Abstract
Calcium-stimulated nuclear factor of activated T cells (NFAT) transcription activity at the interleukin-2 promoter is negatively regulated by cyclic AMP (cAMP). This effect of cAMP is mediated, in part, by protein kinase A phosphorylation of NFAT. The mechanism of regulation involves the creation of a phosphorylation-dependent binding site for 14-3-3. Decreased NFAT phosphorylation caused by the calcium-stimulated phosphatase calcineurin, or mutation of the PKA phosphorylation sites, disrupted 14-3-3 binding and increased NFAT transcription activity. In contrast, NFAT phosphorylation caused by cAMP increased 14-3-3 binding and reduced NFAT transcription activity. The regulated interaction between NFAT and 14-3-3 provides a mechanism for the integration of calcium and cAMP signaling pathways.
Nuclear factor of activated T cells (NFAT) represents a group of transcription factors that are implicated in the expression of Fas ligand and several cytokine genes, including those encoding interleukin 2 (IL-2), IL-4, and tumor necrosis factor alpha (18, 37, 47, 70). Four members of the NFAT group (NFAT1/NFATC2/NFATp, NFAT2/NFATC1/NFATc, NFAT3, and NFAT4/NFATC3) have been identified, and multiple alternatively spliced isoforms have been described (25, 27, 45, 48, 56). Recent studies have confirmed that NFAT plays a critical role in the expression of IL-2 and IL-4 by T cells (12, 18, 26, 77). However, roles for NFAT in other biological processes have also been reported. For example, NFAT4 is required for normal thymocyte development (57), NFATc participates in heart valve development during embryogenesis (16, 61), NFAT3 interacts with GATA4 and induces heart hypertrophy (50), and NFAT activity controls gene expression in different skeletal muscle fiber types (1, 11).
NFAT is present in the cytoplasm of resting cells (reviewed in references 15 and 62). Upon T-cell activation, a sustained increase in intracellular calcium activates the phosphatase calcineurin (72). Calcineurin binds to a docking site in the NH2-terminal region of NFAT (3), dephosphorylates phosphoserine residues located in the NFAT homology domain, and induces translocation of NFAT from the cytoplasm into the nucleus (14, 29, 68). Recently, several protein kinases that phosphorylate sites located in the NFAT homology domain have been identified; these include glycogen synthase kinase 3, casein kinase 1α, and c-Jun N-terminal kinase (5, 13, 83). The nuclear localization of NFAT is therefore regulated by the opposing actions of protein kinases and the phosphatase calcineurin.
NFAT was identified as the molecular target for cyclosporin A (CsA), an immunosuppressive drug that has allowed important advances in transplantation surgery (20, 66). CsA inhibits calcineurin activity and thus blocks nuclear translocation of NFAT, and consequently IL-2 secretion, by activated T cells (14). In addition, stimuli that activate protein kinase A (PKA), including prostaglandin E2, histamine, epinephrine, and immunosuppressive retroviral peptides, inhibit IL-2 gene expression (22, 31, 75). Conversely, suppression of cyclic AMP (cAMP) signaling is required for T-cell activation (39). The mechanism of inhibition appears to be mediated by the transcriptional induction of phosphodiesterase 7 following T-cell costimulation. Increased phosphodiesterase 7 expression is associated with decreased levels of cAMP and increased IL-2 production (39). Thus, the cAMP signaling pathway represents a physiologically relevant regulatory mechanism that controls IL-2 expression by T cells.
The mechanism by which cAMP regulates IL-2 gene expression has not been clearly defined. It is likely that the effects of cAMP are mediated by activation of PKA since transfection studies demonstrate that the expression of the PKA catalytic subunit suppresses IL-2 promoter activity. Several potential targets of PKA signaling have been identified. For example, cAMP inhibits T-cell receptor tyrosine phosphorylation and down-regulates c-Jun N-terminal kinase activity (28, 58). In addition, several transcription factors, including inducible cyclic AMP early repressor (ICER) (6), NF-κB (54), and NFAT, have been identified as potential mediators of negative regulation by cAMP (reviewed in reference 62). Interestingly, overexpression of NFAT antagonizes the inhibitory effect of PKA on IL-2 gene expression (73). Together, these actions of PKA appear to inhibit IL-2 expression. Nevertheless, the mechanisms that account for the effect of PKA remain unclear.
The purpose of this study was to examine the effect of PKA on negative regulation of NFAT-mediated signal transduction. We show that PKA phosphorylates NFAT and inhibits NFAT transcription activity. Mutational analysis demonstrated that these sites of NFAT phosphorylation by PKA are required for regulation of NFAT transcription activity. NFAT phosphorylation by PKA creates binding sites for 14-3-3. We propose that the PKA-regulated formation of NFAT–14-3-3 complexes may, in part, mediate the effects of cAMP on NFAT transcription activity.
MATERIALS AND METHODS
Cell culture and reagents.
BHK fibroblasts, Jurkat T cells, and COS cells were cultured in minimal essential medium, RPMI 1640, and Dulbecco modified Eagle medium, respectively, supplemented with 10% fetal calf serum, 2 mM l-glutamine, penicillin (100 U/ml), and streptomycin (100 μg/ml) (Life Technologies Inc.). BHK and COS cells were transfected by using Lipofectamine (Life Technologies). Ionomycin, phorbol myristate acetate, dibutyryl cAMP, calcineurin, calmodulin, 4′,6′-diamidino-2-phenylindole, H7, 3-isobutyl-1-methylxanthine (IBMX), and monoclonal antibody M2 were obtained from Sigma. Calcineurin inhibitory peptide and CsA were obtained from Calbiochem.
Expression vectors.
Plasmids used for expression of Raf-1 (76), NFAT (27), calcineurin (60), PKA catalytic subunit (46), and 14-3-3 (7, 41) have been described elsewhere. Wild-type and mutated NFAT constructs were prepared by PCR, sequenced, and cloned in pGEX-3X (Amersham Pharmacia Biotech Inc.) and pCDNA3 (Invitrogen Inc.).
Luciferase reporter gene assays.
An NFAT expression vector (0.3 μg) was cotransfected with an NFAT-luciferase reporter plasmid (0.2 μg) and the control plasmid pRSV β-galactosidase (0.2 μg) (13). The PKA catalytic subunit expression vector plasmid (0.1 μg) was cotransfected as indicated. Luciferase and β-galactosidase activity were measured 48 h after transfection (13). The data are presented as relative luciferase activity, calculated as the ratio of the activity of luciferase to the activity of β-galactosidase (mean ± standard deviation [n = 3]).
Protein interaction assays.
Cell extracts were prepared with Triton lysis buffer (20 mM Tris [pH 7.4], 137 mM NaCl, 2 mM EDTA, 1% Triton X-100, 25 mM β-glycerophosphate, 1 mM sodium vanadate, 2 mM sodium pyrophosphate, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride, 10 μg of leupeptin per ml). COS cells were harvested 48 h after transfection. Jurkat cells (2 × 107) were treated (20 min) without or with 2 μM ionomycin or with 500 μM dibutyryl cAMP plus 50 μM IBMX or 100 ng of CsA per ml. The cells were harvested in 400 μl of two-times-concentrated Triton lysis buffer and centrifuged, and the lysate was diluted by adding 400 μl of ice-cold water. Immunoprecipitation was performed with either monoclonal antibody M2 or an NFATp antibody (Upstate Biotechnology Inc.). 14-3-3 in the immunoprecipitates was detected by immunoblot analysis using a pan-14-3-3 antibody (SC-629; Santa Cruz Inc.).
Binding assays were performed by incubating cell extracts in Triton lysis buffer with 5 μg of recombinant glutathione S-transferase (GST)–14-3-3 fusion protein bound to 20 μl of glutathione-Sepharose beads (Amersham Pharmacia Biotech Inc.) for 5 h at 4°C. After three washes with Triton lysis buffer, the bound proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and electrotransferred to a polyvinylidene membrane (Millipore). Immunoblot analysis was performed with the anti-NFATp antibody 67.1 (23) or an antibody to Raf-1 (76) and visualized by enhanced chemiluminescence (Kirkegaard & Perry Laboratories).
In vitro translation.
T7-coupled in vitro transcription-translation (Promega Inc.) was performed in the presence of [35S]methionine (New England Nuclear Inc.). Translated NFAT was used in binding assays, and bound NFAT was visualized by autoradiography and quantitated by PhosphorImager (Molecular Dynamics Inc.) analysis. Dephosphorylation of NFAT was performed in calcineurin reaction buffer (50 mM HEPES [pH 7.4], 2 mM MnCl2, 0.5 mM EDTA, 15 mM 2-mercaptoethanol, 0.1 mg of bovine serum albumin per ml) in the presence of purified calmodulin (250 U) and calcineurin (2.5 and 5 U). Calcineurin inhibitory peptide (10 μg) was preincubated with calcineurin for 10 min at 4°C before incubation with in vitro-translated NFAT protein.
Protein kinase assays.
Phosphorylation of NFAT (1 μg) by purified PKA catalytic subunit (4 U) was performed in kinase reaction buffer (25 mM HEPES [pH 7.4], 25 mM β-glycerophosphate, 25 mM MgCl2, 2 mM dithiothreitol, 0.1 mM sodium vanadate) in the presence of 50 μM [γ-32P]ATP.
Phosphopeptide analysis.
COS cells were transfected with a Flag epitope-tagged NFAT expression vector (6 μg) without and with a PKA expression vector (2 μg) and incubated for 24 h. The transfected cells were then incubated with [32P]phosphate (1 mCi/ml) for 5 h. The NFAT proteins were isolated by immunoprecipitation with anti-Flag monoclonal antibody M2. Immunoprecipitates were separated by SDS-PAGE, electrotransferred to a polyvinylidene difluoride membrane (Millipore), and visualized by autoradiography. The band containing 32P-labeled NFAT was excised from the membrane and digested with trypsin, and the peptides obtained were examined by phosphopeptide mapping (8). Phosphopeptide maps of NFAT phosphorylated in vivo were compared with maps of NFAT phosphorylated by PKA in vitro.
Immunofluorescence analysis.
BHK cells were transfected by using Lipofectamine (Gibco-BRL) and an expression plasmid for Flag-tagged NFAT3 (0.3 μg), PKA (0.1 μg), or calcineurin (0.2 μg). NFAT was detected by immunofluorescence analysis with anti-Flag monoclonal antibody M2 (13). The secondary antibody was Texas red-conjugated anti-mouse immunoglobulin antibody (1:100; Jackson Immunoresearch), and nuclei were visualized with 4′,6′-diamidino-2-phenylindole.
RESULTS
Phosphorylation by PKA inhibits NFAT transcription activity.
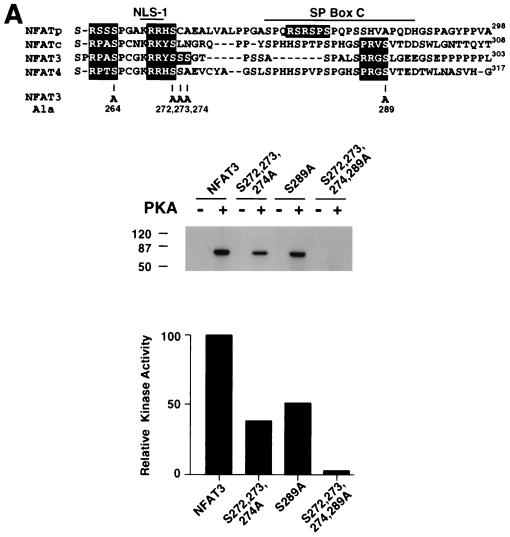
NFAT is implicated as an effector molecule that mediates the negative regulatory action of PKA on IL-2 gene expression. We tested whether NFAT is a substrate for PKA (Fig. 1A). In vitro protein kinase assays using purified PKA catalytic subunit demonstrated that NFAT3 is phosphorylated by PKA (Fig. 1A). Deletion analysis identified the NH2-terminal region of NFAT3 as a substrate for PKA. Sequence analysis indicated that there are several potential PKA phosphorylation sites in this region: Ser-264, Ser-272, Ser-273, and Ser-289 (Fig. 1A). These Ser residues are conserved in other members of the NFAT group (Fig. 1A). We performed site-directed mutagenesis to replace these potential phosphorylation sites with Ala, creating [Ala272,273,274] NFAT. Mutation at Ser-272, Ser-273, and Ser-274 reduced PKA phosphorylation of NFAT3 (Fig. 1A). A similar reduction in PKA phosphorylation was caused by mutation at Ser-289 (Fig. 1A). In contrast, replacement of Ser-272, Ser-273, Ser-274, and Ser-289 with Ala eliminated phosphorylation of NFAT3 by PKA (Fig. 1A). This observation indicated that Ser-264 was not phosphorylated by PKA. Control experiments demonstrated that phosphorylation of CREB by PKA in the same assay was not affected by these NFAT3 mutations (data not shown). Together, these data demonstrate that the PKA sites on NFAT3 are phosphorylated in vitro.
FIG. 1.
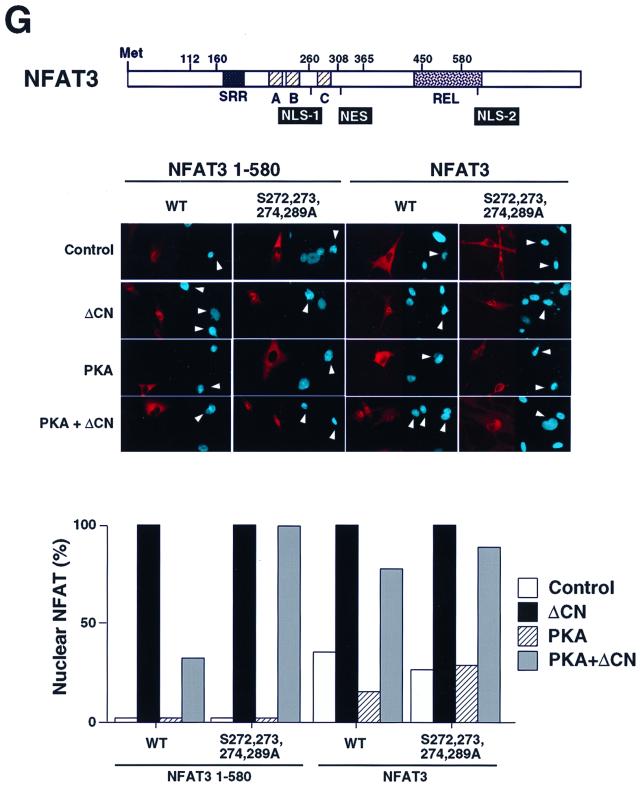
Phosphorylation by PKA inhibits NFAT transcription activity. (A) Mutational analysis of NFAT3 phosphorylation by PKA. The primary sequence of the NFAT homology domain of NFAT3 is compared with sequences of NFATp (NFAT1), NFATc (NFAT2), and NFAT4 (upper panel). Potential PKA phosphorylation sites are highlighted with filled boxes. NLS-1 and the conserved SP box C are indicated. Recombinant NFAT3 (1 μg) was phosphorylated by purified PKA catalytic subunit in the presence of [γ-32P]ATP (middle panel; sizes are indicated in kilodaltons). Phosphorylated NFAT3 was detected by autoradiography and quantitated by PhosphorImager analysis (lower panel). (B) Replacement of Ser272, Ser273, Ser274, and Ser289 with Ala increases the electrophoretic mobility of NFAT3 during SDS-PAGE. Epitope-tagged wild-type and mutated [Ala272,273,274,289] NFAT3 were expressed in COS cells without (Control) and with activated calcineurin (ΔCN). NFAT3 proteins were detected in immunoblot analysis by using monoclonal antibody M2. (C) NFAT3 is phosphorylated by PKA in vivo. Epitope-tagged wild-type and mutated [Ala272,273,274,289] NFAT3 were expressed in COS cells without (Control) and with PKA. The cells were labeled with [32P]phosphate, and the NFAT3 proteins were isolated by immunoprecipitation. The phosphorylation of NFAT3 was examined by tryptic phosphopeptide mapping. (D) PKA phosphorylates NFAT3 on Ser272 and Ser289 in vivo and in vitro. Recombinant NFAT3 (1 μg) was phosphorylated by purified PKA in the presence of [γ-32P]ATP (upper left panel). The effect of replacement of Ser272 and Ser289 with Ala was examined. The in vitro phosphorylated wild-type NFAT3 was examined by tryptic phosphopeptide mapping (upper right panel). The phosphorylation of NFAT3 in vivo was also examined by phosphopeptide mapping of NFAT3 co-expressed with PKA in COS cells (middle and lower panels). The effect of replacement of Ser272, Ser273, Ser274 or Ser289 with Ala was examined. Electrophoresis and chromatography are the horizontal and vertical dimensions of the phosphopeptide maps, respectively. (E) Measurement of NFAT3 transcription activity in BHK cells. The effect of NFAT3 expression was examined in cotransfection assays using an NFAT-luciferase reporter plasmid. The cells were treated without (Untreated) and with ionomycin (2 μM) and PMA (100 nM) (I+P) for 16 h. The data are presented as relative luciferase activity (see Materials and Methods). (F) Mutation of the PKA phosphorylation sites blocks PKA-mediated inhibition of NFAT3 transcription activity. Wild-type and mutated [Ala272,273,274,289] NFAT3 were expressed together with an NFAT-luciferase reporter plasmid in BHK cells. The effect of treatment with dibutyryl cAMP (500 μM) and IBMX (50 μM) or coexpression without (Control) and with PKA on cells treated (24 h) with ionomycin (2 μM) and PMA (100 nM) is presented. (G) Effect of PKA phosphorylation on the nuclear accumulation of NFAT3. The structure of NFAT3 is illustrated schematically to show the serine-rich region (SRR), NLS-1 and NLS-2, NES, the conserved SP boxes (A, B, and C), and the Rel domain. Epitope-tagged NFAT3 proteins were expressed in BHK cells and detected by immunofluorescence microscopy using monoclonal antibody M2. The subcellular distribution of wild-type (WT) and truncated NFAT3 (residues 1 to 580) was examined. The effect of mutation of the PKA phosphorylation sites and coexpression with PKA catalytic subunit or ΔCn is presented. The percentage of cells with NFAT3 in nucleus (n = 100) is presented, and images of representative cells are illustrated. The nuclei (blue) of transfected cells expressing NFAT3 (red) are indicated with arrowheads.
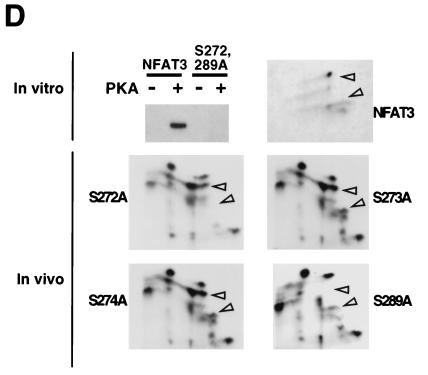
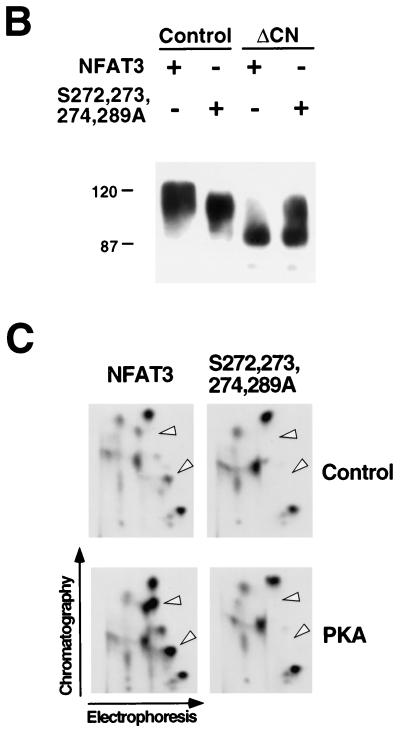
Dephosphorylation of NFAT causes increased electrophoretic mobility during SDS-PAGE (15, 62). We therefore examined the electrophoretic mobility of wild-type and mutated [Ala272,273,274,289] NFAT3. Immunoblot analysis demonstrated that the mutated [Ala272,273,274,289] NFAT3 exhibited increased electrophoretic mobility compared to wild-type NFAT3 (Fig. 1B). Activated calcineurin dephosphorylated both wild-type and mutated [Ala272,273,274,289] NFAT3. Importantly, dephosphorylated wild-type and mutated [Ala272,273,274,289] NFAT3 showed similar electrophoretic mobilities (Fig. 1B). These data suggest that the PKA sites contribute to the phosphorylation of NFAT3 in vivo. To test this hypothesis directly, we examined the in vivo phosphorylation of NFAT3 in cells labeled with [32P]phosphate (Fig. 1C). Tryptic phosphopeptide mapping demonstrated that coexpression of the PKA catalytic subunit increased the number of phosphopeptides in wild-type NFAT3 (arrowheads in Fig. 1C). These PKA-dependent phosphopeptides were absent in maps of the mutated [Ala272,273,274,289] NFAT3 that was not phosphorylated by PKA in vitro. Together, these data establish that NFAT3 is phosphorylated in vitro and in vivo by PKA.
Replacement of Ser-272, Ser-273, Ser-274, and Ser-289 of NFAT3 eliminated phosphorylation by PKA in vitro (Fig. 1A). Interestingly, Ser-272 and Ser-289 are conserved in other members of the NFAT group of transcription factors (Fig. 1A). We therefore postulated that Ser-272 and Ser-289 might represent the sites of NFAT3 phosphorylation by PKA. To test this hypothesis, we tested the effect of the replacement of Ser-272 and Ser-289 with Ala residues. Mutation at Ser-272 and Ser-289 was sufficient to prevent phosphorylation of NFAT3 by PKA (Fig. 1D). These data indicate that Ser-272 and Ser-289 are phosphorylated by PKA. To test whether these sites are phosphorylated in vivo, we examined the effect of individual point mutations at each of these sites on NFAT3 phosphorylation (Fig. 1D). Phosphopeptide mapping demonstrated that mutations at Ser-273 and Ser-274 did not cause marked changes in NFAT phosphorylation in vivo. In contrast, mutations at Ser-272 and Ser-289 caused the loss of phosphopeptides (arrowheads in Fig. 1D). These phosphopeptides correspond to peptides observed in maps of NFAT3 phosphorylated by PKA in vitro. Together, these data demonstrate that Ser-272 and Ser-289 represent the major sites of NFAT3 phosphorylation by PKA in vivo and in vitro (Fig. 1D).
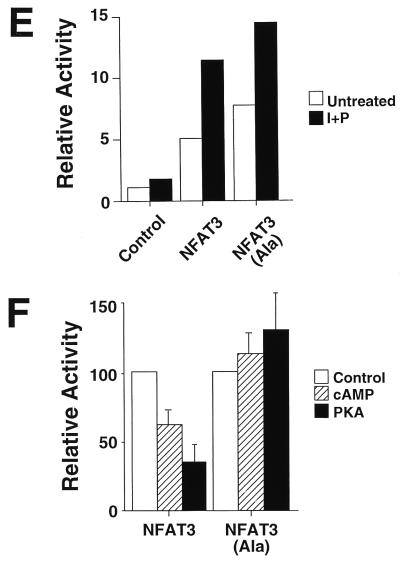
To test the effect of PKA phosphorylation on NFAT function, we examined the transcription activity of wild-type and phosphorylation-defective NFAT3. Transcription activity was measured by using an NFAT-luciferase reporter plasmid containing three copies of an NFAT–AP-1 composite element derived from the IL-2 promoter. Both wild-type and mutated NFAT3 caused increased reporter gene expression (Fig. 1E). Treatment of the cells with dibutyryl cAMP or coexpression of PKA catalytic subunit caused inhibition of transcription activity mediated by wild-type NFAT3 (Fig. 1F). In contrast, transcription activity mediated by the mutated phosphorylation-defective NFAT3 was not inhibited under these conditions (Fig. 1F). These data suggest that PKA down-regulates NFAT transcription activity by a mechanism that requires NFAT phosphorylation.
Phosphorylation is an important mechanism that regulates the subcellular distribution of NFAT (15, 62). Two functionally redundant nuclear localization sequences (NLS) in NFAT have been identified (4, 44). These correspond to NFAT3 residues 268 to 270 (NLS-1) and 672 to 675 (NLS-2) (Fig. 1F). Intramolecular interaction with phosphoserine residues located in the NFAT NH2 terminus may control the function of NLS-2 (5). However, the mechanism that regulates NLS-1 is unclear. Interestingly, sites of PKA phosphorylation are located adjacent to NLS-1. To test whether phosphorylation at these sites alters NLS-1 function, we examined the effect of PKA on the subcellular distribution of a truncated NFAT molecule (residues 1 to 580) that contains NLS-1, but not NLS-2, by immunofluorescence analysis (Fig. 1G). The truncated NFAT3 molecule was found in the cytosol, and this localization was not affected by PKA (Fig. 1G). Activated calcineurin induced NFAT3 nuclear accumulation, which was blocked by PKA (Fig. 1G). Replacement of the PKA phosphorylation sites with Ala residues eliminated the effect of PKA to oppose calcineurin-stimulated nuclear accumulation (Fig. 1G). These data indicate that PKA phosphorylation may regulate the function of NLS-1.
Both NLS-1 and NLS-2 are implicated in the mechanism of NFAT nuclear accumulation (4, 44). To test whether PKA was able to regulate NFAT subcellular distribution we used full-length NFAT3, which contains both NLS-1 and NLS-2. Immunofluorescence analysis demonstrated that both wild-type and mutated phosphorylation-defective NFAT3 were located in the cytosol (Fig. 1G). Calcineurin-stimulated nuclear accumulation was observed in experiments using both wild-type and mutated phosphorylation-defective NFAT3 (Fig. 1G). No effect of PKA on the subcellular distribution of these NFAT3 molecules was observed (Fig. 1G). Together, these data indicate that while PKA may regulate the function of NLS-1, PKA signaling was not sufficient to alter the nuclear accumulation of wild-type NFAT. This is probably because PKA does not regulate NLS-2. PKA can therefore inhibit NFAT transcription activity independently of changes in the subcellular distribution of NFAT. Electrophoretic mobility shift assays indicated that treatment with dibutyryl cAMP did not inhibit NFAT DNA binding activity (data not shown). Thus, PKA may exert a direct action on NFAT transcription activity. This effect of PKA requires the sites of NFAT phosphorylation by PKA.
Phosphorylation-dependent interaction of NFAT with 14-3-3.
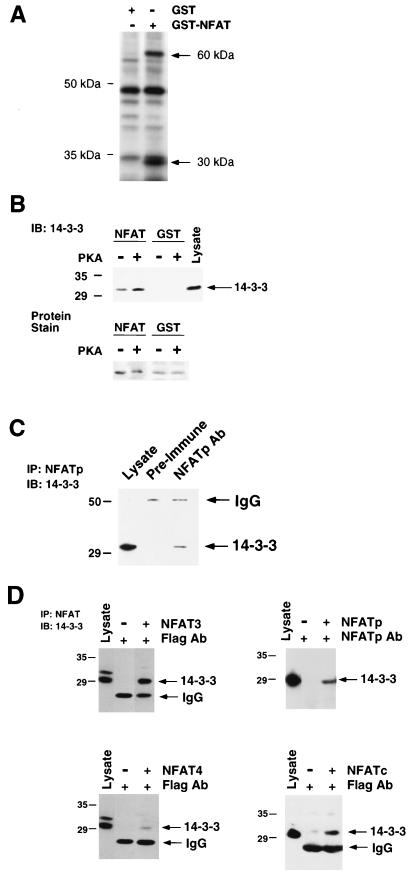
The sites of PKA phosphorylation are located within the conserved NFAT homology region (Fig. 1A). This region is postulated to form a complex subdomain upon phosphorylation that masks important NFAT regulatory elements, such as NLS-1 (62). Formation of this subdomain may involve both intra- and intermolecular interactions. To test whether PKA phosphorylation altered intermolecular interactions, we used NFAT as an affinity matrix to purify molecules that bind the conserved NFAT homology region. Two proteins (30 and 60 kDa) were found to bind GST-NFAT3 but not GST (Fig. 2A). The 60-kDa protein may correspond to the catalytic subunit of calcineurin, which is known to bind to the NFAT homology domain (3, 21). Indeed, immunoblot analysis confirmed that calcineurin bound to GST-NFAT3 (data not shown). In contrast, the identity of the 30-kDa protein that bound NFAT3 was unclear. We suspected that this 30-kDa protein might be 14-3-3 because of the similar mass and the reported function of 14-3-3 as a phosphoprotein ligand (53). To test this hypothesis, we performed immunoblot analysis using an antibody to 14-3-3. These experiments demonstrated that the NH2-terminal region of NFAT3 bound 14-3-3 (Fig. 2B).
FIG. 2.
NFAT interacts with 14-3-3. (A) Immobilized GST and GST-NFAT3 (residues 1 to 308) were incubated with extracts prepared from [35S]methionine-labeled BHK cells. Bound proteins were separated by SDS-PAGE and detected by autoradiography. (B) Recombinant GST and GST-NFAT3 (residues 1 to 308) were phosphorylated by incubation with 1 mM ATP and purified PKA catalytic subunit. Immobilized GST and GST-NFAT3 were incubated with extracts prepared from Jurkat T cells. Bound 14-3-3 was detected by immunoblot (IB) analysis. The GST and GST-NFAT3 fusion proteins were detected by staining with Coomassie blue. (C) NFATp interacts with 14-3-3 in vivo. NFATp was immunoprecipitated from Jurkat T cell extracts by using a rabbit antibody to NFATp. Preimmune antibody was used as a control. 14-3-3 in the immunoprecipitates (IP) was detected by immunoblot (IB) analysis. (D) NFAT proteins bind 14-3-3. NFAT3, NFAT4, NFATp and NFATc were expressed in COS cells. NFATp was immunoprecipitated with an antibody to NFATp. Epitope-tagged NFAT3, NFAT4, and NFATc were immunoprecipitated with the anti-Flag monoclonal antibody (Ab) M2. 14-3-3 in the cells lysate and in the immunoprecipitates (IP) was detected by immunoblot (IB) analysis. Extracts prepared from mock-transfected cells were used as a control. IgG, immunoglobulin G.
NFAT was first characterized as a transcription factor in T cells (67). We therefore examined the interaction of endogenous NFAT with 14-3-3 in T cells by coimmunoprecipitation analysis. Jurkat T cells, which express predominantly the NFATp isoform, were used for these assays. Immunoblot analysis demonstrated the presence of 14-3-3 in NFATp immunoprecipitates (Fig. 2C). These data confirm the conclusion that NFAT interacts with 14-3-3. Since the NH2-terminal region of NFAT3 is conserved in the NFAT group of transcription factors, we also examined whether other members of the NFAT group interacted with 14-3-3. Coimmunoprecipitation assays demonstrated that 14-3-3 was detected in NFAT3, NFAT4, NFATp, and NFATc immunoprecipitates (Fig. 2D). These data indicate that interaction with 14-3-3 is a common property of NFAT transcription factors.
PKA phosphorylation sites mediate the interaction of NFAT with 14-3-3.
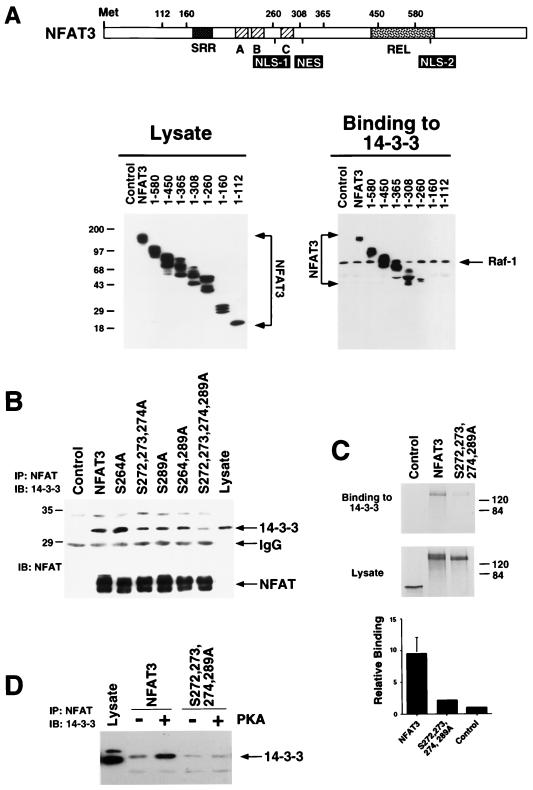
To delineate the 14-3-3 binding site on NFAT, we examined the interaction between various NFAT3 deletion mutants and 14-3-3. Immunoblot analysis demonstrated similar levels of expression of the NFAT proteins (Fig. 3A). The interaction between 14-3-3 and Raf-1 was not affected by the various NFAT3 proteins (Fig. 3A). In contrast, deletion of NFAT3 sequences caused marked changes in the binding of NFAT3 to 14-3-3. Progressive COOH-terminal truncations of NFAT3 demonstrated that the Rel domain was not required for 14-3-3 binding. Further deletion to remove the NFAT homology domain abolished the interaction with 14-3-3 (Fig. 3A). A fragment of the NFAT homology domain (NFAT3 residues 260 to 308) encompassed the major binding sites for 14-3-3. This region includes the sites of PKA phosphorylation on NFAT3. We tested whether these phosphorylation sites are relevant to the interaction of NFAT3 with 14-3-3. Coimmunoprecipitation assays demonstrated that replacement of the PKA phosphorylation sites with Ala markedly reduced the coimmunoprecipitation of NFAT with 14-3-3 (Fig. 3B). These data indicate that the PKA phosphorylation sites of NFAT3 are important for the interaction with 14-3-3. To quantitate the contribution of the PKA phosphorylation sites to 14-3-3 binding, we examined the interaction of in vitro-translated [35S]methionine-labeled NFAT3 to recombinant 14-3-3. We found that wild-type NFAT3 bound to 14-3-3 approximately five times as strongly as the mutated phosphorylation-defective NFAT3 (Fig. 3C).
FIG. 3.
Identification of the 14-3-3 binding site on NFAT. (A) Deletion analysis of NFAT3. The structure of NFAT3 is illustrated schematically. Flag-tagged NFAT3 proteins corresponding to residues 1 to 902, 1 to 580, 1 to 450, 1 to 365, 1 to 308, 1 to 260, 1 to 160, and 1 to 112 were expressed in COS cells and detected by immunoblot analysis of cell lysates with monoclonal antibody M2. The binding of NFAT3 and Raf-1 to immobilized GST–14-3-3τ was examined. Bound NFAT3 and Raf-1 were detected by immunoblot analysis. (B) Replacement of Ser-272, Ser-273, Ser-274, and Ser-289 with Ala decreases NFAT3 binding to 14-3-3. Epitope-tagged wild-type and mutated NFAT3 were expressed in COS cells and detected by immunoblot (IB) analysis of cell lysates with monoclonal antibody M2 (lower panel). The NFAT3 proteins were immunoprecipitated, and 14-3-3 present in the immunoprecipitates (IP) was detected by immunoblot analysis (upper panel; sizes are indicated in kilodaltons. (C) Immobilized GST–14-3-3τ was incubated with [35S]methionine-labeled wild-type and mutated [Ala272,273,274,289] NFAT3 prepared by in vitro translation. Control experiments were performed with in vitro-translated luciferase. Proteins in the lysate and bound to the immobilized GST–14-3-3τ were detected by autoradiography and quantitated by PhosphorImager analysis. (D) Epitope-tagged wild-type and mutated [Ala272,273,274,289] NFAT3 were expressed in COS cells without (Control) and with an expression vector for the PKA catalytic subunit (PKA). NFAT3 proteins were immunoprecipitated, and 14-3-3 present in the immunoprecipitates (IP) was detected by immunoblot analysis (IB).
Phosphorylation of NFAT3 by PKA increased 14-3-3 binding in vitro (Fig. 2B). We therefore tested whether PKA increased the binding of 14-3-3 to the mutated phosphorylation-defective NFAT3. Coexpression of the PKA catalytic subunit with wild-type NFAT3 increased 14-3-3 binding (Fig. 3D). Replacement of the PKA phosphorylation sites with Ala markedly reduced 14-3-3 binding. Importantly, coexpression of the PKA catalytic subunit did not increase 14-3-3 binding to the phosphorylation-defective NFAT3. These data indicate that the PKA phosphorylation sites on NFAT3 are required for the PKA-stimulated interaction of NFAT3 with 14-3-3.
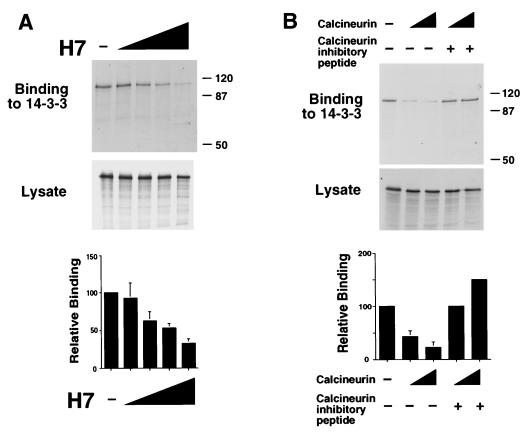
The binding of 14-3-3 can be phosphorylation dependent (53, 79). We therefore examined whether phosphorylation contributes to the interaction of NFAT with 14-3-3. NFAT3 prepared by in vitro translation binds to 14-3-3 (Fig. 3C), suggesting either that phosphorylation is not required or that the translated NFAT3 is phosphorylated in the reticulocyte lysate. To test the latter hypothesis, we investigated the effect of the protein kinase inhibitor H7. Similar amounts of in vitro-translated NFAT3 were obtained in the absence and presence of H7 (Fig. 4A). However, the presence of H7 during in vitro translation caused a marked dose-dependent inhibition of NFAT3 binding to 14-3-3 (Fig. 4A). Similarly, treatment of in vitro-translated NFAT3 with the phosphatase calcineurin caused inhibition of NFAT3 binding to 14-3-3 (Fig. 4B). This effect of calcineurin was blocked by a specific peptide inhibitor of calcineurin activity. These data indicate that phosphorylation increases NFAT interaction with 14-3-3. This conclusion was confirmed by the observation that NFAT3 phosphorylation by PKA increases binding to 14-3-3 both in vitro (Fig. 2A) and in vivo (Fig. 3D).
FIG. 4.
Phosphorylation increases NFAT binding to 14-3-3. (A) [35S]methionine-labeled NFAT3 was prepared by in vitro translation in the presence of various concentrations of the protein kinase inhibitor H7 (0, 1, 10, 100, and 1,000 μM). The amount of NFAT3 in the lysate was detected after SDS-PAGE by autoradiography (middle panel). The binding of NFAT3 to immobilized GST–14-3-3τ was examined by autoradiography (upper panel; sizes are indicated in kilodalton) and was quantitated by PhosphorImager analysis (lower panel). (B) Calcineurin inhibits NFAT3 binding to 14-3-3. [35S]methionine-labeled NFAT3 prepared by in vitro translation was incubated with calcineurin (2.5 and 5 U) and calcineurin inhibitor peptide (10 μg). The amount of NFAT3 in the lysate was detected after SDS-PAGE by autoradiography (middle panel). The binding of NFAT3 to immobilized GST–14-3-3τ was examined by autoradiography (upper panel) and was quantitated by PhosphorImager analysis (lower panel).
To test whether 14-3-3 can mediate the inhibitory effect of PKA phosphorylation on NFAT transcription activity, we investigated the effect of 14-3-3 overexpression. Since 14-3-3 is an abundant cellular protein, a marked effect of ectopic 14-3-3 expression was not anticipated. However, the expression of 14-3-3τ did cause a moderate (30%) reduction of the transcription activity of wild-type NFAT3 but not the mutated phosphorylation defective NFAT3 that lacks PKA phosphorylation sites (data not shown). These data are consistent with the hypothesis that 14-3-3 binding might mediate the effect of PKA on NFAT3 activity.
Regulation of NFAT binding to 14-3-3 by cAMP and calcium.
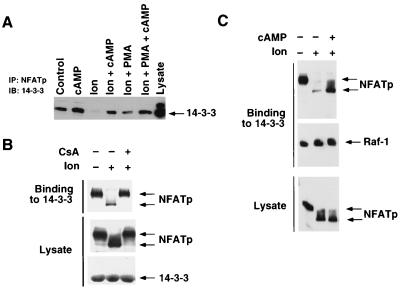
The ability of calcineurin to inhibit NFAT binding to 14-3-3 (Fig. 4B) suggests that calcium signaling may be a negative regulator of the interaction between NFAT and 14-3-3. In contrast, PKA appears to be a positive regulator of NFAT binding to 14-3-3 (Fig. 3D). The opposite effects of calcium and cAMP on NFAT–14-3-3 complexes suggests that these signaling pathways may have an antagonistic relationship in vivo. To test this hypothesis, we examined the effect of calcium and cAMP on the formation of endogenous NFATp–14-3-3 complexes in Jurkat T cells (Fig. 5A). Dibutytryl cAMP and ionomycin were used to activate the PKA and calcium signaling pathways, respectively. Coimmunoprecipitation assays demonstrated that 14-3-3 coprecipitated with NFATp. Treatment with ionomycin caused a marked reduction in NFATp–14-3-3 complex formation. This effect of ionomycin was attenuated by treatment of the cells with dibutyryl cAMP (Fig. 5A). Control experiments demonstrated that calcium and cAMP did not affect the binding of Raf-1 to 14-3-3 (data not shown). These data indicate that the cAMP signaling pathway can antagonize the effect of calcium on NFATp–14-3-3 complex formation.
FIG. 5.
Calcium and cAMP regulate NFAT binding to 14-3-3. (A) Jurkat T cells were incubated without and with dibutyryl cAMP (500 μM) plus IBMX (50 μM) for 20 min prior to treatment with ionomycin (2 μM, 20 min) or with ionomycin plus PMA (100 nM, 20 min). NFATp proteins were immunoprecipitated, and 14-3-3 present in the immunoprecipitates (IP) was detected by immunoblot analysis (IB). (B) Jurkat T cells were incubated without and with CsA (100 ng/ml, 20 min) prior to treatment with ionomycin (2 μM, 20 min). Extracts prepared from the Jurkat T cells were incubated with immobilized GST–14-3-3τ. Bound NFATp and Raf-1 were detected by immunoblot analysis. The presence of equal amounts of NFATp in the lysates was confirmed by immunoblot analysis. The amount of GST–14-3-3τ was confirmed by Coomassie blue staining. (C) Jurkat T cells were incubated without and with dibutyryl cAMP (500 μM) plus IBMX (50 μM) for 20 min prior to treatment with ionomycin (2 μM, 20 min). Extracts prepared from the Jurkat T cells were incubated with immobilized GST–14-3-3τ. Bound NFATp and Raf-1 were detected by immunoblot analysis. The presence of equal amounts of NFATp in the lysates was confirmed by immunoblot analysis.
The ability of ionomycin to inhibit NFATp–14-3-3 complex formation was further examined in in vitro binding assays (Fig. 5B). Western blot analysis of extracts prepared from Jurkat T cells demonstrated that ionomycin caused a marked increase in the electrophoretic mobility of NFATp during SDS-PAGE. This increased mobility has been reported to be caused by calcineurin-mediated dephosphorylation (15, 62). Binding assays demonstrated that ionomycin treatment of Jurkat cells inhibited the interaction of NFATp with 14-3-3 in vitro (Fig. 5B). This ability of ionomycin to increase the electrophoretic mobility of NFATp and to inhibit binding to 14-3-3 was blocked if the cells were incubated with the immunosuppressive drug CsA, which inhibits the phosphatase activity of calcineurin (Fig. 5B). The effect of ionomycin on NFATp–14-3-3 binding was also inhibited when the Jurkat T cells were incubated with dibutyryl cAMP (Fig. 5C). Control experiments demonstrated that dibutyryl cAMP and ionomycin did not alter the interaction of 14-3-3 with the Raf-1 protein kinase. In contrast to CsA, dibutyryl cAMP did not cause a marked decrease in NFATp electrophoretic mobility (Fig. 5C). These data indicate that calcineurin inhibition does not mediate the effect of dibutyryl cAMP. In contrast, these data are consistent with the hypothesis that the phosphorylation of NFATp on a limited number of sites accounts for the regulation of NFATp/14-3-3 complex formation by the cAMP signaling pathway.
DISCUSSION
14-3-3 was first described as a dimeric protein in the brain (51). Subsequent studies demonstrated that 14-3-3 includes a group of proteins that are highly conserved and ubiquitously expressed (reviewed in references 2 and 63). The general mechanism of action of 14-3-3 proteins appears to involve the formation of complexes with target proteins. In some instances, such complexes depend on the phosphorylation of the 14-3-3 binding partner. A general consensus sequence that mediates phosphorylation-dependent binding to 14-3-3 has been defined as RSXSpXP (53, 79). Nevertheless, other sequences containing phosphoserine have been shown to bind 14-3-3 (17, 36, 42, 81). Structural studies demonstrate the presence of a basic pocket within a conserved groove in 14-3-3 (40, 78). This basic pocket was postulated to interact with phosphoserine and to provide a specificity determinant for ligand binding by 14-3-3 (53). This hypothesis was confirmed by structural studies of 14-3-3 bound to phosphopeptides (64, 79). However, additional interactions between the phosphopeptide ligand and residues present in the 14-3-3 groove also contribute to binding specificity (64, 79).
Recent studies have established many functions for the interaction of phosphoproteins with 14-3-3. Examples include the observations that (i) 14-3-3 is required for the stabilization of Raf-1 in its active conformation (19, 65, 71, 74), (ii) the binding of 14-3-3 to Bad prevents interaction with Bcl-xL and plays a key role in cell survival (80), (iii) the binding of 14-3-3 to Cdc25C is required for checkpoint control of the cell cycle (59), and (iv) the binding of 14-3-3 to a Forkhead transcription factor causes nuclear export and contributes to PKB-mediated cell survival (9). Here we report the identification of the transcription factor NFAT as a ligand for 14-3-3.
PKA and calcineurin regulate NFAT binding to 14-3-3.
Members of the NFAT group of transcription factors bind to 14-3-3. This binding is increased by PKA in vitro (Fig. 2B) and in vivo (Fig. 3D). Conversely, the binding of NFAT to 14-3-3 is inhibited by the phosphatase calcineurin in vitro (Fig. 4B) and by calcium in vivo (Fig. 5). Replacement of the PKA phosphorylation sites with Ala blocked the inhibitory effect of PKA on both NFAT transcription activity (Fig. 1) and the binding of NFAT to 14-3-3 (Fig. 3). NFAT binding to 14-3-3 therefore negatively correlates with NFAT transcription activity. Thus, 14-3-3 may function to suppress NFAT activity. This conclusion is further supported by the observation that overexpression of 14-3-3 inhibits IL-2 gene expression (49). Interestingly, PKA can induce 14-3-3 binding to NFAT in the presence of activated calcineurin. Thus, cAMP signaling may act dominantly with respect to signaling by calcineurin.
The sites of phosphorylation-dependent binding of 14-3-3 to NFAT correspond to PKA sites located in the NH2-terminal region of NFAT. These sites are located immediately adjacent to NLS-1 of NFAT. It is therefore likely that the binding of 14-3-3 to NFAT would cause sequestration and inactivation of NLS-1. This might cause redistribution of NFAT from the nucleus to the cytoplasm and therefore inhibition of NFAT-mediated transcription. The mechanism of nuclear export may include sequestration of NLS-1, but it is also possible that other processes are involved. Several possible mechanisms have been established in previous studies. For example, the yeast transcription factor Pho4 interacts in a phosphorylation-dependent manner with the nuclear export receptor Msn5 (30, 35). A second example is provided by the observation that 14-3-3 proteins contain a conserved nuclear export signal (NES) that may mediate CRM1-dependent translocation of 14-3-3 from the nucleus to the cytoplasm (9, 43). Thus, the binding of 14-3-3 to NFAT may cause both the sequestration of NLS-1 and the attachment of an NES. The NES of 14-3-3 may augment the action of an NES present in the NFAT NH2-terminal region that allows rapid CRM1-dependent nuclear export (32, 33, 82).
Sequestration of NFAT NLS-1 by an NES-bearing 14-3-3 protein provides an extremely elegant mechanism by which 14-3-3 could regulate the subcellular localization of NFAT transcription factors and thereby inhibit NFAT transcription activity. Indeed, some evidence for this potential mechanism was obtained. Studies of a truncated NFAT3 molecule (residues 1 to 580) that contains NLS-1 but not NLS-2 demonstrated that calcineurin-induced nuclear translocation of NFAT was opposed by PKA and that this effect of PKA was blocked by mutation of the NFAT phosphorylation sites. However, full-length NFAT3, which contains both NLS-1 and NLS-2, exhibited calcineurin-induced nuclear translocation that was not inhibited by PKA. These data indicate that while 14-3-3 has the potential to regulate the subcellular distribution of NFAT, 14-3-3 by itself was insufficient to cause redistribution of nuclear NFAT to the cytoplasm. It is known that the mode of 14-3-3 binding to phosphoproteins may influence the function of the 14-3-3 NES (64). Thus, the difference observed between the regulation of the full-length and truncated NFAT3 molecules may reflect a difference in the mode of 14-3-3 binding to these phosphoproteins.
The observation that PKA does not alter the subcellular distribution of full-length NFAT indicates that export of NFAT from the nucleus does not account for the effect of PKA to inhibit NFAT transcription activity at the IL-2 promoter. Furthermore, we did not detect PKA-stimulated changes in NFAT DNA binding activity. It is therefore possible that PKA exerts a more direct effect on NFAT transcription activity and that the formation of NFAT complexes with 14-3-3 contributes to the altered function of NFAT following PKA activation. It is likely that NFAT function is altered rather than simply inhibited because while PKA inhibits IL-2 expression, PKA causes increased expression of IL-4 and IL-5 (52). The mechanism by which PKA differentially regulates the NFAT-mediated expression of IL-2 compared with IL-4 and IL-5 is unclear. It is possible that the NFAT complex with 14-3-3 contributes to this differential regulation. Evidence in favor of this hypothesis was obtained from the observation that the overexpression of 14-3-3 inhibits transcription activity at the IL-2 promoter but increases transcription activity at the IL-4 promoter (49). The molecular basis for the differential effect of 14-3-3 on NFAT-mediated transcription is unclear. However, we can speculate that NFAT complexes may differentially collaborate with other transcription factors that are relevant to each promoter, including AP-1 at the IL-2 promoter (15, 62), GATA-3 at the IL-5 promoter (69), and c-Maf or JunB at the IL-4 promoter (24, 38).
Specificity of NFAT interaction with 14-3-3.
Binding to 14-3-3 was detected in coimmunoprecipitation assays using members of the NFAT group, including NFATp, NFATc, NFAT3, and NFAT4 (Fig. 2D). Interaction with 14-3-3 therefore appears to be a common property of NFAT transcription factors. The 14-3-3 group of proteins includes seven members (2, 63). Some of these 14-3-3 isoforms are expressed selectively in certain tissues. For example, 14-3-3τ is expressed in T cells (55). It is possible that NFAT binds differentially to these 14-3-3 proteins. Preliminary studies designed to investigate 14-3-3 binding specificity indicated that the amount of NFATp binding to 14-3-3τ was approximately five times greater than binding to 14-3-3ζ, while an equal amount of Raf-1 was found to bind each 14-3-3 isoform (data not shown). Whether this difference in binding activity in vitro is relevant in vivo is unclear. Further studies will be required to resolve this question. We note that previous studies have not established biochemical differences in the ligand binding properties of 14-3-3 proteins (79). However, such differences are likely since genetically nonredundant functions for specific 14-3-3 genes have been identified in Drosophila (10, 34).
The specificity of NFAT interaction with 14-3-3 is determined, in part, by the requirement of NFAT phosphorylation. The sites of NFAT phosphorylation that contribute to the interaction with 14-3-3 correspond to PKA sites. Indeed, it is likely that PKA is a physiologically relevant protein kinase since dibutyryl cAMP increases NFAT binding to 14-3-3 in calcium-stimulated cells (Fig. 5). However, the sites of PKA phosphorylation may also be phosphorylated by other protein kinases with similar substrate specificity, including calmodulin-dependent protein kinases, PKB, and PKC. Thus, the regulation of NFAT interaction with 14-3-3 may be a site of integration of multiple signaling pathways that ensures correct responses to complex biological stimuli.
ACKNOWLEDGMENTS
We thank A. Altman, T. Hoey, Y.-C. Liu, R. Maurer, A. Rao, and T. Soderling for providing reagents; T. Barrett and M. Sharma for technical assistance; and K. Gemme for administrative assistance.
C.-W. Chow is an Arthritis Foundation fellow. This work was supported in part by grants CA65861 and CA72009 from the National Cancer Institute. R.J.D. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Abbott K L, Friday B B, Thaloor D, Murphy T J, Pavlath G K. Activation and cellular localization of the cyclosporine A-sensitive transcription factor NF-AT in skeletal muscle cells. Mol Biol Cell. 1998;9:2905–2916. doi: 10.1091/mbc.9.10.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aitken A. 14-3-3 proteins on the MAP. Trends Biochem Sci. 1995;20:95–97. doi: 10.1016/s0968-0004(00)88971-9. [DOI] [PubMed] [Google Scholar]
- 3.Aramburu J, Garcia-Cozar F, Raghavan A, Okamura H, Rao A, Hogan P G. Selective inhibition of NFAT activation by a peptide spanning the calcineurin targeting site of NFAT. Mol Cell. 1998;1:627–637. doi: 10.1016/s1097-2765(00)80063-5. [DOI] [PubMed] [Google Scholar]
- 4.Beals C R, Clipstone N A, Ho S N, Crabtree G R. Nuclear localization of NF-ATc by a calcineurin-dependent, cyclosporin- sensitive intramolecular interaction. Genes Dev. 1997;11:824–834. doi: 10.1101/gad.11.7.824. [DOI] [PubMed] [Google Scholar]
- 5.Beals C R, Sheridan C M, Turck C W, Gardner P, Crabtree G R. Nuclear export of NF-ATc enhanced by glycogen synthase kinase-3. Science. 1997;275:1930–1934. doi: 10.1126/science.275.5308.1930. [DOI] [PubMed] [Google Scholar]
- 6.Bodor J, Habener J F. Role of transcriptional repressor ICER in cyclic AMP-mediated attenuation of cytokine gene expression in human thymocytes. J Biol Chem. 1998;273:9544–9551. doi: 10.1074/jbc.273.16.9544. [DOI] [PubMed] [Google Scholar]
- 7.Bonnefoy-Berard N, Liu Y C, von Willebrand M, Sung A, Elly C, Mustelin T, Yoshida H, Ishizaka K, Altman A. Inhibition of phosphatidylinositol 3-kinase activity by association with 14-3-3 proteins in T cells. Proc Natl Acad Sci USA. 1995;92:10142–10146. doi: 10.1073/pnas.92.22.10142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyle W J, van der Geer P, Hunter T. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional thin layer cellulose plates. Methods Enzymol. 1991;201:110–149. doi: 10.1016/0076-6879(91)01013-r. [DOI] [PubMed] [Google Scholar]
- 9.Brunet A, Bonni A, Zigmond M J, Lin M Z, Juo P, Hu L S, Anderson M J, Arden K C, Blenis J, Greenberg M E. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 10.Chang H C, Rubin G M. 14-3-3 epsilon positively regulates Ras-mediated signaling in Drosophila. Genes Dev. 1997;11:1132–1139. doi: 10.1101/gad.11.9.1132. [DOI] [PubMed] [Google Scholar]
- 11.Chin E R, Olson E N, Richardson J A, Yang Q, Humphries C, Shelton J M, Wu H, Zhu W, Bassel-Duby R, Williams R S. A calcineurin-dependent transcriptional pathway controls skeletal muscle fiber type. Genes Dev. 1998;12:2499–2509. doi: 10.1101/gad.12.16.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chow C-W, Rincon M, Davis R J. Requirement of transcription factor NFAT for interleukin 2 expression. Mol Cell Biol. 1999;19:2300–2307. doi: 10.1128/mcb.19.3.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chow C W, Rincon M, Cavanagh J, Dickens M, Davis R J. Nuclear accumulation of NFAT4 opposed by the JNK signal transduction pathway. Science. 1997;278:1638–1641. doi: 10.1126/science.278.5343.1638. [DOI] [PubMed] [Google Scholar]
- 14.Clipstone N A, Crabtree G R. Identification of calcineurin as a key signalling enzyme in T-lymphocyte activation. Nature. 1992;357:695–697. doi: 10.1038/357695a0. [DOI] [PubMed] [Google Scholar]
- 15.Crabtree G R, Clipstone N A. Signal transmission between the plasma membrane and nucleus of T lymphocytes. Annu Rev Biochem. 1994;63:1045–1083. doi: 10.1146/annurev.bi.63.070194.005145. [DOI] [PubMed] [Google Scholar]
- 16.de la Pompa J L, Timmerman L A, Takimoto H, Yoshida H, Elia A J, Samper E, Potter J, Wakeham A, Marengere L, Langille B L, Crabtree G R, Mak T W. Role of the NF-ATc transcription factor in morphogenesis of cardiac valves and septum. Nature. 1998;392:182–186. doi: 10.1038/32419. [DOI] [PubMed] [Google Scholar]
- 17.Du X, Fox J E, Pei S. Identification of a binding sequence for the 14-3-3 protein within the cytoplasmic domain of the adhesion receptor, platelet glycoprotein Ib alpha. J Biol Chem. 1996;271:7362–7367. doi: 10.1074/jbc.271.13.7362. [DOI] [PubMed] [Google Scholar]
- 18.Durand D B, Shaw J-P, Bush M R, Repogla R E, Belagaje R, Crabtree G R. Characterization of antigen receptor response elements within the interleukin-2 enhancer. Mol Cell Biol. 1988;8:1715–1724. doi: 10.1128/mcb.8.4.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fantl W J, Muslin A J, Kikuchi A, Martin J A, MacNicol A M, Gross R W, Williams L T. Activation of Raf-1 by 14-3-3 proteins. Nature. 1994;371:612–614. doi: 10.1038/371612a0. [DOI] [PubMed] [Google Scholar]
- 20.Flanagan W M, Corthesy B, Bram R J, Crabtree G R. Nuclear association of a T-cell transcription factor blocked by FK-506 and cyclosporin A. Nature. 1991;352:803–807. doi: 10.1038/352803a0. [DOI] [PubMed] [Google Scholar]
- 21.Garcia-Cozar F J, Okamura H, Aramburu J F, Shaw K T Y, Pelletier L, Showalter R, Villafranca E, Rao A. Two-site interaction of nuclear factor of activated T cells with activated calcineurin. J Biol Chem. 1998;273:23877–23883. doi: 10.1074/jbc.273.37.23877. [DOI] [PubMed] [Google Scholar]
- 22.Haraguchi S, Good R A, Day N K. Immunosuppressive retroviral peptides: cAMP and cytokine patterns. Immunol Today. 1995;16:595–603. doi: 10.1016/0167-5699(95)80083-2. [DOI] [PubMed] [Google Scholar]
- 23.Ho A M, Jain J, Rao A, Hogan P G. Expression of the transcription factor NFATp in a neuronal cell line and in the murine nervous system. J Biol Chem. 1994;269:28181–28186. [PubMed] [Google Scholar]
- 24.Ho I C, Hodge M R, Rooney J W, Glimcher L H. The proto-oncogene c-maf is responsible for tissue-specific expression of interleukin-4. Cell. 1996;85:973–983. doi: 10.1016/s0092-8674(00)81299-4. [DOI] [PubMed] [Google Scholar]
- 25.Ho S N, Thomas D J, Timmerman L A, Li X, Francke U, Crabtree G R. NFATc3, a lymphoid-specific NFATc family member that is calcium-regulated and exhibits distinct DNA binding specificity. J Biol Chem. 1995;270:19898–19907. doi: 10.1074/jbc.270.34.19898. [DOI] [PubMed] [Google Scholar]
- 26.Hodge M R, Ranger A M, de la Brousse C F, Hoey T, Grusby M J, Glimcher L H. Hyperproliferation and dysregulation of IL-4 expression in NF-ATp-deficient mice. Immunity. 1996;4:397–405. doi: 10.1016/s1074-7613(00)80253-8. [DOI] [PubMed] [Google Scholar]
- 27.Hoey T, Sun Y L, Williamson K, Xu X. Isolation of two new members of the NF-AT gene family and functional characterization of the NF-AT proteins. Immunity. 1995;2:461–472. doi: 10.1016/1074-7613(95)90027-6. [DOI] [PubMed] [Google Scholar]
- 28.Hsueh Y-P, Lai M-Z. c-Jun N-terminal kinase but not mitogen-activated protein kinase is sensitive to cAMP inhibition in T lymphocytes. J Biol Chem. 1995;270:18094–18098. doi: 10.1074/jbc.270.30.18094. [DOI] [PubMed] [Google Scholar]
- 29.Jain J, McCaffrey P G, Miner Z, Kerppola T K, Lambert J N, Verdine G L, Curran T, Rao A. The T-cell transcription factor NFATp is a substrate for calcineurin and interacts with Fos and Jun. Nature. 1993;365:352–355. doi: 10.1038/365352a0. [DOI] [PubMed] [Google Scholar]
- 30.Kaffman A, Rank N M, O'Neill E M, Huang L S, O'Shea E K. The receptor Msn5 exports the phosphorylated transcription factor Pho4 out of the nucleus. Nature. 1998;396:482–486. doi: 10.1038/24898. [DOI] [PubMed] [Google Scholar]
- 31.Kammer G M. The adenylate cyclase-cAMP-protein kinase A pathway and regulation of the immune response. Immunol Today. 1988;9:222–229. doi: 10.1016/0167-5699(88)91220-0. [DOI] [PubMed] [Google Scholar]
- 32.Kehlenbach R H, Dickmanns A, Gerace L. Nucleocytoplasmic shuttling factors including Ran and CRM1 mediate nuclear export of NFAT in vitro. J Cell Biol. 1998;141:863–874. doi: 10.1083/jcb.141.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klemm J D, Beals C R, Crabtree G R. Rapid targeting of nuclear proteins to the cytoplasm. Curr Biol. 1997;7:638–644. doi: 10.1016/s0960-9822(06)00290-9. [DOI] [PubMed] [Google Scholar]
- 34.Kockel L, Vorbruggen G, Jackle H, Mlodzik M, Bohmann D. Requirement for Drosophila 14-3-3 zeta in Raf-dependent photoreceptor development. Genes Dev. 1997;11:1140–1147. doi: 10.1101/gad.11.9.1140. [DOI] [PubMed] [Google Scholar]
- 35.Komeili A, O'Shea E K. Roles of phosphorylation sites in regulating activity of the transcription factor Pho4. Science. 1999;284:977–980. doi: 10.1126/science.284.5416.977. [DOI] [PubMed] [Google Scholar]
- 36.Ku N O, Liao J, Omary M B. Phosphorylation of human keratin 18 serine 33 regulates binding to 14-3-3 proteins. EMBO J. 1998;17:1892–1906. doi: 10.1093/emboj/17.7.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Latinis K M, Norian L A, Eliason S L, Koretzky G A. Two NFAT transcription factor binding sites participate in the regulation of CD95 (Fas) ligand expression in activated human T cells. J Biol Chem. 1997;272:31427–31434. doi: 10.1074/jbc.272.50.31427. [DOI] [PubMed] [Google Scholar]
- 38.Li B, Tournier C, Davis R J, Flavell R A. Regulation of IL-4 expression by the transcription factor JunB during T helper cell differentiation. EMBO J. 1999;18:420–432. doi: 10.1093/emboj/18.2.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li L, Yee C, Beavo J A. CD3- and CD28-dependent induction of PDE7 required for T cell activation. Science. 1999;283:848–851. doi: 10.1126/science.283.5403.848. [DOI] [PubMed] [Google Scholar]
- 40.Liu D, Bienkowska J, Petosa C, Collier R J, Fu H, Liddington R. Crystal structure of the zeta isoform of the 14-3-3 protein. Nature. 1995;376:191–194. doi: 10.1038/376191a0. [DOI] [PubMed] [Google Scholar]
- 41.Liu Y C, Elly C, Yoshida H, Bonnefoy-Berard N, Altman A. Activation-modulated association of 14-3-3 proteins with Cbl in T cells. J Biol Chem. 1996;271:14591–14595. doi: 10.1074/jbc.271.24.14591. [DOI] [PubMed] [Google Scholar]
- 42.Liu Y C, Liu Y, Elly C, Yoshida H, Lipkowitz S, Altman A. Serine phosphorylation of Cbl induced by phorbol ester enhances its association with 14-3-3 proteins in T cells via a novel serine-rich 14- 3-3-binding motif. J Biol Chem. 1997;272:9979–9985. doi: 10.1074/jbc.272.15.9979. [DOI] [PubMed] [Google Scholar]
- 43.Lopez-Girona A, Furnari B, Mondesert O, Russell P. Nuclear localization of Cdc25 is regulated by DNA damage and a 14-3-3 protein. Nature. 1999;397:172–175. doi: 10.1038/16488. [DOI] [PubMed] [Google Scholar]
- 44.Luo C, Shaw K T, Raghavan A, Aramburu J, Garcia-Cozar F, Perrino B A, Hogan P G, Rao A. Interaction of calcineurin with a domain of the transcription factor NFAT1 that controls nuclear import. Proc Natl Acad Sci USA. 1996;93:8907–8912. doi: 10.1073/pnas.93.17.8907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Masuda E S, Naito Y, Tokumitsu H, Campbell D, Saito F, Hannum C, Arai K, Arai N. NFATx, a novel member of the nuclear factor of activated T cells family that is expressed predominantly in the thymus. Mol Cell Biol. 1995;15:2697–2706. doi: 10.1128/mcb.15.5.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maurer R A. Both isoforms of the cAMP-dependent protein kinase catalytic subunit can activate transcription of the prolactin gene. J Biol Chem. 1989;264:6870–6873. [PubMed] [Google Scholar]
- 47.McCaffrey P G, Goldfeld A E, Rao A. The role of NFATp in cyclosporin A-sensitive tumor necrosis factor-alpha gene transcription. J Biol Chem. 1994;269:30445–30450. [PubMed] [Google Scholar]
- 48.McCaffrey P G, Luo C, Kerppola T K, Jain J, Badalian T M, Ho A M, Burgeon E, Lane W S, Lambert J N, Curran T, et al. Isolation of the cyclosporin-sensitive T cell transcription factor NFATp. Science. 1993;262:750–754. doi: 10.1126/science.8235597. [DOI] [PubMed] [Google Scholar]
- 49.Meller N, Liu Y C, Collins T L, Bonnefoy-Berard N, Baier G, Isakov N, Altman A. Direct interaction between protein kinase Cθ (PKCθ) and 14-3-3τ in T cells: 14-3-3 overexpression results in inhibition of PKCθ translocation and function. Mol Cell Biol. 1996;16:5782–5791. doi: 10.1128/mcb.16.10.5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Molkentin J D, Lu J R, Antos C L, Markham B, Richardson J, Robbins J, Grant S R, Olson E N. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell. 1998;93:215–228. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moore B W, Perez V J, editors. Specific acidic proteins of the nervous system. Englewood Cliffs, N.J: Prentice-Hall; 1967. [Google Scholar]
- 52.Munoz E, Zubiaga A M, Merrow M, Sauter N P, Huber B T. Cholera toxin discriminates between T helper 1 and 2 cells in T cell receptor-mediated activation: role of cAMP in T cell proliferation. J Exp Med. 1990;172:95–103. doi: 10.1084/jem.172.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muslin A J, Tanner J W, Allen P M, Shaw A S. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell. 1996;84:889–897. doi: 10.1016/s0092-8674(00)81067-3. [DOI] [PubMed] [Google Scholar]
- 54.Neumann M, Grieshammer T, Chuvpilo S, Kneitz B, Lohoff M, Schimpl A, Franza B R, Jr, Serfling E. RelA/p65 is a molecular target for the immunosuppressive action of protein kinase A. EMBO J. 1995;14:1991–2004. doi: 10.1002/j.1460-2075.1995.tb07191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nielsen P J. Primary structure of a human protein kinase regulator protein. Biochim Biophys Acta. 1991;1088:425–428. doi: 10.1016/0167-4781(91)90136-a. [DOI] [PubMed] [Google Scholar]
- 56.Northrop J P, Ho S N, Chen L, Thomas D J, Timmerman L A, Nolan G P, Admon A, Crabtree G R. NF-AT components define a family of transcription factors targeted in T-cell activation. Nature. 1994;369:497–502. doi: 10.1038/369497a0. [DOI] [PubMed] [Google Scholar]
- 57.Oukka M, Ho I C, de la Brousse F C, Hoey T, Grusby M J, Glimcher L H. The transcription factor NFAT4 is involved in the generation and survival of T cells. Immunity. 1998;9:295–304. doi: 10.1016/s1074-7613(00)80612-3. [DOI] [PubMed] [Google Scholar]
- 58.Patel M D, Samelson L E, Klausner R D. Multiple kinases and signal transduction: phosphorylation of the T cell antigen receptor complex. J Biol Chem. 1987;262:5831–5838. [PubMed] [Google Scholar]
- 59.Peng C Y, Graves P R, Thoma R S, Wu Z, Shaw A S, Piwnica-Worms H. Mitotic and G2 checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science. 1997;277:1501–1505. doi: 10.1126/science.277.5331.1501. [DOI] [PubMed] [Google Scholar]
- 60.Perrino B A, Fong Y L, Bricky D A, Saitoh Y, Ushio Y, Fukunaga K, Miyamoto E, Soderling T R. Characterization of the phosphatase activity of a baculovirus-expressed calcineurin A isoform. J Biol Chem. 1992;267:15965–15969. [PubMed] [Google Scholar]
- 61.Ranger A M, Grusby M J, Hodge M R, Gravallese E M, de la Brousse F C, Hoey T, Mickanin C, Baldwin H S, Glimcher L H. The transcription factor NF-ATc is essential for cardiac valve formation. Nature. 1998;392:186–190. doi: 10.1038/32426. [DOI] [PubMed] [Google Scholar]
- 62.Rao A, Luo C, Hogan P G. Transcription factors of the NFAT family: regulation and function. Annu Rev Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 63.Reuther G W, Pendergast A M. The roles of 14-3-3 proteins in signal transduction. Vitam Horm. 1996;52:149–175. doi: 10.1016/s0083-6729(08)60410-0. [DOI] [PubMed] [Google Scholar]
- 64.Rittinger K, Budman J, Xu J, Volinia S, Cantley L C, Smerdon S J, Gamblin S J, Yaffe M B. Structural analysis of 14-3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14-3-3 in ligand binding. Mol Cell. 1999;4:1–20. doi: 10.1016/s1097-2765(00)80363-9. [DOI] [PubMed] [Google Scholar]
- 65.Roy S, McPherson R A, Apolloni A, Yan J, Lane A, Clyde-Smith J, Hancock J F. 14-3-3 facilitates Ras-dependent Raf-1 activation in vitro and in vivo. Mol Cell Biol. 1998;18:3947–3955. doi: 10.1128/mcb.18.7.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ruhlmann A, Nordheim A. Effects of the immunosuppressive drugs CsA and FK506 on intracellular signalling and gene regulation. Immunobiology. 1997;198:192–206. doi: 10.1016/S0171-2985(97)80040-X. [DOI] [PubMed] [Google Scholar]
- 67.Shaw J P, Utz P J, Durand D B, Toole J J, Emmel E A, Crabtree G R. Identification of a putative regulator of early T cell activation genes. Science. 1988;241:202–205. [PubMed] [Google Scholar]
- 68.Shibasaki F, Price E R, Milan D, McKeon F. Role of kinases and phosphatase calcineurin in the nuclear shuttling of transcription factor NFAT4. Nature. 1996;382:370–373. doi: 10.1038/382370a0. [DOI] [PubMed] [Google Scholar]
- 69.Siegel M D, Zhang D H, Ray P, Ray A. Activation of the interleukin-5 promoter by cAMP in murine EL-4 cells requires the GATA-3 and CLE0 elements. J Biol Chem. 1995;270:24548–24555. doi: 10.1074/jbc.270.41.24548. [DOI] [PubMed] [Google Scholar]
- 70.Szabo S J, Gold J S, Murphy T L, Murphy K M. Identification of cis-acting regulatory elements controlling interleukin-4 gene expression in T cells: roles for NF-Y and NF-ATc. Mol Cell Biol. 1993;13:4793–4805. doi: 10.1128/mcb.13.8.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thorson J A, Yu L W, Hsu A L, Shih N Y, Graves P R, Tanner J W, Allen P M, Piwnica-Worms H, Shaw A S. 14-3-3 proteins are required for maintenance of Raf-1 phosphorylation and kinase activity. Mol Cell Biol. 1998;18:5229–5238. doi: 10.1128/mcb.18.9.5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Timmerman L A, Clipstone N A, Ho S N, Northrop J P, Crabtree G R. Rapid shuttling of NF-AT in discrimination of Ca2+ signals and immunosuppression. Nature. 1996;383:837–840. doi: 10.1038/383837a0. [DOI] [PubMed] [Google Scholar]
- 73.Tsuruta L, Lee H J, Masuda E S, Koyano-Nakagawa N, Arai N, Arai K, Yokota T. Cyclic AMP inhibits expression of the IL-2 gene through the nuclear factor of activated T cells (NF-AT) site, and transfection of NF-AT cDNAs abrogates the sensitivity of EL-4 cells to cyclic AMP. J Immunol. 1995;154:5255–5264. [PubMed] [Google Scholar]
- 74.Tzivion G, Luo Z, Avruch J. A dimeric 14-3-3 protein is an essential cofactor for Raf kinase activity. Nature. 1998;394:88–92. doi: 10.1038/27938. [DOI] [PubMed] [Google Scholar]
- 75.Uotila P. The role of cyclic AMP and oxygen intermediates in the inhibition of cellular immunity in cancer. Cancer Immunol Immunother. 1996;43:1–9. doi: 10.1007/BF03354243. [DOI] [PubMed] [Google Scholar]
- 76.Wartmann M, Davis R. The native structure of the activated Raf protein kinase is a membrane-bound multi-subunit complex. J Biol Chem. 1994;269:6695–6701. [PubMed] [Google Scholar]
- 77.Xanthoudakis S, Viola J P, Shaw K T, Luo C, Wallace J D, Bozza P T, Luk D C, Curran T, Rao A. An enhanced immune response in mice lacking the transcription factor NFAT1. Science. 1996;272:892–895. doi: 10.1126/science.272.5263.892. [DOI] [PubMed] [Google Scholar]
- 78.Xiao B, Smerdon S J, Jones D H, Dodson G G, Soneji Y, Aitken A, Gamblin S J. Structure of a 14-3-3 protein and implications for coordination of multiple signalling pathways. Nature. 1995;376:188–191. doi: 10.1038/376188a0. [DOI] [PubMed] [Google Scholar]
- 79.Yaffe M B, Rittinger K, Volinia S, Caron P R, Aitken A, Leffers H, Gamblin S J, Smerdon S J, Cantley L C. The structural basis for 14-3-3:phosphopeptide binding specificity. Cell. 1997;91:961–971. doi: 10.1016/s0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]
- 80.Zha J, Harada H, Yang E, Jockel J, Korsmeyer S J. Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-X(L) Cell. 1996;87:619–628. doi: 10.1016/s0092-8674(00)81382-3. [DOI] [PubMed] [Google Scholar]
- 81.Zhang S H, Kobayashi R, Graves P R, Piwnica-Worms H, Tonks N K. Serine phosphorylation-dependent association of the band 4.1-related protein-tyrosine phosphatase PTPH1 with 14-3-3beta protein. J Biol Chem. 1997;272:27281–27287. doi: 10.1074/jbc.272.43.27281. [DOI] [PubMed] [Google Scholar]
- 82.Zhu J, McKeon F. NF-AT activation requires suppression of Crm1-dependent export by calcineurin. Nature. 1999;398:256–260. doi: 10.1038/18473. [DOI] [PubMed] [Google Scholar]
- 83.Zhu J, Shibasaki F, Price R, Guillemot J C, Yano T, Dotsch V, Wagner G, Ferrara P, McKeon F. Intramolecular masking of nuclear import signal on NF-AT4 by casein kinase I and MEKK1. Cell. 1998;93:851–861. doi: 10.1016/s0092-8674(00)81445-2. [DOI] [PubMed] [Google Scholar]