Abstract
The treatment of viral infections remains challenging, in particular in the face of emerging pathogens. Broad-spectrum antiviral drugs could potentially be used as a first line of defense. The RNA-dependent RNA polymerase (RdRp) of RNA viruses serves as a logical target for drug discovery and development efforts. Herein we discuss compounds that target RdRp of poliovirus, hepatitis C virus, influenza viruses, respiratory syncytial virus, and the growing data on coronaviruses. We focus on nucleotide analogs and mechanisms of action and resistance.
Keywords: Nucleotide analogs, Antivirals, RNA virus, Viral polymerase inhibitor, Respiratory syncytial virus, Poliovirus, Influenza virus, Coronavirus, Hepatitis C virus
1. Introduction
The fight against emerging and existing viral infections is necessarily multifactorial. The economic and personal burden of viral infections is well understood and established, especially in the case of seasonally fluctuating or chronic illnesses [1], [2], [3], [4], [5], [6], [7], [8], [9], [10]. Lessons from both recent and ongoing outbreaks of previously uncharacterized viruses highlight the need for development of an array of countermeasures, both to curb ongoing situations as well as to prepare for future emergences of pathogens [11], [12], [13], [14], [15], [16]. Most distressing is that, despite several recent outbreaks, the world was overall underprepared for handling a novel coronavirus. One of the potential tools in handling viral outbreaks are broad spectrum antiviral drugs capable of managing an array of viral infections. However, most of the approved direct acting antivirals are selective, with a mechanism of action tailored to a given protein target. Herein we focus on viral RNA-dependent RNA polymerases (RdRp) as a primary target for nucleotide analog inhibitors using poliovirus (PV), hepatitis C virus (HCV), influenza viruses, respiratory syncytial virus (RSV), and coronaviruses such as SARS-CoV-2 as highlighted examples. We describe important principles of viral RdRp inhibition, defined mechanisms of action (MOA), drug resistance, and discuss the potential for the development of nucleotide analogs with a broader spectrum of activities.
Several barriers exist along the road of antiviral development. If a considerable overlap between a compound's given effects is seen between host and viral proteins, treatment may unsurprisingly cause more harm than good. Additionally, the degree of similarity between viral targets can define if any given compound is broad in its spectrum of activity. However, compounds that are more broad spectrum may also begin to affect similar host proteins [17], [18]. Viruses can be classified based upon their genome structure and replication strategy (Fig. 1 ), with significantly different genes encoded from one family to another. Even among RNA viruses, only a single gene product is shared between all known members, the RdRp, responsible for copying viral genomic information [19], [20]. Fundamentally, genome replication of an RNA virus requires a virally encoded RdRp, which is therefore a logical target when designing antiviral compounds. Additionally, the production of RNA from an RNA template is not a catalytic activity found in human polymerases, limiting the potential to affect any native cellular processes when targeting the viral protein [19]. However, the structure of RdRp enzymes or enzyme complexes can differ significantly among RNA viruses.
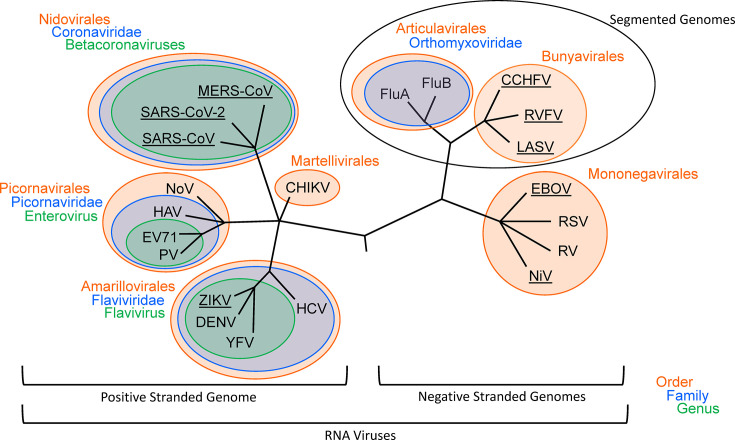
Fig. 1.
Phylogenetic tree outlining relatedness between pathogenic RNA viruses. Relations are as outlined by the International Committee on Taxonomy of Viruses (ICTV). Groupings are shown at the levels of Orders, Families, and Genera. Viruses found on the WHO R&D Blueprint priority list are underlined. Abbreviations are as follows: FluA, influenza A virus; FluB, influenza B virus; CCHFV, Crimean-Congo hemorrhagic fever virus; RVFV, Rift Valley fever virus; LASV, Lassa virus; EBOV, Ebola virus; RSV, respiratory syncytial virus; RV, rabies virus; MERS-CoV, Middle East respiratory syndrome coronavirus; SARS-CoV, severe acute respiratory syndrome coronavirus; NoV, norovirus; PV, poliovirus; HAV, hepatitis A virus; ZIKV, Zika virus; DENV, dengue virus; HCV, hepatitis C virus; EV71, enterovirus 71; YFV, yellow fever virus; CHIKV, chikungunya virus; NiV, Nipah virus.
A defining feature of almost all RNA viruses is their relatively high error rate when compared to DNA viruses, partially due to a “sloppy” RdRp and generally a lack of proofreading capability [21], [22]. There are few exceptions, including coronaviruses, that possess a proofreading 3′ to 5′ exonuclease activity [23]. A high error rate drives up the number of mutations per round of genome replication, allowing for high degrees of diversity and in turn adaptability. This increase in genetic diversity sets the stage for when antiviral treatment is applied, as resistant variants may emerge [24]. Application of drug treatment changes the selective pressures taking place between competing genotypes: where the wild type virus would have a fitness advantage without treatment, with treatment, mutants with a lower fitness at baseline that are less affected by a given drug are then able to propagate [24]. On the other hand, resistance is also important in understanding the mechanism of action of a given drug. Specific mutations give insight on specific drug-protein interactions, indicating which residues may be required for a molecule's activity. A useful step in the rational design of antivirals is therefore the determination of mechanisms of action as well as identifying possible resistance mutations.
2. Viral RNA-dependent RNA polymerases
The RNA-dependent RNA polymerase is responsible for viral genomic replication and transcription, making it a key player in the viral lifecycle [25]. Given the universal requirements for RdRps across all RNA viruses, there are not only important similarities but also differences in structure and function of these enzymes [19]. Despite how these proteins are often defined by the polymerase activity found in the conserved RdRp domain, there may also be additional functional domains which are responsible for other catalytic activities [26], [27], [28], [29]. The tertiary structure typically found in the core RdRp domain is commonly likened to a right hand, with distinct structurally conserved subdomains for the fingers, thumb, and palm, all constituting a common catalytic site of RNA synthesis [25], [30], [31], [32]. Within the RdRp domain is a set of seven (A through G) sequence motifs [32]. These motifs play a direct role in the catalytic activity of the RdRp domain, generating a strong selection for sequence and structural conservation (Fig. 2 ) [32]. The RdRp motifs themselves can be found separated in the primary structure, with their residues found in both the finger and palm subdomains, with the palm housing the majority of the motifs (A through E) involved in NTP substrate and catalytic metal coordination for catalysis [32]. The thumb subdomain, conversely, lacks consistently observed RdRp motifs, but is commonly implicated in holding and positioning the nucleic acid substrate during catalysis [32].
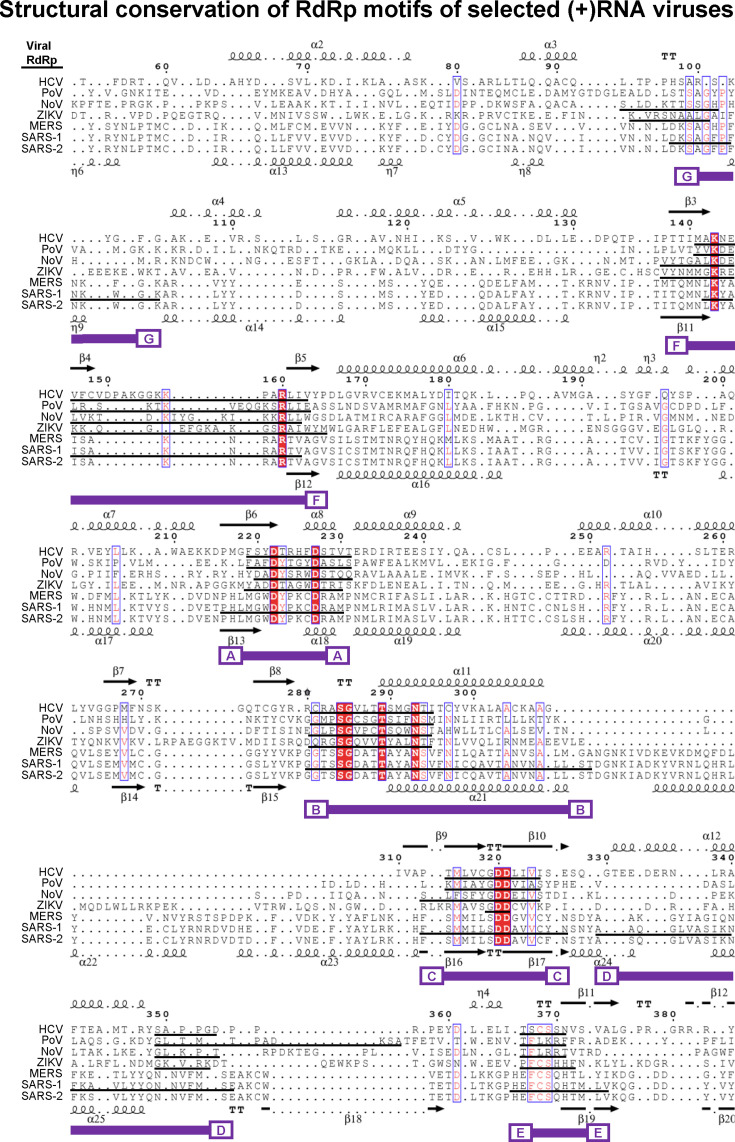
Fig. 2.
Structure-based alignment of select positive-sense genome RdRps. Sequences are aligned based on structural identity, with secondary structural elements of HCV (top) and SARS-2 (bottom) indicated. The following pdb files were used to generate the structure-based alignment: 4wta (HCV), 3o19 (PoV), 3h5y (noV), 5u0b (ZIKV), 6nur (SARS-CoV), and 6yyt (SARS-CoV-2). MERS nsp12 structural model was generated in SWISS-MODEL homology modelling pipeline [32a]. Due to the large nature of the proteins, segments containing RdRp motifs were aligned, rather than the entire protein. Conserved sections containing RdRp motifs A through G are indicated underneath in purple. Black underlined sequences correspond to motif sequences as defined in the literature. The structural alignments were conducted using Chimera UCSF software and visualized in Espript 3.0 [277], [278].
The fingers of most RdRps contain motifs F and G with motif F residues forming the NTP entry channel. Motif G residues guide single-stranded RNA template toward the active site while the N-terminal region of the motif G facilitates the exit of the double-stranded RNA from the active site [33]. In some, the “fingertips,” extended β strands, come in direct contact with the thumb, enclosing the active site and RdRp catalytic residues [25], [32]. Significant changes can be associated with distinct conformations, indicating that the dynamic spatial relationship between individual subdomains and domains is required for function [32].
Regarding specific motifs, foremost is the triad of motif A, C, and D, which are considered critical for polymerase activity [25], [30], [32]. The mechanism of NTP incorporation into the growing RNA strand is metal ion-dependent, with two divalent metal ions coordinated by aspartate residues found in both motif A and C [25], [30], [32]. The same residues also are involved in positioning incoming NTPs [25]. Motif D also participates in the polymerase mechanism—the conserved lysine or histidine directly acts as a general acid, interacting with the PPi leaving group via deprotonation [25], [34], [35].
Motif B plays a significant role in discriminating between dNTPs and NTPs, ensuring the correct substrate is incorporated into the growing RNA strand [31]. Motif E also plays a similar role in facilitating interactions between substrates by positioning the 3′ group of the primer for further extension [25]. Motifs F and G are the only motifs found within the finger subdomains [32]. Motif F plays a role in manipulating incoming NTPs likely via interactions with the template, whereas the role of motif G is less conserved [25], [32]. In some RdRps, motif G's role in binding the template and coordinating polymerase translocation has been suggested [36].
3. Picornaviruses
Picornaviruses represent a diverse family of small, nonenveloped positive-sense RNA viruses with genomes less than 10 kb in size, many of which are known to cause human disease [37], [38]. Significant picornaviral pathogens include poliovirus (PV), hepatitis A virus (HAV), coxsackievirus, human rhinovirus, foot-and-mouth disease virus (FMDV), several numbered enteroviruses, and norovirus [39], [40], [41], [42], [43], [44]. Pathogenic members of this family typically infect intestinal tissues, causing gastrointestinal symptoms [39], [40], [41], [42], [43], [44]. Some demonstrate the ability to spread elsewhere, as with HAV and poliovirus, spreading to the liver and neuronal tissues, respectively [39], [42]. Work surrounding PV, the prototypical picornavirus, stretches back close to a century [45]. The severity of the disease poses a powerful scientific impetus—while most cases resolve as acute gastrointestinal infections, a minority of cases occur such that the virus infects neuronal tissue, leading to temporary or permanent paralysis [46], [47].
Efforts toward managing PV have centered around vaccination. The first PV vaccine, developed by Jonas Salk and adopted in 1955, was based on inactivated viral particles [48]. The vaccine was soon field tested nationally in the US on a massive scale and was found to be effective [49], [50]. Soon after, an orally administered variant based on a live attenuated strain of the virus was developed by Albert Sabin [48], [50]. Due to several advantages, vaccination efforts have mainly relied on the oral Sabin vaccine [51]. However, one key difference is the capability of Sabin's vaccine to revert attenuation and cause disease like the wild type of the virus [52], [53], [54]. Interestingly, as we approach global eradication of PV, a significant concern is the potential for outbreaks triggered by the orally administered vaccine and elimination of already circulating revertant vaccine strains [53], [55].
Due to the many years spent working toward its eradication, several aspects have been intensely studied. The polymerase of PV, 3Dpol serves as a model of positive-stranded monomeric RdRps, with the first incomplete and later complete structures nearing two decades in age [56], [57]. Typical of many viral RdRps, 3Dpol shows a core fold resembling a right-hand, complete with palm, finger, and thumb subdomains [57], [58]. Found within the RdRp core as well are the expected polymerase motifs [58], [59]. Of particular note is the template entry channel found within the fingers, a highly dynamic region that was initially unresolved in the original structure [60]. Like many RNA viruses, the mutational rate of PV is remarkably high [22], [61]. With a body of work pointing out that polymerase fidelity itself not solely responsible, one likely source is the lack of proofreading capability present in 3Dpol [62], [63]. Consequently, the high rate allows for the formation of “quasispecies” due to the variety of viral progeny, leading to a diverse genetic “cloud” of viruses present, even in a single infected individual [64].
Perhaps due to the success of the PV vaccine over the past 70 years, there are currently no approved antiviral treatments for PV [65], [66]. However, there does still exist a need due to circumstances where the use of a vaccine has been limited [67]. Backing this is a significant body of work examining the effects of antivirals on PV, much of which has informed our understanding of basic RNA virus biology: RdRp nucleoside inhibitors (NIs), RdRp template channel-interacting compounds, and capsid-binding compounds have shown inhibitory activity against PV [68], [69], [70], [71]. Of specific note is the well studied nucleoside compound ribavirin (RBV), a broad spectrum guanosine-like analog [72]. Possibly explaining the broad spectrum of activity are the several mechanistic activities shown by RBV, including modulating cellular GTP concentrations, modulating host T-cell responses, inhibiting RNA capping, and directly interacting with viral RdRps by either affecting enzyme activity or inducing lethal mutagenesis of viral genomes through G to A and C to U transition mutations [72], [73]. Mechanistic studies performed with PV and HCV demonstrate the ability of RBV to be incorporated at sites templated by cytidine or uridine residues, and when found in the template, allow for incorporation of cytidine or uridine [74], [75]. One likely explanation is the rotational flexibility of the carbon-carbon bond connecting the amide group of the molecule to the 5 atom heterocycle base mimic [76]. Depending on the rotation of this bond, RBV can mimic the base pairing characteristics of either adenosine or guanosine (Fig. 3 ) [74]. Of significance is the direct activity of RBV upon PV 3Dpol, where the increase in mutational rate drives the RdRp beyond the tolerable threshold into error catastrophe, rendering the produced genome nonviable [77].
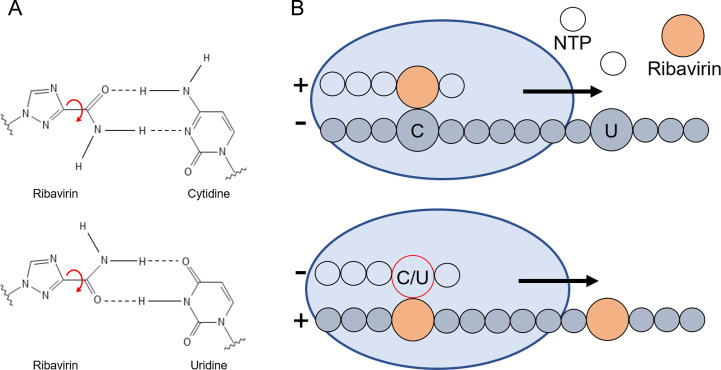
Fig. 3.
Mechanism of action of ribavirin on poliovirus 3Dpol. (A) Base pairing capability of ribavirin with cytidine and uridine. Based on the rotation of the bond between the 5-carbon heterocycle and the amide group, ribavirin is capable of base pairing with either uridine or cytidine. (B) Cartoon depiction of ribavirin incorporation and mutagenesis in resulting rounds of replication. Ribavirin can be incorporated at both cytidine and uridine-templated positions. When the ribavirin-containing strand is then used as a template, cytidine or uridine may be added against ribavirin-templated positions [74].
Panel (A): Adapted from S. Crotty, et al., The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen, Nat. Med. 6 (12) (2000) 1375–1379.
Perhaps the strongest evidence for lethal mutagenesis as a possible antiviral mechanism is the emergence of a RBV resistance-conferring mutation that counteracts these effects. In PV 3Dpol, the mutation G64S emerges and increases polymerase fidelity, lowering the overall mutational load below that of the tolerable threshold [78]. While this mutation in isolation may seem beneficial to the virus, it is associated with lower fitness [79]. For some time after the initial discovery, the mutation was believed to result in lower fitness as a direct result of the lower mutational rate in the absence of RBV, presumably by decreasing quasispecies diversity [64], [79]. Recent work suggested that the decreased fitness of PV 3Dpol G64S is due to a slower, more careful polymerase rather than decreased genetic variety [80], [81]. Later work by Kempf et al. described how L420, a thumb motif residue, was critical in allowing for WT levels of recombination, with L420A heavily lowering the frequency of recombination (Fig. 4 ) [82]. Recent work has solidified the connection between recombination and RBV resistance: While the G64S mutant does demonstrate a higher fidelity over WT, its RBV resistance is dependent on polymerase recombination, with L420A abrogating its resistant phenotype [83].
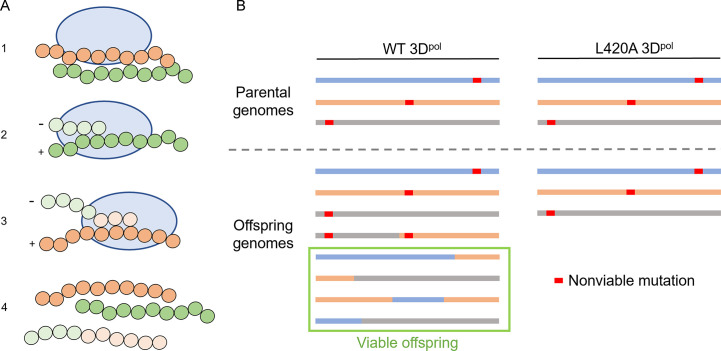
Fig. 4.
Outline of poliovirus recombination. (A) Stepwise outline of the generation of a recombinant genome from two parental genomes. (1) A cell is infected with two genomes harboring sequence differences. (2) The RdRp of poliovirus, 3Dpol, begins to replicate one of the genomes. (3) 3Dpol transfers over to a new template from a differing genome without aborting replication, producing a genomic copy that contains sequences from both templates. (4) The final product yields a new genome distinct from the parental material. (B) Mechanism of recombination-mediated avoidance of lethal mutagenesis as compared between wild type and a recombination-deficient mutant (L420A) as suggested by Kempf et al. [83]. Despite only having initially nonviable genomes, wild type virus can establish entirely viable genomes via recombination between nonviable material. With a mutant polymerase that exhibits greatly reduced recombination rates, the generation of viable offspring is limited.
Panel (A): Adapted from B.J. Kempf, et al., Picornavirus RNA recombination counteracts error catastrophe, J. Virol. 93 (14) (2019).
Similar results have been observed in related picornaviruses and with similar compounds: experiments performed with FMDV demonstrate RBV resistance mutations, some also located in the template channel, and some associated with increased fidelity [84], [85], [86], [87], [88], [89]. The same high fidelity, RBV resistance phenotype has been observed in enterovirus 71 [90]. Work again with FMDV examining 5-fluorouracil also showed potential for mutagenic compounds other than RBV, with similar high-fidelity mutants being identified [91], [92]. The observation that N-6-substituted purine analogs also demonstrate broad spectrum mutagenic effect but are attenuated by PV 3Dpol G64S further suggests a common mechanism of RBV [93], [94].
Recent work, leveraging advances in assay capabilities, has also highlighted a class of compounds with a previously unknown mechanism of action against PV (and later against coronaviruses, below) [95]. Using magnetic tweezers capable of measuring the force of tension across an RNA template between a magnetic bead and a glass coverslip, Dulin et al. demonstrated that pyrazine-carboxamide pseudobase containing nucleotide analogs such as favipiravir and the related T-1106 can act to induce backtracking, causing the polymerase to slide backward and pause, eventually leading to inhibition [95]. Using susceptible and resistant mutants to ribavirin, which contains a similar pseudobase to the above, they demonstrate nonoverlapping mechanisms between ribavirin and their compounds capable of inducing backtracking.
4. Hepatitis C virus
Hepatitis viruses represent a phylogenetically diverse grouping, unified by their ability to infect the liver [96], [97], [98], [99]. Hepatitis B, C and D viruses exist as significant pathogens due to their ability to manifest chronic infections which, if untreated, may lead to cirrhosis, liver failure, and liver cancer [96], [97], [98], [99]. Despite its discovery close to 30 years ago, vaccine development for hepatitis C virus (HCV) has so far been unsuccessful, with no approved or effective vaccine for any genotype to date [100]. While the most recent attempts have established a response and result in lowered viral load, they do not protect from chronic disease [101]. Thankfully, the standard of care for HCV has progressed to a broadly successful clinical treatment, demonstrating how incremental steps forward, pharmacologically, may lead to an effective treatment [102]. With over 150 million people worldwide infected chronically, understanding mechanisms of resistance tied to successful compounds allows for us to preserve such treatments as we progress [103].
HCV contains a positive-sense, single-stranded RNA genome of ~ 9700 bases, producing all its proteins via a single open reading frame [104]. Currently there are seven circulating HCV genotypes (1–7), with approximately 30% of the genome differing between genotypes [105], [106]. Genotype 1 constitutes close to half of all HCV cases globally, and historically has been the most resistant to treatments [107], [108]. As genotypes themselves differ significantly, the barrier may be larger or smaller depending on those differences, allowing certain genotypes to establish resistance with fewer mutations [109]. Additional factors such as the generation of quasispecies in infected individuals as well as intrinsic bias in mutational rates also contribute to the generation of resistance associated mutants, with some being detectable at baseline prior to treatment [110], [111], [112], [113].
A major antiviral target of HCV is the RdRp, nonstructural protein 5B (NS5B). The structure of this monomeric polymerase was solved near the end of the millennium independently by several groups [114], [115], [116]. These structures and later work demonstrated NS5B contained several unique motifs such as a thumb subdomain β-loop insertion and C-terminal membrane anchor, in addition to the expected RdRp motifs and right-handed tertiary structure [117]. The enclosed nature of NS5B was also remarked upon, with the active site of the RdRp enveloped in the finger and thumb subdomains, including the mentioned thumb β-loop [117]. Rearrangements during initiation of polymerase activity by these components has been shown to be indispensable for efficient activity, outlining a requirement for dynamic conformational shifts [118], [119].
Early treatments for HCV infection centered around the use of RBV alongside pegylated interferon alpha, which remained the standard of care into the early 2010s [120], [121]. RBV has many purported mechanisms, with no one clear effect driving its efficacy against HCV [122]. Evidence does exist to indicate that RBV acts directly upon the polymerase as a mutagenic agent, and/or as an inhibitor of RNA synthesis, similar to its effects on poliovirus, however the lack of concrete resistance-associated mutations in NS5B point to other mechanisms [74], [75], [121], [123], [124].
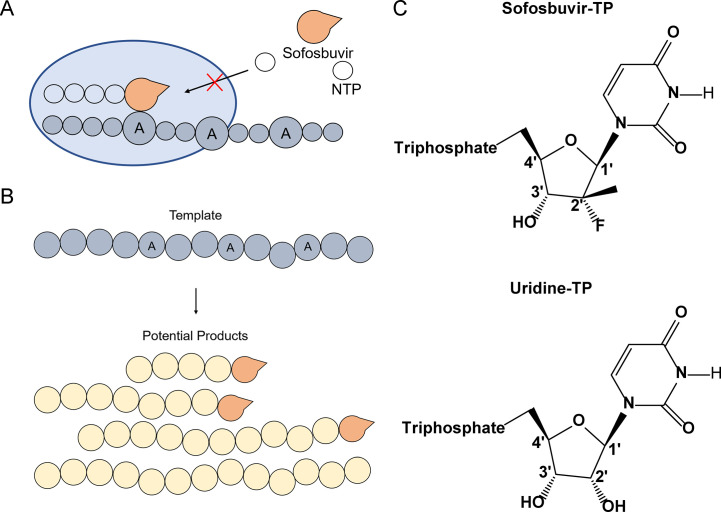
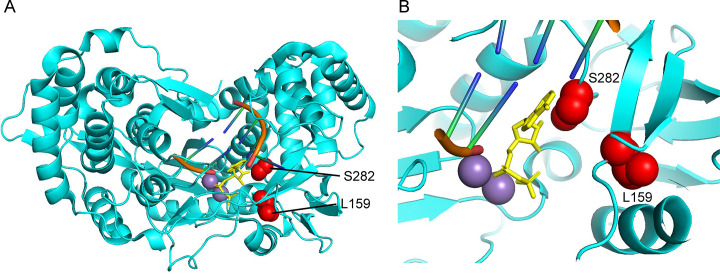
A major step forward in the treatment of HCV was the development of sofosbuvir (SOF), a highly effective uridine analog approved for HCV treatment in 2013 [125], [126]. Mechanistically, SOF-triphosphate acts as a substrate of NS5B, leading to its incorporation into the growing RNA genome after modification from its prodrug to its active form, eventually resulting in chain termination (Fig. 5 ) [127], [128]. Theories primarily supported by structural data suggest that SOF, once incorporated, spatially blocks the attack of additional nucleotides, effectively blocking further elongation [117]. Data from similar molecules suggests this steric clash occurs between incoming nucleotides and the bulky 2′-methyl group [129]. Notably, SOF was observed to be an effective treatment regardless of HCV genotype [130], [131]. While resistance mutations have been noted for SOF, they arise in surprisingly low abundance and are typically not observable at baseline prior to treatment [111], [132], [133], [134]. The primary mutation associated with SOF resistance, S282T, is found within the active site of NS5B (Fig. 6 ) [117], [131]. The mutation would be expected to cause a conflict between the side chain of T282 and SOF, limiting SOF incorporation [117]. Potentially due to the nature of the mutation and its proximity to the active site, S282T is associated with significant loss of fitness in the virus, with resistant genotypes lacking the ability to persist in individuals after SOF treatment [132], [135]. Other mutations have been associated with SOF resistance. L159F, also located in the vicinity of the active site, has been implicated with S282 due to their close structural proximity and the emergence of L159F mutations among low responding SOF patients [136], [137].
Fig. 5.
Mechanism of action of sofosbuvir (SOF) against HCV NS5B. (A) Incorporation of SOF into the growing RNA strand prevents additional NTP incorporation. Sofosbuvir can be incorporated at positions templated by adenosine, mimicking the natural substrate, uridine [127], [128]. After incorporation, due to steric clashes between SOF and incoming NTPs, no further elongation is possible [117]. (B) Schematic of potential products of chain termination as caused by SOF incorporation. At each position with a template adenosine, SOF incorporation may result in chain termination, rendering that round of replication or transcription nonviable. (C) Structure comparison of SOF (uridine analog) and uridine. Of note is the fluoro- and methyl-group moieties found on the 2′ ribose carbon of sofosbuvir.
Fig. 6.
Structural insights into sofosbuvir resistance mutations found in hepatitis C virus NS5B (4WTJ). (A) Overview of NS5B structure with template and elongating RNA strands (orange), incoming ADP (yellow), manganese ions (purple), and two amino acid residues for which sofosbuvir resistance mutations exist for (red, S282T, L259F). (B) Zoomed view of the spatial position of S282, L159, and the incoming ADP nucleotide. As suggested by Appleby et al., due to the proximity of residue 282 to incoming nucleotides, the T282 mutant may sterically clash with the 2′ methyl group found on the functional sofosbuvir molecule, prohibiting incorporation [117]. Structures oriented in PyMol, PDB: 4WTJ.
In addition to compounds that mimic natural substrates of NS5B, several nonnucleoside inhibitors (NNIs) that act upon the RdRp allosterically have been developed and explored. These compounds bind to surface pockets found on NS5B and are categorized based on their site of binding and the mutations associated with that site [138]. Currently there are five binding sites that have been identified and targeted: two thumb sites (T1 and T2), two palm sites (P1 and P2), and one site associated with the β-loop (B1) [139], [140]. Structural work examining the conformational landscape of NS5B suggests that NNIs may share a uniform mechanism, blocking the transitions between the initiation and elongation associated conformations, inhibiting polymerase activity [141], [142]. When compared to NIs, the barrier for resistance to develop is much lower [143], [144]. Indeed, many NNI compounds are genotype restricted, with some resistance mutations commonly found at baseline [143], [144].
The standard of care for treating chronic HCV infection has evolved greatly over time, starting with RBV and pegylated interferon alpha treatments [122]. Limitations of such a treatment centered on rates of treatment success around 50% and significant side effects suffered by patients [122]. Current standard of care is determined depending on several factors, including HCV genotype, if the patient is treatment naive, the degree of liver cirrhosis, the presence of any chronic liver disease, and the presence of chronic viral coinfections [145]. Thanks to the degree of therapeutic development since the days of RBV/interferon treatment, response rates with directly acting antivirals can approach nearly 100% [146], [147], [148].
5. Respiratory syncytial virus
Respiratory syncytial virus (RSV) belongs to the Mononegavirales order of nonsegmented negative strand RNA viruses and is a member of Pneumoviridae family of pneumoviruses [149]. An approximately 15 kb single-stranded RSV genome coding for 11 proteins is encapsidated with the viral nucleoprotein and is associated with the viral RNA polymerase complex inside an enveloped viral particle [149].
RSV infection is omnipresent in children as the leading cause of acute lower respiratory infections in infants and a life-threatening pathogen to the elderly and immunocompromised [150], [151], [152]. Prophylactic developments were severely slowed due to complications in clinical trials of an early vaccine, with no vaccine currently available today [153], [154]. Palivizumab, a humanized monoclonal antibody against the F glycoprotein, was approved for prophylaxis of RSV infections in 1998, however, it has recently been reported that its effect on RSV transmission rates is limited [155]. The only antiviral agent that is approved for treatment of RSV infections is ribavirin (RBV), a broad-spectrum antiviral nucleoside analog—unfortunately, the efficacy of RBV when used to treat RSV is poor [156], [157], [158].
Considering the recent successes with anti-HIV, -HCV, and -SARS-CoV-2 antivirals targeting viral nucleic acid polymerases, RSV RNA polymerase represents an attractive target for the development of novel nucleoside analogs. The functional core of the RSV RNA polymerase consists of two subunits, a ~ 250 kDa large polymerase (L) and a tetramer of the ~ 27 kDa phosphoprotein cofactor (P), both of which are present in the viral particle [159], [160], [161]. Recent cryo-EM structures revealed that the RSV RNA polymerase exists as a complex of one L subunit with P tetramer adopting an asymmetric tentacular arrangement [160], [161]. L protein is a multifunctional enzyme catalyzing RNA transcription, capping and polyadenylation of transcripts, as well as genome replication each mediated by the L specific subdomains [161].
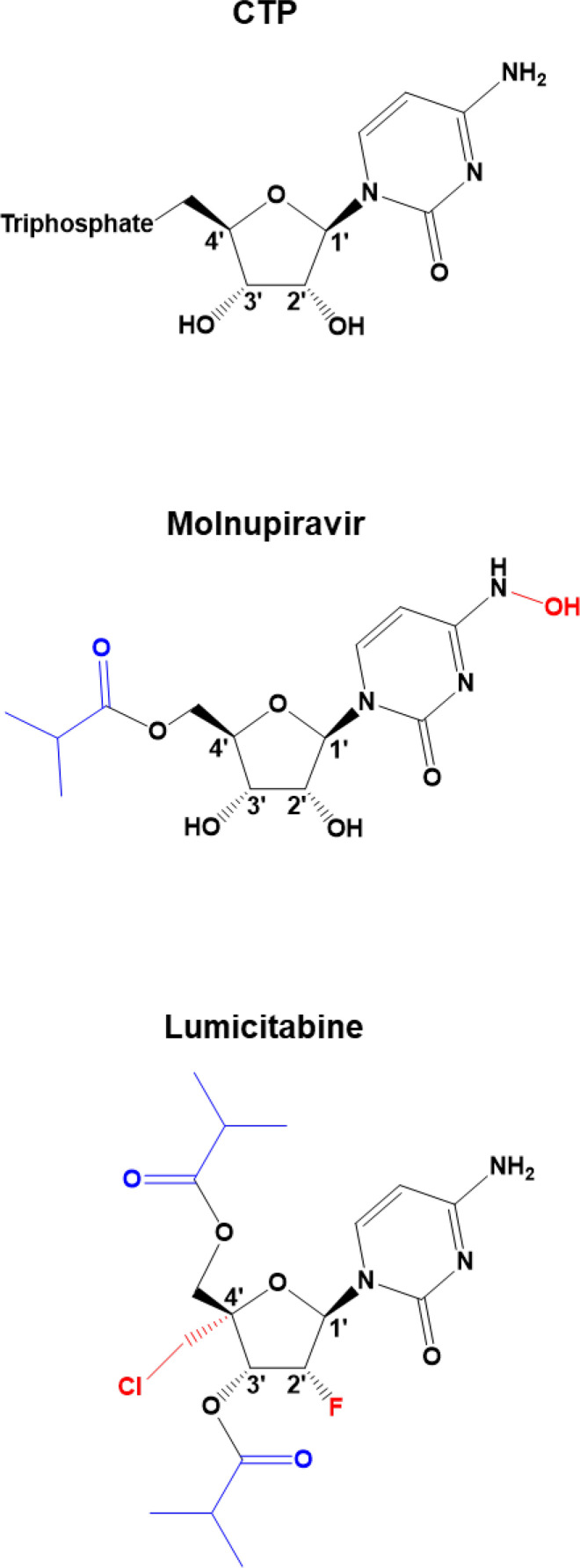
RNA transcription and genome replication is conducted by the RdRp domain of the L protein, which contains the highly conserved motifs [32]. While both nucleoside and nonnucleoside inhibitors have been in the development over the recent years [162], two cytidine nucleoside analogs are of particular interest: molnupiravir (EIDD-2108) and lumicitabine (ALS-8176) (Fig. 7 ): molnupiravir has a modification on the base moiety, while lumicitabine contains a sugar moiety modified at 2′ and 4′ positions [151], [163]. Both compounds are intracellularly metabolized to their triphosphate forms to compete with their natural counterpart CTP for nucleotide incorporation by viral RNA polymerases [163], [164].
Fig. 7.
Chemical structures of CTP analogs. Features highlighted in red illustrate the portion of the molecules that mediate the inhibitory effect on the viral RNA synthesis. Features highlighted in blue illustrate modifications that increase bioavailability of the analog.
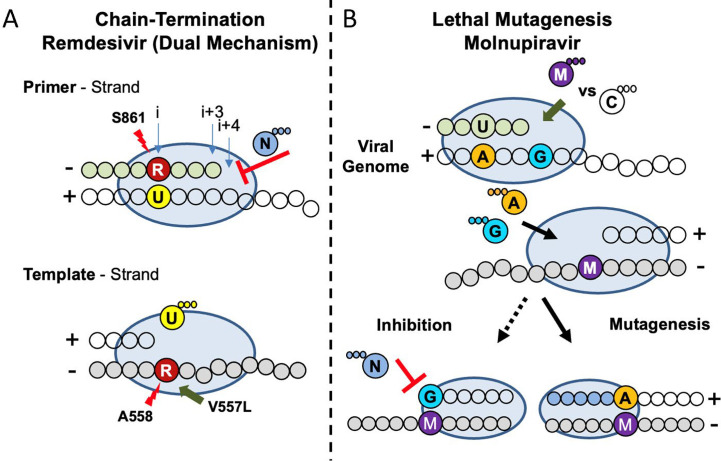
Molnupiravir is a broad-spectrum antiviral that is currently undergoing clinical trials against SARS-CoV-2 infection. It has been shown to act as a mutagen, promoting G-to-A and C-to-U transition mutations in coronaviruses and influenza virus [163]. Interestingly, in the context of RSV only A-to-G transition mutations have been observed [163]. While the mechanism of action (MOA) of molnupiravir against SARS-CoV-2 has been recently elucidated (Fig. 8B) [165], [166], its MOA against RSV remains unknown.
Fig. 8.
Model of the mechanism of action of RDV and molnupiravir against SARS-CoV-2 RdRp. (A) RDV functions as a chain terminator preventing RNA synthesis during both primer and template strand synthesis. (Top) Incorporation of RDV-MP (red) by the viral polymerase (blue oval) opposite UMP in the template (yellow/clear bubbles) results in a steric clash with the residue S861 and incomplete inhibition. (Bottom) The primer is eventually extended and is later used as a template. A clash between RDV-MP and the backbone of A558 is responsible for the reduction in UTP (U) binding. V557L, previously identified in vitro, can compensate for this clash promoting low level resistance. (B) Molnupiravir incorporation results in mutagenesis promoting incorrect base-pairing. (Top) Molnupiravir (purple) primarily base-pairs with GMP (blue), once extended the subsequent template strand promotes ATP (orange) binding opposite molnupiravir explaining lethal mutagenesis observed in cell culture as a result of A-to-G and U-to-C transitions. Biochemical evidence showed evidence of GTP (blue) binding opposite molnupiravir in the template, but this resulted in minimum inhibition and extension of this product would not result in a mismatch.
Adapted from C.J. Gordon, et al., Molnupiravir promotes SARS-CoV-2 mutagenesis via the RNA template, J. Biol. Chem. (2021) 100770. E.P. Tchesnokov, et al., Template-dependent inhibition of coronavirus RNA-dependent RNA polymerase by remdesivir reveals a second mechanism of action, J. Biol. Chem. 295 (47) (2020) 16156–16165.
Lumicitabine is the only RSV-specific nucleoside inhibitor that has reached the stage of clinical trials. ALS-8176 is an improved bioavailability version of the parent drug ALS-8112, which was found to have inhibitory effects on RSV replication in vitro [167]. ALS-8112 is a cytidine analog with sugar moiety modifications, with the triphosphate version (ALS-8112-TP) being recognized by the RSV RNA polymerase as a substrate for RNA synthesis, albeit about 10-fold less than the natural counterpart CTP [167]. Incorporation of the compound into the extended RNA primer caused immediate chain termination due to the inability to incorporate subsequent nucleotides [168]. In addition, the compound was not RSV strain specific [168]. Lumicitabine and ALS-8112 resistance-conferring mutations have been characterized and often found together in replicon assays: M628L, A789V, L795I, and I796V located in the conserved motif B of the RdRp domain [168].
When analyzed in isolation, A789V was the only mutation capable of conferring resistance but was still unable to match the phenotype of the quadruple mutant [169]. Structurally, A789V may be significant due to its position near the catalytic motif C as well as the residue D700 in the motif A, however the overall protein structure suggests that all the mutations together may play a role in modifying the active site and facilitating nucleoside discrimination [160], [161].
Early clinical data with Lumicitabine indicated that it was highly efficacious in decreasing viral load and speeding viral clearance with the additional observation of no resistance mutants arising because of treatment [169], [170]. Unfortunately, further clinical trials were stopped due to treatment-related neutrophil abnormalities [171].
6. Coronaviruses
The Coronaviridae are a diverse family of positive-sense, single stranded RNA viruses with a genome approximately 30 kb in size—the largest genome of any known RNA virus [172], [173]. Coronaviridae are comprised of four genera: alphacoronavirus, betacoronavirus, gammacoronavirus, and deltacoronavirus. Alpha- and betacoronaviruses are only capable of infecting mammals, while the host range for gamma- and deltacoronavirus also includes avian species [174], [175]. There are seven known coronaviruses capable of causing disease in humans predominantly as a result of infection in the upper respiratory and gastrointestinal tract [174], [176]. Consequential outcomes due to coronavirus infection vary from the common cold to severe respiratory damage that can ultimately be fatal [176], [177]. The emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [178], the causative agent of COVID-19, put coronaviruses in the global spotlight. However, previous outbreaks of SARS-CoV and Middle East respiratory syndrome CoV (MERS-CoV) foreshadowed the pathogenic potential possessed by coronaviruses [179], [180].
One outcome of the COVID-19 has been the display of technological advancements resulting in a rapid and expansive pipeline of vaccine candidates [181]. As of June 2021, three vaccines have received Emergency Use Authorization (EUA) from the Food and Drug Administration (FDA). Pfizer-BioNTech [182] and Moderna [183] are mRNA vaccines which demonstrated an efficacy 95% and 94% respectively in protecting against COVID-19 infection [184], [185]. Johnson & Johnson, an adenovirus vector that is unable to replicate, received EUA after exhibiting an efficacy rate of 66.1% during ongoing phase 3 trials [186], [187]. AstraZeneca another adenovirus vaccine, has received approval in Europe and Canada based on the pooled results of four independent clinical trials, efficacy was determined to be approximately 70% [188], [189]. Conversely to vaccine development, antibody cocktails also received EUA for the treatment of outpatients [190], and the nucleotide analog remdesivir has been approved for COVID-19 treatment [191].
Upon host cell entry, ORF1a and ORF1ab of the viral genome is translated and a − 1 ribosomal frameshift enables the production of two distinct polyproteins, 1a and 1ab [192]. The proteolytic processing of 1a and 1ab result in two polyproteins comprised of nonstructural protein (nsp) 1-11 and 1-16 respectively [193], [194]. Posttranslational assembly results in the formation of a replicase transcriptase complex (RTC), driving RNA synthesis and supporting viral propagation [193], [195]. The large genome puts an extended pressure on the replication machinery during RNA synthesis, thus coronaviruses require higher replication fidelity as compared to other RNA viruses [196], [197]. Two crucial subunits within the RTC responsible for fidelity are nsp12 and -14, the RdRp and the 3′ to 5′ exonuclease (ExoN), respectively [194], [196], [197]. The RdRp drives RNA synthesis while the ExoN functions as a proof-reader repairing errors in nucleotide incorporation [196], [197]. The vital role of the RdRp and its highly conserved nature among coronaviruses makes it a logical target for therapeutics to prevent viral propagation [195], [198]. The proofreading capability of the ExoN presents a plausible evolutionary development to support the replication of their large genome [199]. Consequently, the excision of incorporated nucleoside analogs by the ExoN adds an additional layer of complexity for therapeutic development [199], [200].
Additional components crucial to RNA synthesis by the RdRp are nsp7 and -8, often implicated in processivity to support the replication of their large genome [21], [201], [202], [203]. The exact role of nsp7 and -8 remains relatively undefined, however biochemical and structural data suggests nsp8 could serve to function as a primase or a 3′-terminal adenylyltransferase [204], [205], [206], [207]. The structure of a RdRp complex with nsp7 and -8 has been solved for of SARS-CoV and SARS-CoV-2 identifying a stoichiometric ratio of 1:2:1 for nsp7, -8, and -12 respectively [33], [207], [208], [209], [210].
The antiviral activity of nucleoside analogs ribavirin [211], [212], BCX4430 [213], gemcitabine hydrochloride [214], and acyclovir fleximer [215] have been investigated in vitro against SARS-CoV and MERS-CoV, while mizoribine was tested solely against SARS-CoV [216]. A shared characteristic among these therapeutics is a low antiviral effect, indicating the potential to treat coronavirus infection is low. In addition to being readily excised, ribavirin's rate of incorporation was significantly lower than its natural counterpart GTP (Km = 136 μM vs 1.5 μM) [200]. The poor incorporation efficiency of ribavirin observed biochemically coincides with a modest antiviral effect in cell culture studies previously mentioned. Taken together, the requirements for a nucleotide analog to elicit an antiviral effect is twofold, relying on efficient incorporation while going unrecognized by the ExoN to avoid excision.
Favipiravir has demonstrated an antiviral effect against SARS-CoV-2 in vitro with an inhibitory concentration in the high micromolar range [217]. The mechanism of favipiravir remains ambiguous, but previous biochemical, cell culture and structural work suggests lethal mutagenesis as the main MOA [217], [218]. Biochemical studies investigating incorporation efficiency of favipiravir by the SARS-CoV-2 RdRp are contradictory [217], [218], [219], [220], furthermore it appears to be ill-equipped to outcompete its natural counterpart for incorporation [220]. One possible explanation may reside in recent data showing that the SARS-CoV-2 RdRp is capable of backtracking [221]. As favipiravir and related compounds have been shown to trigger backtracking and inhibition upon incorporation upon incorporation by the PV RdRp 3Dpol [95], a similar mechanism may be at play during incorporation by the SARS-CoV-2 RdRp.
Remdesivir (RDV, formerly GS-5734) is a nucleotide analog that, when metabolized to its triphosphate form (RDV-TP), mimics the natural nucleotide adenosine triphosphate (ATP) with 1′-C-nucleoside bond and a 1′-cyano-substitution [222]. RDV is broad-spectrum in nature showing an antiviral effect in vitro and in vivo against Nipah virus (NiV), respiratory syncytial virus (RSV), and Ebola virus (EBOV) [222], [223], [224], [225]. Biochemical assays demonstrated RDV-TP can compete with its natural counterpart, ATP, for incorporation by the RdRp of Ebola resulting in delayed chain-termination [226]. Moreover, the therapeutic potential of RDV was tested in a randomized, controlled trial in 2019 during the Ebola outbreak in the Democratic Republic of the Congo (DRC), but it was ultimately determined two different antibody therapies proved to be more effective [227].
RDV has undergone extensive investigation in its ability to inhibit the RdRp of CoV on route to being the first FDA approved antiviral treatment for COVID-19 [191]. Approval was in large part due to a randomized clinical trial by the National Institute of Allergy and Infectious Disease (NIAID), concluding RDV treatment reduced median time to recover by 5 days [228]. However, another randomized clinical trial performed by the World Health Organization (WHO) did not arrive to the same conclusions [229]. This inconsistency could be linked to discrepancies in experimental design, both of which failed to measure the viral load [230]. Initial investigation demonstrated an EC50 in the submicromolar range [231], [232], and reduced the viral load of SARS-CoV and MERS-CoV in a murine model [232], [233]. The efficacy of RDV was demonstrated further when treatment of MERS-CoV infection in a rhesus macaque model decreased viral replication [234]. The potency of RDV as an antiviral against SARS-CoV-2 was demonstrated in human airways epithelial (HAE) lung cells as well as in mice infected with a SARS-CoV expressing the polymerase of SARS-CoV-2 [235]. RNA synthesis assays determined the SARS-CoV-2 RdRp complex incorporated RDV-TP approximately three times more efficient than its natural counterpart ATP [220]. Following incorporation of RDV-TP in the nascent RNA strand RNA synthesis arrest occurs following the incorporation of three nucleotides (i + 3), demonstrating the first MOA delayed chain termination (Fig. 8A, top) [220]. A biochemical mutagenesis assessment revealed that delayed chain termination is a result of a steric clash with the 1′-cyano-group of the incorporated RDV-TP and the hydroxyl group of S861 [236]. Moreover, increased nucleotide concentrations can diminish the intermediate i + 3 product suggesting newly synthesized negative sense copies of the viral genome contain an incorporated RDV (RDV-MP) [236]. This presents the second mechanism, template-dependent inhibition (Fig. 8A, bottom) [236]. When RDV-MP is embedded in the template strand, incorporation of complementary UTP is severely compromised requiring an increased concentration to promote RNA synthesis beyond the RDV-MP blockade [236]. This obstruction can be attributed to a clash between the backbone of residue A558 with the SARS-CoV-2 RdRp and the 1′-cyano-group of RDV-MP [236].
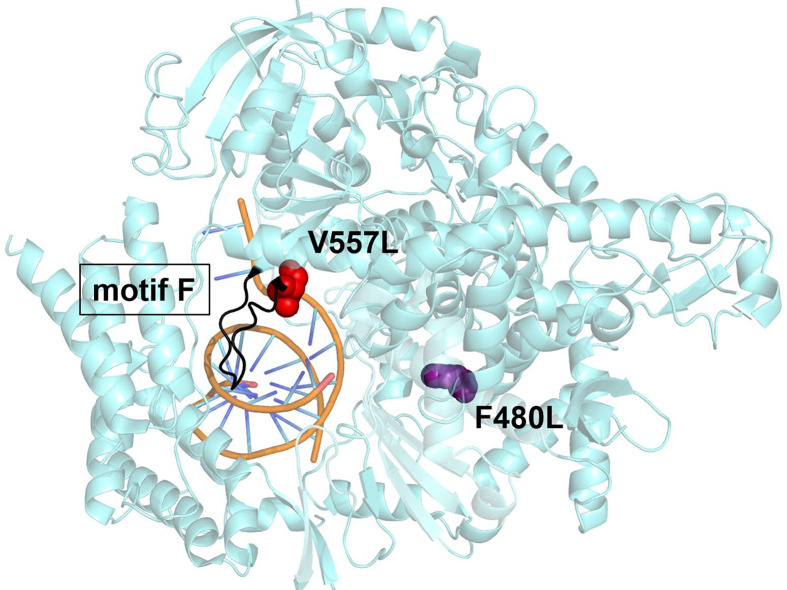
Serial passages of β-CoV model murine hepatitis virus (MHV) with increasing concentrations of the RDV resulted in a resistant phenotype with two amino acid mutations (F476L and V553L) within the RdRp [231]. These residues are conserved within the CoV RdRp, and when aligned, correspond to F480 and V557 of SARS-CoV, and F481 and V558 of MERS-CoV (Fig. 9 ) [231]. Induced mutagenesis in this fashion further solidifies that the mode of inhibition by RDV is imposed on the RdRp of CoV [231]. V557L is positioned in motif F while F480L is not associated with any structural motif [198], [231]. Both residues are within the fingers subdomain of the CoV RdRp responsible in forming the passage that supports incoming nucleotides [231], [235]. The adjacent location of V557 to A558, the residue likely responsible for template-dependent inhibition [236], presents an interesting dynamic for a potential mechanism of resistance. When studied biochemically, the single mutation, V557L, within the SARS-CoV-2 RdRp conferred the low-level resistance previously observed in cell culture [236]. The mechanism of resistance was specific to RDV-MP in the template as decreased UTP concentration was needed to promote RNA synthesis beyond RDV-MP [236]. V557L had no effect on RDV-TP incorporation efficiency or delayed chain termination [236]. RDV resistance studies with EBOV identified the mutation F548S conferring low level resistance [237]. F548 resides in motif F, although its location is not identical to V557, this demonstrates a potential unifying broad spectrum mechanism of RDV as it targets structurally equivalent regions within the RdRp [237].
Fig. 9.
Structural representation of the SARS-CoV-2 RdRp (nsp12) containing the resistance conferring mutations, 7BV2. Two RDV resistant mutations selected for in vitro with the coronavirus model murine hepatitis virus (MHV) align to V557L (red) and F480L (purple) in SARS-CoV-2. Both residues are located in the finger subdomain, V557L is associated with structural motif F (black). This motif is implicated with nucleotide incorporation and contacting the 5′ end of the template. Computational and biochemical studies have associated A558 with template-dependent mechanism of RDV, neighboring V557L counteracts this effect resulting in low-level resistance. The distal location of F480L from the active site and V557L suggest it does not have a direct role in RDV resistance as it relates to RNA synthesis. Structures oriented in PyMol, PDB: 7BV2 [208].
Adapted from M.L. Agostini, et al., Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease, mBio 9 (2) (2018) e00221-00218.
Four additional mutations were identified in the RDV resistant CoV strain none of which were located in the ExoN [231]. Deletion of the ExoN resulted in an increased sensitivity to RDV, indicating nsp14 could participate in the excision of the incorporated RDV [231]. A mechanism of action for the possible excision of RDV by the ExoN has yet to be described in the literature. However, the efficiency of RDV incorporation as well as potential protection due to delay-chain termination could explain why the ExoN is unable completely alleviate RDV induced inhibition [220], [238].
Molnupiravir, the prodrug of β-d-N4-hydroxycytidine (NHC), when metabolized to its triphosphate form (NHC-TP) is an effective substrate for the CoV RdRp [165], [166]. The potency of molnupiravir is exhibited by the submicromolar EC50 values determined against MERS-CoV, SARS-CoV, and SARS-CoV-2 in a variety of cell types [233]. CoV Cell culture data revealed the likely mechanism of molnupiravir is lethal mutagenesis, promoting A-to-G and C-to-U transitions [233], [239]. This is a result of NHC-TP functioning as a tautomer existing as both an N-hydroxylamine and an oxime [240]. RNA synthesis assays revealed NHC-TP as a substrate for the CoV RdRp, which is incorporated most efficiently in its hydroxylamine form as a CTP analog opposite a template G [165], [166]. Incorporated NHC-MP does not result in chain termination; therefore NHC-MP will be present in subsequently generated copies of the CoV genome [165], [166]. Additional biochemical experiments revealed GTP and ATP incorporation is promoted opposite the templated NHC-MP [165], [166]. This suggests NHC-MP in the template exists in both tautomeric forms acting as CMP or UMP forming a solidified mutagenic mechanism of inhibition by molnupiravir (Fig. 8B) [165], [166].
Sequential passaging of MHV and MERS-CoV in the presence NHC revealed a high genetic barrier for the antiviral resistance [239]. Cross-resistance to NHC was examined with the previously mentioned RDV resistant MHV strain (F476L and V553L) [233]. Interestingly, the known RDV resistant strain of MHV displayed an increased sensitivity to NHC [239]. As opposed to RDV, the ExoN deletion does not impact the antiviral potency of molnupiravir, suggesting NHC-MP is resistant to excision [239]. A biochemical mechanism elucidating the relationship between the CoV ExoN and molnupiravir remains unanswered. The potential inability of the ExoN to excise NHC-MP could be due to RNA synthesis remaining unimpeded following NHC-TP incorporation, going unrecognized, and therefore avoiding excision [165], [166].
7. Influenza viruses
Influenza is a severe, contagious respiratory illness caused by infection with influenza viruses. Influenza viruses are segmented negative sense RNA viruses with ~ 14 kb genomes that together comprise six of the eight members of the family Orthomyxoviridae [241]. Four genera of influenza, influenza A, B, C, and D have been identified, two of which, A and B, cause significant morbidity and mortality worldwide [241], [242]. The WHO reports that influenza A and B together account for up to 5 million severe illnesses and 650,000 deaths annually [243]. This accounts for a total economic burden of $87 billion per year in the United States alone, despite the availability of effective vaccines and antivirals against both influenza A and B [242], [244].
Vaccination is widely considered to be the most effective way to prevent influenza infection [245]. Disease resulting from influenza infection is predominantly the result of infection with either influenza A or B [245]. Therefore, the currently available influenza vaccines consist of a mixture of A and B strains which is determined annually based on global surveillance of the circulating strains [245], [246]. Two types of influenza virus vaccine, live attenuated and inactivated vaccine, are currently in use [245]. However, despite the availability of these vaccines influenza still causes significant morbidity and mortality necessitating the development of antivirals against influenza [247].
Antivirals represent an important tool for the treatment and management of influenza infection. Historically, influenza virus infection has been treated using M2 ion channel and neuraminidase inhibitors [248]. Unfortunately, in recent seasons > 99% of circulating influenza strains have been resistant to M2 ion channel inhibitors, limiting treatment options [249]. This has necessitated the development of antivirals with novel mechanisms of action. To this end, inhibitors of the viral RdRp have been developed.
The structures of the RNA dependent RNA polymerases (RdRp) of influenza A and B have been solved [250], [251]. These structures revealed that the influenza RdRp is as a multifunctional heterotrimer consisting of three proteins PA, PB1, and PB2 [250], [251]. PA encodes an endonuclease activity, PB1 encodes the RdRp activity, while PB2 contains 7-methylguanylate cap binding activities [250], [251]. As each of these activities are distinct and indispensable, the viral RdRp serves as an attractive drug target. Influenza performs transcription via a well described process known as cap snatching [251], [252], [253], [254], [255], [256], [257]. Briefly, PB2 binds host mRNAs via a 5′ m7GTP cap binding domain, bringing them into proximity to PA. The endonuclease domain within PA cleaves them, producing short, capped primers, which are subsequently extended by the RdRp activity of PB1 to produce capped viral transcripts.
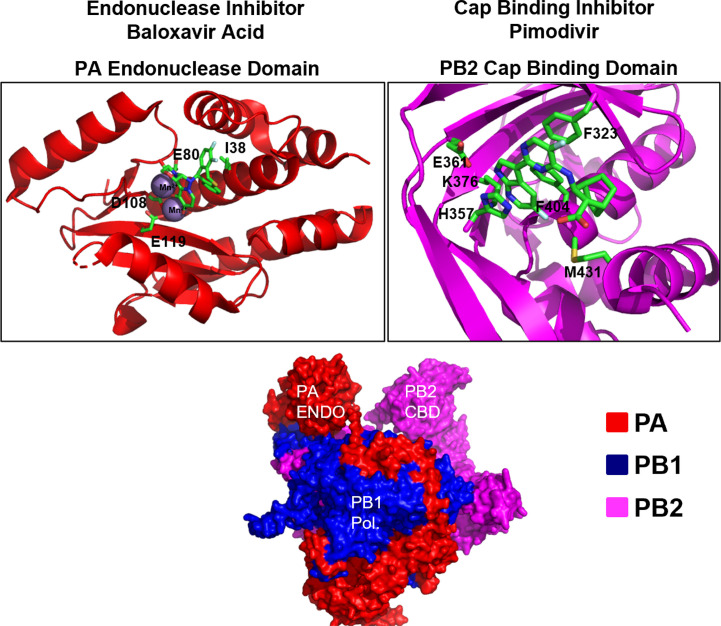
Each aspect of this process has currently been targeted for drug development and compounds targeting each component of the polymerase complex are currently available (Fig. 10 ). For example, the first-in-class PA endonuclease inhibitor baloxavir marboxil (BXM) was approved by the FDA in December of 2018 for the treatment of uncomplicated influenza in adults [258]. A single dose of BXM results in the alleviation of influenza symptoms approximately 23–28 h faster than placebo [259]. This is likely because baloxavir is a highly potent, tight-binding, PA endonuclease inhibitor that is capable of inhibiting influenza viruses in the low nanomolar range [258], [260]. BXM is a prodrug, the active form of which, baloxavir acid (BXA), binds to divalent metal ions in the PA endonuclease active site preventing catalysis [261]. Baloxavir is active against all known genera of influenza including viruses resistant to neuraminidase inhibitors [258], [262]. This, in addition to the fact that BXM can be administered orally as a single dose make BXM highly attractive for clinical use [259]. However, soon after the discovery of BXM resistant strains emerged. The majority of described resistance mutations are substitutions in the I38 position, with I38T/M/F resulting in a 2- to 50-fold increase in EC50 with I38T having the largest effect [259], [261]. It has been postulated that this reduced susceptibility is due to the fact that I38T may reduce van der Waals contacts with the inhibitor [261]. I38T has also been shown to confer resistance to baloxavir in in vitro biochemical assays [260], [261]. It has been reported that these mutations have a significant adverse effect on viral fitness so it is unknown how prevalent they may become should the use of BXM become widespread [261]. However, the development of widespread resistance may be possible as the emergence and transmission viruses containing BXM resistance mutations has been documented in clinical trials [259], [263].
Fig. 10.
Structural overview of inhibitors targeting the influenza RdRp. The influenza RdRp is a multifunctional heterotrimer consisting of three subunits: PA, PB1, and PB2. PA encodes the cap snatching endonuclease, PB1 encodes RdRp activity, and PB2 encodes a m7G-cap binding activity. The binding sites of inhibitors targeting each subunit and selected residues involved in inhibitor binding and resistance are highlighted. Structures adapted from the following and oriented using PyMol: FluA-polymerase heterotrimer 4WSB [251], PA endo with Baloxavir, 6FS6 [261], PB2 with pimodivir 4P1U [273]. Structural snapshots for favipiravir and molnupiravir are not shown as structures of the polymerase bound to these molecules are currently unavailable.
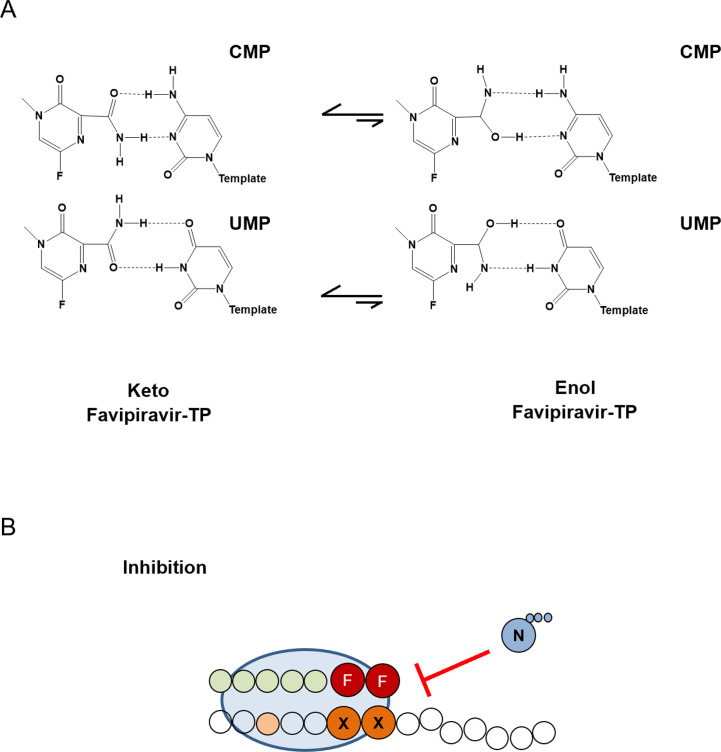
The RNA synthesis activity of the polymerase has been targeted using nucleotide-base moiety analogs such as favipiravir and NIs as molnupiravir (NHC) [163], [264], [265]. To date, however, the only analog approved for the treatment of influenza infection is favipiravir, which gained approval in Japan for the treatment of pandemic influenza [265]. Favipiravir is a nucleotide base-moiety analog which, following ribosylation and triphosphorylation by the cellular machinery functions as an inhibitor of the influenza polymerase [266]. Favipiravir has been evaluated in clinical trials and, while the exact mechanism of action requires clarification, it is thought to function through the induction of lethal mutagenesis, although limited biochemical studies have suggested that chain termination can occur after two successive incorporations (Fig. 11 ) [267], [268], [269]. While favipiravir can tautomerize, the keto tautomer is the most stable form at biological conditions and is therefore responsible for the biological activities of the compound [240]. Specifically, favipiravir functions as either an ATP or GTP analog, the successive incorporation of which leads to chain termination (Fig. 11) [268]. Resistance to favipiravir has been generated in vitro [270]. The PB1 K229R mutation located within the conserved F motif of the RdRp has been shown to confer resistance to favipiravir at the cost of viral fitness [270]. However, this loss of fitness is restored by the PA 653L mutation [270]. Interestingly, resistance to favipiravir has never emerged in clinical trials suggesting that the fitness cost associated with these mutations is likely too great [269].
Fig. 11.
Mechanism of inhibition of the influenza RdRp by favipiravir. (A) Base pairing of the keto and enol forms of favipiravir with CMP and UMP. (B) Two consecutive incorporations of favipiravir leads to chain termination, preventing successful polymerase activity. X, G or A nucleotides; N, any nucleotide; F, favipiravir.
Panel (A): Adapted from N.R. Jena, Role of different tautomers in the base-pairing abilities of some of the vital antiviral drugs used against COVID-19, Phys. Chem. Chem. Phys. 22 (48) (2020) 28115–28122. Panel (B): Mechanism adapted from Z. Jin, et al., The ambiguous base-pairing and high substrate efficiency of T-705 (favipiravir) ribofuranosyl 5'-triphosphate towards influenza A virus polymerase, PLoS One 8 (7) (2013) e68347.
Several unpublished clinical studies on the efficacy of favipiravir for the treatment of uncomplicated influenza have been performed with variable results [271]. Studies have reported anywhere between a 6 and 14 h decrease in time to influenza symptom resolution vs placebo [271]. This moderate effect, when combined with the fact that teratogenicity has been observed in multiple animal models at doses similar to those used in treatment, limit the utility of favipiravir in the treatment of uncomplicated influenza [271]. However, due to its activity against pandemic influenza and highly pathogenic Avian influenza strains, favipiravir remains clinically significant [272].
Molnupiravir (NHC), a NI targeting the RdRp activity of PB1, has also been evaluated for efficacy against influenza [163], [264]. NHC has shown efficacy against influenza A and B in cell culture, mouse, and ferret models of influenza infection with inhibitory concentrations in the nanomolar range [163], [264]. Increased frequencies of G to A and C to U transversion mutations were observed suggesting that NHC inhibits influenza via a mechanism involving lethal mutagenesis [163], [264]. This is similar to the mechanism which has been recently described for inhibition of SARS-CoV-2 and discussed earlier in this chapter [165]. While efficacy has yet to be demonstrated in clinical trials, given the fact that orally bioavailable prodrugs are available, NHC is a promising candidate for further development [163], [264].
The cap binding subunit of the influenza polymerase, PB2, has been targeted by pimodivir. Pimodivir binds to the cap binding domain of PB2 at nanomolar concentrations preventing the binding of capped host mRNAs. This, in turn, prevents cap-snatching and viral transcription [273]. However, pimodivir is only active against influenza A viruses, likely owing to significant amino acid sequence divergence in the cap binding domain of PB2 between different genera of influenza [273]. Resistance to pimodivir has been reported with the Q306H, S324I, S324N, S324R, F404Y, and N510T mutations conferring a greater than 10-fold decrease in susceptibility [274]. Furthermore, resistant strains of influenza containing PB2 substitutions have also emerged in clinical trials examining the efficacy of pimodivir [271], [275]. The most common of these is the PB2 M431I variant which has been shown to confer a 57-fold reduction in pimodivir susceptibility in cell culture [271], [275]. However, interestingly, in patients treated with a combination of a neuraminidase inhibitor (Tamiflu) and pimodivir the emergence of resistant strains was greatly reduced [271], [276].
References
- 1.Keech M., Beardsworth P. The impact of influenza on working days lost: a review of the literature. Pharmacoeconomics. 2008;26(11):911–924. doi: 10.2165/00019053-200826110-00004. [DOI] [PubMed] [Google Scholar]
- 2.Nair H., et al. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet. 2010;375(9725):1545–1555. doi: 10.1016/S0140-6736(10)60206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nair H., et al. Global burden of respiratory infections due to seasonal influenza in young children: a systematic review and meta-analysis. Lancet. 2011;378(9807):1917–1930. doi: 10.1016/S0140-6736(11)61051-9. [DOI] [PubMed] [Google Scholar]
- 4.Kirk M.D., et al. World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: a data synthesis. PLoS Med. 2015;12(12) doi: 10.1371/journal.pmed.1001921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahmed S.M., et al. Global prevalence of norovirus in cases of gastroenteritis: a systematic review and meta-analysis. Lancet Infect. Dis. 2014;14(8):725–730. doi: 10.1016/S1473-3099(14)70767-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Messina J.P., et al. Global distribution and prevalence of hepatitis C virus genotypes. Hepatology. 2015;61(1):77–87. doi: 10.1002/hep.27259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perz J.F., et al. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J. Hepatol. 2006;45(4):529–538. doi: 10.1016/j.jhep.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 8.Bhatt S., et al. The global distribution and burden of dengue. Nature. 2013;496(7446):504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bertino J.S. Cost burden of viral respiratory infections: issues for formulary decision makers. Am. J. Med. 2002;112(Suppl. 6A):42S–49S. doi: 10.1016/s0002-9343(01)01063-4. [DOI] [PubMed] [Google Scholar]
- 10.Stanaway J.D., et al. The global burden of viral hepatitis from 1990 to 2013: findings from the Global Burden of Disease Study 2013. Lancet. 2016;388(10049):1081–1088. doi: 10.1016/S0140-6736(16)30579-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abd El-Aziz T.M., Stockand J.D. Recent progress and challenges in drug development against COVID-19 coronavirus (SARS-CoV-2)—an update on the status. Infect. Genet. Evol. 2020;83 doi: 10.1016/j.meegid.2020.104327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cui J., et al. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17(3):181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Musso D., et al. Zika virus infection—after the pandemic. N. Engl. J. Med. 2019;381(15):1444–1457. doi: 10.1056/NEJMra1808246. [DOI] [PubMed] [Google Scholar]
- 14.Wikan N., Smith D.R. Zika virus: history of a newly emerging arbovirus. Lancet Infect. Dis. 2016;16(7):e119–e126. doi: 10.1016/S1473-3099(16)30010-X. [DOI] [PubMed] [Google Scholar]
- 15.Malvy D., et al. Ebola virus disease. Lancet. 2019;393(10174):936–948. doi: 10.1016/S0140-6736(18)33132-5. [DOI] [PubMed] [Google Scholar]
- 16.Grubaugh N.D., et al. Tracking virus outbreaks in the twenty-first century. Nat. Microbiol. 2019;4(1):10–19. doi: 10.1038/s41564-018-0296-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adalja A., Inglesby T. Broad-spectrum antiviral agents: a crucial pandemic tool. Expert Rev. Anti Infect. Ther. 2019;17(7):467–470. doi: 10.1080/14787210.2019.1635009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andersen P.I., et al. Discovery and development of safe-in-man broad-spectrum antiviral agents. Int. J. Infect. Dis. 2020;93:268–276. doi: 10.1016/j.ijid.2020.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolf Y.I., et al. Origins and evolution of the global RNA virome. mBio. 2018;9(6) doi: 10.1128/mBio.02329-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koonin E.V., et al. Global organization and proposed megataxonomy of the virus world. Microbiol. Mol. Biol. Rev. 2020;84(2) doi: 10.1128/MMBR.00061-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith E.C., Denison M.R. Coronaviruses as DNA wannabes: a new model for the regulation of RNA virus replication fidelity. PLoS Pathog. 2013;9(12) doi: 10.1371/journal.ppat.1003760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duffy S. Why are RNA virus mutation rates so damn high? PLoS Biol. 2018;16(8) doi: 10.1371/journal.pbio.3000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Denison M.R., et al. Coronaviruses: an RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. 2011;8(2):270–279. doi: 10.4161/rna.8.2.15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Irwin K.K., et al. Antiviral drug resistance as an adaptive process. Virus Evol. 2016;2(1) doi: 10.1093/ve/vew014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Venkataraman S., et al. RNA dependent RNA polymerases: insights from structure, function and evolution. Viruses. 2018;10(2) doi: 10.3390/v10020076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scholte F.E.M., et al. Crimean-Congo hemorrhagic fever virus suppresses innate immune responses via a ubiquitin and ISG15 specific protease. Cell Rep. 2017;20(10):2396–2407. doi: 10.1016/j.celrep.2017.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Vlugt C., et al. Insight into influenza: a virus cap-snatching. Viruses. 2018;10(11) doi: 10.3390/v10110641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J., et al. Amino acid residues within conserved domain VI of the vesicular stomatitis virus large polymerase protein essential for mRNA cap methyltransferase activity. J. Virol. 2005;79(21):13373–13384. doi: 10.1128/JVI.79.21.13373-13384.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogino T., et al. Histidine-mediated RNA transfer to GDP for unique mRNA capping by vesicular stomatitis virus RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 2010;107(8):3463–3468. doi: 10.1073/pnas.0913083107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ng K.K., et al. Structure-function relationships among RNA-dependent RNA polymerases. Curr. Top. Microbiol. Immunol. 2008;320:137–156. doi: 10.1007/978-3-540-75157-1_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jia H., Gong P. A structure-function diversity survey of the RNA-dependent RNA polymerases from the positive-strand RNA viruses. Front. Microbiol. 2019;10:1945. doi: 10.3389/fmicb.2019.01945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.te Velthuis A.J. Common and unique features of viral RNA-dependent polymerases. Cell. Mol. Life Sci. 2014;71(22):4403–4420. doi: 10.1007/s00018-014-1695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]; [32a] Bienert S., Waterhouse A., de Beer T.A.P., Tauriello G., Studer G., Bordoli L., Schwede T. The SWISS-MODEL Repository—new features and functionality. Nucleic Acids Res. 2017;45(D1):D313–D319. doi: 10.1093/nar/gkw1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10(1):2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Castro C., et al. Two proton transfers in the transition state for nucleotidyl transfer catalyzed by RNA- and DNA-dependent RNA and DNA polymerases. Proc. Natl. Acad. Sci. U. S. A. 2007;104(11):4267–4272. doi: 10.1073/pnas.0608952104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Castro C., et al. Nucleic acid polymerases use a general acid for nucleotidyl transfer. Nat. Struct. Mol. Biol. 2009;16(2):212–218. doi: 10.1038/nsmb.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu J., et al. A structural overview of RNA-dependent RNA polymerases from the Flaviviridae family. Int. J. Mol. Sci. 2015;16(6):12943–12957. doi: 10.3390/ijms160612943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zell R., et al. ICTV virus taxonomy profile: Picornaviridae. J. Gen. Virol. 2017;98(10):2421–2422. doi: 10.1099/jgv.0.000911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang P., et al. Picornavirus morphogenesis. Microbiol. Mol. Biol. Rev. 2014;78(3):418–437. doi: 10.1128/MMBR.00012-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whitton J.L., et al. Host and virus determinants of picornavirus pathogenesis and tropism. Nat. Rev. Microbiol. 2005;3(10):765–776. doi: 10.1038/nrmicro1284. [DOI] [PubMed] [Google Scholar]
- 40.Grubman M.J., Baxt B. Foot-and-mouth disease. Clin. Microbiol. Rev. 2004;17(2):465–493. doi: 10.1128/CMR.17.2.465-493.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lindesmith L., et al. Human susceptibility and resistance to Norwalk virus infection. Nat. Med. 2003;9(5):548–553. doi: 10.1038/nm860. [DOI] [PubMed] [Google Scholar]
- 42.Lemon S.M., et al. Type A viral hepatitis: a summary and update on the molecular virology, epidemiology, pathogenesis and prevention. J. Hepatol. 2017 doi: 10.1016/j.jhep.2017.08.034. [DOI] [PubMed] [Google Scholar]
- 43.Heikkinen T., Jarvinen A. The common cold. Lancet. 2003;361(9351):51–59. doi: 10.1016/S0140-6736(03)12162-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muehlenbachs A., et al. Tissue tropism, pathology and pathogenesis of enterovirus infection. J. Pathol. 2015;235(2):217–228. doi: 10.1002/path.4438. [DOI] [PubMed] [Google Scholar]
- 45.Racaniello V.R. One hundred years of poliovirus pathogenesis. Virology. 2006;344(1):9–16. doi: 10.1016/j.virol.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 46.Daley J.K., et al. Poliovirus replication and spread in primary neuron cultures. Virology. 2005;340(1):10–20. doi: 10.1016/j.virol.2005.05.032. [DOI] [PubMed] [Google Scholar]
- 47.Huang H.I., Shih S.R. Neurotropic enterovirus infections in the central nervous system. Viruses. 2015;7(11):6051–6066. doi: 10.3390/v7112920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chumakov K., et al. Vaccination against polio should not be stopped. Nat. Rev. Microbiol. 2007;5(12):952–958. doi: 10.1038/nrmicro1769. [DOI] [PubMed] [Google Scholar]
- 49.Meldrum M. "A calculated risk”: the Salk polio vaccine field trials of 1954. BMJ. 1998;317(7167):1233–1236. doi: 10.1136/bmj.317.7167.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pearce J.M. Salk and Sabin: poliomyelitis immunisation. J. Neurol. Neurosurg. Psychiatry. 2004;75(11):1552. doi: 10.1136/jnnp.2003.028530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bandyopadhyay A.S., et al. Polio vaccination: past, present and future. Future Microbiol. 2015;10(5):791–808. doi: 10.2217/fmb.15.19. [DOI] [PubMed] [Google Scholar]
- 52.Dunn G., et al. Virus excretion and mutation by infants following primary vaccination with live oral poliovaccine from two sources. J. Med. Virol. 1990;32(2):92–95. doi: 10.1002/jmv.1890320205. [DOI] [PubMed] [Google Scholar]
- 53.Famulare M., et al. Sabin vaccine reversion in the field: a comprehensive analysis of Sabin-like poliovirus isolates in Nigeria. J. Virol. 2016;90(1):317–331. doi: 10.1128/JVI.01532-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cann A.J., et al. Reversion to neurovirulence of the live-attenuated Sabin type 3 oral poliovirus vaccine. Nucleic Acids Res. 1984;12(20):7787–7792. doi: 10.1093/nar/12.20.7787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kew O.M., et al. Vaccine-derived polioviruses and the endgame strategy for global polio eradication. Annu. Rev. Microbiol. 2005;59:587–635. doi: 10.1146/annurev.micro.58.030603.123625. [DOI] [PubMed] [Google Scholar]
- 56.Hansen J.L., et al. Structure of the RNA-dependent RNA polymerase of poliovirus. Structure. 1997;5(8):1109–1122. doi: 10.1016/s0969-2126(97)00261-x. [DOI] [PubMed] [Google Scholar]
- 57.Thompson A.A., Peersen O.B. Structural basis for proteolysis-dependent activation of the poliovirus RNA-dependent RNA polymerase. EMBO J. 2004;23(17):3462–3471. doi: 10.1038/sj.emboj.7600357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kok C.C., McMinn P.C. Picornavirus RNA-dependent RNA polymerase. Int. J. Biochem. Cell Biol. 2009;41(3):498–502. doi: 10.1016/j.biocel.2008.03.019. [DOI] [PubMed] [Google Scholar]
- 59.Gohara D.W., et al. Poliovirus RNA-dependent RNA polymerase (3Dpol): structural, biochemical, and biological analysis of conserved structural motifs A and B. J. Biol. Chem. 2000;275(33):25523–25532. doi: 10.1074/jbc.M002671200. [DOI] [PubMed] [Google Scholar]
- 60.Kortus M.G., et al. A template RNA entry channel in the fingers domain of the poliovirus polymerase. J. Mol. Biol. 2012;417(4):263–278. doi: 10.1016/j.jmb.2012.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Domingo E., et al. Basic concepts in RNA virus evolution. FASEB J. 1996;10(8):859–864. doi: 10.1096/fasebj.10.8.8666162. [DOI] [PubMed] [Google Scholar]
- 62.Arnold J.J., Cameron C.E. Poliovirus RNA-dependent RNA polymerase (3Dpol): pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mg2 + Biochemistry. 2004;43(18):5126–5137. doi: 10.1021/bi035212y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Freistadt M.S., et al. Biochemical characterization of the fidelity of poliovirus RNA-dependent RNA polymerase. Virol. J. 2007;4:44. doi: 10.1186/1743-422X-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vignuzzi M., et al. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature. 2006;439(7074):344–348. doi: 10.1038/nature04388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Minor P.D. The polio-eradication programme and issues of the end game. J. Gen. Virol. 2012;93(Pt. 3):457–474. doi: 10.1099/vir.0.036988-0. [DOI] [PubMed] [Google Scholar]
- 66.John T.J., Vashishtha V.M. Eradicating poliomyelitis: India's journey from hyperendemic to polio-free status. Indian J. Med. Res. 2013;137(5):881–894. [PMC free article] [PubMed] [Google Scholar]
- 67.Collett M.S., et al. A case for developing antiviral drugs against polio. Antiviral Res. 2008;79(3):179–187. doi: 10.1016/j.antiviral.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 68.Collett M.S., et al. Antiviral activity of pocapavir in a randomized, blinded, placebo-controlled human oral poliovirus vaccine challenge model. J. Infect. Dis. 2017;215(3):335–343. doi: 10.1093/infdis/jiw542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rodriguez P.L., Carrasco L. Gliotoxin: inhibitor of poliovirus RNA synthesis that blocks the viral RNA polymerase 3Dpol. J. Virol. 1992;66(4):1971–1976. doi: 10.1128/jvi.66.4.1971-1976.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Harki D.A., et al. Synthesis and antiviral activity of 5-substituted cytidine analogues: identification of a potent inhibitor of viral RNA-dependent RNA polymerases. J. Med. Chem. 2006;49(21):6166–6169. doi: 10.1021/JM060872x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van der Linden L., et al. The RNA template channel of the RNA-dependent RNA polymerase as a target for development of antiviral therapy of multiple genera within a virus family. PLoS Pathog. 2015;11(3) doi: 10.1371/journal.ppat.1004733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Beaucourt S., Vignuzzi M. Ribavirin: a drug active against many viruses with multiple effects on virus replication and propagation. Molecular basis of ribavirin resistance. Curr. Opin. Virol. 2014;8:10–15. doi: 10.1016/j.coviro.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Graci J.D., Cameron C.E. Mechanisms of action of ribavirin against distinct viruses. Rev. Med. Virol. 2006;16(1):37–48. doi: 10.1002/rmv.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Crotty S., et al. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat. Med. 2000;6(12):1375–1379. doi: 10.1038/82191. [DOI] [PubMed] [Google Scholar]
- 75.Maag D., et al. Hepatitis C virus RNA-dependent RNA polymerase (NS5B) as a mediator of the antiviral activity of ribavirin. J. Biol. Chem. 2001;276(49):46094–46098. doi: 10.1074/jbc.C100349200. [DOI] [PubMed] [Google Scholar]
- 76.Wu J.Z., et al. Ribavirin, viramidine and adenosine-deaminase-catalysed drug activation: implication for nucleoside prodrug design. J. Antimicrob. Chemother. 2003;52(4):543–546. doi: 10.1093/jac/dkg405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vignuzzi M., et al. Ribavirin and lethal mutagenesis of poliovirus: molecular mechanisms, resistance and biological implications. Virus Res. 2005;107(2):173–181. doi: 10.1016/j.virusres.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 78.Pfeiffer J.K., Kirkegaard K. A single mutation in poliovirus RNA-dependent RNA polymerase confers resistance to mutagenic nucleotide analogs via increased fidelity. Proc. Natl. Acad. Sci. U. S. A. 2003;100(12):7289–7294. doi: 10.1073/pnas.1232294100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pfeiffer J.K., Kirkegaard K. Increased fidelity reduces poliovirus fitness and virulence under selective pressure in mice. PLoS Pathog. 2005;1(2) doi: 10.1371/journal.ppat.0010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fitzsimmons W.J., et al. A speed-fidelity trade-off determines the mutation rate and virulence of an RNA virus. PLoS Biol. 2018;16(6) doi: 10.1371/journal.pbio.2006459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Campagnola G., et al. Structure-function relationships underlying the replication fidelity of viral RNA-dependent RNA polymerases. J. Virol. 2015;89(1):275–286. doi: 10.1128/JVI.01574-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kempf B.J., et al. Poliovirus polymerase Leu420 facilitates RNA recombination and ribavirin resistance. J. Virol. 2016;90(19):8410–8421. doi: 10.1128/JVI.00078-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kempf B.J., et al. Picornavirus RNA recombination counteracts error catastrophe. J. Virol. 2019;93(14) doi: 10.1128/JVI.00652-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sierra M., et al. Foot-and-mouth disease virus mutant with decreased sensitivity to ribavirin: implications for error catastrophe. J. Virol. 2007;81(4):2012–2024. doi: 10.1128/JVI.01606-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ferrer-Orta C., et al. Structure of foot-and-mouth disease virus mutant polymerases with reduced sensitivity to ribavirin. J. Virol. 2010;84(12):6188–6199. doi: 10.1128/JVI.02420-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Agudo R., et al. A multi-step process of viral adaptation to a mutagenic nucleoside analogue by modulation of transition types leads to extinction-escape. PLoS Pathog. 2010;6(8) doi: 10.1371/journal.ppat.1001072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Arias A., et al. Determinants of RNA-dependent RNA polymerase (in)fidelity revealed by kinetic analysis of the polymerase encoded by a foot-and-mouth disease virus mutant with reduced sensitivity to ribavirin. J. Virol. 2008;82(24):12346–12355. doi: 10.1128/JVI.01297-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zeng J., et al. Ribavirin-resistant variants of foot-and-mouth disease virus: the effect of restricted quasispecies diversity on viral virulence. J. Virol. 2014;88(8):4008–4020. doi: 10.1128/JVI.03594-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Meng T., Kwang J. Attenuation of human enterovirus 71 high-replication-fidelity variants in AG129 mice. J. Virol. 2014;88(10):5803–5815. doi: 10.1128/JVI.00289-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sadeghipour S., et al. Ribavirin-resistant mutants of human enterovirus 71 express a high replication fidelity phenotype during growth in cell culture. J. Virol. 2013;87(3):1759–1769. doi: 10.1128/JVI.02139-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Agudo R., et al. Molecular characterization of a dual inhibitory and mutagenic activity of 5-fluorouridine triphosphate on viral RNA synthesis. Implications for lethal mutagenesis. J. Mol. Biol. 2008;382(3):652–666. doi: 10.1016/j.jmb.2008.07.033. [DOI] [PubMed] [Google Scholar]
- 92.Zeng J., et al. An increased replication fidelity mutant of foot-and-mouth disease virus retains fitness in vitro and virulence in vivo. Antiviral Res. 2013;100(1):1–7. doi: 10.1016/j.antiviral.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 93.Graci J.D., et al. Lethal mutagenesis of picornaviruses with N-6-modified purine nucleoside analogues. Antimicrob. Agents Chemother. 2008;52(3):971–979. doi: 10.1128/AAC.01056-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Levi L.I., et al. Fidelity variants of RNA dependent RNA polymerases uncover an indirect, mutagenic activity of amiloride compounds. PLoS Pathog. 2010;6(10) doi: 10.1371/journal.ppat.1001163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dulin D., et al. Signatures of nucleotide analog incorporation by an RNA-dependent RNA polymerase revealed using high-throughput magnetic tweezers. Cell Rep. 2017;21(4):1063–1076. doi: 10.1016/j.celrep.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kamar N., et al. Hepatitis E virus infection. Nat. Rev. Dis. Primers. 2017;3:17086. doi: 10.1038/nrdp.2017.86. [DOI] [PubMed] [Google Scholar]
- 97.Tang L.S.Y., et al. Chronic hepatitis B infection: a review. JAMA. 2018;319(17):1802–1813. doi: 10.1001/jama.2018.3795. [DOI] [PubMed] [Google Scholar]
- 98.Hernaez R., et al. Acute-on-chronic liver failure: an update. Gut. 2017;66(3):541–553. doi: 10.1136/gutjnl-2016-312670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Do A., Reau N.S. Chronic viral hepatitis: current management and future directions. Hepatol. Commun. 2020;4(3):329–341. doi: 10.1002/hep4.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.The Lancet Gastroenterology Hepatology The hunt for a vaccine for hepatitis C virus continues. Lancet Gastroenterol. Hepatol. 2021;6(4):253. doi: 10.1016/S2468-1253(21)00073-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Page K., et al. Randomized trial of a vaccine regimen to prevent chronic HCV infection. N. Engl. J. Med. 2021;384(6):541–549. doi: 10.1056/NEJMoa2023345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kish T., et al. Hepatitis C in a new era: a review of current therapies. P. T. 2017;42(5):316–329. [PMC free article] [PubMed] [Google Scholar]
- 103.Thrift A.P., et al. Global epidemiology and burden of HCV infection and HCV-related disease. Nat. Rev. Gastroenterol. Hepatol. 2017;14(2):122–132. doi: 10.1038/nrgastro.2016.176. [DOI] [PubMed] [Google Scholar]
- 104.Mauger D.M., et al. Functionally conserved architecture of hepatitis C virus RNA genomes. Proc. Natl. Acad. Sci. U. S. A. 2015;112(12):3692–3697. doi: 10.1073/pnas.1416266112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Smith D.B., et al. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology. 2014;59(1):318–327. doi: 10.1002/hep.26744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Simmonds P., et al. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology. 2005;42(4):962–973. doi: 10.1002/hep.20819. [DOI] [PubMed] [Google Scholar]
- 107.Hadziyannis S.J., et al. Peginterferon-alpha2a and ribavirin combination therapy in chronic hepatitis C: a randomized study of treatment duration and ribavirin dose. Ann. Intern. Med. 2004;140(5):346–355. doi: 10.7326/0003-4819-140-5-200403020-00010. [DOI] [PubMed] [Google Scholar]
- 108.Kieffer T.L., et al. Viral resistance to specifically targeted antiviral therapies for hepatitis C (STAT-Cs) J. Antimicrob. Chemother. 2010;65(2):202–212. doi: 10.1093/jac/dkp388. [DOI] [PubMed] [Google Scholar]
- 109.Cento V., et al. HCV genotypes are differently prone to the development of resistance to linear and macrocyclic protease inhibitors. PLoS One. 2012;7(7) doi: 10.1371/journal.pone.0039652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Powdrill M.H., et al. Contribution of a mutational bias in hepatitis C virus replication to the genetic barrier in the development of drug resistance. Proc. Natl. Acad. Sci. U. S. A. 2011;108(51):20509–20513. doi: 10.1073/pnas.1105797108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sarrazin C., et al. Prevalence of resistance-associated substitutions in HCV NS5A, NS5B, or NS3 and outcomes of treatment with ledipasvir and sofosbuvir. Gastroenterology. 2016;151(3):501–512. doi: 10.1053/j.gastro.2016.06.002. e501. [DOI] [PubMed] [Google Scholar]
- 112.Kuntzen T., et al. Naturally occurring dominant resistance mutations to hepatitis C virus protease and polymerase inhibitors in treatment-naive patients. Hepatology. 2008;48(6):1769–1778. doi: 10.1002/hep.22549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Farci P., et al. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science. 2000;288(5464):339–344. doi: 10.1126/science.288.5464.339. [DOI] [PubMed] [Google Scholar]
- 114.Lesburg C.A., et al. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 1999;6(10):937–943. doi: 10.1038/13305. [DOI] [PubMed] [Google Scholar]
- 115.Bressanelli S., et al. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 1999;96(23):13034–13039. doi: 10.1073/pnas.96.23.13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ago H., et al. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Structure. 1999;7(11):1417–1426. doi: 10.1016/s0969-2126(00)80031-3. [DOI] [PubMed] [Google Scholar]
- 117.Appleby T.C., et al. Viral replication. Structural basis for RNA replication by the hepatitis C virus polymerase. Science. 2015;347(6223):771–775. doi: 10.1126/science.1259210. [DOI] [PubMed] [Google Scholar]
- 118.Mosley R.T., et al. Structure of hepatitis C virus polymerase in complex with primer-template RNA. J. Virol. 2012;86(12):6503–6511. doi: 10.1128/JVI.00386-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hong Z., et al. A novel mechanism to ensure terminal initiation by hepatitis C virus NS5B polymerase. Virology. 2001;285(1):6–11. doi: 10.1006/viro.2001.0948. [DOI] [PubMed] [Google Scholar]
- 120.Fried M.W., et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 2002;347(13):975–982. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- 121.Sarrazin C., Zeuzem S. Resistance to direct antiviral agents in patients with hepatitis C virus infection. Gastroenterology. 2010;138(2):447–462. doi: 10.1053/j.gastro.2009.11.055. [DOI] [PubMed] [Google Scholar]
- 122.Feld J.J., Hoofnagle J.H. Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature. 2005;436(7053):967–972. doi: 10.1038/nature04082. [DOI] [PubMed] [Google Scholar]
- 123.Cuevas J.M., et al. Effect of ribavirin on the mutation rate and spectrum of hepatitis C virus in vivo. J. Virol. 2009;83(11):5760–5764. doi: 10.1128/JVI.00201-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ortega-Prieto A.M., et al. Extinction of hepatitis C virus by ribavirin in hepatoma cells involves lethal mutagenesis. PLoS One. 2013;8(8) doi: 10.1371/journal.pone.0071039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Poordad F., et al. Exploratory study of oral combination antiviral therapy for hepatitis C. N. Engl. J. Med. 2013;368(1):45–53. doi: 10.1056/NEJMoa1208809. [DOI] [PubMed] [Google Scholar]
- 126.Gane E.J., et al. Nucleotide polymerase inhibitor sofosbuvir plus ribavirin for hepatitis C. N. Engl. J. Med. 2013;368(1):34–44. doi: 10.1056/NEJMoa1208953. [DOI] [PubMed] [Google Scholar]
- 127.Bhatia H.K., et al. Sofosbuvir: a novel treatment option for chronic hepatitis C infection. J. Pharmacol. Pharmacother. 2014;5(4):278–284. doi: 10.4103/0976-500X.142464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Fung A., et al. Efficiency of incorporation and chain termination determines the inhibition potency of 2'-modified nucleotide analogs against hepatitis C virus polymerase. Antimicrob. Agents Chemother. 2014;58(7):3636–3645. doi: 10.1128/AAC.02666-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ma H., et al. Characterization of the metabolic activation of hepatitis C virus nucleoside inhibitor beta-D-2'-deoxy-2'-fluoro-2'-C-methylcytidine (PSI-6130) and identification of a novel active 5'-triphosphate species. J. Biol. Chem. 2007;282(41):29812–29820. doi: 10.1074/jbc.M705274200. [DOI] [PubMed] [Google Scholar]
- 130.Lawitz E., et al. Sofosbuvir for previously untreated chronic hepatitis C infection. N. Engl. J. Med. 2013;368(20):1878–1887. doi: 10.1056/NEJMoa1214853. [DOI] [PubMed] [Google Scholar]
- 131.Lam A.M., et al. Genotype and subtype profiling of PSI-7977 as a nucleotide inhibitor of hepatitis C virus. Antimicrob. Agents Chemother. 2012;56(6):3359–3368. doi: 10.1128/AAC.00054-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Svarovskaia E.S., et al. Infrequent development of resistance in genotype 1-6 hepatitis C virus-infected subjects treated with sofosbuvir in phase 2 and 3 clinical trials. Clin. Infect. Dis. 2014;59(12):1666–1674. doi: 10.1093/cid/ciu697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dietz J., et al. Patterns of resistance-associated substitutions in patients with chronic HCV infection following treatment with direct-acting antivirals. Gastroenterology. 2018;154(4) doi: 10.1053/j.gastro.2017.11.007. 976–988.e4. [DOI] [PubMed] [Google Scholar]
- 134.Gane E.J., et al. The emergence of NS5B resistance associated substitution S282T after sofosbuvir-based treatment. Hepatol. Commun. 2017;1(6):538–549. doi: 10.1002/hep4.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hedskog C., et al. Evolution of the HCV viral population from a patient with S282T detected at relapse after sofosbuvir monotherapy. J. Viral Hepat. 2015;22(11):871–881. doi: 10.1111/jvh.12405. [DOI] [PubMed] [Google Scholar]
- 136.Donaldson E.F., et al. Clinical evidence and bioinformatics characterization of potential hepatitis C virus resistance pathways for sofosbuvir. Hepatology. 2015;61(1):56–65. doi: 10.1002/hep.27375. [DOI] [PubMed] [Google Scholar]
- 137.Svarovskaia E.S., et al. L159F and V321A sofosbuvir-associated hepatitis C virus NS5B substitutions. J. Infect. Dis. 2016;213(8):1240–1247. doi: 10.1093/infdis/jiv564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Patil V.M., et al. Current perspective of HCV NS5B inhibitors: a review. Curr. Med. Chem. 2011;18(36):5564–5597. doi: 10.2174/092986711798347234. [DOI] [PubMed] [Google Scholar]
- 139.Eltahla A.A., et al. Inhibitors of the hepatitis C virus polymerase; mode of action and resistance. Viruses. 2015;7(10):5206–5224. doi: 10.3390/v7102868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Membreno F.E., Lawitz E.J. The HCV NS5B nucleoside and non-nucleoside inhibitors. Clin. Liver Dis. 2011;15(3):611–626. doi: 10.1016/j.cld.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 141.Caillet-Saguy C., et al. An objective assessment of conformational variability in complexes of hepatitis C virus polymerase with non-nucleoside inhibitors. J. Mol. Biol. 2011;414(3):370–384. doi: 10.1016/j.jmb.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 142.Davis B.C., Thorpe I.F. Thumb inhibitor binding eliminates functionally important dynamics in the hepatitis C virus RNA polymerase. Proteins. 2013;81(1):40–52. doi: 10.1002/prot.24154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Le Guillou-Guillemette H., et al. Genetic diversity of the hepatitis C virus: impact and issues in the antiviral therapy. World J. Gastroenterol. 2007;13(17):2416–2426. doi: 10.3748/wjg.v13.i17.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Di Maio V.C., et al. Hepatitis C virus genetic variability and the presence of NS5B resistance-associated mutations as natural polymorphisms in selected genotypes could affect the response to NS5B inhibitors. Antimicrob. Agents Chemother. 2014;58(5):2781–2797. doi: 10.1128/AAC.02386-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.AASLD-IDSA HCV Guidance Panel Hepatitis C guidance 2018 update: AASLD-IDSA recommendations for testing, managing, and treating hepatitis C virus infection. Clin. Infect. Dis. 2018;67(10):1477–1492. doi: 10.1093/cid/ciy585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Asahina Y., et al. Ledipasvir-sofosbuvir for treating Japanese patients with chronic hepatitis C virus genotype 2 infection. Liver Int. 2018;38(9):1552–1561. doi: 10.1111/liv.13685. [DOI] [PubMed] [Google Scholar]
- 147.Berg T., et al. Real-world effectiveness and safety of glecaprevir/pibrentasvir for the treatment of chronic hepatitis C infection: data from the German Hepatitis C-Registry. Aliment. Pharmacol. Ther. 2019;49(8):1052–1059. doi: 10.1111/apt.15222. [DOI] [PubMed] [Google Scholar]
- 148.Chiu W.N., et al. Real-world effectiveness of glecaprevir/pibrentasvir and ledipasvir/sofosbuvir for mixed genotype hepatitis C infection: a multicenter pooled analysis in Taiwan. J. Viral Hepat. 2020 doi: 10.1111/jvh.13305. [DOI] [PubMed] [Google Scholar]
- 149.Collins P.L., et al. Respiratory syncytial virus: virology, reverse genetics, and pathogenesis of disease. Curr. Top. Microbiol. Immunol. 2013;372:3–38. doi: 10.1007/978-3-642-38919-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hall C.B., et al. The burden of respiratory syncytial virus infection in young children. N. Engl. J. Med. 2009;360(6):588–598. doi: 10.1056/NEJMoa0804877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Griffiths C., et al. Respiratory syncytial virus: infection, detection, and new options for prevention and treatment. Clin. Microbiol. Rev. 2017;30(1):277–319. doi: 10.1128/CMR.00010-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tin Tin Htar M., et al. The burden of respiratory syncytial virus in adults: a systematic review and meta-analysis. Epidemiol. Infect. 2020;148 doi: 10.1017/S0950268820000400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Acosta P.L., et al. Brief history and characterization of enhanced respiratory syncytial virus disease. Clin. Vaccine Immunol. 2015;23(3):189–195. doi: 10.1128/CVI.00609-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Elawar F., et al. Pharmacological targets and emerging treatments for respiratory syncytial virus bronchiolitis. Pharmacol. Ther. 2021;220:107712. doi: 10.1016/j.pharmthera.2020.107712. [DOI] [PubMed] [Google Scholar]
- 155.Robinson J.L., et al. Preventing hospitalizations for respiratory syncytial virus infection. Paediatr. Child Health. 2015;20(6):321–333. doi: 10.1093/pch/20.6.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Turner T.L., et al. Respiratory syncytial virus: current and emerging treatment options. Clinicoecon. Outcomes Res. 2014;6:217–225. doi: 10.2147/CEOR.S60710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Simoes E.A.F., et al. Past, present and future approaches to the prevention and treatment of respiratory syncytial virus infection in children. Infect. Dis. Ther. 2018;7(1):87–120. doi: 10.1007/s40121-018-0188-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Law B.J., et al. Does ribavirin impact on the hospital course of children with respiratory syncytial virus (RSV) infection? An analysis using the pediatric investigators collaborative network on infections in Canada (PICNIC) RSV database. Pediatrics. 1997;99(3) doi: 10.1542/peds.99.3.e7. [DOI] [PubMed] [Google Scholar]
- 159.Simabuco F.M., et al. Structural analysis of human respiratory syncytial virus p protein: identification of intrinsically disordered domains. Braz. J. Microbiol. 2011;42(1):340–345. doi: 10.1590/S1517-83822011000100043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gilman M.S.A., et al. Structure of the respiratory syncytial virus polymerase complex. Cell. 2019;179(1) doi: 10.1016/j.cell.2019.08.014. 193–204.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Cao D., et al. Cryo-EM structure of the respiratory syncytial virus RNA polymerase. Nat. Commun. 2020;11(1):368. doi: 10.1038/s41467-019-14246-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Nicholson E.G., Munoz F.M. A review of therapeutics in clinical development for respiratory syncytial virus and influenza in children. Clin. Ther. 2018;40(8):1268–1281. doi: 10.1016/j.clinthera.2018.06.014. [DOI] [PubMed] [Google Scholar]
- 163.Yoon J.J., et al. Orally efficacious broad-spectrum ribonucleoside analog inhibitor of influenza and respiratory syncytial viruses. Antimicrob. Agents Chemother. 2018;62(8) doi: 10.1128/AAC.00766-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Patel K., et al. Respiratory syncytial virus—a dynamics and the effects of lumicitabine, a nucleoside viral replication inhibitor, in experimentally infected humans. J. Antimicrob. Chemother. 2019;74(2):442–452. doi: 10.1093/jac/dky415. [DOI] [PubMed] [Google Scholar]
- 165.Gordon C.J., et al. Molnupiravir promotes SARS-CoV-2 mutagenesis via the RNA template. J. Biol. Chem. 2021:100770. doi: 10.1016/j.jbc.2021.100770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kabinger F., et al. Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis. Nat. Struct. Mol. Biol. 2021 doi: 10.1038/s41594-021-00651-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wang G., et al. Discovery of 4'-chloromethyl-2'-deoxy-3',5'-di-O-isobutyryl-2'-fluorocytidine (ALS-8176), a first-in-class RSV polymerase inhibitor for treatment of human respiratory syncytial virus infection. J. Med. Chem. 2015;58(4):1862–1878. doi: 10.1021/jm5017279. [DOI] [PubMed] [Google Scholar]
- 168.Deval J., et al. Molecular basis for the selective inhibition of respiratory syncytial virus RNA polymerase by 2'-fluoro-4'-chloromethyl-cytidine triphosphate. PLoS Pathog. 2015;11(6) doi: 10.1371/journal.ppat.1004995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Deval J., et al. Biochemical effect of resistance mutations against synergistic inhibitors of RSV RNA polymerase. PLoS One. 2016;11(5) doi: 10.1371/journal.pone.0154097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.DeVincenzo J.P., et al. Activity of oral ALS-008176 in a respiratory syncytial virus challenge study. N. Engl. J. Med. 2015;373(21):2048–2058. doi: 10.1056/NEJMoa1413275. [DOI] [PubMed] [Google Scholar]
- 171.Beigel J.H., et al. Advances in respiratory virus therapeutics—a meeting report from the 6th isirv Antiviral Group conference. Antiviral Res. 2019;167:45–67. doi: 10.1016/j.antiviral.2019.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Gorbalenya A.E., et al. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117(1):17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Stern D.F., Kennedy S.I. Coronavirus multiplication strategy. I. Identification and characterization of virus-specified RNA. J. Virol. 1980;34(3):665–674. doi: 10.1128/jvi.34.3.665-674.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Corman V.M., et al. Hosts and sources of endemic human coronaviruses. Adv. Virus Res. 2018;100:163–188. doi: 10.1016/bs.aivir.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5(4):536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Su S., et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24(6):490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Huang C., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wu F., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Zaki A.M., et al. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367(19):1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 180.Drosten C., et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 181.Forni G., et al. COVID-19 vaccines: where we stand and challenges ahead. Cell Death Differ. 2021;28(2):626–639. doi: 10.1038/s41418-020-00720-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.US Food and Drug Administration . US Food and Drug Administration; Silver Spring, MD: 2020. Pfizer Letter of Authorization. [Google Scholar]
- 183.US Food and Drug Administration . US Food and Drug Administration; Silver Spring, MD: 2020. Moderna COVID-19 Vaccine Letter of Authorization. [Google Scholar]
- 184.Polack F.P., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383(27):2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Baden L.R., et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021;384(5):403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.US Food and Drug Administration . US Food and Drug Administration; Silver Spring, MD: 2020. Janssen COVID-19 Vaccine Emergency Use Authorization Letter of Authorization. [Google Scholar]
- 187.Sadoff J., et al. Interim results of a phase 1-2a trial of Ad26.COV2.S Covid-19 vaccine. N. Engl. J. Med. 2021;384(19):1824–1835. doi: 10.1056/NEJMoa2034201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Government of Canada . 2021. Recalls and Safety Alerts: Authorization of AstraZeneca COVID-19 Vaccine With English-Only Vial and Carton Labels. [Google Scholar]
- 189.Voysey M., et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397(10269):99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.US Food and Drug Administration . US Food and Drug Administration; Silver Spring, MD: 2020. Fact Sheet for Health Care Providers Emergency Use Authorization (EUA) of Casirivimab and Imdevimab. [Google Scholar]
- 191.US Food and Drug Administration . US Food and Drug Administration; Silver Spring, MD: 2020. Fact Sheet for Health Care Providers Emergency Use Authorization (EUA) of Remdesivir (GS-5734™) [Google Scholar]
- 192.Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7(6):439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.da Costa V.G., et al. The emergence of SARS, MERS and novel SARS-2 coronaviruses in the 21st century. Arch. Virol. 2020 doi: 10.1007/s00705-020-04628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Ziebuhr J. The coronavirus replicase. Curr. Top. Microbiol. Immunol. 2005;287:57–94. doi: 10.1007/3-540-26765-4_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Snijder E.J., et al. The nonstructural proteins directing coronavirus RNA synthesis and processing. Adv. Virus Res. 2016;96:59–126. doi: 10.1016/bs.aivir.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Eckerle L.D., et al. Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 2010;6(5) doi: 10.1371/journal.ppat.1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Smith E.C., et al. Coronaviruses lacking exoribonuclease activity are susceptible to lethal mutagenesis: evidence for proofreading and potential therapeutics. PLoS Pathog. 2013;9(8) doi: 10.1371/journal.ppat.1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Shannon A., et al. Remdesivir and SARS-CoV-2: structural requirements at both nsp12 RdRp and nsp14 exonuclease active-sites. Antiviral Res. 2020;178:104793. doi: 10.1016/j.antiviral.2020.104793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Snijder E.J., et al. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331(5):991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Ferron F., et al. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc. Natl. Acad. Sci. U. S. A. 2018;115(2):E162–E171. doi: 10.1073/pnas.1718806115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Subissi L., et al. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. U. S. A. 2014;111(37):E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Zhai Y., et al. Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nat. Struct. Mol. Biol. 2005;12(11):980–986. doi: 10.1038/nsmb999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Peng Q., et al. Structural and biochemical characterization of the nsp12-nsp7-nsp8 core polymerase complex from SARS-CoV-2. Cell Rep. 2020;31(11):107774. doi: 10.1016/j.celrep.2020.107774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Tvarogova J., et al. Identification and characterization of a human coronavirus 229E nonstructural protein 8-associated RNA 3'-terminal adenylyltransferase activity. J. Virol. 2019;93(12) doi: 10.1128/JVI.00291-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Imbert I., et al. A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. EMBO J. 2006;25(20):4933–4942. doi: 10.1038/sj.emboj.7601368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.te Velthuis A.J., et al. The SARS-coronavirus nsp7 + nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension. Nucleic Acids Res. 2012;40(4):1737–1747. doi: 10.1093/nar/gkr893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Hillen H.S., et al. Structure of replicating SARS-CoV-2 polymerase. Nature. 2020 doi: 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- 208.Yin W., et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020 doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Gao Y., et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368(6492):779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Wang Q., et al. Structural basis for RNA replication by the SARS-CoV-2 polymerase. Cell. 2020 doi: 10.1016/j.cell.2020.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Chan J.F., et al. Broad-spectrum antivirals for the emerging Middle East respiratory syndrome coronavirus. J. Infect. 2013;67(6):606–616. doi: 10.1016/j.jinf.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Chen F., et al. In vitro susceptibility of 10 clinical isolates of SARS coronavirus to selected antiviral compounds. J. Clin. Virol. 2004;31(1):69–75. doi: 10.1016/j.jcv.2004.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Warren T.K., et al. Protection against filovirus diseases by a novel broad-spectrum nucleoside analogue BCX4430. Nature. 2014;508(7496):402–405. doi: 10.1038/nature13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Dyall J., et al. Repurposing of clinically developed drugs for treatment of Middle East respiratory syndrome coronavirus infection. Antimicrob. Agents Chemother. 2014;58(8):4885–4893. doi: 10.1128/AAC.03036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Peters H.L., et al. Design, synthesis and evaluation of a series of acyclic fleximer nucleoside analogues with anti-coronavirus activity. Bioorg. Med. Chem. Lett. 2015;25(15):2923–2926. doi: 10.1016/j.bmcl.2015.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Saijo M., et al. Inhibitory effect of mizoribine and ribavirin on the replication of severe acute respiratory syndrome (SARS)-associated coronavirus. Antiviral Res. 2005;66(2–3):159–163. doi: 10.1016/j.antiviral.2005.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Shannon A., et al. Rapid incorporation of favipiravir by the fast and permissive viral RNA polymerase complex results in SARS-CoV-2 lethal mutagenesis. Nat. Commun. 2020;11(1):4682. doi: 10.1038/s41467-020-18463-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Peng Q., et al. Structural basis of SARS-CoV-2 polymerase inhibition by favipiravir. Innovation (N. Y.) 2021;2(1) doi: 10.1016/j.xinn.2021.100080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Naydenova K., et al. Structure of the SARS-CoV-2 RNA-dependent RNA polymerase in the presence of favipiravir-RTP. Proc. Natl. Acad. Sci. U. S. A. 2021;118(7) doi: 10.1073/pnas.2021946118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Gordon C.J., et al. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020;295(20):6785–6797. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Bera S.C., et al. The nucleotide addition cycle of the SARS-CoV-2 polymerase. bioRxiv. 2021 doi: 10.1016/j.celrep.2021.109650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Siegel D., et al. Discovery and synthesis of a phosphoramidate prodrug of a pyrrolo[2,1-f][triazin-4-amino] adenine C-nucleoside (GS-5734) for the treatment of Ebola and emerging viruses. J. Med. Chem. 2017;60(5):1648–1661. doi: 10.1021/acs.jmedchem.6b01594. [DOI] [PubMed] [Google Scholar]
- 223.Warren T.K., et al. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531(7594):381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Lo M.K., et al. Remdesivir (GS-5734) protects African green monkeys from Nipah virus challenge. Sci. Transl. Med. 2019;11(494) doi: 10.1126/scitranslmed.aau9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Lo M.K., et al. GS-5734 and its parent nucleoside analog inhibit filo-, pneumo-, and paramyxoviruses. Sci. Rep. 2017;7:43395. doi: 10.1038/srep43395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Tchesnokov E.P., et al. Mechanism of inhibition of Ebola virus RNA-dependent RNA polymerase by remdesivir. Viruses. 2019;11(4) doi: 10.3390/v11040326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Mulangu S., et al. A randomized, controlled trial of Ebola virus disease therapeutics. N. Engl. J. Med. 2019;381(24):2293–2303. doi: 10.1056/NEJMoa1910993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Beigel J.H., et al. Remdesivir for the treatment of Covid-19—final report. N. Engl. J. Med. 2020;383(19):1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.WHO Solidarity Trial Consortium, et al. Repurposed antiviral drugs for COVID-19-interim WHO solidarity trial results. N. Engl. J. Med. 2021;384:497–511. doi: 10.1056/NEJMoa2023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Gotte M. Remdesivir for the treatment of Covid-19: the value of biochemical studies. Curr. Opin. Virol. 2021;49:81–85. doi: 10.1016/j.coviro.2021.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Agostini M.L., et al. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio. 2018;9(2) doi: 10.1128/mBio.00221-18. e00221-00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Sheahan T.P., et al. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 2017;9(396) doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Sheahan T.P., et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020;12(541) doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.de Wit E., et al. Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc. Natl. Acad. Sci. U. S. A. 2020;117(12):6771–6776. doi: 10.1073/pnas.1922083117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Pruijssers A.J., et al. Remdesivir potently inhibits SARS-CoV-2 in human lung cells and chimeric SARS-CoV expressing the SARS-CoV-2 RNA polymerase in mice. bioRxiv. 2020 doi: 10.1016/j.celrep.2020.107940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Tchesnokov E.P., et al. Template-dependent inhibition of coronavirus RNA-dependent RNA polymerase by remdesivir reveals a second mechanism of action. J. Biol. Chem. 2020;295(47):16156–16165. doi: 10.1074/jbc.AC120.015720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Lo M.K., et al. Remdesivir targets a structurally analogous region of the Ebola virus and SARS-CoV-2 polymerases. Proc. Natl. Acad. Sci. U. S. A. 2020;117(43):26946–26954. doi: 10.1073/pnas.2012294117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Gordon C.J., et al. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J. Biol. Chem. 2020;295(15):4773–4779. doi: 10.1074/jbc.AC120.013056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Agostini M.L., et al. Small-molecule antiviral beta-d-N (4)-hydroxycytidine inhibits a proofreading-intact coronavirus with a high genetic barrier to resistance. J. Virol. 2019;93(24) doi: 10.1128/JVI.01348-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Jena N.R. Role of different tautomers in the base-pairing abilities of some of the vital antiviral drugs used against COVID-19. Phys. Chem. Chem. Phys. 2020;22(48):28115–28122. doi: 10.1039/d0cp05297c. [DOI] [PubMed] [Google Scholar]
- 241.Bouvier N.M., Palese P. The biology of influenza viruses. Vaccine. 2008;26(Suppl. 4):D49–D53. doi: 10.1016/j.vaccine.2008.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Molinari N.A., et al. The annual impact of seasonal influenza in the US: measuring disease burden and costs. Vaccine. 2007;25(27):5086–5096. doi: 10.1016/j.vaccine.2007.03.046. [DOI] [PubMed] [Google Scholar]
- 243.World Health Organization . 2019. Global Influenza Strategy 2019–2030. [Google Scholar]
- 244.Plotkin S.A. Vaccines: past, present and future. Nat. Med. 2005;11(4 Suppl):S5–11. doi: 10.1038/nm1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Mameli C., et al. Influenza vaccination: effectiveness, indications, and limits in the pediatric population. Front. Pediatr. 2019;7:317. doi: 10.3389/fped.2019.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Wei C.-J., et al. Next-generation influenza vaccines: opportunities and challenges. Nat. Rev. Drug Discov. 2020;19(4):239–252. doi: 10.1038/s41573-019-0056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Iuliano A.D., et al. Estimates of global seasonal influenza-associated respiratory mortality: a modelling study. Lancet. 2018;391(10127):1285–1300. doi: 10.1016/S0140-6736(17)33293-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Gubareva L.V., et al. Influenza virus neuraminidase inhibitors. Lancet. 2000;355(9206):827–835. doi: 10.1016/S0140-6736(99)11433-8. [DOI] [PubMed] [Google Scholar]
- 249.CDC . 2020. Influenza Antiviral Medications: Summary for Clinicians. [Google Scholar]
- 250.Reich S., et al. Structural insight into cap-snatching and RNA synthesis by influenza polymerase. Nature. 2014;516(7531):361–366. doi: 10.1038/nature14009. [DOI] [PubMed] [Google Scholar]
- 251.Pflug A., et al. Structure of influenza A polymerase bound to the viral RNA promoter. Nature. 2014;516(7531):355–360. doi: 10.1038/nature14008. [DOI] [PubMed] [Google Scholar]
- 252.te Velthuis A.J., Fodor E. Influenza virus RNA polymerase: insights into the mechanisms of viral RNA synthesis. Nat. Rev. Microbiol. 2016;14(8):479. doi: 10.1038/nrmicro.2016.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Dou D., et al. Influenza A virus cell entry, replication, virion assembly and movement. Front. Immunol. 2018;9:1581. doi: 10.3389/fimmu.2018.01581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Dias A., et al. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature. 2009;458(7240):914–918. doi: 10.1038/nature07745. [DOI] [PubMed] [Google Scholar]
- 255.Guilligay D., et al. The structural basis for cap binding by influenza virus polymerase subunit PB2. Nat. Struct. Mol. Biol. 2008;15(5):500. doi: 10.1038/nsmb.1421. [DOI] [PubMed] [Google Scholar]
- 256.Yuan P., et al. Crystal structure of an avian influenza polymerase PA(N) reveals an endonuclease active site. Nature. 2009;458(7240):909–913. doi: 10.1038/nature07720. [DOI] [PubMed] [Google Scholar]
- 257.Fodor E., te Velthuis A.J. Structure and function of the influenza virus transcription and replication machinery. Cold Spring Harb. Perspect. Med. 2019 doi: 10.1101/cshperspect.a038398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Noshi T., et al. In vitro characterization of baloxavir acid, a first-in-class cap-dependent endonuclease inhibitor of the influenza virus polymerase PA subunit. Antiviral Res. 2018;160:109–117. doi: 10.1016/j.antiviral.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 259.Hayden F.G., et al. Baloxavir marboxil for uncomplicated influenza in adults and adolescents. N. Engl. J. Med. 2018;379(10):913–923. doi: 10.1056/NEJMoa1716197. [DOI] [PubMed] [Google Scholar]
- 260.Todd B., et al. The active form of the influenza cap-snatching endonuclease inhibitor baloxavir marboxil is a tight binding inhibitor. J. Biol. Chem. 2021:100486. doi: 10.1016/j.jbc.2021.100486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Omoto S., et al. Characterization of influenza virus variants induced by treatment with the endonuclease inhibitor baloxavir marboxil. Sci. Rep. 2018;8(1):9633. doi: 10.1038/s41598-018-27890-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Mishin V.P., et al. Susceptibility of influenza A, B, C, and D viruses to baloxavir. Emerg. Infect. Dis. 2019;25(10):1969. doi: 10.3201/eid2510.190607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Takashita E., et al. Human-to-human transmission of influenza A (H3N2) virus with reduced susceptibility to baloxavir, Japan, February 2019. Emerg. Infect. Dis. 2019;25(11):2108. doi: 10.3201/eid2511.190757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Toots M., et al. Characterization of orally efficacious influenza drug with high resistance barrier in ferrets and human airway epithelia. Sci. Transl. Med. 2019;11(515) doi: 10.1126/scitranslmed.aax5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Delang L., et al. Favipiravir as a potential countermeasure against neglected and emerging RNA viruses. Antiviral Res. 2018;153:85–94. doi: 10.1016/j.antiviral.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 266.Furuta Y., et al. Mechanism of action of T-705 against influenza virus. Antimicrob. Agents Chemother. 2005;49(3):981–986. doi: 10.1128/AAC.49.3.981-986.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Baranovich T., et al. T-705 (favipiravir) induces lethal mutagenesis in influenza A H1N1 viruses in vitro. J. Virol. 2013;87(7):3741–3751. doi: 10.1128/JVI.02346-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Jin Z., et al. The ambiguous base-pairing and high substrate efficiency of T-705 (favipiravir) ribofuranosyl 5'-triphosphate towards influenza A virus polymerase. PLoS One. 2013;8(7) doi: 10.1371/journal.pone.0068347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Takashita E., et al. Antiviral susceptibility of influenza viruses isolated from patients pre-and post-administration of favipiravir. Antiviral Res. 2016;132:170–177. doi: 10.1016/j.antiviral.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 270.Goldhill D.H., et al. The mechanism of resistance to favipiravir in influenza. Proc. Natl. Acad. Sci. U. S. A. 2018;115(45):11613–11618. doi: 10.1073/pnas.1811345115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Hayden F.G., Shindo N. Influenza virus polymerase inhibitors in clinical development. Curr. Opin. Infect. Dis. 2019;32(2):176–186. doi: 10.1097/QCO.0000000000000532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Shiraki K., Daikoku T. Favipiravir, an anti-influenza drug against life-threatening RNA virus infections. Pharmacol. Ther. 2020;209:107512. doi: 10.1016/j.pharmthera.2020.107512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Clark M.P., et al. Discovery of a novel, first-in-class, orally bioavailable azaindole inhibitor (VX-787) of influenza PB2. J. Med. Chem. 2014;57(15):6668–6678. doi: 10.1021/jm5007275. [DOI] [PubMed] [Google Scholar]
- 274.Byrn R.A., et al. Preclinical activity of VX-787, a first-in-class, orally bioavailable inhibitor of the influenza virus polymerase PB2 subunit. Antimicrob. Agents Chemother. 2015;59(3):1569–1582. doi: 10.1128/AAC.04623-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Trevejo J.M., et al. Pimodivir treatment in adult volunteers experimentally inoculated with live influenza virus: a phase IIa, randomized, double-blind, placebo-controlled study. Antivir. Ther. 2018;23(4):335–344. doi: 10.3851/IMP3212. [DOI] [PubMed] [Google Scholar]
- 276.Finberg R.W., et al. Phase 2b study of pimodivir (JNJ-63623872) as monotherapy or in combination with oseltamivir for treatment of acute uncomplicated seasonal influenza A: TOPAZ trial. J. Infect. Dis. 2019;219(7):1026–1034. doi: 10.1093/infdis/jiy547. [DOI] [PubMed] [Google Scholar]
- 277.Robert X., Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42(Web Server issue):W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Pettersen E.F., et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]