Abstract
We report on the influence of ~22 million variants on 731 immune cell traits in a cohort of 3,757 Sardinians. We detected 122 significant (P < 1.28 × 10−11) independent association signals for 459 cell traits at 70 loci (53 of them novel) identifying several molecules and mechanisms involved in cell regulation. Furthermore, 53 signals at 36 loci overlapped with previously reported disease-associated signals, predominantly for autoimmune disorders, highlighting intermediate phenotypes in pathogenesis. Collectively, our findings illustrate complex genetic regulation of immune cells with highly selective effects on autoimmune disease risk at the cell-subtype level. These results identify drug-targetable pathways informing the design of more specific treatments for autoimmune diseases.
Assessing the impact of natural genetic variation on quantitative and discrete immune-related traits provides a powerful route to better understand immune system function and dysfunction.
We thus previously performed a genome-wide association study (GWAS) on 272 blood immune-cell-related traits profiled by flow cytometry in 1,629 general population individuals from the founder population of Sardinia1. We found significant association signals at 13 loci, 3 of them coinciding with autoimmune disease risk (‘coincident associations’), revealing possible contributions from these loci to disease pathophysiology. The approach differed from comparisons of immune cell traits in case–control cohorts, which can be influenced by disease process and therapy.
Using a similar approach, four subsequent GWASs analyzed up to 1,000 individuals and detected 28 additional loci associated with immune cell traits2–5. All of these studies provided mechanistic clues about the genetic regulation of immune cells but had some limitations, including relatively small sample sizes and number of cell traits tested by GWAS, and only partial coverage of genetic variation. Furthermore, our previous analysis1 and another study3 did not consider median fluorescence intensities (MFIs) reflecting the levels of cell surface antigens, and a third study considered proportions of cells but not absolute cell counts2.
Interpretation of the coincident associations detected thus far has been hampered by the observation that a variant associated with a clinical outcome often influences multiple immune phenotypes (‘pleiotropy’), and it is not always clear which specific immune phenotype is truly implicated in the disease process. Moreover, the true disease-related immune phenotype may not even have been measured in previous studies, supporting the need to analyze by GWAS even larger numbers of immune traits. This could lead to the identification of multiple independent genetic associations with effects on the same pairs of immune trait and disease risk (‘multiple coincident associations’), giving further support to a possible causal relationship between levels of that immune cell type and that particular disease.
To overcome previous limitations, we have performed a new GWAS with almost three times the number of immune cell traits and more than double the number of individuals previously studied. We have also improved considerably the information content of the genetic map, interrogating ~22 million genetic variants derived from our population-specific whole-genome sequencing effort6. The enhanced power of the study increased considerably the number of reported associations with immune cell traits and coincident associations with disease risk, revealing numerous potential druggable protein targets for autoimmune diseases.
Results
Heritability and genetic associations of immune cell traits.
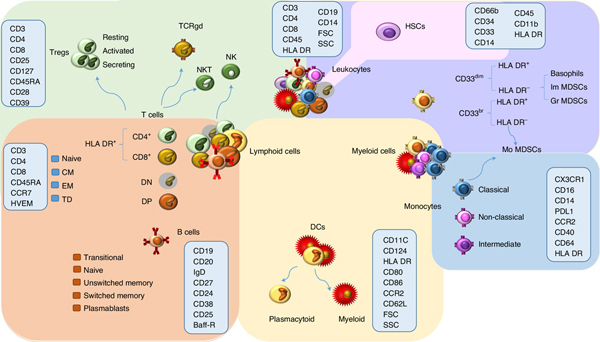
We profiled by flow cytometry 539 immune traits, including 118 absolute cell counts, 389 MFIs of surface antigens and 32 morphological parameters. In addition, we considered 192 relative counts (ratios between cell levels) for a total of 731 cell traits assessed in a general population cohort of 3,757 Sardinians (Fig. 1, Extended Data Figs. 1–6, Supplementary Table 1 and Supplementary Information).
Fig. 1 |. Schematic representation of the main leukocyte subsets assessed by GWAS.
The background colors indicate different cell groupings: T-cell populations are indicated in green, B cells in orange, DCs in yellow, monocytes in blue, other myeloid cells in violet, and hematopoietic stem cells in pink. The markers assessed for MFI are indicated within the light blue rectangles. TCRgd, gamma delta T cells; NK, natural killer cells; NKT, NK T cells; DN, CD4− CD8− T cells; DP, CD4+ CD8+ T cells; CM, central memory; EM, effector memory; HSCs, hematopoietic stem cells; Im MDSCs, immature myeloid-derived suppressor cells; Gr MDSCs, granulocytic MDSCs; Mo MDSCs, monocytic MDSCs; FSC, forward scatter; SSC, side scatter.
Using our unique family-based cohort of individuals with varying degrees of genetic relatedness (Methods), we first provided robust estimates of the proportion of phenotypic variation in immune traits attributable to inherited variation (‘heritability’). Narrow-sense heritability of the assessed traits—which captures the proportion of phenotypic variation due to additive genetic effects—had a median value of 37.0% (Supplementary Information and Supplementary Table 2).
Focusing on cell counts, lymphoid cells were more heritable than myeloid cells (median values, 37.7% versus 32.3%, respectively), which roughly overlap with higher heritability of adaptive immunity compared to innate immunity (37.7% versus 32.6%, respectively). Among lymphoid cell counts, CD4 T cells were the most heritable followed by B-cell and CD8 T-cell traits (42.6%; 38.9%; 33.8%, respectively). The maturation stages of T- and B-cell counts had a clearer heritability pattern: naive cells were more heritable than memory cells (naive T: 47.0% versus memory T: 37.1%; naive B: 44.7% versus memory B: 38.9%). This pattern is most likely due to the fact that memory cells are more strongly influenced by previous encounters with environmental exposures. In fact, the terminally differentiated (TD) T cells, which are the last maturation state of memory T cells, were among the least heritable T-cell subsets (29.3% for TD CD4 and 30.6% for TD CD8 T cells).
From these findings, we inferred that the cell types with the highest heritability, which roughly correspond to adaptive immunity, are those with more sophisticated functions—and are also the last to appear during ontogenesis and after hematopoietic stem cell transplantation7,8.
Overall, our results are in line with those obtained in previous studies on immune traits9, in which, although the importance of environmental exposure in immune system modulation was emphasized, a mean heritability comparable to ours was obtained.
To identify the genetic variation accounting for the inherited component of the 731 immunophenotypes, we next performed a GWAS, testing 20,143,392 SNPs and 1,688,858 indels, either genotyped with high-density arrays or imputed through our Sardinian sequence-based reference panel of 3,514 individuals6.
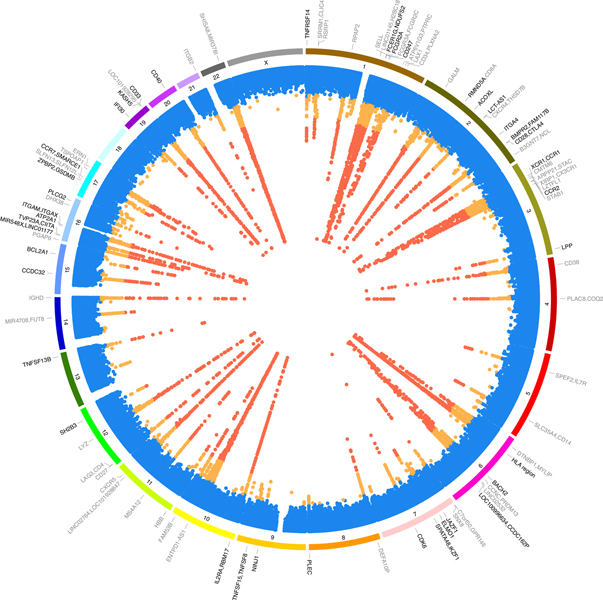
At the genome-wide significant threshold of P < 1.28 × 10−11—based on our empirical genome-wide P value (P = 6.9 × 10−9)6 corrected for 539 independent traits tested (Methods)—we found 122 independent associations (111 outside the HLA region). These signals were localized at 70 loci, 53 of them novel, and resulted in a total of 763 trait–variant associations (Fig. 2). Of the 122 association signals, 49 (40%) were purely cis (locally acting) associations, 57 (47%) were trans (acting at distant sites), and 16 (13%) were both cis and trans.
Fig. 2 |. Genetic associations of the immune traits assessed.
The innermost track shows the lowest association P value among all traits for each locus. Significant variants (P < 1.28 × 10−11) are in red, suggestive variants (P value ranging from 5 × 10−8 to 1.28 × 10−11) in orange, and non-significant variants in blue. The middle track shows the chromosome number. The outermost track indicates the genes nearest to signals with P < 1.28 × 10−11. Genes showing coincident genetic association with diseases are shown in bold font. The coincident associated genes are indicated even if in some instances their P values range from 5 × 10−8 to 1.28 × 10−11. The summary statistics were obtained using a linear mixed association model. The significance threshold was calculated by applying a Bonferroni correction to the empirical significance threshold (P = 6.9 × 10−9)6 considering 539 independent tests.
At a more relaxed standard GWAS threshold of P < 5 × 10−8, we identified an additional 198 independent associations at 154 loci (152 of them novel; Supplementary Table 3A–C).
Identification of candidate causal variants and genes.
To identify candidate causal variants driving the observed association profiles, we determined for each association signal the so-called ‘credible set’, or the minimum set of variants with a 95% summed posterior probability of including the causal variant (Supplementary Table 3D,E)10.
Among the 24 signals with only 1 likely causal variant in their credible set, 6 were driven by protein-altering variants (25%), 8 by variants within regulatory regions, and the remaining 10 were variants without an obvious functional impact (synonymous, intronic or intergenic), although 8 of these were annotated as molecular quantitative trait loci (QTLs) in the LinDA QTL Catalog (http://linda.irgb.cnr.it/, Methods and Supplementary Table 4A).
Using a combination of criteria (Methods), we also detected candidate causal genes for the 122 independent signals. Importantly, 83 of 122 signals overlapped (linkage disequilibrium (LD) with r2 > 0.7 between lead variants) with expression QTLs (eQTLs), which overall were significantly enriched among our signals (odds ratio (OR) = 16.54, P = 1.88 × 10−42, Fisher’s exact test). Likewise, 64 of the 198 association signals with 1.28 × 10−11 < P < 5 × 10−8 (suggestive set) showed significant enrichment for eQTLs (OR = 4.87, P = 1.39 × 10−19), consistent with a number of these signals representing genuine associations (Supplementary Table 4B,C).
Coincident associations with disease risk.
GWASs performed thus far have identified thousands of signals associated with immune-related diseases, but the underlying mechanisms and the specific immune cell subtypes involved remain largely unknown or speculative. To overcome this gap in knowledge, a useful approach is to identify overlaps (‘coincident associations’) between disease risk and blood immune-cell-level association signals, pointing to intermediate quantitative phenotypes that bridge the mechanistic lacunae between genetic variation and disease endpoints1,11.
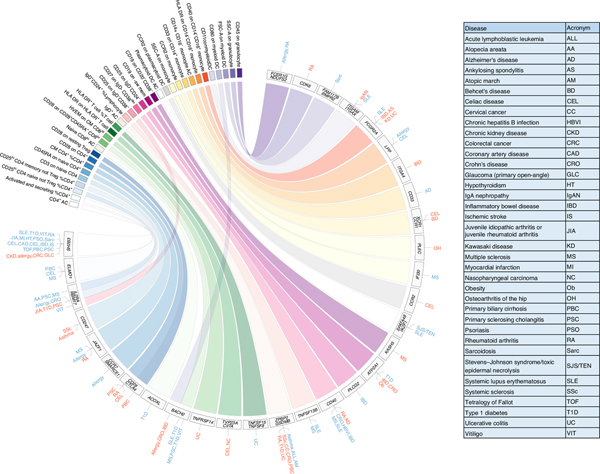
We found that 53 independent association signals with immune cell traits at 36 loci (of which 27 are newly identified) overlapped with reported GWAS associations with disease risk (lead variants with r2 > 0.7; Fig. 3, Supplementary Fig. 1 and Supplementary Table 5A). The association profiles were also assessed for colocalization (posterior probability > 0.8) with disease GWAS with available genome-wide summary statistics (Methods and Supplementary Table 5B).
Fig. 3 |. Coincident genetic association between immune traits and diseases.
The circular plot shows the top immune traits in colored rectangles and the genes nearest the associated signals in gray rectangles. The pathologies linked to the immune traits via coincident genetic associations (P < 5.0 × 10−8) are listed radially to the associated gene. The pathologies for which the protective allele correlates with a reduction of an immune trait level are in blue, whereas those for which the protective allele correlates with an increase are in red. The HLA region is not included. The pathology acronyms are detailed in the table. The summary statistics for the immune traits were obtained using a linear mixed association model.
Furthermore, some of the associations with immune traits we report here were driven by genetic variants far more common in Sardinia than elsewhere and may thus have been missed by disease GWASs in other populations. For instance, among the 122 independent significant signals, 16 (not showing overlap with disease in published GWASs) have a low frequency in Europeans from the 1000 Genome Project (<2%) but at least twofold higher frequency in Sardinians (Supplementary Table 3A,F and Supplementary Information).
Among the 53 coincident association signals, 32 (60.4%) act in trans, 16 (30.2%) in cis, and 5 (9.4%) both in cis and trans (Supplementary Table 5A). Overlaps include signals involved in the risk of one or, more frequently, multiple diseases, and the direction of the effects on risk can be opposite for different diseases. Furthermore, in some instances, multiple independent coincident association signals coherently pointed to the same cell trait and disease, further supporting the causal involvement of that cell trait in that disease (Table 1 and Supplementary Table 6A).
Table 1 |.
Multiple coincident associations between immune traits and diseases
| Immune trait association | Disease association | Correlation bw trait and disease | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Trait name | Trait rsID | Derived/ancestral allele | Derived allele effect | s.e. | P value | Disease | Disease rsID | Effect/other allele | OR | P value | Reference | r 2 | Derived allele/corresponding allele in disease | Effects of derived allele/corresponding allele in disease |
| FCGR2A | CD27 on IgD−CD38dim | rs1801274 | G/A | −0.204 | 0.03 | 1.14 × 10−14 | CRO | rs1801274 | G/A | 0.93 | 1.53 × 10−7 | Ref. 17 | 1.00 | G/G | −/− |
| IBD | 0.89 | 2.00 × 10−38 | |||||||||||||
| CD40 | rs1883832 | T/C | 0.640 | 0.03 | 6.21 × 10−147 | CRO | rs6074022 | C/T | 1.10 | 3.00 × 10−12 | Ref. 33 | 0.98 | T/C | +/+ | |
| IBD | 1.08 | 8.00 × 10−11 | |||||||||||||
| FCGR2A | CD27 on sw mem | rs1801274 | G/A | −0.293 | 0.03 | 2.48 × 10−28 | CRO | rs1801274 | G/A | 0.93 | 1.53 × 10−7 | Ref. 17 | 1.00 | G/G | −/− |
| IBD | 0.89 | 2.00 × 10−38 | |||||||||||||
| CD40 | rs1883832 | T/C | 0.585 | 0.03 | 2.53 × 10−118 | CRO | rs6074022 | C/T | 1.10 | 3.00 × 10−12 | Ref. 33 | 0.98 | T/C | +/+ | |
| IBD | 1.08 | 8.00 × 10−11 | |||||||||||||
| CD28, CTLA4 | CD28 on CD4+ | rs1973872 | T/G | −0.386 | 0.03 | 2.24 × 10−42 | MS | rs6435203 | G/A | 0.93 | 1.00 × 10−7 | Ref. 16 | 0.83 | T/G | −/− |
| BACH2 | rs72928038 | A/G | 0.247 | 0.03 | 4.43 × 10−13 | rs72928038 | A/G | 1.14 | 1.50 × 10−15 | 1.00 | A/A | +/+ | |||
| BACH2 | CD28 on CD28+CD45RA+CD8br | rs619192 | C/T | −0.199 | 0.03 | 2.97 × 10−10 | T1D | rs597325 | A/G | 0.85 | 3.38 × 10−10 | Ref. 37 | 0.98 | C/A | −/− |
| rs72928038 | A/G | 0.589 | 0.03 | 5.13 × 10−67 | rs72928038 | A/G | 1.20 | 6.40 × 10−14 | Ref. 23 | 1.00 | A/A | +/+ | |||
| FCGR2A | CD64 on CD14+CD16+ monocyte | rs1801274 | G/A | 0.383 | 0.03 | 1.86 × 10−48 | SLE | rs1801274 | G/A | 1.16 | 1.04 × 10−12 | Ref. 52 | 1.00 | G/G | +/+ |
| ATXN2, BRAP | rs11065979 | T/C | 0.150 | 0.03 | 6.46 × 10−9 | rs653178 | C/T | 1.14 | 7.00 × 10−9 | Ref. 15 | 0.97 | T/C | +/+ | ||
| TNFSF15, TNFSF8 | HLA DR+ T cell %T cell | rs7032773 | T/C | −0.147 | 0.03 | 1.46 × 10−9 | UC | rs726657 | T/C | 0.89 | 2.26 × 10−14 | Ref. 22 | 0.91 | T/T | −/− |
| CIITA | rs3087456 | A/G | −0.461 | 0.03 | 3.08 × 10−70 | rs4781011 | G/T | 0.87 | 3.00 × 10−6 | Ref. 53 | 0.94 | A/G | −/− | ||
| LCT-AS1 | pDC AC | rs2164210 | C/T | 0.194 | 0.03 | 2.12 × 10−13 | SLE | rs12615624 | A/G | 1.11 | 4.00 × 10−7 | Ref. 15 | 0.76 | C/A | +/+ |
| SPATA48, IKZF1 | rs876039 | C/G | −0.197 | 0.03 | 9.51 × 10−13 | rs4917014 | G/T | 0.72 | 3.00 × 10−23 | Ref. 12 | 0.99 | C/G | −/− | ||
| TNFSF13B | Unsw mem %lymphocyte | rs200748895 | A/GCTGT | 0.209 | 0.03 | 1.49 × 10−14 | MS | rs200748895 | A/GCTGT | 1.27 | 1.23 × 10−9 | Ref. 11 | 1.00 | A/A | +/+ |
| SLE | 1.44 | 6.74 × 10−10 | |||||||||||||
| NCOA5, CD40 | rs6032660 | G/A | 0.153 | 0.03 | 7.20 × 10−9 | MS | rs4810485 | T/G | 1.11 | 7.70 × 10−16 | Ref. 16 | 0.98 | G/T | +/+ | |
| SLE | 1.43 | 1.00 × 10−8 | Ref. 15 | ||||||||||||
Columns indicate: annotated gene name(s) (column 1); association statistics (obtained using a linear mixed model accounting for population structure and hidden relatedness) for the immune traits including trait name, its associated marker, corresponding derived and ancestral alleles, effect size for the derived allele expressed in standard deviation units, its standard error (s.e.) and P value (columns 2–6). Association statistics for pathologies obtained from published papers and comprising disease name, its associated marker, corresponding effect and non-effect alleles, OR for the effect allele and P value (columns 7–11); disease association reference papers (column 12). Correlation between trait-associated and disease-associated markers including LD (r2) between immune trait variant and disease associated variant (column 13), derived immune trait allele and corresponding allele of associated disease variant (column 14), and their corresponding direction of effect. Specifically, ‘+’ indicates increased trait levels or predisposition for the disease, whereas ‘−’ indicates decreased trait levels or protection for disease (column 15). AC, cell absolute count; other acronyms are as in Fig. 3.
A more comprehensive treatment of the observed coincident associations is provided in Supplementary Information. Below are details for some examples with especially strong biological and clinical implications. Unless otherwise specified, we refer to the effects due to the derived (mutated) allele.
Genetic regulation of plasmacytoid cells.
A signal led by rs876039[C] in the SPATA48–IKZF1 intergenic region decreased the level of plasmacytoid dendritic cells (pDCs; effect −0.20 s.d., P = 9.51 × 10−13) and colocalized with decreased risk for systemic lupus erythematosus (SLE)12. Promoter capture Hi-C data suggest enhancer–promoter contacts between DNA sequence variation marked by this signal and the IKZF1 gene13, which encodes for the eponymous transcription factor. Furthermore, this signal coincided with several eQTLs acting in trans14 (Supplementary Table 4A), suggesting an even more complex regulation that could likely be mediated by IKZF1 action. Interestingly, another independent signal, localized in LCT-AS1, decreased the levels of pDCs (ancestral allele rs2164210[T], effect −0.20 s.d., P = 2.12 × 10−13). This signal overlapped with associations decreasing risk for SLE12,15. The potential causal role of the downregulation of pDCs in inherited protection from SLE is further supported by Mendelian randomization (MR) analyses (Methods, Supplementary Information and Supplementary Table 6) and may have implications for SLE therapy.
Genetic regulation of CD40 and CD27 in B-cell involvement in autoimmunity.
A signal led by rs1883832[T] in the 5′ untranslated region of CD40 increases in trans the expression of CD27 on memory-B-cell subsets (effect +0.64 s.d., P = 6.21 × 10−147) and decreases the level of a specific B-cell subset that does not express CD27 (IgD−CD27−; effect −0.39 s.d., P = 7.20 × 10−50). The signal overlaps with reported increased risk of multiple sclerosis (MS), inflammatory bowel disease (IBD), Crohn’s disease, SLE and chronic hepatitis infection, but with reduced risk of rheumatoid arthritis (RA) and Kawasaki disease16–20 (Fig. 3).
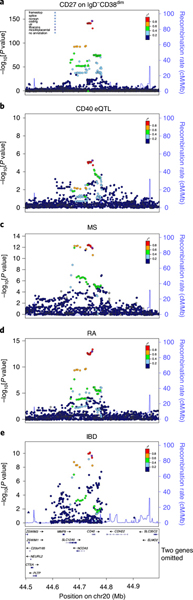
As CD40 was not included in our original panel to assess B cells, we then measured both CD40 and CD27 on B-cell subsets in an additional group of 999 individuals and found that rs1883832[T] was indeed associated with decreased CD40 expression as well as increased CD27 expression on B-cell subsets (Supplementary Table 7A). Notably, CD40 is consistently downregulated by rs1883832[T] in all B-cell subsets (both naive and memory), with the strongest effect in IgD−CD27− B cells (effect −0.73 s.d.; P = 4.55 × 10−66). This variant lies in the Kozak consensus sequence of the CD40 gene and might thereby directly affect ribosome binding and translation of CD40 messenger RNA. Furthermore, it lies in a region recognized by numerous transcription factors, suggesting that variants in that region could also influence CD40 gene transcription. Consistent with that possibility, the signal led by rs1883832[T] overlaps with a cis eQTL decreasing CD40 mRNA (Fig. 4 and Supplementary Table 4A).
Fig. 4 |. Regional association plots in the CD40 region.
The significance of the association (−log10[P value]; left y axis) for each trait is plotted relative to the genomic positions on the hg19/GRCh37 genomic build (x axis). The symbols reflect genomic functional annotations. SNPs are colored to reflect their LD with rs1883832 (indicated with a purple dot). a, Expression of CD27 on IgD−CD38dim cells. The P values were obtained using a linear mixed association model. b, CD40 expression on leukocytes calculated using RNA-seq data from Pala et al.54. c–e, Association profiles for the autoimmune diseases MS (c), RA (d) and IBD (e). The data plotted in c–e are from published results16,19,55. Genes, position of exons and direction of transcription are noted below e. The plots were drawn using the standalone version of LocusZoom56.
MR analyses (Methods) supported a unidirectional negative effect of CD40 on CD27 at the protein level (P = 1.75 × 10−5; Supplementary Table 7B,C), contributing to the inverse correlation observed between the two proteins on memory B cells (Spearman correlation ρ = −0.198, P = 2.53 × 10−10). We then assessed whether the association in trans of rs1883832 with CD27 levels on memory B cells was completely mediated by the CD40 protein or whether it was also due to horizontal (independent) pleiotropy. Our results show that the effect of this variant on CD27 is only 2% mediated by CD40 (Methods and Supplementary Table 7D). We thus hypothesize that the signal in the CD40 region has two cis-acting effects: one leading to direct downregulation of the corresponding protein, which in turn directly contributes to a relatively modest extent to the observed increase of CD27, while the second, mediated by a nearby but still undetermined gene, accounts for most of the increase of CD27 due to variation in the CD40 gene region. This latter mechanism is consistent with interactions between rs1883832 (and/or other variants in LD with it) with regulatory elements affecting expression in nearby genes (see https://genome.ucsc.edu/ and https://www.chicp.org).
Further MR analyses (Methods and Supplementary Tables 6 and 7E) suggested that decreased CD40 leads to increased risk for MS, whereas decreased CD27 leads to increased risk for Crohn’s disease. A causal role of increased CD27 expression on memory B cells in increased risk for Crohn’s disease is further supported by another independent and coherent coincident association led by the ancestral allele rs1801274[A], encoding His, in the FCGR2A gene (effect +0.29 s.d., P = 2.48 × 10−28; Supplementary Table 6A)17.
Complex coincident associations.
In some cases, the observed associations show particularly complex effects such as the presence of multiple independent signals in one or more gene regions that influence the expression of a given surface marker in different cell subtypes with distinct consequences on disease risk. Such complex regulation is exemplified below by variation at the IL2RA locus, affecting the expression of CD25, and by variation at the CD28–CTLA4 and BACH2 loci, affecting the expression of CD28 and CD80.
IL2RA, a complex autoimmunity locus.
IL2RA (CD25), encoding the alpha chain of the IL-2 receptor, contains seven independent signals, four of them also associated with diseases. As previously reported1, one signal driven by rs61839660[T], indicated as the candidate causal variant by fine-mapping analyses, increased the expression of CD25 on CD25hi memory helper T cells (effect +0.61 s.d., P = 6.70 × 10−28) as well as the levels of these cells (effect +0.62 s.d., P = 2.78 × 10−28) and was also associated with increased risk for allergic disease, Crohn’s disease and SLE, but decreased risk for type 1 diabetes (T1D), juvenile RA and primary sclerosing cholangitis15,21–24.
The second signal, led by rs10905719[A], increased CD25 expression on naive helper T cells (effect +0.23 s.d., P = 2.56 × 10−14) and predisposed to primary sclerosing cholangitis, alopecia areata, psoriasis and MS25–28.
The third signal, driven by rs706779[C], was associated with lower CD25 on IgD−CD38− B cells (effect −0.29 s.d., P = 6.64 × 10−31) and decreased risk for vitiligo and autoimmune thyroiditis29,30. The fourth signal, led by rs41294937[C], was associated with increased expression of CD25 on B cells, especially IgD−CD38dim (effect +0.57 s.d., P = 9.24 × 10−48), and overlapped with increased risk for asthma and hay fever21.
Three other signals regulated the levels or expression of CD25 in subsets of B and T cells, but have thus far not been correlated with any disease risk association (Supplementary Table 3A).
Overall, our correlations show the influence of the expression of CD25 in specific cell subtypes, including non-T cells, in predisposition to or protection from different autoimmune diseases.
Genetic regulation of CD28 and CD80 and autoimmunity risk.
CD28 is a key co-stimulatory molecule expressed on T cells, binding CD80 and CD86 on antigen-presenting cells and promoting T-cell activation. In the CTLA4–CD28 gene region, two partially correlated (r2 = 0.36) cis-acting signals, led by rs1973872[T] and rs3116493[G] (ancestral alleles), reduced CD28 levels on a number of T-cell subsets, especially CD4+ T cells and CD39+ activated regulatory T cells (Tregs; effect −0.39 s.d., P = 2.24 × 10−42 and effect −0.30 s.d., P = 8.44 × 10−30, respectively) and overlapped with predisposition to celiac disease, ulcerative colitis and IBD, but with protection against MS16,19,22,25,31–33. A further independent signal led by rs4675369[G] in the same gene region reduced CD28 expression in resting Tregs (effect −0.21 s.d., P = 2.23 × 10−10) and coincided with reported predisposition to primary biliary cholangitis34.
The expression of CD28 on T cells was also affected by genetic variation in the BACH2 gene region. Here a single fine-mapped variant, rs72928038[A], associated with increased CD28 expression on CD45RA+ cells—both CD8+ (effect +0.59 s.d., P = 5.13 × 10−67) and CD4+ (effect +0.53 s.d., P = 1.61 × 10−55) subsets—and overlapped with increased risk for MS, T1D, autoimmune thyroiditis and vitiligo16,22,23,30,35,36. After conditional analysis, a second signal, led by the ancestral allele rs619192[T], was found positively associated with the same cell traits (effect +0.20 s.d., P = 2.97 × 10−10) and overlapped with increased risk for T1D and SLE37,38. Notably, both the CD28–CTLA4 and BACH2 gene regions were associated with expression of CD28 on CD4+ T cells and overlapped with risk for MS16, further supporting the predisposing role of increased CD28 levels in MS (related MR analyses are in Supplementary Information and Supplementary Table 6). Adding an additional layer of complexity, a third signal in BACH2, led by rs12199079[G], increased CD80 on myeloid DCs (effect +0.23 s.d., P = 7.54 × 10−16) and overlapped with decreased risk for allergic disease, Crohn’s disease and IBD17,21. These findings link genetic variation in BACH2 to CD28 and CD80 regulation and add to the reported transcriptional activity of BACH2 a pivotal role in directing T-cell fate toward inflammatory or regulatory status39.
Druggability and therapeutic implications of findings.
To further assess the potential therapeutic utility of our findings, we explored the druggability of the implicated proteins (Table 2 and Supplementary Table 8A). We considered as a druggable target any protein: whose expression (here measured as MFI) was influenced in cis or in trans by the associated signals (pQTLs) underlying coincident associations; whose pharmacological modulation might reproduce a therapeutic protective effect on disease risk40,41; and for which an approved or investigational drug was available. Of the 29 proteins identified through pQTL associations, 24 were classified as drug targets in the Pharmaprojects database from Citeline (https://pharma.id.informa.com; Supplementary Table 8A,B).
Table 2 |.
Selected targets for therapeutic modulation
| Cell trait name | Biomarkers to ensure cell-subtype specificity | Primary drug target | Disease | Proposed therapeutic modulation of primary drug target |
|---|---|---|---|---|
| HVEM on T cell (especially memory) | CD3, CD45RO | HVEM | UC, IBD | Activation |
| CD28 on CD4+ | CD4, CD3, CD28 | CD28 | MS | Inhibition |
| UC | Activation | |||
| CD28 on CD28+CD45RA+ T cells (mainly CD8br) | CD45RA, CD8, CD3, CD28 | CD28 | MS, thyroiditis, T1D, VIT | Inhibition |
| CD80 on myeloid DC | CD62L, CD33, CD11c, CD141 or CD1c | CD80 | CRO, IBD, Asthma | Activation |
| pDC AC | CD123/CLEC4C | CD123/CLEC4C | SLE | Inhibition |
| CD80 on pDC | CD80/CLEC4C | CD80 | SLE | Inhibition |
| CD62L on pDC | CD62L, CD123, CLEC4C | CD62L | SLE | Inhibition |
| CD25 on CD45RA+ CD4 not Treg | CD25, CD127, CD45RA, CD4, CD3 | CD25 | PSC, AA, MS | Inhibition |
| CD25 on CD45RA− CD4 not Treg | CD25, CD127, CD45RO, CD4, CD3 | CD25 | Allergy, CRO | Inhibition |
| PSC, T1D | Activation | |||
| B cell % lymphocyte (especially IgD+CD24+ %lymphocyte) | IgD, CD24, CD19/CD20 | IgD, CD24, CD19/CD20 | MS, SLE | Inhibition |
| CD33 on myeloid cell (especially CD14+ monocyte) | CD14, HLA DR | CD33 | AD | Inhibition |
| CD27 on memory B cell (especially IgD−CD38dim) | CD27, CD19/CD20 | CD27 | HBVI, CRO, IBD, MS, SLE | Inhibition |
| RA, KD | Activation | |||
| CD40 on B cell (especially IgD−CD27−) | CD19/CD20 | CD40 | RA, KD | Inhibition |
| HBVI, CRO, IBD, MS, SLE | Activation | |||
| IgD−CD27− %B cell | CD19/CD20 | CD19/CD20 | RA, KD | Inhibition |
| HBVI, CRO, IBD, MS, SLE | Activation |
The table lists the cell trait name, specifying the primary drug target and the biomarkers used to ensure the specificity of the cell subtype. The correlated pathologies and the proposed therapeutic direction of modulation are also indicated. In the second column, a comma indicates the simultaneous need for specific markers; ‘or’ the mutually exclusive presence of two markers; a slash symbolizes the presence of at least one of the two indicated markers. Complete results are reported in Supplementary Table 8A. The disease acronyms are those of Fig. 3.
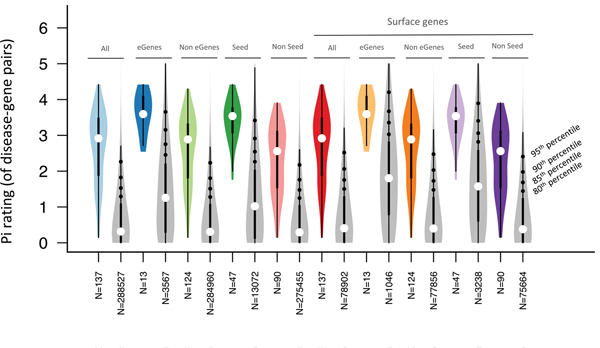
Using a recently developed drug target prioritization score (Pi rating)42, we then found that overall the 29 proteins (Supplementary Table 8B) are highly scored to be pharmacologically relevant (Fig. 5). Specifically, we retrieved the Pi ratings of 137 gene–disease pairs (IgD, HLA, CD45RA and CD3 have been excluded because the Pi rating was not available) resulting from our coincident association data, and found that their median is higher than the 95th percentile of all the disease–gene pair ratings in the Pi database (Fig. 5). We observed a high Pi rating for gene–disease pairs regardless of whether they were supported or not by genetic associations in the Pi-score database (Extended Data Fig. 7), demonstrating that our results were not biased by the fact that both our protein selection pipeline and the Pi-score computation leverage genetic information.
Fig. 5 |. Drug target prioritization (priority index, Pi) score of our drug target candidates.
The red lines indicate the Pi rating of our drug target candidates for the respective diseases (listed in Supplementary Table 8A,B). The gray violin plots indicate the distribution of the Pi rating, for the respective diseases, of all the genes for which a Pi rating has been computed (number of genes indicated in parenthesis). The gray lines indicate the 90th and the 95th percentiles of the Pi rating values. IgD, HLA, CD45RA and CD3 have been excluded because the Pi rating was not available. Gene aliases: TNFRSF13C (BAFF-R), ITGAM (CD11b), ITGAX (CD11c), IL3RA (CD123), MS4A1 (CD20), IL2RA (CD25), PTPRC (CD45), SELL (CD62L), FCGR1A (CD64), CD8A (CD8), TNFRSF14 (HVEM), FCGR2A (CD32). The acronyms are as in Fig. 3; ATD, autoimmune thyroiditis; ALG, allergy; ASM, asthma. The boxplot inside the violin plot reports a circle indicating the median value, with the box limits indicating the upper and the lower quartiles. The whisker at the upper side of the box extends to the minimum between the interquartile range × 1.5 and the overall maximum value of the data. The whisker at the bottom side of the box extends to the maximum between the interquartile range × 1.5 and the overall minimum value of the data.
As a proof of concept of our approach to select drug targets, the two signals in the SPATA48–IKZF1 intergenic region and in LCT-AS1—associated with decreasing levels of pDCs and decreased risk for SLE12,15—point to the downregulation of pDCs as a promising therapeutic route for SLE. Proteasomal degradation of the protein target IKZF1, also known as IKAROS, enhanced by the antitumoral drug lenalidomide, decreases pDC numbers in vivo43 and has been shown to be efficacious in treating patients with refractory cutaneous lupus erythematosus44. Furthermore, another existing route to SLE therapy is based on the downregulation of pDCs through the inhibition of the protein product of the gene BDCA2 (blood DC antigen 2), whose expression is regulated in trans by the IKZF1 signal overlapping with SLE protection (Supplementary Table 4). The BDCA2 product, also known as CLEC4C, is a pDC-specific receptor that following engagement inhibits the production of type I interferons (IFN-1), implicated in the pathogenesis of SLE43,45. An anti-BDCA2 monoclonal antibody, BIIB059, indeed suppresses the ability of human pDCs to produce IFN-146–48 and is currently in a phase 2 trial for SLE therapy (NCT02847598).
On the basis of our data, some drugs already approved or under clinical investigation against single target antigens could be repurposed to meet new needs. For instance, the anti-CD28 drug lulizumab pegol, developed for SLE and Sjogren’s syndrome therapy, could potentially be repositioned for MS, given the above-mentioned coincident association signals affecting increased expression of CD28 on both CD4 and CD8 T cells and MS risk.
However, on the basis of our findings, drugs directed against a single target antigen may not be an optimal therapeutic choice, especially when the antigen is expressed in different cell subtypes with opposing direction of effect on different disease risk. In such situations, it seems advisable to target more than one antigen to ensure cell-subtype specificity. For instance, CD25 needs to be modulated differently in diverse T- and B-cell subsets on the basis of the associated disease. Thus, our data suggest a possible explanation for why some trials have not been successful, and predict the efficacy of other drugs currently under clinical trial. Of note, trials of daclizumab, an CD25 antagonist, for T1D were ceased, and its efficacy was likely limited49 because, as our genetic data suggest, in this disease CD25 should be activated instead of inhibited—and in a specific T-cell subset (Supplementary Table 8A and Table 2).
Discussion
By combining high-resolution immune and genetic profiling in a Sardinian cohort, we have greatly increased the number of known genetic variants affecting the regulation of immune cell types. Our findings have linked specific cells to immune-related disorders such as MS, T1D, RA, IBD, asthma and Kawasaki disease. The analysis of quantitative immune traits has also facilitated fine-mapping of many overlapping disease association signals to one or a few variants. Furthermore, in several instances we have identified candidate causal genes and established the direction of effect of the underlying disease associations via overlapping eQTLs and pQTLs.
In particular, the assessment of pQTLs in numerous cell subtypes reported here overcomes limitations such as the restricted availability of RNA-seq data from cell subtypes, and post-transcriptional effects in gene expression that cannot be detected through eQTL analysis. The unbiased genetic approach and the large number of coincident associations detected help to reveal or validate therapeutic target proteins controlling disease-related intermediate phenotypes. Candidate targets include co-stimulatory molecules and cell surface receptors that control key immune participants in the pathogenesis of disease. Many of these targets, including CD27 and CD80, were detected only through trans pQTL associations, which can uncover molecules whose relation to disease would otherwise remain hidden.
Furthermore, in some cases, MR analyses and multiple independent signals that associated the same cell trait and disease risk provided additional support for the causal involvement of some targets in a given disease (Table 1 and Supplementary Table 6A).
However, the complex genetic regulation of immune cell levels impacting immune-related diseases also suggests that their therapeutic modulation may be just as complex. Indeed, we report many examples, including effects on CD28, CD27, CD25 and CD40 that strongly support the dependence of the efficacy of new therapies on the direction and extent of modulation of the targets in specific cell subtypes. For instance, multiple coincident associations in both the CD28–CTLA4 and BACH2 gene regions increased the expression of CD28 on CD4+ T cells and overlapped with higher risk for MS16, consistent with inhibition of CD28 on this cell subtype as a potential therapeutic route in MS. Likewise, two signals in the CD40 and FCGR2A gene regions were associated with higher CD27 expression on memory-B-cell subsets and overlapped with signals increasing risk for IBD, especially Crohn’s disease17, thus supporting therapeutic inhibition of CD27 on B cells in these diseases. In another instance, two signals in the TNFSF13B and CD40 genes (Table 1 and Supplementary Table 7B) support a causal role of unswitched memory B cells (IgD+CD27+) in MS and SLE pathogenesis, pointing to a targeted inhibition/depletion of this B-cell subtype as a therapy of these diseases. Instead, current anti-CD20 monoclonal antibodies approved or in clinical trials for MS and SLE50 are based on a broad depletion of B cells.
Thus, in contrast to classical autoimmune disease treatments directed against individual protein targets, future therapies of greater efficacy and safety may more usefully co-target two or more proteins to discriminate a particular cell subtype51 or could be based on the targeted delivery of a drug to a specific cell type—for example, by poly-specific monoclonal antibodies or other carriers for small molecules.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41588-020-0684-4.
Methods
The SardiNIA dataset.
The SardiNIA project is a longitudinal study57 comprising 6,602 general population individuals (57% females, 43% males), ranging from 18 to 102 years, native of the central east coast of Sardinia, Italy. As detailed below, all volunteers are deeply genetically characterized and 3,757 of them are immune profiled. All participants signed informed consent to study protocols approved by the Sardinian Regional Ethics Committee (protocol no. 2171/CE).
Flow cytometric measurements.
Donors’ peripheral blood was collected in heparin tubes, and then antibody-stained and processed for flow cytometry. Data were acquired with two standardized BD FACSCanto II flow cytometers and analyzed by BD FACSDiva software (BD Biosciences). The cell populations were manually gated by the same specialist to increase consistent processing of the data. To minimize time-dependent artifacts, cell phenotyping was performed in the same recruitment center within 2 h after blood collection and using erythrocyte-lysed fresh samples. BD Lyotube technology and a lyse–wash protocol were used for all panels except for TBNK; for that panel, a lyse–no-wash protocol was applied to obtain more precise cell concentrations. Samples were stained with the following antibody panels.
TBNK panel.
Leukocytes (CD45+) were divided into granulocytes, monocytes and lymphocytes on the basis of morphological parameters (forward scatter (FSC)-A and side scatter (SSC)-A, which are proportional to the size and intracellular complexity of cells, respectively). Lymphocytes were divided into CD3+, corresponding to T cells, and CD3−, including B cells (CD19+) and natural killer cells (CD16+ or CD56+). T lymphocytes were split into six subsets based on the expression of the CD4 and CD8 markers: CD4−CD8− (DN), CD4−CD8dim (CD8dim), CD4−CD8bright (CD8br), CD4+CD8br (DP), CD4+CD8dim, CD4+CD8− (CD4+). The HLA DR positivity of CD4+ and CD8br T cells and NK cells was considered as an activation marker. The gamma–delta antigen was also measured on T cells. To get absolute cell counts, BD TruCount absolute counting tubes were used. The concentrations of T cells, B cells and monocytes from this panel were used to calculate the absolute counts in the other antibody panels assessed (except for the circulating DC panel in which liquid counting beads were included).
Treg panel.
Tregs were identified on the basis of high expression of CD25 and low expression of CD127 surface antigens (CD25hiCD127lo) and further subdivided into activated (CD25+++CD45RA−), resting (CD25++CD45RA+) and secreting (CD25++CD45RA−). Tregs were subtracted from CD4+CD25hi T cells, resulting in the CD25hiCD4+ not Treg population (CD25hiCD127hi), which was further subdivided on the basis of CD45RA expression. In this panel, the CD8 T cells were divided according to their expression of CD28 and CD45RA antigens. The high positivity for CD25 and the negativity for CD127 on CD28− CD8 cells were also measured. Finally, Treg subsets, CD4+ and CD8+ T cells were also subdivided on the basis of the expression of CD39.
Maturation stages of T-cell panel.
The maturation status of CD4+, CD8br and CD4−CD8− T lymphocytes was assessed on the basis of the expression of CD45RA and CCR7. Naive (CD45RA+CCR7+), central memory (CCR7+CD45RA−), effector memory (CD45RA−CCR7−) and TD (CCR7−CD45RA+) maturation stages were identified.
DC panel.
DCs were identified on the basis of their positivity for HLA DR and their negativity for the lineage cocktail (Lin) targeting CD3, CD14, CD16, CD19, CD20 and CD56 markers. The DCs were subdivided into myeloid (CD11c+) and plasmacytoid (CD123+) cells. Their maturation and activation status were ascertained by the adhesion molecule CD62L and the co-stimulatory molecules CD80 and CD86. In addition, monocytes were morphologically ascertained and analyzed for HLA DR, CD62L and CD11c expression. The absolute number of cells was estimated by adding BD Liquid Counting Beads to the samples.
B-cell panel.
Total B cells were identified as CD19-positive and were further subdivided using several classification approaches. CD24 versus CD38 classification identified transitional (CD24+CD38hi), memory (CD24+CD38−/dim) and naive mature (CD24−CD38−/dim) subsets. CD27 versus IgD classification discriminated switched memory (CD27+IgD−), unswitched memory (CD27+IgD+), naive (CD27−IgD+) and CD27−IgD− B cells. IgD versus CD38 classification, also known as Bm1–Bm5 classification, distinguished six B cell subsets: Bm1 (IgD+CD38−) mainly virgin naive cells; Bm2 (IgD+CD38dim) activated naive cells; Bm2′ (IgD+CD38br) pre-germinal center cells; Bm3–Bm4 (IgD−CD38br) centroblasts and centrocytes present in germinal center cells but very low/absent in blood; early Bm5 (IgD−CD38dim); and late Bm5 (IgD−CD38−) memory cells. CD24 versus CD27 classification identified CD24+CD27+ memory cells. IgD versus CD24 classification subdivided B cells into four subsets: IgD+CD24+; IgD−CD24+; IgD−CD24; and IgD+CD24−. CD20 versus CD38 discriminated plasma blasts/plasma cells (as CD20−CD38hi) and CD20−CD38− cells.
Monocyte panel.
Monocytes were identified on the basis of morphological parameters and HLA DR positivity. Monocytes were then subdivided into classical (CD14+CD16−), non-classical (CD14−CD16+) and intermediate (CD14+CD16+). Each subset was assessed for CD40, CD64, CCR2, CX3CR1 and PD-L1 expression level.
Myeloid cell panel.
The fluorescent intercalator 7-aminoactinomycin D was used to recognize and exclude dead cells. In parallel, a cocktail including CD19, CD20 and CD3 antibodies was used to remove lymphoid cells. The resulting myeloid-enriched cells were subdivided on the basis of CD14 high positivity (corresponding to classical monocytes) and into five subsets based on CD33 and HLA DR expression. CD11b and CD66b antibodies were used for additional sub-characterization. The CD33dim HLA DR− were subdivided into: granulocytic myeloid-derived suppressor cells (MDSCs), based on the high positivity for CD66b cells; immature MDSCs, which are negative for CD11b; and basophils, which are positive for CD11b. Monocytic MDSCs were identified on the basis of their high positivity for CD14 and CD33 and weak positivity for HLA DR. Finally, hematopoietic stem cells were identified as CD34+CD45dim.
Four of these seven panels (that is, TBNK, Treg, maturation stages of T cell and cDC) have been described previously in a smaller set of individuals1, but without assessing MFIs or morphological parameters (FSC and SSC). As the name suggests, MFI represents the median expression level of a fluorescent-conjugated antibody bound to a cell and is proportional to the median amount of antigen expressed in that cell. To control for batch effects in MFIs due to variability in antibody lots and any seasonal shifts, the distribution was normalized for overall and daily changes as in Steri et al.11. Briefly, values for each trait were normalized by calculating the cohort means of all the samples and the daily means of the samples analyzed in the same day. Each MFI value was then multiplied by the ratio between cohort mean and daily means to compensate for daily fluctuations. The normalization was calculated independently for each MFI trait. Morphological parameters were assessed by light scattering measured by two optical detectors. One detector measured scatter along the path of the laser, namely FSC, primarily due to light diffraction around the cell, and is proportional to the diameter and the size of the cell. The other detector measured scatter in a perpendicular direction relative to the laser, namely SSC, caused by the refraction or reflection of the interface between the laser and intracellular structures, and provided information about the internal complexity (that is, granularity) of a cell.
Measures to ensure reproducibility of measurements have already been described1. Briefly, the internal parameters of the two FACS CantoII analyzers used to measure immune traits were adjusted daily by standardized fluorescent beads to check and correct for laser wear and fluidic instability. Actual cell counts were validated weekly by processing stabilized blood samples with characterized cell concentrations. To directly assess reproducibility, we repeated the FACS measurements in 35 participants for the TBNK, Treg, maturation stages of T cell and DC panels as previously described1, in 87 individuals for the monocyte panel and in 91 volunteers for the B cell panel. Reproducibility was assessed between paired samples using a two-sided Pearson’s product moment correlation coefficient on the inverse-normalized traits, finding overall high reproducibility.
Statistical and bioinformatic analyses.
Heritability estimation.
Broad heritability (the ratio of the total genetic variance to the phenotypic variance of a trait) and narrow heritability (the proportion of broad heritability due to additive effects of genes on phenotype) were estimated according to models previously described1. Among the 3,757 immunoprofiled individuals, 3,371 were grouped into 847 multigenerational families (ranging from 1 to 5 generations, average 2.66), comprising 2,405 sib pairs (including 4 monozygotic twins), 79 half-sib pairs, 2,258 cousins pairs, 1,587 parent–child pairs, 88 grandparent–grandchild pairs and 2,997 avuncular pairs. Heritability estimates were obtained with the poly-0.5.1 software57; all models were adjusted for age and sex, and traits were first normalized using inverse-normal transformation on the same traits or, when necessary, on the covariate-adjusted residuals. Statistical significance of the difference in the average heritability among the defined trait categories was evaluated by non-parametric tests (a two-sided Wilcoxon signed-rank test for pairwise comparisons and a Kruskal–Wallis test for multiple comparisons), with P < 0.05 considered as significant.
Bivariate analysis.
Phenotypic and genetic correlations were estimated for MFIs and cell counts/ratios separately. Phenotypic correlations were calculated for trait pairs using the Spearman coefficient in R (version 3.5.3). Genetic correlations between trait pairs, proportional to the cross trait–cross individual additive genetic covariance, were obtained using poly-0.5.1 software, as previously described1. A hierarchical clustering based on estimated correlation coefficients was also performed using the heatmap.2() function in R with default settings (Euclidean distance and the complete clustering method).
Genotyping and imputation.
All the genetic analyses were performed using a genetic map based on 6,602 samples genotyped with 4 Illumina arrays (OmniExpress, ImmunoChip, Cardio-MetaboChip and ExomeChip) as previously described6. Imputation was performed on a genome-wide scale using a Sardinian sequence-based reference panel of 3,514 individuals and the software Minimac58 on pre-phased genotypes6. After imputation, only markers with imputation quality (RSQR) > 0.3 for estimated minor allele frequency (MAF) ≥ 1% or > 0.6 for MAF < 1% were retained for association analyses59, yielding ~22 million variants (20,143,392 SNPs and 1,688,858 indels) useful for analyses.
Association analyses.
Genome-wide association analysis for each quantitative immune trait was carried out using the q.emmax (quantitative EMMAX–Efficient Mixed Model Association eXpedited) function included in EPACTS −3.2.6 (https://genome.sph.umich.edu/wiki/EPACTS). The method implemented in this software accounts for a wide range of sample structures, such as cryptic relatedness and population stratification, by applying a linear mixed model adjusted for a genomic-based kinship matrix obtained from quality-checked genotyped autosomal SNPs with MAF > 1%6. All assessed traits were normalized with inverse-normal transformation and adjusted for sex, age and age2 as covariates. To evaluate any significant deviation of summary association statistics from the null distribution, the genomic inflation factor was calculated: lambda values ranged from 0.95 to 1.035 (median = 0.994, mean = 0.995, s.d. = 0.017). Genomic correction was then not applied.
To adjust for multiple testing, Bonferroni correction was applied to the empirical significance threshold (P = 6.9 × 10−9)6, taking into account the total number of absolute cell counts, MFIs and morphological parameters assessed here (N = 539) to establish a final threshold of P < 1.28 × 10−11. Conditional analyses were performed for each trait that reached standard genome-wide significance (P < 5 × 10−8) by adding the top associated variant as a covariate to the model adjusted for sex, age and age2.
Specific analyses in the FGCR2A locus.
Specific association analyses in the FGCR2A locus on 117 volunteers were carried out comparing the two homozygous genotypes using a non-parametric model (two-sided Wilcoxon signed-rank test) on the raw phenotypes.
Definition of signals using LD.
To establish ‘independent variants’ for each signal that reached the standard threshold for genome-wide significance, the clumping function in PLINK60 and LD patterns in the SardiNIA genetic map were used. In particular, the options --clump-p1 0.00000005 --clump-p2 0.00000005 --clump-r2 0.1 --clump-kb 500 were used to clump into single-group variants with P < 5 × 10−8 and r2 > 0.1 in a 1-megabase window. The same clumping procedure was applied to define ‘sentinel’ variants using r2 > 0.7 at each gene region (‘locus’).
Allele frequency differentiation and positive selection.
Analysis of allelic differentiation was carried out on associated variants and on data from the 1000 Genomes Project Phase 3 (http://www.internationalgenome.org/category/phase-3/).
We identified highly differentiated variants by calculating the difference in allelic frequency between Sardinians and Europeans (SardiNIA MAF − 1000G-EUR AF = ΔAF) with respect to the minor allele in the Sardinian data.
For a subset of variants, we also investigated evidence of positive selection by using standard statistical tests based on allelic frequency (population-branch statistic) and haplotype diversity (integrated haplotype score, cross-population extended haplotype homozygosity and the number of segregating sites by length), as previously described11. For each variant, we calculated a ‘genomic percentile’, ranking each statistic value with that obtained for a set of variants sampled across the genome and matched for MAF in Sardinians, local recombination rates and levels of background selection11.
Replication and validation of findings.
For each association at P < 5 × 10−8, we considered a signal replicated if the top variant or its proxy with r2 > 0.8 was already reported as significantly associated with a related trait in GWASs from Ferreira et al.61, Roederer et al.2, Aguirre-Gamboa et al.3, Patin et al.4 or Lagou et al.5. Furthermore, for each genome-wide significant association (P < 1.28 × 10−11), we considered a signal validated if the top variant was genotyped or in LD with a genotyped proxy (r2 > 0.80) with a P < 1.28 × 10−11.
Fine-mapping and credible sets.
Credible set analyses were performed for 584 trait–locus associations having at least one SNP with P < 1.28 × 10−11. Credible sets were obtained using FINEMAP v1.310 and setting at most ten causal variants for each association profile (–n-causal-snps 10). These variants were annotated with VEP (https://www.ensembl.org/Tools/VEP) to evaluate which signals contain any variant(s) with strong predicted functional impact in their credible sets.
Definition of causal genes at associated loci.
To infer candidate causal genes underlying association signals, we used a combination of criteria that included: lead variant, or one variant in the credible set, with a clear functional impact (for example, a biochemically relevant coding variant) located in a specific gene; overlap with molecular QTLs in LD with r2 ≥ 0.7 (based on the Sardinian genetic map) assessed using the LinDA Catalog (Version 20190109), which included eQTLs as well as QTLs for other molecular traits such as alternative splicing, polyA usage, histone modifications and DNA methylation; overlap with a cis pQTL for levels of the measured surface protein.
Enrichment of molecular QTLs in immune trait association signals.
To test for the enrichment of molecular QTLs in immune trait association signals, we used a set of background variants determined as follows. We considered each lead variant of the LD-independent immune trait loci, and we randomly selected 50 variants matched for MAF (±0.05), imputation quality (±0.05), distance from the nearest transcriptional start site (±1 megabase) and LD-independent (between each other and with respect to the immune trait loci lead variants). The LD-independent loci have been chosen in the LD-clumped Sardinian genetic map (PLINK version 1.90_b3.38, with parameters --clump-r2 0.1 --clump-kb 1,500) and excluding sex chromosomes and the HLA region (chr6: 27000000–33000000). The positions of the resulting background variants were checked for any molecular QTLs with r2 ≥ 0.7 (in the Sardinian genetic map) in the LinDA molecular QTL Catalog. The Fisher exact test was used to test for enrichment of molecular QTLs in the LD-independent immune trait loci. Only molecular QTLs having more than 10 LD overlapping hits among the LD-independent immune loci were considered for enrichment.
Coincident associations of immune traits and disease.
To identify associations overlapping with previously described complex disease associations (‘coincident associations’), both the GWAS Catalog (https://www.ebi.ac.uk/gwas/home, version v1.0, Ensembl release version e93, date 2018–08-28) and the ImmunoBase (https://genetics.opentargets.org/immunobase, version 12-May-2016) websites were screened for variants in LD (r2 ≥ 0.7) with the top associations for any of the assessed traits. For a primary group of diseases (RA19, SLE62, MS16, T1D63, Crohn’s disease55, ulcerative colitis55, IBD55, allergy21), we performed colocalization analyses using the coloc v2.3–1 package64 with default settings. Trait and disease were considered colocalized if the probability of colocalization (coloc_P) ≥ 0.8. Colocalizations with coloc_P ≤ 0.2 were considered as not colocalized. Intermediate values (0.2 < coloc_P < 0.8) were called ‘uncertain’. To establish the direction of a genetic signal effect on traits and diseases, we considered as the ‘effect allele’ the ‘derived’ allele for variants associated with immune parameters, and the allele corresponding by LD (r2 ≥ 0.7) to the derived immune trait allele for variants associated with diseases. For each associated variant, we inferred the ancestral/derived allele by comparing Sardinian sequences with the ancestral sequences for Homo sapiens (GRCh37) extracted from the alignment set ensembl_compara_59@ens-livemirror:3306 (using 6 primates EPO (474)).
The confidence in the ancestral call was determined according to the 1000 Genomes Project pipeline (described in ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/phase1/analysis_results/supporting/ancestral_alignments/human_ancestor_GRCh37_e59.README).
MR.
To assess the causal role of the immune traits (‘exposures’) on the identified coincident diseases (‘outcomes’), we applied a two-sample multivariable MR approach on seven clinical outcomes detailed in Supplementary Table 6A showing multiple coincident associations.
For each multiple coincident association, we prioritized the seven most representative phenotypes by selecting, for each coincident disease, the trait showing the most significant P value at one of the associated coincident loci (Supplementary Table 6B).
We then identified the instrumental variable (IV) models by extracting from our GWAS: variants with P < 1 × 10−5; independent variants (that is, variants showing a LD r2 < 0.1 in a 500-kilobase window, with r2 calculated in our SardiNIA genetic map using PLINK v1.9060); when a variant was not included in the outcome GWAS, we replaced it with its best proxy in 1000G-EUR data (r2 > 0.8); when a proxy was not available, the variant was excluded from the analyses. For each selected IV, we extracted its summary statistics, in particular effects and standard errors, to be used in MR analyses.
To guarantee the robustness of the association of the IVs with exposure and ensure the statistical significance of the IV models, we evaluated the F-statistic according to the following formula:
where n is the number of individuals analyzed in the exposure GWAS, k is the number of variants included in the IV model and R2 is the variance explained by the genetic instrument(s). For each variant, R2 was calculated as
with β being the estimated effect of the variant on the exposure and var(exposure) being the variance of the tested exposure. For multivariable IV models, R2 was calculated on single-regression-model fitting all of the IVs. An F-statistic > 10 was required to define an IV model as significantly informative in MR analyses65.
To perform MR analyses, we applied different methods: the random-effects inverse-variance-weighted (IVW) method, weighted median (WM), MR-Egger and MR-PRESSO66. All of the analyses were carried out with the Two-SampleMR version 0.4.22 and the MR-PRESSO version 1.0 packages implemented in R (https://cran.r-project.org/).
We estimated the heterogeneity for the IVW and MR-Egger methods using the mr_heterogeneity() function, and when the Cochran’s Q statistic indicated a significant value, IVW was replaced by WM. Similarly, we tested for MR-Egger intercept pleiotropy with the mr_pleiotropy_test() function. When MR-Egger showed significant heterogeneity and/or pleiotropy and the number of IVs was >3, we applied MR-PRESSO to identify and discard significant pleiotropic IVs (P < 0.05), defined as ‘outliers’; the global P value of the model, referring to the hypothesis of no directional pleiotropy, raw and outlier-corrected causal estimations were reported; again, if the number of filtered IVs was >3, IWV, WM and MR-Egger were carried out on outlier-corrected models.
A significance threshold of P < 7.1 × 10−3 was established on the basis of the seven exposures analyzed. MR results were considered robust if confirmed by at least two different methods. When reported in the text, the P values refer to the WM test, with outliers removed if necessary.
Analyses in the CD40 locus.
To establish the relative causal impact of CD40 and CD27 protein levels in the CD40 locus association, a series of analyses were performed to: understand whether the expression of CD27 is regulated by rs1883832 or is mediated by CD40 protein levels; quantify the amount of rs1883832 effect on CD27 protein levels mediated by CD40 protein levels; understand whether the association with autoimmune disease at this locus is mediated by CD40 or CD27 expression.
First, bidirectional one-sample multivariable MR analyses were applied to check whether rs1883832 showed a horizontal pleiotropic effect on both CD40 and CD27 protein levels, or CD40 protein levels mediate CD27 protein levels. We extracted instrumental variables associated with CD40 and with CD27 expression on IgD−CD38dim B cells as already described. We then applied two different MR methods: the Wald ratio test (implemented in the TwoSampleMR version 0.4.22 package in R) on rs1883832 as a single-variant instrument; the two-stage least squares regression method using the tsls() function of the sem version 3.1–9 package in R67. In particular, we tested three different models with two-stage least squares: a model including only rs1883832, to compare results with the Wald ratio test; a model including all of the selected IVs; a model including all IVs, except pleiotropic variants according to the MR-PRESSO test (only rs1883832 was excluded). A Bonferroni correction was applied considering the four tests performed (two traits analyzed in two different variant settings), and a significance threshold of P < 1.25 × 10−2 was established.
Second, the proportion of the rs1883832 effect on CD27 mediated by the CD40 protein was estimated using nested linear regression models as follows: first, we evaluated a baseline model regressing out rs1883832 on CD27 protein levels (model 1); then a second model regressing out the rs1883832 effect adjusted for the CD40 protein level effect (model 2); finally, a third model regressing out the rs1883832 effect adjusted for the effect on CD40 expression of all IVs used in MR (model 3). In all models, CD27 and CD40 protein levels were inverse-normalized and adjusted for sex, age and age2 before the analyses. A likelihood ratio test was applied to compare nested models. We calculated the proportion of the SNP effect on CD27 mediated by the CD40 protein as the difference of the R2-adjusted between model 1 and model 2, and between model 1 and model 3, obtaining comparable results.
Finally, we also carried out two-sample MR analyses with CD40 and with CD27 expression on IgD−CD38dim B cells as exposures, and MS and RA as outcomes, using the same procedure described above. A multiple-testing-adjusted significance threshold of P < 1.25 × 10−2 has been considered (two traits in two diseases).
Selection of drug targets and their prioritization score.
To investigate the potential for drug discovery of the observed coincident associations between immune-related trait and disease risk, we applied a set of rules to define optimal candidates for pharmaceutical development40,41.
Overall, we ranked our targets into three broad categories: biological targets for which MFIs and expression data indicated a loss/reduction of function associated with disease protection (or a gain of function associated with disease predisposition), thus supporting possible therapeutic inhibition; biological targets for which MFIs and expression data indicated a gain of function associated with disease protection, thus supporting possible therapeutic activation of the gene product; more complex situations where the same biological target was associated with more than one disease but with discordant directions of effects.
We used the priority index (PI) to score each gene as a potential drug target for a specific disease42.
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Full GWAS summary statistics have been deposited in the GWAS Catalog with accession numbers from GCST0001391 (https://www.ebi.ac.uk/gwas/studies/GCST0001391) to GCST0002121 (https://www.ebi.ac.uk/gwas/studies/GCST0002121). The accession number for each trait is reported in Supplementary Table 1B.
Disease summary statistics used to identify coincident associations were obtained from the GWAS Catalog (https://www.ebi.ac.uk/gwas/home, version v1.0, Ensembl release version e93, date 2018–08-28) and from ImmunoBase (https://genetics.opentargets.org/immunobase, version 12-May-2016). Summary statistics for colocalization analyses were downloaded from the respective web pages: RA (http://plaza.umin.ac.jp/~yokada/datasource/files/GWASMetaResults/RA_GWASmeta_European_v2.txt.gz); T1D (https://datadryad.org/stash/dataset/doi:10.5061/dryad.ns8q3); MS (http://imsgc.net/publications/); SLE (http://insidegen.com/insidegen-LUPUS-data.html); allergy (https://genepi.qimr.edu.au/staff/manuelf/gwas_results/SHARE-without23andMe.LDSCORE-GC.SE-META.v0.gz); IBD, Crohn’s disease and ulcerative colitis (ftp://ftp.sanger.ac.uk/pub/project/humgen/summary_statistics/human/2016-11-07/). Molecular QTLs are from the LinDA QTL Catalog (http://linda.irgb.cnr.it/, version 20190109). Source data are provided with this paper.
Extended Data
Extended Data Fig. 1 |. Flow cytometry gating strategy of TBNK, regulatory T cell, maturation stages of T cell, and dendritic cell antibody panel.
TBNK panel. a, Lymphocytes (violet) and granulocytes (blue). b, CD14+ monocytes (light blue). c, HLA DR++CD14+ monocytes. d, CD3+ (T cells, purple) and CD3− (green) lymphocytes. e, B and NK cells are CD19+ and CD16/CD56+, respectively. f, HLA DR+ NK cells. g, T cells are divided based on CD4 and CD8 expression. h, TCR-ϒδ+ T cells. i, NKT are CD3+ and CD16/CD56+. j, HLA DR+ T cells. HLA DR+CD4 and HLA DR+CD8 subsets are obtained by intersecting HLA DR+ T cell with CD4+ and CD8br lymphocytes. Regulatory T cell panel. a, CD4+ (blue) and CD8+ (violet) lymphocytes. b, CD4+ Tregs (green) are CD25high CD127low. c, Resting (CD45RA+CD25++, pink), activated (CD45RA-CD25+++, orange) and secreting (CD45RA-CD25++, purple) CD4+ Tregs. D-E) CD25hiCD4+ lymphocytes are divided based on CD45RA expression. F) CD4-CD8− T cells (DN, black) are divided in CD28+ and CD28−. G-H-I-J-K) CD39 expression on Treg subsets, CD4 and CD8br T cells, respectively. L) CD8br cells division based on CD45RA vs CD28 expression. M-N) CD25++CD28-CD8br and CD127-CD28-CD8br identification. Maturation stages of T cell panel. a, CD4+ (blue), CD8br (violet) and CD4-CD8− (black) T cells are analyzed for CD45RA vs CCR7 (plots B-C-D, respectively) identifying naïve (CCR7+CD45RA+), central memory (CM, CCR7+CD45RA−), effector memory (EM, CCR7-CD45RA−), and terminally differentiated (TD, CCR7-CD45RA+) subsets. Dendritic cell antibody panel. a, Monocytes (pink). b, c, DCs are Lineage (Lin) negative and HLA DR+. d, Myeloid (green) and plasmacytoid (violet) DCs are CD11c+ and CD123+, respectively. e, f, CD86 and CD62L expression on cDC. G-H-I) CD11c, CD62L and HLA DR expression on monocytes.
Extended Data Fig. 2 |. Flow cytometry gating strategy of B cell, monocyte, myeloid cell antibody panel.
B cell panel. a-b, Lymphocyte (red). c, B lymphocytes (violet) are CD19+. d, IgD+ B cells. B cells classification based on e) CD24 vs CD38; f) CD27 vs IgD; g) IgD vs CD38; h) IgD vs CD24; i) CD24+CD27+ memory B cells. j, Plasma blasts/plasma cells (PB/PC) are CD20-CD38hi B cells. Monocyte panel. A-B-C) Monocyte (blue). D) Monocytes division into CD14+CD16− (classical), CD14-CD16+ (non-classical) and CD14+CD16+ (intermediate). Myeloid panel. A-B-C) Lympho-monocytes (red). d, Viable and myeloid-enriched cells (green) are obtained excluding lymphoid cells, which are lineage1 (Lin1) positive, and dead cells, which are 7-aminoactinomycin-D (7AAD) positive. e, Hematopoietic stem cells (HSC). f, CD14 vs CD66b expression and g) CD33 vs HLA DR expression on myeloid-enriched cells. The intersection of CD33dim/br HLA DRdim/− cells in g) with CD14+ monocytes (orange) in f) results in monocytic myeloid-derived dendritic cells (Mo MDSC). h, The deletion of CD14+ monocytes (orange) from cells in g) discriminates five subsets using CD33 vs HLA DR markers. i, CD66b+ cells were excluded from the CD33dim HLA DR− cells (blue) and j) the resulting CD33dimHLA DR-CD66− population was further divided into basophils and immature MDSC (Im MDSC) based on CD45 and CD11b expression. k, CD33br HLA DR+ cells (black) division into CD14 dim and CD14−. l, CD11b expression on CD33dim HLA DR+ cells (purple). Intersection of CD33dim HLA DR− in h) with CD66b++ cells in f) corresponds to granulocytic myeloid-derived dendritic cells (Gr MDSC).
Extended Data Fig. 3 |. Phenotypic correlation among expression level of surface markers.
Heatmap of phenotypic correlations for MFI pairs calculated using the Spearman coefficient. Dendrograms represent the clustering: short branches indicate strong phenotypic correlation between traits, whereas long branches weak correlation. Color gradations represent the correlation strength, with red indicating direct correlation (from 0 to +1) and blue inverse correlation (from 0 to −1).
Extended Data Fig. 4 |. Genetic correlation among expression level of surface markers.
Heatmap of genetic correlations for MFIs pairs calculated as previously described1. The description of the figure is as for Extended Data Fig. 3.
Extended Data Fig. 5 |. Phenotypic correlation among cell levels.
Heatmap of phenotypic correlations for cell counts and T/B and CD4/CD8br ratios, calculated using the Spearman coefficient. The description of the figure is as for Extended Data Fig. 3.
Extended Data Fig. 6 |. Genetic correlation among absolute cell counts.
Heatmap of genetic correlations for cell counts and T/B and CD4/CD8br ratios, calculated as previously described1. The description of the figure is as for Extended Data Fig. 3.
Extended Data Fig. 7 |. Drug target prioritization (Priority index, Pi) score of our drug targets candidates segmented by gene categories.
It is shown the distributions of the Pi-rating (computed in Fang et al., 42) of our candidate genes (colored boxplots) segmented for different gene categories (“All” genes, “eGenes”, “Seed” genes and cell surface genes) with the relative background distributions (grey boxplots, that is that consider all the genes belonging to the respective category). eGenes means that the gene has an eQTL colocalizing with the disease; seed gene means that the genes have a genetic link to the disease (by eQTL, gene proximity or chromatin conformation) as defined in Fang et al., 42. The boxplot inside the violin plot reports a white circle indicating the median value, with the box limits indicating the upper and the lower quartiles. The whisker at the upper side of the box extends to the minimum between the interquartile range (IQR) x 1.5 and the overall maximum value of the data. The whisker at the bottom side of the box extends to the maximum between IQR x 1.5 and the overall minimum value of the data.
Supplementary Material
Acknowledgements
We thank all of the volunteers who generously participated in this study; S. Naitza and J. Todd for helpful suggestions; the Consortium ‘Sa Corona Arrubia della Marmilla’ for making available equipment and scientific instruments; and the IMSGC and WTCCC2 consortia for access to the summary statistics from the key disease GWAS. We acknowledge support by grants (2011/R/13 and 2015/R/09, to F.C.) from the Italian Foundation for Multiple Sclerosis; contracts (HHSN271201600005C, to F.C.) from the Intramural Research Program of the National Institute on Aging, National Institutes of Health (NIH); a grant (FaReBio2011 ‘Farmaci e Reti Biotecnologiche di Qualità’, to F.C.) from the Italian Ministry of Economy and Finance; a grant (633964, to F.C.) from the Horizon 2020 Research and Innovation Program of the European Union; grants (U1301.2015/AI.1157. BE Prat. 2015–1651, to F.C. and U965.2014/AI.847.MGB Prat. 2014.0597, to M.S.) from Fondazione di Sardegna; grants (‘Centro per la Ricerca di Nuovi Farmaci per Malattie Rare, Trascurate e della Povertà’ and ‘Progetto Collezione di Composti Chimici ed Attività di Screening’ to F.C.) from Ministero dell’Istruzione, dell’Università e della Ricerca.
Footnotes
Competing interests
The authors declare no competing interests.
Additional information
Extended data is available for this paper at https://doi.org/10.1038/s41588-020-0684-4.
Supplementary information is available for this paper at https://doi.org/10.1038/s41588-020-0684-4.
Reprints and permissions information is available at www.nature.com/reprints.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Orrù V et al. Genetic variants regulating immune cell levels in health and disease. Cell 155, 242–256 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roederer M et al. The genetic architecture of the human immune system: a bioresource for autoimmunity and disease pathogenesis. Cell 161, 387–403 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aguirre-Gamboa R et al. Differential effects of environmental and genetic factors on T and B cell immune traits. Cell Rep. 17, 2474–2487 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patin E et al. Natural variation in the parameters of innate immune cells is preferentially driven by genetic factors. Nat. Immunol. 19, 302–314 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Lagou V et al. Genetic architecture of adaptive immune system identifies key immune regulators. Cell Rep. 25, 798–810 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sidore C et al. Genome sequencing elucidates Sardinian genetic architecture and augments association analyses for lipid and blood inflammatory markers. Nat. Genet. 47, 1272–1281 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang YJ, Zhao XY & Huang XJ Immune reconstitution after haploidentical hematopoietic stem cell transplantation. Biol. Blood Marrow Transplant. 20, 440–449 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Ogonek J et al. Immune reconstitution after allogeneic hematopoietic stem cell transplantation. Front. Immunol. 17, 507 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brodin P et al. Variation in the human immune system is largely driven by non-heritable influences. Cell 160, 37–47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benner C et al. FINEMAP: efficient variable selection using summary data from genome-wide association studies. Bioinformatics 32, 1493–1501 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steri M et al. Overexpression of the cytokine BAFF and autoimmunity risk. N. Engl. J. Med. 376, 1615–1626 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han JW et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat. Genet. 41, 1234–1237 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Javierre BM et al. Lineage-specific genome architecture links enhancers and non-coding disease variants to target gene promoters. Cell 167, 1369–1384 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Westra HJ et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 45, 1238–1243 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langefeld CD et al. Transancestral mapping and genetic load in systemic lupus erythematosus. Nat. Commun. 8, 16021 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.International Multiple Sclerosis Genetics Consortium (IMSGC) et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat. Genet. 45, 1353–1360 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jostins L et al. Host–microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang DK et al. Genetic variants in five novel loci including CFB and CD40 predispose to chronic hepatitis B. Hepatology 62, 118–128 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Okada Y et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 506, 376–381 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee YC et al. Two new susceptibility loci for Kawasaki disease identified through genome-wide association analysis. Nat. Genet. 44, 522–525 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Ferreira MA et al. Shared genetic origin of asthma, hay fever and eczema elucidates allergic disease biology. Nat. Genet. 49, 1752–1757 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ellinghaus D et al. Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat. Genet. 48, 510–518 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Onengut-Gumuscu S et al. Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat. Genet. 47, 381–386 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hinks A et al. Dense genotyping of immune-related disease regions identifies 14 new susceptibility loci for juvenile idiopathic arthritis. Nat. Genet. 45, 664–669 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu JZ et al. Dense genotyping of immune-related disease regions identifies nine new risk loci for primary sclerosing cholangitis. Nat. Genet. 45, 670–675 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petukhova L et al. Genome-wide association study in alopecia areata implicates both innate and adaptive immunity. Nature 466, 113–117 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Betz RC et al. Genome-wide meta-analysis in alopecia areata resolves HLA associations and reveals two new susceptibility loci. Nat. Commun. 6, 5966 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.International Multiple Sclerosis Genetics Consortium (IMSGC) et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 476, 214–219 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin Y et al. Variant of TYR and autoimmunity susceptibility loci in generalized vitiligo. N. Engl. J. Med. 362, 1686–1697 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cooper JD et al. Seven newly identified loci for autoimmune thyroid disease. Hum. Mol. Genet. 21, 5202–5208 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trynka G et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat. Genet. 43, 1193–1201 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eyre S et al. High-density genetic mapping identifies new susceptibility loci for rheumatoid arthritis. Nat. Genet. 44, 1336–1340 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu JZ et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 47, 979–986 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qiu F et al. A genome-wide association study identifies six novel risk loci for primary biliary cholangitis. Nat. Commun. 20, 14828 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fortune MD et al. Statistical colocalization of genetic risk variants for related autoimmune diseases in the context of common controls. Nat. Genet. 47, 839–846 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin Y et al. Genome-wide association studies of autoimmune vitiligo identify 23 new risk loci and highlight key pathways and regulatory variants. Nat. Genet. 48, 1418–1424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bradfield JP et al. A genome-wide meta-analysis of six type 1 diabetes cohorts identifies multiple associated loci. PLoS Genet. 7, e1002293 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morris DE et al. Genome-wide association meta-analysis in Chinese and European individuals identifies ten new loci associated with systemic lupus erythematosus. Nat. Genet. 48, 940–946 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roychoudhuri R et al. BACH2 represses effector programs to stabilize T(reg)-mediated immune homeostasis. Nature 498, 506–510 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Plenge RM, Scolnick EM & Altshuler D Validating therapeutic targets through human genetics. Nat. Rev. Drug Discov. 12, 581–594 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Floris M, Olla S, Schlessinger D & Cucca F Genetic-driven druggable target identification and validation. Trends Genet. 34, 558–570 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fang H et al. A genetics-led approach defines the drug target landscape of 30 immune-related traits. Nat. Genet. 51, 1082–1091 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cytlak U et al. Ikaros family zinc finger 1 regulates dendritic cell development and function in humans. Nat. Commun. 9, 1239 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Presto JK & Werth VP Cutaneous lupus erythematosus: current treatment options. Curr. Treat. Options Rheumatol. 2, 36–48 (2016). [Google Scholar]
- 45.Kim JM, Park SH, Kim HY & Kwok SK A plasmacytoid dendritic cells-type I interferon axis is critically implicated in the pathogenesis of systemic lupus erythematosus. Int. J. Mol. Sci. 16, 14158–14170 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dzionek A BDCA-2, a novel plasmacytoid dendritic cell-specific type II C-type lectin, mediates antigen capture and is a potent inhibitor of interferon alpha/beta induction. J. Exp. Med. 194, 1823–1834 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blomberg S, Eloranta ML, Magnusson M, Alm GV & Rönnblom L Expression of the markers BDCA-2 and BDCA-4 and production of interferon-alpha by plasmacytoid dendritic cells in systemic lupus erythematosus. Arthritis Rheumatol. 48, 2524–2532 (2003). [DOI] [PubMed] [Google Scholar]
- 48.Cao W et al. BDCA2/Fc epsilon RI gamma complex signals through a novel BCR-like pathway in human plasmacytoid dendritic cells. PLoS Biol. 5, e248 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gottlieb PA Failure to preserve β-cell function with mycophenolate mofetil and daclizumab combined therapy in patients with new- onset type 1 diabetes. Diabetes Care 33, 826–832 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Du FH et al. Next-generation anti-CD20 monoclonal antibodies in autoimmune disease treatment. Autoimmun. Highlights 8, 12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Labrijn AF, Janmaat ML, Reichert JM & Parren PWHI Bispecific antibodies: a mechanistic review of the pipeline. Nat. Rev. Drug Discov. 18, 585–608 (2019). [DOI] [PubMed] [Google Scholar]
- 52.Gateva V et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat. Genet. 41, 1228–1233 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McGovern D et al. Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nat. Genet. 42, 332–337 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pala M et al. Population- and individual-specific regulatory variation in Sardinia. Nat. Genet. 49, 700–707 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Lange KM et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat. Genet. 49, 256–261 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pruim RJ et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26, 2336–2337 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pilia G et al. Heritability of cardiovascular and personality traits in 6,148 Sardinians. PLoS Genet. 2, e132 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Das S et al. Next-generation genotype imputation service and methods. Nat. Genet. 48, 1284–1287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pistis G et al. Rare variant genotype imputation with thousands of study-specific whole-genome sequences: implications for cost-effective study designs. Eur. J. Hum. Genet. 23, 975–983 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Purcell S et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ferreira MA et al. Quantitative trait loci for CD4:CD8 lymphocyte ratio are associated with risk of type 1 diabetes and HIV-1 immune control. Am. J. Hum. Genet. 86, 88–92 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bentham J et al. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat. Genet. 47, 1457–1464 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Censing JC et al. Childhood adiposity and risk of type 1 diabetes: A Mendelian randomization study. PLoS Med. 14, e1002362 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Giambartolomei C et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 10, e1004383 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Palmer TM et al. Instrumental variable estimation of causal risk ratios and causal odds ratios in Mendelian randomization analyses. Am. J. Epidemiol. 173, 1392–1403 (2011). [DOI] [PubMed] [Google Scholar]
- 66.Hemani G et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife 7, e34408 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Teumer A Common methods for performing Mendelian randomization. Front. Cardiovasc. Med. 5, 51 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Full GWAS summary statistics have been deposited in the GWAS Catalog with accession numbers from GCST0001391 (https://www.ebi.ac.uk/gwas/studies/GCST0001391) to GCST0002121 (https://www.ebi.ac.uk/gwas/studies/GCST0002121). The accession number for each trait is reported in Supplementary Table 1B.
Disease summary statistics used to identify coincident associations were obtained from the GWAS Catalog (https://www.ebi.ac.uk/gwas/home, version v1.0, Ensembl release version e93, date 2018–08-28) and from ImmunoBase (https://genetics.opentargets.org/immunobase, version 12-May-2016). Summary statistics for colocalization analyses were downloaded from the respective web pages: RA (http://plaza.umin.ac.jp/~yokada/datasource/files/GWASMetaResults/RA_GWASmeta_European_v2.txt.gz); T1D (https://datadryad.org/stash/dataset/doi:10.5061/dryad.ns8q3); MS (http://imsgc.net/publications/); SLE (http://insidegen.com/insidegen-LUPUS-data.html); allergy (https://genepi.qimr.edu.au/staff/manuelf/gwas_results/SHARE-without23andMe.LDSCORE-GC.SE-META.v0.gz); IBD, Crohn’s disease and ulcerative colitis (ftp://ftp.sanger.ac.uk/pub/project/humgen/summary_statistics/human/2016-11-07/). Molecular QTLs are from the LinDA QTL Catalog (http://linda.irgb.cnr.it/, version 20190109). Source data are provided with this paper.