Abstract
Plants transitioned from an aquatic to a terrestrial lifestyle during their evolution. On land, fluctuations on water availability in the environment became one of the major problems they encountered. The appearance of morpho‐physiological adaptations to cope with and tolerate water loss from the cells was undeniably useful to survive on dry land. Some of these adaptations, such as carbon concentrating mechanisms (CCMs), desiccation tolerance (DT) and root impermeabilization, appeared in multiple plant lineages. Despite being crucial for evolution on land, it has been unclear how these adaptations convergently evolved in the various plant lineages. Recent advances on whole genome and transcriptome sequencing are revealing that co‐option of genes and gene regulatory networks (GRNs) is a common feature underlying the convergent evolution of these adaptations. In this review, we address how the study of CCMs and DT has provided insight into convergent evolution of GRNs underlying plant adaptation to dry environments, and how these insights could be applied to currently emerging understanding of evolution of root impermeabilization through different barrier cell types. We discuss examples of co‐option, conservation and innovation of genes and GRNs at the cell, tissue and organ levels revealed by recent phylogenomic (comparative genomic) and comparative transcriptomic studies.
Keywords: apoplastic barriers, comparative genomics, desiccation tolerance, exodermis
Short abstract
In this review, the authors highlight recent research on signatures of convergent evolution of regulatory networks underlying carbon concentrating mechanisms such as C4 and CAM photosynthesis, desiccation tolerance in seeds and resurrection plants and impermeabilization of root exodermis.
1. INTRODUCTION
The transition of an ancestor aquatic green algae to a terrestrial environment, termed terrestrialization, was a major event in the evolution and diversification of the land plant flora. About 500 Ma after the first ancestor colonized the land, the appearance of a multitude of morpho‐physiological adaptations allowed plants to cope with several problems related to terrestrial life, such as water scarcity (Becker & Marin, 2009; Delaux et al., 2012; Kenrick & Crane, 1997; Wodniok et al., 2011). Some of the major adaptations to terrestrial lifestyle include modification of the life cycle, divergence of the plant body into roots and shoots, the appearance of complex phenolic compounds (e.g., lignin and flavonoids), vascularization and the development of specialized cells (such as stomata) (Delaux et al., 2012). As they colonized land, exposure to high radiations and drought became a recurring problem encountered by multiple plant lineages, and common adaptations at the cell, tissue and organ levels emerged in diverging plant clades. It was until recently unknown how these morpho‐physiological adaptations evolved on a genetic level in phylogenetically distant plant species.
Recent advances on whole genome and transcriptome sequencing are now providing evidence that co‐option of genes and gene regulatory networks (GRNs) underlies the appearance of cell‐, organ‐ and tissue‐level adaptations to dry environments. For example, carbon concentrating mechanisms (CCMs) allow photosynthesis under high‐light and low‐water availability conditions and involve either temporal or spatial separation of the initial carbon fixing from the photosynthetic carbon fixing via anatomical adaptations (Edwards & Ogburn, 2012). Studies have shown that all of the enzymes necessary for the temporally separated Crassulacean acid metabolism (CAM) and the spatially separated C4 metabolic pathways are present in all plants and function in other processes (Burgess et al., 2016; Christin et al., 2013; Dunning, Moreno‐Villena, et al., 2019; Heyduk et al., 2019; Rondeau et al., 2005; Yang et al., 2017). The co‐option of these enzymes for CCMs was based on regulatory co‐option and neofunctionalization of pre‐existing genes, including those involved in C3 photosynthesis, and rewiring of ancestral GRNs (Figure 1) (Ming et al., 2015; Yang et al., 2017).
FIGURE 1.
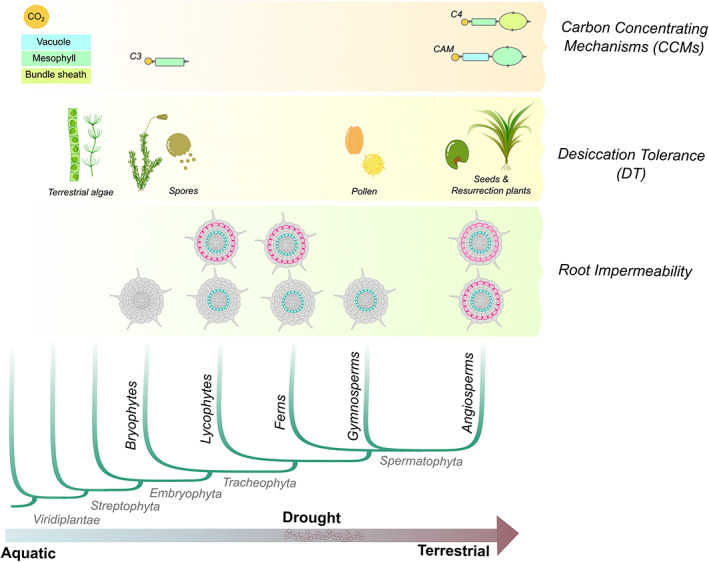
Overview of the evolution of plant adaptations to dry environments. The recurring appearance of carbon concentrating mechanisms (CCMs), desiccation tolerance (DT) and root impermeability in Viridiplantae lineages are shown. Root impermeability shows endodermis (yellow), exodermis with suberin only (bright turquoise) and exodermis with suberin and lignin (blue)
Another clear example of plant adaptation to dry environments is desiccation tolerance (DT), the ability to survive extreme drying and remain alive in the dry state (Alpert, 2000; Leprince & Buitink, 2010; Oliver et al., 2000). It has been long hypothesized that DT mechanisms present in the vegetative body of ancestral land plants became confined in small reproductive organs, such as seeds, during the evolution of tracheophytes (Figure 1) (Alpert, 2000; Oliver et al., 2000). These DT mechanisms of seeds were then co‐opted in the vegetative body of angiosperm ‘resurrection plants’ to be able to survive in extremely dry environments (Artur et al., 2019; Farrant & Moore, 2011). The co‐option of DT genes and GRNs has been recently assessed in the genome and transcriptome of resurrection plants (Costa et al., 2017; Giarola et al., 2017; VanBuren et al., 2017; VanBuren et al., 2018; VanBuren et al., 2019). Comparative genomic studies are also revealing that similar gene families and GRNs have expanded and/or being rewired similarly in multiple resurrection plants (Artur et al., 2019; Oliver et al., 2020; VanBuren et al., 2019).
The production of hydrophobic extracellular biopolymers (such as lignin, cutin and suberin) is also an important adaptation to survive on dry land that appeared in specific cells, tissues and organs of diverging plant clades. Comparative genome studies have shown that the ancestral green algae and red algae were already able to produce ‘lignin‐like’ compounds (Delwiche et al., 1989; Labeeuw et al., 2015; Martone et al., 2009), but lycophytes and spermatophytes seem to have independently developed the ability to produce monomers for lignin (Renault et al., 2017; Weng et al., 2010; Weng et al., 2008). Cutin and suberin seem to have also independently evolved in different plant clades, as homologs of genes encoding enzymes necessary for the biosynthesis of their precursors were absent in ancestral non‐angiosperm species (Cannell et al., 2020; Philippe et al., 2020; Pollard et al., 2008). The co‐option of GRNs for the biosynthesis of these biopolymers in specific root cell types has likely contributed to plant plasticity on dry land. For example, suberin is known to dynamically form barrier for water movement in root endodermis, and it also contributes to plant acclimation to drought when deposited in the exodermis (Ejiri & Shiono, 2019; Enstone et al., 2002; Kreszies et al., 2020; Líška et al., 2016; Reinhardt & Rost, 1995; Taleisnik et al., 1999). The evolution of exodermis remains poorly understood. A suberized exodermis possibly first appeared in early land plants (lycophytes). Its plasticity may have involved the co‐option of regulatory genes from the endodermis and may have convergently occurred in some flowering plant lineages (Angiosperms) submitted to constant environmental water fluctuations (Figure 1) (Perumalla et al., 1990). Due to the coexistence of endodermis and exodermis in roots, separation of these cell types for gene expression analyses has been required to understand which genes and GRNs are active in each cell type and to assess co‐option and innovation in exodermis GRN evolution (Kajala et al., 2021; Shiono et al., 2014).
In addition to CCMs, DT and root impermeabilization, there are many other adaptations to dry environments that have evolved convergently, such as succulence (Griffiths & Males, 2017), leaf shape (Nicotra et al., 2011; Peguero‐Pina et al., 2020) and shallow root system architecture (Griffiths & Males, 2017). However, in this review, we focus on the current knowledge about genes and GRNs recently found to be involved with the convergent evolution of CCMs, DT and root impermeabilization in different plant clades. Using these three examples, we provide insight into the current state of understanding of the following gaps in knowledge:
How do GRNs convergently evolve to adapt to dry environments?
Are the same genes and GRNs co‐opted or repurposed during independent evolutions?
What are the shared ancestral GRNs for these traits? What makes ancestral GRNs predisposed for convergent co‐option or repurposing?
What are the different, not shared, pathways taken during independent evolutions?
We will also address how functional genomics can help to generate and to test the novel hypotheses about the evolution and function of these genes.
2. CONVERGENT GRN EVOLUTION AT THE SPATIAL–TEMPORAL LEVEL: CARBON CONCENTRATING MECHANISMS
Dry habitats are often also sunny, placing further specific restraints on plant physiology and specifically photosynthesis: high‐light intensity and high temperature. As plants acclimate to drought by restricting their water loss through closing their stomata, they also restrict CO2 uptake, reducing the available CO2 for photosynthesis. High light becomes damaging if the energy obtained from light absorption is not used by the photosynthetic electron transport chain due to the absence of CO2. Simultaneously, high temperature reduces the solubility of gases, so that RuBisCO activity shifts away from carboxylation towards more oxygenation, increasing photorespiration and leading to wasted energy (Edwards, 2019). To adapt to these constraints, CCMs have evolved in numerous lineages to enable efficient photosynthesis in dry, hot and high‐light environments while improving water‐use efficiency.
CCMs work by separating the initial carbon fixing away from the photosynthetic carbon fixing (Calvin cycle) either temporally across the diurnal cycle or spatially across different cell types or compartments. Both the temporal separation, as seen in CAM, and spatial separation, as seen in C4 photosynthesis, have evolved independently in over 60 lineages (Edwards & Ogburn, 2012). The evolutionary paths to both CAM and C4 photosynthesis have been recently reviewed and discussed in great detail (Bräutigam et al., 2017; Chen et al., 2020; Edwards, 2019; Heyduk et al., 2019; Niklaus & Kelly, 2019; Schlüter & Weber, 2020; Sedelnikova et al., 2018), and in short, both require two main aspects: (a) an anatomical adaptation and (b) co‐option of the carbon concentrating metabolic pathway to the correct spatiotemporal location. The main anatomical adaptation for CAM is enlarged storage vacuole to store the malate synthesized during the night and enable the day–night CCM (Luttge, 1987). In many C4 plants, the spatial separation is across two cell types, mesophyll and bundle sheath, and to achieve this, C4 leaves adapt with so‐called Kranz anatomy with enlarged bundle sheath cells with increased plastid numbers and increased vein density (Haberlandt, 1904). The regulation of Kranz anatomy is proving to be a complex process (Sedelnikova et al., 2018), and although it has readily evolved convergently in some plant clades, it is starting to appear that not all plant clades are pre‐conditioned for the C4 photosynthesis to evolve (Edwards & Ogburn, 2012). Conversely to complex leaf anatomy and its regulation, all the enzymes required for both the CAM and C4 metabolic pathways, such as phosphoenolpyruvate carboxylase and malate dehydrogenase, are present in all plants serving other functions. To co‐opt these enzymes for CCMs, the expression and regulatory patterns have evolved to be spatially and temporally specific (Brown et al., 2011; Burgess et al., 2016; Christin et al., 2013; Dunning, Moreno‐Villena, et al., 2019; Gowik et al., 2004; Heyduk, Ray, et al., 2019; Kajala et al., 2012; Ming et al., 2015; Rondeau et al., 2005; Schulze et al., 2013; Williams et al., 2016; Yang et al., 2017). Whole genome sequencing has enabled a level of understanding of how these CCMs evolved in plants. The first C4 (Sorghum bicolor) (Paterson et al., 2009) and CAM (Phalaenopsis equestris) (Cai et al., 2015) genomes provided insights about redirection of genes involved in C3 photosynthesis and expansion of ancient and recent gene families. Transcriptomic approaches are also offering new evidence about convergent evolution of genes and regulatory pathways underlying these CCMs. For example, the genome and temporal transcriptome sequencing of the CAM species Kalanchoë fedtschenkoi revealed that the independent emergences of CAM from C3 have been based on rewiring of diel gene expression patterns along with protein sequence mutations (Yang et al., 2017). Furthermore, the pineapple genome (Ananas comosus [L.] Merr.), another CAM species, indicated that the transition from C3 to CAM was based on regulatory neofunctionalization of pre‐existing genes and regulation of circadian clock components through evolution of novel cis‐regulatory elements (Ming et al., 2015).
To resolve how gene expression patterns and GRNs have evolved in convergent C4 lineages, comparative leaf transcriptomics have been utilized, including comparisons of C3, C4 and intermediate C3‐C4 leaves, developmental gradients, specific cell types and environmental cues (Aubry et al., 2014; Bräutigam et al., 2011; Burgess et al., 2016; Dunning et al., 2017; Gowik et al., 2004; Li et al., 2010). From these comparisons, shared routes to C4 are starting to emerge. Transcriptomic comparison across the monocot–dicot divide revealed deep evolutionary conservation of C4 leaf development pathways and that certain homologous cell type–specific regulators were co‐opted during the independent evolutions of C4 photosynthesis (Aubry et al., 2014). Not only is it possible to co‐opt the same orthologues as another species, but it is also possible to co‐opt the very gene from another species. Transcriptomics of closely related Alloteropsis grasses revealed that recurrent evolution of C4 among the group was enabled by co‐opting genes across species boundaries, specifically by introgressions of the C4 components (Dunning et al., 2017). Subsequently, it was discovered that grasses have utilized lateral gene transfer also to speed up C4 evolution (Dunning, Olofsson, et al., 2019; Phansopa et al., 2020). Furthermore, the understanding of C4 enzymes' regulatory networks in ancestral C3 state was elucidated also by a transcriptomics approach: comparison of how light and chloroplasts regulate C4 enzymes in closely related C3 and C4 plants. This linked the C4 enzymes into a pre‐existing C3 regulatory network, explaining the readiness of C4 to evolve at the molecular level (Burgess et al., 2016). Regardless of these examples of GRN readiness for C4 evolution, or ‘genetic potentiation’, the C4 lineages are not evenly distributed across the angiosperms (Edwards et al., 2010). This can be due either to the different lineages facing different evolutionary pressures or differences in either genetic or anatomic potentiation (Heyduk, Moreno‐Villena, et al., 2019) and remains a question to be addressed in the future.
Counter‐intuitively, C4 and CAM can exist in the same leaf. A recent transcriptomic approach was taken to dissect the behaviour of both C4 and drought‐induced CAM in the same plant, Portulaca oleracea, offering insight on how the regulatory networks of shared enzymes might be able to coexist while responding to different environmental and temporal cues (Ferrari et al., 2020). With more C4 and CAM genomes and transcriptomes becoming recently available (Phytozome v.12.1, Goodstein et al. (2012)), the door is open for better understanding of convergent evolution of CCMs and other parallel drought adaptations in these species.
3. DESICCATION TOLERANCE GRN IN SEEDS AND RESURRECTION PLANTS
DT, the ability to survive the loss of almost all cellular water without irreversible damage, appeared in the plant lineage during terrestrialization of their ancestor streptophyte algae (Figure 1) (Alpert, 2000; Leprince & Buitink, 2010; Oliver et al., 2000; Terlova et al., 2021; Wodniok et al., 2011). DT refers to survival after extreme dehydration (below 0.3 g H2O/g dry weight) where the cellular metabolic activity nearly stops, entering into a state of ‘anhydrobiosis’ (Hoekstra et al., 2001).
At least six independent major clades of green algae were able to colonize dry environments and to display this remarkable ability to withstand extreme dehydration (Lewis & McCourt, 2004; Terlova et al., 2021). The initial appearance of DT features in the ancestor green algae was a crucial step for plant radiation on land (Oliver et al., 2000; Rensing et al., 2008; Wodniok et al., 2011). Vegetative DT is nowadays documented in about 68 bryophytes, 10 ferns and 10 angiosperms families (Artur, Costa, et al., 2019; Marks et al., 2021; Oliver et al., 2000; Oliver et al., 2020). DT has been retained in reproductive structures (such as spores, pollen and seeds) of most land plant lineages, suggesting that virtually all plants display the genetic potential to become DT but are limited by morpho‐physiological constraints (Alpert, 2000; Farrant & Moore, 2011; Marks, Farrant, et al., 2021; Oliver et al., 2000). It has been proposed that vegetative DT in Angiosperm resurrection plants re‐evolved by re‐directing common regulatory pathways from their reproductive structures (Costa et al., 2017; Farrant & Moore, 2011; Oliver et al., 2000). In fact, common morpho‐physiological and biochemical signatures of DT have been found among evolutionarily distant resurrection plants and between desiccation‐tolerant vegetative and reproductive structures. One example is the ability that certain groups of resurrection plants have to display leaf curling, rolling or folding, which provide protection against photo‐damage during dehydration (‘homoichlorophylly’), while other groups undergo chlorophyll breakdown, chloroplast disassembly and synthesis of anthocyanin (‘poikilochlorophylly’), a common mechanism involved in seed acquisition of DT (Alpert, 2000; Artur, Zhao, et al., 2019; Charuvi et al., 2019; Dekkers et al., 2015; Radermacher et al., 2019).
Recent developments in sequencing technologies are now also facilitating the assessment of genomic and regulatory adaptations underlying the origin and convergent evolution of DT in plants. In the past 5 years, at least eight whole genomes and several transcriptomes of desiccation‐tolerant plant species from distinct phylogenetic groups became available (Artur, Costa, et al., 2019; Oliver et al., 2020). Comparison between desiccation‐tolerant and desiccation‐sensitive genomes and transcriptomes is revealing loss and repurposing of genes associated with the aquatic lifestyle of the ancestor green algae and the expansion of gene families and refinement of gene expression necessary for survival on dry environments (Khraiwesh et al., 2015; Marks et al., 2021; Peredo & Cardon, 2020; Rensing et al., 2008; VanBuren et al., 2019; Xu et al., 2018). The latter is clearly exemplified by expansion of late embryogenesis abundant proteins (LEAs) and early‐light induced proteins (ELIPs) families in resurrection plants (Costa et al., 2017; Khraiwesh et al., 2015; Rensing et al., 2008; VanBuren et al., 2019; Xu et al., 2018). LEA genes are expressed during orthodox seed maturation, and their corresponding proteins accumulate when the seeds start to dry (Dure et al., 1989; Dure III et al., 1980; Galau et al., 1986; Verdier et al., 2013). These proteins are intrinsically disordered and can undergo disorder to‐order transitions, what contribute to stabilization of membranes, organelles and the cytoplasm (Artur, Rienstra, et al., 2019; Buitink & Leprince, 2004; Crowe et al., 1992; Wise & Tunnacliffe, 2004). A few LEA families were found to be commonly expanded in resurrection plant genomes, what indicates that specific LEAs contributed to the convergent evolution of DT in these species (Artur, Zhao, et al., 2019; Costa et al., 2017; VanBuren et al., 2017). Similarly, ELIPs, known to protect the cells against photo‐oxidative damage under high‐light intensities (Hutin et al., 2003), have undergone a massive proliferation as tandem duplications in the genome of resurrection plants (VanBuren et al., 2019). Altogether, these studies are shedding light on common patterns of gene family expansion associated with convergent evolution of DT in resurrection plants.
Usually, angiosperm resurrection plants respond to vegetative desiccation by inducing the expression of regulatory pathways typically related to seed DT (Costa et al., 2017; Giarola et al., 2017; Pardo et al., 2020; VanBuren et al., 2017). A recent study has shown, however, that despite conserved seed regulatory networks being activated in vegetative tissues of the poikilochlorophyllous resurrection plant Xerophyta humilis, the master transcription factors (TFs) upstream of these pathways in seeds are not activated in vegetative tissues (Lyall et al., 2020). This finding brings novel hypotheses about the evolution of DT. For example, it is likely that the activation of components of seed DT in vegetative tissues involved the appearance of alternative TFs that have evolved in a similar fashion in different resurrection plant genomes. A comparative genome and transcriptome study have recently shown that seed dehydration‐related genes shared similar expression patterns among desiccation‐tolerant and ‐sensitive grass species during drought; however, subsets of seed‐specific genes were identified as expressing only in desiccation‐tolerant grasses (Pardo et al., 2020). With more seed and resurrection plant genomes and transcriptomes becoming available, more information will be provided about the identity of the genes and pathways that underlie the convergent evolution of DT in different plant species and organs.
4. ROOT CELL TYPE GRN ADAPTATIONS TO DROUGHT: IMPERMEABILIZATION OF ENDODERMIS AND EXODERMIS
The evolution of hydrophobic extracellular biopolymers was a critical innovation for plant terrestrialization, contributing to permeability and water transport control. Some of these compounds include complex phenolic‐derived polymers such as lignin, cutin and suberin (Niklas et al., 2017). Lignin generally works in the reinforcement of secondary cell walls; cutin is commonly found as part of the impermeabilizing cuticle of aerial parts and was recently reported as being also part of the root cap cuticle, and suberin is found in many tissues including specialized root cells, tubers, fruit skin and seed coat (Berhin et al., 2019; Niklas et al., 2017; Philippe et al., 2020; Renault et al., 2017). In general, these compounds function in mechanical support, defence against pathogens and herbivores and in the control of the movement of water, nutrient and gases (Niklas et al., 2017; Philippe et al., 2020; Renault et al., 2017; Pei Wang et al., 2020).
Convergent evolution has also played an important role in the appearance and shaping of the biosynthetic pathways of some of these biopolymers across diverging plant taxa. For example, the ancestral green algae and red algae have the ability to produce ‘lignin‐like’ compounds, leading to the hypothesis that ancient biosynthesis pathways have been rewired in the vascular plant lineage (Delwiche et al., 1989; Labeeuw et al., 2015; Martone et al., 2009). Furthermore, lycophytes and spermatophytes have independently developed the ability to produce monomers necessary for lignin biosynthesis and assembly (Renault et al., 2017; Weng et al., 2008; Weng et al., 2010). It is possible that convergent evolution also played a role on the appearance of cutin and suberin across distantly related plant species. Both cutin and suberin share similar initial biosynthetic steps and lipid precursors; however, different enzymes belonging to the same superfamily and with different mechanisms of action work on their modification and assembly (Philippe et al., 2020; Pollard et al., 2008). A recent large‐scale comparative genomic study has revealed that the ability to synthesize precursor molecules of cutin and suberin emerged prior the evolution of land plants; however, the subsequent steps of the biosynthetic pathway may have evolved independently across land plants (Cannell et al., 2020). Still, little is known about how the other parts of the pathway, such as polymerization and distribution in plant cells, evolved in land plants (Niklas et al., 2017; Philippe et al., 2020; Pollard et al., 2008).
The diversification of biosynthetic pathways leading to the production of impermeabilizing hydrophobic compounds has also contributed to the evolution of distinct cell types important for adaptation to terrestrial life and for resilience to drought. For example, the roots of all vascular plants contain an endodermis surrounding the vascular tissues (Doblas et al., 2017; Enstone et al., 2002). The endodermis cell layer forms a diffusion barrier for water, gases and nutrients due to the presence of two cell wall modifications: the Casparian strip and suberin lamella (Barberon et al., 2016; Doblas et al., 2017; Seago Jr. & Fernando, 2013; Vishwanath et al., 2015). Casparian strip is composed of lignin, deposited in the walls of endodermal cells at their junctions, dividing the layer into outward and inward polarities and forming an effective barrier to the apoplastic movement of molecules into the stele and preventing their backflow (Barberon, 2017; Enstone et al., 2002; D. Roppolo et al., 2014; Daniele Roppolo et al., 2011). The suberin lamella is a secondary cell wall modification deposited in the inner surface of the primary cell walls, usually after the Casparian strip is formed in the mature endodermis (Barberon, 2017; Enstone et al., 2002). Different from Casparian strip, the suberin lamella may not form in every root nor in every endodermal cell (the so‐called passage cells) (Andersen et al., 2018; Barberon et al., 2016; Enstone et al., 2002; Holbein et al., 2021). Despite debate in the past years, the role of suberin lamella as an apoplastic barrier for water and nutrient uptake from the apoplast to the endodermis cytoplasm has been demonstrated (Barberon et al., 2016; Ranathunge & Schreiber, 2011; Peng Wang et al., 2019).
The roots of several species also develop an exodermis below the epidermis, which is a specialized type of hypodermis with Casparian bands and suberin lamellae depositions (Enstone et al., 2002; Perumalla et al., 1990). The exodermis functions as a dynamic barrier not only against water loss under drought and salinity but also against loss of oxygen under anoxic conditions, against penetration of ions and heavy metals and against pathogen infections (Aloni et al., 1998; Damus et al., 1997; Ejiri & Shiono, 2019; Enstone et al., 2002; Líška et al., 2016; Namyslov et al., 2020; Ranathunge et al., 2011; Tylová et al., 2017). At the same time, the development of exodermis barriers has its downside as it may impair the uptake of nutrients and interaction with beneficial microbes (Kamula et al., 1994). To cope with this problem, many plant species developed the ability to induce an exodermis dynamically in response to abiotic stresses, such as drought (Enstone et al., 2002; Kreszies et al., 2020; Líška et al., 2016; Reinhardt & Rost, 1995; Taleisnik et al., 1999). Interestingly, the development of the exodermis may vary among closely related species displaying distinct stress response phenotypes (Ejiri & Shiono, 2019), indicating that this cell type contributes to plant plasticity and acclimation and may also help plants to adapt and colonize dry environments.
Regardless of its adaptive role, the evolution of exodermis in plants still remains untangled. Perumalla et al. (1990) surveyed 181 species from 53 families of plants from different ecological groups (hydrophytic, mesophytic and xerophytic) to determine the presence of hypodermis with Casparian bands (exodermis). As the majority (156) of the species assessed presented an exodermis with suberin only (hypodermis) or with both suberin and lignin, the authors hypothesized that the presence of a modified hypodermis is ancestral to flowering plants and has been retained in many species (Perumalla et al., 1990). Furthermore, the authors found that festucoid grasses lack Casparian bands despite presenting cells with similar shape and packing as species with hypodermal Casparian bands, leading to the hypothesis that their recent ancestor may have lost the trait. Interestingly, in seminal roots of modern cultivars of barley (a festucoid species), the exodermis fails to develop even upon severe osmotic stress (Kreszies et al., 2019), while in wild barley, the exodermis is induced in response to osmotic stress (Kreszies et al., 2020). On the other hand, in other crop grasses (non‐festucoid), such as rice and maize, an exodermis is present and develops faster in response to stress (Ranathunge et al., 2016; Schreiber et al., 2005).
Understanding how the exodermis evolved in plants can help in the identification of the underlying regulatory networks responsible for its induction in response to drought. To obtain more knowledge about the evolution of exodermis, we compiled the current information about their presence in plant species based on literature search (Figure 2) (Bani et al., 2018; Barrios‐Masias et al., 2015; Barykina & Kramina, 2006; M. Brundrett et al., 1990; Calvo‐Polanco et al., 2014; Damus et al., 1997; Demchenko et al., 2004; Eissenstat & Achor, 1999; Ejiri & Shiono, 2019; Enstone et al., 2002; Ghanati et al., 2005; Kosma et al., 2015; Liu et al., 2019; Perumalla et al., 1990; Ranathunge et al., 2017; Reinhardt & Rost, 1995; Ron et al., 2013; Schreiber et al., 2005; Schreiber et al., 1999; Shiono & Yamada, 2014; Thomas et al., 2007; Zhang et al., 2018). Based on this analysis, the exodermis with suberin first appeared in early land plants (lycophytes), but it is missing from other seedless vascular plants and all but one gymnosperm (Damus et al., 1997). Interestingly, four species in the lycophyte genus Selaginella contain exodermis with lignified Casparian strips (Damus et al., 1997). Most flowering plants contain an exodermis with suberization only (hypodermis), while a lignified exodermis appears in about a third of the species. The scattered appearances of the exodermal lignification indicate that it has evolved independently multiple times, suggesting a high evolutionary pressure and pre‐conditioning for the characteristic to arise. Species with no exodermis have been identified in seven clades (in purple, Figure 2), and the most parsimonious explanation for presence/absence of exodermis is the loss of the cell type in these lineages. However, the evolutionary hypotheses are restricted by the sparse sampling in families of interest. This is highlighted by the relevant literature containing contradictions (e.g., pea Bani et al. (2018); Perumalla et al. (1990); Taleisnik et al. (1999)), likely due to the dynamic nature of the exodermis.
FIGURE 2.
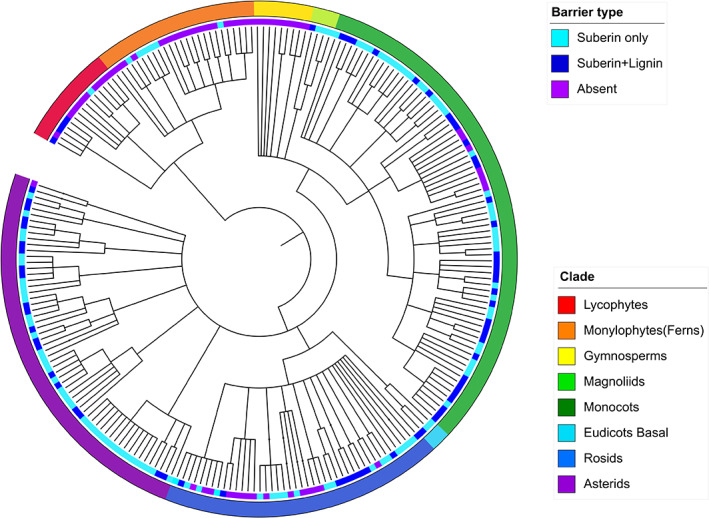
Exodermis (hypodermis) evolution across land plants. The inner ring represents the exodermis phenotype and the outer ring, the major clades. The phylogenetic tree was generated using NCBI Common Tree (www.ncbi.nlm.nih.gov/Taxonomy/CommonTree/wwwcmt.cgi) and edited using iTOL (itol.embl.de/)
Evolutionary studies focussed on characterizing the exodermis, for example, by staining suberin and lignin or using barrier property assays (Table S1) will contribute with important information about how this cell type has appeared or disappeared multiple times across the plant lineages. Coupling that with comparative genomics and transcriptomics of phylogenetically close species (e.g., from the same family) but with different phenotypes (e.g., non‐exodermal, constitutive and stress‐inducible exodermis) will be key to identify the origin of the regulatory networks and how master regulators underlying exodermis development and suberization in response to drought evolved. It is possible that the sporadic appearance of exodermis during plant evolution was possible through rewiring regulatory networks of Casparian strips and suberin lamellae formation in endodermis or similar lignin and suberin biosynthetic pathways from other cell types. The coexistence of endodermis and exodermis in roots has complicated transcriptomic approaches, and to investigate the exodermis gene expression and GRNs in individual species, methods such as laser capture microdissection and translating ribosome affinity purification (TRAP) have been used (Kajala et al., 2021; Shiono et al., 2014). The exodermis‐specific gene expression patterns and GRNs are a foundation for understanding the genetic underpinnings of the cell type and enable investigating its evolutionary paths.
Recent studies are showing the importance of distinct clades of the MYB TF family as conserved regulators of suberin deposition in response to osmotic stress in different cell types and in phylogenetically distant plants, linking their evolution with colonization of dry terrestrial environments by early land plants (Capote et al., 2018; Cohen et al., 2020; Gou et al., 2017; Kajala et al., 2021; Kosma et al., 2014; Lashbrooke et al., 2016; Legay et al., 2016; Shukla et al., 2021; To et al., 2020; Wei et al., 2020). Two of the possible scenarios are: (a) osmotic stress–inducible regulation of suberization diversified from pre‐existing developmental pathways and (b) the regulation of suberization in response to drought was re‐activated as plants colonized drier environments. Taking that into account, further evolutionary/phylogenetic study of exodermis is needed and selection of a good clade(s) to dissect the gain/loss events is key for understanding exodermis development and how it evolves so readily.
5. CONCLUSIONS AND PERSPECTIVES
The appearance and diversification of adaptations to dry environments and their underlying GRNs have been tightly shaped at cell, tissue and organismal levels during evolution. These adaptations were crucial for plants to colonize dry lands and survive variations on water availability in the environment. Carbon concentration mechanisms, DT and root impermeabilization are among the main plant adaptations to dry environments and share a similar pattern of convergent evolution in phylogenetically distant organisms. However, we are only starting to understand the evolutionary paths of the GRNs underpinning these adaptations in individual lineages. More comparative research between independent evolutions will inform us of the possible GRN evolutionary paths, including finding out if the same molecular and regulatory changes have occurred in independent lineages.
Detailed physiological and histological studies combined with advanced genome and transcriptome sequencing techniques such as TRAP‐seq and single‐cell sequencing (Heiman et al., 2014; Reynoso et al., 2015; Thellmann et al., 2020) will allow us to assess how these adaptations evolved through millions of years. This information in combination with novel methods to simulate drought conditions (e.g., Marchin et al. (2019)) and technologies for plant genetic modification (e.g., CRISPR/Cas9) will have the power to help on the improvement of plant performance in dry environments, especially in the current scenario of increasing climate change. These data will also provide knowledge about key TFs working upstream of GRNs controlling DT pathways, which are of potential interest for engineering more drought‐tolerant crops.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supporting information
Table S1. Histochemical tests for barrier cell‐type identification
ACKNOWLEDGEMENTS
KK was supported by Marie Skłodowska‐Curie Re‐integration fellowship #790057.
Artur, M. A. S. , & Kajala, K. (2021). Convergent evolution of gene regulatory networks underlying plant adaptations to dry environments. Plant, Cell & Environment, 44(10), 3211–3222. 10.1111/pce.14143
Funding information Marie Skłodowska‐Curie re‐integration fellowship, Grant/Award Number: 790057
Contributor Information
Mariana A. S. Artur, Email: mariana.silvaartur@wur.nl.
Kaisa Kajala, Email: k.kajala@uu.nl.
REFERENCES
- Aloni, R. , Enstone, D. E. , & Peterson, C. A. (1998). Indirect evidence for bulk water flow in root cortical cell walls of three dicotyledonous species. Planta, 207(1), 1–7. [Google Scholar]
- Alpert, P. (2000). The discovery, scope, and puzzle of desiccation tolerance in plants. Plant Ecology, 151(1), 5–17. [Google Scholar]
- Andersen, T. G. , Naseer, S. , Ursache, R. , Wybouw, B. , Smet, W. , De Rybel, B. , … Geldner, N. (2018). Diffusible repression of cytokinin signalling produces endodermal symmetry and passage cells. Nature, 555(7697), 529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artur, M. A. S. , Costa, M. C. D. , Farrant, J. M. , & Hilhorst, H. W. M. (2019). Genome‐level responses to the environment: Plant desiccation tolerance. Emerging Topics in Life Sciences, 3(2), 153–163. [DOI] [PubMed] [Google Scholar]
- Artur, M. A. S. , Rienstra, J. , Dennis, T. J. , Farrant, J. M. , Ligterink, W. , & Hilhorst, H. (2019). Structural plasticity of intrinsically disordered LEA proteins from Xerophyta schlechteri provides protection in vitro and in vivo. Frontiers in Plant Science, 10, 1272. 10.3389/fpls.2019.01272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artur, M. A. S. , Zhao, T. , Ligterink, W. , Schranz, E. , & Hilhorst, H. W. M. (2019). Dissecting the genomic diversification of late embryogenesis abundant (LEA) protein gene families in plants. Genome Biology and Evolution, 11(2), 459–471. 10.1093/gbe/evy248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubry, S. , Kelly, S. , Kümpers, B. M. , Smith‐Unna, R. D. , & Hibberd, J. M. (2014). Deep evolutionary comparison of gene expression identifies parallel recruitment of trans‐factors in two independent origins of C 4 photosynthesis. PLoS Genetics, 10(6), e1004365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bani, M. , Pérez‐De‐Luque, A. , Rubiales, D. , & Rispail, N. (2018). Physical and chemical barriers in root tissues contribute to quantitative resistance to Fusarium oxysporum f. sp. pisi in pea. Frontiers in Plant Science, 9, 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberon, M. (2017). The endodermis as a checkpoint for nutrients. The New Phytologist, 213(4), 1604–1610. 10.1111/nph.14140 [DOI] [PubMed] [Google Scholar]
- Barberon, M. , Vermeer, J. E. , De Bellis, D. , Wang, P. , Naseer, S. , Andersen, T. G. , … Geldner, N. (2016). Adaptation of root function by nutrient‐induced plasticity of endodermal differentiation. Cell, 164(3), 447–459. 10.1016/j.cell.2015.12.021 [DOI] [PubMed] [Google Scholar]
- Barrios‐Masias, F. , Knipfer, T. , & McElrone, A. (2015). Differential responses of grapevine rootstocks to water stress are associated with adjustments in fine root hydraulic physiology and suberization. Journal of Experimental Botany, 66(19), 6069–6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barykina, R. P. , & Kramina, T. E. (2006). A comparative morphological and anatomical study of the model legume Lotus japonicus and related species. Wulfenia, 13, 33–56. [Google Scholar]
- Becker, B. , & Marin, B. (2009). Streptophyte algae and the origin of embryophytes. Annals of Botany, 103(7), 999–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berhin, A. , de Bellis, D. , Franke, R. B. , Buono, R. A. , Nowack, M. K. , & Nawrath, C. (2019). The root cap cuticle: A cell wall structure for seedling establishment and lateral root formation. Cell, 176(6), 1367–1378 e1368. 10.1016/j.cell.2019.01.005 [DOI] [PubMed] [Google Scholar]
- Bräutigam, A. , Kajala, K. , Wullenweber, J. , Sommer, M. , Gagneul, D. , Weber, K. L. , … Weber, A. P. M. (2011). An mRNA blueprint for C4 photosynthesis derived from comparative transcriptomics of closely related C3 and C4 species. Plant Physiology, 155(1), 142–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräutigam, A. , Schlüter, U. , Eisenhut, M. , & Gowik, U. (2017). On the evolutionary origin of CAM photosynthesis. Plant Physiology, 174(2), 473–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, N. J. , Newell, C. A. , Stanley, S. , Chen, J. E. , Perrin, A. J. , Kajala, K. , & Hibberd, J. M. (2011). Independent and parallel recruitment of preexisting mechanisms underlying C4 photosynthesis. Science, 331(6023), 1436–1439. [DOI] [PubMed] [Google Scholar]
- Brundrett, M. , Murase, G. , & Kendrick, B. (1990). Comparative anatomy of roots and mycorrhizae of common Ontario trees. Canadian Journal of Botany, 68(3), 551–578. [Google Scholar]
- Brundrett, M. C. , Enstone, D. E. , & Peterson, C. A. (1988). A berberine‐aniline blue fluorescent staining procedure for suberin, lignin, and callose in plant tissue. Protoplasma, 146(2), 133–142. [Google Scholar]
- Brundrett, M. C. , Kendrick, B. , & Peterson, C. A. (1991). Efficient lipid staining in plant material with Sudan Red 7B or Fluoral Yellow 088 in polyethylene glycol‐glycerol. Biotechnic & Histochemistry, 66(3), 111–116. [DOI] [PubMed] [Google Scholar]
- Buitink, J. , & Leprince, O. (2004). Glass formation in plant anhydrobiotes: Survival in the dry state. Cryobiology, 48(3), 215–228. [DOI] [PubMed] [Google Scholar]
- Burgess, S. J. , Granero‐Moya, I. , Grangé‐Guermente, M. J. , Boursnell, C. , Terry, M. J. , & Hibberd, J. M. (2016). Ancestral light and chloroplast regulation form the foundations for C 4 gene expression. Nature Plants, 2(11), 1–6. [DOI] [PubMed] [Google Scholar]
- Cai, J. , Liu, X. , Vanneste, K. , Proost, S. , Tsai, W.‐C. , Liu, K.‐W. , … Liu, Z.‐J. (2015). The genome sequence of the orchid Phalaenopsis equestris. Nature Genetics, 47(1), 65–72. [DOI] [PubMed] [Google Scholar]
- Calvo‐Polanco, M. , Sánchez‐Romera, B. , & Aroca, R. (2014). Mild salt stress conditions induce different responses in root hydraulic conductivity of Phaseolus vulgaris over‐time. PLoS One, 9(3), e90631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannell, N. , Emms, D. M. , Hetherington, A. J. , MacKay, J. , Kelly, S. , Dolan, L. , & Sweetlove, L. J. (2020). Multiple metabolic innovations and losses are associated with major transitions in land plant evolution. Current Biology, 30(10), 1783–1800 e1711. 10.1016/j.cub.2020.02.086 [DOI] [PubMed] [Google Scholar]
- Capote, T. , Barbosa, P. , Usié, A. , Ramos, A. M. , Inácio, V. , Ordás, R. , … Morais‐Cecílio, L. (2018). ChIP‐Seq reveals that QsMYB1 directly targets genes involved in lignin and suberin biosynthesis pathways in cork oak (Quercus suber). BMC Plant Biology, 18(1), 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charuvi, D. , Nevo, R. , Aviv‐Sharon, E. , Gal, A. , Kiss, V. , Shimoni, E. , … Reich, Z. (2019). Chloroplast breakdown during dehydration of a homoiochlorophyllous resurrection plant proceeds via senescence‐like processes. Environmental and Experimental Botany, 157, 100–111. [Google Scholar]
- Chen, L.‐Y. , Xin, Y. , Wai, C. M. , Liu, J. , & Ming, R. (2020). The role of cis‐elements in the evolution of crassulacean acid metabolism photosynthesis. Horticulture Research, 7(1), 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christin, P.‐A. , Boxall, S. F. , Gregory, R. , Edwards, E. J. , Hartwell, J. , & Osborne, C. P. (2013). Parallel recruitment of multiple genes into C4 photosynthesis. Genome Biology and Evolution, 5(11), 2174–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, H. , Fedyuk, V. , Wang, C. , Wu, S. , & Aharoni, A. (2020). SUBERMAN regulates developmental suberization of the Arabidopsis root endodermis. The Plant Journal, 102(3), 431–447. [DOI] [PubMed] [Google Scholar]
- Costa, M. D. , Artur, M. A. , Maia, J. , Jonkheer, E. , Derks, M. F. , Nijveen, H. , Williams Brett, Sagadevan G. Mundree , José M. Jiménez‐Gómez , Hesselink Thamara, Schijlen Elio G. W. M., Ligterink Wilco, Melvin J. Oliver , Jill M. Farrant , Hilhorst, H. W. (2017). A footprint of desiccation tolerance in the genome of Xerophyta viscosa . Nature Plants , 3, 17038. doi: 10.1038/nplants.2017.38 [DOI] [PubMed] [Google Scholar]
- Crowe, J. H. , Hoekstra, F. A. , & Crowe, L. M. (1992). Anhydrobiosis. Annual Review of Physiology, 54, 579–599. 10.1146/annurev.ph.54.030192.003051 [DOI] [PubMed] [Google Scholar]
- Damus, M. , Peterson, R. , Enstone, D. E. , & Peterson, C. A. (1997). Modifications of cortical cell walls in roots of seedless vascular plants. Botanica Acta, 110(2), 190–195. [Google Scholar]
- Dekkers, B. J. , Costa, M. C. D. , Maia, J. , Bentsink, L. , Ligterink, W. , & Hilhorst, H. W. (2015). Acquisition and loss of desiccation tolerance in seeds: From experimental model to biological relevance. Planta, 241(3), 563–577. [DOI] [PubMed] [Google Scholar]
- Delaux, P.‐M. , Nanda, A. K. , Mathé, C. , Sejalon‐Delmas, N. , & Dunand, C. (2012). Molecular and biochemical aspects of plant terrestrialization. Perspectives in Plant Ecology, Evolution and Systematics, 14(1), 49–59. [Google Scholar]
- Delwiche, C. F. , Graham, L. E. , & Thomson, N. (1989). Lignin‐like compounds and sporopollenin coleochaete, an algal model for land plant ancestry. Science, 245(4916), 399–401. 10.1126/science.245.4916.399 [DOI] [PubMed] [Google Scholar]
- Demchenko, K. , Winzer, T. , Stougaard, J. , Parniske, M. , & Pawlowski, K. (2004). Distinct roles of Lotus japonicus SYMRK and SYM15 in root colonization and arbuscule formation. New Phytologist, 163(2), 381–392. [DOI] [PubMed] [Google Scholar]
- Doblas, V. G. , Geldner, N. , & Barberon, M. (2017). The endodermis, a tightly controlled barrier for nutrients. Current Opinion in Plant Biology, 39, 136–143. 10.1016/j.pbi.2017.06.010 [DOI] [PubMed] [Google Scholar]
- Dunning, L. T. , Lundgren, M. R. , Moreno‐Villena, J. J. , Namaganda, M. , Edwards, E. J. , Nosil, P. , … Christin, P. A. (2017). Introgression and repeated co‐option facilitated the recurrent emergence of C4 photosynthesis among close relatives. Evolution, 71(6), 1541–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning, L. T. , Moreno‐Villena, J. J. , Lundgren, M. R. , Dionora, J. , Salazar, P. , Adams, C. , … Grundy, I. M. (2019). Key changes in gene expression identified for different stages of C4 evolution in Alloteropsis semialata . Journal of Experimental Botany, 70(12), 3255–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning, L. T. , Olofsson, J. K. , Parisod, C. , Choudhury, R. R. , Moreno‐Villena, J. J. , Yang, Y. , … Bennetzen, J. L. (2019). Lateral transfers of large DNA fragments spread functional genes among grasses. Proceedings of the National Academy of Sciences, 116(10), 4416–4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dure, L. S., III , Galau, G. A. , & Greenway, S. (1980). Changing protein patterns during cotton cotyledon embryogenesis and germination as shown by in vivo and in vitro synthesis. Israel Journal of Botany, 29(1–4), 293–306. [Google Scholar]
- Dure, L. , Crouch, M. , Harada, J. , Ho, T.‐H. D. , Mundy, J. , Quatrano, R. , Thomas, T. , & Sung, Z. (1989). Common amino acid sequence domains among the LEA proteins of higher plants. Plant Molecular Biology, 12(5), 475–486. [DOI] [PubMed] [Google Scholar]
- Edwards, E. J. (2019). Evolutionary trajectories, accessibility and other metaphors: The case of C4 and CAM photosynthesis. New Phytologist, 223(4), 1742–1755. [DOI] [PubMed] [Google Scholar]
- Edwards, E. J. , & Ogburn, R. M. (2012). Angiosperm responses to a low‐CO2 world: CAM and C4 photosynthesis as parallel evolutionary trajectories. International Journal of Plant Sciences, 173(6), 724–733. [Google Scholar]
- Edwards, E. J. , Osborne, C. P. , Strömberg, C. A. , Smith, S. A. , & Consortium, C. G. (2010). The origins of C4 grasslands: Integrating evolutionary and ecosystem science. Science, 328(5978), 587–591. [DOI] [PubMed] [Google Scholar]
- Eissenstat, D. , & Achor, D. (1999). Anatomical characteristics of roots of citrus rootstocks that vary in specific root length. New Phytologist, 141(2), 309–321. [DOI] [PubMed] [Google Scholar]
- Ejiri, M. , & Shiono, K. (2019). Prevention of radial oxygen loss is associated with exodermal suberin along adventitious roots of annual wild species of Echinochloa. Frontiers in Plant Science, 10, 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enstone, D. E. , Peterson, C. A. , & Ma, F. (2002). Root endodermis and exodermis: Structure, function, and responses to the environment. Journal of Plant Growth Regulation, 21(4), 335–351. [Google Scholar]
- Farrant, J. M. , & Moore, J. P. (2011). Programming desiccation‐tolerance: From plants to seeds to resurrection plants. Current Opinion in Plant Biology, 14(3), 340–345. [DOI] [PubMed] [Google Scholar]
- Ferrari, R. C. , Bittencourt, P. P. , Rodrigues, M. A. , Moreno‐Villena, J. J. , Alves, F. R. , Gastaldi, V. D. , … Freschi, L. (2020). C4 and crassulacean acid metabolism within a single leaf: Deciphering key components behind a rare photosynthetic adaptation. New Phytologist, 225(4), 1699–1714. [DOI] [PubMed] [Google Scholar]
- Galau, G. A. , Hughes, D. W. , & Dure, L. (1986). Abscisic acid induction of cloned cotton late embryogenesis‐abundant (lea) mRNAs. Plant Molecular Biology, 7(3), 155–170. [DOI] [PubMed] [Google Scholar]
- Ghanati, F. , Morita, A. , & Yokota, H. (2005). Deposition of suberin in roots of soybean induced by excess boron. Plant Science, 168(2), 397–405. [Google Scholar]
- Giarola, V. , Hou, Q. , & Bartels, D. (2017). Angiosperm plant desiccation tolerance: Hints from transcriptomics and genome sequencing. Trends in Plant Science, 22(8), 705–717. 10.1016/j.tplants.2017.05.007 [DOI] [PubMed] [Google Scholar]
- Goodstein, D. M. , Shu, S. , Howson, R. , Neupane, R. , Hayes, R. D. , Fazo, J. , … Rokhsar, D. S. (2012). Phytozome: A comparative platform for green plant genomics. Nucleic Acids Research, 40(D1), D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou, M. , Hou, G. , Yang, H. , Zhang, X. , Cai, Y. , Kai, G. , & Liu, C.‐J. (2017). The MYB107 transcription factor positively regulates suberin biosynthesis. Plant Physiology, 173(2), 1045–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowik, U. , Burscheidt, J. , Akyildiz, M. , Schlue, U. , Koczor, M. , Streubel, M. , & Westhoff, P. (2004). Cis‐regulatory elements for mesophyll‐specific gene expression in the C4 plant Flaveria trinervia, the promoter of the C4 phosphoenolpyruvate carboxylase gene. The Plant Cell, 16(5), 1077–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths, H. , & Males, J. (2017). Succulent plants. Current Biology, 27(17), R890–R896. [DOI] [PubMed] [Google Scholar]
- Haberlandt, G. (1904). Physiologische pflanzenanatomie. Leipzig: Wilhelm Engelman. [Google Scholar]
- Heiman, M. , Kulicke, R. , Fenster, R. J. , Greengard, P. , & Heintz, N. (2014). Cell type–specific mRNA purification by translating ribosome affinity purification (TRAP). Nature Protocols, 9(6), 1282–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyduk, K. , Moreno‐Villena, J. J. , Gilman, I. S. , Christin, P.‐A. , & Edwards, E. J. (2019). The genetics of convergent evolution: Insights from plant photosynthesis. Nature Reviews Genetics, 20(8), 485–493. [DOI] [PubMed] [Google Scholar]
- Heyduk, K. , Ray, J. N. , Ayyampalayam, S. , Moledina, N. , Borland, A. , Harding, S. A. , … Leebens‐Mack, J. (2019). Shared expression of crassulacean acid metabolism (CAM) genes pre‐dates the origin of CAM in the genus Yucca. Journal of Experimental Botany, 70(22), 6597–6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra, F. A. , Golovina, E. A. , & Buitink, J. (2001). Mechanisms of plant desiccation tolerance. Trends in Plant Science, 6(9), 431–438. [DOI] [PubMed] [Google Scholar]
- Holbein, J. , Shen, D. , & Andersen, T. G. (2021). The endodermal passage cell–just another brick in the wall? New Phytologist, 230, 1321–1328. [DOI] [PubMed] [Google Scholar]
- Hutin, C. , Nussaume, L. , Moise, N. , Moya, I. , Kloppstech, K. , & Havaux, M. (2003). Early light‐induced proteins protect Arabidopsis from photooxidative stress. Proceedings of the National Academy of Sciences of the United States of America, 100(8), 4921–4926. 10.1073/pnas.0736939100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, W. (1962). Botanical histochemistry.,(W. Freeman & Co. [Google Scholar]
- Johansen, D. A. (1940). Plant microtechnique (p. 530). New York, London: McGraw‐Hill Book Company Inc. [Google Scholar]
- Kajala, K. , Brown, N. J. , Williams, B. P. , Borrill, P. , Taylor, L. E. , & Hibberd, J. M. (2012). Multiple Arabidopsis genes primed for recruitment into C4 photosynthesis. The Plant Journal, 69(1), 47–56. [DOI] [PubMed] [Google Scholar]
- Kajala, K. , Gouran, M. , Shaar‐Moshe, L. , Mason, G. A. , Rodriguez‐Medina, J. , Kawa, D. , … Brady, S. M. (2021). Innovation, conservation, and repurposing of gene function in root cell type development. Cell, 184, 3333–3348.e19. [DOI] [PubMed] [Google Scholar]
- Kamula, S. , Peterson, C. , & Mayfield, C. (1994). The plasmalemma surface area exposed to the soil solution is markedly reduced by maturation of the exodermis and death of the epidermis in onion roots. Plant, Cell & Environment, 17(11), 1183–1193. [Google Scholar]
- Kenrick, P. , & Crane, P. R. (1997). The origin and early evolution of plants on land. Nature, 389(6646), 33–39. 10.1038/37918. [DOI] [Google Scholar]
- Khraiwesh, B. , Qudeimat, E. , Thimma, M. , Chaiboonchoe, A. , Jijakli, K. , Alzahmi, A. , … Salehi‐Ashtiani, K. (2015). Genome‐wide expression analysis offers new insights into the origin and evolution of Physcomitrella patens stress response. Scientific Reports, 5, 17434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosma, D. K. , Murmu, J. , Razeq, F. M. , Santos, P. , Bourgault, R. , Molina, I. , & Rowland, O. (2014). At MYB 41 activates ectopic suberin synthesis and assembly in multiple plant species and cell types. The Plant Journal, 80(2), 216–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosma, D. K. , Rice, A. , & Pollard, M. (2015). Analysis of aliphatic waxes associated with root periderm or exodermis from eleven plant species. Phytochemistry, 117, 351–362. [DOI] [PubMed] [Google Scholar]
- Kreszies, T. , Eggels, S. , Kreszies, V. , Osthoff, A. , Shellakkutti, N. , Baldauf, J. A. , … Schreiber, L. (2020). Seminal roots of wild and cultivated barley differentially respond to osmotic stress in gene expression, suberization, and hydraulic conductivity. Plant, Cell & Environment, 43(2), 344–357. 10.1111/pce.13675 [DOI] [PubMed] [Google Scholar]
- Kreszies, T. , Shellakkutti, N. , Osthoff, A. , Yu, P. , Baldauf, J. A. , Zeisler‐Diehl, V. V. , … Schreiber, L. (2019). Osmotic stress enhances suberization of apoplastic barriers in barley seminal roots: Analysis of chemical, transcriptomic and physiological responses. The New Phytologist, 221(1), 180–194. 10.1111/nph.15351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labeeuw, L. , Martone, P. T. , Boucher, Y. , & Case, R. J. (2015). Ancient origin of the biosynthesis of lignin precursors. Biology Direct, 10, 23. 10.1186/s13062-015-0052-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lashbrooke, J. , Cohen, H. , Levy‐Samocha, D. , Tzfadia, O. , Panizel, I. , Zeisler, V. , … Aharoni, A. (2016). MYB107 and MYB9 homologs regulate suberin deposition in angiosperms. The Plant Cell, 28(9), 2097–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legay, S. , Guerriero, G. , André, C. , Guignard, C. , Cocco, E. , Charton, S. , … Hausman, J. F. (2016). MdMyb93 is a regulator of suberin deposition in russeted apple fruit skins. New Phytologist, 212(4), 977–991. [DOI] [PubMed] [Google Scholar]
- Leprince, O. , & Buitink, J. (2010). Desiccation tolerance: From genomics to the field. Plant Science, 179(6), 554–564. [Google Scholar]
- Lewis, L. A. , & McCourt, R. M. (2004). Green algae and the origin of land plants. American Journal of Botany, 91(10), 1535–1556. [DOI] [PubMed] [Google Scholar]
- Li, P. , Ponnala, L. , Gandotra, N. , Wang, L. , Si, Y. , Tausta, S. L. , … Brutnell, T. P. (2010). The developmental dynamics of the maize leaf transcriptome. Nature Genetics, 42(12), 1060–1067. [DOI] [PubMed] [Google Scholar]
- Líška, D. , Martinka, M. , Kohanová, J. , & Lux, A. (2016). Asymmetrical development of root endodermis and exodermis in reaction to abiotic stresses. Annals of Botany, 118(4), 667–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, G. , Stirnemann, M. , Gübeli, C. , Egloff, S. , Courty, P.‐E. , Aubry, S. , … Borghi, L. (2019). Strigolactones play an important role in shaping exodermal morphology via a KAI2‐dependent pathway. Iscience, 17, 144–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttge, U. (1987). Carbon dioxide and water demand: Crassulacean acid metabolism (CAM), a versatile ecological adaptation exemplifying the need for integration in ecophysiological work. New Phytologist, 106, 593–629. [DOI] [PubMed] [Google Scholar]
- Lyall, R. , Schlebusch, S. A. , Proctor, J. , Prag, M. , Hussey, S. G. , Ingle, R. A. , & Illing, N. (2020). Vegetative desiccation tolerance in the resurrection plant Xerophyta humilis has not evolved through reactivation of the seed canonical LAFL regulatory network. The Plant Journal, 101(6), 1349–1367. 10.1111/tpj.14596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchin, R. M. , Ossola, A. , Leishman, M. R. , & Ellsworth, D. S. (2019). A simple method for simulating drought effects on plants. Frontiers in Plant Science, 10, 1715. 10.3389/fpls.2019.01715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks, R. A. , Farrant, J. M. , Nicholas McLetchie, D. , & VanBuren, R. (2021). Unexplored dimensions of variability in vegetative desiccation tolerance. American Journal of Botany, 108, 346–358. [DOI] [PubMed] [Google Scholar]
- Marks, R. A. , Smith, J. J. , VanBuren, R. , & McLetchie, D. N. (2021). Expression dynamics of dehydration tolerance in the tropical plant Marchantia inflexa. The Plant Journal, 105(1), 209–222. [DOI] [PubMed] [Google Scholar]
- Martone, P. T. , Estevez, J. M. , Lu, F. , Ruel, K. , Denny, M. W. , Somerville, C. , & Ralph, J. (2009). Discovery of lignin in seaweed reveals convergent evolution of cell‐wall architecture. Current Biology, 19(2), 169–175. 10.1016/j.cub.2008.12.031 [DOI] [PubMed] [Google Scholar]
- Ming, R. , VanBuren, R. , Wai, C. M. , Tang, H. , Schatz, M. C. , Bowers, J. E. , … Yu, Q. (2015). The pineapple genome and the evolution of CAM photosynthesis. Nature Genetics, 47(12), 1435–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namyslov, J. , Bauriedlová, Z. , Janoušková, J. , Soukup, A. , & Tylová, E. (2020). Exodermis and endodermis respond to nutrient deficiency in nutrient‐specific and localized manner. Plants, 9(2), 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicotra, A. B. , Leigh, A. , Boyce, C. K. , Jones, C. S. , Niklas, K. J. , Royer, D. L. , & Tsukaya, H. (2011). The evolution and functional significance of leaf shape in the angiosperms. Functional Plant Biology, 38(7), 535–552. [DOI] [PubMed] [Google Scholar]
- Niklas, K. J. , Cobb, E. D. , & Matas, A. J. (2017). The evolution of hydrophobic cell wall biopolymers: From algae to angiosperms. Journal of Experimental Botany, 68(19), 5261–5269. 10.1093/jxb/erx215 [DOI] [PubMed] [Google Scholar]
- Niklaus, M. , & Kelly, S. (2019). The molecular evolution of C4 photosynthesis: Opportunities for understanding and improving the world's most productive plants. Journal of Experimental Botany, 70(3), 795–804. [DOI] [PubMed] [Google Scholar]
- Oliver, M. J. , Farrant, J. M. , Hilhorst, H. W. , Mundree, S. , Williams, B. , & Bewley, J. D. (2020). Desiccation tolerance: Avoiding cellular damage during drying and rehydration. Annual Review of Plant Biology, 71, 435–460. [DOI] [PubMed] [Google Scholar]
- Oliver, M. J. , Tuba, Z. , & Mishler, B. D. (2000). The evolution of vegetative desiccation tolerance in land plants. Plant Ecology, 151(1), 85–100. [Google Scholar]
- Pardo, J. , Man Wai, C. , Chay, H. , Madden, C. F. , Hilhorst, H. W. M. , Farrant, J. M. , & VanBuren, R. (2020). Intertwined signatures of desiccation and drought tolerance in grasses. Proceedings of the National Academy of Sciences of the United States of America, 117(18), 10079–10088. 10.1073/pnas.2001928117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson, A. H. , Bowers, J. E. , Bruggmann, R. , Dubchak, I. , Grimwood, J. , Gundlach, H. , … Rokhsar, D. S. (2009). The Sorghum bicolor genome and the diversification of grasses. Nature, 457(7229), 551–556. [DOI] [PubMed] [Google Scholar]
- Pecková, E. , Tylová, E. , & Soukup, A. (2016). Tracing root permeability: Comparison of tracer methods. Biologia Plantarum, 60(4), 695–705. [Google Scholar]
- Peguero‐Pina, J. J. , Vilagrosa, A. , Alonso‐Forn, D. , Ferrio, J. P. , Sancho‐Knapik, D. , & Gil‐Pelegrín, E. (2020). Living in drylands: Functional adaptations of trees and shrubs to cope with high temperatures and water scarcity. Forests, 11(10), 1028. [Google Scholar]
- Peredo, E. L. , & Cardon, Z. G. (2020). Shared up‐regulation and contrasting down‐regulation of gene expression distinguish desiccation‐tolerant from intolerant green algae. Proceedings of the National Academy of Sciences, 117(29), 17438–17445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perumalla, C. J. , Peterson, C. A. , & Enstone, D. E. (1990). A survey of angiosperm species to detect hypodermal Casparian bands. I. Roots with a uniseriate hypodermis and epidermis. Botanical Journal of the Linnean Society, 103(2), 93–112. [Google Scholar]
- Phansopa, C. , Dunning, L. T. , Reid, J. D. , & Christin, P.‐A. (2020). Lateral gene transfer acts as an evolutionary shortcut to efficient C4 biochemistry. Molecular Biology and Evolution, 37(11), 3094–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe, G. , Sorensen, I. , Jiao, C. , Sun, X. , Fei, Z. , Domozych, D. S. , & Rose, J. K. (2020). Cutin and suberin: Assembly and origins of specialized lipidic cell wall scaffolds. Current Opinion in Plant Biology, 55, 11–20. 10.1016/j.pbi.2020.01.008 [DOI] [PubMed] [Google Scholar]
- Pollard, M. , Beisson, F. , Li, Y. , & Ohlrogge, J. B. (2008). Building lipid barriers: Biosynthesis of cutin and suberin. Trends in Plant Science, 13(5), 236–246. 10.1016/j.tplants.2008.03.003 [DOI] [PubMed] [Google Scholar]
- Radermacher, A. L. , du Toit, S. F. , & Farrant, J. M. (2019). Desiccation‐driven senescence in the resurrection plant Xerophyta schlechteri (baker) NL Menezes: Comparison of anatomical, ultrastructural, and metabolic responses between senescent and non‐senescent tissues. Frontiers in Plant Science, 10, 1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranathunge, K. , Kim, Y. X. , Wassmann, F. , Kreszies, T. , Zeisler, V. , & Schreiber, L. (2017). The composite water and solute transport of barley (Hordeum vulgare) roots: Effect of suberized barriers. Annals of Botany, 119(4), 629–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranathunge, K. , Lin, J. , Steudle, E. , & Schreiber, L. (2011). Stagnant deoxygenated growth enhances root suberization and lignifications, but differentially affects water and NaCl permeabilities in rice (Oryza sativa L.) roots. Plant, Cell & Environment, 34(8), 1223–1240. [DOI] [PubMed] [Google Scholar]
- Ranathunge, K. , & Schreiber, L. (2011). Water and solute permeabilities of Arabidopsis roots in relation to the amount and composition of aliphatic suberin. Journal of Experimental Botany, 62(6), 1961–1974. 10.1093/jxb/erq389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranathunge, K. , Schreiber, L. , Bi, Y. M. , & Rothstein, S. J. (2016). Ammonium‐induced architectural and anatomical changes with altered suberin and lignin levels significantly change water and solute permeabilities of rice (Oryza sativa L.) roots. Planta, 243(1), 231–249. 10.1007/s00425-015-2406-1 [DOI] [PubMed] [Google Scholar]
- Reinhardt, D. , & Rost, T. (1995). Salinity accelerates endodermal development and induces an exodermis in cotton seedling roots. Environmental and Experimental Botany, 35(4), 563–574. [Google Scholar]
- Renault, H. , Alber, A. , Horst, N. A. , Basilio Lopes, A. , Fich, E. A. , Kriegshauser, L. , … Werck‐Reichhart, D. (2017). A phenol‐enriched cuticle is ancestral to lignin evolution in land plants. Nature Communications, 8, 14713. 10.1038/ncomms14713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing, S. A. , Lang, D. , Zimmer, A. D. , Terry, A. , Salamov, A. , Shapiro, H. , … Boore, J. L. (2008). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science, 319(5859), 64–69. [DOI] [PubMed] [Google Scholar]
- Reynoso, M. A. , Juntawong, P. , Lancia, M. , Blanco, F. A. , Bailey‐Serres, J. , & Zanetti, M. E. (2015). Translating ribosome affinity purification (TRAP) followed by RNA sequencing technology (TRAP‐SEQ) for quantitative assessment of plant translatomes. In Plant functional genomics (pp. 185–207). New York: Springer. [DOI] [PubMed] [Google Scholar]
- Ron, M. , Dorrity, M. W. , de Lucas, M. , Toal, T. , Hernandez, R. I. , Little, S. A. , … Brady, S. M. (2013). Identification of novel loci regulating interspecific variation in root morphology and cellular development in tomato. Plant Physiology, 162(2), 755–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondeau, P. , Rouch, C. , & Besnard, G. (2005). NADP‐malate dehydrogenase gene evolution in Andropogoneae (Poaceae): Gene duplication followed by sub‐functionalization. Annals of Botany, 96(7), 1307–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roppolo, D. , Boeckmann, B. , Pfister, A. , Boutet, E. , Rubio, M. C. , Denervaud‐Tendon, V. , … Geldner, N. (2014). Functional and evolutionary analysis of the Casparian strip membrane domain protein family. Plant Physiology, 165(4), 1709–1722. 10.1104/pp.114.239137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roppolo, D. , De Rybel, B. , Tendon, V. D. , Pfister, A. , Alassimone, J. , Vermeer, J. E. , … Geldner, N. (2011). A novel protein family mediates Casparian strip formation in the endodermis. Nature, 473(7347), 380–383. [DOI] [PubMed] [Google Scholar]
- Schlüter, U. , & Weber, A. P. (2020). Regulation and evolution of C4 photosynthesis. Annual Review of Plant Biology, 71, 183–215. [DOI] [PubMed] [Google Scholar]
- Schreiber, L. , Franke, R. , & Hartmann, K. (2005). Effects of NO 3 deficiency and NaCl stress on suberin deposition in rhizo‐and hypodermal (RHCW) and endodermal cell walls (ECW) of castor bean (Ricinus communis L.) roots. Plant and Soil, 269(1–2), 333–339. [Google Scholar]
- Schreiber, L. , Franke, R. , Hartmann, K. D. , Ranathunge, K. , & Steudle, E. (2005). The chemical composition of suberin in apoplastic barriers affects radial hydraulic conductivity differently in the roots of rice (Oryza sativa L. cv. IR64) and corn (Zea mays L. cv. Helix). Journal of Experimental Botany, 56(415), 1427–1436. 10.1093/jxb/eri144 [DOI] [PubMed] [Google Scholar]
- Schreiber, L. , Hartmann, K. , Skrabs, M. , & Zeier, J. (1999). Apoplastic barriers in roots: Chemical composition of endodermal and hypodermal cell walls. Journal of Experimental Botany, 50(337), 1267–1280. [Google Scholar]
- Schulze, S. , Mallmann, J. , Burscheidt, J. , Koczor, M. , Streubel, M. , Bauwe, H. , … Westhoff, P. (2013). Evolution of C4 photosynthesis in the genus Flaveria: Establishment of a photorespiratory CO2 pump. The Plant Cell, 25(7), 2522–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seago, J. L., Jr. , & Fernando, D. D. (2013). Anatomical aspects of angiosperm root evolution. Annals of Botany, 112(2), 223–238. 10.1093/aob/mcs266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedelnikova, O. V. , Hughes, T. E. , & Langdale, J. A. (2018). Understanding the genetic basis of C4 Kranz anatomy with a view to engineering C3 crops. Annual Review of Genetics, 52, 249–270. [DOI] [PubMed] [Google Scholar]
- Shiono, K. , & Yamada, S. (2014). Waterlogging tolerance and capacity for oxygen transport in Brachypodium distachyon (Bd21). Plant Root, 8, 5–12. [Google Scholar]
- Shiono, K. , Yamauchi, T. , Yamazaki, S. , Mohanty, B. , Malik, A. I. , Nagamura, Y. , … Nakazono, M. (2014). Microarray analysis of laser‐microdissected tissues indicates the biosynthesis of suberin in the outer part of roots during formation of a barrier to radial oxygen loss in rice (Oryza sativa). Journal of Experimental Botany, 65(17), 4795–4806. [DOI] [PubMed] [Google Scholar]
- Shukla, V. , Han, J.‐P. , Cléard, F. , Legendre‐Lefebvre, L. , Gully, K. , Flis, P. , … Barberon, M. (2021). Suberin plasticity to developmental and exogenous cues is regulated by a set of MYB transcription factors. BioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taleisnik, E. , Peyrano, G. , Cordoba, A. , & Arias, C. (1999). Water retention capacity in root segments differing in the degree of exodermis development. Annals of Botany, 83(1), 19–27. [Google Scholar]
- Terlova, E. F. , Holzinger, A. , & Lewis, L. A. (2021). Terrestrial green algae show higher tolerance to dehydration than do their aquatic sister‐species. Microbial Ecology, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thellmann, M. , Andersen, T. G. , & Vermeer, J. E. (2020). Translating ribosome affinity purification (trap) to investigate Arabidopsis thaliana root development at a cell type‐specific scale. Journal of Visualized Experiments (Jove), 159, e60919. [DOI] [PubMed] [Google Scholar]
- Thomas, R. , Fang, X. , Ranathunge, K. , Anderson, T. R. , Peterson, C. A. , & Bernards, M. A. (2007). Soybean root suberin: Anatomical distribution, chemical composition, and relationship to partial resistance to Phytophthora sojae. Plant Physiology, 144(1), 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To, A. , Joubès, J. , Thueux, J. , Kazaz, S. , Lepiniec, L. , & Baud, S. (2020). AtMYB92 enhances fatty acid synthesis and suberin deposition in leaves of Nicotiana benthamiana . The Plant Journal, 103(2), 660–676. [DOI] [PubMed] [Google Scholar]
- Tylová, E. , Pecková, E. , Blascheová, Z. , & Soukup, A. (2017). Casparian bands and suberin lamellae in exodermis of lateral roots: An important trait of roots system response to abiotic stress factors. Annals of Botany, 120(1), 71–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBuren, R. , Man Wai, C. , Pardo, J. , Giarola, V. , Ambrosini, S. , Song, X. , & Bartels, D. (2018). Desiccation tolerance evolved through gene duplication and network rewiring in Lindernia. Plant Cell, 30(12), 2943–2958. 10.1105/tpc.18.00517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBuren, R. , Pardo, J. , Man Wai, C. , Evans, S. , & Bartels, D. (2019). Massive tandem proliferation of ELIPs supports convergent evolution of desiccation tolerance across land plants. Plant Physiology, 179(3), 1040–1049. 10.1104/pp.18.01420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBuren, R. , Wai, C. M. , Zhang, Q. , Song, X. , Edger, P. P. , Bryant, D. , … Bartels, D. (2017). Seed desiccation mechanisms co‐opted for vegetative desiccation in the resurrection grass Oropetium thomaeum . Plant, Cell & Environment, 40(10), 2292–2306. 10.1111/pce.13027 [DOI] [PubMed] [Google Scholar]
- Verdier, J. , Lalanne, D. , Pelletier, S. , Torres‐Jerez, I. , Righetti, K. , Bandyopadhyay, K. , … Buitink, J. (2013). A regulatory network‐based approach dissects late maturation processes related to the acquisition of desiccation tolerance and longevity of Medicago truncatula seeds. Plant Physiology, 163(2), 757–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishwanath, S. J. , Delude, C. , Domergue, F. , & Rowland, O. (2015). Suberin: Biosynthesis, regulation, and polymer assembly of a protective extracellular barrier. Plant Cell Reports, 34(4), 573–586. 10.1007/s00299-014-1727-z [DOI] [PubMed] [Google Scholar]
- Wang, P. , Calvo‐Polanco, M. , Reyt, G. , Barberon, M. , Champeyroux, C. , Santoni, V. , … Salt, D. E. (2019). Surveillance of cell wall diffusion barrier integrity modulates water and solute transport in plants. Scientific Reports, 9(1), 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P. , Wang, C.‐M. , Gao, L. , Cui, Y.‐N. , Yang, H.‐L. , de Silva, N. D. , … Wang, S.‐M. (2020). Aliphatic suberin confers salt tolerance to Arabidopsis by limiting Na+ influx, K+ efflux and water backflow. Plant and Soil, 448, 1–18. [Google Scholar]
- Wei, X. , Lu, W. , Mao, L. , Han, X. , Wei, X. , Zhao, X. , … Xu, C. (2020). ABF2 and MYB transcription factors dominate feruloyl transferase FHT gene involved in ABA‐mediated wound suberization of kiwifruit. Journal of Experimental Botany, 71, 305–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng, J. K. , Akiyama, T. , Bonawitz, N. D. , Li, X. , Ralph, J. , & Chapple, C. (2010). Convergent evolution of syringyl lignin biosynthesis via distinct pathways in the lycophyte Selaginella and flowering plants. Plant Cell, 22(4), 1033–1045. 10.1105/tpc.109.073528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng, J. K. , Li, X. , Stout, J. , & Chapple, C. (2008). Independent origins of syringyl lignin in vascular plants. Proceedings of the National Academy of Sciences of the United States of America, 105(22), 7887–7892. 10.1073/pnas.0801696105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, B. P. , Burgess, S. J. , Reyna‐Llorens, I. , Knerova, J. , Aubry, S. , Stanley, S. , & Hibberd, J. M. (2016). An untranslated cis‐element regulates the accumulation of multiple C4 enzymes in Gynandropsis gynandra mesophyll cells. The Plant Cell, 28(2), 454–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise, M. J. , & Tunnacliffe, A. (2004). POPP the question: What do LEA proteins do? Trends in Plant Science, 9(1), 13–17. 10.1016/j.tplants.2003.10.012 [DOI] [PubMed] [Google Scholar]
- Wodniok, S. , Brinkmann, H. , Glöckner, G. , Heidel, A. J. , Philippe, H. , Melkonian, M. , & Becker, B. (2011). Origin of land plants: Do conjugating green algae hold the key? BMC Evolutionary Biology, 11(1), 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Z. , Xin, T. , Bartels, D. , Li, Y. , Gu, W. , Yao, H. , … Chen, S. (2018). Genome analysis of the ancient tracheophyte Selaginella tamariscina reveals evolutionary features relevant to the acquisition of desiccation tolerance. Molecular Plant, 11(7), 983–994. [DOI] [PubMed] [Google Scholar]
- Yang, X. , Hu, R. , Yin, H. , Jenkins, J. , Shu, S. , Tang, H. , … Tuskan, G. A. (2017). The Kalanchoë genome provides insights into convergent evolution and building blocks of crassulacean acid metabolism. Nature Communications, 8(1), 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Yang, C. , & Seago, J. L., Jr. (2018). Anatomical and histochemical traits of roots and stems of Artemisia lavandulaefolia and A. selengensis (Asteraceae) in the Jianghan Floodplain, China. Flora, 239, 87–97. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Histochemical tests for barrier cell‐type identification


