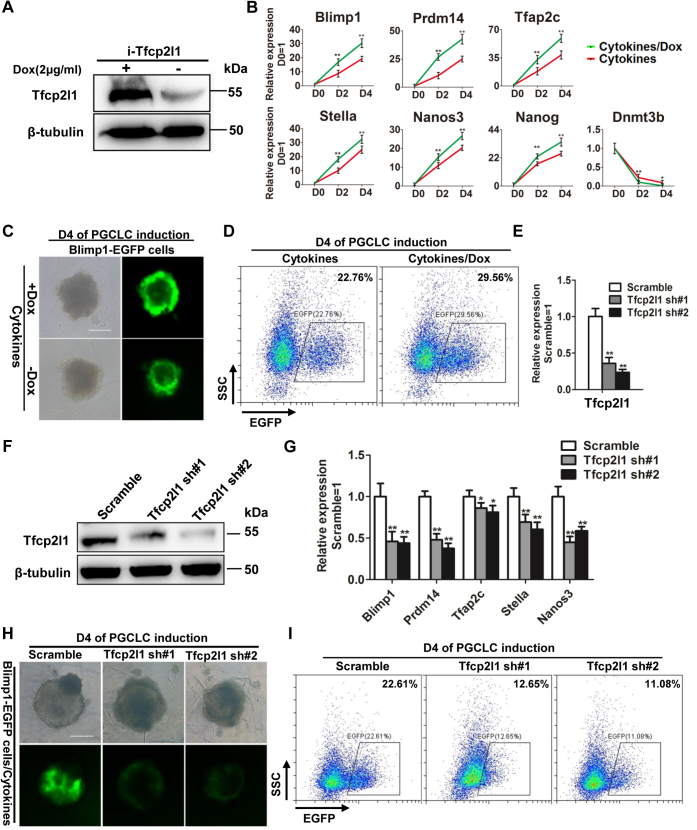
To investigate the function of Tfcp2l1 in PGCLC generation, we used a mouse ESC line harboring a doxycycline (DOX)-inducible Tfcp2l1 (i-Tfcp2l1) transgene (21), in which Tfcp2l1 expression could be efficiently induced after the addition of DOX (Fig. 2A). i-Tfcp2l1 ESCs were first differentiated into EpiLCs, and then BMP4, LIF, SCF, and EGF were further supplemented to stimulate PGCLC formation in the presence or absence of DOX for 4 days. The expression levels of PGC markers were assessed by qRT-PCR and FACS, and the results showed that DOX treatment increased key genes of PGC specification, Blimp1, Prdm14, Tfap2c, Nanos3, Stella, and Nanog (Fig. 2, B–D), indicating that upregulation of Tfcp2l1 is capable of promoting mouse ESC differentiation into PGCLCs. Next, to evaluate whether Tfcp2l1 is necessary for PGCLC generation, we designed two shRNAs specific for mouse Tfcp2l1 mRNA (Tfcp2l1 shRNA) with a lentivirus system. Compared with scramble control cells, Tfcp2l1 transcripts were knocked down by approximately 70 to 80% at the mRNA and protein levels in 46C mouse ESCs after infection with Tfcp2l1 shRNA lentiviruses (Fig. 2, E and F). Tfcp2l1–shRNA mouse ESCs-derived PGCLCs exhibited lower levels of the PGC markers Blimp1, Prdm14, Tfap2c, Stella, and Nanos3 than scramble control cells (Fig. 2, G–I), suggesting that downregulation of Tfcp2l1 limits the ability of mouse ESCs to convert into PGCLCs. Taken together, these data demonstrated that Tfcp2l1 is important for PGCLC specification.
Figure 2.
Tfcp2l1 is important for the generation of PGCLCs from mouse ESCs.A, Western blot analysis of Tfcp2l1 protein in i-Tfcp2l1 ESCs in the presence or absence of doxycycline (DOX) for 24 h. β-Tubulin was used as a loading control. B, qRT-PCR analysis of the expression levels of PGC markers under the treatment of PGCLC-inductive cytokines with or without DOX. Data are the mean ± SD (N = 3 biological replicates). ∗p < 0.05, ∗∗p < 0.01 versus cytokines. C, in Blimp1-EGFP transfected i-Tfcp2l1 ESCs, the fluorescence intensity of PGCLCs was induced by BMP4, LIF, SCF, and EGF in the presence or absence of 2 μg/ml DOX. The scale bar represents 100 μm. D, FACS analysis of Blimp1-EGFP on day 4 in i-Tfcp2l1 cell–derived PGCLCs in the presence or absence of DOX. E, qRT-PCR analysis of mouse Tfcp2l1 expression in 46C mouse ESCs infected with scramble or Tfcp2l1 shRNA lentiviruses. Data are the mean ± SD (N = 3 biological replicates). ∗∗p < 0.01 versus scramble. F, Western blot analysis of the Tfcp2l1 protein levels in scramble and Tfcp2l1 shRNA 46C mouse ESCs. G, qRT-PCR analysis of mouse PGC genes in scramble and Tfcp2l1 shRNA PGCLCs induced by BMP4, LIF, SCF, and EGF. Data are the mean ± SD (N = 3 biological replicates). ∗p < 0.05, ∗∗p < 0.01 versus scramble. H, green fluorescence changes after 4 days of PGCLC induction in Blimp1-EGFP cells infected with scramble or mouse Tfcp2l1 shRNA lentiviruses. The scale bar represents 100 μm. I, FACS analysis of Blimp1-EGFP in day 4 embryoids infected with scramble or Tfcp2l1 shRNA lentiviruses in the presence of PGCLC-inductive cytokines. Blimp1, B lymphocyte–induced maturation protein-1; BMP, bone morphogenetic protein; ESCs, embryonic stem cells; FACS, fluorescence-activated cell sorting; LIF, leukemia inhibitory factor; PGCLCs, PGC-like cells; PGCs, primordial germ cells; SCF, stem cell factor; Tfcp2l1, transcription factor CP2-like 1.
Tfcp2l1 promotes PGCLC formation partially through Prdm14
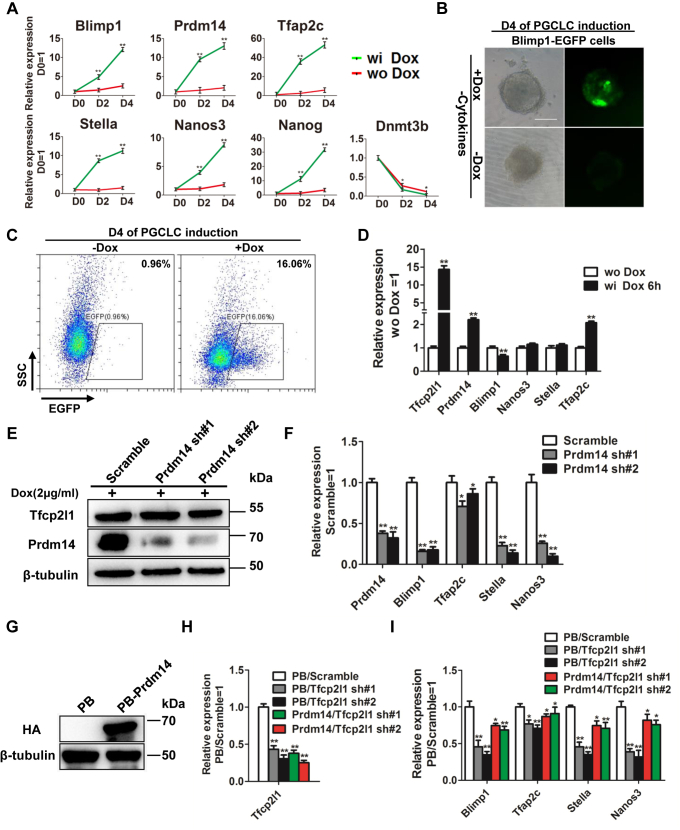
Given that Tfcp2l1 is one of the key targets of the LIF/Stat3 signaling pathway (21, 25) and LIF combined with BMP4 is also able to induce PGCLCs (14), we then treated i-Tfcp2l1 ESCs with BMP4 in the presence of DOX or LIF. BMP4 and DOX increased PGCLC marker levels, but the efficiency was lower than those of BMP4 and LIF (Fig. S2, A and B). To investigate whether overexpression of Tfcp2l1 alone can promote mouse ESC differentiation into PGCLCs, DOX was added after i-Tfcp2l1 ESCs were transformed into EpiLCs. The results showed that DOX treatment only upregulated the expression of PGCLC marker genes (Fig. 3, A–C). To further investigate the mechanism by which Tfcp2l1 induces PGCLCs, we treated i-Tfcp2l1 cells with DOX for a short period of time to detect the expression of PGC marker genes, and the results showed that 6 h of DOX treatment could increase the expression of Prdm14 and Tfap2c (Fig. 3D). The previous literature reported that overexpression of Tfap2c alone cannot reach the state of PGCLCs from EpiLCs, whereas ectopic expression of the Prdm14 gene alone suffices for the induction of the PGC state in EpiLCs (15). Thus, we next wanted to examine whether the function of Tfcp2l1 depends on Prdm14. First, we constructed a Prdm14–shRNA lentivirus system and infected i-Tfcp2l1 EpiLCs. After 4 days, qRT-PCR analysis results showed that Prdm14 expression was efficiently decreased and that downregulation of Prdm14 gene expression was able to reduce the expression of PGCLC marker genes induced by DOX compared with the scramble control group (Fig. 3, E and F). Shortly thereafter, we generated a mouse ESC line that overexpressed the HA-tagged mouse Prdm14 gene using a PiggyBac vector (PB-Prdm14) in which Prdm14 expression was efficiently enhanced (Fig. 3G). Empty vector control (PB) and PB-Prdm14 EpiLCs were infected with Tfcp2l1–shRNA lentivirus (Fig. 3H). After switching to cytokine-induced condition medium, Prdm14 upregulation was capable of preventing the PGCLC differentiation defect caused by Tfcp2l1 gene downregulation (Fig. 3I). Taken together, these results indicate that Tfcp2l1 partially depends on the Prdm14 gene to promote mouse ESC differentiation into PGCLCs.
Figure 3.
Prdm14 mediates the effect of Tfcp2l1 during mouse PGCLC specification.A, qRT-PCR analysis of the expression levels of PGC markers in i-Tfcp2l1 ESCs differentiated into EpiLCs and then cultured in GK15 medium in the presence or absence of 2 μg/ml DOX. Data are the mean ± SD (N = 3 biological replicates). ∗p < 0.05, ∗∗p < 0.01 versus wo DOX. B, the fluorescence intensity of Blimp1-EGFP transfected i-Tfcp2l1 ESCs cultured in GK15 medium in the absence of PGCLC-inductive cytokines and treated with or without DOX. The scale bar represents 100 μm. C, FACS analysis of Blimp1-EGFP on day 4 i-Tfcp2l1 embryoids in the presence or absence of DOX without PGCLC-inductive cytokines. D, qRT-PCR analysis of the expression levels of Tfcp2l1, Prdm14, Blimp1, Nanos3, Stella, and Tfap2c in i-Tfcp2l1 ESCs treated with or without DOX for 6 h. Data are the mean ± SD (N = 3 biological replicates). ∗∗p < 0.01 versus wo DOX. E, Western blot analysis of Tfcp2l1 and Prdm14 protein levels in i-Tfcp2l1 ESCs infected with scramble or Prdm14 shRNA lentiviruses in the presence of DOX. F, qRT-PCR analysis of PGC markers in i-Tfcp2l1 PGCLCs infected with scramble or Prdm14 shRNA lentiviruses in the presence of DOX. Data are the mean ± SD (N = 3 biological replicates). ∗p < 0.05, ∗∗p < 0.01 versus scramble. G, Western blot analysis of HA in 46C mouse ESCs overexpressing HA (PB) or HA-tagged mouse Prdm14 (PB-Prdm14) and cultured in LIF/serum conditions. H, qRT-PCR analysis of Tfcp2l1 expression levels in PB and PB-Prdm14 mouse ESCs infected with scramble or Tfcp2l1 shRNA lentiviruses. Data are the mean ± SD (N = 3 biological replicates). ∗∗p < 0.01 versus PB/scramble. I, qRT-PCR analysis of PGC genes in PB and PB-Prdm14 ESCs infected with scramble or Tfcp2l1 shRNA lentiviruses and converted into PGCLCs under PGCLC-inductive cytokines for 4 days. Data are the mean ± SD (N = 3 biological replicates). ∗p < 0.05, ∗∗p < 0.01 versus PB/scramble. Blimp1, B lymphocyte–induced maturation protein-1; ESCs, embryonic stem cells; i-Tfcp2l1, inducible Tfcp2l1; LIF, leukemia inhibitory factor; PB, PiggyBac; PGCLCs, PGC-like cells; PGCs, primordial germ cells; Prdm14, PR domain zinc finger protein 14; qRT-PCR, quantitative real-time PCR; Tfcp2l1, transcription factor CP2-like 1.
Prdm14 is a direct target of Tfcp2l1
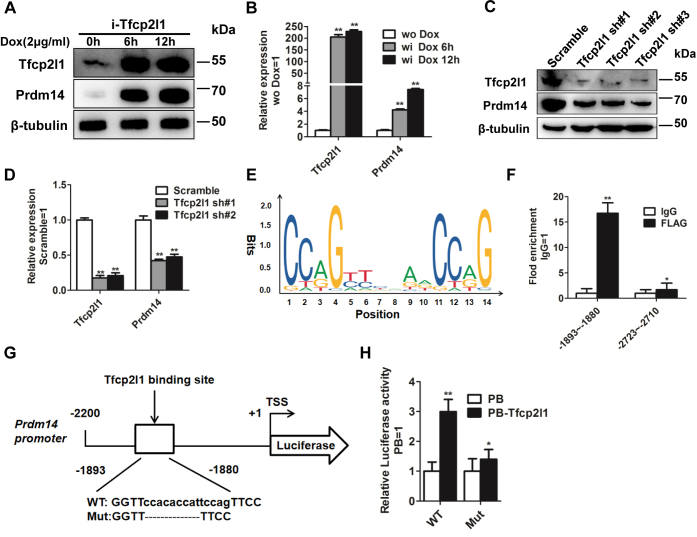
To investigate whether Tfcp2l1 directly regulates the expression level of Prdm14 in mouse ESCs, we used three different approaches. First, we examined Prdm14 transcription under different Tfcp2l1 expression levels by using i-Tfcp2l1 ESCs at the protein and mRNA levels. Prdm14 had a similar expression pattern to Tfcp2l1 after treatment with DOX for 6 h and 12 h (Fig. 4, A and B). Second, to detect whether knockdown of Tfcp2l1 has a negative effect on Prdm14 expression, we infected 46C mouse ESCs with Tfcp2l1–shRNA lentiviruses (Tfcp2l1 sh#1, Tfcp2l1 sh#2, and Tfcp2l1 sh#3). As expected, the transcripts of Prdm14 decreased in Tfcp2l1–shRNA cells but not in scramble control cells (Fig. 4, C and D). Third, to examine whether Tfcp2l1 directly binds to the Prdm14 gene locus, we analyzed the Tfcp2l1-binding consensus motifs using the JASPAR CORE database and predicted two potential binding sites within Prdm14 promoter regions: motif 1 is located at −1893∼−1880, and motif 2 is located at −2723∼−2710 (Fig. 4E). We then performed chromatin immunoprecipitation (ChIP) in FLAG-tagged Tfcp2l1 (PB-Tfcp2l1)-overexpressing 46C mouse ESCs with an anti-FLAG antibody affinity gel and discovered high enrichment binding within motif 1 (Fig. 4F). To further confirm that Tfcp2l1 is a functional activator of the Prdm14 promoter, the core base sequences of motif 1 were depleted and theoretically did not bind to the Tfcp2l1 protein (Fig. 4G). The Prdm14 promoter sequences comprising WT or mutated motif 1 were inserted into pGL6 to drive the expression of luciferase (pGL6-Prdm14). Subsequently, Tfcp2l1 was cotransfected into 46C mouse ESCs with pGL6-Prdm14. After 48 h, these cells were collected and lysed, and 2.14-fold upregulation was observed in WT promoter activity relative to the mutant sequence (Fig. 4H). These results collectively indicate that Tfcp2l1 directly binds to and activates Prdm14.
Figure 4.
Tfcp2l1 stimulates Prdm14 transcription in mouse ESCs.A, Western blot analysis of Tfcp2l1 and Prdm14 protein in i-Tfcp2l1 ESCs treated with DOX for 6 h and 12 h. B, qRT-PCR analysis of Tfcp2l1 and Prdm14 expression in i-Tfcp2l1 ESCs treated with DOX. Data are the mean ± SD (N = 3 biological replicates). ∗∗p < 0.01 versus without DOX. C, Western blot analysis of Tfcp2l1 and Prdm14 protein levels in 46C mouse ESCs infected with scramble or mouse Tfcp2l1 shRNA lentiviruses. D, qRT-PCR analysis of Tfcp2l1 and Prdm14 expression in Tfcp2l1 shRNA mouse ESCs. Data are the mean ± SD (N = 3 biological replicates). ∗∗p < 0.01 versus scramble. E, predicted consensus binding motif of Tfcp2l1 target loci from the JASPAR CORE database. F, ChIP–qRT-PCR analysis of the fold enrichment in the indicated regions of the Prdm14 promoter. Data are the mean ± SD (N = 3 biological replicates). ∗p < 0.05, ∗∗p < 0.01 versus IgG. G, the positions of one putative Tfcp2l1-binding site in the Prdm14 promoter and the corresponding mutant sequence. H, luciferase activity analysis of PB or PB-Tfcp2l1 cells overexpressing the WT or mutant (Mut) Prdm14 promoter reporter plasmids. Data are the mean ± SD (N = 3 biological replicates). ∗p < 0.05, ∗∗p < 0.01 versus PB. ChIP, chromatin immunoprecipitation; ESCs, embryonic stem cells; Prdm14, PR domain zinc finger protein 14; Tfcp2l1, transcription factor CP2-like 1; TSS, transcription start site.
Given that both Tfcp2l1 and Prdm14 have the ability to sustain the undifferentiated state of mouse ESCs in the absence of LIF, which is able to maintain naïve pluripotency by triggering Stat3 phosphorylation (Fig. S3, A–D) (21, 25), we examined whether Prdm14 is essential for Tfcp2l1 function. PB-Tfcp2l1 mouse ESCs infected with scramble or Prdm14–shRNA lentiviruses were cultured in serum-containing medium in the absence of LIF (Fig. S3, E and F). After 7 days, most of these cells retained typical ESC morphology and positive alkaline phosphatase activity, a marker of mouse ESCs (Fig. S3G). Meanwhile, PB-Prdm14 46C mouse ESCs infected with scramble or Tfcp2l1–shRNA lentivirus were still alkaline phosphatase positive (Fig. S3, H–J). These data suggest that Prdm14 is an important but not the key target of Tfcp2l1 in sustaining mouse ESC stemness.
Tfcp2l1 expression is upregulated during human PGC specification in vivo and in vitro
To determine whether Tfcp2l1 favors human PGCLC specification, we first detected its transcript in nonhuman primate PGCs. PGCs isolated from cynomolgus monkeys at days 43, 50, and 51 (corresponding to E10.5–E13.5 in mice) showed higher levels of Tfcp2l1 and the PGC markers Blimp1 and Tfap2c than cynomolgus monkey ESCs (GSE67259) (18) (Fig. 5A). We also observed obvious upregulation of PGC genes and Tfcp2l1 in human cells at week 5.5 to week 9 (SRA: SRP057098) (26) (Fig. 5B). To gain further insights into the function of Tfcp2l1 in promoting human PGCLC specification, we cultured human iPSCs in activin A, CHIR99021, and Y27632 conditions to induce iMeLCs according to a previous report (18) (Fig. 5, C and D). PGCLCs were then efficiently induced by transferring iMeLCs into KnockOut Serum Replacement (KSR)-containing medium supplemented with BMP4, LIF, SCF, and EGF, characterized by the upregulation of the PGC markers, Stella, Tfap2c, Nanos3, and Blimp1 (Fig. 5, E and F). Similar to a previous study (18), the mesoderm marker gene EOMES was highly expressed in iMeLCs, but its expression was decreased in iPSCs and PGCLCs (Fig. 5F). Interestingly, the transcription of Prdm14 decreased in iMeLCs and PGCLCs (Fig. 5F). Tfcp2l1 expression decreased when human iPSCs became iMeLCs but increased markedly upon PGCLC formation (Fig. 5F). A similar expression pattern was observed in H9 ESCs (Fig. S4A). Overall, the data not only reveal that Tfcp2l1 is highly elevated in human PGCLCs but also imply that Tfcp2l1 may be a potential driving factor for the generation of PGCs.
Figure 5.
Tfcp2l1 is strongly expressed in primate PGCs.A, heatmap showing the expression of Blimp1, Tfcp2l1, and Tfap2c during cynomolgus monkey PGC development. B, heatmap showing the expression pattern of the indicated genes during human PGC development. C, schematic diagram of human iPSCs transforming into PGCLCs. D, phenotype of human iPSCs and iMeLCs. The scale bar represents 100 μm. E, immunofluorescence staining of Blimp1 (green) and Tfap2c (red) in human PGCLCs. The nuclei were counterstained with Hoechst 33342 (Hoechst). The scale bar represents 100 μm. F, qRT-PCR analysis of the expression levels of PGC markers in human iPSCs transformed into PGCLCs in the presence or absence of PGCLC-inductive cytokines. Data are the mean ± SD (three independent experiments). ∗p < 0.05, ∗∗p < 0.01 versus without cytokines. Blimp1, B lymphocyte–induced maturation protein-1; cyESC, cynomolgus monkey ESCs; hPGC Wk9 F, week 9 female human PGCs; hPGC Wk9 M, week 9 male human PGCs; iPSCs, induced pluripotent stem cells; PGCd43, PGC on day 43; PGCLCs, PGC-like cells; PGCs, primordial germ cells; Tfap2c, transcription factor AP-2 gamma; Tfcp2l1, transcription factor CP2-like 1.
Tfcp2l1 facilitates human iPSC differentiation into PGCLCs
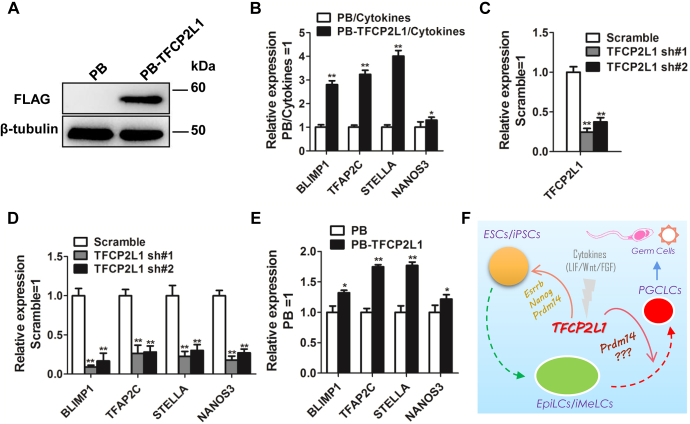
To validate the above hypothesis, we transduced FLAG-tagged human Tfcp2l1 into human iPSCs with a PB system in which Tfcp2l1 was successfully upregulated (PB-TFCP2L1) (Fig. 6A). To evaluate whether enforced Tfcp2l1 expression facilitates PGCLC emergence, we differentiated PB control and PB-TFCP2L1 human iPSCs into iMeLCs and swiftly induced PGCLCs by changing the medium into KSR-containing medium supplemented with BMP4, LIF, SCF, and EGF. After 4 days, we extracted RNA from these cells and found that PB-TFCP2L1 cells expressed higher levels of the PGC markers Blimp1, Stella, Tfap2c, and Nanos3 than PB cells (Fig. 6B). The same results were also observed in H9 human ESCs (Fig. S4, B and C). Next, to detect the function of Tfcp2l1 in the generation of PGCLCs, we suppressed Tfcp2l1 transcription with RNA interference. As shown in Figure 6C, human Tfcp2l1 expression was successfully downregulated (Fig. 6C). Meanwhile, Tfcp2l1–shRNA decreased PGCLC marker levels in the presence of PGCLC-inductive cytokines (Fig. 6D). Similar phenotypes could also be obtained in H9 ESCs (Fig. S4, D and E). These results suggest a crucial role of Tfcp2l1 during human PGCLC specification. As overexpression of Tfcp2l1 only can promote mouse PGCLC generation (Fig. 3A), to test whether TFCP2L1 has similar effect in human iPSCs, we finally differentiated PB and PB-TFCP2L1 human iPSCs into iMeLCs and then transferred these cells into differentiation medium for 4 days. qRT-PCR detection showed that PB-TFCP2L1 cells expressed higher levels of PGC markers than PB cells (Fig. 6E). Similar results were also observed in H9 ESCs (Fig. S4F). Overall, these data indicate that exogenous expression of Tfcp2l1 favors PGCLC generation from human pluripotent stem cells.
Figure 6.
Tfcp2l1 increases human PGCLC generation efficiency.A, Western blot analysis of FLAG in human iPSCs overexpressing the FLAG-tagged human Tfcp2l1 gene. B, qRT-PCR analysis of the expression levels of several core PGC genes in PB and PB-TFCP2L1 human iPSCs cultured in PGCLC-indicative cytokines. Data are the mean ± SD (N = 3 biological replicates). ∗p < 0.05, ∗∗p < 0.01 versus PB/cytokines. C, qRT-PCR analysis of the expression levels of human Tfcp2l1 genes in human iPSCs infected with Tfcp2l1 shRNA lentiviruses. Data are the mean ± SD (N = 3 biological replicates). ∗∗p < 0.01 versus scramble. D, qRT-PCR analysis of PGC markers in human iPSCs infected with scramble or human Tfcp2l1 shRNA lentiviruses and induced into PGCLCs under PGCLC-inductive cytokines for 4 days. Data are the mean ± SD (N = 3 biological replicates). ∗∗p < 0.01 versus scramble. E, qRT-PCR analysis of the expression levels of PGC genes in PB and PB-TFCP2L1 human iPSCs transformed into iMeLCs and then cultured in GK15 medium for 4 days. Data are the mean ± SD (N = 3 biological replicates). ∗∗p < 0.01 versus PB. F, schematic diagram of the positive role of Tfcp2l1 in pluripotent stem cell maintenance and PGCLC specification. iMeLCs, incipient mesoderm-like cells; iPSCs, induced pluripotent stem cells; PB, PiggyBac; PGCLCs, PGC-like cells; PGCs, primordial germ cells; qRT-PCR, quantitative real-time PCR; Tfcp2l1, transcription factor CP2-like 1.
Discussion
In this study, we found that the expression of Tfcp2l1 increased during ESC differentiation into PGCLCs. Therefore, upregulation of Tfcp2l1 is able to induce PGCLC generation, whereas downregulation of its transcript inhibits PGCLC formation in mouse and human pluripotent stem cells. Further mechanistic study showed that Tfcp2l1 might function by directly increasing the transcription of the germ cell marker Prdm14 in mouse ESCs, as knockdown of Prdm14 would impair PGCLC formation mediated by Tfcp2l1. Therefore, our data demonstrated a critical role for Tfcp2l1 in germ cell specification (Fig. 6F).
The aforementioned function of Tfcp2l1 can be explained. First, although Tfcp2l1 transcript is reported to be enriched in mouse ESCs and kidney tissues, it is also highly expressed in PGCs and testis samples (27, 28, 29). Second, Tfcp2l1 is a converged target of three self-renewal–associated pathways, the LIF/Stat3, Wnt/β-catenin, and fibroblast growth factor/extracellular signal-regulated kinase signaling pathways, in mouse ESCs (21, 25). Activation of either LIF/Stat3 or Wnt/β-catenin signaling or inhibition of MEK can induce Tfcp2l1 expression (21, 25, 30). Therefore, forced expression of Tfcp2l1 can substitute for each to promote the maintenance of mouse ESC identity (21, 25). Notably, only knockdown of Tfcp2l1 was able to suppress the function of Stat3 among the reported Stat3 target genes (21, 23, 25). LIF is an important cytokine that induces mouse and human PGCLC specification (14, 18, 31, 32). Moreover, activation of Wnt/β-catenin signaling by Wnt3 and inhibition of ERK by a MEK inhibitor can also promote PGC formation (8, 33). Third, in addition to Prdm14, Tfcp2l1 increases the expression levels of many genes that harbor the function of enhancing PGCLC generation. For instance, upregulation of Nanog can markedly increase the efficiency of PGCLC specification in EpiLCs after overexpression (20). In contrast, PGCLC differentiation is impaired when the Nanog gene is knocked out, showing decreased proliferation and increased apoptosis (20, 34). However, induced expression of estrogen related receptor beta (Esrrb) can restore PGCLC numbers in Nanog-null mouse ESCs (34). Notably, both Nanog and Esrrb are the downstream targets of Tfcp2l1 (21, 25, 35). Fourth, Tfcp2l1 physically interacts with Oct4 in mouse ESCs (36). Oct4 is an essential gene that has been recognized as fundamental in the maintenance of the state of ESCs and PGCs (19). Loss of Oct4 function leads to the differentiation of ESCs and the apoptosis of PGCs (19). Recently, a study reported that Tfcp2l1 is dispensable for the generation of PGCLCs from human ESCs (27). However, depletion of the Tfcp2l1 gene could slightly reduce the efficiency of the differentiation of human ESCs into PGCLCs revealed by their FACS results (27). This result is consistent with our observation, although we used different human pluripotent stem cell lines and PGCLC-inductive methods. Future studies on genetically modifying the locus of Tfcp2l1 in animal models to validate its impact on germline development are needed.
Another important finding of this study is that we identified Prdm14 as a direct target gene of Tfcp2l1 for the first time. Prdm14 is a key factor for the PGC development of different species (6, 37, 38). Simultaneous overexpression of Prdm14 alone or of three germline genes, Blimp1, Prdm14, and Tfap2c, could induce germline induction in mouse ESCs. Prdm14 KO mice are sterile because of complete lack of germ cells in both females and males (6, 15), suggesting a central role of Prdm14 in the mouse PGC regulatory network. Prdm14 shares many similar functional features with Tfcp2l1 during early development. For example, both are highly expressed in the pluripotent inner cell mass and have the ability to promote ESC self-renewal (21, 25, 39), whereas loss of each gene destabilizes ESCs and leads to the emergence of lineage markers (21, 22, 25, 40, 41), but these cells can be maintained indefinitely in naïve conditions (21, 40, 42, 43). In addition, the expression of both proteins ceases in postimplantation epiblast cells; overexpression of each protein alone can convert EpiSCs into naïve pluripotent cells (21, 25, 44), and the reprogramming efficiency can be significantly increased in combination with Klf2 (23, 44). However, it is worth noting that Prdm14 cannot mediate the self-renewal–promoting effect of Tfcp2l1 (Fig. S3, E–J). We previously reported that Esrrb is the major target of Tfcp2l1 in mouse ESCs (35). Interestingly, Tfcp2l1 acts during human PGCLC specification, and possibly not through PRDM14, because their expression patterns in this process are different (Fig. 5F). This is understandable because the current research results around PRDM14 in the human PGCLC specification are uncertain. A previous knockdown experiment suggested that PRDM14 might be dispensable for PGCLC generation (45), while a recent study used inducible degrons for more rapid and comprehensive PRDM14 depletion and found reduced specification efficiency in human ESC-derived PGCLCs (38). In addition, the sets of targets regulated by PRDM14 in mice and humans are vastly different, and PRDM14 alone cannot induce human PGCLCs, unlike in mice (38). These different results may be related to the status of mouse and human pluripotent stem cells, the latter being similar to mouse postimplantation EpiSCs in growth requirements, morphology, clonogenicity, and gene expression patterns (11, 16, 17). Therefore, human PGC specification depends on the expression of SOX17, an endoderm marker that is dispensable for mouse PGC development (46). Interestingly, SOX2 is crucial for mouse, but not for human PGCs (46, 47, 48). These reports combined with our observations suggest that there must be other targets that mediate Tfcp2l1 in inducing the PGCLC state. Identification of these genes in the future may be an effective way to understand the similarities and differences between mouse and human PGC development.
In summary, we revealed a novel and conserved function of Tfcp2l1 in mouse and human PGCLC specification. Meanwhile, we preliminarily demonstrated that Tfcp2l1 exerts this function partially by upregulating the expression of the Prdm14 gene in mouse ESCs. A detailed analysis of the molecular mechanisms involved in the function of Tfcp2l1 will provide new insights into the full understanding of the regulatory circuitry that induces PGC specification across species, which will eventually contribute to the study of the early development of reproduction and the treatment of infertility.
Experimental procedures
Cell culture
The 46C mouse ESCs were kindly provided by Qi-Long Ying (The University of Southern California) and were cultured in 0.1% gelatin-coated dishes at 37 °C in 5% CO2. The conventional cell culture conditions were Dulbecco's modified Eagle's medium (Biological Industries) supplemented with 10% fetal bovine serum (FND500, ExCell Bio), 1× MEM nonessential amino acids (N1250, Solarbio), 1× penicillin/streptomycin (P1400, Solarbio), 0.1 mM β-mercaptoethanol (M3148, Sigma), and LIF (made in house). Human transgene-free iPSCs were kindly provided by Nuwacell Ltd (ZSSY-001) and were cultured in ncTarget (RP01020, Nuwacell Ltd). Cells were dissociated using EDTA solution (RP01007, Nuwacell Ltd) every 3 to 5 days.
Plasmid construction
The coding regions of the mouse and human Tfcp2l1 and mouse Prdm14 genes were inserted into PB transposon vectors carrying FLAG or HA tags. The targeting sequences designed for decreasing mouse or human Tfcp2l1 or mouse Prdm14 transcript were cloned into pLKO.1-TRC (#10878, Addgene). The sequences are listed in Tables S1 and S2. For construction of Blimp1 promoter-mediated EGFP expression, the promoter sequence of Blimp1 (from −1000 to +50) was inserted into pEGFP-N1 to replace the original CMV promotor, which induces EGFP expression in the pEGFP-N1 plasmid.
Induction of mouse and human PGCLCs
We mainly referred to two previous studies for the induction protocols (14, 18). Briefly, for mouse PGCLC formation, 1 × 105 46C ESCs were seeded in fibronectin bovine plasma (16.7 μl/ml, F1141-5MG, Sigma) coated plate and were cultured in activin A (20 ng/ml, C678, Novoprotein) and basic fibroblast growth factor (12 ng/ml, C044, Novoprotein) containing medium to induce EpiLCs. The medium was changed every day. After 2 days, 3 × 105 EpiLCs were incubated in GK15 medium, consisting of Glasgow's minimal essential medium (11710035, Gibco), 15% KSR (10828028, Invitrogen), 1× penicillin/streptomycin (P1400, Solarbio), 1× MEM nonessential amino acids, 0.1 mM β-mercaptoethanol, and 1 mM sodium pyruvate (N1250, Solarbio), to generate PGCLCs in the presence of BMP4 (500 ng/ml, 315-27-10, PeproTech), LIF (1000 U/ml, Millipore), SCF (100 ng/ml, AF-250-03, PeproTech), and EGF (50 ng/ml, AF-100-15, PeproTech).
For human PGCLC induction, 1 × 105 human iPSCs were differentiated into iMeLCs with 50 ng/ml activin A, 3 mM CHIR99021, and 10 mM ROCK inhibitor (Y27632). iMeLCs (3 × 105) were then incubated in GK15 medium supplemented with BMP2 (200 ng/ml, C012, Novoprotein) or BMP4 (500 ng/ml, 120-05-5, PeproTech), LIF (1000 U/ml, Millipore), SCF (100 ng/ml, C034, Novoprotein), EGF (50 ng/ml, AF-100-15, PeproTech), and 10 mM ROCK inhibitor for 4 days.
Western blot
Western blotting was performed according to the conventional protocol. Briefly, the cells were lysed on ice with radioimmunoprecipitation assay buffer (P0013B, Beyotime Biotechnology). The extracted proteins were separated on a 10% PAGE gel and electrotransferred onto a polyvinylidene fluoride membrane. The membranes were incubated with specific primary antibodies for investigation. The antibodies used are FLAG (1:1000, GNI4110-FG-S, GNI), HA (1:1000, GNI4110-HA-S, GNI, 1:1000), β-tubulin mouse monoclonal antibody (1:2000, 200608, Zen-Bio), Prdm14 (1:1000, D121722, BBI), and Tfcp2l1 (1:1000, AF5726, R&D systems).
Immunofluorescence staining
The cells were fixed in 4% paraformaldehyde for 20 min and then incubated in the blocking buffer (PBS containing 5% BSA and 0.2% Triton X-100) for 2 h. After washing three times with PBS, the cells were incubated overnight at 4 °C under the corresponding antibody. Then, after washing with PBS, cells were incubated with the secondary antibody at 37 °C for 1 h. Finally, the cells were photographed with a Leica DMI8 microscope. The antibodies used are Tfcp2l1(AF5726, 1:100, R&D systems), Stella (ab19878, 1:100, Abcam), Tfap2c (sc-12762, 1:100, Santa Cruz), and Blimp1 (sc-47732, 1:100, Santa Cruz).
FACS
Blimp1-EGFP-expressing PGCLCs were digested into single cells with Solase solution (RP01021, Nuwacell) for 15 min. Cells were resuspended in 500 μl of precooled D-PBS, and then the EGFP fluorescence intensity was measured using a CytoFLEX analytical flow cytometer (Beckman).
qRT-PCR
The total RNA was extracted using the EZ-10 RNA extraction kit (B610583, BBI). cDNA was synthesized from 1 μg of total RNA using the reverse transcription kit (with dsDNase) (BL699A, Biosharp). Finally, qRT-PCR was performed using Hieff qPCR SYBR Green Master Mix (No Rox) (11201ES03, YEASEN) in a PikoReal real-time PCR machine. The relative expression level was determined by the 2-ΔCq method and normalized to human β-actin or mouse Rpl19 expression. The primers used are listed in Table S3.
ChIP assay
ChIP experiments were carried out using a ChIP Analysis Kit (P2078, Beyotime Biotechnology) according to the manufacturer's instructions. The FLAG antibody was used for immunoprecipitation, and immunoglobulin G was used as a negative control. The ChIP enrichment was confirmed by qRT-PCR. Table S4 lists the sequences and positions of primers for cloning the promoter regions of Prdm14.
Luciferase assay
The promoter sequence of Prdm14 (from −2200 to +1) was cloned into the pGL6 plasmid (pGL6-Prdm14). The WT and mutant pGL6-Prdm14 plasmids were cotransfected into 46C mouse ESCs using the Tfcp2l1-overexpressing and Renilla-luciferase constructs. After 48 h, the luciferase activity was measured using a TransDetect Double-Luciferase Reporter Assay Kit (FR201, TransGen Biotech).
Cell transfection and virus production
For gene overexpression, 2 μg of PB and 2 μg of transposon plasmids were transduced into cells using the Hieff Trans Liposomal Transfection Reagent (40802ES03, YEASEN) according to the manufacturer's instructions. For lentivirus production, 2 μg of pLKO.1, 0.75 μg of VSV-G, and 1.25 μg of psPAX2 were transfected into 293FT cells together. The supernatant containing the secreted viruses was then collected and added to the culture medium. After 2 days, puromycin or blasticidin S was added to screen the positive cells.
Statistical analysis
All data are reported as the mean ± SD. Student's t test is used to determine the significance of the comparison difference using GraphPad Prism 8 software. Values with p < 0.05 are considered statistically significant.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank everyone in the Ye lab for technical help. This work was supported by the Natural Science Foundation of Anhui Province [1908085J13], the Anhui Provincial Key Research and Development Plan [202104b11020026], the University Synergy Innovation Program of Anhui Province [GXXT-2020-064], and the Open Fund for Discipline Construction, Institute of Physical Science and Information Technology, Anhui University [S01003106] and receives funding support from the Department of Education of Anhui Province and the Department of Human Resources and Social Security of Anhui Province [gxyqZD2020001 and 2020H210].
Author contributions
M. Z. and J. J. conceptualization; M. Z., J. J., Xiaoxiao Wang, X. Z., and Y. Z. investigation; M. Z. writing–original draft; Y. L., Xin Wang, X. L., and Q. B. methodology; Y. Z. validation; Y. L., Xin Wang, X. L., and Q. B. formal analysis, Y. L., Xin Wang, X. L., and Q. B. visualization; Y. L., Xin Wang, X. L., and Q. B. resources; S.-D. Y. supervision; S.-D. Y. writing–review and editing.
Edited by Qi-Qun Tang
Supporting information
References
- 1.Ginsburg M., Snow M.H., McLaren A. Primordial germ cells in the mouse embryo during gastrulation. Development. 1990;110:521–528. doi: 10.1242/dev.110.2.521. [DOI] [PubMed] [Google Scholar]
- 2.Lawson K.A., Dunn N.R., Roelen B.A., Zeinstra L.M., Davis A.M., Wright C.V., Korving J.P., Hogan B.L. Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev. 1999;13:424–436. doi: 10.1101/gad.13.4.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ying Y., Zhao G.Q. Cooperation of endoderm-derived BMP2 and extraembryonic ectoderm-derived BMP4 in primordial germ cell generation in the mouse. Dev. Biol. 2001;232:484–492. doi: 10.1006/dbio.2001.0173. [DOI] [PubMed] [Google Scholar]
- 4.Ohinata Y., Ohta H., Shigeta M., Yamanaka K., Wakayama T., Saitou M. A signaling principle for the specification of the germ cell lineage in mice. Cell. 2009;137:571–584. doi: 10.1016/j.cell.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 5.Ohinata Y., Payer B., O'Carroll D., Ancelin K., Ono Y., Sano M., Barton S.C., Obukhanych T., Nussenzweig M., Tarakhovsky A., Saitou M., Surani M.A. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature. 2005;436:207–213. doi: 10.1038/nature03813. [DOI] [PubMed] [Google Scholar]
- 6.Yamaji M., Seki Y., Kurimoto K., Yabuta Y., Yuasa M., Shigeta M., Yamanaka K., Ohinata Y., Saitou M. Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat. Genet. 2008;40:1016–1022. doi: 10.1038/ng.186. [DOI] [PubMed] [Google Scholar]
- 7.Weber S., Eckert D., Nettersheim D., Gillis A.J., Schafer S., Kuckenberg P., Ehlermann J., Werling U., Biermann K., Looijenga L.H., Schorle H. Critical function of AP-2 gamma/TCFAP2C in mouse embryonic germ cell maintenance. Biol. Reprod. 2010;82:214–223. doi: 10.1095/biolreprod.109.078717. [DOI] [PubMed] [Google Scholar]
- 8.Aramaki S., Hayashi K., Kurimoto K., Ohta H., Yabuta Y., Iwanari H., Mochizuki Y., Hamakubo T., Kato Y., Shirahige K., Saitou M. A mesodermal factor, T, specifies mouse germ cell fate by directly activating germline determinants. Dev. Cell. 2013;27:516–529. doi: 10.1016/j.devcel.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 9.Evans M.J., Kaufman M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 10.Martin G.R. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. U. S. A. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 13.Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R., Slukvin I.I., Thomson J.A. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 14.Hayashi K., Ohta H., Kurimoto K., Aramaki S., Saitou M. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell. 2011;146:519–532. doi: 10.1016/j.cell.2011.06.052. [DOI] [PubMed] [Google Scholar]
- 15.Nakaki F., Hayashi K., Ohta H., Kurimoto K., Yabuta Y., Saitou M. Induction of mouse germ-cell fate by transcription factors in vitro. Nature. 2013;501:222–226. doi: 10.1038/nature12417. [DOI] [PubMed] [Google Scholar]
- 16.Brons I.G., Smithers L.E., Trotter M.W., Rugg-Gunn P., Sun B., Chuva de Sousa Lopes S.M., Howlett S.K., Clarkson A., Ahrlund-Richter L., Pedersen R.A., Vallier L. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 17.Tesar P.J., Chenoweth J.G., Brook F.A., Davies T.J., Evans E.P., Mack D.L., Gardner R.L., McKay R.D. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 18.Sasaki K., Yokobayashi S., Nakamura T., Okamoto I., Yabuta Y., Kurimoto K., Ohta H., Moritoki Y., Iwatani C., Tsuchiya H., Nakamura S., Sekiguchi K., Sakuma T., Yamamoto T., Mori T. Robust in vitro induction of human germ cell fate from pluripotent stem cells. Cell Stem Cell. 2015;17:178–194. doi: 10.1016/j.stem.2015.06.014. [DOI] [PubMed] [Google Scholar]
- 19.Kehler J., Tolkunova E., Koschorz B., Pesce M., Gentile L., Boiani M., Lomeli H., Nagy A., McLaughlin K.J., Scholer H.R., Tomilin A. Oct4 is required for primordial germ cell survival. EMBO Rep. 2004;5:1078–1083. doi: 10.1038/sj.embor.7400279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murakami K., Gunesdogan U., Zylicz J.J., Tang W.W.C., Sengupta R., Kobayashi T., Kim S., Butler R., Dietmann S., Surani M.A. NANOG alone induces germ cells in primed epiblast in vitro by activation of enhancers. Nature. 2016;529:403–407. doi: 10.1038/nature16480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye S., Li P., Tong C., Ying Q.L. Embryonic stem cell self-renewal pathways converge on the transcription factor Tfcp2l1. EMBO J. 2013;32:2548–2560. doi: 10.1038/emboj.2013.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun H., You Y., Guo M., Wang X., Zhang Y., Ye S. Tfcp2l1 safeguards the maintenance of human embryonic stem cell self-renewal. J. Cell Physiol. 2018;233:6944–6951. doi: 10.1002/jcp.26483. [DOI] [PubMed] [Google Scholar]
- 23.Qiu D., Ye S., Ruiz B., Zhou X., Liu D., Zhang Q., Ying Q.L. Klf2 and Tfcp2l1, two Wnt/beta-catenin targets, act synergistically to induce and maintain naive pluripotency. Stem Cell Rep. 2015;5:314–322. doi: 10.1016/j.stemcr.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mise N., Fuchikami T., Sugimoto M., Kobayakawa S., Ike F., Ogawa T., Tada T., Kanaya S., Noce T., Abe K. Differences and similarities in the developmental status of embryo-derived stem cells and primordial germ cells revealed by global expression profiling. Genes Cells. 2008;13:863–877. doi: 10.1111/j.1365-2443.2008.01211.x. [DOI] [PubMed] [Google Scholar]
- 25.Martello G., Bertone P., Smith A. Identification of the missing pluripotency mediator downstream of leukaemia inhibitory factor. EMBO J. 2013;32:2561–2574. doi: 10.1038/emboj.2013.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang W.W., Dietmann S., Irie N., Leitch H.G., Floros V.I., Bradshaw C.R., Hackett J.A., Chinnery P.F., Surani M.A. A unique gene regulatory network resets the human germline epigenome for development. Cell. 2015;161:1453–1467. doi: 10.1016/j.cell.2015.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hancock G.V., Liu W., Peretz L., Chen D., Gell J.J., Collier A.J., Zamudio J.R., Plath K., Clark A.T. Divergent roles for KLF4 and TFCP2L1 in naive ground state pluripotency and human primordial germ cell development. Stem Cell Res. 2021;55:102493. doi: 10.1016/j.scr.2021.102493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aguilar A. Development: Tfcp2l1 drives Notch signalling and epithelial diversity in the collecting duct. Nat. Rev. Nephrol. 2017;13:445. doi: 10.1038/nrneph.2017.93. [DOI] [PubMed] [Google Scholar]
- 29.Werth M., Schmidt-Ott K.M., Leete T., Qiu A., Hinze C., Viltard M., Paragas N., Shawber C.J., Yu W., Lee P., Chen X., Sarkar A., Mu W., Rittenberg A., Lin C.S. Transcription factor TFCP2L1 patterns cells in the mouse kidney collecting ducts. Elife. 2017;6 doi: 10.7554/eLife.24265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martello G., Sugimoto T., Diamanti E., Joshi A., Hannah R., Ohtsuka S., Gottgens B., Niwa H., Smith A. Esrrb is a pivotal target of the Gsk3/Tcf3 axis regulating embryonic stem cell self-renewal. Cell Stem Cell. 2012;11:491–504. doi: 10.1016/j.stem.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith A.G., Heath J.K., Donaldson D.D., Wong G.G., Moreau J., Stahl M., Rogers D. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature. 1988;336:688–690. doi: 10.1038/336688a0. [DOI] [PubMed] [Google Scholar]
- 32.Williams R.L., Hilton D.J., Pease S., Willson T.A., Stewart C.L., Gearing D.P., Wagner E.F., Metcalf D., Nicola N.A., Gough N.M. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature. 1988;336:684–687. doi: 10.1038/336684a0. [DOI] [PubMed] [Google Scholar]
- 33.Kimura T., Kaga Y., Ohta H., Odamoto M., Sekita Y., Li K., Yamano N., Fujikawa K., Isotani A., Sasaki N., Toyoda M., Hayashi K., Okabe M., Shinohara T., Saitou M. Induction of primordial germ cell-like cells from mouse embryonic stem cells by ERK signal inhibition. Stem Cells. 2014;32:2668–2678. doi: 10.1002/stem.1781. [DOI] [PubMed] [Google Scholar]
- 34.Zhang M., Leitch H.G., Tang W.W.C., Festuccia N., Hall-Ponsele E., Nichols J., Surani M.A., Smith A., Chambers I. Esrrb complementation rescues development of nanog-null germ cells. Cell Rep. 2018;22:332–339. doi: 10.1016/j.celrep.2017.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X., Wang X., Zhang S., Sun H., Li S., Ding H., You Y., Zhang X., Ye S.D. The transcription factor TFCP2L1 induces expression of distinct target genes and promotes self-renewal of mouse and human embryonic stem cells. J. Biol. Chem. 2019;294:6007–6016. doi: 10.1074/jbc.RA118.006341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van den Berg D.L., Snoek T., Mullin N.P., Yates A., Bezstarosti K., Demmers J., Chambers I., Poot R.A. An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell. 2010;6:369–381. doi: 10.1016/j.stem.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kobayashi T., Kobayashi H., Goto T., Takashima T., Oikawa M., Ikeda H., Terada R., Yoshida F., Sanbo M., Nakauchi H., Kurimoto K., Hirabayashi M. Germline development in rat revealed by visualization and deletion of Prdm14. Development. 2020;147 doi: 10.1242/dev.183798. [DOI] [PubMed] [Google Scholar]
- 38.Sybirna A., Tang W.W.C., Pierson Smela M., Dietmann S., Gruhn W.H., Brosh R., Surani M.A. A critical role of PRDM14 in human primordial germ cell fate revealed by inducible degrons. Nat. Commun. 2020;11:1282. doi: 10.1038/s41467-020-15042-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burton A., Muller J., Tu S., Padilla-Longoria P., Guccione E., Torres-Padilla M.E. Single-cell profiling of epigenetic modifiers identifies PRDM14 as an inducer of cell fate in the mammalian embryo. Cell Rep. 2013;5:687–701. doi: 10.1016/j.celrep.2013.09.044. [DOI] [PubMed] [Google Scholar]
- 40.Ma Z., Swigut T., Valouev A., Rada-Iglesias A., Wysocka J. Sequence-specific regulator Prdm14 safeguards mouse ESCs from entering extraembryonic endoderm fates. Nat. Struct. Mol. Biol. 2011;18:120–127. doi: 10.1038/nsmb.2000. [DOI] [PubMed] [Google Scholar]
- 41.Tsuneyoshi N., Sumi T., Onda H., Nojima H., Nakatsuji N., Suemori H. PRDM14 suppresses expression of differentiation marker genes in human embryonic stem cells. Biochem. Biophys. Res. Commun. 2008;367:899–905. doi: 10.1016/j.bbrc.2007.12.189. [DOI] [PubMed] [Google Scholar]
- 42.Yamaji M., Ueda J., Hayashi K., Ohta H., Yabuta Y., Kurimoto K., Nakato R., Yamada Y., Shirahige K., Saitou M. PRDM14 ensures naive pluripotency through dual regulation of signaling and epigenetic pathways in mouse embryonic stem cells. Cell Stem Cell. 2013;12:368–382. doi: 10.1016/j.stem.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 43.Ying Q.L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gillich A., Bao S., Grabole N., Hayashi K., Trotter M.W., Pasque V., Magnusdottir E., Surani M.A. Epiblast stem cell-based system reveals reprogramming synergy of germline factors. Cell Stem Cell. 2012;10:425–439. doi: 10.1016/j.stem.2012.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sugawa F., Arauzo-Bravo M.J., Yoon J., Kim K.P., Aramaki S., Wu G., Stehling M., Psathaki O.E., Hubner K., Scholer H.R. Human primordial germ cell commitment in vitro associates with a unique PRDM14 expression profile. EMBO J. 2015;34:1009–1024. doi: 10.15252/embj.201488049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Irie N., Weinberger L., Tang W.W., Kobayashi T., Viukov S., Manor Y.S., Dietmann S., Hanna J.H., Surani M.A. SOX17 is a critical specifier of human primordial germ cell fate. Cell. 2015;160:253–268. doi: 10.1016/j.cell.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Jong J., Stoop H., Gillis A.J., van Gurp R.J., van de Geijn G.J., Boer M., Hersmus R., Saunders P.T., Anderson R.A., Oosterhuis J.W., Looijenga L.H. Differential expression of SOX17 and SOX2 in germ cells and stem cells has biological and clinical implications. J. Pathol. 2008;215:21–30. doi: 10.1002/path.2332. [DOI] [PubMed] [Google Scholar]
- 48.Campolo F., Gori M., Favaro R., Nicolis S., Pellegrini M., Botti F., Rossi P., Jannini E.A., Dolci S. Essential role of Sox2 for the establishment and maintenance of the germ cell line. Stem Cells. 2013;31:1408–1421. doi: 10.1002/stem.1392. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.