Abstract
Aims
In‐vitro/In‐vivo evaluation of cholesterol‐lowering probiotic strain Lactobacillus paracasei DTA81 and the possible connection with the gut microbiota modulation.
Methods and Results
In the present study, strain DTA81 has been evaluated for the possible influence on blood lipid and glucose concentrations, modulation of the immune system, gastrointestinal survivability and modulation of gut microbiota in BALB/c mice receiving a high‐fat diet. After 6 weeks of treatment, a significant reduction of total cholesterol and fasting blood sugar (FBS) among animals treated with L. paracasei DTA81 has been recorded. Comparison of colon tissue levels of different cytokines revealed a significant reduction of the inflammatory cytokine interleukin‐6. The comparison of gut microbiota using the 16S rRNA approach indicated that the treatment with L. paracasei DTA81 significantly increased the taxa Bacteroidetes and Coprococcus. Moreover, the genome of DTA81 was sequenced for the in‐silico assessment, and the analysis indicated the presence of cholesterol assimilation‐related genes as well as the absence of negative traits such as transmissible antibiotic resistance genes, plasmids and prophage regions.
Conclusion
The outcome of this study revealed the in‐vitro and in‐vivo properties of L. paracasei DTA81 and the possible mechanism between consumption of this strain, the abundance of Bacteriodetes/Coprococcus taxa, immunomodulatory activity and the subsequent reduction of cholesterol/FBS in BALB/c mice.
Significance and Impact of the Study
Lactobacillus paracasei DTA81 as a non‐pharmacological potential probiotic supplement can influence metabolic homeostasis in individuals, particularly those adopting high‐fat diets, and it can contribute to reduce coronary heart disease.
Keywords: 16S rRNA, genome sequencing, gut microbiota, immunomodulatory effect, probiotic, total cholesterol‐lowering
Introduction
According to the last definition by the FAO/WHO (Food and Agriculture Organization/World Health Organization), probiotics are ‘live microorganisms which when administered in adequate amounts confer a health benefit on the host’ (Hill et al. 2014). Probiotic foods represent a relevant portion of functional foods available on the market worldwide, projected to reach a value of US $ 46·55 billion by 2020 (Singh et al. 2018). During the past years, the probiotic potential of many lactic acid bacteria has been studied since they are generally recognized as safe micro‐organisms and can, therefore, be safely used in food preparations (Tarrah et al. 2020a, 2020b). Probiotic consumption can benefit human health by modulating the immune system, affecting the gut microbial composition and by producing antimicrobial substances that can contribute to the reduction of deleterious bacteria and promote stability of beneficial microbes (Magnusson and Schnürer 2001; Baker et al. 2009; Amund 2016). Many studies revealed the influence of gut microbiota on metabolic disorders and obesity in humans (Crovesy et al. 2020; Wang et al. 2020). A recent study indicated that energy homeostasis and metabolism of the human can be directly influenced by the gut microbiota (Cani et al. 2019). Indeed, new studies report that transferring the gut microbiota from obese to germ‐free mice can result in a higher weight increase in comparison to the transfer of gut microbiota from a lean mouse (Rosenbaum et al. 2015; Cani et al. 2019). Nowadays, hypercholesterolaemia is reported to be a common human disorder, which is mostly related to cardiovascular disease (CVD) and coronary heart disease (CHD) (Dunn‐Emke et al. 2001). Many in‐vitro and in‐vivo studies recently reported that probiotics such as some Lactobacillus and Bifidobacterium can have beneficial effects on serum lipid profiles (He et al. 2017; Mo et al. 2019). Probiotics can also reduce blood cholesterol in different ways, by utilizing prebiotics to produce short‐chain fatty acids (SCFAs) in the human gut that can further inhibit hepatic cholesterol synthesis and will result in a reduction of blood lipids or by assimilating cholesterol directly and reduce its presence in the human gut (Pereira and Gibson 2002). Therefore, several probiotic bacteria have been proposed and used as food supplements to reduce the rate of hypercholesterolaemia in humans (Marchesi et al. 2016). On the other side, it has been proven that probiotic strains can modulate the human immune system in different ways. The expression of cytokines in the human body has been the most frequent approach to describe the immunomodulatory effect of probiotics. In Wang et al.'s (2015) study, treated people with Bifidobacterium bifidum, Bifidobacterium catenulatum, Bifidobacterium longum and Lactobacillus plantarum showed a decrease in serum levels of pro‐inflammatory cytokines TNF‐α, IL‐5 and IL‐6, while levels of serum of IL‐10 significantly increased. In a previous in‐vitro study (Tarrah et al. 2019), we have characterized the probiotic potential of strain Lactobacillus paracasei DTA81 which was found to possess interesting properties. In particular, DTA81 revealed a strong adherence ability to human cell lines. Therefore, this strain was used in this study to be further investigated regarding its immunomodulatory effects, metabolic alteration and possible gut microbiota modulation effect, using in‐vitro and in‐vivo approaches.
Materials and methods
Bacterial strain and growth conditions
Lactobacillus paracasei DTA81 (Guerra et al. 2018) was routinely grown using MRS medium (Sigma, MO) at 37°C for 24 h. For in‐vivo assays, overnight cultures were centrifuged at 5000 g for 5 min, washed two times with sterile PBS (NaCl 8·0 g l−1, KCl 0·2 g l−1, Na2HPO4 1·44 g l−1, KH2PO4 0·24 g l−1, pH 7·4) and resuspended in skim milk (10%) to a final concentration of about 1010 CFU per ml.
Measurement of cholesterol assimilation by L. paracasei DTA81
Initially, 1% overnight culture was incubated in MRS broth (Sigma) containing 0·30% ox gall bile salt (Sigma) and 100 µg ml−1 filter‐sterilized cholesterol (Cholesterol–methyl‐β‐cyclodextrin, Sigma) for 24 h at 37°C. Tubes were then centrifuged at 5500 g for 15 min at 4°C, and 1 ml of supernatant was collected for measurement of residual cholesterol using a colorimetric method (Miremadi et al. 2014). Cholesterol concentration was measured using a standard curve from 0 to 100 µg ml−1. The experiment was repeated two times with three replicates each. The ability of L. paracasei DTA81 to assimilate cholesterol was calculated as a percentage of cholesterol removal after 24 h.
Animals
In all, 24 male BALB/c mice (4‐week old) were obtained from the Animal House at the Biological Sciences Center of the Universidade Federal de Viçosa. All mice were housed (four animals per cage) in a controlled environment: temperature 22°C, humidity 55 ± 5%, 12 h light/dark cycle and received food (Nuvilab, São Paulo, Brazil) and sterilized tap water ad libitum except at sampling time when the access to food was restricted. Mice body weight and food consumption were recorded weekly and daily respectively. The animal study design was approved by the animal ethics committee at the Universidade Federal de Viçosa (CEUA/UFV, protocol nº 15/2020) and it was in accordance with the National Research Council guide for the care and use of laboratory animals (Clark et al. 1997).
Diets and experimental design
All animals were fed for a week (week 0) with a conventional diet (CD). Then they were randomly divided considering the body weight and GTT (glucose tolerance test) into three experimental groups (8 animals per group). The first one received the CD (CD group), the second one was fed with a high‐fat diet (HFD group) and the third one received HFD + L. paracasei DTA81 (DTA81 group). The composition of CD and HFD diets is reported in Table 1 (Reeves et al. 1993; Zhao et al. 2019).
Table 1.
Composition of basal diets for conventional and high‐fat diet (g/100 g)
| Ingredient | Conventional diet | High‐fat diet |
|---|---|---|
| Corn starch | 46.56 | – |
| Fat (lard) | – | 31.7 |
| Casein | 14 | 25.8 |
| Dextrinized starch | 15.5 | 16.2 |
| Sucrose | 10 | 8.9 |
| Soybean oil | 4 | 3.2 |
| Microfine cellulose | 5 | 6.5 |
| Mineral mix | 3.5 | 1.3 |
| Vitamin mix | 1 | 1.3 |
| L‐cystine | 0.18 | 0.39 |
| Choline bitartrate | 0.25 | 0.3 |
| Potassium citrate | – | 2.1 |
| Calcium phosphate | – | 1.7 |
| Calcium carbonate | – | 0.7 |
| Energy density (kcal g−1) | 3.76 | 5.17 |
The potential probiotic strain was administered daily for 6 weeks (from weeks 1 to 6) via a unique oral administration by gavage of 100 μl, equivalent to approx. 109 CFU dispersed in 10% skim milk. During the same period, the remaining groups received the same amount of skim milk without cells. Then, the animals were anesthetized using ketamine (Imalgène, 200 mg kg−1, Sigma) and Rompun (Xylasine, 20 mg kg−1, Sigma) diluted in NaCl 0·9%, the mice were sacrificed by cervical dislocation, and the blood samples were collected from the retro‐orbital sinus for the biochemical analysis.
Oral glucose tolerance test
The oral glucose tolerance test (OGTT) was performed at the end of week 0 and 6 (end of the experiment), according to Ferrere et al. (2016), with some modifications. A solution of 0·2% d‐glucose was given to each animal via gavage after overnight (12 h) fasting conditions, then blood was collected from the tail after 0, 30, 60 and 120 min. Glucose concentration in the blood serum was measured by putting a blood drop on a Comfort Curve Strips (F. Hoffmann–La Roche, Basel, Switzerland) that was then inserted into an ACCU‐CHEK Advantage Glucometer (Roche, Basel, Switzerland) and the OGT was determined according to Cardoso et al. (2011).
Viable bacteria enumeration after transit through the GIT
The survival of L. paracasei DTA81 after transit through the GIT was evaluated at the end of week 3 and at the end of the study (end of week 6). Three mice from each group (from different cages) were randomly selected and their faeces were collected, weighed resuspended in 10 ml of sterilized PBS and serially diluted using the same solution. Then they were plated on MRS medium supplemented with kanamycin (64 μg ml−1; Sigma) and incubated at 37°C for 48 h. Resistance of L. paracasei DTA81 to kanamycin had been determined in a previous study (Tarrah et al. 2019). After incubation, colony forming units were counted and reported per gram of wet faeces. Besides, five colonies were also randomly taken from plates and investigated by Gram staining, catalase and oxidase tests. The same mice were used for this evaluation at week 3 and at week 6.
Determination of the lipid profile and transaminases
Blood samples collected from sacrificed animals were centrifuged at 700 g for 10 min to obtain the serums and immediately examined for total cholesterol, high‐density lipoprotein (HDL), triglyceride, glutamate‐oxaloacetate transaminase (GOT), and glutamate‐pyruvate transaminase (GPT) using Bioclin kits (Diagnostics, Belo Horizonte, Brazil) and an auto‐analyzer equipment (Analyzer BS‐200; Mindray, Shenzhen, China). Low‐density lipoprotein (LDL) was calculated according to the method of Friedwald et al. (Friedwald 1972).
Immunomodulatory effects on colon tissue
The commercial BD CBA Human Th1/Th2/Th17 Cytokine Kit II (BD Biosciences, San Jose, CA) and a BD FACSVers flow cytometer were used to quantitatively measure interleukin‐2 (IL‐2), interleukin‐4 (IL‐4), interleukin‐6 (IL‐6), interleukin‐10 (IL‐10), interleukin‐17 (IL‐17), tumour necrosis factor (TNF) and interferon‐γ (IFN‐γ) protein levels in mice colon samples. To perform local inflammatory cytokine analysis, colon tissue (100–200 mg) was ground using a tissue homogenizer (IKA WORKS GMBH & CO, Staufen, Germany, model T10 basic), washed in cold phosphate‐buffered saline (pH 7·0) and centrifuged (10 000 g , for 10 min at 4°C). Finally, the supernatant was collected and stored at −80°C for further analysis. The samples were prepared according to the manufacturer’s instruction before being analysed by flow cytometry.
16S rRNA gene amplicon target sequencing
Four mice from each group (from different cages) were randomly chosen at the beginning and at the end of the experiment and their faeces were collected in a separated clean cage for each mouse and the samples were immediately stored at −80°C for further analysis. Later, 0·25 g of collected faeces was weighed and total DNA was extracted using the DNeasy PowerSoil Microbial Kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. The quality and quantity of the extracted DNA were assessed by the Spark 10 M spectrophotometer (Tecan Trading AG, Männedorf, Switzerland). The V3–V5 regions of the 16S rRNA gene were PCR amplified and sequenced using an Illumina MiSeq desktop sequencer (Eurofins Genomics Germany GmbH, Ebersberg, Germany) producing 300 bp paired‐end (PE) reads.
Genome sequencing and genomic analysis of L. paracasei DTA81
Lactobacillus paracasei DTA81 was grown overnight in MRS broth at 37°C for 24 h and genomic DNA was isolated using the DNeasy PowerSoil Microbial Kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. The assessment of isolated DNA quality and quantity was done using Spark 10M spectrophotometer (Tecan Trading AG, Männedorf, Switzerland). Lactobacillus paracasei DTA81 genome was sequenced using the paired‐end sequencing technology with NextSeq500 Illumina sequencer at the Interdepartmental Center for Research on Innovative Biotechnology CRIBI (CRIBI, Padova, Italy).
Bioinformatic analyses
The raw data of 16S rRNA gene sequencing were imported and analysed with the CLC Genomics Workbench software v.12.0.2 (Qiagen, Hilden, Germany) using the Microbial genomics module plugin as described by da Silva Duarte et al. (2020). In summary, quality filtering, operational taxonomic unit (OTU) clustering, taxonomical assignment (Greengenes v13_8 database), alpha‐ and beta‐diversity indices calculation were performed with default parameters. Raw reads were deposited in the Sequence Read Archive (SRA) database (http://www.ncbi.nlm.nih.gov/sra) under the BioProject ‘PRJNA638135’.
The Shannon and Chao 1 indices were compared among the experimental groups (CD, DTA81 and HFD) using ANOVA and Kruskal–Wallis tests, followed by Tukey's and Dunn’s post hoc tests, correspondingly. The Welch’s t test was chosen to identify significant differences inside the same group (intra‐group comparison) between the time‐points ‘t0’ and ‘t1’. Both statistical analyses were conducted using GraphPad Prism software (version 7, GraphPad Software, Inc., San Diego, CA). For beta‐diversity analysis, gut microbial dissimilarities among the groups were calculated by permutational multivariate analysis of variance (PERMANOVA) with 999 permutations using Unweighted and Weighted UniFrac diversity metrics. Principal coordinate analysis (PCoA) was chosen as the ordination method to explore and visualize the data.
After data inspection, data filtering (low count and low variance filters) and normalization (cumulative sum scaling), differential abundance analysis were carried with MicrobiomeAnalyst (Dhariwal et al. 2017) considering three taxonomic levels (phylum, family and genus) by applying the linear discriminant analysis (LDA) effect size (LEfSe) function setting up FDR‐adjusted p value (q value) cutoff of 0·05 and log LDA score of 2·0. Pearson correlation with a complete linkage method was chosen to cluster the samples based on taxon abundance in a heat map graph generated with Heatmapper (Babicki et al. 2016).
Regarding DTA81 genome analysis, de novo assembly of the raw reads was done using Velvet algorithm package, ver. 1.1.04 setting on parameters as (min read length: 15, min average quality of read: 20 and min adapter match: 15) (Larsen et al. 2012). Rapid Annotation using Subsystems Technology (RAST) was also used for gene prediction and annotation (Aziz et al. 2008).
The PATRIC 3.6.3 server (Wattam et al. 2017) was used to construct a graphical genome map/annotation after scaffolding the related contigs using the Medusa web server (Bosi et al. 2015) and L. paracasei ATCC 334 as the reference genome.
The presence of prophage regions was predicted using the PHASTER server (Arndt et al. 2016). The detection of plasmid and transmissible antibiotic resistance genes was assessed using PlasmidFinder 2.0 and ResFinder 3.2 servers, respectively (Zankari et al. 2012; Carattoli et al. 2014).
This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession JAAVWK000000000. The version described in this paper is version JAAVWK010000000.
Statistical analyses
Data were analysed using one‐way analysis of variance (ANOVA). Tukey’s test was used as post hoc analysis by the GraphPad Prism software (version 7, GraphPad Software, Inc., San Diego, CA). In general, results were considered significantly different when P values were lower than 0·05. The number of asterisks is used to indicate the level of confidence of the statistical analyses results: *P < 0·05, **P < 0·01, ***P < 0·001.
Results
In‐vitro cholesterol assimilation
Cholesterol reduction by L. paracasei DTA81 grown in MRS supplemented with 0·3% ox bile was measured in‐vitro. DTA81 was able to reduce 16·16 ± 2·0% of the cholesterol presents in the medium after 24 h.
Weight gain and oral glucose tolerance
At the end of the experimental period (6 weeks), mice treated with DTA81 did not evidence a significant difference in weight gain (Fig. 1a); the weekly monitoring of animal weight (Fig. 1b) showed a ponderal increase of HFD during the last 2 weeks while DTA81 group had the same trend of CD. The influence of DTA81 supplementation on plasma glucose is indicated in Fig. 1c,d. Regarding the fasting blood sugar (FBS), a significant glucose reduction was recorded in the animals treated with L. paracasei DTA81 compared with the CD and HFD groups (P < 0·05) (Fig. 1c). However, after receiving the glucose, no significant difference was recorded regarding the glucose tolerance among the groups, neither at the beginning nor at the end of the experiments (Fig. 1d).
Figure 1.
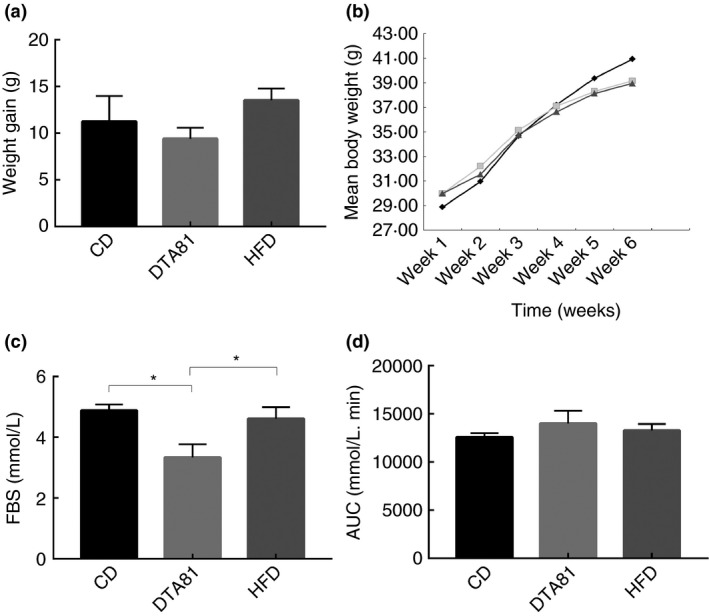
Effect of probiotic consumption on body weight, fasting blood sugar (FBS) and glucose tolerance test. (a) Mean body weight; (b) weight gain at the end of the experiment; (c) fasting blood sugar (FBS); (d) glucose tolerance test. Results are expressed as means ± SEM (n = 8). The number of asterisks is used to indicate the level of confidence of the statistical analyses results: *statistically significant P < 0·05, ( ) HFD; (
) HFD; ( ) DTA81; (
) DTA81; ( ) CD.
) CD.
Determination of the lipid profile and transaminases
Results of mice blood biochemical analyses at the end of the study (week 6) are reported in Fig. 2. Total cholesterol (TC), HDL and LDL evidenced a significant (P < 0·01) reduction in the DTA81 group compared with the HFD group, whereas triglycerides, GOT and GPT did not show significant differences between any group. In addition, a significant difference (P < 0·05) was detected for TC and HDL between groups HFD and CD while no statistically significant difference was seen for LDL.
Figure 2.
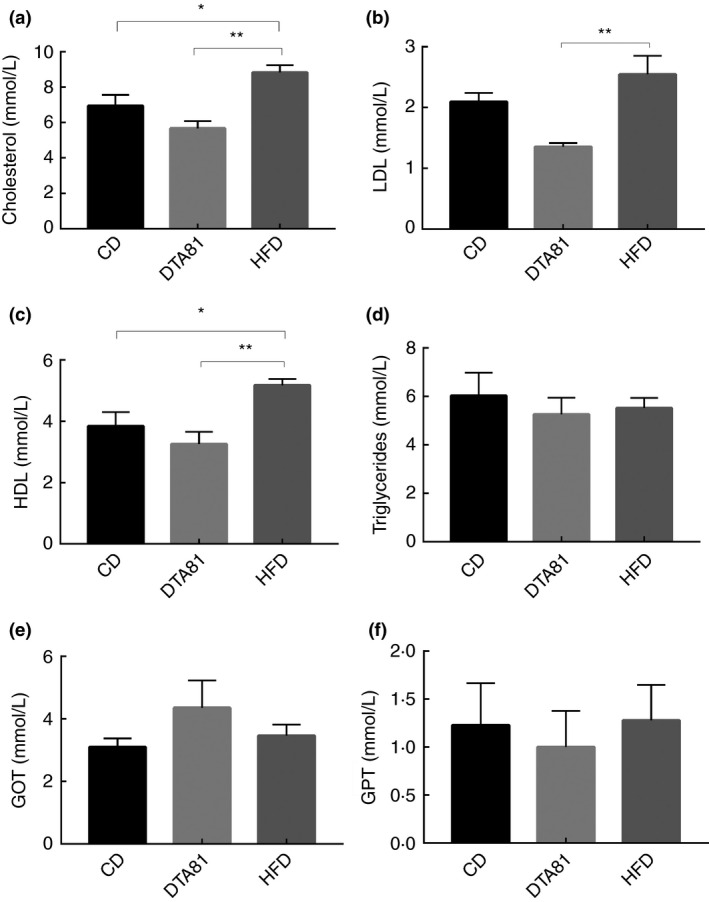
Effect of different treatments on blood parameters. (a) Total cholesterol; (b) high‐density lipoprotein (HDL); (c) low‐density lipoprotein (LDL); (d) triglyceride; (e) glutamate‐oxaloacetate transaminase (GOT); (f) glutamate‐pyruvate transaminase (GPT); Results are expressed as means ± SEM (n = 8). The number of asterisks is used to indicate the level of confidence of the statistical analyses results: *statistically significant P < 0·05, **statistically significant P < 0·01.
Immunomodulatory activity in the colon tissue
Comparison of colon tissue levels of Th1 (IL2, TNF and IFN‐γ), Th2 (IL4, IL6 and IL10) and Th17 (IL‐17) at the end of the experiment (week 6) is presented in Fig. 3. The outcome revealed a significant reduction of IL‐6 and IL‐10 in the group treated with L. paracasei DTA81 when compared with CD and HFD groups. However, the level of cytokines IL‐2, IL‐4, IL‐17, TNF and INF‐γ did not show any significant difference between DTA81 and HFD or CD groups, except for INF‐γ that showed a significant difference with the CD group.
Figure 3.
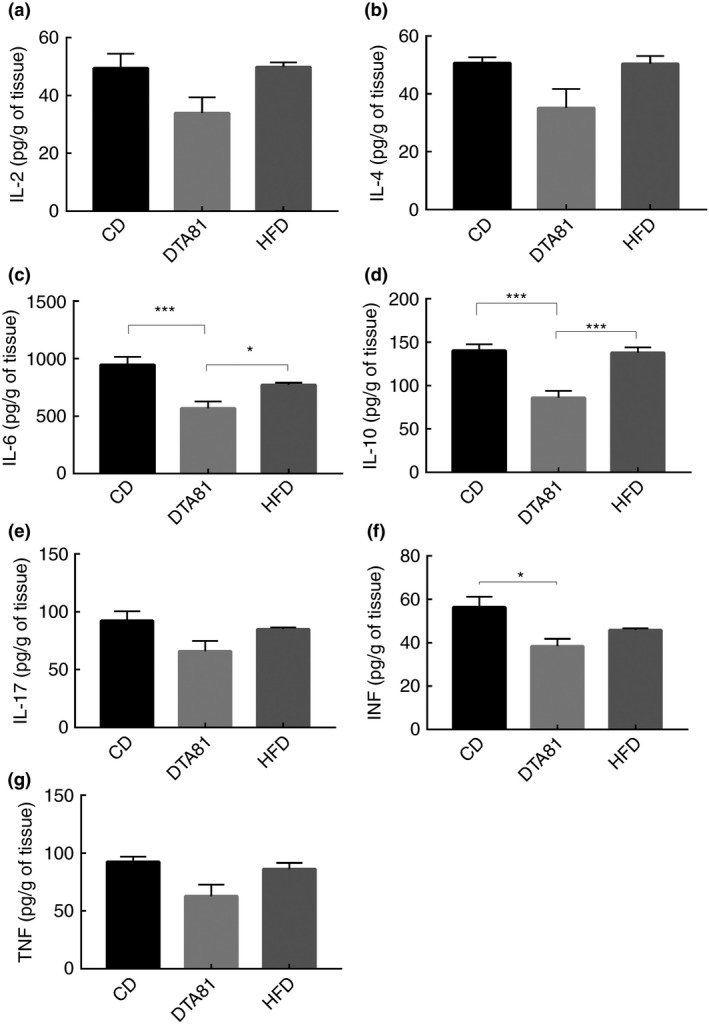
Effect of different treatments on local cytokines. (a) Interleukin‐2; (b) interleukin‐4; (c) interleukin‐6; (d) interleukin‐10; (e) interleukin‐17; (f) interferon gamma; (g) tumour necrosis factor. Results are expressed as means ± SEM (n = 8). The number of asterisks is used to indicate the level of confidence of the statistical analyses results: *statistically significant P < 0·05, ***statistically significant P < 0·001.
16S rRNA sequencing analysis of the gut microbiota
A high‐throughput 16S rRNA sequencing was performed to assess possible changes in the gut microbiota composition caused by the administration of high‐fat diet or by L. paracasei DTA81 supplementation. A total of 536 330 reads from mice faecal microbiota were analysed with a mean of 22 621 (±8·191) sequences for each sample (8 samples for each experimental group; n = 24). Shannon index and phylogenetic diversity curves (Fig. S1) confirmed that the total number of reads obtained from 16S rRNA gene amplicon sequencing covered most of the microbial diversity and that the majority of bacterial phylotypes inside each group were sampled.
At the end of the experimental period (t1), a significant reduction in faecal microbial diversity was observed inside all groups (Fig. 4a), as evidenced by Shannon's diversity index, compared to the same groups at the beginning of the experiment (week 0; t0). A reduced Chao1 value (richness diversity index) was also detected at the end of the experimental period in faecal samples of all animals regardless the experimental group. However, only animals of the HFD group displayed a significant reduction in diversity compared to the same group at the beginning of the trial (Fig. 4b).
Figure 4.
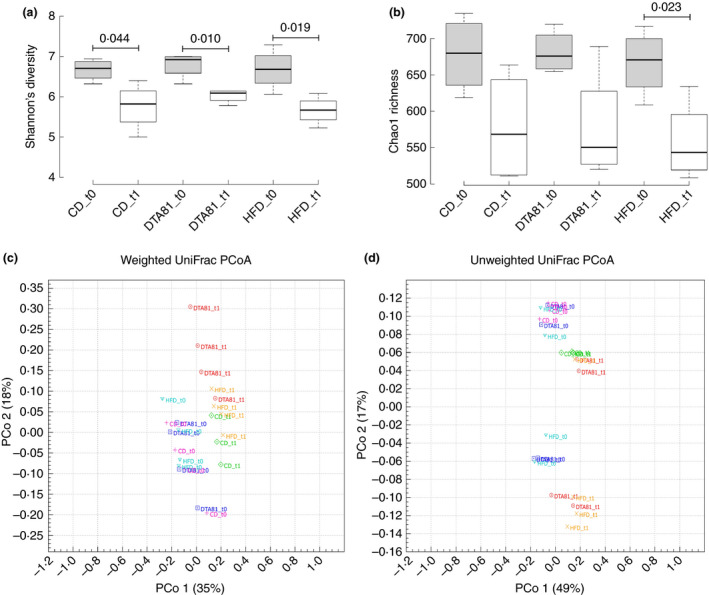
Alpha‐ and beta‐diversity analysis of faecal samples. Box and whisker plots comparing the alpha‐diversity indices Shannon diversity (a) and Chao1 richness (b) among the three groups (CD, DTA81 and HFD) before (t0) and after (t) their respective intervention. Horizontal bold lines show the median values. The bottom and top of the boxes show the 25th and the 75th percentiles respectively. The whiskers extend up to the most extreme points within 1·5 times the interquartile ranges (IQR). Level of significance: P ≤ 0·05. Principal coordinate analysis (PCoA) based on Weighted (c) and Unweighted (d) UniFrac distances for CD, DTA81 and HFD in two different time‐points (t0 and t1). PERMANOVA with 999 permutations was used to detect significant microbial community structure dissimilarity in the different experimental groups enrolled in this study. , ( ) HFD_t0, (
) HFD_t0, ( ) DTA81_t0, (
) DTA81_t0, ( ) CD_t1, (
) CD_t1, ( ) HFD t1, (
) HFD t1, ( ) DTA81 t1, (
) DTA81 t1, ( ) CD_t0.
) CD_t0.
The investigation of community structures of faecal samples of BALB/c mice was done using the phylogenetic distance‐based measurements weighted and unweighted UniFrac. As shown in Fig. 4c,d, scatter plot of the PCoA using both distance metrics revealed a significant microbial shift (PERMANOVA weighted UniFrac: P = 0·001, Pseudo‐f statistic = 4·87; PERMANOVA unweighted UniFrac: P = 0·001, Pseudo‐f statistic = 2·24) after the experimental period in all groups enrolled in this study, although a more pronounced difference was observed when weighted UniFrac distance metric was considered, which accounts for the relative abundance of OTUs. Supplementation of strain DTA81 significantly changed the intestinal microbial composition when compared to the groups receiving a CD (PERMANOVA weighted UniFrac: P = 0·029, Pseudo‐f statistic = 4·49; PERMANOVA unweighted UniFrac: P = 0·031, Pseudo‐f statistic = 1·50) and an HFD (PERMANOVA unweighted UniFrac: P = 0·027, Pseudo‐f statistic = 1·60). A significant difference in terms of beta‐diversity of the intestinal microbiota was also observed between CD and HFD groups (PERMANOVA weighted UniFrac: P = 0·023, Pseudo‐f statistic = 3·11; PERMANOVA unweighted UniFrac: P = 0·025, Pseudo‐f statistic = 1·60). In total, weighted and unweighted UniFrac components (PCoA 1 and PCoA 2) accounted, respectively, for 53 and 66% of the total variance.
The relative distributions of bacteria at the phylum, family and genus level identified by 16S rRNA gene amplicon sequencing are reported in Fig. S2. Prevalent phyla considering all groups were Firmicutes (65%), Proteobacteria (16%) and Bacteroidetes (6%). At t1, the consumption of a high‐fat diet (HFD group) significantly increased Firmicutes (LDA = 3·9), Actinobacteria (LDA = 3·3), Deferribacteres (LDA = 2·8) and Spirochaetes (LDA = 2·2), whereas the supplementation of DTA81 significantly increased Bacteroidetes (LDA = 3·1) (Fig. 5). Considered important in the development of obesity, the Firmicutes/Bacteroidetes (F/B) ratio was calculated and results show that the potential probiotic intervention improved on average the proportion F/B (7·8 ± 4·65) when compared to the groups that received a CD (11·11 ± 3·95) or a high‐fat diet (18·71 ± 12·65).
Figure 5.
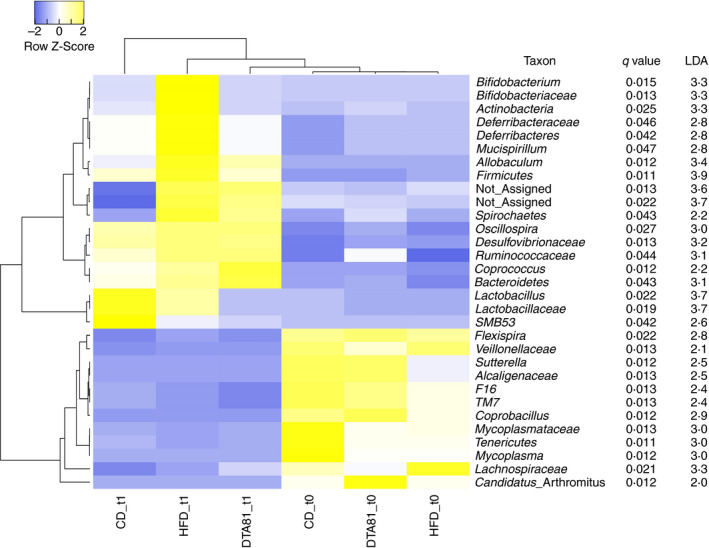
Heat map based on linear discriminant analysis effect size (LEfSe) at phylum, family and genus levels. An FDR adjusted P value (or q value) less than 5% and an LDA score greater than 2 were used to determine taxa that are significantly enriched among the groups CD, DTA81 and HFD before (t0) and after (t1) the experimental period. In the heat map, an enrichment trend is reported in yellow while depletion is represented in blue.
At family level, Lachnospiraceae, Ruminococcaceae, Erysipelotrichaceae, Lactobacillaceae and Desulfovibrionaceae were the top five taxa (Fig. S2b). At the end of the experimental period, major changes were observed mainly in samples from the HFD group, where four families (Bifidobacteriaceae, LDA = 3·3; Desulfovibrionaceae, LDA = 3·2; Ruminococcaceae, LDA = 3·1; Deferribacteraceae, LDA = 2·8) resulted overrepresented in this group (Fig. 5). Only the family Lactobacillaceae increased in the CD group (LDA = 3·7), while in the DTA81 group there were no statistically significant biomarkers (LDA score <2 and/or q value >0·05) after the potential probiotic supplementation.
With regard to the genera observed in faecal samples across the groups, Lactobacillus, Oscillospira, Allobaculum, Helicobacter and Ruminococcus were identified as the top five taxa (Fig. S2c). Linear discriminant effect size (LEfSe) analysis revealed an enrichment of 13 biomarkers (Fig. 5), among which a higher relative abundance of the genera Oscillospira (LDA = 3·0) and Coprococcus (LDA = 2·2) were associated with L. paracasei DTA81 administration. In stool samples of mice of the CD group, an enrichment of the genera Lactobacillus (LDA = 3·7) and SMB53 (LDA = 2·6) was observed at the end of the experimental period. Lastly, the HFD was associated with a higher proportion of Allobaculum (LDA = 3·4), Bifidobacterium (LDA = 3·3) and Mucispirillum (LDA = 2·8) in faeces samples.
L. paracasei DTA81 cell enumeration after mice GI transit
Survival of L. paracasei DTA81 after passage through mice GIT was evaluated after 21 (week 3) and 42 (week 6) days. The strain was administered to the animals at the dose of approx. 109 CFU per day and viable cells were then enumerated from collected faeces. After week 3, potential probiotic‐treated animals revealed 8·50 ± 0·16 log CFU per g of wet faeces while no colony was recovered from CDC and HFD groups. After week 6, no significant difference in the number of retrieved cells (8·67 ± 0·23 log CFU per g) was recorded.
Genome sequencing and genomic analysis of L. paracasei DTA81
Sequencing of L. paracasei DTA81 genome produced 1 443 422 reads with an average size of 150·5 bp. The assembled genome of L. paracasei DTA81 produced 39 scaffolds, giving a genome size of 3·00 Mb with a GC content of 46·1% (Table 2). RASTtk server predicted a total number of 3077 protein‐coding sequences (CDSs) classified into 236 different subsystems. The largest part of this subsystem is allocated to the carbohydrate metabolism (22·03%) followed by amino acids and derivatives (13·45%) and protein metabolism (11·86%). A total number of 59 structural RNAs including 3 complete rRNAs (5S, 16S and 23S) and 56 tRNAs were predicted, too. The average nucleotide identity (ANI) of 98·7% with the closest neighbour (L. paracasei ATCC 334) allowed a precise taxonomical placement of strain DTA81 inside the species L. paracasei. In Fig. 6, a circular graphical map of the distribution of the genome annotations of L. paracasei DTA81 is provided which indicates the location of CDS on the forward and reverse strands, RNA genes, antimicrobial resistance genes and virulence factors. A deep analysis within the genome of DTA81 has also revealed the presence of cholesterol assimilation‐related genes namely, ccpA, fba, lbpg_rs09895, lbpg_rs11190 and lbpg_rs10085 (Table 3) (Lee et al. 2010). Finally, the absence of mobile elements such as prophage regions, acquired antibiotic resistance genes and plasmid sequences on DTA81 genome was confirmed using PHASTER, PlasmidFinder and ResFinder servers respectively.
Table 2.
Main characteristics of L. paracasei DTA81 genome
| Feature | Value |
|---|---|
| Genome size | 3 002 945 |
| G+C content (%) | 46.2 |
| Contig N50 | 156 284 |
| Contig L50 | 7 |
| Number of scaffolds | 39 |
| Number of protein coding sequences (CDSs) | 3077 |
| Number of rRNAs | 3 |
| Number of tRNAs | 56 |
| Number of genes related to Virulence, disease, and defense | 0 |
Figure 6.
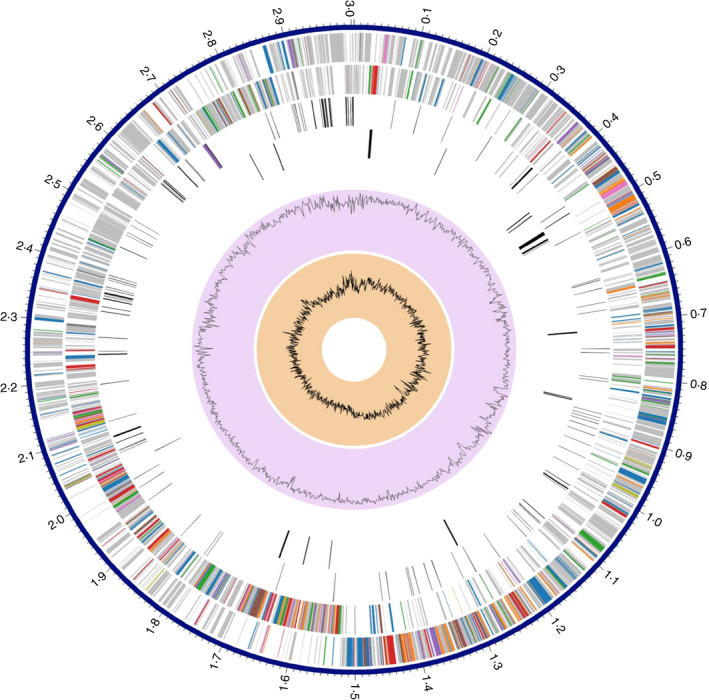
Circular graphical display of the distribution of the genome map and annotations. This includes, from outer to inner rings, CDS on the forward strand, CDS on the reverse strand, RNA genes, CDS with homology to known antimicrobial resistance genes, CDS with homology to known virulence factors, GC content and GC skew. The colours of the CDS on the forward and reverse strand indicate the subsystem that these genes belong to. ( ) Metabolism, (
) Metabolism, ( ) Protein processing, (
) Protein processing, ( ) Stress response, Defence, Virulence, (
) Stress response, Defence, Virulence, ( ) DNA processing, (
) DNA processing, ( ) Energy, (
) Energy, ( ) Cellular processing, (
) Cellular processing, ( ) RNA processing, (
) RNA processing, ( ) Membrane transport, (
) Membrane transport, ( ) Cell envelope, (
) Cell envelope, ( ) Regulation and cell signalling, (
) Regulation and cell signalling, ( ) Miscellaneous.
) Miscellaneous.
Table 3.
Identification of cholesterol assimilation‐related genes in Lactobacillus paracasei DTA81
| Gene | Gene description | Organism | Identity score (%) | Accession number |
|---|---|---|---|---|
| ccpA | Catabolite control protein A | L. paracasei AO356 | 100.00 | CP025499.1 |
| fba | Class II fructose‐1,6‐bisphosphate aldolase | L. paracasei N1115 | 100.00 | CP007122.1 |
| lbpg_rs09895 | Glycogen phosphorylase | L. paracasei AO356 | 100.00 | CP025499.1 |
| lbpg_rs11190 | FMN‐binding protein | L. paracasei W56 | 100.00 | HE970764.1 |
| lbpg_rs10085 | MFS transporter | L. paracasei N1115 | 100.00 | CP007122.1 |
Discussion
It has been reported that the presence of probiotics inside the gut microbiota can be considered as a potential therapeutic strategy for some metabolic disorders such as hyperglycaemia, hypercholesterolemia and obesity (Janssen and Kersten 2015).
In this study, mice treated with DTA81 did not evidence a significant difference in weight gain. It should be noted that the use of a high‐fat diet does not always cause changes of total body weight, and metabolic changes related to obesity, such as hyperlipidaemia, hyperglycaemia, diabetes and low‐grade inflammation, should also be observed. In a recently published study, the GTT was recorded significantly higher in BALB/c mice that had received a high‐fat diet compared to BALC/c mice that received a control diet. This indicates that the high‐fat diet interfered with the glucose metabolism in these mice; besides, serum levels of total cholesterol, LDL‐c, triglycerides/HDL‐c ratio, liver and adipose tissue were higher in BALC/c mice that had received the high‐fat diet. However, there was no change in body weight (Li et al. 2020).
After 6 weeks of treatment, a significant fasting blood glucose reduction was recorded in the animals treated with L. paracasei DTA81 compared with the CD and HFD groups. The reduction of fasting blood glucose by DTA81 compared to the group receiving a CD is very interesting since the CD (control) contains higher carbohydrate concentrations than that of the high‐fat diet; therefore, our findings demonstrate the possible benefit of DTA81 in this condition too. The mechanism of glucose‐lowering by probiotics is not well understood, but probiotics can modulate the human immune system which can influence glucose metabolism. Moreover, probiotics can reduce the inflammatory cytokines and regulate the immune system (de LeBlanc and Perdigón 2010).
Laitinen et al. (2008) demonstrated that the immunomodulatory effect of probiotics can lead to glucose reduction. In our study, L. paracasei DTA81, with its fasting blood‐sugar‐lowering activity, induced significantly lower values for interleukin 6 and 10.
Probiotics can also lower blood cholesterol in different ways, indirectly by fermentation of prebiotics and consequent production of SCFA in the human gut that can further inhibit hepatic cholesterol synthesis and will result in a reduction of blood lipids (Ashaolu et al. 2020). Alternatively, probiotics can assimilate cholesterol directly, thus eliminating it from the human gut (Pereira and Gibson 2002; Pan et al. 2011; Öner et al. 2014). In their study, Shimizu et al. (2015) reported that consumption of probiotics by elderly and hypercholesterolaemic patients could be more effective than in youngsters and individuals with normal lipid levels. Besides, probiotics can reduce cholesterol levels by assimilating and entrapping this molecule into bacterial membranes (Castorena‐Alba et al. 2018; Bhat and Bajaj 2020). There are numerous species of Lactobacillus and Bifidobacterium that have shown cholesterol assimilation in in‐vitro experiments and it has been reported that this ability is strictly strain‐dependent (Costabile et al. 2017; Castorena‐Alba et al. 2018). Taking into account the published data in the literature, cholesterol assimilation by probiotic strains can range from 0·86% to more than 40%; however, we usually do not see the same reduction when we use probiotics in the in‐vivo conditions, which can be due to the effect of gastrointestinal conditions of the strains, microbial competition, colonization on the epithelial cells, etc. (Belviso et al. 2009; Tokatlı et al. 2015; Castorena‐Alba et al. 2018). The outcome of our study indicates that consumption of L. paracasei DTA81 can lead in mice to a statistically significant reduction of total cholesterol and LDL and HDL, which appears very interesting and useful in people who suffer from CVD and CHD. This ability by L. paracasei DTA81 could be connected to the presence of cholesterol assimilation‐related genes, coding membrane‐associated proteins that can adhere to the cholesterol molecule and further incorporate it inside the cell (Lee et al. 2010). Besides, in our previous study (Tarrah et al. 2019), DTA81 had revealed a great adhesion capability to human cell lines, which could play a significant role in its potential beneficial activity.
Gut microbiota dysbiosis can also promote the occurrence of metabolic syndromes in diverse ways, such as low‐grade inflammation, through increased production of bacterial lipopolysaccharide. In the present study, we detected an increase in Bacteroidetes and Coprococcus after 6 weeks of treatment with strain DTA81. The phylum Bacteroidetes belongs to Gram‐negative bacteria that normally colonize the human lower gastrointestinal tract during infancy, due to the abundance of non‐digestible oligosaccharides in mother’s milk which support their growth (Marcobal et al. 2011). Since colonization, they play an essential role by breaking down complex sugars and degrading proteins in the human gut, as well as by exerting an immunomodulatory effect (Rakoff‐Nahoum et al. 2004; Rajilić‐Stojanović and de Vos 2014). Another important function of this bacterial group in the human gut is related to the deconjugation of bile acids which is linked to cholesterol‐lowering activity (Narushima et al. 2006; Leitch et al. 2007). Also, it has been reported that, due to the broad metabolic potential of Bacteroidetes, their reduced abundance could be linked to obesity in humans (Ley 2010). On the other side, it was proven that the increase in the genus Coprococcus, an anaerobic genus which is normally present in the human faecal microbiota, can have an anti‐carcinogenic and anti‐inflammatory effect in the human gut, due to butyric acid production (Hamer et al. 2008; Ai et al. 2019).
As determinants and modulators of immune pathology, cytokines play a key regulatory role among the many components of the animal immune system (Lin and Karin 2007). A broad spectrum of cells such as fibroblasts, endothelial cells, neuronal cells, macrophages and mast cells can produce cytokines; however, production is mainly dependent from the differentiation state of T cells, which can be divided into three different types according to the pattern of cytokine production (Saito et al. 2010; Yang et al. 2017). Among the cytokines considered in the present study, IL‐2, TNF‐α and IFN‐γ are produced by T helper 1 cells and play an important role in the cell‐mediated immune response. By contrast, IL‐4, IL‐6 and IL‐10 are secreted by T helper 2 cells and enhance humoral immunity (Kikuchi and Crystal 2001). Moreover, we have studied the IL‐17 secreted by T helper 17 cells which is involved in allergic responses by inducing and mediating the proinflammatory responses (Korn et al. 2009). Overproduction or inappropriate production of certain cytokines by the body can result in inflammatory diseases.
Occasionally, it has been reported that the insertion of external bacterial cells inside the human body leads to inappropriate production of certain cytokines which can cause inflammatory diseases (Percoco et al. 2013; Cattaneo et al. 2017). In our study, the group of mice treated with L. paracasei DTA81 showed lower average values for IL‐6 and IL‐10 when compared to CD and HFD groups. Cytokine IL‐6 is locally produced in response to infection or injury and delivered to other body parts by the bloodstream, activating immunological defences. IL‐6 also stimulates intestinal epithelial proliferation and repair after injury (Kuhn et al. 2014); however, excessive or prolonged production of IL‐6 is involved in various diseases (Narazaki and Kishimoto 2018). Several studies have reported that IL‐6 is a pro‐inflammatory cytokine that is detected in higher amounts in obese individuals and contributes to the occurrence of type 2 diabetes, insulin resistance and CVDs (Higa and Panee 2011; Shi et al. 2019). The IL‐6 level observed in this work does not differ between CD and HFD groups and can be explained by the reduced duration of the experiment when compared to other studies in the literature assessing the immunomodulatory effect of probiotics (8–14 weeks) (Sheil et al. 2006; Antunes et al. 2020). Moreover, it can be related to a potential pro‐inflammatory property of the commercial diet, which has approximately 56% carbohydrate, a component that at high concentrations induces inflammation in skeletal muscle (Antunes et al. 2020). As regards the reduction of IL‐10, it could be explained considering the strong positive correlation between IL‐6 and IL‐10 normally found in the human body, which contributes to homeostasis maintenance (Dizdarević‐Hudić et al. 2009; Sapan et al. 2016). The IL‐10 level was reduced by DTA81 intake, and a significant difference was observed compared to the other two groups. Although the increase in IL‐10 level is referred to as the immunomodulatory effect of probiotics, this is not the unique form of probiotics action, which depends on the bacterial strain, concentration and administration method. The reduction level of IL‐10 may decrease the immunostimulatory effect of this cytokine in innate immunity (de Moreno de LeBlanc et al. 2011).
It is well known that probiotic traits are strain‐specific and this gives strong motivation to keep seeking new potentially better strains (Senok et al. 2005). In our study, L. paracasei DTA81 indicated a good resistance to the gastrointestinal environment in‐vivo. The resistance of lactobacilli to the harsh GIT conditions reported previously (Noriega et al. 2004; Burns et al. 2010) seems to be linked to the preservation of cell internal pH, functionality and integrity of cell membrane, and to the existence of bile salt efflux pumps (Bustos et al. 2011; Wu et al. 2012, 2014).
The outcomes of this study revealed the possible mechanism that started from the gut microbiota regulation by L. paracasei DTA81 with increasing the abundance of Bacteriodetes and Coprococcus taxa followed by a significant reduction of inflammatory cytokine interleukin 6 which has subsequently led to a decrement of FBS. On the other side, total cholesterol reduction in the group treated with DTA81 could be possibly related to the above‐mentioned gut microbiota modulation as well as the direct cholesterol assimilation by L. paracasei DTA81. Overall, considering the results of this study alongside the previous findings, L. paracasei DTA81 has a great potential to be used as a commercial promising probiotic with great influence on metabolic homeostasis in individuals, particularly those adopting high‐fat diets.
Conflict of Interest
No conflict of interest declared.
Author Contributions
Conceptualization: A.T., B.C.S.C. and V.S.D.; investigation: A.T., B.C.S.C., V.S.D., R.S.D., S.P., L.L.O.; data curation: A.T. and V.S.D.; writing—original draft preparation: A.T.; writing—review and editing: A.T. and A.G.; supervision: A.G., S.O.P., M.C.G.P., L.L.O.; funding acquisition: A.G., V.C., S. O. P. and M.C.G.P.; All authors have read and agreed to the published version of the manuscript.
Supporting information
Figure S1. Rarefaction curves of Shannon entropy (a) and phylogenetic diversity (b) of stool samples before (t0) and after (t1) 6 weeks of experimental period.
Figure S2. Relative abundance of bacterial phyla (a), top‐20 families (b), and top‐20 genera (c) identified in feces samples of BALB/c mice in groups CD, DTA81 and HFD before (t0) and after (t1) the experimental period.
Acknowledgements
We are grateful to the Núcleo de Análise de Biomoléculas and Núcleo de Microscopia e Microanálise of the Universidade Federal de Viçosa for providing the facilities to conduct the experiments. This work was supported in part by the ‘Ministero dell’Università e della Ricerca Scientifica’. We also acknowledge the financial support of the following Brazilian agencies: Fundação de Amparo à Pesquisa do Estado de Minas Gerais (Fapemig), Coordenacão de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Financiadora de Estudos e Projetos (Finep), Sistema Nacional de Laboratórios em Nanotecnologias (SisNANO)/Ministério da ciência, tecnologia e Informação (MCTI).
References
- Ai, D. , Pan, H. , Li, X. , Gao, Y. , Liu, G. and Xia, L.C. (2019) Identifying gut microbiota associated with colorectal cancer using a zero‐inflated lognormal model. Front Microbiol 10, 826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amund, O.D. (2016) Exploring the relationship between exposure to technological and gastrointestinal stress and probiotic functional properties of lactobacilli and bifidobacteria. Can. J. Microbiol. 62, 715–725. [DOI] [PubMed] [Google Scholar]
- Antunes, M.M. , Godoy, G. , de Almeida‐Souza, C.B. , da Rocha, B.A. , da Silva‐Santi, L.G. , Masi, L.N. , Carbonera, F. , Visentainer, J.V. et al. (2020) A high‐carbohydrate diet induces greater inflammation than a high‐fat diet in mouse skeletal muscle. Brazilian J. Med. Biol Res 53.e9039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt, D. , Grant, J.R. , Marcu, A. , Sajed, T. , Pon, A. , Liang, Y. and Wishart, D.S. (2016) PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, 1–6. 10.1093/nar/gkw387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashaolu, T.J. , Ashaolu, J.O. and Adeyeye, S.A.O. (2020) Fermentation of prebiotics by human colonic microbiota in vitro and short‐chain fatty acids production: a critical review. Microbiol J. Appl, 130, 677–687. [DOI] [PubMed] [Google Scholar]
- Aziz, R.K. , Bartels, D. , Best, A.A. , DeJongh, M. , Disz, T. , Edwards, R.A. , Formsma, K. , Gerdes, S. et al. (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genom 9, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babicki, S. , Arndt, D. , Marcu, A. , Liang, Y. , Grant, J.R. , Maciejewski, A. and Wishart, D.S. (2016) Heatmapper: web‐enabled heat mapping for all. Nucleic Acids Res. 44, W147–W153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, H.C. , Tran, D.N. and Thomas, L.V. (2009) Health benefits of probiotics for the elderly: a review. J. Foodserv 20, 250–262. [Google Scholar]
- Belviso, S. , Giordano, M. , Dolci, P. and Zeppa, G. (2009) In vitro cholesterol‐lowering activity of Lactobacillus plantarum and Lactobacillus paracasei strains isolated from the Italian Castelmagno PDO cheese. Dairy Sci Technol 89, 169–176. [Google Scholar]
- Bhat, B. and Bajaj, B.K. (2020) Multifarious cholesterol lowering potential of lactic acid bacteria equipped with desired probiotic functional attributes. 3 Biotech 10, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosi, E. , Donati, B. , Galardini, M. , Brunetti, S. , Sagot, M.‐F. , Lió, P. , Crescenzi, P. , Fani, R. et al. (2015) MeDuSa: a multi‐draft based scaffolder. Bioinformatics 31, 2443–2451. [DOI] [PubMed] [Google Scholar]
- Burns, P. , Sánchez, B. , Vinderola, G. , Ruas‐Madiedo, P. , Ruiz, L. , Margolles, A. , Reinheimer, J. and Clara, G. (2010) Inside the adaptation process of Lactobacillus delbrueckii subsp. lactis to bile. Int J Food Microbiol 142, 132–141. [DOI] [PubMed] [Google Scholar]
- Bustos, A.Y. , Raya, R. , de Valdez, G.F. and Taranto, M.P. (2011) Efflux of bile acids in Lactobacillus reuteri is mediated by ATP. Biotechnol Lett. https://doi.org/10.1007/s10529‐011‐0696‐333 2265–2269. [DOI] [PubMed] [Google Scholar]
- Cani, P.D. , Van Hul, M. , Lefort, C. , Depommier, C. , Rastelli, M. and Everard, A. (2019) Microbial regulation of organismal energy homeostasis. Nat Metab 1, 34–46. [DOI] [PubMed] [Google Scholar]
- Carattoli, A. , Zankari, E. , García‐Fernández, A. , Larsen, M.V. , Lund, O. , Villa, L. , Aarestrup, F.M. and Hasman, H. (2014) In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58, 3895–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso, F.C. , Sears, W. , LeBlanc, S.J. and Drackley, J.K. (2011) Comparison of 3 methods for analyzing areas under the curve for glucose and nonesterified fatty acids concentrations following epinephrine challenge in dairy cows. J. Dairy Sci 94, 6111–6115. [DOI] [PubMed] [Google Scholar]
- Castorena‐Alba, M.M. , Vázquez‐Rodríguez, J.A. , López‐Cabanillas Lomelí, M. and González‐Martínez, B.E. (2018) Cholesterol assimilation, acid and bile survival of probiotic bacteria isolated from food and reference strains. CyTA‐J Food 16, 36–41. [Google Scholar]
- Cattaneo, A. , Cattane, N. , Galluzzi, S. , Provasi, S. , Lopizzo, N. , Festari, C. , Ferrari, C. , Guerra, U.P. et al. (2017) Association of brain amyloidosis with pro‐inflammatory gut bacterial taxa and peripheral inflammation markers in cognitively impaired elderly. Neurobiol Aging 49, 60–68. [DOI] [PubMed] [Google Scholar]
- Clark, J.D. , Gebhart, G.F. , Gonder, J.C. , Keeling, M.E. and Kohn, D.F. (1997) The 1996 guide for the care and use of laboratory animals. ILAR J 38, 41–48. [DOI] [PubMed] [Google Scholar]
- Costabile, A. , Buttarazzi, I. , Kolida, S. , Quercia, S. , Baldini, J. , Swann, J.R. , Brigidi, P. and Gibson, G.R. (2017) An in vivo assessment of the cholesterol‐lowering efficacy of Lactobacillus plantarum ECGC 13110402 in normal to mildly hypercholesterolaemic adults. PLoS One 12, e0187964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crovesy, L. , Masterson, D. and Rosado, E.L. (2020) Profile of the gut microbiota of adults with obesity: a systematic review. Eur J Clin Nutr 74, 1251–1262. [DOI] [PubMed] [Google Scholar]
- Dhariwal, A. , Chong, J. , Habib, S. , King, I.L. , Agellon, L.B. and Xia, J. (2017) MicrobiomeAnalyst: a web‐based tool for comprehensive statistical, visual and meta‐analysis of microbiome data. Nucleic Acids Res 45, W180–W188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dizdarević‐Hudić, L. , Kušljugić, Z. , Baraković, F. , Brkić, S. , Sabitović, D. , Jahić, E. , Isabegović, M. , Smajić, E. et al. (2009) Correlation between interleukin 6 and interleukin 10 in acute myocardial infarction. Bosn J Basic Med Sci 9, 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn‐Emke, S. , Weidner, G. and Ornish, D. (2001) Benefits of a low‐fat plant‐based diet. Obesity 9, 731. [DOI] [PubMed] [Google Scholar]
- Ferrere, G. , Leroux, A. , Wrzosek, L. , Puchois, V. , Gaudin, F. , Ciocan, D. , Renoud, M.‐L. , Naveau, S. et al. (2016) Activation of kupffer cells is associated with a specific dysbiosis induced by fructose or high fat diet in mice. PLoS One 11, e0146177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedwald, W.T. (1972) Estimation of the concentration of low density lipoprotein cholesterol in plasma without use of the preparative ultracentrifuge. Clin Chem 18, 499–502. [PubMed] [Google Scholar]
- Guerra, A.F. , Lemos Junior, W.J.F. , dos Santos, G.O. , Andrighetto, C. , Gianomini, A. , Corich, V. , Luchese, R.H. , Guerra, A.F. et al. (2018) Lactobacillus paracasei probiotic properties and survivability under stress‐induced by processing and storage of ice cream bar or ice‐lolly. Ciência Rural 48, 10.1590/0103-8478cr20170601 [DOI] [Google Scholar]
- Hamer, H.M. , Jonkers, D. , Venema, K. , Vanhoutvin, S. , Troost, F.J. and Brummer, R. (2008) The role of butyrate on colonic function. Aliment Pharmacol Ther 27, 104–119. [DOI] [PubMed] [Google Scholar]
- He, J. , Zhang, F. and Han, Y. (2017) Effect of probiotics on lipid profiles and blood pressure in patients with type 2 diabetes: a meta‐analysis of RCTs. Medicine (Baltimore) 96.e9166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higa, J.K. and Panee, J. (2011) Bamboo extract reduces interleukin 6 (IL‐6) overproduction under lipotoxic conditions through inhibiting the activation of NF‐κB and AP‐1 pathways. Cytokine 55, 18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, C. , Guarner, F. , Reid, G. , Gibson, G.R. , Merenstein, D.J. , Pot, B. , Morelli, L. , Canani, R.B. et al. (2014) Expert consensus document: The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol 11, 506. [DOI] [PubMed] [Google Scholar]
- Janssen, A.W.F. and Kersten, S. (2015) The role of the gut microbiota in metabolic health. FASEB J 29, 3111–3123. [DOI] [PubMed] [Google Scholar]
- Kikuchi, T. and Crystal, R.G. (2001) Antigen‐pulsed dendritic cells expressing macrophage‐derived chemokine elicit Th2 responses and promote specific humoral immunity. J Clin Invest 108, 917–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn, T. , Bettelli, E. , Oukka, M. and Kuchroo, V.K. (2009) IL‐17 and Th17 Cells. Annu Rev Immunol 27, 485–517. [DOI] [PubMed] [Google Scholar]
- Kuhn, K.A. , Manieri, N.A. , Liu, T.‐C. and Stappenbeck, T.S. (2014) IL‐6 stimulates intestinal epithelial proliferation and repair after injury. PLoS One 9, e114195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laitinen, K. , Poussa, T. and Isolauri, E. (2008) Probiotics and dietary counselling contribute to glucose regulation during and after pregnancy: a randomised controlled trial. Br J Nutr 101, 1679–1687. [DOI] [PubMed] [Google Scholar]
- Larsen, M.V. , Cosentino, S. , Rasmussen, S. , Friis, C. , Hasman, H. , Marvig, R.L. , Jelsbak, L. , Sicheritz‐Pontén, T. et al. (2012) Multilocus sequence typing of total‐genome‐sequenced bacteria. J Clin Microbiol 50, 1355–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de LeBlanc, A.M. and Perdigón, G. (2010) The application of probiotic fermented milks in cancer and intestinal inflammation. Proc Nutr Soc 69, 421–428. [DOI] [PubMed] [Google Scholar]
- Lee, J. , Kim, Y. , Yun, H.S. , Kim, J.G. , Oh, S. and Kim, S.H. (2010) Genetic and proteomic analysis of factors affecting serum cholesterol reduction by Lactobacillus acidophilus A4. Appl Environ Microbiol 76, 4829–4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch, E.C.M. , Walker, A.W. , Duncan, S.H. , Holtrop, G. and Flint, H.J. (2007) Selective colonization of insoluble substrates by human faecal bacteria. Environ Microbiol 9, 667–679. [DOI] [PubMed] [Google Scholar]
- Ley, R.E. (2010) Obesity and the human microbiome. Curr Opin Gastroenterol 26, 5–11. [DOI] [PubMed] [Google Scholar]
- Li, J. , Wu, H. , Liu, Y. and Yang, L. (2020) High fat diet induced obesity model using four strains of mice: Kunming, C57BL/6. BALB/c and ICR. Exp Anim 69, 326–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, W.‐W. and Karin, M. (2007) A cytokine‐mediated link between innate immunity, inflammation, and cancer. J Clin Invest 117, 1175–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson, J. and Schnürer, J. (2001) Lactobacillus coryniformis subsp. coryniformis strain Si3 produces a broad‐spectrum proteinaceous antifungal compound. Appl Environ Microbiol 67, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchesi, J.R. , Adams, D.H. , Fava, F. , Hermes, G.D.A. , Hirschfield, G.M. , Hold, G. , Quraishi, M.N. , Kinross, J. et al. (2016) The gut microbiota and host health: a new clinical frontier. Gut 65, 330–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcobal, A. , Barboza, M. , Sonnenburg, E.D. , Pudlo, N. , Martens, E.C. , Desai, P. , Lebrilla, C.B. , Weimer, B.C. et al. (2011) Bacteroides in the infant gut consume milk oligosaccharides via mucus‐utilization pathways. Cell Host Microbe 10, 507–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miremadi, F. , Ayyash, M. , Sherkat, F. and Stojanovska, L. (2014) Cholesterol reduction mechanisms and fatty acid composition of cellular membranes of probiotic Lactobacilli and Bifidobacteria. J Funct Foods 9, 295–305. 10.1016/J.JFF.2014.05.002 [DOI] [Google Scholar]
- Mo, R. , Zhang, X. and Yang, Y. (2019) Effect of probiotics on lipid profiles in hypercholesterolaemic adults: A meta‐analysis of randomized controlled trials. Med Clínica (English Ed.) 152, 473–481. [DOI] [PubMed] [Google Scholar]
- de Moreno de LeBlanc, A. , Del Carmen, S. , Zurita‐Turk, M. , Santos Rocha, C. , Van de Guchte, M. , Azevedo, V. , Miyoshi, A. and LeBlanc, J.G. (2011) Importance of IL‐10 modulation by probiotic microorganisms in gastrointestinal inflammatory diseases. ISRN Gastroenterol, 2011 892971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narazaki, M. and Kishimoto, T. (2018) The two‐faced cytokine IL‐6 in host defense and diseases. Int J Mol Sci 19, 3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narushima, S. , Itoh, K. , Miyamoto, Y. , Park, S. , Nagata, K. , Kuruma, K. and Uchida, K. (2006) Deoxycholic acid formation in gnotobiotic mice associated with human intestinal bacteria. Lipids 41, 835–843. [DOI] [PubMed] [Google Scholar]
- Noriega, L. , Gueimonde, M. , Sánchez, B. , Margolles, A. and de los Reyes‐Gavilán, C.G. (2004) Effect of the adaptation to high bile salts concentrations on glycosidic activity, survival at low pH and cross‐resistance to bile salts in Bifidobacterium. Int J Food Microbiol 94, 79–86. [DOI] [PubMed] [Google Scholar]
- Öner, Ö. , Aslim, B. and Aydaş, S.B. (2014) Mechanisms of cholesterol‐lowering effects of lactobacilli and bifidobacteria strains as potential probiotics with their bsh gene analysis. J Mol Microbiol Biotechnol 24, 12–18. [DOI] [PubMed] [Google Scholar]
- Pan, D.D. , Zeng, X.Q. and Yan, Y.T. (2011) Characterisation of Lactobacillus fermentum SM‐7 isolated from koumiss, a potential probiotic bacterium with cholesterol‐lowering effects. J Sci Food Agric 91, 512–518. [DOI] [PubMed] [Google Scholar]
- Percoco, G. , Merle, C. , Jaouen, T. , Ramdani, Y. , Bénard, M. , Hillion, M. , Mijouin, L. , Lati, E. et al. (2013) Antimicrobial peptides and pro‐inflammatory cytokines are differentially regulated across epidermal layers following bacterial stimuli. Exp Dermatol 22, 800–806. [DOI] [PubMed] [Google Scholar]
- Pereira, D.I.A. and Gibson, G.R. (2002) Effects of consumption of probiotics and prebiotics on serum lipid levels in humans. Crit Rev Biochem Mol Biol 37, 259–281. [DOI] [PubMed] [Google Scholar]
- Rajilić‐Stojanović, M. and de Vos, W.M. (2014) The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol Rev 38, 996–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakoff‐Nahoum, S. , Paglino, J. , Eslami‐Varzaneh, F. , Edberg, S. and Medzhitov, R. (2004) Recognition of commensal microflora by toll‐like receptors is required for intestinal homeostasis. Cell 118, 229–241. [DOI] [PubMed] [Google Scholar]
- Reeves, P.G. , Nielsen, F.H. and Fahey, G.C. Jr . (1993) AIN‐93 purified diets for laboratory rodents: final report of the American Institute of Nutrition ad hoc writing committee on the reformulation of the AIN‐76A rodent diet. J. Nutr 123, 1939–51. [DOI] [PubMed] [Google Scholar]
- Rosenbaum, M. , Knight, R. and Leibel, R.L. (2015) The gut microbiota in human energy homeostasis and obesity. Trends Endocrinol Metab 26, 493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, S. , Nakashima, A. , Shima, T. and Ito, M. (2010) Th1/Th2/Th17 and regulatory T‐cell paradigm in pregnancy. Am J Reprod Immunol 63, 601–610. [DOI] [PubMed] [Google Scholar]
- Sapan, H.B. , Paturusi, I. , Jusuf, I. , Patellongi, I. , Massi, M.N. , Pusponegoro, A.D. , Arief, S.K. , Labeda, I. et al. (2016) Pattern of cytokine (IL‐6 and IL‐10) level as inflammation and anti‐inflammation mediator of multiple organ dysfunction syndrome (MODS) in polytrauma. Int J Burns Trauma 6, 37. [PMC free article] [PubMed] [Google Scholar]
- Senok, A.C. , Ismaeel, A.Y. and Botta, G.A. (2005) Probiotics: facts and myths. Clin Microbiol Infect 11, 958–966. [DOI] [PubMed] [Google Scholar]
- Sheil, B. , MacSharry, J. ., O’Callaghan, L. , O’Riordan, A. , Waters, A. , Morgan, J. , Collins, J.K. , O’Mahony, L. et al. (2006) Role of interleukin (IL‐10) in probiotic‐mediated immune modulation: an assessment in wild‐type and IL‐10 knock‐out mice. Clin Exp Immunol 144, 273–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, J. , Fan, J. , Su, Q. and Yang, Z. (2019) Cytokines and abnormal glucose and lipid metabolism in type 2 diabetes. Front Endocrinol 10, 703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu, M. , Hashiguchi, M. , Shiga, T. , Tamura, H. and Mochizuki, M. (2015) Meta‐analysis: effects of probiotic supplementation on lipid profiles in normal to mildly hypercholesterolemic individuals. PLoS One 10, e0139795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Duarte, V. , Carlot, M. , Pakroo, S. , Tarrah, A. , Lombardi, A. , Santiago, H. , Corich, V. and Giacomini, A. (2020) Comparative evaluation of cheese whey microbial composition from four Italian cheese factories by viable counts and 16S rRNA gene amplicon sequencing. Int Dairy J 104, 104656. [Google Scholar]
- Singh, N. , Singh, J. and Singh, K. (2018) Small at size, big at impact: Microorganisms for sustainable development. In Microbial Bioprospecting for Sustainable Development ed. Singh, J. , Sharma, D. , Kumar, G. and Sharma, N.R. pp. 3–28. Singapore: Springer Singapore. 10.1007/978-981-13-0053-0_1 [DOI] [Google Scholar]
- Tarrah, A. , Pakroo, S. , Corich, V. and Giacomini, A. (2020b) Whole‑genome sequence and comparative genome analysis of Lactobacillus paracasei DTA93, a promising probiotic lactic acid bacterium. Arch Microbiol, 202, 1997–2003. [DOI] [PubMed] [Google Scholar]
- Tarrah, A. , da Silva Duarte, V. , de Castilhos, J. , Pakroo, S. , Junior, W.J.F.L. , Luchese, R.H. , Guerra, A.F. , Rossi, R.C. et al. (2019) Probiotic potential and biofilm inhibitory activity of Lactobacillus casei group strains isolated from infant feces. J Funct Foods 54, 489–497. [Google Scholar]
- Tarrah, A. , da Silva Duarte, V. , Pakroo, S. , Corich, V. and Giacomini, A. (2020a) Genomic and phenotypic assessments of safety and probiotic properties of Streptococcus macedonicus strains of dairy origin. Food Res Int 130, 108931. [DOI] [PubMed] [Google Scholar]
- Tokatlı, M. , Gülgör, G. , Bağder Elmacı, S. , Arslankoz İşleyen, N. and Özçelik, F. (2015) In vitro properties of potential probiotic indigenous lactic acid bacteria originating from traditional pickles. Biomed Res Int 2015 315819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, I.‐K. , Wu, Y.‐Y. , Yang, Y.‐F. , Ting, I.‐W. , Lin, C.‐C. , Yen, T.‐H. , Chen, J.‐H. , Wang, C.‐H. et al. (2015) The effect of probiotics on serum levels of cytokine and endotoxin in peritoneal dialysis patients: a randomised, double‐blind, placebo‐controlled trial. Benef Microbes 6, 423–430. [DOI] [PubMed] [Google Scholar]
- Wang, P. , Li, D. , Ke, W. , Liang, D. , Hu, X. and Chen, F. (2020) Resveratrol‐induced gut microbiota reduces obesity in high‐fat diet‐fed mice. Int J Obes 44, 213–225. [DOI] [PubMed] [Google Scholar]
- Wattam, A.R. , Davis, J.J. , Assaf, R. , Boisvert, S. , Brettin, T. , Bun, C. , Conrad, N. , Dietrich, E.M. et al. (2017) Improvements to PATRIC, the all‐bacterial bioinformatics database and analysis resource center. Nucleic Acids Res. 45, D535–D542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C. , He, G. and Zhang, J. (2014) Physiological and proteomic analysis of Lactobacillus casei in response to acid adaptation. J Ind Microbiol Biotechnol 41, 1533–1540. 10.1007/s10295-014-1487-3 [DOI] [PubMed] [Google Scholar]
- Wu, C. , Zhang, J. , Wang, M. , Du, G. and Chen, J. (2012) Lactobacillus casei combats acid stress by maintaining cell membrane functionality. J Ind Microbiol Biotechnol 39, 1031–1039. 10.1007/s10295-012-1104-2 [DOI] [PubMed] [Google Scholar]
- Yang, F. , Wang, D. , Li, Y. , Sang, L. , Zhu, J. , Wang, J. , Wei, B. , Lu, C. and et al. (2017) Th1/Th2 balance and Th17/Treg‐mediated immunity in relation to murine resistance to dextran sulfate‐induced colitis. J Immunol Res, 2017 7047201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zankari, E. , Hasman, H. , Cosentino, S. , Vestergaard, M. , Rasmussen, S. , Lund, O. , Aarestrup, F.M. and Larsen, M.V. (2012) Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67, 2640–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, L. , Wang, Y. , Zhang, G. , Zhang, T. , Lou, J. and Liu, J. (2019) L‐Arabinose elicits gut‐derived hydrogen production and ameliorates metabolic syndrome in C57BL/6J mice on high‐fat‐diet. Nutrients 11, 3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Rarefaction curves of Shannon entropy (a) and phylogenetic diversity (b) of stool samples before (t0) and after (t1) 6 weeks of experimental period.
Figure S2. Relative abundance of bacterial phyla (a), top‐20 families (b), and top‐20 genera (c) identified in feces samples of BALB/c mice in groups CD, DTA81 and HFD before (t0) and after (t1) the experimental period.


