Abstract
Background
Biliary tract cancers (BTCs) are rare and highly heterogenous malignant neoplasms. Because obtaining BTC tissues is challenging, the purpose of this study was to explore the potential roles of bile as a liquid biopsy medium in patients with BTC.
Patients and methods
Sixty-nine consecutive patients with suspected BTC were prospectively enrolled in this study. Capture-based targeted sequencing was performed on tumor tissues, whole blood cells, plasma, and bile samples using a large panel consisting of 520 cancer-related genes.
Results
Of the 28 patients enrolled in this cohort, tumor tissues were available in eight patients, and plasma and bile were available in 28 patients. Somatic mutations were detected in 100% (8/8), 71.4% (20/28), and 53.6% (15/28) of samples comprising tumor tissue DNA, bile cell-free DNA (cfDNA), and plasma cfDNA, respectively. Bile cfDNA showed a significantly higher maximum allele frequency than plasma cfDNA (P = 0.0032). There were 56.2% of somatic single-nucleotide variant (SNVs)/insertions and deletions (indels) shared between bile and plasma cfDNA. When considering the genetic profiles of tumor tissues as the gold standard, the by-variant sensitivity and positive predictive value for SNVs/indels in bile cfDNA positive for somatic mutations were both 95.5%. The overall concordance for SNVs/indels in bile was significantly higher than that in plasma (99.1% versus 78.3%, P < 0.0001). Moreover, the sensitivity of CA 19-9 combined with bile cfDNA achieved 96.4% in BTC diagnosis.
Conclusion
We demonstrated that bile cfDNA was superior to plasma cfDNA in the detection of tumor-related genomic alterations. Bile cfDNA as a minimally invasive liquid biopsy medium might be a supplemental approach to confirm BTC diagnosis.
Key words: cell-free nucleic acids, bile, sequence analysis, DNA, liquid biopsy, biliary tract neoplasms
Highlights
-
•
This is the first study to compare somatic mutation profiling in tumor tissues, bile and plasma samples in BTC patients.
-
•
Our findings demonstrated that bile cfDNA outperformed plasma cfDNA in detecting somatic mutations.
-
•
Bile cfDNA might be a feasible liquid biopsy medium for BTC diagnosis.
Introduction
Biliary tract cancers (BTCs) are rare and a heterogenous group of neoplasms, which are anatomically classified into gallbladder cancer (GBC), intrahepatic cholangiocarcinoma (iCCA), extrahepatic cholangiocarcinoma (eCCA) with the further classification of perihilar cholangiocarcinoma (pCCA) and distal cholangiocarcinoma (dCCA), and carcinoma of ampulla of Vater (CAV) with distinct epidemiological and molecular pathological processes.1,2
The prognosis of patients with BTC remains poor with a 5-year survival rate of 7%-15% because most patients with BTC are diagnosed with advanced disease and those patients who undergo surgery frequently experience a recurrent disease.3,4 Treatment options for BTC are limited. Chemotherapy remains the current standard of care in both adjuvant and advanced disease setting.5,6 In recent years, identification of mutations driving oncogenesis and development of targeted therapy and immunotherapy provide new opportunities for treating metastatic BTC.7 The U.S. Food and Drug Administration has approved neurotrophic receptor tyrosine kinase (NTRK) inhibitors (entrectinib and larotrectinib) and the programmed cell death protein 1 (PD-1) inhibitor pembrolizumab for treatment of patients with solid tumor who carry NTRK fusions and microsatellite instability (MSI)-high/mismatch repair-deficient, respectively.8,9 The National Comprehensive Cancer Network (NCCN) guidelines currently recommend ivosidenib and pemigatinib for treatment of cholangiocarcinoma patients who harbor isocitrate dehydrogenase 1 (IDH1) mutations and fibroblast growth factor receptor 2 (FGFR2) fusions, respectively.9,10 Therefore molecular testing of tumor tissues could be a crucial approach to guide precision therapy for BTC in the future. However, obtaining BTC tissues is challenging because BTC frequently infiltrates bile duct wall and the tumors are commonly adjacent to large blood vessels and vital organs including liver and pancreas,11 and potentially devastating complications involving biliary hemorrhaging and bile duct perforation.12, 13, 14
Liquid biopsy, specifically cell-free DNA (cfDNA) analysis, as a minimally invasive approach, has shown promising results within several clinical applications for patients with insufficient tumor tissue, including mutation profiling, treatment monitoring, and cancer detection.15 Plasma cfDNA is the most commonly studied cancer noninvasive biomarker due to its remarkable advantages over traditional methods, including invasiveness, no intratumor heterogeneity, and short turnaround time,16 which has been approved for clinical applications in several malignancies.17 However, the clinical applications of plasma cfDNA for mutation testing is limited by the anatomical location of the tumors.18 Previous studies have demonstrated that other biofluids, such as cerebrospinal fluid, pleural effusion, ascites, and urine, are superior to plasma in the detection of tumor evolution and identification of resistance mechanism in several special malignancies.19, 20, 21
For BTC, bile sample can be minimally invasive and is obtained by percutaneous transhepatic cholangial drainage (PTCD). It has been reported that bile contains tumor-associated peptides, microRNAs, and cfDNA.22,23 Bile cfDNA as a novel and powerful liquid biopsy for detecting somatic variants by using a customized panel comprising 150 cancer-related genes in 10 BTC patients has been reported.24 However, the difference in somatic mutation profiling between bile and plasma in BTC patients has not been documented. Furthermore, the question of whether bile cfDNA is superior to plasma cfDNA in detecting tumor-related somatic alterations remains elusive.
In this study, we investigated the genomic profiling of tumor tissues, plasma samples, and bile samples using a large panel consisting of 520 cancer-related genes in BTC patients. We also explored the potential roles of bile cfDNA as a source of liquid biopsy in BTC patients.
Methods and materials
Patients
A total of 69 consecutive patients with suspected BTC were prospectively enrolled between April 2018 and April 2020 at Guangdong Provincial People’s Hospital in this study. Those patients who met the following inclusion criteria were screened for further analysis: (i) With BTC diagnosis confirmed by pathological or cytological examinations; (ii) without contraindications for PTCD; (iii) without other malignant tumors. The study was approved by the Ethics Committee of Guangdong Provincial People’s Hospital. Informed consent was obtained from each patient for the use of their tumor, peripheral blood, and bile samples.
Sample collection
Tumor tissues, peripheral blood, and bile samples were obtained from BTC patients. Formalin-fixed, paraffin-embedded (FFPE) sections of tumor tissues were obtained from BTC patients who underwent surgical resection or percutaneous biopsy. In addition, hematoxylin and eosin staining of each sample was evaluated by an experienced pathologist to confirm the tumor content. Eight milliliters of peripheral blood of each patient was collected and centrifuged at 1800g for 10 min at 4°C within 2 h after blood collection to separate plasma and white blood cells (WBCs). Plasma samples were subsequently transferred into a new tube for extraction of cfDNA. WBC sediments were used for genomic DNA extraction as the germline controls. Eight milliliters of bile of each patient who underwent PTCD was collected in an ethylene diamine tetraacetic acid-coated tube. Bile samples were then centrifuged at 1800g for 15 min within 2 h after bile collection. The supernatants were transferred into a new tube and centrifuged at 1800g for 10 min. The supernatants were subsequently collected and examined under a light microscope to ensure that there were no residual cells or debris. Samples were then aliquoted and stored at –80°C.
Tissue DNA and cfDNA extraction
Tissue DNA was extracted with a QIAamp DNA FFPE tissue kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. cfDNA was extracted from each plasma/bile sample using the QIAamp Circulating Nucleic Acid Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. The concentration of tissue DNA and cfDNA was measured by Qubit 2.0 Fluorometer with a Qubit double-stranded DNA assay kit (Life Technologies, Carlsbad, CA, USA).
NGS library preparation and capture-based targeted sequencing
Tissue DNA, plasma cfDNA, and bile cfDNA were fragmented by Covaris M220 focused-ultrasonicator (Covaris, Inc., Woburn, MA, USA) followed by end repair, phosphorylation, dA addition, and adaptor ligation for library construction, respectively.
Tissue, plasma, and bile samples were subjected to capture-based targeted sequencing using a panel consisting of 520 cancer-related genes (including whole exons of 312 genes and exons, introns, and promoter regions of the remaining 208 genes), spanning 1.7 Mb of human genome (OncoScreen Plus; Burning Rock Biotech, Guangzhou, China). DNA was hybridized with the capture probe baits, selected with magnetic beads, and PCR amplified. A bioanalyzer high-sensitivity DNA assay was then performed to assess the quality and size of the fragments and indexed samples were sequenced on a Nextseq500 sequencer (Illumina, Inc., San Diego, CA, USA) with paired-end reads. WBCs were used for filtering germline mutations. The mean sequencing coverage for tumor tissue, WBC, plasma, and bile was 1217×, 6864×, 139 13×, and 5863×, respectively.
Sequence data analysis
The raw sequencing data were preprocessed using Trimmomatic 0.36 for trimming adaptor and low-quality reads. Preprocessed sequencing data were mapped to the human genome (hg19) using Burrows-Wheeler Aligner 0.7.10. Variant calling and annotation were performed using GATK 3.2, MuTect, and VarScan. Plasma/bile samples were compared against paired WBCs to identify somatic variants. According to the ExAC, 1000 Genomes, dbSNP, ESP6500SI-V2 database, variants with population frequency >0.1% were grouped as single-nucleotide polymorphisms and excluded from further analysis. Remaining variants were annotated with ANNOVAR and SnpEff version 3.6. Copy number variations (CNVs) were detected by in-house analysis scripts based on the depth of coverage data of capture intervals as previously described.25 DNA translocation analysis was performed using both Tophat2 and Factera 1.4.3. The allele frequency of mutations was calculated. The MSI status of tumor samples was determined based on a read count distribution-based method.26,27
Tumor mutation burden
Tumor mutation burden (TMB) was defined as the somatic single-nucleotide variants (SNVs) and small insertions and deletions (indels; fusions and CNVs were excluded) locating at the coding region and its 20-bp upstream/downstream region. TMB was calculated according the following equation:
Statistical analysis
Differences between the two groups were accessed by two-tailed t-test for continuous variables with normal distribution or by Mann-Whitney U test for continuous variables with non-normal distribution or by Fisher’s exact test for discontinuous variables. P < 0.05 was considered to be statistically significant. All statistical analyses were performed in R 3.3.3 (R Foundation, Vienna, Austria).
Results
Patient characteristics
From April 2018 to April 2020, 69 consecutive patients with suspected BTC were prospectively enrolled in this study. Twenty-eight patients with BTC confirmed by pathological or cytological examinations who had no contraindications for PTCD (Supplementary Figure S1, available at https://doi.org/10.1016/j.esmoop.2021.100275) were screened for further analyses, including one patient with GBC (3.6%), eight with iCCA (28.6%), eight with dCCA (28.6%), seven with pCCA (25.0%), and four with CAV (14.3%). The median age of 28 patients including 11 (39.3%) males was 61.8 years (range 38-85 years). Twenty-six patients (92.9%) were diagnosed with adenocarcinomas and two patients (7.1%) had mucinous carcinomas. According to the eighth edition of the American Joint Committee on Cancer (AJCC) Tumor, Node, Metastasis staging system, 2, 4, 9, and 13 patients were distributed in stage I, stage II, stage III, and stage IV, respectively. The clinical characteristics for the cohort are summarized in Supplementary Table S1, available at https://doi.org/10.1016/j.esmoop.2021.100275.
Mutational profiling in tumor DNA, plasma cfDNA, and bile cfDNA samples
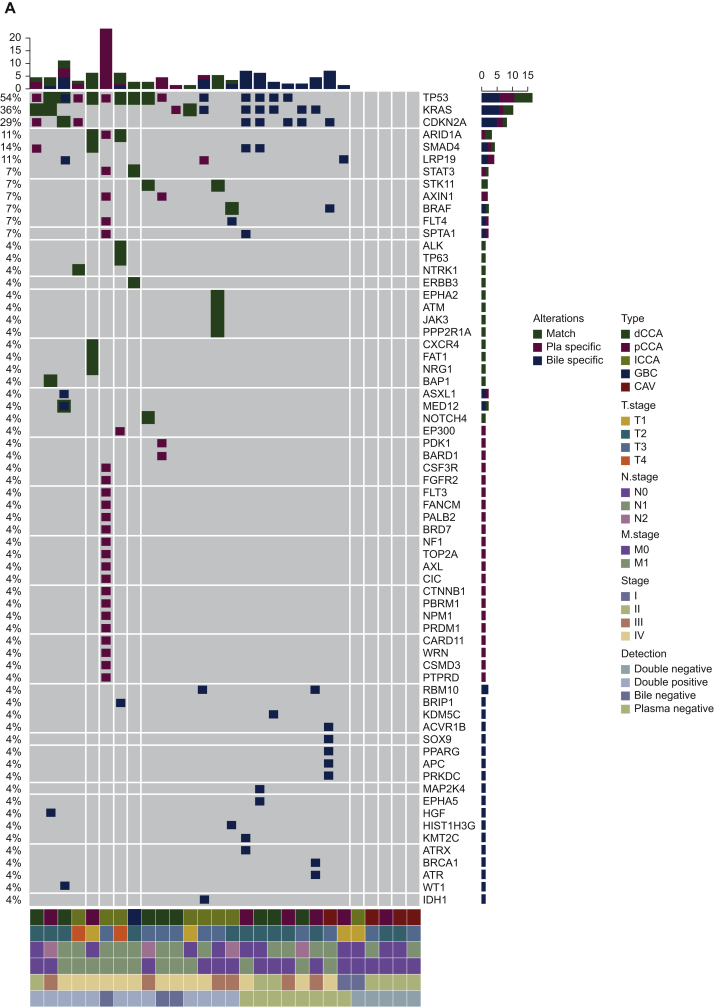
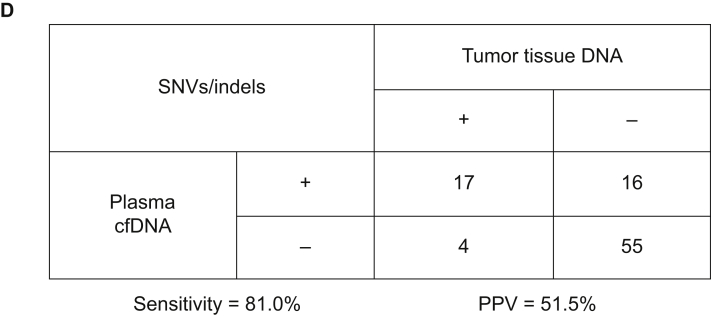
Somatic mutational profiles were respectively obtained from tumor tissue samples of eight patients, plasma and bile samples of 28 patients. Collectively, 72 genomic alterations spanning 31 genes were identified in eight tumor tissues, including 44 SNVs, one indel, and 27 CNVs. TP53, KRAS, CDKN2A, LRP1B, SPTA1, and SMAD4 were the most commonly mutated genes identified in tumor tissues, occurring in 62% (n = 5), 38% (n = 3), 38% (n = 3), 25% (n = 2), and 25% (n = 2) of patients, respectively (Supplementary Figure S2A, available at https://doi.org/10.1016/j.esmoop.2021.100275). Seventy-three SNVs, two fusions, one indel, and one CNV were identified in 28 plasma cfDNA samples. The most commonly mutated genes in plasma cfDNA were TP53, KRAS, CDKN2A, and ARID1A, occurring in 36% (n = 10), 14% (n = 4), 11% (n = 3), and 11% (n = 3) of patients, respectively (Supplementary Figure S2B, available at https://doi.org/10.1016/j.esmoop.2021.100275). Seventy-eight SNVs, two fusions, one indel, and nine CNVs were identified in 28 bile cfDNA samples. TP53, KRAS, CDKN2A, and SMAD4, occurring in 39% (n = 11), 32% (n = 9), 21% (n = 6), and 11% (n = 3) of patients, were the most commonly mutated genes in bile cfDNA samples (Supplementary Figure S2C, available at https://doi.org/10.1016/j.esmoop.2021.100275). All patients had microsatellite stable tumors. None of the patients harbored POLE/POLD1 mutations.
The number of SNVs/indels in plasma and matched bile samples in each patient is shown in Figure 1A. Among those mutations, 10 (17.5%) were plasma specific, 15 (26.3%) were bile specific, and 32 (56.2%) were shared between the two media (Figure 1B).
Figure 1.
Distribution of somatic mutations in plasma and bile samples.
(A) Heatmap indicating the mutations detected in bile and matched plasma samples. Dark green indicates mutations detected from both sources, maroon indicates mutations that were present only in the plasma samples, and dark blue indicates mutations present only in the bile samples. (B) Venn diagram showing the overlap of genetic mutations in plasma and bile samples obtained from 28 patients with BTC.
BTC, biliary tract cancer; CAV, carcinoma of ampullar of Vater; dCCA, distal cholangiocarcinoma; GBC, gallbladder cancer; iCCA, intrahepatic cholangiocarcinoma; pCCA, perihilar cholangiocarcinoma; Pla, plasma.
The overall mutation detection rate in tissue, plasma, and bile samples
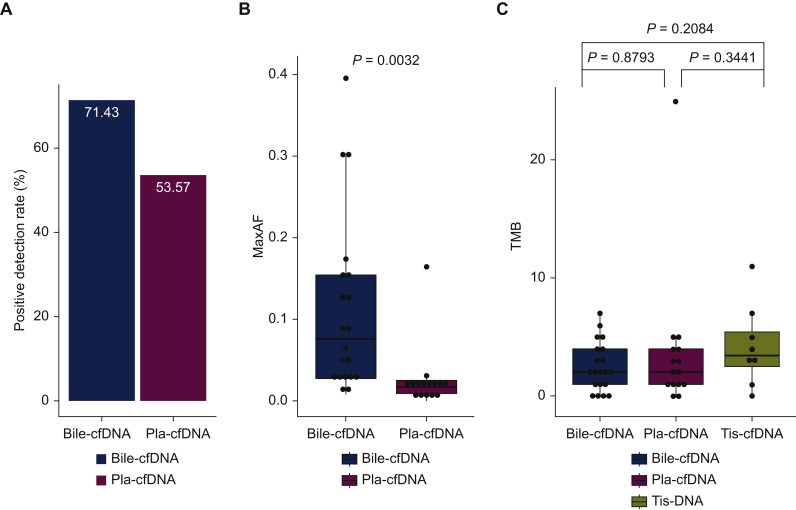
Somatic mutations were detected in 100% (8/8) of tumors. Of the 28 plasma samples, 13 (46.4%) had no genomic alterations, achieving a mutation detection rate of 53.6% (15/28; Figure 2A). Of the 28 bile samples, eight (28.6%) had no genomic alterations, achieving a mutation detection rate of 71.4% (20/28; Figure 2A). The median maximum allele fractions (maxAFs) of tumor tissues, plasma samples, and bile samples were 24.3%, 1.9%, and 7.7%, respectively. Moreover, the average maxAF of bile cfDNA was significantly higher than that of plasma cfDNA (P = 0.0032; Figure 2B). The median TMB of bile was 1.99 (range 0-6.98) mutations/Mb, which was comparable with that of tumor tissue (3.49 mutations/Mb; range 0-10.97) and plasma (1.99 mutations/Mb; range 0-24.93), respectively (Figure 2C).
Figure 2.
The difference in (A) somatic mutation detection rate, (B) maxAF, and (C) TMB between bile cfDNA and plasma cfDNA samples.
cfDNA, cell-free DNA; maxAF, maximum allele fractions; Pla, plasma; Tis, tumor tissue; TMB, tumor mutation burden.
Association between bile cfDNA detection rate and clinical features
Next, we investigated associations between bile cfDNA detection rates and patients’ clinical features, including stage and tumor location. The bile cfDNA detection rate in GBC, pCCA, dCCA, iCCA, and CAV were 100% (1/1), 85.7% (6/7), 75.0% (6/8), 75.0% (6/8), and 25.0% (1/4), respectively (Supplementary Figure S3A, available at https://doi.org/10.1016/j.esmoop.2021.100275). Patients with pCCA and dCCA were grouped into the eCCA subgroup for further analysis. Bile cfDNA detection rate in patients with iCCA and eCCA was comparable (75% versus 80%, P > 0.99, Supplementary Figure S3B, available at https://doi.org/10.1016/j.esmoop.2021.100275). eCCA had a trend of higher bile cfDNA detection rate compared with CAV (80.0% versus 25.0%, P = 0.07; Supplementary Figure S3B, available at https://doi.org/10.1016/j.esmoop.2021.100275). The bile cfDNA mutation detection rates in stage I, II, III, and IV were 50.0% (1/2), 42.9% (3/7), 100% (6/6), and 76.9% (10/13), respectively (Supplementary Figure S3C, available at https://doi.org/10.1016/j.esmoop.2021.100275). Because of the limited number of patients, patients with stage I and stage II and those with stage III and stage IV were grouped together for subsequent analysis, respectively. Patients with stage III/IV disease had a marginally significantly higher bile cfDNA mutation detection rate than those with stage I/II disease (84.2% versus 44.4%, P = 0.07; Supplementary Figure S3D, available at https://doi.org/10.1016/j.esmoop.2021.100275).
Bile cfDNA outperforms plasma cfDNA in detecting somatic mutations
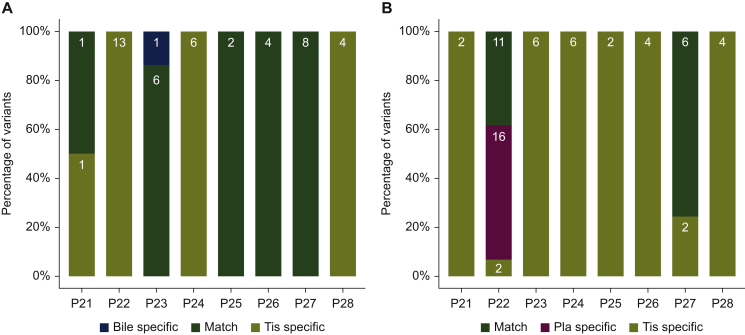
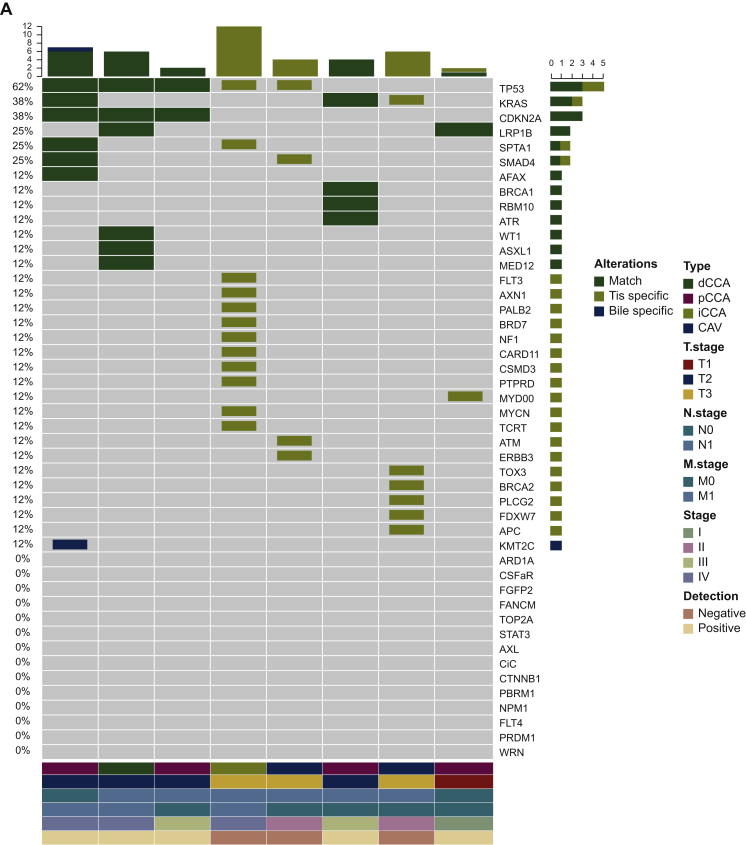
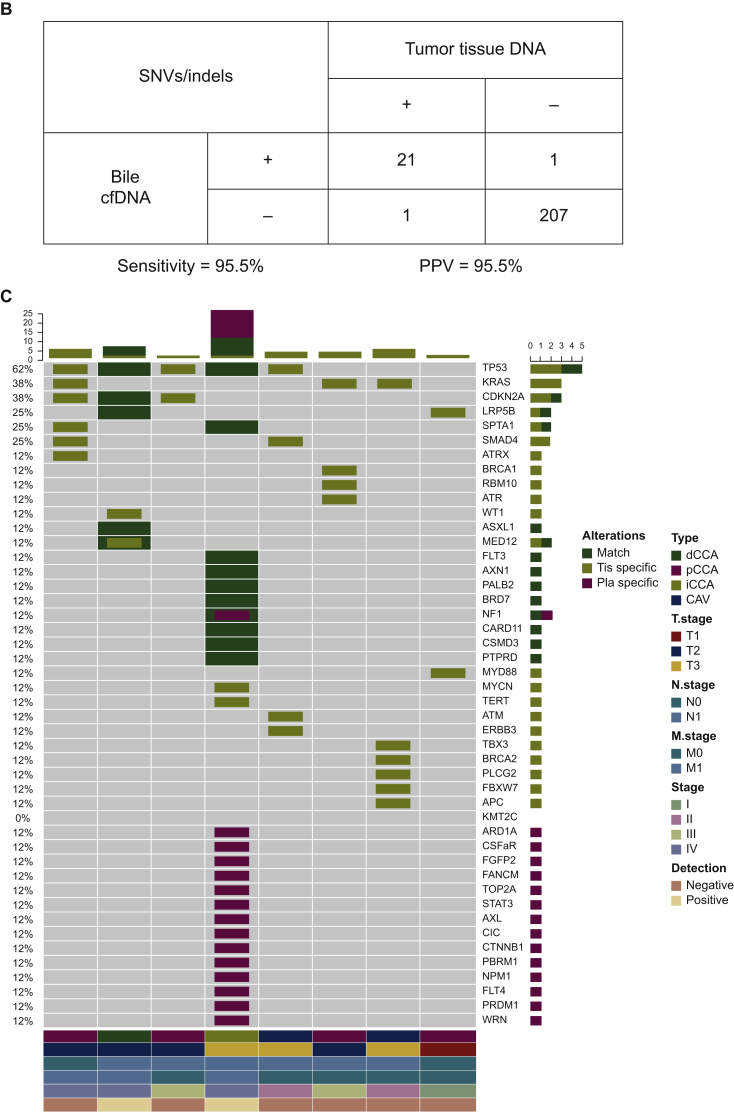
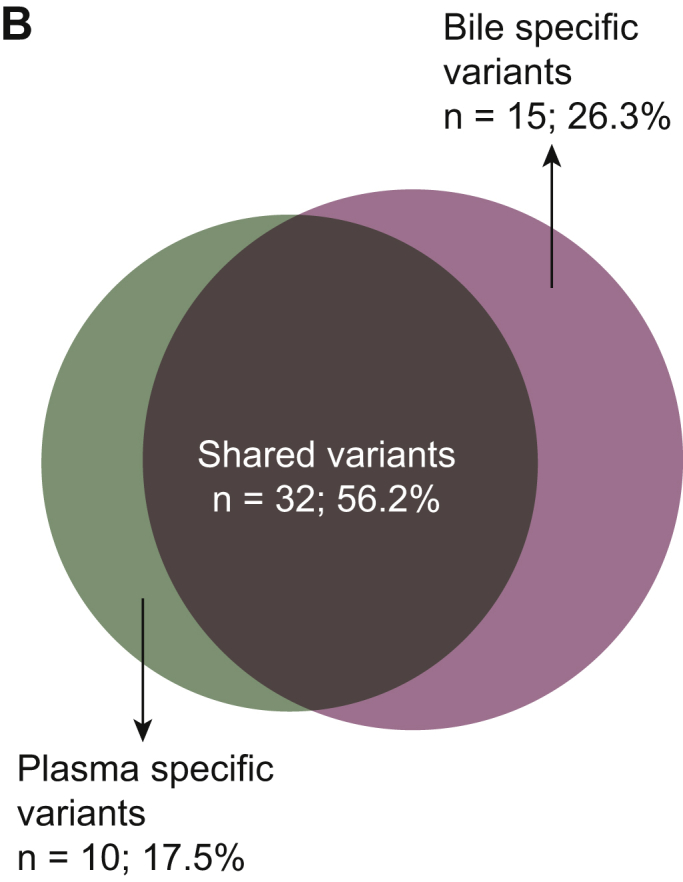
Next, the concordance of utilizing capture-based targeted sequencing to detect SNVs/indels between bile/plasma samples and tumor tissues was analyzed. Tumor tissues, plasma samples, and bile samples were available from eight patients. When tumor tissues were used as the reference, five patients harboring somatic mutations were identified in bile cfDNA with a by-patient sensitivity of 62.5% (5/8, Figure 3A), while only two patients harboring somatic mutations were identified in plasma cfDNA with a by-patient sensitivity of 25.0% (Figure 3B). By-variant sensitivity is commonly used to evaluate the concordance of genomic profiling between different biopsy methods. Next, we demonstrated the by-variant concordance for SNVs/indels in bile/plasma cfDNA. When the genetic profiles of tumor tissues were considered the gold standard, the by-variant sensitivity and positive predictive value (PPV) for SNVs/indels in bile samples were both 95.5% in five patients who had NGS results showing positivity for somatic alterations in bile (Figure 4A). The overall concordance for SNVs/indels in bile cfDNA was 99.1% (228/230). The by-variant sensitivity and PPV for SNVs/indels in plasma samples were 81.0% and 51.5% (Figure 4B) in two patients who had NGS results showing positivity for somatic alterations in plasma, respectively. The overall concordance for SNVs/indels in plasma was 78.3% (72/92). Furthermore, the overall concordance for SNVs/indels in bile was significantly higher than that in plasma (P < 0.0001).
Figure 3.
The distribution of somatic mutations detected in eight patients with BTC who had available tumor tissue, plasma, and bile samples.
(A) The distribution of matched, bile-specific, and tissue-specific mutations in eight patients. (B) The distribution of matched, plasma-specific, and tissue-specific mutations in eight patients.
BTC, biliary tract cancer; cfDNA, cell-free DNA; Pla, plasma; Tis, tumor tissue.
Figure 4.
Somatic mutations identified in bile, plasma, and tissue samples.
(A) Heatmap showing somatic mutations identified in bile and matched tissue samples. Dark green indicates mutations identified in both sources, olive green indicates mutations present only in the tissue samples, and dark blue indicates mutations present only in the bile samples. (B) The by-variant sensitivity and PPV of utilizing capture-based targeted sequencing to identify SNVs/indels in bile cfDNA were calculated, with tissue samples used as references. (C) Heatmap displaying somatic mutations identified in plasma and matched tissue samples. Dark green indicates mutations identified in both sources, olive green indicates mutations present only in the tissue samples, and maroon indicates mutations present only in the plasma samples. (D) The by-variant sensitivity and PPV of utilizing capture-based targeted sequencing to identify SNVs/indels in plasma cfDNA were calculated, with tissue samples used as references.
CAV, carcinoma of ampullar of Vater; cfDNA, cell-free DNA; dCCA, distal cholangiocarcinoma; iCCA, intrahepatic cholangiocarcinoma; indels, insertions and deletions; pCCA, perihilar cholangiocarcinoma; Pla, plasma; PPV, positive predictive value; SNVs, single-nucleotide variants; Tis, tumor tissue.
Bile cfDNA as a supplemental tool for BTC diagnosis
In this study, all 28 patients with BTC diagnosis were confirmed by pathological or cytological examinations. All 28 patients underwent imaging using abdominal/pelvis computed tomography, positron emission tomography–computed tomography, or magnetic resonance cholangiopancreatography (Figure 5). The presence of a mass on imaging, cancer antigen 19-9 (CA 19-9) level >100 U/ml, carcinoembryonic antigen (CEA) level >5 ng/ml, bile cfDNA positive for somatic mutations, or plasma cfDNA positive for somatic mutations was suggestive of BTC in this work. When pathological or cytological examination report was used as the gold standard for BTC diagnosis, the sensitivity of imaging, CA 19-9, CEA, bile cfDNA, and plasma cfDNA were 60.7%, 82.1%, 60.7%, 71.4%, and 53.6%, respectively (Supplementary Table S2, available at https://doi.org/10.1016/j.esmoop.2021.100275). The sensitivity of CA 19-9 combined with plasma-cfDNA was 85.7% (Figure 5, Supplementary Table S2, available at https://doi.org/10.1016/j.esmoop.2021.100275). Furthermore, the sensitivity of imaging combined with bile cfDNA, and CA 19-9 combined with bile cfDNA increased to 89.3% and 96.4%, respectively (Figure 5, Supplementary Table S2, available at https://doi.org/10.1016/j.esmoop.2021.100275).
Figure 5.
The performance of bile cfDNA in BTC diagnosis.
BTC, biliary tract cancer; CA 19-9, cancer antigen 19-9; CEA, carcinoembryonic antigen; cfDNA, cell-free DNA.
Discussion
Our study is the first to compare somatic mutation profiling in tumor tissues, plasma samples, and bile samples in patients with BTC. Our findings showed that bile cfDNA outperformed plasma cfDNA in detecting somatic mutations, suggesting the importance of bile cfDNA as a liquid biopsy medium for BTC diagnosis.
In this work, 96.3% (26/27) of CCA patients had adenocarcinoma, which was consistent with the prior studies indicating that >90% of CCA patients have adenocarcinoma.28 The genomic features of BTC have been investigated in several previous studies.5,24,29 The most frequently mutated genes are TP53, CDKN2A/B, and KRAS in primary iCCA tissues.5 Similar results were observed in the present work. We found that TP53, CDKN2A, and KRAS were also the most frequently mutated genes in both bile and plasma samples obtained from BTC patients. IDH1 mutations and FGFR2 fusions occur in 13% and 8%-14% of patients with iCCA in the Western population, respectively.30, 31, 32, 33 In this work, FGFR2 fusions were not observed, which might be attributed to the relatively small sample size of this study. Only one IDH1 point mutation (p.R132H) was identified in this study, which was present in bile cfDNA rather than in plasma cfDNA. These data suggested that the patient might benefit from the IDH1 inhibitor ivosidenib and bile cfDNA might be a feasible tool for identifying patients who could receive targeted therapy. A large-cohort study of patients harboring IDH1 mutations or FGFR2 fusions is warranted to validate these results. The comprehensive genomic profiling of Chinese BTC patients has not been well elucidated and is needed to be investigated in large cohorts. Furthermore, NTRK fusions occurring in 0.75% of patients with BTC,34 were not observed in this cohort.
Somatic mutations were detected in 71.4% of bile cfDNA samples, whereas these were identified in only 53.6% of plasma cfDNA samples in this study. A significantly higher median maxAF was observed in bile cfDNA than that in plasma cfDNA. These findings suggested that more tumor-derived DNA fragments are released into bile compared with plasma in patients with BTC. We also revealed that bile cfDNA detection rate was associated with AJCC stage and tumor location. Patients with stage III/IV disease had a significantly higher mutation detection rate in bile cfDNA than those with stage I/II disease in this work. CAV showed the lowest bile cfDNA detection rate (25.0%) compared with iCCA and eCCA, which might be related to its distinct anatomical location. CAV frequently causes biliary obstruction in BTC patients.12 cfDNA is not commonly released into bile in CAV patients except for those patients having a tumor invading the entire wall of bile duct. Moreover, the anatomic location of sampling bile is commonly located above the tumor, which results in a low abundance of bile cfDNA in patients with CAV.
Our study revealed that bile cfDNA samples positive for somatic mutations had a high by-variant sensitivity and PPV for SNVs/indels (both 95.5%), when genomic profiles of tumor tissues were regarded as references. Similar results were also documented in a previous report.24 However, plasma samples positive for somatic mutations revealed a compromised by-variant sensitivity (81.0%) and PPV (51.5%) for SNVs/indels. In addition, bile cfDNA showed a significantly higher overall concordance rate for SNVs/indels than plasma cfDNA. These findings indicated that bile cfDNA outperformed plasma cfDNA in detecting SNVs/indels in BTC patients. Currently available serum biomarkers for BTC, such as CEA and CA 19-9, lack sensitivity and specificity and ultimate diagnosis still requires invasive procedures for histological confirmation. A feasible and reliable biopsy strategy thus remains an unmet need for patients who cannot provide sufficient tumor tissues to confirm diagnosis. The potential roles of bile cfDNA within clinical applications have not been previously documented. This study demonstrated that the sensitivity of CA 19-9 was 82.1% in BTC diagnosis, which was comparable with that of CA 19-9 combined with plasma cfDNA. The sensitivity of CA 19-9 in conjunction with bile cfDNA achieved 96.4%. We also found that the combination of bile cfDNA and CA 19-9 could diagnose 27 cases out of 28, which were also the same ones detected by the combination of imaging and CA 19-9 plus CEA. Our proof-of-principle study suggests that bile cfDNA as a minimally invasive liquid biopsy medium might be a reliable supplemental tool for BTC diagnosis. The diagnostic value of bile cfDNA should be further validated in a large cohort of BTC patients.
There were several limitations in this study. First, the sample size was small. This study presented a proof of concept of the use of bile cfDNA as a further supplemental tool for BTC diagnosis. A large cohort is needed to investigate the feasibility of bile cfDNA in BTC diagnosis. Second, the specificity of bile cfDNA in BTC diagnosis was not explored because obtaining bile from healthy individuals or patients with benign disease affecting the biliary tree is challenging in clinical practice. Third, the potential roles of bile cfDNA in other clinical applications, including treatment monitoring and early screening, need to be explored in further studies.
In conclusion, bile cfDNA outperformed plasma cfDNA in the detection of somatic mutations in patients with BTC. Bile cfDNA might be a reliable liquid biopsy in the diagnosis of BTC and it might be a feasible supplemental tool to confirm BTC diagnosis.
Funding
This work was supported by the National Natural Science Foundation of China [Grant No. 82072637].
Disclosure
The authors declare that they have no conflicts of interests.
Data sharing
Data are not publicly available.
Ethics approval
This study was approved by the Ethics Committee of the Guangdong Provincial People’s Hospital.
Consent to participate
Informed consent was obtained from all patients for the use of their and tumor tissue, peripheral blood, and bile samples.
Consent for publication
Obtained.
Code availability
Not applicable.
Contributor Information
X.M. Chen, Email: cjr.chenxiaoming@vip.163.com.
Z.J. Zhou, Email: zhzejian@126.com.
Supplementary data
Schematic design of our study.
BTC, biliary tract cancer; cfDNA, cell-free DNA; PTCD, percutaneous transhepatic cholangial drainage; cfDNA, cell-free DNA.
Heatmap of somatic SNVs/Indels indels identified in eight tissue samples (A), 28 plasma cfDNA samples (B), and 28 bile cfDNA samples (C).
CAV, carcinoma of ampullar of Vater; cfDNA, cell-free DNA; dCCA, distal cholangiocarcinoma; SNVs, single nucleotide variants; Indels, insertions and deletions; cfDNA, cell-free DNA; GBC, gallbladder cancer; iCCA, intrahepatic cholangiocarcinoma; Iindels, insertions and deletions; pCCA, perihilar cholangiocarcinoma; dCCA, distal cholangiocarcinoma; SNVs, single -nucleotide variants; CAV, carcinoma of ampullar of Vater.
Potential effect of tumor stage and location the bile cfDNA mutation detection rate.
(A) The distribution of patients positive or negative for somatic mutations in bile cfDNA with different location of the primary tumor. (B) The difference of percentage of patients positive for somatic mutations in bile cfDNA with different location of the primary tumor. (C) The distribution of patients positive or negative for somatic mutations in bile cfDNA with different tumor stage. (D) The difference of percentage of patients positive for somatic mutations in bile cfDNA with different tumor stage.
BTC, biliary tract cancer; CAV, carcinoma of ampullar of Vater; cfDNA, cell-free DNA; dCCA, distal cholangiocarcinoma; eCCA, extrahepatic cholangiocarcinoma; GBC, gallbladder cancer; iCCA, intrahepatic cholangiocarcinoma; eCCA, extrahepatic cholangiocarcinoma; pCCA, perihilar cholangiocarcinoma; dCCA, distal cholangiocarcinoma; CAV, carcinoma of ampullar of Vater.
References
- 1.Ciombor K.K., Goff L.W. Advances in the management of biliary tract cancers. Clin Adv Hematol Oncol. 2013;11:28–34. [PMC free article] [PubMed] [Google Scholar]
- 2.Saha S.K., Zhu A.X., Fuchs C.S., Brooks G.A. Forty-year trends in cholangiocarcinoma Incidence in the U.S.: intrahepatic disease on the rise. Oncologist. 2016;21:594–599. doi: 10.1634/theoncologist.2015-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lamarca A., Barriuso J., McNamara M.G., Valle J.W. Molecular targeted therapies: ready for “prime time” in biliary tract cancer. J Hepatol. 2020;73:170–185. doi: 10.1016/j.jhep.2020.03.007. [DOI] [PubMed] [Google Scholar]
- 4.Valle J., Wasan H., Palmer D.H. Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N Engl J Med. 2010;362:1273–1281. doi: 10.1056/NEJMoa0908721. [DOI] [PubMed] [Google Scholar]
- 5.Banales J.M., Marin J.J.G., Lamarca A. Cholangiocarcinoma 2020: the next horizon in mechanisms and management. Nat Rev Gastroenterol Hepatol. 2020;17:557–588. doi: 10.1038/s41575-020-0310-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goetze T.O. Gallbladder carcinoma: prognostic factors and therapeutic options. World J Gastroenterol. 2015;21:12211–12217. doi: 10.3748/wjg.v21.i43.12211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oneda E., Abu Hilal M., Zaniboni A. Biliary tract cancer: current medical treatment strategies. Cancers (Basel) 2020;12:1237. doi: 10.3390/cancers12051237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marabelle A., Le D.T., Ascierto P.A. Efficacy of pembrolizumab in patients with noncolorectal high microsatellite instability/mismatch repair-deficient cancer: results from the phase II KEYNOTE-158 study. J Clin Oncol. 2020;38:1–10. doi: 10.1200/JCO.19.02105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abou-Alfa G.K., Sahai V., Hollebecque A. Pemigatinib for previously treated, locally advanced or metastatic cholangiocarcinoma: a multicentre, open-label, phase 2 study. Lancet Oncol. 2020;21:671–684. doi: 10.1016/S1470-2045(20)30109-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abou-Alfa G.K., Macarulla T., Javle M.M. Ivosidenib in IDH1-mutant, chemotherapy-refractory cholangiocarcinoma (ClarIDHy): a multicentre, randomised, double-blind, placebo-controlled, phase 3 study. Lancet Oncol. 2020;21:796–807. doi: 10.1016/S1470-2045(20)30157-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mizuno T., Ebata T., Yokoyama Y. Combined vascular resection for locally advanced perihilar cholangiocarcinoma. Ann Surg. 2020 doi: 10.1097/SLA.0000000000004322. In press. [DOI] [PubMed] [Google Scholar]
- 12.Wang A.Y., Yachimski P.S. Endoscopic management of pancreatobiliary neoplasms. Gastroenterology. 2018;154:1947–1963. doi: 10.1053/j.gastro.2017.11.295. [DOI] [PubMed] [Google Scholar]
- 13.Heimbach J.K., Sanchez W., Rosen C.B., Gores G.J. Trans-peritoneal fine needle aspiration biopsy of hilar cholangiocarcinoma is associated with disease dissemination. HPB (Oxford) 2011;13:356–360. doi: 10.1111/j.1477-2574.2011.00298.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kato K., Sugimori S., Kakiya Y. Brushing the distal biliary stricture in the surrounding of the papilla increased the risk of post-endoscopic retrograde cholangiopancreatography pancreatitis: a retrospective study using propensity score analysis. United European Gastroenterol J. 2017;5:1015–1023. doi: 10.1177/2050640617694279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corcoran R.B., Chabner B.A. Application of cell-free DNA analysis to cancer treatment. N Engl J Med. 2018;379:1754–1765. doi: 10.1056/NEJMra1706174. [DOI] [PubMed] [Google Scholar]
- 16.Sun K., Jiang P., Cheng S.H. Orientation-aware plasma cell-free DNA fragmentation analysis in open chromatin regions informs tissue of origin. Genome Res. 2019;29:418–427. doi: 10.1101/gr.242719.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z., Cheng Y., An T. Detection of EGFR mutations in plasma circulating tumour DNA as a selection criterion for first-line gefitinib treatment in patients with advanced lung adenocarcinoma (BENEFIT): a phase 2, single-arm, multicentre clinical trial. Lancet Respir Med. 2018;6:681–690. doi: 10.1016/S2213-2600(18)30264-9. [DOI] [PubMed] [Google Scholar]
- 18.Merker J.D., Oxnard G.R., Compton C. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J Clin Oncol. 2018;36:1631–1641. doi: 10.1200/JCO.2017.76.8671. [DOI] [PubMed] [Google Scholar]
- 19.Li Y.S., Jiang B.Y., Yang J.J. Unique genetic profiles from cerebrospinal fluid cell-free DNA in leptomeningeal metastases of EGFR-mutant non-small-cell lung cancer: a new medium of liquid biopsy. Ann Oncol. 2018;29:945–952. doi: 10.1093/annonc/mdy009. [DOI] [PubMed] [Google Scholar]
- 20.Tong L., Ding N., Tong X. Tumor-derived DNA from pleural effusion supernatant as a promising alternative to tumor tissue in genomic profiling of advanced lung cancer. Theranostics. 2019;9:5532–5541. doi: 10.7150/thno.34070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujii T., Barzi A., Sartore-Bianchi A. Mutation-enrichment next-generation sequencing for quantitative detection of KRAS mutations in urine cell-free DNA from patients with advanced cancers. Clin Cancer Res. 2017;23:3657–3666. doi: 10.1158/1078-0432.CCR-16-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Voigtländer T., Metzger J., Husi H. Bile and urine peptide marker profiles: access keys to molecular pathways and biological processes in cholangiocarcinoma. J Biomed Sci. 2020;27:13. doi: 10.1186/s12929-019-0599-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L., Masica D., Ishida M. Human bile contains microRNA-laden extracellular vesicles that can be used for cholangiocarcinoma diagnosis. Hepatology. 2014;60:896–907. doi: 10.1002/hep.27050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen N., Zhang D., Yin L. Bile cell-free DNA as a novel and powerful liquid biopsy for detecting somatic variants in biliary tract cancer. Oncol Rep. 2019;42:549–560. doi: 10.3892/or.2019.7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ying S., Ke H., Ding Y. Unique genomic profiles obtained from cerebrospinal fluid cell-free DNA of non-small cell lung cancer patients with leptomeningeal metastases. Cancer Biol Ther. 2019;20:562–570. doi: 10.1080/15384047.2018.1538614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cai Z., Wang Z., Liu C. Detection of microsatellite instability from circulating tumor DNA by targeted deep sequencing. J Mol Diagn. 2020;22:860–870. doi: 10.1016/j.jmoldx.2020.04.210. [DOI] [PubMed] [Google Scholar]
- 27.Zhu L., Huang Y., Fang X. A novel and reliable method to detect microsatellite instability in colorectal cancer by next-generation sequencing. J Mol Diagn. 2018;20:225–231. doi: 10.1016/j.jmoldx.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 28.Lim J.H. Cholangiocarcinoma: morphologic classification according to growth pattern and imaging findings. AJR Am J Roentgenol. 2003;181:819–827. doi: 10.2214/ajr.181.3.1810819. [DOI] [PubMed] [Google Scholar]
- 29.Tian W., Hu W., Shi X. Comprehensive genomic profile of cholangiocarcinomas in China. Oncol Lett. 2020;19:3101–3110. doi: 10.3892/ol.2020.11429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borger D.R., Tanabe K.K., Fan K.C. Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist. 2012;17:72–79. doi: 10.1634/theoncologist.2011-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang P., Dong Q., Zhang C. Mutations in isocitrate dehydrogenase 1 and 2 occur frequently in intrahepatic cholangiocarcinomas and share hypermethylation targets with glioblastomas. Oncogene. 2013;32:3091–3100. doi: 10.1038/onc.2012.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graham R.P., Barr Fritcher E.G., Pestova E. Fibroblast growth factor receptor 2 translocations in intrahepatic cholangiocarcinoma. Hum Pathol. 2014;45:1630–1638. doi: 10.1016/j.humpath.2014.03.014. [DOI] [PubMed] [Google Scholar]
- 33.Arai Y., Totoki Y., Hosoda F. Fibroblast growth factor receptor 2 tyrosine kinase fusions define a unique molecular subtype of cholangiocarcinoma. Hepatology. 2014;59:1427–1434. doi: 10.1002/hep.26890. [DOI] [PubMed] [Google Scholar]
- 34.Demols A., Rocq L., Charry M. NTRK gene fusions in biliary tract cancers. J Clin Oncol. 2020;38:574. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic design of our study.
BTC, biliary tract cancer; cfDNA, cell-free DNA; PTCD, percutaneous transhepatic cholangial drainage; cfDNA, cell-free DNA.
Heatmap of somatic SNVs/Indels indels identified in eight tissue samples (A), 28 plasma cfDNA samples (B), and 28 bile cfDNA samples (C).
CAV, carcinoma of ampullar of Vater; cfDNA, cell-free DNA; dCCA, distal cholangiocarcinoma; SNVs, single nucleotide variants; Indels, insertions and deletions; cfDNA, cell-free DNA; GBC, gallbladder cancer; iCCA, intrahepatic cholangiocarcinoma; Iindels, insertions and deletions; pCCA, perihilar cholangiocarcinoma; dCCA, distal cholangiocarcinoma; SNVs, single -nucleotide variants; CAV, carcinoma of ampullar of Vater.
Potential effect of tumor stage and location the bile cfDNA mutation detection rate.
(A) The distribution of patients positive or negative for somatic mutations in bile cfDNA with different location of the primary tumor. (B) The difference of percentage of patients positive for somatic mutations in bile cfDNA with different location of the primary tumor. (C) The distribution of patients positive or negative for somatic mutations in bile cfDNA with different tumor stage. (D) The difference of percentage of patients positive for somatic mutations in bile cfDNA with different tumor stage.
BTC, biliary tract cancer; CAV, carcinoma of ampullar of Vater; cfDNA, cell-free DNA; dCCA, distal cholangiocarcinoma; eCCA, extrahepatic cholangiocarcinoma; GBC, gallbladder cancer; iCCA, intrahepatic cholangiocarcinoma; eCCA, extrahepatic cholangiocarcinoma; pCCA, perihilar cholangiocarcinoma; dCCA, distal cholangiocarcinoma; CAV, carcinoma of ampullar of Vater.