Key Points
Question
Are clonal hematopoiesis of indeterminate potential (CHIP) variants detected in peripheral blood cells (PBCs) before treatment with rucaparib associated with the risk of therapy-related myeloid neoplasms (t-MNs) after rucaparib treatment in patients with high-grade ovarian cancer?
Findings
In this genetic association study, the prevalence of pretreatment TP53-variant CHIP with a variant allele frequency of 1% or higher in PBCs was significantly higher among patients with high-grade ovarian cancer who developed t-MNs after rucaparib therapy vs those who did not and was associated with longer prior exposure to platinum therapy.
Meaning
The results of this study suggest that pretreatment TP53-variant CHIP detected at a variant allele frequency of 1% or higher in PBCs may be associated with development of t-MNs after rucaparib treatment.
Abstract
Importance
A total of 1% to 3% of patients treated with a poly(adenosine diphosphate–ribose) polymerase inhibitor for high-grade ovarian cancer (HGOC) develop therapy-related myeloid neoplasms (t-MNs), which are rare but often fatal conditions. Although the cause of these t-MNs is unknown, clonal hematopoiesis of indeterminate potential (CHIP) variants can increase the risk of primary myeloid malignant neoplasms and are more frequent among patients with solid tumors.
Objectives
To examine whether preexisting CHIP variants are associated with the development of t-MNs after rucaparib treatment and how these CHIP variants are affected by treatment.
Design, Setting, and Participants
This retrospective genetic association study used peripheral blood cell (PBC) samples collected before rucaparib treatment from patients in the multicenter, single-arm ARIEL2 (Study of Rucaparib in Patients With Platinum-Sensitive, Relapsed, High-Grade Epithelial Ovarian, Fallopian Tube, or Primary Peritoneal Cancer) (n = 491; between October 30, 2013, and August 9, 2016) and the multicenter, placebo-controlled, double-blind ARIEL3 (Study of Rucaparib as Switch Maintenance Following Platinum-Based Chemotherapy in Patients With Platinum-Sensitive, High-Grade Serous or Endometrioid Epithelial Ovarian, Primary Peritoneal or Fallopian Tube Cancer) (n = 561; between April 7, 2014, and July 19, 2016), which tested rucaparib as HGOC therapy in the treatment and maintenance settings, respectively. The follow-up data cutoff date was September 1, 2019. Of 1052 patients in ARIEL2 and ARIEL3, PBC samples from 20 patients who developed t-MNs (cases) and 44 randomly selected patients who did not (controls) were analyzed for the presence of CHIP variants using targeted next-generation sequencing. Additional longitudinal analysis was performed on available ARIEL2 samples collected during treatment and at the end of treatment.
Main Outcomes and Measures
Enrichment analysis of preexisting variants in 10 predefined CHIP-associated genes in cases relative to controls; association with clinical correlates.
Results
Among 1052 patients (mean [SE] age, 61.7 [0.3] years) enrolled and dosed in ARIEL2 and ARIEL3, 22 (2.1%) developed t-MNs. The t-MNs were associated with longer overall exposure to prior platinum therapies (13.2 vs 9.0 months in ARIEL2, P = .04; 12.4 vs 9.6 months in ARIEL3, P = .003). The presence of homologous recombination repair gene variants in the tumor, either germline or somatic, was associated with increased prevalence of t-MNs (15 [4.1%] of 369 patients with HGOC associated with an HRR gene variant vs 7 [1.0%] of 683 patients with wild-type HGOC, P = .002). The prevalence of preexisting CHIP variants in TP53 but not other CHIP-associated genes at a variant allele frequency of 1% or greater was significantly higher in PBCs from cases vs controls (9 [45.0%] of 20 cases vs 6 [13.6%] of 44 controls, P = .009). TP53 CHIP was associated with longer prior exposure to platinum (mean 14.0 months of 15 TP53 CHIP cases vs 11.1 months of 49 non-TP53 CHIP cases; P = .02). Longitudinal analysis showed that preexisting TP53 CHIP variants expanded in patients who developed t-MNs.
Conclusions and Relevance
The findings of this genetic association study suggest that preexisting TP53 CHIP variants may be associated with t-MNs after rucaparib treatment.
This genetic association study examines whether patients who harbor specific clonal hematopoiesis of indeterminate potential variants before rucaparib treatment may be at an increased risk of developing therapy-related myeloid neoplasms.
Introduction
Therapy-related secondary myeloid neoplasms (t-MNs), including myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML), are severe, often fatal diseases that occur in a small fraction of patients with solid tumors, including approximately 0.3% of ovarian cancer survivors, who have received cytotoxic treatment.1,2,3,4,5 Therapy-related secondary myeloid neoplasms account for 10% to 20% of MDS and AML cases and are characterized by complex karyotypes and frequent TP53 (OMIM 191170) alterations.1,6,7 Resistance to standard therapies and poor outcomes are common, with 5-year survival rates of less than 10%.1,2
Clonal hematopoiesis of indeterminate potential (CHIP) is conventionally defined as somatic abnormalities in genes associated with myeloid malignant neoplasms at a variant allele frequency (VAF) of 2% or greater detected in the peripheral blood cells (PBCs) of healthy individuals.8,9,10,11 With increasingly sensitive targeted and whole-genome and exome sequencing technologies, the definition of CHIP is evolving to encompass lower-VAF events.12 Common variants in CHIP include epigenetic modulators (DNMT3A [OMIM 602769], TET2 [OMIM 612839], and ASXL1 [OMIM 612990]), DNA damage repair factors (TP53 and PPM1D [OMIM 605100]), splicing factors (SF3B1 [OMIM 605590] and SRSF2 [OMIM 600813]), and others.8,9,10 CHIP prevalence increases with age, correlates with exposure to mutagenic substances (such as smoking),10,11,13 and is a known risk factor for the development of primary myeloid malignant neoplasms.10,11
The prevalence of CHIP in patients with solid tumors is higher than in the general population.13,14,15 CHIP variants in DNA damage repair genes, including TP53, specifically provide fitness advantage and preferentially expand during platinum treatment, which is commonly used in ovarian cancer.16 The presence of CHIP in patients with solid tumors has been linked to increased risk of subsequently developing t-MNs.14,16,17
Rucaparib is a small-molecule poly(adenosine diphosphate–ribose) polymerase inhibitor (PARPi) approved for the treatment of advanced high-grade ovarian cancer (HGOC) and castration-resistant prostate cancer.18 Therapy-related secondary myeloid neoplasms have been observed in PARPi HGOC trials, with a prevalence of 1% to 3%.19,20,21,22 A recent meta-analysis22 of multiple trials suggests an increased risk of secondary myeloid neoplasms in PARPi-treated patients. A higher number of prior courses of chemotherapy was also linked to increased risk of developing t-MNs during or after PARPi treatment, suggesting that cumulative prior chemotherapy exposure may be another relevant risk factor.21 Beyond this, little is known about what factors contribute to t-MNs in PARPi-treated patients. We hypothesized that patients who harbor specific CHIP variants before rucaparib treatment may be at an increased risk of developing t-MNs. To address this hypothesis, we performed a retrospective genetic association study that analyzed PBC samples collected from patients enrolled in the ARIEL2 (Study of Rucaparib in Patients With Platinum-Sensitive, Relapsed, High-Grade Epithelial Ovarian, Fallopian Tube, or Primary Peritoneal Cancer) and ARIEL3 (Study of Rucaparib as Switch Maintenance Following Platinum-Based Chemotherapy in Patients With Platinum-Sensitive, High-Grade Serous or Endometrioid Epithelial Ovarian, Primary Peritoneal or Fallopian Tube Cancer) clinical trials that evaluated rucaparib as an active and maintenance treatment for HGOC, respectively.
Methods
Participants
In this retrospective genetic association study, t-MN cases and controls were identified within ARIEL2, a 2-part, single-arm, open-label, phase 2 study of rucaparib in the treatment of relapsed HGOC23 that enrolled 491 patients between October 30, 2013, and August 9, 2016, who received treatment and ARIEL3, a phase 3, double-blind, placebo-controlled study of rucaparib for the maintenance treatment of platinum-responsive HGOC24 that enrolled 561 patients between April 7, 2014, and July 19, 2016, who received treatment (eMethods in the Supplement). The follow-up data cutoff date for the current analysis was September 1, 2019. The trials were approved by national or local institutional review boards and carried out in accordance with the Declaration of Helsinki and Good Clinical Practice Guidelines of the International Council on Harmonisation. Patients provided written informed consent before participation, which included consent to use their samples for translational research. The study followed the Strengthening the Reporting of Genetic Association Studies (STREGA) reporting guideline.
t-MN Reporting Process
Clinical sites that participated in ARIEL2 and ARIEL3 were instructed to report t-MN cases to Clovis Oncology regardless of causality as serious adverse events or adverse events of special interest within 24 hours of becoming aware of events (eMethods in the Supplement).
Samples and Molecular Analysis
Preservation of PBC samples and DNA extraction details are provided in the eMethods in the Supplement. DNA was sequenced using the FoundationOne Heme targeted next-generation sequencing (NGS) assay (Foundation Medicine). Matched tumor tissues were sequenced using the FoundationOne CDx assay (Foundation Medicine). Cell-free DNA (cfDNA) plasma sequencing was performed using the FoundationOne Liquid CDx (Foundation Medicine) or Guardant360 (Guardian Health) panels.
CHIP Definition
CHIP variants were predefined as loss of function somatic alterations (VAF ≤40%) in DNMT3A, TET2, ASXL1, TP53, JAK2 (OMIM 147796), SF3B1, GNB1 (OMIM 139380), CBL (OMIM 165360), SRSF1 (OMIM 600812), and GNAS (OMIM 139320).10 Other alterations were reported separately. The limit of detection of the assay was 1%.
Statistical Analysis
Expected prevalence of CHIP was estimated to be 55% in cases and 20% in controls based on prior publications.13,14,17 Given that 20 of 22 ARIEL2 and ARIEL3 t-MN cases had PBC samples available, we estimated that 45 controls (control-case ratio of approximately 2:1) would be sufficient to detect this difference with approximately 80% power and an α error rate of .025 (1-sided, greater in cases). In all, 17 ARIEL2 and 28 ARIEL3 controls were randomly selected, and 44 were successfully sequenced. All comparisons presented are exploratory because they were not prespecified; multiple comparison adjustments were not performed. Prism software, version 9 (GraphPad) was used for statistical tests. Tests were 1-sided or 2-sided as indicated; P < .05 was considered to be statistically significant.
Results
t-MN Prevalence in the ARIEL2 and ARIEL3 Clinical Trials
Among 1052 patients (mean [SE] age, 61.7 [0.3] years) enrolled in ARIEL2 and ARIEL3, 22 (2.1%) developed t-MNs. In ARIEL2, 6 of 491 patients (1.2%) developed t-MNs. In ARIEL3, 16 of 561 patients (2.9%) developed t-MNs: 13 of 372 (3.5%) in the rucaparib arm and 3 of 189 (1.6%) in the placebo arm (P = .28) (Table 1). The time from start of treatment to t-MN diagnosis was similar in the rucaparib and placebo arms (eFigure 1 in the Supplement). Patients in the rucaparib arm of ARIEL3 who developed t-MNs received treatment longer than patients in the same arm who did not develop secondary malignant neoplasms (median, 25.0 vs 8.0 months; P = .003) (Table 1); the finding was similar in ARIEL2 but did not reach statistical significance (median, 14.7 vs 4.4 months; P = .16) (Table 1). A total of 19 of the 22 patients who developed t-MNs received rucaparib while in the trial (3 were in the placebo arm of ARIEL3); of those, 7 had treatment-emergent t-MNs (diagnosed within 28 days of last dose). Of the remaining 12 long-term follow-up cases, 8 were diagnosed without any additional anticancer therapy exposure, whereas 4 received additional treatment after rucaparib, including PARPi- and platinum-based therapies (eTable 1 in the Supplement). The ARIEL2 and ARIEL3 t-MNs were characterized by complex karyotypes with multiple chromosomal abnormalities, typical of t-MNs1,6 (eTable 2 in the Supplement).
Table 1. ARIEL2 and ARIEL3 Patient Characteristics in Patients With and Without t-MNsa.
| Characteristic | ARIEL2 | ARIEL3 | ||||
|---|---|---|---|---|---|---|
| Participants, No. (%) | P valueb | Participants, No. (%) | P valueb | |||
| t-MN (n = 6) | No t-MN (n = 485) | t-MN (n = 16) | No t-MN (n = 545) | |||
| Age at start of trial, median (range), y | 70.5 (58-76) | 63 (31-91) | .09 | 58.5 (45-83) | 61 (36-85) | .90 |
| Smoking status | ||||||
| Ever | 0 | 152 (31.3) | .24 | 6 (37.5) | 183 (33.6) | .91 |
| Never | 6 (100) | 327 (67.4) | 10 (62.5) | 359 (65.9) | ||
| Missing | 0 | 6 (1.2) | 0 | 3 (0.6) | ||
| Histologic subtype | ||||||
| Serous | 6 (100) | 460 (94.8) | >.99 | 15 (93.8) | 519 (95.2) | .55 |
| Endometrioid, mixed, or other | 0 | 25 (5.2) | 1 (6.3) | 26 (4.8) | ||
| ECOG performance status | ||||||
| 0 | 2 (33.3) | 266 (54.8) | .42 | 10 (62.5) | 403 (73.9) | .39 |
| ≥1 | 4 (66.7) | 219 (45.2) | 6 (37.5) | 142 (26.1) | ||
| No. of prior courses of chemotherapyc | ||||||
| 1-2 | 0 | 173 (35.7) | .09 | 7 (43.8) | 346 (63.5) | .12 |
| ≥3 | 6 (100) | 312 (64.3) | 9 (56.3) | 199 (36.5) | ||
| No. of prior courses of platinum | ||||||
| 1-2 | 2 (33.3) | 317 (65.4) | .19 | 8 (50.0) | 352 (64.6) | .29 |
| ≥3 | 4 (66.7) | 168 (34.6) | 8 (50.0) | 193 (35.4) | ||
| Total prior time receiving platinum, median (range), mo | 13.2 (10.9-17.4) | 9.0 (1.4-43.7) | .04 | 12.4 (6.0-27.9) | 9.6 (4.8-31.8) | .003 |
| Treatment | ||||||
| Rucaparib | 6 (100) | 485 (100) | NA | 13 (81.3) | 359 (65.9) | .28 |
| Placebo | NA | NA | 3 (18.8) | 186 (34.1) | ||
| Time undergoing treatment, median (range), mo | ||||||
| Rucaparib | 14.7 (0.9-23.9) | 4.4 (0.1-57.6) | .16 | 25.0 (8.7-52.2) | 8.0 (0-67.4) | .003 |
| Placebo | NA | NA | NA | 10.6 (1.9-11.0) | 5.5 (0-67.9) | .82 |
Abbreviations: ARIEL2, Study of Rucaparib in Patients With Platinum-Sensitive, Relapsed, High-Grade Epithelial Ovarian, Fallopian Tube, or Primary Peritoneal Cancer; ARIEL3, Study of Rucaparib as Switch Maintenance Following Platinum-Based Chemotherapy in Patients With Platinum-Sensitive, High-Grade Serous or Endometrioid Epithelial Ovarian, Primary Peritoneal or Fallopian Tube Cancer; ECOG, Eastern Cooperative Oncology Group; NA, not available; t-MN, therapy-related myeloid neoplasm.
No multiple hypothesis testing correction was performed. Comparisons are exploratory in nature.
P values for age at start of treatment and time undergoing treatment are based on 2-sided t tests. P values for smoking status are based on the χ2 test. All others are based on the Fisher exact test.
All ARIEL3 patients had 2 or more prior courses of chemotherapy per enrollment requirement.
Next, we considered baseline characteristics that may be associated with an increased risk of t-MNs. No differences were found in age, smoking status, Eastern Cooperative Oncology Group performance status, or histologic findings between patients who developed t-MNs and those who did not (Table 1). A higher fraction of patients who developed t-MNs vs the unaffected population had 3 or more prior courses of chemotherapy in general (6 [100%] vs 312 [64.3%] [P = .09] in ARIEL2 and 9 [56.3%] vs 199 [36.5%] [P = .12] in ARIEL3) and 3 or more prior courses of platinum therapy specifically (4 [66.7%] vs 168 [34.6%] [P = .19] in ARIEL2 and 8 [50.0%] vs 193 [35.4%] [P = .29] in ARIEL3), but the findings were not statistically significant (Table 1). However, patients with t-MNs received prior platinum treatments for a significantly longer period (median, 13.2 vs 9.0 months in ARIEL2, P = .04; median, 12.4 vs 9.6 months in ARIEL3, P = .003), suggesting that longer prior exposure to platinum increases the risk of developing secondary malignant neoplasms after rucaparib.
We also explored whether there was an association between the presence of a homologous recombination repair (HRR) gene variant (BRCA1 [OMIM 113705], BRCA2 [OMIM 600185], RAD51C [OMIM 602774], or RAD51D [OMIM 602954]) in the tumor and the emergence of t-MNs. Indeed, the prevalence of t-MNs was higher in patients with HGOC that harbored a variant in 1 of these genes (15 of 369 [4.1%]) compared with those with wild-type HGOC (7 of 683 [1.0%], P = .002) (eTable 3 in the Supplement). This association, however, was not driven by inherited predisposition because t-MN rates were similar among patients with germline vs somatic alterations (3.8% vs 3.9%) (eTable 3 in the Supplement). Instead, we observed that patients with HGOC that contained variants in these 4 HRR genes were significantly more likely to have had 3 or more courses of prior chemotherapy (55.8% vs 46.9%, P = .007), to have had 3 or more courses of prior platinum therapy (42.5% vs 31.6%, P < .001), and to have received prior platinum treatment for longer (10.4 vs 9.0 months, P < .001) (eTable 4 in the Supplement), suggesting that more extensive exposure to prior platinum treatment contributed to their development of t-MNs.
Prevalence of Pretreatment CHIP in ARIEL2 and ARIEL3 Patients
Next, we hypothesized that CHIP prevalence at enrollment (pretreatment) may be higher in patients who developed t-MNs vs those who did not. We performed targeted NGS variant and copy number analysis on PBC samples from ARIEL2 and ARIEL3 patients who did (cases) or did not (controls) develop t-MNs. A total of 44 control (all before treatment) and 20 t-MN PBC samples (19 patients before treatment and 1 patient at day 1 of cycle 4 [83 days of treatment]) were successfully sequenced. Sequenced t-MN cases and controls were confirmed to be balanced on age, number of prior courses of chemotherapy, smoking status, and BRCA status (eTable 5 in the Supplement).
We identified variants in 4 of the 10 predefined CHIP-associated genes: TP53, DNMT3A, TET2, and ASXL1 (Table 2). All detected CHIP variants had a VAF of 1% or greater (the limit of detection of the assay). At least 1 CHIP event occurred in 9 of 20 patients (45.0%) who developed t-MNs compared with 11 of 44 patients (25.0%) who did not (odds ratio [OR], 2.5; 95% CI, 0.8-7.9; P = .10). Variants were detected in more than 1 gene in 4 of 20 cases (20.0%) compared with 2 of 44 controls (4.5%) (OR, 5.3; 95% CI, 1.1-28.8; P = .07) (Table 3). We ruled out an HGOC origin for all TP53 CHIP variants (eTable 6 in the Supplement); in all cases, the TP53 variant with the highest VAF in the HGOC was different from that in the PBCs. Finally, we compared the PBC findings with pretreatment cfDNA NGS data available for a subset of ARIEL2 patients (4 cases and 3 controls). All alterations detected in PBCs were also found in plasma, suggesting that CHIP variants can be incidental findings in cfDNA analysis (Table 2).
Table 2. All Variants in Predefined Clonal Hematopoiesis of Indeterminate Potential–Associated Genes Detected in Peripheral Blood Cells From Cases and Controls and Their Presence in Matched High-grade Ovarian Cancer (HGOC) and Cell-Free DNA Samples.
| Patient No. | Study | Treatment arm (ARIEL3 only) | Age, y | No. of prior courses of chemotherapy | No. of prior courses of platinum | Ever smoker | Time point | Variant detected (VAF, %)a | Variant detected in the HGOC (VAF in tumor, %) | Variant detected in plasma before treatment (VAF in plasma, %)b |
|---|---|---|---|---|---|---|---|---|---|---|
| t-MN cases | ||||||||||
| 1 | ARIEL2 | NA | 71 | 4 | 3 | No | During treatment (cycle 4 day 1) | DNMT3A p.R736H (1.3) | No | Not in panel |
| TP53 p.E56* (1.0) | No | Yes (0.4) | ||||||||
| TP53 p.R273C (2.1) | No | Yes (0.6) | ||||||||
| TP53 p.Y220C (1.8) | No | Yes (0.6) | ||||||||
| 2 | ARIEL2 | NA | 58 | 3 | 2 | No | Before treatment | TP53 p.K164E (1.3) | No | Yes (1.2) |
| TP53 p.S240fs*24 / c.716_717insT (9.4) | No | Yes (19.0) | ||||||||
| 3 | ARIEL2 | NA | 75 | 4 | 4 | No | Before treatment | TP53 p.M246K (4.5) | No | Yes (3.1) |
| 4 | ARIEL2 | NA | 76 | 3 | 3 | No | Before treatment | TET2 p.T229fs*25 / c.685_686insA (21.1) | No | Yes (23.6) |
| TP53 p.Y220C (13.0) | Yes (5) | Yes (14.5) | ||||||||
| DNMT3A splice site c.1429 + 1G>T (22.6) | Yes (6) | Yes (27.8) | ||||||||
| 7 | ARIEL3 | Rucaparib | 83 | 4 | 4 | Yes | Before treatment | DNMT3A p.R635W (1.2) | No | NA |
| TP53 p.I251S (1.4) | No | |||||||||
| TP53 p.R273C (1.2) | No | |||||||||
| 8 | ARIEL3 | Rucaparib | 63 | 2 | 2 | Yes | Before treatment | DNMT3A p.R882H (8.2) | No | NA |
| TP53 p.K132R (14.2) | No | |||||||||
| TP53 p.R337C (35.9) | No | |||||||||
| 9 | ARIEL3 | Placebo | 60 | 3 | 3 | Yes | Before treatment | TP53 p.N239D (1.5) | No | NA |
| 10 | ARIEL3 | Rucaparib | 57 | 5 | 4 | No | Before treatment | TP53 p.R273G (5.0) | No | NA |
| 11 | ARIEL3 | Rucaparib | 60 | 3 | 3 | No | Before treatment | TP53 p.V272M (2.0) | No | NA |
| Non–t-MN controls | ||||||||||
| 1 | ARIEL2 | NA | 51 | 3 | 3 | No | Before treatment | TP53 p.H178P (1.6) | NA | NA |
| 2 | ARIEL2 | NA | 41 | 3 | 2 | No | Before treatment | TP53 p.I251N (2.2) | No | Yes (0.6) |
| 3 | ARIEL2 | NA | 68 | 2 | 2 | Yes | Before treatment | TP53 p.V173M (2.7) | No | Yes (1.2) |
| TP53 p.V274D (16.9) | No | Yes (16.5) | ||||||||
| 4 | ARIEL2 | NA | 58 | 3 | 2 | No | Before treatment | DNMT3A p.P904L (1.7) | NA | NA |
| 5 | ARIEL2 | NA | 82 | 3 | 2 | No | Before treatment | DNMT3A p.R882C (6.4) | Yes (2) | Yes (3.4) |
| ASXL1 p.G642* (2.4) | No | Yes (0.6) | ||||||||
| 18 | ARIEL3 | Placebo | 59 | 3 | 3 | Yes | Before treatment | DNMT3A p.R749C (18.6) | No | NA |
| TP53 p.R175H (5.7) | No | |||||||||
| TP53 p.R273H (6.3) | No | |||||||||
| TP53 p.Y220C (16.4) | No | |||||||||
| 19 | ARIEL3 | Placebo | 63 | 5 | 4 | Yes | Before treatment | TP53 p.R175H (40.7) | No | NA |
| 20 | ARIEL3 | Placebo | 63 | 2 | 2 | No | Before treatment | TP53 p.R248Q (1.4) | No | NA |
| 21 | ARIEL3 | Rucaparib | 58 | 2 | 2 | Yes | Before treatment | DNMT3A p.G543C (3) | No | NA |
| 22 | ARIEL3 | Rucaparib | 72 | 3 | 3 | Yes | Before treatment | DNMT3A splice site c.1015-2A>C (6.7) | No | NA |
| 23 | ARIEL3 | Rucaparib | 58 | 2 | 2 | Yes | Before treatment | TET2 p.Q644* (2.8) | Yes (2) | NA |
Abbreviations: ARIEL2, Study of Rucaparib in Patients With Platinum-Sensitive, Relapsed, High-Grade Epithelial Ovarian, Fallopian Tube, or Primary Peritoneal Cancer; ARIEL3, Study of Rucaparib as Switch Maintenance Following Platinum-Based Chemotherapy in Patients With Platinum-Sensitive, High-Grade Serous or Endometrioid Epithelial Ovarian, Primary Peritoneal or Fallopian Tube Cancer; NA, not available; t-MN, therapy-related myeloid neoplasm; VAF, variant allele frequency.
Limit of detection of assay was 1%.
All plasma data are based on FoundationOne Liquid CDx, except case 1, which is based on the Guardant360 test.
Table 3. Number of Participants With Variants in Predefined CHIP-Associated Genes Detected Before Treatment or in Early Treatment in t-MN Cases and Controlsa.
| CHIP-associated gene | Participants, No. (%) | OR (95% CI) | P value | |
|---|---|---|---|---|
| t-MN (n = 20) | Controls (n = 44) | |||
| TP53 | 9 (45.0) | 6 (13.6) | 5.2 (1.6-16.0) | .009 |
| DNMT3A | 4 (20.0) | 5 (11.4) | 2.0 (0.5-7.9) | .29 |
| TET2 | 1 (5.0) | 1 (2.3) | 2.3 (0.1-43.8) | .53 |
| ASXL1 | 0 | 1 (2.3) | NA | .69 |
| At least 1 CHIP event | 9 (45.0) | 11 (25.0) | 2.5 (0.8-7.9) | .10 |
| CHIP in multiple genes | 4 (20.0) | 2 (4.5) | 5.3 (1.1-28.8) | .07 |
Abbreviations: CHIP, clonal hematopoiesis of indeterminate potential; NA, not available; OR, odds ratio; t-MN, therapy-related myeloid neoplasm.
CHIP prevalence is based on variants with a variant allele frequency of 1% or greater, which was the limit of detection of the assay. P values are based on a 1-sided Fisher exact test. No multiple hypothesis testing correction was performed. Comparisons are exploratory in nature.
Association of Pretreatment TP53 CHIP With Increased Risk of Developing t-MNs
We observed no differences in the prevalence of DNMT3A, TET2, and ASXL1 CHIP between cases and controls (Table 3). In contrast, deleterious alterations in TP53, the most common CHIP-associated variant in this cohort, were significantly more frequent in the t-MN cases (9 of 20 [45.0%]) than in controls (6 of 44 [13.6%]; OR, 5.2; 95% CI, 1.6-16.0; P = .009) (Table 3). TP53 CHIP variant VAFs were not significantly different between cases and controls (eFigure 2A in the Supplement). Moreover, TP53 variant VAFs at pretreatment did not affect the time from start of treatment to t-MN diagnosis (eFigure 2B in the Supplement).
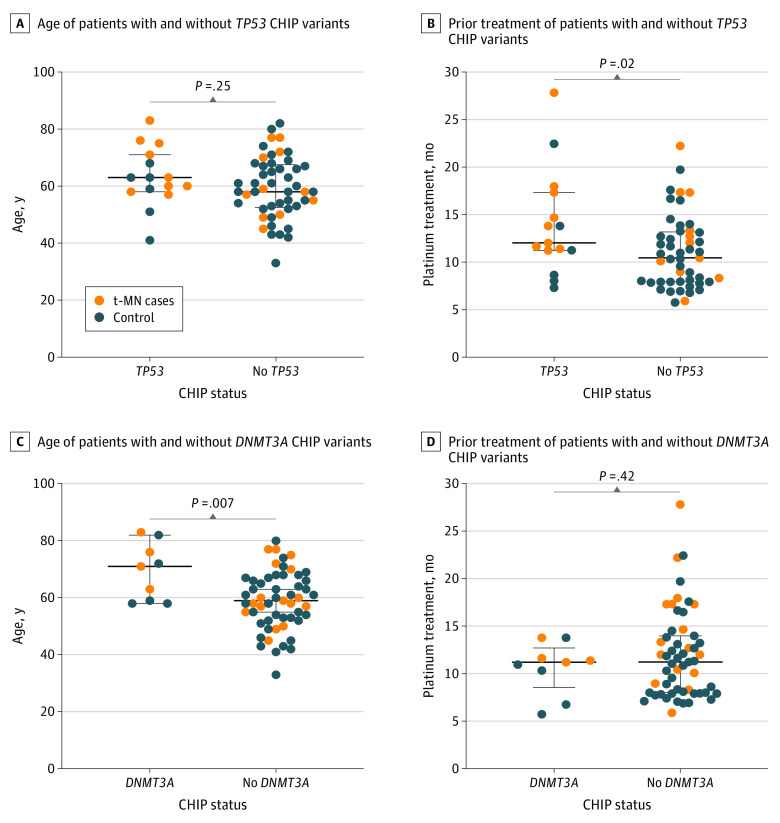
Patients with TP53-variant CHIP, including those who developed t-MNs, did not have a significant difference in age compared with the rest of the patients (Figure 1A), but they had a significantly longer prior exposure to platinum-based therapy (mean 14.0 months of 15 TP53 CHIP cases vs 11.1 months of 49 non-TP53 CHIP cases; P = .02) (Figure 1B). In contrast, patients with DNMT3A CHIP were significantly older than those with no DNMT3A variants (mean age, 69 years among 9 DNMT3A CHIP cases vs 59 years among 55 non-DNMT3A CHIP cases P = .007) (Figure 1C) but had no association with prior platinum exposure (Figure 1D).
Figure 1. Association of TP53 and DNMT3A Clonal Hematopoiesis of Indeterminate Potential (CHIP) Variants With Patient Age and Prior Exposure to Platinum.
A, Median age of sequenced patients with and without TP53 CHIP variants detected before treatment. B, Median length of exposure to prior platinum treatment of sequenced patients with and without TP53 CHIP variants detected before treatment. C, Median age of sequenced patients with and without DNMT3A CHIP variants detected before treatment. D, Median length of exposure to prior platinum treatment of sequenced patients with and without DNMT3A CHIP variants detected before treatment. P values are based on a 2-sided t test. No multiple hypothesis testing correction was performed. Comparisons are exploratory in nature. Error bars indicate IQR. t-MN indicates therapy-related malignant neoplasm.
A total of 21 of the 23 TP53 CHIP variants (91%) were missense variants in the DNA-binding domain. All patients with TP53 CHIP had at least 1 such variant (15 of 15 [100%]) (Table 2). By contrast, only 8 of 15 (53.0%) of the TP53 variants detected in the matched tumor samples were missense variants (P = .02) (eTables 6 and 7 in the Supplement).
In addition to alterations in the 10 predefined genes, we also identified a highly expanded activating NRAS G12D variant (OMIM 164790) (VAF, 37.2%) and a number of copy number events (high copy amplification of PIM1 [OMIM 164960]; losses in the ephrin receptors EPHA3 [OMIM 179611], EPHA5 [OMIM 600004], and EPHA7 [OMIM 602190]; and loss in the tumor suppressor LRP1B [OMIM 608766]) in 2 ARIEL3 patients who subsequently developed AML (eTable 8 in the Supplement). Two ARIEL2 patients who developed t-MNs harbored low-VAF ATM variants (OMIM 607585) (eTable 8 in the Supplement).
Expansion of Pretreatment TP53 Variants in CHIP During Rucaparib Treatment
To explore the association of rucaparib with CHIP variants, we sequenced PBC samples collected during and after treatment that were available for 5 of 6 ARIEL2 patients who developed t-MNs (samples during and after treatment were not collected as part of ARIEL3) (Figure 2A). No additional genes, predefined or not, were altered in any of the posttreatment samples analyzed. All 4 patients who had a TP53 CHIP alteration before treatment or early during treatment experienced a VAF increase for at least 1 of the detected variants, suggesting that the expansion of some preexisting clones likely contributes to the development of t-MNs (Figure 2B; eFigure 3 in the Supplement). Case 2, in which TP53 S240fs*24/716_717insT expanded from 9.4% before treatment to 41.6% after treatment in PBCs, had the same variant identified at 39% VAF in a bone marrow aspirate collected at the time of MDS diagnosis by independent targeted NGS (Figure 2A). This finding links the preexisting TP53 variant with the secondary malignant neoplasm developed after treatment.
Figure 2. Expansion of Preexisting TP53 Clonal Hematopoiesis of Indeterminate Potential (CHIP) Variants During Rucaparib Treatment in Patients Who Developed Therapy-Related Myeloid Neoplasms (t-MNs) After Treatment.
A, Detailed patient histories during and after being enrolled in the ARIEL2 (Study of Rucaparib in Patients With Platinum-Sensitive, Relapsed, High-Grade Epithelial Ovarian, Fallopian Tube, or Primary Peritoneal Cancer) trial, including timeline of start on treatment, last dose, TP53 CHIP variant detection, and t-MN diagnosis. Bars without an arrow end indicate patient death. B, Kinetics of TP53 CHIP variant allele frequency (VAF) expansion during the time between the pretreatment/early treatment period and posttreatment sample collection. HRR indicates homologous recombination repair; PARP, poly(adenosine diphosphate–ribose) polymerase; PBC, peripheral blood cell.
Case 1 involved the simultaneous increase in 2 TP53 variants (Figure 2B; eFigure 3 in the Supplement). The parallel increase and similar VAFs suggest that the 2 variants are found within the same clone and may together result in the biallelic inactivation of the gene. Indeed, a copy number analysis detected no loss of heterozygosity (LOH) at the TP53 locus in the posttreatment sample of this patient, which would be expected with biallelic TP53 variants (eTable 9 in the Supplement). In contrast, case 2 and case 3, both of which had an increase in a single TP53 variant, had accompanying LOH after treatment (eTable 9 in the Supplement). Although based on a small sample size, these data suggest that biallelic inactivation is required for the development of t-MNs in patients with TP53-variant CHIP. In support of this hypothesis, copy number analysis was also successful in a pretreatment sample of a patient who did not develop t-MNs but harbored a high-VAF TP53 CHIP event (40.7% R175H in control 19) (Table 2). The analysis indicated no LOH at the TP53 locus, suggesting that, in the absence of LOH, a TP53-variant clone can reach substantial expansion without becoming malignant.
The rate of TP53 VAF increase during rucaparib treatment was different in the 4 patients and was independent of pretreatment VAF (Figure 2B). In 2 instances, we also noticed the loss of a CHIP event detected at pretreatment (TP53 R273C/DNMT3A R736G in case 1 and TP53 K164E in case 2) (eFigure 3 in the Supplement). We found only minor to no changes in TP53 CHIP variant VAFs after treatment in 3 ARIEL2 control patients who did not develop t-MNs during the follow-up (eFigure 4 in the Supplement).
An increase in VAF of non-TP53 CHIP variants was observed in only 1 patient (case 4); the rates of increase of TET2 and DNMT3A VAF in this case were similar to that of the TP53 variant, raising the possibility that all 3 variants were found within the same clone. Two patients with longitudinal data had ATM CHIP variants, neither of whom experienced VAF change during treatment (eFigure 3 in the Supplement).
Discussion
This genetic association study found that the presence of TP53 CHIP variants before initiating rucaparib treatment was significantly associated with development of t-MNs after rucaparib treatment. Although t-MNs have been observed after PARPi treatment, little is known about factors that increase the risk of developing these serious adverse events.
Pretreatment TP53 CHIP VAF, which estimates the relative fraction of the CHIP clone among PBCs, was not different in cases vs controls; it also did not influence the time from start of rucaparib treatment to t-MN diagnosis. We also noted different kinetics of TP53 CHIP expansion during rucaparib treatment as well as the loss of some TP53 alterations in patients who developed t-MNs and little change in the TP53 CHIP VAF in patients who did not. These observations indicate that additional factors, beyond rucaparib treatment, determine whether and how fast a TP53-mutant clone expands or gains malignant potential. One relevant aspect may be the biallelic inactivation of the gene. Understanding the additional variables that contribute to malignant neoplasms would be essential for determining a patient’s risk of developing t-MNs after PARPi treatment.
A recent report16 specifically linked the presence of TP53 CHIP variants to platinum therapy and found that TP53-mutant clones display increased fitness and preferentially expand during such treatment. Furthermore, TP53 alterations are more common in t-MNs compared with primary myeloid malignant neoplasms.7,25 In the SOLO2 (Olaparib Treatment in BRCA Mutated Ovarian Cancer Patients After Complete or Partial Response to Platinum Chemotherapy) study of olaparib maintenance treatment, prevalence of t-MNs increased with the number of prior courses of platinum therapy.21 In agreement with these observations, we noted an association between TP53 CHIP events and longer overall prior exposure to platinum therapy in ARIEL2 and ARIEL3. Longer exposure to prior platinum was significantly associated with t-MNs, as was longer exposure to rucaparib and the presence of a germline or somatic variant in BRCA1, BRCA2, RAD51C, or RAD51D. Of importance, these clinical and molecular characteristics are not independent; for example, the presence of HRR gene variants is associated with platinum sensitivity, and both are associated with PARPi sensitivity. Because these variables are highly correlated, it is hard to extrapolate how much each of them individually contributes to the overall risk of t-MNs.
Limitations
This study has limitations, including the retrospective design. Prospective cohort studies are needed to confirm the risk conferred by preexisting TP53 CHIP variants for t-MNs after PARPi therapy, to define the natural history of TP53 CHIP variants in HGOC survivors both receiving and not receiving therapy and to evaluate the contribution of other risk factors, including the time undergoing treatment and presence of HRR gene variants.
As plasma sequencing of cfDNA becomes more widely used in cancer treatment and monitoring, the presence and particularly the expansion of TP53 CHIP variants over time may be an additional finding that could inform clinicians about individual patient risks of developing a secondary malignant neoplasm. Because our current analysis focused on CHIP variants with a VAF of 1% or greater in PBCs, the significance of CHIP events with lower frequencies that may be identified with more sensitive liquid biopsy technologies remains unclear. Our results also underscore the importance of ascertaining the tumor or CHIP origin of variants detected in plasma, especially in the case of TP53, which is 1 of the most altered genes in both hematopoietic malignant neoplasms and solid tumors.
Conclusions
This study found that pretreatment TP53 CHIP variants present at a VAF of 1% or greater are a risk factor for developing secondary t-MNs after rucaparib treatments. Poly(adenosine diphosphate–ribose) polymerase inhibitors are increasingly being used earlier in the treatment of HGOC, at which point TP53 CHIP variants are less frequent and the consequent risk of t-MNs is lower, which may lead to an improved benefit-risk ratio. How different therapies affect the expansion of preexisting TP53 CHIP clones and the benefit of screening for them require further research with careful longitudinal monitoring.
eMethods. Supplemental Methods
eTable 1. Characteristics of All t-MN Cases in ARIEL2 and ARIEL3, Including Age, HRR Gene Variant Status, Number of Prior Chemotherapies, Presence of TP53 CHIP, Time From Last Dose to t-MN Diagnosis, Reason for Treatment Discontinuation in Trial and Anti-cancer Therapies Received Between Last Dose on Trial and t-MN Diagnosis, if Applicable
eTable 2. Cytogenetic Findings for t-MN Cases in ARIEL2 and ARIEL3
eTable 3. Prevalence of t-MN in Patients With HGOC Positive (Variant) or Negative (Wild Type) for a BRCA1, BRCA2, RAD51C, or RAD51D Alteration
eTable 4. Exposure to Prior Treatment of Patients With HGOC Positive or Negative for a BRCA1, BRCA2, RAD51C, or RAD51D Alteration
eTable 5. Demographics, Clinical, and Molecular Characteristics of Sequenced Cases and Controls
eTable 6. TP53 Variants Detected in HGOC Tumor Tissue in Patients With TP53 CHIP
eTable 7. Types of TP53 Variants Detected in PBCs and in Matched HGOC Tumor Tissue
eTable 8. Alterations in Nonpredefined CHIP-Associated Genes Detected in Pretreatment Blood Samples
eTable 9. TP53 LOH Status in Posttreatment Samples From Patients Who Developed t-MN
eFigure 1. Time From Start of Treatment to t-MN Diagnosis in Rucaparib- and Placebo-Treated Patients From the ARIEL3 Trial
eFigure 2. Relationship Between Pretreatment TP53 CHIP VAF and the Development of t-MN
eFigure 3. Changes in VAF of Different Genes Detected in Pre/On-treatment and End of Treatment PBC Samples in ARIEL2 Patients Who Developed t-MN
eFigure 4. Pretreatment and Posttreatment VAF of Preexisting TP53 CHIP in Patients Who Did Not Develop t-MN
References
- 1.McNerney ME, Godley LA, Le Beau MM. Therapy-related myeloid neoplasms: when genetics and environment collide. Nat Rev Cancer. 2017;17(9):513-527. doi: 10.1038/nrc.2017.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fianchi L, Criscuolo M, Fabiani E, et al. Therapy-related myeloid neoplasms: clinical perspectives. Onco Targets Ther. 2018;11:5909-5915. doi: 10.2147/OTT.S101333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Travis LB, Holowaty EJ, Bergfeldt K, et al. Risk of leukemia after platinum-based chemotherapy for ovarian cancer. N Engl J Med. 1999;340(5):351-357. doi: 10.1056/NEJM199902043400504 [DOI] [PubMed] [Google Scholar]
- 4.Morton LM, Dores GM, Schonfeld SJ, et al. Association of chemotherapy for solid tumors with development of therapy-related myelodysplastic syndrome or acute myeloid leukemia in the modern era. JAMA Oncol. 2019;5(3):318-325. doi: 10.1001/jamaoncol.2018.5625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shenolikar R, Durden E, Meyer N, Lenhart G, Moore K. Incidence of secondary myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML) in patients with ovarian or breast cancer in a real-world setting in the United States. Gynecol Oncol. 2018;151(2):190-195. doi: 10.1016/j.ygyno.2018.09.003 [DOI] [PubMed] [Google Scholar]
- 6.Sperling AS, Gibson CJ, Ebert BL. The genetics of myelodysplastic syndrome: from clonal haematopoiesis to secondary leukaemia. Nat Rev Cancer. 2017;17(1):5-19. doi: 10.1038/nrc.2016.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ok CY, Patel KP, Garcia-Manero G, et al. TP53 mutation characteristics in therapy-related myelodysplastic syndromes and acute myeloid leukemia is similar to de novo diseases. J Hematol Oncol. 2015;8:45. doi: 10.1186/s13045-015-0139-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steensma DP. Clinical consequences of clonal hematopoiesis of indeterminate potential. Hematology Am Soc Hematol Educ Program. 2018;2018(1):264-269. doi: 10.1182/asheducation-2018.1.264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaiswal S, Ebert BL. Clonal hematopoiesis in human aging and disease. Science. 2019;366(6465):eaan4673. doi: 10.1126/science.aan4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaiswal S, Fontanillas P, Flannick J, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371(26):2488-2498. doi: 10.1056/NEJMoa1408617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Genovese G, Kähler AK, Handsaker RE, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371(26):2477-2487. doi: 10.1056/NEJMoa1409405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Razavi P, Li BT, Brown DN, et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat Med. 2019;25(12):1928-1937. doi: 10.1038/s41591-019-0652-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coombs CC, Zehir A, Devlin SM, et al. Therapy-related clonal hematopoiesis in patients with non-hematologic cancers is common and associated with adverse clinical outcomes. Cell Stem Cell. 2017;21(3):374-382.e4. doi: 10.1016/j.stem.2017.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gillis NK, Ball M, Zhang Q, et al. Clonal haemopoiesis and therapy-related myeloid malignancies in elderly patients: a proof-of-concept, case-control study. Lancet Oncol. 2017;18(1):112-121. doi: 10.1016/S1470-2045(16)30627-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swisher EM, Harrell MI, Norquist BM, et al. Somatic mosaic mutations in PPM1D and TP53 in the blood of women with ovarian carcinoma. JAMA Oncol. 2016;2(3):370-372. doi: 10.1001/jamaoncol.2015.6053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bolton KL, Ptashkin RN, Gao T, et al. Cancer therapy shapes the fitness landscape of clonal hematopoiesis. Nat Genet. 2020;52(11):1219-1226. doi: 10.1038/s41588-020-00710-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takahashi K, Wang F, Kantarjian H, et al. Preleukaemic clonal haemopoiesis and risk of therapy-related myeloid neoplasms: a case-control study. Lancet Oncol. 2017;18(1):100-111. doi: 10.1016/S1470-2045(16)30626-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rubraca (rucaparib) tablets. Prescribing information. Clovis Oncology Inc; 2020.
- 19.Kim G, Ison G, McKee AE, et al. FDA approval summary: olaparib monotherapy in patients with deleterious germline BRCA-mutated advanced ovarian cancer treated with three or more lines of chemotherapy. Clin Cancer Res. 2015;21(19):4257-4261. doi: 10.1158/1078-0432.CCR-15-0887 [DOI] [PubMed] [Google Scholar]
- 20.Master SR, Mansour RP. Myelodysplastic syndrome and acute myeloid leukemia as side effect of PARP inhibitors. J Clin Oncol. 2020;38(suppl 15):3601. doi: 10.1200/JCO.2020.38.15_suppl.3601 [DOI] [Google Scholar]
- 21.Korach J, Turner S, Milenkova T, et al. Incidence of myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML) in patients (pts) with a germline (g) BRCA mutation (m) and platinum-sensitive relapsed ovarian cancer (PSR OC) receiving maintenance olaparib in SOLO2: impact of prior lines of platinum therapy. J Clin Oncol. 2018;36(suppl 15):5548. doi: 10.1200/JCO.2018.36.15_suppl.5548 [DOI] [Google Scholar]
- 22.Morice PM, Leary A, Dolladille C, et al. Myelodysplastic syndrome and acute myeloid leukaemia in patients treated with PARP inhibitors: a safety meta-analysis of randomised controlled trials and a retrospective study of the WHO pharmacovigilance database. Lancet Haematol. 2021;8(2):e122-e134. doi: 10.1016/S2352-3026(20)30360-4 [DOI] [PubMed] [Google Scholar]
- 23.Swisher EM, Lin KK, Oza AM, et al. Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 Part 1): an international, multicentre, open-label, phase 2 trial. Lancet Oncol. 2017;18(1):75-87. doi: 10.1016/S1470-2045(16)30559-9 [DOI] [PubMed] [Google Scholar]
- 24.Coleman RL, Oza AM, Lorusso D, et al. ; ARIEL3 investigators . Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;390(10106):1949-1961. doi: 10.1016/S0140-6736(17)32440-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong TN, Ramsingh G, Young AL, et al. Role of TP53 mutations in the origin and evolution of therapy-related acute myeloid leukaemia. Nature. 2015;518(7540):552-555. doi: 10.1038/nature13968 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eMethods. Supplemental Methods
eTable 1. Characteristics of All t-MN Cases in ARIEL2 and ARIEL3, Including Age, HRR Gene Variant Status, Number of Prior Chemotherapies, Presence of TP53 CHIP, Time From Last Dose to t-MN Diagnosis, Reason for Treatment Discontinuation in Trial and Anti-cancer Therapies Received Between Last Dose on Trial and t-MN Diagnosis, if Applicable
eTable 2. Cytogenetic Findings for t-MN Cases in ARIEL2 and ARIEL3
eTable 3. Prevalence of t-MN in Patients With HGOC Positive (Variant) or Negative (Wild Type) for a BRCA1, BRCA2, RAD51C, or RAD51D Alteration
eTable 4. Exposure to Prior Treatment of Patients With HGOC Positive or Negative for a BRCA1, BRCA2, RAD51C, or RAD51D Alteration
eTable 5. Demographics, Clinical, and Molecular Characteristics of Sequenced Cases and Controls
eTable 6. TP53 Variants Detected in HGOC Tumor Tissue in Patients With TP53 CHIP
eTable 7. Types of TP53 Variants Detected in PBCs and in Matched HGOC Tumor Tissue
eTable 8. Alterations in Nonpredefined CHIP-Associated Genes Detected in Pretreatment Blood Samples
eTable 9. TP53 LOH Status in Posttreatment Samples From Patients Who Developed t-MN
eFigure 1. Time From Start of Treatment to t-MN Diagnosis in Rucaparib- and Placebo-Treated Patients From the ARIEL3 Trial
eFigure 2. Relationship Between Pretreatment TP53 CHIP VAF and the Development of t-MN
eFigure 3. Changes in VAF of Different Genes Detected in Pre/On-treatment and End of Treatment PBC Samples in ARIEL2 Patients Who Developed t-MN
eFigure 4. Pretreatment and Posttreatment VAF of Preexisting TP53 CHIP in Patients Who Did Not Develop t-MN