Abstract
Background
Bacterial infections are usually suspected in infertile couples seeking IVF with no clear understanding of the microbial compositions present in the seminal fluids and vaginal niche of the patients. We used next-generation sequencing technology to correlate microbiota compositions with IVF clinical outcomes.
Methods
Thirty-six couples were recruited to provide seminal fluids and vaginal swabs. Bacterial DNA was extracted, and V4 region of the 16S rRNA was amplified and sequenced in a pair-end configuration on the Illumina MiSeq platform rendering 2 × 150 bp sequences. Microbial taxonomy to species level was generated using the Greengenes database. Linear discriminant analysis (LDA) effect size (LEfSe) was used to identify biologically and statistically significant differences in relative abundance.
Results
Seminal fluid microbiota compositions had lower bacterial concentrations compared with the vagina, but species diversity was significantly higher in seminal fluid samples. Azoospermic subjects had more relative abundance of Mycoplasma and Ureaplasma. In Normospermic semen, Lactobacillus (43.86%) was the most abundant, followed by Gardnerella (25.45%), while the corresponding vaginal samples, Lactobacillus (61.74%) was the most abundant, followed by Prevotella (6.07%) and Gardnerella (5.86%).
Conclusions
Semen samples with positive IVF were significantly colonized by Lactobacillus jensenii (P=0.002), Faecalibacterium (P=0.042) and significantly less colonized by Proteobacteria, Prevotella, Bacteroides, and lower Firmicutes/Bacteroidetes ratio compared with semen samples with negative IVF. Vaginal samples with positive IVF clinical outcome were significantly colonized by Lactobacillus gasseri, less colonized by Bacteroides and Lactobacillus iners. This study has opened a window of possibility for Lactobacillus replenishments in men and women before IVF treatment.
Keywords: seminal fluid, microbiome, in vitro fertilization - pregnancy, infertility, bacteria infections, vagina, 16S rRNA, sequencing
Introduction
Bacterial infections affecting the reproductive tracts of males and/or females with infertility have been documented in previous studies using culture methods (Eggert-Kruse et al., 1995; Gdoura et al., 2008). Some bacteria, fungi, viruses, and parasites are known to interfere with reproductive functions in both male and females of reproductive age, and infections of the genitourinary tract account for about 15% of male infertility cases. More than a few bacteria, including Lactobacillus iners, Gardnerella vaginalis, Escherichia faecalis, E. coli, and Staphylococcus aureus, have been found to be associated with male infertility as demonstrated using polymerase chain reaction (PCR) (Franasiak and Scott, 2015). Bacterial vaginosis (BV) has been found to be a major risk factor for infertility (Al-Awadhi et al., 2013). Some specific bacteria incriminated in BV, such as Atopobium vaginae, Ureaplasma vaginae, U. parvum, U. urealyticum, Gardnerella vaginalis, and reduced abundance of Lactobacillus species, are associated with infertility in women (Onemu and Ibeh, 2001). Women with endometrial dysbiosis have also been found to experience implantation failure leading to infertility (Momoh et al., 2007). While vaginal microbiota is normally under the influence of oestrogen (Ibadin and Kai, 2008), endometrial microbiota is not affected by hormonal fluctuations (Ekhaise and Richard, 2001).
There are several studies confirming that a vaginal microbiota replete with relative abundances of Lactobacillus species, devoid of bacterial vaginosis, leads to more positive IVF clinical outcomes (Nwadioha et al., 2016).
Besides other sexually transmitted infections, genital mycoplasmas are associated with poor reproductive health of women including but not limited to endometritis, cervicitis and pelvic inflammatory disease, and adverse pregnancy outcomes (Lis et al., 2015; Taylor et al., 2018). The pregnancy success rate of the various assisted reproductive health care such as in vitro fertilization (IVF) tend to be reduced as a result of prior Mycoplasma colonization of the female and male genital tract (Günyeli et al., 2011). One wonders whether genital Mycoplasma was the only pathogen associated with poor pregnancy outcome, although several studies have shown that Mycoplasma species can attach to spermatozoa and remain adherent to spermatozoa after assisted reproductive treatment washing procedures (Ibadin and Osemwenkha, 2013; Ahmadi et al., 2018).
Clinical studies have shown that bacterial contamination of the embryo transfer catheter has significant negative effect on the clinical pregnancy rates (Ikechebelu, 2003 ). Approximately 35% of infertile women are afflicted with post-inflammatory changes of the oviduct or surrounding peritoneum that interfere with tubal-ovarian function mostly as a result of infection and are likely to develop ectopic (tubal) pregnancy (Swift and Liu, 2014).
In Africa, especially Nigeria, little is known about the bacterial communities found in the seminal fluids of men seeking reproductive health care with next-generation sequencing technology. A recent pilot study of seminal fluid in a tertiary hospital revealed varying bacterial community diversities that are unique in each sample in contrast to culture-dependent methods (Ndiokwere et al., 2019). We have also previously shown that women with bacterial vaginosis (BV) had varying proportions of diverse bacteria including Lactobacillus species in all BV subjects, but the total number of all the BV-associated microbes (Gardnerella, Prevotella, Magasphaera, and others) outnumbered Lactobacillus genera (Anukam et al., 2019). In the present study, next-generation 16S rRNA gene sequencing method was used to compare seminal bacterial composition in couples seeking reproductive health care IVF. In addition, the study delineated semen quality, bacterial functional gene predictions, and correlated microbiota composition with clinical outcome of the IVF-assisted reproductive care.
Materials and Methods
Ethical Approval
The study was approved by the Ethics Review Committee on Human Research from Nnamdi Azikiwe University Teaching Hospital (Ref # NAUTH/CS/66/VOL11/175/2018/111).
Participation in the study was voluntary. Informed written consent was obtained from the patients. All methods were performed in accordance with the relevant guidelines and regulations.
Selection Criteria
Couples seeking reproductive health care at Nnamdi Azikiwe University Teaching Hospital (NAUTH), Nnewi campus, Anambra State, Nigeria, were referred to Life Fertility Center, Nnewi, for IVF/Embryo Transfer for self-cycle with cases of primary or secondary infertility after 1–12 years of uninterrupted sexual intercourse with partner.
Collection of Specimens
Two high vaginal swabs were collected by a qualified gynecologist with a non-lubricated sterile disposable plastic speculum. One of the swabs was agitated into a tube containing buffer for DNA preservation at ambient temperature, and the other was used for microscopy to detect leukocytes. Each semen sample was produced by masturbation after 5 days of sexual intercourse abstinence, and on the same day vaginal sample was collected.
Semen Analysis
The semen quality of the patients was analyzed with Semen Quality Analyzer-Vision (SQA-V) Gold (Medical Electronic Systems, USA), following the manufacturer’s procedural instructions.
Extraction of Bacterial DNA From Vaginal Swabs/Semen Samples and Sequencing of the Amplified 16S rRNA Region
Bacterial DNA was extracted from the vaginal swabs/semen samples using a protocol developed by uBiome Inc. Briefly, samples were lysed using bead-beating, and DNA was extracted in a class 1,000 clean room by a guanidine thiocyanate silica column-based purification method using a liquid-handling robot. PCR amplification of the 16S rRNA genes was performed with primers containing universal primers amplifying the V4 region (515F: GTGCCAGCMGCCGCGGTAA and 806R: GGACTACHVGGGTWTCTAAT). In addition, the primers contained Illumina tags and barcodes. Samples were barcoded with a unique combination of forward and reverse indexes allowing for simultaneous processing of multiple samples. PCR products were pooled, column-purified, and size-selected through microfluidic DNA fractionation. Consolidated libraries were quantified by quantitative real-time PCR using the Kapa Bio-Rad iCycler qPCR kit on a BioRad MyiQ before loading into the sequencer. Sequencing was performed in a pair-end modality on the Illumina NextSeq 500 platform rendering 2 × 150 bp pair-end sequences (Almonacid et al., 2017).
16S rRNA Sequence Analysis
Raw sequence reads were demultiplexed using Illumina’s BCL2FASTQ algorithm. Reads were filtered using an average Q-score >30. The paired-end sequence FASTQ reads were imported into MG-RAST pipeline for quality check (QC). EzBiocloud Microbiome Taxonomic Profile (MTP) pipeline (Yoon et al., 2017) was employed for alpha and beta diversity estimation using PKSSU4.0 version database and Open reference UCLUST_MC2 for OTUs picking at 97% cutoff. Sequences were prescreened using QIIME-UCLUST algorithms for at least 97% identity to ribosomal sequences from the RNA databases (Quast et al., 2013). Rarefication to 1,000 reads per sample was employed to calculate microbial diversity. Alpha diversity was calculated for species richness by Abundance Coverage Estimate (ACE), Chao1 and Jackknife method, while diversity indexes were calculated by Shannon, Non-parametric Shannon, and Simpson index. Principal coordinate analysis (PCoA) with Jensen-Shannon divergence distance metrices were used to evaluate beta diversity between vaginal and semen samples (Koren et al., 2013). Linear discriminant analysis (LDA) effect size (LEfSe) (Segata et al., 2011) was used to identify biologically and statistically significant differences in the OTU relative abundance. Phylogenetics Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) was used to predict the metabolic function of the metagenomes from the 16S rRNA gene dataset (Langille et al., 2013) with reference to Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthologs categorizations (Kanehisa et al., 2014).
Availability of Data and Materials
The datasets used and or analyzed in the current study are available from the corresponding author on reasonable request.
Results
Demographic Information, IVF Clinical Outcome, and Semen Quality
As shown in Table 1A / Table 1B , of the 36 men that were examined for semen quality and that had result for 16S rRNA gene sequencing, 11 were clinically diagnosed as having secondary infertility with duration of infertility ranging from 1 to 13 years, while 25 men were diagnosed with primary infertility, had duration of infertility from 1 to 8 years. The semen characteristics of the subjects were as follows: 11 samples were assigned as normospermia [>15×10 (6)], 7 had oligospermia [<15×10 (6)], 7 had azoospermia, 10 had asthenozoospermia, while 1 sample was classified as tetratozoospermia. The number of semen samples that had leukocytes (pyospermia) were 11, while 1 sample was assigned as oligoasthenozoospermia.
Table 1A.
Demographic information and IVF clinical outcome.
| Sample No. | Female age (years) | Clinical pregnancy outcome | Scan result | Sex of the baby | Live delivery | Male age (years) | Duration of marriage (years) | Duration of infertility (years) | Type of infertility |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 26–30 | Negative | – | - | 31–35 | 6 | 6 | Primary | |
| 2 | 36–40 | Negative | – | - | 41–45 | 10 | 8 | Secondary | |
| 3 | 36–40 | Negative | – | - | 46–50 | 6 | 6 | Primary | |
| 4 | 31–35 | Negative | – | - | 36–40 | 10 | 10 | Primary | |
| 5 | 26–30 | Negative | – | - | 36–40 | 5 | 5 | Primary | |
| 6 | 26–30 | Negative | – | - | 26–30 | 4 | 4 | Primary | |
| 7 | 36–40 | POSITIVE | Singleton | Male | YES | 46–50 | 9 | 8 | Secondary |
| 8 | 31–35 | Negative | – | – | 31–35 | 3 | 3 | Primary | |
| 9 | 21–25 | Negative | – | – | 26–30 | 4 | 4 | Primary | |
| 10 | 36–40 | POSITIVE | Singleton | Female | YES | 36–40 | 5 | 5 | Primary |
| 11 | 41–45 | POSITIVE | Twins | Male, Female | YES | 46–50 | 3 | 2 | Secondary |
| 12 | 36–40 | Negative | – | – | 41–45 | 4 | 4 | Primary | |
| 13 | 26–30 | Negative | – | – | 26–30 | 1 | 1 | Primary | |
| 14 | 31–35 | POSITIVE | Singleton | Male | YES | 36–40 | 2 | 2 | Primary |
| 15 | 26–30 | Negative | – | – | 26–30 | 5 | 3 | Secondary | |
| 16 | 26–30 | POSITIVE | Singleton | Male | YES | 31–35 | 2 | 2 | Primary |
| 17 | 26–30 | POSITIVE | Singleton | Male | YES | 31–35 | 3 | 3 | Primary |
| 18 | 41–45 | Negative | – | – | 46–50 | 7 | 6 | Secondary | |
| 19 | 31–35 | POSITIVE | Twins | Male, Female | YES | 41–45 | 2 | 2 | Primary |
| 20 | 26–30 | Negative | – | – | 31–35 | 6 | 6 | Primary | |
| 21 | 36–40 | Negative | – | – | 46–50 | 9 | 9 | Secondary | |
| 22 | 31–35 | Negative | – | – | 41–45 | 4 | 1 | Secondary | |
| 23 | 36–40 | POSITIVE | Twins | Male, Female | YES | 46–50 | 12 | 6 | Secondary |
| 24 | 31–36 | Negative | – | - | 31–35 | 5 | 5 | Primary | |
| 25 | 26–30 | POSITIVE | Singleton | Female | No (Misca) | 31–35 | 3 | 3 | Primary |
| 26 | 41–45 | Negative | – | – | 51–55 | 2 | 2 | Primary | |
| 27 | 31–35 | POSITIVE | Singleton | Male | YES | 36–40 | 6 | 6 | Primary |
| 28 | 31–35 | Negative | – | – | 31–35 | 2 | 2 | Primary | |
| 29 | 31–35 | Negative | – | – | 31–35 | 4 | 4 | Primary | |
| 30 | 31–35 | Negative | – | – | 36–40 | 5 | 4 | Secondary | |
| 31 | 46–50 | Negative | – | – | 56–60 | 13 | 13 | Primary | |
| 32 | 36–40 | Negative | – | – | 46–50 | 6 | 6 | Primary | |
| 33 | 41–45 | POSITIVE | Singleton | Male | YES | 51–55 | 5 | 5 | Primary |
| 34 | 36–40 | Negative | – | – | 51–55 | 10 | 7 | Secondary | |
| 35 | 31–35 | POSITIVE | Singleton | Female | YES | 41–45 | 4 | 1 | Secondary |
| 36 | 36–40 | Negative | – | - | 36–40 | 6 | 6 | Primary |
Table 1B.
Semen Characteristics.
| Sample # | Days of abstinence | Viscosity | Liquefaction | Volume (ml) | Progressive motility (%) | Non-progressive motility (%) | Non-motile (%) | Total motility (%) | Velocity (mic/s) | Sperm conc (m/ml) | Total sperm/volume (million) | Presence of pus cells |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 3 | NORMAL | NORMAL | 2 | 9 | 3 | 88 | 12 | 32 | 88 | 176.1 | + |
| 2 | 5 | NORMAL | NORMAL | 2.5 | 5 | 2 | 93 | 7 | 27 | 22.2 | 55.4 | NIL |
| 3 | 4 | ABNORMAL | ABNORMAL | 1.2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2+ |
| 4 | 3 | NORMAL | NORMAL | 2.1 | 2 | 9 | 89 | 11 | 0 | 2 | 0 | NIL |
| 5 | 3 | NORMAL | NORMAL | 1.2 | 27 | 6 | 67 | 33 | 41 | 120.5 | 144.6 | NIL |
| 6 | 4 | ABNORMAL | ABNORMAL | 1.2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2+ |
| 7 | 3 | NORMAL | NORMAL | 2.8 | 37 | 10 | 53 | 47 | 38 | 77.4 | 216.7 | NIL |
| 8 | 3 | NORMAL | NORMAL | 1 | 44 | 8 | 48 | 52 | 43 | 72 | 72 | NIL |
| 9 | 4 | NORMAL | NORMAL | 3 | 22 | 10 | 68 | 32 | 30 | 33.8 | 101.4 | NIL |
| 10 | 5 | NORMAL | NORMAL | 1.5 | 39 | 8 | 53 | 47 | 42 | 194.5 | 291.8 | NIL |
| 11 | 3 | NORMAL | NORMAL | 2.3 | 51 | 12 | 37 | 63 | 39 | 52.7 | 121.3 | NIL |
| 12 | 5 | NORMAL | NORMAL | 0.8 | 17 | 3 | 80 | 20 | 46 | 207.6 | 166.1 | NIL |
| 13 | 4 | NORMAL | NORMAL | 2 | 52 | 20 | 28 | 72 | 32 | 6.4 | 12.8 | NIL |
| 14 | 4 | ABNORMAL | ABNORMAL | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3+ |
| 15 | 3 | NORMAL | NORMAL | 3.2 | 0 | 1 | 99 | 1 | 0 | 16 | 51.1 | NIL |
| 16 | 5 | NORMAL | NORMAL | 2.2 | 35 | 8 | 57 | 43 | 42 | 77.2 | 169.8 | NIL |
| 17 | 3 | NORMAL | NORMAL | 1.5 | 11 | 7 | 82 | 18 | 25 | 66.4 | 99.7 | NIL |
| 18 | 3 | ABNORMAL | ABNORMAL | 2.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2+ |
| 19 | 4 | NORMAL | NORMAL | 1.5 | 30 | 6 | 62 | 38 | 43 | 163.3 | 244.9 | NIL |
| 20 | 3 | ABNORMAL | ABNORMAL | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2+ |
| 21 | 4 | NORMAL | NORMAL | 3.2 | 46 | 9 | 40 | 60 | 44 | 50.3 | 160.9 | NIL |
| 22 | 5 | ABNORMAL | ABNORMAL | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2+ |
| 23 | 3 | NORMAL | NORMAL | 2 | 50 | 4 | 84 | 16 | 34 | 157 | 314 | 2+ |
| 24 | 3 | NORMAL | NORMAL | 3.2 | 47 | 10 | 40 | 60 | 45 | 61.6 | 197.2 | NIL |
| 25 | 3 | ABNORMAL | ABNORMAL | 2.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2+ |
| 26 | 4 | NORMAL | NORMAL | 2 | 5 | 24 | 29 | 29 | 5 | 4.7 | 9.4 | NIL |
| 27 | 3 | NORMAL | NORMAL | 2.4 | 3 | 2 | 95 | 5 | 22 | 75.3 | 180 | NIL |
| 28 | 4 | NORMAL | NORMAL | 2 | 40 | 14 | 46 | 54 | 34 | 44.6 | 89.2 | NIL |
| 29 | 3 | NORMAL | NORMAL | 1 | 3 | 2 | 95 | 5 | 22 | 72.2 | 72.2 | NIL |
| 30 | 5 | NORMAL | NORMAL | 3 | 8 | 19 | 73 | 27 | 10 | 5.6 | 16.9 | 3+ |
| 31 | 3 | NORMAL | NORMAL | 3.2 | 1 | 11 | 88 | 12 | 4 | 11.3 | 36 | NIL |
| 32 | 4 | ABNORMAL | ABNORMAL | 2.1 | 39 | 22 | 39 | 61 | 26 | 5.1 | 10.8 | 2+ |
| 33 | 5 | NORMAL | NORMAL | 1.5 | 27 | 6 | 67 | 33 | 40 | 77.6 | 116.4 | NIL |
| 34 | 4 | NORMAL | NORMAL | 1 | 20 | 5 | 75 | 25 | 37 | 134.1 | 134.1 | NIL |
| 35 | 3 | NORMAL | NORMAL | 2.5 | 29 | 9 | 62 | 38 | 34 | 80.8 | 97 | NIL |
| 36 | 5 | NORMAL | NORMAL | 3 | 44 | 7 | 49 | 51 | 45 | 83.8 | 251.5 | NIL |
Key: NIL represents absence of pus cells.
Semen and Vagina Microbiome Compositions
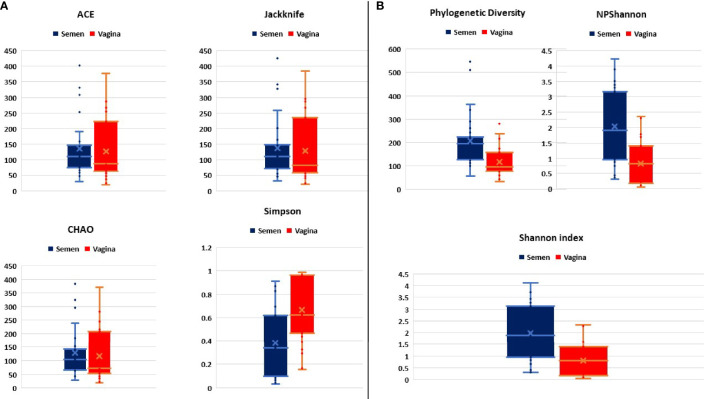
The first objective was to determine whether seminal fluid microbiota differ substantially with the vagina. In this regard, the alpha diversity that estimates the species richness typified by ACE, CHAO Jackknife, Shannon, and Simpson shows that seminal fluid microbiota composition is less in species richness (lower bacterial concentrations) compared with the vagina as shown in Figure 1A .
Figure 1.
(A, B) Comparative alpha diversities between semen and vagina showing species richness typified by ACE, CHAO Jackknife, Shannon, and Simpson index.
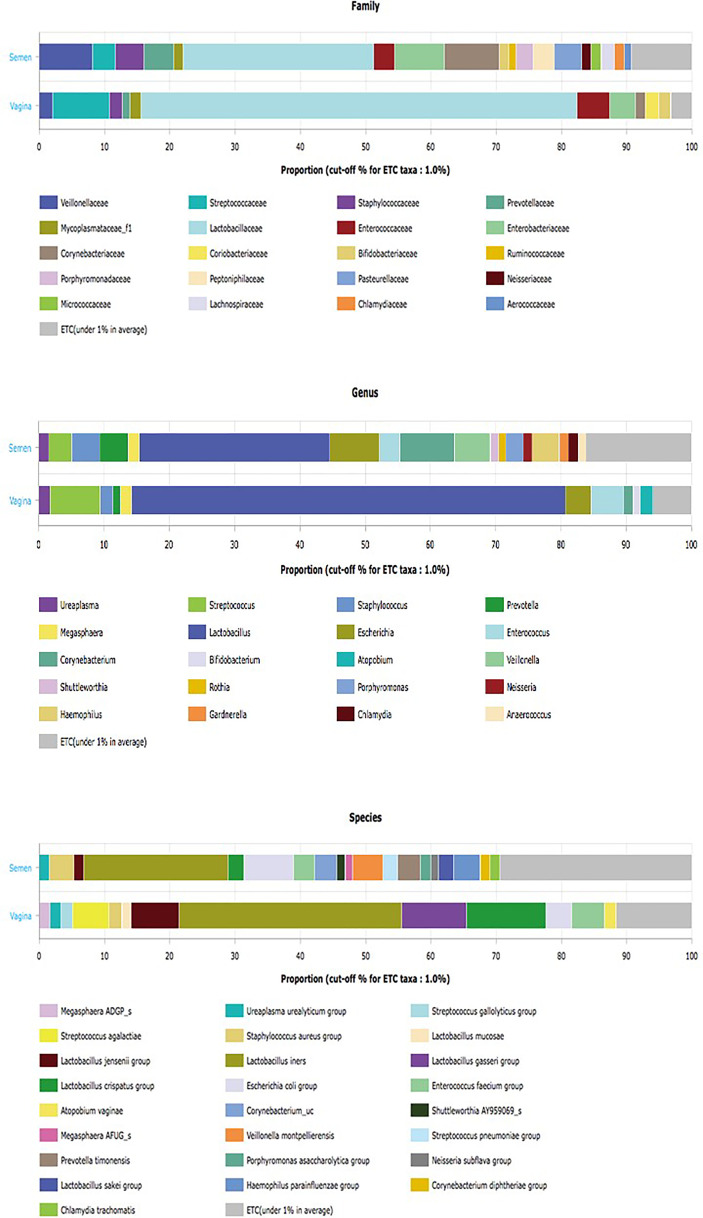
However, species diversity was significantly higher in seminal fluid samples represented in Figure 1B . The Principal Coordinate Analysis (PCoA) with Bray-Curtis metrices confirmed the differences in bacterial community diversities between semen and vagina as shown in Supplementary Figure S1 . The EzBiocloud Microbiome Taxonomic Profile (MTP) pipeline was able to provide distinct taxonomic categories (Family, Genus, and Species) at 1% cutoff between semen and vagina as shown in Figure 2 .
Figure 2.
EzBiocloud Microbiome Taxonomic Profile (MTP) pipeline showing distinct taxonomic categories (Family, Genus, and Species) at 1% cutoff between semen and vagina.
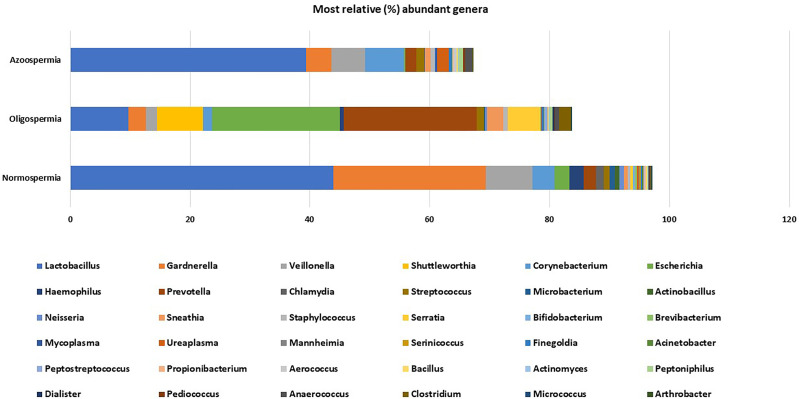
The rarefaction curve showing the number of reads between the semen samples and vaginal microbiota is presented in Supplementary Figure S2 . For comparative purposes, we selected the corresponding semen and vagina bacterial communities in line with the three semen categories. At the genera taxonomic level, out of 621 genera identified, 6.8% were exclusive to normospermia, 20.5% to oligospermia, and 2.9% exclusive to azoospermia, while 41.5% were common to all the categories. When the genera taxa are examined with the three semen categories, Mycoplasma and Ureaplasma occurred more in relative abundance in azoospermic subjects ( Figure 3 ).
Figure 3.
Most relative (%) genera abundance between the semen categories.
At the species taxonomic level, out of 1,384 species identified, 10.3% were exclusive to normospermia, 26.4% to oligospermia, and 6.8% exclusive to azoospermia, while 27% were common to all the categories as shown in Supplementary Figure S3 .
Microbiota Compositions From Normospermia Couples
Twenty-four phyla were identified from subjects with normospermia, while 22 were identified from the corresponding vagina samples. Firmicutes accounted for 54.47 vs 75.89% relative abundance, followed by Actinobacteria (32.26 vs 8.72%), Proteobacteria (8.60 vs 7.20%), Bacteroidetes (2.29 vs 5.95%), Chlamydiae (1.32 vs 0.0003%), Fusobacteria (0.67 vs 2.09%), Tenericutes (0.30 vs 0.09%), and others represented in Supplementary Figure S4 . At the genera taxonomic level, 451 genera were identified in normospermia samples, while 331 genera were found in the corresponding females. Interestingly, 282 genera were shared between couples. Lactobacillus (43.86%) was the most abundant genera in semen/vagina, followed by Gardnerella (25.45%), Veillonella (7.78%), Corynebacterium (3.73%), Escherichia (2.47%), Haemophilus (2.36%), Prevotella (2.03%), and others, while in the corresponding vagina samples, Lactobacillus (61.74%) was the most abundant genera, followed by Prevotella (6.07%), Gardnerella (5.86%), Streptococcus (5.84%), Escherichia (5.40%), Megasphaera (4.51%), Sneathia (2.13%), and others represented in Supplementary Figure S5 .
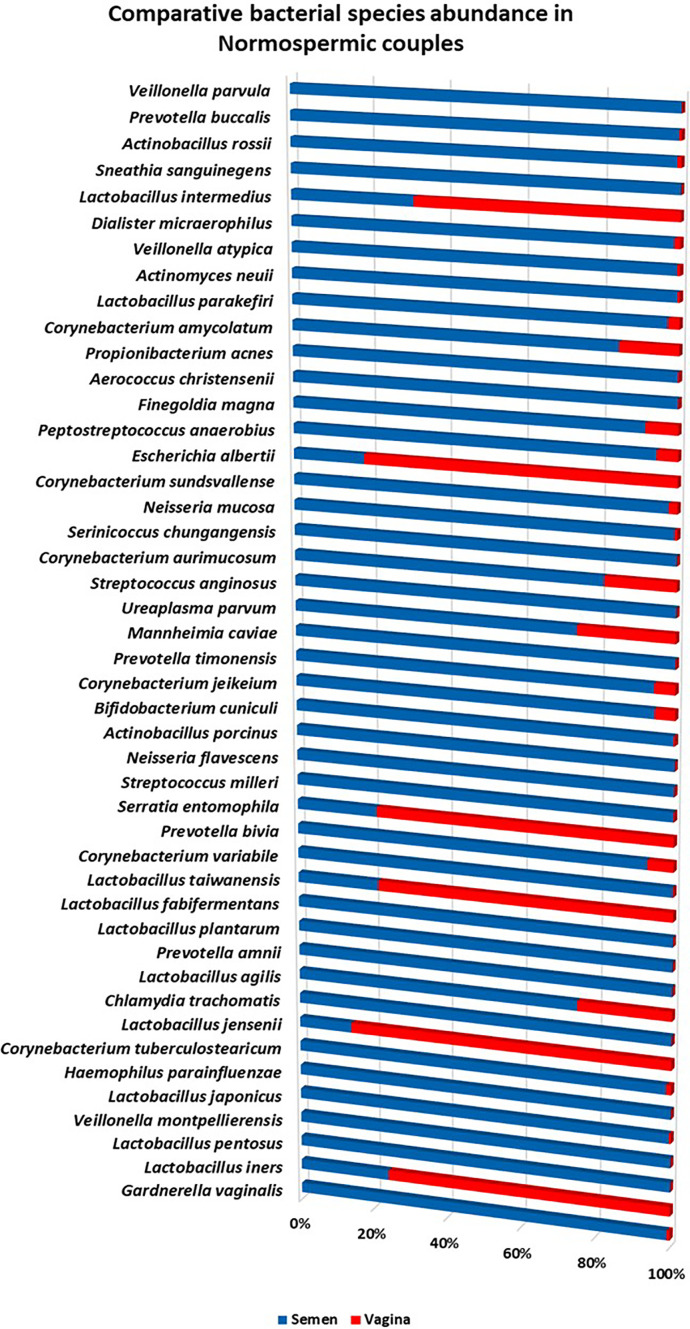
At the species taxonomic level, 848 species were identified in normospermia samples, while 585 species were found in the corresponding vagina samples. Gardnerella vaginalis (31.93%) was the most abundant species identified in 10/12 of the semen samples, followed by Lactobacillus iners 8/12 (15.56%), Lactobacillus pentosus 1/12 (12.39%), Veillonella montpellierensis 8/12 (9.61%), Lactobacillus japonicus 3/12 (4.29%), Haemophilus parainfluenzae 9/12 (2.91%), Corynebacterium tuberculostearicum 11/12 (1.75%), Lactobacillus jensenii 3/12 (1.73%), and others. The corresponding vaginal samples had Lactobacillus iners 9/12 (49.06%) as the most abundant species, followed by Lactobacillus jensenii 2/12 (10.60%), Peptostreptococcus stomatis 6/12 (6.92%), Actinocatenispora silicis 5/12 (6.19%), Pasteurella pneumotropica 2/12 (4.09%), Actinomyces naturae 4/12 (3.32%), Lactobacillus taiwanensis 10/12 (2.78%), Peptoniphilus asaccharolyticus 8/12 (2.69%) and others as shown in Figure 4 .
Figure 4.
Comparative relative (%) species abundance in normospermia couples.
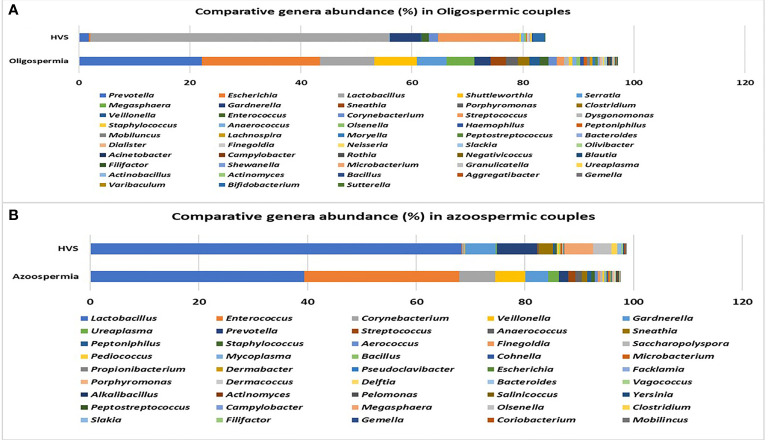
Microbiota Compositions From Oligospermia Couples
Among the oligospermic couples, 555 genera were identified in semen samples, while 403 genera were found in the corresponding vaginal samples. Prevotella (22.13%) was the most abundant genus in oligospermic semen, followed by Escherichia (21.33%), Lactobacillus (9.73%), Shuttleworthia (7.67%), Serratia (5.33%), Megasphaera (5.04%), Gardnerella (2.85%), Sneathia (2.79%), Porphyromonas (2.22%), and others. The corresponding vaginal samples had Lactobacillus (53.90%) as the most abundant genus, followed by Streptococcus (14.68%), Gardnerella (5.48%), and others as shown in Figure 5A .
Figure 5.
(A, B) Comparative relative (%) genera abundance in oligospermia and azoospermia couples.
Microbiota Compositions From Azoospermia Couples
The results from azoospermic couples show that 342 genera colonized the semen samples, while 309 genera were found in the corresponding vaginal samples. Lactobacillus (39.38%) was the most abundant genus in azoospermic men, followed by Enterococcus (28.52%), Corynebacterium (6.58%), Veillonella (5.53%), Gardnerella (4.25%), Ureaplasma (1.91%), Prevotella (1.82%), and others. The corresponding vaginal samples were colonized mostly by Lactobacillus (68.38%), Prevotella (7.37%), Gardnerella (5.69%), Megasphaera (5.23%), Olsenella (3.41%), Sneathia (2.69%), and others represented in Figure 5B .
Microbiota Compositions From Semen Samples With Leukocytes
We compared the microbiota compositions of nine semen samples with leukocytes (2+ to 3++) and nine semen samples without the presence of leukocytes. Semen samples with leukocytes tend to be significantly less colonized by Lactobacillus reuteri group, Faecalibacterium, and more inhabited by Bacteroides and Prevotella ( Supplementary Figure S6 ). The corresponding microbiota at genera level from vaginal samples is presented in Supplementary Figure S7 showing Lactobacillus and Gardnerella as the most relative abundance. At the species taxonomic level, semen samples with leukocytes had more relative abundance of Lactobacillus iners and Enterococcus faecium compared with semen without leukocytes as shown in Supplementary Figure S8 . The corresponding female partners had Lactobacillus iners as the most abundant species as shown in Supplementary Figure S9 . Bacterial metabolic functional genes that were downregulated in the semen samples with leukocytes include but not limited to hemoglobin/transferrin/lactoferrin receptor protein, MFS transporter, OPA family, sugar phosphate sensor protein UhpC as presented in Supplementary Table S1 .
Microbiota Compositions in Couples With Positive and Negative IVF Clinical Outcome
This study compared the relative abundance of the microbiota in 12 couples with positive IVF clinical outcome and 24 couples with unsuccessful or negative IVF clinical outcome as shown in Tables 1A , 1B .
Table 2.
LefSe comparison of the taxonomic biomarkers in the seminal fluid of men that had positive IVF clinical outcome and seminal fluid of those with negative IVF clinical outcome.
| Taxon name | LDA effect size | p-value | p-value (FDR) | Semen positive IVF clinical outcome | Semen negative IVF clinical outcome |
|---|---|---|---|---|---|
| Cutibacterium | 3.50568 | 0.00034 | 0.00034 | 0.76044 | 0.12494 |
| Cutibacterium acnes group | 3.45684 | 0.00067 | 0.00067 | 0.66262 | 0.09221 |
| Propionibacteriales | 3.48056 | 0.00068 | 0.00068 | 0.82203 | 0.23874 |
| Propionibacteriaceae | 3.49112 | 0.00068 | 0.00068 | 0.80718 | 0.21312 |
| Salinifilum | 3.70772 | 0.01579 | 0.01584 | 1.02006 | 0.00000 |
| Leptotrichiaceae | 3.12386 | 0.02997 | 0.03009 | 0.00000 | 0.26525 |
| Facklamia hominis group | 2.60266 | 0.02997 | 0.03011 | 0.00000 | 0.07949 |
| Lactobacillus_uc | 2.72540 | 0.03372 | 0.03390 | 0.01318 | 0.11892 |
| Actinobacteria_c | 4.76455 | 0.03899 | 0.03923 | 22.04688 | 10.41718 |
| Micrococcales | 2.99379 | 0.04751 | 0.04783 | 3.00235 | 2.80542 |
| Streptomycetales | 2.92289 | 0.04825 | 0.04861 | 0.16874 | 0.00170 |
| Streptomycetaceae | 2.92289 | 0.04825 | 0.04865 | 0.16874 | 0.00170 |
| Streptomyces | 2.92289 | 0.04825 | 0.04868 | 0.16874 | 0.00170 |
| Cutibacterium avidum | 2.44565 | 0.04825 | 0.04871 | 0.05682 | 0.00568 |
| Saccharibacteria_TM7 | 2.20252 | 0.04825 | 0.04874 | 0.00000 | 0.03151 |
| Saccharimonas_c | 2.20252 | 0.04825 | 0.04878 | 0.00000 | 0.03151 |
| Saccharimonas_o | 2.20252 | 0.04825 | 0.04881 | 0.00000 | 0.03151 |
| Saccharimonas_f | 2.20252 | 0.04825 | 0.04885 | 0.00000 | 0.03151 |
| Prevotella_uc | 2.60734 | 0.04825 | 0.04888 | 0.00000 | 0.08053 |
| Actinomyces europaeus group | 2.45905 | 0.04825 | 0.04891 | 0.00000 | 0.05622 |
| Streptococcus_uc | 2.02046 | 0.04825 | 0.04895 | 0.00000 | 0.01467 |
FDR, False Discovery Rate; LDA, Linear Discriminant Analysis.
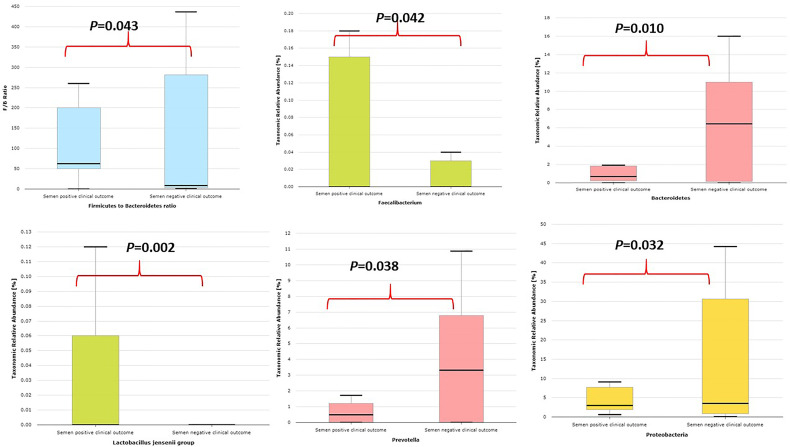
Semen samples with positive IVF clinical outcome have less alpha diversity as typified by Shannon index and phylogenetic diversity ( Supplementary Figure S10 ) and are significantly colonized by Lactobacillus jensenii group and Faecalibacterium and significantly less colonized by Proteobacteria taxa, Prevotella, Bacteroidetes taxa, and Bacteroides and lower Firmicutes/Bacteroidetes ratio compared with semen samples with negative IVF clinical outcome as shown in Figure 6 .
Figure 6.
Comparative relative abundance of some selected taxa showing significant difference between semen samples with positive IVF clinical outcome and samples with negative IVF clinical outcome.
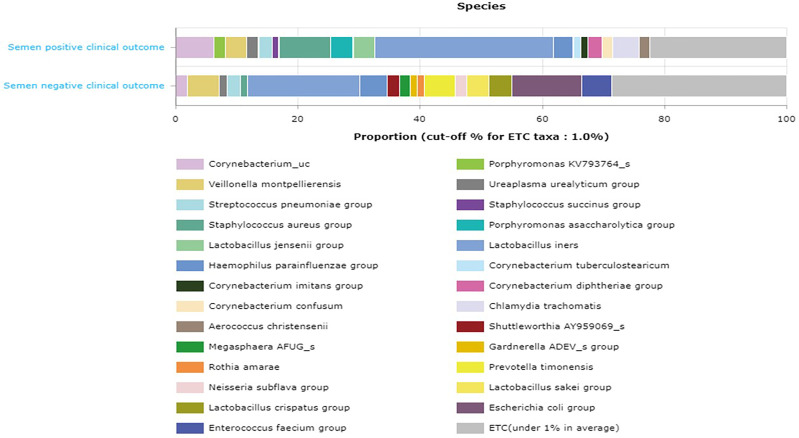
The comparative proportion of the relative abundance at the species taxonomic level is represented in Figure 7 .
Figure 7.
Comparative proportion (cutoff 1.0%) of the relative abundance at the species taxonomic level between semen samples with positive IVF clinical outcome and samples with negative IVF clinical outcome.
LefSe comparison of the taxonomic microbiota biomarkers in the seminal fluid of men that had positive IVF clinical outcome and seminal fluid of those with negative IVF clinical outcome is presented in Table 2 .
In addition, the LefSe comparison of the taxonomic microbiota biomarkers for positive IVF clinical outcome for the couples is presented in Supplementary Table S2 .
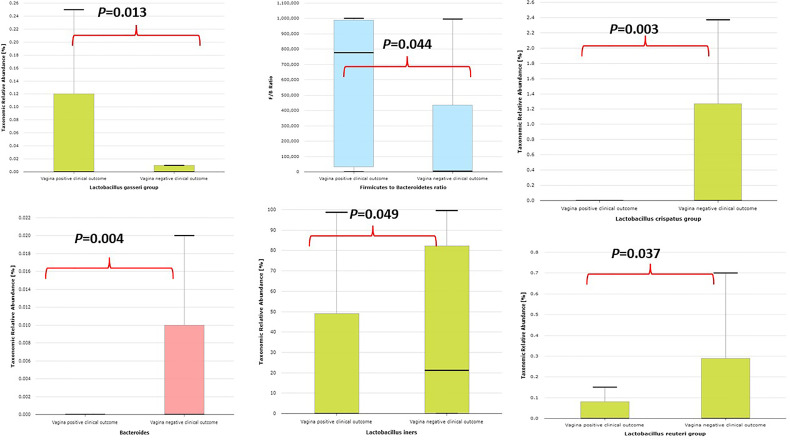
Vaginal samples with positive IVF clinical outcome are significantly colonized by Lactobacillus gasseri group, higher Firmicutes/Bacteroidetes ratio, and significantly less colonized by Bacteroides, Bacteroidetes taxa, Lactobacillus reuteri group, Lactobacillus iners, and Lactobacillus crispatus as represented in Figure 8 .
Figure 8.
Comparative relative abundance of some selected taxa showing significant difference between vaginal samples with positive IVF clinical outcome and vaginal samples with negative IVF clinical outcome.
LefSe comparison of the taxonomic microbiota biomarkers in the vaginal samples of women that had positive IVF clinical outcome and vaginal samples of those with negative IVF clinical outcome is presented in Table 3 .
Table 3.
LefSe comparison of the taxonomic biomarkers in the vaginal swabs of women that had positive IVF clinical outcome and vaginal swabs of women with negative IVF clinical outcome.
| Taxon name | LDA effect size | p-value | p-value (FDR) | Vagina positive IVF clinical outcome | Vagina negative IVF clinical outcome |
|---|---|---|---|---|---|
| Bacteroidetes | 3.95843 | 0.00783 | 0.00783 | 0.01044 | 1.82702 |
| Bacteroidia | 3.95832 | 0.00889 | 0.00890 | 0.00978 | 1.82624 |
| Bacteroidales | 3.95832 | 0.00889 | 0.00891 | 0.00978 | 1.82624 |
| Prevotellaceae | 3.95382 | 0.01060 | 0.01064 | 0.00267 | 1.80030 |
| Prevotella | 3.95366 | 0.01060 | 0.01066 | 0.00267 | 1.79968 |
| Citrobacter | 2.17062 | 0.01579 | 0.01589 | 0.02942 | 0.00000 |
| Citrobacter koseri | 2.17062 | 0.01579 | 0.01591 | 0.02942 | 0.00000 |
| Cutibacterium acnes group | 3.04732 | 0.01822 | 0.01839 | 0.00000 | 0.22235 |
| Enterobacterales_uc | 2.34217 | 0.01864 | 0.01884 | 0.04385 | 0.00008 |
| Prevotella timonensis | 3.79064 | 0.01866 | 0.01888 | 0.00224 | 1.23701 |
| Cutibacterium | 3.06412 | 0.02560 | 0.02594 | 0.00038 | 0.23154 |
| Streptococcus agalactiae | 4.38488 | 0.03698 | 0.03767 | 8.66950 | 3.81779 |
| Veillonella | 2.28915 | 0.04785 | 0.04881 | 0.02708 | 0.06579 |
| Fusobacteria | 2.40327 | 0.04825 | 0.04928 | 0.00000 | 0.05037 |
| Fusobacteria_c | 2.40327 | 0.04825 | 0.04935 | 0.00000 | 0.05037 |
| Fusobacteriales | 2.40327 | 0.04825 | 0.04942 | 0.00000 | 0.05037 |
| Mobiluncus | 2.52442 | 0.04825 | 0.04962 | 0.00000 | 0.06573 |
| Mobiluncus curtisii group | 2.51681 | 0.04825 | 0.04968 | 0.00000 | 0.06468 |
| Veillonella dispar | 2.37510 | 0.04825 | 0.04975 | 0.00000 | 0.04684 |
FDR, False Discovery Rate; LDA, Linear Discriminant Analysis.
The results from PICRUSt indicated that some bacterial metabolic functional genes were upregulated in the semen of men who had positive IVF clinical outcome. For example, bacterial metabolic functional gene orthologs for phenylalanyl-tRNA synthetase beta chain was significantly upregulated (P=0.0231) with LDA effect size of 2.0847, when compared with bacterial metabolic genes from seminal fluid of men with negative IVF clinical outcome. Other several bacterial metabolic functional gene orthologs that were significantly upregulated include but not limited to methionyl aminopeptidase (P=0.0231), peptide/nickel transport system permease protein (P=0.0132), chaperonin GroEL (P=0.0286), glucose-6-phosphate 1-dehydrogenase (P=0.0374), mycothione reductase (P=0.0318), multicomponent Na+:H+ antiporter subunits D, E, G, A, F, and C ( Supplementary Table S3 ).
Similarly, bacterial metabolic functional genes in the vagina of women with positive IVF clinical outcome were upregulated. Notably, iron/zinc/manganese/copper transport system permease protein (P=0.0326) and diphosphoinositol-polyphosphate diphosphatase (P=0.0369) had a twofold increase with positive IVF clinical outcome when compared with negative IVF clinical outcome ( Table 4 ).
Table 4.
Bacterial metabolic functional gene orthologs in the vagina of women with positive IVF clinical outcome and women with negative IVF clinical outcome.
| Gene Ortholog | Definition | LDA effect size | p-value | p-value (FDR) | Vagina positive IVF clinical outcome | Vagina negative IVF clinical outcome |
|---|---|---|---|---|---|---|
| K21572 | starch-binding outer membrane protein, SusD/RagB family | 2.053646 | 0.006891 | 0.006898 | 0.000183145 | 0.022612675 |
| K21573 | TonB-dependent starch-binding outer membrane protein SusC | 1.836336 | 0.006891 | 0.006899 | 0.000123226 | 0.013643597 |
| K01277 | dipeptidyl-peptidase III | 1.106637 | 0.007833 | 0.007843 | 1.51541E-05 | 0.00237178 |
| K00895 | diphosphate-dependent phosphofructokinase | 0.934877 | 0.010441 | 0.010463 | 5.83556E-05 | 0.001579854 |
| K16363 | UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase/3-hydroxyacyl-[acyl-carrier-protein] dehydratase | 0.903303 | 0.024279 | 0.024405 | 1.06454E-05 | 0.001411429 |
| K11705 | iron/zinc/manganese/copper transport system permease protein | 1.112538 | 0.032613 | 0.032839 | 0.004644898 | 0.002253295 |
| K07766 | diphosphoinositol-polyphosphate diphosphatase | 1.112288 | 0.036982 | 0.037275 | 0.004642652 | 0.002252543 |
FDR, False Discovery Rate; LDA, Linear Discriminant Analysis.
Discussion
To our knowledge, this is the first study from Nigeria that utilized next-generation sequencing (NGS) technology to determine the microbiota compositions of the seminal fluids and vaginal swabs from couples seeking assisted reproductive health care. This study showed a higher percentage of primary infertility (69.0%) when compared with subjects who had secondary infertility (31.0%). The high rate of primary infertility in this study was in agreement with the previous results of Ikechebelu et al. (Ikechebelu, 2003; Ikechebelu et al., 2003).
Clinical pregnancy determined with ultrasound scan showed 33% (12/36) positive IVF outcomes, which led to 91.6% (11/12) live delivery, and only 8.3% (1/12) clinical pregnancy loss (miscarriage) occurred. This clinical pregnancy rate (33%) is similar to our previous study (Ikechebelu et al., 2016) and very close to the overall pregnancy rate (36%) as reported by Haahr et al. (2019). Among couples diagnosed with secondary infertility, only 11% (4/36) had positive clinical IVF outcome, compared with 22% (8/36) of couples with primary infertility. The 16S rRNA results, when taken together, revealed that the semen microbiota is highly polymicrobial, typified by alpha diversity indexes such as Shannon index and phylogenetic diversity, but low in species concentrations, as shown in a similar study by Mandar et al. (2015). This finding was consistent with our previous pilot study of seminal fluids in a tertiary hospital that revealed varying bacterial diversities that are unique in each sample in contrast to culture-dependent methods (Ndiokwere et al., 2019). The origin of these diverse bacteria in seminal fluids is still not fully determined, but interestingly, most of the bacterial species are closely related to human vaginal microbes (Ravel et al., 2010; Anukam et al., 2019; Okoli et al., 2019), urine microbiota (Nelson et al., 2010), and urethra (Riemersma et al., 2003). In contrast, the vaginal microbiota had higher species concentrations with less bacterial diversity. The family taxa Lactobacillaceae and the Lactobacillus genus were significantly higher in the vagina than in the semen (61.74 vs 43.86%), which is consistent with several studies showing that the healthy vagina is colonized by Lactobacilli that help to fend off pathogenic microbes by increasing the pH and preventing urogenital infections (Amabebe and Anumba, 2018). However, previous studies have shown that Lactobacillus is part of a normal microbiota of the seminal fluid in healthy subjects (Ivanov et al., 2009 ). It is noteworthy that couples shared many of the predominant genera (56%) and some species (41%) in their reproductive tracts, such as Gardnerella vaginalis, Lactobacillus iners, Lactobacillus japonicus, Lactobacillus jensenii, and Lactobacillus agilis, which suggests that a healthy vagina or vagina with dysbiosis could influence the reproductive tract microbiota composition of the sexual partner, and vice versa. Similar observations have been documented on the skin microbiome of cohabiting couples (Ross et al., 2017). Another interesting finding showed that couples have the same genera but different species, though this is not surprising as the physiological condition of the vagina is acidic while semen is alkaline. This study shows that Gardnerella (25.45 vs 5.86%) and Veillonella (7.78 vs 0.04%) were more in abundance in the seminal fluids with normospermia than the corresponding vagina microbiota, although Gardnerella vaginalis has severally been associated with bacterial vaginosis (Anukam et al., 2019). The physiological role of Gardnerella vaginalis in normozoospermic healthy subjects is yet to be determined as Weng et al. (2014) found that Lactobacillus, Gardnerella, Propionibacterium, and Atopobium were relatively more in abundance and significantly present in the normal semen samples. In this study, we found that relative signatures of bacterial communities could be used to disentangle semen categories. Men seeking reproductive health care in the tested population, though found to be normozoospermic, tend to have more Lactobacillus >Gardnerella >Veillonella >Corynebacterium, while the female partners have more Lactobacillus >Prevotella >Gardnerella >Streptococcus. This suggests that the source of their infertility could probably be more than altered bacterial communities. A similar finding was reported by Hou et al. ( Hou et al., 2013) showing that infertile subjects did not have altered or unusual semen bacterial communities compared to normal sperm donors. The men categorized as oligospermia tend to have more Prevotella >Escherichia > Lactobacillus >Shuttleworthia >Serratia >Megasphaera >Gardnerella >Sneathia, and their female partners tend to have more Lactobacillus >Streptococcus >Gardnerella >Lactococcus >Bifidobacterium >Prevotella. It appears that oligospermia men may be under the influence of these pathogens that overwhelm Lactobacillus’s protective activities. The factors responsible for these microbial community differences observed in oligospermic couples are yet to be delineated. In contrast, azoospermia men in these cohorts were observed to have more Lactobacillus >Enterococcus >Corynebacterium >Veillonella >Gardnerella >Ureaplasma >Prevotella, while their female partners have more Lactobacillus >Prevotella >Gardnerella >Megasphaera >Olsenella >Sneathia >Peptoniphillus, and other BV-associated bacteria. Previous studies have reported that several bacteria, including Lactobacillus iners, Gardnerella vaginalis, Escherichia faecalis, E. coli, and Staphylococcus aureus, are associated with male infertility (Franasiak and Scott, 2015; Javurek et al., 2016). These bacterial taxa are common to both semen and vagina. The caveat is that they occur at different proportions in the semen and vaginal niche. The differences in these bacterial communities in these partners may be due to sexual intercourse, episodes of receptive oral sex, and anal sex before vaginal intercourse, which has been reported to influence vaginal and genital tract microbiota in infertile couples (Beigi et al., 2005). The presence of leukocytospermia or pyospermia may have been triggered by the presence of Gardnerella, Prevotella, and other BV-associated bacteria, which leads to alteration in the partner’s vaginal microbiota, and BV has been associated with a 40% increase in the risk of preterm birth (Hillier et al., 1995) and infertility (Al-Awadhi et al., 2013). The occurrence of BV-related microbiota in semen suggests a possible reservoir and supports the concept of sexual transmission of BV, besides semen’s alkaline properties, which may alter the acidic pH of the vagina, leading to BV (Gallo et al., 2011). An earlier study examined adherence, biofilm formation, and cytotoxicity in vitro for G. vaginalis strains isolated from women with BV as well as other BV-associated bacteria, including Atopobium, Prevotella, and Mobiluncus (Patterson et al., 2010). In terms of the impact of BV on IVF, several authors have observed high rates of BV on women with reproductive IVF failure and adverse pregnancy outcome (Spandorfer et al., 2001; Wilson et al., 2002; Wittemer et al., 2004), which corroborates with the findings in this study. Surprisingly, in this study, Lactobacillus crispatus was found to be less colonized in women with positive IVF clinical pregnancy outcome in contrast to the work of Haahr et al. (2019). Instead, Lactobacillus gasseri was significantly associated with positive IVF clinical outcome. The negative IVF clinical outcome observed in these cohorts of men and women may have been due to the induction of inflammatory response that inhibited the sperms from fertilizing the ovum. Although, Vergaro et al. (2019) showed that vaginal microbiota profile at the time of embryo transfer does not affect the live birth rate in IVF cycles, however, the study has been challenged as it was ladened with poor diagnosis and flawed conclusions (Haahr et al., 2019). The microbiota biomarkers identified in women with negative IVF clinical outcomes in this study point towards an infectious perturbation of the IVF process, which corroborates the findings of Haahr et al. (2016). For example, in this study, the significant increase of Bacteroidales, Prevotellaceae, Mobiluncus curtisii, and Cutibacterium acnes in the vagina of those with negative IVF clinical outcome lends credence to previous observations on causes of IVF failure (Onemu and Ibeh, 2001; Al-Awadhi et al., 2013). Cutibacterium acnes are involved in the inflammation of the skin by secreting lipase enzymes, which metabolize sebum into free fatty acids, but its activity in the vagina of women with negative IVF clinical outcomes is yet to be determined. A recent study has demonstrated that IVF does not occur in a sterile environment, and the presence of Staphylococcus sp. and Alphaproteobacteria is associated with clinical indicators such as sperm and embryo quality (Štšepetova et al., 2020).
Transferrin/lactoferrin receptor proteins were significantly upregulated in the bacterial metabolic genes found in semen samples without leukocytes and with positive IVF clinical outcomes when compared with those that had negative IVF clinical outcomes. It should be noted that lactoferrin is a member of the iron-binding transferrin proteins known to have antimicrobial properties and plays a significant role in the mucosal immune response. In addition, lactoferrin aids neutrophils by regulating hydroxyl radical production and participates in the secretion of IgA antibodies (Spik and Montreuil, 1983). It is very interesting to observe that some bacterial metabolic functional genes were upregulated in the vagina of women with positive IVF outcomes. For example, metal ions transport system permease protein such as iron/zinc/manganese/copper were upregulated. These permease systems are required in many biological processes as components of metalloproteins and serve as cofactors or structural elements for enzymes. It should be noted that some bacteria employ a variety of metal uptake and export mechanisms to regulate metal homeostasis by numerous transcriptional regulators (Porcheron et al., 2013).
Conclusions
The fact that semen samples of men with positive IVF clinical outcomes were significantly colonized by Lactobacillus jensenii group and Faecalibacterium and significantly less colonized by Proteobacteria taxa, Prevotella, Bacteroidetes taxa suggests an association between semen and vaginal microbiota correlation. Lactobacillus gasseri was significantly associated with positive IVF clinical outcome in women. In addition, this study has opened a window of the possibility of using clinically tested probiotics therapy for men and women before IVF treatment. The need for implementing this approach has been advocated (Verstraelen and Senok, 2005; Garcia-Velasco et al., 2017).
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: the 72 raw sequence reads (FASTQ files) have been deposited on the Sequence Read Archive (SRA) of the National Center for Biotechnology Information (NCBI) with Project Accession Number PRJNA762524 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA762524).
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Review Committee on Human Research from Nnamdi Azikiwe University Teaching Hospital (Ref. # NAUTH/CS/66/VOL11/175/2018/111). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
KCA designed the study and sourced for funding; SIO and KCA were responsible for taxonomic data organization, initial analysis and manuscript drafting; KCA was responsible for bioinformatic analysis; JII and SIO were involved in clinical evaluation and the collection of samples; KCA, JII and NRA were the principal investigators and participated in the final design of the study, coordination and drafting the manuscript. All authors contributed to the interpretation of data and made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
This study was partly funded by uBiome Inc. San Francisco, USA, under an academic consortium grant-in-kind award to KA and Uzobiogene Genomics, London, Ontario, Canada. However, uBiome did not participate in the design of the study, collection, analysis, interpretation of data and in writing the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to acknowledge uBiome Inc., San Francisco, California, USA, for assisting in the 16S rRNA metagenomics sequencing under an academic consortium grant-in-kind award to KA.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.709372/full#supplementary-material
Principal Coordinate Analysis (PCoA) with the Bray-Curtis clustering in bacterial community diversities between semen and vagina.
The rarefaction curve showing the number of reads between semen samples and vaginal samples.
Venn diagram showing number of taxa exclusive to normospermia, oligospermia, azoospermia, and taxa that were common to the semen categories.
Comparative stacked relative (%) abundance of taxonomic phyla between semen and vagina in normospermia.
Comparative stacked relative (%) abundance of taxonomic genera between semen and vagina in normospermia.
Comparative relative abundance of some selected taxa showing significant difference between semen samples without leukocytes and semen samples leukoocytes.
Relative (%) abundance of taxonomic genera in the vaginal samples of the corresponding pyospermia samples.
Comparative relative (%) abundance of some species taxa between semen samples with leukocytes and semen without leukocytes.
Relative (%) abundance of taxonomic species in the vaginal samples of the corresponding pyospermia samples.
Comparative alpha diversities between semen samples with positive IVF clinical outcome and semen samples with negative IVF clinical outcome.
References
- Ahmadi M. H., Mirasalehian A., Gilani M. A. S., Bahador A., Talebi M. (2018). Improvement of Semen Parameters After Antibiotic Therapy in Asymptomatic Infertile Men Infected With Mycoplasma Genitalium . Infection 46 (1), 31–38. doi: 10.1007/s15010-017-1075-3 [DOI] [PubMed] [Google Scholar]
- Al-Awadhi H., Dashti N., Khanafer M., Al-Mailem D., Ali N., Radwan S. (2013). Bias Problems in Culture-Independent Analysis of Environmental Bacterial Communities: A Representative Study on Hydrocarbonoclastic Bacteria. Springerplus 2, 369. doi: 10.1186/2193-1801-2-369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almonacid D. E., Kraal L., Ossandon F. J., Budovskaya Y. V., Cardenas J. P., Bik E. M., et al. (2017). 16s rRNA Gene Sequencing and Healthy Reference Ranges for 28 Clinically Relevant Microbial Taxa From the Human Gut Microbiome. PloS One 12 (5), e0176555. doi: 10.1371/journal.pone.0176555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amabebe E., Anumba D. O. C. (2018). The Vaginal Microenvironment: The Physiologic Role of Lactobacilli. Front. Med. 5, 181. doi: 10.3389/fmed.2018.00181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anukam K. C., Agbakoba N. R., Okoli A. C., Oguejiofor C. B. (2019). Vaginal Bacteriome of Nigerian Women in Health and Disease: A Study With 16S rRNA Metagenomics. Trop. J. Obstet. Gynaecol. 36, 96−104. doi: 10.4103/TJOG.TJOG_67_18 [DOI] [Google Scholar]
- Beigi R. H., Wiesenfeld H. C., Hillier S. L., Straw T., Krohn M. A. (2005). Factors Associated With Absence of H2O2-Producing Lactobacillus Among Women With Bacterial Vaginosis. J. Infect. Dis. 191, 924–929. doi: 10.1086/428288 [DOI] [PubMed] [Google Scholar]
- Eggert-Kruse W., Rohr G., Strock W., Pohl S., Schwalbach B., Runnebaum B. (1995). Anaerobes in Ejaculates of Subfertile Men. Hum. Reprod. Update 1, 462–478. doi: 10.1093/humupd/1.5.462 [DOI] [PubMed] [Google Scholar]
- Ekhaise F. O., Richard F. R. (2001). Common Bacterial Isolates Associated With Semen of Men Attending the Fertility Clinic of the University of Teaching Hospital (U.B.T.H) Benin City, Nigeria. Afr J. Microbiol. 5 (22), 3805–3809. [Google Scholar]
- Franasiak J. M., Scott R. T., Jr (2015). Reproductive Tract Microbiome in Assisted Reproductive Technologies. Fertil Steril 104, 1364−1371. doi: 10.1016/j.fertnstert.2015.10.012 [DOI] [PubMed] [Google Scholar]
- Gallo M. F., Warner L., King C. C., Sobel J. D., Klein R. S., Cu-Uvin S., et al. (2011). Association Between Semen Exposure and Incident Bacterial Vaginosis. Infect. Dis. Obstet Gynecol 2011, 842652. doi: 10.1155/2011/842652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Velasco J. A., Menabrito M., Catalan I. B. (2017). What Fertility Specialists Should Know About the Vaginal Microbiome: A Review. Reprod. BioMed. Online 35, 103–112. doi: 10.1016/j.rbmo.2017.04.005 [DOI] [PubMed] [Google Scholar]
- Gdoura R., Kchaou W., Ammar-Keskes L., Chakroun N., Sellemi A., Znazen A., et al. (2008). Assessment of Chlamydia Trachomatis, Ureaplasma Urealyticum, Ureaplasma Parvum, Mycoplasma Hominis, and Mycoplasma Genitalium in Semen and First Void Urine Specimens of Asymptomatic Male Partners of Infertile Couples. J. Androl. 29, 198–206. doi: 10.2164/jandrol.107.003566 [DOI] [PubMed] [Google Scholar]
- Günyeli İ, Abike F., Dünder İ, Aslan C., Tapısız ÖL, Temizkan O. (2011). Chlamydia, Mycoplasma and Ureaplasma Infections in Infertile Couples and Effects of These Infections on Fertility. Arch. Gynecol. Obstet. 283, 379–385. doi: 10.1007/s00404-010-1726-4 [DOI] [PubMed] [Google Scholar]
- Haahr T., Humaidan P., Elbaek H. O., Alsbjerg B., Laursen R. J., Rygaard K., et al. (2019). Vaginal Microbiota and In Vitro Fertilization Outcomes: Development of a Simple Diagnostic Tool to Predict Patients at Risk of a Poor Reproductive Outcome. J. Infect. Dis. 219 (11), 1809–1817. doi: 10.1093/infdis/jiy744 [DOI] [PubMed] [Google Scholar]
- Haahr T., Jensen J. S., Humaidan P. (2019). Vaginal Microbiota and IVF Outcomes: Poor Diagnosis Results in Flawed Conclusions. Repro BioMed. Online 39 (1), 178. doi: 10.1016/j.rbmo.2019.04.006 [DOI] [PubMed] [Google Scholar]
- Haahr T., Jensen J. S., Thomsen L., Duus L., Rygaard K., Humaidan P. (2016). Abnormal Vaginal Microbiota may be Associated With Poor Reproductive Outcomes: A Prospective Study in IVF Patients. Hum. Reprod. Oxf Engl. 31, 795–803. doi: 10.1093/humrep/dew026 [DOI] [PubMed] [Google Scholar]
- Hillier S. L., Nugent R. P., Eschenbach D. A., Krohn M. A., Gibbs R. S., Martin D. H., et al. (1995). Association Between Bacterial Vaginosis and Preterm Delivery of a Low-Birth-Weight Infant. The Vaginal Infections and Prematurity Study Group. N Engl. J. Med. 333 (26), 1737–1742. doi: 10.1056/NEJM199512283332604 [DOI] [PubMed] [Google Scholar]
- Hou D., Zhou X., Zhong X., Settles M., Herring J., Wang L., et al. (2013). Microbiota of the Seminal Fluid From Healthy and Infertile Men. Fertil Steril 100 (5), 1261–1269. doi: 10.1016/j.fertnstert.2013.07.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibadin O., Kai I. N. (2008). Bacteriospermia and Sperm Quality in Infertile Male Patient at University of Benin Teaching Hospital, Benin City, Nigeria. Malaysian J. Microbiol. 4 (2), 65–67. [Google Scholar]
- Ibadin K. O., Osemwenkha K. P. (2013). Serodynamics of Chlamydia Trachomatis Infection in Women With In-Vitro Fertilization-Embryo Transfer Failures. J. Infertl. Reprod. Biol. 1 (4), 69–73. [Google Scholar]
- Ikechebelu J. I. (2003). Assisted Reproduction Techniques (ART): The State of Art in Nigeria. J. Clin. Med. 8 (1), 1–6. [Google Scholar]
- Ikechebelu J. I., Adimma J. I. B., Orie E. F., Ikegwuonu S. O. (2003). High Prevalence of Male Infertility in Southeastern, Nigeria. J. Obstet. Gynaecol. 23, 657–659. doi: 10.1080/01443610310001604475 [DOI] [PubMed] [Google Scholar]
- Ikechebelu J. I., Eleje G. U., Ibadin K., Joe-Ikechebelu N. N., Nwaefulu K., Okwelogu I. S. (2016). Outcome of in-Vitro Fertilization Procedure at a Private Fertility Center in Nnewi, South-East Nigeria. Afr. J. Infert. Assisted Conception 1 (1), 2–5. doi: 10.4103/2468-8452.189960 [DOI] [Google Scholar]
- Ivanov I. B., Kuzmin M. D., Gritsenko V. A. (2009). Microflora of the Seminal Fluid of Healthy Men and Men Suffering From Chronic Prostatitis Syndrome. Int. J. Androl. 32, 462–467. doi: 10.1111/j.1365-2605.2008.00878.x [DOI] [PubMed] [Google Scholar]
- Javurek A. B., Spollen W. G., Ali A. M., Johnson S. A., Lubahn D. B., Bivens N. J., et al. (2016). Discovery of a Novel Seminal Fluid Microbiome and Influence of Estrogen Receptor Alpha Genetic Status. Sci. Rep. 6, 23027. doi: 10.1038/srep23027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Sato Y., Kawashima M., Furumichi M., Tanabe M. (2014). Data, Information, Knowledge and Principle: Back to Metabolism in KEGG. Nucleic Acids Res. 42 (D1), D199–D205. doi: 10.1093/nar/gkt1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren O., Knights D., Gonzalez A., Waldron L., Segata N., Knight R., et al. (2013). A Guide to Enterotypes Across the Human Body: Meta-Analysis of Microbial Community Structures in Human Microbiome Datasets. PloS Comput. Biol. 9 (1), e1002863. doi: 10.1371/journal.pcbi.1002863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille M. G., Zaneveld J., Caporaso J. G., McDonald D., Knights D., Reyes J. A., et al. (2013). Predictive Functional Profiling of Microbial Communities Using 16S rRNA Marker Gene Sequences. Nat. Biotechnol. 31, 814–821. doi: 10.1038/nbt.2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lis R., Rowhani-Rahbar A., Manhart L. E. (2015). Mycoplasma Genitalium Infection and Female Reproductive Tract Disease: A Meta-Analysis. Clin. Infect. Dis. 61 (3), 418–426. doi: 10.1093/cid/civ312 [DOI] [PubMed] [Google Scholar]
- Mandar R., Punab M., Borovkova N., Lapp E., Kiiker R., Korrovits P., et al. (2015). Complementary Seminovaginal Microbiome in Couples. Res. Microbiol. 166, 440–447. doi: 10.1016/j.resmic.2015.03.009 [DOI] [PubMed] [Google Scholar]
- Momoh A. R., Odike M. A., Samuel S. O., Momoh A. A., Okolo P. O. (2007). Infertility in Male: Risk Factors, Causes and Management-A Review. Benin J. Post Grad Med. 9 (1), 22–27. [Google Scholar]
- Ndiokwere C., Olise N. A., Nmewurum V., Omoregie R., Agbakoba N. R., Anukam K. C. (2019). 16s rRNA Metagenomics of Seminal Fluids From Medical Microbiology Laboratory in a Tertiary Hospital, Southern Nigeria. J. Med. Lab. Sci. 29 (3), 86–109. doi: 10.5281/zenodo.4007804 [DOI] [Google Scholar]
- Nelson D. E., van der Pol B., Dong Q., Revanna K. V., Fan B., Easwaran S., et al. (2010). Characteristic Male Urine Microbiomes Associate With Asymptomatic Sexually Transmitted Infection. PloS One 5, e14116. doi: 10.1371/journal.pone.0014116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwadioha S. I., Odimayo M. S., Jombo G. T. A. (2016). Microbiologic Review of Seminal Fluids in a Nigerian Tertiary Health Centre. Arch. Clin. Microbiol. 7, 4. [Google Scholar]
- Okoli A. C., Agbakoba N. R., Ezeanya C. C., Oguejiofor C. B., Anukam K. C. (2019). Comparative Abundance and Functional Biomarkers of the Vaginal and Gut Microbiome of Nigerian Women With Bacterial Vaginosis: A Study With 16S rRNA Metagenomics. J. Med. Lab. Sci. 29, 1–26. doi: 10.5281/zenodo.4010799 [DOI] [Google Scholar]
- Onemu S. O., Ibeh I. N. (2001). Studies on the Significance of Positive Bacterial Semen Cultures in Male Fertility in Nigeria. Int. J. Fertil Womens Med. 46 (4), 210–214. [PubMed] [Google Scholar]
- Patterson J. L., Stull-Lane A., Girerd P. H., Jefferson K. K. (2010). Analysis of Adherence, Biofilm Formation and Cytotoxicity Suggests a Greater Virulence Potential of Gardnerella Vaginalis Relative to Other Bacterial-Vaginosis Associated Anaerobes. Microbiol. 156, 392–399. doi: 10.1099/mic.0.034280-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcheron G., Garénaux A., Proulx J., Sabri M., Dozois C. M. (2013). Iron, Copper, Zinc, and Manganese Transport and Regulation in Pathogenic Enterobacteria: Correlations Between Strains, Site of Infection and the Relative Importance of the Different Metal Transport Systems for Virulence. Front. Cell Infect. Microbiol. 5, 90. doi: 10.3389/fcimb.2013.00090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. (2013). The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Webbased Tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravel J., Gajer P., Abdo Z., Schneider G. M., Koenig S. S., McCulle S. L., et al. (2010). Vaginal Microbiome of Reproductive-Age Women. Proc. Natl. Acad. Sci. U.S.A. 108 (Suppl 1), 4680–4687. doi: 10.1073/pnas.1002611107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemersma W. A., van der Schee C. J., van der Meijden W. I., Verbrugh H. A., van Belkum A. (2003). Microbial Population Diversity in the Urethras of Healthy Males and Males Suffering From non-Chlamydial, Nongonococcal Urethritis. J. Clin. Microbiol. 41, 1977–1986. doi: 10.1128/JCM.41.5.1977-1986.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross A. A., Doxey A. C., Neufeld J. D. (2017). The Skin Microbiome of Cohabiting Couples. mSystems 2, e00043–e00017. doi: 10.1128/mSystems.00043-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W. S., et al. (2011). Metagenomic Biomarker Discovery and Explanation. Gen. Biol. 12 (6), R60. doi: 10.1186/gb-2011-12-6-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spandorfer S. D., Neuer A., Giraldo P. C., Rosenwaks Z., Witkin S. S. (2001). Relationship of Abnormal Vaginal Flora, Proinflammatory Cytokines and Idiopathic Infertility in Women Undergoing IVF. J. Reprod. Med. 46 (9), 806–810. [PubMed] [Google Scholar]
- Spik G., Montreuil J. (1983). The Role of Lactotransferrin in the Molecular Mechanisms of Antimicrobial Defense. Bull. Eur. Physiopathol. Respir. 19 (2), 123–130. [PubMed] [Google Scholar]
- Štšepetova J., Baranova J., Simm J., Parm U., Roop T., Sokmann S., et al. (2020). The Complex Microbiome From Native Semen to Embryo Culture Environment in Human In Vitro Fertilization Procedure. Reprod. Biol. Endocrinol. 18, 3. doi: 10.1186/s12958-019-0562-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift B. E., Liu K. E. (2014). The Effect of Age, Ethnicity, and Level of Education on Fertility Awareness and Duration of Infertility. J. Obstet Gynaecol Can. 36 (11), 990–996. doi: 10.1016/S1701-2163(15)30412-6 [DOI] [PubMed] [Google Scholar]
- Taylor B. D., Zheng X., O’Connell C. M., Wiesenfeld H. C., Hillier S. L., Darville T. (2018). Risk Factors for Mycoplasma Genitalium Endometritis and Incident Infection: A Secondary Data Analysis of the T-Cell Response Against Chlamydia (TRAC) Study. Sex Trans. Dis. 2 (2), 405–410. doi: 10.1136/sextrans-2017-053376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergaro P., Tiscornia G., Barragán M., García D., Rodriguez A., Santaló J., et al. (2019). Vaginal Microbiota Profile at the Time of Embryo Transfer Does Not Affect Live Birth Rate in IVF Cycles With Donated Oocytes. Reprod. BioMed. Online 38 (6), 883–891. doi: 10.1016/j.rbmo.2018.12.019 [DOI] [PubMed] [Google Scholar]
- Verstraelen H., Senok A. C. (2005). Vaginal Lactobacilli, Probiotics and IVF. Reprod. BioMed. Online 11 (6), 674–675. doi: 10.1016/S1472-6483(10)61683-5 [DOI] [PubMed] [Google Scholar]
- Weng S.-L., Chiu C.-M., Lin F.-M., Huang W.-C., Liang C., Yang T., et al. (2014). Bacterial Communities in Semen From Men of Infertile Couples: Metagenomic Sequencing Reveals Relationships of Seminal Microbiota to Semen Quality. PloS One 9 (10), e110152. doi: 10.1371/journal.pone.0110152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J. D., Ralph S. G., Rutherford A. J. (2002). Rates of Bacterial Vaginosis in Women Undergoing In Vitro Fertilisation for Different Types of Infertility. BJOG 109 (6), 714–717. doi: 10.1111/j.1471-0528.2002.01297.x [DOI] [PubMed] [Google Scholar]
- Wittemer C., Bettahar-Lebugle K., Ohl J., Rongieres C., Viville S., Nisand I. (2004). Abnormal Bacterial Colonisation of the Vagina and Implantation During Assisted Reproduction. Gynecol. Obstet. Fertil. 32, 135e9. doi: 10.1016/j.gyobfe.2003.11.017 [DOI] [PubMed] [Google Scholar]
- Yoon S. H., Ha S. M., Kwon S., Lim J., Kim Y., Seo H., et al. (2017). Introducing EzBioCloud: A Taxonomically United Database of 16S rRNA and Whole Genome Assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617. doi: 10.1099/ijsem.0.001755 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Principal Coordinate Analysis (PCoA) with the Bray-Curtis clustering in bacterial community diversities between semen and vagina.
The rarefaction curve showing the number of reads between semen samples and vaginal samples.
Venn diagram showing number of taxa exclusive to normospermia, oligospermia, azoospermia, and taxa that were common to the semen categories.
Comparative stacked relative (%) abundance of taxonomic phyla between semen and vagina in normospermia.
Comparative stacked relative (%) abundance of taxonomic genera between semen and vagina in normospermia.
Comparative relative abundance of some selected taxa showing significant difference between semen samples without leukocytes and semen samples leukoocytes.
Relative (%) abundance of taxonomic genera in the vaginal samples of the corresponding pyospermia samples.
Comparative relative (%) abundance of some species taxa between semen samples with leukocytes and semen without leukocytes.
Relative (%) abundance of taxonomic species in the vaginal samples of the corresponding pyospermia samples.
Comparative alpha diversities between semen samples with positive IVF clinical outcome and semen samples with negative IVF clinical outcome.
Data Availability Statement
The datasets used and or analyzed in the current study are available from the corresponding author on reasonable request.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: the 72 raw sequence reads (FASTQ files) have been deposited on the Sequence Read Archive (SRA) of the National Center for Biotechnology Information (NCBI) with Project Accession Number PRJNA762524 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA762524).