Abstract
Human IQGAP1 is a widely expressed 190-kDa Cdc42-, Rac1-, and calmodulin-binding protein that interacts with F-actin in vivo and that can cross-link F-actin microfilaments in vitro. Recent results have implicated IQGAP1 as a component of pathways via which Cdc42 or Rac1 modulates cadherin-based cell adhesion (S. Kuroda et al., Science 281:832–835, 1998), whereas yeast IQGAP-related proteins have been found to play essential roles during cytokinesis. To identify critical in vivo functions of IQGAP1, we generated deficient mice by gene targeting. We demonstrate that IQGAP1 null mutants arise at normal frequency and show no obvious defects during development or for most of their adult life. Loss of IQGAP1 also does not affect tumor development or tumor progression, but mutant mice exhibit a significant (P < 0.0001) increase in late-onset gastric hyperplasia relative to wild-type animals of the same genetic background. While we cannot exclude that functional redundancy with IQGAP2 contributes to the lack of developmental phenotypes, the restricted expression pattern of IQGAP2 is not obviously altered in adult IQGAP1 mutant mice. Thus, IQGAP1 does not serve any essential nonredundant functions during murine development but may serve to maintain the integrity of the gastric mucosa in older animals.
Members of the Rho family of Ras-related GTPases, including RhoA to -C, Rac1 to -3, and Cdc42, function in a wide variety of biological processes, at least in part by directing the formation of specific F-actin structures and by controlling the activities of several transcription factors (12, 29). To determine how Rho family members mediate these and other effects, several groups have identified numerous Rho GTPase-binding putative effector proteins. Among such proteins, mammalian IQGAP1 has attracted special attention because it harbors an N-terminal calponin-related F-actin binding domain and interacts with Cdc42 and Rac1 by means of a C-terminal segment related to Ras GTPase-activating proteins (RasGAPs) (5, 13, 19, 23). Supporting a role in Cdc42 or Rac1-induced F-actin rearrangements, IQGAP1 forms complexes with F-actin in vivo and can cross-link actin microfilaments in vitro (1, 8, 9).
We identified human IQGAP1 as an abundant and widely expressed 190-kDa RasGAP-related protein (31) and subsequently characterized IQGAP2 as a 62% identical 180-kDa protein expressed in the liver and in several liver-derived cell lines (5). As is common for signaling proteins, both IQGAPs include several putative or confirmed protein interaction domains, including a calponin-related F-actin binding segment, a WW domain, and four calmodulin-binding IQ motifs. Both IQGAPs also harbor several segments with extensive coiled-coil potential and five (IQGAP2) or six (IQGAP1) so-called IQGAP repeats of unknown function (5).
Within their RasGAP-related segments, both IQGAPs lack an arginine finger residue that is essential for GAP activity (26), which may explain why neither IQGAP showed in vitro GAP activity toward Ras or several related GTPases (5, 13). Rather, IQGAP1 and IQGAP2 interacted with Cdc42 and Rac1 and inhibited their intrinsic and RhoGAP-stimulated GTPase activities in a dose-dependent manner (5, 13, 19, 23). Thus, rather than acting as GAPs, IQGAPs may maintain Cdc42 and Rac1 in their active GTP-bound state. IQGAP1 may also provide a link between Cdc42 and Ca2+-mediated signals, because addition of Ca2+/calmodulin abolished the interaction between IQGAP1 and Cdc42 in vitro (14, 16). Thus, signals that increase intracellular Ca2+ may dissociate IQGAP-GTPase complexes, allowing the GTPase to interact with other effectors.
Clues to the function(s) of putative signaling proteins can often be obtained by analyzing their expression patterns or subcellular localization. Results of such studies indicate that IQGAP1 is widely expressed and may exist in several subcellular pools, perhaps depending on the cell type. Thus, IQGAP1 localizes to lamellipodia and membrane ruffles in fibroblasts and other cell types (1, 19), colocalizes with Cdc42 and F-actin to the Golgi membrane in Cdc42-overexpressing CHO cells (22), and resides at adherens junctions in confluent epithelial cells (19). This latter finding is especially intriguing, given that Rho, Rac, and Cdc42 have been implicated in the establishment and the maintenance of cadherin-based cell contacts (3, 4, 18, 28).
Results of recent cell transfection studies have implicated IQGAP1 as a potential component of pathways via which Cdc42 and Rac1 modulate cadherin-based cell adhesion (10, 20). Thus, IQGAP1 localized to adherens junctions in E-cadherin-expressing murine L cells (EL cells) and coprecipitated with β-catenin and E-cadherin from EL cell extracts. Moreover, overexpressed IQGAP1 dissociated α-catenin from EL cell junctions and reduced their adhesion (20). Subsequent in vitro binding studies suggested that IQGAP1 may modulate cadherin-based adhesion by acting as a competitive inhibitor of β-catenin–α-catenin complex formation (10). However, IQGAP1 may also have roles beyond modulating cadherin function, since the protein is also abundant in nonadherent peripheral blood myeloid and lymphoid cells. It is interesting in this respect that others have detected mammalian IQGAPs in F-actin-containing complexes associated with the Golgi apparatus (22) and that IQGAP-related Iqg1p/Cyk1p from budding yeast and Rng2p from fission yeast play essential roles during cytokinesis (6, 7, 27). Thus, to shed light on critical in vivo functions for IQGAP1, we generated deficient mice by gene targeting. Arguing against any essential nonredundant functions, we report that mice that completely lack IQGAP1 develop normally, show no obvious alteration in their tumor spectrum, and exhibit no consistent defects beyond an approximately fivefold increase in late-onset gastric hyperplasia.
MATERIALS AND METHODS
Gene targeting and genotyping.
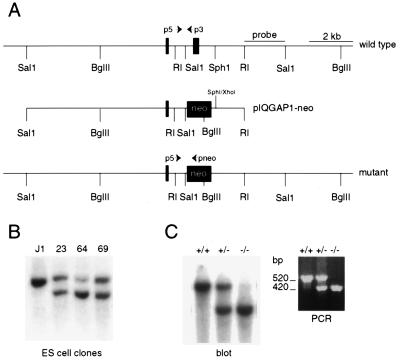
Genomic clones representing the mouse Iqgap1 gene were isolated from a J1 embryonic stem (ES) cell phage library. Restriction maps of clones that hybridized to an IQGAP1-RasGAP-related domain (GRD) cDNA fragment were prepared, and the locations of two 85- and 233-bp exons were established by sequence analysis. A targeting vector was made by inserting an approximately 3-kb SalI-EcoRI fragment harboring the 233-bp exon into pBluescript. This clone (pIQG1SE) was modified by replacing a 1.9-kb SalI-SphI fragment containing the 233-bp exon with the phosphoglycerate kinase-neomycin phosphotransferase resistance gene (PGK-neo) cassette in the opposite transcriptional orientation to generate pIQG1SE-neo. The region of homology upstream of the PGK-neo cassette was extended by inserting a 9-kb genomic SalI fragment containing the 85-bp exon into the SalI site of pIQG1SE-neo, to generate targeting construct pIQGAP1-neo (Fig. 1A). This vector was linearized and electroporated into J1 ES cells, and DNA prepared from neomycin-resistant ES cell clones was digested with BglII and probed with the 2-kb genomic EcoRI-SalI fragment identified in Fig. 1A. ES cell lines 23 and 64 were used to generate chimeric mice as described elsewhere (21). Progeny of the chimeric mice was initially genotyped by blotting BglII-digested tail DNA. Subsequent mice were genotyped by a PCR-based assay, using the p3 (5′ CCT GCT GAC AGG TCA ATG AT 3′), p5 (5′ TTG CAG TCT GTG GCA TGT G 3′), and pneo (5′ CCT GCT CTT TAC TGA AGG CT 3′) primers, the approximate locations of which are indicated in Fig. 1A. Tail DNA was prepared by standard procedures and dissolved in 100 μl of 10 mM Tris HCl (pH 7.5)–1 mM EDTA. PCR amplification was performed in a reaction volume of 20 μl containing 2 μl of tail DNA, 2 μl of 10× reaction buffer, 2 μl of 2.5 mM deoxynucleoside triphosphates, 1 μl of 25 mM MgCl2, 40 pM pneo, 20 pM p3, 20 pM p5, and 1 μl of Taq polymerase. Samples were denatured for 7 min at 95°C, followed by 35 cycles of amplification (1 min at 95°C, 1 min at 55°C, and 1 min at 72°C). The wild-type and mutant alleles give rise to PCR-amplified fragments of approximately 520 and 420 bp, respectively (Fig. 1C).
FIG. 1.
Generation of IQGAP1-deficient mice. (A) Restriction map of murine IQGAP1 gene segment showing the locations of two GRD exons (top), structure of the pIQGAP1-neo targeting construct (middle), and structure of the mutant allele after homologous recombination (bottom). The approximate locations of three primers (p3, p5, and pneo) used in PCR-based genotyping are indicated. The indicated 2-kb EcoRI (RI)-SalI fragment was initially used in DNA blot-based genotyping. (B) BglII digest of DNA isolated from neomycin-resistant ES cell clones probed with the EcoRI-SalI fragment indicated in panel A. Untransfected J1 ES cells show only an approximately 15-kb BglII fragment, whereas 3 of approximately 40 tested ES cell clones showed an additional 9-kb fragment as evidence of homologous recombination. (C) Mouse tail DNA was genotyped either by a DNA blot assay (left) or by PCR using the P3, p5 and pneo primers (right). See Materials and Methods for details.
Immunoblot analysis.
A rabbit polyclonal antiserum (CSH-183) against a keyhole limpet hemocyanin-conjugated IQGAP1 peptide (H-Cys-Leu-Leu-Asn-Lys-Lys-Phe-Tyr-Gly-Lys-OH) was a kind gift from Nouria Hernandez. A rabbit serum against an N-terminal IQGAP1 fusion protein was kindly donated by Kozo Kaibuchi (19). A monoclonal antibody made against a human IQGAP1-GRD fusion protein that cross-reacts with murine IQGAP1 was bought from Transduction Laboratories. Additional antipeptide sera have been described previously (5).
Miscellaneous.
Stomachs were opened along the greater curvature, and the absence or presence of generalized hyperplasia or localized polyps was scored by visual inspection using a binocular microscope. To provide a more quantitative assessment, transverse sections of the glandular stomachs were prepared and bilateral (relative to the midline) mucosal thickness was measured under a microscope. Full-thickness dermal wound healing of three Iqgap1−/− and three control +/+ mice was assayed as described elsewhere (17). Three-color (CD43/CD45R/immunoglobulin M) and four-color flow cytometric analyses (CD45R/CD43/CD24/BP-1) of B-lymphocyte populations in bone marrow were performed as described previously (30).
RESULTS AND DISCUSSION
IQGAP1 and IQGAP2 are 62% identical at the protein sequence level, and each binds Cdc42, Rac1, calmodulin, and F-actin (5, 13, 22). However, whereas IQGAP1 is widely expressed (31), we previously reported that in a survey of adult mouse tissues IQGAP2 appeared to be largely liver specific (5). Thus, to shed light on the in vivo function(s) of IQGAPs, we set out to generate mice deficient for IQGAP1. To inactivate the murine Iqgap1 gene, we isolated genomic clones representing the GRD from a J1 ES cell lambda phage library, subcloned the inserts into a plasmid vector, and established the locations of two consecutive 85- and 233-bp GRD exons by sequence analysis (Fig. 1A). Recently, the sequence of an 87,034-bp human chromosome 15q26.1 phage artificial chromosome clone was reported to include several regions of homology to IQGAP1 (GenBank accession no. AC004587). Computer analysis of this clone confirmed the presence of 36 IQGAP1 exons, representing the cDNA from codon 53 to the poly(A) addition site. Exons 26 and 27 in the PAC clone code for amino acids 1158 to 1185 and 1186 to 1263 of human IQGAP1 and correspond to the murine exons identified by us. The protein segment predicted by the 233-bp murine exon includes the NLLYYRYMNPAIVAP RasGAP signature motif (2). We predicted that deleting this exon would, minimally, disrupt the interaction between IQGAP1 and its GTPase substrates.
A targeting vector was made by replacing the 233-bp exon with a PGK-neo cassette in the opposite transcriptional orientation (see Fig. 1A and Materials and Methods for details). This construct was electroporated into J1 ES cells, and several neomycin-resistant clones showing evidence of homologous recombination were identified (Fig. 1B). Upon confirmation of the structure of the targeted gene by further restriction mapping, two independent ES cell clones were introduced into C57BL/6 blastocysts. Chimeric mice that transmitted the mutant allele to their progeny were obtained and bred with C57BL/6 and 129 animals in order to obtain heterozygous Iqgap1 mutant mice in mixed 129/BL6 and pure 129 genetic backgrounds (Fig. 1C).
Heterozygous Iqgap1 mutant mice representing both ES cell clones were mated to determine the effects of homozygous loss of IQGAP1. Among 165 progeny from 23 heterozygous crosses genotyped at around 3 weeks of age, we identified 46 +/+, 75 +/−, and 44 −/− animals, representing no significant deviation from the expected 1:2:1 Mendelian ratio. Thus, homozygosity for the mutant Iqgap1 allele does not confer a detectable disadvantage during murine development or early life.
The Iqgap1 insertion-deletion mutation generated by us is likely to represent a null allele. Thus, a 5′ Iqgap1 cDNA probe detected an abundant approximately 8-kb mRNA in total RNA from several wild-type mouse tissues, but no mRNA of this or any other size was detected in the same tissues of homozygous mutant animals (not shown). Moreover, blots of adult tissue or embryonic day 17 total embryo extracts probed with antibodies against N-terminal, central, or C-terminal IQGAP1 epitopes (see Materials and Methods for details) showed a prominent 190-kDa protein in +/+ and +/− but not −/− tissues (Fig. 2A).
FIG. 2.
Immunoblots showing absent IQGAP1 and unaltered IQGAP2 expression in homozygous mutant mice. (A) Blot containing extracts of the indicated tissues from IQGAP1 +/+ and −/− mice, probed with a monoclonal antibody that detects murine IQGAP1 but not IQGAP2. The 97-kDa protein seen in wild-type but not mutant spleen is a proteolytic degradation product of IQGAP1. (B) Blot containing liver and lung extracts from +/+, +/−, and −/− mice, probed with a polyclonal serum against an epitope that is conserved between IQGAP1 and IQGAP2.
To determine whether the restricted expression pattern of IQGAP2 is altered in IQGAP1-deficient mice, we probed adult tissue extracts with an antibody against a C-terminal epitope that is conserved between IQGAP1 and IQGAP2. Figure 2B shows that IQGAP2 expression, at least at this level of analysis, is not altered in IQGAP1-deficient mice. Thus, functional redundancy with IQGAP2 is unlikely to mask essential roles for IQGAP1 in several adult mouse tissues.
RNA and protein blots show that IQGAP1 is highly expressed in mouse placenta and in the bodies of embryonic day 17 embryos (reference 31 and results not shown). Moreover, multiple IQGAP1 and IQGAP2 entries in the dbEST database derive from cDNA libraries representing various stages of mouse embryonic development. Thus, while IQGAP1 and IQGAP2 are likely to be coexpressed in embryos, we have been unable to establish whether the two proteins overlap in their embryonic expression patterns. For this reason, we cannot exclude that functional redundancy between IQGAP1 and IQGAP2 contributes to the lack of developmental phenotypes in IQGAP1 deficient mice.
IQGAP1 and IQGAP2 show very limited overlap in their adult expression patterns (5). Thus, to determine whether loss of IQGAP1 causes abnormalities in adult tissues, we sacrificed several +/+, +/−, and −/− mice at approximately 5 months of age. No defects were detected in a comprehensive survey of tissues from these animals. Lung, kidney, and placenta produce little or no IQGAP2 and express particularly high levels of IQGAP1 (5, 31). However, near-term placentas from Iqgap1−/− mothers appeared normal, and no abnormalities were detected in kidney sections stained for aquaporin-2, GP330/megalin, or the H+-ATPase. IQGAP1 is also expressed at a low level in primary mouse embryo fibroblasts and is abundant in mouse skin (not shown), but we detected no abnormalities in motility assays using low-passage-number Iqgap1−/− embryo fibroblasts. Furthermore, dorsal wound healing was not obviously impaired in the mutant mice, and Iqgap1−/− primary keratinocytes underwent normal Ca2+-induced stratification. Likewise, flow cytometric analysis of bone marrow revealed no obvious abnormalities in B-lymphocyte populations (results not shown; see Materials and Methods for details). Thus, loss of IQGAP1 does not cause any obvious anatomical or functional defects in many tissues that normally express this protein.
Loss of E-cadherin is common in malignant carcinomas and plays a causative role in the progression of benign adenoma to invasive adenocarcinoma in a transgenic model of murine pancreatic cancer (24). Thus, we argued that loss of the proposed cadherin-modulating function of IQGAP1 might alter the incidence or the severity of spontaneous tumors. Among the most frequent tumors in 129 or mixed 129 × C57/BL6 mice are lung adenoma or adenocarcinoma, hepatoma, lymphoma, and histiocytic sarcoma. To determine whether these or other cancers showed altered incidences in Iqgap1-deficient mice, we monitored 47 +/+, 86 +/−, and 168 −/− mice over an approximately 2-year period. Mice that developed visible or palpable masses or other disease symptoms were sacrificed, while other animals were analyzed for tumors after being sacrificed for other purposes. Although 55% of wild-type animals remained free of obvious benign or malignant tumors at an average age of 20 months, compared to 36 to 37% tumor-free survival for similarly aged +/− or −/− mice, only minor differences in the incidences of specific tumors were apparent among the three genotypes (Table 1). Moreover, histological analysis failed to reveal any tendency toward a more aggressive phenotype in IQGAP1-deficient lung and other tumors. Thus, loss of IQGAP1 does not cause a major increase in the incidence of spontaneous tumors and does not alter the progression of spontaneous tumors in a large cohort of mice.
TABLE 1.
Tumor incidence in wild-type and Iqgap1 mutant mice
| Tumor type | No. with tumor (% with tumor; avg day of detection)
|
||
|---|---|---|---|
| +/+ (n = 47) | +/− (n = 86) | −/− (n = 168) | |
| None | 26 (55.3; 598) | 31 (36.0; 597) | 63 (37.5; 542) |
| Lung adenoma/adenocarcinoma | 11 (23.4; 606) | 24 (27.9; 653) | 49 (29.2; 588) |
| Hepatoma/HCC | 9 (19.1; 640) | 11 (12.6; 663) | 26 (15.4; 568) |
| Lymphoma | 4 (8.5; 647) | 14 (16.3; 615) | 6 (3.6; 553) |
| Histiocytic sarcoma | 2 (4.3; 600) | 9 (10.3; 577) | 11 (6.5; 546) |
| Hemangioma/hemangiosacroma | 0 | 2 (2.3) | 6 (3.5; 576) |
| Harderian gland adenoma | 0 | 4 (4.6) | 3 (1.8) |
| Sarcoma (other) | 0 | 3 (3.4) | 0 |
| Othersa | 0 | 4 (4.6) | 3 (1.8) |
| Avg age (with tumors) in days | 598 (623) | 612 (626) | 562 (573) |
In +/− mice, included one leiomyoma, one mammary gland fibroadenoma, one nerve sheath tumor, and one pheochromocytoma. Other tumors in −/− mice included two leiomyomas and one pituitary adenoma.
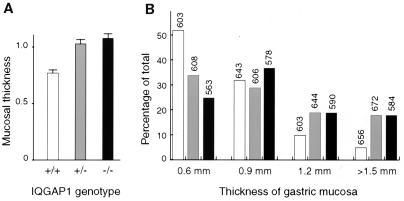
The E-cadherin gene has been implicated as a tumor suppressor in familial gastric carcinoma (11). Intriguingly, whereas Iqgap1-deficient mice showed no stomach defects at 5 months of age, at 18 months both heterozygous and homozygous mutants showed a marked increase in gastric hyperplasia relative to wild-type mice of the same genetic background (Fig. 3 and 4). Thus, in a comprehensive survey of their tissues, 1 of 13 +/+ mice (8%), 10 of 27 +/− mice (37%), and 23 of 51 −/− animals (45%) showed unilateral or bilateral hyperplasia or polyps of the gastric mucosa. To provide a more quantitative assessment of this phenotype, we prepared sections of the glandular stomachs of a larger series of 40 +/+, 47 +/−, and 80 −/− randomly chosen animals and measured bilateral mucosal thickness. Results demonstrate that both heterozygous and homozygous Iqgap1 mutants exhibit a highly significant increase in average gastric mucosal thickness, compared to wild-type mice of the same genetic background (Mann Whitney test P value < 0.0001; Fig. 3A). The gastric lesions presented either as bilateral hyperplasia, or as localized polyps (Fig. 4B and 4C). Many lesions in mice older than approximately 15 months included eosinophilic inclusions and areas of dysplasia (Fig. 4D), but no carcinoma in situ or invasive stomach carcinoma was apparent in any animal. As is not unusual for late-onset defects, heterozygous and homozygous mutants showed similar levels of gastric hyperplasia (Fig. 3). Attempts to demonstrate loss of the wild-type Iqgap1 allele in the heterozygous stomachs were not successful. However, supporting the view that additional genetic hits occur in the heterozygous animals, the gastric lesions in the homozygous mutants were detected at an earlier average age (Fig. 3B).
FIG. 3.
Gastric hyperplasia in IQGAP1-deficient mice. Bilateral mucosal thickness was measured in transverse sections of the glandular stomachs. A total of 79 +/+, 94 +/−, and 160 −/− measurements were used to generate the graphs. (A) Average mucosal thickness in millimeters plus the standard error of the mean. (B) Distribution of mucosal thickness measurements among +/+, +/−, and −/− mice. The number above each column gives the average age of mice in days.
FIG. 4.
Hemotoxylin-eosin-stained glandular stomach sections showing mucosal hyperplasia and dysplasia. (A) Normal −/− stomach (0.6-mm mucosal thickness); (B and C) gastric polyps in −/− animals (A, B, and C at 2.5× magnification); (D) Dysplastic region within a −/− polyp at 10× magnification.
Beyond gastric lesions, 2 of 51 comprehensively analyzed −/− mice showed atrophy of one kidney, with five males showing either testicular atrophy (two cases) or testicular mineralization. Moreover, a gallbladder polyp was found in one homozygous mutant and colonic hyperplasia was noted in two. Although these lesions are uncommon, they do occur as normal background defects in mice of this genetic background. Thus, hyperplasia of the gastric mucosa is the only statistically significant defect in mice lacking IQGAP1, and it was found in mice of either mixed C57/BL6 × 129 or pure 129 genetic background.
The lack of obvious developmental defects suggests that IQGAP1 does not play essential nonredundant roles during the intricate tissue remodeling events that characterize murine embryogenesis. Loss of IQGAP1 also does not obviously affect tumorigenesis in adult animals. Thus, while these results argue against a critical function for IQGAP1 in regulating cadherin-based cell adhesion, it is interesting that germ line E-cadherin mutations are specifically associated with familial gastric carcinoma in humans (11). Thus, gastric mucosal cells may be especially sensitive to defects in cadherin-based adhesion. However, the gastric hyperplasia in mice lacking IQGAP1 may also reflect a defect in Cdc42 or Rac1 signals, because NF-κB is activated by Rho family GTPases (25) and because constitutive activation of murine NF-κB2 causes severe gastric hyperplasia leading to perinatal lethality (15). Further genetic and biochemical studies are required to determine the mechanism responsible for the gastric lesions in IQGAP1 mutant animals and to reveal the extent to which IQGAP1 and IQGAP2 serve redundant functions during development.
ACKNOWLEDGMENTS
The first two authors contributed equally to this work.
This work was supported by Public Health Service grant CA70294 from the National Cancer Institute to A.B.
We are grateful to Dennis Brown, Annaiah Cariappa, Nouria Hernandez, Kozo Kaibuchi, En Li, Ivan Stamenkovic, Robert Tyszkowski, Chun-xun Zeng, Janice Williams, and Monique DiSanto for reagents or technical assistance. We also thank the MGH Gene Knockout Facility for services provided and Jeff Settleman for comments on the manuscript.
REFERENCES
- 1.Bashour A M, Fullerton A T, Hart M J, Bloom G S. IQGAP1, a Rac- and Cdc42-binding protein, directly binds and cross-links microfilaments. J Cell Biol. 1997;137:1555–1566. doi: 10.1083/jcb.137.7.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boguski M S, McCormick F. Proteins regulating Ras and its relatives. Nature. 1993;366:643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- 3.Braga V M, Del Maschio A, Machesky L, Dejana E. Regulation of cadherin function by Rho and Rac: modulation by junction maturation and cellular context. Mol Biol Cell. 1999;10:9–22. doi: 10.1091/mbc.10.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braga V M, Machesky L M, Hall A, Hotchin N A. The small GTPases Rho and Rac are required for the establishment of cadherin-dependent cell-cell contacts. J Cell Biol. 1997;137:1421–1431. doi: 10.1083/jcb.137.6.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brill S, Li S, Lyman C W, Church D M, Wasmuth J J, Weissbach L, Bernards A, Snijders A J. The Ras GTPase-activating-protein-related human protein IQGAP2 harbors a potential actin binding domain and interacts with calmodulin and Rho family GTPases. Mol Cell Biol. 1996;16:4869–4878. doi: 10.1128/mcb.16.9.4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eng K, Naqvi N I, Wong K C, Balasubramanian M K. Rng2p, a protein required for cytokinesis in fission yeast, is a component of the actomyosin ring and the spindle pole body. Curr Biol. 1998;8:611–621. doi: 10.1016/s0960-9822(98)70248-9. [DOI] [PubMed] [Google Scholar]
- 7.Epp J A, Chant J. An IQGAP-related protein controls actin-ring formation and cytokinesis in yeast. Curr Biol. 1997;7:921–929. doi: 10.1016/s0960-9822(06)00411-8. [DOI] [PubMed] [Google Scholar]
- 8.Erickson J W, Cerione R A, Hart M J. Identification of an actin cytoskeletal complex that includes IQGAP and the Cdc42 GTPase. J Biol Chem. 1997;272:24443–24447. doi: 10.1074/jbc.272.39.24443. [DOI] [PubMed] [Google Scholar]
- 9.Fukata M, Kuroda S, Fujii K, Nakamura T, Shoji I, Matsuura Y, Okawa K, Iwamatsu A, Kikuchi A, Kaibuchi K. Regulation of cross-linking of actin filament by IQGAP1, a target for Cdc42. J Biol Chem. 1997;272:29579–29583. doi: 10.1074/jbc.272.47.29579. [DOI] [PubMed] [Google Scholar]
- 10.Fukata M, Kuroda S, Nakagawa M, Kawajiri A, Itoh N, Shoji I, Matsuura Y, Yonehara S, Fujisawa H, Kikuchi A, Kaibuchi K. Cdc42 and rac1 regulate the interaction of IQGAP1 with beta-catenin. J Biol Chem. 1999;274:26044–26050. doi: 10.1074/jbc.274.37.26044. [DOI] [PubMed] [Google Scholar]
- 11.Guilford P, Hopkins J, Harraway J, McLeod M, McLeod N, Harawira P, Taite H, Scoular R, Miller A, Reeve A E. E-cadherin germline mutations in familial gastric cancer. Nature. 1998;392:402–405. doi: 10.1038/32918. [DOI] [PubMed] [Google Scholar]
- 12.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 13.Hart M J, Callow M G, Souza B, Polakis P. IQGAP1, a calmodulin-binding protein with a rasGAP-related domain, is a potential effector for cdc42Hs. EMBO J. 1996;15:2997–3005. [PMC free article] [PubMed] [Google Scholar]
- 14.Ho Y D, Joyal J L, Li Z, Sacks D B. IQGAP1 integrates Ca2+/calmodulin and Cdc42 signaling. J Biol Chem. 1999;274:464–470. doi: 10.1074/jbc.274.1.464. [DOI] [PubMed] [Google Scholar]
- 15.Ishikawa H, Carrasco D, Claudio E, Ryseck R P, Bravo R. Gastric hyperplasia and increased proliferative responses of lymphocytes in mice lacking the COOH-terminal ankyrin domain of NF-kappaB2. J Exp Med. 1997;186:999–1014. doi: 10.1084/jem.186.7.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joyal J L, Annan R S, Ho Y D, Huddleston M E, Carr S A, Hart M J, Sacks D B. Calmodulin modulates the interaction between IQGAP1 and Cdc42. Identification of IQGAP1 by nanoelectrospray tandem mass spectrometry. J Biol Chem. 1997;272:15419–15425. doi: 10.1074/jbc.272.24.15419. [DOI] [PubMed] [Google Scholar]
- 17.Kaya G, Rodriguez I, Jorcano J L, Vassalli P, Stamenkovic I. Selective suppression of CD44 in keratinocytes of mice bearing an antisense CD44 transgene driven by a tissue-specific promoter disrupts hyaluronate metabolism in the skin and impairs keratinocyte proliferation. Genes Dev. 1997;11:996–1007. doi: 10.1101/gad.11.8.996. [DOI] [PubMed] [Google Scholar]
- 18.Kuroda S, Fukata M, Fujii K, Nakamura T, Izawa I, Kaibuchi K. Regulation of cell-cell adhesion of MDCK cells by Cdc42 and Rac1 small GTPases. Biochem Biophys Res Commun. 1997;240:430–435. doi: 10.1006/bbrc.1997.7675. [DOI] [PubMed] [Google Scholar]
- 19.Kuroda S, Fukata M, Kobayashi K, Nakafuku M, Nomura N, Iwamatsu A, Kaibuchi K. Identification of IQGAP as a putative target for the small GTPases, Cdc42 and Rac1. J Biol Chem. 1996;271:23363–23367. doi: 10.1074/jbc.271.38.23363. [DOI] [PubMed] [Google Scholar]
- 20.Kuroda S, Fukata M, Nakagawa M, Fujii K, Nakamura T, Ookubo T, Izawa I, Nagase T, Nomura N, Tani H, Shoji I, Matsuura Y, Yonehara S, Kaibuchi K. Role of IQGAP1, a target of the small GTPases Cdc42 and Rac1, in regulation of E-cadherin-mediated cell-cell adhesion. Science. 1998;281:832–835. doi: 10.1126/science.281.5378.832. [DOI] [PubMed] [Google Scholar]
- 21.Li E, Bestor T H, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 22.McCallum S J, Erickson J W, Cerione R A. Characterization of the association of the actin-binding protein, IQGAP, and activated Cdc42 with Golgi membranes. J Biol Chem. 1998;273:22537–22544. doi: 10.1074/jbc.273.35.22537. [DOI] [PubMed] [Google Scholar]
- 23.McCallum S J, Wu W J, Cerione R A. Identification of a putative effector for Cdc42Hs with high sequence similarity to the RasGAP-related protein IQGAP1 and a Cdc42Hs binding partner with similarity to IQGAP2. J Biol Chem. 1996;271:21732–21737. doi: 10.1074/jbc.271.36.21732. [DOI] [PubMed] [Google Scholar]
- 24.Perl A K, Wilgenbus P, Dahl U, Semb H, Christofori G. A causal role for E-cadherin in the transition from adenoma to carcinoma. Nature. 1998;392:190–193. doi: 10.1038/32433. [DOI] [PubMed] [Google Scholar]
- 25.Perona R, Montaner S, Saniger L, Sanchez-Perez I, Bravo R, Lacal J C. Activation of the nuclear factor-kappaB by Rho, CDC42, and Rac-1 proteins. Genes Dev. 1997;11:463–475. doi: 10.1101/gad.11.4.463. [DOI] [PubMed] [Google Scholar]
- 26.Scheffzek K, Ahmadian M R, Wittinghofer A. GTPase-activating proteins: helping hands to complement an active site. Trends Biochem Sci. 1998;23:257–262. doi: 10.1016/s0968-0004(98)01224-9. [DOI] [PubMed] [Google Scholar]
- 27.Shannon K B, Li R. The multiple roles of Cyk1p in the assembly and function of the actomyosin ring in budding yeast. Mol Biol Cell. 1999;10:283–296. doi: 10.1091/mbc.10.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takaishi K, Sasaki T, Kotani H, Nishioka H, Takai Y. Regulation of cell-cell adhesion by rac and rho small G proteins in MDCK cells. J Cell Biol. 1997;139:1047–59. doi: 10.1083/jcb.139.4.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Aelst L, D'Souza-Schorey C. Rho GTPases and signaling networks. Genes Dev. 1997;11:2295–2322. doi: 10.1101/gad.11.18.2295. [DOI] [PubMed] [Google Scholar]
- 30.Wang J H, Avitahl N, Cariappa A, Friedrich C, Ikeda T, Renold A, Andrikopoulos K, Liang L, Pillai S, Morgan B A, Georgopoulos K. Aiolos regulates B cell activation and maturation to effector state. Immunity. 1998;9:543–553. doi: 10.1016/s1074-7613(00)80637-8. [DOI] [PubMed] [Google Scholar]
- 31.Weissbach L, Settleman J, Kalady M F, Snijders A J, Murthy A E, Yan Y X, Bernards A. Identification of a human rasGAP-related protein containing calmodulin-binding motifs. J Biol Chem. 1994;269:20517–20521. [PubMed] [Google Scholar]