Recent epidemiological studies of dissemination of vanA-type glycopeptide resistance in enterococci indicated localization of the vanA gene cluster on plasmids smaller than 100 kbp (1, 4, 6) or on the chromosome (2, 3, 6). However, until now the integration of the vanA cluster into a particular chromosomal SmaI fragment of a vancomycin-resistant Enterococcus (VRE) strain has been unambigiously demonstrated only in one case (2). Many authors interpreted their findings of lack of plasmid isolation and hybridization of a labelled vanA gene probe to fragments resolved in pulsed-field gel electrophoresis (PFGE) as chromosomal localization of the vanA cluster (3, 4, 6).
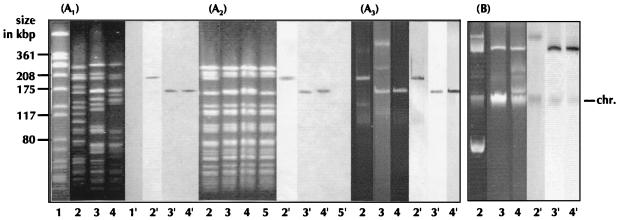
For 18 of 26 VRE (Enterococcus faecium) strains from Germany analyzed by PFGE, the vanA gene clusters were documented to be located on macrorestriction fragments (125 to 208 kbp) which hybridized with a vanA probe (8) (Fig. 1A1). The glycopeptide resistance determinant was transferred to a recipient strain by filter mating. Macrorestriction analysis (MRA) of the transconjugant DNA and corresponding Southern hybridizations localized the vanA gene on the same-sized fragments as in the donor strains (Fig. 1A2).
FIG. 1.
(A) Macrorestriction patterns in PFGE (left) and corresponding Southern blots (right) hybridized with a digoxigenin-labelled vanA gene probe resulting from SmaI-digested cell DNA of VRE (A1), SmaI-digested cell DNA of transconjugants of VRE (A2), and SmaI-digested plasmid DNA of VRE (A3). (B) Nondigested plasmids of VRE in 0.8% agarose gel (left) and corresponding Southern blots (right) hybridized with a digoxigenin-labelled vanA gene probe. Lanes: 1, Staphylococcus aureus NCTC 8325 (molecular weight standard for PFGE); 2, strain U200; 3, strain AW5; 4, strain AW9; 5, recipient strain 64/3. chr., chromosome.
The plasmid isolation method of Woodford et al. (10) was modified as follows: lysozyme treatment was extended to 60 min, ethanol precipitation of DNA at −20°C was extended to overnight, and gentle handling was employed without vortexing, stirring, or filtering of the probes. Plasmid DNA from donor and transconjugant strains was demonstrated in 0.8% agarose gels (Fig. 1B). When plasmid DNA was digested with SmaI and resolved by PFGE, plasmid bands (125 to 200 kbp) could be detected (Fig. 1A3), whereas no bands appeared without SmaI digestion (data not shown; nondigested plasmids do not appear as fragments on PFGE).
For epidemiological studies of dissemination of the glycopeptide resistance determinant in enterococci, the localization of the corresponding gene cluster is of particular interest. Mobile vanA gene clusters were found on conjugative plasmids as well as on conjugative chromosomal elements. Integration of conjugative chromosomal gene clusters into a recipient chromosome can easily be detected by MRA using PFGE (7), which leads to a differentiation between chromosomal and plasmid-encoded glycopeptide resistance. For large resistance plasmids (>100 kb), gentle isolation methodologies without any vortexing, stirring, or filtering steps are recommended.
Based on the recommendations for determining clonal identity by means of PFGE, macrorestriction patterns differing in more than three fragments are considered to indicate separate clones (5). As described recently, VRE can alter their van genotype during an outbreak (9), possessing different van plasmids. For the large van plasmids described here, a 2-fragment shift would already result from plasmid exchange, without any other alteration(s) in the chromosome. These considerations must be kept in mind when comparing MRA results in outbreaks.
REFERENCES
- 1.Clark N C, Cooksey R C, Hill B C, Swenson J M, Tenover F C. Characterization of glycopeptide-resistant enterococci from U.S. hospitals. Antimicrob Agents Chemother. 1993;37:2311–2317. doi: 10.1128/aac.37.11.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Handwerger S, Skoble J. Identification of chromosomal mobile element conferring high-level vancomycin resistance in Enterococcus faecium. Antimicrob Agents Chemother. 1995;39:2446–2453. doi: 10.1128/aac.39.11.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jensen L B, Ahrens P, Dons L, Jones R N, Hammerun A, Aarestrup F M. Molecular analysis of Tn1546 in Enterococcus faecium isolated from animals and humans. J Clin Microbiol. 1998;36:437–442. doi: 10.1128/jcm.36.2.437-442.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mato R, de Lencastre H, Roberts R B, Tomasz A. Multiplicity of genetic backgrounds among vancomycin-resistant Enterococcus faecium isolates recovered from an outbreak in a New York City hospital. Microb Drug Resist. 1996;2:309–317. doi: 10.1089/mdr.1996.2.309. [DOI] [PubMed] [Google Scholar]
- 5.Tenover F C, Arbeit R D, Goering R V, Mickelson P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thal L, Donabedian S, Robinson-Dunn B, Chow J W, Dembry L, Clewell D B, Alshab D, Zervos M J. Molecular analysis of glycopeptide-resistant Enterococcus faecium isolates collected from Michigan hospitals over a 6-year period. J Clin Microbiol. 1998;36:3303–3308. doi: 10.1128/jcm.36.11.3303-3308.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thal L, Silverman J, Donabedian S, Zervos M. The effect of Tn916 insertions on contour-clamped homogeneous electrophoresis patterns of Enterococcus faecalis. J Clin Microbiol. 1997;35:969–972. doi: 10.1128/jcm.35.4.969-972.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Werner G, Klare I, Witte W. Arrangement of the vanA gene cluster in enterococci of different ecological origin. FEMS Microbiol Lett. 1997;155:55–61. doi: 10.1111/j.1574-6968.1997.tb12685.x. [DOI] [PubMed] [Google Scholar]
- 9.Woodford N, Chadwick P R, Morrison D, Cookson B D. Strains of glycopeptide-resistant Enterococcus faecium can alter their van genotypes during an outbreak. J Clin Microbiol. 1997;35:2966–2968. doi: 10.1128/jcm.35.11.2966-2968.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woodford N, Morrison D, Cookson B, George R C. Comparison of high-level gentamicin-resistant Enterococcus faecium isolates from different continents. Antimicrob Agents Chemother. 1993;37:681–684. doi: 10.1128/aac.37.4.681. [DOI] [PMC free article] [PubMed] [Google Scholar]