Abstract
The large tumor suppressor homolog 2 (LATS2), one of the central regulators of the Hippo/MST signaling pathway, plays an inhibitory role in ovarian function and different organ development and growth in mammals. However, the exact roles and molecular regulatory mechanisms of LATS2 in chicken granulosa cell (GC) proliferation, differentiation, and steroidogenesis required for ovarian follicle growth, development, and follicular selection remain poorly understood. This study demonstrated that the LATS2 protein was predominantly localized in the oocytes and undifferentiated GCs of various-sized prehierarchical follicles of the hen ovary. Expression levels of LATS2 mRNA were significantly higher in the smaller follicles (from 1 mm to 5.9 mm in diameter) and the GCs than in the larger follicles (6–6.9 mm in diameter up to F1). Moreover, we found that high levels of LATS2 suppressed the GC proliferation and the mRNA and protein expression of the genes serving as the biomarkers of follicle selection, GC differentiation, and steroidogenesis in the GCs, including FSHR, STAR, CYP11A1, ESR1, and ESR2. Interestingly, the LATS2 significantly downregulated SAV1 and YAP1 transcripts but upregulated the expression of STK3, STK4, TEAD1, and TEAD3 mRNA. Our study provided evidences that STK3/4-LATS2-YAP1 not only acts as a suppressor of cell proliferation and follicle selection but also LATS2 may serve as an enhancer in cell proliferation and follicle selection through the YAP1-LATS2 and the LATS2-STK3/4 feedback loops by promoting the expression of TEAD1/3 but inhibiting the expression of SAV1 transcripts in the prehierarchical follicle development of hen ovary. Taken together, the present study initially revealed the pivotal role and molecular mechanism of LATS2 in the regulation of hen prehierarchical follicle development by controlling GC proliferation, differentiation, steroidogenesis, and follicle selection via the Hippo/MST signaling pathway.
Key words: chicken, LATS2, prehierarchical follicle, Hippo pathway, ovary development
ABBREVIATIONS: LATS2, large tumor suppressor homolog 2; MST, mammalian Ste20-like kinase; GCs, granulosa cells; FSH, follicle-stimulating hormone; FSHR, FSH receptor; STAR, steroidogenic acute regulatory protein; CYP11A1, cytochrome P450 family 11 subfamilies A member 1; ESR1, estrogen receptor α; ESR2, estrogen receptor β; STK3, serine/threonine kinase 3; STK4, serine/threonine kinase 4; SAV1, Salvador homolog 1; LATS1, large tumor suppressor homolog 1; YAP1, Yes-associated protein 1; TAZ, transcriptional coactivator with PDZ binding motif; MOB1, MOB family member 1; OGSCs, ovarian germline stem cells; HRP, horseradish peroxidase; HA, hemagglutinin epitope; PF, prehierarchical follicles; TEAD1, TEA domain transcription factor 1; TEAD3, TEA domain transcription factor 3; TEA, transcriptional enhancer factor
INTRODUCTION
Egg production is the most desirable economic trait in laying hens (Kim et al., 2004; Qin et al., 2015). It mainly depends upon ovarian follicle recruitment, selection, development, and a well-organized preovulatory hierarchy (Johnson and Woods, 2009; Son et al., 2011; Johnson, 2012; Johnson, 2015). The ovarian follicle growth and development in chicken is a highly intricate and coordinated process involving a multitude of biological events controlled by reproductive hormones in the ovary (Gilchrist et al., 2006; Woodruff and Shea, 2007; Xu et al., 2018a). A wide variety of local intraovarian factors, such as steroidogenic acute regulatory protein (StAR) and cytochrome P450 family 11 subfamilies A member 1 (CYP11A1/P450scc), various members of gonadotropins, gonadal steroid hormones and their receptors, such as follicle-stimulating hormone (FSH) and FSH receptor (FSHR), estrogen and their receptor isoforms, ESR1/ERα and ESR2/ERβ, are implicated in follicle development, selection, differentiation, and steroidogenesis (Lin et al., 1995; Pollack et al., 1997; Bentsi-Barnes et al., 2010; Johnson, 2015; Oh et al., 2017). Furthermore, various cell signaling transduction pathways are also involved in ovarian follicle development and ovarian function. In recent years, the Hippo/MST (mammalian Ste20-like kinase) signaling pathway emerges as a crucial pathway regulating ovarian functions (Kawamura et al., 2013; Xiang et al., 2015; Lyu et al., 2016; Ye et al., 2017).
The core members of the Hippo pathway in chicken comprises of 2 upstream MST homologous named serine/threonine kinase 3 (STK3, named MST2 in mammals) and STK4 (MST1 in mammals) (Lyu et al., 2016), along with a scaffolding protein Salvador homolog 1 (SAV1), 2 serine/threonine (Ser/Thr) protein kinases, LATS1 (large tumor suppressor homolog 1) and LATS2. The Hippo pathway interacts with MOB1/2 proteins, which regulate transcriptional coactivators, Yes-associated protein 1 (YAP1, also named YAP65), and with a PDZ-binding motif (TAZ) (Zhao et al., 2008a; Kawamura et al., 2013; Xiang et al., 2015; Lyu et al., 2016). In mammals, SAV1 recruits LATS to the MST and then promotes LATS phosphorylation mediated by the MST1 and MST2 (Wu et al., 2003; Chan et al., 2005); the activated LATS kinases then phosphorylate YAP and TAZ (Wang et al., 2009). In the absence of the inhibitory phosphorylation for the YAP1 and TAZ by LATS1/2 in the cytoplasm, these transcription factors move into the nucleus to trigger the transcription of genes that enhance cell proliferation (Vassilev et al., 2001; Camargo et al., 2007; Zhao et al., 2008b; Huntoon et al., 2010). These studies indicated that the Hippo/MST signaling pathway, a highly conserved cell signaling system, exists in most multicellular organisms and regulates cell proliferation, differentiation, and apoptosis in chicken and mammals (Kawamura et al., 2013; Xiang et al., 2015; Ye et al., 2017).
The LATS1 and LATS2, two central regulators of the Hippo/MST signaling pathway, regulate the ovarian growth, development, and functions in mammals (Dong et al., 2007; Huntoon et al., 2010; Tsoi et al., 2019). Inactivation of Lats1/2 in murine granulosa cells (GCs) resulted in a loss of granulosa cell morphology, function, and gene expression (Tsoi et al., 2019). Furthermore, our previous study revealed that Hippo/MST signaling regulates chicken prehierarchical follicle development, and the phosphorylation of LATS1 is induced by the kinases STK3 and STK4 in vitro, which is promoted by the scaffolding protein SAV1 (Lyu et al., 2016). Consistent with the roles of LATS1 in germ cell proliferation and apoptosis of the follicles, the paralogous kinase LATS2 was suggested to function in cell proliferation and differentiation in the ovary as reported in other organs (Dong et al., 2007; An et al., 2013). Moreover, the elevated LATS2 kinase activity phosphorylates YAP1 and inhibits the transcriptional coactivity of YAP1 (Camargo et al., 2007; Hoa et al., 2016). All these studies indicated that the LATS2 is strongly associated with ovarian follicle growth and development through granulosa cell proliferation, differentiation, and apoptosis (Kawamura et al., 2013; Xiang et al., 2015; Hoa et al., 2016; Ye et al., 2017). However, the exact functions and molecular mechanisms of kinase LATS2 in ovarian follicle growth, development, and follicular selection, involved in GCs proliferation, differentiation, and steroidogenesis in chicken, remained unknown.
To comprehensively understand the functions of the Hippo/MST signaling pathway in ovarian follicle development, we aimed to explore the potential upstream kinases and coactivator (STK3, STK4, and SAV1) of the LATS2 and its downstream effectors (YAP1, TEAD1, and TEAD3) in the GCs in vitro, and the roles and molecular mechanism of LATS2 protein in GC proliferation, differentiation, steroidogenesis, and follicle selection during the prehierarchical follicular development in hen ovary.
MATERIALS AND METHODS
Ethics Statement
All procedures performed in animals were approved by the Institutional Animal Care and Use Committee (IACUC) of Jilin Agricultural University (Changchun, China). The animal experiment was carried out in compliance with the ARRIVE guidelines (Percie et al., 2020). Hens were sacrificed before removing organs following the IACUC guidelines for experimental animals (Permission No. GR (J) 19-030). Euthanasia of the chickens is fully compliant with the Chinese applicable laws and regulations concerning the care and use of laboratory animals, which was issued based on the Regulations for the Administration Affairs Concerning Experimental Animals of the State Council of the People's Republic of China (2017 Revision). All of our efforts have been made to minimize the suffering of the animals.
Animals and Sampling
A commercial strain of Lohmann Brown laying hens was reared in laying batteries according to the standard husbandry practices as previously reported (Xu et al., 2018a). Twenty birds were obtained from the population and euthanized at 21 wk of age. Various sized ovarian follicles and GCs from the prehierarchical follicles (1–3.9 mm, 4–4.9 mm, 5–5.9 mm, 6–6.9 mm, 7–8 mm in diameters) and preovulatory follicles (F6 - F1) were isolated from the hen ovaries, and then cultured as previously described (Johnson et al., 1996, 2002). A representative portion of each ovary was sampled and immediately snapped frozen in liquid nitrogen, and stored at −80°C until use. Another equal part of the ovarian tissue was fixed in 4% neutral-buffered formalin at 4°C. GCs from the follicles were collected as previously reported (Qin et al., 2020). In brief, after follicles were cleaned with ice-cold 0.9% NaCl solution, follicular walls were dissected with sterile scalpel blades or needles following washed with preheated culture media (DMEM/F12 supplemented with 1% penicillin-streptomycin mixture) to remove oocyte contents. Granulosa cell layers were separated by gently shaking or peeling off from the inverted follicular tissue while theca layers remained within the residual follicular walls. The collected granulosa cells were disaggregated at 37 °C for 15 min under continuous agitation in 5 mL of digesting solution containing 1 mg/mL of trypsin type II in DPBS for culture and subsequent experimental purposes.
Immunohistochemistry
Tissue slides of the ovarian prehierarchical follicles were prepared, pretreated, blocked, and immunohistochemical staining was performed as previously described (Lyu et al., 2016). The slides were incubated with mouse anti-LATS2 of chicken (1:500, Sangon Co, Shanghai, China) overnight at 4°C, and then incubated at room temperature for 1 h with goat anti-mouse secondary antibody (1:1,000) labeled with horseradish peroxidase (HRP). Negative control sections were treated with the same concentration of normal mouse serum for 30 min instead of the primary antibody. Images of the sections were taken using a JNOEC XS-213 biological microscope (Jiangnan Optics & Electronics Co., Ltd. Nanjing, China).
Quantitative Real-Time Reverse Transcriptase -PCR
To determine target genes' expression, real-time quantitative reverse transcriptase PCR (RT-qPCR) was conducted as previously described (Xu et al., 2018a). The primers used for the LATS2 gene: forward 5’-TCTTCCAACAGCAAGCACAC-3’ and reverse 5’- AAGCTCCAGTCTGATCCACC-3’. The 18S rRNA gene was used as an internal control: forward 5’-TAGTTGGTGGAGCGATTTGTCT-3’ and reverse 5’- CGGACATCTAAGGGCATCACA-3’. The other primers utilized for amplifying the genes, including FSHR, STAR, CYP11A1, ESR1, and ESR2, are listed in Table 1. Using the 2−ΔΔCt method, mRNA expression was determined and further normalized against 18S rRNA.
Table 1.
Primer pairs designed for the quantitative real-time PCR analysis.
| Gene | Forward primer (5′ - 3′) | Reverse primer (5′ - 3′) | Accession No. | Size |
|---|---|---|---|---|
| LATS2 | TCTTCCAACAGCAAGCACAC | AAGCTCCAGTCTGATCCACC | XM_015279299.2 | 169 bp |
| STK3 | CATGCACGAACCTTACCACA | CCATCGCATCCAGGATAGGT | NM_001031337.2 | 206 bp |
| STK4 | GTGTCTGATATTATTCGGTTAC | ATGTCCCTCCGTGTTTAG | NM_001030853.1 | 159 bp |
| SAV1 | ACCACACTGCTGAGATTCCA | TCGTACAGCTTCACAATGCG | XM_015276749.2 | 169 bp |
| YAP1 | GCCAAAGTGCTCCAGTGAAA | CATCGTTGGAAGCTGGCTAC | NM_205243.1 | 197 bp |
| TEAD1 | TGCAAGGTTTGAGAATGGCC | AGCCATGCAGAGTAGGGTTT | NM_001199405.1 | 190 bp |
| TEAD3 | GCGGATCTGAACAGCACAAT | TCCGGGAGATGCTTCAGTTT | XM_015298756.2 | 248 bp |
| FSHR | TCCTGTGCTAACCCTTTCCTCTA | AACCAGTGAATAAATAGTCCCATC | NM_205079.1 | 207 bp |
| STAR | AGCAGATGGGCGACTGGAAC | GGGAGCACCGAACACTCACAA | NM_204686.2 | 147 bp |
| CYP11A1 | TCCGCTTTGCCTTGGAGTCTGTG | ATGAGGGTGACGGCGTCGATGAA | NM_001001756.1 | 112 bp |
| ESR1 | CAGATGGTCAGTGCCTTGCT | CGCCAGACTAAGCCGATCAT | NM_205183.2 | 242 bp |
| ESR2 | GATTACATCTGCCCAGCTACCAA | TACCCACAGCGTTCTCTTCTTGA | NM_204794.2 | 134 bp |
| 18SrRNA | TAGTTGGTGGAGCGATTTGTCT | CGGACATCTAAGGGCATCACA | AF173612.1 | 169 bp |
Cell Culture
The GCs culture from hen prehierarchical follicles (6–8 mm in diameter) was performed as previously described, along with some modification in our laboratory (Johnson et al., 2002; Qin et al., 2020). Briefly, fresh granulosa cells isolated from the follicles were dispersed using a Pasteur pipette. After that, the granulosa cell pellets were resuspended in the culture medium (medium M199 containing 1.1 g of 4-(2-Hydroxyethyl) piperazin-1-ylethanesulphonic acid (HEPES), 10% of newborn calf serum, 100 IU/mL of penicillin, 75 IU/mL of streptomycin, and 10 ng/mL of insulin), and viable cells number was determined using a hemocytometer. GCs were incubated in a humidified atmosphere with 5% CO2 and 95% air at 41 °C. The specificity of the granulosa cells was identified by the H & E staining and fluorescence staining analysis (Xu et al., 2018a).
Construction of Recombinant Plasmids and Cells Transfection
After chicken LATS2 cDNA sequence (GenBank accession: XM_015279299.2) was amplified from a chicken cDNA library by RT-PCR, and cloned into the pUC57-Simple plasmid (Sangon Biotech Co., Ltd., Shanghai, China) by using the specific primers for the LATS2 gene (see Supplementary Table S1 online): forward 5’-CCGGAATTCGCCACCATGAGGCCAAAGACTTTTCC-3’ and reverse 5’-CCGCTCGAGCGGTTACACATATACAGGTTGAC-3’, and then subcloned into a pYr-adshuttle-4 expression vector, which contains an N-terminal hemagglutinin epitope (HA) tag (Wuhan Biobuffer Biotech Service Co. Ltd., China) to generate pYr-adshuttle-4-LATS2 expression construct (Xu et al., 2018a,b).
Transfection of LATS2 expression by using the recombinant plasmid vector pYr-adshuttle-4-LATS2 was performed as previously reported (Lyu et al., 2016). Briefly, the GCs were transfected with the plasmid pYr-adshuttle-4-LATS2 and pYr-adshuttle-4 blank vector using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Cells (1 × 105 cells/well in a 24-well plate) were cultured in a basal medium containing 1 μL/mL Polybrene (hexadimethrine bromide, Sigma) and incubated at 37 °C with 5% CO2. After 24 h of culture, the granulosa cells were collected and then lysed for immunoblot analysis and RT-qPCR analysis.
Transfection of siRNA
Specific siRNAs targeting the LATS2 gene were designed using an InvivoGen siRNA Wizard v3.1. All designed siRNA sequences were blasted against the chicken genome database to eliminate the cross-silence phenomenon with non-target genes. A most effective LATS2 specific siRNA was further screened by RT-qPCR and Western blotting: 5′-GCAGCUGUUGAAGUGAAUATT-3′. Scrambled siRNA was used as the negative control siRNA: 5′-UUCUCCGAACGUGUCACGUTT-3′. As mentioned above, GCs were plated in 24-well plates, and the siRNAs were transfected into the cultured cells using Lipofectamine 2000 (Invitrogen, Carlsbad) according to the manufacturer's instructions as we previously reported (Xu et al., 2018a,b). The other siRNA sequences utilized to interfere with the targeted TEAD1, TEAD3, and YAP1 genes were listed in Supplementary Table S2 online.
Western Blotting
Western blot analysis for LATS2, YAP1, FSHR, STAR, CYP11A1, ESR1, and ESR2 was performed using total cellular extracts as previously described (Xu et al., 2018a,b). Briefly, equal amounts of protein were separated by 10% (w/v) SDS-polyacrylamide gel under reducing conditions and electrotransferred to Protran nitrocellulose membranes (Whatman, Dassel, Germany). After electrophoresis of the protein samples in a mini gel apparatus, a prestained protein molecular weight marker was loaded to locate/monitor the target proteins in the electrophoresis (SDS-PAGE). At the approximated protein size position, the gel was directly cut and transferred to the nitrocellulose membrane for western blotting. The affinity-purified antibodies for LATS2 and the other antibodies were used (Supplementary Table S3). The horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG secondary antibody was incubated for 2 h at room temperature. Blots were subsequently performed with ECL western blotting agent (Rockford, IL) for 5 min and exposed to X-ray film for 1 to 5 min. The signals were detected using the ECL Plus Western blotting detection system according to the manufacturer's instructions. Anti-β-actin (dilution 1:1,000, Boster, Wuhan, China) antibody acted as a loading control. As a result, the molecular size of the ladders was not observed in the original blots.
EdU Cell Proliferation Assay
Following the transfection of the cells with LATS2 overexpression or knockdown, a variation in cell proliferation was determined by EdU (5′-Ethynyl-2′- deoxyuridine) incorporation assay using the Cell-Light EdU imaging kit (RiboBio, Guangzhou, China) according to the manufacturer's protocol. Briefly, the control and transfected cells were seeded at a density of 1 × 105 cells/well in a 96-well flat-bottom plate and cultured for 24 h. Then, cells were then incubated with 50 nM of EdU for an additional 2 h at 37°C. Cells were fixed with 4% formaldehyde for 15 min at room temperature and washed with glycine (2 mg/mL) for 5 min in a decolorization shaker, 0.5% Triton X-100 was added for 10 min, and then cells were washed thrice with PBS. Next, the cells were incubated with 1 × Apollo reaction cocktail (100 μL/well) for 30 min. Finally, DNA was stained with 10 μg/mL of Hoechst 33342 stain (100 μL/well) for 20 min and visualized with fluorescence microscopy (Olympus, Tokyo, Japan). Each experiment was performed in triplicate and repeated 5 times. The number of EdU-positive cells was expressed as a percentage and calculated relative to the total number of cells (positive cells/total cells in one field) as previously described (Xu et al., 2018a,b). The ratio of positive cells was calculated from the average of the 5 group values.
Statistical Analysis
Statistical analysis was performed using the SPSS12.0 software package (Lyu et al., 2016). All the experiments were repeated at least 3 times using different batches of sampled birds. The data were collected and analyzed with a one-way ANOVA and Tukey's multiple-comparison test when more than 2 groups were involved or using a Student's t test when treatment and control groups were compared after confirming normal distributions for parametric analysis. P < 0.01 or P < 0.05 was considered to be statistically significant.
RESULTS
Expression of LATS2 Gene in the Ovarian Prehierarchical Follicles
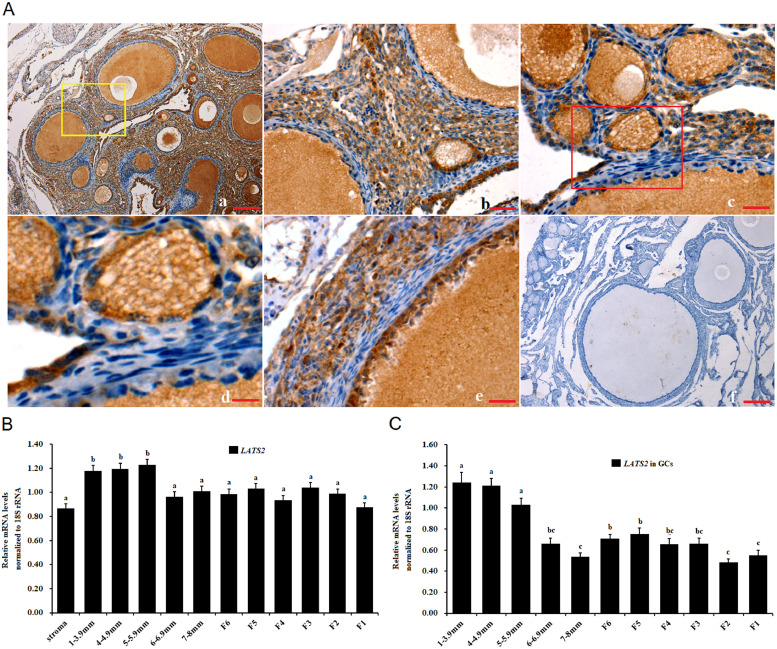
To determine the localization of LATS2 kinase in various sized ovarian prehierarchical follicles (PF), immunohistochemistry was performed. LATS2 protein was predominantly localized in the undifferentiated GCs and oocytes in the PFs (60 μm to 8 mm in diameter) as well as primordial follicles (smaller than 20 μm in diameter) and the primary follicles of 30 to 90 μm in diameter (Figure 1A). Furthermore, a strong expression intensity of LATS2 was observed in GCs and oocytes of all the follicles. LATS2 protein was also expressed in the ovarian stroma (mainly containing stromal cells, reticular, and collagen fibers) surrounded the follicles at the various development stages. Moreover, a higher expression of LATS2 transcript was detected in the smaller follicles (from 1 mm to 5.9 mm in diameter) and their GCs than the larger follicles (from 6 mm in diameter up to F1) and the surrounding GCs. The endogenous expression level of LATS2 mRNA kept stable in the larger follicles (>5.9 mm in diameter) of the chicken ovary (Figure 1B). However, the expression patterns of LATS2 mRNA in the GCs from the smallest prehierarchical follicles (1 mm to 3.9 mm in diameter) up to the largest follicles F1 appear different from those in the whole follicles (Figure 1C).
Figure 1.
Expression of LATS2 mRNA and protein in the ovarian follicles and their granulosa cells. (A) Immunolocalization of LATS2 protein was performed in the ovarian prehierarchical follicle. Panel a, intense staining detected in granulosa cells (GCs) and oocytes (OC) of the variously sized prehierarchical follicles as well as in ovarian stroma ( × 10). Panel b, paralleling the image indicated with the yellow box of panel a ( × 40); Panel c, a larger PF follicle with two or three layers of GCs ( × 100); Panel d, paralleling the image marked in the red box of panel c ( × 200); Panel e, the larger PF follicle with multiple layers of GCs and thecal cells ( × 100); Panel f, one of the negative controls for LATS2. The negative control sections were treated with the same concentration of normal mouse serum; no specific staining was observed ( × 10). Five birds were sampled for immunohistochemical analysis, and representative microscopic fields were screened. Scale bars, 200 μm (a and f); 50 μm (b); 20 μm (c and e); 10 μm (d). (B) Quantification of LATS2 transcripts in the various-sized ovarian follicles. The ovarian stroma was mainly containing stromal cells, reticular, and collagen fibers that extensively dispersed in the follicles. The prehierarchical follicles were grouped in diameters from 1 to 3.9 mm, 4–4.9 mm, 5–5.9 mm, 6–6.9 mm, and 7–8 mm, and hierarchical follicles were classified as F6, F5, F4, F3, F2, and F1). (C) The expression of LATS2 transcripts was determined in the GCs from the prehierarchical and hierarchical follicles (n = 10). Data presented are mean ± SEM, and normalized with 18S rRNA. Bars with different superscripts show significantly different at P < 0.01.
Overexpressing LATS2 Downregulates FSHR, STAR, CYP11A1, ESR1, and ESR2 Expressions and Represses GC Proliferation
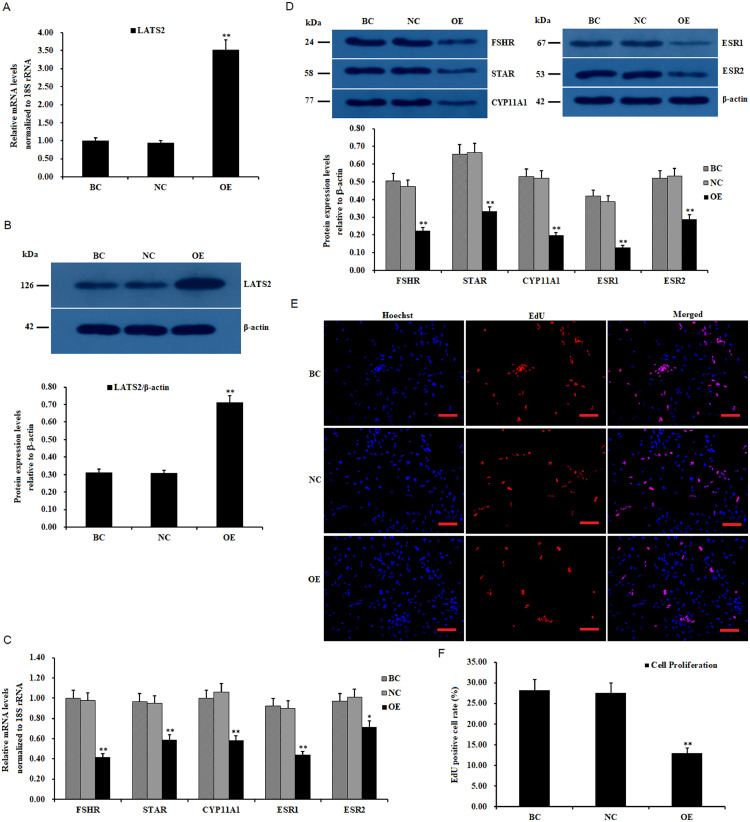
To delineate the physiological role of LATS2 in ovarian follicle development, a pYr-adshuttle-4-LATS2 expression vector was prepared and transfected into the cultured GCs. The regulation of LATS2 on the expression of the genes (FSHR, STAR, CYP11A1, ESR1, and ESR2) and GC proliferation was analyzed. As shown in Figures 2A–2D, a markedly enhanced LATS2 mRNA expression and protein were observed in the cells at 24 h of the post-transfection pYr-adshuttle-4-LATS2 vector (P < 0.01). The overexpression of LATS2 significantly downregulated the expression of FSHR, STAR, CYP11A1, ESR, and ESR2 mRNA and proteins in the cells (P < 0.01). Furthermore, cell proliferation ratios of the GCs were markedly decreased as compared to the negative control (P < 0.01; Figures 2E and 2F). It indicated that LATS2 plays an inhibitory role in GC proliferation and control follicle selection, GC differentiation, and steroidogenesis by regulating the expression of FSHR, STAR, CYP11A1, ESR1, and ESR2 in the prehierarchal follicle development.
Figure 2.
Inhibitory effects of LATS2 overexpression on FSHR, STAR, CYP11A1, ESR1and ESR2 mRNA, and granulosa cell (GC) proliferation. (A) GCs of the prehierarchical follicles (6–8 mm in diameter) were cultured and transfected with reconstructed pYr-adshuttle-4-LATS2 vector (OE group), pYr-adshuttle-4 empty vector (negative control) and no plasmid (blank control), respectively. The expression of LATS2 with or without transfection of the pYr-adshuttle-4-LATS2 vector for 24 h was evaluated by RT-qPCR. (B) Expression of LATS2 protein with or without transfection of the pYr-adshuttle-4-LATS2 vector was examined by western blotting. The β-actin was utilized as the loading control. (C) Effects of LATS2 overexpression on the expression of FSHR, STAR, CYP11A1, ESR1, and ESR2 mRNA in the granulosa cells. (D) The influence of LATS2 overexpression on the expression of FSHR, STAR, CYP11A1, ESR1, and ESR2 proteins. (E) The results of overexpressing LATS2 on GC proliferation were analyzed using EdU cell proliferation assay. All cell nuclei show blue fluorescence indicating Hoechst 33342 staining; the EdU-labeled cells exhibited red fluorescence showing the proliferated cells with newly synthesized DNA (original magnification × 20). Scale bars, 25 μm. (F) Quantification of cell proliferation percentage after GCs being transfected with the pYr-adshuttle-4-LATS2 vector. Values were indicated as mean ± SEM, and bars with superscript symbol present the significant difference compared with control groups at **P < 0.01, * P < 0.05.
Confirmation of the Inhibitory Effects of LATS2 on FSHR, STAR, CYP11A1, ESR1, and ESR2 Expressions, and GC Proliferation
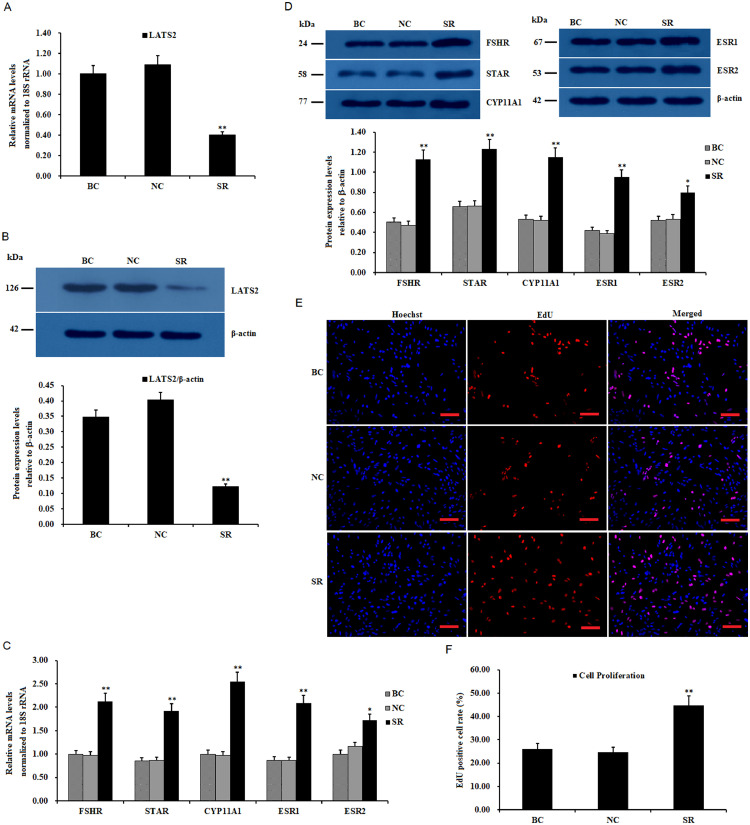
To further confirm the suppressive effects of LATS2 on GC proliferation, the expression of LATS2 was knocked down in the GCs using LATS2-specific siRNA transfection. As shown in Figures 3A–3D, the expression levels of FSHR, STAR, CYP11A1, ESR1, and ESR2 mRNA and proteins were remarkably upregulated in the cells after being transfected with the LATS2 siRNA (P < 0.01). Furthermore, the knockdown of LATS2 expression significantly enhanced GC proliferation compared to the negative control (P < 0.01; Figures 3E and 3F).
Figure 3.
Effects of LATS2 knockdown on the expression of FSHR, STAR, CYP11A1, ESR1, and ESR2, and granulosa cell (GC) proliferation. (A) GCs were transfected with LATS2-specific siRNA (SR group), scrambled siRNA (negative control, NC), and no siRNA as a vehicle (blank control, BC). The LATS2 mRNA expression in the GCs was analyzed using RT-qPCR. The values on the bar graphs are the mean ± SEM (n = 10) from a representative experiment. (B) Expression of LATS2 protein in the GCs with or without interference using the siRNA was analyzed by western blotting. The β-actin was used as the loading control. (C) The influence of LATS2 knockdown on FSHR, STAR, CYP11A1, ESR1, and ESR2 mRNA abundances in the GCs. (D) The impact of silencing of LATS2 on FSHR, STAR, CYP11A1, ESR1, and ESR2 protein expression. (E) All cell nuclei display blue fluorescence indicative of Hoechst 33342 staining; the EdU-labeled cells showed red fluorescence implying the proliferated cells with the newly synthesized DNA (original magnification × 20). Scale bars, 25 μm. (F) Quantification of cell proliferation percentage after granulosa cells transfected with the LATS2-specific siRNA. Values were represented as mean ± SEM, and bars with superscript symbols indicate a significant difference compared with control groups at **P < 0.01, * P < 0.05.
Regulations of LATS2 on STK3, STK4, SAV1, YAP1, TEAD1, and TEAD3 mRNA Expressions
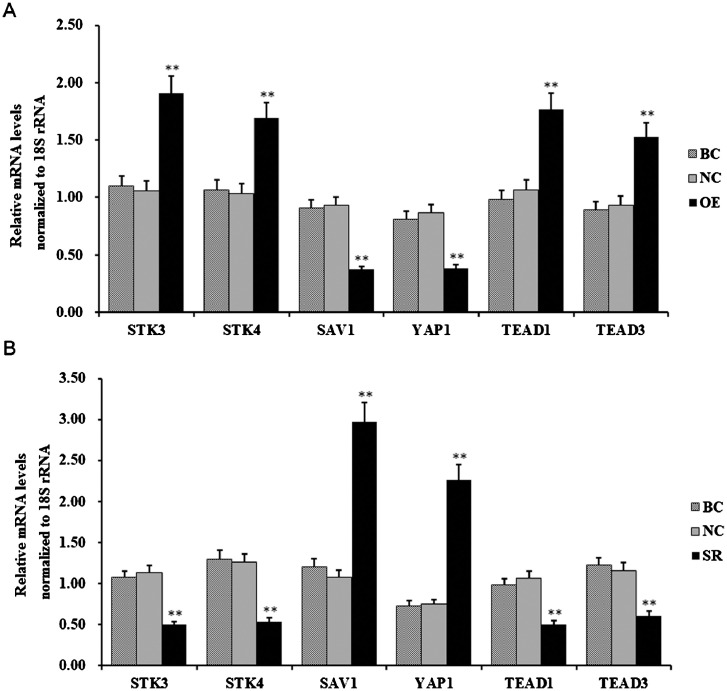
To explore the regulatory mechanism of LATS2 through mediating the Hippo/MST signaling cascades in ovarian follicular growth and development, effects of LATS2 on the expressions of its upstream kinases and coactivator (STK3, STK4, and SAV1), and downstream effectors (YAP1, TEAD1, and TEAD3) were detected in the GCs transfected with the pYr-adshuttle-4-LATS2 vector or LATS2-specific siRNA. As shown in Figure 4, overexpression of LATS2 remarkably upregulated STK3, STK4, TEAD1, and TEAD3, while significantly downregulates SAV1 and YAP1 (P < 0.01). Conversely, knocking down of LATS2 notably downregulated the expression of STK3, STK4, TEAD1, and TEAD3, but significantly upregulated the expression of SAV1 and YAP1 (P < 0.01). Unexpectedly, the results indicated that LATS2-STK3/4 feedback loops might inversely modulate the activities of STK3 and STK4.
Figure 4.
LATS2 regulates the expressions of STK3, STK4, SAV1, YAP1, TEAD1, and TEAD3 transcripts. (A) Effects of LATS2 overexpression on the expression of STK3, STK4, SAV1, YAP1, TEAD1, and TEAD3 in the granulosa cells (GCs). (B) Effects of LATS2 knockdown on the expression of STK3, STK4, SAV1, YAP1, TEAD1, and TEAD3 mRNA. Values were represented as mean ± SEM, and bars with superscript symbols indicate a significant difference compared with control groups at **P < 0.01.
Roles of TEAD1 and TEAD3 in Mediating the Regulation of LATS2 in GC Proliferation, Differentiation, and Follicle Selection
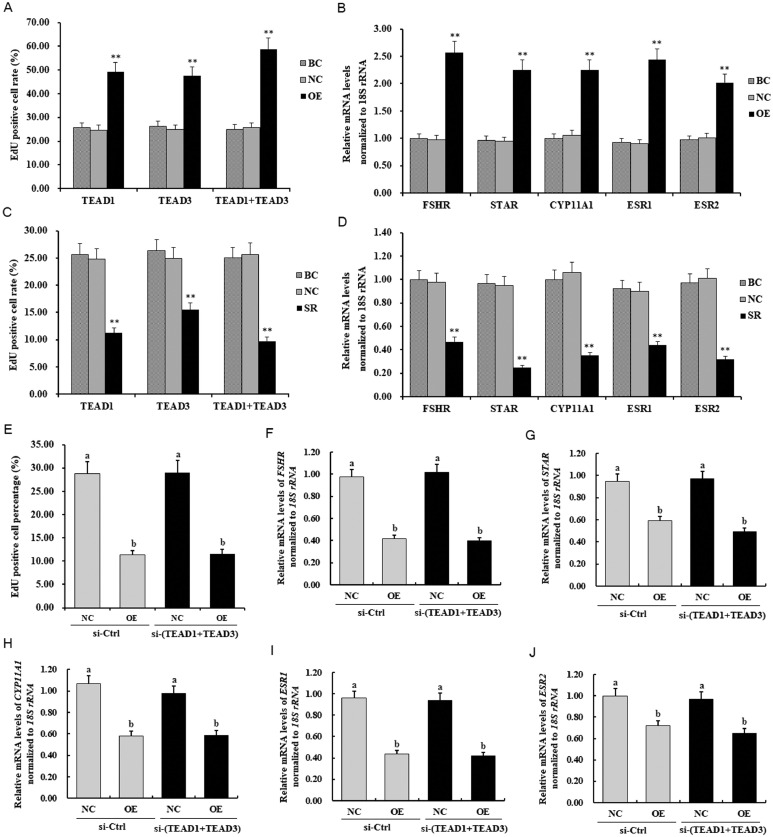
To investigate the roles of TEAD1 and TEAD3 in the regulation of GC proliferation, overexpression of TEAD1 and TEAD3 were carried out by transfecting the GCs with pYr-adshuttle-4-TEAD1 or/and TEAD3 vectors (Supplementary Figure S1). As shown in Figure 5, overexpression of TEAD1 and/or TEAD3 resulted in a significant increase in GC proliferation and remarkably enhanced the mRNA expression levels of FSHR, STAR, CYP11A1, ESR1, and ESR2 (P < 0.01, Figures 5A and 5B). Conversely, specific siRNA-mediated knockdown of TEAD1 and TEAD3 in the GCs resulted in a significant reduction in GC proliferation and a sharp decline in the expression of FSHR, STAR, CYP11A1, ESR1, and ESR2 transcripts (P < 0.01, Figures 5C and 5D). These results demonstrate that TEAD1 and TEAD3 promote cell proliferation, differentiation, steroidogenesis, and follicle selection.
Figure 5.
Roles of TEAD1 and TEAD3 in the LATS2-induced inhibitory regulation of granulosa cell (GC) proliferation and expression of FSHR, STAR, CYP11A1, ESR1, and ESR2 transcripts. (A) GCs were transfected with reconstructed pYr-adshuttle-4-TEAD1, pYr-adshuttle-4-TEAD3, and pYr-adshuttle-4-TEAD1 and pYr-adshuttle-4-TEAD3 vectors (OE group), pYr-adshuttle-4 empty vector (negative control, NC) and no plasmid (blank control, BC). GCs were transfected with pYr-adshuttle-4-TEAD1, pYr-adshuttle-4-TEAD3, and pYr-adshuttle-4-TEAD1 and pYr-adshuttle-4-TEAD3 vectors for 24 h, and cell proliferation was detected by EdU incorporation assay. (B) Effects of TEAD1 and TEAD3 overexpression on FSHR, STAR, CYP11A1, ESR1, and ESR2 mRNA abundances in the GCs from prehierarchichal follicles (6–8 mm in diameter) was analyzed by RT-qPCR. (C) GCs were administrated with TEAD1/3-specific siRNA (SR group), scrambled siRNA (negative control, NC), and no siRNA (blank control, BC). The cell proliferation transfected with TEAD1 siRNA, TEAD3 siRNA, and TEAD1 and TEAD3 siRNA for 24 h were detected by EdU incorporation assay. (D) Abundances of FSHR, STAR, CYP11A1, ESR1, and ESR2 mRNA expression impacted by TEAD1 and TEAD3 knockdown in the GCs. (E–J) Results of TEAD1 and TEAD3 knockdown by using specific siRNA in the GCs with the LATS2 overexpression. NC, the granulosa cells were transfected with pYr-adshuttle-4 empty vector; OE, the cells were transfected with reconstructed pYr-adshuttle-4-LATS2. Values were represented as mean ± SEM, and bars with superscript symbols indicate a significant difference compared with control groups at ** P < 0.01, * P < 0.05.
To further understand the role of TEAD1 and TEAD3 in the LATS2-induced inhibitory effect on granulosa cell proliferation, TEAD1 and TEAD3 expressions were knocked down using specific siRNA in the GCs with the LATS2 overexpression (Supplementary Figure S2). Knockdown of TEAD1 and TEAD3 did not influence the repressive effects of LATS2 on GC proliferation, and the expression levels of FSHR, STAR, CYP11A1, ESR1, and ESR2 transcripts (P < 0.01, Figures 5E–5J). These results suggested that TEAD1 and TEAD3 did not directly mediate the effects of LATS2 on GC proliferation, differentiation, steroidogenesis, and follicle selection in the prehierarchical follicles. Therefore, TEAD1 and TEAD3 as the candidates for mediating the inhibitory effect on granulosa cell proliferation were excluded.
Transduction of YAP1 in the Regulation of LATS2 in GC Proliferation, Differentiation, and Follicle Selection
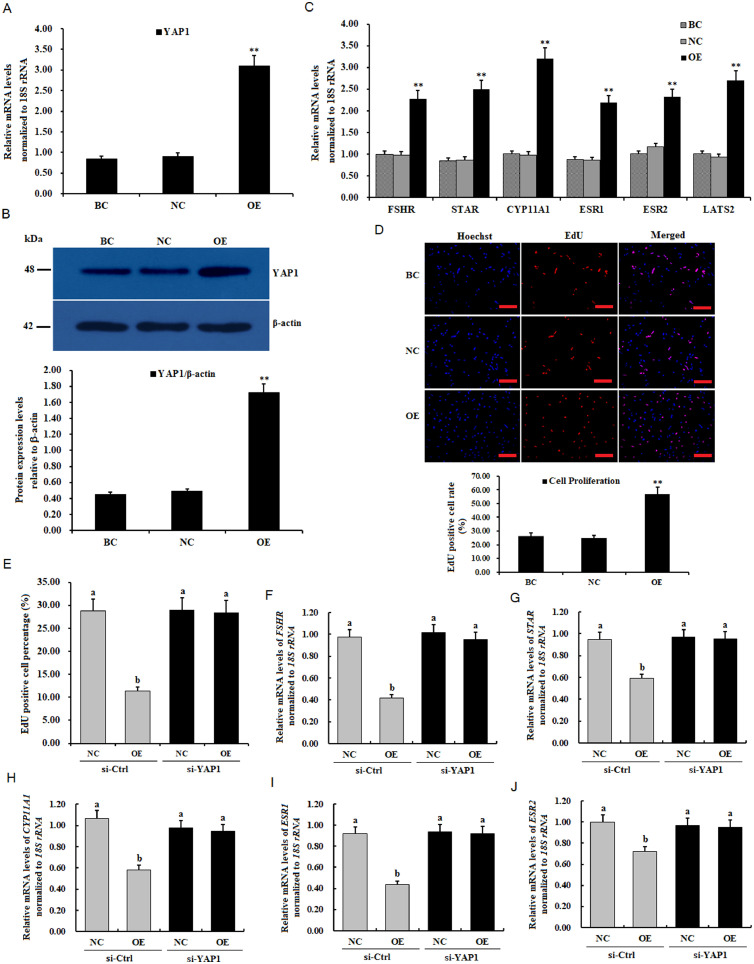
To explore the functions of YAP1 in ovarian follicle development via the Hippo pathway, overexpression of YAP1 was performed by the transfection of a pYr-adshuttle-4-YAP1 vector into the GCs. As shown in Figures 6A–6D, the expression levels of FSHR, STAR, CYP11A1, ESR1, and ESR2 mRNA were significantly elevated (P < 0.01). Furthermore, the proliferation ratio of the GCs was notably enhanced as compared to the negative control (P < 0.01). Thus, it indicated that YAP1 exerts a stimulatory effect on ovarian follicle growth and development. However, incredibly, the LATS2 transcript was also remarkably increased through YAP1 overexpression (P < 0.01). It suggested that LATS2 may form a feedback loop with YAP1 (i.e., YAP1-LATS2 loop) in the GCs to inversely enhance the negative regulation of LATS2 on the promotive role of YAP1 in ovary follicle development.
Figure 6.
The YAP1 transduction affects the LATS2-induced inhibition in granulosa cell (GC) proliferation. GCs were transfected using the reconstructed pYr-adshuttle-4-YAP1 vector (OE group), pYr-adshuttle-4 empty vector (negative control), and no plasmid (blank control) for 24 h. (A) The expression of YAP1 mRNA with or without transfection of the pYr-adshuttle-4-YAP1 vector by RT-qPCR. (B) Expression levels of YAP1 protein with or without transfection of the pYr-adshuttle-4-YAP1 vector by western blotting. The β-actin was used as the loading control. (C) Abundances of FSHR, STAR, CYP11A1, ESR1, and ESR2 mRNA expression affected by YAP1 overexpression in the GCs. (D) The effects of YAP1 overexpression on granulosa cell proliferation. Scale bars: 50 μm. (E–J) Knockdown of YAP1 abolished the inhibitions of LATS2 on GC proliferation and the mRNA expression levels of FSHR, STAR, CYP11A1, ESR1, and ESR2. NC, the granulosa cells were transfected with pYr-adshuttle-4 empty vector; OE, the cells were transfected with reconstructed pYr-adshuttle-4-LATS2. For each group, bars with superscript symbol shows significant difference compared to the control group ** P < 0.01, * P < 0.05.
In an attempt to further confirm whether LATS2-YAP1 is an actual pathway in the LATS2-induced inhibitory effect on granulosa cell proliferation, endogenous YAP1 was knocked down by using specific siRNA in the GCs with LATS2 overexpression. As shown in Figures 6E–6J, the knockdown of YAP1 completely abolished the inhibitory effects of LATS2 on GC proliferation (Supplementary Figure S3), and the expression of FSHR, STAR, CYP11A1, ESR1, and ESR2 transcripts (P < 0.01). This result suggested that YAP1 serves as a downstream effector through the LATS2-YAP1 pathway and plays a crucial role in the transduction of the LATS2 effects on GC proliferation, differentiation, steroidogenesis, and follicle selection via the Hippo pathway in the prehierarchical follicles. Therefore, current results provide evidence that LATS2 is involved in ovarian follicle growth and development not only by LATS2-YAP1 transduction but also by YAP1-LATS2 loop feedback in the Hippo/MST signaling pathway, as shown in Figure 7.
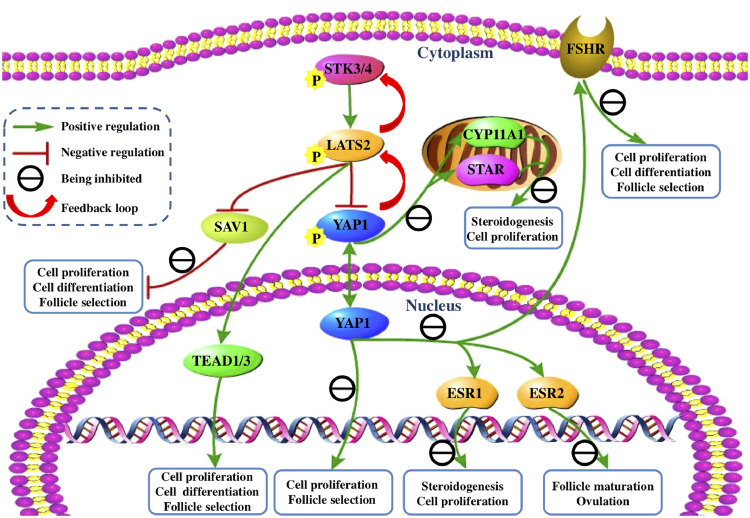
Figure 7.
A schematic representation of the roles and molecular mechanism of LATS2 in the homeostasis-maintaining for granulosa cell proliferation, differentiation, steroidogenesis, and follicle selection via the Hippo/MST signaling pathway.
DISCUSSION
Previous studies have reported that the Hippo/MST pathway's key members coordinate the GC proliferation, GC differentiation, oocyte maturation, follicular atresia, and other physiological processes in the mammalian ovaries (Kawamura et al., 2013; Cheng et al., 2015; Tsoi et al., 2019). This pathway is essential in controlling ovarian cell proliferation for the growth and development of the follicles (Xu et al., 2018b). The uncontrolled GC proliferation and reduced cell differentiation and/or death were substantiated to result in hyperplasia of the granulosa cells layer and GC tumors (Fu et al., 2014). In mammals, LATS2 and its paralogue LATS1 are activated by cell-cell interaction and repress cell proliferation and organ size (Huntoon et al., 2010, Furth and Aylon, 2017). Moreover, the LATS2 and LATS1 also act independently in the Hippo signaling pathway, playing multiple biological roles in cell proliferation, cell migration, apoptosis, transcriptional regulation, and maintaining genetic stability (Tsoi et al., 2019). Our current data revealed that the overexpressed LATS2 directly decremented GC proliferation by downregulating the abundance of FSHR, STAR, and CYP11A1, ESR1, and ESR2 transcript and protein in GCs of prehierarchical follicles of 6 mm up to 8 mm in diameters. During the follicle selection, FSH is required for ovarian follicle maturation and initiation of steroidogenesis in the ovarian GCs (Li and Johnson, 1993). FSHR is fundamental for GC proliferation and differentiation through FSH (Kossowska-Tomaszczuk et al., 2013). In the ovarian follicle growth and development, the increased level of FSHR transcript in the GCs of prehierarchical follicles (6–8 mm in diameter) has been recognized as an indicator to initiate follicle selection, by which one or a limited number of the undifferentiated follicle(s) commence final differentiation and maturation in preparation for ovulation in chicken (Johnson, 2012; Ocón-Grove et al., 2012; Xu et al., 2018a). The STAR and CYP11A1 are the critical regulatory factors involved in the rate-limiting step of ovarian steroidogenesis and promote GC differentiation (Lin et al., 1995; Pollack et al., 1997; Bentsi-Barnes et al., 2010). The latest study has shown that ESR1 mediates the estrogen-induced negative regulation of steroidogenesis and proliferation (Oh et al., 2017), while ESR2 regulates essential genes in GCs required for follicle maturation and ovulation in rats (Khristi et al., 2018). The overexpression of ESR2 triggered the deregulation of genes associated with apoptosis, growth factors, and others leading to endometriosis (Abu-Asab et al., 2011). Based on our present study and previously published reports in other species, LATS2 may play an essential role in controlling the selection of dominant prehierarchical follicles into a differentiation stage of follicular hierarchy and enhancing GC proliferation and steroidogenesis during ovarian development. On the contrary, our data indicated that the GC proliferation index was significantly magnified, in which the expression of genes serving as biomarkers of follicle selection, granulosa cell differentiation, and steroidogenesis (Xu et al., 2018b; Tyasi et al., 2020), including FSHR, StAR, CYP11A1, ESR1, and ESR2 (their functions and biological roles were clarified as aforementioned, respectively), were notably elevated by knockdown of LAST2. The LATS2 knockdown result supports the hypothesis that LATS2 plays a suppressive role in ovarian follicle development, and it may hinder GC proliferation, differentiation, steroidogenesis, and follicle selection.
To further investigate the involvement of LATS2 in ovarian follicle development via the Hippo pathway, expression of the critical components, including STK3, STK4, SAV1, YAP1, TEAD1, and TEAD3 transcripts were analyzed in the GCs with LATS2 overexpression and knockdown. Interestingly, our study provided evidence that LATS2 negatively regulated the expression of SAV1 and YAP1 transcripts but positively regulated STK3, STK4, TEAD1, and TEAD3 transcripts.
It is well-known that YAP1 is the major downstream effector of the Hippo pathway (Strano et al., 2005; Harvey et al., 2013). In our previous study, a relatively higher expression of YAP1 transcript was observed in the ovarian prehierarchical follicles (1 mm up to 8 mm in diameter), and YAP1 protein was mainly expressed in the GCs and oocytes (Zhang et al., 2019). Our current data displays that YAP1 promotes GC proliferation, differentiation, steroidogenesis, and follicle selection by enhancing the expression of FSHR, STAR, CYP11A1, ESR1, and ESR2 transcripts. These results suggest that YAP plays a vital role in initiating cell proliferation and steroidogenesis, as previously reported in human granulosa cell tumors (Fu et al., 2014). However, the expression of YAP1 was inhibited by LATS2. Knocking down of endogenous YAP1 in the GCs with LATS2 overexpression has completely abolished the inhibitory effects of LATS2 on GC proliferation and the expression FSHR, STAR, CYP11A1, ESR1, and ESR2 mRNA that directly affect follicle selection, granulosa cell differentiation, and steroidogenesis. Therefore, the present evidence directly substantiated that the LATS2-YAP1 pathway transduces the inhibitory LATS2 effect on the ovary follicles via the Hippo/MST signaling system. However, unexpectedly, the expression of LATS2 transcript is also positively regulated by its effector YAP1. So, we speculated that the inhibition of LATS2 on the expression of YAP1 might be modulated by a YAP1-LATS2 feedback loop to inversely strengthen the negative regulation of LATS2 on the YAP1 stimulatory roles in ovary follicle development. Although there is little information about this activity's underlying mechanisms, the recent study reported that the YAP1-LATS2 feedback loop forms a homeostatic rheostat for dictating senescence or malignant fate in cultured human ovarian surface epithelial cells (He et al., 2019). Moreover, the latest study reported that knockout of Lats1 and/or Lats2 in murine granulosa cells either in vitro or in vivo has caused the loss of granulosa cell function and expression of genes associated with granulosa cell differentiation, including Wnt4, Fshr, Lhcgr, and Cyp19a1, and concomitantly accompanied with an increase in total YAP levels (Tsoi et al., 2019), which indirectly support our findings.
SAV1, a tumor suppressor in mammals and Drosophila, serves as a core partner of the Ser/Thr protein kinase MSTs (Xiang et al., 2015; Tapon et al., 2020). The previous studies have reported that SAV1 recruits LATS to the MST to increase LATS phosphorylation mediated by MST1 and MST2 (Wu et al., 2003; Chan et al., 2005). Furthermore, our previous work has shown that SAV1 plays a suppressive role in prehierarchical follicles development in hen by inhibiting GC proliferation, differentiation, and follicle selection (Lyu et al., 2016). However, current data show that LATS2 acts to prohibit the inhibitory effects of SAV1 on follicles development. This revelation suggests that LATS2 may also serve as an attenuator to reduce the oversuppression of SAV1 via the Hippo/MST signaling on follicles development. These data conclude that LATS2 plays a role in maintaining the homeostasis in the follicle growth by coordinating the involved positively and negatively regulatory elements of the pathway.
Our previous study in the chicken ovary, along with the data reported in humans and mice, the Hippo/MST pathway consists of the STK3 and STK4 kinases, their cofactor Salvador and LATS1, and LATS2 (Chan et al., 2005; Lyu et al., 2016). The STK3 and STK4 serve as an upstream kinase of the pathway; the phosphorylation levels of LATS2 were induced by the kinase of STK4 or STK3 in vitro (Lyu et al., 2016). The phosphorylated LATS then phosphorylates and inactivates the downstream transcriptional coactivator YAP1, which is responsible for expressing multiple apoptosis-related genes (Zhao et al., 2007; Li et al., 2012). However, the present result reveals that LATS2 overexpression upregulates the STK4 and STK3 mRNA transcription, which indicated that the LATS2-STK3/4 feedback loop might inversely modulate the activities of STK3 and STK4. Thus, LATS2 may play a pivotal role in this pathway by providing feedback to its upstream kinases STK3/4 to attenuate or enhance the downstream effectors in maintaining the homeostasis with the Hippo/MST family members implicated in the cell process. Additionally, our present study showed that TEAD1 and TEAD3 promoted GC proliferation and enhanced mRNA expression levels of FSHR, STAR, CYP11A1, ESR1, and ESR2. However, the LATS2 overexpression resulted in the significantly enhanced expression levels of TEAD1 and TEAD3 transcript. Therefore, TEAD1 and TEAD3 as the potential effectors to mediate the suppression of LATS2 on the ovarian follicles have been excluded. On the other hand, this result also provides additional evidence that LATS2 serves as a negative regulator and may act as a positive modulator in ovarian follicle development.
CONCLUSIONS
In summary, the present study initially revealed the pivotal role and regulatory mechanism of LATS2 in the ovarian prehierarchical follicle development by regulating GC proliferation, differentiation, steroidogenesis, and follicle selection via the Hippo/MST signaling pathway. Then, unexpectedly, our study provides evidence that STK3/4-LATS2-YAP1 not only acts as a suppressor of cell proliferation and follicle selection but also LATS2 may serve as an enhancer in cell proliferation and follicle selection by the YAP1-LATS2 and the LATS2-STK3/4 feedback loops through promoting the TEAD1/3 but inhibiting the SAV1 transcripts in the follicle development of hen ovary (Figure 7).
Acknowledgments
ACKNOWLEDGMENTS
This work was supported by grants from the National Natural Science Foundation of China (Grant No.31672407 and 31902145); the Project of Science and Technology Development Plan of Jilin Province (Grant No. 20170101019JC); the Project of Education Development Plan of Jilin Province (Grant No. JJKH20180646KJ); and the China Agriculture Research System of MOF and MARA (Grant No. CARS-41-Z03)
Authors' contributions: XS and XTN contributed equally to this manuscript. XS and XTN mainly performed the experiments; NQ, XSS, JHZ and CM analyzed the data, and helped perform some experiments; RFX conceived, designed the experiments, and wrote the manuscript; BM designed the experiments and revised the manuscript. All authors read and approved the final manuscript.
DISCLOSURES
The authors declare that they have no competing interests.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.psj.2021.101454.
Appendix. Supplementary materials
Supplementary Figure S1. Overexpression of TEAD1 and TEAD3 in the GCs. (A) The expression of TEAD1 with or without transfection of the pYr-adshuttle-4-TEAD1 vector for 24 h was evaluated by RT-qPCR. OE, the GCs were transfected with pYr-adshuttle-4-TEAD1 vector; BC, no transfection; NC, transfected with pYr-adshuttle-4 empty vector. (B) Expression of TEAD1 protein with or without transfection of the pYr-adshuttle-4-TEAD1 vector was examined by western blotting. (C) The expression of TEAD3 with or without transfection of the pYr-adshuttle-4-TEAD3 vector for 24 h was evaluated by RT-qPCR. OE, the GCs were transfected with pYr-adshuttle-4-TEAD3 vector; BC, no transfection; NC, transfected with pYr-adshuttle-4 empty vector. (D) Expression of TEAD3 protein with or without transfection of the pYr-adshuttle-4-TEAD3 vector was examined by western blotting. The β-actin was utilized as the loading control.
Supplementary Figure S2. Knockdowns of TEAD1 and TEAD3 in the GCs. (A) The TEAD1 mRNA expression in the GCs was analyzed using RT-qPCR. GCs were transfected with TEAD1-specific siRNA (SR group), scrambled siRNA (negative control, NC), and no siRNA as a vehicle (blank control, BC). (B) Expression of TEAD1 protein in the GCs with or without interference using the siRNA was analyzed by western blotting. (C) The TEAD3 mRNA expression in the GCs was analyzed using RT-qPCR. GCs were transfected with TEAD3-specific siRNA (SR group), scrambled siRNA (negative control, NC), and no siRNA as a vehicle (blank control, BC). (D) Expression of TEAD3 protein in the GCs with or without interference using the siRNA was analyzed by western blotting. The β-actin was used as the loading control.
Supplementary Figure S3. Effects of YAP1 knockdown on cell proliferation in the GCs with the LATS2 overexpression. (A) Effects of LATS2 overexpression on GC proliferation. NC, the granulosa cells were transfected with pYr-adshuttle-4 empty vector; OE, the cells were transfected with reconstructed pYr-adshuttle-4-LATS2. (B) Effects of YAP1 knockdown on cell proliferation in the GCs with the LATS2 overexpression. NC, the granulosa cells were transfected with pYr-adshuttle-4 empty vector, but no transfection of YAP1 specific siRNA; OE, the cells were transfected with reconstructed pYr-adshuttle-4-LATS2 in the GCs with transfection of YAP1 specific siRNA. Scale bars, 50 μm.
REFERENCES
- Abu-Asab M., Zhang M., Amini D., Abu-Asab N., Amri H. Endometriosis gene expression heterogeneity and biosignature: a phylogenetic analysis. Obstet. Gynecol. Int. 2011;2011 doi: 10.1155/2011/719059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An Y., Kang Q., Zhao Y., Hu X., Li N. Lats2 modulates adipocyte proliferation and differentiation via hippo signaling. PLoS One. 2013;8:e72042. doi: 10.1371/journal.pone.0072042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsi-Barnes I.K., Kuo F.T., Barlow G.M., Pisarska M.D. Human forkhead L2 represses key genes in granulosa cell differentiation including aromatase, P450scc, and cyclin D2. Fertil. Steril. 2010;94:353–356. doi: 10.1016/j.fertnstert.2009.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo F.D., Gokhale S., Johnnidis J.B., Fu D., Bell G.W., Jaenisch R., Brummelkamp T.R. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr. Biol. 2007;17:2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Chan E.H., Nousiainen M., Chalamalasetty R.B., Schäfer A., Nigg E.A., Silljé H.H. The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene. 2005;24:2076–2086. doi: 10.1038/sj.onc.1208445. [DOI] [PubMed] [Google Scholar]
- Cheng Y., Feng Y., Jansson L., Sato Y., Deguchi M., Kawamura K., Hsueh A.J. Actin polymerization-enhancing drugs promote ovarian follicle growth mediated by the Hippo signaling effector YAP. FASEB J. 2015;29:2423–2430. doi: 10.1096/fj.14-267856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., Feldmann G., Huang J., Wu S., Zhang N., Comerford S.A., Gayyed M.F., Anders R.A., Maitra A., Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu D., Lv X., Hua G., He C., Dong J., Lele S.M., Li D.W., Zhai Q., Davis J.S., Wang C. YAP regulates cell proliferation, migration, and steroidogenesis in adult granulosa cell tumors. Endocr. Relat. Cancer. 2014;21:297–310. doi: 10.1530/ERC-13-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furth N., Aylon Y. The LATS1 and LATS2 tumor suppressors: beyond the Hippo pathway. Cell Death Differ. 2017;24:1488–1501. doi: 10.1038/cdd.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist R.B., Ritter L.J., Myllymaa S., Kaivo-Oja N., Dragovic R.A., Hickey T.E., Ritvos O., Mottershead D.G. Molecular basis of oocyte-paracrine signalling that promotes granulosa cell proliferation. J. Cell Sci. 2006;119:3811–3821. doi: 10.1242/jcs.03105. [DOI] [PubMed] [Google Scholar]
- Harvey K.F., Zhang X., Thomas D.M. The Hippo pathway and human cancer. Nat. Rev. Cancer. 2013;13:246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- He C., Lv X., Huang C., Hua G., Ma B., Chen X., Angeletti P.C., Dong J., Zhou J., Wang Z., Rueda B.R., Davis J.S., Wang C. YAP1-LATS2 feedback loop dictates senescent or malignant cell fate to maintain tissue homeostasis. EMBO Rep. 2019;20:e44948. doi: 10.15252/embr.201744948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoa L., Kulaberoglu Y., Gundogdu R., Cook D., Mavis M., Gomez M., Gomez V., Hergovich A. The characterisation of LATS2 kinase regulation in Hippo-YAP signalling. Cell Signal. 2016;28:488–497. doi: 10.1016/j.cellsig.2016.02.012. [DOI] [PubMed] [Google Scholar]
- Huntoon C.J., Nye M.D., Geng L., Peterson K.L., Flatten K.S., Haluska P., Kaufmann S.H., Karnitz L.M. Heat shock protein 90 inhibition depletes LATS1 and LATS2, two regulators of the mammalian hippo tumor suppressor pathway. Cancer Res. 2010;70:8642–8650. doi: 10.1158/0008-5472.CAN-10-1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A.L., Bridgham J.T., Witty J.P., Tilly J.L. Susceptibility of avian ovarian granulosa cells to apoptosis is dependent upon stage of follicle development and is related to endogenous levels of bcl-xlong gene expression. Endocrinology. 1996;137:2059–2066. doi: 10.1210/endo.137.5.8612548. [DOI] [PubMed] [Google Scholar]
- Johnson A.L., Solovieva E.V., Bridgham J.T. Relationship between steroidogenic acute regulatory protein expression and progesterone production in hen granulosa cells during follicle development. Biol. Reprod. 2002;67:1313–1320. doi: 10.1095/biolreprod67.4.1313. [DOI] [PubMed] [Google Scholar]
- Johnson A.L., Woods D.C. Dynamics of avian ovarian follicle development: cellular mechanisms of granulosa cell differentiation. Gen. Comp. Endocrinol. 2009;163:12–17. doi: 10.1016/j.ygcen.2008.11.012. [DOI] [PubMed] [Google Scholar]
- Johnson A.L. Ovarian follicle selection and granulosa cell differentiation. Poult. Sci. 2015;94:781–785. doi: 10.3382/ps/peu008. [DOI] [PubMed] [Google Scholar]
- Johnson P.A. Follicle selection in the avian ovary. Reprod. Domest. Anim. 2012;47:283–287. doi: 10.1111/j.1439-0531.2012.02087.x. [DOI] [PubMed] [Google Scholar]
- Kawamura K., Cheng Y., Suzuki N., Deguchi M., Sato Y., Takae S., Ho C.H., Kawamura N., Tamura M., Hashimoto S., Sugishita Y., Morimoto Y., Hosoi Y., Yoshioka N., Ishizuka B., Hsueh A.J. Hippo signaling disruption and Akt stimulation of ovarian follicles for infertility treatment. Proc. Natl. Acad. Sci. U. S. A. 2013;110:17474–17479. doi: 10.1073/pnas.1312830110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khristi V., Chakravarthi V.P., Singh P., Ghosh S., Pramanik A., Ratri A., Borosha S., Roby K.F., Wolfe M.W., Rumi M.A.K. ESR2 regulates granulosa cell genes essential for follicle maturation and ovulation. Mol. Cell Endocrinol. 2018;474:214–226. doi: 10.1016/j.mce.2018.03.012. [DOI] [PubMed] [Google Scholar]
- Kim M.H., Seo D.S., Ko Y. Relationship between egg productivity and insulin-like growth factor-I genotypes in Korean native Ogol chickens. Poult. Sci. 2004;83:1203–1208. doi: 10.1093/ps/83.7.1203. [DOI] [PubMed] [Google Scholar]
- Kossowska-Tomaszczuk K., De Geyter C. Cells with stem cell characteristics in somatic compartments of the ovary. Biomed. Res. Int. 2013;2013 doi: 10.1155/2013/310859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Zhao H., Zhao X., Zhang B., Cui L., Shi Y., Li G., Wang P., Chen Z.J. Identification of YAP1 as a novel susceptibility gene for polycystic ovary syndrome. J. Med. Genet. 2012;49:254–257. doi: 10.1136/jmedgenet-2011-100727. [DOI] [PubMed] [Google Scholar]
- Li Z., Johnson A.L. Regulation of P450 cholesterol side-chain cleavage messenger ribonucleic acid expression and progesterone production in hen granulosa cells. Biol. Reprod. 1993;49:463–469. doi: 10.1095/biolreprod49.3.463. [DOI] [PubMed] [Google Scholar]
- Lin D., Sugawara T., 3rd S.J.F., Clark B.J., Stocco D.M., Saenger P., Rogol A., Miller W.L. Role of steroidogenic acute regulatory protein in adrenal and gonadal steroidogenesis. Science. 1995;267:1828–1831. doi: 10.1126/science.7892608. [DOI] [PubMed] [Google Scholar]
- Lyu Z., Qin N., Tyasi T.L., Zhu H., Liu D., Yuan S., Xu R. The Hippo/MST pathway member SAV1 plays a suppressive role in development of the prehierarchical follicles in hen ovary. PLoS One. 2016;11 doi: 10.1371/journal.pone.0160896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocón-Grove O.M., Poole D.H., Johnson A.L. Bone morphogenetic protein 6 promotes FSH receptor and anti-Müllerian hormone mRNA expression in granulosa cells from hen prehierarchal follicles. Reproduction. 2012;143:825–833. doi: 10.1530/REP-11-0271. [DOI] [PubMed] [Google Scholar]
- Oh Y.S., Koh I.K., Choi B., Gye M.C. ESR1 inhibits hCG-induced steroidogenesis and proliferation of progenitor Leydig cells in mice. Sci. Rep. 2017;7:43459. doi: 10.1038/srep43459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percie du Sert N., Hurst V., Ahluwalia A., Alam S., Avey M.T., Baker M., Browne W.J., Clark A., Cuthill I.C., Dirnagl U., Emerson M., Garner P., Holgate S.T., Howells D.W., Karp N.A., Lazic S.E., Lidster K., MacCallum C.J., Macleod M., Pearl E.J., Petersen O.H., Rawle F., Reynolds P., Rooney K., Sena E.S., Silberberg S.D., Steckler T., Würbel H. The ARRIVE guidelines 2.0: updated guidelines for reporting animal research. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack S.E., Furth E.E., Kallen C.B., Arakane F., Kiriakidou M., Kozarsky K.F., Strauss J.F., 3rd Localization of the steroidogenic acute regulatory protein in human tissues. J. Clin. Endocrinol. Metab. 1997;82:4243–4251. doi: 10.1210/jcem.82.12.4445. [DOI] [PubMed] [Google Scholar]
- Qin N., Liu Q., Zhang Y.Y., Fan X.C., Xu X.X., Lv Z.C., Wei M.L., Jing Y., Mu F., Xu R.F. Association of novel polymorphisms of forkhead box L2 and growth differentiation factor-9 genes with egg production traits in local Chinese Dagu hens. Poult. Sci. 2015;94:88–95. doi: 10.3382/ps/peu023. [DOI] [PubMed] [Google Scholar]
- Qin N., Tyasi T.L., Sun X., Chen X., Zhu H., Zhao J., Xu R. Determination of the roles of GREM1 gene in granulosa cell proliferation and steroidogenesis of hen ovarian prehierarchical follicles. Theriogenology. 2020;151:28–40. doi: 10.1016/j.theriogenology.2020.03.030. [DOI] [PubMed] [Google Scholar]
- Son W.Y., Das M., Shalom-Paz E., Holzer H. Mechanisms of follicle selection and development. Minerva Ginecol. 2011;63:89–102. [PubMed] [Google Scholar]
- Strano S., Monti O., Pediconi N., Baccarini A., Fontemaggi G., Lapi E., Mantovani F., Damalas A., Citro G., Sacchi A., Del Sal G., Levrero M., Blandino G. The transcriptional co-activator Yes-associated protein drives p73 gene-target specificity in response to DNA damage. Mol. Cell. 2005;18:447–459. doi: 10.1016/j.molcel.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Tapon N., Harvey K.F., Bell D.W., Wahrer D.C., Schiripo T.A., Haber D., Hariharan I.K. Salvador promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2020;110:467–478. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- Tsoi M., Morin M., Rico C., Johnson R.L., Paquet M., Gévry N., Boerboom D. Lats1 and Lats2 are required for ovarian granulosa cell fate maintenance. FASEB J. 2019;33:10819–10832. doi: 10.1096/fj.201900609R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyasi T.L., Sun X., Shan X., Liswaniso S., Chimbaka I.M., Qin N., Xu R. Effects of RAC1 on proliferation of hen ovarian prehierarchical follicle granulosa cells. Animals (Basel). 2020;10:1589. doi: 10.3390/ani10091589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev A., Kaneko K.J., Shu H., Zhao Y., DePamphilis M.L. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 2001;15:1229–1241. doi: 10.1101/gad.888601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Degerny C., Xu M., Yang X.J. YAP, TAZ, and Yorkie: a conserved family of signal-responsive transcriptional coregulators in animal development and human disease. Biochem. Cell Biol. 2009;87:77–91. doi: 10.1139/O08-114. [DOI] [PubMed] [Google Scholar]
- Woodruff T.K., Shea L.D. The role of the extracellular matrix in ovarian follicle development. Reprod. Sci. 2007;14:6–10. doi: 10.1177/1933719107309818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Huang J., Dong J., Pan D. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114:445–456. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- Xiang C., Li J., Hu L., Huang J., Luo T., Zhong Z., Zheng Y., Zheng L. Hippo signaling pathway reveals a spatio-temporal correlation with the size of primordial follicle pool in mice. Cell Physiol. Biochem. 2015;35:957–968. doi: 10.1159/000369752. [DOI] [PubMed] [Google Scholar]
- Xu R., Qin N., Xu X., Sun X., Chen X., Zhao J. Inhibitory effect of SLIT2 on granulosa cell proliferation mediated by the CDC42-PAKs-ERK1/2 MAPK pathway in the prehierarchical follicles of the chicken ovary. Sci. Rep. 2018;8:9168. doi: 10.1038/s41598-018-27601-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R., Qin N., Xu X., Sun X., Chen X., Zhao J. Implication of SLIT3-ROBO1/ROBO2 in granulosa cell proliferation, differentiation and follicle selection in the prehierarchical follicles of hen ovary. Cell Biol. Int. 2018;42:1643–1657. doi: 10.1002/cbin.11063. [DOI] [PubMed] [Google Scholar]
- Ye H., Li X., Zheng T., Hu C., Pan Z., Huang J., Li J., Li W., Zheng Y. The Hippo signaling pathway regulates ovarian function via the proliferation of ovarian germline stem cells. Cell Physiol. Biochem. 2017;41:1051–1062. doi: 10.1159/000464113. [DOI] [PubMed] [Google Scholar]
- Zhang F., Qin N., Sun X., Niu X., Liu D., Xu R. Analysis of spatio-temporal expression of YAP1 gene in the developing follicles of chicken ovaries. J. Jilin Agric. Univ. 2019;41:71–76. [Google Scholar]
- Zhao B., Lei Q.Y., Guan K.L. The Hippo-YAP pathway: new connections between regulation of organ size and cancer. Curr. Opin. Cell Biol. 2008;20:638–646. doi: 10.1016/j.ceb.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Wei X., Li W., Udan R.S., Yang Q., Kim J., Xie J., Ikenoue T., Yu J., Li L., Zheng P., Ye K., Chinnaiyan A., Halder G., Lai Z.C., Guan K.L. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Ye X., Yu J., Li L., Li W., Li S., Yu J., Lin J.D., Wang C.Y., Chinnaiyan A.M., Lai Z.C., Guan K.L. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. Overexpression of TEAD1 and TEAD3 in the GCs. (A) The expression of TEAD1 with or without transfection of the pYr-adshuttle-4-TEAD1 vector for 24 h was evaluated by RT-qPCR. OE, the GCs were transfected with pYr-adshuttle-4-TEAD1 vector; BC, no transfection; NC, transfected with pYr-adshuttle-4 empty vector. (B) Expression of TEAD1 protein with or without transfection of the pYr-adshuttle-4-TEAD1 vector was examined by western blotting. (C) The expression of TEAD3 with or without transfection of the pYr-adshuttle-4-TEAD3 vector for 24 h was evaluated by RT-qPCR. OE, the GCs were transfected with pYr-adshuttle-4-TEAD3 vector; BC, no transfection; NC, transfected with pYr-adshuttle-4 empty vector. (D) Expression of TEAD3 protein with or without transfection of the pYr-adshuttle-4-TEAD3 vector was examined by western blotting. The β-actin was utilized as the loading control.
Supplementary Figure S2. Knockdowns of TEAD1 and TEAD3 in the GCs. (A) The TEAD1 mRNA expression in the GCs was analyzed using RT-qPCR. GCs were transfected with TEAD1-specific siRNA (SR group), scrambled siRNA (negative control, NC), and no siRNA as a vehicle (blank control, BC). (B) Expression of TEAD1 protein in the GCs with or without interference using the siRNA was analyzed by western blotting. (C) The TEAD3 mRNA expression in the GCs was analyzed using RT-qPCR. GCs were transfected with TEAD3-specific siRNA (SR group), scrambled siRNA (negative control, NC), and no siRNA as a vehicle (blank control, BC). (D) Expression of TEAD3 protein in the GCs with or without interference using the siRNA was analyzed by western blotting. The β-actin was used as the loading control.
Supplementary Figure S3. Effects of YAP1 knockdown on cell proliferation in the GCs with the LATS2 overexpression. (A) Effects of LATS2 overexpression on GC proliferation. NC, the granulosa cells were transfected with pYr-adshuttle-4 empty vector; OE, the cells were transfected with reconstructed pYr-adshuttle-4-LATS2. (B) Effects of YAP1 knockdown on cell proliferation in the GCs with the LATS2 overexpression. NC, the granulosa cells were transfected with pYr-adshuttle-4 empty vector, but no transfection of YAP1 specific siRNA; OE, the cells were transfected with reconstructed pYr-adshuttle-4-LATS2 in the GCs with transfection of YAP1 specific siRNA. Scale bars, 50 μm.