Abstract
Different forms of photoreceptor degeneration cause blindness. Retinal degeneration-3 protein (RD3) deficiency in photoreceptors leads to recessive congenital blindness. We proposed that aberrant activation of the retinal membrane guanylyl cyclase (RetGC) by its calcium-sensor proteins (guanylyl cyclase–activating protein [GCAP]) causes this retinal degeneration and that RD3 protects photoreceptors by preventing such activation. We here present in vivo evidence that RD3 protects photoreceptors by suppressing activation of both RetGC1 and RetGC2 isozymes. We further suggested that insufficient inhibition of RetGC by RD3 could contribute to some dominant forms of retinal degeneration. The R838S substitution in RetGC1 that causes autosomal-dominant cone–rod dystrophy 6, not only impedes deceleration of RetGC1 activity by Ca2+GCAPs but also elevates this isozyme's resistance to inhibition by RD3. We found that RD3 prolongs the survival of photoreceptors in transgenic mice harboring human R838S RetGC1 (R838S+). Overexpression of GFP-tagged human RD3 did not improve the calcium sensitivity of cGMP production in R838S+ retinas but slowed the progression of retinal blindness and photoreceptor degeneration. Fluorescence of the GFP-tagged RD3 in the retina only partially overlapped with immunofluorescence of RetGC1 or GCAP1, indicating that RD3 separates from the enzyme before the RetGC1:GCAP1 complex is formed in the photoreceptor outer segment. Most importantly, our in vivo results indicate that, in addition to the abnormal Ca2+ sensitivity of R838S RetGC1 in the outer segment, the mutated RetGC1 becomes resistant to inhibition by RD3 in a different cellular compartment(s) and suggest that RD3 overexpression could be utilized to reduce the severity of cone–rod dystrophy 6 pathology.
Keywords: cyclic GMP, eye, calcium-binding proteins, GCAP, guanylate cyclase (guanylyl cyclase), GUCY2D, photoreceptor, RetGC, retinal degeneration, RD3
Abbreviations: CNG, cyclic nucleotide–gated channel; CORD6, cone–rod dystrophy 6; ERG, electroretinography; GCAP, guanylyl cyclase–activating protein; HEK293, human embryonic kidney cells 293; LCA12, Leber's congenital amaurosis type 12; OCT, optical coherence tomography; ONL, outer nuclear layer; RD3, retinal degeneration-3 protein; RetGC, the retinal membrane guanylyl cyclase; ROS, rod outer segment; TTBS, Tris-buffered saline containing 0.5% Tween-20
Cyclic GMP mediates rod and cone responses to light by regulating cGMP-gated channels in the photoreceptor outer segment, which open in the dark but close when illumination activates rapid cGMP hydrolysis by the rhodopsin–transducin–phosphodiesterase-6 cascade (reviewed in Refs. (1, 2, 3, 4)). Negative Ca2+ feedback controls the rate at which the retinal membrane guanylyl cyclase (RetGC) isozymes, RetGC1 (GUCY2D) and RetGC2 (GUCY2F) (5, 6, 7), produce cGMP in photoreceptor outer segments: when the influx of Ca2+ via the cyclic nucleotide–gated (CNG) channels stops, the catalytic activity of RetGC becomes accelerated by the Mg2+-liganded form of guanylyl cyclase–activating proteins (GCAPs), the Ca2+/Mg2+ sensor proteins (reviewed in Refs. (1, 4, 8, 9)). Activation of RetGC continues until cGMP reopens the CNG channels during the recovery phase of the response and resumes the influx of Na+ and Ca2+. Consequently, GCAPs bind Ca2+ and decelerate RetGC to prevent excessive production of cGMP and avoid opening more CNG channels. Some mutations in the human GUCY2D gene coding for RetGC1 cause inherited dominant retinopathies by deregulating Ca2+ sensitivity of RetGC1 (reviewed in Refs. (10, 11, 12)). In particular, various substitutions of Arg838 in the RetGC1 dimerization domain frequently cause autosomal dominant cone–rod dystrophy 6 (CORD6) (10, 11, 12). Several GUCY2D CORD6-linked mutations (13, 14), including R838S, have been biochemically characterized in vitro (13, 14, 15, 16) and in vivo (17, 18), demonstrating that in transgenic mice the mutated RetGC1 requires higher free Ca2+ concentrations to decelerate the cyclase activity and therefore abnormally elevates cGMP production and Ca2+ influx in the dark, leading to progressive photoreceptor degeneration.
Such Arg838 substitutions increase RetGC1 affinity for Mg2+GCAP, the activator form of GCAP, over the inhibitory form, Ca2+GCAP (13, 14, 15, 16). This alters two main biochemical aspects of the guanylyl cyclase regulation in photoreceptors. First, deceleration of the mutated RetGC1 now requires higher than normal free Ca2+ concentrations, which in turn elevates cGMP production and the influx of Ca2+ (16, 17). The apoptotic changes in photoreceptors triggered by the elevated levels of cGMP and Ca2+ in the outer segment can be therefore regarded as a “phototransduction disease” caused by deregulated Ca2+ feedback (17). Second, the increased affinity for Mg2+GCAP makes the mutated RetGC1 more resistant to inhibition by RD3 protein (16, 18).
RD3 was first identified as the product of a gene disrupted by mutations causing recessive degenerative retinal blindness in humans (Leber's congenital amaurosis type 12 [LCA12]) and in rd3 mouse strain (Rd3−/−) (19). RD3 is a low-abundance protein required for RetGC to be effectively transferred from the photoreceptor inner segment to the outer segment (20, 21, 22). RD3 also strongly inhibits RetGC basal activity and activation by GCAPs (23, 24). The in vivo evidence suggests that aberrant guanylyl cyclase activation by GCAPs in the absence of RD3 provokes the apoptotic process (25, 26), but this does not fully exclude the possibility that GCAPs mediate the apoptotic process via some target(s) other than RetGC (27). Therefore, we tested here the effects of deleting RetGC1 or RetGC2 in Rd3−/− retinas. We reasoned that if GCAPs mediated photoreceptor degeneration independently of RetGC, then the reduction of RetGC content would not affect, or possibly even increase, the progression of photoreceptor loss. Conversely, if the main cause of the Rd3−/− photoreceptor degeneration were their inability to suppress guanylyl cyclase, then Rd3−/−RetGC1−/− and/or Rd3−/−RetGC2−/− photoreceptors would degenerate more slowly than Rd3−/−.
The main hypothesis addressed in this study is that the lower susceptibility of the CORD6 RetGC1 variants to inhibition by RD3 (23, 24) contributes to the progression of this dominant retinal dystrophy. This hypothesis emerged from in vitro studies showing that GCAP1-activated R838S RetGC1 requires higher concentrations of RD3 to decelerate its activity. We reasoned that if the increased resistance to inhibition by RD3 aggravates the pathology caused by the Arg838 substitution in RetGC1, then increased expression of RD3 could slow the pace of degeneration.
We here present the evidence that (i) the removal of RetGC1 or RetGC2 delays Rd3−/− photoreceptor degeneration, thus confirming that RD3 protects photoreceptors mainly by suppressing aberrant RetGC activity and (ii) transgenic overexpression of RD3 in mice harboring R838S RetGC1 (R838S+) prolongs the survival of R838S+ photoreceptors. Our findings indicate that RD3 is involved in protecting photoreceptors from two different types of retinal degeneration—the recessive, such as LCA12, and the dominant, such as CORD6.
Results
Deletion of guanylyl cyclase isozymes RetGC1 or RetGC2 slows degeneration of Rd3−/− photoreceptors
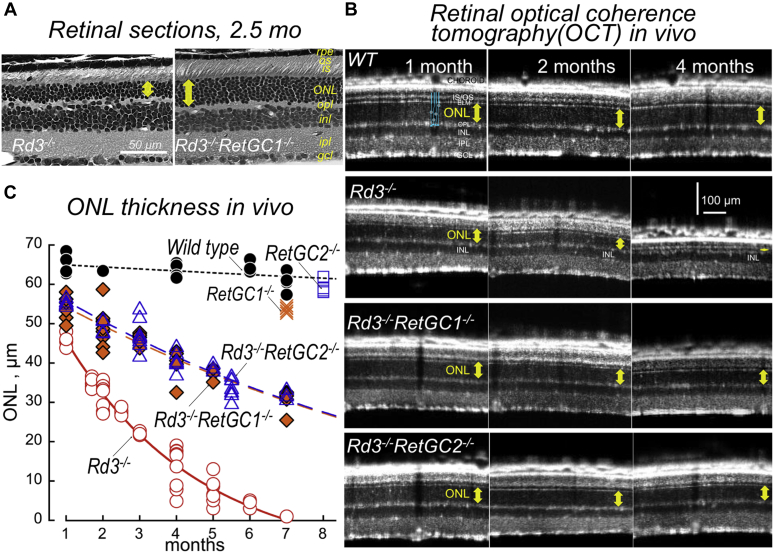
In wildtype (Rd3+/+) mice, simultaneous deletion of both RetGC1 and RetGC2 completely disables cGMP production, eliminates the photoreceptor function, and causes a slowly progressing degeneration of photoreceptors (25, 28). We reasoned that studying the en masse degenerating RetGC1−/−RetGC2−/− double-knockout photoreceptors lacking RD3 would make it rather cumbersome to isolate the potential contribution of RetGCs to the degeneration caused by the RD3 deficiency per se. In contrast, the individual gene knockouts, RetGC1−/− or RetGC2−/−, only reduce the levels of cGMP synthesis and alter photoresponses but do not entail massive photoreceptor degeneration (28, 29). So the Rd3−/− degeneration in RetGC1−/− or RetGC2−/− background would be caused by RD3 deficiency rather than the RetGC isozymes deficiency, making it possible to assess any potential role of RetGC activity in Rd3−/− degeneration. We therefore produced Rd3−/− in either RetGC1−/− or RetGC2−/−, both congenic with the C57B6 strain background (24). The reduction of the outer nuclear layer (ONL) of photoreceptors assessed by post-mortem morphological analysis and optical coherence tomography (OCT) in vivo showed that the rapid loss of the Rd3−/− ONL was dramatically delayed after deletion of RetGC1 or RetGC2 (Fig. 1, A and B). In all combinations, including Rd3−/−, the ONL was significantly thinner than in the wildtype (ANOVA, F = 184, Scheffé post hoc test p < 0.0001, confidence level 99% hereafter), but whereas only 20% normal nuclei count remained after 4 months in Rd3−/− retinas (Fig. 1, B and C), the respective 63% and 64% remained in Rd3−/−RetGC1−/− and Rd3−/−RetGC2−/−, both significantly higher than in Rd3−/− (p < 0.0001). This result conforms to the hypothesis that the primary cause of Rd3−/− photoreceptor rapid degeneration is the remaining RetGC activity unsuppressed by RD3.
Figure 1.
Deletion of RetGC isozymes slows degeneration of Rd3−/−photoreceptors.A, representative post-mortem retinal morphology in Rd3−/− (left) and Rd3−/−RetGC1−/− (right) littermates at 2.5 months of age. Photoreceptor outer nuclei layer (ONL) thickness hereafter is marked by the yellow arrow; other histological layers hereafter are as follows: GCL, ganglion cell layer; INL, inner nuclear layer; IPL, inner plexiform layer; IS, inner segments; OPL, outer plexiform layer; OS, rod outer segments; RPE, retinal pigment epithelium. B, representative in vivo OCT of the retina at 1, 2, and 4 months of age in (top to bottom) wildtype, Rd3−/−, Rd3−/−RetGC1−/−, and Rd3−/−RetGC2−/− mice; note the markedly improved Rd3−/− ONL thickness after deletion of either RetGC isozyme at 2 and 4 months; the location of photoreceptors is schematically shown in blue; bars in either direction—100 μm. C, progressing reduction of ONL thickness measured in vivo using OCT: wildtype (•), Rd3−/− (◯), RetGC1−/− (✕), RetGC2−/− (□), Rd3−/−RetGC1−/− (◆), and Rd3−/−RetGC2−/−(△); each data point is an average of ten measurements of ONL in retinal OCT sections per different mouse of indicated genotype; data fitted assuming exponential decay; note the much slower progression of the ONL loss in Rd3−/− after deletion of RetGC1 or RetGC2. OCT, optical coherence tomography; RetGC, the retinal membrane guanylyl cyclase.
Overexpression of RD3 does not alter the abnormal regulation of RetGC1 in R838S+ retinas
In order to test the hypothesis that the reduced sensitivity of R838S RetGC1 to inhibition by RD3 contributes to photoreceptor death in R838S+ rods, we overexpressed a human RD3 tagged with GFP at the C terminus in mouse line 362, in which rods transgenically express a human R838S RetGC1 (16, 17) and therefore undergo rapid degeneration. We reasoned that the increase of RD3 concentrations in R838S+ rods could compensate for the reduction of the R838S RetGC1/GCAP1 sensitivity to RD3 (23, 24). To enable monitoring the localization of the expressed RD3, we used RD3GFP that was previously shown to inhibit RetGC activity similarly to untagged RD3 and to fully rescue Rd3−/− rods from degeneration in vivo (25).
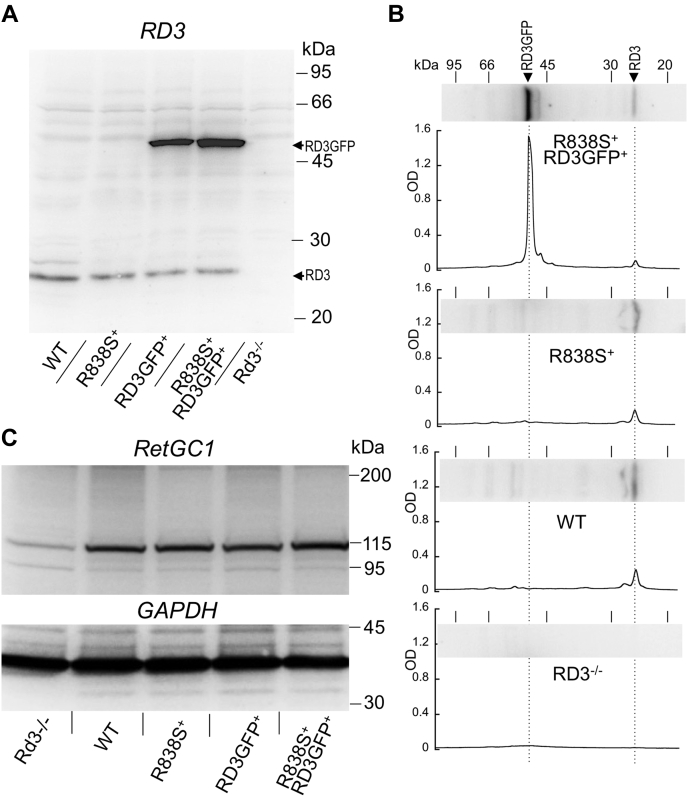
The overexpression of RD3GFP (Fig. 2) did not significantly affect regulation of the guanylyl cyclase activity in R838S+ retinas. The levels of RetGC expression in R838S+ and R838S+RD3GFP+ were similar, whereas the RD3 content was markedly increased in R838S+RD3GFP+ as compared with wildtype or R838S+. The expressed RD3GFP was of human origin, which could potentially reduce its reactivity toward the antimouse RD3 antibody. But even assuming that its immunoreactivity is similar to that of endogenous mouse RD3 and further taking into account the molecular mass being nearly twice that of the endogenous RD3 (Fig. 2B), the total RD3 content, the endogenous plus the transgenically expressed, in R838S+RD3GFP+ was at least fourfold to fivefold higher than normal (possibly even higher if the affinity of the antibody for the human RD3GFP was lower).
Figure 2.
Overexpression of RD3 in R838S+retinas.A and B, immunoblotting of the retinas from indicated genotypes were probed by antimouse RD3 antibody. B, the relative intensity of the RD3GFP versus the endogenous RD3 images on densitometry scans of the endogenous RD3 and the transgenically expressed human RD3GFP in (bottom to top) Rd3−/− (as negative control), wildtype, R838S+, and R838S+RD3GFP+ (all tracks in panel B are from the same blot). C, immunoblotting from the retinas of indicated genotypes probed by anti-RetGC1 antibody (top) or anti-GAPDH (bottom) antibody. RD3, retinal degeneration-3 protein.
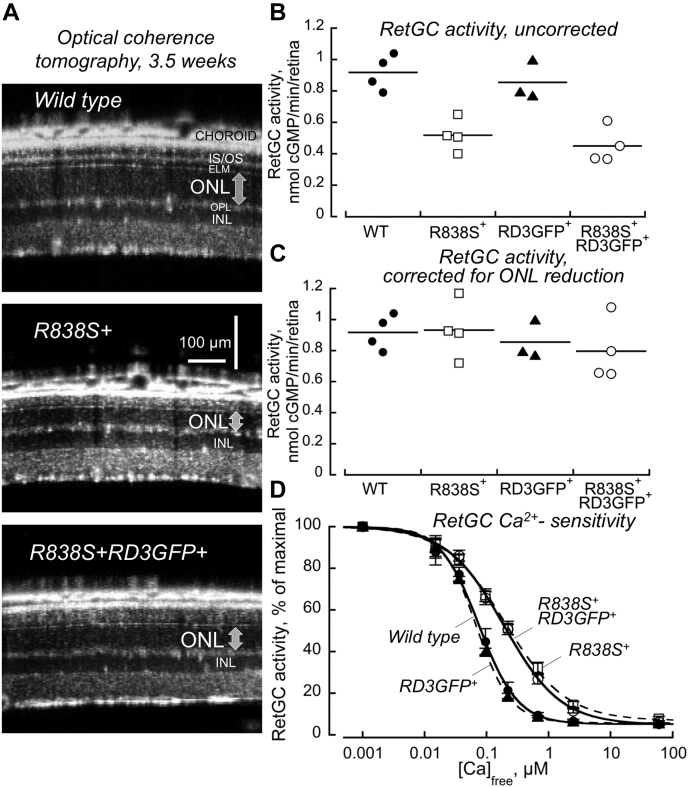
Also, the overexpression of RD3 in R838S+ RD3GFP+ did not critically affect overall RetGC activity or its calcium sensitivity (Fig. 3, A–C). After 3 weeks of age, the early stage of degeneration only partially reducing the photoreceptor content, the bulk activity of the photoreceptor guanylyl cyclase in R838S+ and R838S+RD3GFP+ retinas appeared lower than normal. However, it became similar to the wildtype after being corrected for the partial reduction of the photoreceptor mass as compared with wildtype by measuring the ONL thickness in vivo (Fig. 3, A and C). Overexpression of RD3GFP in wildtype retinas did not significantly alter the normal GCAP-dependent Ca2+ sensitivity of the cGMP production ([Ca]1/2, mean ± SD): 0.067 ± 0.003 μM versus 0.076 ± 0.012 μM (p = 0.25, Student's t test) (Fig. 3D). Notably, the abnormal Ca2+ sensitivity of RetGC in R838S+RD3GFP+ retinas also remained identical to its abnormal Ca2+ sensitivity in R838S+ (Fig. 3D). The [Ca]1/2 for deceleration of RetGC activity, 0.191 ± 0.021 and 0.203 ± 0.026 μM, respectively, was both markedly different from the wildtype (p < 0.0008) but virtually indistinguishable from each other (p = 0.50).
Figure 3.
Overexpression of RD3 does not affect photoreceptor guanylyl cyclase activity in R838S+retinas.A, OCT in vivo scans comparing the photoreceptor nuclei mass in wildtype (top) degenerating R838S+ and R838S+RD3GFP+ at 3.5 weeks of age; the arrow marks photoreceptor ONL thickness. B and C, RetGC activity measured in the presence of 1 mM EGTA in the retinas from 3.5-week-old mice before (B) or after (C) correction for the loss of photoreceptor ONL layer: wildtype (•), R838S+(□), RD3GFP+ (▲), and R838S+RD3GFP+(◯). D, normalized Ca2+ sensitivity of RetGC activity in wildtype (•), R838S+(□), RD3GFP+ (▲), and R838S+RD3GFP+(◯) retinas. See Experimental procedures section for details. OCT, optical coherence tomography; ONL, outer nuclear layer; RD3, retinal degeneration-3 protein; RetGC, the retinal membrane guanylyl cyclase.
Overexpression of RD3 delays degeneration of R838S+ rods
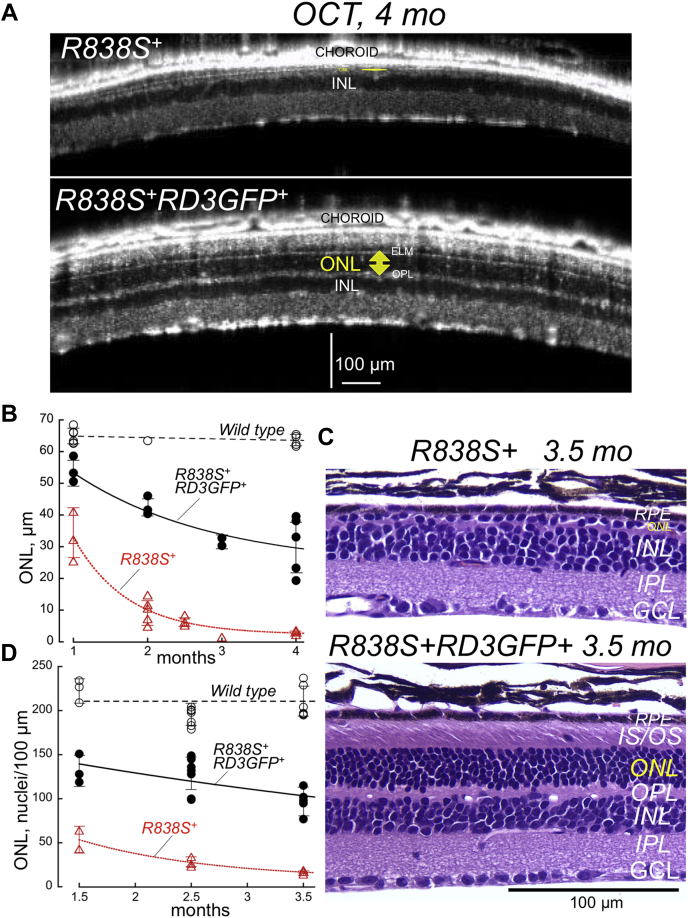
Both R838S+ and R838S+RD3GFP+ developed photoreceptor degeneration. Yet, despite retaining the same abnormal Ca2+ sensitivity of RetGC as in their parental R838S+ line, the degenerative changes of retinal morphology in R838S+RD3GFP+ mice developed much more slowly than in R838S+ siblings (Fig. 4). The in vivo OCT scans revealed markedly improved ONL thickness after 4 months (31 ± 9 versus 3 ± 1 μm in R838S+; Student’s t test, p = 0.0023) (Fig. 4A). In contrast to R838S+, where the photoreceptor layer completely degenerated within 3.5 months, the ONL thickness in R838S+RD3GFP+ gradually declined to 50% wildtype at the end of the 4-month period (Fig. 4B). Likewise, in a post-mortem histological analysis of the retinas (Fig. 4, C and D), photoreceptor nuclei count per 100 μm in the R838S+RD3GFP+ retinas was lower than in wildtype (97 ± 14, n = 3 versus 210 ± 17, n = 6; p < 0.0001), yet dramatically improved compared with R838S+ (17 ± 3, n = 5; p = 0.0001). The remaining 49% of the photoreceptors in R838S+RD3GFP+ retained recognizable outer segment layer, whereas no recognizable photoreceptor structures remained in R838S+ (Fig. 4D).
Figure 4.
RD3 overexpression partially rescues Rd3−/−rod degeneration.A, representative retinal OCT images of R838S+ and R838S+RD3GFP+ littermates at 4 months of age; note the complete lack of photoreceptors in R838S+ and the preservation of identifiable ONL in R838S+RD3GFP+. B, quantitation of the ONL in wildtype (◯), R838S+ (△), and R838S+RD3GFP+ (•) by OCT in vivo at different ages; error bars represent SD. C, representative post-mortem retinal morphology in histological sections from R838S+ and R838S+RD3GFP+ littermates at 3.5 months of age; note the nearly complete lack of identifiable photoreceptor nuclei in R838S+ and their abundant presence in R838S+RD3GFP+ along with the identifiable histological layer of rod inner and outer segments. D, linear density of photoreceptor nuclei in wildtype (◯), R838S+ (△), and R838S+RD3GFP+ (•) counted using histological sections. OCT, optical coherence tomography; ONL, outer nuclear layer; RD3, retinal degeneration-3 protein.
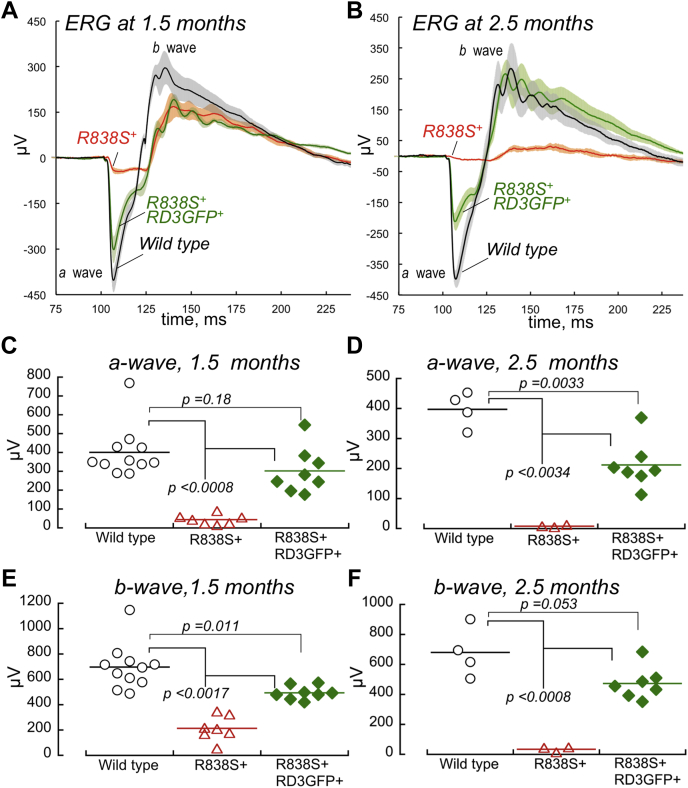
RD3GFP overexpression delays the loss of electroretinography responses in R838S+ retinas
Retinal function assessed by dark-adapted electroretinography (ERG) was also preserved in R838S+RD3GFP+ much better than in R838S+ (Fig. 5). The ERG a-wave (negative voltage deflection produced by hyperpolarization of photoreceptors in response to a bright flash) was strongly suppressed at 1.5 months of age (44 ± 26 μV; mean ± SD) (Fig. 5A) and virtually undetectable (<10 μV) at 2.5 months of age in R838S+ (Fig. 5B) because of rapidly progressing en masse loss of photoreceptors. In comparison with R838S+, the ERG a-wave amplitudes in R838S+RD3GFP+ were much larger both at 1.5 months (302 ± 120 μV, p = 0.0008) (Fig. 5, A and C) and at 2.5 months (212 ± 79 μV; p < 0.0001) (Fig. 5, B and D) of age. They were only slightly lower in R838S+RD3GFP+ than the wildtype (401 ± 135 μV; p = 0.18) at 1.5 months (Fig. 5, A and C) but more significantly (p = 0.0033) reduced (212 ± 79 μV) than the 397 ± 58 μV in the wildtype at 2.5 months (Fig. 5B), as R838S+RD3GFP+ photoreceptors continued to gradually degenerate, albeit much slower than in R838S+. The b-wave generated by inner retina neurons receiving input from photoreceptors (positive voltage deflection measured from the bottom of the a-wave), despite being lower than in wildtype, was also drastically improved in R838S+RD3GFP+ as compared with R838S+, both at 1.5 (Fig. 5E) and 2.5 months (Fig. 5F).
Figure 5.
RD3 overexpression offsets the loss of rod function in R838S+.A and B, dark-adapted ERG (mean ± SEM error bars) recorded in wildtype (black traces, gray error bars), R838S+ (red traces, pale orange error bars), and R838S+RD3GFP+ (green traces, pale green error bars) mice at 1.5 months (A) and 2.5 months (B) of age in response to a 1-ms 505-nm flash of ∼1 × 106 photons/rod delivered at 100 ms on the timescale. C and D, the a-wave amplitudes in wildtype (◯), R838S+ (△), and R838S+RD3GFP+ (◆) mice at 1.5 months of age (C) and 2.5 months of age (D). E and F, the respective b-wave amplitudes at 1.5 months of age (E) and 2.5 months of age (F). ERG, electroretinography; RD3, retinal degeneration-3 protein.
RD3 is displaced from the complex with RetGC1 in the outer segment
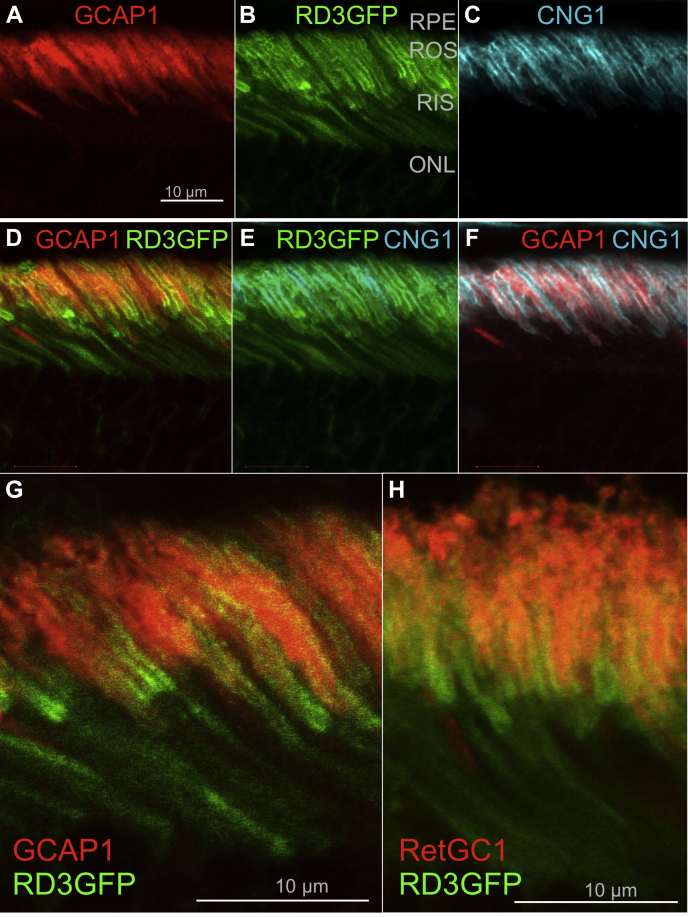
RD3 and GCAP regulate RetGC activity in opposite directions: GCAP activates RetGC, and RD3 inhibits it in a manner indicating that it disables the guanylyl cyclase stimulation by competing with GCAP (23, 24, 25). Conversely, GCAP would have to take over the RetGC regulation in the outer segment, otherwise Ca2+ feedback on the cyclase would be disabled (23). Therefore, we tested RD3GFP versus GCAP1 and RetGC1 localization in R838S+ and R838S+RD3GFP+ retinas (Fig. 6).
Figure 6.
RD3GFP separates from the RetGC1 and GCAP1 accumulated in ROS.A–F, immunofluorescence in 3.5-week-old RD3GFP+ retinas probed with anti-GCAP1 rabbit polyclonal antibody (red) (A, D, and F) or anti-CNG1 mouse polyclonal antibody (pseudocolored cyan; C, E, and F) in comparison with RD3GFP fluorescence (green; B, D, E, and F); the bar represents 10 μm. G and H, separation of RD3GFP from GCAP1 (G) and RetGC1 (H) immunofluorescence in R838S+RD3GFP+ ROS. The R838S+RD3GFP+ retina sections were probed by rabbit polyclonal anti-GCAP1 (red, G) or RetGC1 (red, H) antibody. Note the similarity between the GCAP1 and RetGC1 localization inside ROS, different in the both cases from the localization of green RD3GFP fluorescence, which is concentrated in the inner segment and near the base of the ROS, partially extending along the plasma membrane but not to the inside of the ROS. CNG, cyclic nucleotide–gated; GCAP, guanylyl cyclase–activating protein; RD3, retinal degeneration-3 protein; RetGC, the retinal membrane guanylyl cyclase; ROS, rod outer segment.
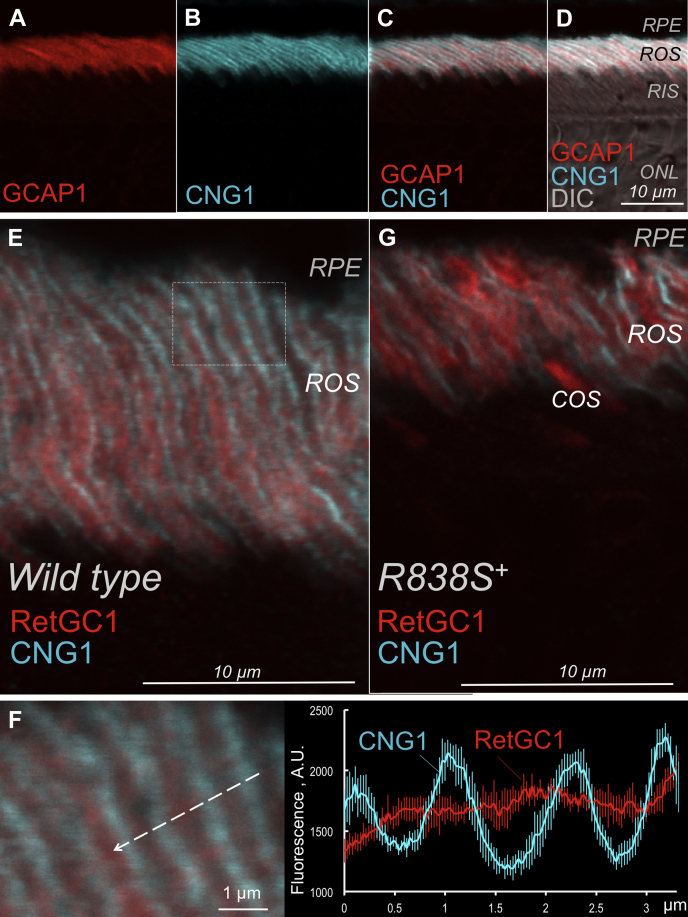
Whereas the anti-CNG1α fluorescence exclusively marking the rod outer segment (ROS) plasma membrane was distributed along the entire ROS length, RD3GFP fluorescence only partially overlapped with the anti-CNG1α in both RD3GFP+ and R838S+RD3GFP+ (Figs. 6 and 7). The bulk of the green RD3GFP fluorescence was observed in proximity to the lower portion of the ROS only partially spreading toward the ROS plasma membrane (Fig. 6): the brightest fluorescence accumulated near the base of the outer segment and was also spread in the inner segment/myoid part of rods (Fig. 6). RetGC1 and GCAP1 immunofluorescence signals (Figs. 6 and 7) did not overlap with CNG1α either (Pearson's correlation coefficient <0.5), but in contrast to RD3GFP, they were both detectable inside R838S+, R838S+RD3GFP+, and wildtype ROS (Fig. 7, E and F), showing a pattern more consistent with the predominant localization of the cyclase in photoreceptor discs (30) rather than the ROS plasma membrane.
Figure 7.
GCAP1 accumulates with RetGC inside wildtype and R838S+ROS.A–D, immunofluorescence in the wildtype retinas probed with anti-GCAP1 rabbit polyclonal antibody (red) (A, C, and D) and anti-CNG1 mouse polyclonal antibody (cyan) (B, C, and D); the merged anti-GCAP1 and anti-CNG1 fluorescence (C and D) were superimposed on DIC image (D). E–G, anti-RetGC1 (AlexaFluor 543, red) and anti-CNG1 (AlexaFluor 647, pseudocolored cyan) immunofluorescence in photoreceptors of 3.5-week-old wildtype (E and F) and R838S+ (G) mice. In F (left panel), the magnified region of the wildtype retina (marked with the rectangle in E) was scanned along the dashed line; the distribution of the respective fluorescence intensities is shown in the right panel. Pearson's correlation coefficient for the two fluorochromes in (E) was 0.31 ± 0.25 (n = 8). See Experimental procedures section for details. CNG, cyclic nucleotide–gated; DIC, differential interference contrast image; GCAP, guanylyl cyclase–activating protein; RetGC, the retinal membrane guanylyl cyclase; ROS, rod outer segment.
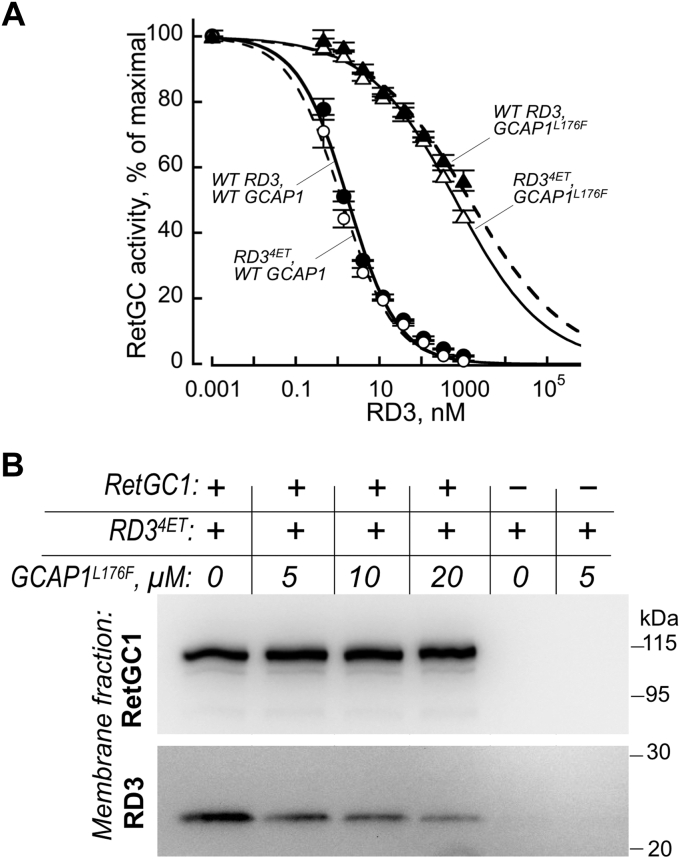
We hypothesized that the different compartmentalizations of GCAP and RD3 in photoreceptors result from displacement of RD3 by GCAP from the complex with RetGC, presumably before the guanylyl cyclase reaches its destination in the ROS photoreceptor discs. However, the content of RD3 in the retina is too low (20) to allow for a reliable direct assessment of its complex with the RetGC and possible RD3 displacement by GCAPs. Therefore, to assess whether such displacement is mechanistically possible, we used reconstitution of the recombinant RD3 with RetGC1 expressed in human embryonic kidney 293 (HEK293) cells membranes and tested their binding using cosedimentation after ultracentrifugation (Fig. 8). We found that the micromolar affinity of the RetGC isozymes for GCAPs (Ref. (31) and Fig. 8A) is insufficiently strong to allow a stable GCAP1:RetGC1 membrane complex to be isolated in this type of experiment. In contrast, the apparent RetGC affinity for RD3 is at least 100-fold higher, and the RD3:RetGC complex is more stable (20, 23, 24). However, the very poor solubility of RD3 heterologously expressed in bacterial and human cells (23, 32, 33) presents a major challenge because precipitated RD3 nonspecifically contaminates the fraction of the RetGC1-containing membranes. To overcome that obstacle, we produced an RD3 variant (RD34ET) in which Glu replaced Arg in positions 154, 156, 158, and 167, and the last 18 C-terminal residues were deleted. RD34ET fully retained the inhibitory activity of wildtype RD3 in vitro (Fig. 8A), but, unlike wildtype RD3, remained in a soluble fraction after ultracentrifugation. The much lower apparent affinity of GCAP1 for RetGC1 than in RD3 presents another major challenge for the binding experiments, requiring unrealistically high concentrations of GCAP1 in the assay. To mitigate this, we used L176F GCAP1, mutation that increases RetGC1 affinity for GCAP1 approximately fivefold (34, 35).
Figure 8.
GCAP can displace RD3 from RetGC1.A, inhibition of RetGC1 activity by RD3 in vitro. The RetGC1 activity in HEK293 membranes activated by 1.5 μM wildtype Mg2+ GCAP1 (●, ◯) or Mg2+ GCAP1L176F (▲, △) was assayed in the presence of increasing concentrations of wildtype RD3 (•, ▲) or RD34ET (◯, △). B, cosedimentation assay of RD34ET binding to RetGC1 using ultracentrifugation. Immunoblots of HEK293 membrane pellets, either containing (+) or not containing (−) expressed RetGC1, were obtained by ultracentrifugation after preincubation with 50 nM RD34ET in the presence of indicated concentrations of L176F GCAP1. The immunoblot was probed by anti-RetGC1 (top) or anti-RD3 (bottom) antibody as described in the Experimental procedures section. GCAP, guanylyl cyclase–activating protein; HEK293, human embryonic kidney 293 cells; RD3, retinal degeneration-3 protein; RetGC, the retinal membrane guanylyl cyclase.
Confirming the specificity of the binding of RD3s with the RetGC1, in the centrifugation assay, the soluble RD34ET cosedimented with the HEK293 membranes containing RetGC1 but not with HEK293 membranes lacking the expressed guanylyl cyclase (Fig. 8B). There was clear evidence that RD3 was bound to the RetGC1-containing membranes in the absence of GCAP1, yet it was displaced from the membrane complex with RetGC1 in a dose-dependent manner in the presence of high concentrations of GCAP1.
Discussion
Two RetGC isozymes mediate Rd3−/− photoreceptor degeneration
RD3 inhibits the RetGC1 and RetGC2 basal activity and also prevents RetGC activation by GCAPs (23, 24) (Fig. 9A). Studying murine models in vivo (24, 25) indicated that activation of RetGC by GCAPs, most likely in the inner segment, was the major reason that Rd3−/− photoreceptors begin to rapidly degenerate soon after completing their differentiation (36) (Fig. 9B). Deletion of the two GCAP isoforms dramatically rescued Rd3−/− photoreceptors from degeneration (25, 26, 27). Yet the possibility that the apoptotic effect of GCAPs in Rd3−/− photoreceptors was propagated not via aberrant activation of the guanylyl cyclase but through some other process (27) could not be fully excluded. The most interesting alternative possibility that has been suggested is that GCAP2 causes photoreceptor death by activating in the inner segment an apoptotic pathway(s) in a way that is unrelated to RetGC regulation (27). In our recent study (26), we observed that deletion of GCAP2 indeed led to partial rescue, but it was less effective than deletion of GCAP1 and especially less effective than simultaneous deletion of both GCAPs. The results of our present study demonstrate that removal of either RetGC isozyme markedly reduces the rate of Rd3−/− photoreceptor degeneration (Fig. 1), similarly to the previously documented deletion of GCAPs (25, 26, 27). Taken together, these results do not support the hypothesis that GCAPs in the absence of RD3 facilitate apoptosis primarily through a process unrelated to activation of RetGC. Instead, the data support the hypothesis that RD3 protects photoreceptors by suppressing aberrant RetGC1 and RetGC2 activities stimulated by GCAPs (Fig. 9A) and that the lack of such suppression is the primary cause of Rd3−/− photoreceptor death.
Figure 9.
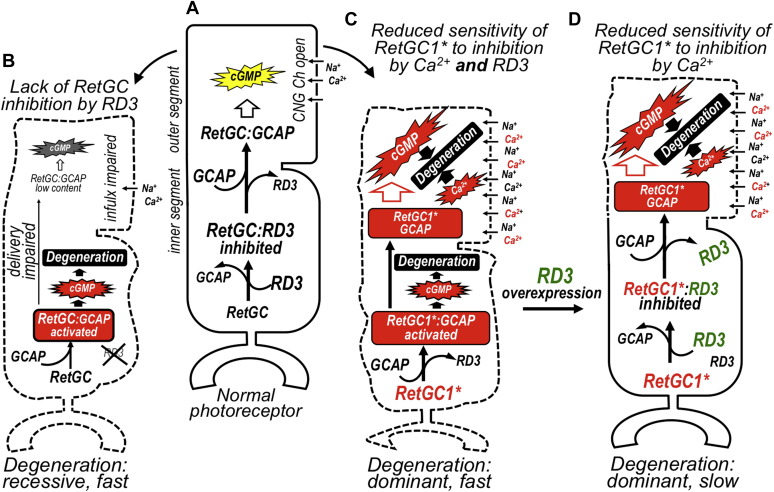
The putative role of RD3 in preventing two different types of rapid photoreceptor degeneration.A, in normal photoreceptors, RD3 plays a dual role: (i) it facilitates RetGC delivery to the outer segment and (ii) protects the cells against aberrant activation of guanylyl cyclase in the inner segment until the enzyme reaches its destination in the outer segment, where GCAPs displace RD3 and control RetGC activity via Ca2+ feedback. B, in RD3-deficient photoreceptors such as in rd3 mice and LCA12 patients, the RetGC content sharply declines and the production of cGMP in the outer segment becomes insufficient to maintain the normal inward current through CNG channels, which reduces responses to light and possibly provokes a slow component of the degenerative process. The remaining RetGC is no longer protected against aberrant activation by GCAPs, and such activation causes rapid degeneration of the RD3-deficient photoreceptors. C, in photoreceptors expressing GUCY2D dominant gain-of-function mutations (RetGC1∗), such as CORD6-linked mutation R838S, the higher affinity of the RetGC1∗ for GCAP causes a “phototransduction disease” by deregulating the suppression of the RetGC1∗ activity by Ca2+ in the outer segment, which in turn causes excessive cGMP production and abnormal elevation of Ca2+ influx in the dark, promoting the apoptotic process. The increased affinity of RetGC1∗ for GCAP also hinders RetGC1∗ silencing by RD3. The aberrant stimulation of the cyclase, possibly in the inner segment, activates the secondary pathway of degeneration, similarly to that in RD3-deficient photoreceptors. D, the increase of RD3 levels by overexpressing RD3GFP enables RD3 to compete with GCAPs for RetGC1∗ more effectively and offsets the secondary, RD3-related, pathway of the apoptosis. Therefore, the dominant photoreceptor degeneration driven now only by the primary apoptotic pathway of deregulated Ca2+ feedback in R838S+ outer segment becomes slower. CNG, cyclic nucleotide–gated; CORD6, cone–rod dystrophy 6; GCAP, guanylyl cyclase–activating protein; RD3, retinal degeneration-3 protein; RetGC, the retinal membrane guanylyl cyclase.
The strongly defined protective effect of deleting RetGC2 was somewhat surprising. Based on the biochemical evidence, RetGC2 is mostly an ancillary isozyme in mouse rods, accounting for ∼30% of the total guanylyl cyclase activity (31). Therefore, one may expect that RetGC2, which is regulated in vivo almost exclusively by GCAP2 (37), would contribute less to the progression of Rd3−/− photoreceptor death than RetGC1. However, the preservation of Rd3−/− photoreceptors after deletion of RetGC2 was similar to that of RetGC1 (Fig. 1, B and C). One possible reason is that RetGC2 in mouse photoreceptors has a higher basal activity than RetGC1 (31). Consequently, the contribution of RetGC2 to the apoptotic process triggered by RD3 deficiency can be higher than its contribution to the total GCAP-stimulated RetGC activity.
Another component causing major dysfunction of Rd3−/− photoreceptors is the severe reduction of RetGC content (20, 21, 25), resulting in suppression of their photoresponses even before the en masse degeneration occurs (Fig. 5 and Refs. (19, 24, 25)). Even though the low guanylyl cyclase activity per se is not the main reason for the rapidly progressing Rd3−/− degeneration (24), mouse photoreceptors completely lacking RetGC activity are known to slowly degenerate (24, 28). Hence, the decrease in RetGC activity could conceivably facilitate apoptosis via drastically lowering cGMP and Ca2+ levels in the Rd3−/− outer segment. More definitive evaluation of such a possibility will require deeper insight into the as-yet insufficiently understood role of RD3 in facilitating RetGC delivery to the outer segment.
The protective function of RD3 in two different types of photoreceptor degeneration
Deregulation of RetGC1 makes GUCY2D CORD6 primarily a “phototransduction disease,” altering the negative Ca2+ feedback on RetGC (17). Various mutations linked to CORD6 cause substitutions or Arg838 in a human RetGC1 dimerization domain (10, 11, 12, 13, 14). The dimerization domain of RetGC1 is an important structural part of the cyclase enzyme regulated by GCAPs (32, 38). The coiled–coil interactions altered by the Arg838 substitutions elevate the affinity of RetGC1 for Mg2+GCAP1 (13, 14, 15, 18). As a result, higher concentrations of Ca2+ are needed to drive RetGC1 into a complex with Ca2+GCAP1 in order to decelerate the cyclase activity so that the normal levels of Ca2+ in photoreceptors become insufficient for properly suppressing cGMP production in the dark (16, 17).
The two main apoptotic triggers are considered to be the increased rate of cGMP production in the outer segment and the elevated influx of Ca2+ in the dark via cGMP-gated CNG channels (39, 40). Both changes occur in the R838S+ mouse rods in vivo (17). However, in a previous study (16, 26), we observed that another, more recently recognized aspect of the RetGC regulation—its inhibition by RD3—may contribute to the disease (Fig. 3). The higher affinity of the mutated cyclase for GCAP also makes it more difficult for RD3 to counteract the cyclase activation. Hence, the normal concentrations of RD3 in CORD6 photoreceptors may become insufficient to prevent the aberrant RetGC1 activation by GCAP in the inner segment and hence aggravate the apoptotic process (Fig. 9C). If this hypothesis were correct, then one would expect that increasing concentrations of RD3 would reduce the pace of degeneration in an animal model harboring a human RetGC1 with a CORD6 mutation, and in fact, this protective effect of RD3 overexpression in R8383S+ rods was evident in the present study (Figs. 4 and 5).
It is also important to note that despite being evident, the rescue of R838S+RD3GFP+ photoreceptors was not complete and the photoreceptors continued to degenerate, albeit at a much slower pace than in R838S+ littermates. This indicates that the two components in the R838S+ degeneration (Fig. 9C) propagate the apoptotic process via different pathways—one triggers the degenerative “phototransduction disease” by deregulating Ca2+ feedback in the outer segment, and the other by unprotecting the mutant RetGC1 against aberrant activation by GCAPs, either in the inner segment or in a “wrong” part of the outer segment before RetGC1 compartmentalizes with GCAP (Figs. 6 and 7).
The results of our present study (Fig. 8) are consistent with kinetic data (23, 24) indicating that GCAPs displace RD3 from RetGC to enable the Ca2+-sensitive regulation of the enzyme in the outer segment. However, it evidently requires very high local concentrations of GCAP to displace RD3 from the complex with the cyclase (Fig. 8), despite the higher than normal affinity for RetGC1 of L176F GCAP1 used in this experiment. It therefore remains unclear how such displacement occurs upon transition of the enzyme from the inner segment to the photoreceptor disc membrane. It is possible that the RetGC1:RD3 complex moves from the inner segment through a compartment containing a high local concentration of GCAP, but it is also possible that RD3, RetGC, and/or GCAP upon entering the outer segment undergo modification(s) that reduce(s) RetGC affinity for RD3 and increases its affinity for GCAP1. These possibilities remain to be explored in future studies.
Expression of RD3 (41) or RD3GFP (25) in Rd3−/− photoreceptors restores their function and prevents degeneration in a mouse rd3 model of the human recessive LCA12. Importantly, the results of our present study also indicate that overexpression of RD3 should be further explored as a possible approach to support the survival of photoreceptors expressing human GUCY2D with Arg838 substitutions. Furthermore, some other types of dominant retinal degeneration originate from mutations in a human GUCA1A gene coding for GCAP1 (reviewed in Refs. (9, 12)), and in many cases, such mutations deregulate Ca2+ sensitivity of guanylyl cyclase in a manner similar to the Arg838 mutations in RetGC1. In at least some of these cases, the affinity of Mg2+GCAP1 for RetGC1 can increase and sensitivity to inhibition by RD3 can decrease (42). Additional studies could help shed light on whether degeneration of photoreceptors caused by the dominant mutations in GCAP1 can be partially rescued by overexpression of RD3.
Experimental procedures
Animals
All experiments involving animals were conducted in accordance with the Public Health Service guidelines and approved by the Salus University Institutional Animal Care and Use Committee. The wildtype C57B6J and rd3/rd3 mouse strains were purchased from JAX Research/Jackson's Laboratory. R838S+ mice (line 362) and mice overexpressing RD3GFP (line 932) were produced as described previously (16, 25). Mice of other parental genotypes used in this study were kindly provided by other investigators: RetGC1−/− line (29)—by Dr David Garbers (University of Texas), and RetGC2−/− (28)—by Dr Wolfgang Baehr (University of Utah). All mouse lines used in this study were made congenic to the C57B6 background by repetitive breeding for over ten generations prior to conducting the experiments (24). The RetGC1−/− or RetGC2−/− mice were bred for several generations to Rd3−/− in C57B6 background to first produce Rd3−/−RetGC1+/− or Rd3−/−RetGC2+/−, then the Rd3−/−RetGC1+/− and Rd3−/−RetGC2+/− were bred to produce Rd3−/−RetGC1−/− or Rd3−/−RetGC2−/−, and those were subsequently bred for the experiments described in Figure 1. Age-matched Rd3−/− mice of the parental rd3/rd3 strain were used for comparison. The animals were genotyped using PCR utilizing the respective genotype-specific primers and, in the case of Rd3−/− background, DNA sequencing of the PCR fragments as described previously in full detail (24, 25, 31). The R838S+RD3GFP+ mice were produced by breeding heterozygous transgenic mouse line 932 (25) expressing human RD3GFP (RD3GFP+) to a heterozygous transgenic R838S+ mouse line 362 (16, 17) expressing the human R838S RetGC1. In the subsequent experiments, the R838S+RD3GFP+ mice were compared with their R838S+ littermates. Mice were fed the same diet and were housed in the same temperature- and humidity-controlled environment using 12 h/12 h light/dark cycle.
RD3 expression and purification
Recombinant human RD3 was expressed from a pET11d vector in a BL21(DE3) Codon Plus Escherichia coli strain (Stratagene/Agilent Technologies) induced by isopropyl-β-d-thiogalactopyranoside, extracted from the inclusion bodies and purified by salt precipitation and dialysis as previously described (23, 24, 33). RD34ET was in addition purified using gel filtration on Sephacryl S-200 column.
GCAP1 expression and purification
Myristoylated bovine GCAP1 for the in vitro assays was expressed from pET11d vector in a BLR(DE3) E. coli strain (both originated from Novagen/Calbiochem) harboring a pBB131 plasmid coding for a yeast N-myristoyl transferase and purified by calcium precipitation, butyl-Sepharose and Sephacryl S-100 chromatography using previously described procedures (43, 44, 45). The purity of GCAP1 preparations estimated by SDS gel electrophoresis was ≥90%.
RetGC1 expression and guanylyl cyclase activity assays
Human recombinant RetGC1 was expressed from a modified Invitrogen pRCCMV vector in HEK293 cells transfected using calcium phosphate precipitation method, and the membrane fraction containing the expressed RetGC1 was isolated as previously described (15). The guanylyl cyclase activity was assayed as previously described in detail (15) with modification described (24). In brief, the assay mixture (25 μl) containing HEK293 membranes, 30 mM Mops–KOH (pH 7.2), 60 mM KCl, 4 mM NaCl, 1 mM DTT, 2 mM Ca2+/Mg2+/EGTA buffers, 0.9 mM free Mg2+, 0.3 mM ATP, 4 mM cGMP, 1 mM GTP, and 1 μCi of [α-32P]GTP, 100 μM zaprinast and dipyridamole, and 10 mM creatine phosphate/0.5 unit of creatine phosphokinase (Sigma–Aldrich) was incubated at 30 °C for 30 min, and the reaction was stopped by heat inactivation at 95° for 2 min. The resultant [32P]cGMP product was separated by TLC using fluorescently backed polyethyleneimine cellulose plates (Merck) developed in 0.2 M lithium chloride, cut from the plate, and eluted with 2 M lithium chloride in scintillation vials, and the radioactivity was counted using liquid scintillation. Mouse retinas for RetGC activity measurements were excised from the dark-adapted 3.5-week-old mice using infrared illumination (Kodak number 11 infrared filters) and a dissecting microscope fitted with an Excalibur infrared goggles, wrapped in aluminum foil, frozen in liquid nitrogen, and stored at −70 °C prior to their use in the cyclase activity assays, also conducted under infrared illumination. The incubation time for the reaction in that case was 12 min. The assay contained [3H]cGMP internal standard to ensure the lack of the cGMP product hydrolysis by retinal phosphodiesterase. Ca2+/EGTA buffers at 0.9 mM free Mg2+ were prepared using Tsien and Pozzan method (46) and verified by fluorescent indicator dyes as previously described in detail (44). Data fitting was performed using Synergy KaleidaGraph 4 software.
ERG
Mice were dark-adapted overnight, their pupils were dilated by applying 1% tropicamide and 2.5% phenylephrine ophthalmic eye drops under dim red safelight illumination, and the mice were dark adapted for another 10 min. Full-field ERG in mice anesthetized by inhalation of 1.7 to 1.9% isoflurane (VEDCO)/air mix delivered by a Kent Scientific SomnoSuite setup at 50 ml/min was performed in the dark as previously described in detail (24, 25) using a Phoenix Research Laboratories Ganzfeld ERG2 instrument. A 505-nm 1-ms light pulse (∼1 × 106 photoisomerizations/rod) was delivered through the infrared camera–guided corneal electrode/LED light source of the instrument.
Retinal post-mortem morphology
Mice were anesthetized with a lethal dose of ketamine/xylazine injection, perfused through the heart with PBS, and then with 2.5% glutaraldehyde in PBS. The eyes were surgically removed and fixed overnight in 2.5% glutaraldehyde/2.5% formaldehyde in PBS (Electron Microscopy Sciences) at 4 °C. The fixed eyes were washed in PBS, soaked in PBS overnight, processed for paraffin embedding, sectioned at 5 μm, and stained with hematoxylin/eosin (AML Laboratories). The retinal sections were photographed using an Olympus BX21 microscope fitted with an Olympus MagnaFire camera, and then the identifiable photoreceptor nuclei per 100 μm in the ONL between the optic nerve and the periphery were counted and averaged from 425-μm frames.
In vivo OCT
Mice were anesthetized using intraperitoneal injection of 20 mg/kg ketamine and 8 mg/kg xylazine. The pupils were dilated by applying 1% tropicamide and 2.5% phenylephrine ophthalmic eye drops ∼10 min before the scan. The B-scans of the retinas were acquired using an IIScience spectral domain OCT camera calibrated by the manufacturer at 2.47 μm/pixel axial scale and 3.5 μm/pixel lateral scale resolution and averaged from 200 to 400 frames. The thickness of the ONL layer was measured between the outer plexiform and the external limiting membrane reflective layers (47, 48). The OCT scans are presented with the choroid on top of the image and the ganglion cell layer on the bottom of the image.
Antibodies
Rabbit RD3 polyclonal antibody 10929 was produced against purified recombinant mouse RD3 expressed in E. coli as described (24); anti-GCAP1 and anti-RetGC1 rabbit polyclonal (RRID: AB_2877058) antibodies were characterized previously (17, 18, 24, 49); rabbit polyclonal anti-GAPDH antibody was purchased from Invitrogen/Thermo Fisher Scientific (catalog no. PA1987). Polyclonal mouse antibody against CNG1 alpha subunit was produced in mice immunized by ∼22-kDa recombinant fragment, Ser493–Asp684, of a mouse CNG1α subunit expressed in E. coli from pET15b vector with a 6-His tag at the NH2 terminus and purified using affinity chromatography on a Ni column; for immunization, the antigen was injected subcutaneously, first using a complete Freund’s adjuvant and then incomplete Freund’s adjuvant (both from Thermo Fisher Scientific) for three subsequent booster injections delivered ca. 3 weeks apart. The blood was collected from mice anesthetized with a lethal injection of ketamine/xylazine and supplemented with 5 mM EDTA to prevent coagulation; the plasma was then separated from blood cells by centrifugation for 5 min at 5000g, 4 °C, and then twice for 10 min at 10,000g, 4 °C; the clear supernatant was aliquoted and frozen in liquid nitrogen and stored at −70 °C.
Membrane binding assay
A 50-μl assay mixture containing HEK293 membranes in 30 mM Mops/KOH, pH 7.2, 60 mM KCl, 5 mM NaCl, 1 mM DTT, 0.1 mM ATP, 2 mM MgCl2, 0.4 mg/ml bovine serum albumin, 2 mM EGTA, inhibitors of proteases, 50 nM hRD34ET, and various concentrations of L176F bGCAP1D6S was assembled on ice and then incubated for 15 min at room temperature. The assay mixture was loaded on top of 0.5 ml of cold 20% sucrose in 5 mM Tris, pH 7.5 in a centrifuge tube for a Beckman Optima TLS rotor, and centrifuged for 10 min at 40,000 rpm at 4 °C. First, the top 0.2 ml of supernatant was removed, then 0.4 ml of 5 mM Tris was carefully added on top of the remaining sucrose layer, and after that, all the supernatants were carefully aspirated, from top to bottom, to prevent nonspecific contamination of the pellet by RD3. The pellet was resuspended in 40 μl SDS-PAGE sample buffer, heated for 3 min at 100 °C and subjected to SDS-PAAG and Western immunoblotting using anti-RetGC1 and anti-RD3 antibodies.
Immunoblotting
HEK293 membranes were subjected to Western immunoblotting after dissolving them in a Laemmli SDS sample buffer. The retinas from mice aged 3.5 weeks were excised in 20 μl of 10 mM Tris–HCl, pH 7.4, containing 1:100 dilution of a Millipore–Sigma protease inhibitor cocktail and homogenized for protein extraction in an Abcam radioimmunoprecipitation assay mixture of detergents containing the protease inhibitors, 300 μl/6 retinas. After 30 min of extraction on ice, the insoluble material was removed by centrifugation at 14,000g for 10 min, 4 °C, and the protein extract was mixed with equal volume of 2× Laemmli SDS sample buffer (Millipore–Sigma) and subjected to electrophoresis in linear or gradient polyacrylamide gels containing 0.1% SDS. Following the electrophoresis, the proteins were transferred overnight at 50 V constant voltage to Immobilon P membrane (Millipore) at 18 °C using Tris–glycine transfer buffer (Invitrogen/Thermo Fisher Scientific). The membranes were transiently stained by Ponceau S (Millipore–Sigma) dye solution in 1% acetic acid to mark the positions of molecular mass markers, destained by series of washes in water and Tris-buffered saline (Thermo Fisher Scientific) containing 0.5% Tween-20 (TTBS), blocked by SuperBlock (Thermo Fisher Scientific) solution in TTBS, probed by primary antibody for 1 h at room temperature, washed three times in TTBS and probed with secondary antibody for 1 h at room temperature. The secondary antibody was removed by washing three times for 15 min in TTBS and twice in Tris-buffered saline. The luminescence signal was developed using peroxidase-conjugated secondary polyclonal IgG (Cappel/MP Biomedical) and a Pierce SuperSignal Femto substrate kit (Thermo Fisher Scientific). The images were acquired and processed using a Fotodyne Luminous FX imager, ImageJ (National Institutes of Health) software; unmodified images were subjected to densitometry using Bio-Rad Multi-Analyst software.
Confocal microscopy
Mice were euthanized by lethal injection of ketamine/xylazine and perfused with formaldehyde fixative solution, then the enucleated eyes were dissected using cryomicrotome, and the sections were mounted for microscopy as previously described (16, 25). The sections were washed three times in PBS containing 0.1 M glycine (pH 7.4), blocked for 1 h at 30 °C with the same solution containing 5% bovine serum albumin and 0.1% Triton X-100, incubated overnight at 4 °C and then 1 h at room temperature with the primary antibody, then washed with PBS solution three times for 15 min each, incubated with 1:400 diluted goat or donkey anti-rabbit and rabbit antimouse secondary antibody conjugated with AlexaFluor 568 or AlexaFluor 647 (Thermo Fisher Scientific), and washed four times for 15 min with PBS at room temperature. Confocal images were acquired using an Olympus FV1000 Spectral instrument controlled by FluoView FV10-ASW software, collecting the emitted fluorescence of different wavelengths in a sequential mode. Where indicated, the fluorescence was superimposed on a differential interference contrast image. The far-red fluorescence of AlexaFluor 647 in the images was assigned cyan pseudocolor. No changes were made to the original images, except for gamma correction applied to the whole image for better clarity in print.
Statistics
Statistical significance of the differences was tested by ANOVA/Scheffe post hoc (confidence level 99%; alpha 0.01) multiple-pairs comparison or Student's t test (unpaired/unequal variance), using Synergy KaleidaGraph 4 software.
Data availability
The data referred to in this article are contained within the article. Unprocessed data can be available from the corresponding author (adizhoor@salus.edu) upon reasonable request.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
This work was supported by the National Institutes of Health grant EY11522 from National Eye Institute and by a CURE Formula grant from Pennsylvania Department of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions
A. M. D. conceptualization; A. M. D. and I. V. P. methodology; A. M. D. and I. V. P. formal analysis; A. M. D., E. V. O., and I. V. P. investigation; A. M. D. and I. V. P. writing – original draft; A. M. D. supervision.
Edited by Henrik G. Dohlman
References
- 1.Koch K.-W., Dell'Orco D. Protein and signaling networks in vertebrate photoreceptor cells. Front. Mol. Neurosci. 2015;8:67. doi: 10.3389/fnmol.2015.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fu Y., Yau K.-W. Phototransduction in mouse rods and cones. Pflugers Arch. 2007;454:805–819. doi: 10.1007/s00424-006-0194-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arshavsky V.Y., Lamb T.D., Pugh E.N., Jr. G proteins and phototransduction. Annu. Rev. Physiol. 2002;64:153–187. doi: 10.1146/annurev.physiol.64.082701.102229. [DOI] [PubMed] [Google Scholar]
- 4.Pugh E.N., Jr., Nikonov S., Lamb T.D. Molecular mechanisms of vertebrate photoreceptor light adaptation. Curr. Opin. Neurobiol. 1999;9:410–418. doi: 10.1016/S0959-4388(99)80062-2. [DOI] [PubMed] [Google Scholar]
- 5.Dizhoor A.M., Lowe D.G., Olshevskaya E.V., Laura R.P., Hurley J.B. The human photoreceptor membrane guanylyl cyclase, RetGC, is present in outer segments and is regulated by calcium and a soluble activator. Neuron. 1994;12:1345–1352. doi: 10.1016/0896-6273(94)90449-9. [DOI] [PubMed] [Google Scholar]
- 6.Lowe D.G., Dizhoor A.M., Liu K., Gu Q., Spencer M., Laura R., Lu L., Hurley J.B. Cloning and expression of a second photoreceptor-specific membrane retina guanylyl cyclase (RetGC), RetGC-2. Proc. Natl. Acad. Sci. U. S. A. 1995;92:5535–5539. doi: 10.1073/pnas.92.12.5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang R.B., Foster D.C., Garbers D.L., Fulle H.J. Two membrane forms of guanylyl cyclase found in the eye. Proc. Natl. Acad. Sci. U. S. A. 1995;92:602–606. doi: 10.1073/pnas.92.2.602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dizhoor A.M., Olshevskaya E.V., Peshenko I.V. Mg2+/Ca2+ cation binding cycle of guanylyl cyclase activating proteins (GCAPs): Role in regulation of photoreceptor guanylyl cyclase. Mol. Cell. Biochem. 2010;334:117–124. doi: 10.1007/s11010-009-0328-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dizhoor A.M., Peshenko I.V. Regulation of retinal membrane guanylyl cyclase (RetGC) by negative calcium feedback and RD3 protein. Pflugers Arch. 2021;473:1393–1410. doi: 10.1007/s00424-021-02523-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunt D.M., Buch P., Michaelides M. Guanylate cyclases and associated activator proteins in retinal disease. Mol. Cell. Biochem. 2010;334:157–168. doi: 10.1007/s11010-009-0331-y. [DOI] [PubMed] [Google Scholar]
- 11.Downes S.M., Payne A.M., Kelsel R.E., Fitzke F.W., Holder G.E., Hunt D.M., Moore A.T., Bird A.C. Autosomal dominant dystrophy with mutations in the guanylate cyclase 2D gene encoding retinal guanylate cyclase-1. Arch. Ophthalmol. 2001;119:1667–1673. doi: 10.1001/archopht.119.11.1667. [DOI] [PubMed] [Google Scholar]
- 12.Sharon D., Wimberg H., Kinarty Y., Koch K.W. Genotype-functional-phenotype correlations in photoreceptor guanylate cyclase (GC-E) encoded by GUCY2D. Prog. Retin. Eye Res. 2018;63:69–91. doi: 10.1016/j.preteyeres.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 13.Tucker C.L., Woodcock S.C., Kelsell R.E., Ramamurthy V., Hunt D.M., Hurley J.B. Biochemical analysis of a dimerization domain mutation in RetGC-1 associated with dominant cone-rod dystrophy. Proc. Natl. Acad. Sci. U. S. A. 1999;96:9039–9044. doi: 10.1073/pnas.96.16.9039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramamurthy V., Tucker C., Wilkie S.E., Daggett V., Hunt D.M., Hurley J.B. Interactions within the coiled-coil domain of RetGC-1 guanylyl cyclase are optimized for regulation rather than for high affinity. J. Biol. Chem. 2001;276:26218–26229. doi: 10.1074/jbc.M010495200. [DOI] [PubMed] [Google Scholar]
- 15.Peshenko I.V., Moiseyev G.P., Olshevskaya E.V., Dizhoor A.M. Factors that determine Ca2+ sensitivity of photoreceptor guanylyl cyclase. Kinetic analysis of the interaction between the Ca2+-bound and the Ca2+-free guanylyl cyclase activating proteins (GCAPs) and recombinant photoreceptor guanylyl cyclase 1 (RetGC-1) Biochemistry. 2004;43:13796–13804. doi: 10.1021/bi048943m. [DOI] [PubMed] [Google Scholar]
- 16.Dizhoor A.M., Olshevskaya E.V., Peshenko I.V. The R838S mutation in retinal guanylyl cyclase 1 (RetGC1) alters calcium sensitivity of cGMP synthesis in the retina and causes blindness in transgenic mice. J. Biol. Chem. 2016;291:24504–24516. doi: 10.1074/jbc.M116.755553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sato S., Peshenko I.V., Olshevskaya E.V., Kefalov V.J., Dizhoor A.M. GUCY2D cone-rod dystrophy-6 is a ‘phototransduction disease’ triggered by abnormal calcium feedback on retinal membrane guanylyl cyclase 1. J. Neurosci. 2018;38:2990–3000. doi: 10.1523/JNEUROSCI.2985-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peshenko I.V., Olshevskaya E.,V., Dizhoor A.M. GUCY2D mutations in retinal guanylyl cyclase 1 provide biochemical reasons for dominant cone-rod dystrophy but not for stationary night blindness. J. Biol. Chem. 2020;295:18301–18315. doi: 10.1074/jbc.RA120.015553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedman J.S., Chang B., Kannabiran C., Chakarova C., Singh H.P., Jalali S., Hawes N.L., Branham K., Othman M., Filippova E., Thompson D.A., Webster A.R., Andreasson S., Jacobson S.G., Bhattacharya S.S. Premature truncation of a novel protein, RD3, exhibiting subnuclear localization is associated with retinal degeneration. Am. J. Hum. Genet. 2006;79:1059–1070. doi: 10.1086/510021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Azadi S., Molday L.L., Molday R.S. RD3, the protein associated with Leber congenital amaurosis type 12, is required for guanylate cyclase trafficking in photoreceptor cells. Proc. Natl. Acad. Sci. U. S. A. 2010;107:21158–21163. doi: 10.1073/pnas.1010460107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zulliger R., Naash M.I., Rajala R.V., Molday R.S., Azadi S. Impaired association of retinal degeneration-3 with guanylate cyclase-1 and guanylate cyclase-activating protein-1 leads to Leber congenital amaurosis-1. J. Biol. Chem. 2015;290:3488–3499. doi: 10.1074/jbc.M114.616656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Molday L.L., Jefferies T., Molday R.S. Insights into the role of RD3 in guanylate cyclase trafficking, photoreceptor degeneration, and Leber congenital amaurosis. Front. Mol. Neurosci. 2014;7:44. doi: 10.3389/fnmol.2014.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peshenko I.V., Olshevskaya E.V., Azadi S., Molday L.L., Molday R.S., Dizhoor A.M. Retinal degeneration 3 (RD3) protein inhibits catalytic activity of retinal membrane guanylyl cyclase (RetGC) and its stimulation by activating proteins. Biochemistry. 2011;50:9511–9519. doi: 10.1021/bi201342b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peshenko I.V., Olshevskaya E.V., Dizhoor A.M. Functional study and mapping sites for interaction with the target enzyme in retinal degeneration 3 (RD3) protein. J. Biol. Chem. 2016;291:19713–19723. doi: 10.1074/jbc.M116.742288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dizhoor A.M., Olshevskaya E.V., Peshenko I.V. Retinal guanylyl cyclase activation by calcium sensor proteins mediates photoreceptor degeneration in an rd3 mouse model of congenital human blindness. J. Biol. Chem. 2019;294:13729–13739. doi: 10.1074/jbc.RA119.009948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dizhoor A.M., Olshevskaya E.V., Peshenko I.V. Retinal degeneration-3 protein promotes photoreceptor survival by suppressing activation of guanylyl cyclase rather than accelerating GMP recycling. J. Biol. Chem. 2021;296:100362. doi: 10.1016/j.jbc.2021.100362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plana-Bonamaisó A., López-Begines S., Andilla J., Fidalgo M.J., Loza-Alvarez P., Estanyol J.M., Villa P., Méndez A. GCAP neuronal calcium sensor proteins mediate photoreceptor cell death in the rd3 mouse model of LCA12 congenital blindness by involving endoplasmic reticulum stress. Cell Death Dis. 2020;24:62. doi: 10.1038/s41419-020-2255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baehr W., Karan S., Maeda T., Luo D.G., Li S., Bronson J.D., Watt C.B., Yau K.W., Frederick J.M., Palczewski K. The function of guanylate cyclase 1 and guanylate cyclase 2 in rod and cone photoreceptors. J. Biol. Chem. 2007;282:8837–8847. doi: 10.1074/jbc.M610369200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang R.B., Robinson S.W., Xiong W.H., Yau K.W., Birch D.G., Garbers D.L. Disruption of a retinal guanylyl cyclase gene leads to cone-specific dystrophy and paradoxical rod behavior. J. Neurosci. 1999;19:5889–5897. doi: 10.1523/JNEUROSCI.19-14-05889.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nemet I., Tian G., Imanishi Y. Organization of cGMP sensing structures on the rod photoreceptor outer segment plasma membrane. Channels (Austin) 2014;8:528–535. doi: 10.4161/19336950.2014.973776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peshenko I.V., Olshevskaya E.V., Savchenko A.B., Karan S., Palczewski K., Baehr W., Dizhoor A.M. Enzymatic properties and regulation of the native isozymes of retinal membrane guanylyl cyclase (RetGC) from mouse photoreceptors. Biochemistry. 2011;50:5590–5600. doi: 10.1021/bi200491b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peshenko I.V., Olshevskaya E.V., Dizhoor A.M. Dimerization domain of retinal membrane guanylyl cyclase 1 (RetGC1) is an essential part of guanylyl cyclase-activating protein (GCAP) binding interface. J. Biol. Chem. 2015;290:19584–19596. doi: 10.1074/jbc.M115.661371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peshenko I.V., Yu Q., Lim S., Cudia D., Dizhoor A.M., Ames J.B. Retinal degeneration 3 (RD3) protein, a retinal guanylyl cyclase regulator, forms a monomeric and elongated four-helix bundle. J. Biol. Chem. 2019;294:2318–2328. doi: 10.1074/jbc.RA118.006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peshenko I.V., Olshevskaya E.V., Lim S., Ames J.B., Dizhoor A.M. Calcium-myristoyl Tug is a new mechanism for intramolecular tuning of calcium sensitivity and target enzyme interaction for guanylyl cyclase-activating protein 1: Dynamic connection between N-fatty acyl group and EF-hand controls calcium sensitivity. J. Biol. Chem. 2012;287:13972–13984. doi: 10.1074/jbc.M112.341883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vocke F., Weisschuh N., Marino V., Malfatti S., Jacobson S.G., Reiff C.M., Dell'Orco D., Koch K.W. Dysfunction of cGMP signalling in photoreceptors by a macular dystrophy-related mutation in the calcium sensor GCAP1. Hum. Mol. Genet. 2017;26:133–144. doi: 10.1093/hmg/ddw374. [DOI] [PubMed] [Google Scholar]
- 36.Chang B., Heckenlively J.R., Hawes N.L., Roderick T.H. New mouse primary retinal degeneration (rd-3) Genomics. 1993;16:45–49. doi: 10.1006/geno.1993.1138. [DOI] [PubMed] [Google Scholar]
- 37.Olshevskaya E.V., Peshenko I.V., Savchenko A.B., Dizhoor A.M. Retinal guanylyl cyclase isozyme 1 is the preferential in vivo target for constitutively active GCAP1 mutants causing congenital degeneration of photoreceptors. J. Neurosci. 2012;32:7208–7217. doi: 10.1523/JNEUROSCI.0976-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peshenko I.V., Olshevskaya E.V., Dizhoor A.M. Evaluating the role of retinal membrane guanylyl cyclase 1 (RetGC1) domains in binding guanylyl cyclase-activating proteins (GCAPs) J. Biol. Chem. 2015;290:6913–6924. doi: 10.1074/jbc.M114.629642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu J., Morris L., Thapa A., Ma H., Michalakis S., Biel M., Baehr W., Peshenko I.V., Dizhoor A.M., Ding X.-Q. cGMP accumulation causes photoreceptor degeneration in CNG channel deficiency: Evidence of cGMP cytotoxicity independently of enhanced CNG channel function. J. Neurosci. 2013;33:14939–14948. doi: 10.1523/JNEUROSCI.0909-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang T., Tsang S.H., Chen J. Two pathways of rod photoreceptor cell death induced by elevated cGMP. Hum. Mol. Genet. 2017;26:2299–2306. doi: 10.1093/hmg/ddx121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Molday L.L., Djajadi H., Yan P., Szczygiel L., Boye S.L., Chiodo V.A., Gregory-Evans K., Sarunic M.V., Hauswirth W.W., Molday R.S. RD3 gene delivery restores guanylate cyclase localization and rescues photoreceptors in the Rd3 mouse model of Leber congenital amaurosis 12. Hum. Mol. Genet. 2013;22:3894–3905. doi: 10.1093/hmg/ddt244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peshenko I.V., Cideciyan A.V., Sumaroka A., Olshevskaya E.V., Scholten A., Abbas S., Koch K.-W., Jacobson S.G., Dizhoor A.M. A Gly86Arg mutation in the calcium-sensor protein GCAP1 alters regulation of retinal guanylyl cyclase and causes dominant cone-rod degeneration. J. Biol. Chem. 2019;294:3476–3488. doi: 10.1074/jbc.RA118.006180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peshenko I.V., Olshevskaya E.V., Lim S., Ames J.B., Dizhoor A.M. Identification of target binding site in photoreceptor guanylyl cyclase activating protein 1 (GCAP1) J. Biol. Chem. 2014;289:10140–10154. doi: 10.1074/jbc.M113.540716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peshenko I.V., Dizhoor A.M. Ca2+ and Mg2+ binding properties of GCAP-1. Evidence that Mg2+-bound form is the physiological activator of photoreceptor guanylyl cyclase. J. Biol. Chem. 2006;281:23830–23841. doi: 10.1074/jbc.M600257200. [DOI] [PubMed] [Google Scholar]
- 45.Peshenko I.V., Olshevskaya E.V., Dizhoor A.M. Binding of guanylyl cyclase activating protein 1(GCAP1) to retinal guanylyl cyclase (RetGC1): The role of individual EF-hands. J. Biol. Chem. 2008;283:21747–21757. doi: 10.1074/jbc.M801899200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsien R., Pozzan T. Measurement of cytosolic free Ca2+ with quin2. Methods Enzymol. 1989;172:230–262. doi: 10.1016/s0076-6879(89)72017-6. [DOI] [PubMed] [Google Scholar]
- 47.Huber G., Beck S.C., Grimm C., Sahaboglu-Tekgoz A., Paquet-Durand F., Wenzel A., Humphries P., Redmond T.M., Seeliger M.W., Fischer M.D. Spectral domain optical coherence tomography in mouse models of retinal degeneration. Invest. Ophthalmol. Vis. Sci. 2009;50:5888–5895. doi: 10.1167/iovs.09-3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang P., Zawadzki R.J., Goswami M., Nguyen P.T., Yarov-Yarovoy V., Burns M.E., Pugh E.N., Jr. In vivo optophysiology reveals that G-protein activation triggers osmotic swelling and increased light scattering of rod photoreceptors. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E2937–E2946. doi: 10.1073/pnas.1620572114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Makino C.L., Peshenko I.V., Wen X.H., Olshevskaya E.V., Barrett R., Dizhoor A.M. A role for GCAP2 in regulating the photoresponse. Guanylyl cyclase activation and rod electrophysiology in GUCA1B knock-out mice. J. Biol. Chem. 2008;283:29135–29143. doi: 10.1074/jbc.M804445200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data referred to in this article are contained within the article. Unprocessed data can be available from the corresponding author (adizhoor@salus.edu) upon reasonable request.