Abstract
The koala biovar of Chlamydia pneumoniae was identified in lung tissue from a sick, free-ranging giant barred frog (Mixophyes iteratus) by using electron microscopy, C. pneumoniae-specific fluorescent-antibody staining, cell culture, and sequencing of the ompA, ompB and 16S rRNA genes. This is the first report of a chlamydial strain infecting both a homeotherm and a poikilotherm and only the fourth host (in addition to humans, koalas, and horses) to be naturally infected with this species of Chlamydia. The frog had severe, chronic, mononuclear pneumonia and nonregenerative anemia and pancytopenia.
Chlamydia species are important pathogens infecting birds, humans, and a wide variety of other mammals (6, 18). Until 1988, there were only two species recognized in the genus, Chlamydia trachomatis and Chlamydia psittaci. In 1988, members of the TWAR group, formerly C. psittaci, were assigned to the new species Chlamydia pneumoniae (2), and then in 1992, the ruminant strains of C. psittaci were assigned to the fourth chlamydial species, Chlamydia pecorum (4). Members of the species C. pneumoniae are recognized as important human pathogens causing respiratory infections which may be asymptomatic or result in sinusitis, bronchitis, or pneumonia (7). In addition, they have recently been linked to coronary heart disease (1, 20). Apart from humans, the only other hosts naturally infected by C. pneumoniae are koalas (6, 24) and horses (based on a single report [22]).
Chlamydia infections have been reported previously in captive amphibians, causing moderate to high mortality rates in various species, including African clawed frogs (Xenopus laevis) in the United States (10, 17, 26), eyelash leaf frogs (Ceratobatrachus guentheri) in Canada (9), and Bufo maculatum and a Pachytriton sp. in Germany (16). There have also been a few case reports of Chlamydia infections in captive and wild reptiles (8, 12, 13). In all cases, however, the chlamydial species was either unknown or assumed to be C. psittaci. Here we report the first isolation of Chlamydia from a frog in Australia and demonstrate that it is identical to the C. pneumoniae strain that infects koalas.
A male, 60-g, giant barred frog (Mixophyes iteratus) with a snout-to-vent length of 7.8 cm, free-ranging in the Orara East State forest, New South Wales, was observed to be behaving abnormally, i.e., sitting unprotected during the day and in a lethargic state. At the laboratory the next day, the frog was found to be in poor nutritional condition and moribund and died soon after arrival. Hematological analysis of heparinized blood collected just before death revealed nonregenerative anemia (packed cell volume [PCV] = 18%) and a low total protein level (15 g/liter). The aspartate aminotransferase level was greatly increased, to 6,080 U/liter. The total white cell count was very low (0.36 × 109/liter), and examination of a Giemsa-stained blood smear showed that 85% of the leukocytes were large lymphocytes or monocytes that were difficult to differentiate. No neutrophils were detected.
During postmortem, organ samples were collected into 10% buffered neutral formalin, dehydrated, and embedded in paraffin wax or were frozen at −80°C for bacterial and chlamydial culture. Upon gross examination, the lungs were thickened and did not fully collapse after sectioning, and the spleen appeared small. Six-micrometer-thick histological sections were cut and stained with hematoxylin and eosin (H&E), Gram stain, or Perl’s stain (3). On histological examination, the spleen and bone marrow were found to be depleted of hemopoietic cells, although no cause of the immunosuppression was apparent. Severe chronic mononuclear pneumonia with marked thickening of the septae due to the inflammation was present, and the airspaces contained masses of free monocytes, lymphocytes, erythrocytes, and plasma cells (Fig. 1). Many mononuclear cells had swollen cytoplasms with fine basophilic stippling (Fig. 2). A large proportion of renal tubular epithelial cells contained round intracytoplasmic deposits of light brown pigment which did not stain for iron with Perl’s stain.
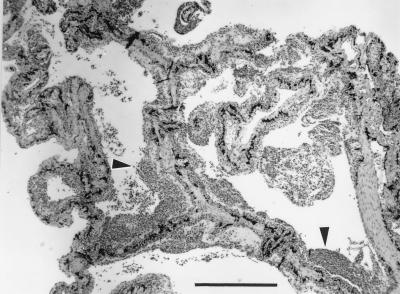
FIG. 1.
Histological section of lung tissue from a giant barred frog with severe, chronic, mononuclear pneumonia. The septae, which are normally covered by a thin epithelium, are here markedly thickened by a layer of mononuclear inflammation (arrowheads). H&E stain. Bar, 500 μm.
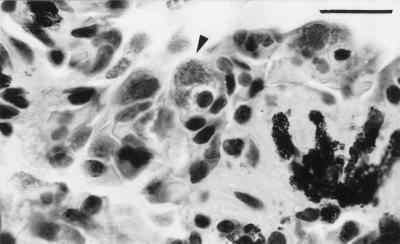
FIG. 2.
Histological section of lung tissue with an infected mononuclear cell (arrowhead). It is important to note the swollen cytoplasm containing chlamydial organisms, visible as fine stippling. H&E stain. Bar, 20 μm.
For electron microscopy, formalin-fixed lung tissue was postfixed in 2.5% glutaraldehyde and then 1% osmium tetroxide, dehydrated, embedded in Spurr’s resin, sectioned at 70 nm, stained with lead citrate and uranyl acetate, and examined with a Hitachi 7000 transmission electron microscope. Typical chlamydial particles were frequently observed within membrane-bound inclusions in the cytoplasm of mononuclear inflammatory cells (Fig. 3) and also occasionally in alveolar epithelial cells. The chlamydial particles included dense elementary bodies (314 ± 39 nm in diameter [mean ± standard deviation; n = 12]), intermediate bodies (371 ± 44 nm [n = 12]), and numerous dividing reticulate bodies (537 ± 130 nm [n = 12]) (Fig. 4). The round elementary bodies had eccentric nuclei and a narrow or nonexistent periplasmic space. Mitochondria were not associated with the inclusion membrane. These ultrastructural characteristics are consistent with those of C. pneumoniae (14).
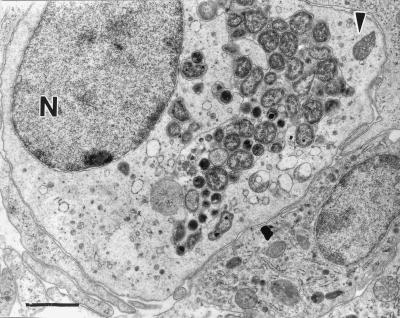
FIG. 3.
Transmission electron micrograph of infected mononuclear cell with chlamydial particles at various stages present within a membrane-bound cytoplasmic inclusion. N, nucleus. The arrowhead indicates a mitochondrion. Bar, 1,500 nm.
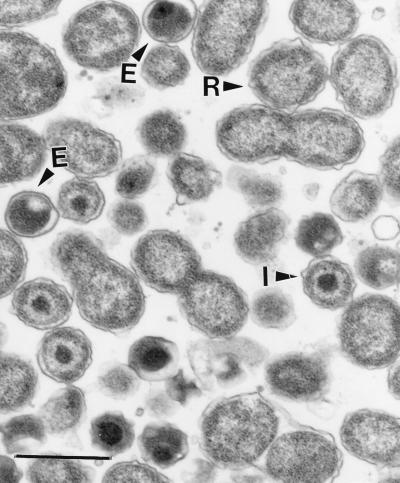
FIG. 4.
Transmission electron micrograph of chlamydial particles at various stages in lung tissue. It is important to note the large reticulate bodies (R), which undergo binary fission, intermediate bodies (I), and condensed elementary bodies (E). Bar, 700 nm.
Cultures of bacteria from frozen lung, liver, and kidney samples were attempted on horse blood agar and MacConkey agar incubated at 37°C. A heavy growth of Flavobacterium species was obtained from lung sample cultures, and a much lighter growth was observed on cultures of bacteria from kidney and liver samples.
The presence of a Chlamydia sp. in lung tissue was initially confirmed by a strong reaction with the genus-specific Clearview lipopolysaccharide (LPS) antigen detection assay (Oxoid). The chlamydial species was identified as C. pneumoniae by positive staining of an impression smear with the Cellabs (Sydney, Australia) Chlamydia-TWAR fluorescent monoclonal antibody. C. pneumoniae was subsequently cultured from the frozen lung tissue in HEp-2 cells by using the polyethylene glycol pretreatment method (23). Infection levels were observed to be high in this cell line (with more than 75% of cells containing inclusions) when stained with either genus-specific (Cellabs Chlamydia-LPS monoclonal) or C. pneumoniae species-specific (Cellabs Chlamydia-TWAR monoclonal) antibodies.
The genus, species, and biovar designations of the frog isolate were confirmed by sequence analysis of the chlamydial 16S rRNA gene (235-bp fragment, positions 566 to 801) (19), the ompB gene (392-bp fragment) (25), and the ompA gene (279-bp fragment, variable domain 4 region) (24). The 16S rRNA gene sequence of the frog isolate was identical to both the horse N16 and koala strain sequences but was different by one nucleotide from the sequence of human strain TW-183. ompB sequence analysis indicated that the frog isolate was identical to the koala biovar of C. pneumoniae but its sequence was different by two nucleotides from that of the horse N16 isolate and by four nucleotides from that of human strain TW-183. ompA variable domain 4 sequence analysis indicated that the frog isolate was again identical to the koala biovar of C. pneumoniae but its sequence differed by 25 nucleotides from that of the horse N16 isolate and by 5 nucleotides from that of human strain TW-183.
This case report is significant not only for our understanding of frog diseases but also for expanding the range of hosts infected with C. pneumoniae. We suspect that the Chlamydia infection in the frog in this study was an opportunistic infection, secondary to immunosuppression of unknown cause. Previous reports of outbreaks of Chlamydia infection in captive amphibians have involved a number of animals, usually with fulminant, multisystemic infections, which is in contrast to the chronic pathology present in the free-ranging giant barred frog of the present study, where lesions were confined to the lung. C. pneumoniae in the other hosts that it infects, namely, humans, horses, and koalas, also usually causes respiratory disease. This might suggest that this species has a tropism for epithelial cells lining the respiratory tract. The human strains of C. pneumoniae have also recently been shown to be able to infect and grow in both peripheral blood and alveolar macrophages (15) as well as vascular endothelium and arterial smooth muscle cells (1).
The finding that the sequence of this frog isolate is identical to that of the koala biovar of C. pneumoniae raises interesting questions. We are confident that the gene analysis is reliable and does not represent laboratory cross-contamination. At this stage, however, we are unsure of the source of infection, although the Orara East State forest where the frog was found contains a significant population of koalas and reports indicate that C. pneumoniae infection is common in most Australian koala populations (11, 24). Mixophyes iteratus is a ground-dwelling fossorial frog with opportunity to come into indirect contact with koalas, who regularly walk on the ground to move between food trees. Despite recent investigations into diseases of wild amphibians in Australia (21), Chlamydia strains have not been identified previously in native frogs or introduced cane toads, although specific chlamydial tests for asymptomatic carriers were not done. Several studies have reported the very high genetic similarity of human C. pneumoniae strains (5) and also the clonality of koala C. pneumoniae strains (24), suggesting a possible recent divergence of these biovars. It is possible that the infection of a wild frog described in this report was an isolated incident, or alternatively, increased testing may show that amphibians are commonly infected with C. pneumoniae and that they are a natural reservoir for this species.
Nucleotide sequence accession numbers.
The ompA variable domain 4, ompB, and 16S rRNA gene sequences have been deposited in the GenBank database under accession no. AF102830, AF102831, and AF102832, respectively.
Acknowledgments
We thank Julia Hammond and Trevor Taylor for assistance with bacteriology, Michael Mahony for collecting the frog, Bruce Parry for performing the hematology, Megan Braun and Terry Wise for cutting sections, and Alex Hyatt and Peter Hooper for advice.
Lee Berger was funded by Environment Australia.
REFERENCES
- 1.Campbell L A, Kuo C-C, Grayston J T. Chlamydia pneumoniae and cardiovascular disease. Emerg Infect Dis. 1998;4:571–579. doi: 10.3201/eid0404.980407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cox R, Kuo C-C, Grayston T, Campbell L A. Deoxyribonucleic acid relatedness of Chlamydia sp. strain TWAR to Chlamydia trachomatis and Chlamydia psittaci. Int J Syst Bacteriol. 1988;38:265–268. [Google Scholar]
- 3.Drury R A, Wallington E A. Carleton’s histological technique. Oxford, United Kingdom: Oxford University Press; 1980. p. 264. [Google Scholar]
- 4.Fukushi H, Hirarai K. Proposal of Chlamydia pecorum sp. nov. for Chlamydia strains derived from ruminants. Int J Syst Bacteriol. 1992;42:306–308. doi: 10.1099/00207713-42-2-306. [DOI] [PubMed] [Google Scholar]
- 5.Gaydos C A, Quinn T C, Bobo L D, Eiden J J. Similarity of Chlamydia pneumoniae strains in the variable domain IV region of the major outer membrane protein gene. Infect Immun. 1992;60:5319–5323. doi: 10.1128/iai.60.12.5319-5323.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glassick T, Giffard P, Timms P. Outer membrane protein 2 gene sequences indicate that Chlamydia pecorum and Chlamydia pneumoniae cause infections in koalas. Syst Appl Microbiol. 1996;19:457–464. [Google Scholar]
- 7.Grayston J T, Campbell L E, Kuo C, Mordhorst C H, Saikku P, Thom D H, Wang S. A new respiratory tract pathogen: Chlamydia pneumoniae strain TWAR. J Infect Dis. 1990;161:618–625. doi: 10.1093/infdis/161.4.618. [DOI] [PubMed] [Google Scholar]
- 8.Homer B L, Jacobson E R, Schumacher J, Scherba G. Chlamydiosis in mariculture-reared green sea turtles (Chelonia mydas) Vet Pathol. 1994;31:1–7. doi: 10.1177/030098589403100101. [DOI] [PubMed] [Google Scholar]
- 9.Honeyman V L, Mehran K G, Barker I K, Crawshaw G J. Proceedings of the Joint Meeting of the American Association of Zoo Veterinarians and the American Association of Wildlife Veterinarians. 1992. Bordetella septicaemia and chlamydiosis in eyelash leaf frogs (Ceratobatrachus guentheri) p. 168. [Google Scholar]
- 10.Howerth E W. Pathology of naturally occurring chlamydiosis in African clawed frogs (Xenopus laevis) Vet Pathol. 1984;21:28–32. doi: 10.1177/030098588402100105. [DOI] [PubMed] [Google Scholar]
- 11.Jackson M, White N, Giffard P, Timms P. Epizootiology of Chlamydia infections in two free-range koala populations. Vet Microbiol. 1999;65:255–264. doi: 10.1016/s0378-1135(98)00302-2. [DOI] [PubMed] [Google Scholar]
- 12.Jacobsen E R, Gaskin J M, Mansell J. Chlamydial infection in puff adders (Bitis arietans) J Zoo Wildl Med. 1989;20:364–369. [Google Scholar]
- 13.Jacobsen E R, Telford S R. Chlamydial and poxvirus infections of circulating monocytes of a flap necked chameleon (Chamaeleo dilepis) J Wildl Dis. 1990;26:572–577. doi: 10.7589/0090-3558-26.4.572. [DOI] [PubMed] [Google Scholar]
- 14.Miyashita N, Kanamoto Y, Matsumoto A. The morphology of Chlamydia pneumoniae. J Med Microbiol. 1993;38:418–425. doi: 10.1099/00222615-38-6-418. [DOI] [PubMed] [Google Scholar]
- 15.Moazed T, Kuo C-C, Grayston T, Campbell L A. Evidence of systemic dissemination of Chlamydia pneumoniae via macrophages in the mouse. J Infect Dis. 1998;177:1322–1325. doi: 10.1086/515280. [DOI] [PubMed] [Google Scholar]
- 16.Mutschmann C, Tierarztpraxis F. Detection of Chlamydia psitacci in amphibians using an immunofluorescence test (IFT) Berl Muench Tieraerztl Wochenschr. 1998;111:187–189. [PubMed] [Google Scholar]
- 17.Newcomer C E, Anver M R, Simmons J L, Wilcke B W, Nace G W. Spontaneous and experimental infections of Xenopus laevis with Chlamydia psittaci. Lab Anim Sci. 1982;32:680–686. [PubMed] [Google Scholar]
- 18.Peeling R W, Brunham R C. Chlamydiae as pathogens: new species and new issues. Emerg Infect Dis. 1996;2:307–319. doi: 10.3201/eid0204.960406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pettersson B, Anderssonn A, Leitner T, Olsivik Ø, Uhlen M, Storey C, Black C M. Evolutionary relationships among members of the genus Chlamydia based on 16S ribosomal DNA analysis. J Bacteriol. 1997;179:4195–4205. doi: 10.1128/jb.179.13.4195-4205.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saikku P, Mattila K, Nieminen M, Huttunen J, Leinonen M, Ekamn M-R, Makela P, Valtonen V. Serological evidence of an association of a novel Chlamydia TWAR with chronic coronary heart disease and acute myocardial infarction. Lancet. 1988;29:983. doi: 10.1016/s0140-6736(88)90741-6. [DOI] [PubMed] [Google Scholar]
- 21.Speare, R., and L. Berger. Unpublished data.
- 22.Storey C, Lusher M, Yates P, Richmond S. Evidence of Chlamydia pneumoniae of non-human origin. J Gen Microbiol. 1993;139:2621–2626. doi: 10.1099/00221287-139-11-2621. [DOI] [PubMed] [Google Scholar]
- 23.Tjhie J H T, Roosendaal R, MacLaren D M, Vandenbroucke-Grauls C M J E. Improvement of growth of Chlamydia pneumoniae on HEp-2 cells by pretreatment with polyethylene glycol in combination with additional centrifugation and extension of culture time. J Clin Microbiol. 1997;35:1883–1884. doi: 10.1128/jcm.35.7.1883-1884.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wardrop S, Fowler A, O’Callaghan P, Giffard P, Timms P. Characterization of the koala biovar of Chlamydia pneumoniae at four gene loci—ompAVD4, ompB, 16S rRNA, groESL. Syst Appl Microbiol. 1999;22:22–27. doi: 10.1016/S0723-2020(99)80024-1. [DOI] [PubMed] [Google Scholar]
- 25.Watson M W, Lambden P R, Clarke I N. Genetic diversity of human infection by amplification of the chlamydial 60-kilodalton cysteine-rich outer membrane protein gene. J Clin Microbiol. 1991;29:1188–1193. doi: 10.1128/jcm.29.6.1188-1193.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilcke B W, Newcomer C E, Anver M R, Simmons J L, Nace G W. Isolation of Chlamydia psittaci from naturally infected African clawed frogs (Xenopus laevis) Infect Immun. 1983;41:789–794. doi: 10.1128/iai.41.2.789-794.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]