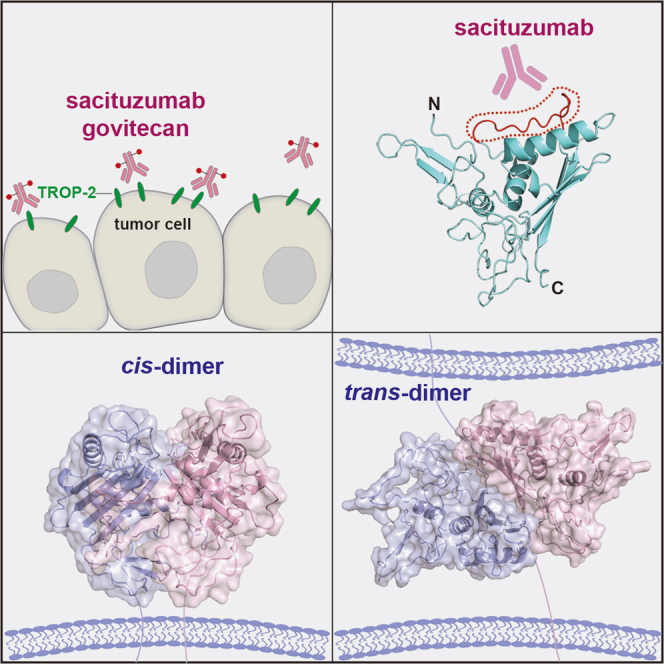
Summary
Human trophoblast cell surface antigen 2 (TROP-2) is an important target of tumor therapy, and antibody-drug conjugates with sacituzumab targeting TROP-2 have been approved for the treatment of triple-negative breast cancer. Here, we report the crystal structures of TROP-2-ECD, which can be either cis- or trans-dimers depending on which distinct but overlapping interfaces is used to engage with monomers. The cis- or trans-tetrameric forms of TROP-2 can also be assembled with a non-overlapping interface with either cis- or trans-dimerization, suggesting that cis- and trans-dimers cluster on the cell surface. The binding site of sacituzumab on TROP-2 is mapped to be located on a stretched polypeptide in CPD (Q237-Q252), which is not involved in either cis- or trans-interactions. The present findings will improve understanding of the molecular assembly of TROP-2 on tumor cells and shed light on future design of biologics for tumor therapy.
Subject areas: Immunology, Structural biology, Cancer
Graphical abstract
Highlights
-
•
TROP-2-ECD assembles into cis- or trans-dimer
-
•
cis- or trans-dimeric TROP-2-ECD can be further cross-linked
-
•
Sacituzumab binds to the stretched polypeptide in CPD not involved in TROP-2 assembly
Immunology; Structural biology; Cancer
Introduction
The GA733 gene family comprises human trophoblast cell surface antigen 2 (TROP-2, GA733-1) and epithelial cell adhesion molecule (EpCAM, TROP-1, GA733-2). The upregulated expression of these two molecules is associated with the prognosis of various tumors (Wen et al., 2018; Zeng et al., 2016). Also known as tumor-associated calcium signal transducer 2, membrane component 1 surface marker 1 (M1S1), or epithelial glycoprotein 1 (EGP1), TROP-2 was first identified as a surface marker of trophoblasts (Lipinski et al., 1981). As the only homolog of TROP-2, EpCAM has been widely applied in the development of tumor-targeting strategies, such as antibody-drug conjugates (ADCs), chimeric antigen receptor (CAR) T cell therapy, and antibody-redirected T cell engagers (Eyvazil et al., 2018; Simon et al., 2013). Catumaxomab, which targets EpCAM and CD3, is the first clinically approved bispecific antibody, and it has ushered in a new era for the developing bispecific antibodies (Ruf et al., 2010).
The cell surface glycoprotein TROP-2 plays important roles in signal transduction, tumor progression, stem cell development, and autosomal recessive diseases (Goldenberg et al., 2018). The US Food and Drug Administration (FDA) has granted accelerated approval to the first TROP-2-directed ADC drug, sacituzumab govitecan, for treating triple-negative breast cancer (TNBC) (Fenn and Kalinsky, 2019). Sacituzumab govitecan ADC consists of a humanized anti-TROP-2 monoclonal antibody, sacituzumab (hRS7), coupled to the topoisomerase-I inhibitor, 7-ethyl-10-hydroxycamptothecin (SN-38). This agent has shown promising therapeutic effects against cancers, including TNBC and non-small cell lung cancer (Bardia et al., 2017; Heist et al., 2017). Furthermore, a bispecific antibody, which was designed to target TROP-2 expressed on tumor cells and to attract T cells with the anti-CD3 antibody for tumor cell killing, substantially suppresses tumors (Chang et al., 2017). A study of the action of TROP-2/PD-L1 bispecific CAR-T cells generated promising results against gastric cancer (Zhao et al., 2019).
As a type I transmembrane glycoprotein, TROP-2 consists of 323 amino acids with a hydrophobic leader peptide, an extracellular domain, a single transmembrane domain, and a cytoplasmic tail (Linnenbach et al., 1989; Pavšič et al., 2015). The extracellular domain of TROP-2 contains a thyroglobulin type-1 (TY) repeat domain (Linnenbach et al., 1989), suggesting that it inhibits cysteine proteases to protect tumor cells from their own secreted cathepsins during metastasis (Baeuerle and Gires, 2007; Mihelic and Turk, 2007). Various human epithelial breast, uterine, and ovarian tumors overexpress TROP-2, whereas normal epithelial cells express substantially less (Ning et al., 2013; Peng et al., 2019; Varughese et al., 2011). The overexpression of TROP-2 is associated with a poor prognosis in several types of cancers, and TROP-2 can promote tumor growth and metastasis via different signaling pathways (Fong et al., 2008; Liu et al., 2013; Peng et al., 2019; Zeng et al., 2016). The mechanism through which TROP-2 promotes tumor metastasis might be via modulating integrin β-1 functions to regulate cell adhesion and motility (Trerotola et al., 2013). A mutation in the TROP-2 gene also leads to the decreased expression of claudin 1 and 7, thus altering the formation of tight junctions between adjacent cells (Nakatsukasa et al., 2010). However, the molecular assembly of TROP-2 on the cell surface and the intercellular assembly of adjacent cells are not yet fully understood but are believed to be critical for understanding the functions of TROP-2 and drug design.
The only known counterpart of TROP-2 in the GA733 gene family is EpCAM, a cell surface protein that is involved in homotypic cell-cell adhesion via cis and/or trans oligomerization (Trebak et al., 2001). The EpCAM structure is a self-assembled cis-dimer (Pavšič et al., 2014). Structural and functional findings of molecules involved in cell adhesion, for example, nectin and nectin-like molecules that play central roles in cell adhesion, suggest a mechanism through which the cis-homodimerization of a receptor on a cell surface is followed by the formation of a trans-dimer between juxtaposed cells (Stengel et al., 2012). However, Gaber et al. reported that no inter-cellular homo-oligomers of EpCAM were detectable, suggesting EpCAM may not function as a homophilic cell adhesion molecule (Gaber et al., 2018).
Recent studies have demonstrated that TROP-2 molecules on the cell membrane are clustered, especially in tumor cells (Fu et al., 2020). The present study determined the crystal structures of the TROP-2 ectodomain and found that it comprised either trans- or cis-dimers, or even tetramers, indicating the oligomeric complexity of this molecule. We also defined the region for sacituzumab binding. These data provide clues to understanding the role of TROP-2 in tumorigenesis and metastasis.
Results
Oligomeric states and overall structure of TROP-2-ECD
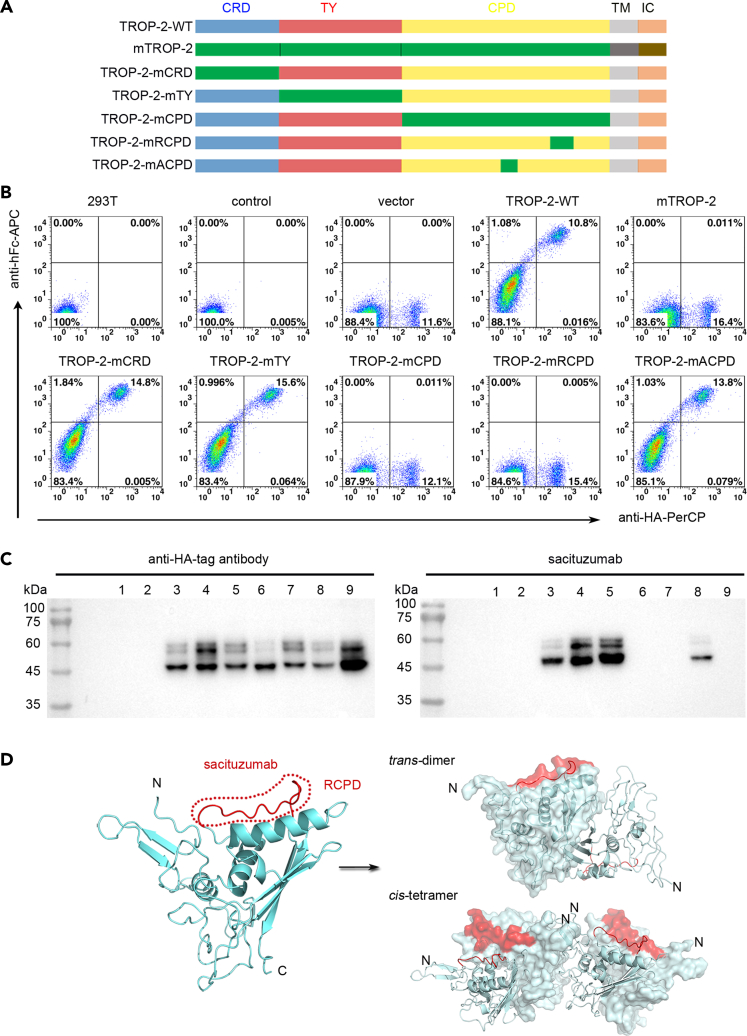
The oligomeric states of TROP-2 are not fully understood, although it has been proposed that TROP-2 may exist as a dimer (Vidmar et al., 2013). Here, the TROP-2 ectodomain (TROP-2-ECD) proteins were expressed in HEK-293T or insect cells, then self-assembled oligomers were initially analyzed by chemical cross-linking. The molecular weight of TROP-2-ECD proteins determined by SDS-PAGE was substantially greater when expressed in 293T (45 kD) than in insect (35 kD) cells, indicating more glycosylation modifications (Figures S1A and S1B). Subjecting the protein to PNGase treatment resulted in a substantially decreased molecular weight. The molecular weight of TROP-2-ECD post PNGase treatment was still higher than the prediction of 28 kDa, possibly due to incomplete removal of glycan by PNGase (Figure 1A). The extracellular domain of TROP-2 contains four potential N-glycosylation sites, N33Q, N120Q, N168Q, and N208Q, in which mutations led to a substantial reduction in the molecular weight of TROP-2-ECD compared with the wild type (WT), confirming N-glycosylation at these sites (Figure 1B).
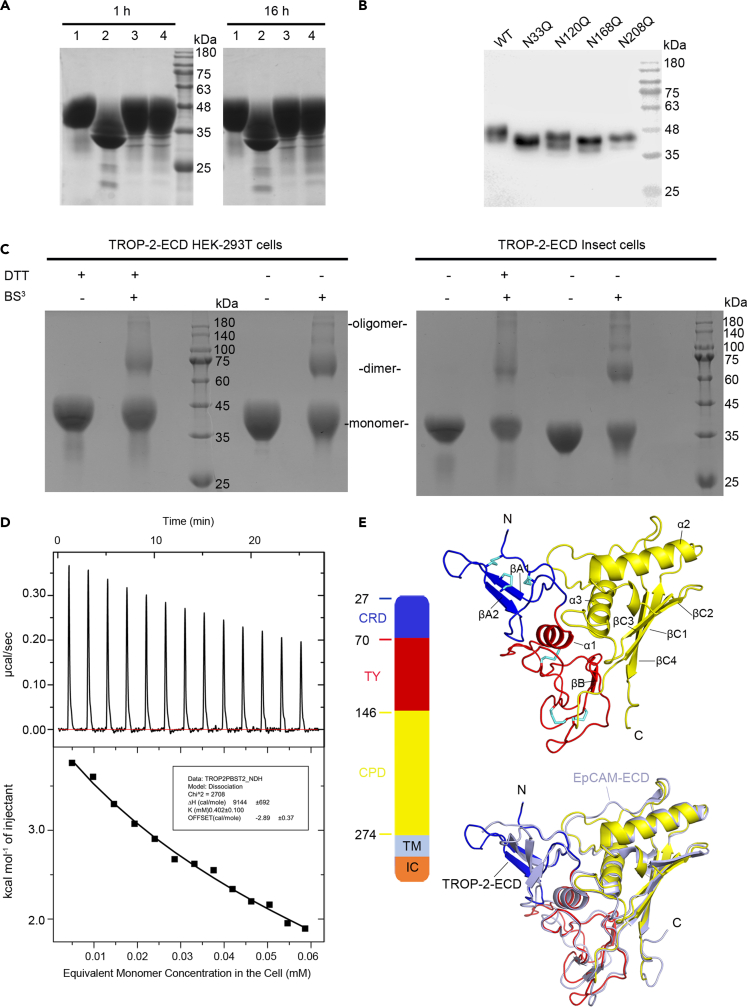
Figure 1.
Oligomeric state and overall subunit structure of TROP-2-ECD
(A) Coomassie Blue staining SDS-PAGE analysis of TROP-2-ECD proteins treated with deglycosylase PNGase or Endo H. Lane 1 and lane 2 represent buffer and PNGase-treated TROP-2-ECD protein in the pH 8.0 buffer, respectively. Lane 3 and lane 4 represent Endo H and buffer-treated TROP-2-ECD protein in the pH 6.0 buffer, respectively. TROP-2-ECD was incubated with deglycosylase at 4°C for either 1 or 16 h as indicated.
(B) A western blot analysis of wild type and potential N-glycosylation site-mutated TROP-2-ECD protein (N33Q, N120Q, N168Q, and N208Q).
(C) Coomassie Blue staining SDS-PAGE analysis of cross-linked states of TROP-2-ECD proteins expressed from HEK-293T cells (left) or insect cells (right) under reducing (with DTT) or non-reducing (without DTT) conditions. The TROP-2-ECD monomer can be cross-linked by the addition of BS3 (bis (sulfosuccinimidyl) suberate) and presented as dimer or higher levels of oligomers.
(D) ITC dissociation model shows the heat changes of decomposition of the substrate, which indicates that TROP-2-ECD exists in dimeric form at least.
(E) Schematic diagram of TROP-2-ECD (left). TROP-2-ECD is composed of an ectodomain (27–274) consisting of a cysteine-rich domain (CRD), TY domain, and cysteine-poor domain (CPD), and followed by transmembrane (TM) and cytoplasmic region. The cartoon represents an ectodomain of TROP-2, including CRD (blue), TY domain (red), and CPD (yellow), with all disulfide bonds shown as sticks (cyan). Secondary structure elements are labeled by the order in which they appear along the peptide chain from N to C terminus. The cartoon representation below is a comparison of the structural differences between the TROP-2 subunit and the EpCAM subunit (gray), with RMSD at 0.780 Å over 216 Cα atoms. See also Figures S1, S2 and S3.
The findings of reducing and non-reducing SDS-PAGE showed that a proportion of TROP-2-ECD molecules in solution assumed various oligomeric forms such as dimers, tetramers, or even higher oligomers in the presence of the cross-linker BS3 (Figure 1C). The ratios of oligomeric TROP-2-ECD proteins expressed in HEK-293T or insect cells were similar. Self-assembled TROP-2-ECD oligomers were analyzed using isothermal titration calorimetry (ITC) with a dimeric dissociation model to determine monomer-oligomer equilibrium. The results showed that TROP-2-ECD was oligomeric at high concentrations, whereas it dissociated into monomers with serial dilutions (dissociation constant (K) = 0.402 ± 0.100 mM and ΔH = 9144 ± 692 cal/mol) (Figure 1D). Taken together, these results suggested that the ectodomain of TROP-2 forms dimers or higher-level architectures. The proteolytic cleavage of TROP-2 at R87-T88 promotes cancer cell growth (Trerotola et al., 2021). Here, truncated TROP-2-ΔQ31-T88 (also ectodomain of TROP-2) proteins were expressed in HEK-293T cells to determine whether oligomeric states of TROP-2 change after proteolytic cleavage. Monomer- and disulfide-bond-mediated dimeric proteins were generated (Figure S2A). Cross-linking revealed a multimeric assembly of monomeric TROP-2-ΔQ31-T88 that resembled that of WT TROP-2, whereas dimeric TROP-2-ΔQ31-T88 still formed oligomers (Figure S2B). These results indicated that proteolytically processed TROP-2 induces a profound rearrangement of its own structure but maintained the ability to form oligomeric assemblies.
The characteristics of TROP-2-ECD oligomers were investigated by crystal screening TROP-2-ECD proteins expressed in 293T or insect cells (Figures S1A and S1B). Two sets of X-ray diffraction data were collected with TROP-2-ECD proteins expressed in either 293T cells or insect cells, and both structures were determined at 3.2 Å (Table S1). The structure of each subunit was highly conserved (Figures S1C–S1E). The TROP-2-ECD subunit comprises an N-terminal cysteine-rich domain (CRD) and a TY domain, followed by a C-terminal cysteine-poor domain (CPD) (Figure 1E). The overall structure of TROP-2-ECD is similar to that of EpCAM-ECD, which shares ∼50% amino acid sequence similarity with its ectodomains (Figures 1E and S3) (Pavšič et al., 2014). Superimposing the TROP-2-ECD structure on that of EpCAM-ECD yielded a root-mean-square deviation (RMSD) of 0.780 Å over 216 Cα atoms, indicating high similarity between them. Structural analysis revealed that the Cys34-Cys53, Cys36-Cys66, Cys44-Cys55 disulfide bonds stabilized the conformation of the CRD domain of TROP-2 and that one of these bonds linked two β-sheets (Cys44-Cys55) (Figure 1E). The TY domain contained a thyroglobulin type-1 repeat module, consisting of cysteine-rich motifs with evolutionarily conserved cysteine residues as in EpCAM-ECD, and the conserved sequence motifs QC and CWCV (Figure S3) (Mihelic and Turk, 2007). The disulfide bonds, Cys73-Cys108, Cys119-Cys125, and Cys127-Cys145, stabilized the TY domain (Figure 1E). The C-terminal CPD domain contained two α-helices (α2, α3) and a βC sheet comprising four β ribbons (βC1-4), and its structural stability was maintained by a hydrophobic interaction network. Of note, the long loop connecting α3 and βC4 constituted a stretched polypeptide chain in parallel with the α2 helix that substantially differed from the structure of EpCAM-ECD. Taken together, the structural features of the three domains of TROP-2-ECD were similar to those of EpCAM-ECD.
Cis- and trans-dimers of TROP-2-ECD
The dimeric conformations distinctly differed between TROP-2-ECD proteins expressed in the two types of host cells. The dimeric structure of TROP-2-ECD proteins expressed in 293T cells comprised two TROP-2-ECD monomers that were organized inversely and symmetrically along with a βC sheet (Figure 2A). The interface of this TROP-2-ECD dimerization was distinct from that of the EpCAM-ECD cis-dimer, and the two C-termini of the TROP-2-ECD monomers are on opposite sides (Pavšič et al., 2014). Therefore, we postulated that this dimer would be in a trans conformation, indicating the intercellular assembly of TROP-2-ECD molecules from two adjacent cells.
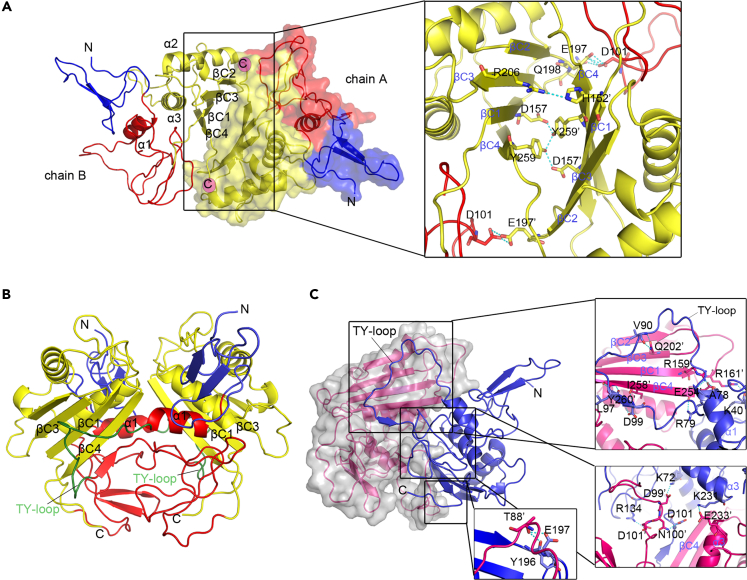
Figure 2.
Characteristics of trans- and cis-dimer TROP-2-ECD
(A) The structure of the TROP-2-ECD trans-dimer is shown as cartoon and surface representation, with colors consistent with Figure 3C. The enlarged area shows the details of the interaction interface.
(B) Cartoon representation of bilateral cis-dimer of TROP-2-ECD and the color of subunits are consistent with Figure 3C except for the TY-loop (green).
(C) One subunit in the cis-dimer structure presents a translucent surface format, and the other subunit is the cartoon representation. The enlarged upper right and the bottom left box indicate the main interaction interface of the two subunits containing βC sheet and TY-loop. Residues that participate in hydrogen bond interactions are shown as sticks and labeled. Hydrogen bonds are shown as dashed cyan lines. See also Figure S1, Tables S1, S2 and S3.
The concave βC sheet of the CPD domain of the two monomers predominantly mediated formation of the trans-dimer, which covered a surface area of 914 Å2 (Figure 2A). Two sets of symmetrical amino acids formed several hydrogen bond interactions between the two monomers. Among them, R206 from the βC3 strand formed hydrogen bond interactions with H152′ from the top of βC1 of the other monomer. D157 in βC1 formed a symmetrical hydrogen bond interaction with Y259′ from the βC4 strand. In addition, the hydrogen bonds formed by D101 and E197′ also contribute to the formation of a stable trans-dimer conformation (Figure 2A, right) (Table S2). The trans-dimeric architecture suggested that TROP-2-ECD molecules between juxtaposed cells mediate intercellular signal transduction or cell-cell communication through trans interaction.
The assembly of TROP-2-ECD proteins expressed in insect cells was distinctly dimeric and similar to the structure of the EpCAM-ECD cis-dimer (PDB:4MZV) (Figure 2B). Formation of the TROP-2-ECD cis-dimer is prominently mediated by the TY and CPD domains, in which the main interacting region is buckled by the TY-loop of one monomer to the βC sheet of the other. The cis-dimer covered a surface area of 2,397.9 Å2, which was ∼2-fold that of the trans-dimer, as described above. Amino acids from the TY-loop and its adjacent α1 helix and βB (A78, R79, T88′, V90, L97, and D99) formed a complicated hydrogen bond interaction network with residues from the βC sheet of the other monomer (R159′, E254′, Y196′, E197′, Q202′, Y260′, and I258′) (Figure 2C) (Table S3). In addition, amino acid residues from adjacent regions to the TY-loop and βB sheet also formed hydrogen bond interactions between K40, K72, D101, R134, and K231 from one monomer and R161′, D99′, N100′, D101′, and E233′ in the other, which helped to stabilize the cis-dimeric structure (Figure 2C bottom right). Sequence alignment revealed that 59% of the residues from TROP-2, involved in the formation of a hydrogen bond interaction network in the cis-dimer, were conserved along with those from EpCAM, indicating a similar cis assembly mode between these homologs (Figure S3).
Cross-linking of TROP-2-ECD
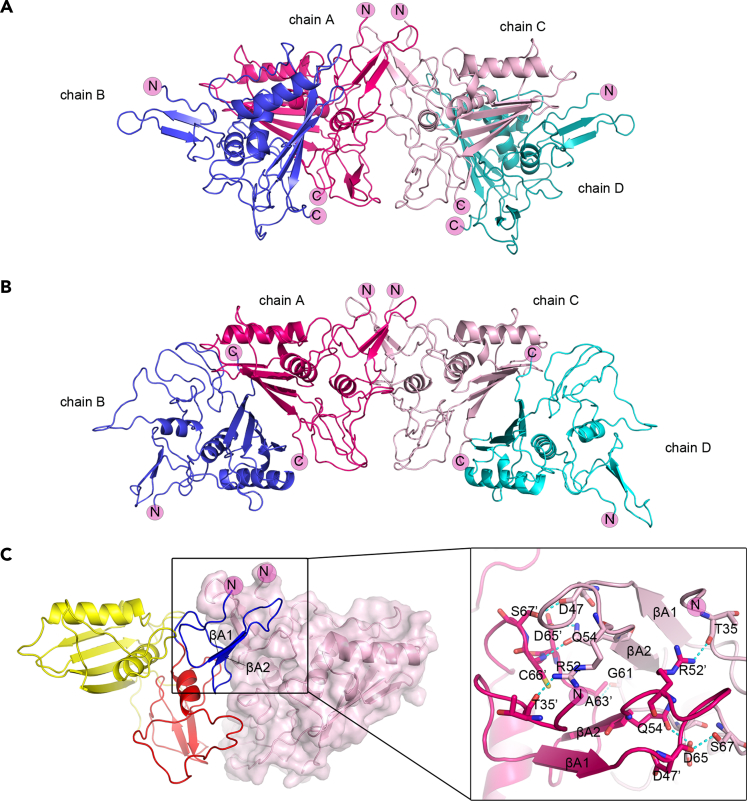
TROP-2 molecules on the cell membrane exhibited in a clustered form, especially on tumor cell membranes (Fu et al., 2020). A tetrameric assembly of TROP-2-ECD, which is composed of two symmetrical cis-dimers, is found in the structure determined with TROP-2-ECD proteins expressed in insect cells (Figure 3A). Tetramerization of the two cis-dimers was mediated by the N-terminal CRD domain, which is distinct from the interface of either cis- or trans-dimers, and covered a surface area of 850 Å2. The homolateral distribution of the C terminus of the four TROP-2-ECD monomers in the tetrameric assembly indicated a proximal location on the cytomembrane, whereas the N terminus was distally located (Figure 3A). Therefore, we postulated that this would be a cis-tetramer, indicating that cis-tetramerization of TROP-2 molecules occurs on the same cell surface. Moreover, a TROP-2-ECD tetramer could also be observed in the structure of TROP-2-ECD trans-dimer after simple symmetrical operation (Figure 3B). Consistent with the structure of cis-tetramer, the tetramerization of the two trans-dimers was also mediated by the N-terminal CRD domain. Specifically, the amino acids from the CRD of one monomer (T35′, D47′, R52′, Q54′, A63′, S67′, D65′, C66′) participated in hydrogen bond interactions with residues from the CRD (R52, S67, T35, G61, D65, D47, Q54) of the other monomer (Figure 3C) (Table S4). Considering that the tetramerization interface mediated by the N-terminal CRD is located away from the cis-dimer and trans-dimer, TROP-2 may extend to form cross-linked clusters on the cell surface.
Figure 3.
Structure of TROP-2-ECD tetramer
(A) The structure of TROP-2-ECD cis-tetramer shown as cartoon representation; each subunit is represented by a different color (hot pink, blue, light pink, teal).
(B) Cartoon representation shows a trans-tetramer of TROP-2-ECD trans-dimer after simple symmetrical operation. The two trans-dimers could be further cross-linked with the N-terminal CRD domain of each of one subunit from the trans-dimer, with colors the same as in Figure 3A.
(C) Two subunits in the middle that cross-linked the two cis-dimers of TROP-2-ECD via the N-terminal CRD domain were presented with one molecule displayed in cartoon representation and the other displayed in the surface format. The enlarged part shows the details of the interaction of the interface of cis-tetramer via the N-terminal CRD. Residues involved in the hydrogen bond interactions are displayed as sticks and labeled, and hydrogen bonds are shown as dashed cyan lines. See also Figures S1, S4 and Tables S1, S3, S4.
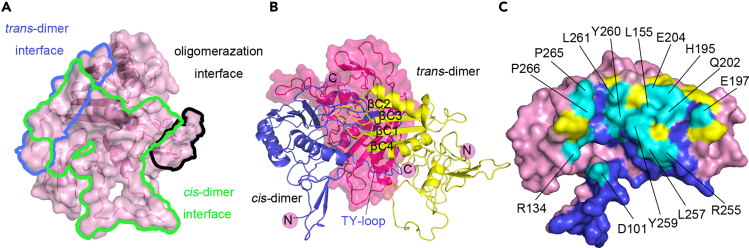
The three interfaces that form trans-dimers, cis-dimers, or N-terminal CRD-mediated tetramerization were further compared (Figure 4A). The formation of trans- or cis-dimers induced mutual stereo hindrance, and the surfaces where they formed substantially overlapped, indicating that formation of the trans-dimer would interrupt that of the cis-dimer, and vice versa (Figures 4B and 4C). The overlapping binding sites are located in a region consisting of residues near the βC sheet (P266, P265, D262, L261, Y260, L155, E204, H195, Q202, Y259) and residues on the TY-loop (V98, D99, N100), which lead to spatial steric hindrance between trans- and cis-dimers (Figure 4C). In contrast, the interface that cross-linked the cis-dimer or trans-dimer to form a tetramer did not overlap with that of trans- or cis-dimers, indicating that tetramers or even higher levels of cis-trans assemblies cluster between adjacent cells. Therefore, the N-terminal domain could serve as cross-linking region to mediate clustering of cis- or trans-dimers of TROP-2-ECD. More complicated clusters might also be formed with both cis- and trans-interacting complexes between adjacent cells (Figure S4). We speculate that these assemblies mediate cellular adhesion or signal transmission between two adjacent cells and that extended TROP-2 clustering may have determined the signaling strength.
Figure 4.
Three contacting interfaces
(A) The translucent surface representation shows a subunit TROP-2-ECD with surface areas of the interacting surfaces of trans-dimer, bilateral cis-dimer, and the interface of cis-tetramer via the N-terminal CRD circled by lines with blue, green, and black, respectively.
(B) Superimposition of TROP-2-ECD cis- and trans-dimeric structure of TROP-2-ECD to illustrate the stereospecific hindrance between cis- and trans-dimerization.
(C) Competitive binding surfaces between cis- and trans-dimeric TROP-2-ECD. Residues involved only in cis-dimer interactions are colored in blue, whereas the residues that are only contacted in trans-dimer are colored in yellow. The amino acid residues involved in the interacting interfaces of both cis- and trans-dimer are indicated in cyan and labeled.
Sacituzumab binds at a site in TROP-2 far from cis- or trans-interface
We investigated the binding site of sacituzumab, which is the antibody used for the ADC drug approved by the US FDA, and whether the binding of sacituzumab would interrupt the cis- or trans-assembly of TROP-2.
Sacituzumab binding to TROP-2 variants was assessed by flow cytometry and western blotting. The results of both analyses showed that sacituzumab did not bind to mouse (m) TROP-2 (Figures 5A–5C). Therefore, we substituted each of the three domains of human TROP-2 with those of a mouse homolog to define the binding epitope of sacituzumab (Figure 5A). An N-terminal hemagglutinin (HA) tag was added to each of the WT or mutated TROP-2 constructs to verify their expression in HEK-293T cells. The domain-substituted full-length TROP-2 constructs were transduced into HEK-293T cells, which were then stained with sacituzumab or anti-HA antibodies and analyzed by flow cytometry and western blotting. Binding was detected by flow cytometry in 293T cells transiently expressing the mouse CRD domain-substituted TROP-2 (TROP-2-mCRD), mouse TY domain-substituted TROP-2 (TROP-2-mTY), or mouse CPD domain-substituted TROP-2 (TROP-2-mCPD), and stained with sacituzumab (Figure 5A). The substitutions prevented sacituzumab from binding TROP-2-mCPD, but not TROP-2-mCRD and TROP-2-mTY (Figure 5B). Binding epitopes were further mapped by substituting two exposed loops in the CPD domain from mTROP-2, the stretched polypeptide in parallel with the α2 helix like the ridge on CPD (Q237-Q252, mRCPD), and the loop connecting the βC3 and α3 helix-like arch on the side of CPD (T209-G215, mACPD) that substantially varied between human and mouse TROP-2 (Figure S5). We found that TROP-2-mACPD did not affect binding with sacituzumab, whereas substitution of the stretched polypeptide Q237-Q252 (TROP-2-mRCPD) abolished sacituzumab binding, suggesting that sacituzumab binds to the Q237-Q252 polypeptide (Figure 5B). Western blots showed that sacituzumab bound to denatured TROP-2 proteins, indicating that the binding site is a linear epitope in TROP-2. Westerns blots of TROP-2 variants were similar to those of flow cytometry in which sacituzumab did not bind to the substituted TROP-2-mCPD and TROP-2-mRCPD (Figure 5C). Therefore, sacituzumab binds to a linear epitope within the RCPD of TROP-2. Further analysis revealed that the RCPD region for sacituzumab binding was located far from the trans-interface and was not involved in formation of the cis-assemblies (Figure 5D). Therefore, sacituzumab binding might not interrupt the assembly of TROP-2 molecules on the cell surface.
Figure 5.
Binding analysis of sacituzumab
(A) Schematic diagram of domain- or loop-substituted TROP-2 constructs with that from mouse homolog.
(B) Flow cytometry analyses of the key domain of interaction between TROP-2 and sacituzumab. The HA-tagged proteins of wild-type (TROP-2-WT), mouse TROP-2 (mTROP-2), mouse CRD-substituted (TROP-2-mCRD), mouse TY-substituted (TROP-2-mTY), mouse CPD-substituted (TROP-2-mCPD), and two exposed loops of CPD substituted (TROP-2-mRCPD and TROP-2-mACPD) TROP-2 were expressed on HEK-293T cells and incubated with sacituzumab, respectively. The PerCP-conjugated anti-HA tag antibody was used to detect the expression of TROP-2 on the surface of HEK-293T cells, whereas APC-conjugated anti-human IgG antibody was used to detect the binding of sacituzumab with TROP-2 on HEK-293T cells. Untransfected HEK-293T cells and TROP2-WT expressing HEK-293T cells stained with isotype control antibody were used as negative controls.
(C) Western blot analysis of the key domain responsible for the interaction between TROP-2 and sacituzumab. The left panel represents the detection of different variants of TROP-2 expression on HEK-293T cells with anti-HA-tag antibody (left). The right panel represents the detection of the binding of sacituzumab to different TROP-2 variants as indicated using sacituzumab as detection antibody. Lanes 1 to 9 represent untransfected HEK-293T cells, HEK-293T cells transfected with empty vector, TROP-2-WT, TROP-2-mCRD, TROP-2-mTY, TROP-2-mCPD, TROP-2-mRCPD, TROP-2-mACPD and mTROP-2, respectively.
(D) The predicted binding region of sacituzumab on TROP-2 based on the flow cytometry-based binding assay, with RCPD loop highlighted in red (left). The location of RCPD in TROP-2 cis- or trans-assembly is indicated (right). See also Figure S5.
Discussion
The molecular assembly of TROP-2 and EpCAM is considered critical for modulating cell-cell communication, cell adhesion, and tumor metastasis. The present study found that TROP-2-ECD oligomers were present in solutions containing the chemical cross-linker BS3, indicating the existence of varied assemblies of TROP-2-ECD. The structures determined here also support the existence of cis- or trans-dimeric TROP-2-ECD, or TROP-2-ECD tetramer. The previously reported structure of EpCAM-ECD was demonstrated to be cis-dimer, which was mainly mediated by the TY-loop, and the cis assembly mode was conserved with the cis-dimeric TROP-2-ECD structure found herein.
Since TROP-2 is a highly glycosylated protein, glycosylation status of the two preparations of TROP-2-ECD was confirmed by PNGase treatment as well as mutational analysis. The results indicated that all of the four potential N-glycosylation sites in TROP-2-ECD protein were glycosylated. The known structures of EpCAM-ECD and TROP-2-ECD cis-dimers were both defined from these proteins expressed in insect cells. The lower molecular weight determined by SDS-PAGE indicated that the TROP-2-ECD proteins seemed less glycosylated when expressed in insect cells than in 293T cells. Therefore, less glycosylated TROP-2-ECD or EpCAM-ECD proteins might be more prone to forming cis-dimeric assemblies. However, cis- and trans-dimer proteins are difficult to discriminate with each other in solution and might be at equilibrium.
Pavšič et al. proposed a trans-interaction model of EpCAM based on the structure of the EpCAM cis-dimer, which utilizes the same interface as cis-interaction (Pavšič et al., 2014). However, the trans-dimeric assembly of TROP-2-ECD was distinct in the present study, and it was mediated by the βC sheet of the C-terminal CPD domain. The surface area covered by this trans-dimer was much smaller, and fewer hydrogen bond interactions were involved in this interface compared with the cis-dimer, suggesting that interactions of TROP-2-ECD cis-dimerization might be much more stable than those of trans-dimerization. The interface of the trans-dimer substantially overlapped that of the cis-dimer, indicating competition between TROP-2-ECD trans- and cis-dimers. Considering the high similarity of the conformations and amino acid sequences of TROP-2 and EpCAM monomers, we speculated that EpCAM trans-dimerization occurs via a similar type of trans-assembly, although there is no direct experimental evidence to show that EpCAM can form trans-dimer. Bidirectional signaling into juxtaposed cells or cell adhesion might be mediated by cis-dimerization of the receptor/ligand on the same cell surface followed by trans-dimerization between juxtaposed cells using either identical or distinct interfaces (Stengel et al., 2012). This process would require rearrangement and breakup of the cis-interaction. However, the mechanism of the formation of trans-interaction of TROP-2 is not yet well understood, especially considering that the present structural findings indicated better stability for cis-than trans-interaction. Other influential factors, including lipid composition and the engagement of other proteins during cell-cell contact, might also play critical roles in cis or trans interactions of these molecules.
We also found that the structures of homolateral cis-tetrameric TROP-2-ECD assembly and trans-tetrameric TROP-2-ECD assembly are both mediated by the N-terminal CRD domain. The tetramerization interface mediated by the N-terminal CRD is distinct from the trans- or cis-dimerization interface, indicating that trans-and cis-interacting TROP-2 molecules are cross-linked on the cell surface. If so, cross-linked TROP-2 molecules on tumor cell surfaces and a clustered synapse formed within the interface of adjacent cells might be critical for signal transduction between juxtaposed tumor cells. Of note, EpCAM also forms tetramers in solution with a reversible dimer-tetramer association of moderate affinity (Kd ∼ 10 μM), whereas the affinity of monomer-dimer association (Kd ∼ 10 nM) is 1,000-fold higher (Trebak et al., 2001). The present finding shows that tetramerized TROP-2-ECD mediated via the N-terminal CRD covered a much smaller surface area than the dimer and fewer hydrogen bond interactions occurred within the tetramerization interface. However, there is no structural evidence that EpCAM could form tetramer. Moreover, Gaber et al. have reported multiple experimental data to show that EpCAM does not form higher-order homo-oligomers and may not be involved in cell-cell adhesion (Gaber et al., 2018). Therefore, TROP-2 and EpCAM may adopt distinct assemblies to mediate their function.
Tumor-specific proteolytic cleavage of TROP-2 occurs at the conserved cleavage site R87-T88 of the TY loop, which is similar to that of EpCAM at the typical dibasic site R80-R81 (Kamble et al., 2020; Pavšič et al., 2014; Trerotola et al., 2021; Wu et al., 2017). Proteolytic cleavage activates the cell-growth stimulatory properties of TROP-2 (Trerotola et al., 2021). The cleavage of EpCAM at R80-R81 of the TY loop interrupts the dimeric forms of EpCAM (Pavšič et al., 2014). However, cross-linking analysis of truncated TROP-2-ΔQ31-R88 protein indicated that the dimeric assemblies were not interrupted. Moreover, Q31-R88 depletion yielded a TROP-2-ECD dimer mediated by a disulfide bond. Such disulfide bond-mediated dimeric proteins might correlate with free C108, which forms disulfide bonds with C73 in wild-type TROP-2-ECD, after the N-terminal region is removed. Therefore, proteolytically processed TROP-2 might induce profound rearrangement of the TROP-2 structure, and the assembly mode of this tumor-specific truncated form awaits further investigation.
We found that sacituzumab mainly binds to the Q237-Q252 polypeptide of TROP-2 by substituting the domains or exposed loops of human TROP-2 with that of a mouse homolog. Like the structure of EpCAM-ECD, the cis-dimer of TROP-2-ECD forms a central groove edged by the Q237-Q252 polypeptides. Structural analysis revealed that this region was not involved in the formation of either cis or trans interactions. This indicated that sacituzumab might not block the formation of a TROP-2 cis- or trans-assembly, although the possibility of binding-induced stereo hindrance could not be excluded. Sacituzumab alone does not effectively suppress tumors in vivo, which also indicates that sacituzumab does not inhibit the function of TROP-2 during tumor metastasis (Han et al., 2020). Therefore, the tumor suppressive effects of sacituzumab govitecan are mainly induced by SN-38 after binding to TROP-2 expressed in tumor cells. However, whether blocking TROP-2 assembly affects its biological function requires further investigation.
In summary, we found that TROP-2 assumes various oligomeric forms and the structures of TROP-2-ECD revealed the assembly of both cis- and trans-dimeric TROP-2 molecules at distinct but overlapping interfaces. The structures of cis- or trans-tetrameric TROP-2-ECD via the N-terminal CRD have a non-overlapping tetramerization interface with either cis- or trans-dimers, suggesting that cis- and trans-TROP-2 molecules are cross-linked on the cell surface. The binding site of sacituzumab was located on a stretched polypeptide in the CPD domain that was not involved in the generation of either cis- or trans-interactions. Considering the critical role of TROP-2 in tumor metastasis, the structural basis of cis- or trans-interactions of TROP-2 presented herein deepens the understanding of its roles in cell-cell communication and should facilitate the design of biologics for tumor therapy.
Limitations of the study
Although in structural biology we have found that TROP-2-ECD forms trans- and cis-assemblies, whether TROP-2 is expressed in these formats on the cell surface needs to be further verified by additional experiments. Based on the region where sacituzumab binds to TROP-2, we speculate that sacituzumab may prevent TROP-2 from forming a cis- or trans-dimer, but this speculation also needs to be supported by further experimental data. The influential factors of the formation of varied TROP-2 assemblies and subsequent effects on its biological function remain undetermined.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| PerCP-conjugated anti-HA tag antibody | Abcam | Cat# ab1117491 |
| APC-conjugated anti-human IgG Fc | Biolegend | Cat# 410712 RRID:AB_2565790 |
| anti-HA-tag antibody | EasyBio | Cat# BE2007 |
| Goat anti-mouse IgG(H+L)-HRP | Gene-Protein Link | Cat# P03S01M |
| HRP-conjugated goat anti-human IgG(H+L) | Proteintech | Cat# SA00001-17 RRID:AB_2890979 |
| anti-His-tag mAB-HRP-DirecT A | MBL | Cat# D291-7; RRID: AB_10694870 |
| Bacterial and virus strains | ||
| Escherichia coli (E.coli) strain T1 | TransGen | Cat# CD501-2 |
| Escherichia coli (E.coli) strain DH10Bac | BioMed | BC112 |
| Escherichia coli (E. coli) strain BL21 (DE3) | Novagen | Cat# 69450 |
| Chemicals, peptides, and recombinant proteins | ||
| BS3 (bis (sulfosuccinimidyl) suberate) | Thermo | Cat# 21580 |
| EndoH | This paper | N/A |
| PNGase | This paper | N/A |
| TROP-2-ECD-293T | This paper | N/A |
| TROP-2-ECD-insect | This paper | N/A |
| sacituzumab | This paper | N/A |
| Critical commercial assays | ||
| Crystallization kits | Hampton Research and Molecular Dimensions and Wizard Classic |
http://www.hamptonresearch.com; https://www.moleculardimensions.com/;https://rigakureagents.com/ |
| HisTrap excel 5mL column | GE Healthcare | Cat# 17371205 |
| RESOURCETM S 6 mL column | GE Healthcare | Cat# 17118001 |
| Hiload Superdex® 200 16/600 pg column | GE Healthcare | Cat# 2898933 |
| Superdex® 200 Increase 10/300 GL column | GE Healthcare | Cat# 28990944 |
| HiTrap Protein A HP | GE Healthcare | Cat# 17040303 |
| Deposited data | ||
| TROP-2-ECD-293T | This paper | PDB: 7E5M |
| TROP-2-ECD-insect | This paper | PDB: 7E5N |
| Experimental models: cell lines | ||
| HEK-293T cells | ATCC | CRL-3216; RRID:CVCL_0063 |
| High Five cells | Invitrogen | B85502 |
| Sf9 cells | Invitrogen | B82501 |
| Recombinant DNA | ||
| pFastBac1-TROP-2-ECD-insect | This paper | N/A |
| pCAGGS | MiaoLingPlasmid | Cat# P0165 |
| pCAGGS-TROP-2-ECD-293T | This paper | N/A |
| pCAGGS-TROP-2-N33Q | This paper | N/A |
| pCAGGS-TROP-2-N120Q | This paper | N/A |
| pCAGGS-TROP-2-N168Q | This paper | N/A |
| pCAGGS-TROP-2-N208Q | This paper | N/A |
| pCAGGS-TROP-2-WT | This paper | N/A |
| pCAGGS-mTROP-2 | This paper | N/A |
| pCAGGS-TROP-2-mCRD | This paper | N/A |
| pCAGGS-TROP-2-mTY | This paper | N/A |
| pCAGGS-TROP-2-mCPD | This paper | N/A |
| pCAGGS-TROP-2-mRCPD | This paper | N/A |
| pCAGGS-TROP-2-mACPD | This paper | N/A |
| pCAGGS- sacituzumab-H-chain | This paper | N/A |
| pCAGGS- sacituzumab-L-chain | This paper | N/A |
| pET28a-PNGase | This paper | N/A |
| pET28a-ENdoH | This paper | N/A |
| Software and algorithms | ||
| PyMOL software | Molecular Graphics System, Version 1.8 Schrö dinger |
https://pymol.org/2/ RRID:SCR_000305 |
| FlowJo 7.6.1 | FLOWJO | https://www.flowjo.com/solutions/flowjo/downloads |
| Phenix | Adams et al., 2010 |
http://www2.mrclmb.cam.ac.uk/Personal/pemsley/coot/; RRID:SCR_014222 |
| Coot | Emsley and Cowtan, 2004 |
http://www.phenixonline.org/; RRID:SCR_014224 |
| CCP4 | Collaborative Computational Project |
https://www.ccp4.ac.uk/ RRID:SCR_007255 |
| MolProbity | Chen et al., 2010 | N/A |
| Origin | OriginLab |
https://www.originlab.com/ RRID:SCR_002815 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Shuguang Tan (tansg@im.ac.cn).
Materials availability
Plasmids and all unique reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Experimental model and subject details
Cells
Human embryonic kidney 293T (HEK293T) cells from ATCC were cultured in Dulbecco's Modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS) in 37°C incubator containing 5% CO2. High Five cells and Sf9 cells from Invitrogen Life were cultured in Insect-XPRESS medium in 27°C incubator.
Microbe strains
E. coli strain T1, BL21 (DE3), DH10Bac were cultured in LB with corresponding antibiotics after transfected with recombinant plasmid or bacmid in 37°C incubator.
Method details
Protein expression and purification
The DNA sequences encoding the ectodomains of TROP-2 (H27 to R247) (UniProtKB-P09758) were cloned into pCAGGS expression vector with EcoRI and BglII restriction sites followed by a C-terminal 6 × His affinity tag to facilitate protein purification. An insect cell expression plasmid was also constructed with the DNA sequences encoding the ectodomains of TROP-2 (H27 to R247) cloned into the baculovirus transfer vector pFastBac1 (Invitrogen), in-frame with an N-terminal gp67 signal peptide for secretion and also a 6xHis affinity tag at the C-terminus for purification, respectively.
TROP-2-ECD proteins were expressed from either HEK-293T cells (ATCC) with pCAGGS (Addgene) expressing construct or insect cells with pFastBac1expressing construct. For 293T cells expressing, the pCAGGS plasmids carrying TROP-2-ECD (H27 to R247) were transiently transfected into 293T cells. The cells were cultured at 37°C in Dulbecco's Modified Eagle medium (DMEM) supplemented containing 10% fetal bovine serum (FBS) with 5% CO2, and supernatants were collected 96h after transfection. and then captured cell culture supernatant by immobilized metal affinity chromatography (IMAC) with a HisTrap excel 5mL column (GE Healthcare), and then purified by ion-exchange chromatography using a RESOURCETM S 6 mL column (GE Healthcare), the final purification was carried out by size exclusion chromatography on Hiload Superdex 200 16/600 pg column (GE Healthcare) equilibrated with a buffer containing 20 mM Tris-HCl and150 mM NaCl (pH 8.0).
The Bac-to-Bac baculovirus expression system (Invitrogen) was used to express soluble TROP-2-ECD proteins constructed in pFastBac1 vector. Transfection of bacmids and virus amplification were operated based on the Bac-to-Bac baculovirus expression system manual, and Hi5 cells suspension cultures were infected with high-titer recombinant baculovirus to express soluble proteins. The supernatant was collected and the protein was purified by sequentially His-Trap HP column (GE Healthcare) and size exclusion chromatography on Hiload Superdex 200 16/600 pg column (GE Healthcare) in a buffer containing 20 mM Tris and 50 mM NaCl (pH 8.0).
The truncated TROP-2-ΔQ31-T88 proteins were constructed and expressed from HEK-293T cells. The DNA sequences encoding TROP-2-ΔQ31- T88 were cloned into pCAGGS expression vector followed by a C-terminal 6 × His affinity tag. The method of expression and purification of TROP-2-ΔQ31-T88 protein was the same as that of TROP-2-ECD protein produced by HEK-293T cells, and was finally purified by Superdex 200 Increase 10/300 GL column (GE Healthcare) in PBS buffer.
The full length heavy chain and light chain gene of sacituzumab were cloned into the pCAGGS vector individually with EcoR I and Xho I sites and then co-transfected into 293T cells. The culture medium was changed to fresh DMEM without FBS 6 h after transfection. The supernatants were collected on the 3rd day and 7th day after transfection, and the proteins were purified with protein A column (GE Healthcare).
The DNA sequences encoding PNGase (A41-N354) (UniProtKB-P21163) and EndoH (A43-P313) (UniProtKB - P04067) were cloned into pET28a expression vector and transformed into E. coli strain BL21 (DE3) for protein expression.
Deglycosylation
TROP-2-ECD proteins expressed by 293T cells was treated with deglycosylase PNGase or EndoH, respectively, and the mass ratio of TROP-2 protein to enzyme is 100:1. The TROP-2 was incubated with deglycosylase at 4°C for 1 hour or 16 hours, and then analysis by Coomassie Blue staining SDS-PAGE. The PNGase treated TROP-2 protein was incubated in a buffer containing 20 mM Tris and 150 mM NaCl (pH 8.0), and the Endo H treated TROP-2 protein was incubated in a buffer containing 20 mM MES and 30 mM NaCl (pH 6.0).
Chemical crosslinking
The oligomeric states of TROP-2-ECD or TROP-2-ΔQ31-T88 were analyzed with BS3. To avoid moisture condensation onto the product, the vial was equilibrated to room temperature before opening and then dissolved BS3 in water to 6.25 mM, and used immediately. TROP-2-ECD proteins expressed from insect cells or 293T cells were adjusted to a concentration of 3 mg/mL, and TROP-2-ΔQ31-T88 proteins expressed from 293T cells were adjusted to a concentration of 1 mg/mL, and both incubated with a 50-fold molar excess of the crosslinker on ice for 2 hours. The reaction was quenched by adding Quenching Buffer to a final concentration of 50 mM glycine for 15 minutes at room temperature. Reaction mixtures were analyzed on 12% SDS–PAGE under reducing conditions with DTT and nonreducing conditions without DTT.
Isothermal titration calorimetry (ITC)
The TROP-2-ECD proteins expressed from insect cells were prepared in PBST buffer containing 0.02% Tween® 20. ITC experiments were performed with an ITC200 microcalorimeter (MicroCal). TROP-2-ECD proteins were loaded into the syringe, and PBST buffer was added to the cell. The start concentration of the TROP-2-ECD in the syringe was 340 μM. Reactions were run by performing 13 injections of TROP-2-ECD into the cell at 25°C with each injection of 3μL. Data were fit using the dissociation model in Origin (OriginLab).
Crystallization, data collection, and structure determination
TROP-2-ECD proteins expressed from 293T cells or insect cells were both crystallized via the sitting-drop vapor diffusion method at 18°C by mixing 1 μL protein with 1μL reservoir solution. Crystals with TROP-2-ECD proteins obtained from 293T cells were grown in a reservoir solution of 0.2 M potassium iodide, 0.1 M MES, pH 6.5, and 25% w/v PEG 4000, while crystals with TROP-2 proteins obtained from insect cells were grown in a reservoir solution of 100 mM Tris base/Hydrochloric acid pH 7.0, 200 mM Sodium chloride and 1000 mM Sodium citrate tribasic. The crystals were cryoprotected in their reservoir solution with the addition of 20% (vol/vol) glycerol before being flash-cooled in liquid nitrogen. The X-ray diffraction data were collected at the Shanghai Synchrotron Radiation Facilities (SSRF: 17U/19U). All diffraction data sets were processed with HKL-2000 software (Otwinowski and Minor, 1997). The TROP-2-ECD structures were solved by molecular replacement using phase with the reported structure of the EpCAM (PDB code: 4MZV) as a search model (Collaborative Computational Project, 1994; Read, 2001). Subsequent model building and refinement were performed using Coot and Phenix to refine the results, respectively (Adams et al., 2010; Emsley and Cowtan, 2004). The stereochemical qualities of the final model were assessed with MolProbity (Chen et al., 2010). All structural images were made using PyMOL (http://www.pymol.org).
Flow cytometry based binding analysis
To define the binding region of sacituzumab on TROP-2, we constructed four different full-length TROP-2 mutants that replaced the CRD (H27-L69), TY (T70-C145), CPD (D146-T274) and two loops of CPD domain (RCPD, Q237-Q252; ACPD, T209-G215) of human TROP-2 with that of murine homolog, mCRD (Q25-L63), mTY (T64-C139), mCPD (D140-G270), mRCPD (M231-H246) and mACPD (A203-R209), respectively, together with full length mTROP-2. The full-length TROP-2, TROP-2-mCRD, TROP-2-mTY TROP-2-mCPD, TROP-2-mRCPD and mACPD substituted mutants were cloned into the pCAGGS vector with N-terminal HA tag, and then transfected into the 293T cell line.
For cell staining, the cells expressing different TROP-2 constructs were washed twice and then suspended in PBS with 4% FBS and incubated with 5μg/mL sacituzumab for 1h on ice. Untransfected 293T cells and TROP-2-WT expressing 293T cells incubated with isotype controlled antibody were enrolled as negative controls. Then the cells were rinsed and further stained with PerCP-conjugated anti-HA tag antibody (Abcam) and APC-conjugated anti-human IgG antibody (Biolegend) for another 30 min on ice. After washing, the cells were analyzed by flow cytometry (BD Canto). The figures were generated with FlowJo 7.6.
Western blotting
Western blot analysis of the key domain responsible for the interaction between TROP-2 and sacituzumab. The full-length TROP-2, mouse TROP-2, TROP-2-mCRD, and TROP-2-mTY, TROP-2-mCPD, TROP-2-mRCPD and TROP-2-mACPD mutants were transiently transfected separately into 293T cells. 48 hours after transfection, the cells were collected and lysed, and then the samples were analyzed by western blotting with anti-HA-tag antibody (EasyBio) or sacituzumab. Goat anti-mouse (Gene-Protein Link) or anti-human IgG-horseradish peroxidase (HRP) (Proteintech) conjugated antibodies were used as the secondary antibodies.
Each of the four potential N-glycosylation sites in the ectodomains of TROP-2 (H27 to R247) were mutated to Q (N33Q, N120Q, N168Q and N208Q). The DNA sequence were cloned into pCAGGS expression vector with a C-terminal 6 × His affinity tag for western blot analysis. 293T cells were transiently transfected with N-glycosylation site mutated TROP-2-ECD (N33Q, N120Q, N168Q and N208Q). After 48 hours, culture supernatants were collected. Protein samples were analyzed by western blotting with anti-His-tag -HRP-Direct antibody.
Quantification and statistical analysis
This manuscript does not include quantification or statistical analysis.
Acknowledgments
This study was supported by National Major Science & Technology Major Project (2018ZX10302302-001-002, 2018ZX10101004-001-003), Strategic Priority Research Program of Chinese Academy of Sciences (CAS) (XDB29040201), and the Chair Professor Grant (CPG) 2019-00019-FHS. We thank the staff of BL17U and BL19U beamline at the Shanghai Synchrotron Radiation Facility for assistance with data collection. We also thank Yuanyuan Chen, Junying Jia, Bingxue Zhou, and Zhenwei Yang from the Institute of Biophysics, CAS, for their technical support in the ITC and FACS assay.
Author contributions
M.S. and M.J. performed experiments; S.T., M.S., and G.F.G. analyzed the data and wrote the paper; M.S., S.T., and G.F.G. designed the experiments; Y.C. and J.Q. solved the structure; H.Z. and M.J. analyzed the data.
Declaration of interests
The authors declare no competing interests.
Published: October 22, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103190.
Contributor Information
George F. Gao, Email: gaof@im.ac.cn.
Shuguang Tan, Email: tansg@im.ac.cn.
Supplemental information
Data and code availability
All the atomic coordinates and diffraction data reported in this paper have been uploaded to Protein Data Bank (PDB) with the following accession codes: PDB: 7E5M (crystal structure of TROP-2-ECD trans-dimer) and PDB: 7E5N (crystal structure of TROP-2-ECD cis-tetramer). This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Struct. Biol. 2010;66:213–221. doi: 10.1107/s0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeuerle P.A., Gires O. EpCAM (CD326) finding its role in cancer (vol 96, pg 417, 2007) Br. J. Cancer. 2007;96:1491. doi: 10.1038/sj.bjc.6603758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardia A., Mayer I.A., Diamond J.R., Moroose R.L., Isakoff S.J., Starodub A.N., Shah N.C., O'Shaughnessy J., Kalinsky K., Guarino M. Efficacy and safety of anti-Trop-2 antibody drug conjugate sacituzumab govitecan (IMMU-132) in heavily pretreated patients with metastatic triple-negative breast cancer. J. Clin. Oncol. 2017;2017:2141–2148. doi: 10.1200/JCO.2016.70.8297.2017.2.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.-H., Wang Y., Li R., Rossi D.L., Liu D., Rossi E.A., Cardillo T.M., Goldenberg D.M. Combination therapy with bispecific antibodies and PD-1 blockade enhances the antitumor potency of t cells. Cancer Res. 2017;77:5384–5394. doi: 10.1158/0008-5472.Can-16-3431. [DOI] [PubMed] [Google Scholar]
- Chen V.B., Arendall W.B., Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D-struct. Biol. 2010;66:12–21. doi: 10.1107/s0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project The CCP4 Suite: programs for protein crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D Struct. Biol. 2004;60:2126–2132. doi: 10.1107/s0907444904019158. [DOI] [PubMed] [Google Scholar]
- Eyvazil S., Farajnia S., Dastmalchi S., Kanipour F., Zarredar H., Bandehpour M. Antibody based EpCAM targeted therapy of cancer, review and update. Curr. Cancer Drug Targets. 2018;18:857–868. doi: 10.2174/1568009618666180102102311. [DOI] [PubMed] [Google Scholar]
- Fenn K.M., Kalinsky K. Sacituzumab govitecan: antibody-drug conjugate in triple-negative breast cancer and other solid tumors. Drugs Today. 2019;55:575–585. doi: 10.1358/dot.2018.55.9.3039669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong D., Moser P., Krammel C., Gostner J.M., Margreiter R., Mitterer M., Gastl G., Spizzo G. High expression of TROP2 correlates with poor prognosis in pancreatic cancer. Br. J. Cancer. 2008;99:1290–1295. doi: 10.1038/sj.bjc.6604677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Jing Y., Gao J., Li Z., Wang H., Cai M., Tong T. Variation of Trop2 on non-small-cell lung cancer and normal cell membranes revealed by super-resolution fluorescence imaging. Talanta. 2020;207:120312. doi: 10.1016/j.talanta.2019.120312. [DOI] [PubMed] [Google Scholar]
- Gaber A., Kim S.J., Kaake R.M., Benčina M., Krogan N., Šali A., Pavšič M., Lenarčič B. EpCAM homo-oligomerization is not the basis for its role in cell-cell adhesion. Sci. Rep. 2018;8:13269. doi: 10.1038/s41598-018-31482-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldenberg D.M., Stein R., Sharkey R.M. The emergence of trophoblast cell-surface antigen 2 (TROP-2) as a novel cancer target. Oncotarget. 2018;9:28989–29006. doi: 10.18632/oncotarget.25615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C., Perrone E., Zeybek B., Bellone S., Tymon-Rosario J., Altwerger G., Menderes G., Feinberg J., Haines K., Karger M.E.M. In vitro and in vivo activity of sacituzumab govitecan, an antibody-drug conjugate targeting trophoblast cell-surface antigen 2 (Trop-2) in uterine serous carcinoma. Gynecol. Oncol. 2020;156:430–438. doi: 10.1016/j.ygyno.2019.11.018. [DOI] [PubMed] [Google Scholar]
- Heist R.S., Guarino M.J., Masters G., Purcell W.T., Starodub A.N., Horn L., Scheff R.J., Bardia A., Messersmith W.A., Berlin J. Therapy of advanced non-small-cell lung cancer with an SN-38-Anti-Trop-2 drug conjugate, Sacituzumab Govitecan. J. Clin. Oncol. 2017;35:2790–2797. doi: 10.1200/jco.2016.72.1894. [DOI] [PubMed] [Google Scholar]
- Kamble P.R., Rane S., Breed A.A., Joseph S., Mahale S.D., Pathak B.R. Proteolytic cleavage of Trop2 at Arg87 is mediated by matriptase and regulated by Val194. FEBS Lett. 2020;594:3156–3169. doi: 10.1002/1873-3468.13899. [DOI] [PubMed] [Google Scholar]
- Linnenbach A.J., Wojcierowski J., Wu S.A., Pyrc J.J., Ross A.H., Dietzschold B., Speicher D., Koprowski H. Sequence investigation of the major gastrointestinal tumor-associated antigen gene family, GA733. Proc. Natl. Acad. Sci. U S A. 1989;86:27–31. doi: 10.1073/pnas.86.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski M., Parks D.R., Rouse R.V., Herzenberg L.A. Human trophoblast cell-surface antigens defined by monoclonal-antibodies. Proc. Natl. Acad. Sci. U S A. 1981;78:5147–5150. doi: 10.1073/pnas.78.8.5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Liu Y.Y., Bao X.X., Tian J.G., Liu Y., Yang X.S. Overexpression of TROP2 predicts poor prognosis of patients with cervical cancer and promotes the proliferation and invasion of cervical cancer cells by regulating ERK signaling pathway. PLoS ONE. 2013;8:e75864. doi: 10.1371/journal.pone.0075864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihelic M., Turk D. Two decades of thyroglobulin type-1 domain research. Biol. Chem. 2007;388:1123–1130. doi: 10.1515/bc.2007.155. [DOI] [PubMed] [Google Scholar]
- Nakatsukasa M., Kawasaki S., Yamasaki K., Fukuoka H., Matsuda A., Tsujikawa M., Tanioka H., Nagata-Takaoka M., Hamuro J., Kinoshita S. Tumor-associated calcium signal transducer 2 is required for the proper subcellular localization of claudin 1 and 7 implications in the pathogenesis of gelatinous drop-like corneal dystrophy. Am. J. Pathol. 2010;177:1344–1355. doi: 10.2353/ajpath.2010.100149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning S.L., Liang N., Liu B., Chen X., Pang Q., Xin T. TROP2 expression and its correlation with tumor proliferation and angiogenesis in human gliomas. Neurol. Sci. 2013;34:1745–1750. doi: 10.1007/s10072-013-1326-8. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/s0076-6879(97)76066-x. [DOI] [PubMed] [Google Scholar]
- Pavšič M., Gunčar G., Djinović-Carugo K., Lenarčič B. Crystal structure and its bearing towards an understanding of key biological functions of EpCAM. Nat. Commun. 2014;5:4764. doi: 10.1038/ncomms5764. [DOI] [PubMed] [Google Scholar]
- Pavšič M., Ilc G., Vidmar T., Plavec J., Lenarčič B. The cytosolic tail of the tumor marker protein Trop2--a structural switch triggered by phosphorylation. Sci. Rep. 2015;5:10324. doi: 10.1038/srep10324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J.H., Ou Q.J., Deng Y.X., Xiao B.Y., Zhang L., Li J.B., Li Y., Wan D.S., Lu Z.H., Fang Y.J. TROP2 overexpression in colorectal liver oligometastases is associated with poor prognosis after liver resection. Ther. Adv. Med. Oncol. 2019;11:1–13. doi: 10.1177/1758835919897543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read R.J. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr. Sect. D Struct. Biol. 2001;57:1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- Ruf P., Kluge M., Jager M., Burges A., Volovat C., Heiss M.M., Hess J., Wimberger P., Brandt B., Lindhofer H. Pharmacokinetics, immunogenicity and bioactivity of the therapeutic antibody catumaxomab intraperitoneally administered to cancer patients. Br. J. Clin. Pharmacol. 2010;69:617–625. doi: 10.1111/j.1365-2125.2010.03635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M., Stefan N., Pluckthun A., Zangemeister-Wittke U. Epithelial cell adhesion molecule-targeted drug delivery for cancer therapy. Expert Opin. Drug Deliv. 2013;10:451–468. doi: 10.1517/17425247.2013.759938. [DOI] [PubMed] [Google Scholar]
- Stengel K.F., Harden-Bowles K., Yu X., Rouge L., Yin J., Comps-Agrar L., Wiesmann C., Bazan J.F., Eaton D.L., Grogan J.L. Structure of TIGIT immunoreceptor bound to poliovirus receptor reveals a cell-cell adhesion and signaling mechanism that requires cis-trans receptor clustering. Proc. Natl. Acad. Sci. U S A. 2012;109:5399–5404. doi: 10.1073/pnas.1120606109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trebak M., Begg G.E., Chong J.M., Kanazireva E.V., Herlyn D., Speicher D.W. Oligomeric state of the colon carcinoma-associated glycoprotein GA733-2 (Ep-CAM/EGP40) and its role in GA733-mediated homotypic cell-cell adhesion. J. Biol. Chem. 2001;276:2299–2309. doi: 10.1074/jbc.M004770200. [DOI] [PubMed] [Google Scholar]
- Trerotola M., Guerra E., Ali Z., Aloisi A.L., Ceci M., Simeone P., Acciarito A., Zanna P., Vacca G., D'Amore A. Trop-2 cleavage by ADAM10 is an activator switch for cancer growth and metastasis. Neoplasia. 2021;23:415–428. doi: 10.1016/j.neo.2021.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trerotola M., Jernigan D.L., Liu Q., Siddiqui J., Fatatis A., Languino L.R. Trop-2 promotes prostate cancer metastasis by modulating beta(1) integrin functions. Cancer Res. 2013;73:3155–3167. doi: 10.1158/0008-5472.Can-12-3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varughese J., Cocco E., Bellone S., Bellone M., Todeschini P., Carrara L., Schwartz P.E., Rutherford T.J., Pecorelli S., Santin A.D. High-grade, chemotherapy-resistant primary ovarian carcinoma cell lines overexpress human trophoblast cell-surface marker (Trop-2) and are highly sensitive to immunotherapy with hRS7, a humanized monoclonal anti-Trop-2 antibody. Gynecol. Oncol. 2011;122:171–177. doi: 10.1016/j.ygyno.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidmar T., Pavsic M., Lenarcic B. Biochemical and preliminary X-ray characterization of the tumor-associated calcium signal transducer 2 (Trop2) ectodomain. Prot. Exp. Pur. 2013;91:69–76. doi: 10.1016/j.pep.2013.07.006. [DOI] [PubMed] [Google Scholar]
- Wen K.C., Sung P.L., Chou Y.T., Pan C.M., Wang P.H., Lee O.K.S., Wu C.W. The role of EpCAM in tumor progression and the clinical prognosis of endometrial carcinoma. Gynecol. Oncol. 2018;148:383–392. doi: 10.1016/j.ygyno.2017.11.033. [DOI] [PubMed] [Google Scholar]
- Wu C.-J., Feng X., Lu M., Morimura S., Udey M.C. Matriptase-mediated cleavage of EpCAM destabilizes claudins and dysregulates intestinal epithelial homeostasis. J. Clin. Invest. 2017;127:623–634. doi: 10.1172/JCI88428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng P., Chen M.-B., Zhou L.-N., Tang M., Liu C.-Y., Lu P.-H. Impact of TROP2 expression on prognosis in solid tumors: a systematic review and meta-analysis. Sci. Rep. 2016;6:33658. doi: 10.1038/srep33658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Jia L., Zhang M., Huang X., Qian P., Tang Q., Zhu J., Feng Z. The killing effect of novel bi-specific Trop2/PD-L1 CAR-T cell targeted gastric cancer. Am. J. Cancer Res. 2019;9:1846–1856. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the atomic coordinates and diffraction data reported in this paper have been uploaded to Protein Data Bank (PDB) with the following accession codes: PDB: 7E5M (crystal structure of TROP-2-ECD trans-dimer) and PDB: 7E5N (crystal structure of TROP-2-ECD cis-tetramer). This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.