Abstract
A region located at kbp −3.9 to −2.6 5′ to the first hematopoietic exon of the GATA-1 gene is necessary to recapitulate gene expression in both the primitive and definitive erythroid lineages. In transfection analyses, this region activated reporter gene expression from an artificial promoter in a position- and orientation-independent manner, indicating that the region functions as the GATA-1 gene hematopoietic enhancer (G1HE). However, when analyzed in transgenic embryos in vivo, G1HE activity was orientation dependent and also required the presence of the endogenous GATA-1 gene hematopoietic promoter. To define the boundaries of G1HE, a series of deletion constructs were prepared and tested in transfection and transgenic mice analyses. We show that G1HE contains a 149-bp core region which is critical for GATA-1 gene expression in both primitive and definitive erythroid cells but that expression in megakaryocytes requires the core plus additional sequences from G1HE. This core region contains one GATA, one GAT, and two E boxes. Mutational analyses revealed that only the GATA box is critical for gene-regulatory activity. Importantly, G1HE was active in SCL−/− embryos. These results thus demonstrate the presence of a critical network of GATA factors and GATA binding sites that controls the expression of this gene.
GATA-1 gene expression is essential for hematopoietic cell differentiation (reviewed in reference 33). The transcription factor GATA-1 is expressed in erythroid cells, megakaryocytes, eosinophils, and mast cells (9, 20, 36), as well as in Sertoli cells in the testis (6, 35). Two promoters, or first exons, exist in the GATA-1 gene (6). The distal (IT) promoter specifies the expression of the GATA-1 gene in Sertoli cells, whereas the proximal (IE) promoter, located between the IT exon and the common coding exons, directs GATA-1 gene expression in the hematopoietic lineages (6). Gene ablation experiments of GATA-1 demonstrate that GATA-1 is required for the differentiation of erythroid cells and also for platelet formation in the final stage of megakaryopoiesis (18, 23–26). GATA-1 is also important for the formation of connective tissue type mast cells (4).
We previously identified critical regulatory regions for expression of the GATA-1 gene in erythroid-lineage cells with a β-galactosidase (lacZ) reporter gene in a transgenic-mouse analysis (17). Transgenic mouse lines bearing a DNA fragment beginning at kbp −3.9 5′ to the IE exon and extending through the second exon, when fused to a lacZ gene (IE3.9intLacZ), recapitulated GATA-1 gene expression in both primitive and definitive erythroid cells. LacZ activity was abolished in transgenic mouse lines bearing a smaller fragment extending from kbp −2.6 (5′ to IE) through the second exon (IE2.6intLacZ), indicating that the 1.3-kbp region acts as an upstream activating element (UE) (17). UE contains a region corresponding to a DNase I-hypersensitive site 1 (21). In transgenic mice with a 3.9-kbp fragment, including UE but lacking the first intron (IE3.9LacZ), the LacZ reporter was expressed only in primitive erythroid cells, not in definitive erythroid cells. As before, deletion of UE from the construct (IE2.6LacZ) ablated this primitive cell-specific LacZ expression. These results suggest the existence of at least two regulatory regions in the GATA-1 gene, i.e., UE and the first intron element (17). In a rescue experiment with our GATA-1 gene knockdown mouse (26), we have demonstrated that the DNA fragment extending from UE through the second exon is sufficient to recapitulate the physiological level of GATA-1 gene expression (S. Takahashi and M. Yamamoto, unpublished data). Thus the UE fragment is one of the regions required for complete GATA-1 gene regulatory activity.
Upstream activating sequences such as UE frequently contain enhancers that can activate transcription from core promoters in a position- and orientation-independent manner and can activate transcription from heterologous promoters (1, 2). These characteristics of enhancers have been established in vitro, in transfection analyses with tissue culture cells (for example, see reference 12). We speculated, however, that novel characteristics of enhancers would be revealed when these elements were tested in vivo in transgenic-mouse assays, leading to the development of new concepts in gene regulation. Indeed, in this study we compared the activity of UE in transgenic mice and in a transfection assay with K562 cells. UE was found to satisfy the classic criteria of an enhancer in the transfection assay and consequently was renamed the GATA-1 gene hematopoietic enhancer (G1HE). However, when G1HE was integrated into the mouse genome, its activity was more restricted. We also performed a detailed dissection of G1HE to delineate a core region, using both the reporter transfection and transgenic-mouse assays, and assessed the importance of each cis-acting element in the core region for stage- and lineage-specific expression of GATA-1. The results of these analyses demonstrate that a network of GATA factors regulates the expression of the GATA-1 gene during hematopoietic cell differentiation, through the GATA box in G1HE, and that G1HE consists of two elements which determine erythroid or megakaryocyte lineage specificity.
MATERIALS AND METHODS
Construction of plasmids and generation of transgenic mice.
Various reporter genes were constructed by using restriction enzyme sites in the GATA-1 gene regulatory regions. The lacZ gene in pSVβ (Clontech) was used as a reporter gene for the transgenic-mouse analysis. An EcoRI site in the first intron was deleted (delE) from pIE3.9intLacZ (17). A series of G1HE deletion constructs and all the point mutant constructs were made with the delE construct. To make pHE-SV40-LacZ and pHE-TK-LacZ, a genomic BamHI (kbp −3.9)-EcoRI (kbp −2.6) restriction fragment (G1HE) was inserted into pSVβ and pTKβ (Clontech), respectively. The firefly luciferase (LUC) gene was used as a reporter gene for the transfection analysis. To make pHE-SV40 and inverted HE-SV40, the BamHI-EcoRI fragment was inserted into the pGL3 promoter vector in both orientations (Promega). Transgenic mice were generated by standard methods (5).
Transfection analysis.
LUC reporter plasmids were transfected into K562 cells (5 × 106 cells/sample) by the DEAE-dextran procedure as described previously (7), and the cells were grown for 24 h. In each experiment, plasmid (10 μg) was transfected in triplicate. Preparation of cell lysates and measurement of LUC activity were carried out with a LUC assay kit (TOYO INK) as specified by the supplier. LUC activity was normalized by the transfection efficiency, determined by using control sea pansy luciferase activity. Cotransfection of pEF-SP (13) did not interfere with the activity of our reporter constructs.
Analysis of transgenic mouse embryos and yolk sacs.
For whole-mount 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal) staining, embryos at embryonic day 8.5 (E8.5) or E9.5 were fixed at room temperature for 30 min in 1% formaldehyde–0.2% glutaraldehyde–0.02% Nonidet P-40 in phosphate-buffered saline (PBS, pH 7.3). After being washed with PBS, the embryos were incubated overnight at 37°C in PBS containing 2 mM MgCl2, 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6 and 1 mg of X-Gal per ml. For section staining, embryos at E15.0 were fixed and incubated overnight at 4°C in PBS–20% sucrose. Samples were embedded in Tissue-Tek OCT compound (Sakura Finetechnical, Tokyo, Japan) and rapidly frozen. Cryosections were stained at 37°C for 8 h with X-Gal. Genomic DNA was purified from the yolk sacs and embryos, and integration of transgenes was verified by PCR with a set of primers (primers 2 and 3) described previously (17). Another set of primers, beta-gal1 and beta-gal2, corresponding to the sequences in the lacZ gene, were also used; their sequences are 5′-ACCGACTACACAAATCAGCG-3′ and 5′-CAACCACCGCACGATAGAGA-3′, respectively.
Preparation of nuclear extract.
Mouse erythroleukemia (MEL) cells were used for detection of DNA binding proteins. Also, GATA-1, GATA-2, or GATA-3 proteins were individually overexpressed in 293T cells. To prepare nuclear extract, MEL cells were washed twice and collected by centrifugation to measure the packed-cell volume (PCV). The cells were then washed twice in 5 PCV of buffer A (10 mM HEPES [pH 8.0], 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol, protease inhibitors), resuspended in buffer A (3 PCV), and incubated on ice for 15 min. They were then homogenized. Nuclei were collected by centrifugation, and the packed-nucleus volume (PNV) was measured. The nuclei were resuspended in 2 PNV of buffer C (20 mM HEPES [pH 8.0], 1.5 mM MgCl2, 420 mM KCl, 0.2 mM EDTA, 25% [vol/vol] glycerol, 0.5 mM DTT, protease inhibitors). The suspension was mixed gently for 30 min at 4°C and centrifuged, and aliquots of the supernatant were frozen immediately in liquid nitrogen and stored.
EGMSA.
Electrophoretic gel mobility shift assay (EGMSA) was performed as described previously (32, 34). MEL and 293T cell nuclear extracts were incubated at room temperature with a 32P-labeled consensus E-box–GATA oligonucleotide (9-bp spacing) (32), GGE, GGmE, or GGEm probe (see Fig. 7A). Competitor oligonucleotides or specific antibodies raised against GATA-1 (N6) or E2A (SC-349X Santa Cruz) were added to the reaction mixture 15 min after the start of incubation. After an additional 15-min incubation, DNA-protein complexes were separated from the free probe by nondenaturing polyacrylamide gel electrophoresis (4% polyacrylamide) and analyzed by autoradiography.
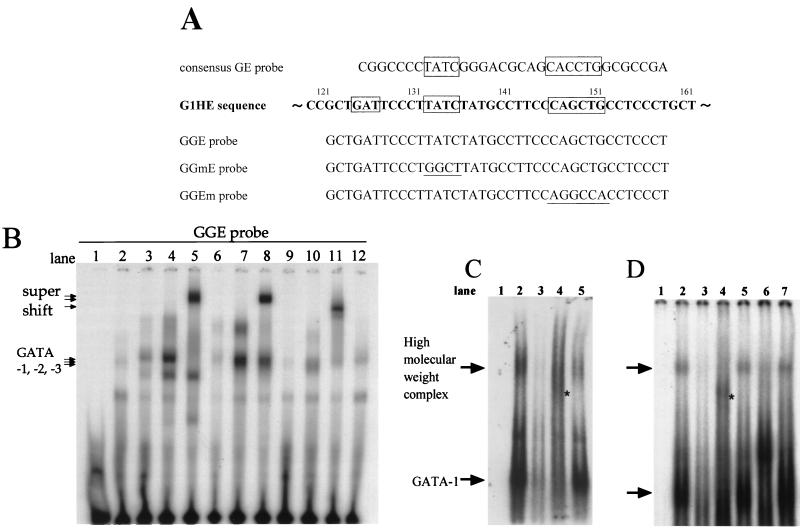
FIG. 7.
GATA factors can bind to the core GATA sequence of G1HE. (A) Sequences of the consensus GE probe (9-bp spacing) (32) and GGE probe of G1HE. The G1HE sequence is shown in bold type. The GAT, GATA, and E-box sequences are boxed. The GGmE probe indicates the GATA motif mutant of GGE probe, whereas the GGEm probe indicates the E-box mutant of the GGE probe. Underlines show mutated sequences. (B) GATA-1, GATA-2, and GATA-3 can bind to the GGE probe. Nuclear extracts from 293T cells transfected with GATA-1 (lanes 3 to 5), GATA-2 (lanes 6 to 8), or GATA-3 (lanes 9 to 11) expression plasmids were examined. Lanes 3, 6, and 9 show shifted bands with 0.2 μl of each extract, while lanes 4, 7, and 10 show bands with 1.0 μl of each extract. The bands in the latter condition were supershifted with specific monoclonal antibodies against each GATA factor (lane 5, anti-GATA-1; lane 8, anti-GATA-2; lane 11, anti-GATA-3). Lane 1 contains probe alone. Lanes 2 and 12 show binding of 293T cell and MEL cell nuclear extract, respectively, to the GGE probe. Of the hematopoietic GATA factors, MEL cells express predominantly GATA-1, so that lane 12 serves as a marker lane for GATA-1. (C) EGMSA with MEL cell nuclear extract. The labeled consensus GE probe was incubated alone (lane 1) or with MEL cell nuclear extract (lanes 2 to 5) for 15 min. Then cold consensus GE oligonucleotide (lane 3), anti-GATA-1 antibody (lane 4), or anti-E2A antibody (lane 5) was added to the reaction mixture. An asterisk in lane 4 marks a band that was supershifted by the anti-GATA-1 antibody. (D) Lanes 1 to 5 show EGMSA with the GGE probe. Labeled GGE probe was incubated alone (lane 1) or with MEL cell nuclear extract (lanes 2 to 5) for 15 min. Then a 100-fold excess of cold GGE probe (lane 3), anti-GATA-1 antibody (lane 4), or anti-E2A antibody (lane 5) was added to the reaction mixture. Note that the high-molecular-weight complex was not affected by the addition of anti-E2A antibody. GGmE probe (lane 6) or GGEm probe (lane 7) was used in the reaction instead of GGE probe. An asterisk in lane 4 shows a band that was supershifted by the anti-GATA-1 antibody.
Analysis of SCL homozygous mutant mice.
Transgenic mice bearing pIE3.9intGFP were newly prepared. SCL knockout mutant mice (22) were obtained from Jackson Laboratory. The heterozygous SCL mutant mice were mated with GFP transgenic mice. The SCL−/+::GFP+ mice were then intercrossed to generate mice with the SCL−/−::GFP+ genotype. Because the SCL homozygous mutant mice die by E10.5, embryos were analyzed at E9.5. For genotype screening, the neomycin resistance gene and wild-type allele were analyzed by PCR. The SCL mutant allele was amplified with a set of primers for the neo gene (27). The PCR conditions were denaturation for 30 s at 94°C, annealing for 30 s at 55°C, and extension for 30 s at 72°C (30 cycles). Amplification of wild-type allele was performed with primers TALS1 (5′-CACCAGACAAGAAACTAAGC-3′) and TALAS1 (5′-ATAGGAAGGCAAGTCTCAGT-3′). The PCR conditions were denaturation for 30 s at 94°C, annealing for 30 s at 56°C, and extension for 1 min at 72°C (35 cycles). The GFP gene was amplified with primers GFPS (5′-AGCAAGGGCGAGGAGCTGTTCACC-3′) and GFPAS (5′-TGCCGTCGTCCTTGAAGAAGATG-3′). The PCR conditions were the same as for neo gene amplification (30 cycles).
RESULTS
G1HE functions in an orientation-dependent manner in an F0 assay.
To examine whether UE of the GATA-1 gene (17) actually fulfills the criteria for an enhancer, we fused the 1.3-kbp UE fragment to the simian virus 40 (SV40) promoter and LUC gene. This reporter construct was transfected into K562 cells. The addition of the UE fragment stimulated expression from this reporter 16-fold (data not shown). When UE was tested in the opposite orientation from that of the normal gene, it activated LUC reporter gene expression eightfold. Thus, UE can enhance reporter gene transcription from a heterologous promoter regardless of orientation and distance, satisfying the classical criteria for an enhancer; it is subsequently referred to as G1HE (see above).
To examine whether G1HE also functions as an enhancer in vivo after integration into chromatin, we used a transgenic “blue” mouse system with the lacZ reporter gene. We previously detected G1HE activity by comparing transgenic mouse lines carrying the IE3.9intLacZ construct with lines containing IE2.6intLacZ, which lacks G1HE (17). Whereas erythroid cell-specific expression of the reporter was reproduced in the former mice, there was no significant blue staining in E8.5 yolk sac cells or E12.5 fetal liver cells of the latter transgenic mouse embryos. Since making a large number of transgenic mouse lines was technically not feasible, we examined transgenic founder mice (F0 or G0 for generation 0) to analyze lacZ reporter gene expression during primitive-stage hematopoiesis in the yolk sac. The yolk sacs of mouse embryos at E8.5 or E9.5 were stained directly with X-Gal. A typical whole-mount staining pattern of IE3.9intLacZ at E8.5 is shown in Fig. 1 (17). Intense blue staining was seen in the transgenic-mouse yolk sacs (Fig. 1A and data not shown), whereas normal yolk sacs and embryos contained essentially no positively stained cells under these assay conditions (Fig. 1B). Only embryos with more than 10 blue-stained blood islands in the yolk sac were scored positive.
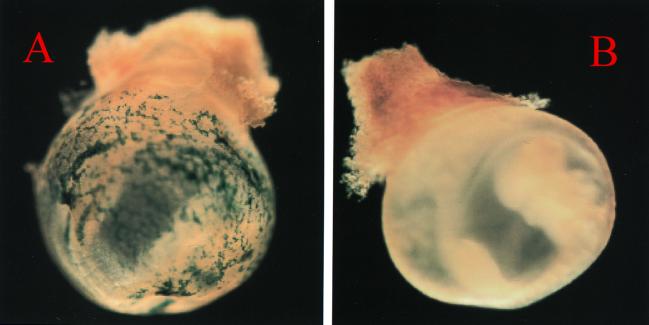
FIG. 1.
Whole-mount LacZ staining of an E8.5 yolk sac of a IE3.9intLacZ transgenic mouse. (A) Typical positively stained yolk sac blood islands, in which the blue area indicates LacZ activity. Primitive erythroid cell-specific expression of LacZ activity is observed. (B) A transgene-negative littermate.
In the IE3.9intLacZ transgenic F0 embryos, six of nine yolk sacs were positive (Fig. 2A). Position effects due to the integration site are commonly observed in transgenic-mouse analyses and are probably responsible for the three negative cases. This result is in agreement with our previous transgenic-mouse analysis (see above). When G1HE was moved 2 kbp 5′ of the original position, two of nine yolk sacs still stained positively (Fig. 2A). Similarly, six out of six yolk sacs were positive when G1HE was moved 500 bp closer to the reporter gene. While the magnitude of the stimulation varies, G1HE can obviously activate transcription from multiple locations. The position independence of G1HE clearly indicated that this element itself is sufficient for enhancer activity.
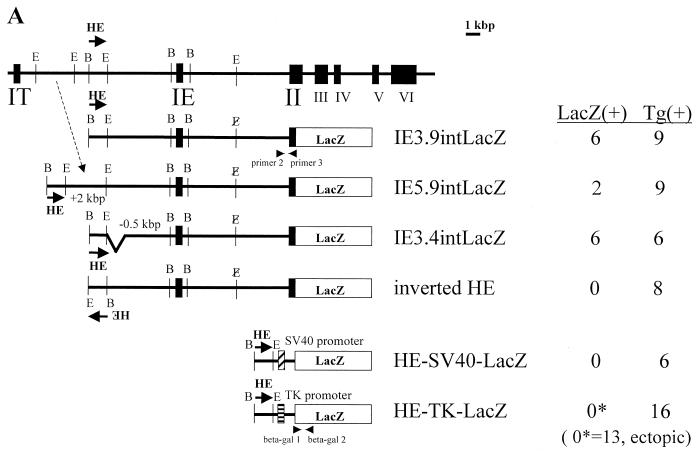
FIG. 2.
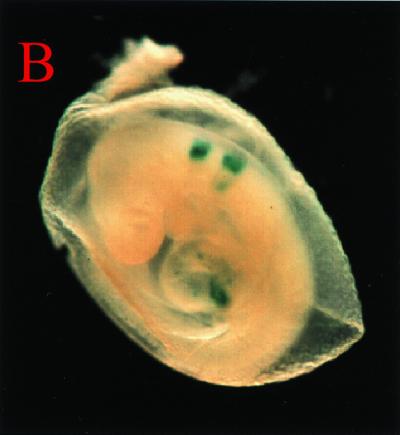
Functional analysis of G1HE activity by a transgenic mouse assay. (A) The mouse GATA-1 locus is shown at the top; exons are depicted as solid boxes, and G1HE is shown as HE. Abbreviations of the restriction enzyme sites: B, BamHI; E, EcoRI. Arrows indicate whether G1HE is cloned in the sense or antisense orientation relative to the promoter. A reporter lacZ gene was fused to exon II of the GATA-1 gene. In the IE3.9intLacZ, IE5.9intLacZ, IE3.4intLacZ, and inverted HE constructs, an EcoRI site in intron I was deleted (delE). The number of embryos staining positive for LacZ [LacZ(+)] and the total number of transgenic embryos [Tg(+)] are shown. (B) Ectopic LacZ expression in an embryo with a HE-TK-lacZ transgene. In this embryo, LacZ activity was observed in the rhombomere and heart.
We then tested a construct in which G1HE was inverted. In contrast to results obtained with the parent IE3.9intLacZ, no positively stained yolk sacs were observed with this construct in a total of eight samples (Fig. 2A). This indicates that G1HE works in vivo in an orientation-dependent manner. In addition, the G1HE fragment (normal orientation) failed to activate transcription from the SV40 or thymidine kinase promoters, since no positively stained yolk sacs were observed. Since G1HE is able to activate SV40 promoter in K562 cells, the transgenic-mouse assay detects a fundamentally different gene regulatory activity from that detected by the transient-transfection assay. The mice carrying the thymidine kinase promoter construct showed ectopic blue staining at high frequency (13 of 16 [81.3%]). However, the staining patterns differed among embryos, and the blood islands remained completely unstained. For instance, one F0 embryo showed rhombomere staining but no yolk sac blood cell staining (Fig. 2B) (see Discussion). We conclude that these signals were all ectopic. Thus, G1HE is not sufficient to confer tissue specificity on the GATA-1 gene in vivo, but additional regulatory sequences are necessary. The orientation dependence and promoter selectivity of G1HE were observed only in transgenic mice, not in transient-transfection analyses.
A core sequence exists near the 5′ end of G1HE.
The 1,174-bp nucleotide sequence of G1HE is shown in Fig. 3, and potential transcription factor binding sites are underlined. We have numbered the first nucleotide of the BamHI site +1, and this is the 5′ end of the fragment. This sequence has been deposited in DDBJ/GenBank Database under accession no. AB000965 and agrees in general with the partial sequence reported by Ronchi et al. (21). The differences between the two sequences probably reflect variation of mouse strains.
FIG. 3.
Structure of the G1HE region. The sequence is numbered from the BamHI site at kbp −3.9, which is the 5′ end of G1HE. Binding sites for transcription factors are underlined. Note that E box 1, GAT, GATA, and E box 2 are mutated in the later analysis.
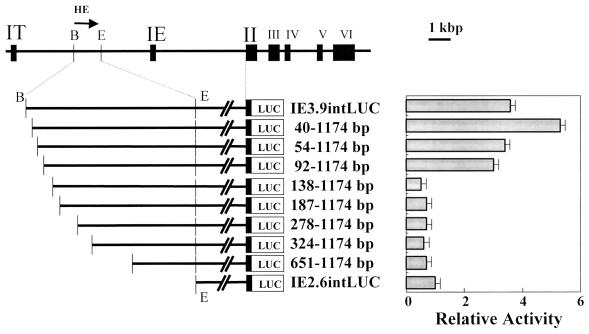
To identify a core region, we prepared various truncated forms of G1HE. We first inserted these into IE2.6intLUC and analyzed the mutants by transfection into K562 cells. When 137 bp was deleted from the 5′ end of G1HE (BamHI site), the LUC activity was notably reduced to the level of the IE2.6intLUC construct, indicating that this 137-bp sequence contains part of the core (Fig. 4). Because a 91-bp deletion mutant showed markedly more LUC activity than the 137-bp deletion mutant did, at least one important cis-acting element appeared to be located between nucleotides (nt) 92 and 137. Similar results were observed when the same series of deletions was tested in a vector with an SV40 promoter-driven reporter gene (data not shown).
FIG. 4.
Deletion analysis of G1HE in a transfection assay with K562 cells. LUC activity of pIE2.6intLUC was set to 1, and the relative LUC activities of other constructs are shown.
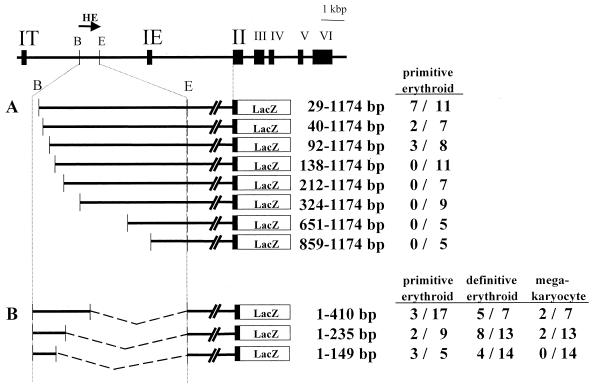
We then tested the G1HE deletion mutants in a transgenic-mouse assay. The yolk sac expression of LacZ in E8.5 or E9.5 transgenic F0 embryos was examined and scored as above. Approximately 65% of embryos harboring the reporter plasmid with complete G1HE (Fig. 2) or with nt 29 through 1174 (Fig. 5A) (6 of 9 and 7 of 11, respectively), showed positively stained yolk sacs. Similarly, 33% of embryos with nt 40 to 1174 and 92 to 1174 constructs (2 of 7 and 3 of 8, respectively) were positive (Fig. 5A). In contrast, no positively stained yolk sacs were observed in embryos with nt 138 to 1174 or shorter constructs. These results coincide very well with those of the transfection assay and confirm the importance of the region between nt 92 and 137. In addition, the decrease in the frequency of positive embryos from 65% to 33% suggests that the region between nt 29 and 92 may also contain some important regulatory elements (see below).
FIG. 5.
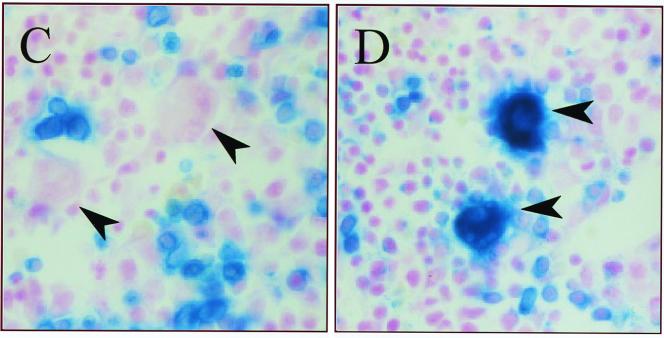
Deletion analysis of G1HE activity in a transgenic-mouse assay. (A and B) A series of 5′ deletion constructs (A) and a series of 3′ deletion constructs (B) were prepared and analyzed. F0 embryos were analyzed at E8.5 (or E9.5) or E14.5 (or E15.5) for LacZ activity in primitive erythroid cells and definitive erythroid cells/megakaryocytes, respectively. The numbers are the number of embryos staining positive for LacZ/the total number of transgenic embryos. (C and D) E14.5 livers of transgenic-mouse embryos bearing 3′ deletion constructs were stained with X-Gal. Embryonic livers with the 149-bp core region (C) and the 235-bp fragment (D) of GIHE, respectively, are shown. Arrowheads indicate megakaryocytes. Magnification, ×140.
Meanwhile, we made three reporter constructs that truncate G1HE from the 3′ end. The smallest construct contained only the 5′ 149 bp of G1HE adjacent to the IE2.6intLacZ reporter gene (Fig. 5B). This construct consequently lacked most of the G1HE sequence. When tested in the transgenic-mouse F0 assay, the 149-bp fragment alone could activate transcription of the lacZ reporter gene in yolk sac hematopoietic cells. Thus, the 149-bp region contains the core enhancer sequence of G1HE, which is sufficient to direct specific GATA-1 gene expression in primitive erythroid cells.
We also carried out similar F0 experiments for E14.5 or E15.5 embryos to determine the activity of G1HE in definitive erythroid cells and in megakaryocytes. As expected, the core 149-bp region stimulated expression of the lacZ reporter gene in definitive erythroid cells (Fig. 5B). Surprisingly, however, the 149-bp region failed to induce LacZ activity in megakaryocytes (Fig. 5C). In contrast, a longer region of G1HE (1 to 235 bp) enhanced LacZ expression both in definitive erythroid cells and in megakaryocytes (Fig. 5D). These results suggest that in addition to the 149-bp core region, G1HE contains another regulatory region that specifies GATA-1 gene expression in the megakaryocyte lineage.
The consensus GATA box is essential for G1HE activity.
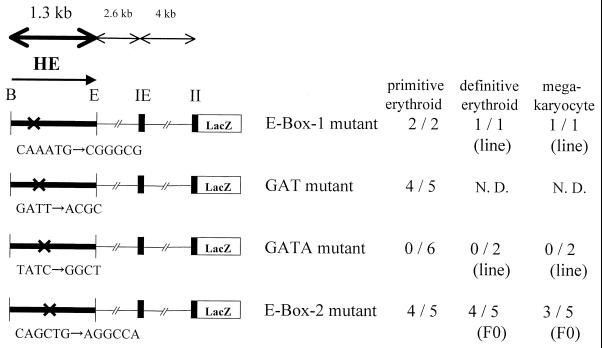
In the region between nt 92 and 137, there is a GATnnnnTTATCT (the underline indicates GATA) sequence (Fig. 3). An E box (E box 2) resides 10 bp 3′ to this GATA box. This sequence arrangement is close to the binding site for a model transcription complex, which includes the GATA-1, SCL, Lmo2, E2A, and Ldb1 proteins (32). If a similar protein complex was formed on the GATA and E boxes of G1HE, it might play an important role in GATA-1 gene expression. To test the functional importance of the GATA-box–E-box combination in vivo, we prepared several lacZ transgene constructs that contain mutations in each motif and tested their expression during both primitive and definitive hematopoiesis. While mutations in the atypical GAT and in E box 2 did not affect the enhancer activity at all, no positively stained erythroid cells and megakaryocytes were observed in the yolk sacs or fetal livers of the embryos containing mutations in the consensus GATA box (Fig. 6). These results clearly demonstrate that only the GATA box, and not E box 2, is essential for G1HE activity in both primitive and definitive hematopoiesis.
FIG. 6.
A GATA site in the core of G1HE is essential for the GATA-1 gene expression in primitive erythroid cells, definitive erythroid cells, and megakaryocytes. There are two E boxes, an atypical GAT, and a consensus GATA box in the core of G1HE. Mutations in each cis-acting element were tested in the transgenic-mouse system. F0 indicates the transient transgenic assay, and “line” indicates the F1 or F2 assay.
We also noticed another E box (E box 1; CAAATG) between nt 29 and 39 of G1HE. Deletion of this region reduced transcriptional activity in the transgenic-mouse analysis (see above). We therefore mutated the E-box 1 sequence and examined this transgene construct similarly. No decrease in the frequency of the LacZ-positive embryos was observed (Fig. 6), indicating that E box 1 is also dispensable for reporter gene expression in this in vivo analysis.
GATA factors bind to the GATA box.
To identify the molecule(s) binding to the consensus GATA box in G1HE, EGMSA was performed. To this end, we prepared three probes from the G1HE core region that contain various combinations of the GATA boxes and the E box (Fig. 7A). These are GGE (wild-type sequence), GGmE (mutation in the second GATA sequence), and GGEm (mutation in E-box-2). We first tested three hematopoietic GATA factors for binding to the wild-type GGE probe. GATA-1, GATA-2, and GATA-3 proteins were expressed individually in 293T cells, and nuclear extracts were prepared from these cells. All three hematopoietic GATA factors were found to bind the GGE probe (Fig. 7B). Supershift experiments with antibodies confirmed the identity of each GATA factor. These analyses thus demonstrated that GATA-1, GATA-2, or GATA-3 could occupy the GATA box in the core of G1HE.
We also prepared a probe with the consensus GATA-box–E-box motif (consensus GE), which was shown to mediate the formation of a high-molecular-weight complex (Fig. 7A) (32). In good agreement with the previous analysis (32), a GATA-1 band was observed with the consensus GE probe and nuclear extract from MEL cells (Fig. 7C, lanes 2 and 5). Considering the results shown in Fig. 7B, this band is most likely to represent monomeric GATA-1 binding to the consensus GE probe. Upon overexposure of the gel, we also detected a high-molecular-weight complex (Fig. 7C, lane 2), of much reduced intensity relative to the GATA-1 band. Both the GATA-1 and high-molecular-weight bands were abolished in the presence of an excess of cold consensus GE competitor (Fig. 7C, lane 3), indicating specific binding. Inclusion of anti-GATA-1 antibody in the reaction mixture resulted in a large reduction of the high-molecular-weight complex, as well as in the appearance of a supershift of the GATA-1 band (Fig. 7C, lane 4). Similarly, addition of anti-E2A antibody destroyed the high-molecular-weight complex with the consensus GE probe (Fig. 7C, lane 5), indicating that this complex contains both GATA-1 and E2A.
With the GGE probe, we also detected a high-molecular-weight band (Fig. 7D, lane 2), which was competed with a 100-fold excess of cold GGE (lane 3). The high-molecular-weight complex was disrupted specifically with an anti-GATA-1 antibody (lane 4) but not with anti-E2A antibody (lane 5). Importantly, whereas the amount of high-molecular-weight complex was markedly decreased with the GGmE probe (a GATA site mutant probe [lane 6]), the complex was formed normally with the GGEm probe (an E box 2 mutant probe [lane 7]). These results suggest that the high-molecular-weight complex with the GGE probe is distinct from that formed on the consensus GE probe, in that E2A may not be a major constituent of the GGE complex. The observation that the E box 2 in G1HE is not important for the high-molecular-weight complex formation in gel shift analyses is consistent with the results of transgenic-mouse analyses (see above).
G1HE is active in SCL-null primitive erythroid cells.
The results thus far suggest that the consensus GATA box at nt 133 is essential for the G1HE activity whereas the E box 2 at nt 147 is not. This conclusion is somewhat unexpected, since SCL, one of the E-box binding transcription factors, is indispensable for hematopoiesis. GATA-1 expression could not be detected in SCL-deficient embryos (19). However, SCL may regulate GATA-1 gene expression through interactions with GATA factors or with other transcription factors, rather than by direct binding to the GATA-1 gene promoter/enhancer. In this scenario, SCL does not necessarily bind the G1HE E box directly but can still regulate GATA-1 gene expression. This possibility was tested by mating heterozygous SCL knockout mice (22) with IE3.9intGFP transgenic mice. The latter transgenic mice express green fluorescence protein (GFP) reporter gene in yolk sac hematopoietic cells under G1HE regulation.
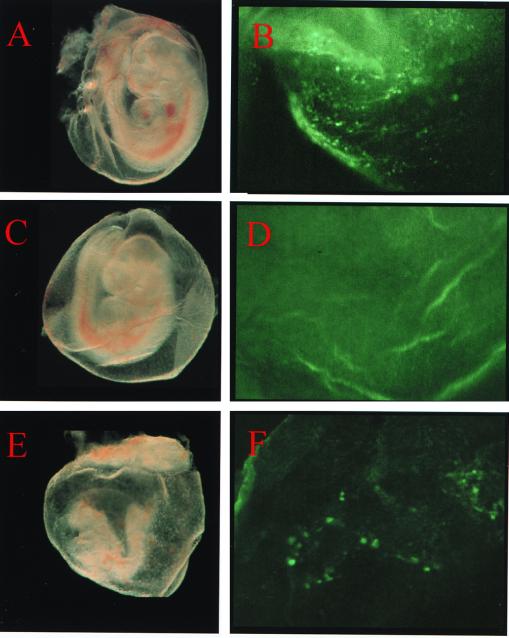
Yolk sac hematopoietic cells in the SCL+/−::GFP+ E9.5 embryos show clear green fluorescence (Fig. 8B; panel A shows the yolk sac and embryo proper), indicating that G1HE is active in primitive erythroid cells. The yolk sacs of wild-type littermates with the SCL+/+::GFP− genotype did not contain any green fluorescence-positive cells (Fig. 8C and D). The SCL−/−::GFP+ embryos were very small and were not well developed (Fig. 8E). However, although the number was significantly decreased, some green fluorescence-positive cells were clearly visible in the yolk sac (Fig. 8F). This result was quite reproducible. We found green fluorescence-positive cells in five SCL−/−::GFP+ yolk sacs from three independent litters. This result indicates that G1HE can support the initiation of transcription from the GATA-1 promoter without SCL, so that SCL is not the essential constituent of GATA-1 gene transcription initiation.
FIG. 8.
G1HE can activate transcription in SCL-null primitive erythroid-cell progenitors. (A and B) Appearance of SCL heterozygous and IE3.9intGFP transgene-positive (SCL+/−::GFP+) E9.5 embryo (A) and yolk sac (B). Green fluorescence-positive cells were seen in the yolk sac. (C and D) A wild-type littermate without the GFP transgene (SCL+/+::GFP−) did not show any GFP-positive cells. (E and F) An SCL−/−::GFP+ embryo (E) is severely anemic and has developmental defects, but a small number of GFP-positive cells are clearly visible in the yolk sac (F).
DISCUSSION
The GATA-1 gene hematopoietic enhancer, G1HE, is a powerful activator of transcription in a transgenic-mouse assay in vivo (17). In this study, we have demonstrated that G1HE acts as a classic enhancer element in a reporter transfection assay. Results of a deletion analysis with both the transfection and transgenic-mouse systems indicate that the core 149-bp region within G1HE contains a strong GATA-1 gene regulatory activity for the erythroid lineage. A consensus GATA box, an atypical GAT box, and two E boxes reside within the core region. Mutational analyses of these cis-acting elements have revealed that while the consensus GATA box is critical for the activity of G1HE in both primitive and definitive hematopoiesis, the other elements are not. Hematopoietic GATA factors GATA-1, GATA-2, and GATA-3 were all demonstrated to bind to the consensus GATA box of G1HE. This result is quite intriguing, since duplicated GATA boxes have been found in the IT promoter (16) and the upstream region of the IE promoter (3, 15, 29). These GATA boxes are required for reporter gene expression in transfection analyses. Taken together, these results demonstrate that a network of GATA factors and binding sites regulate GATA-1 gene expression in hematopoietic cells and suggest that GATA-1 and/or other GATA factors work as key regulators in this network through direct interaction with the cis-acting GATA boxes in multiple regulatory regions. This network seems to be the molecular basis for the erythroid cell-specific expression of the GATA-1 gene.
We also found that while the core 149-bp region of G1HE was sufficient to drive lacZ reporter gene expression in erythroid cells, the region was inadequate to generate reporter gene expression in megakaryocytes of E15 fetal liver. To attain megakaryocytic expression of the reporter gene, a slightly longer region of G1HE (i.e., bp 1 to 235) was required. Since disruption of the GATA box at nt 133 in the G1HE reporter resulted in loss of megakaryocytic expression of the reporter gene, the core region was also required for the enhancer activity of G1HE in the fetal liver megakaryocytes. These results thus indicate that G1HE consists of multiple cis-acting elements and that these elements have distinct functions in erythroid and megakaryocytic lineage-specific gene expression.
G1HE can activate transcription in a position-independent manner from a cognate promoter in both the transfection and transgenic-mouse assays. Unlike in the transfection assay, however, the presence of the IE promoter was necessary for the reporter genes to recapitulate the endogenous GATA-1 gene expression profile in the transgenic-mouse assay. Whereas G1HE activated the SV40 promoter in the transfection analysis, it could not do so in the transgenic mice. These discrepancies in G1HE function between the transfection and transgenic-mouse assays may be due to the transgene configuration. In the reporter transfection assay, the reporter gene exists transiently in the nucleus as an episome outside the host genome. In contrast, the reporter gene is integrated into the transgenic mouse genome, where chromatin structure and the influences of regulatory sequences surrounding the integration site can affect reporter gene expression. Thus, the transgenic-mouse assay is a more stringent test for correct gene-regulatory activity than is the DNA transfection assay.
While inversion of G1HE relative to the IE promoter had only a quantitative effect on reporter gene transcription in the transfection assay, proper orientation of G1HE is absolutely required for its activity in the transgenic mice. We do not know the basis for the strict orientation dependence of the G1HE activity in vivo. One plausible explanation is that the three-dimensional structure of the protein complex interacting with the core enhancer (and/or promoter) region may be crucial for transcriptional activation in a chromatin configuration. In fact, a high-molecular-weight complex containing GATA-1 was detected in nuclear extracts of MEL cells with the GGE probe of G1HE. An alternative explanation is to assume that transcription initiation at the GATA-1 locus is regulated by a tracking mechanism and that the inverted G1HE may disturb the synthesis of long RNA transcripts initiated from far upstream. These issues remain to be resolved.
A transcription factor complex containing GATA-1, SCL, and E2A assembled by Lmo2 and Lbd1 was proposed to associate with DNA through an E box and then to be stabilized by interaction with a GATA box in an in vitro reconstitution analysis (32). Importantly, in G1HE, E box 2 resides 10 bp 3′ to the consensus GATA box, similar to the arrangement of the DNA binding site used in the reconstitution assay. Because this model has never been tested in vivo, we examined the activity of each cis element in G1HE in detail. While mutation of E box 2 did not affect the enhancer activity at all, mutations in the consensus GATA box severely affected reporter gene expression in yolk sacs, definitive erythrocytes, and megakaryocytes, demonstrating that the GATA box is essential for the activity of G1HE but E box 2 is not. In addition, a high-molecular-weight complex was detected with the GGE probe and the GGEm (E-box-mutated) probe in MEL cell nuclear extracts, while its level was markedly decreased when the GGmE (GATA box-mutated) probe was used. Our results also suggest that the high-molecular-weight complex generated with the GGE probe may be distinct from the complex formed with the consensus GE probe, in that E2A is not involved in the GGE-based high-molecular-weight complex. Thus, GATA-1 or other hematopoietic GATA factors bind to the GATA box in G1HE and serve as an anchoring factor, as well as contributing to the formation of the high-molecular-weight protein complex which regulates GATA-1 gene expression.
An attractive model proposes that GATA-2 binds the GATA motif in the early stage of differentiation whereas GATA-1 replaces GATA-2 in the late stage. Three lines of evidence support this notion. First, we previously demonstrated that G1HE is active in the GATA-1 knockdown environment (26), suggesting that some other GATA factors can substitute at the GATA box. Second, our recent experiment shows that the GATA-1 knockdown mice can be rescued by GATA-2, and the GATA box in G1HE is indeed occupied by GATA-2 in the mouse cells (Takahashi and Yamamoto, unpublished). This indicates that other GATA factors can actually substitute for GATA-1 in vivo. Third, GATA-1, GATA-2, and GATA-3 can bind to the GATA boxes in the G1HE core (C. D. Trainor, unpublished observation; see above); these sequences actually form a GATA-pal configuration similar to other double GATA sites, to which both GATA-1 and GATA-2 can bind (28). It has also been shown that the expression of GATA-2 precedes that of GATA-1 in the hematopoietic lineage (8, 11, 14, 30).
SCL-null mice were reported to die by E10.5 due to severe anemia (19). mRNAs for GATA-1 and EKLF were not detected in E9.5 yolk sacs and embryos, suggesting that SCL is necessary for GATA-1 gene expression. These findings thus appear inconsistent with the results of our E-box mutation. To resolve this discrepancy, we carried out an additional experiment, in which SCL−/− mice were crossed with IE3.9intGFP transgenic mice. The latter mice express GFP reporter in yolk sac hematopoietic cells under the regulatory influence of G1HE. The results unequivocally demonstrated that GFP reporter is expressed in yolk sac cells in the absence of SCL, thus excluding the possibility that SCL is essential for initiation of G1HE activity. This data is in good agreement with the recent rescue experiment of SCL−/− embryos by using a transgene that expresses SCL under the influence of regulatory sequences from the GATA-1 gene (31). In the latter mouse embryos, the GATA-1 gene regulatory region drove the expression of SCL cDNA in the absence of endogenous SCL protein, proving that SCL is not required for transcription from the GATA-1 enhancer/promoter.
The reason for the marked decrease in the number of GFP-positive cells in the yolk sacs of SCL−/−::GFP+ embryos is not clear. One plausible explanation is that SCL is essential for the growth and/or differentiation of early primitive myeloerythroid progenitors, so that without SCL, the hematopoietic compartment cannot expand in the yolk sac. If this is the case, the GFP-positive cells may correspond to the yolk sac progenitors of hematopoiesis. However, it is technically not feasible to analyze the properties of the GFP-positive cells in SCL−/−::GFP+ embryos, and this point remains to be addressed.
Recently, the zebrafish GATA-1 gene-regulatory region was examined in a transgenic-fish assay, and distal double GATA sites were found to promote and maintain GATA-1 transcription (10). In the zebrafish GATA-1 gene, a proximal CACCC box is also critical for the initiation of GATA-1 gene expression in hematopoietic cells. However, since the zebrafish GATA-1 gene sequence has diverged substantially from those of the mouse and human GATA-1 genes, the cis-acting regulatory elements are difficult to compare. For instance, we previously identified a regulatory region in the first intron that specifies GATA-1 gene expression in definitive erythroid cells (17), but this intronic regulatory element does not appear to be conserved in the zebrafish GATA-1 gene.
Our preliminary data suggests that the erythroid-cell specificity of the GATA-1 gene also depends on the contribution of a regulatory element in the upstream promoter region (S. Nishimura, S. Takahashi, and M. Yamamoto, unpublished observation). Identification of cis-acting elements that organize the lineage specificity of GATA-1 gene expression and elucidation of the intricate relationships among the factors interacting with these elements are apparently the focus of our future research.
ACKNOWLEDGMENTS
We thank N. Kajiwara, N. Kaneko, Y. Kikuchi, N. Kasai, K.-C. Lim, J. Ohta, H. Motohashi, F. Sugiyama, N. Suzuki, and K. Yagami for help and discussion.
This work was supported in part by Grants-in-Aids from the Ministry of Education, Science, Sports and Culture, Core Research for Evolutional Science and Technology (CREST), the Japanese Society for Promotion of Sciences (JSPS), and NIH.
REFERENCES
- 1.Blackwood E M, Kadonaga J T. Going the distance: a current view of enhancer action. Science. 1998;281:60–63. doi: 10.1126/science.281.5373.60. [DOI] [PubMed] [Google Scholar]
- 2.Groudine M, Peretz M, Weintraub H. Transcriptional regulation of hemoglobin switching on chicken embryos. Mol Cell Biol. 1981;1:281–288. doi: 10.1128/mcb.1.3.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hannon R, Evans T, Felsenfeld G, Gould H. Structure and promoter activity of the gene for the erythroid transcription factor GATA-1. Proc Natl Acad Sci USA. 1991;88:3004–3008. doi: 10.1073/pnas.88.8.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harigae H, Takahashi S, Suwabe N, Ohtsu H, Gu L, Yang Z, Tsai F Y, Kitamura Y, Engel J D, Yamamoto M. Differential roles of GATA-1 and GATA-2 in growth and differentiation of mast cells. Genes Cells. 1998;3:39–50. doi: 10.1046/j.1365-2443.1998.00166.x. [DOI] [PubMed] [Google Scholar]
- 5.Hogan B, Costantini F, Lacy E. Manipulating the mouse embryo: a laboratory manual. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- 6.Ito E, Toki T, Ishihara H, Ohtani H, Gu L, Yokoyama M, Engel J D, Yamamoto M. Erythroid transcription factor GATA-1 is abundantly transcribed in mouse testis. Nature. 1993;362:466–468. doi: 10.1038/362466a0. [DOI] [PubMed] [Google Scholar]
- 7.Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel J D, Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leonard M, Brice M, Engel J D, Papayannopoulou T. Dynamics of GATA transcription factor expression during erythroid differentiation. Blood. 1993;82:1071–1079. [PubMed] [Google Scholar]
- 9.Martin D I K, Zon L I, Mutter G, Orkin S H. Expression of an erythroid transcription factor in megakaryocytic and mast cell lineages. Nature. 1990;344:444–447. doi: 10.1038/344444a0. [DOI] [PubMed] [Google Scholar]
- 10.Meng A, Tang H, Yuan B, Ong B A, Long Q, Lin S. Positive and negative cis-acting elements are required for hematopoietic expression of zebrafish GATA-1. Blood. 1999;93:500–508. [PubMed] [Google Scholar]
- 11.Minegishi N, Ohta J, Yamagiwa H, Suzuki N, Kawauchi S, Zhou Y, Takahashi S, Hayashi N, Engel J D, Yamamoto M. The mouse GATA-2 gene is expressed in the para-aortic splanchinopleura and aorta, gonads and mesonephros region. Blood. 1999;93:4196–4207. [PubMed] [Google Scholar]
- 12.Moreau P, Hen R, Wasylyk B, Everett R, Gaub M P, Chambon P. The SV40 72 base repair repeat has a striking effect on gene expression both in SV40 and other chimeric recombinants. Nucleic Acids Res. 1981;9:6047–6068. doi: 10.1093/nar/9.22.6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muto A, Hoshino H, Madisen L, Yanai N, Obinata M, Karasuyama H, Hayashi N, Nakauchi H, Yamamoto M, Groudine M, Igarashi K. Identification of Bach2 as a B-cell-specific partner for small Maf proteins that negatively regulate the immunoglobulin heavy chain gene 3′ enhancer. EMBO J. 1998;17:5734–5743. doi: 10.1093/emboj/17.19.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagai T, Harigae H, Ishihara H, Motohashi H, Minegishi N, Tsuchiya S, Hayashi N, Gu L, Andres B, Engel J D, Yamamoto M. Transcription factor GATA-2 is expressed in erythroid, early myeloid, and CD34+ human leukemia-derived cell lines. Blood. 1994;84:1074–1084. [PubMed] [Google Scholar]
- 15.Nicolis S, Bertini C, Ronchi A, Crotta S, Lanfranco L, Moroni E, Gilioni B, Ottolenghi S. An erythroid specific enhancer upstream to the gene encoding the cell-type specific transcription factor GATA-1. Nucleic Acids Res. 1991;19:5285–5291. doi: 10.1093/nar/19.19.5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Onodera K, Yomogida K, Suwabe N, Takahashi S, Muraosa Y, Hayashi N, Ito E, Gu L, Rassoulzadegan M, Engel J D, Yamamoto M. Conserved structure, regulatory elements, and transcriptional regulation from the GATA-1 gene testis promoter. J Biochem. 1997;121:251–263. doi: 10.1093/oxfordjournals.jbchem.a021581. [DOI] [PubMed] [Google Scholar]
- 17.Onodera K, Takahashi S, Nishimura S, Ohta J, Motohashi H, Yomogida K, Hayashi N, Engel J D, Yamamoto M. GATA-1 transcription is controlled by distinct regulatory mechanisms during primitive and definitive erythropoiesis. Proc Natl Acad Sci USA. 1997;94:4487–4492. doi: 10.1073/pnas.94.9.4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pevny L, D'Agati C V, Simon M C, Orkin S H, Costantini F. Development of hematopoietic cells lacking transcription factor GATA-1. Development. 1995;121:163–172. doi: 10.1242/dev.121.1.163. [DOI] [PubMed] [Google Scholar]
- 19.Robb L, Lyons I, Li R, Hartley L, Kntgen F, Harvey R P, Metcalf D, Begley C G. Absence of yolk sac hematopoiesis from mice with a targeted disruption of the scl gene. Proc Natl Acad Sci USA. 1995;92:7075–7079. doi: 10.1073/pnas.92.15.7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Romeo P-H, Prandini M H, Joulin V, Mignotte V, Prenant M, Vainchenker W, Marguerie G, Uzan G. Megakaryocytic and erythrocytic lineages share specific transcription factors. Nature. 1990;344:447–449. doi: 10.1038/344447a0. [DOI] [PubMed] [Google Scholar]
- 21.Ronchi A, Cir M, Cairns L, Basilico L, Corbella P, Ricciardi-Castagnoli P, Cross M, Ghysdael J, Ottolenghi S. Molecular heterogeneity of regulatory elements of the mouse GATA-1 gene. Genes Funct. 1997;1:245–258. doi: 10.1046/j.1365-4624.1997.00021.x. [DOI] [PubMed] [Google Scholar]
- 22.Shivdasani R A, Mayer E L, Orkin S H. Absence of blood formation in mice lacking the T-cell leukemia oncoprotein tal-1/SCL. Nature. 1995;373:432–434. doi: 10.1038/373432a0. [DOI] [PubMed] [Google Scholar]
- 23.Shivdasani R A, Fujiwara Y, MacDevit M A, Orkin S H. A lineage-selective knockout establishes the critical role of transcription factor GATA-1 in megakaryocyte growth and platelet development. EMBO J. 1997;16:3965–3973. doi: 10.1093/emboj/16.13.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simon M C, Pevny L, Wiles M V, Keller G, Costantini F, Orkin S H. Rescue of erythroid development in gene targeted GATA-1-mouse embryonic stem cell. Nat Genet. 1992;1:92–97. doi: 10.1038/ng0592-92. [DOI] [PubMed] [Google Scholar]
- 25.Suwabe N, Takahashi S, Nakano T, Yamamoto M. GATA-1 regulates growth and differentiation of erythroid lineage cells during in vitro ES cell differentiation. Blood. 1998;92:4108–4118. [PubMed] [Google Scholar]
- 26.Takahashi S, Onodera K, Motohashi H, Suwabe N, Hayashi N, Yanai N, Nabeshima Y, Yamamoto M. Arrest in primitive erythroid cell development caused by promoter-specific disruption of the GATA-1 gene. J Biol Chem. 1997;272:12611–12615. doi: 10.1074/jbc.272.19.12611. [DOI] [PubMed] [Google Scholar]
- 27.Takahashi S, Komeno T, Suwabe N, Yoh K, Nakajima O, Nishimura S, Kuroha T, Nagasawa T, Yamamoto M. Role of GATA-1 in proliferation and differentiation of definitive erythroid and megakaryocytic cells in vivo. Blood. 1998;92:434–442. [PubMed] [Google Scholar]
- 28.Trainor C D, Omichinski J G, Vandergon T L, Gronenborn A M, Clore G M, Felsenfeld G. A palindromic regulatory site within vertebrate GATA-1 promoters requires both zinc fingers of the GATA-1 DNA-binding domain for high-affinity interaction. Mol Cell Biol. 1996;16:2238–2247. doi: 10.1128/mcb.16.5.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsai S-F, Strauss E, Orkin S H. Functional analysis and in vivo footprinting implicate the erythroid transcription factor GATA-1 as a positive regulator of its own promoter. Genes Dev. 1991;5:919–931. doi: 10.1101/gad.5.6.919. [DOI] [PubMed] [Google Scholar]
- 30.Tsai F-Y, Keller G, Kuo F C, Weiss M, Chen J, Rosenblatt M, Alt F W, Orkin S H. An early hematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 1994;371:221–226. doi: 10.1038/371221a0. [DOI] [PubMed] [Google Scholar]
- 31.Visvader J E, Fujiwara Y, Orkin S H. Unsuspected role for the T-cell leukemia protein SCL/tal-1 in vascular development. Genes Dev. 1998;12:473–479. doi: 10.1101/gad.12.4.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wadman I A, Osada H, Grts G G, Agulnick A D, Westphal H, Foster A, Rabbitts T H. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamamoto M, Takahashi S, Onodera K, Muraosa Y, Engel J D. Upstream and downstream of erythroid transcription factor GATA-1. Genes Cells. 1997;2:107–115. doi: 10.1046/j.1365-2443.1997.1080305.x. [DOI] [PubMed] [Google Scholar]
- 34.Yamamoto M, Ko L J, Leonard M W, Beug H, Orkin S H, Engel J D. Activity and expression of the NF-E1 multigene family. Genes Dev. 1990;4:1650–1662. doi: 10.1101/gad.4.10.1650. [DOI] [PubMed] [Google Scholar]
- 35.Yomogida K, Ohtani H, Harigae H, Ito E, Nishimune Y, Engel J D, Yamamoto M. Developmental stage- and spermatogenic cycle-specific expression of transcription factor GATA-1 in mouse Sertoli cells. Development. 1994;120:1759–1766. doi: 10.1242/dev.120.7.1759. [DOI] [PubMed] [Google Scholar]
- 36.Zon L I, Yamaguchi Y, Yee K, Albee E A, Kimura A, Bennett J C, Orkin S H. Expression of mRNA for the GATA-binding proteins in human eosinophils and basophils: potential role in gene transcription. Blood. 1993;81:3234–3241. [PubMed] [Google Scholar]