Abstract
Background
Vibrio spp. are aquatic bacteria that prefer warm seawater with moderate salinity. In humans, they can cause gastroenteritis, wound infections, and ear infections. During the summers of 2018 and 2019, unprecedented high sea surface temperatures were recorded in the German Baltic Sea.
Aim
We aimed to describe the clinical course and microbiological characteristics of Vibrio infections in Germany in 2018 and 2019.
Methods
We performed an observational retrospective multi-centre cohort study of patients diagnosed with domestically-acquired Vibrio infections in Germany in 2018 and 2019. Demographic, clinical, and microbiological data were assessed, and isolates were subjected to whole genome sequencing and antimicrobial susceptibility testing.
Results
Of the 63 patients with Vibrio infections, most contracted the virus between June and September, primarily in the Baltic Sea: 44 (70%) were male and the median age was 65 years (range: 2–93 years). Thirty-eight patients presented with wound infections, 16 with ear infections, six with gastroenteritis, two with pneumonia (after seawater aspiration) and one with primary septicaemia. The majority of infections were attributed to V. cholerae (non–O1/non-O139) (n = 30; 48%) or V. vulnificus (n = 22; 38%). Phylogenetic analyses of 12 available isolates showed clusters of three identical strains of V. vulnificus, which caused wound infections, suggesting that some clonal lines can spread across the Baltic Sea.
Conclusions
During the summers of 2018 and 2019, severe heatwaves facilitated increased numbers of Vibrio infections in Germany. Since climate change is likely to favour the proliferation of these bacteria, a further increase in Vibrio-associated diseases is expected.
Keywords: Vibriosis, Vibrio vulnificus, Vibrio cholerae, Vibrio parahaemolyticus, Baltic Sea, climate change
Introduction
The genus Vibrio, which belongs to the family Vibrionaceae and the class Gammaproteobacteria, includes many species that are potential human pathogens [1]. V. cholerae is a highly diverse species that consists of more than 200 serogroups [2]. Strains within the serogroups O1 and O139 produce cholera toxin and are the causative agents of endemic and epidemic cholera, which represent an important cause of morbidity and mortality in countries with inadequate access to clean water and sanitation facilities [3]. V. cholerae strains not included in these serogroups as well as other Vibrio spp. are referred to as non-cholera Vibrio spp. and are ubiquitous aquatic bacteria with a worldwide distribution, especially in warm estuarine and marine ecosystems [2]. These halophilic bacteria prefer low to moderate salinity (less than 25 parts per thousand (ppt) NaCl) [4]. The abundance of Vibrio spp. in marine and estuarine waters closely corresponds with the sea surface temperatures (SSTs) since they proliferate in warm water [5]. Thus, regional variations in environmental conditions are paramount importance in understanding the ecology of Vibrio spp.
Human infections with non-cholera Vibrio spp. can manifest as wound infections, ear infections, gastroenteritis, and primary septicaemia and have been predominantly reported in tropical and subtropical regions [6]. In Europe, cases are rare, and infections associated with the Mediterranean Sea [7-9], the Atlantic Ocean [10-12], or the Baltic Sea [13,14] have only been sporadically reported. However, a rapidly warming marine environment accompanied by an increase in extreme weather events such as heatwaves has resulted in unprecedented peak SSTs favouring the spread of Vibrio spp. worldwide. Recently, larger Vibrio spp. outbreaks have been reported in temperate regions such as Spain [15], Sweden, and Finland [16]. In Germany, Vibrio spp. other than toxigenic V. cholerae strains were not classified as reportable pathogenic agents before March 2020; to date, only a few cases have been reported and few case series of autochthonous infections have been published [17-20]. Over the last decades, resistance to various antibiotics, including to ampicillins, tetracyclines, and carbapenems, has emerged among Vibrio spp [21,22]. As a result of resistant isolates, the monitoring of antibiotic profiles of clinical and environmental Vibrio spp. has become of increasing importance. However, information on the number of human cases and the abundance of Vibrio spp. in coastal waters is scarce in most other European countries.
When we became aware of a high number of human infections with virulent Vibrio strains contracted in the Baltic Sea during the summer months of 2018 and 2019 (data not shown), we decided to conduct an observational retrospective multi-centre cohort study of domestically-acquired Vibrio infections. This study describes the epidemiology and the clinical impact of those infections and assesses antibiotic resistance patterns and phylogenetic relationships among clinical isolates. To determine the association between Vibrio infections and SST anomalies, we analysed the degree of warming in the south-western Baltic Sea, where the majority of the infections occurred.
Methods
Study population
Although non-cholera Vibrio spp. were not notifiable pathogenic agents in Germany before March 2020, clinical isolates of V. cholerae could be submitted to the Unit of Highly Pathogenic Microorganism (ZBS 2) at the Center for Biological Threats and Special Pathogens at the Robert Koch Institute (RKI), Germany’s national public health institute, for confirmation and serotyping. In addition, clinical isolates of other virulent Vibrio spp. could be sent for further analysis to the consultant laboratory for Vibrio spp. at the Federal Institute for Risk Assessment (BfR).
To identify as many patients with Vibrio-associated diseases as possible, the first author contacted the RKI, the BfR, and hospitals and microbiological laboratories within 20 km of the German North Sea or Baltic Sea coastline. Patients with Vibrio infections and a recent history of foreign travel who presumably did not acquire those infections in Germany were not included in the analysis. We (TTB, LB, HR) developed a standardised case report form, which was used to obtain demographic and clinical characteristics of patients from the treating hospitals and physicians.
Laboratory investigations
Antibiotic susceptibility testing and whole genome sequencing (WGS) of V. cholerae isolates initially submitted to the Unit of Highly Pathogenic Microorganisms (ZBS 2) at the Centre for Biological Threats and Special Pathogens at the RKI by microbiological laboratories or hospitals were performed directly at the RKI. All other isolates were analysed at the Institute of Medical Microbiology, Virology and Hygiene at the University Medical Center Hamburg-Eppendorf.
Species isolates were confirmed by matrix-assisted laser desorption ionization/time-of-flight mass spectrometry (MALDI-TOF) fingerprinting using a biotyper instrument (Bruker Daltonics, Bremen, Germany). To verify a V. cholerae species-specific sequence of sodB gene (superoxide dismutase), multiplex real-time PCR (5´ nuclease assay) was performed on all suspected isolates. The same assay confirmed the lack of ctxA, the encoding cholera toxin produced by toxigenic V. cholerae. In addition, conventional PCRs for the determination of serotype serogroup-specific gene rfb were performed to confirm the identification of non-O:1/non-O:139 V. cholerae strains. Primers and probes were used as previously described [23].
Antibiotic susceptibility testing
Available Vibrio isolates were subjected to antimicrobial susceptibility testing (AST) using disk diffusion and classified according to Clinical and Laboratory Standards Institute (CLSI) breakpoints [24]. Briefly, Vibrio isolates were grown on Colombia sheep blood agar at 37 °C for 18 hours. A colony suspension equivalent to 0.5 McFarland was prepared in 0.85% NaCl, streaked on Müller Hinton agar (MHA) plates, and incubated with respective antimicrobial agent disks at 35 ± 2 °C for 18 hours. Sixteen types of antibiotics disks (Oxoid by Thermo Fischer Scientific, Waltham, USA) were used: ampicillin (10 μg), amoxicillin-clavulanic acid (20/10 μg), piperacillin-tazobactam (100/10 μg), amikacin (30 μg), cefepime (30 μg), cefuroxime (30 μg), cefotaxime (30 μg), ceftazidime (30 μg), gentamicin (10 μg), imipenem (10 μg), meropenem (10 μg), ciprofloxacin (5 μg), levofloxacin (5 μg), chloramphenicol (30 μg), sulfamethoxazole/trimethoprim (1.25/23.75 μg), and tetracycline (30 μg). Escherichia coli ATCC 25922 was included as a positive control in each test. After incubation, the inhibition zone was measured and interpreted based on guidelines of CLSI M45-A2 [24]. Results for disk diffusion testing are shown in Supplementary Table S1.
Whole genome sequencing
WGS was performed retrospectively with an Illumina NextSeq instrument (Illumina, San Diego, USA) using 2 x 150 bp paired-end chemistry. Sequencing reads have been deposited in NCBI’s small reads archive (BioProject: PRJNA723758). Processing of WGS data for further phylogenetic analysis was performed at the Institute of Medical Microbiology, Virology and Hygiene at the University Medical Center Hamburg-Eppendorf. Reads were assembled with shovill 1.0.4 (https://github.com/tseemann/shovill) and spades 3.13.1 [25] and annotated with prokka 1.13.3 [26]. An average nt identity over 80% with genomes of the respective reference genome strains (Vibrio fluvialis_ASM155841v2; Vibrio parahaemolyticus_ASM19609v1; Vibrio vulnificus_ASM221513v1; Vibrio cholera ASM674v1; and Vibrio alginolyticus ASM35417v2) confirmed the identified species [27]. Sequencing reads were initially mapped to the respective reference genome and variants were called using snippy (https://github.com/tseemann/snippy). An alignment of core genome single nucleotide polymorphisms (SNPs) was produced in snippy to infer a phylogeny. The phylogenetic tree was constructed with Molecular Evolutionary Genetic Analysis (MEGA) X version 10.2.0 [28] using a neighbour-joining algorithm [29]. Possible relationships between clusters, potential genotypic, and phenotypic traits associated with pathogenicity and source of isolation were investigated. The virulence gene profile was determined with ABRicate (https://github.com/tseemann/abricate) and an additional blast-search against the virulence factor database [30].
Climatological analyses
SST data were originally derived using the near-infrared channel of the space-borne Advanced Very High Resolution Radiometer (AVHRR) satellite. We used the Reynolds Optimum Interpolation Sea Surface Temperature (OISST) data set with a spatial resolution of ca 25 km. The daily data were obtained from National Oceanic and Atmospheric Administration’s (NOAA) National Center for Environmental Information (NCEI) [31]. The annual climatological cycle from 1982 to 2001 was calculated using the OISST as a reference. SST anomalies were calculated from the higher resolving Moderate Resolution Imaging Spectroradiometer (MODIS) SST data, which have been available since 2002 from the MODIS sensors onboard the EOS-AQUA and Terra satellites. The MODIS Aqua SST data (version 2019 Reprocessing, NASA OB.DAAC, Greenbelt, USA) were provided with a time variable by the Integrated Climate Data Center (ICDC) at the University of Hamburg, Germany. The MODIS sensors offer a spatial resolution of 4.6 km; however, because the sensors cannot penetrate clouds, the data coverage is reduced. The annual climatology from the OISST data was interpolated to the higher resolution of the MODIS data and subtracted from the MODIS SST time series. In addition, a daily MODIS SST spatial average was calculated for the German coastlines where the infections in our study population occurred. The trend in SSTs was calculated from the low-passed SSTs (19 months) by a simple linear regression using standard statistics. Only values with a significance of p < 0.05 were considered.
Ethical statement
The study was reviewed and approved by the Ethics Committee of the Medical Council of Hamburg (PV7066) and written informed consent was waived because of the retrospective design.
Results
Patients and course of disease
Of the 63 patients with domestically-acquired Vibrio infections, 44 (70%) were male and the median age for all patients was 65 years (range: 2–93 years). Reliable anamnestic information about the precise place of infection was available for 36 patients, all of whom contracted Vibrio infections after recreational exposure to the open Baltic Sea or its estuaries (n = 34) or after consumption of shrimp caught in the North Sea or the Baltic Sea (n = 2). Nine patients were diagnosed while hospitalised near the Baltic Sea, so it is highly likely that their infections occurred during exposure to the Baltic Sea. For the remaining 18 patients, the place of infection is not known. A total of 38 patients (60%) presented with wound infections, 16 (25%) with ear infections, and six (10%) with gastroenteritis (Table 1).
Table 1. Characteristics of patients with domestically-acquired Vibrio spp. infections by age group, Germany, 2018 and 2019 (n = 63).
| Characteristic | Total (n = 63) |
Wound infections (n = 38) | Ear infections (n = 16) | Gastroenteritis (n = 6) |
Pneumonia (n = 2) |
Primary septicaemia (n = 1) |
|---|---|---|---|---|---|---|
| Age in years, Median (range) | 65 (2–93) | 69 (7–93) | 13 (3–78) | 60 (2–88) | 73 (68–78) | 88 |
| Sex | ||||||
| Male | 44 | 26 | 10 | 5 | 2 | 1 |
| Female | 19 | 12 | 6 | 1 | 0 | 0 |
| Vibrio spp. | ||||||
|
V. cholerae
(non-O1/non-O139) |
30 | 9 | 15 | 5 | 0 | 1 |
| V. vulnificus a | 24 | 22 | 0 | 0 | 2 | 0 |
| V. parahaemolyticus a | 5 | 4 | 1 | 0 | 0 | 0 |
| V. fluvialis | 3 | 2 | 0 | 1 | 0 | 0 |
| V. alginolyticus | 2 | 2 | 0 | 0 | 0 | 0 |
| Comorbidities | ||||||
| Information available | 41 | 33 | 5 | 1 | 2 | 0 |
| Chronic disease | 25 | 22 | 0 | 1 | 2 | 0 |
| Immunosuppressionb | 8 | 6 | 0 | 1 | 1 | 0 |
| Course of disease | ||||||
| Information available (n) | 41 | 33 | 5 | 1 | 2 | 0 |
| Antibiotic treatment | 37 | 32 | 2 | 1 | 2 | 0 |
| Hospital admission | 35 | 31 | 1 | 1 | 2 | 0 |
| Intensive care unit admission | 21 | 18 | 0 | 1 | 2 | 0 |
| Death | 8 | 6 | 0 | 1 | 1 | 0 |
a One patient was co-infected with V. vulnificus and V. parahaemolyticus, which were detected in wounds as well as in blood cultures.
b Immunosuppression was the result of immunosuppressive therapy, underlying haematological malignancies, or splenectomy.
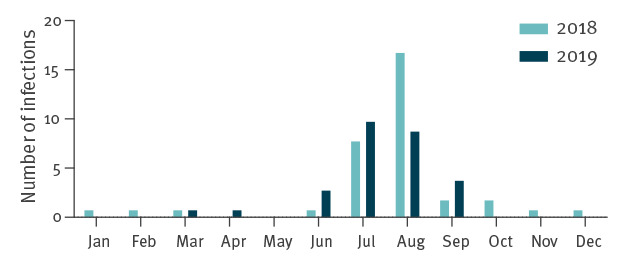
The vast majority of infections occurred in the summer months between June and September (n = 54; 86%) (Figure 1). In addition, nine infections with V. cholerae (non-O1/non-O139) occurred between October and April. Among these cases, detailed information on the route of transmission was only available for one patient, who contracted a wound infection after bathing in the Baltic Sea in October. However, no foreign travel was documented for these cases, so it can be assumed that these sporadic infections were acquired in Germany.
Figure 1.
Monthly distribution of patients with domestically-acquired Vibrio spp. infections, Germany 2018 and 2019
Infections spiked during summer heat waves in the Baltic Sea area.
Among patients who contracted wound infections, the majority (n = 32; 84%) had traumatic injuries either before or during contact with contaminated seawater. Another patient had superficial skin lesions as the result of mosquito bites, which served as an entry point for the Vibrio spp. In addition, five patients had underlying chronic skin diseases, including diabetic foot syndrome, lymphoedema, and atopic dermatitis, which predisposed them for wound infections.
Twenty-one patients with wound infections and one patient with gastroenteritis developed septicaemia, suggesting systemic disease progression. In addition, two patients developed pneumonia after seawater aspiration, and one patient presented with primary septicaemia without a known primary focus of infection. Wound infections and primary septicaemia mainly occurred in elderly patients (median age 69 and 78 years, respectively), and ear infections mainly occurred in younger patients (median age 13 years).
Information about the medical history and the course of disease was available for 65% (n = 41) of all patients. The most prevalent comorbidities were cardiovascular diseases (n = 25) and diabetes mellitus (n = 8). Eight patients were immunocompromised because of immunosuppressive therapy, underlying haematological malignancies, or splenectomy. The vast majority of patients had received antibiotic therapy (n = 37) and required hospitalisation (n = 35) with a mean time of hospitalisation of 18 days. Admission to an intensive care unit (ICU) was necessary for 21 patients. Eight patients died of the infection (Table 2).
Table 2. Deaths from Vibrio spp. infections, Germany, 2018 and 2019 (n = 8).
| Year | Age group in years | Vibrio spp. | Clinical manifestation | Chronic disease | Immuno-suppression |
|---|---|---|---|---|---|
| 2018 | 80−89 | V. vulnificus | Wound infection | yes | yes |
| 2018 | 70−79 | V. vulnificus | Wound infection | yes | no |
| 2018 | 70−79 | V. vulnificus | Wound infection | yes | yes |
| 2019 | 80−89 |
V. cholerae
(non-O1/non-O139) |
Gastroenteritis after eating crab meat | yes | yes |
| 2019 | 80−89 | V. vulnificus | Wound infection | yes | no |
| 2019 | 60−69 | V. vulnificus | Pneumogenic sepsis after drowning accident | no | no |
| 2019 | >90 | V. vulnificus | Wound infection | yes | no |
| 2019 | 50−59 | V. vulnificus | Wound infection | yes | no |
All but one of these eight patients were male, and the median age was 80 years (range: 56–93 years). Six patients died of wound infections from V. vulnificus, one patient died of gastroenteritis from V. cholerae (non-O1/non-O139) after eating crab meat, and one patient died of pneumonia from V. vulnificus after aspirating seawater. Of the patients who died of infection, three were immunocompromised and all but one had chronic diseases.
Detected clinical Vibrio spp. isolates
Most infections were either attributed to V. cholerae (non–O1/non-O139) (n = 30; 48%) or V. vulnificus (n = 24; 38%) (Table 1). Less frequently, V. parahaemolyticus (n = 5; 8%), V. fluvialis (n = 3; 5%), and V. alginolyticus (n = 2; 3%) were detected.
Ear infections were primarily caused by V. cholerae (non–O1/non-O139) (n = 15/16), and wound infections were primarily caused by V. vulnificus (n = 22/38). One patient was infected with both V. vulnificus and V. parahaemolyticus; both species were detected in the wounds and in the blood cultures. One patient was diagnosed with polymicrobial ear infections with V. parahaemolyticus and Shewanella putrefaciens, and one patient was diagnosed with V. cholerae, Staphylococcus aureus, Pseudomonas stutzeri, and E. coli.
A total of 48 clinical Vibrio spp. isolates (V. cholerae (non–O1/non-O139), n = 29; V. vulnificus, n = 12; V. parahaemolyticus, n = 3; V. alginolyticus, n = 2; and V. fluvialis, n = 1) were available for further analysis and subsequently subjected to AST and WGS. One V. vulnificus isolate was only available for AST but not WGS, and one V. vulnificus isolate was only available for WGS but not for AST.
Antimicrobial susceptibilities
Most of the 16 antibiotics tested in this study – e.g., tetracycline, third-generation cephalosporins (cefotaxime and ceftazidime), aminoglycosides (gentamicin and amikacin), and fluoroquinolones (ciprofloxacin and levofloxacin) – are recommended by the United States Centers for Disease Control and Prevention (CDC) for the treatment of Vibrio spp. infections [32]. Based on the AST results, the highest rate of phenotypic antimicrobial resistance of all Vibrio ssp. isolates in our study were detected for ampicillin (n = 6) and amikacin (n = 2). Low rates of resistance were found for gentamicin (n = 1) and amoxicillin-clavulanic acid (n = 1). However, all isolates were susceptible to cefepime, cefotaxime, ceftazidime, cefuroxime, chloramphenicol, ciprofloxacin, gentamicin, imipenem, meropenem, levofloxacin, and trimethoprim-sulfamethoxazole (Supplementary Table S1).
Resistance gene profiling identified resistance genes in 72% (34/47) of all Vibrio ssp. isolates (Table 3). In all but one ampicillin-resistant isolate, blaCARB genes encoding for a beta-lactamase were detected (blaCARB-7; n = 2). One of these isolates showed additional resistance to amoxicillin-clavulanic acid. Gentamicin-resistance is mostly conferred by aminoglycoside-modifying enzymes. However, no corresponding resistance gene was found for gentamicin-resistant and gentamicin-intermediate resistant isolates.
Table 3. Phenotypic antibiotic susceptibilities and associated genetic resistance markers in isolates from patients with Vibrio spp. infections, Germany, 2018 and 2019 (n = 48 isolates).
| Antibiotic | Phenotypic susceptibility (n = 47) | Resistance detected by WGS (n = 47) | |||||
|---|---|---|---|---|---|---|---|
| Resistant | Intermediate | Gene or mutation | Positive | ||||
| n | % | n | % | n | % | ||
| AMK | 2 | 4.3 | 11 | 23.4 | NA | 0 | 0 |
| AMP | 6 | 12.8 | 3 | 6.4 | blaCARB-7 | 2 | 4.3 |
| AMC | 1 | 2.1 | 2 | 4.3 | blaCARB-18, blaCARB-21, blaCARB-26, blaCARB-30, blaCARB-42 | 7 | 14.9 |
| MEM | 0 | 0 | 0 | 0 | varG | 15 | 31.9 |
| TET | 0 | 0 | 0 | 0 | tet 34, tet 35 | 18 | 38.3 |
| GEN | 1 | 2.1 | 0 | 0 | NA | 0 | 0 |
AMC: amoxicillin-clavulanic acid; AMK: amikacin; AMP: ampicillin; GEN: gentamicin; MEM: meropenem; NA: not applicable; TET: tetracycline; WGS: whole genome sequencing.
Genomical analysis showed that varG, which encodes a metallo-beta-lactamase, was present in 31% (n = 15/29) of V. cholerae (non–O1/O139) isolates. VarG has beta-lactamase activity against penicillins, cephalosporins, and carbapenems, with the highest activity against meropenem [33], but no meropenem resistance was detected in our study isolates. Two V. cholerae (non–O1/non-O139) isolates additionally carried variants of blaCARB –7, a determinant of beta-lactam resistance.
The prevalence of antibiotic resistance genes tet34 and tet35 indicates intrinsic resistance to tetracycline, and these genes are present in all V. vulnificus and V. parahaemolyticus strains [34]. The function of tet34 is associated with xanthine-guanine phosphoribosyltransferase (XPRT)-like activity and has been shown to be present in certain Vibrio spp [35]. However, the phenotype of antimicrobial resistance indicates that all strains are sensitive to tetracycline, suggesting that tet34 is potentially non-functional [36].
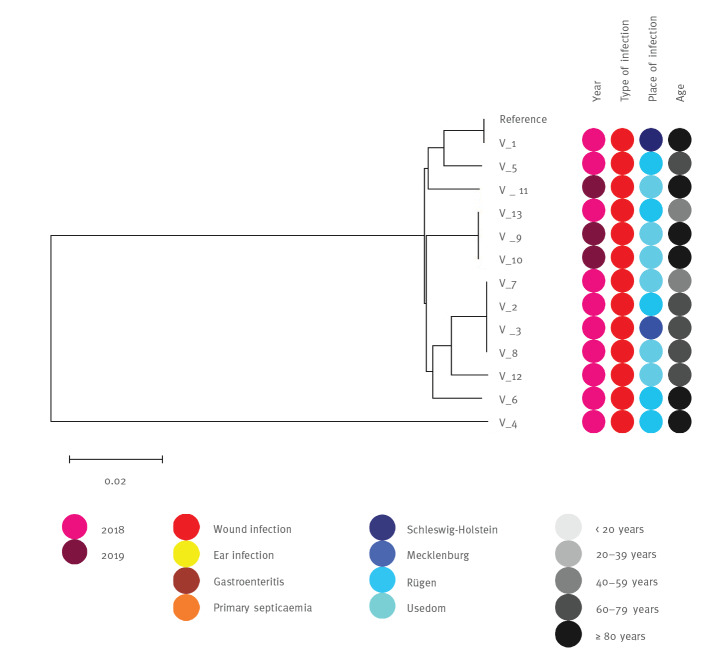
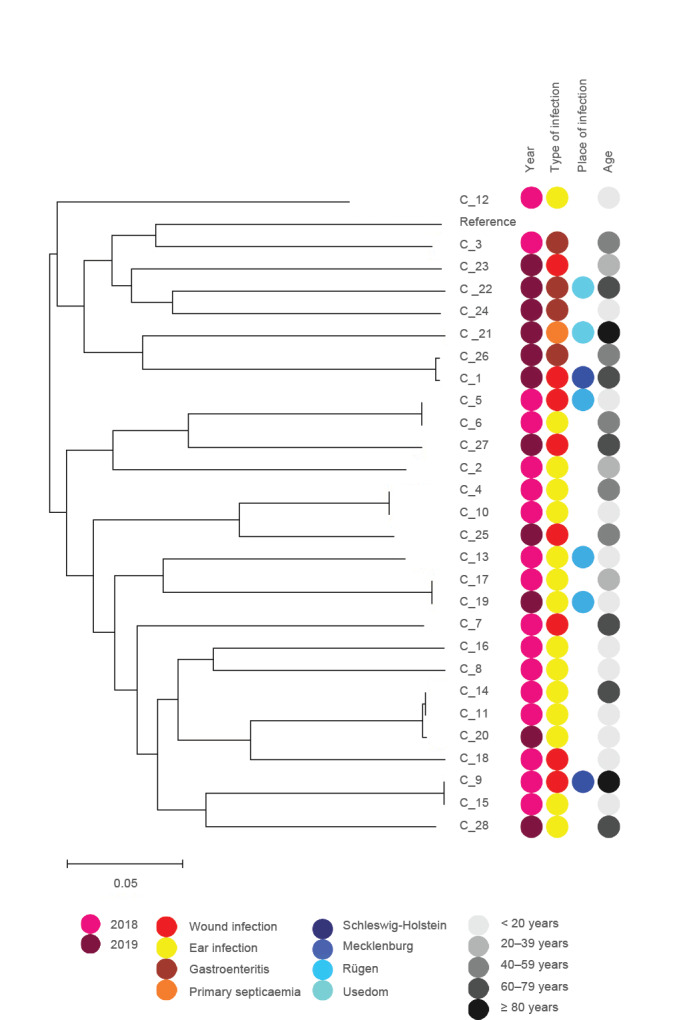
Phylogenetic data
The genetic diversity of V. vulnificus isolates (n = 12) and V. cholera (non–O1/non-O139) isolates (n = 29) was evaluated using a phylogenomic tree based on single nucleotide polymorphisms (SNPs) (Figure 2 and 3). Despite the genomic divergence among clusters, no distinct patterns linking strain phylogeny, isolation source, or virulent capabilities were apparent. As the isolates within the same clade were closely related, we identified isolates that were identical in the detected SNPs, but mostly with different isolation sources. Virulence gene profiling did not reveal atypical virulence factors (e.g., toxins) in our study isolates (Supplementary Table S2).
Figure 2.
Maximum likelihood trees showing phylogenetic relationships of 13 Vibrio vulnificus genomes based on SNP analysis
SNP: single nucleotide polymorphism.
The V. vulnificus ASM221513v1 sequence was used as reference. Bar, 2% estimated sequence divergence.
Figure 3.
Maximum likelihood trees showing phylogenetic relationships of 28 Vibrio cholerae genomes based on SNP analysis
SNP: single nucleotide polymorphism.
The V. cholerae ASM674v1 sequence was used as reference. Bar, 5% estimated sequence divergence.
Climatological data
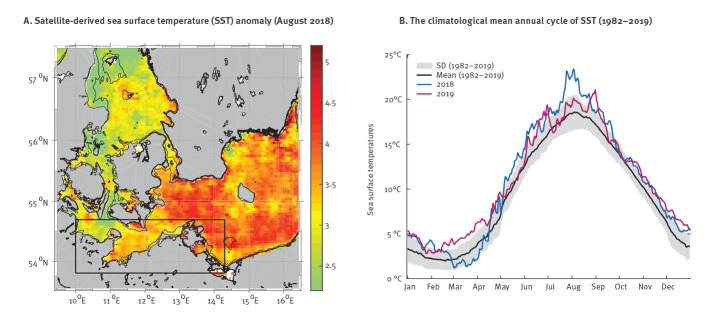
Between 1982 and 2019, the SST warming trend in the south-western Baltic Sea (Box in Figure 4A) was 0.56 °C per decade. During this period, the Baltic Sea reached the highest SST in August 2018 (Figure 4A). The positive SST anomaly reached 2 °C throughout the south-western Baltic Sea. In the central-southern Baltic Sea and in some coastal areas, especially the eastern coasts with shallow areas, the positive SST anomaly even reached 5 °C. Although the SSTs in 2019 were not as high as in the summer of 2018, positive SST anomalies of up to 4 °C were observed in 2019, primarily in June and September (Figure 4B).
Figure 4.
Sea surface temperatures in the south-eastern Baltic Sea in 2018 and 2019
4A: Satellite-derived sea surface temperature (SST) anomaly in August 2018 with a reference to the climatological mean between 1982 and 2001. The thin black line shows the water depth of 10 m and reveals the shallow coastal areas. 4B: The climatological mean annual cycle of SST (black) based on data between 1982 and 2019 and averaged within the black rectangular box in 4A with the standard deviation underlaid as a grey shadow. The annual SSTs between 2018 and 2019 are shown in blue and pink as indicated in the legend.
Discussion
Here, we present a large case series of domestically-acquired Vibrio infections in Germany. Consistent with previous studies [37,38], we demonstrate the broad clinical spectrum and various disease manifestations of Vibrio infections, ranging from mild gastroenteritis or ear infections to fulminant wound infections and primary septicaemia.
Also in line with previous findings, we observed a striking male predominance among patients with Vibrio infections [16,37]. While this may be attributed to sex differences in predisposing comorbidities and the choice of recreational activities, it has also been suggested that female sex hormones may have a protective effect against severe V. vulnificus infections [39]. Our study further emphasises that Vibrio infections occur across all age groups, but different disease manifestations have different age distributions. While ear infections most commonly affected children, wound infections and primary septicaemia were predominantly detected in elderly patients. The high prevalence of immunocompromising conditions and the chronic medical conditions among patients in our study cohort in general and in those who died of the infection in particular suggest these conditions may be determinants of disease severity in patients who contract Vibrio infections [37].
Importantly, recreational exposure to the Baltic Sea and its estuaries was responsible for the majority of infections in our study cohort. As in other studies, this finding demonstrates that most infections with Vibrio spp. involve either exposure of previously contracted wounds to seawater or injuries acquired during water-based activities [9,37]. The vast majority of Vibrio infections (n = 54; 86%) occurred between June and September: when water-associated recreational activities increase, high SSTs are responsible for an elevated abundance of Vibrio spp. in coastal waters. Sporadic infections outside these months were all caused by V. cholerae (non-O1/non-O139), which has a higher probability of occurrence at relatively low water temperatures compared to other Vibrio spp [4,40].
As the result of global warming, the SSTs in the Baltic Sea are expected to rise between 4 and 5 °C over the next several decades [41], which will likely lead to an increase of virulent Vibrio strains in coastal waters. On an interannual scale, the main driving force behind the Baltic Sea warming is the varying ambient temperature; less important are the intensity of solar radiation and the varying wind forces [42]. On a seasonal scale, the SSTs in the Baltic Sea are characterised by an average amplitude of 16 to 18 °C between summer and winter temperatures, which is higher in shallow bays and coastal regions as bathymetry prevents mixing and circulation of the water [43]. In addition, the rise of sea levels is expected to cause more coastal flooding, which will expand estuarine and brackish water environments suitable for Vibrio spp. growth [44]. Recently, early warning systems have been established that can forecast the environmental suitability of coastal waters for Vibrio spp. growth using remotely sensed SSTs and sea surface salinity measurements [40]. These systems can be used to alert healthcare providers and individuals at risk. Importantly, infection risk from Vibrio spp. does not correlate with faecal indicator organisms and therefore common water surveillance practices cannot predict infection risk from Vibrio spp [45]. To prevent severe infections, patients with open wounds or chronic skin diseases, especially those with underlying immunocompromising conditions, should be advised to avoid contact with seawater in the Baltic and Northern Sea area when water temperatures exceed 20 °C. In addition, vaccine candidates are being developed that may prevent Vibrio spp. infections in vulnerable populations [46].
Given the potentially fulminant disease course, patients with a presumptive diagnosis of severe Vibrio-associated diseases such as wound infections or primary septicaemia should promptly receive appropriate antibiotics for optimal disease outcomes. Recommended empiric treatment regimens include third-generation cephalosporins and either tetracyclines or fluoroquinolones [47]. In this study, the proportion of resistance among Vibrio spp. was comparable to a similar survey conducted on environmental isolates from marine coastal waters in Germany [22]. Using AST and WGS, we were able to show that empirical therapy would have been effective in all investigated cases. Resistance gene profiling identified some beta-lactamase and tetracycline inactivation enzyme genes, but no specific antimicrobial resistance patterns among Vibrio spp. isolates were detected that could pose a public health risk. Studies have shown that some strain subgroups are more likely to cause disease in humans than other subgroups, which is consistent with phylogenetic studies that show evidence of clustering of human isolates [48]. According to the SNP analysis, the strains showed three identical clusters for V. vulnificus, which caused wound infections. These clusters were found in three areas of the Baltic Sea coast, suggesting that clonal lines can spread across the Baltic Sea. However, because the number of reported Vibrio-associated diseases was relatively low, we could perform a phylogenetic analysis only between a small number of V. vulnificus and V. cholerae strains. Nevertheless, the results of the present survey provide an important resource for future prospective studies that focus on the emergence of Vibrio spp. infections contracted in the Baltic Sea and its estuaries.
Our study is subject to several limitations. First, it is not always possible to retrospectively differentiate bacterial colonisation from infection in patients with Vibrio spp. detected in wound or ear swabs after recent contact with seawater, especially in individuals with more than one potentially pathogenic agent. However, in patients with a typical clinical presentation and detection of human pathogenic Vibrio spp., the bacterium is highly likely the causative pathogen. Second, since non-cholera Vibrio spp. were not notifiable in Germany in 2018 and 2019, we aimed to contact all hospitals and microbiological laboratories near the German Baltic Sea and Northern Sea; however, this data collection approach meant that Vibrio infections detected elsewhere were probably not captured. In addition, patients with self-limiting and transient infections may not have undergone microbiological testing or sought medical attention. Although this selection bias may result in overestimation of disease severity in our study cohort, the actual clinical burden associated with Vibrio infections may be grossly underestimated due to under-reporting and underdiagnosis. Third, we were not able to obtain reliable information on comorbidities, disease course, and place of infection from all patients. These limitations underscore the need for mandatory standardised surveillance systems for Vibrio spp. infections [2,6,49]. In Germany, a mandatory notification for Vibrio spp. infections has been in place since March 2020. In 2020, only 13 Vibrio spp. infections were reported to the RKI. This relatively low number of cases was probably the result of restrictions imposed during the coronavirus disease (COVID-19) pandemic. In the future, this mandatory surveillance may help capture comprehensive clinical and microbiological data. These data are paramount for obtaining the precise epidemiological and clinical impact of domestically-acquired Vibrio infections, identifying vulnerable populations, and guiding future public health preparedness activities.
Conclusion
Our study suggests that severe heatwaves during the summer of 2018 and 2019 were responsible for at least 63 Vibrio spp. infections contracted in the German Baltic Sea and its estuaries. Since global warming is predicted to favour the proliferation of these bacteria in the aquatic environment and demographic change will likely increase the number of vulnerable individuals, a rise in severe Vibrio-associated diseases may be expected in the future. Although the burden of disease from Vibrio spp. infections is currently relatively low, the potentially fulminant disease course in patients with wound infections and primary septicaemia underlines the need to protect vulnerable population groups.
Acknowledgements
Funding statement: TTB is funded by the German Center for Infection Research (DZIF).
Supplementary Data
German Vibrio Study Group
Veronika Balau (MVZ IMD Labor Greifswald GmbH, Greifswald, Germany); Elsa Baufeld (Friedrich Loeffler-Institute of Medical Microbiology, University Hospital Greifswald, Germany); Simone Brechmann (MVZ Diamedis, Bielefeld, Germany); Lutz Briedigkeit (Institute for Laboratory and Transfusion Medicine, Helios Kliniken Schwerin, Schwerin, Germany); Stephan Diedrich (Department of General, Visceral, Thoracic and Vascular Surgery, Universitätsmedizin Greifswald, Greifswald, Germany); Ulrike Ebert (Kreiskrankenhaus Frankenberg, Frankenberg (Eder), Germany); Helmut Fickenscher (Institute for Infection Medicine, University of Kiel and University Hospital Schleswig-Holstein, Kiel, Germany); Bojana Grgic (Department of Hygiene and Infection Prevention, Helios Kliniken Schwerin, Schwerin, Germany); Claus-Dieter Heidecke (Department of General, Visceral, Thoracic and Vascular Surgery, University Medicine of Greifswald, Greifswald, Germany); Peter Hinz (Department of Trauma and Orthopaedic Surgery, Clinic of Surgery, University Medicine Greifswald, Greifswald, Germany); Ada Hoffmann (Department of Infectious Diseases and Microbiology, University Hospital Schleswig-Holstein, Lübeck, Germany); Meike Holbe (Department of Paediatrics, Medical University of Lübeck, Lübeck, Germany); Ralf Ignatius (MVZ Labor 28 GmbH, Berlin, Germany, and Charité, Institute of Microbiology, Infectious Diseases and Immunology, Berlin, Germany); Olaf Kaup (Microbiologic Laboratory, Klinikum Bielefeld Rosenhöhe, Bielefeld, Germany); Martin Kern (IMD Labor Oderland GmbH, Frankfurt (Oder), Germany); Martina Kerwat (Institute of Medical Microbiology and Hygiene, Philipps University Marburg, Marburg, Germany); Ingo Klempien (Department of Clinical Hygiene and Infectious Diseases, Helios Hanseklinikum Stralsund, Stralsund, Germany); Georg Lamprecht (Department of Medicine II, Division of Gastroenterology and Endocrinology, University Medical Center Rostock, Rostock, Germany); Astrid Meerbach (MVZ Labor Dr Reising-Ackermann und Kollegen, Leipzig, Germany); Alexander Mischnik (Department of Infectious Diseases and Microbiology, University of Lübeck, Lübeck, Germany); Andreas Podbielski (Institute of Medical Microbiology, Virology and Hygiene, University Hospital Rostock, Rostock, Germany); Stephan Schaefer (MVZ Labor Limbach Vorpommern-Rügen GmbH, Stralsund, Germany); Roman Schwarz (Labor Mönchengladbach, MVZ Dr. Stein + Kollegen, Mönchengladbach, Germany); Eckhard Strauch (Bundesinstitut für Risikobewertung (BfR) - German Federal Institute for Risk Assessment, Berlin, Germany; Philipp Warnke (Institute of Medical Microbiology, Virology and Hygiene, University Hospital Rostock, Rostock, Germany); Simone Weikert-Asbeck (Department of Infectious Diseases and Microbiology, University of Lübeck, Lübeck, Germany); Maria Witte (Department of General, Thoracic, Vascular, and Transplantation Surgery, Rostock University Medical Center, Rostock, Germany); Waleed Zaki (Berlin, Germany).
Conflict of interest: None declared.
Authors’ contributions: TTB, LB, SD, SS and HR initiated the study. MSM, DJ, OD, JB, KB, AK, MC and MA contributed to the acquisition and interpretation of data. All German Vibrio Study Group members contributed to acquisition of data. TTB, LB, MSM and HR wrote the first draft manuscript. All authors, including the German Vibrio Study Group members, critically revised the manuscript and approved the final version.
References
- 1.Farmer JJ. The Family Vibrionaceae. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, editors. The prokaryotes: Volume 6: proteobacteria: gamma subclass. New York, NY: Springer New York; 2006. p. 495-507. [Google Scholar]
- 2. Baker-Austin C, Trinanes J, Gonzalez-Escalona N, Martinez-Urtaza J. Non-cholera Vibrios: the microbial barometer of climate change. Trends Microbiol. 2017;25(1):76-84. 10.1016/j.tim.2016.09.008 [DOI] [PubMed] [Google Scholar]
- 3. Clemens JD, Nair GB, Ahmed T, Qadri F, Holmgren J. Cholera. Lancet. 2017;390(10101):1539-49. 10.1016/S0140-6736(17)30559-7 [DOI] [PubMed] [Google Scholar]
- 4.Muhling BA, Jacobs J, Stock CA, Gaitan CF, Saba VS. Projections of the future occurrence, distribution, and seasonality of three Vibrio species in the Chesapeake Bay under a high-emission climate change scenario. Geohealth. 2017;1(7):278-96. [DOI] [PMC free article] [PubMed]
- 5. Baker-Austin C, Trinanes JA, Taylor NGH, Hartnell R, Siitonen A, Martinez-Urtaza J. Emerging Vibrio risk at high latitudes in response to ocean warming. Nat Clim Chang. 2012;3(1):73-7. 10.1038/nclimate1628 [DOI] [Google Scholar]
- 6. Baker-Austin C, Oliver JD, Alam M, Ali A, Waldor MK, Qadri F, et al. Vibrio spp. infections. Nat Rev Dis Primers. 2018;4(1):8. 10.1038/s41572-018-0005-8 [DOI] [PubMed] [Google Scholar]
- 7. Marinello S, Marini G, Parisi G, Gottardello L, Rossi L, Besutti V, et al. Vibrio cholerae non-O1, non-O139 bacteraemia associated with pneumonia, Italy 2016. Infection. 2017;45(2):237-40. 10.1007/s15010-016-0961-4 [DOI] [PubMed] [Google Scholar]
- 8. Thodis E, Kriki P, Kakagia D, Passadakis P, Theodoridis M, Mourvati E, et al. Rigorous Vibrio vulnificus soft tissue infection of the lower leg in a renal transplant patient managed by vacuum therapy and autologous growth factors. J Cutan Med Surg. 2009;13(4):209-14. 10.2310/7750.2008.08033 [DOI] [PubMed] [Google Scholar]
- 9. Brehm TT, Berneking L, Rohde H, Chistner M, Schlickewei C, Sena Martins M, et al. Wound infection with Vibrio harveyi following a traumatic leg amputation after a motorboat propeller injury in Mallorca, Spain: a case report and review of literature. BMC Infect Dis. 2020;20(1):104. 10.1186/s12879-020-4789-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Reilly GD, Reilly CA, Smith EG, Baker-Austin C. Vibrio alginolyticus-associated wound infection acquired in British waters, Guernsey, July 2011. Euro Surveill. 2011;16(42):19994. 10.2807/ese.16.42.19994-en [DOI] [PubMed] [Google Scholar]
- 11. Engel MF, Muijsken MA, Mooi-Kokenberg E, Kuijper EJ, van Westerloo DJ. Vibrio cholerae non-O1 bacteraemia: description of three cases in the Netherlands and a literature review. Euro Surveill. 2016;21(15). 10.2807/1560-7917.ES.2016.21.15.30197 [DOI] [PubMed] [Google Scholar]
- 12. De Keukeleire S, Hoste P, Crivits M, Hammami N, Piette A. Atypical manifestation of Vibrio cholerae: fear the water! Acta Clin Belg. 2018;73(6):462-4. 10.1080/17843286.2018.1483563 [DOI] [PubMed] [Google Scholar]
- 13. Stypulkowska-Misiurewicz H, Pancer K, Roszkowiak A. Two unrelated cases of septicaemia due to Vibrio cholerae non-O1, non-O139 in Poland, July and August 2006. Euro Surveill. 2006;11(11):E061130.2. [DOI] [PubMed] [Google Scholar]
- 14. Dalsgaard A, Frimodt-Møller N, Bruun B, Høi L, Larsen JL. Clinical manifestations and molecular epidemiology of Vibrio vulnificus infections in Denmark. Eur J Clin Microbiol Infect Dis. 1996;15(3):227-32. 10.1007/BF01591359 [DOI] [PubMed] [Google Scholar]
- 15. Martinez-Urtaza J, Trinanes J, Abanto M, Lozano-Leon A, Llovo-Taboada J, Garcia-Campello M, et al. Epidemic dynamics of Vibrio parahaemolyticus illness in a hotspot of disease emergence, Galicia, Spain. Emerg Infect Dis. 2018;24(5):852-9. 10.3201/eid2405.171700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Baker-Austin C, Trinanes JA, Salmenlinna S, Löfdahl M, Siitonen A, Taylor NG, et al. Heat wave-associated Vibriosis, Sweden and Finland, 2014. Emerg Infect Dis. 2016;22(7):1216-20. 10.3201/eid2207.151996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Frank C, Littman M, Alpers K, Hallauer J. Vibrio vulnificus wound infections after contact with the Baltic Sea, Germany. Euro Surveill. 2006;11(8):E060817.1. 10.2807/esw.11.33.03024-en [DOI] [PubMed] [Google Scholar]
- 18. Hoyer J, Engelmann E, Liehr RM, Distler A, Hahn H, Shimada T. Septic shock due to Vibrio vulnificus serogroup 04 wound infection acquired from the Baltic Sea. Eur J Clin Microbiol Infect Dis. 1995;14(11):1016-8. 10.1007/BF01691388 [DOI] [PubMed] [Google Scholar]
- 19. Kuhnt-Lenz K, Krengel S, Fetscher S, Heer-Sonderhoff A, Solbach W. Sepsis with bullous necrotizing skin lesions due to vibrio vulnificus acquired through recreational activities in the Baltic Sea. Eur J Clin Microbiol Infect Dis. 2004;23(1):49-52. 10.1007/s10096-003-1056-6 [DOI] [PubMed] [Google Scholar]
- 20. Ruppert J, Panzig B, Guertler L, Hinz P, Schwesinger G, Felix SB, et al. Two cases of severe sepsis due to Vibrio vulnificus wound infection acquired in the Baltic Sea. Eur J Clin Microbiol Infect Dis. 2004;23(12):912-5. 10.1007/s10096-004-1241-2 [DOI] [PubMed] [Google Scholar]
- 21. Lee LH, Ab Mutalib NS, Law JW, Wong SH, Letchumanan V. Discovery on antibiotic resistance patterns of Vibrio parahaemolyticus in Selangor reveals carbapenemase producing Vibrio parahaemolyticus in marine and freshwater fish. Front Microbiol. 2018;9:2513. 10.3389/fmicb.2018.02513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bier N, Schwartz K, Guerra B, Strauch E. Survey on antimicrobial resistance patterns in Vibrio vulnificus and Vibrio cholerae non-O1/non-O139 in Germany reveals carbapenemase-producing Vibrio cholerae in coastal waters. Front Microbiol. 2015;6:1179. 10.3389/fmicb.2015.01179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dupke S, Akinsinde KA, Grunow R, Iwalokun BA, Olukoya DK, Oluwadun A, et al. Characterization of Vibrio cholerae strains isolated from the Nigerian cholera outbreak in 2010. J Clin Microbiol. 2016;54(10):2618-21. 10.1128/JCM.01467-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clinical and Laboratory Standards Institute (CLSI). Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria. Guideline - Second Edition. Pennsylvania: CLSI. 2010;30(18). M45-A2. [Google Scholar]
- 25. Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455-77. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068-9. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- 27. Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, et al. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol. 2017;67(5):1613-7. 10.1099/ijsem.0.001755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725-9. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406-25. [DOI] [PubMed] [Google Scholar]
- 30. Chen L, Yang J, Yu J, Yao Z, Sun L, Shen Y, et al. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 2005;33(Database issue):D325-8. 10.1093/nar/gki008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.National Centers for Environmental Information. National Oceanic and Atmospheric Administration (NCEI.NOAA). NOAA 0.25-degree daily optimum interpolation sea surface temperature (OISST), version 2.1. Washington: NCEI.NOAA. [Accessed: 6 Nov 2020]. Available from: https://www.ncei.noaa.gov/metadata/geoportal/rest/metadata/item/gov.noaa.ncdc:C01606/html
- 32. Shaw KS, Rosenberg Goldstein RE, He X, Jacobs JM, Crump BC, Sapkota AR. Antimicrobial susceptibility of Vibrio vulnificus and Vibrio parahaemolyticus recovered from recreational and commercial areas of Chesapeake Bay and Maryland Coastal Bays. PLoS One. 2014;9(2):e89616. 10.1371/journal.pone.0089616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lin HV, Massam-Wu T, Lin CP, Wang YA, Shen YC, Lu WJ, et al. The Vibrio cholerae var regulon encodes a metallo-β-lactamase and an antibiotic efflux pump, which are regulated by VarR, a LysR-type transcription factor. PLoS One. 2017;12(9):e0184255. 10.1371/journal.pone.0184255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. López-Pérez M, Jayakumar JM, Haro-Moreno JM, Zaragoza-Solas A, Reddi G, Rodriguez-Valera F, et al. Evolutionary model of cluster divergence of the emergent marine pathogen Vibrio vulnificus: from genotype to ecotype. MBio. 2019;10(1):e02852-18. 10.1128/mBio.02852-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nonaka L, Suzuki S. New Mg2+-dependent oxytetracycline resistance determinant tet 34 in Vibrio isolates from marine fish intestinal contents. Antimicrob Agents Chemother. 2002;46(5):1550-2. 10.1128/AAC.46.5.1550-1552.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang Z, Yu YX, Jiang Y, Wang YG, Liao MJ, Rong XJ, et al. First report of isolation and complete genome of Vibrio rotiferianus strain SSVR1601 from cage-cultured black rockfish (Sebastes schlegelii) associated with skin ulcer. J Fish Dis. 2019;42(5):623-30. 10.1111/jfd.12963 [DOI] [PubMed] [Google Scholar]
- 37. Dechet AM, Yu PA, Koram N, Painter J. Nonfoodborne Vibrio infections: an important cause of morbidity and mortality in the United States, 1997-2006. Clin Infect Dis. 2008;46(7):970-6. 10.1086/529148 [DOI] [PubMed] [Google Scholar]
- 38. Heng S-P, Letchumanan V, Deng C-Y, Ab Mutalib N-S, Khan TM, Chuah L-H, et al. Vibrio vulnificus: an environmental and clinical burden. Front Microbiol. 2017;8:997. 10.3389/fmicb.2017.00997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Merkel SM, Alexander S, Zufall E, Oliver JD, Huet-Hudson YM. Essential role for estrogen in protection against Vibrio vulnificus-induced endotoxic shock. Infect Immun. 2001;69(10):6119-22. 10.1128/IAI.69.10.6119-6122.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hirk S, Huhulescu S, Allerberger F, Lepuschitz S, Rehak S, Weil S, et al. Necrotizing fasciitis due to Vibrio cholerae non-O1/non-O139 after exposure to Austrian bathing sites. Wien Klin Wochenschr. 2016;128(3-4):141-5. 10.1007/s00508-015-0944-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Semenza JC, Trinanes J, Lohr W, Sudre B, Löfdahl M, Martinez-Urtaza J, et al. Environmental suitability of Vibrio infections in a warming climate: an early warning system. Environ Health Perspect. 2017;125(10):107004. 10.1289/EHP2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kniebusch M, Meier HEM, Neumann T, Börgel F. Temperature variability of the Baltic Sea since 1850 and attribution to atmospheric forcing variables. J Geophys Res Oceans. 2019;124(6):4168-87. 10.1029/2018JC013948 [DOI] [Google Scholar]
- 43. Stramska M, Białogrodzka J. Spatial and temporal variability of sea surface temperature in the Baltic Sea based on 32-years (1982-2013) of satellite data. Oceanologia. 2015;57(3):223-35. 10.1016/j.oceano.2015.04.004 [DOI] [Google Scholar]
- 44. Semenza JC, Herbst S, Rechenburg A, Suk JE, Höser C, Schreiber C, et al. Climate change impact assessment of food- and waterborne diseases. Crit Rev Environ Sci Technol. 2012;42(8):857-90. 10.1080/10643389.2010.534706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Böer SI, Heinemeyer EA, Luden K, Erler R, Gerdts G, Janssen F, et al. Temporal and spatial distribution patterns of potentially pathogenic Vibrio spp. at recreational beaches of the German north sea. Microb Ecol. 2013;65(4):1052-67. 10.1007/s00248-013-0221-4 [DOI] [PubMed] [Google Scholar]
- 46. Lee TH, Kim MH, Lee CS, Lee JH, Rhee JH, Chung KM. Protection against Vibrio vulnificus infection by active and passive immunization with the C-terminal region of the RtxA1/MARTXVv protein. Vaccine. 2014;32(2):271-6. 10.1016/j.vaccine.2013.11.019 [DOI] [PubMed] [Google Scholar]
- 47. Stevens DL, Bisno AL, Chambers HF, Dellinger EP, Goldstein EJ, Gorbach SL, et al. Infectious Diseases Society of America . Practice guidelines for the diagnosis and management of skin and soft tissue infections: 2014 update by the Infectious Diseases Society of America. Clin Infect Dis. 2014;59(2):e10-52. 10.1093/cid/ciu296 [DOI] [PubMed] [Google Scholar]
- 48. Roig FJ, González-Candelas F, Sanjuán E, Fouz B, Feil EJ, Llorens C, et al. Phylogeny of Vibrio vulnificus from the analysis of the core-genome: implications for intra-species taxonomy. Front Microbiol. 2018;8:2613. 10.3389/fmicb.2017.02613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Newton A, Kendall M, Vugia DJ, Henao OL, Mahon BE. Increasing rates of vibriosis in the United States, 1996-2010: review of surveillance data from 2 systems. Clin Infect Dis. 2012;54 Suppl 5(0 5):S391-5. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.