Abstract
Background and Aim
This study aims to construct a strategy that uses assistance from artificial intelligence (AI) to assist radiologists in the identification of malignant versus benign focal liver lesions (FLLs) using contrast‐enhanced ultrasound (CEUS).
Methods
A training set (patients = 363) and a testing set (patients = 211) were collected from our institute. On four‐phase CEUS images in the training set, a composite deep learning architecture was trained and tuned for differentiating malignant and benign FLLs. In the test dataset, AI performance was evaluated by comparison with radiologists with varied levels of experience. Based on the comparison, an AI assistance strategy was constructed, and its usefulness in reducing CEUS interobserver heterogeneity was further tested.
Results
In the test set, to identify malignant versus benign FLLs, AI achieved an area under the curve of 0.934 (95% CI 0.890–0.978) with an accuracy of 91.0%. Comparing with radiologists reviewing videos along with complementary patient information, AI outperformed residents (82.9–84.4%, P = 0.038) and matched the performance of experts (87.2–88.2%, P = 0.438). Due to the higher positive predictive value (PPV) (AI: 95.6% vs residents: 88.6–89.7%, P = 0.056), an AI strategy was defined to improve the malignant diagnosis. With the assistance of AI, radiologists exhibited a sensitivity improvement of 97.0–99.4% (P < 0.05) and an accuracy of 91.0–92.9% (P = 0.008–0.189), which was comparable with that of the experts (P = 0.904).
Conclusions
The CEUS‐based AI strategy improved the performance of residents and reduced CEUS's interobserver heterogeneity in the differentiation of benign and malignant FLLs.
Keywords: artificial intelligence, diagnosis, computer‐assisted, liver neoplasms, ultrasonography
Introduction
The worldwide incidence of focal liver lesions (FLLs) is increasing and is accompanied by an increase in the prevalence of hepatocellular carcinoma, intrahepatic cholangiocarcinoma, and metastasis from colorectal cancer. 1 , 2 The noninvasive differentiation of malignant lesions from benign lesions is a key diagnostic process before treatment and routinely relies on computed tomography (CT) and magnetic resonance (MR). Unfortunately, the reported error rate of FLL characterization varies from 11% to 33%. 3 , 4
Compared with CT and MR, contrast‐enhanced ultrasound (CEUS) has the advantages of allowing real‐time scanning and providing dynamic perfusion information with fewer application limitations. 5 , 6 CEUS has been widely used in Europe and Asia, especially in China, where members of the population tend to have lower body mass index values. A meta‐analysis demonstrated that compared with CT and MR, CEUS has an equivalent diagnostic sensitivity (Se) (87% vs 86% and 75%, respectively) and specificity (Sp) (91% vs 88% and 82%, respectively). 7 The main controversy regarding CEUS is its poor generalizability in the reading of real‐time videos between different readers. Regarding the differentiation of diagnoses of hepatocellular carcinoma from target FLLs, the Se of CEUS varied from 84% to 95%, and the Sp varied from 25% to 77% among different radiologists at a single center. 8 However, at different centers, the Se varied from 52% to 98%, and the Sp varied from 71% to 100%. 7 To date, no solution to this critical issue has been proposed.
Artificial intelligence (AI) in medicine has been widely explored. Specifically, deep learning has been reported to achieve excellent performance on images of breast cancer, 9 pulmonary diseases, 10 , 11 diabetic retinopathy, 12 , 13 and dermatoma, 14 , 15 even outperforming human experts. 16 , 17 Deep learning models can learn the most predictive features directly from raw image pixels and avoid the subjective feature engineering required in conventional machine learning, 18 making them independent of prior human knowledge and capable of a high degree of fault tolerance. 19 Hwang et al. 20 reported a deep‐learning‐based algorithm that could detect major thoracic diseases from chest radiographs, and good validation results were achieved across five external test datasets with an area under the curve (AUC) of 0.973–1.000. These findings indicate that deep learning offers inherently good generalizability across different radiologists at different centers; thus, this methodology has the potential to overcome the disadvantage of the poor generalizability of CEUS.
Previous studies applied machine learning algorithms to CT/MRI/CEUS images to characterize FLL, 21 , 22 , 23 , 24 but none of these algorithms were targeted at reducing imaging interobserver heterogeneity or reported how these algorithms could interact with radiologists and improve diagnosing accuracy (ACC). In this study, we aimed to construct a deep learning model based on CEUS video analysis for the differentiation of benign and malignant FLLs. The performance of AI was compared with that of radiologists with varied experiences. The influence of the AI‐radiologist interaction on performance improvement was assessed, focusing on AI's potential to reduce interobserver heterogeneity.
Methods
Study design and participants
This retrospective study was approved by the ICE for Clinical Research and Animal Trials of the First Affiliated Hospital of Sun Yat‐sen University (No. [2015]106). Informed consent from patients was waived given the retrospective nature of the study. Patients who underwent CEUS examination for FLL characterization met the inclusion criteria. Cases were excluded if they met the following criteria: (i) patients who received pre‐imaging treatment with surgery, trans‐arterial chemoembolization, ablation, systemic chemotherapy, or catheterization; (ii) cases with simple cystic lesions that were not indicated for CEUS examination; (iii) images with greater than 1/3 of the target lesion covered by an acoustic shadow; (iv) cases with missing images of any needed phase; and (v) cases who could not be given a definite diagnosis based on the reference standard. As shown in Table 1, two datasets were collected from the hospital: a development set of 363 patients obtained from January 2014 to May 2015 and a test set of 211 patients obtained from June 2015 to December 2015.
Table 1.
Baseline characteristics of the included datasets
| Data sets | Development set | Testing set | P | |
|---|---|---|---|---|
| Reference standard | Malignant, No. | 281 | 164 | 0.984 |
| Benign, No. | 82 | 47 | ||
| Gender | Male, No. | 273 | 152 | 0.457 |
| Female, No. | 90 | 59 | ||
| Age | Mean ± SD, year | 52.64 ± 13.77 | 54.30 ± 12.58 | 0.151 |
| Lesion size | Mean ± SD, cm | 5.10 ± 3.27 | 4.74 ± 4.05 | 0.245 |
| No. of images | 614,728 (augmented) | 616 | ‐ | |
| Ultrasound devices | Types, No. | 5 | 6 | ‐ |
| CEUS examiners | No. | 10 | 11 | ‐ |
CEUS, contrast‐enhanced ultrasound; No., number; SD, standard deviation.
The reference diagnoses for malignant lesions, such as hepatocellular carcinoma and liver metastasis, were obtained by pathology. For benign lesions, such as hemangiomas and focal nodular hyperplasia, we used typical characteristics on contrast‐enhanced ultrasonography (CEUS) and at least 12 months of follow‐up without progression as standard criteria. For abscesses, the diagnosis was obtained by successful suction of pus or lesion shrinkage after anti‐infection treatment. For other benign and malignant lesion categories, pathology was needed for diagnosis confirmation.
Contrast‐enhanced ultrasonography examination
Contrast‐enhanced ultrasonography systems used were listed in Appendix A. First, the target lesions were detected and assessed by the unenhanced sonography. Second, patients intravenously received a bolus injection of 2.4 mL (up to 3 mL) SonoVue (Bracco) via the antecubital vein followed by 5 mL of 0.9% normal saline solution. Third, CEUS of the largest tumor cross‐section within 6 min was recorded as the arterial, portal venous, and delayed phases at 0–30 s, 31–120 s, and 121–360 s after injection in separate clips with varied time duration. The images and video clips were stored in the Digital Imaging and Communications in Medicine (DICOM) format.
Data preparation
All patients' CEUS examinations, pathological results, and clinical information, which included age, gender, alpha‐fetoprotein, hepatitis, liver cirrhosis, and history of malignancy, were collected from the automatic storage and retrieval system in the hospital. Cases were deidentified before further processing.
The results of the CEUS examinations were stored as plain scans and video clips of enhanced phases in DICOM format. Videos were converted into consecutive frames using the native function of MicroDicom DICOM viewer 2.8.3. Based on the 2012 version of the Guidelines and Good Clinical Practice Recommendations for CEUS in the Liver, 25 plain scans and enhanced frames were extracted from specified time durations of CEUS video clips. In total, 32 (1 unenhanced, 15 arterial, 15 portal, and 1 delayed phase images) or 46 (1 unenhanced, 15 arterial, 15 portal, and 15 delayed phase images) representative frames were manually selected from each case (Appendix B). For the test datasets, four representative frames per case (one from each phase) were randomly selected. The frames were preprocessed into a square image containing the lesion and a perilesional area that was 1–2 cm in diameter. The preprocessed images were saved in an 8‐bit JPEG format. Finally, 14 296 original frames for AI development and 844 frames for testing were included in this study. Gold standard labels for the images of each case were assigned based on the reference diagnosis.
Artificial intelligence development: deep learning model
Network architectures
Network architectures and the flowchart of AI development are presented in Figure 1. Microsoft's residual neural network architecture (ResNet), which is regarded as a 4th‐generation convolutional neural network, was used for deep learning model training (Appendix C). 26 Four 152‐layer ResNet branches on four‐phase images were trained independently while fused by a max‐pooling layer and a fully connected layer to obtain the final output. Given the limited data available for training, we applied a transfer learning algorithm that preserved most parts of the network (152‐layer ResNet) that had already been trained on a large dataset (ImageNet) and retrained the weights of the fully connected layer with random initialization on the target dataset (our training set). 27
Figure 1.
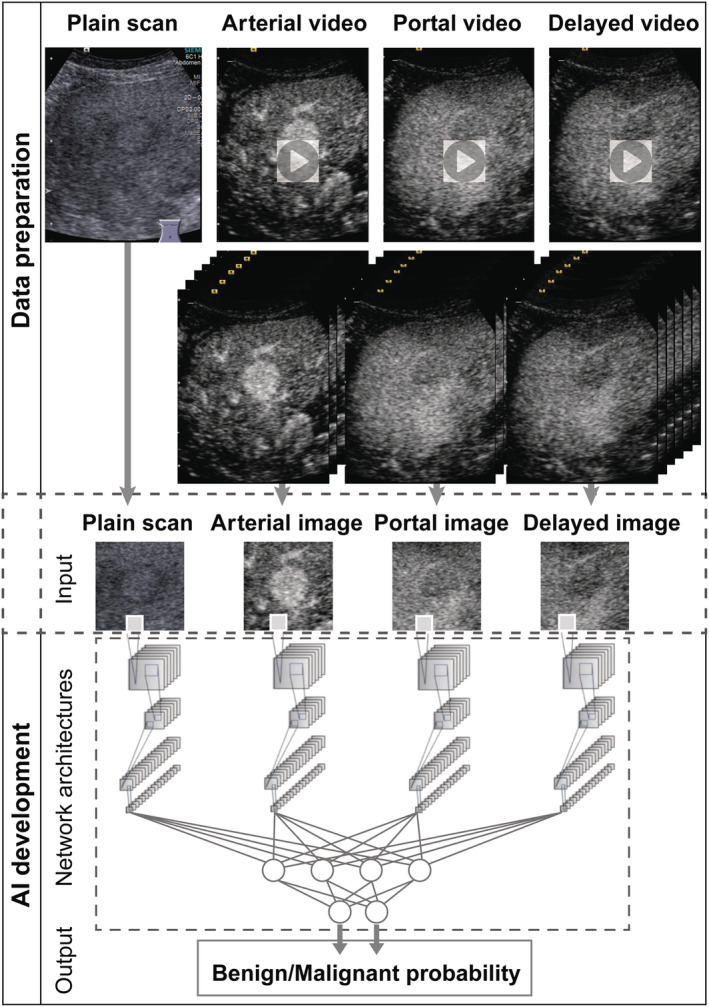
Flowchart of data preparation and AI development. Data preparation consisted of data collection, decomposition of video clips into frames, frame selection, and image cropping into square four‐phase AI inputs. AI development consisted of input, network architectures, and output.
Input and output
The input images were resized to a resolution of 224 × 224 pixels. To improve the model's generalizability, we applied an augmentation procedure to enrich the data diversity 28 ; this augmentation was based on algorithms via brightness changes, contrast adjustment, rotation, parallel shifting, and simple combinations thereof to mimic the data diversity observed in clinical practice (Appendix D). Through augmentation, 43 images (including the original image) were generated from a single image. The augmentation procedure generated 614 728 images for AI training. The four‐phase images were input to the corresponding four branches of the 152‐layer ResNet. The output for each case was the risk probability of malignancy with values ranging from 0 to 1 and the initial diagnosis of benign or malignant.
Training protocol
Training was performed on a workstation with a GeForce GTX 1080 Ti graphics processing unit (NVIDIA), a Core i7‐6700 K (Intel) central processing unit, and 64 GB of random‐access memory. Python 3.5 (https://www.python.org) and the Torch (http://torch.ch) framework for neural networks were used for this purpose. Augmentation was performed using the Python imaging library of Pillow 3.3.1 (https://pypi.python.org/pypi/Pillow/3.3.1). During training, the dataset was randomly divided into a training set (80%) and a tuning set (20%). Detailed training configuration can be found in Appendix E.
Artificial intelligence performance and comparison with radiologists
Performance of the artificial intelligence model versus radiologists
By applying the AI to the test set, each case was evaluated by the same input method as used in the training process, and output was presented as a risk probability of malignancy for each case and the corresponding diagnosis of benign or malignant. For radiologists reading CEUS, the diagnosis was referenced to the Guidelines and Good Clinical Practice Recommendations for CEUS in the Liver (Update 2012). 29 For lesions in the noncirrhotic liver, those with arterial hyper‐enhancement and late hypo‐enhancement tend to be malignant. Otherwise, lesions tend to be benign. For lesions in the cirrhotic liver, sustained hyper‐ or iso‐arterial and late enhancement indicate benign features; otherwise, lesions are considered malignant. Clinical information, such as medical history and blood test, aided in diagnosing.
Four radiologists (two residents and two experts with 2, 3, 6, and 8 years of experience with hepatic CEUS, separately) who were blinded to the final diagnoses and did not participate in the data preparation work reviewed the cases in random order. The radiologists independently reviewed the CEUS videos along with the patients' clinical information. The performance was evaluated in terms ACC, and the diagnostic tests were assessed based on Se, Sp, positive predictive value (PPV), and negative predictive value (NPV).
Performance of radiologists alone versus radiologists with artificial intelligence assistance
By comparing the performance of the AI with that of the radiologists, an AI assistance strategy was developed based on AI's advantage in the diagnostic PPV or NPV, which suggested a more reliable diagnosis of malignancy or benignity. After an additional 1‐month interval, the radiologists reviewed the CEUS cases again with AI assistance. By assistance, the AI results provided a strong reference in cases of conflict with the radiologists' diagnoses, and the radiologists made the final decision of whether to modify the diagnosis or adhere to the initial diagnosis. Comparisons were drawn between the radiologists alone and the AI‐assisted radiologist performance.
Statistical methods
The performances of the radiologists and AI were mainly evaluated in terms of the AUC, ACC, Se, Sp, PPV, NPV, and error rates. R software (version 3.4.1; https://www.r‐project.org) was used for statistical analysis. Results with two‐sided P‐values of less than 0.05 were considered to indicate a statistically significant difference. Detailed statistical methods can be found in Appendix F.
Results
Performance of the artificial intelligence model versus radiologists
On the test set, the AI achieved an AUC of 0.934 (95% CI 0.890–0.978) and an ACC of 91.0% (95% CI 87.1–94.9%). Radiologists had an ACC varied from 82.0% to 86.7% (P = 0.116) (Table 2, Fig. 2a). In particular, the residents achieved similar Se compared with the experts (88.4–89.6% vs 88.4–90.2%, P = 0.380) but showed a deficiency in Sp (59.6–63.8% vs 72.3–80.9%, P = 0.034) (Fig. 2b).
Table 2.
Detailed performance comparison between the AI and the four radiologists on the testing set
| Statistics | ACC | Se | Sp | PPV | NPV |
|---|---|---|---|---|---|
| AI | 0.910 (0.871, 0.949) | 0.927 (0.887, 0.967) | 0.851 (0.749, 0.953) | 0.956 (0.924, 0.988) | 0.769 (0.655, 0.884) |
| Expert1 | 0.867 (0.822, 0.913) | 0.884 (0.835, 0.933) | 0.809 (0.696, 0.921) | 0.942 (0.905, 0.979) | 0.667 (0.544, 0.789) |
| Expert2 | 0.863 (0.816, 0.909) | 0.902 (0.857, 0.948) | 0.723 (0.596, 0.851) | 0.919 (0.877, 0.961) | 0.680 (0.551, 0.809) |
| Resident1 | 0.839 (0.789, 0.888) | 0.896 (0.850, 0.943) | 0.638 (0.501, 0.776) | 0.896 (0.850, 0.943) | 0.638 (0.501, 0.776) |
| Resident2 | 0.820 (0.768, 0.872) | 0.884 (0.835, 0.933) | 0.596 (0.455, 0.736) | 0.884 (0.835, 0.933) | 0.596 (0.455, 0.736) |
| P (AI vs Experts) | 0.256 | 0.419 | 0.297 | 0.385 | 0.453 |
| P (AI vs Residents) | 0.021 * | 0.406 | 0.016 * | 0.052 | 0.157 |
ACC, accuracy; Se, sensitivity; Sp, specificity. Bold fonts indicate the best performance per column.
Statistically significant (P < 0.05).
Figure 2.
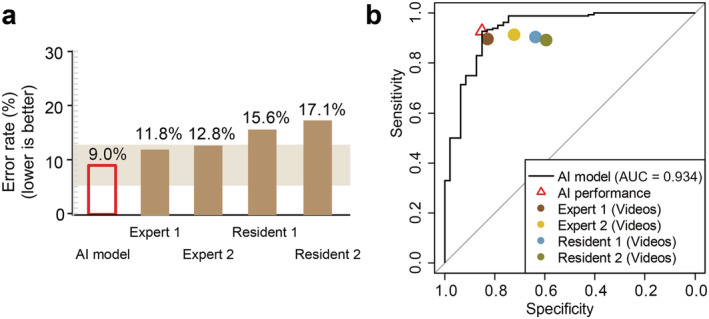
Performance comparison between AI and radiologists. (a) Error rate (1‐accuracy) comparison between AI and radiologists. (b) Detailed comparison of diagnostic sensitivity and specificity between AI and the radiologists.
By comparison, AI outperformed residents (AUC: 82.9–84.4%, P = 0.038; ACC: 91.0% vs 86.3–86.7%, P = 0.256) and matched experts (AUC: 87.2–88.2%, P = 0.438; ACC: 91.0% vs 82.0–83.9%, P = 0.021). Specifically, AI achieved a higher PPV than the residents (95.6% vs 88.4–89.6%, P = 0.052) but comparable with experts (95.6% vs 91.9–94.2%, P = 0.385). NPV of the AI was higher than all four radiologists but not significantly (76.9% vs 59.6–68.0%, P = 0.157–0.453). This indicated that AI is more reliable diagnosis of malignancy than benignity (Table 2, Fig. 2).
Performance of radiologists alone versus radiologists with artificial intelligence assistance
The higher diagnostic PPV of AI suggested a more reliable diagnosis of malignancy. The AI strategy was defined to improve the true malignant rate, especially for residents. When a radiologist made a diagnosis that conflicted with AI's malignant prediction, a strong recommendation to modify his or her diagnosis was suggested. While when the radiologist's diagnosis conflicted with AI's benign prediction, the suggestion for diagnosis modification was general.
Compared with radiologists alone, radiologists with AI assistance achieved 7.4–11.0% (P < 0.001–0.015) improved sensitivity for both residents and experts, 21.1–37.3% (P = 0.001–0.031) NPV improvement and 5.1–9.9% (P = 0.004–0.080) improved accuracy. Expert 1 experienced a 4.3% reduced Sp (P = 0.801) and 0.7% decreased PPV (P = 0.998) (Table 3, Fig. 3). With AI assistance, interobserver performance between residents and experts was comparable based on ACC (91.0–92.9%, P = 0.904), Se (97.0–99.4%, P = 0.360), Sp (66.0–76.6%, P = 0.671), PPV (91.1–93.5%, P = 0.818), and NPV (86.8–96.9%, P = 0.460) (Table 4, Fig. 3).
Table 3.
Performance comparison of the four radiologists between radiologist‐alone and AI assisted radiologists on the testing set
| Statistics | ACC | Se | Sp | PPV | NPV | |
|---|---|---|---|---|---|---|
| Expert 1 | Alone/AI assisted | 0.867/0.924 | 0.884/0.970 | 0.809/0.766 | 0.942/0.935 | 0.667/0.878 |
| P | 0.080 | 0.006 * | 0.801 | 0.998 | 0.031 * | |
| Expert 2 | Alone/AI assisted | 0.863/0.929 | 0.902/0.982 | 0.723/0.745 | 0.919/0.931 | 0.680/0.921 |
| P | 0.038 * | 0.005 * | 1.000 | 0.852 | 0.014 * | |
| Resident 1 | Alone/AI assisted | 0.839/0.910 | 0.896/0.970 | 0.638/0.702 | 0.896/0.919 | 0.638/0.868 |
| P | 0.040 * | 0.015 * | 0.661 | 0.594 | 0.031 * | |
| Resident 2 | Alone/AI assisted | 0.820/0.919 | 0.884/0.994 | 0.596/0.660 | 0.884/0.911 | 0.596/0.969 |
| P | ‐ | 0.004 * | <0.001 * | 0.670 | 0.528 | 0.001 * |
ACC, accuracy; Se, sensitivity; Sp, specificity. Bold fonts indicate the best performance per column.
Statistically significant (P < 0.05).
Figure 3.
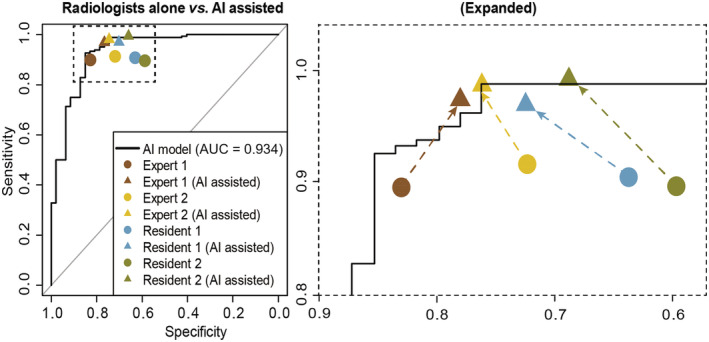
Performance validation of the strategy of AI assistance in the testing dataset. Performance comparison between radiologists with AI assistance and radiologists alone.
Table 4.
Performance comparison between the four radiologists with AI assistance on the testing set
| Statistics | ACC | Se | Sp | PPV | NPV |
|---|---|---|---|---|---|
| Expert1 | 0.924 (0.888, 0.960) | 0.970 (0.943, 0.996) | 0.766 (0.645, 0.887) | 0.935 (0.898, 0.972) | 0.878 (0.778, 0.978) |
| Expert2 | 0.929 (0.894, 0.964) | 0.982 (0.961, 1.000) | 0.745 (0.620, 0.869) | 0.931 (0.893, 0.968) | 0.921 (0.835, 1.000) |
| Resident1 | 0.910 (0.871, 0.949) | 0.970 (0.943, 0.996) | 0.702 (0.571, 0.833) | 0.919 (0.878, 0.960) | 0.868 (0.761, 0.976) |
| Resident2 | 0.919 (0.883, 0.956) | 0.994 (0.982, 1.000) | 0.660 (0.524, 0.795) | 0.911 (0.869, 0.952) | 0.969 (0.908, 1.000) |
| P | 0.904 | 0.360 | 0.671 | 0.818 | 0.460 |
ACC, accuracy; Se, sensitivity; Sp, specificity. Bold fonts indicate the best performance per column.
Discussion
In this study, we constructed a CEUS‐based AI for FLL differentiation between benignity and malignancy, which significantly outperformed resident radiologists and matched the performance of experts who had access to complementary clinical information on patients. Considering the advantage of AI's high diagnostic PPV compared with radiologists, the strategy of AI assistance was developed to improve their true malignancy rate. For the independent testing set, radiologists with AI assistance exhibited improved performance especially for residents who reached the expert level; thus, interobserver heterogeneity was reduced.
Contrast‐enhanced ultrasound is complementary to and even substitutable for CT and MR in the characterization of FLLs, and the main advantages include the increased temporal resolution of CEUS videos and their ability to show detailed blood perfusion morphology. CEUS videos provide time‐sequence information on dynamic blood perfusion, enabling the differentiation of focal nodular hyperplasia from atypical hepatocellular carcinoma. 8 In addition to these visible features, potential pixel‐based “features” of time‐sequence information may be recognizable with the aid of deep learning techniques. By applying multiphase video‐based images and a deep neural network for model development, the advantages of CEUS could be optimally exploited. Our model achieved a tested AUC of 93.4% and ACC of 91.0%. Compared with models trained on single‐frame images, our model outperformed or matched the previously reported performances of AI‐CT (ACC: 82–90%), 24 and AI‐MRI (ACC: 88.0–91.9%). 30 , 31 For AI‐US, our study reported the largest sample size with an independent test dataset. 32 , 33 , 34 Compared with a previous AI‐US study with an independent test dataset, 35 our AI model exhibited better performance (AUC: 93.4% vs 88.1%).
For multiphase imaging analysis, an architecture based on multiple ResNet branches was designed. ResNet was the first network architecture to outperform human experts in the ImageNet Large Scale Visual Recognition Challenge. Its pixel‐based convolution and backpropagation design for automatic weight optimization make it powerful in recognizing the distinguishing features of different categories. 26 An ACC of 96.4% has been achieved with the use of ResNet in colonoscopy video analysis for polyp detection in a study by Urban et al. 36 Our ResNet‐based and video‐based AI model achieved an Se of 92.7% and an Sp of 85.1% for FLL differentiation on the test dataset. Its performance was comparable with or even better than the previously reported performance of non‐AI CT (Se: 89%, Sp: 94%) and MR (Se: 83%, Sp: 75%). 7 A CEUS‐based AI model was also reported in a recent study of FLL differentiation 34 ; however, that study used machine learning algorithms based on manually extracted features for model development. That study reported an Se of 83.3% and an Sp of 62.7%, and these values are lower than those obtained with our deep learning model.
For the application of AI in clinical practice, the authority of decision‐making should remain under radiologists' supervision. In this study, we proposed a man–AI interaction strategy for FLL diagnosis, which improved residents' performance and reduced interobserver heterogeneity associated with CEUS. Because they lack clinical experience, residents can be less confident in their diagnoses, especially for benign lesions in high‐risk liver background, leading to a low Sp. In this study, our residents achieved similar Se results but significantly lower Sp results compared with experts (59.6–63.8% vs 72.3–80.9%, P = 0.034). By contrast, the AI had a similar PPV and Sp compared with the best‐performing expert and outperformed residents. Therefore, the strategy of AI assistance was designed to compensate for the residents' deficiency in Sp by referring to a more reliable diagnosis of malignancy. In the test procedure, radiologists were informed of AI's high confidence in malignancy diagnoses (comparable PPV with experts) and low confidence of benignity diagnoses (no better NPV than residents). This information gave the radiologists evidence for their choice, as they can modify their diagnosis or not when it conflicts with the diagnosis provided by AI. In the previously reported studies, this specific man–AI interaction strategy was always missed. 33 , 37 As shown in the testing dataset, our strategy was proven to be effective. This AI system may also be helpful in radiology resident training programs and radiologist training at less developed centers.
This study has several limitations. First, we used only image data for AI training, thus neglecting potentially important information, such as patients' clinical information related to alpha‐fetoprotein, hepatitis, and liver cirrhosis. Although this limitation can be compensated by the intended purpose of the AI, that is, to provide assistance for radiologists, a comprehensive AI model integrating CEUS and complementary patient information could enable a further breakthrough in FLL differentiation. Second, although our study reported the largest cohort to date compared with previous studies on CEUS, the sample size was still small considering the deep learning nature of this study. Although transfer learning allows the development of an accurate model with a relatively small training dataset, the model performance will still be inferior to that of a model trained from a random initialization on an extremely large dataset. 12 Future studies using a much larger dataset that ideally includes data from multiple centers for training may further improve the AI model's performance.
Conclusion
In summary, we developed CEUS‐based AI for differentiating between benign and malignant FLLs, which outperformed our radiologists. Consequently, a clinically applicable strategy of AI assistance was developed, which improved the performance of residents to the expert level and thus reduced interobserver heterogeneity associated with CEUS.
Acknowledgments
We thank employees of General Electric China Technology Center Xin Li for his statistical work of this study and Ting‐Fan Wu, Ling Chen, and Xin‐Rong Wang for their helpful and valuable statistical assistance. We thank the American Journal Experts for their language editing service (Certificate Verification Key: C84B‐2306‐027C‐D842‐FD1P).
Appendix A. CEUS systems used in the study
All CEUS examinations were performed by radiologists with at least 4 years of experience in hepatic CEUS, using an Acuson Sequoia 512 scanner (Siemens Medical Solutions, Mountain View, CA, USA) equipped with a 4V1 vector transducer (frequency range 1.0–4.0 MHz) with contrast pulse sequencing (CPS, Mechanic Index from 0.15 to 0.21) or an Aplio 500 or Aplio XV (Toshiba Medical Systems, Tokyo, Japan) scanner equipped with a 375BT convex transducer (frequency range 1.9–6.0 MHz) with contrast harmonic imaging (CHI, Mechanic Index from 0.05 to 0.10).
Appendix B. Frame selection criteria
The frames were chosen in accordance with the following criteria: (i) images with at least 1 cm of perilesional hepatic parenchyma; (ii) the lesion should remain close to the same location in the tri‐phase frames; (iii) images without or with less than 1/3 of the target lesion covered by an acoustic shadow.
Appendix C. 152‐layer ResNet architecture
The 152‐layer ResNet architecture is separated into five parts: conv1, conv2_x, conv3_x, conv4_x, and conv5_x. Not including conv1, the remaining parts consist of 3, 8, 36, and 3 building blocks, respectively, with three convolutional layers (1 × 1, 3 × 3, and 1 × 1) in each block. With the 1 layer (7 × 7) in conv1 and the fully connected layer, there are a total of 152 layers in the network.
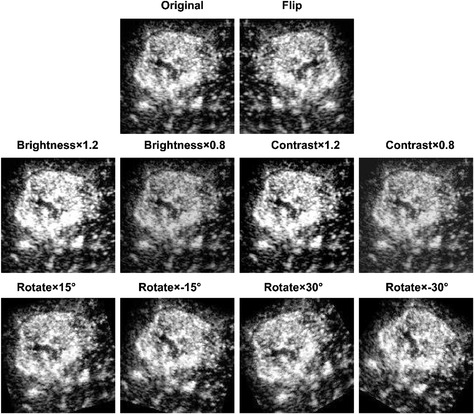
Appendix D. Examples of image augmentation
The augmentation was based on algorithms via brightness changes (brighter by 1.2‐fold or darker by 0.2), contrast adjustment (1.2‐fold increased or 0.2‐fold reduced), rotation (by −30, −15, 15, and 30 degrees), parallel shifting (horizontally), and simple combinations thereof to mimic the data diversity observed in clinical practice.
Appendix E. Detailed training configuration
The images of the training set were fed into the network. The output probability of the malignancy or benignity of each image was automatically compared with the reference label, and the error between them was backpropagated for weight optimization. The weights in the fully connected layer were further updated by applying the network to the test set. Ten epochs (iterations through the entire dataset), a constant learning rate of 0.001, batch normalization [1], minibatch gradient descent, and a γ value of 0.1 were applied. Early termination of the training process was applied when no further improvement in loss and accuracy on the tuning set was achieved in at least 500 iterations.
Appendix F. Detailed statistical methods
ROC curves were plotted using the “pROC” package by plotting the Se against the Sp for a varying predicted probability threshold, and the AUC values were calculated accordingly. For the calculation of the ACC, Se, Sp, PPV, and NPV metrics, the “confusion matrix” function of the “caret” package was used, and the error rate was calculated as 1‐ACC. For the comparison of the statistical metrics among the AI, the radiologists, and the radiologists with AI assistance, Wilcoxon rank test was applied for AUC comparison, and χ 2 test was applied for ACC, Se, Sp, PPV, and NPV.
Hu, H.‐T. , Wang, W. , Chen, L.‐D. , Ruan, S.‐M. , Chen, S.‐L. , Li, X. , Lu, M.‐D. , Xie, X.‐Y. , and Kuang, M. (2021) Artificial intelligence assists identifying malignant versus benign liver lesions using contrast‐enhanced ultrasound. Journal of Gastroenterology and Hepatology, 36: 2875–2883. 10.1111/jgh.15522.
Declaration of conflict of interest: The funding source had no involvement in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript. The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Financial support: This work is supported by the National Natural Science Foundation of China (no. 81971630 and 81701701).
Contributor Information
Xiao‐Yan Xie, Email: xiexyan@mail.sysu.edu.cn.
Ming Kuang, Email: kuangm@mail.sysu.edu.cn.
References
- 1. Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet (London, England) 2018; 391: 1301–1314. [DOI] [PubMed] [Google Scholar]
- 2. Rizvi S, Gores GJ. Pathogenesis, diagnosis, and management of cholangiocarcinoma. Gastroenterology 2013; 145: 1215–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sangiovanni A, Manini MA, Iavarone M et al. The diagnostic and economic impact of contrast imaging techniques in the diagnosis of small hepatocellular carcinoma in cirrhosis. Gut 2010; 59: 638–644. [DOI] [PubMed] [Google Scholar]
- 4. Roberts LR, Sirlin CB, Zaiem F et al. Imaging for the diagnosis of hepatocellular carcinoma: a systematic review and meta‐analysis. Hepatology (Baltimore, Md) 2018; 67: 401–421. [DOI] [PubMed] [Google Scholar]
- 5. Wang W, Chen LD, Lu MD et al. Contrast‐enhanced ultrasound features of histologically proven focal nodular hyperplasia: diagnostic performance compared with contrast‐enhanced CT. Eur. Radiol. 2013; 23: 2546–2554. [DOI] [PubMed] [Google Scholar]
- 6. Xie XH, Xu HX, Xie XY et al. Differential diagnosis between benign and malignant gallbladder diseases with real‐time contrast‐enhanced ultrasound. Eur. Radiol. 2010; 20: 239–248. [DOI] [PubMed] [Google Scholar]
- 7. Chou R, Cuevas C, Fu R et al. Imaging techniques for the diagnosis of hepatocellular carcinoma: a systematic review and meta‐analysis. Ann. Intern. Med. 2015; 162: 697–711. [DOI] [PubMed] [Google Scholar]
- 8. Li W, Wang W, Liu GJ et al. Differentiation of atypical hepatocellular carcinoma from focal nodular hyperplasia: diagnostic performance of contrast‐enhanced us and microflow imaging. Radiology 2015; 275: 870–879. [DOI] [PubMed] [Google Scholar]
- 9. Antropova N, Huynh BQ, Giger ML. A deep feature fusion methodology for breast cancer diagnosis demonstrated on three imaging modality datasets. Med. Phys. 2017; 44: 5162–5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lakhani P, Sundaram B. Deep learning at chest radiography: automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology 2017; 284: 574–582. [DOI] [PubMed] [Google Scholar]
- 11. Coudray N, Ocampo PS, Sakellaropoulos T et al. Classification and mutation prediction from non‐small cell lung cancer histopathology images using deep learning. Nat. Med. 2018; 24: 1559–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kermany DS, Goldbaum M, Cai W et al. Identifying medical diagnoses and treatable diseases by image‐based deep learning. Cell 2018; 172: 1122–1131 e9. [DOI] [PubMed] [Google Scholar]
- 13. Gulshan V, Peng L, Coram M et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 2016; 316: 2402–2410. [DOI] [PubMed] [Google Scholar]
- 14. Han SS, Kim MS, Lim W, Park GH, Park I, Chang SE. Classification of the clinical images for benign and malignant cutaneous tumors using a deep learning algorithm. J. Invest. Dermatol. 2018; 138: 1529–1538. [DOI] [PubMed] [Google Scholar]
- 15. Esteva A, Kuprel B, Novoa RA et al. Dermatologist‐level classification of skin cancer with deep neural networks. Nature 2017; 542: 115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liang H, Tsui BY, Ni H et al. Evaluation and accurate diagnoses of pediatric diseases using artificial intelligence. Nat. Med. 2019; 25: 433–438. [DOI] [PubMed] [Google Scholar]
- 17. Haenssle HA, Fink C, Schneiderbauer R et al. Man against machine: diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Ann. Oncol. 2018; 29: 1836–1842. [DOI] [PubMed] [Google Scholar]
- 18. LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015; 521: 436–444. [DOI] [PubMed] [Google Scholar]
- 19. Litjens G, Kooi T, Bejnordi B et al. A survey on deep learning in medical image analysis. Med. Image Anal. 2017; 42: 60–88. [DOI] [PubMed] [Google Scholar]
- 20. Hannun AY, Rajpurkar P, Haghpanahi M et al. Cardiologist‐level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat. Med. 2019; 25: 65–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gatos I, Tsantis S, Karamesini M et al. Focal liver lesions segmentation and classification in nonenhanced T2‐weighted MRI. Med. Phys. 2017; 44: 3695–3705. [DOI] [PubMed] [Google Scholar]
- 22. Gatos I, Tsantis S, Spiliopoulos S et al. A new automated quantification algorithm for the detection and evaluation of focal liver lesions with contrast‐enhanced ultrasound. Med. Phys. 2015; 42: 3948–3959. [DOI] [PubMed] [Google Scholar]
- 23. Guo LH, Wang D, Qian YY et al. A two‐stage multi‐view learning framework based computer‐aided diagnosis of liver tumors with contrast enhanced ultrasound images. Eur. Radiol. 2018; 69: 343–354. [DOI] [PubMed] [Google Scholar]
- 24. Yasaka K, Akai H, Abe O, Kiryu S. Deep learning with convolutional neural network for differentiation of liver masses at dynamic contrast‐enhanced CT: a preliminary study. Radiology 2018; 286: 887–896. [DOI] [PubMed] [Google Scholar]
- 25. Nam JG, Park S, Hwang EJ et al. Development and validation of deep learning‐based automatic detection algorithm for malignant pulmonary nodules on chest radiographs. Radiology 2018; 290: 218–228. [DOI] [PubMed] [Google Scholar]
- 26. He K, Zhang X, Ren S, Sun J. Delving deep into rectifiers: surpassing human‐level performance on ImageNet classification. In: IEEE International Conference on Computer Vision (ICCV). Santiago, Chile, 2015; 1026–1034. [Google Scholar]
- 27. Meyer A, Zverinski D, Pfahringer B et al. Machine learning for real‐time prediction of complications in critical care: a retrospective study. Lancet Respir. Med. 2018; 6: 905–914. [DOI] [PubMed] [Google Scholar]
- 28. Ramachandram D, Taylor GW. Deep multimodal learning: a survey on recent advances and trends. IEEE Signal Proc. Mag. 2017; 34: 96–108. [Google Scholar]
- 29. Claudon M, Dietrich CF, Choi BI et al. Guidelines and good clinical practice recommendations for contrast enhanced ultrasound (CEUS) in the liver—update 2012: a WFUMB‐EFSUMB initiative in cooperation with representatives of AFSUMB, AIUM, ASUM, FLAUS and ICUS. Ultraschall in der Medizin 2013; 34: 11–29. [DOI] [PubMed] [Google Scholar]
- 30. Hamm CA, Wang CJ, Savic LJ et al. Deep learning for liver tumor diagnosis part I: development of a convolutional neural network classifier for multi‐phasic MRI. Eur. Radiol. 2019; 29: 3338–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang CJ, Hamm CA, Savic LJ et al. Deep learning for liver tumor diagnosis part II: convolutional neural network interpretation using radiologic imaging features. Eur. Radiol. 2019; 29: 3348–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hwang YN, Lee JH, Kim GY, Jiang YY, Kim SM. Classification of focal liver lesions on ultrasound images by extracting hybrid textural features and using an artificial neural network. Biomed. Mater. Eng. 2015; 26: S1599–S1611. [DOI] [PubMed] [Google Scholar]
- 33. Moga TV, Popescu A, Sporea I et al. Is contrast enhanced ultrasonography a useful tool in a beginner's hand? How much can a computer assisted diagnosis prototype help in characterizing the malignancy of focal liver lesions? Med. Ultrason. 2017; 19: 252–258. [DOI] [PubMed] [Google Scholar]
- 34. Ta CN, Kono Y, Eghtedari M et al. Focal liver lesions: computer‐aided diagnosis by using contrast‐enhanced US cine recordings. Radiology 2018; 286: 1062–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schmauch B, Herent P, Jehanno P et al. Diagnosis of focal liver lesions from ultrasound using deep learning. Diagn. Interv. Imaging 2019; 100: 227–233. [DOI] [PubMed] [Google Scholar]
- 36. Urban G, Tripathi P, Alkayali T et al. Deep learning localizes and identifies polyps in real time with 96% accuracy in screening colonoscopy. Gastroenterology 2018; 155: 1069–78.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sim Y, Chung MJ, Kotter E et al. Deep convolutional neural network‐based software improves radiologist detection of malignant lung nodules on chest radiographs. Radiology 2020; 294: 199–209. [DOI] [PubMed] [Google Scholar]


