Abstract
The CD8αβ heterodimer plays a crucial role in the stabilization between major histocompatibility complex class I molecules (MHC-I) and the T cell receptor (TCR). The interaction between CD8 and MHC-I can be regulated by posttranslational modifications, which are proposed to play an important role in the development of CD8 T cells. One modification that has been proposed to control CD8 coreceptor function is ribosylation. Utilizing NAD+, the ecto-enzyme adenosine diphosphate (ADP) ribosyl transferase 2.2 (ART2.2) catalyzes the addition of ADP-ribosyl groups onto arginine residues of CD8α or β chains and alters the interaction between the MHC and TCR complexes. To date, only interactions between modified CD8 and classical MHC-I (MHC-Ia), have been investigated and the interaction with non-classical MHC (MHC-Ib) has not been explored. Here, we show that ADP-ribosylation of CD8 facilitates the binding of the liver-restricted nonclassical MHC, H2-Q10, independent of the associated TCR or presented peptide, and propose that this highly regulated binding imposes an additional inhibitory leash on the activation of CD8-expressing cells in the presence of NAD+. These findings highlight additional important roles for nonclassical MHC-I in the regulation of immune responses.
Keywords: MHC-Ib, non-classical MHC, H2-Q10, ribosylation, CD8αβ
Abbreviations: ADP, adenosine diphosphate; APC, antigen-presenting cell; ART2.2, ADP ribosyl transferase 2.2; FACS, flow cytometry; GPI, glycosylphosphatidylinositol; MHC-I, major histocompatibility complex class I; NAD, nicotinamide adenine dinucleotide; TCR, T cell receptor
The CD8 glycoprotein enhances T cell activation by interacting with peptide bound major histocompatibility class I molecules (pMHC-I) on antigen-presenting cells (APC) (1). The CD8 coreceptor is expressed as a disulfide-linked dimer of an alpha (α) and beta (β) domain and/or an α−chain homodimer, with the vast majority of peripheral CD8+ T cells expressing the CD8αβ heterodimer (2). Upon T cell activation, the T cell receptor (TCR) binds to pMHC-I and CD8 binding follows, leading to a TCR-pMHC-I-CD8 complex. CD8 interacts with the Src tyrosine kinase Lck, resulting in phosphorylation of CD3 in the TCR-CD3 complex, ultimately resulting in T cell activation and proliferation (3).
Posttranslational modification represents an additional regulatory checkpoint in T cell activation. During thymic maturation, the CD8 receptor on developing T cells exhibits promiscuous binding to MHC-I; however, as the CD8+ T cell matures, CD8 undergoes glycosylation and sialic acid capping, abolishing nonspecific binding and enforcing specific binding to cognate TCR (4, 5). In addition, upon activation CD8 undergoes a reduction in sialylation of N-glycans, resulting in a reduction in binding to MHC-I (6). Furthermore, distinct glycosylation patterns in different organs may influence local T cell function in those organs. For example, deficiencies in O-linked glycosylation have been associated with impaired differentiation of regulatory T cells in the liver, resulting in increased liver injury in a mouse model of autoimmune hepatitis (7).
Ribosylation of CD8 is another important posttranslational modification that can control T cell function. Ribosylation occurs when extracellular soluble nicotinamide adenine dinucleotide (NAD+) acts as a substrate for the glycosylphosphatidylinositol (GPI)-anchored cell surface enzyme ADP-ribosyltransferase 2 (ART2.2), resulting in the transfer of ADP-ribose to arginine residues. NAD+ is released by damaged or stressed cells, acting as a danger signal, and also has the ability to induce apoptosis via the ribosylation of the purinoceptor P2X7 (8, 9, 10, 11). ART2.2 is expressed by mature T cells and is cleaved from the cell surface upon activation of T cells (12, 13, 14, 15). While the cleaved form of ART2.2 is still enzymatically active and may act as an intercellular signal, the shedding of ART2.2 reduces the sensitivity of the cell to NAD+ and reduces subsequent ribosylation (16). As such, the ribosylation of CD8 is entirely dependent upon the enzymatic activity of ART2.2 and results in decreased T cell cytotoxicity (15). Interestingly, there are tissue-specific patterns of ribosylation, with the liver showing increased modification of Arg residues contained in membrane and extracellular proteins compared with other tissues (17). In addition, tissue-resident T cells (Trm) in the liver have been reported to express high levels of ART2.2 and P2X7, and that blockade of ART2.2 can reverse the effects of ADP-ribosylation induced cellular damage (18).
To date, only interactions between modified CD8 and classical MHC-I (MHC-Ia), have been investigated and the interaction with nonclassical MHC (MHC-Ib) has not been explored. MHC-Ib molecules are distinguished from MHC-Ia by their limited polymorphism and often restricted tissue distribution, such as the H2-Q10 molecule that is highly restricted to the liver. We and others have observed that MHC-Ib family members can interact with CD8αα homodimers (19) but MHC-Ib has not been reported to recognize CD8αβ (19). Here, we found that the liver-restricted MHC-Ib molecule H2-Q10 binds liver-derived CD8 T cells following the ribosylation of the CD8αβ heterodimer. Therefore, we hypothesized that upon cellular damage or stress, the release of NAD+ reduces the interaction between the CD8 and the TCR:MHC-Ia interaction, but provides a specific mechanism for binding to H2-Q10.
Results
High expression of H2-Q10 in the liver correlates with binding to TCRαβ CD8 T cells
MHC-Ib proteins generally display organ-specific expression levels and H2-Q10 had previously been reported to be restricted to the liver (20), a finding we confirmed in this study (Fig. 1A). Given our previous studies showing that H2-Q10 bound the natural killer (NK) cell receptor Ly49C (21) and CD8αα (19), we analyzed the binding of H2-Q10 to cells expressing these receptors within the bone marrow, spleen, lung, and liver. H2-Q10 tetramers did not bind to NK cells from any of the organs tested, likely due to the binding of Ly49C to MHC-Ia in cis (21) (Fig. 1B). In contrast, CD8αα-expressing γδ T cells from all organs bound to H2-Q10 tetramers (Fig. 1C). Strikingly, H2-Q10 tetramers only bound to CD8 αβ T cells isolated from the liver, demonstrating a tissue-specific interaction (Fig. 1D).
Figure 1.
H2-Q10 is highly expressed in the liver, recognizes hepatic αβ/γδ T cells irrespective of the organ origin, and increases upon aging.A, real-time PCR detection demonstrates that H2-Q10 is highly expressed by hepatocytes in contrast to low or absence of expression in the bone marrow, spleen, and lung of mice at 7 weeks of age. Data are from ≥9 samples prepared in isolation ∗∗∗∗p < 0.0001. Binding of H2-Q10 tetramer to NK cells (B), γδ T cells (C) and αβ T cells (D) isolated from 7 week old C57BL/6 mice. Histograms are representative of two independent experiments using four mice per experiment (n = 8). All histograms have been offset and are modal. The controls are unstained cells. E, real-time PCR detection demonstrates that H2-Q10 expression in hepatocytes increases upon aging, in contrast to low expression in the bone marrow, spleen, and lung of mice at 1, 3, 5, and 7 weeks of age. Data are from nine samples prepared in isolation ∗∗p = 0.0022. F, total bone marrow, spleen, lung, or liver was processed into single cell suspensions and H2-Q10 tetramer binding was determined by flow cytometry (FACS). Histograms are representative of two independent experiments using five mice per experiment (n = 10). G, NK and CD8 αβ T cells were isolated from the liver via cell sorting at 1, 3, 5, and 7 weeks of age, and H2-Q10 tetramer binding was determined by FACS. Histograms are representative of two independent experiments using five mice per experiment (n = 10). ∗∗∗∗p ≤ 0.0001.
Age-associated changes of H2-Q10 expression and binding to CD8αβ T cells
Given that previous studies have shown developmental regulation of CD8-MHC binding (4), and that the expression of H2-Q10 in the liver increases with age (22, 23), we determined whether there was any difference in the ability of H2-Q10 to bind CD8αβ T cells isolated from different aged mice. In line with previous studies, we found an increase in the expression of H2-Q10 RNA in the liver that occurred between 3 and 7 weeks of age (Fig. 1E). We also observed a significant increase in the number and frequency of TCRαβ CD8 T cells in the liver that bound H2-Q10 tetramers at the same time points (Fig. 1F), whereas the tetramer did not bind to NK cells at any age (Fig. 1G). Moreover, the H2-Q10 tetramer did not bind to TCRαβ CD8 T cells isolated from the bone marrow, spleen, or lung at any age (Fig. 1E). Thus, age-related correlation exists between the increase in H2-Q10 expression in the liver and its ability to bind to liver-derived CD8αβ T cells.
Binding to TCRαβ CD8 T cells in the liver is specific to H2-Q10, TCR-independent, not peptide specific, and involves the CD8β chain
Having demonstrated that the H2-Q10 tetramer binds to TCRαβ CD8 T cells in the liver and that this is dependent upon a liver specific factor, we next sought to determine the specificity of this interaction. We generated H2-Q10 heavy chains and murine β2-microglobulin and refolded in the presence of either riboforin, proteosome peptide or TCR Vβ chain peptides. The purified protein preparation (Fig. 2A) depicts a single band of H2-Q10 monomer, with almost complete biotinylation of the H2-Q10 protein, indicated by the shift upon addition of streptavidin. We stained liver CD8 T cells with MHC-Ia and MHC-Ib tetramers to determine if this binding was specific to H2-Q10. These studies showed that only H2-Q10 tetramers bound to CD8αβ T cells and not other MHC-Ib (H2-Q9, -M3) or MHC-Ia (H-2Kb, -2Db, -2Ld) tetramers (Fig. 2B). In addition, binding of H2-Q10 tetramer to liver-derived CD8αβ T cells was independent of a specific peptide loaded into H2-Q10 (Fig. 2C). Moreover, identical levels of H2-Q10 tetramer binding were observed for liver-derived CD8αβ T cells isolated from wild-type mice and mice where the majority of CD8αβ T cells are specific for ovalbumin (OT-I mice), indicating that tetramer binding was independent of the specificity of the TCR (Fig. 2D). We next determined the role of the alpha and beta domains of CD8 in this interaction. Incubating H2-Q10 tetramers with a CD8α antibody known to block CD8+ T cell activation (24, 25) had no effect on binding, while incubation with antibodies that prevented binding to both CD8α and CD8β completely abolished tetramer binding (Fig. 2E). Moreover, the H2-Q10 tetramer failed to bind to liver-derived CD8αβ T cells isolated from CD8β deficient mice (Fig. 2F). These data demonstrate that the binding of H2-Q10 tetramer to CD8αβ T cells in the liver is H2-Q10 specific, independent of TCR specificity, not peptide-specific, but dependent on the presence of the CD8β chain.
Figure 2.
H2-Q10 binding to CD8αβ T cells in the liver is specific to H2-Q10, independentofTCR andisnot peptide specific.A, 10% native PAGE gel with a single band of H2-Q10 monomer refolded with VF9 or HGT peptides. Almost complete biotinylation is indicated by the shift upon the addition of streptavidin (wells 2 and 4). B, binding of MHC-Ia tetramers H-2Ld, H-2Db, and H-2Kb and MHC-Ib tetramers H2-M3, H2-Q9 and H2-Q10 to liver CD8αβ cells isolated from WT mice. C, liver CD8αβ cells stained with H2-Q10 tetramers folded with HGT, VGI, or TGT peptide. D, Liver CD8αβ T cells were isolated from either WT or OT-I transgenic mice and binding of H2-Q10 tetramer determined to each genotype. E, competition assays were performed using the H2-Q10 tetramer alongside antibodies against CD8α and CD8β on liver CD8αβ cells. F, CD8 cells were isolated from the liver from either WT or CD8β−/− mice and binding of H2-Q10 tetramer determined to each genotype.
Liver CD8 T cells do not have a distinct pattern of glycosylation
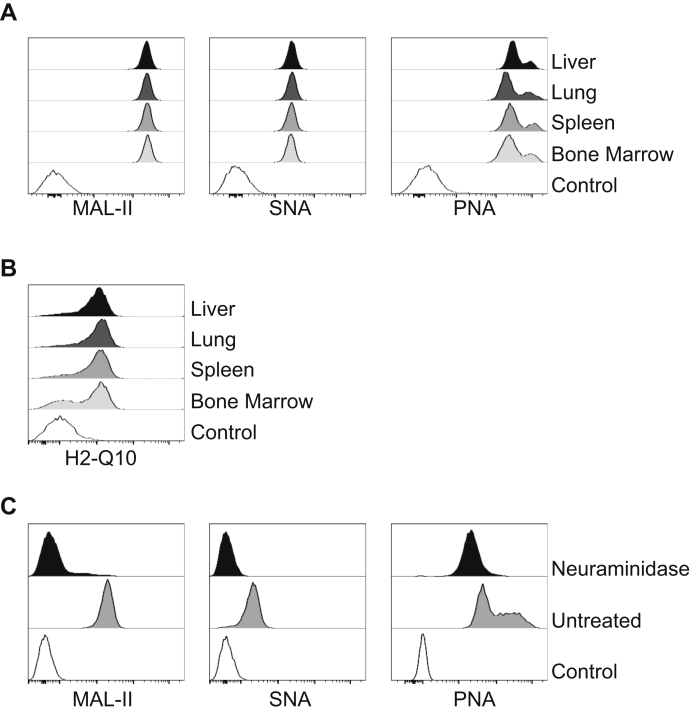
Our previous results demonstrated that H2-Q10 binds to CD8αα expressing γδ T cells as they have lower levels of sialic acids when compared with αβT cells (19). Furthermore, we demonstrated that H2-Q10 is able to bind to CD8αα heterodimers when sialic acids are removed (19). Hence, we determined whether the sialic acid profile of liver TCRαβ CD8 T cells was different from that of TCRαβ CD8 T cells isolated from other organs. Sialylation profiling indicated equivalent levels of α2,3 and α2,6 linked sialic acids, reflected by the comparable binding of the lectins maackia amurensis lectin II and sambucus nigra lectin, respectively, on CD8αβ T cells isolated from all organs (Fig. 3A). Moreover, equivalent levels of peanut agglutinin binding indicated similar proportions of hypo-sialylated O-linked glycans on CD8αβ T cells isolated from all organs (Fig. 3A). Furthermore, removal of terminal sialic acid residues following treatment with neuraminidase (Fig. 3C) resulted in binding of H2-Q10 to CD8αβ T cells, independent of the organs from which they were harvested (Fig. 3B). Thus, in contrast to the interaction with CD8αα, the binding of H2-Q10 tetramer to liver-derived CD8αβ T cells does not appear to be controlled by the surface levels of sialic acids.
Figure 3.
Glycosylation patterns of liver CD8αβ cells.A, single cell suspensions were isolated from the liver, lung, spleen, and bone marrow before cell sorting on CD8αβ cells (CD3+ CD8a+ TCRαβ+) and staining with the lectins maackia amurensis lectin II, sambucus nigra lectin and peanut agglutinin. Histograms are representative of two independent experiments using five mice per experiment (n = 10). CD8αβ cells isolated from organs were treated with 5 μg/ml Neuraminidase for 20 min and H2-Q10 binding (B) and glycosylation profile via lectin binding (C) was observed. Histograms are representative of two independent experiments using five mice per experiment (n = 10).
Soluble factors released by the liver parenchyma control H2-Q10 binding to TCRαβ CD8 T cells
Isolation of leukocytes from the liver involves separation of parenchymal and nonparenchymal cells using Percoll. We investigated whether a factor released from the liver during enrichment of leukocytes was involved in the binding of H2-Q10 to CD8αβ T cells. We isolated splenocytes from wild-type (CD45.2) mice and determined H2-Q10 tetramer binding in the absence or presence of liver cells isolated by mechanical disruption from congenic mice (CD45.1). Analysis of splenocytes in the absence of liver preparations demonstrated no binding of H2-Q10 to TCRαβ CD8 T cells, (Fig. 4i). In contrast, there was an increase in H2-Q10 tetramer binding to CD45.2 splenocytes when in the presence of liver preparations from CD45.1 mice (Fig. 4ii). These data demonstrate that H2-Q10 tetramer binding to TCRαβ CD8 T cells in the liver is a function of a soluble factor released upon disruption of the liver.
Figure 4.
H2-Q10 binding to CD8αβ cells is controlled by liver-soluble factors. Splenocytes were isolated from CD45.2 mice and either stained with H2-Q10 tetramer directly (i) or processed through Percoll in the presence of disrupted liver from CD45.1 mice. In this case, cells were divided into (ii) CD45.2 positive (splenocytes) or (iii) negative (liver leukocytes), and stained with H2-Q10 tetramer. Histograms are representative of two independent experiments using five mice per experiment (n = 10).
NAD and ART2.2 control binding of H2-Q10 to CD8β
Our results indicated that H2-Q10 binding to CD8αβ T cells was not dependent on sialylation but instead was dependent on a soluble liver-derived factor. Therefore, we hypothesized that these findings were attributed to ribosylation of CD8. Intriguingly, the H2-Q10 tetramer showed binding to splenocytes in the presence NAD, the substrate for ribosylation, in a dose-dependent manner, an interaction that was specific for H2-Q10 and not observed with any other MHC-I (Fig. 5A). Next, we generated CHO cell lines expressing CD8αα or CD8αβ cotransfected with or without with ART2.2, the enzyme that catalyzes ribosylation of Arg residues (26). CHO cells expressing CD8αα or CD8αβ in the absence of ART2.2 did not bind any tetramers, as depicted by “untreated” samples (Fig. 5A). In contrast, H2-Q10 tetramers specifically bound to CHO cells coexpressing ART 2.2 and CD8αβ but not CD8αα (Fig. 5B). This indicates that both the substrate and the enzyme for ribosylation are required to facilitate binding of H2-Q10 to CD8αβ cells.
Figure 5.
H2-Q10 binding to CD8αβ is reliant upon NAD treatment and the CD8 beta chain.A, splenocytes were isolated and treated with increasing concentrations of NAD+ for 30 min at 37 °C. Following treatment binding of MHC-Ia tetramers H-2Ld, H-2Db, and H-2Kb and the MHC-Ib tetramers H2-M3, H2-Q9 and H2-Q10 binding to CD8αβ cells were determined by FACS. Histograms are representative of two independent experiments using five mice per experiment (n = 10). B, CHO cells were generated, which expressed the ART2.2 ectoenzyme alongside either CD8αα or CD8αβ. Tetramer binding to these cells was determined via FACS. Histograms are representative of three independent experiments (n = 3).
H2-Q10 does not compete with H-2Kb for CD8αβ binding
Having shown that NAD treatment of cells expressing CD8αβ in the presence of ART2.2 increases binding of H2-Q10, along with previous studies having observed that H-2Kb binding is impaired in the presence of NAD+ (15), we sought to determine if H2-Q10 binding competes with H-2Kb binding. For these experiments we used the CHO CD8αβ ART2.2 cells and treated with increasing doses of NAD+. To investigate competition, we tested H-2Kb tetramer binding in both the absence and presence of the H2-Q10 tetramer (Fig. 6). Interestingly, the binding of the H-2Kb is unaffected by the presence of H2-Q10. To confirm that this observation was not peptide-specific, we also tested binding of H-2Kb refolded with three additional peptides and saw similar results (Fig. 6), Therefore, these data indicate that H2-Q10 tetramer binds to a different region of CD8αβ, and this does not compete with H-2Kb binding.
Figure 6.
H2-Q10 binding does not compete with H-2Kb binding to CD8αβ. CHO ART2.2-CD8αβ cells were treated with increasing doses of NAD+ for 30 min at 37 °C before incubating with either H-2Kb tetramer (folded with SIINFEKL, SIITFEKL, SIIQFEKL, or SIIGFEKL) or with H-2Kb tetramer and H2-Q10 tetramer together. Histograms are representative of three independent experiments (n = 3).
Identification of binding motifs of H2-Q10 to CD8αβ cells suggests NAD treatment exposes CD8α to H2-Q10
To determine the effect of NAD+ on the CD8β chain, we isolated splenocytes from WT mice and isolated CD8+ T cells, which express the CD8αβ heterodimer. Upon treatment with NAD+, we identified that the levels of CD8β chain being expressed by cells are reduced (Fig. 7A). Due to this, we queried whether ribosylation of the beta chain was not leading to direct binding of H2-Q10, but instead it was revealing key residues on the alpha chain that then allowed H2-Q10 to bind. To address this hypothesis, we mutated key residues on the H2-Q10 tetramer that have been previous identified to interact with the alpha chain of CD8, and using these, we tested binding to ribosylated CD8 (Fig. 7B). As previously shown, binding of native H2-Q10 tetramer increased upon increasing doses of NAD+; however, the single point mutant 194 showed significant reduction in binding, and this loss of binding was enhanced when examining the double mutant of 187/194. Therefore, these data suggest that ribosylation of the CD8 beta chain exposes the alpha chain, thereby increasing the ability of H2-Q10 to bind.
Figure 7.
Identification of binding motifs of H2-Q10 to CD8αβ cells. CHO ART2.2 CD8αβ cells were treated with increasing concentrations of NAD+ before (A) levels of CD8 alpha and beta chain were determined via FACS (n = 3) or (B) stained with H2-Q10 tetramers with point mutations on the putative binding sites between CD8α and MHC-Ib (n = 3).
Discussion
The MHC-Ib molecules are an underresearched family of proteins, whose importance in regulating the immune response is only just coming to light. Out of the extended family of MHC-Ib in the mouse, only a handful have identified binding partners, and there remains uncertainties surrounding their physiological relevance (21). The binding partners identified for MHC-Ib to date include TCR (27, 28), NK cell receptors Ly49A (29) and Ly49C (21), and CD8 (30). MHC-Ia are recognised ligands for the CD8αβ heterodimer; however, how they bind is dependent upon posttranslational modifications. CD8 preferentially binds MHC-Ia in trans; however, when the CD8β chain undergoes posttranslational modification and acquires O-linked glycans near positively charged residues, this favors binding of the MHC-Ia to CD8 in cis, which in turn alters the response of the CD8+ T cell (31). Unlike CD8αα, the CD8αβ heterodimer binds MHC-Ia independently of the β2M loop and relies on the β chain to orient the CD8αβ heterodimer (32). However, MHC-Ib has previously only been identified to bind to the CD8αα homodimer (19, 33). We recently observed that the liver-specific MHC-Ib, H2-Q10, was a high-affinity ligand for CD8αα, predominantly expressed by liver γδ T cells. Unlike MHC-Ia, where early expression in the thymus is essential for the development of CD8+ T cells, our recent study identifying that H2-Q10 recognizes that γδ T cells in the periphery help rationalize why H2-Q10 expression peaks at a later timepoint (19). γδ T cells do not mature within the thymus, but instead in the periphery, with large populations found within the gut and liver, as well as other peripheral organs, and therefore they require interaction with MHC-Ib later than intrathymically developing cells (34, 35). Herein, we show that the ability of γδ T cells to recognize H2-Q10 is unaffected by the age of these cells, instead the point of control being the induction of H2-Q10 expression by hepatocytes at 5 weeks of age. Moreover, we find that H2-Q10 is also a ligand for CD8αβ heterodimers and may play a role in the immune response following cellular damage or stress. During cell damage, NAD+ release occurs, and via the enzyme ART2.2, this leads to the subsequent ribosylation of the CD8β chain on T cells, thereby reducing binding of MHC-Ia (15). It has been proposed that this mechanism serves to curtail the CD8αβ T cell response to prevent immune-mediated damage. However, we show that ribosylation of CD8 also induces binding of the nonclassical MHC-I receptor H2-Q10. We show that binding is controlled by key residues of H2-Q10 that interact with CD8αα, suggesting that ribosylation of the beta chain exposes the CD8α chain, allowing for H2-Q10 to interact. It is possible that the binding of H2-Q10 serves to further dampen down the immune response of CD8αβ T cells in the liver, either by blocking binding of MHC-Ia or potentially through preventing efficient signaling. Indeed, the liver is well recognized as a site of immune tolerance, evidenced by the observation that liver allografts are often not rejected despite mismatched MHC-Ia (36). Notably, although CD8α enhances the response of CD8αβ T cells by lipid raft partitioning of the TCR, it is the beta chain of CD8 that leads to efficient and effective Lck activity and downstream signaling (1, 37). Moreover, a recent study demonstrated that CD8 T cells are inherently more autoreactive than CD4 T cells due to an increase in stoichiometry of the interaction between CD8 and Lck, which does not occur for CD4 (38). Therefore, the interaction between H2-Q10 and ribosylated CD8 may contribute to mechanisms of immune tolerance in the liver.
The other interesting finding is the observation of the developmentally regulated increase in H2-Q10 in mice, coincident with the increased binding of H2-Q10 to liver CD8αβ T cells. Notably, ART2.2 is also developmentally regulated, with a dramatic increase in splenocytes expressing the enzyme between 3 and 7 weeks of age (39). Although the expression of ART2.2 was not examined in this study, it coincides with the timepoints in which we observed increased H2-Q10 binding. Future work aimed at defining the developmental regulation of ART2.2 in the liver will confirm these findings.
Collectively, in this study we identified soluble NAD+ in the liver mediating the binding of the MHC-Ib molecule H2-Q10 to CD8αβ T cells. Upon release from cells, NAD+ ribosylate CD8, which results in a reduction in MHC-Ia binding and an increase in H2-Q10 binding. Future work will be aimed at uncovering the functional consequence and developmental regulation of this interaction.
Experimental Procedures
Mice
C57BL/6, CD45.1, and CD45.2 mice were from Alfred Medical Research and Education Precinct (AMREP) Animal services. C57BL/6 CD8−/− mice were obtained from Professor Dan Littman (New York University School of Medicine). OT-I mice were provided by Nicole La Gruta (Monash University). All mice were used between 6 and 8 weeks of age. All experiments were in accordance with the animal ethics guidelines of the National Health and Medical Research Council of Australia. All experiments were approved by the AMREP Animal Ethics Committee.
Generation of tetramers
cDNA encoding the soluble regions of H2-Q10, H2-T3/TL, H2-Q9 and H2-Q10 and H-2Kb, H-2Db, and H-2Ld were generated by Genscript and cloned into a pUC57 vector. These were subcloned into a pET-30-based vector that allowed for an in-frame fusion of a substrate peptide for the biotin ligase enzyme BirA. The heavy chains of MHC-Ia and MHC-Ib proteins and mouse β2-microglobulin were expressed separately in E. coli, purified from inclusion bodies and refolded in the presence of either ribophorin (VGITNVDL), TCR Vβ chain (TGTETLYF), or proteosome peptide (HGTTTLAF) for H2-Q10. Q10 mutant tetramers were generated either point mutations (N187A or N194A), a double mutation (N187A, N194A), or a full α3 domain swap to that of H-2Kb (H2-Q10α3). H2-Q10 monomers were purified by anion exchange and gel filtration chromatography prior to biotinylation with BirA, as previously described (21). Monomers were conjugated into tetramers by the addition of streptavidin-PE or -APC. Additional monomers were folded with LemA (fMIGWII) for H2-M3, VP2.139 (HALNVVHDW) for H2-Q9, motif peptide (AGPARAAAL) for H-2Dd, and OVA (SIINFEKL), (SIIQFEKL), (SIIGFEKL), or (SIINFEKL) for H-2Kb.
Generation of CHO ART2.2 CD8αα/CD8αβ cells lines
The full-length sequences of ART2.2, CD8α, or CD8β were cloned separately into MSCV plasmids, and retroviral supernatants containing either ART2.2 and CD8α or ART2.2, CD8α, and CD8β were generated using 293T cells and standard calcium phosphate transfection protocols. Cells were transfected with plasmid DNA and pCL-Ampho retrovirus packaging vector, harvested at 48 or 72 h posttransfection, and retroviral supernatants were used to transduce CHO cells on retronectin (TaKaRa Bio) precoated plates (BD Bioscience) (30). CHO cells infected with retrovirus were sorted on the basis of GFP, mCherry, or cell surface protein expression. Populations were sorted by flow cytometry (FACS) until stable via surface expression of the recombinant receptors.
Real-time PCR (RT-PCR)
Total RNA was isolated from organs either using the RNEasy kit (Qiagen), or TRI Reagent RT (Merck) and complementary DNA (cDNA) synthesized from 1 μg total RNA using an iScript cDNA Synthesis Kit (Bio-Rad). Complementary DNA was amplified using QuantiNova SYBR Green mix (Qiagen) and the Quantstudio 6 qPCR (Applied Biosystems) with a two-step protocol including an annealing/extension temperature of 60 °C for all primer pairs. Primer sequences are as follows: H2-Q10 Fwd 3′ – 5′ GCAGAGTTTCCATGTGAGCCT, Rev 3′ – 5′ GGACCCCACTTTACAGCCATAC, GAPDH Fwd 3′ – 5′ TGGCCTTCCGTGTTCCTAC, Rev 3′ – 5′ GAGTTGCTGTTGAAGTCGCA.
Sorting of liver populations into hepatocyte and nonhepatocyte populations
Livers were perfused with ice-cold PBS and dissected whole. They were subsequently dissociated in the presence of 100 U mL−1 collagenase at 37 °C for 30 min. Single-cell suspensions were then filtered through 100 μM sieve and centrifuged twice using ice-cold PBS. Hepatocytes and nonhepatocytes were subsequently sorted on the basis of size and complexity (FSChi v SSChi for hepatocytes and FSClo v SSClo/hi for nonhepatocytes).
Flow cytometry
Flow cytometric analysis of organs
Organs were collected from WT, CD45.1, CD45.2, OT-I, or CD8β-deficient mice and single-cell suspensions prepared using standard protocols. Following removal of red blood cells using ACK, nonspecific receptors were blocked with monoclonal antibody (mAb) 2.4G2, before cells (1–5 × 106) were stained with mAb to mouse NKp46 (29A1.4; BioLegend), CD3 (17A2; BD Biosciences), TCRγδ (GL3, BioLegend), TCRαβ (H57-597, BioLegend), CD8α (53-6.7; BD Biosciences), CD8β (H35-17.2; BD Biosciences). Alternatively cells were stained with the lectins peanut agglutinin (Vector Laboratories), sambucus nigra lectin (Vector Laboratories), or maackia amurensis lectin II (Vector Laboratories) before detecting using Streptavidin (BD Biosciences). Cells stained with tetramers were fixed in 2% paraformaldehyde for 15 min and washed twice with FACS buffer (1% FCS/PBS) before being resuspended in FACS buffer. All other FACS combinations were acquired unfixed. For acquisition, events were electronically gated on FSC-A versus FSC-H (singlets), followed by FSC-A and SSC-A (to exclude doublets and debris). Among the remaining population, at least 5000 electronic events of interest were collected using an LSR-II or X- 20 Fortessa (BD Biosciences).
Flow cytometric analysis of CHO ART2.2 CD8αα/αβ cells
CHO ART2.2-CD8αα and CHO ART2.2-CD8αβ cells were routinely cultured and liberated from the tissue culture flask with the use of TrypLE Express (Thermo Fisher Scientific). The cells were stained with the antibodies CD8α or CD8β. Cells stained with tetramers were fixed in 2% paraformaldehyde and washed twice with FACS buffer (1% FCS/PBS) before being resuspended in FACS buffer. All other FACS combinations were acquired unfixed. For acquisition, events were electronically gated on FSC-A versus FSC-H (singlets), followed by FSC-A and SSC-A (to exclude doublets and debris). Among the remaining population at least 10,000 electronic events of interest were collected using an LSR-II or X- 20 Fortessa (BD Biosciences).
NAD+ and neuraminidase treatment of cells
Single cell suspensions from organs or CHO ART2.2 cell lines were treated with 0 μM up to 20 μM β-nicotinamide adenine dinucleotide sodium salt (NAD; Merck) for 30 min at 37 °C in the presence of 5% CO2. Alternatively, cells were treated with 5ug/ml neuraminidase (Vibrio cholera) for 20 min (Sigma Aldrich). After incubation, cells were washed with PBS once before staining with the relevant antibodies or tetramers. Cells stained with tetramers were fixed in 2% paraformaldehyde and washed twice with FACS buffer (1% FCS/PBS) before being resuspended in FACS buffer. All other FACS combinations were acquired unfixed.
Data availability
All data are contained within the manuscript.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank the members of the Alfred Medical Research and Education Precinct (AMREP) Flow Cytometry unit for expert assistance. We also thank the Monash Animal Research Platform (MARP) and AMREP Animal Services (AAS) for husbandry.
Author contributions
K. J. G., D. M. A., and L. C. S., conceptualization; K. J. G., A. N., and D. M. A., data curation; K. J. G. and D. M. A., formal analysis; D. M. A. and L. C. S., funding acquisition; K. J. G., A. N., and D. M. A., investigation; K. J. G., A. N., and D. M. A., methodology; K. J. G. and D. M. A., project administration; D. M. A. and L. C. S., resources; D. M. A., supervision; K. J. G., A. N., and D. M. A., validation; K. J. G. and D. M. A., visualization; K. J. G., writing–original draft; K. J. G., D. M. A., and L. C. S., writing–review and editing.
Funding and additional information
This work was supported by an NHMRC project grant (APP1141950) for D. M. A. and L. C. S. and a Career Development Fellowship (CDF) for D. M. A. K. J. G. was supported by a Monash University Post-Doctoral Award.
Edited by Peter Cresswell
Footnotes
Present address for Katharine Jennifer Goodall: oNKo-innate Pty Ltd, Monash Biomedicine Discovery Institute, Clayton, Australia.
Present address for Angela Nguyen: Biomedicine Discovery Institute, Department of Biochemistry and Molecular Biology, Monash University, Clayton, Australia.
Present address for Daniel Mark Andrews: Bioproperties Pty Ltd, RMIT University Bundoora Campus, Bundoora, Victoria, Australia.
Present address for Lucy Catherine Sullivan: South Australian Transplantation and Immunogenetics Service, Australian Red Cross Lifeblood, North Adelaide, South Australia, Australia.
References
- 1.Cheroutre H., Lambolez F. Doubting the TCR coreceptor function of CD8alphaalpha. Immunity. 2008;28:149–159. doi: 10.1016/j.immuni.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 2.Zloza A., Sullivan Y.B., Connick E., Landay A.L., Al-Harthi L. CD8+ T cells that express CD4 on their surface (CD4dimCD8bright T cells) recognize an antigen-specific target, are detected in vivo, and can be productively infected by T-tropic HIV. Blood. 2003;102:2156–2164. doi: 10.1182/blood-2002-07-1972. [DOI] [PubMed] [Google Scholar]
- 3.Wei Q., Brzostek J., Sankaran S., Casas J., Hew L.S., Yap J., Zhao X., Wojciech L., Gascoigne N.R.J. Lck bound to coreceptor is less active than free Lck. Proc. Natl. Acad. Sci. U. S. A. 2020;117:15809–15817. doi: 10.1073/pnas.1913334117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daniels M.A., Devine L., Miller J.D., Moser J.M., Lukacher A.E., Altman J.D., Kavathas P., Hogquist K.A., Jameson S.C. CD8 binding to MHC class I molecules is influenced by T cell maturation and glycosylation. Immunity. 2001;15:1051–1061. doi: 10.1016/s1074-7613(01)00252-7. [DOI] [PubMed] [Google Scholar]
- 5.Kao C., Sandau M.M., Daniels M.A., Jameson S.C. The sialyltransferase ST3Gal-I is not required for regulation of CD8-class I MHC binding during T cell development. J. Immunol. 2006;176:7421–7430. doi: 10.4049/jimmunol.176.12.7421. [DOI] [PubMed] [Google Scholar]
- 6.Comelli E.M., Sutton-Smith M., Yan Q., Amado M., Panico M., Gilmartin T., Whisenant T., Lanigan C.M., Head S.R., Goldberg D., Morris H.R., Dell A., Paulson J.C. Activation of murine CD4+ and CD8+ T lymphocytes leads to dramatic remodeling of N-linked glycans. J. Immunol. 2006;177:2431–2440. doi: 10.4049/jimmunol.177.4.2431. [DOI] [PubMed] [Google Scholar]
- 7.Hao X., Li Y., Wang J., Ma J., Zhao S., Ye X., He L., Yang J., Gao M., Xiao F., Wei H. Deficient O-GlcNAc glycosylation impairs regulatory T cell differentiation and notch signaling in autoimmune hepatitis. Front. Immunol. 2018;9:2089. doi: 10.3389/fimmu.2018.02089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haag F., Adriouch S., Brass A., Jung C., Moller S., Scheuplein F., Bannas P., Seman M., Koch-Nolte F. Extracellular NAD and ATP: Partners in immune cell modulation. Purinergic Signal. 2007;3:71–81. doi: 10.1007/s11302-006-9038-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruzzone S., Guida L., Zocchi E., Franco L., De Flora A. Connexin 43 hemi channels mediate Ca2+-regulated transmembrane NAD+ fluxes in intact cells. FASEB J. 2001;15:10–12. doi: 10.1096/fj.00-0566fje. [DOI] [PubMed] [Google Scholar]
- 10.Adriouch S., Hubert S., Pechberty S., Koch-Nolte F., Haag F., Seman M. NAD+ released during inflammation participates in T cell homeostasis by inducing ART2-mediated death of naive T cells in vivo. J. Immunol. 2007;179:186–194. doi: 10.4049/jimmunol.179.1.186. [DOI] [PubMed] [Google Scholar]
- 11.Scheuplein F., Schwarz N., Adriouch S., Krebs C., Bannas P., Rissiek B., Seman M., Haag F., Koch-Nolte F. NAD+ and ATP released from injured cells induce P2X7-dependent shedding of CD62L and externalization of phosphatidylserine by murine T cells. J. Immunol. 2009;182:2898–2908. doi: 10.4049/jimmunol.0801711. [DOI] [PubMed] [Google Scholar]
- 12.Wang J., Nemoto E., Kots A.Y., Kaslow H.R., Dennert G. Regulation of cytotoxic T cells by ecto-nicotinamide adenine dinucleotide (NAD) correlates with cell surface GPI-anchored/arginine ADP-ribosyltransferase. J. Immunol. 1994;153:4048–4058. [PubMed] [Google Scholar]
- 13.Okamoto S., Azhipa O., Yu Y., Russo E., Dennert G. Expression of ADP-ribosyltransferase on normal T lymphocytes and effects of nicotinamide adenine dinucleotide on their function. J. Immunol. 1998;160:4190–4198. [PubMed] [Google Scholar]
- 14.Wang J., Nemoto E., Dennert G. Regulation of CTL by ecto-nictinamide adenine dinucleotide (NAD) involves ADP-ribosylation of a p56lck-associated protein. J. Immunol. 1996;156:2819–2827. [PubMed] [Google Scholar]
- 15.Lischke T., Schumacher V., Wesolowski J., Hurwitz R., Haag F., Koch-Nolte F., Mittrucker H.W. CD8-beta ADP-ribosylation affects CD8(+) T-cell function. Eur. J. Immunol. 2013;43:1828–1838. doi: 10.1002/eji.201243231. [DOI] [PubMed] [Google Scholar]
- 16.Kahl S., Nissen M., Girisch R., Duffy T., Leiter E.H., Haag F., Koch-Nolte F. Metalloprotease-mediated shedding of enzymatically active mouse ecto-ADP-ribosyltransferase ART2.2 upon T cell activation. J. Immunol. 2000;165:4463–4469. doi: 10.4049/jimmunol.165.8.4463. [DOI] [PubMed] [Google Scholar]
- 17.Martello R., Leutert M., Jungmichel S., Bilan V., Larsen S.C., Young C., Hottiger M.O., Nielsen M.L. Proteome-wide identification of the endogenous ADP-ribosylome of mammalian cells and tissue. Nat. Commun. 2016;7:12917. doi: 10.1038/ncomms12917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rissiek B., Lukowiak M., Raczkowski F., Magnus T., Mittrucker H.W., Koch-Nolte F. In vivo blockade of murine ARTC2.2 during cell preparation preserves the vitality and function of liver tissue-resident memory T cells. Front. Immunol. 2018;9:1580. doi: 10.3389/fimmu.2018.01580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodall K.J., Nguyen A., Matsumoto A., McMullen J.R., Eckle S.B., Bertolino P., Sullivan L.C., Andrews D.M. Multiple receptors converge on H2-Q10 to regulate NK and gammadeltaT-cell development. Immunol. Cell Biol. 2019;97:326–339. doi: 10.1111/imcb.12222. [DOI] [PubMed] [Google Scholar]
- 20.Cosman D., Kress M., Khoury G., Jay G. Tissue-specific expression of an unusual H-2 (class I)-related gene. Proc. Natl. Acad. Sci. U. S. A. 1982;79:4947–4951. doi: 10.1073/pnas.79.16.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sullivan L.C., Berry R., Sosnin N., Widjaja J.M., Deuss F.A., Balaji G.R., LaGruta N.L., Mirams M., Trapani J.A., Rossjohn J., Brooks A.G., Andrews D.M. Recognition of the major histocompatibility complex (MHC) class Ib molecule H2-Q10 by the natural killer cell receptor Ly49C. J. Biol. Chem. 2016;291:18740–18752. doi: 10.1074/jbc.M116.737130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lew A.M., Maloy W.L., Coligan J.E. Characteristics of the expression of the murine soluble class I molecule (Q10) J. Immunol. 1986;136:254–258. [PubMed] [Google Scholar]
- 23.Melo-Lima B.L., Evangelista A.F., de Magalhaes D.A., Passos G.A., Moreau P., Donadi E.A. Differential transcript profiles of MHC class Ib(Qa-1, Qa-2, and Qa-10) and Aire genes during the ontogeny of thymus and other tissues. J. Immunol. Res. 2014;2014:159247. doi: 10.1155/2014/159247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takahashi K., Nakata M., Tanaka T., Adachi H., Nakauchi H., Yagita H., Okumura K. CD4 and CD8 regulate interleukin 2 responses of T cells. Proc. Natl. Acad. Sci. U. S. A. 1992;89:5557–5561. doi: 10.1073/pnas.89.12.5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson P. A human homolog of the mouse CD8 molecule, Lyt-3: Genomic sequence and expression. Immunogenetics. 1987;26:174–177. doi: 10.1007/BF00365908. [DOI] [PubMed] [Google Scholar]
- 26.Laing S., Unger M., Koch-Nolte F., Haag F. ADP-ribosylation of arginine. Amino Acids. 2011;41:257–269. doi: 10.1007/s00726-010-0676-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berg R.E., Princiotta M.F., Irion S., Moticka J.A., Dahl K.R., Staerz U.D. Positive selection of an H2-M3 restricted T cell receptor. Immunity. 1999;11:33–43. doi: 10.1016/s1074-7613(00)80079-5. [DOI] [PubMed] [Google Scholar]
- 28.Sullivan B.A., Reed-Loisel L.M., Kersh G.J., Jensen P.E. Homeostatic proliferation of a Qa-1b-restricted T cell: A distinction between the ligands required for positive selection and for proliferation in lymphopenic hosts. J. Immunol. 2004;173:6065–6071. doi: 10.4049/jimmunol.173.10.6065. [DOI] [PubMed] [Google Scholar]
- 29.Andrews D.M., Sullivan L.C., Baschuk N., Chan C.J., Berry R., Cotterell C.L., Lin J., Halse H., Watt S.V., Poursine-Laurent J., Wang C.R., Scalzo A.A., Yokoyama W.M., Rossjohn J., Brooks A.G. Recognition of the nonclassical MHC class I molecule H2-M3 by the receptor Ly49A regulates the licensing and activation of NK cells. Nat. Immunol. 2012;13:1171–1177. doi: 10.1038/ni.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goodall K.J., Nguyen A., McKenzie C., Eckle S.B.G., Sullivan L.C., Andrews D.M. The murine CD94/NKG2 ligand, Qa-1(b), is a high-affinity, functional ligand for the CD8alphaalpha homodimer. J. Biol. Chem. 2020;295:3239–3246. doi: 10.1074/jbc.RA119.010509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y., Cuendet M.A., Goffin L., Sachl R., Cebecauer M., Cariolato L., Guillaume P., Reichenbach P., Irving M., Coukos G., Luescher I.F. CD8 binding of MHC-peptide complexes in cis or trans regulates CD8(+) T-cell responses. J. Mol. Biol. 2019;431:4941–4958. doi: 10.1016/j.jmb.2019.10.019. [DOI] [PubMed] [Google Scholar]
- 32.Wang R., Natarajan K., Margulies D.H. Structural basis of the CD8 alpha beta/MHC class I interaction: Focused recognition orients CD8 beta to a T cell proximal position. J. Immunol. 2009;183:2554–2564. doi: 10.4049/jimmunol.0901276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.La Gruta N.L., Gras S., Daley S.R., Thomas P.G., Rossjohn J. Understanding the drivers of MHC restriction of T cell receptors. Nat. Rev. Immunol. 2018;18:467–478. doi: 10.1038/s41577-018-0007-5. [DOI] [PubMed] [Google Scholar]
- 34.Fahl S.P., Coffey F., Wiest D.L. Origins of gammadelta T cell effector subsets: A riddle wrapped in an enigma. J. Immunol. 2014;193:4289–4294. doi: 10.4049/jimmunol.1401813. [DOI] [PubMed] [Google Scholar]
- 35.Sato K., Ohtsuka K., Watanabe H., Asakura H., Abo T. Detailed characterization of gamma delta T cells within the organs in mice: Classification into three groups. Immunology. 1993;80:380–387. [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang Y., Que W., Zhu P., Li X.K. The role of diverse liver cells in liver transplantation tolerance. Front. Immunol. 2020;11:1203. doi: 10.3389/fimmu.2020.01203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Irie H.Y., Mong M.S., Itano A., Crooks M.E., Littman D.R., Burakoff S.J., Robey E. The cytoplasmic domain of CD8 beta regulates Lck kinase activation and CD8 T cell development. J. Immunol. 1998;161:183–191. [PubMed] [Google Scholar]
- 38.Horkova V., Drobek A., Mueller D., Gubser C., Niederlova V., Wyss L., King C.G., Zehn D., Stepanek O. Dynamics of the coreceptor-LCK interactions during T cell development shape the self-reactivity of peripheral CD4 and CD8 T cells. Cell Rep. 2020;30:1504–15014.e7. doi: 10.1016/j.celrep.2020.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koch-Nolte F., Duffy T., Nissen M., Kahl S., Killeen N., Ablamunits V., Haag F., Leiter E. A new monoclonal antibody detects a developmentally regulated mouse ecto-ADP-ribosyltransferase on T cells: Subset distribution, inbred strain variation, and modulation upon T cell activation. J. Immunol. 1999;163:6014–6022. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are contained within the manuscript.