Abstract
Background
Diagnostic tests for allergy rely on detecting allergen‐specific IgE. Component‐resolved diagnostics incorporate multiple defined allergen components to improve the quality of diagnosis and patient care.
Objective
To develop a new approach for determining sensitization to specific allergen components that utilizes fluorescent protein tetramers for direct staining of IgE on blood basophils by flow cytometry.
Methods
Recombinant forms of Lol p 1 and Lol p 5 proteins from ryegrass pollen (RGP) and Api m 1 from honeybee venom (BV) were produced, biotinylated, and tetramerized with streptavidin‐fluorochrome conjugates. Blood samples from 50 RGP‐allergic, 41 BV‐allergic, and 26 controls were incubated with fluorescent protein tetramers for flow cytometric evaluation of basophil allergen binding and activation.
Results
Allergen tetramers bound to and activated basophils from relevant allergic patients but not controls. Direct fluorescence staining of Api m 1 and Lol p 1 tetramers had greater positive predictive values than basophil activation for BV and RGP allergy, respectively, as defined with receiver operator characteristics (ROC) curves. Staining intensities of allergen tetramers correlated with allergen‐specific IgE levels in serum. Inclusion of multiple allergens coupled with distinct fluorochromes in a single‐tube assay enabled rapid detection of sensitization to both Lol p 1 and Lol p 5 in RGP‐allergic patients and discriminated between controls, BV‐allergic, and RGP‐allergic patients.
Conclusion
Our novel flow cytometric assay, termed CytoBas, enables rapid and reliable detection of clinically relevant allergic sensitization. The intensity of fluorescent allergen tetramer staining of basophils has a high positive predictive value for disease, and the assay can be multiplexed for a component‐resolved and differential diagnostic test for allergy.
Keywords: allergen tetramers, basophils, component‐resolved diagnostics, flow cytometry, surface IgE
Fluorescent recombinant allergen tetramers specifically bind to and activate basophils from patients allergic to bee venom (Api m 1) and ryegrass pollen (Lol p 1 and Lol p 5). Staining intensities of recombinant Api m 1 and Lol p 1 tetramers have greater positive predictive values for allergy than basophil activation. A single flow cytometric stain with multiple allergen tetramers can be applied as a rapid component‐resolved and differential diagnostic test for allergy. Abbreviation: Strep, streptavidin

Abbreviations
- AIT
allergen‐specific immunotherapy
- Anti‐hIgE
anti‐human IgE
- AU
arbitrary units
- AUC
area under the curve
- BAT
basophil activation test
- BV
bee venom
- MFI
median fluorescence intensity
- PBMC
peripheral blood mononuclear cells
- RGP
ryegrass pollen
- ROC
receiver operator characteristics
- SPT
skin prick test
1. INTRODUCTION
Diagnosis of IgE‐mediated allergy is based on the patient's clinical symptoms and medical history, confirmed with laboratory tests. 1 The latter are becoming increasingly important for patient management, as well as for selection for and monitoring of allergen‐specific immunotherapy (AIT). Currently, skin prick testing (SPT) and detection of serum‐specific IgE using allergen extracts are standard methods to differentiate sensitized from non‐sensitized individuals. 2 While serum‐specific IgE detection is highly sensitive, the use of allergen extracts can yield positive results due to recognition by cross‐reactive antibodies of components that are not the drivers of disease. 3 , 4 , 5 , 6 , 7 , 8
Basophil activation tests (BAT), that is in vitro exposure of blood basophils to increasing doses of allergen, 9 have the advantage of detecting functional IgE that on binding allergen can directly result in degranulation and development of allergic symptoms. 10 IgE bound to FcεRI is cross‐linked by allergen and triggers signaling and basophil degranulation. Resultant surface expression of CD63 is a reliable indicator of the sensitivity and reactivity of basophils to an allergen. 9 BAT is a functional assay with advantages over serology for specific IgE in that it can more accurately diagnose allergies and monitor responses to immunotherapy. 10 However, BAT requires in vitro stimulation of multiple blood samples with serial dilutions of allergen as well as positive and negative controls, which has thus far hampered large‐scale diagnostic implementation. 8
A different approach for detection of relevant allergen sensitization is the use of molecular allergen components that are either purified from allergen extracts or produced recombinantly. Detection of IgE reactivity to major allergen components has the potential to be highly sensitive with high specificity through omission of cross‐reactive components that are less relevant for disease. 11 , 12 , 13 For example, sensitization to Der p 1 and/or Der p 2, the major house dust mite (HDM) allergens, is a better positive predictor for HDM‐driven asthma than whole HDM extract. 14 , 15 Furthermore, sensitization to Ara h 2, a major peanut allergen, is more specific for systemic allergic responses than whole peanut extract. 16 , 17 Consequently, sensitization patterns to molecular allergen components can be relevant for patient management and treatment. 18 , 19 The major ryegrass pollen (RGP) allergens Lol p 1 and Lol p 5 are each detected by serum IgE of 80–90% of RGP‐allergic individuals. 20 , 21 , 22 , 23 Importantly, Lol p 5 sensitization is associated with a higher risk of thunderstorm asthma, 24 , 25 and would be a potential indicator for AIT. 26
The major allergen component of BV is Api m 1, whereby 78–90% of people allergic to BV have Api m 1‐specific IgE in serum. 27 , 28 BV extracts used in AIT for BV allergy typically contain Api m 1 but may lack another key major allergen associated with systemic reactions, Api m 10. Patients with high sensitization to Api m 10 will not benefit from treatment with such currently available SCIT preparations, highlighting the importance of resolving both disease and treatment at the component level for accurate prescription of AIT. 5 , 19 , 29
Despite our understanding of these allergen components, the use of component‐resolved diagnostics has not become routine or standard practice. At present, these may be applied as follow‐up tests to confirm or refute an initial test using whole allergen extract. 17 Ideally, a single laboratory test would enable differential detection of allergen sensitization and stratification of patients into clinical risk groups. This would constitute a multiplex assay that can incorporate a relevant set of allergen components for, for example, aeroallergens, food allergens, or stinging insect allergens.
We have generated fluorescently labeled tetramers of the major allergen components Api m 1, Lol p 1, and Lol p 5 to explore component‐resolved diagnostics using direct staining of blood basophils in a single test—CytoBas.
2. METHODS
2.1. Study participants
Allergic subjects were recruited from the Allergy Clinics of The Alfred and Box Hill Hospitals, Melbourne, Victoria, Australia (Alfred Ethics Committee projects 509/11 and 514/13). Forty‐one BV‐allergic patients (18–68 years; 32% female) were diagnosed on the basis of a systemic allergic response to a bee sting and serum BV‐specific IgE of ≥0.35kUA/L (ImmunoCAP, Phadia, Uppsala, Sweden). Fifty RGP‐allergic patients (18–65 years; 52% female) were included with moderate to severe seasonal allergic rhinitis with or without asthma, and serum RGP‐specific IgE of ≥0.35kUA/L (ImmunoCAP). Blood from RGP‐allergic subjects was collected in April/May outside of the Australian grass pollen season (September‐December). Twenty‐six control subjects (22–58 years; 50% female) had no clinical history of BV and/or RGP allergy as appropriate for the assay, and no detectable specific IgE to the relevant allergen (Monash University project 2016–0289). Controls comprised both allergic (including 8 HDM SPT positive, 5 tree pollen SPT positive) and non‐allergic subjects. Exclusion criteria were immunodeficiency, AIT within the last five years, and treatment with continuous oral corticosteroids and/or β‐blockers. The use of symptomatic medications (incl. antihistamines and topical corticosteroids) for allergic rhinitis was permitted. The study was conducted according to the principles of the Declaration of Helsinki, and written informed consent from each participant was obtained prior to inclusion.
2.2. Blood sampling and ELISA
Heparinized blood samples were processed within 24 hours of collection for basophil activation, PBMC and serum isolation, and storage. Serum RGP‐specific IgE and BV‐specific IgE levels were measured by ImmunoCAP using allergen extracts as per manufacturer's instructions at the Alfred Pathology Services (Alfred Hospital, Melbourne, Australia). Serum Lol p 1‐ and Lol p 5‐specific IgE were measured by a semi‐quantitative in‐house ELISA as described previously. 24 , 30 , 31 Briefly, ELISA plate wells were coated with recombinant monomeric, non‐biotinylated Lol p 1 (MyBiosource, San Diego, CA, US) or Lol p 5 (see below), blocked with 5% skim milk powder in PBS, and incubated with serial dilutions of serum samples. Separate wells were incubated with a range of concentrations of purified recombinant human IgE (clone AbD18705; Bio‐Rad, Puchheim, Germany) to generate a standard curve for quantification of IgE in serum. Bound IgE was detected using polyclonal rabbit anti‐hIgE (Agilent, Santa Clara, CA, US) followed by polyclonal goat anti‐rabbit HRP (Promega, Madison, WI, US). ELISA was developed using TMB (Thermo Fisher Scientific, Waltham, MA, US) before the reaction was stopped with 1 M HCl and absorbance measured at OD 450 nm on a Multiskan Microplate Spectrophotometer (Thermo Fisher Scientific). Wells without allergens were used to determine background values that were subtracted from allergen‐specific IgE values and results are expressed in arbitrary units (AU).
2.3. Recombinant allergen production
For recombinant protein production, the protein sequences of Api m 1.0101, Lol p 1.0101, and Lol p 5.0101 were obtained from the Allergen Nomenclature website (allergen.org). 32 , 33 All three constructs were generated with the Api m 1 N‐terminal leader sequence for secretion, as well as a 6‐His tag for purification and a BirA tag for biotinylation (Figure 1A). To prevent unwanted effects of catalytic activity, mutations were introduced in Api m 1 (H67Q) and Lol p 1 (H104 V), as published previously. 34 , 35 All constructs were codon‐optimized for Spodoptera frugiperda and cloned into the pFastBac vector (Thermo Fisher Scientific), prior to incorporation into a Bacmid for baculovirus production. Bacmids were transfected into Sf21 cells, which were subsequently cultured at 27°C. Supernatants from infected Sf21 cultures were clarified by centrifugation, and the 6‐His tagged proteins were purified through retention on a cobalt column. Supernatants were gravity‐fed through a 25 ml column packed with 4 ml Talon NTA‐cobalt‐agarose beads (Clontech, Mountain View, CA, US). Beads were washed with PBS and allergens eluted with PBS, pH 8.5, containing 200 mM imidazole. Eluate was dialyzed against 10 mM TRIS, pH 7.5. The purified recombinant proteins were incubated overnight at RT with BirA enzyme for targeted biotinylation (2.5 µg/ml BirA in 10 mM TRIS containing 62.5 mM Bicine‐HCl, 12.5 mM ATP, 12.5 mM MgOAc, 62.5 µM D‐biotin). Following subsequent dialysis against PBS, tetramerization was performed with fluorochrome‐conjugated streptavidin (PE, APC, and BV711 conjugates; all from BD Biosciences, San Jose, CA, US) at a 4:1 molar ratio of allergen:streptavidin.
FIGURE 1.
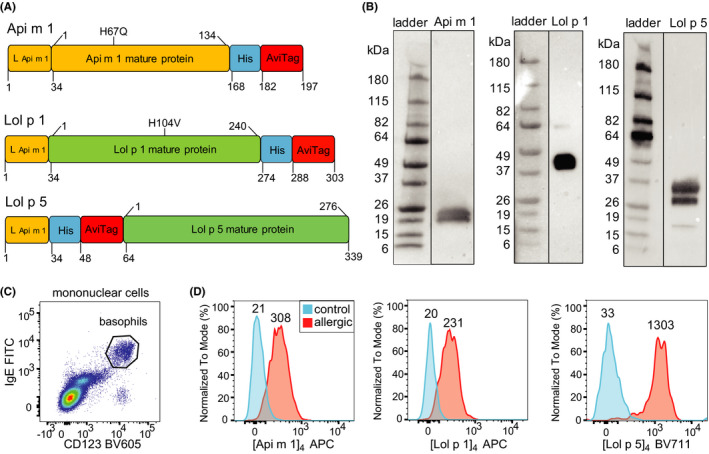
Detection of allergen sensitization by quantifying binding of fluorescent allergen components of BV and RGP to basophils. A) Schematic diagram of DNA constructs for generation of the recombinant allergens Api m 1, Lol p 1, and Lol p 5. Recombinant allergens contained mutations to render the catalytic site inactive (Api m 1 H67Q, Lol p 1 H104V). Leader peptide and spacer (L); 6‐histidine tag (His); avidin tag (AviTag). B) Western blots of recombinant allergens detected with anti‐His antibody. C) Representative flow plot for detection of circulating basophils (CD123+IgE+) within gated mononuclear cells (Figure S1). D) Histograms depicting representative fluorescent staining of Api m 1, Lol p 1, and Lol p 5 tetramers on basophils from relevant allergic patients (red) and controls (blue). Values above the histograms represent median fluorescence intensity values of the population.
2.4. Western blotting
Baculovirus‐infected Sf21 supernatants were mixed with 6X non‐reducing buffer (0.1 M Tris‐HCl, pH 6.8, 0.2% bromophenol blue, and 20% glycerol). Protein samples were then loaded onto 4–15% Mini‐PROTEAN® TGX Stain‐Free gels (Bio‐Rad) and separated at 200 V for 30 mins. Proteins were transferred onto PVDF membranes (Bio‐Rad) using the Trans‐Blot Turbo Transfer System (Bio‐Rad). Membranes were probed with mouse anti‐His (clone 27471001, GE Healthcare, Chicago, IL, US) followed by goat anti‐mouse IgG HRP (clone NA9310 N; Cell Signaling Technology, Danvers, MA, US). PVDF membranes were developed using Amersham ECL Western Blotting Detection Reagent (GE Healthcare), and chemiluminescence was detected using a ChemiDoc Imager (Bio‐Rad).
2.5. Basophil activation test
To prime circulating basophils, whole blood was incubated for 10 mins at 37°C in stimulation buffer (Hepes 20 mM, NaCl 133 mM, KCl 5 mM, CaCl2 7 mM, CaCl2 3.5 mM, BSA 1 mg/ml, rIL‐3 2 ng/ml, Heparin 20 µl/ml, pH 7.4). Basophils were incubated with RGP extract (0.001 – 1 µg/ml; Stallergenes Greer, London, UK), BV extract (0.01, 0.1, 1 µg/ml; HollisterStier Allergy, Spokane, WA, US), fluorescent allergen tetramers (0.01, 0.1,1 µg/ml), or streptavidin conjugate only (1 µg/ml) for 20 mins at 37°C. In parallel, stimulations with rabbit anti‐human IgE (0.1–10 µg/ml; Agilent) or fMLP (8 nM; Sigma, St. Louis, MO, US) were performed to control for FcεRI‐dependent and FcεRI‐independent degranulation, respectively. 36 Activation was stopped by incubating on ice for 5 mins. Cells were washed with cold wash buffer (Hepes 20 mM, NaCl 133 mM, KCl 5 mM, EDTA 0.27 mM, pH 7.3) prior to flow cytometry.
2.6. Flow cytometry
Flow cytometry was performed on blood cells after one of three types of processing: whole blood, washed blood, or PBMC. Whole blood was directly incubated with fluorescent antibodies and allergen tetramers followed by red blood cell lysis with NH4Cl. Washed blood was first subjected to 2 PBS washes prior to incubation with fluorescent reagents. Post‐Ficoll PBMC were either used fresh or stored in liquid nitrogen with 10% DMSO prior to incubation with fluorescent reagents and flow cytometry. To acid strip IgE from the surface of basophils, fresh PBMC were twice incubated in acetate buffer (0.05 M acetate, 0.085 M NaCl, 0.01 M EDTA, 0.03% BSA, pH 4) for 5 min at 4°C, followed by neutralization with 0.2% BSA in PBS as previously described. 37
Flow cytometry on whole or washed blood samples used for BAT with a single allergen tetramer was performed with CD123‐BV605 (6H6; BioLegend, San Diego, CA, US), CD63‐PE (H5C6; BD Biosciences), and either IgE‐FITC (polyclonal, Thermo Fisher Scientific) or HLA‐DR‐APC (L243; BioLegend). Flow cytometry on thawed or fresh PBMC for multiplex allergen staining was performed with CD123‐BV605, IgE‐FITC, and HLA‐DR‐APC for 15 min at RT in the dark. 7AAD was added to samples immediately prior to flow cytometry to distinguish between live and dead cells (Figure S1). Cell acquisition was performed across two instruments (4‐laserBD LSRII and 5‐laserBD LSRFortessa X‐20) with a nearly identical set‐up for the shared 4 lasers. Instrument set‐up and calibration were performed using standardized EuroFlow SOPs as described in detail previously. 38 , 39
2.7. Statistical analysis
All flow cytometry data were analyzed with FACS DIVA v8.0.1 (BD Biosciences) and FlowJo v10 software packages (FlowJo LLC, Ashland, OR, US). Statistical analyses were performed using GraphPad Prism (v8.4.1): the non‐parametric Wilcoxon signed‐rank test for paired data and the non‐parametric Mann‐Whitney U‐test for unpaired data. Receiver operating characteristic (ROC) curves were used to determine specificity and sensitivity of BAT or CytoBas for distinguishing between control and allergic patients. For all tests, p < 0.05 was considered significant.
3. RESULTS
3.1. Design and production of recombinant allergen tetramers for CytoBas
Immunodominant allergen components of BV and RGP were selected for generation of recombinant allergen tetramers (Api m 1, Lol p 1, and Lol p 5). 20 , 21 , 22 , 27 , 28 The full‐length Api m 1 protein was produced with the native leader sequence, and C‐terminal 6‐His and BirA tags for purification and biotinylation (Figure 1A). The construct contained a H67Q mutation to prevent catalytic activity without affecting IgE reactivity. 34 Similarly, the Lol p 1 and Lol p 5 constructs were generated with the Api m 1 leader sequence to target these for secretion. Lol p 1 catalytic activity was abolished by a H104V mutation. 35 Purified allergens were probed by Western blot using an anti‐6‐His antibody and detected at expected sizes with typical diversity in glycosylation density (Figure 1B). Recombinant allergens were biotinylated and tetramerized with a streptavidin‐APC conjugate to generate Api m 1, Lol p 1, and Lol p 5 tetramers.
The ability of allergen tetramers to detect specific IgE on basophils using flow cytometry was assessed using whole blood samples of allergic patients and controls. Following 15 min incubation with the relevant allergen tetramer and mAbs, binding of allergen to CD123+IgE+ basophils was examined (Figure 1C). The Api m 1 tetramer ([Api m 1]4‐APC) specifically stained basophils from a BV‐allergic individual and not a control (Figure 1D). Similarly, [Lol p 1]4‐APC and [Lol p 5]4‐APC each positively stained basophils from an RGP‐allergic patient, and not a control. Thus, these recombinant allergen tetramers can be used to detect allergen sensitization by binding specific IgE on the surface of basophils using flow cytometry (CytoBas).
3.2. Recombinant Api m 1 tetramer staining detects functional allergic sensitization to bee venom
To determine whether positive staining of basophils with [Api m 1]4 indicates functional allergic sensitization to BV, blood samples from 41 BV‐allergic patients and 24 controls were incubated for 20 min at 37°C with increasing amounts of [Api m 1]4‐APC or, as a control, 1 µg/ml streptavidin‐APC. Samples were then stained with mAbs to detect basophils, and within this population, expression of CD63 was assessed as a marker of activation and degranulation. There was a dose‐dependent increase in staining of basophils following incubation with [Api m 1]4‐APC in all 41 patients with BV allergy (Figure 2A). In contrast, there was no increase in staining with [Api m 1]4‐APC over the streptavidin‐APC reagent on basophils from the control cohort (Figure 2A). Thus, the specific binding of allergen tetramer to basophils of a BV‐allergic patient was accompanied by a dose‐dependent increase in the frequency of basophils with surface expression of CD63 (Figure 2B). The frequency of CD63+basophils following incubation with [Api m 1]4‐APC was negligible for control subjects. Similarly, parallel incubation of blood samples with BV extract resulted in CD63 expression only on basophils of BV‐allergic patients and not controls (Figure 2C). Basophils from all BV‐allergic and control subjects expressed surface CD63 following incubation with positive controls anti‐IgE and/or fMLP, indicating that basophils in all blood samples were functionally capable of degranulation (Suppl. Figure S2A, B). Thus, the CytoBas approach with [Api m 1]4 has the capacity to detect relevant and functional allergic sensitization to BV.
FIGURE 2.
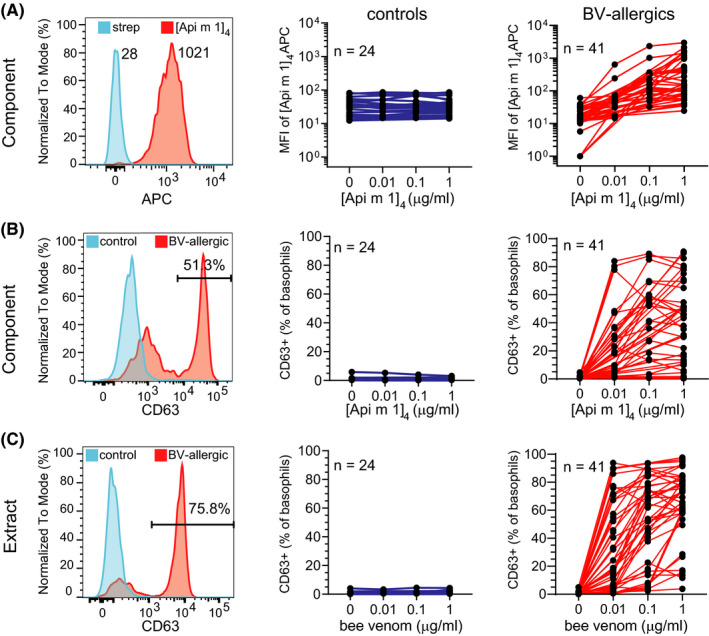
Recombinant Api m 1 binds basophils from BV‐allergic patients in a dose‐dependent manner and induces basophil activation. A) Representative staining of basophils from blood of a BV‐allergic patient stimulated with [Api m 1]4‐APC (red) or streptavidin‐APC (blue)—left plot. Median fluorescence intensity (MFI) of basophils from controls (blue; middle) and BV‐allergic subjects (red; right) following incubation with 1 µg/ml streptavidin‐APC (“0”) or 0.01, 0.1, and 1 µg/ml [Api m 1]4‐APC. B) Representative histogram of CD63 staining on basophils (left), and frequencies of basophils from controls (middle) and BV‐allergic patients (right) expressing CD63 following stimulation with streptavidin‐APC or increasing concentrations of [Api m 1]4‐APC. C) Representative histogram of CD63 staining on basophils (left), and frequencies of basophils from controls (middle) and BV‐allergic patients (right) expressing CD63 following stimulation with increasing concentrations of BV extract.
3.3. Recombinant Lol p 1 tetramer staining detects functional allergic sensitization to ryegrass pollen
The ability of [Lol p 1]4 to detect functional allergen sensitization was examined using blood samples from 50 RGP‐allergic patients and 20 controls. Following incubation with increasing amounts of [Lol p 1]4‐APC or control streptavidin‐APC, basophils of patients and controls were examined for allergen binding (Figure 3A) and activation (CD63 positivity; Figure 3B). Incubation with [Lol p 1]4‐APC resulted in a dose‐dependent increase in signal observed in all 50 patients with RGP allergy (Figure 3A). The increase in staining intensity indicated functional allergen binding, as this was accompanied by increased frequencies of CD63+ basophils in the patient samples (Figure 3B). In contrast, basophils in the control cohort did not show any increase of allergen staining over the strep‐APC reagent (Figure 3A). There was negligible CD63 expression on basophils of controls. Similarly, parallel incubations of blood with RGP extract only resulted in a significant increase in the fraction of CD63 expressing basophils of patients and not controls (Figure 3C). Activation by anti‐IgE resulted in large fractions of basophils expressing CD63 for most patients and controls. Activation with fMLP resulted in a large fraction of basophils expressing CD63 in all patients, indicating that these cells were functionally capable of degranulation (Figure S3A, B). Thus, as for BV allergy, the CytoBas approach can be utilized for RGP allergy with a major recombinant allergen component, Lol p 1, to detect relevant and functional allergic sensitization.
FIGURE 3.
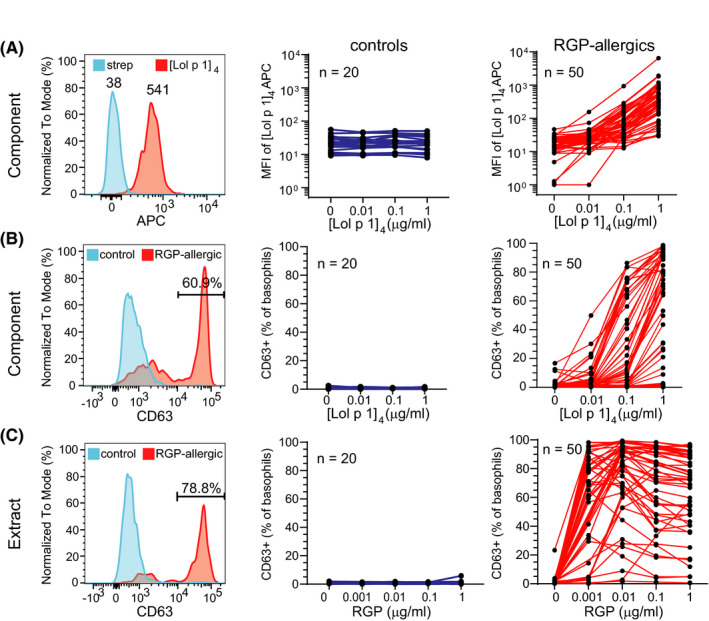
Recombinant Lol p 1 binds basophils from RGP‐allergic patients in a dose‐dependent manner and induces basophil activation. A) Left plot, representative staining of basophils from blood of a RGP‐allergic patient stimulated with [Lol p 1]4‐APC (red) or streptavidin‐APC (blue). Median fluorescence intensity (MFI) of basophils from controls (blue; middle) and RGP‐allergic subjects (red; right) following incubation with 1 µg/ml streptavidin‐APC (“0”) or 0.01, 0.1, and 1 µg/ml [Lol p 1]4‐APC.B) Representative histogram of CD63 staining on basophils (left), and frequencies of basophils from controls (middle) and RGP‐allergic patients (right) expressing CD63 following stimulation with streptavidin‐APC or increasing concentrations of [Lol p 1]4. C) Representative histogram of CD63 staining on basophils (left), and frequencies of basophils from controls (middle) and RGP‐allergic patients (right) expressing CD63 following stimulation with increasing concentrations of RGP extract.
3.4. Allergen tetramers bind the total basophil population in sensitized individuals irrespective of activation
Conventional understanding of basophils capturing polyclonal IgE via their FcεRI suggests that all basophils of an allergic patient have the potential to bind IgE with specificity to an allergen molecule. However, typically only a fraction of the basophils will degranulate and express CD63 following stimulation with that allergen. To examine whether the CD63+ basophils differed from CD63– basophils in terms of allergen‐binding specificity and capability, the staining intensities of the allergen tetramers were re‐examined following stratification of basophils based on CD63 expression after in vitro stimulation (Figures 2B, 3B). Both the CD63– and the CD63+basophils from all 41 BV‐allergic individuals showed strong staining with [Api m 1]4‐APC as compared to streptavidin‐APC. Importantly, the CD63– and CD63+fractions bound similar amounts of Api m 1 (Figure S4A). Similarly, [Lol p 1]4‐APC showed high degrees of binding to both CD63+ and CD63– basophils from the 50 RGP‐allergic individuals, albeit with slightly but significantly lower levels on the CD63– subset (Figure S4B). Thus, in sensitized individuals, all basophils may have surface‐bound allergen‐specific IgE, but not all will degranulate upon binding to allergen.
3.5. Greater diagnostic ability of basophil staining than activation with recombinant allergen tetramers
The BV‐allergic and RGP‐allergic individuals in this study were diagnosed based on their clinical history, SPT, and/or allergen‐specific serum IgE, as per clinical standards. To compare the predictive values of the BAT and CytoBas for allergy, ROC of their outcomes were determined. For each individual, the highest CD63+frequency was selected from the serial dilutions of allergen tetramer (Figure 2B; Figure 3B). CD63 positivity following stimulation with [Api m 1]4 was<10% for all control subjects and >10% in 40 of 41 BV‐allergic individuals (Figure 4A). Incubation with [Lol p 1]4 resulted in <5% of basophils becoming CD63+in all controls and >5% in 42/50 RGP‐allergic patients (Figure 4B).
FIGURE 4.
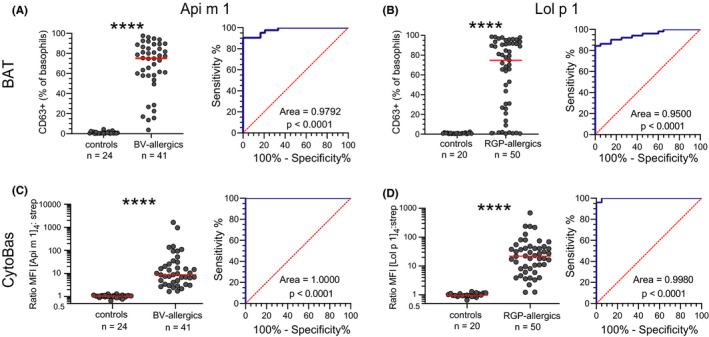
Sensitivity and specificity of BAT and CytoBas for allergy. A) Frequencies of CD63+ basophils following in vitro stimulation with [Api m 1]4 (left) and receiver operator characteristics (ROC) curve of CD63 positivity (right). For each individual, the highest frequency was selected from the serial dilutions in Figure 2B. B) Frequencies of CD63+ basophils following in vitro stimulation with [Lol p 1]4 and ROC curve of CD63 positivity (right). For each individual, the highest frequency was selected from the serial dilutions in Figure 3B. C) Staining intensities as ratio of [Api m 1]4‐APC/streptavidin‐APC on basophils (left) and ROC curve of [Api m 1]4 staining intensity (right). D) Staining intensities as ratio of [Lol p 1]4‐APC/streptavidin‐APC on basophils (left) and ROC curve of [Lol p 1]4 staining intensity (right). Statistics for CD63+ frequencies and allergen tetramer stains, Mann‐Whitney U test; for ROC curves, Wilson/Brown method to test whether the confidence level of the outcome distribution is greater than 95%. **** p < 0.0001.
Staining intensities with the allergen tetramers were determined by calculating the ratio of MFI from staining with 1 µg/ml tetramer‐APC over 1 µg/ml streptavidin‐APC. This MFI ratio was <2 for all control subjects stained with [Api m 1]4‐APC, whereas all 41 patients with BV allergy had a ratio >2 (Figure 4C). Furthermore, the MFI ratios for all patients were higher than for any of the controls, resulting in an area under the curve (AUC) in the ROC curve of 1.0000. This was slightly higher than the AUC for the ROC curve of the [Api m 1]4 BAT (0.9792). Similarly, all control subjects showed a ratio <2 for [Lol p 1]4‐APC staining, whereas 48/50 RGP‐allergic patients had a ratio >2 (Figure 4D). As a result, the AUC for the ROC curve (0.9980) was substantially higher than that of the ROC curve for the BAT of [Lol p 1]4 (0.9500). Basophils of control subjects with sensitization to HDM or tree pollens did not display staining with the BV and RGP allergen tetramers. In conclusion, the CytoBas approach with single allergens showed high specificity and sensitivity for BV (Api m 1) and RGP (Lol p 1) allergies with improved performance over BAT.
To investigate the impact of serum allergen‐specific immunoglobulin on allergen tetramer staining, we washed blood to remove serum prior to conducting CytoBas. Removing serum had negligible effect on [Api m 1]4‐ APC or [Lol p 1]4‐ APC staining intensities of basophils from BV‐ or RGP‐allergic subjects, respectively (Figure S5). Serum allergen‐specific immunoglobulin, therefore, has minimal effect on basophil staining with allergen tetramers.
3.6. Component‐resolved differential diagnosis approach with a single flow cytometry assay
To enhance the component‐resolved diagnostic performance of our CytoBas technique for RGP allergy, multiplex flow cytometry was conducted with [Lol p 1]4‐APC and [Lol p 5]4‐BV711 tetramers in a single tube. Basophils in thawed PBMC from an allergic patient were successfully discriminated from those of a control by a higher MFI for one or both allergen tetramers (Figure 5A–C). Multiplex flow cytometry on thawed PBMC with both RGP allergen tetramers in a single tube exhibited clear resolution between a series of RGP‐allergic patients and controls (Figure 5D).
FIGURE 5.
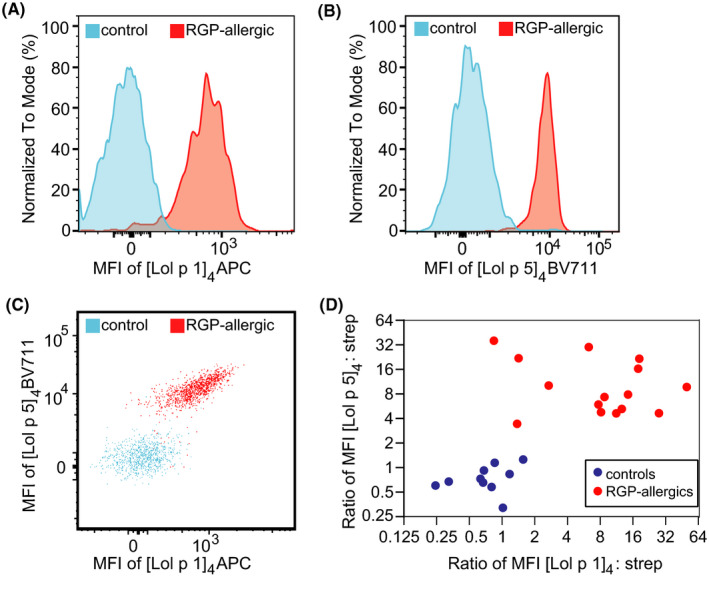
Multiplex flow cytometry using multiple allergen tetramers in a single tube as a component‐resolved diagnostic test for allergy. A, B) Representative histograms of basophils from thawed PBMC of a control (blue) or an RGP‐allergic patient (red) stained with Lol p 1 and Lol p 5 tetramers in a single tube. C) 2D dot plot of Lol p 1 and Lol p 5 tetramer staining of the basophils from the control and patient in panels A and B. D) Scatterplot depicting MFI ratios of allergen:streptavidin for [Lol p 1]4‐APC vs strep‐APC and [Lol p 5]4‐BV711 vs strep‐BV711 on basophils from controls (blue, n = 10) and RGP‐allergic subjects (red, n=15) stained with both allergen tetramers or both streptavidin conjugates in a single tube.
To test for discrimination between multiple allergic sensitizations, Api m 1, Lol p 1, and Lol p 5 tetramers, each labeled with a different fluorochrome, were included simultaneously in a single multiplex flow cytometric test. Since the MFI of the allergen tetramers on pDCs was shown to be similar to the MFI of streptavidin control conjugates on basophils, pDCs provided a suitable background control in blood samples for multiplex flow cytometry (Figure S6). Basophils from thawed PBMC of RGP‐allergic patients demonstrated selective binding to only [Lol p 1]4 or [Lol p 5]4 tetramers and not [Api m 1]4, while basophils from a BV‐allergic patient only bound [Api m 1]4 (Figure 6A). The MFI of staining on basophils for each of the three allergens correlated strongly with serum allergen‐specific IgE (each p < 0.0001; Figure 6B‐D). This is in contrast to the frequency of activated (CD63+) basophils stimulated with Api m 1 and Lol p 1 tetramers, which showed a poorer correlation with Api m 1‐specific IgE (r2 = 0.03561, p = 0.2436) or Lol p 1‐specific IgE (r2 = 0.1313, p = 0.0114), respectively. To further investigate the impact of surface IgE on allergen staining, we acid‐stripped IgE from the surface of basophils before conducting multiplex flow cytometry with [Api m 1]4‐ PE, [Lol p 1]4‐ BV711, and [Lol p 5]4‐ BV480 on PBMC from subjects with BV allergy, RGP allergy, or controls. Acid‐stripping successfully removed IgE from the basophil surface (Figure S7), and in parallel, detection of allergen tetramer staining was lost (Figure S7). The CytoBas technique, therefore, detects allergen sensitization with greater accuracy than basophil activation. Taken together, these data demonstrate that a single flow cytometric assay with multiple allergen tetramers can provide a rapid component‐resolved and differential diagnostic test for allergy.
FIGURE 6.
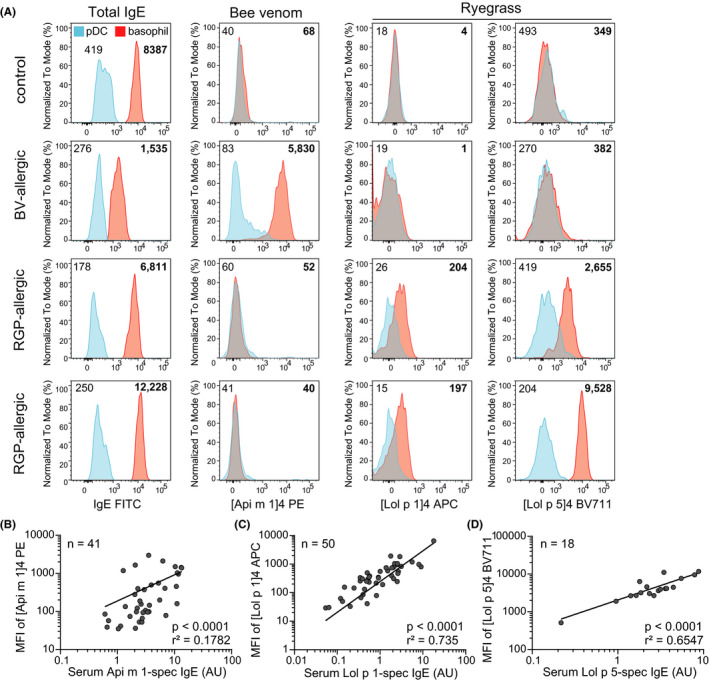
Multiplex flow cytometry distinguishes between different allergic sensitizations with a single tube. A) Representative histograms of [Lol p 1]4, [Lol p 5]4, or [Api m 1]4 MFI on pDCs (blue) and basophils (red) from thawed PBMC of controls, BV‐allergic and RGP‐allergic patients. Numbers accompanying histograms denote MFI (non‐bolded indicate pDCs; bolded indicate basophils). Scatterplots depicting correlation between serum levels of allergen‐specific IgE and MFI of allergen tetramers on basophils stained with B) Api m 1 C) Lol p 1 and D) Lol p 5 allergen tetramers. AU, arbitrary units. Significance and r‐squared (r2) of correlation determined by linear regression.
4. DISCUSSION
We describe here a novel methodology for a component‐resolved diagnostic test for allergy using multiplex flow cytometry involving direct staining of basophils with recombinant allergen tetramers—CytoBas. This flow cytometric approach enabled rapid detection with high sensitivity and specificity of BV allergy with Api m 1 and of RGP allergy with Lol p 1. Combined staining of Api m 1 and Lol p 1 reliably enabled differential detection of allergen sensitization, and inclusion of both Lol p 1 and Lol p 5 facilitated quantification of the relative sensitization to distinct components of RGP in a single‐tube test. The combination of multiple allergens from a single species provided complete discrimination between RGP‐allergic patients and controls, highlighting the utility of multiplex CytoBas in diagnosing allergies that exhibit sensitization to multiple allergen components.
The allergens in our study were recombinantly produced proteins. This approach facilitates the introduction of mutations that disrupted enzymatic function and the addition of protein tags for biotinylation and purification. Api m 1, Lol p 1, and Lol p 5 contain glycan structures. 40 , 41 Because the glycosylation patterns of proteins in invertebrates and plants are more similar to each other and differ strongly from mammals, 42 , 43 , 44 we here produced their recombinant forms in the Sf21 insect cell line. Similar to a previous report utilizing Sf9 cells, 45 we obtained two main glycosylated products of the recombinant Api m 1, fitting with the native paucimannosidic type of glycosylation. 44 Both recombinant RGP allergens had greater sizes than predicted for non‐glycosylated variants, and showed high sensitivity and specificity for detection of RGP allergy. The high sensitivity and specificity of our recombinant allergen components for confirmation of BV and RGP allergies indicate that our approach preserves the major epitopes for IgE binding. The fact that invertebrate‐ and plant‐derived proteins can be generated as such in an insect cell line, as confirmed here, demonstrates the potential to apply our CytoBas assay for detection of sensitization to a huge diversity of food‐, aero‐, and stinging insect‐derived allergens.
Our CytoBas study exploits the concept that basophils present high levels of IgE on their surface through FcεRI binding, and that this IgE is polyclonal and reflective of the specificities produced in an individual. This is the same concept that is utilized for BAT, even though there is typically a fraction of cells that does not degranulate and express CD63 after allergen stimulation. Our direct staining approach showed that in the case of allergen sensitization, all basophils become positive with the population showing a Gaussian distribution of staining intensity. The median staining intensities correlated strongly with allergen‐specific serum IgE levels, and so could be used in a similar manner for quantitating the response. Rather than using absolute MFI values, we propose to calculate the staining index over a known negative cell population in the same tube, that is, plasmacytoid dendritic cells (Figure 6) to obtain a normalized value for each individual.
RGP‐allergic patients with Lol p 1 and Lol p 5 co‐sensitization showed double‐positive basophil populations. This is in contrast to a recent study where distinct basophil subsets were positive for either recombinant Api m 1 or recombinant Api m 2 conjugated to fluorescent quantum dots (Qdots). 46 The reason for this difference is unclear, as it is unlikely that within an individual, distinct basophil populations will present unique allergen specificities. Therefore, it will be important for applications of multicolor flow cytometry to incorporate appropriate controls and establish standardized instrument settings for reproducible and robust measurements. 38 , 39
Another novel observation from this study is that degranulated basophils (CD63+) bind allergen tetramers with similar intensity as basophils that have not degranulated (CD63–) in the same assay. Typically, the proportion of basophils that degranulate in a BAT forms a bell‐shaped curve with increasing concentrations of allergen. The maximum frequency of CD63+cells will differ between individuals, and the dose‐response curve is thought to be affected by the affinity of IgE to the antigen, the density of the epitope‐specific IgE on the cell surface, and the functional capacity of the basophil itself. 47 , 48 , 49 Our observation argues against differences in basophils from the same individual with regards to IgE affinity or density of epitope‐specific IgE. Rather, intrinsic characteristics of the basophil, for example, signaling threshold, degranulation capacity, and maturity, could underlie the differences between those cells that do degranulate and the fraction of cells that does not.
Our allergens were tetramerized using streptavidin‐fluorochrome conjugates. Incubation of basophils with the streptavidin‐fluorochrome conjugates only did not stain basophils of any control or allergic subject. Thus, these conjugates were not recognized by IgE and did not result in false‐positive signals. A previous study has reported that activated basophils may have the capacity to bind streptavidin via positively charged molecules that are expressed on the basophil surface upon degranulation. 50 This could potentially lead to non‐specific binding of allergen tetramers to degranulated basophils. In our experiments, we did not observe such an effect and the fluorescence intensities of our allergen tetramers were similar between basophils that had degranulated and those that had not. Still, it is recommended for multiparameter analysis of allergen binding to perform incubations for staining at room temperature or even at 4ºC to minimize the potential for activation and degranulation of basophils that might mediate non‐specific binding of streptavidin.
The grass pollen‐allergic patients in our study were sampled outside of the pollen season to avoid any confounding effects of pollen exposure on basophil activation or binding to grass pollen allergens. During the pollen season, serum‐specific IgE as well as FcεRI‐IgE complexes on the surface of basophils tend to be increased. 51 , 52 , 53 While increased surface IgE on basophils potentially increases the sensitivity of CytoBas, serum IgE could block binding. Similarly, allergen‐specific IgG has been observed to block binding of IgE to allergen in plate and chip‐based methods of detecting allergen‐specific IgE. 54 , 55 While the effects of allergen‐specific Ig in serum will need to be examined experimentally, it is recommended to perform CytoBas on washed cells to maximize the sensitivity of the assay by avoiding potential blocking effects of serum IgE or IgG.
Using a multiparameter staining approach with three allergen components, we demonstrated that CytoBas enabled detection of sensitization to multiple components of the same allergen (RGP; Lol p 1 and Lol p 5) as well as differential sensitization to distinct allergens (BV; Api m 1 vs RGP; Lol p 1 and/or Lol p 5). This differential and component‐resolved analysis through staining of basophils has several advantages over BAT: 1) The CytoBas assay only requires direct staining with allergen tetramers and mAbs without the need for in vitro stimulation, making it less dependent on maintaining basophil function ex vivo, less labor‐intensive and easier to standardize; 2) Sensitization to multiple different allergen components can be assessed in a single CytoBas staining. This contrasts with BATs for which each single component has to be evaluated separately and ideally in multiple dilution steps; 3) CytoBas had a greater sensitivity and specificity than BAT for detection of BV and RGP allergy; 4) Basophil staining with recombinant allergen tetramers is possible on fresh whole blood samples (Figures 2 and 3), as well as on cryopreserved PBMC (Figures 5 and 6) enabling more flexibility for transport or batch analysis of clinical samples than is currently possible with the BAT, which requires processing of fresh blood within 24 hrs 56 ; 5) In up to 7% of individuals, basophils do not activate upon FcεRI‐mediated stimulation, 57 , 58 , 59 and these cannot be evaluated using BAT. As antigen binding occurs irrespective of activation, the CytoBas does not have this limitation.
A current limitation of CytoBas is the availability of recombinant allergens with the correct modifications for tetramerization with fluorescently labeled streptavidin. However, with the potential for use of insect cells to generate plant and invertebrate proteins, and the vast experience with recombinant proteins in serology‐based component‐resolved diagnostics, this will only be a temporary hurdle. Furthermore, an important consideration in generating new allergen tetramers will be the fluorochrome selection. In the current study, [Lol p 5]4‐BV711 exhibited approximately one log higher MFI than [Lol p 1]4‐APC despite prior reports of only slightly higher serum IgE titers for Lol p 5 compared to Lol p 1. 24 This is likely due to the greater staining index of the BV711 fluorochrome than that of APC. As each recombinant protein is likely to behave differently in the CytoBas assay, it will be important to experimentally determine the optimal fluorochrome choice for maximum positive discriminative capacity.
CytoBas with Lol p 1 tetramers exhibited greater sensitivity and specificity than previously reported for clinical history, SPT, and allergen‐specific IgE in allergic rhinitis patients sensitized to pollen aeroallergens. 60 This positive predictive value was defined with a control group that had no symptoms and was not sensitized to RGP as defined by SPT or BAT. Potentially, individuals without symptoms can be sensitized to RGP. Therefore, similar to other laboratory tests for allergen sensitization, CytoBas results should be interpreted in the clinical context.
With the potential for multiplex analysis in a single assay, CytoBas has the potential advantage over serology‐based IgE tests by being a faster diagnostic assay and to simultaneously detect functional allergen‐specific IgE bound to the surface of basophil effector cells, rather than detecting allergen‐specific IgE in serum. Theoretically, a combination of 2–4 major allergen components could be applied to a) detect allergen reactivity in a highly sensitive manner; and b) detect specific sensitization and risk of systemic response, without the need for repeat tests that are currently recommended for, for example, peanut allergy. 17 In the current study, it was not explored whether CytoBas also has the potential to monitor immunological changes induced by allergen immunotherapy. We did not observe an effect of removing serum on allergen staining on patient basophils at diagnosis. AIT monitoring studies might, therefore, require staining with lower concentrations of fluorescent tetramers than used here for diagnosis (1 µg/ml) to maximize the effect of AIT‐induced increases of serum allergen‐specific serum Ig on CytoBas staining intensities. In practice, the composition of a CytoBas assay and its performance will need to be tested and compared to the standard diagnostic workflow for each diagnostic process. The availability of high‐end clinical cytometers that allow 10 or more components to be multiplexed in one assay provides the potential for CytoBas to provide a rapid component‐resolved diagnostic test for allergy while avoiding allergen challenge.
CONFLICT OF INTEREST
MCvZ, ROH, and PMH are inventors on a patent application related to this work. All the other authors declare that they have no relevant conflicts of interest.
Supporting information
Supplementary Material
ACKNOWLEDGMENTS
The authors gratefully acknowledge Kirsten Deckert, Sara Bullen, Anita Hazard, Monique Dols, and Anna Mackay for collection of clinical data and blood samples from patients, and Adam Nelson and Rosalyn Cao for technical support. We thank Dr. Lucy Sullivan, Ms Sandra Verschoor, and Prof Joseph Trapani for sharing of reagents, and the AMREP Flow Cytometry Core Facility team for technical assistance.
Funding information
The studies were supported financially by a NHMRC Senior Research Fellowship GNT1117687 to MCvZ, a Central Clinical School Early Career Fellowship to CIM, and by NHMRC project grant GNT145303 to PMH, REO’H, BDW.
REFERENCES
- 1. van Zelm MC, McKenzie CI, Varese N, Rolland JM, O'Hehir RE. Recent developments and highlights in immune monitoring of allergen immunotherapy. Allergy 2019;74(12):2342‐2354. [DOI] [PubMed] [Google Scholar]
- 2. Valenta R, Karaulov A, Niederberger V, et al. Allergen extracts for in vivo diagnosis and treatment of allergy: is there a future? J Allergy Clin Immunol Pract. 2018;6(6):1845‐1855.e1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Carnés J, Iraola V, Cho SH, Esch RE. Mite allergen extracts and clinical practice. Ann Allergy Asthma Immunol 2017;118(3):249‐256. [DOI] [PubMed] [Google Scholar]
- 4. Codina R, Lockey RF. Pollen used to produce allergen extracts. Ann Allergy Asthma Immunol 2017;118(2):148‐153. [DOI] [PubMed] [Google Scholar]
- 5. Frick M, Fischer J, Helbling A, et al. Predominant Api m 10 sensitization as risk factor for treatment failure in honey bee venom immunotherapy. J Allergy Clin Immunol Pract. 2016;138(6):1663‐1671.e9. [DOI] [PubMed] [Google Scholar]
- 6. Rolland JM, Varese NP, Abramovitch JB, et al. Effect of heat processing on IgE reactivity and cross‐reactivity of tropomyosin and other allergens of Asia‐pacific Mollusc species: identification of novel Sydney rock oyster tropomyosin Sac g 1. Mol Nutr Food Res 2018;62(14):1800148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vieths S, Hoffmann A, Holzhauser T, Muller U, Reindl J, Haustein D. Factors influencing the quality of food extracts for in vitro and in vivo diagnosis. Allergy 1998;53(46 Suppl):65‐71. [DOI] [PubMed] [Google Scholar]
- 8. Uyttebroek AP, Sabato V, Faber MA, et al. Basophil activation tests: time for a reconsideration. Exp Rev Clin Immunol 2014;10(10):1325‐1335. [DOI] [PubMed] [Google Scholar]
- 9. MacGlashan DW. Basophil activation testing. J Allergy Clin Immunol Pract. 2013;132(4):777‐787. [DOI] [PubMed] [Google Scholar]
- 10. Hoffmann HJ, Santos AF, Mayorga C, et al. The clinical utility of basophil activation testing in diagnosis and monitoring of allergic disease. Allergy 2015;70(11):1393‐1405. [DOI] [PubMed] [Google Scholar]
- 11. Bilò MB, Ollert M, Blank S. The role of component‐resolved diagnosis in Hymenoptera venom allergy. Curr Opin Allergy Clin Immunol 2019;19(6). [DOI] [PubMed] [Google Scholar]
- 12. Luengo O, Cardona V. Component resolved diagnosis: when should it be used? Clinical and Translational Allergy 2014;4(1):28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stringari G, Tripodi S, Caffarelli C, et al. The effect of component‐resolved diagnosis on specific immunotherapy prescription in children with hay fever. J Allergy Clin Immunol. 2014;134(1):75‐81.e72. [DOI] [PubMed] [Google Scholar]
- 14. Huang HJ, Resch‐Marat Y, Rodriguez‐Dominguez A, et al. Underestimation of house dust mite‐specific IgE with extract‐based ImmunoCAPs compared with molecular ImmunoCAPs. J Allergy Clin Immunol. 2018;142(5):1656‐1659.e9. [DOI] [PubMed] [Google Scholar]
- 15. Huang Y, Wang C, Lin X, et al. Association between component‐resolved diagnosis of house dust mite and efficacy of allergen immunotherapy in allergic rhinitis patients. Clin Transl Allergy. 2019;9:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dang TD, Tang M, Choo S, et al. Increasing the accuracy of peanut allergy diagnosis by using Ara h 2. J Allergy Clin Immunol. 2012;129(4):1056‐1063. [DOI] [PubMed] [Google Scholar]
- 17. Lieberman JA, Glaumann S, Batelson S, Borres MP, Sampson HA, Nilsson C. The utility of peanut components in the diagnosis of IgE‐mediated peanut allergy among distinct populations. J Allergy Clin Immunol Pract. 2013;1(1):75‐82. [DOI] [PubMed] [Google Scholar]
- 18. Treudler R, Simon JC. Overview of component resolved diagnostics. Curr Allergy Asthma Rep. 2013;13(1):110‐117. [DOI] [PubMed] [Google Scholar]
- 19. Köhler J, Blank S, Müller S, et al. Component resolution reveals additional major allergens in patients with honeybee venom allergy. J Allergy Clin Immunol. 2014;133(5):1383‐1389.e1386. [DOI] [PubMed] [Google Scholar]
- 20. Matthiesen F, Løwenstein H. Group V allergens in grass pollens. II. Investigation of group V allergens in pollens from 10 grasses. Clin Exp Allergy 1991;21(3):309‐320. [DOI] [PubMed] [Google Scholar]
- 21. Ong EK, Knox RB, Singh MB. Mapping of the antigenic and allergenic epitopes of Lol p VB using gene fragmentation. Mol Immunol 1995;32(4):295‐302. [DOI] [PubMed] [Google Scholar]
- 22. Tamborini E, Faccini S, Lidholm J, et al. Biochemical and immunological characterization of recombinant allergen Lol p 1. Eur J Biochem 1997;249(3):886‐894. [DOI] [PubMed] [Google Scholar]
- 23. Niederberger V, Laffer S, Froschl R, et al. IgE antibodies to recombinant pollen allergens (Phl p 1, Phl p 2, Phl p 5, and Bet v 2) account for a high percentage of grass pollen‐specific IgE. J Allergy Clin Immunol. 1998;101(2 Pt 1):258‐264. [DOI] [PubMed] [Google Scholar]
- 24. Hew M, Lee J, Varese N, et al. Epidemic thunderstorm asthma susceptibility from sensitization to ryegrass (Lolium perenne) pollen and major allergen Lol p 5. Allergy 2020;75(9):2369‐2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Knox RB. Grass pollen, thunderstorms and asthma. Clin Exp Allergy. 1993;23(5):354‐359. [DOI] [PubMed] [Google Scholar]
- 26. O'Hehir RE, Varese NP, Deckert K, et al. Epidemic thunderstorm asthma protection with five‐grass pollen tablet sublingual immunotherapy: a clinical trial. Am J Respir Crit Care Med. 2018;198(1):126‐128. [DOI] [PubMed] [Google Scholar]
- 27. Müller U, Schmid‐Grendelmeier P, Hausmann O, Helbling A. IgE to recombinant allergens Api m 1, Ves v 1, and Ves v 5 distinguish double sensitization from crossreaction in venom allergy. Allergy 2012;67(8):1069‐1073. [DOI] [PubMed] [Google Scholar]
- 28. Müller UR, Johansen N, Petersen AB, Fromberg‐Nielsen J, Haeberli G. Hymenoptera venom allergy: analysis of double positivity to honey bee and Vespula venom by estimation of IgE antibodies to species‐specific major allergens Api m1 and Ves v5. Allergy 2009;64(4):543‐548. [DOI] [PubMed] [Google Scholar]
- 29. Jakob T, Müller U, Helbling A, Spillner E. Component resolved diagnostics for hymenoptera venom allergy. Curr Opin Allergy Clin Immunol 2017;17(5):363‐372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Whitrow MJ, Moore VM, Rumbold AR, Davies MJ. Effect of supplemental folic acid in pregnancy on childhood asthma: a prospective birth cohort study. Am J Epidemiol 2009;170(12):1486‐1493. [DOI] [PubMed] [Google Scholar]
- 31. Heeringa JJ, McKenzie CI, Varese N, et al. Induction of IgG2 and IgG4 B‐cell memory following sublingual immunotherapy for ryegrass pollen allergy. Allergy 2020;75(5):1121‐1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chan SK, Pomes A, Hilger C, et al. Keeping allergen names clear and defined. Front Immunol. 2019;10:2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pomes A, Davies JM, Gadermaier G, et al. WHO/IUIS allergen nomenclature: providing a common language. Mol Immunol. 2018;100:3‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Förster E, Dudlerb T, Gmachl M, Aberer W, Urbanek R, Suter M. Natural and recombinant enzymatically active or inactive bee venom phospholipase A2 has the same potency to release histamine from basophils in patients with Hymenoptera allergy. J Allergy Clin Immunol. 1995;95(6):1229‐1235. [DOI] [PubMed] [Google Scholar]
- 35. Grobe K, Pöppelmann M, Becker W‐M, Petersen A. Properties of group I allergens from grass pollen and their relation to cathepsin B, a member of the C1 family of cysteine proteinases. Eur J Biochem 2002;269(8):2083‐2092. [DOI] [PubMed] [Google Scholar]
- 36. Apostolou E, Deckert K, Puy R, et al. Anaphylaxis to Gelofusine confirmed by in vitro basophil activation test: a case series. Anaesthesia 2006;61(3):264‐268. [DOI] [PubMed] [Google Scholar]
- 37. de Leon MP, Drew AC, Glaspole IN, Suphioglu C, Rolland JM, O'Hehir RE. Functional analysis of cross‐reactive immunoglobulin E antibodies: peanut‐specific immunoglobulin E sensitizes basophils to tree nut allergens. Clin Exp Allergy. 2005;35(8):1056‐1064. [DOI] [PubMed] [Google Scholar]
- 38. Kalina T, Flores‐Montero J, van der Velden VHJ, et al. EuroFlow standardization of flow cytometer instrument settings and immunophenotyping protocols. Leukemia 2012;26(9):1986‐2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Edwards ESJ, Bosco JJ, Aui PM, et al. Predominantly antibody‐deficient patients with non‐infectious complications have reduced naive B, Treg, Th17, and Tfh17 Cells. Front Immunol. 2019;10:2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Altmann F. The role of protein glycosylation in allergy. Int Arch Allergy Immunol 2007;142(2):99‐115. [DOI] [PubMed] [Google Scholar]
- 41. Halim A, Carlsson MC, Madsen CB, et al. Glycoproteomic analysis of seven major allergenic proteins reveals novel post‐translational modifications. Mol Cell Proteomics. 2015;14(1):191‐204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Strasser R. Plant protein glycosylation. Glycobiology 2016;26(9):926‐939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rendj D, Wilson IBH, Paschinger K. The Glycosylation Capacity of Insect Cells. 2008.
- 44. Shi X, Jarvis DL. Protein N‐glycosylation in the baculovirus‐insect cell system. Curr Drug Targets. 2007;8(10):1116‐1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Blank S, Michel Y, Seismann H, et al. Evaluation of different glycoforms of honeybee venom major allergen phospholipase A2 (Api m 1) produced in insect cells. Protein Pept Lett. 2011;18(4):415‐422. [DOI] [PubMed] [Google Scholar]
- 46. Koren A, Lunder M, Molek P, et al. Fluorescent labeling of major honeybee allergens Api m 1 and Api m 2 with quantum dots and the development of a multiplex basophil activation test. Allergy 2020;75(7):1753‐1756. [DOI] [PubMed] [Google Scholar]
- 47. Chirumbolo S, Vella A, Ortolani R, et al. Differential response of human basophil activation markers: a multi‐parameter flow cytometry approach. Clin Mol Allergy. 2008;6:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Prussin C, Metcalfe DD. 5. IgE, mast cells, basophils, and eosinophils. J Allergy Clin Immunol. 2006;117(2):S450‐S456. [DOI] [PubMed] [Google Scholar]
- 49. Knol EF. Requirements for effective IgE cross‐linking on mast cells and basophils. Mol Nutr Food Res 2006;50(7):620‐624. [DOI] [PubMed] [Google Scholar]
- 50. Mukai K, Chinthrajah RS, Nadeau KC, Tsai M, Gaudenzio N, Galli SJ. A new fluorescent‐avidin–based method for quantifying basophil activation in whole blood. J Allergy Clin Immunol. 2017;140(4):1202‐1206.e1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Carlsson M, Thorell L, Sjölander A, Larsson‐Faria S. Variability of total and free IgE levels and IgE receptor expression in allergic subjects in and out of pollen season. Scand J Immunol 2015;81(4):240‐248. [DOI] [PubMed] [Google Scholar]
- 52. Stone KD, Prussin C, Metcalfe DD. IgE, mast cells, basophils, and eosinophils. J Allergy Clin Immunol 2010;125(2 Suppl 2):S73‐S80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sihra BS, Kon OM, Grant JA, Kay AB. Expression of high‐affinity IgE receptors (Fc epsilon RI) on peripheral blood basophils, monocytes, and eosinophils in atopic and nonatopic subjects: relationship to total serum IgE concentrations. J Allergy Clin Immunol. 1997;99(5):699‐706. [DOI] [PubMed] [Google Scholar]
- 54. Fall BI, Nießner R. Detection of known allergen‐specific IgE antibodies by immunological methods. In: Bilitewski U, ed. Microchip Methods in Diagnostics. Totowa, NJ: Humana Press; 2009: pp 107‐122. [DOI] [PubMed] [Google Scholar]
- 55. Lupinek C, Wollmann E, Baar A, et al. Advances in allergen‐microarray technology for diagnosis and monitoring of allergy: the MeDALL allergen‐chip. Methods 2014;66(1):106‐119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sturm GJ, Kranzelbinder B, Sturm EM, Heinemann A, Groselj‐Strele A, Aberer W. The basophil activation test in the diagnosis of allergy: technical issues and critical factors. Allergy 2009;64(9):1319‐1326. [DOI] [PubMed] [Google Scholar]
- 57. Knol EF, Mul FP, Kuijpers TW, Verhoeven AJ, Roos D. Intracellular events in anti‐IgE nonreleasing human basophils. J Allergy Clin Immunol. 1992;90(1):92‐103. [DOI] [PubMed] [Google Scholar]
- 58. Nguyen KL, Gillis S, MacGlashan DW Jr. A comparative study of releasing and nonreleasing human basophils: nonreleasing basophils lack an early component of the signal transduction pathway that follows IgE cross‐linking. J Allergy Clin Immunol. 1990;85(6):1020‐1029. [DOI] [PubMed] [Google Scholar]
- 59. Santos AF, Alpan O, Hoffmann H‐J. Basophil activation test: mechanisms and considerations for use in clinical trials and clinical practice. Allergy. 2021. Accepted Author Manuscript. [DOI] [PubMed] [Google Scholar]
- 60. Wagner N, Rudert M. Sensitivity and specificity of standardised allergen extracts in skin prick test for diagnoses of IgE‐mediated respiratory allergies. Clin Translat Allergy. 2019;9(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


